ABSTRACT
Insecticide resistance among mosquito species is now a pervasive phenomenon that threatens to jeopardize global malaria vector control efforts. Evidence of links between the mosquito microbiota and insecticide resistance is emerging, with significant enrichment of insecticide degrading bacteria and enzymes in resistant populations. Using 16S rRNA amplicon sequencing, we characterized and compared the microbiota of Anopheles coluzzii in relation to their deltamethrin resistance and exposure profiles. Comparisons between 2- and 3-day-old deltamethrin-resistant and -susceptible mosquitoes demonstrated significant differences in microbiota diversity. Ochrobactrum, Lysinibacillus, and Stenotrophomonas genera, each of which comprised insecticide-degrading species, were significantly enriched in resistant mosquitoes. Susceptible mosquitoes had a significant reduction in alpha diversity compared to resistant individuals, with Asaia and Serratia dominating microbial profiles. There was no significant difference in deltamethrin-exposed and -unexposed 5- to 6-day-old individuals, suggesting that insecticide exposure had minimal impact on microbial composition. Serratia and Asaia were also dominant in 5- to 6-day-old mosquitoes, which had reduced microbial diversity compared to 2- to 3-day-old mosquitoes. Our findings revealed significant alterations of Anopheles coluzzii microbiota associated with deltamethrin resistance, highlighting the potential for identification of novel microbial markers for insecticide resistance surveillance. qPCR detection of Serratia and Asaia was consistent with 16S rRNA sequencing, suggesting that population-level field screening of bacterial microbiota may be feasibly integrated into wider resistance monitoring, if reliable and reproducible markers associated with phenotype can be identified.
IMPORTANCE Control of insecticide-resistant vector populations remains a significant challenge to global malaria control and while substantial progress has been made elucidating key target site mutations, overexpressed detoxification enzymes and alternate gene families, the contribution of the mosquito microbiota to phenotypic insecticide resistance has been largely overlooked. We focused on determining the effects of deltamethrin resistance intensity on Anopheles coluzzii microbiota and identifying any microbial taxa associated with phenotype. We demonstrated a significant reduction in microbial diversity between deltamethrin-resistant and -susceptible mosquitoes. Insecticide degrading bacterial species belonging to Ochrobactrum, Lysinibacillus, and Stenotrophomonas genera were significantly enriched in resistant mosquitoes, while Asaia and Serratia dominated microbial profiles of susceptible individuals. Our results revealed significant alterations of Anopheles coluzzii microbiota associated with deltamethrin resistance, highlighting the potential for identification of novel microbial markers for surveillance and opportunities for designing innovative control techniques to prevent the further evolution and spread of insecticide resistance.
KEYWORDS: Anopheles coluzzii, insecticide resistance, microbiota, deltamethrin, malaria, Côte d’Ivoire, Asaia, Serratia
INTRODUCTION
Malaria remains a considerable public health problem with an estimated 229 million cases worldwide, including 409,000 deaths in 2019 alone (1). Malaria mortality has fallen since 2010, largely due to the scale-up of treatment, diagnostics, and insecticide-based vector control interventions, principally long-lasting insecticidal nets (LLINs) and indoor residual spraying. However, global gains in malaria control have begun to stall (2). Insecticide resistance among major malaria vector species is now a pervasive phenomenon, affecting more than 90% of countries with ongoing transmission (2). Of particular concern is the continued spread of resistance to pyrethroids, which were until recently, the only class of insecticide recommended for use in LLINs. Pyrethroids are still a crucial component of next-generation LLINs (3), and resistance may severely threaten the long-term effectiveness of contemporary vector control programs.
Control of insecticide-resistant vector populations is predicated on a clear understanding of the complex interplay between molecular mechanisms and fitness costs which contribute to mosquito behavior, phenotype and vectorial capacity, the genetic and local environmental factors driving ongoing resistance selection, and the implications of resistance for intervention operational efficacy. Substantial progress has been made elucidating key target site mutations (4–7), overexpression of detoxification enzymes (8–12) and alternate gene families and pathways (13–17), all of which play important roles in resistance modulation. Furthermore, the recent publication of genome data for more than 1,000 Anopheles (An.) gambiae sensu lato (s.l.) has illustrated the considerable genetic diversity among natural vector populations, raising concerns for the rapid evolution and spread of novel resistance mechanisms (18, 19).
In addition to host-mediated resistance mechanisms, evidence is emerging that changes in mosquito microbiota may confer resistance to certain insecticides. The mosquito microbiota is a heterogenous and variable network of microorganisms, comprising the bacterial, archaeal, viral, fungal, and other eukaryotic microbial communities which inhabit the mosquito cuticle and internal structures such as the midgut, salivary glands, and ovaries. Constituents of the microbiota can be either inherited from mother to offspring (20) or acquired from the environment, predominantly the larval habitat (21). Characterization of the microbiota in mosquitoes has shown varied phenotypic impacts on the host species, including on fitness (22), blood feeding (23), fecundity (24), immunity (25, 26), pathogen infection (27–33), and transmission (34). There is increasing interest in investigating symbionts of mosquito vectors because they may offer unique transmission-blocking opportunities. Similarly, studies on the role played by mosquito symbionts in insecticide resistance may offer a better understanding of the underlying mechanisms and the potential for designing innovative control techniques (35) and developing new insecticide resistance monitoring tools.
The interaction between insecticide resistance and arthropod microbiota has been examined principally in agricultural pest species. Chlorpyrifos (organophosphate)- and fipronil (phenylpyrazole)-resistant strains of the Diamondback moth, Plutella xylostella were shown to have a higher proportion of Lactobacillales, Pseudomonadales, and Xanthomonadales bacteria (36). Furthermore, the bean bug Riptortus pedestris and allied stinkbug species harbor symbiotic Burkholderia bacteria which degrade fenitrothion (organophosphate) and are present in greater abundance when this insecticide is applied to their habitat (37). As advanced molecular technologies become increasingly accessible, research in this area is expanding to disease vectors, with recent studies on several mosquito species. Whole-metagenome sequencing of microbiota from wild-caught fenitrothion-resistant and -susceptible An. albimanus mosquitoes showed distinct differences between these two groups (38). Fenitrothion-resistant mosquitoes had significant enrichment of organophosphate degrading bacteria and enzymes such as hydrolases, carboxylesterases, and phosphomonoesterases. Resistant mosquitoes also had lower bacterial diversity, with an overabundance of Klebsiella spp. and a reduction in the relative abundance of Enterobacter spp. It was suggested that selection for organophosphate-degrading bacteria may have developed alongside resistance, potentially in response to prior insecticide exposure (38). F1 progeny of field-caught An. albimanus exposed to the pyrethroids alpha-cypermethrin and permethrin had significantly greater abundance of bacteria from the genus Pseudomonas, of which several strains have been shown to metabolize pyrethroids, and from the genus Pantoea (39), which had previously been identified in insecticide-resistant mosquitoes (38). Pseudomonas, alongside Clostridium and Rhizobium species, were also implicated in lambda-cyhalothrin (pyrethroid) resistance in wild populations of Aedes aegypti (Ae. aegypti) from Colombia (40). Addition of tetracycline to temephos-resistant (organophosphate) strains of An. stephensi destroyed the bacterial component of the microbiota and significantly reduced the activity of three main resistance enzymes: α esterase, glutathione S-transferase, and acetylcholinesterase, restoring mosquito susceptibility (41). Similarly, sterilization of An. arabiensis gut microbiota by antibiotics resulted in a decreased tolerance to deltamethrin (pyrethroid) and malathion (organophosphate) (42).
To date, information on field populations of the An. gambiae complex, the main malaria vectors in sub-Saharan Africa, is limited to recent reports of significant enrichment of known pyrethroid degrading taxa (Sphingobacterium, Lysinibacillus, and Streptococcus) in permethrin-resistant An. gambiae sensu stricto (s.s.) from Kenya (43). To address this deficit, we comparatively characterized the bacterial microbiota of An. coluzzii, collected from an area of high pyrethroid resistance in Côte d’Ivoire. We specifically focused on determining the effects of deltamethrin resistance intensity on host microbiota and identifying any microbial taxa associated with resistance phenotypes.
RESULTS
Species identification and deltamethrin resistance profiles.
In total, 580 blood-fed An. gambiae s.l. were collected from Agboville using human landing catches (HLCs) during the rainy season in July 2019. Of these, 245 (42%) laid eggs via forced oviposition. Following larval development, 1,015 F1 An. gambiae s.l. pupae were identified as female and tested in deltamethrin resistance intensity assays as 2- to 3-day-old adults. Individuals were classified as susceptible if they were knocked down after exposure to 1× deltamethrin, resistant if they survived 60 min (2 to 3 days old) or 72 h (5 to 6 days old) postexposure to 1×, 5×, or 10× deltamethrin, or controls if they were unexposed to insecticide (comprising a mix of age-matched individuals of unknown phenotype). A total of 380 mosquitoes were randomly selected for DNA extraction, across all exposure and time groups, with 338 individuals identified as An. coluzzii (78.3%). From the remaining individuals, 31 were An. gambiae s.s. (8.1%), 10 failed to amplify (2.6%), and one individual was an An. gambiae s.s.–An. coluzzii hybrid (0.26%). Table S1 summarizes the number of mosquitoes selected for DNA extraction, pooling, and sequencing.
Sequencing metrics.
A total of 1,156,076 reverse reads were obtained from sequencing. Quality control and denoising resulted in 2,999 unique amplicon sequence variants (ASVs), 878,155 in total. Filtering of ASVs associated with water and ethanol blanks, low-frequency ASVs, and ASVs not classified to the phylum level resulted in 210 unique ASVs, totaling 556,254 across 94 pools of mosquitoes. Table S3 summarizes the number of sequences processed per sample and the number of reads remaining after denoising and filtering.
Susceptible An. coluzzii had microbiota which were significantly different to, and less diverse than, resistant mosquitoes.
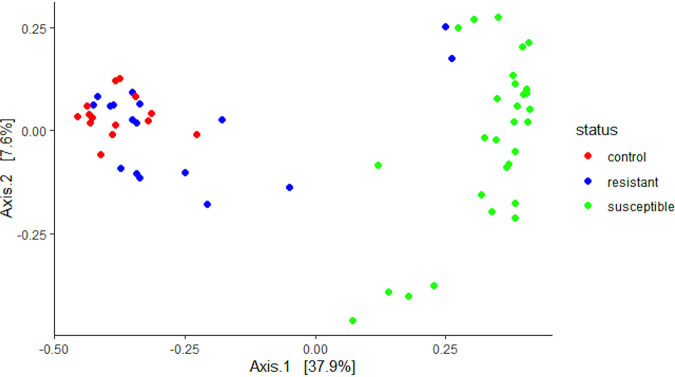
Comparison of the Bray-Curtis dissimilarity index using pairwise PERMANOVA with 999 permutations showed significant differences in bacterial composition between microbiota of 2- to 3-day-old deltamethrin-resistant and -susceptible An. coluzzii (pseudo-F = 19.44, P = 0.0015). Principal coordinate analysis (PCoA) visualizations showed the microbiota of susceptible mosquitoes clustered away from resistant and control mosquitoes (Fig. 1), indicating that the microbiota of susceptible mosquitoes were more similar to each other than to resistant and control mosquitoes.
FIG 1.
PCoA plot showing Bray-Curtis distance of microbiota between resistant, susceptible, and control F1 2- to 3-day-old An. coluzzii adults. Each point represents the bacterial composition of a pool of three mosquitoes of the same resistance phenotype. There was a distinct separation between resistant/control and susceptible mosquitoes, which was shown to be a significant difference using a pairwise PERMANOVA (999 permutations) (pseudo-F = 19.44, P = 0.0015).
Susceptible mosquitoes had significantly lower Shannon and Faith phylogenetic diversity indices (see Fig. S2) than resistant (Shannon: H = 13.91, q = 0.0003, Faith: H = 6.68, q = 0.01) and control (Shannon: H = 22.6 q = 0.000006, Faith: H = 16.6, q = 0.0001) mosquitoes of the same age, indicating that the susceptible group had reduced microbial diversity. There was no significant difference in alpha or beta diversity in deltamethrin-exposed and unexposed 5- to 6-day-old mosquitoes (Shannon: H = 5.12, q = 0.02, Faith: H = 0.27, q = 0.6, Bray-Curtis: pseudo-F = 1.61, q = 0.17), suggesting that insecticide exposure during the CDC bottle bioassays had minimal impact on microbial composition. There was no significant difference in alpha or beta diversity between mosquitoes exposed to 5 or 10 times the diagnostic dose of deltamethrin (Shannon: H = 0.05, q = 0.81, Faith: H = 0.68, q = 0.41, Bray-Curtis: pseudo-F = 0.96, q = 0.503).
Serratia and Asaia dominated in older and younger susceptible An. coluzzii.
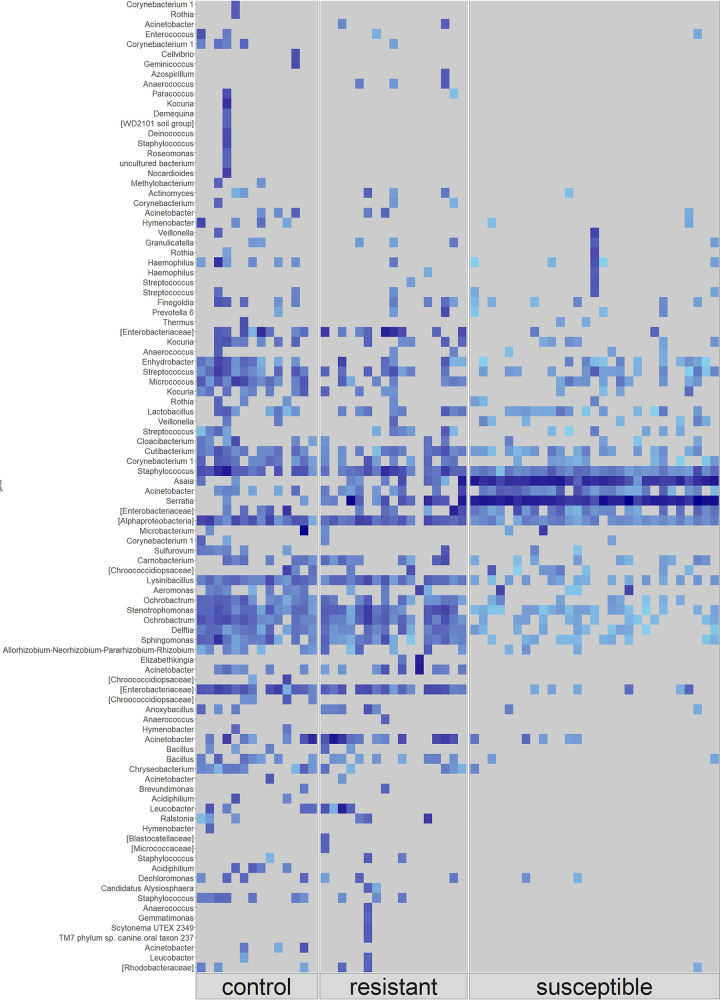
There were significant differences in the microbiota of 2- to 3- and 5- to 6-day old mosquitoes (pseudo-F = 11.34, q = 0.001). Following taxonomic annotation of ASVs to the genus or lowest possible taxonomic level, 114 and 57 bacterial taxa were detected in 2- to 3-day-old and 5- to 6-day-old mosquitoes, respectively. The less diverse 5- to 6-day-old microbiota was predominantly comprised of ASVs assigned to the genera Serratia (75.5%) and Asaia (13.6%) (see Fig. S3). In 2- to 3-day-old mosquitoes, microbial composition varied by resistance phenotype. Control mosquitoes had the highest number of taxa present (n = 97), followed by resistant (n = 90) and susceptible (n = 66). A total of 20 taxa were unique to control mosquitoes, and 15 were unique to resistant mosquitoes. No taxa were unique to the susceptible group of mosquitoes, and 60 taxa were common to all groups (Fig. 2).
FIG 2.
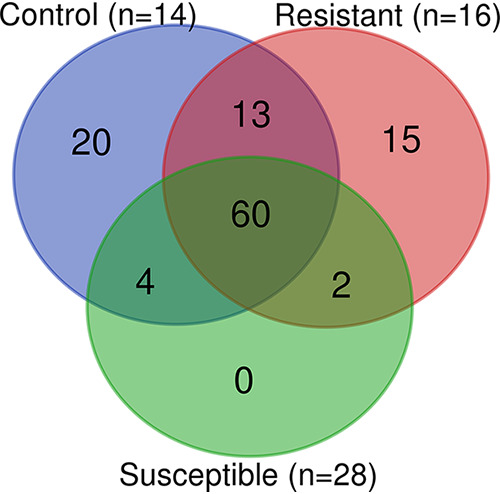
Venn diagram showing number of bacterial taxa unique to or shared between pools of 2- to 3-day-old resistant, susceptible, or control mosquitoes. Taxa were identified to genus level or lowest possible taxonomic rank. n = number of pools (each pool consists of three mosquitoes of the same age and phenotype).
In control mosquitoes, an unclassified species within the Enterobacteriaceae family (15.24%), Acinetobacter (8.83%) and Staphylococcus (8.29%) were most abundant, while Enterobacteriaceae (15.12%), Acinetobacter (14.26%), and Serratia (11.8%) were the most abundant in resistant mosquitoes. In susceptible mosquitoes, Serratia (56.4%) and Asaia (30.92%) were the dominant genera, with Acinetobacter (1.96%), Enterobacteriaceae (1.57%), and Staphylococcus (1.4%) present at low abundance (Fig. 3). The remaining 61 taxa were present at an abundance of <1% of total ASVs present (see Fig. S3 and Table S4 in the supplemental material). Differential rankings confirmed that Asaia and Serratia were significantly associated with susceptibility and that Stenotrophomonas, Ochrobactrum, Lysinibacillus, and Alphaproteobacteria were significantly associated with phenotypic resistance.
FIG 3.
Raw frequency of ASVs from the microbiota of control (n = 14), resistant (n = 16), and susceptible (n = 28) F1 2- to 3-day-old An. coluzzii adults. Each column represents a pool of three mosquitoes of the same phenotype. ASVs were annotated to genus level or lowest possible taxonomic level (in square brackets). Only taxonomically annotated ASVs with a frequency of >150 are shown. Light blue indicates a low frequency of ASVs present, while darker blue indicates a higher frequency. Gray indicates ASV was not present in that pool.
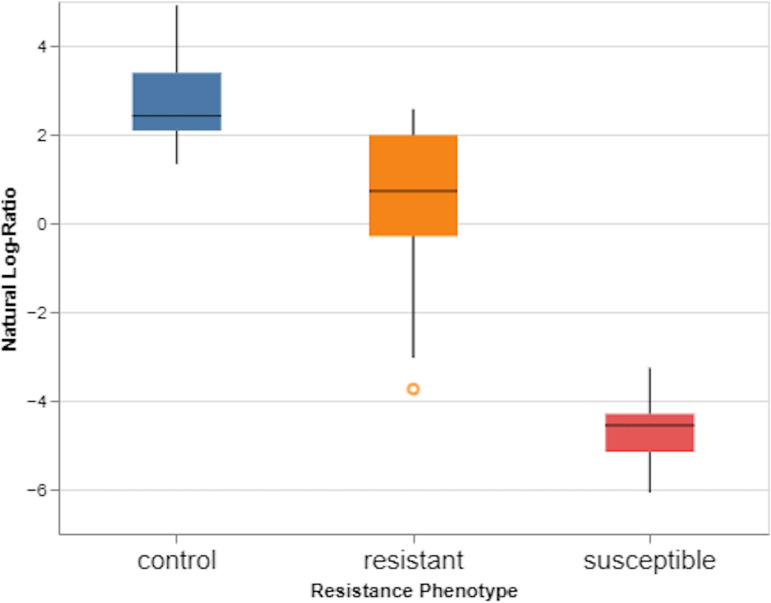
Songbird was used to identify taxa which were differentially abundant in 2- to 3-day-old resistant, susceptible, or control mosquitoes. Evaluation of our Songbird model with resistance phenotype as the variable, against a baseline model with no variable resulted in a pseudo Q-squared value of 0.42, indicating that the model had not been overfit and that roughly 42% of variation in the model was predicted by resistance phenotype (see Fig. S4). There were significant differences in the log ratios of highest to lowest ranked taxa between resistant and susceptible microbiota (Fig. 4; see also Table S5), suggesting that the highest ranked taxa were significantly overabundant in resistant microbiota and that the lowest ranked taxa were significantly overabundant in susceptible microbiota.
FIG 4.
Log ratios of 10% highest ranked features to 10% lowest ranked features in control, resistant, and susceptible 2- to 3-day-old F1 An. coluzzii. Susceptible mosquitoes had a significantly lower ratio than control or resistant mosquitoes indicating that the lowest ranked features were overabundant in the susceptible group, while the highest ranked features were overabundant in either resistant or control mosquitoes.
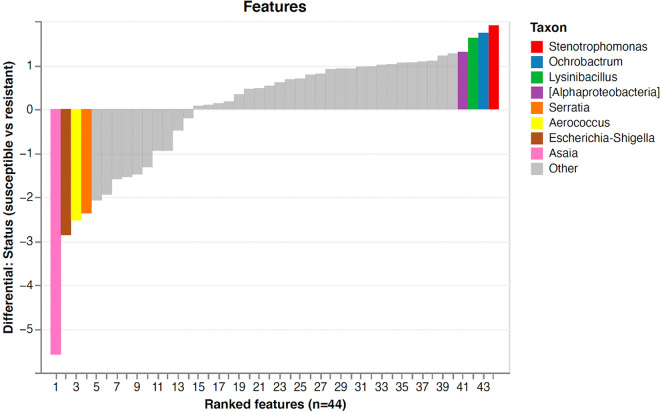
Stenotrophomonas, Ochrobactrum, Lysinibacillus, and Alphaproteobacteria (highest ranked) were most strongly associated with insecticide resistance, while Serratia, Aerococcus, E. shigella, and Asaia (lowest ranked) were most strongly associated with insecticide susceptibility (Fig. 5). Comparing log ratios of control and susceptible pools indicated that Rhodococcus, Sphingomonas, Haemophilus, and E. shigella were most strongly associated with controls, while an uncultured Chroocooccidiopsaceae, Serratia, an unclassified member of Enterobacteriaceae, and Asaia were most strongly associated with susceptible mosquitoes (see Table S5).
FIG 5.
Sorted differential ranks of features associated with a resistant or a susceptible phenotype in 2- to 3-day-old An. coluzzii. The highest 10% and lowest 10% of ranked features are shown, colored by their corresponding assigned taxon. Taxa are shown to genus or to the lowest possible taxonomic level (square brackets).
These results were confirmed by the ANCOM method. Serratia (W = 208) and Asaia (W = 208) were significantly overabundant in susceptible mosquitoes relative to resistant and controls, while Ochrobactrum (W = 199), Lysinibacillus (W = 188), and Enterobacteriaceae (W = 201) were overabundant in resistant and control mosquitoes (see Fig. S5).
Increased abundance of Serratia and Asaia species in susceptible individuals confirmed by qPCR.
Quantitative PCR (qPCR) assays confirmed that Serratia was significantly overabundant in 2- to 3-day-old susceptible mosquitoes compared to deltamethrin-resistant (5× P = 0.028; 10× P = 0.002) and control (P = 0.02) (average ΔCT values for susceptible, −8.4 [95% CI = −9.0 to –7.74]; 5×, –7.43 [–7.9 to –6.9]; 10×, –6.3 [–7.2 to –5.4]; and control, –7.2 [–7.9 to –6.6]) mosquitoes. Asaia was also significantly overabundant in 2- to 3-day-old susceptible mosquitoes compared to resistant (5×, P = <0.001; 10×, P < 0.001) and control (P < 0.001) (average ΔCT values for susceptible, –7.6 [95% CI = –8.5 to –6.8]; 5×, 0.8 [–1.6 to 3.2]; 10×, 5.1 [3.0 to 7.1]; and control, 4.5 [3.3 to 5.7]). Five- to six-day-old mosquitoes also had increased abundance of both bacterial species (average ΔCT value for 5- to 6-day-old 5×, Serratia, –8.2 [95% CI = –8.9 to –7.4] and Asaia, –1.3 [–5.6 to 2.9]; 5- to 6-day-old 10×, Serratia, –7.5 [–7.9 to –7.1] and Asaia, 5.4 [4.6 to 6.3]; 5- to 6-day-old control Serratia, –8.7 [–9.6 to –7.8] and Asaia, –0.68 [–3.0 to 1.7]).
DISCUSSION
There is increasing evidence for an association between insecticide resistance phenotype (38, 41, 43) and Anopheles spp. microbiota. This study revealed distinct differences between the microbiota of deltamethrin-resistant and -susceptible An. coluzzii, with significant enrichment of insecticide degrading taxa in resistant individuals and an overabundance of Serratia and Asaia taxa in susceptible individuals. This population of field-caught An. gambiae s.l. from Agboville, Southeast Côte d’Ivoire, has previously been characterized as intensely resistant to pyrethroids, with average vector mortalities of 14.56% (95% CI = 8.92 to 22.88%), 61.62% (95% CI = 51.58 to 70.75%), and 73.79% (95% CI = 64.35 to 81.45%) to 1, 5, and 10 times the diagnostic doses of deltamethrin, respectively, and pyrethroid resistance associated with overexpression of CYP450 enzymes (CYP6P4, CYP6Z1, and CYP6P3) (44).
Our study demonstrated significant differences in alpha and beta diversity between deltamethrin-resistant and -susceptible An. coluzzii. Resistant mosquitoes harbored a wider variety of microbial taxa and had more microbial diversity both within and between themselves. Susceptible mosquitoes had fewer bacterial taxa and were far more homogenous, with Serratia and Asaia dominating in all samples. Previous studies have demonstrated differences between the microbiota of insecticide-resistant and -susceptible An. stephensi (38), An. arabiensis (42), and An. gambiae s.s. (43). This study is the first to detect an increase in alpha diversity in resistant mosquitoes, with other studies reporting no difference (39, 43) or a decrease (45). There are multiple potential reasons for the decreased microbial diversity previously identified, including processing of individuals rather than pooled mosquitoes, host species-specific differences, and variability among geographical collection sites (45). Furthermore, field-caught mosquitoes may be of unknown age and physiological status; prior environmental insecticide exposure might also be responsible for reducing overall diversity, as bacteria with the ability to metabolize insecticides have greater access to compounds for growth and reproduction and can outcompete other species (38). In our study, larvae were raised in distilled water, adults were age standardized, unable to mate, or blood-feed, and had no insecticide exposure prior to resistance assays. We therefore consider this inherited microbiota to be linked to the resistance status of the host, with mosquitoes characterized by a wider range of bacteria having an increased chance of harboring an insecticide degrading strain. Raising in sterile cups under identical rearing conditions reduces the environmental impact on the microbiome (45). In other organisms, particularly coprophagic animals such as rodents, individuals residing in the same cage have increasingly homogenous fecal microbiomes over time (46). It is plausible that the cage effect may have influenced the mosquito microbiota in this study, particularly during larval development where bacteria may be transferred between larvae via breeding water (21). The cage effect has not been widely studied in mosquitoes; however, future research should control for this in downstream analysis to minimize any environmental confounding.
Our study identified significant enrichment of several insecticide-degrading taxa in resistant compared to susceptible mosquitoes: Ochrobactrum, Lysinibacillus, and Stenotrophomonas. Ochrobactrum spp. have been isolated from contaminated soil and shown to degrade a variety of insecticides, including pyrethroids (47, 48) and organophosphates (49, 50). Similarly, Lysinibacillus spp. derived from soil and sewage can metabolize deltamethrin (51) and cyfluthrin (52), while Stenotrophomonas in the microflora of cockroaches living in pesticide-treated environments can degrade endosulfan in vitro (53). Elevated expression of xenobiotic degrading genes (51) and enzymes (38) may be contributing to the insecticide degrading properties of these bacteria, as well as the direct degradation of pesticides. Future work could attempt to isolate these bacteria from the mosquito microbiota and perform in vitro insecticide degradation assays, which would provide more evidence for bacterial degradation of pesticides, thus establishing a causal relationship between the microbiota and resistance phenotype.
While certain species of bacteria can confer insecticide resistance to the host, others may influence susceptibility. Indigenous gut bacteria have been implicated in the susceptibility of the gypsy moth, L. dispar, to the insecticidal toxin Bacillus thuringiensis. Treatment of larvae with antibiotics eliminated gut microbiota and subsequently reduced mortality to B. thuringiensis; susceptibility was restored upon oral administration of Enterobacter sp., a Gram-negative bacterium widely present in the L. dispar gut (54). In our study, Serratia and Asaia were found to be significantly overabundant in susceptible mosquitoes. While there are no prior reports of an association of these species with mosquito insecticide resistance phenotype, when the relative abundance of Serratia sp. in the gut of the diamondback moth was increased, susceptibility to chlorpyrifos significantly increased (55). Serratia marcescens plays a role in the susceptibility of field-caught Aedes to dengue virus infection by secreting SmEnhancin, an enzyme which digests gut epithelia mucins, enabling the virus to penetrate the gut (35). The Bel protein, similar to SmEnhancin and produced by the bacteria B. thuringiensis, has been shown to significantly increase the toxicity of Cry1Ac toxin in the cotton bollworm larvae, Helicoverpa armigera, by perforating the midgut peritrophic matrix and degrading the insect intestinal mucin, enabling the toxin to reach the target epithelial membrane (56). A similar mechanism may occur in these mosquitoes, whereby proteins produced by Serratia spp. increase the permeability of the internal organs to deltamethrin, enabling it to reach its target in the mosquito nervous system.
Insecticide resistance status has also been shown to influence P. falciparum development, with pyrethroid exposure adversely affecting L1014S-kdr-resistant An. gambiae s.s. in Uganda (57). Other studies have reported potentiation of An. gambiae vector competence in individuals carrying the kdr mutation (58). Elucidating the impact of mosquito host microbiota composition and molecular and metabolic resistance mechanisms on parasite infection dynamics is crucial for the design of novel transmission-blocking strategies. As well as being associated with resistance, Asaia and Serratia sp. have also both previously been implicated in the modulation of Anopheles vector competence. Asaia spp. have been shown to activate antimicrobial peptide expression in An. stephensi (59), while some strains of S. marcescens isolated from An. sinensis can inhibit Plasmodium development by altering the immunity-related effector genes TEP1 and FBN9 (27). Serratia spp. may also directly inhibit malaria parasite development by secretion of serralysin proteins and prodigiosin, which can have a pathogen-killing effect in vitro (60); the latter can also act as a larvicidal agent against Ae. aegypti and An. stephensi (61). By comparison, the presence of a dominant commensal Enterobacteriaceae has been positively correlated with Plasmodium infection (33).
No significant difference in the microbiota of deltamethrin-exposed and -unexposed mosquitoes was observed at 60 min or 72 h postexposure. One hour is likely insufficient time for the microbial composition to significantly shift in response to insecticide, given the relatively slow rate of bacterial growth, and the fact that any bacteria killed by insecticide exposure would still have been present during microbiota extraction. At 72 h, differences in the microbiota of exposed and unexposed mosquitoes, if present, should have been apparent. The lack of difference observed may in part be due to the low sample size of the 5-day-old 10×-resistant group and may also reflect the short insecticide contact time. Bioassays are a single exposure at a lethal dose in a sterile environment used to determine resistance phenotype (62). In the wild, multiple insecticide exposures are likely to happen at sublethal doses at both the larval stage, as habitats are contaminated with agricultural pesticides, and the adult stage, as there is frequent interaction with treated surfaces or materials indoors (63). Bioassays may therefore not induce the same shifts in microbiota as insecticide exposure in the wild. Furthermore, as mosquitoes acquire most of their microbiota from the aquatic environment at the larval stage (21) and may obtain insecticide metabolizing bacteria at this stage, studying the effects of deltamethrin exposure on larvae, or adults which were exposed as larvae, may be more informative.
Our results demonstrated a significant relative reduction in alpha diversity in resistant and control 5- to 6-day-old mosquitoes compared to the 2- to 3-day-old group, as expected based on prior reports that microbial diversity declines with age (64). Older mosquitoes also had lower relative abundances of Ochrobactrum, Lysinibacillus, and Stenotrophomonas, the insecticide-degrading species shown to be significantly enriched in resistant 2- to 3-day-old individuals. Serratia and Asaia, the species associated with susceptibility, were present in increased abundance in the older age group. It has been widely reported that insecticide resistance declines with age (65–70). The shift in microbiota may also be a contributing factor, and further research is warranted to determine the association between the microbiota, resistance phenotype, and age.
In our study, qPCR detection of Serratia and Asaia was consistent with the 16S rRNA sequencing data, with susceptible mosquitoes having significantly lower ΔCT values than resistant or control individuals. Population-level field screening using qPCR as a less-expensive, faster, and more-feasible option than amplicon sequencing should be considered for integration in wider insecticide resistance monitoring, if reliable and reproducible bacterial markers associated with phenotype can be identified.
Conclusions.
Insecticide susceptibility is influenced by a range of diverse factors, including host genetics, detoxification systems, and behavior, as well as the mosquito microbiota. We report significant differences in the microbiota of deltamethrin-resistant An. coluzzii and have identified several bacterial species that were associated with either resistance or susceptibility to the host and therefore may represent important markers of resistance phenotype. The role of bacteria in determining resistance phenotype is highly complex and specific to the host and bacterial species and insecticide and likely involves multiple, parallel mechanisms, including direct degradation of insecticide, an altered host immune system, and changes to the midgut. In addition, these interactions may have important implications for host species fitness, vector competence, and pathogen development and transmission. Further investigation into the mechanisms of microbiota-mediated susceptibility is necessary since this may provide opportunities for preventing or reducing insecticide resistance, which is crucial to maintain gains in malaria vector control.
MATERIALS AND METHODS
Mosquito collections and mass rearing.
This study was conducted in Agboville (GPS: 5°55′21″N 4°13′13″W), Agnéby-Tiassa region, southeast Côte d’Ivoire. The location was chosen because of its high mosquito densities, malaria prevalence (26% in children <5 years in 2016 [71]), and intense deltamethrin resistance (44). The main industry is agriculture, with livestock such as cows, goats, and chickens living close to households and cultivation of crops, including bananas, cocoa, and rice (72).
Sampling was conducted between 5 July and 26 July 2019, coinciding with the long rainy season (May-November) and peak malaria transmission. Adult mosquitoes were collected using HLCs, inside and outside households from 18:00 h to 06:00 h. Fieldworkers used individual hemolysis tubes to collect host-seeking mosquitoes, which were transported each morning to the Centre Suisse de Recherche Scientifique en Côte d’Ivoire (CSRS) in Abidjan. Blood-fed mosquitoes, morphologically identified as female An. gambiae s.l. (73), were transferred to cages with 10% sugar solution and left for 2 to 3 days to become fully gravid.
A total of 580 fully gravid females were used for forced oviposition. Oviposition was achieved by placing a single gravid mosquito into a 1.5-ml Eppendorf tube, half filled with damp cotton wool, with small holes in the tube cap for ventilation (74). Mosquitoes were held under standard insectary conditions (25°C, 70% humidity, and a 12-h light-dark cycle) until eggs were laid or adult death. Eggs were removed daily and placed into sterile paper cups containing distilled water and NISHIKOI (Nishikoi, United Kingdom) (75) staple fish food pellets. Emergent larvae were reared in 50-cm washing-up bowls in distilled water under the same insectary conditions. Pupae were removed daily and separated by sex with the aid of a stereomicroscope. Female pupae were put in a clean plastic cup with distilled water and placed in a cage for eclosion, while male pupae were discarded. Adults were housed in cages in an incubator (26.6°C, 70% humidity) with a 12-h light-dark cycle and given unlimited access to 10% glucose solution. The cages were checked to ensure that only virgin females were used in bioassays, since mating can potentially introduce changes to the microbiome (20). Care was also taken to ensure that no mosquito obtained a blood meal during handling, since this can significantly decrease bacterial diversity in the gut (64).
Determining deltamethrin resistance status of adult F1 progeny of field-caught An. gambiae s.l.
Deltamethrin resistance was characterized using Centers for Disease Control (CDC) bottle bioassays (62), with some modifications. Two- to three-day-old virgin F1 females were exposed to 1, 5, or 10 times the diagnostic dose of deltamethrin (12.5 μg/bottle) for 30 min. Stock solutions of deltamethrin were prepared using 100% ethanol as the solvent. Per bioassay, multiple 250-ml Wheaton bottles and their lids were coated with 1 ml of stock solution and left to dry in a dark storage area to avoid exposure to UV light. A control bottle, treated with 1 ml of ethanol, was assayed in parallel. Prior to bioassay testing, approximately 20 to 25 mosquitoes were aspirated into holding cups. After 1 to 2 h of acclimatization, they were introduced into each test or control bottle.
Knockdown was scored at 0, 15, and 30 min. A subset of mosquitoes that were alive at 60 min was held for 72 h, with mortality recorded every 24 h. These mosquitoes were housed in paper cups in the insectary, with unlimited access to sterile 10% glucose made with distilled water. Mosquitoes were counted as dead if they were unable to stand as per WHO criteria (62).
At the end of the bioassay and subsequent holding time, mosquitoes were classified as susceptible if they were knocked down following exposure to 1× deltamethrin; resistant if they survived 60 min or 72 h postexposure to 1×, 5×, or 10× deltamethrin; or controls if they were in the ethanol-coated bottle. Specimens were separated into their respective phenotype and concentration/time group and stored at −70°C.
DNA extraction.
DNA was extracted from 380 mosquitoes that had been categorized as resistant, susceptible, or unexposed to deltamethrin. Individuals were homogenized in a Qiagen TissueLyser II with sterilized 5-mm stainless steel beads for 5 min at 30 Hz and incubated overnight at 56°C. DNA was extracted using a Qiagen DNeasy 96 blood and tissue kit (Qiagen, UK) according to the manufacturer’s protocol (76) with DNA eluted in 45 μl of buffer AE. Extracted DNA was stored at −70°C.
Four blank extraction controls were processed alongside mosquitoes: three blanks containing RNase-free water as the extraction template and one blank containing the 70% ethanol used for reagent dilution and sterilization of instruments. All steps were performed under sterile conditions, with tweezers and other instruments being rinsed with 70% ethanol in between handling each mosquito, to avoid microbial or DNA contamination.
PCR for mosquito species identification.
Individual mosquitoes were identified to species level according to Santolamazza et al. (77). PCRs contained 2 μl of 10 μM forward primer (5′-TCGCCTTAGACCTTGCGTTA-3′), 2 μl of 10 μM reverse primer (5′-CGCTTCAAGAATTCGAGATAC-3′), 1 μl of extracted DNA, and 10 μl of Hot Start Taq 2X Master Mix (New England Biolabs, UK) for a final reaction volume of 20 μl. Prepared reactions were run on a Bio-Rad T100 thermal cycler with the following conditions: 10 min denaturation time at 94°C, followed by 35 amplification cycles of 94°C for 30 s, 54°C for 30 s, and 72°C for 60 s, followed by a final extension at 72°C for 10 min. PCR products were visualized on 2% E-gel agarose gels in an Invitrogen E-gel iBase real-time transilluminator. A Quick-Load 100-bp DNA ladder (New England Biolabs) was used to determine band size. Amplified PCR products of 479 or 249 bp were indicative of An. coluzzii or An. gambiae s.s., respectively. As the dominant species, only An. coluzzii individuals of the same age and resistance phenotype were selected and pooled for 16S rRNA sequencing.
16S rRNA gene amplicon sequencing.
DNA concentration from each mosquito was measured using an Invitrogen Qubit 4 fluorometer (Thermo Fisher Scientific, USA). Pools were prepared by combining equal concentrations of DNA from three mosquitoes of the same phenotype/deltamethrin concentration/time group to give 100 ng in a final volume of 20 μl (see Table S1 in the supplemental material). Two negative controls, one comprised of a pool of the three RNase-free water blanks mentioned above and the other a 70% ethanol blank, were processed in parallel.
The microbial composition of the microbiome was determined by amplification of the V3-V4 region of the 16S rRNA gene, using the primers 5′-CCTACGGGNGGCWGCAG-3′ and 5′-GGACTACHVGGGTATCTAATCC-3′. PCRs were prepared in a 25 μl reaction volume, comprising 12.5 μl of KAPA HiFi Hot Start ReadyMix PCR kit (78) (Roche, Switzerland), 0.5 μl of forward and reverse primers (10 μM), and 12.5 ng of DNA. The following PCR cycling was used: 95°C for 3 min and 35 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s, followed by a final extension at 72°C for 5 min. The resulting amplified PCR products were purified with AMPure XP beads (Beckman Coulter, UK) at 1× sample volume.
Next, an index PCR was performed using 5 μl of purified PCR products, 5 μl of Nextera XT Index 1 Primers (N7XX), 5 μl of Nextera XT Index 2 Primers (S5XX) (both from the Nextera XT Index kit; Illumina, USA), 10 μl PCR grade water and 25 μl of KAPA HiFi Hot Start Ready Mix (Roche, Switzerland). The following PCR cycling was used: 95°C for 3 min and 12 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s, followed by a final extension at 72°C for 5 min. The final library was purified with AMPure XP beads, at a 1.12× sample volume, before quantification.
Sequencing was performed on an Illumina MiSeq platform. Libraries were sequenced as 250-bp paired-end reads. Sequences were demultiplexed and filtered for read quality using Bcl2Fastq conversion software (Illumina, Inc.). In total, 1,156,076 sequences were generated in the FASTQPhred33 format44.
Data cleaning and filtering.
Sequencing data were imported into the “Quantitative Insights Into Microbial Ecology” pipeline, version 2020.8 (Qiime2) (79), and primary analysis was performed on the reverse reads, as the quality of the forward reads was not sufficient for merging (see Fig. S1). Sequencing primers and adapters were removed using the “cutadapt” plugin (80) with an error rate of 10%. The divisive amplicon denoising algorithm (DADA2) plugin (81) was used to “denoise” sequencing reads, removing phiX reads and chimeric sequences, to produce high-resolution ASVs (82). DADA2 was run using the denoise-single command, with samples truncated at 206 nucleotides (trunc-len 206), to remove bases with a low-quality score. All other parameters were set to the default. The resulting feature table (83) and sequences were filtered to remove ASVs present in the two blank samples and those with a frequency of below 100 to reduce biases in comparison of diversity indices across groups and especially in differential abundance tests.
Taxonomic annotation.
Taxonomic annotation of ASVs was performed using the -feature-classifier plugin (84), with a Naive-Bayes classifier (85) pretrained on the 16S SILVA reference (99% identity) database version 132. The -extract-reads command was used to trim the reference sequences to span the V3-V4 region (425 bp) of the 16S rRNA gene. Any features not classified to phylum level were also removed, these included hosts’ mitochondrial 16S rRNA genes. The resulting ASV table was exported into R (version 3.6.3) for analysis with the phyloseq package.
Bacterial diversity analysis.
A rooted and unrooted phylogenetic tree was generated using the qiime phylogeny plugin (86–88) and were used to compute alpha and beta diversity metrics using the qiime2-diversity (89) plugin. For alpha diversity metrics, samples were rarefied (90) at a depth of 2,359, where alpha rarefaction curves plateaued, indicating that there was adequate sampling of the microbiota during sequencing. Beta diversity metrics were computed for both rarefied and nonrarefied data, with no significant differences between methods (see Table S2); nonrarefied data are presented here. Mosquitoes 2 to 3 days old and 5 to 6 days old were analyzed separately, since age was shown to significantly impact the bacterial composition of the microbiota. Mosquitoes resistant to 5 and 10 times the diagnostic dose of deltamethrin were analyzed together since the insecticide concentration was not shown to significantly impact the bacterial composition of resistant mosquitoes.
Two methods of alpha diversity were selected: the Shannon diversity Index, which considers the abundance and evenness of ASVs present, and the Faith phylogenetic diversity, a measure of community richness which incorporates phylogenetic relationships between species. Pairwise Kruskal-Wallis comparisons of these alpha diversity indices between groups of insecticide resistance phenotypes were performed, with a Benjamini-Hochberg false discovery rate (FDR) correction for multiple comparisons (91). Significance was set to the FDR adjusted P value, i.e., q < 0.05.
The Bray-Curtis dissimilarity index (92, 93), which measures differences in relative species composition between samples, was chosen as the beta diversity metric. Comparisons of this index between insecticide resistance phenotype groups were conducted using pairwise PERMANOVA tests with 999 permutations (94). Results were visualized using PCoA generated using the phyloseq (95) package. Significance was set to a P value of <0.05.
Determination of association between microbiota composition and insecticide resistance phenotype, and identification of differentially abundant microbial taxa.
Comparison of alpha and beta diversity indices indicated that both insecticide resistance phenotype and mosquito age affected the bacterial composition of An. coluzzii in this study. Following taxonomic annotation of ASVs, multinomial regression and differential abundance analysis was performed using Songbird (96) to determine the microbial taxa that were associated with and differentially abundant across insecticide resistance phenotype for mosquitoes separated by age group. Songbird is a compositionally aware differential abundance method which ranks features based on their log fold change with respect to covariates of interest (96) The following Songbird parameters were used: epochs, 10000; number of random test examples, 15; and differential prior, 0.5. The fit of the model was tested against the null hypothesis (-p-formula “1”). Differential log ratios of features were computed in Qurro (97). We present the highest and lowest 10% ranked features associated with resistance phenotype. The analysis of composition of microbiome method (ANCOM) was used to complement Songbird analysis, and this was computed using the composition plugin (98) with all parameters set to the default. Significance was determined using the automatic cut off for the test statistic, W (98).
Quantitative PCR validation of sequencing data.
The abundance of Serratia spp. and Asaia spp. was assessed using qPCR, relative to the nuclear single-copy An. gambiae s.l. ribosomal protein S7 housekeeping gene (RPS7). Serratia reactions contained 1 μl of 10 μM forward primer (5′-CCGCGAAGGCAAAGTGCACGAACA-3′), 1 μl of 10 μM reverse primer (5′-CTTGGCCAGAAGCGCACCATAG-3′) (99), 2 μl of pooled DNA, and 5 μl LightCycler 480 SYBR green Master Mix (Roche, UK) for a final reaction volume of 10 μl. Prepared reactions were run on an Agilent Technologies Stratagene Mx3005P qPCR system which performed 40 cycles of 95°C for 15 s and 60°C for 1 min, followed by a dissociation curve. Asaia reactions contained 1 μl of 10 μM forward primer (5′-GCGCGTAGGCGGTTTACAC-3′), 1 μl of 10 μM reverse primer (5′-AGCGTCAGTAATGAGCCAGGTT-3′) (100), 2 μl of pooled DNA, and 5 μl of LightCycler 480 SYBR green Master Mix (Roche, UK) for a final reaction volume of 10 μl. Prepared reactions were run on an Agilent Technologies Stratagene Mx3005P qPCR system with the following conditions: 95°C for 15 min and 40 cycles of 95°C for 10 s, 60°C for 10 s, and 72°C for 10 s, followed by a dissociation curve. RSP7 reactions contained 1 μl of 10 μM forward primer (5′-TCCTGGAGCTGGAGATGAAC-3′), 1 μl of 10 μM reverse primer (5′-GACGGGTCTGTACCTTCTGG-3′) (101), 2 μl of pooled DNA, and 5 μl of LightCycler 480 SYBR green Master Mix (Roche, UK) for a final reaction volume of 10 μl. Prepared reactions were run on an Agilent Technologies Stratagene Mx3005P qPCR system with the following conditions: 40 cycles of 95°C for 10 s, 65°C for 60 s, and 97°C for 1 s, followed by a dissociation curve. All samples were run in technical triplicate. Relative bacterial abundance was normalized relative to the endogenous control gene (RPS7). qPCR results were analyzed using the MxPro software (Agilent Technologies).
Ethics approval and consent to participate.
The study protocol was reviewed and approved by the Comite National d’Ethique des Sciences de la Vie et de la Sante (catalog no. 069-19/MSHP/CNESVS-kp) and the institutional review board (IRB) of the London School of Hygiene and Tropical Medicine (no. 16860); all study procedures were performed in accordance with relevant guidelines and regulations. Prior to study initiation, community consent was sought from village leaders and written, informed consent was obtained from the heads of all households selected for participation and from all fieldworkers who performed HLCs. Fieldworkers participating in HLCs were provided with doxycycline malaria prophylaxis.
Data availability.
Sequence data generated by this study is available at Sequence Read Archive (SRA) BioProject PRJNA702915 (accession numbers: SRR13743435 to SRR13743530). All other relevant data are available from the corresponding author upon reasonable request. Codes can be accessed at the public repository Zenodo (http://zenodo.org) under https://doi.org/10.5281/zenodo.5102954.
ACKNOWLEDGMENTS
We thank Fidele Assamoa, Claver Adjobi, and Laurent Didier Dobri, laboratory technicians at Centre Suisse de Recherches Scientifiques en Côte d’Ivoire (CSRS), for their support in mosquito collection and rearing, as well as the chief and population of the village of Aboudé (Agboville) and the entomology fieldworkers of CSRS.
We declare no conflicts of interest.
This study was supported by the Sir Halley Stewart Trust (LAM) and the Wellcome Trust/Royal Society (101285/Z/13/Z to T.W.). C.E. was also supported by the Wellcome Trust (110430/Z/15/Z). B.P. was supported by the Nagasaki University “Doctoral Program for World-leading Innovative and Smart Education” for Global Health (Ministry of Education, Culture, Sports, Science, and Technology of Japan).
N.D., L.A.M., B.P., and T.W. designed the study. B.P., E.C., A.M., and C.E. conducted fieldwork, and B.P., M.K., and L.A.M. undertook mosquito rearing, phenotyping, and preparation of samples for sequencing. Laboratory supervision was provided by L.A.M., C.L.J., and T.W., B.P., N.D., and L.A.M. performed the data analysis, and B.P. and L.A.M. drafted the manuscript. All authors read and approved the final manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Louisa A. Messenger, Email: louisa.messenger@lshtm.ac.uk.
Neha Garg, Georgia Institute of Technology.
REFERENCES
- 1.World Health Organization. 2020. World Malaria Report 2020: 20 years of global progress and challenges. World Health Organization, Geneva, Switzerland. https://www.who.int/teams/global-malaria-programme/reports/world-malaria-report. Accessed 15 December 2020. [Google Scholar]
- 2.World Health Organization. 2019. World malaria report 2019. World Health Organization, Geneva, Switzerland. https://www.who.int/publications/i/item/world-malaria-report-2019. Accessed 26 October 2020. [Google Scholar]
- 3.World Health Organization. 2020. Prequalified vector control products. World Health Organization, Geneva, Switzerland. https://www.who.int/pq-vector-control/prequalified-lists/VCP_PQ-List_26August2020.pdf?ua=1. Accessed 26 October 2020. [Google Scholar]
- 4.Clarkson C, Miles A, Harding N, Weetman D, Kwiatkowski D, Donnelly M. 2018. The genetic architecture of target-site resistance to pyrethroid insecticides in the African malaria vectors Anopheles gambiae and Anopheles coluzzii. bioRxiv 323980. [DOI] [PMC free article] [PubMed]
- 5.Mugenzi LMJ, et al. 2019. cis-Regulatory CYP6P9b P450 variants associated with loss of insecticide-treated bed net efficacy against Anopheles funestus. Nat Commun 10:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Assogba BS, et al. 2015. An ace-1 gene duplication resorbs the fitness cost associated with resistance in Anopheles gambiae, the main malaria mosquito. Sci Rep 5:14529. doi: 10.1038/srep14529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grau-Bové X, et al. 2020. Resistance to pirimiphos-methyl in West African Anopheles is spreading via duplication and introgression of the Ace1 locus. bioRxiv 102343. [DOI] [PMC free article] [PubMed]
- 8.Müller P, Warr E, Stevenson BJ, Pignatelli PM, Morgan JC, Steven A, Yawson AE, Mitchell SN, Ranson H, Hemingway J, Paine MJI, Donnelly MJ. 2008. Field-caught permethrin-resistant Anopheles gambiae overexpress CYP6P3, a P450 that metabolizes pyrethroids. PLoS Genet 4:e1000286. doi: 10.1371/journal.pgen.1000286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stevenson BJ, Bibby J, Pignatelli P, Muangnoicharoen S, O’Neill PM, Lian L-Y, Müller P, Nikou D, Steven A, Hemingway J, Sutcliffe MJ, Paine MJI. 2011. Cytochrome P450 6M2 from the malaria vector Anopheles gambiae metabolizes pyrethroids: sequential metabolism of deltamethrin revealed. Insect Biochem Mol Biol 41:492–502. doi: 10.1016/j.ibmb.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 10.Chiu TL, Wen Z, Rupasinghe SG, Schuler MA. 2008. Comparative molecular modeling of Anopheles gambiae CYP6Z1, a mosquito P450 capable of metabolizing DDT. Proc Natl Acad Sci USA 105:8855–8860. doi: 10.1073/pnas.0709249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ibrahim SS, Riveron JM, Stott R, Irving H, Wondji CS. 2016. The cytochrome P450 CYP6P4 is responsible for the high pyrethroid resistance in knockdown resistance-free Anopheles arabiensis. Insect Biochem Mol Biol 68:23–32. doi: 10.1016/j.ibmb.2015.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riveron JM, Yunta C, Ibrahim SS, Djouaka R, Irving H, Menze BD, Ismail HM, Hemingway J, Ranson H, Albert A, Wondji CS. 2014. A single mutation in the GSTe2 gene allows tracking of metabolically based insecticide resistance in a major malaria vector. Genome Biol 15:R27. doi: 10.1186/gb-2014-15-2-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ingham VA, Pignatelli P, Moore JD, Wagstaff S, Ranson H. 2017. The transcription factor Maf-S regulates metabolic resistance to insecticides in the malaria vector Anopheles gambiae. BMC Genomics 18. doi: 10.1186/s12864-017-4086-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ingham VA, Wagstaff S, Ranson H. 2018. Transcriptomic meta-signatures identified in Anopheles gambiae populations reveal previously undetected insecticide resistance mechanisms. Nat Commun 9:5282. doi: 10.1038/s41467-018-07615-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ingham VA, Anthousi A, Douris V, Harding NJ, Lycett G, Morris M, Vontas J, Ranson H. 2020. A sensory appendage protein protects malaria vectors from pyrethroids. Nature 577:376–380. doi: 10.1038/s41586-019-1864-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Isaacs AT, Mawejje HD, Tomlinson S, Rigden DJ, Donnelly MJ. 2018. Genome-wide transcriptional analyses in Anopheles mosquitoes reveal an unexpected association between salivary gland gene expression and insecticide resistance. BMC Genomics 19:225. doi: 10.1186/s12864-018-4605-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balabanidou V, et al. 2019. Mosquitoes cloak their legs to resist insecticides. Proc Biol Sci 286:2019091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clarkson CS, et al. 2020. Genome variation and population structure among 1142 mosquitoes of the African malaria vector species Anopheles gambiae and Anopheles coluzzii. Genome Res 30:1533–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miles A, et al. 2017. Genetic diversity of the African malaria vector Anopheles gambiae. Nature 552:96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Favia G, Ricci I, Damiani C, Raddadi N, Crotti E, Marzorati M, Rizzi A, Urso R, Brusetti L, Borin S, Mora D, Scuppa P, Pasqualini L, Clementi E, Genchi M, Corona S, Negri I, Grandi G, Alma A, Kramer L, Esposito F, Bandi C, Sacchi L, Daffonchio D. 2007. Bacteria of the genus Asaia stably associate with Anopheles stephensi, an Asian malarial mosquito vector. Proc Natl Acad Sci USA 104:9047–9051. doi: 10.1073/pnas.0610451104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gimonneau G, Tchioffo MT, Abate L, Boissière A, Awono-Ambéné PH, Nsango SE, Christen R, Morlais I. 2014. Composition of Anopheles coluzzii and Anopheles gambiae microbiota from larval to adult stages. Infect Genet Evol 28:715–724. doi: 10.1016/j.meegid.2014.09.029. [DOI] [PubMed] [Google Scholar]
- 22.Ezemuoka LC, Akorli EA, Aboagye-Antwi F, Akorli J. 2020. Mosquito midgut Enterobacter cloacae and Serratia marcescens affect the fitness of adult female Anopheles gambiae s.l. PLoS One 15:e0238931. doi: 10.1371/journal.pone.0238931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kozlova EV, et al. 2020. Microbial interactions in the mosquito gut determine Serratia colonization and blood-feeding propensity. ISME J 15:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaio ADO, Gusmão DS, Santos AV, Berbert-Molina MA, Pimenta PFP, Lemos FJA. 2011. Contribution of midgut bacteria to blood digestion and egg production in Aedes aegypti (diptera: Culicidae) (L). Parasites Vectors 4:105. doi: 10.1186/1756-3305-4-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cappelli A, Damiani C, Mancini MV, Valzano M, Rossi P, Serrao A, Ricci I, Favia G. 2019. Asaia activates immune genes in mosquito eliciting an anti-plasmodium response: implications in malaria control. Front Genet 10:836. doi: 10.3389/fgene.2019.00836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muhammad A, Habineza P, Ji T, Hou Y, Shi Z. 2019. Intestinal microbiota confer protection by priming the immune system of red palm weevil Rhynchophorus ferrugineus Olivier (Coleoptera: Dryophthoridae). Front Physiol 10:1303. doi: 10.3389/fphys.2019.01303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bai L, Wang L, Vega-Rodríguez J, Wang G, Wang S. 2019. A gut symbiotic bacterium Serratia marcescens renders mosquito resistance to Plasmodium infection through activation of mosquito immune responses. Front Microbiol 10:1580. doi: 10.3389/fmicb.2019.01580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jupatanakul N, Sim S, Dimopoulos G. 2014. The insect microbiome modulates vector competence for arboviruses. Viruses 6:4294–4313. doi: 10.3390/v6114294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cirimotich CM, Dong Y, Garver LS, Sim S, Dimopoulos G. 2010. Mosquito immune defenses against Plasmodium infection. Dev Comp Immunol 34–395. doi: 10.1016/j.dci.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dennison NJ, Saraiva RG, Cirimotich CM, Mlambo G, Mongodin EF, Dimopoulos G. 2016. Functional genomic analyses of Enterobacter, Anopheles, and Plasmodium reciprocal interactions that impact vector competence. Malar J 15:425. doi: 10.1186/s12936-016-1468-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gonzalez-Ceron L, Santillan F, Rodriguez MH, Mendez D, Hernandez-Avila JE. 2003. Bacteria in midguts of field-collected Anopheles albimanus block Plasmodium vivax sporogonic development. J Med Entomol 40:371–374. doi: 10.1603/0022-2585-40.3.371. [DOI] [PubMed] [Google Scholar]
- 32.Dong Y, Manfredini F, Dimopoulos G. 2009. Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathog 5. doi: 10.1371/journal.ppat.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boissière A, Tchioffo MT, Bachar D, Abate L, Marie A, Nsango SE, Shahbazkia HR, Awono-Ambene PH, Levashina EA, Christen R, Morlais I. 2012. Midgut microbiota of the malaria mosquito vector Anopheles gambiae and interactions with Plasmodium falciparum infection. PLoS Pathog 8:e1002742. doi: 10.1371/journal.ppat.1002742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gendrin M, et al. 2015. Antibiotics in ingested human blood affect the mosquito microbiota and capacity to transmit malaria. Nat Commun 6:5291. doi: 10.1038/ncomms6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu P, Sun P, Nie K, Zhu Y, Shi M, Xiao C, Liu H, Liu Q, Zhao T, Chen X, Zhou H, Wang P, Cheng G. 2019. A gut commensal bacterium promotes mosquito permissiveness to arboviruses. Cell Host Microbe 25:101–112. doi: 10.1016/j.chom.2018.11.004. [DOI] [PubMed] [Google Scholar]
- 36.Xia X, Zheng D, Zhong H, Qin B, Gurr GM, Vasseur L, Lin H, Bai J, He W, You M. 2013. DNA sequencing reveals the midgut microbiota of diamondback moth, Plutella xylostella (L.) and a possible relationship with insecticide resistance. PLoS One 8:e68852. doi: 10.1371/journal.pone.0068852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kikuchi Y, Hayatsu M, Hosokawa T, Nagayama A, Tago K, Fukatsu T. 2012. Symbiont-mediated insecticide resistance. Proc Natl Acad Sci USA 109:8618–8622. doi: 10.1073/pnas.1200231109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dada N, Sheth M, Liebman K, Pinto J, Lenhart A. 2018. Whole metagenome sequencing reveals links between mosquito microbiota and insecticide resistance in malaria vectors. Sci Rep 8:2084. doi: 10.1038/s41598-018-20367-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dada N, Lol JC, Benedict AC, López F, Sheth M, Dzuris N, Padilla N, Lenhart A. 2019. Pyrethroid exposure alters internal and cuticle surface bacterial communities in Anopheles albimanus. ISME J 13:2447–2464. doi: 10.1038/s41396-019-0445-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arévalo-Cortés A, Mejia-Jaramillo AM, Granada Y, Coatsworth H, Lowenberger C, Triana-Chavez O. 2020. The midgut microbiota of Colombian Aedes aegypti populations with different levels of resistance to the insecticide lambda-cyhalothrin. Insects 11:584. doi: 10.3390/insects11090584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soltani A, Vatandoost H, Oshaghi MA, Enayati AA, Chavshin AR. 2017. The role of midgut symbiotic bacteria in resistance of Anopheles stephensi (Diptera: Culicidae) to organophosphate insecticides. Pathog Glob Health 111:289–296. doi: 10.1080/20477724.2017.1356052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barnard K, Jeanrenaud ACSN, Brooke BD, Oliver SV. 2019. The contribution of gut bacteria to insecticide resistance and the life histories of the major malaria vector Anopheles arabiensis (Diptera: Culicidae). Sci Rep 9:9117. doi: 10.1038/s41598-019-45499-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Omoke D, et al. 2021. Western Kenyan Anopheles gambiae showing intense permethrin resistance harbor distinct microbiota. Malar J 20:77. doi: 10.1186/s12936-021-03606-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meiwald A, et al. 2020. Reduced long-lasting insecticidal net efficacy and pyrethroid insecticide resistance are associated with overexpression of CYP6P4, CYP6P3 and CYP6Z1 in populations of Anopheles coluzzii from South-East Côte d’Ivoire. J Infect Dis doi: 10.1093/infdis/jiaa699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dada N, Jupatanakul N, Minard G, Short S, Akorli J, Villegas LM. 2020. Considerations for mosquito microbiome research from the Mosquito Microbiome Consortium. Microbiome 9:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Knight R, Vrbanac A, Taylor BC, Aksenov A, Callewaert C, Debelius J, Gonzalez A, Kosciolek T, McCall L-I, McDonald D, Melnik AV, Morton JT, Navas J, Quinn RA, Sanders JG, Swafford AD, Thompson LR, Tripathi A, Xu ZZ, Zaneveld JR, Zhu Q, Caporaso JG, Dorrestein PC. 2018. Best practices for analyzing microbiomes. Nat Rev Microbiol 16:410–422. doi: 10.1038/s41579-018-0029-9. [DOI] [PubMed] [Google Scholar]
- 47.Chen S, Hu M, Liu J, Zhong G, Yang L, Rizwan-Ul-Haq M, Han H. 2011. Biodegradation of beta-cypermethrin and 3-phenoxybenzoic acid by a novel Ochrobactrum lupini DG-S-01. J Hazard Mater 187:433–440. doi: 10.1016/j.jhazmat.2011.01.049. [DOI] [PubMed] [Google Scholar]
- 48.Wang B-Z, Ma Y, Zhou W-Y, Zheng J-W, Zhu J-C, He J, Li S-P. 2011. Biodegradation of synthetic pyrethroids by Ochrobactrum tritici strain pyd-1. World J Microbiol Biotechnol 27:2315–2324. doi: 10.1007/s11274-011-0698-2. [DOI] [Google Scholar]
- 49.Talwar MP, Mulla SI, Ninnekar HZ. 2014. Biodegradation of organophosphate pesticide quinalphos by Ochrobactrum sp. strain HZM. J Appl Microbiol 117:1283–1292. doi: 10.1111/jam.12627. [DOI] [PubMed] [Google Scholar]
- 50.Qiu XH, Bai WQ, Zhong QZ, Li M, He FQ, Li BT. 2006. Isolation and characterization of a bacterial strain of the genus Ochrobactrum with methyl parathion mineralizing activity. J Appl Microbiol 101:986–994. doi: 10.1111/j.1365-2672.2006.03016.x. [DOI] [PubMed] [Google Scholar]
- 51.Hao X, Zhang X, Duan B, Huo S, Lin W, Xia X, Liu K. 2018. Screening and genome sequencing of deltamethrin-degrading bacterium ZJ6. Curr Microbiol 75:1468–1476. doi: 10.1007/s00284-018-1546-5. [DOI] [PubMed] [Google Scholar]
- 52.Hu GP, Zhao Y, Song FQ, Liu B, Vasseur L, Douglas C, You MS. 2014. Isolation, identification and cyfluthrin-degrading potential of a novel Lysinibacillus sphaericus strain, FLQ-11-1. Res Microbiol 165:110–118. doi: 10.1016/j.resmic.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 53.Ozdal M, Ozdal OG, Algur OF. 2016. Isolation and characterization of α-endosulfan degrading bacteria from the microflora of cockroaches. Pol J Microbiol 65:63–68. doi: 10.5604/17331331.1197325. [DOI] [PubMed] [Google Scholar]
- 54.Broderick NA, Raffa KF, Handelsman J. 2006. Midgut bacteria required for Bacillus thuringiensis insecticidal activity. Proc Natl Acad Sci USA 103:15196–15199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xia X, Sun B, Gurr GM, Vasseur L, Xue M, You M. 2018. Gut microbiota mediate insecticide resistance in the diamondback moth, Plutella xylostella (L). Front Microbiol 9:25. doi: 10.3389/fmicb.2018.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fang S, Wang L, Guo W, Zhang X, Peng D, Luo C, Yu Z, Sun M. 2009. Bacillus thuringiensis Bel protein enhances the toxicity of Cry1Ac protein to Helicoverpa armigera larvae by degrading insect intestinal mucin. Appl Environ Microbiol 75:5237–5243. doi: 10.1128/AEM.00532-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kristan M, Lines J, Nuwa A, Ntege C, Meek SR, Abeku TA. 2016. Exposure to deltamethrin affects development of Plasmodium falciparum inside wild pyrethroid-resistant Anopheles gambiae s.s. mosquitoes in Uganda. Parasites Vectors 9:100. doi: 10.1186/s13071-016-1384-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alout H, Ndam NT, Sandeu MM, Djégbe I, Chandre F, Dabiré RK, Djogbénou LS, Corbel V, Cohuet A. 2013. Insecticide resistance alleles affect vector competence of Anopheles gambiae s.s. for Plasmodium falciparum field isolates. PLoS One 8:e63849. doi: 10.1371/journal.pone.0063849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Capone A, et al. 2013. Interactions between Asaia, Plasmodium, and Anopheles: new insights into mosquito symbiosis and implications in malaria symbiotic control. Parasites Vectors 6:182. doi: 10.1186/1756-3305-6-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Castro AJ. 1967. Antimalarial activity of prodigiosin. Nature 213:903–904. doi: 10.1038/213903a0. [DOI] [PubMed] [Google Scholar]
- 61.Patil CD, Patil SV, Salunke BK, Salunkhe RB. 2011. Prodigiosin produced by Serratia marcescens NMCC46 as a mosquito larvicidal agent against Aedes aegypti and Anopheles stephensi. Parasitol Res 109:1179–1187. doi: 10.1007/s00436-011-2365-9. [DOI] [PubMed] [Google Scholar]
- 62.Guideline for evaluating insecticide resistance in vectors using the CDC bottle bioassay. https://www.cdc.gov/malaria/resources/pdf/fsp/ir_manual/ir_cdc_bioassay_en.pdf.
- 63.Guedes RNC, Walse SS, Throne JE. 2017. Sublethal exposure, insecticide resistance, and community stress. Curr Opin Insect Sci 21:47–53. doi: 10.1016/j.cois.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 64.Wang Y, Gilbreath TM, Kukutla P, Yan G, Xu J. 2011. Dynamic gut microbiome across life history of the malaria mosquito Anopheles gambiae in Kenya. PLoS One 6:e24767. doi: 10.1371/journal.pone.0024767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rajatileka S, Burhani J, Ranson H. 2011. Mosquito age and susceptibility to insecticides. Trans R Soc Trop Med Hyg 105:247–253. doi: 10.1016/j.trstmh.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 66.Jones CM, Sanou A, Guelbeogo WM, Sagnon N, Johnson PCD, Ranson H. 2012. Aging partially restores the efficacy of malaria vector control in insecticide-resistant populations of Anopheles gambiae s.l. from Burkina Faso. Malar J 11:1–11. doi: 10.1186/1475-2875-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rowland M, Hemingway J. 1987. Changes in malathion resistance with age in Anopheles stephensi from Pakistan,” Pestic. Biochem Physiol 28:239–247. doi: 10.1016/0048-3575(87)90022-8. [DOI] [Google Scholar]
- 68.Lines JD, Nassor NS. 1991. DDT resistance in Anopheles gambiae declines with mosquito age. Med Vet Entomol 5:261–265. doi: 10.1111/j.1365-2915.1991.tb00550.x. [DOI] [PubMed] [Google Scholar]
- 69.Hodjati MH, Curtis CF. 1999. Evaluation of the effect of mosquito age and prior exposure to insecticide on pyrethroid tolerance in Anopheles mosquitoes (Diptera: Culicidae). Bull Entomol Res 89:329–337. doi: 10.1017/S0007485399000462. [DOI] [Google Scholar]
- 70.Collins E, et al. 2019. The relationship between insecticide resistance, mosquito age, and malaria prevalence in Anopheles gambiae s.l. from Guinea. Sci Rep 9:8846. doi: 10.1038/s41598-019-45261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.MPD/INS/MSHP/PNLP/DHS Program/University of Maryland. 2016. Côte d’Ivoire enquête de prévalence parasitaire du paludisme et de l’anémie. University of Maryland, Rockville, MD. [Google Scholar]
- 72.Fodjo BK, Koudou BG, Tia E, Saric J, N’dri PB, Zoh MG, Gba CS, Kropf A, Kesse NB, Chouaïbou MS. 2018. Insecticides resistance status of An. gambiae in areas of varying agrochemical use in Côte D’Ivoire. Biomed Res Int 2018:1–9. doi: 10.1155/2018/2874160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gillies MT, Coetzee M. 1987. A supplement to the Anophelinae of Africa south of the Sahara (Afrotropical Region). South African Institute for Medical Research, Johannesburg, South Africa. [Google Scholar]
- 74.Morgan JC, Irving H, Okedi LM, Steven A, Wondji CS. 2010. Pyrethroid resistance in an Anopheles funestus population from Uganda. PLoS One 5:e11872. doi: 10.1371/journal.pone.0011872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nishikoi Aquaculture, Ltd. 2020. https://www.nishikoi.co.uk/. Accessed 23 November 2020.
- 76.Qiagen. 2020. (EN)—DNeasy blood and tissue handbook. Qiagen. https://www.qiagen.com/gb/resources/resourcedetail?id=68f29296-5a9f-40fa-8b3d-1c148d0b3030&lang=en. Accessed 14 February 2020.
- 77.Santolamazza F, Mancini E, Simard F, Qi Y, Tu Z, della Torre A. 2008. Insertion polymorphisms of SINE200 retrotransposons within speciation islands of Anopheles gambiae molecular forms. Malar J 7:163. doi: 10.1186/1475-2875-7-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Roche. 2020. KAPA HiFi HotStart ReadyMix | Roche Sequencing Store. Roche. https://rochesequencingstore.com/catalog/kapa-hifi-hotstart-readymix/. Accessed 14 February 2020.
- 79.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, et al. 2019. Reproducible, interactive, scalable, and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 81.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Callahan BJ, McMurdie PJ, Holmes SP. 2017. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J 11:2639–2643. doi: 10.1038/ismej.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McDonald D, Clemente JC, Kuczynski J, Rideout JR, Stombaugh J, Wendel D, Wilke A, Huse S, Hufnagle J, Meyer F, Knight R, Caporaso JG. 2012. The Biological Observation Matrix (BIOM) format or: how I learned to stop worrying and love the ome-ome. Gigascience 1:7. doi: 10.1186/2047-217X-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bokulich NA, Kaehler BD, Rideout JR, Dillon M, Bolyen E, Knight R, Huttley GA, Gregory Caporaso J. 2018. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6:90. doi: 10.1186/s40168-018-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pedregosa F, et al. 2011. Scikit-learn: machine learning in Python. J Mach Learn Res 12:2825–2830. [Google Scholar]
- 86.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2: approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–175. In Stackebrandt E, Goodfellow M (ed), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, New York, NY. [Google Scholar]
- 89.Faith DP. 1992. Conservation evaluation and phylogenetic diversity. Biol Conserv 61:1–10. doi: 10.1016/0006-3207(92)91201-3. [DOI] [Google Scholar]
- 90.Weiss S, Xu ZZ, Peddada S, Amir A, Bittinger K, Gonzalez A, Lozupone C, Zaneveld JR, Vázquez-Baeza Y, Birmingham A, Hyde ER, Knight R. 2017. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome 5:27. doi: 10.1186/s40168-017-0237-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kruskal WH, Wallis WA. 1952. Use of ranks in one-criterion variance analysis. J Am Stat Assoc 47:583–621. doi: 10.1080/01621459.1952.10483441. [DOI] [Google Scholar]
- 92.Sorenson T. 1948. A method of establishing groups of equal amplitude in plant sociology based on similarity of species content. K Danske Vide Nskabernes Selsk [Google Scholar]
- 93.Legendre P, Legendre L. 2012. Numerical ecology, p 499, 3rd ed. Elsevier, New York, NY. [Google Scholar]
- 94.Anderson MJ. 2001. A new method for nonparametric multivariate analysis of variance. Austral Ecol 26:32–46. doi: 10.1046/j.1442-9993.2001.01070.x. [DOI] [Google Scholar]
- 95.McMurdie PJ, Holmes S. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Morton JT, et al. 2019. Establishing microbial composition measurement standards with reference frames. Nat Commun 10:2719. doi: 10.1038/s41467-019-10656-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fedarko MW, et al. 2020. Visualizing ‘omic feature rankings and log-ratios using Qurro. NAR Genom Bioinform 2:lqaa023. doi: 10.1093/nargab/lqaa023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mandal S, Van Treuren W, White RA, Eggesbø M, Knight R, Peddada SD. 2015. Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb Ecol Health Dis 26:27663. doi: 10.3402/mehd.v26.27663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhu H, Sun S-J, Dang H-Y. 2008. PCR detection of Serratia spp. using primers targeting pfs and luxS genes involved in AI-2-dependent quorum sensing. Curr Microbiol 57:326–330. doi: 10.1007/s00284-008-9197-6. [DOI] [PubMed] [Google Scholar]
- 100.Jeffries CL, Lawrence GG, Golovko G, Kristan M, Orsborne J, Spence K, Hurn E, Bandibabone J, Tantely LM, Raharimalala FN, Keita K, Camara D, Barry Y, Wat’senga F, Manzambi EZ, Afrane YA, Mohammed AR, Abeku TA, Hedge S, Khanipov K, Pimenova M, Fofanov Y, Boyer S, Irish SR, Hughes GL, Walker T. 2018. Novel Wolbachia strains in Anopheles malaria vectors from Sub-Saharan Africa [version 2; peer review: 3 approved]. Wellcome Open Res 3:113. doi: 10.12688/wellcomeopenres.14765.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ren X, Rasgon JL. 2010. Potential for the Anopheles gambiae densonucleosis virus to act as an ‘evolution-proof’ biopesticide. J Virol 84:7726–7729. doi: 10.1128/JVI.00631-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download SPECTRUM00157-21_Supp_1_seq7.pdf, PDF file, 0.6 MB (606.7KB, pdf)
Supplemental material. Download SPECTRUM00157-21_Supp_2_seq8.xlsx, XLSX file, 0.02 MB (16.2KB, xlsx)
Supplemental material. Download SPECTRUM00157-21_Supp_3_seq9.xlsx, XLSX file, 0.02 MB (17.8KB, xlsx)
Supplemental material. Download SPECTRUM00157-21_Supp_4_seq10.xlsx, XLSX file, 0.01 MB (10.2KB, xlsx)
Supplemental material. Download SPECTRUM00157-21_Supp_5_seq11.xlsx, XLSX file, 0.01 MB (14.6KB, xlsx)
Data Availability Statement
Sequence data generated by this study is available at Sequence Read Archive (SRA) BioProject PRJNA702915 (accession numbers: SRR13743435 to SRR13743530). All other relevant data are available from the corresponding author upon reasonable request. Codes can be accessed at the public repository Zenodo (http://zenodo.org) under https://doi.org/10.5281/zenodo.5102954.