Abstract
Ninety-one isolates of nutritionally variant streptococci (NVS) that were previously isolated from the human mouth were regarded as consisting of 7 Streptococcus defectivus isolates, 78 Streptococcus adjacens isolates, and 6 Gemella morbillorum isolates. However, recent references to the taxonomic reclassification of NVS, from S. defectivus to Abiotrophia defectiva and from S. adjacens to Abiotrophia adiacens, and the newly introduced species Abiotrophia elegans as a third Abiotrophia species, emphasize the need for genetic analyses for identification of NVS. When PCR-restriction fragment length polymorphism (RFLP) and phylogenetic distances were examined based on 16S rRNA gene sequences, the results indicated that 7 of the 91 NVS isolates were closely related to A. elegans. These seven isolates consisted of four isolates previously identified as G. morbillorum and three isolates previously identified as S. adjacens. Two isolates previously identified as G. morbillorum were related to A. adiacens. In biochemical tests, A. elegans and the seven isolates related to it possessed arginine dihydrolase (ADH) activity but the other Abiotrophia species did not. As a result, A. elegans strains comprised 8% of the 91 NVS isolates. Our findings suggest that A. elegans, A. adiacens, and A. defectiva exist in the human mouth in proportions of about 1:11:1 and that A. elegans can be genetically distinguished from the other two Abiotrophia species by PCR-RFLP analysis of 16S rRNA gene sequences and can be biochemically distinguished by ADH activity.
Nutritionally variant streptococci (NVS) can be seen as satellite colonies around other microorganisms and require cysteine or vitamin B6 for growth in complex medium (3, 13). Although such streptococci are responsible for a variety of infectious diseases (13), they have been isolated not only from flora associated with disease but also from normal flora in the form of symbiotic streptococci (5). In particular, occurrence of NVS in the human mouth is typical (6, 9, 10).
With respect to taxonomy, in 1989, Bouvet et al. identified Streptococcus defectivus and Streptococcus adjacens as new species of NVS based on their different biochemical characteristics and DNA homology (2). Then, in 1995, Kawamura et al. proposed a new genus, Abiotrophia, based on the phylogenetic distances of 16S rRNA gene sequences and named two species, Abiotrophia defectiva and Abiotrophia adiacens, based on the species names S. defectivus and S. adjacens, respectively (7). In 1998, Roggenkamp et al. specified the 16S rRNA gene sequences, biochemical characteristics, and growth characteristics for a third Abiotrophia species, Abiotrophia elegans (12). They also showed the differentiation among these three species by PCR amplification with various primers which had sequences found in 16S rRNA genes (11). Thus, at this moment, NVS are regarded as comprising three Abiotrophia species: A. defectiva, A. adiacens, and A. elegans.
In 1996, we reported 91 NVS isolates from the human mouth (6). All of them presented bacteriolytic activity and pink chromophores and required additional vitamin B6 in the medium for their growth. The results of identification with a Rapid ID32 STREP kit showed that these 91 NVS isolates consisted of 7 S. defectivus isolates, 78 S. adjacens isolates, including NMP3, S943-2, and S1052-1, and 6 Gemella morbillorum isolates. Despite this identification, the six isolates previously identified as G. morbillorum, i.e., C9-2, HHC5, HKT1-1, S43-1, TKT2, and YTM1, present an as yet unsolved problem, since G. morbillorum is able to grow without additional vitamin B6 and presents neither bacteriolytic activity nor chromophores (3).
For identification of NVS, recent studies on Abiotrophia species have emphasized the need for genetic analyses, particularly for a PCR assay based on 16S rRNA gene sequences. In order to solve the problem presented above, we reexamined the 91 isolates of NVS by such genetic analyses. The observed characteristics and resulting reidentifications are presented here.
MATERIALS AND METHODS
Strains and culture conditions.
The 91 NVS, which included C9-2, HHC5, YTM1, S43-1, NMP3, S943-2, and S1052-1, were previously isolated from a healthy human mouth in our laboratory (6). A. defectiva ATCC 49176T, A. adiacens ATCC 49175T, and G. morbillorum ATCC 27824T were purchased from the American Type Culture Collection (Manassas, Va.). A. elegans DSM 11693T was purchased from Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (Braunschweig, Germany). Bacteria, with the exception of A. elegans DSM 11693T, were grown in Todd-Hewitt broth (THB; BBL Becton Dickinson and Company, Cockeysville, Md.) containing 0.001% pyridoxal hydrochloride. A. elegans DSM 11693T was grown in THB containing 5% horse serum and 0.01% l-cysteine hydrochloride.
DNA extraction for PCR.
Cells in 1 ml of culture were collected, suspended in 0.2 ml of lysis buffer, and boiled for 3 min based on the method of Watanabe and Frommel (16). After centrifugation, DNA-containing supernatant was obtained.
Primers and PCR.
A pair of primers corresponding to Escherichia coli 16S rRNA gene positions 8 to 27 (5′-AGAGTTTGATCATGGCTCAG-3′) (17) and 1405 to 1391 (5′-ACGGGCGGTGTGTAC-3′) (8) was obtained from Amersham Pharmacia Biotech (Tokyo, Japan). A mixture of DNA extract (0.5 μl), 0.2 μM primers, and Taq DNA polymerase (Premix Taq; Takara Shuzo Co., Ltd., Shiga, Japan) in a 50-μl volume was incubated for 30 cycles of 94°C for 1 min and 64°C for 1 min and for an extension at 64°C for 10 min with a Zymoreactor II thermal cycler (Atto Corporation, Tokyo, Japan).
PCR-restriction fragment length polymorphism (RFLP) analysis.
The PCR product (20 μl) was digested first with KpnI in low-salt buffer, then with HindIII in medium-salt buffer, and finally with PstI in high-salt buffer at 37°C, for 1 h for each digestion (final volume, 30 μl). Five units of each restriction enzyme (Nippon Gene, Osaka, Japan) was used. The triple enzyme digest (12 μl) was analyzed in a 2.5% ethidium bromide-stained agarose gel.
16S rRNA gene sequence of HHC5.
The PCR product was cloned into a pCR2.1 vector (TA cloning kit; Invitrogen Corporation, Carlsbad, Calif.) and cut into two EcoRI fragments. Their single-stranded DNAs were obtained by subcloning into M13mp18 and sequenced by using a Thermo Sequenase premixed cycle sequencing kit (Amersham) with an automatic sequencer (model Hitachi Ltd., SQ-5500; Tokyo, Japan). The HHC5 and the other sequences derived from data deposited with the DNA Data Bank of Japan (DDBJ, Mishima, Japan) were analyzed, and the results were used to construct a phylogenetic tree by means of a DDBJ Super Computer (Fujitsu VPP500) and the program Clustalw (supplied by DDBJ).
DNA-DNA hybridization.
Chromosomal DNA was extracted as described previously without treatment with achromopeptidase (15). The DNA (10 μg) was loaded onto a Hybond-N+ membrane (Amersham Pharmacia Biotech) and hybridized with [∂-32P]dCTP (NEN, Boston, Mass.)-labeled DNA in 25 mM phosphate buffer (pH 6.5) containing 30% formamide, 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 5× Denhardt’s solution, and salmon sperm DNA (0.2 mg/ml) at 42°C for 24 h. The membrane was washed with 2× SSC–0.1% sodium dodecyl sulfate (SDS) at 52°C (15 min) and then with 0.2× SSC–0.1% SDS at room temperature (10 min). Radioactivity of the hybridized DNA was quantified with a BAS 1000 bioimaging analyzer (Fuji Photo Film Co., Tokyo, Japan). Triplicate tests were run for each assay, and the data were normalized with the value for the homologous DNA-DNA hybridization taken as 100%.
Biochemical characterization.
Bacteriolytic activity on Micrococcus luteus was tested as described previously (10). To examine an essential growth factor, cells were cultured in brain heart infusion broth (BHIB), BHIB containing 0.001% pyridoxal hydrochloride (vitamin B6), or BHIB containing 0.01% l-cysteine (12). The activities of 32 enzymes were tested by using a Rapid ID32 STREP kit (BioMérieux S.A., Marcy l’Etoile, France).
DNA sequence data and nucleotide sequence accession numbers.
The sequence data for 16S rRNA genes obtained from DDBJ had the following accession numbers.: A. defectiva ATCC 49176T, D50541 (7); A. adiacens ATCC 49175T, D50540 (7); A. elegans DSM 11693T, AF016390 (12); G. morbillorum ATCC 27824T, L14327 (18); and E. coli, A14565. The sequence of strain HHC5 has been deposited in the DDBJ under accession no. AB022026.
RESULTS
Comparison of PCR-RFLP patterns among NVS, Abiotrophia species, and G. morbillorum.
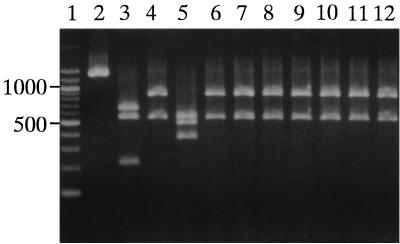
PCR-RFLP analysis of the 16S rRNA gene (Fig. 1) showed that A. defectiva produced one PCR product of 1,400 bp which was not digested by any of the three enzymes. However, A. adiacens and G. morbillorum each produced PCR products which were digested into three fragments of 650, 550, and 210 bp and of 550, 490, and 370 bp, respectively. On the other hand, the new species, A. elegans, produced a PCR product which was digested into two fragments of 860 and 550 bp. Surprisingly, 7 of the 91 clinical isolates produced PCR products which were digested into two fragments of the same sizes as those produced by A. elegans. These results suggested that the seven isolates might be A. elegans.
FIG. 1.
PCR-RFLP analysis of the 16S rRNA genes. Lanes: 1, size marker of 100-bp ladder; 2, A. defectiva ATCC 49176T; 3, A. adiacens ATCC 49175T; 4, A. elegans DSM 11693T; 5, G. morbillorum ATCC 27824T; 6 to 12, NVS isolates C9-2, HHC5, YTM1, S43-1, NMP3, S943-2, and S1052-1, respectively.
This finding was unexpected, since our isolates can grow in the presence of vitamin B6 without l-cysteine in the medium, like A. defectiva and A. adiacens, while the original A. elegans isolate requires specific addition of l-cysteine to the medium for growth (12).
Of the remainder of our isolates, 77 presented three digested fragments similar in length to those produced by A. adiacens and 7 presented nondigested fragments similar to those produced by A. defectiva (data not shown). As a result, the PCR-RFLP analysis of the 16S rRNA genes readily allocated all 91 NVS isolates among the three Abiotrophia species: 7 isolates as A. defectiva, 77 isolates as A. adiacens, and 7 isolates as A. elegans.
The seven isolates of A. defectiva correctly corresponded to the seven isolates previously identified as S. defectivus, while the seven new A. elegans isolates consisted of three previously identified as S. adjacens and four previously identified as G. morbillorum. Thus, the 77 isolates of A. adiacens consisted of 75 previously identified as S. adjacens and 2 previously identified as G. morbillorum.
16S rRNA gene sequence of the isolate HHC5.
The HHC5 sequence, which was typical of the seven isolates described above, was 1,407 bp in length and included the forward and reverse primers in the 5′ and 3′ directions, respectively. The result of a multiple alignment analysis showed that the HHC5 sequence resembled A. elegans more than it did the other two Abiotrophia species and resembled G. morbillorum less than it did the Abiotrophia species. The homologies between HHC5 and each of A. elegans, A. adiacens, A. defectiva, and G. morbillorum were 99, 97, 93, and 86%, respectively.
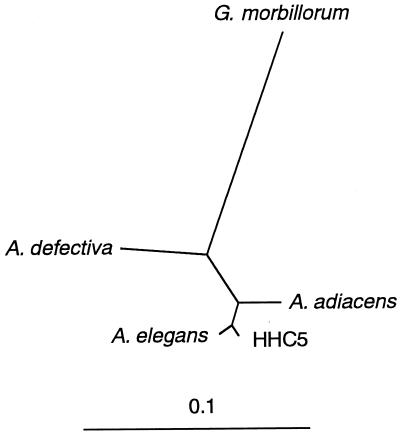
An unrooted phylogenetic tree (Fig. 2) clearly revealed that HHC5 was highly homologous to A. elegans but considerably dissimilar to G. morbillorum, despite the previous identification of HHC5 as G. morbillorum (6).
FIG. 2.
Phylogenetic relationships based on 16S rRNA gene sequences. The line denotes evolutionary distance.
The findings that the phylogenetic distance between A. elegans and HHC5 was the shortest observed and that the homology between the two sequences was significant (99% identity) strongly supported the notion that the seven isolates (Fig. 1, lanes 6 to 12) are A. elegans, despite the different factors required for their growth.
Restriction enzyme sites in 16S rRNA gene.
Agarose gel electrophoretic analyses of the restriction enzyme digests and the data for the DNA sequences revealed that in the 16S rRNA gene A. defectiva did not possess HindIII, PstI, or KpnI sites, A. adiacens possessed HindIII and PstI sites located at positions 214 and 862, respectively, and A. elegans possessed a PstI site located at position 863. Only G. morbillorum possessed a KpnI site at position 492 in addition to a PstI site at position 863.
DNA-DNA hybridization.
As shown in Table 1, chromosomal DNA of HHC5 hybridized with A. elegans DNA at a relatedness of 70%, but it hybridized with A. defectiva, A. adiacens, and G. morbillorum DNAs to a lesser degree.
TABLE 1.
DNA-DNA hybridization
| Species or strain | Relatedness (%)
|
|||
|---|---|---|---|---|
| A. defectiva | A. adiacens | A. elegans | G. morbillorum | |
| A. defectiva | 100 | 6 | 7 | 1 |
| A. adiacens | 6 | 100 | 20 | 3 |
| A. elegans | 6 | 7 | 100 | 1 |
| G. morbillorum | 5 | 4 | 7 | 100 |
| HHC5 | 6 | 14 | 70 | 2 |
Biochemical characteristics.
We found that arginine dihydrolase (ADH) and urease (URE) were critical to distinguishing A. elegans from the other Abiotrophia species. As shown in Table 2, ADH was positive in both A. elegans DSM 11693T and the seven related isolates but was negative in all of the others. This suggested that ADH-positive NVS are A. elegans. On the other hand, URE was negative in A. elegans DSM 11693T and three of the seven related isolates, but a URE-positive isolate of NVS is probably A. elegans, since all of A. defectiva and A. adiacens and their related clinical isolates were URE negative. The other 30 biochemical characteristics tested by the Rapid ID32 STREP kit did not distinguish A. elegans and the seven related isolates from either A. defectiva or A. adiacens.
TABLE 2.
Critical biochemical characteristics specific to A. elegans and seven related isolates
| Bacterial strain or isolate (no. of isolates) | Characteristic (no. of isolates)
|
||||
|---|---|---|---|---|---|
| Lytic enzyme production | Vitamin B6a | l-Cysteineb | Activity
|
||
| ADH | URE | ||||
| A. defectiva ATCC 49176T | + | + | + | − | − |
| Clinical isolates identified as A. defectiva (7) | + (7) | + (7) | + (7) | − (7) | − (7) |
| A. adiacens ATCC 49175T | + | + | + | − | − |
| Clinical isolates identified as A. adiacens (77) | + (77) | + (77) | + (77) | − (77) | − (77) |
| A. elegans DSM 11693T | + | − | + | + | − |
| Clinical isolates identified as A. elegans (7) | + (7) | + (7) | + (7) | + (7) | + (4) |
| G. morbillorum ATCC 27824T | − | −c | −c | − | − |
Ability to grow in vitamin B6-supplemented medium without l-cysteine.
Ability to grow in l-cysteine-supplemented medium without vitamin B6.
G. morbillorum required neither vitamin B6 nor l-cysteine for growth.
Only A. elegans DSM 11693T was unable to grow in the medium supplemented with vitamin B6 in the absence of l-cysteine, but it did grow in the medium supplemented with l-cysteine in the absence of vitamin B6, as previously described by Roggenkamp et al. (12). In contrast, all of the other isolates were able to grow in the medium supplemented with either vitamin B6 or l-cysteine (Table 2). Thus, vitamin B6 and l-cysteine could be substituted for each other as essential growth factors for all NVS tested with the exception of A. elegans DSM 11693T. We were unable to determine if the requirement for l-cysteine as a growth factor was characteristic of only A. elegans DSM 11693T or of A. elegans strains in general.
G. morbillorum did not produce any lytic enzymes and was able to grow in THB containing neither vitamin B6 nor l-cysteine.
DISCUSSION
NVS, initially regarded as consisting of S. adjacens and S. defectivus, are now classified into A. elegans, A. adiacens, and A. defectiva by genetic analyses (7, 9, 11, 12). We reexamined genetically the identification of 91 isolates of NVS. As shown in Fig. 1, the 16S rRNA genes of seven of those isolates presented the same PCR-RFLP pattern as A. elegans DSM 11693T. Sequence analysis of HHC5, the most typical of the seven isolates, also showed that HHC5 was most related to A. elegans (99% identity) and had the shortest phylogenetic distance from it (Fig. 2). These results strongly indicated that the seven isolates were A. elegans. Further results of DNA-DNA hybridization (Table 1) (14) led us to conclude that the seven isolates were A. elegans.
Although HHC5 was previously identified as G. morbillorum (6), the present genetic analyses clearly showed that this isolate was unrelated to G. morbillorum. This finding is reasonable since the fundamental characteristics of HHC5 are different from those of G. morbillorum (Table 2).
The seven isolates genetically related to A. elegans consisted of four isolates previously identified as G. morbillorum and three isolates previously identified as S. adjacens. In addition, two isolates previously identified as G. morbillorum were reidentified as A. adiacens (data not shown), but the seven A. defectiva isolates completely corresponded with the seven isolates previously identified as S. defectivus. These results suggest that the results of genetic analysis and biochemical identification correspond exactly for A. defectiva but not for A. elegans and A. adiacens. They also indicate that A. elegans and A. adiacens isolates are sometimes identified as G. morbillorum isolates based on a biochemical identification system, e.g., the Rapid ID32 STREP kit.
In this experiment, DSM 11693 (12) was used as a type strain of A. elegans; however, only DSM 11693T was unable to grow in the vitamin B6-containing medium without added l-cysteine (Table 2). We are very interested in the requirement for l-cysteine of other A. elegans strains (11).
Roggenkamp et al. demonstrated that the polypeptide profiles differed among A. defectiva, A. adiacens, and A. elegans (12). Further, Collins et al. showed a similarity dendrogram based on whole-cell protein patterns which clearly indicated that A. defectiva, A. adiacens, A. elegans, and G. morbillorum belong to independent clusters (4). The protein profiles resulting from the present SDS-polyacrylamide gel electrophoresis analysis also suggested that HHC5 more closely resembled A. elegans than it did either A. defectiva or A. adiacens (data not shown).
Beighton et al. reported enzymatic activities differentiating S. defectivus from S. adjacens (1), but A. elegans-specific enzyme activities have not yet been described. When we reexamined the biochemical characteristics, ADH activity was demonstrated as the only characteristic specific to A. elegans DSM 11693T and the seven related isolates (Table 2). URE activity was also a critical characteristic in identifying some isolates, as described in the Results section. However, the specificity of ADH and URE activities would have to apply to a large number of A. elegans isolates before we could infer that these characteristics distinguish A. elegans from the other Abiotrophia species.
Our reassignment of seven isolates to the species A. elegans brings the percentage of A. elegans to all isolates of NVS in the human mouth to 7.7%. This proportion is the same as that for A. defectiva. Ohara-Nemoto et al. did not estimate the proportion of A. elegans isolates, but they noted colonization frequencies of 11.8% for A. defectiva and 87.1% for A. adiacens for 92 isolates from normal flora found in the human oral cavity (9). Of our Abiotrophia isolates 84.6% were A. adiacens, which seems to be a reasonable amount. We plan to study the frequencies of A. elegans relative to A. adiacens and A. defectiva in other isolates from normal human flora.
REFERENCES
- 1.Beighton D, Homer K A, Bouvet A, Storey A R. Analysis of enzymatic activities for differentiation of two species of nutritionally variant streptococci, Streptococcus defectivus and Streptococcus adjacens. J Clin Microbiol. 1995;33:1584–1587. doi: 10.1128/jcm.33.6.1584-1587.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bouvet A, Grimont F, Grimont P A D. Streptococcus defectivus sp. nov. and Streptococcus adjacens sp. nov., nutritionally variant streptococci from human clinical specimens. Int J Syst Bacteriol. 1989;39:290–294. doi: 10.1099/00207713-41-4-483. [DOI] [PubMed] [Google Scholar]
- 3.Bouvet A, Villeroy F, Cheng F, Lamesch C, Williamson R, Gutmann L. Characterization of nutritionally variant streptococci by biochemical tests and penicillin-binding proteins. J Clin Microbiol. 1985;22:1030–1034. doi: 10.1128/jcm.22.6.1030-1034.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins M D, Hutson R A, Falsen E, Sjöden B, Facklam R R. Description of Gemella sanguinis sp. nov., isolated from human clinical specimens. J Clin Microbiol. 1998;36:3090–3093. doi: 10.1128/jcm.36.10.3090-3093.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.George R H. The isolation of symbiotic streptococci. J Med Microbiol. 1974;7:77–83. doi: 10.1099/00222615-7-1-77. [DOI] [PubMed] [Google Scholar]
- 6.Kanamoto T, Eifuku-Koreeda H, Inoue M. Isolation and properties of bacteriolytic enzyme-producing cocci from the human mouth. FEMS Microbiol Lett. 1996;144:135–140. doi: 10.1111/j.1574-6968.1996.tb08519.x. [DOI] [PubMed] [Google Scholar]
- 7.Kawamura Y, Hou X-G, Sultana F, Liu S, Yamamoto H, Ezaki T. Transfer of Streptococcus adjacens and Streptococcus defectivus to Abiotrophia gen. nov. as Abiotrophia adiacens comb. nov. and Abiotrophia defectiva comb. nov., respectively. Int J Syst Bacteriol. 1995;45:798–803. doi: 10.1099/00207713-45-4-798. [DOI] [PubMed] [Google Scholar]
- 8.Lane D J, Pace B, Olsen G J, Stahl D A, Sogin M L, Pace N R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci USA. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohara-Nemoto Y, Tajika S, Sasaki M, Kaneko M. Identification of Abiotrophia adiacens and Abiotrophia defectiva by 16S rRNA gene PCR and restriction fragment length polymorphism analysis. J Clin Microbiol. 1997;35:2458–2463. doi: 10.1128/jcm.35.10.2458-2463.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pompei R, Caredda E, Piras V, Serra C, Pintus L. Production of bacteriolytic activity in the oral cavity by nutritionally variant streptococci. J Clin Microbiol. 1990;28:1623–1627. doi: 10.1128/jcm.28.7.1623-1627.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roggenkamp A, Leitritz L, Baus K, Falsen E, Heesemann J. PCR for detection and identification of Abiotrophia spp. J Clin Microbiol. 1998;36:2844–2846. doi: 10.1128/jcm.36.10.2844-2846.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roggenkamp A, Abele-Horn M, Trebesius K H, Tretter U, Autenrieth I B, Heesemann J. Abiotrophia elegans sp. nov., a possible pathogen in patients with culture-negative endocarditis. J Clin Microbiol. 1998;36:100–104. doi: 10.1128/jcm.36.1.100-104.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruoff K L. Nutritionally variant streptococci. Clin Microbiol Rev. 1991;4:184–190. doi: 10.1128/cmr.4.2.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stackebrandt E, Goebel B M. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 15.Taketoshi M, Yakushiji T, Inoue M. Deoxyribonucleic acid relatedness and phenotypic characteristics of oral Streptococcus milleri strains. Microbios. 1993;73:269–280. [PubMed] [Google Scholar]
- 16.Watanabe K, Frommel T O. Detection of Porphyromonas gingivalis in oral plaque samples by use of the polymerase chain reaction. J Dent Res. 1993;72:1040–1044. doi: 10.1177/00220345930720060801. [DOI] [PubMed] [Google Scholar]
- 17.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whitney A M, O’Connor S P. Phylogenetic relationship of Gemella morbillorum to Gemella haemolysans. Int J Syst Bacteriol. 1993;43:832–838. doi: 10.1099/00207713-43-4-832. [DOI] [PubMed] [Google Scholar]