Abstract
Interest in mesenchymal stem cell derived extracellular vesicles (MSC‐EVs) as therapeutic agents has dramatically increased over the last decade. Current approaches to the characterization and quality control of EV‐based therapeutics include particle tracking techniques, Western blotting, and advanced cytometry, but standardized methods are lacking. In this study, we established and verified quartz crystal microbalance (QCM) as highly sensitive label‐free immunosensing technique for characterizing clinically approved umbilical cord MSC‐EVs enriched by tangential flow filtration and ultracentrifugation. Using QCM in conjunction with common characterization methods, we were able to specifically detect EVs via EV (CD9, CD63, CD81) and MSC (CD44, CD49e, CD73) markers. Furthermore, analysis of QCM dissipation versus frequency allowed us to quantitatively determine the ratio of marker‐specific EVs versus non‐vesicular particles (NVPs) – a parameter that cannot be obtained by any other technique so far. Additionally, we characterized the topography and elasticity of these EVs by atomic force microscopy (AFM), enabling us to distinguish between EVs and NVPs in our EV preparations. This measurement modality makes it possible to identify EV sub‐fractions, discriminate between EVs and NVPs, and to characterize EV surface proteins, all with minimal sample preparation and using label‐free measurement devices with low barriers of entry for labs looking to widen their spectrum of characterization techniques. Our combination of QCM with impedance measurement (QCM‐I) and AFM measurements provides a robust multi‐marker approach to the characterization of clinically approved EV therapeutics and opens the door to improved quality control.
Keywords: atomic force microscopy (AFM), extracellular vesicles (EVs), label‐free sensors, quartz crystal microbalance (QCM)
1. INTRODUCTION
The interest in extracellular vesicles (EVs) for therapeutic use or as a medical product has increased massively over the past decade. EVs are membrane surrounded particles that are naturally released by potentially all eukaryotic cell types (Ludwig & Giebel, 2012). Exosomes or small EVs range in size from 30 to 150 nm and carry cell type‐specific proteins, lipids and coding and non‐coding ribonucleic acids. They have been implicated in cellular signalling to regulate biological processes such as immunomodulation and regeneration, but also play a pathophysiological role in cancer, infections, and degenerative diseases such as neurodegeneration (Rohde et al., 2019). EVs obtained from multipotent mesenchymal stem cells (MSC) are of particular interest for therapeutic use (Lener et al., 2015) since they convey anti‐inflammatory (Romanelli et al., 2019) and anti‐apoptotic effects (Terlecki‐Zaniewicz et al., 2018), stimulate angiogenesis (Zhang et al., 2019) and wound healing (Shabbir et al., 2015). A current clinical study investigates the use of MSC‐EVs for the treatment of type I diabetes mellitus (NCT02138331). In an individual treatment attempt, MSC‐EVs were administered in a patient with graft versus host disease (Kordelas et al., 2014). The properties of an EV‐based therapeutic, a product of natural processes, fluctuate widely due to the variable cellular secretome. Furthermore, the amount and composition of EVs that can be produced is impacted by the cell cultivation and expansion conditions as well as by the EV harvesting and isolation techniques, for example, the media supplements used during MSC cultivation (Gimona et al., 2017; Konoshenko et al., 2018). From the cellular secretome, a mixed population of EVs, such as exosomes, microvesicles, shedding vesicles (ectosomes), and microparticles, as well as a number of soluble factors, all of which are also involved in intercellular communication (Gurunathan et al., 2019), can be enriched. Although EVs were used in patients for the first time in 2005 for cancer therapy (Escudier et al., 2005), there are still no routinely established standard techniques for quality control of EV therapeutics for clinical production. The latest minimal information for studies of extracellular vesicles (MISEV) guidelines stated that quantitation and single‐particle characterization should be performed by methods including but not limited to sizing and counting by particle tracking techniques, imaging by electron microscopy, and advanced flow cytometry (Witwer et al., 2019). Current particle quantitation methods such as nanoparticle tracking analysis (NTA), dynamic light scattering (DLS), and resistive pulse sensing (RPS) cannot discriminate EVs from other particles (Giebel & Helmbrecht, 2017). Raman Spectroscopy is capable of determining differences in EV preparations in terms of purity and composition (protein‐to‐lipid and nucleic acids‐to‐lipid ratio) but this technique is not suitable for EV preparations generated by commercial kits based on precipitation or via density gradient purification (Gualerzi et al., 2019). Additionally, single or at least small numbers of EVs from different cellular origin could be distinguished by their molecular composition using synchronized Rayleigh and Raman scattering and laser tweezers Raman spectroscopy (Enciso‐Martinez et al., 2020). However, superposition of various EV components complicates data interpretation, and a complete quantitative spectral interpretation is still in progress (Kruglik et al., 2019). Dual‐wavelength surface plasmon resonance (SPR) (Rupert et al., 2016) provides information about the concentration and size of nanoparticles or the presence of characteristic surface proteins, but cannot distinguish between EVs and other particles (e.g. protein aggregates), which may become co‐enriched during isolation (Lobb et al., 2015). Due to the heterogeneity and small dimensions, EV characterization is still challenging. Atomic force microscopy (AFM) imaging can be used to characterize the impact of EV isolation procedure on EV size distribution, morphology, and mechanical properties (Parisse et al., 2017). Perissinotto et al. analysed EVs with a combination of Fourier Transform Infrared Spectroscopy, Ultraviolet Resonant Raman Spectroscopy, AFM, and Small Angle X‐Ray Scattering to address size, stability and purity of EVs (Perissinotto, 2020). A more advanced method for measuring particle number combines SPR with AFM (Obeid et al. (2017)). The latter presents a promising first strategy to obtain more detailed information about EV properties (Vorselen et al., 2020). Nevertheless, ensuring that the counted particles are indeed EVs and that these EVs are therapeutically active (e.g. as indicated by their surface marker profile and meaningful biochemical or cell‐based assays), in a robust, accessible and economical manner remains unsolved. Quartz crystal microbalance (QCM) is a highly sensitive method able to simultaneously detect biomarker association and the resulting changes in mechanical properties at the binding interface (Suthar et al., 2020). QCM provides highly sensitive mass sensing, based on the piezoelectric effect via changes in the oscillation of a gold‐coated quartz crystal. It is a recognized and established method for the detection and characterization of adsorption processes on surfaces. Changes in frequency directly relate to changes in mass according to the Sauerbrey equation (Sauerbrey, 1959), given thin and rigid adlayers. Soft and thus more dissipative adlayers such as those represented by biological binding processes often require more complex modelling (Reviakine et al., 2011). Specific interactions of ligand‐receptor or antigen‐antibody as well as the behaviour of supramolecular systems such as the adhesion of vesicles and liposomes can be examined with a sensitivity of less than 1 ng/cm2 in real time to determine kinetic rates and affinity constants (Kasper et al., 2016; Strasser et al., 2020). The simultaneous detection of energy loss during the oscillation of the quartz crystal–adsorbate system allows experimental access to interfacial changes that do not alter the mass of the system but do affect its dissipative properties. It enables the differentiation between rigid and soft adsorbates, and—using the state of the art QCM‐D device—has been employed previously to phenotypically subtype exosomes at native concentrations (Suthar et al., 2020). Here we propose a QCM‐I and AFM based protocol to characterize EVs in terms of their elasticity and surface markers, and to distinguish them from NVPs exhibiting the same surface markers, tailored to be robust, reproducible, and easy to integrate in existing laboratory infrastructures.
2. METHODS
2.1. Primary isolation and expansion of human MSCs
Human umbilical cord (UC)‐derived MSCs were isolated as previously described (Pachler, 2017). Immediately after delivery, cords were collected and stored in phosphate buffered saline (PBS) until further processing. Whole cords were washed with PBS to remove contaminating blood cells before the cord stroma was cut into small pieces of 1 – 2 mm3. Pieces were transferred into a culture plate allowing them to dry‐adhere to the plastic surface before adding culture medium based on alpha‐modified minimum essential medium (α‐MEM Sigma‐Aldrich, MO, USA) supplemented with 10% (v/v) pooled human platelet lysate (pHPL) and Dipeptiven (5.5 mg/ml, Fresenius‐Kabi, Germany). Pooled HPL was prepared as previously described (Laner‐Plamberger et al., 2015). In brief, expired platelet concentrates were lysed by several freeze/thaw cycles. Platelet fragments were pelleted by centrifugation (4000 × g, 15 min at room temperature) and aliquots of the supernatant were frozen at ‐30°C until use. After 10 to 12 days, outgrowing UC‐MSC colonies became visible and cord tissue pieces were removed. UC‐derived MSCs were detached enzymatically by addition of TrypLE Select CTS (A12859‐01, Thermo Fisher Scientific, MA, USA), and further expanded in cell factory systems (CF4, Thermo Scientific). Immuno‐phenotyping and viability analysis of MSC was carried out according to the proposed surface marker profile for defining MSC identity as published by the International Society of Cell Therapy (ISCT) in 2006 (Dominici et al., 2006).
2.2. Flow cytometric analysis of UC‐MSCs
To demonstrate the MSC identity of the parental cells at the time of collection of the conditioned medium the immunophenotype and viability analysis was carried out according to the established marker profile for defining MSC identity as published by the ISCT. In brief, collected cells were centrifuged (300 × g for 7 min), resuspended in 5% v/v sheep serum‐containing blocking buffer to reach a concentration of 1.5 × 107cells / ml and incubated for 20 min at +4 °C in the dark. 3 × 105 cells were stained with mouse anti‐human monoclonal antibodies against CD90 (IM1839U, Beckman Coulter, France), CD105 (Life Technologies, Austria), CD14, CD34, CD45, CD73, HLA‐II (DR) (Becton Dickinson), or with corresponding isotype controls (Becton Dickinson) for 25 min at +4 °C in the dark. Samples were washed in PBS, resuspended in 100 μl 7AAD‐containing PBS (1:10 dilution, 0.0005 w/v % final concentration) and stained for 10 min at room temperature. Finally, 400 μl cold PBS was added and the samples were measured immediately using FACSCanto II flow cytometer (Becton Dickinson) until 10.000 events were recorded per staining. Blue (488 nm) and red (633 nm) laser excited fluorescence signals were detected with the following standard light filters: FITC: 530/30 nm; PE: 585/42 nm; APC: 660/20 nm; 7AAD: 670LP. Results were analysed with WinList software 8.0 (Verity Software House, Topsham, USA). FSCA ‐SSCA dot plot analyses were applied for debris exclusion, and a doublet discrimination panel was set on the FSC channel for the detection of height and width of the fluorescence signals. The ratio of the viable cells was determined on [SSC‐7AAD] dot plot.
2.3. Manufacturing and characterization of MSC‐EVs
Batches of EVs from UC‐MSCs were prepared according to Good Manufacturing Practice (GMP) as previously described (Desgeorges et al., 2020; Gimona et al., 2017; Pachler et al., 2017b; Warnecke et al., 2020). In brief, cells were cultured in fibrinogen‐depleted culture medium at 5% CO2 and 37°C. Upon reaching 60%–70% confluence, growth medium was exchanged with EV‐depleted harvest medium containing 5% HPL. After 24 h, conditioned medium was collected, centrifuged at 2.500 × g for 20 min at 18°C and clarified via sterile filtration (0.22 μm). The resulting supernatant was concentrated and buffer‐exchanged to PBS by Tangential Flow Filtration (TFF) and diafiltration, respectively, using a 100 kDa hollow fibre column filter (Spectrum Labs, Greece). Ultimately, EVs were enriched by ultracentrifugation at 120.000 × g for 3 h at 18°C in a Sorvall model WX‐80 using a fixed angle rotor model Fiberlite F37L‐8x100 (Angle = 25°, K Factor = 168). Resulting pellets were washed once with 10 ml PBS and subsequently resuspended in Ringer's Lactate. Resuspended EVs were subsequently centrifuged at 3.000 × g for 10 min at 4°C and the supernatant was sterile filtered (0.22 μm). Individual doses were stored in glass vials at ‐ 80°C and batches were tested for endotoxin levels, bacterial sterility and the presence of mycoplasma.
2.4. Total protein mass determination
Total protein amounts were determined using a QuBit 3.0 Fluorometer instrument (Life Technologies, CA, USA) according to the manufacturer´s instructions.
2.5. Protein profiling
Proteins (IL‐1ß, IL‐6, IL‐8, TNF‐α, MCP‐1, Insulin, Leptin, ß‐NGF, BDNF and IL‐10) from various preparations were analysed using V‐Plex and U‐Plex human multiplex immunoassay kits on the MSD platform (Meso Scale Diagnostics, MD, USA) according to the manufacturer´s instructions.
2.6. Nanoparticle tracking analysis (NTA) in light scatter mode
To determine the size and amount of particles in the individual EV preparations, samples were analysed using a Nanoparticle Tracking Device (ZetaView PMC 110 from Particle Metrix, Germany) in light scatter mode essentially as described (Desgeorges, 2020). Previously frozen EV preparations were used and samples were diluted to a concentration of 4 – 7 × 107 particles/ml in PBS. Prior to NTA analysis, the instrument was calibrated using Yellow/Green‐labelled 100 nm polystyrene standard beads (1:1.000.000 dilution in ddH2O). The minimum brightness was set to 20 arbitrary units (AU), temperature to 21.5°C, shutter to 70 AU, and sensitivity to 85 AU. Subsequently, data for two exposures at 11 measurement positions were acquired per sample. Based on the Stokes‐Einstein equation, particle size was calculated using the ZetaView software (PMX 110: Version 8.4.2).
2.7. MACSPlex surface protein profiling
The bead‐based multiplexed FACS‐based MACSPlex Exosome Kit (Miltenyi Biotec, Germany) is an assay for the analysis of surface markers present on EVs. To characterize the various MSC‐EV preparations we used the MACSPlex kit according to the manufacturer's instructions and following a validated standard operating procedure with 5 × 107 to 5 × 108 total particles as input. Data acquisition was done using a FACS Canto II instrument (BD Biosciences, CA, USA). For additional CD73 analyses an anti‐CD73‐BV421 antibody (BD Biosciences) was used. Data normalization was directed towards CD9/CD63/CD81 APC signal. Isotype control normalization was performed as described (Wiklander et al., 2018).
2.8. Western blotting
EV solutions (10 μl) from two independent batches (7,32 μg protein from 3,63 × 108 and 3,86 μg protein from 2,93 × 108 total particles) were incubated with an equal amount of Laemmli sample buffer (Bio‐Rad Laboratories, Portland, ME, USA) supplemented with 2‐Mercaptoethanol (Bio‐Rad Laboratories) at 95°C for 5 min. EV proteins were then separated on 4%–15% gradient polyacrylamide gels (Bio‐Rad Laboratories) and transferred onto nitrocellulose membranes (Bio‐Rad Laboratories). Precision Plus Protein Dual Colour Standards served as the protein size marker. Recombinant human 5′‐ectonucleotidase (CD73) at 0.4 mg/ml was purchased from Sigma. After electrotransfer nitrocellulose membranes were incubated in Ponceau‐S solution (0.1% in 5% acetic acid; Sigma) for 5 min and rinsed in distilled H2O for 5 min. Membranes were then blocked with 5% non‐fat dry milk in Tris‐buffered saline containing 0.1% Tween‐20 (TBS‐T) for 1 h at room temperature and probed with primary antibody against CD73 (Cell Signaling, 1:1000 in in TBST) and secondary antibody anti rabbit IgG, HRP linked (Cell Signaling, in TBST). Documentation was performed using ChemiDoc with ECL Prime (Amersham).
2.9. CryoEM analysis
UC‐MSC EV samples were deposited on an electron microscopy (EM) grid coated with a perforated carbon film. Samples were quickly frozen in liquid nitrogen‐cooled liquid ethane using a Leica EM‐PC cryo system. EM grids were maintained under liquid nitrogen until use. EM grids transferred to a Tecnai F20 cryo‐electron microscope (FEI, ThermoFisher) operating at 200 kV. Grids were mounted in a Gatan 626 cryo‐holder and images were recorded with a FEI‐Eagle camera by Prof. Alain Brisson (University of Bordeaux, France)
2.10. CD73 activity assay
The enzymatic activity of CD73 in UC‐MSC EV preparations was determined by incubating 10 μl of EVs (representing between 7 × 109 ‐ 2.8 x 1010 particles) in 10 mM HEPES (Sigma H3537) buffer containing 2 mM MgCl2 (Merck Millipore, MA, USA) with 10 μM AMP (Sigma 01930) for 20 min at 37°C. The amount of AMP consumption was detected with the AMP‐Glo Assay Kit (Promega, WI, USA) according to the manufacturer´s protocol and measured with Spark multimode microplate reader (Tecan, Austria). 2 ng rhCD73 (Sigma N1665) was used as positive control and AMP‐CP (Sigma M8386) as CD73 inhibitor.
2.11. Immunomodulation assay
To investigate the immunomodulatory activity of UC‐MSC EV preparations, the capacity to inhibit T cell proliferation in vitro was studied, essentially as described previously (Pachler et al., 2017a). Stimulation of T‐cell proliferation was achieved by incubation with CD3/CD28 antibody beads (Thermo Fisher). Carboxyfluorescein succinimidyl ester (CFSE) pre‐labelled pooled peripheral blood mononuclear cells were stimulated with CD3 and CD28 and co‐cultured with different ratios of UC‐MSC EVs for 72 h essentially as described (Trickett and Kwan, 2003) at a ratio of 1.5 × 105 cells per 1 × 1010 particles. The percentage of inhibition of fluorescently labelled CD3 T‐cell proliferation was analysed by flow cytometry in triplicates.
2.12. Liposome preparation
Liposomes containing biotinylated headgroups were prepared from 1,2‐dioleoyl‐sn‐glycero‐3‐phosphoethanolamine‐N‐(cap biotinyl) (Biotinyl‐cap‐DOPE), 1,2‐dioleoyl‐sn‐glycero‐3‐phosphocholine (DOPC) and 1,2‐dioleoyl‐sn‐glycero‐3‐phospho‐L‐serine (DOPS) in a ratio of 1:7:2 (Karner et al., 2017). In total 5 mg of lipids (all Avanti Polar Lipids, AL, USA) of Biotinyl‐cap‐DOPE:DOPC:DOPS were dissolved in 2 ml of Trichlormethan/Chloroform (Roth, Germany) and mixed with 1 ml of Methanol (Roth, Germany). Lipids were dried using a R‐100 rotary evaporator (Buchi, Germany) at 180 rpm for 30 min. To remove the methanol, the dried lipid mixture was dissolved in 3 ml chloroform and dried again using the rotary evaporator at 180 rpm for 30 min. Afterwards, the lipid mixture was further dried for 1‐2 h with a high vacuum pump DCP 3000 (Vacuubrand, Germany). The dried lipid mixture was dissolved in 500 μl mQ‐H2O by repeated pipetting and transferred into a glass vial capped with a septum. The lipid mixture was sonicated in an ultrasonic bath (Sonorex, Germany) for 10 min until the emulsion got clear. To obtain a final concentration of 2 mg/ml, 2 ml of running buffer (RB) consisting of 10 mM Hepes, 150 mM NaCl, 2 mM CaCl2 (pH 7.4) were added and aliquots of 100 μl were shock‐frozen with liquid nitrogen and stored at ‐80°C until further use.
2.13. QCM measurements
Quartz crystal microbalance (QCM) is an ensemble technique utilizing the inverse piezoelectric effect to measure mass adsorption and changes in fluid viscosity. In QCM, adsorption processes and changes in the medium surrounding the crystal result in in proportional changes in the frequency and energy dissipation of its oscillation (Cheng et al., 2012). Monitoring changes in resonance frequency due to mass deposition on the quartz surface thus yields real‐time traces of association and dissociation of everything from small molecules to entire cells.
All QCM experiments were conducted using a two‐channel QCM‐I system from MicroVacuum Ltd. (Hungary). AT‐cut SiO2‐coated quartz crystals with a diameter of 14.0 mm and a resonance frequency of 5.000 MHz were used (Quartz Pro AB, Sweden). All sensorgrams were recorded on the first, third and fifth harmonic frequencies simultaneously. The data shown relates to the third harmonic. A constant flow rate was provided by a programmable peristaltic pump. Before each set of experiments, the SiO2‐coated QCM crystals were cleaned as described (Strasser et al., 2020). Briefly, chips were immersed in 2% SDS for 30 min, followed by thorough rinsing with mQ−H2O. The chips were then dried in a gentle stream of N2 and activated using air plasma (4 min at 80 W), after which they were ready to be mounted in the measurement chamber. Once securely affixed, the sensor surface was cleaned by pumping 2% SDS through the liquid cell at 250 μl/min for 5 min, followed by mQ−H2O at 250 μl/min for 5 min, and finally RB for equilibration at 50 μl/min. All subsequent injections were performed at 50 μl/min. After approximately 30 min of RB equilibration, 200 μg/ml biotin‐liposome solution was injected until complete bilayer fusion was achieved (approx. 20 min). To ensure complete bilayer coverage without exposed substrate, a 6.25 μg/ml Control Protein (CP) solution was injected. The used CP, C1q (Complement Technology, TX, USA), adheres to activated SiO2 but not to the lipid bilayer. No change in frequency proves that the lipid is closed and that there is no non‐specific protein adsorption. Subsequently the bilayer is loaded with 60 μg/ml streptavidin (Merck Millipore, MA, USA) to saturation. Finally, 10 μg/ml biotinylated antibody are added and bound to the lipid bilayer via streptavidin. Specific EV markers were recruited using biotin anti‐human IgG1 targeting CD9 (312112), CD44 (103003), CD63 (353017), CD73 (344017) and CD81 (349514) (all from BioLegend, CA, US), CD49e (Invitrogen 13‐0496‐80) respectively. Biotin anti‐DNP IgG1 served as negative control (ACROBiosystems, US). Then UC‐MSC EVs can be analysed for the presence of different surface markers by diluting the stock concentration 1:10 with RB and injecting the solution for 5 min to ensure appropriate volume exchange. After this time the flow was stopped for 30 min in order to limit sample consumption to < 300 μl and to allow for stable EV association to the surface. Finally, the sample was washed with RB for 20 min under constant flow to observe any dissociation of the loaded EVs. Finally, the EV recruitment efficiency, i.e. the frequency shift during EV binding normalized to the shift during antibody immobilization, is calculated and compared between markers using MATLAB (MathWorks). Analysis of frequency and dissipation response was performed with the QCM software BioSense, version 3.11. After each measurement, the chip can be regenerated by online pumping 2% SDS for 5 min followed by H2O for 5 min at 250 μl/min through the liquid cell, respectively.
We determined the limit of detection (LOD), limit of quantification (LOQ), and dynamic range of the system towards our EVs by comparing the response at a range of EV concentrations to a control surface. LOD, LOQ, and the dynamic range were determined using CD73 as a model marker. Relative recruitment for 5 × 106, 1 × 107, 2.5 × 107, 5 × 107, 1 × 108, 5 × 108, 2.5 × 109 and 9.5 × 109 particles/ml were recorded in duplicates. The resulting sigmoid curve was fitted with R = Rmax*c/(c+c50) where R is the relative recruitment, Rmax its maximum, c the particle concentration, and c50 the particle concentration eliciting half of Rmax. The required control surface was established by using biotin anti‐DNP IgG1 instead of biotin‐anti‐CD73. Here, triplicate measurements using 5 × 108 particles/ml were performed. For the purposes of this study, LOD and LOQ were defined as the analyte concentration eliciting the control's recruitment efficiency plus three and ten times the standard deviation of the control measurements, respectively (Shrivastava A, 2011). The dynamic range was defined as the concentration range from the LOQ as lower bond and the limit of linearity as upper bond, as estimated from the recruitment efficiency over particle/ml plot.
2.14. AFM
An atomic force microscope (AFM, JPK Nano Wizard4, Germany) mounted on an Olympus IX71 inverted optical microscope was used for sample characterization. MLCT‐F cantilevers with a nominal tip radius of 20 nm and a spring constant of 0.6 N/m (Bruker, Germany) were used for measurements. Cantilevers were calibrated prior to each measurement using the commercial JPK NanoWizard control software V6.1.163. The indentation force was set constant to a value of 0.8 nN for EV‐imaging.
The off‐resonance JPK QI mode was used for topography imaging and mechanical characterization of EVs. In this mode, several sample properties (topography, stiffness, adhesion, elasticity) can be acquired within a single measurement by recording complete force‐distance curves in each pixel. As a result, lateral forces on the sample are eliminated during scanning which minimizes sample deformation and possible displacements for loosely bound substances. All measurements were done in an aqueous environment (PBS).
Glass slides were cleaned by rinsing with mQ‐H2O, Isopropanol (99,9%), mQ‐H2O and Ethanol (96%), and dried with a stream of N2 gas. Glass slides were cleaned in Argon plasma for 15 min at 40 kHz 96 W (Zepto, Diener electronic Germany). Samples were diluted in PBS, until a sparse surface density of ∼2 particles/μm2 was achieved after incubation for 45 min on the cleaned glass slide to ensure proper separation between objects for individual characterization. After incubation, the sample was gently rinsed 3 times with PBS while ensuring continuous liquid coverage of the glass slide to prevent the removal of surface‐bound EVs.
For all measured particles, height, and Young's modulus (elasticity) were determined using JPK data processing software, version 6.1.121. Young's Modulus were obtained by fitting Hertz‐contact model (paraboloid tip shape; tip radius of 20 nm; poison ratio of 0.5) to the force‐distance curves.
3. RESULTS
3.1. Quality control of UC‐MSC EVs according to MISEV guidelines
In this study, UC‐MSC EVs that were manufactured according to the pharmaceutical standards and guided by approved Standard Operating Procedures (SOPs) were enriched from the MSC secretome by Tangential Flow Filtration (TFF) and ultracentrifugation (UC). The MISEV 2018 guidelines state that both, the source and the preparation of EVs, should be described as quantitatively as possible. First, the number of cultured cells at the time of collection must be determined for each experimental use (Thery et al., 2018). Isolated MSCs exhibit a viability of ≥ 90% and represent the typical MSC surface marker profile with ≥ 95% positive for CD29+, CD44+, CD73+, CD90+, CD105+, CD166+ and with ≤ 2% for negative markers CD14, CD19‐, CD34‐, CD45‐ and MHC class II‐ (Table 1, Supplementary Figure S1a). Regarding quantitation and single‐particle characterization of EVs, particle tracking, imaging, and advanced flow cytometry has been recommended (Witwer et al., 2019). Our NTA measurements revealed a particle number of > 5x1010/ml, particle sizes ranging from 80 to 150 nm (Table 1, Figure 1A), and a fraction ≥10‐15% of CD63+, CD81+ and CD73+ EVs. Additionally, a total protein concentration of < 5 mg/ml was determined (Table 1). Analysing these EVs using MACSPlex, we observed a panel of positive (CD9+, CD29+, CD44+, CD49e+, CD63+, CD81+, CD105+, MCSP+) and negative (CD14‐, CD19‐, CD45‐, CD142‐, MHC class I (= HLA_ABC) ‐ and class II (= HLA‐DP, DR, DQ)‐ surface markers that are typical for MSC‐EVs (Table 1, Figure 1B). An additional MACSPlex analysis demonstrates the presence of CD73 on the surface of UC‐MSC EVs and shows that CD73 positive EVs preferentially contain CD44 in addition to the other EV markers (Supplementary Figure S1b). The presence of CD73 on two independent UC‐MSC EV batches was further confirmed by Western blotting (Supplementary Figure S1c). The protein profile of MSC‐EV preparations demonstrates absence/low levels of IL‐1ß, insulin, leptin and IL‐10, intermediate levels of IL‐6, IL‐8, TNF‐α and ß‐NGF and high levels of MCP‐1 and BDNF. (Figure 1C). Cryo‐electron microscopy images of MSC‐EVs (Figure 1D) clearly revealed the presence of EVs (identified through their lipid bilayer membrane) and other particles of approx. similar size (identified through the absence of a lipid bilayer membrane), respectively (Emelyanov et al., 2020). AFM topography images of MSC‐EVs incubated on a flat glass substrate revealed a mixture of various particle sizes (Figure 1E).
TABLE 1.
Multimodal quality control parameters of uc‐msc evs
| Parameter | Release criteria | Method |
|---|---|---|
| Cell count and viability | ≥90% viable cells, cell count determines cell equivalent | 7‐AAD stain; flow cytometry |
| Cell surface marker profile | ≥95% CD29+, CD44+, CD73+, CD90+, CD105+, CD166+ | Multi‐colour flow cytometry |
| ≤2% CD14–, CD19–, CD34–, CD45–, MHC class II– | ||
| Particle size | 80–150 nm | Nanoparticle Tracking Analysis (NTA) |
| Particle number | >5 × 1010/ml | Nanoparticle NTA |
| EV number | Percentage of CD63+, CD81+, CD73+ (≥10%–15%) | Fluorescent NTA |
| EV particle identity | CD9+, CD29+, CD44+, CD49e+, CD63+, CD81+, CD73+, CD105+, MCSP+ | Flow cytometry‐based bead array MACS Plex |
| CD14–, CD19–, CD34–, CD45–, CD142–, MHC class I–, class II– | ||
| Protein concentration | <5 mg/ml | Qubit 3 |
Parental cell characterization, identity, purity and impurity determination of EV preparations is performed for the standard quality release testing of all research scale preparations and for GMP training and GMP clinical runs.
FIGURE 1.
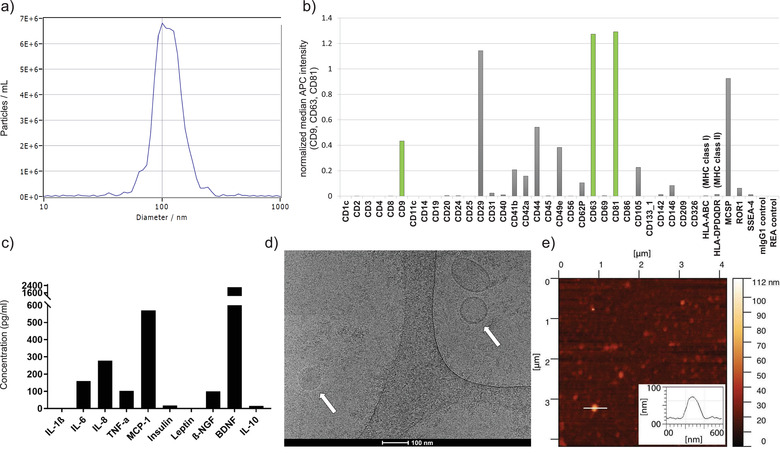
Characterization of UC‐MSC EVs. (A) Size distribution of UC‐MSC EVs by nanoparticle tracking analysis. (B) Surface marker profiling by MACSPlex. The surface profile of one representative UC‐MSC EV batch is shown. (C) UC‐MSC EV protein profile demonstrates absence/low levels of the pro‐inflammatory cytokines IL‐1ß, insulin, leptin and IL‐10, intermediate levels of IL‐6, IL‐8, TNF‐α and ß‐NGF and high levels of MCP‐1 and BDNF. (D) Cryo‐electron microscopy image from a representative batch of MSC‐derived EVs and NVPs, size bar 100 nm. The lipid bilayer surrounding the EV can be recognized (right arrow). Other particles can be identified through the absence of a lipid bilayer (left arrow). (E) AFM topographic image of UC‐MSC EVs incubated on a flat glass substrate and a cross‐section through an EV (inset)
3.2. Functional characterization of UC‐MSC EVs
We have previously shown, that UC‐MSC EVs retain the immunomodulatory potential of their parent MSCs (Pachler et al., 2017a). The precise mechanism underlying the T‐cell proliferation inhibition and potential immunomodulatory activity of UC‐MSC EVs is not completely understood. However, adenosine signalling via the conversion of AMP to adenosine by the GPI‐anchored 5´‐ecto‐nucleotidase CD73 can shape various lymphocyte functions, the majority of effects being described as suppressive, and could be, at least in part, responsible for this activity. We thus investigated if the EV‐associated CD73 is still functional on enriched UC‐MSC EVs after the purification process. As shown in Supplementary Figure S2 UC‐MSC EV preparations actively converted AMP in a biochemical assay in a dose dependent manner. This activity was sensitive to heat and heat‐treated UC‐MSC EV preparations that were neither capable of converting AMP nor of inhibiting T‐cell proliferation in vitro. These findings suggest that the UC‐MSC EVs that were used in this study retained biochemical and biological activity.
3.3. Advanced AFM characterization
AFM is a well‐established technique for high‐resolution imaging of biological structures under physiological conditions (Preiner et al., 2007, 2014, 2015; Strasser et al., 2019). Beyond simple topography (height) images, AFM can be used to determine the mechanical properties of particles (LeClaire, 2020). Figure 2A displays an AFM topography image of UC‐MSC EVs adsorbed to a glass slide. Due to their overall similarity in morphological appearance, distinction between EVs and NVPs based on their height was not possible, as evident from a histogram of 252 individually analysed particles (Figure 2B). However, quantitative analysis of the simultaneously recorded QI™‐mode elasticity map (Figure 2C) revealed two distinct populations of particles that differ in their specific elasticities, as apparent from their Young's modulus depicted in Figure 2D. The histogram is reasonably described by the sum of two Gaussian distributions with means 2.9 ± 2.1 MPa and 14.9 ± 6.5 MPa. Given the area under the respective curves, the first (lower Young's modulus) population includes 158 particles (∼ 63%) and the second (higher Young's modulus) population 94 (∼ 37%). Remarkably, the Young's modulus of the first population overlaps with the value determined for pure vesicles (4.5 ± 2.0 MPa, Supplementary Figure S3a) and is within the range for various different EVs reported in literature (0.2 – 2.7 MPa; (Laney et al., 1997; Liang et al., 2004; Sharma et al., 2011)). The Young's modulus of the second population was significantly higher, comparable to the value determined for large proteins (18.8 ± 10.8 MPa, Supplementary Figure S3b) and within the range of other large proteins reported in literature (∼ 9 MPa; (Perrino and Garcia, 2016)). Strikingly, when treating our UC‐MSC EVs with Triton X‐100 to lyse EVs prior to the QI‐mode characterization, the lower Young's modulus population was no longer detected, while the larger Young's modulus population was still present (15.3 ± 5.2 MPa, Supplementary Figure S3c). This clearly identifies the remaining population as NVPs, and attributes the appearance of the lower Young's modulus population in the untreated sample to the presence of EVs (Supplementary Figure S3c). Together, advanced AFM characterization proves the existence of two distinct particle populations in our UC‐MSC EVs that differ in their Young's modulus and can thus be identified as EVs and proteinaceous NVPs, respectively.
FIGURE 2.
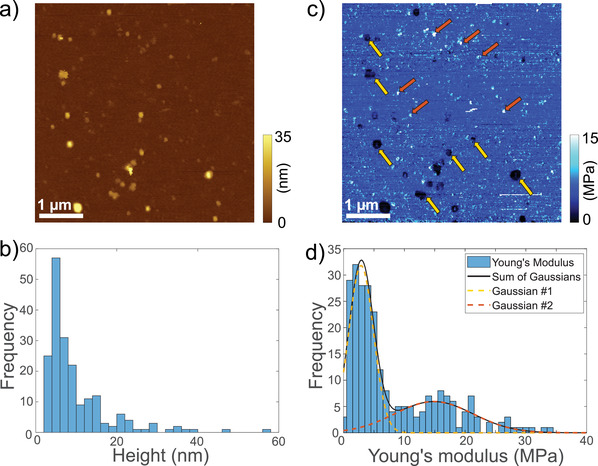
AFM characterization of UC‐MSC EVs incubated on a glass slide. (A) Topography image, showing particles of various sizes. (B) Histogram of particle heights (n = 252) determined through AFM imaging as in (a). (C) Simultaneously recorded QI™‐mode elasticity map of the same sample area as in (a). Arrows indicate particles that likely represent EVs (yellow) or NVPs (brown) based on their Young's modulus. (D) Histogram of Young's modulus determined from simultaneously recorded QI™‐mode elasticity maps. A sum of two Gaussians with means 2.9 ± 2.1 MPa and 14.9 ± 6.5 MPa, and amplitudes 31.8 ± 4.4 (63%) and 5.9 ± 2.2 (37%) reasonably fits the data
3.4. QCM characterization of UC‐MSC EVs
Our results so far describe a biologically active and potentially therapeutically relevant but complex mixture of EVs and NVPs. We thus established a flexible and easy to use QCM‐I protocol for the detection of surface markers on the EV fraction, which utilizes a combination of specific recruitment of marker‐positive particles by antibodies and the distinction of EVs and NVPs (that potentially also carry a marker) through their differing viscoelastic properties. Fluid‐filled vesicles are highly viscoelastic, while protein aggregates are significantly more rigid. Figure 3A provides a schematic illustration of the experimental setup. At first, a lipid bilayer containing 10% biotinylated lipids was established on the QCM sensor chip (Figure 3B). The corresponding characteristic total frequency shift ΔfLipid was ∼25‐30 Hz (Keller & Kasemo, 1998). We employed a 450 kD control protein, which adsorbs to exposed SiO2 but not to lipids (Figure 3C) to check for bilayer integrity and prevent unspecific binding of antibodies and EVs/NVPs later in the experiment. The surface was then loaded with streptavidin and subsequently with the biotinylated IgG of interest (ΔfSA and ΔfIgG respectively, both ∼20 Hz). Finally, the EV/NVP solution was injected, resulting in a maximal association signal Δfmax and, after a dissociation phase, a stable immobilization signal Δfstable. To make our analysis independent of subtle differences in the amount of marker‐specific IgGs bound to our QCM chips, we determined the relative recruitment of EV/NVP material from solution, i.e. we normalized the frequency shift observed during sample application by the frequency shift caused by immobilized IgGs, Δfmax/ΔfIgG. Notably, our experimental design allows for an easy in‐situ reset of the experiment (cf. methods) without removing the senor chip from the instrument, which leads to highly reproducible measurements (Supplementary Figure S4).
FIGURE 3.
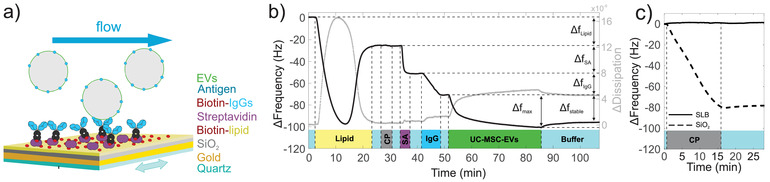
QCM workflow for characterizing EV surface proteins. (A) Schematic of the experimental setup. A quartz crystal is coated with a supported lipid bilayer containing biotin. Streptavidin is added and recruits biotinylated antibodies, which subsequently capture antigen‐carrying EVs. Vesicles, extracellular particles, or proteins without this marker are not detected. (B) Representative QCM sensorgram of an EV characterization experiment. ΔfLipid serves as indication of lipid coverage and quality, as does the subsequent injection of the Control Protein CP. No binding of CP was observed after lipid fusion, demonstrating that a homogenous lipid bilayer without defects or non‐specific protein adsorption has formed. ΔfSA and ΔfIgG denote the amount of bound streptavidin and biotinylated antibodies, respectively. Δfmax is the maximum association observed for the given EV solution, Δfstable indicates the mass remaining after 20 min of dissociation. The presence of a certain substance in the running buffer is indicated by the coloured rectangles at the bottom of the figure. Light blue corresponds to buffer only. (C) CP tightly associates to the bare SiO2 surface of the QCM sensor chip; decrease in frequency shift). A supported lipid bilayer passivates the sensor surface against unspecific adsorption (stable baseline during CP incubation)
Antibodies targeting common EV markers, the tetraspanins CD9, CD63 and CD81, showed significant recruitment of the UC‐MSC EV sample relative to the control, as did the typical MSC markers CD44 and CD73, and the marker typical for UC‐MSC, CD49e (Figure 4A), which is in line with our EV surface marker expression characterized by MACSPlex analysis (Table 1, Figure 1). Our approach additionally provides a semi‐quantitative comparison of marker‐positive particle recruitment: Markers that result in recruitment efficiencies below that of the negative control plus 3 standard deviations (n.c.+3∂) are considered negative, those with values between n.c.+3∂ and n.c.+10∂ as weakly positive, and those above n.c.+10∂ as strongly positive (Figure 4B). Given these parameters, CD9, CD81, CD73, CD63, CD49e, and CD44 are classified as strong positive. No major distinction can be made between relative EV/NVP recruitment with or without 20 min of dissociation (Supplementary Figure S5a and b). This further indicates that indeed the relative abundances of surface markers rather than a potentially different affinity of the marker‐specific IgGs give rise to the respective relative recruitment, thereby justifying the above comparison and classification. On the other hand, direct comparison to the MACSPlex analysis (APC intensity, Figure 1B) is not straightforward since the affinities of detection antibodies are not known (not disclosed by the manufacturer) and might very well be different from the affinities of antibodies used in our QCM platform.
FIGURE 4.
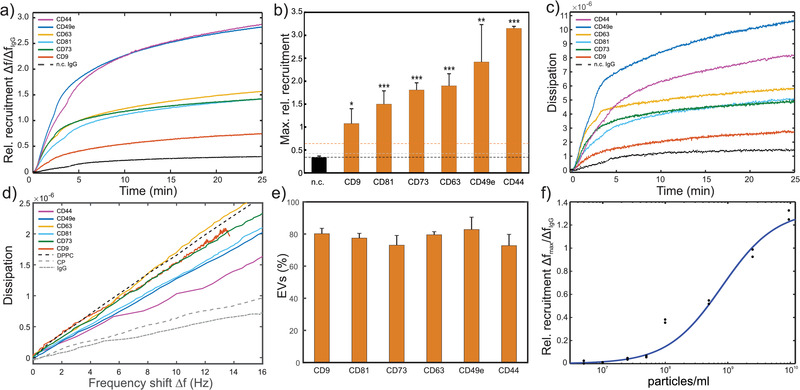
QCM analysis of a CD9, CD44, CD49e, CD63, CD73 and CD81 positive primary UC‐MSC EV preparation. (A) Relative recruitment for the different marker‐specific antibodies (anti‐CD9, anti‐CD44, anti‐CD49e, anti‐CD63, anti‐CD73, and anti‐CD81 antibodies) and a control anti‐DNP IgG (dashed line) calculated from the binding curves (frequency shift) normalized to the total number of immobilized antibodies, respectively. (B) Maximal rel. recruitment levels for each marker. Bars represent means ± standard deviation of experimental replicates. The black dashed line represents the mean of all negative controls (n.c.). Grey and orange dashed lines represent the mean of all negative controls plus 3 and 10 standard deviations, respectively. All data relates to the third harmonic frequency of the 5 MHz quartz crystals used. Statistical analysis was performed using ordinary one‐way Anova, Dunnett's multiple comparisons test. *... P < 0.05; **... P < 0.01; ***...P < 0.001, N = 3‐6. (C) Dissipation monitoring revealed an increase in dissipative energy loss upon sample binding to the sensor surface. (D) Frequency shift vs dissipation signals obtained from (a) and (c) through eliminating time. Additionally, similar analysis of DPPC vesicles and two protein samples (CP and IgG) is displayed. (E) Marker specific EVs included in our UC‐MSC EVs as determined via Equations (3), (4) from the slope of frequency shift vs dissipation curves as in (d). Bars represent means ± standard deviation of experimental replicates (N = 3). (F) Response curve of biotin‐anti‐CD73 IgG functionalized sensors at a range of particle concentrations
QCM‐I dissipation monitoring revealed an increase in energy dissipation during sample association caused by the increase of viscoelastic mass bound to the sensor surface (Figure 4C). To better compare the viscoelastic properties of the respective marker‐specific subpopulations, we plotted the corresponding energy dissipation vs. frequency shift (ΔD vs. Δf; (Saftics et al., 2018; Tagaya, 2015)) curves into Figure 4D and compared these curves to curves obtained for pure lipid vesicles (DPPC) and pure protein samples (IgGs, control protein). All curves exhibit a linear dependence of the dissipation on the frequency, whereas the slopes of the marker‐specific subpopulations lie in between the curves for pure vesicles and pure protein samples, suggesting that the marker‐specific material captured on our QCM chip contains a mixture of EVs and proteinaceous NVPs. Theoretically, such variation in slope of the ΔD vs. Δf curves could be caused by differently sized vesicles predominately recruited by a certain marker. This effect has been analysed in detail in literature for pure vesicles of various diameters (Ferhan et al., 2017). It was found that the slope of ΔD vs. Δf curves monotonically increases with vesicle size, implying a higher increase in energy dissipation per adsorbing vesicle caused by the larger amount of solvent coupled to larger vesicles. In this case, the initial rates of vesicle adsorption (Δf vs. time curves) monotonically decrease with increasing vesicle size, reflecting that the rate of vesicle adsorption is limited by their bulk diffusion, and larger vesicles diffuse more slowly. Since this anti‐correlation between slope of the ΔD vs. Δf curves and the corresponding Δf vs. time curves is not observed in our data (cf. Figure 4D and a) we conclude that the observed differences are not a consequence of differently sized EVs binding to our marker‐specific antibodies, but it is indeed caused by a mixture of marker‐positive EVs and proteinaceous NVPs that bind to our sensor surface with comparable kinetics. This is further supported by the narrow size distribution of particles in our samples (Figure 1) and the finding that EVs and NVPs cannot be distinguished by their size (Figure 2), so that not even the extrema of the distributions could lead to the observed differences, when compared to the vesicle size range investigated in the abovementioned study.
We can now estimate the ratio of EVs to NVPs immobilized on our sensor chip by each marker‐specific antibody. For a two component mixture with its pure constituents characterized by their respective ΔD versus Δf slopes k1,2 , we can write down the slope kmix of the resulting mixture as
| (1) |
with Δf1, 2 being the frequency shift caused by the respective components 1 and 2 within the mixture. Introducing the ratio of wet masses (note that Δf measured in QCM is not only dependent on the dry mass of the respective entity, but also includes solvent molecules coupled to it) α = Δf1 /Δf2 we get
| (2) |
which can be solved for α yielding
| (3) |
Assuming that the viscoelastic properties of pure EVs and NVPs can be approximated by the viscoelasticity of pure vesicles (k1 = slope of DPPC vesicles, Figure 4D) and pure proteins (k2 = average slope of CP and IgG, Figure 4D) as determined via following their binding to bare QCM chips, respectively, we can quantify the wet mass ratio α for each marker by determining the slope kmix of each curve (Supplementary Figure S5c). By correcting for the solvent coupled to NVPs (factor of ∼3 for large proteins (Höök, 2002)), assuming equal particle volume for single EVs and NVPs (supported by our AFM measurements, Figure 2), and modelling EVs as solvent filled spheres, we can estimate the amount of marker specific NVPs per EV in our UC‐MSC EVs, n through
| (4) |
with ρEV and ρNVP being the effective density of EVs (approx. by ρwater = 1.0 g cm‐3) and NVPs (approx. by ρprotein = 1.3 g cm‐3), respectively. Thus, the specifically captured particles on the sensor consist of EVs and NVPs, respectively. For our UC‐MSC EVs, the percentage of captured EVs for each marker is displayed in Figure 4E and amounts between ∼73% (CD73 and CD44) and ∼83% (CD49e) which is close to what have been determined in our advanced AFM characterization (Figure 2), albeit by employing a different, unspecific capturing strategy that cannot distinguish between different markers. We further assessed the sensitivity of the platform by applying a range of particle concentrations to an anti‐CD73‐IgG modified QCM chip (Figure 4F). The system displayed an LOD of 3.1 × 108 particles/ml and an LOQ of 8.2 × 108 particles/ml, resulting in a dynamic range of 3.1 × 108 ‐ 5.5 × 109 particles/ml.
4. DISCUSSION
Both EV based research and clinical applications have seen a surge in interest in recent years. Accordingly, experts in the field have established quality standards to ensure safety and efficacy for patients and enable efficient comparison of data between laboratories. When characterizing EV preparations, it is critical to recognize that even with careful enrichment and purification steps a population of non‐vesicular particles may be enriched alongside the EVs. Separating experimental data from EVs and NVPs may hence be an important prerequisite for the robust characterization of EV functionality, making it necessary to expand the current portfolio of standardized techniques. Here we present such an extended approach that provides data on EV quality and quantity using established techniques and makes it possible to discriminate between EVs and NVPs. We first confirmed the presence of both particle types using QI‐mode AFM imaging and then used this complex mixture in QCM‐I to selectively characterize EV surface proteins and the amount of marker‐positive NVPs by carefully designing, monitoring and analysing the sensor properties. The sensitivity of our method (LOD of 3.1 × 108 particles/ml) is comparable to other QCM‐based approaches (2.9 × 108 particles/ml (Suthar et al., 2020)) and multiplex bead‐based flow cytometry assays (LOD of 5 × 106 – 5 × 107 particles depending on the marker (Wiklander et al., 2018) compared to LOD of 1.2 × 107 particles in 40 μl volume in the sample chamber of our QCM‐I), but more sensitive than commercially available immunoassays (LOD of 100 pg/ml ∼ 9 × 108 particles (CD73) /ml, (Zhang et al., 2014)). To ensure reproducibility and quality of EV‐based therapeutics, establishment of a standard and reliable quality control for the contained particle subtypes is important. For instance, MISEV suggests demonstrating the presence of three different kinds of proteins on EVs, including at least one transmembrane or lipid‐bound extracellular protein (CD9, CD63, CD81), which have been originally established as exosome markers. Methods like ELISA, bead‐based flow cytometry, aptamer‐ and carbon nanotube‐based colorimetric assays, and SPR can be used to quantify the amount of one or more specific molecules in the EV preparation (Witwer et al., 2019), as well as the concentration and size of nanoparticles carrying characteristic surface proteins (Rupert et al., 2016). Moreover, advanced methods such as synchronized Rayleigh and Raman scattering can distinguish EVs from different cellular origins based on their molecular composition (Enciso‐Martinez et al., 2020). QCM‐D was proposed as an attractive method for EV surface marker characterization (Suthar et al., 2020), however, despite the high sensitivity of this technique it comes with a major barrier of entry in terms of instrumentation costs. Additionally, sample preparation methods based on self‐assembled monolayers and covalently immobilized IgGs are challenging to establish, time‐consuming, and limit sensor reusability. To address these issues and to make QCM‐based EV characterization accessible to a wider audience, we have established a label‐free immunosensing method using QCM‐I, based on a modular in‐device functionalization and easy in‐situ regeneration of the sensor surface ensuring optimal experimental reproducibility. Our capturing strategy using QCM‐I was established based on the appropriate lipid composition of 65% DOPC, 25% DOPS and 10% DOPE cap biotin, which is easily produced, reproducible, fast to apply (20 min) and does not involve overnight steps during preparation. We further provide a formalism that can be employed to analyse QCM dissipation vs. frequency curves to determine the fractions of EVs and NVPs making up the overall amount of marker‐specific particles included in a certain EV‐preparation, which cannot be obtained by any other technique to date. Our approach is based on the assumption that the (I) viscoelastic properties of NVPs and EVs may be approximated by the properties of large proteins and pure vesicles, respectively and (II) that the volumes of single NVPs and EVs are approximately equal. While these assumptions appear to be largely plausible and will allow the relative comparison of NVP/EV fractions for different markers, future determination of the exact values for the viscoelastic properties and volumes of NVPs and EVs would further improve the absolute values obtained through our formalism.
Our approach thus represents a flexible (lipids are adaptable to the specific application, IgGs can be exchanged easily), reproducible, economical (chips can be reused, sample consumption is minimized, inexpensive device compared to QCM‐D), and comprehensive way to analyse EVs and their surface proteins. Employing this approach, we have demonstrated the presence of all three EV‐specific transmembrane proteins CD9, CD63, and CD81 in our EV preparation, thus corroborating our MACSPlex results. In terms of functionality, UC‐MSC EVs express several adhesion molecules (CD29, CD44, CD49e and CD73), which facilitate their homing to the injured and inflamed tissues (Harrell et al., 2019). We were able to detect these typical MSC (CD44 and CD73) and UC‐MSC (CD49e) markers for immobilizing UC‐MSC EVs as well. This is in line with our standard characterization protocols using the MACSPlex analysis that reveal the presence of these surface markers in UC‐MSC EV preparations. CD73, a GPI‐anchored 5´‐ecto‐nucleotidase that drives adenosine signalling. CD73 positive MSC exosomes cause the phosphorylation of ERK1/2 and AKT kinases and elicited pro‐survival signalling via these two latter enzymes (Lai et al., 2013). The conversion of AMP to adenosine by CD73 also triggers the immunomodulatory environment via T cells (Antonioli et al., 2019). CD73 could thus play a dual role in mediating immunomodulation, survival and cytoprotection in the target tissue. The integrin α5, known as CD49e, was shown to promote osteogenic differentiation potential of UC‐MSC (Zheng et al., 2018). Thus, we suggest that the association of these markers with functional membranous vesicles in addition to determining the MSC origin of EVs represents a valuable quality characteristic.
In conclusion, our combined approach enabled us to detect and characterize a roster of specific surface markers on membranous vesicles and also on NVPs in a complex and clinically relevant EV preparation. QCM‐I proved especially valuable to this end by not only providing information on the EV recruitment efficiency of a panel of EV markers but also by simultaneously recording the viscoelastic properties of the adsorbates. We believe that the demonstrated quantitative determination of marker‐specific EV versus NVP ratios has a high applicability for providing insight into this complex composition and functionality of EV‐based therapeutics. Finally, our modular and reversible in‐device sensor functionalization makes QCM‐I a highly promising and accessible technique for the specific detection of particle subpopulations.
DISCLOSURE OF INTEREST
The author(s) declare no competing interests, either financial or non‐financial, in the work described.
AUTHOR CONTRIBUTIONS
Eleni Priglinger, Juergen Strasser, Johannes Preiner, Jaroslaw Jacak and Mario Gimona made substantial contributions to conception and design, enquired and drafted the manuscript. Eleni Priglinger, Juergen Strasser, Boris Buchroithner, Florian Weber, Claudia Arzt, Daniela Auer, Johannes Preiner, Eva Grasmann and Dmitry Sivun performed the scientific experiments and analysed data. Susanne Wolbank, Johannes Grillari, Johannes Preiner, Jaroslaw Jacak and Mario Gimona have given final approval and revised the manuscript critically. All authors read and approved the final manuscript.
Supporting information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
ACKNOWLEDGEMENTS
We are indebted to Alain Brisson (Univ. Bordeaux, FRA) for providing cryo‐EM images. This work was supported by the Austrian Forschungsförderungsgesellschaft Coin project “BioCETA” (No. 15379797), the European Fund for Regional Development (EFRE, IWB2020), and the Federal State of Upper Austria. We gratefully acknowledge financial support through Projects from Land Salzburg/IWB/EFRE 2014‐2020 P1812596 “EV‐TT” and Land Salzburg/WISS 2025 20102‐F1900731‐KZP “EV‐TT‐Bpro”.
Priglinger, E. , Strasser, J. , Buchroithner, B. , Weber, F. , Wolbank, S. , Auer, D. , Grasmann, E. , Arzt, C. , Sivun, D. , Grillari, J. , Jacak, J. , Preiner, J. , & Gimona, M. (2021). Label‐free characterization of an extracellular vesicle‐based therapeutic. Journal of Extracellular Vesicles, 10, e12156. 10.1002/jev2.12156
Eleni Priglinger, Juergen Strasser, Johannes Preiner and Mario Gimona contributed equally to this study.
REFERENCES
- Antonioli, L. , Fornai, M. , Blandizzi, C. , Pacher, P. , & Hasko, G. (2019). Adenosine signaling and the immune system: When a lot could be too much. Immunology Letters, 205, 9–15. [DOI] [PubMed] [Google Scholar]
- Cheng, C. I. , Chang, Y. P. , & Chu, Y. H. (2012). Biomolecular interactions and tools for their recognition: Focus on the quartz crystal microbalance and its diverse surface chemistries and applications. Chemical Society Reviews, 41, 1947–1971. [DOI] [PubMed] [Google Scholar]
- Desgeorges, A. , Hollerweger, J. , Lassacher, T. , Rohde, E. , Helmbrecht, C. , & Gimona, M. (2020). Differential fluorescence nanoparticle tracking analysis for enumeration of the extracellular vesicle content in mixed particulate solutions. Methods, 177, 67–73. [DOI] [PubMed] [Google Scholar]
- Dominici, M. , Le Blanc, K. , Mueller, I. , Slaper‐Cortenbach, I. , Marini, F. , Krause, D. , Deans, R. , Keating, A. , Prockop, D. , & Horwitz, E. (2006). Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy, 8, 315–317. [DOI] [PubMed] [Google Scholar]
- Emelyanov, A. , Shtam, T. , Kamyshinsky, R. , Garaeva, L. , Verlov, N. , Miliukhina, I. , Kudrevatykh, A. , Gavrilov, G. , Zabrodskaya, Y. , Pchelina, S. , Emelyanov, A. K.A. , Shtam, T. , Kamyshinsky, R. , Garaeva, L. , Verlov, N. , Miliukhina, I. , Kudrevatykh, A. , Gavrilov, G. , Zabrodskaya, Y. , Pchelina, S. , & Konevega, A. (2020). Cryo‐electron microscopy of extracellular vesicles from cerebrospinal fluid. PloS One, 15, e0227949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enciso‐Martinez, A. , Van Der Pol, E. , Hau, C. M. , Nieuwland, R. , Van Leeuwen, T. G. , Terstappen, L. , & Otto, C. (2020). Label‐free identification and chemical characterisation of single extracellular vesicles and lipoproteins by synchronous Rayleigh and Raman scattering. Journal of Extracellular Vesicles, 9, 1730134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escudier, B. , Dorval, T. , Chaput, N. , Andre, F. , Caby, M. P. , Novault, S. , Flament, C. , Leboulaire, C. , Borg, C. , Amigorena, S. , Boccaccio, C. , Bonnerot, C. , Dhellin, O. , Movassagh, M. , Piperno, S. , Robert, C. , Serra, V. , Valente, N. , Le Pecq, J. B. , … Zitvogel, L. (2005). Vaccination of metastatic melanoma patients with autologous dendritic cell (DC) derived‐exosomes: Results of the first phase I clinical trial. Journal of Translational Medicine, 3, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferhan, A. R. , Jackman, J. A. , & Cho, N. J. (2017). Investigating how vesicle size influences vesicle adsorption on titanium oxide: A competition between steric packing and shape deformation. Physical Chemistry Chemical Physics : PCCP, 19, 2131–2139. [DOI] [PubMed] [Google Scholar]
- Giebel, B. , & Helmbrecht, C. (2017). Methods to analyze EVs. Methods in Molecular Biology, 1545, 1–20. [DOI] [PubMed] [Google Scholar]
- Gimona, M. , Pachler, K. , Laner‐Plamberger, S. , Schallmoser, K. , & Rohde, E. (2017). Manufacturing of Human Extracellular Vesicle‐Based Therapeutics for Clinical Use. International Journal of Molecular Sciences 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualerzi, A. , Kooijmans, S. A. A. , Niada, S. , Picciolini, S. , Brini, A. T. , Camussi, G. , & Bedoni, M. (2019). Raman spectroscopy as a quick tool to assess purity of extracellular vesicle preparations and predict their functionality. Journal of Extracellular Vesicles, 8, 1568780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurunathan, S. , Kang, M. H. , Jeyaraj, M. , Qasim, M. , & Kim, J. H. (2019). Review of the isolation, characterization, biological function, and multifarious therapeutic approaches of exosomes. Cells, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrell, C. R. , Jankovic, M. G. , Fellabaum, C. , Volarevic, A. , Djonov, V. , Arsenijevic, A. , & Volarevic, V. (2019). Molecular mechanisms responsible for anti‐inflammatory and immunosuppressive effects of mesenchymal stem cell‐derived factors. Advances in Experimental Medicine and Biology, 1084, 187–206. [DOI] [PubMed] [Google Scholar]
- Hööka, F. , Vörösb, J. , Rodahla, M. , Kurratb, R. , Bönid, P. , Ramsdene, J. J. , Textorb, M. , Spencerb, N. D. , Tengvallc, P. , Golda, J. , & Kasemo, B. (2002). A comparative study of protein adsorption on titanium oxide surfaces using in situ ellipsometry, optical waveguide lightmode spectroscopy, and quartz crystal microbalance/dissipation. Colloids and Surfaces B: Biointerfaces, 24(2), 155–170. [Google Scholar]
- Karner, A. , Nimmervoll, B. , Plochberger, B. , Klotzsch, E. , Horner, A. , Knyazev, D. G. , Kuttner, R. , Winkler, K. , Winter, L. , Siligan, C. , Ollinger, N. , Pohl, P. , & Preiner, J. (2017). Tuning membrane protein mobility by confinement into nanodomains. Nature Nanotechnology, 12, 260–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasper, M. , Traxler, L. , Salopek, J. , Grabmayr, H. , Ebner, A. , & Kienberger, F. (2016). Broadband 120 MHz Impedance Quartz Crystal Microbalance (QCM) with calibrated resistance and quantitative dissipation for biosensing measurements at higher harmonic frequencies. Biosensors, 6, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller, C. A. , & Kasemo, B. (1998). Surface specific kinetics of lipid vesicle adsorption measured with a quartz crystal microbalance. Biophysical Journal, 75, 1397–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konoshenko, M. Y. , Lekchnov, E. A. , Vlassov, A. V. , & Laktionov, P. P. (2018). Isolation of extracellular vesicles: General methodologies and latest trends. BioMed Research International, 2018, 8545347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordelas, L. , Rebmann, V. , Ludwig, A. K. , Radtke, S. , Ruesing, J. , Doeppner, T. R. , Epple, M. , Horn, P. A. , Beelen, D. W. , & Giebel, B. (2014). MSC‐derived exosomes: A novel tool to treat therapy‐refractory graft‐versus‐host disease. Leukemia, 28, 970–973. [DOI] [PubMed] [Google Scholar]
- Kruglik, S. G. , Royo, F. , Guigner, J. M. , Palomo, L. , Seksek, O. , Turpin, P. Y. , Tatischeff, I. , & Falcon‐Perez, J. M. (2019). Raman tweezers microspectroscopy of circa 100 nm extracellular vesicles. Nanoscale, 11, 1661–1679. [DOI] [PubMed] [Google Scholar]
- Lai, R. C. , Yeo, R. W. , Tan, K. H. , & Lim, S. K. (2013). Mesenchymal stem cell exosome ameliorates reperfusion injury through proteomic complementation. Regenerative Medicine, 8, 197–209. [DOI] [PubMed] [Google Scholar]
- Laner‐Plamberger, S. , Lener, T. , Schmid, D. , Streif, D. A. , Salzer, T. , Oller, M. , Hauser‐Kronberger, C. , Fischer, T. , Jacobs, V. R. , Schallmoser, K. , Gimona, M. , & Rohde, E. (2015). Mechanical fibrinogen‐depletion supports heparin‐free mesenchymal stem cell propagation in human platelet lysate. Journal of Translational Medicine, 13, 354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laney, D. E. , Garcia, R. A. , Parsons, S. M. , & Hansma, H. G. (1997). Changes in the elastic properties of cholinergic synaptic vesicles as measured by atomic force microscopy. Biophysical Journal, 72, 806–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeClaire, M.G. J. , Sharma, S. (2020). A review of the biomechanical properties of single extracellular vesicles. NanoSelect, 17. [Google Scholar]
- Lener, T. , Gimona, M. , Aigner, L. , Borger, V. , Buzas, E. , Camussi, G. , Chaput, N. , Chatterjee, D. , Court, F. A. , Del Portillo, H. A. , O'Driscoll, L. , Fais, S. , Falcon‐Perez, J. M. , Felderhoff‐Mueser, U. , Fraile, L. , Gho, Y. S. , Görgens, A. , Gupta, R. C. , Hendrix, A. , … Bernd, Giebel . (2015). Applying extracellular vesicles based therapeutics in clinical trials ‐ an ISEV position paper. Journal of Extracellular Vesicles, 4, 30087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, X. , Mao, G. , & Simon Ng, K. Y. (2004). Probing small unilamellar EggPC vesicles on mica surface by atomic force microscopy. Colloids and Surfaces B, Biointerfaces, 34, 41–51. [DOI] [PubMed] [Google Scholar]
- Lobb, R. J. , Becker, M. , Wen, S. W. , Wong, C. S. , Wiegmans, A. P. , Leimgruber, A. , & Moller, A. (2015). Optimized exosome isolation protocol for cell culture supernatant and human plasma. Journal of Extracellular Vesicles, 4, 27031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig, A. K. , & Giebel, B. (2012). Exosomes: Small vesicles participating in intercellular communication. The International Journal of Biochemistry & Cell Biology, 44, 11‐15. [DOI] [PubMed] [Google Scholar]
- Obeid, S. , Ceroi, A. , Mourey, G. , Saas, P. , Elie‐Caille, C. , & Boireau, W. (2017). Development of a NanoBioAnalytical platform for "on‐chip" qualification and quantification of platelet‐derived microparticles. Biosensors & Bioelectronics, 93, 250–259. [DOI] [PubMed] [Google Scholar]
- Pachler, K. , Ketterl, N. , Desgeorges, A. , Dunai, Z. A. , Laner‐Plamberger, S. , Streif, D. , Strunk, D. , Rohde, E. , & Gimona, M. (2017a). An in vitro potency assay for monitoring the immunomodulatory potential of stromal cell‐derived extracellular vesicles. International Journal of Molecular Sciences, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachler, K. , Lener, T. , Streif, D. , Dunai, Z. A. , Desgeorges, A. , Feichtner, M. , Oller, M. , Schallmoser, K. , Rohde, E. , & Gimona, M. (2017b). A good manufacturing practice‐grade standard protocol for exclusively human mesenchymal stromal cell‐derived extracellular vesicles. Cytotherapy 19, 458–472. [DOI] [PubMed] [Google Scholar]
- Parisse, P. , Rago, I. , Ulloa Severino, L. , Perissinotto, F. , Ambrosetti, E. , Paoletti, P. , Ricci, M. , Beltrami, A. P. , Cesselli, D. , & Casalis, L. (2017). Atomic force microscopy analysis of extracellular vesicles. European Biophysics Journal : EBJ, 46, 813–820. [DOI] [PubMed] [Google Scholar]
- Perissinotto, F.S. B. , Vaccari, L. , Pachetti, M. , D'Amico, F. , Amneitsch, H. , Sartori, B. , Pachler, K. , Mayr, M. , Gimona, M. , Rohde, E. , Caponnetto, F. , Cesselli, D. , Casalis, L. , & Parisse, P. (2020). Multi‐technique analysis of extracellular vesicles: Not only size matters. Advances in Biomembranes and Lipid Self‐Assembly, 32, 21. [Google Scholar]
- Perrino, A. P. , & Garcia, R. (2016). How soft is a single protein? The stress‐strain curve of antibody pentamers with 5 pN and 50 pm resolutions. Nanoscale, 8, 9151–9158. [DOI] [PubMed] [Google Scholar]
- Preiner, J. , Horner, A. , Karner, A. , Ollinger, N. , Siligan, C. , Pohl, P. , & Hinterdorfer, P. (2015). High‐speed AFM images of thermal motion provide stiffness map of interfacial membrane protein moieties. Nano Letters, 15, 759–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiner, J. , Kodera, N. , Tang, J. , Ebner, A. , Brameshuber, M. , Blaas, D. , Gelbmann, N. , Gruber, H. J. , Ando, T. , & Hinterdorfer, P. (2014). IgGs are made for walking on bacterial and viral surfaces. Nature Communications, 5, 4394. [DOI] [PubMed] [Google Scholar]
- Preiner, J. , Tang, J. , Pastushenko, V. , & Hinterdorfer, P. (2007). Higher harmonic atomic force microscopy: Imaging of biological membranes in liquid. Physical Review Letters, 99, 046102. [DOI] [PubMed] [Google Scholar]
- Reviakine, I. , Johannsmann, D. , & Richter, R. P. (2011). Hearing what you cannot see and visualizing what you hear: Interpreting quartz crystal microbalance data from solvated interfaces. Analytical Chemistry, 83, 8838–8848. [DOI] [PubMed] [Google Scholar]
- Rohde, E. , Pachler, K. , & Gimona, M. (2019). Manufacturing and characterization of extracellular vesicles from umbilical cord‐derived mesenchymal stromal cells for clinical testing. Cytotherapy, 21, 581–592. [DOI] [PubMed] [Google Scholar]
- Romanelli, P. , Bieler, L. , Scharler, C. , Pachler, K. , Kreutzer, C. , Zaunmair, P. , Jakubecova, D. , Mrowetz, H. , Benedetti, B. , Rivera, F. J. , Aigner, L. , Rohde, E. , Gimona, M. , Strunk, D. , & Couillard‐Despres, S. (2019). Extracellular Vesicles Can Deliver Anti‐inflammatory and Anti‐scarring Activities of Mesenchymal Stromal Cells After Spinal Cord Injury. Frontiers in Neurology, 10, 1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupert, D. L. M. , Shelke, G. V. , Emilsson, G. , Claudio, V. , Block, S. , Lasser, C. , Dahlin, A. , Lotvall, J. O. , Bally, M. , Zhdanov, V. P. , & Höök, F. (2016). Dual‐wavelength surface plasmon resonance for determining the size and concentration of sub‐populations of extracellular vesicles. Analytical Chemistry. 88, 9980–9988. [DOI] [PubMed] [Google Scholar]
- Saftics, A. , Prosz, G. A. , Turk, B. , Peter, B. , Kurunczi, S. , & Horvath, R. (2018). In situ viscoelastic properties and chain conformations of heavily hydrated carboxymethyl dextran layers: A comparative study using OWLS and QCM‐I chips coated with waveguide material. Science Reports, 8, 11840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabbir, A. , Cox, A. , Rodriguez‐Menocal, L. , Salgado, M. , & Van Badiavas, E. (2015). Mesenchymal stem cell exosomes induce proliferation and migration of normal and chronic wound fibroblasts, and enhance angiogenesis in vitro. Stem Cells and Development, 24, 1635‐1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, S. , Gillespie, B. M. , Palanisamy, V. , & Gimzewski, J. K. (2011). Quantitative nanostructural and single‐molecule force spectroscopy biomolecular analysis of human‐saliva‐derived exosomes. Langmuir : The ACS Journal of Surfaces and Colloids, 27, 14394–14400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastava, A. G.V. (2011). Methods for the determination of limit of detection and limit of quantitation of the analytical methods. Chronicles of Young Scientists, 2, 5. [Google Scholar]
- Strasser, J. , de Jong, R. N. , Beurskens, F. J. , Schuurman, J. , Parren, P. , Hinterdorfer, P. , & Preiner, J. (2020). Weak fragment crystallizable (Fc) domain interactions drive the dynamic assembly of IgG oligomers upon antigen recognition. ACS Nano, 14, 2739–2750. [DOI] [PubMed] [Google Scholar]
- Strasser, J. , de Jong, R. N. , Beurskens, F. J. , Wang, G. , Heck, A. J. R. , Schuurman, J. , Parren, P. , Hinterdorfer, P. , & Preiner, J. (2019). Unraveling the macromolecular pathways of IgG oligomerization and complement activation on antigenic surfaces. Nano Letters, 19, 4787‐4796. [DOI] [PubMed] [Google Scholar]
- Suthar, J. , Parsons, E. S. , Hoogenboom, B. W. , Williams, G. R. , & Guldin, S. (2020). Acoustic immunosensing of exosomes using a quartz crystal microbalance with dissipation monitoring. Analytical Chemistry, 92, 4082–4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagaya, M. (2015). In situ QCM‐D study of nano‐bio interfaces with enhanced biocompatibility. Polymer Journal, 47, 10. [Google Scholar]
- Terlecki‐Zaniewicz, L. , Lammermann, I. , Latreille, J. , Bobbili, M. R. , Pils, V. , Schosserer, M. , Weinmullner, R. , Dellago, H. , Skalicky, S. , Pum, D. , Almaraz, J. C. H. , Scheideler, M. , Morizot, F. , Hackl, M. , Gruber, F. , & Grillari, J. (2018). Small extracellular vesicles and their miRNA cargo are anti‐apoptotic members of the senescence‐associated secretory phenotype. Aging, 10, 1103‐1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thery, C. , Witwer, K. W. , Aikawa, E. , Alcaraz, M. J. , Anderson, J. D. , Andriantsitohaina, R. , Antoniou, A. , Arab, T. , Archer, F. , Atkin‐Smith, G. K. , Ayre, D. C. , Bach, J. ‐. M. , Bachurski, D. , Baharvand, H. , Balaj, L. , Baldacchino, S. , Bauer, N. N. , Baxter, A. A. , Bebawy, M. , & Zuba‐Surma, E. K. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of Extracellular Vesicles, 7, 1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trickett, A. , & Kwan, Y. L. (2003). T cell stimulation and expansion using anti‐CD3/CD28 beads. Journal of Immunological Methods, 275, 251–255. [DOI] [PubMed] [Google Scholar]
- Vorselen, D. , Piontek, M. C. , Roos, W. H. , & Wuite, G. J. L. (2020). Mechanical characterization of liposomes and extracellular vesicles, a protocol. Frontiers In Molecular Biosciences, 7, 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnecke, A. , Harre, J. , Staecker, H. , Prenzler, N. , Strunk, D. , Couillard‐Despres, S. , Romanelli, P. , Hollerweger, J. , Lassacher, T. , Auer, D. , Pachler, K. , Wietzorrek, G. , Köhl, U. , Lenarz, T. , Schallmoser, K. , Laner‐Plamberger, S. , Falk, C S. , Rohde, E. , & Gimona, M. (2020). Extracellular vesicles from human multipotent stromal cells protect against hearing loss after noise trauma in vivo. Clinical and Translational Medicine, 10, e262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiklander, O. P. B. , Bostancioglu, R. B. , Welsh, J. A. , Zickler, A. M. , Murke, F. , Corso, G. , Felldin, U. , Hagey, D. W. , Evertsson, B. , Liang, X. M. , Gustafsson, M. O. , Mohammad, D. K. , Wiek, C. , Hanenberg, H. , Bremer, M. , Gupta, D. , Björnstedt, M. , Giebel, B. , Nordin, J. Z. , … Görgens, A. (2018). Systematic methodological evaluation of a multiplex bead‐based flow cytometry assay for detection of extracellular vesicle surface signatures. Frontiers in Immunology, 9, 1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witwer, K. W. , Van Balkom, B. W. M. , Bruno, S. , Choo, A. , Dominici, M. , Gimona, M. , Hill, A. F. , De Kleijn, D. , Koh, M. , Lai, R. C. , Mitsialis, S. A. , Ortiz, L. A. , Rohde, E. , Asada, T. , Toh, W. S. , Weiss, D. J. , Zheng, L. , Giebel, B. , & Lim, S. K. (2019). Defining mesenchymal stromal cell (MSC)‐derived small extracellular vesicles for therapeutic applications. Journal of Extracellular Vesicles, 8, 1609206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Hao, Z. , Wang, P. , Xia, Y. , Wu, J. , Xia, D. , Fang, S. , & Xu, S. (2019). Exosomes from human umbilical cord mesenchymal stem cells enhance fracture healing through HIF‐1alpha‐mediated promotion of angiogenesis in a rat model of stabilized fracture. Cell Proliferation, 52, e12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, H. , Li, X. , Chen, Y. , Zhou, R. , Zhao, H. , & Qian, C. (2018). Integrin subunits alphaV and beta3 promote the osteogenic differentiation of umbilical cord blood mesenchymal stem cells. International Journal of Clinical and Experimental Pathology, 11, 2008–2016. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information


