Abstract
Background
Endoscopic and histological remission are both important treatment goals in patients with ulcerative colitis (UC). We aimed to define cellular architecture, expression of molecular markers, and their correlation with endoscopic scores assessed by ultra-high magnification endocytoscopy (ECS) and histological scores.
Methods
Patients with UC (n = 29) were prospectively recruited. The correlation among ECS score (ECSS), Mayo endoscopic score (MES), and histological scores were determined. Area under curve were plotted to determine the best thresholds for ECSS that predicted histological remission by Robarts (RHI) and Nancy Histological Index (NHI).
Soluble analytes relevant to inflammation were measured in serum and mucosal culture supernatants using ProcartaPlex Luminex assays and studied by partial least square discriminant analysis and logistic model. Mucosal RNA sequencing and bioinformatics analysis were performed to define differentially expressed genes/pathways.
Results
Endocytoscope scoring system correlated strongly with RHI (r = 0.89; 95% CI, 0.51–0.98) and NHI (r = 0.86; 95% CI, 0.42–0.98) but correlated poorly with MES (r = 0.28; 95% CI, 0.27–0.70). We identified soluble brain-derived neurotrophic factors (BDNF), macrophage inflammatory proteins (MIP-1 α) and soluble vascular cell adhesion molecule 1 (sVCAM-1) predicted histological remission. Mucosal biopsy cultures also identified sVCAM-1 associated with healed mucosa. RNA-seq analysis identified gene expressions shared between ECSS, RHI, or NHI defined healing. A number of gene expressions and pathways were identified including inflammation and metabolic and tumor suppressors that discriminated healed from nonhealed mucosa.
Conclusions
Endocytoscopy represents an interesting tool that may sit between endoscopy and histology—but closer to the latter—identifying gene expression markers and pathways that are also identified by histology.
Keywords: mucosal healing, histological healing, endocytoscope, noninvasive markers, RNA-sequencing
INTRODUCTION
Achieving mucosal healing (MH) is considered to be a primary target in the treatment of inflammatory bowel disease (IBD).1–3 Mucosal healing increasingly incorporates both endoscopic remission (ER) and histologic remission (HR). Nevertheless, ER does not necessarily correspond to HR.4–6 Indeed, there is an increasing evidence that patients with ER have histological inflammation, accounting for relapse and adverse outcomes at follow-up.7, 8 This discrepancy between HR and ER might be explained by the use of conventional endoscopes that cannot accurately detect subtle inflammation.9, 10 Several studies have shown that HR is associated with reduced steroid use, lower risk of complication, hospitalization, and colorectal cancer development, suggesting it is an important endpoint to achieve—especially in ulcerative colitis (UC).11–14
The advanced endoscopy technologies are getting closer to histology by introducing the new concepts of mucosal and vascular healing patterns.15, 16 We have demonstrated that with widely available advanced technologies such as virtual electronic chromoendoscopy (VEC) and high definition (HD) imaging, endoscopic and histologic scores correlate more strongly and reduced discrepancy between the two, unlike previous publications.17–19 This has also been shown using artificial intelligence approach utilizing deep neural network approach.20, 21 Among these new endoscopic armamentarium, endocytoscopy (ECS; CF- Y-0058-1 prototype, Olympus Japan) is a new technique that provides in vivo microscopic imaging during endoscopy, with ultra-high magnification ranging from 450-fold to 1400-fold, looking at cells and nuclei of mucosal surfaces. Several studies have reported that ECS is a reliable technique to assess precisely ER and HR, potentially reducing the need for biopsy specimens. Notably, biopsies can assess only a limited area, whereas ECS is an optical diagnosis tool, which can sample a wider area in vivo of the colonic mucosa.22 Studies with ECS have shown high reproducibility between endoscopists and demonstrated that it predicts accurately histological inflammation and HR.22–25 Therefore, ECS is a bridge between endoscopy and histology, but further studies are required.
The identification of noninvasive molecular markers to monitor IBD patients by predicting MH and the risk of flare-up is a growing area of interest. Frequent endoscopic examinations are costly and uncomfortable for the patient; therefore, a need exists for blood-based biomarkers that accurately assess MH, which correlates well with endoscopic findings.26 Recently, a commercial panel based on blood levels of 13 proteins, called the endoscopic healing index (EHI), was developed and preliminary evidence supports that it can identify endoscopic healing, favoring the noninvasive monitoring and management of Crohn’s disease (CD) patients.27 Despite the encouraging results of EHI, it did not include histological assessment to confirm MH and has not been reproduced in UC.
Though many studies have explored the gene expression profiles in the context of specific therapies,28, 29 few studies have investigated ultrastructural and molecular mechanisms of intestinal healing beyond absence of inflammation. Whether advanced endoscopic techniques such as ECS and histology share molecular footprints of intestinal restitution and repair requires further evidence. Importantly, MH is not just an absence of inflammation but an active process of restitution and repair at the cellular and molecular level.30
The main objectives of this exploratory study were to understand ER and HR in UC determined by ultra-high magnification in vivo microscopy ECS, determine the correlation between endocytoscopy and histology, and define cellular architecture, expression of molecular and genomic markers and their correlation with endoscopic score defined by ultra-high magnification with histology scores.
MATERIALS AND METHODS
Patients
We conducted a prospective study enrolling 29 consecutive UC patients (18 males, 62%; mean age 41 years ± SD 15) referred for assessing ER achieved after treatment or for surveillance between January 2018 and July 2019 at a tertiary academic center (Queen Elizabeth II Hospital, Birmingham, UK).
We collected demographic data including patient characteristics, clinical data detailing age at diagnosis, disease characteristics (extent of disease, Montreal classification), treatments (corticosteroids, immunosuppressive therapy, biologics), clinical disease activity scores (full and partial Mayo score31), and endoscopic activity scores (Mayo endoscopic score; MES),32 ulcerative colitis endoscopic index (UCEIS),33 and Paddington International virtual chromoendoscopy score (PICaSSO)34–36 (Table 1).
TABLE 1.
Demographic and Clinical Data of IBD Patients Enrolled
| UC | Demographic and Clinical Characteristics |
|---|---|
| Sex | 11 F; 18 M (62%M and 38% F) |
| Mean age ± SD (range) | 41± 15 (20–66) |
| Disease duration median years (range) | 12 (1–38) |
| Localization | |
| Pancolitis (E3) | 25 (86%) |
| Left colitis (E2) | 4 (15%) |
| Proctosigmoiditis (E1) | 0 |
| Mayo endoscopy score (MES) | |
| Mayo 0 | 11 (38%) |
| Mayo 1 | 8 (27.5%) |
| Mayo 2 | 8 (27.5%) |
| Mayo 3 | 2 (7%) |
| Clinical Mayo | |
| Remission < 2 remission | 15 (52%) |
| Mild 2–4 mild activity | 7 (24%) |
| Moderate 5–7 moderate activity | 3 (10%) |
| Severe > 7 severe activity | 4 (14%) |
| Biological therapy | |
| Adalimumab | 3 (10.3%) |
| Infliximab | 1 (3.5%) |
| Vedolizumab | 3 (10.3%) |
| Ustekinumab | 1 (3.5%) |
| No biological therapy | 21 (72.4%) |
| Medication | |
| Mesalazine | 24 (83%) |
| Steroids | 7 (24%) |
| Immunosuppressants | 6 (20%) |
| CRP mean (range) mg/dL | 9 (1–33) |
| FC mean (range) mcg/gram | 656 (30–2363) |
Endoscopic Procedure
All the recruited patients underwent colonoscopies with endocytoscope (ECS; CF- Y-0058-1 prototype Olympus, Japan) with 520-fold magnification to obtain ultra-magnified images and define inflamed and healed areas. For consistency of technique, all of the procedures were performed by one colonoscopist (MI) experienced in ECS and optical diagnosis in IBD to harmonize and accurately assess the grade of inflammation. Additionally, the endoscopic findings were scored by 2 endoscopists during the procedure and also video recorded. The agreed score between the 2 endoscopists was recorded and analyzed.
The colonic mucosa was initially assessed using HD white light endoscopy (HD WLE) and narrow banding imaging (NBI) with and without magnification. The inflammatory activity was scored using the MES, and ER was defined as MES 0.2 Biopsies were taken targeting either the worst affected area of inflammation or the most representative area of endoscopic healing. After washing of the mucosa with water plus simethicone and staining with 10 mL of 1% methylene blue solution, ECS was performed in the same area of the colon where endoscopic activity was scored. The following endoscopic parameters for ECSS were assessed to grade the inflammation (Table 2): (1) the shape of the crypts (normal round, elongated, irregular, necrosis); (2) infiltration of the cell between the crypts (≤50% vs ≥50%); (3) the distance between the crypts (normal, 3 or more crypts in a visual field [VF], ≤2 crypts in a VF, intermediate 2≤ or ≥ 3 crypts in a VF with infiltrating cells in lamina propria (LP), dropout/necrosis); and (4) the visibility of superficial microvessels (not visible vs visible).We set an overall score of 3 to 9, and we defined the following subscores: the shape of crypts (range, 1–3), infiltration between the crypts (range, 1–2), distance between the crypts (range, 1–3), and the visibility of superficial microvessels (range, 0–1). This score was adapted from Nakazato et al by including endoscopic findings representative of disease activity (infiltration of cells, Fig. 1).22
TABLE 2.
Endocytoscopy Scoring System
| Endocytoscopy Items | Score |
|---|---|
| Crypts architecture | |
| Normal, elongated | 1 |
| Irregular | 2 |
| Necrosis | 3 |
| Infiltration of the cell between the crypts | |
| ≤50% | 1 |
| ≥50% | 2 |
| Distance between the crypts | |
| Normal: 3 or more crypts in a VF | 1 |
| Elongated = <2 crypts in a VF | 1 |
| Intermediate = 2 ≤ crypts ≥ 3 in a VF with infiltrating cells in LP | 2 |
| Drop-out /necrosis | 3 |
| Visibility of superficial microvessels | |
| Not visible | 0 |
| Visible | 1 |
| ECS total score | 3–9 |
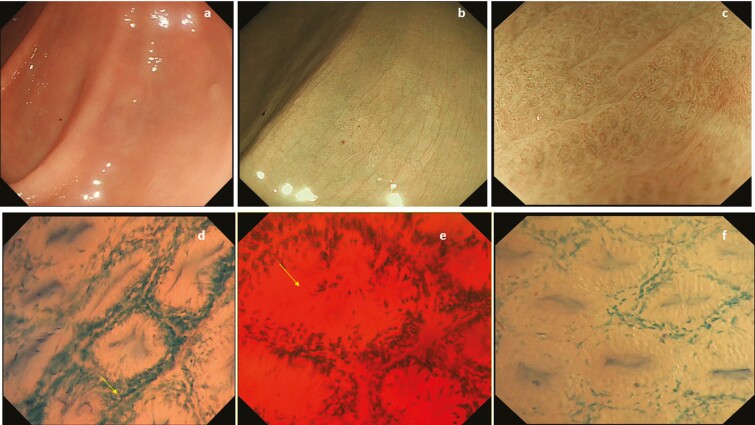
FIGURE 1.
A–C, Endoscopic MH assessed by HD white light endoscopy and NBI with and without magnification showing elongated crypts as observed in endoscopic mucosal healing in UC. D–F, Ultra-high magnification endocytoscope after using dye chromoendoscopy with methylene blue 1% showed increase infiltration with cells between the crypts (arrow) and elongated distorted crypts (arrow) as per histological healing changes.
Pictures and videos collected under HD WLE, NBI, and then ECS from each patient were all recorded and digitally stored for subsequent analyses by using the Olympus recording system IMH-20.
Histological Scoring
Histological assessment of inflammatory activity was scored by an expert pathologist (DZ) blinded to clinical and endoscopic information. The following histological scores were used: NHI score (range, 0–4)37 and Robarts Histological Index (RHI,38 range, 0–33). Histologic remission was defined as NHI ≤137, and RHI ≤3 without neutrophils in the epithelium and lamina propria.39
Analysis of Soluble Markers
Serum collection
Serum samples were collected from all patients at the time of the colonoscopy in BD SST vacutainer tubes and processed within 4 hours. Samples were spun at 1300 g for 10 minutes and serum removed and stored at −80C before analysis.
Culture of mucosal biopsies
Endoscopic biopsies were cultured in 10% fetal bovine serum, (50 U/mL Penicillin/Streptomycin, 200 μM L-Glutamine, 50 μg/mL Gentamicin, 2.5 μg/mL Amphotericin B, 50 μM 2-mercaptoethanol in RPMI 1640) at mass to volume ratio of 20 ug/mL. At 24 hours, supernatants were harvested, centrifuged for 10 minutes at 10000 g and stored at −80C before analysis.
Serum and mucosal biopsy culture supernatants were thawed on ice and analyzed for 56 soluble markers (Table 3) using ProcartaPlex Luminex assays (ThermoFisher Scientific, Waltham, Massachusetts, US) according to manufacturer guidelines. Serum samples were diluted 2-fold for all analytes except for matrix metalloprotein-2 (MMP-2), matrix metalloprotein 9 (MMP-9), Intercellular Adhesion Molecule 1 (ICAM-1), and soluble vascular cell adhesion molecule 1 (sVCAM-1), which were diluted 100-fold. Culture supernatants were used neat.
TABLE 3.
ProcartaPlex Luminex Arrays Used to Analyze Serum and Endoscopy Biopsy Culture Supernatants
| ProcartaPlex Array | Analytes |
|---|---|
| 4 Plex (customized) | MMP-2; MMP-9; sICAM-1; sVCAM-1 |
| 7 Plex (customized) | BLC; IL-12p40; MMP-1; OSM a,b; TNFR1; TNFR2; TREM-1a,b |
| 45 Plex (ProcartaPlex Hu) Cytokine/Chemokine/GF 1 45plex (Cat no. EPX450-12171–901) | BDNFb; Eotaxin/CCL11; EGFb; FGF-2a,b; GM-CSFa; GRO alpha/CXCL1a; HGF; NGF beta a,b; LIF; IFN alpha a,b; IFN gammaa; IL-1 beta a; IL-1 alpha a; IL-1RA a; IL-2 a; IL-4 a,b; IL-5 a,b; IL-6 a; IL-7; IL-8/CXCL8 a; IL-9 a,b; IL-10 a; IL-12 p70 a; IL-13*; IL-15 a; IL-17A a; IL-18 a; IL-21 a,b; IL-22 a; IL-23 a,b; IL-27 a; IL-31 a,b; IP-10/CXCL10; MCP-1/CCL2; MIP-1 alpha/CCL3; MIP-1 beta/CCL4; RANTES/CCL5; SDF-1 alpha/CXCL12; TNF alpha a; TNF beta/LTA a,b; PDGF-BBb; PLGF; SCF; VEGF-A; VEGF-D a,b |
aNot detected in serum
bNot detected in culture supernatant
RNA Extraction, Library Preparation, and Sequencing
Endoscopic biopsies of the colon were stored in RNA later before RNeasy on-column RNA extraction and purification (Qiagen, Venio, Netherlands). RNA was quantified by Qubit (Life Technologies, Carlsbad, California, US) and 0.5 ng used to prepare uniquely indexed cDNA QIAseq UPX 3’ Transcriptome libraries according to manufacturer’s instructions. Libraries were quantified and quality controlled using the QIAseq Library Quant Assay Kit and tapestation analysis and sequenced on the Miseq and Nextseq Illumina platforms to a depth of 1 to 3milion reads/sample. Fastq files were obtained through BaseSpace and reads demultiplexed aligned, quantified, and normalized using the CLC Genomics Workbench (Qiagen, Venio, Netherlands). The RNA seq data was deposited, and the raw data is available at Array Express through accession number E-MATB-9731.
Statistical Analysis and Informatics
Demographic, clinical and endoscopic data
Demographic, clinical, and endoscopic data were collected using the REDCap platform, and the results were transferred to a Microsoft Excel database.
Pearson correlations between MES, ECSS, and histological scores and between Picasso total score, UCEIS, and ECSS were calculated. Very strong correlation was considered as a value of 0.8 to 1.0, strong as 0.6 to 0.79, moderate as 0.40 to 0.59, and weak as 0.2 to 0.39
We determined diagnostic accuracy, sensitivity, and specificity of MES and ECSS assessed by HD-NBI to predict HR. Receiver operating characteristic (ROC) curves were plotted as sensitivity vs specificity to explore the ability of endoscopic scores to predict HR. The area under the receiver operating characteristic curve (AUROC) was determined to accurately identify the cutoff of MES and ECSS that reflect HR.
Cytokine panel analysis from blood samples
A total of 56 soluble analytes relevant to inflammation or shown to be altered in UC were measured in serum (Table 3). Measurements that did not fall within the standard range of the assay were assigned the maximum or lower limits of the assay as appropriate, and analytes that were not detected in at least 40% of patients in 1 group were excluded from analysis to reduce type 1 errors. Those not detected in serum or culture supernatants are marked in Table 3. Detected analytes were used to train a logistic regression model.
Luminex soluble marker analysis from mucosal culture supernatants
Luminex data were initially analyzed using t test across all the comparisons separately. The selected analytes were then included in a regression model utilising partial least square discriminant analysis (PLS-DA). A false discovery rate (FDR) corrected P value (q value) was considered for significance. Significant cytokines/soluble markers were then further analyzed using PLS-DA modeling.40 We used variable importance in projection (VIP) to prioritize the features. Variable importance in projection is a measure of a variable’s importance in the PLS-DA model. It summarizes the contribution a variable makes to the model. Markers with a VIP score of more than 1.5 were used.41 Area under the curve for each comparison was calculated to show the sensitivity and specificity of each combinations of the markers.
RNA sequencing data
Bioinformatics analysis of the RNA sequencing data was performed (AA, GG). The following differential expression procedure was performed for participants classified as healed vs nonhealed according to each of the scoring procedures. Transcripts per million (TPM) normalized values42 were log-transformed with a pseudocount of 1 added. Transcripts with <1 transformed count in any sample were excluded from further analysis, as were transcripts with low variance (defined as less than 10% unique counts across both conditions and greater than a 19:1 ratio of the most frequent count to the second most frequent across both conditions). Differential expression analysis between conditions was conducted with the limma package.43 Library size was estimated using reduced maximum likelihood estimator with 500 iterations. Initial fitting was performed using a robust M-estimation, and moderated test statistics were computed by empirical Bayes. An FDR-corrected P value <0.05 was considered and filtered for further downstream data analysis. Partial least squares discriminant analysis modeling was performed on these genes. A VIP score was used to further filter the genes and measured a variable’s importance in the PLS-DA model. Genes with a VIP score of more than 1 were used for gene expression analysis. To understand the biological significance and pathways, enrichment analysis using EnrichR was also performed.44 Pathway enrichment was done using gene ontology and kyoto encyclopaedia of genes and genomes (KEGG).45 Differentially expressed up and down regulated genes were visualized using volcano plots. We considered Fold change >2.0 and FDR-corrected P value < 0.05 across healed vs nonhealed samples.
The RNA sequencing data were loaded into ArrayExpress (ArrayExpress accession:E-MTAB-9731).
ETHICAL CONSIDERATIONS
The study was approved by the research ethics committee (IRAS ID: 227882), and all patients provided written informed consent. Exclusion criteria included inability to provide consent, coagulation defect, or severe comorbidity.
RESULTS
Patient Characteristics
The demographic characteristics of the patients are summarized in Table 1. Eighty-six percent (25 of 29) of UC patients had pancolitis, and 52% (15 of 29) were in clinical remission according to partial Mayo clinical score. Although, 38% (11 of 29) showed an MES of 0, and 28% (8 of 29) showed an MES of 1; 45% (13 of 29) and 52% (15 of 29) were in histologic remission (HR) according to NHI ≤137 and RHI ≤3,39 respectively.
Correlations of Endocytoscopy and Histology (NHI and RHI) Scores in UC
The ECS total score had very strong correlation with HR as defined by NHI r = 0.86 (95% CI, 0.42–0.98) and RHI r = 0.89 (95% CI, 0.51–0.98). Whereas ECS total score correlated weakly with MES r = 0.28 (95% CI, 0.27–0.70). Mayo endoscopic score correlated strongly with NHI r = 0.73 (95%CI 95% 0.35–090) and RHI r = 0.63 (95% CI, 0.18–0.86), though less strongly than with ECSS. (Table 4)
TABLE 4.
Correlations of ECS With NHI and RHI Score in UC Patients
| Correlations of Endocytoscopy Scores and NHI Score of UC Patients | |
|---|---|
| Crypts architecture | 76.4%; 95% CI, 12.9- 95.4 |
| Infiltration of the cell between the crypts | 66.1%; 95% CI, 8.11–93.2 |
| Distance between the crypts | 86.6%; 95% CI, 41.52–97.5 |
| Visibility of vessels | 75.0%; 95% CI, 9.61–95.2 |
| Endocytoscopy total score | 86.6%; 95% CI, 41.5–97.5 |
| Correlations of Endocytoscopy Scores and RHI Score of UC Patients | |
| Crypts architecture | 66.3%; 95% CI, 7.7–93.2 |
| Infiltration of the cell between the crypts | 82.7%; 95% CI, 29.4–96.7 |
| Distance between the crypts | 89.6%; 95% CI, 52.05–98.1 |
| Visibility of vessels | 86.7%; 95% CI, 41.9–97.6 |
| Endocytoscopy total score | 89.3 %; 95% CI, 50.8–98.0 |
Correlations of Endocytoscopy and Endoscopic PICaSSO and UCEIS Scores in UC
The ECS total score showed a strong correlation with PICaSSO total score r = 0.67 (95% CI, 0.40–0.83) and with UCEIS r = 0.74 (95% CI, 0.51–0.87), better than MES but weaker than HR defined by NHI.
Diagnostic Accuracy of Endocytoscopy Score System (ECSS) in UC Patients for RHI ≦3 and NHI ≦1
Using the defined threshold of HR, we analyzed the diagnostic accuracy of each ECSS endoscopic feature, the total score in the colonic area, and from where biopsies were taken. (Tables 4, 5). A ROC curve was used to calculate the best value of total ECSS to predict HR. The best value of ECSS total score was ≤3 for predicting HR, with RHI ≤3 with no neutrophilis in epithelium/LP, with an AUROC of 0.81 (95% CI, 0.66–0.97), specificity of 0.89 (95% CI, 0.52–1), sensitivity of 0.62 (95% CI, 0.31–0.86), and an accuracy of 0.80 (95% CI, 0.57–0.87). Similarly, an ECSS total score of ≤3 was the best cutoff to predict HR defined as NHI ≤1, with an AUROC of 0.77 (95% CI, 0.59–0.95), specificity of 0.86 (95% CI-43-1), sensitivity of 0.64 % (95% CI, 27–91), and an accuracy of 0.80% (95% CI, 49–90). (Tables 5, 6)
TABLE 5.
Diagnostic Accuracy of the Best Threshold ECSS Total to Predict Histological Healing
| UC | Sensitivity 95% CI | Specificity 95% CI | Accuracy 95% CI | AUROC 95% CI |
|---|---|---|---|---|
| ECS total score ≤3 and RHI ≤3 | 61.5% (31–85) | 88.5 % (52–100) | 79.5% (57–87) | 81.2% (66–97) |
| ECS total score ≤3 and NHI ≤1 | 64% (27–91) | 86% (43–100) | 79.5% (49–90) | 77% (59–95) |
TABLE 6.
Diagnostic Accuracy of the Best Threshold of Each ECS ≤3 Item to Predict Histological Healing in UC Defined by RHI and NHI Scores
| ECS Item in UC | Sensitivity 95% CI | Specificity 95% CI | Accuracy 95% CI | AUROC 95% CI |
|---|---|---|---|---|
| Crypt architecture ≤0 and RHI ≤3 | 46% (11.5–71) | 96% (52–100) | 80% (50–82.5) | 74% (57–91) |
| Infiltration of the cell ≤1 between the crypts and RHI ≤3 | 69% (33–92) | 81.5% (42-–94) | 77.5% (51–85.5) | 75% (60–90) |
| Distance between the crypts ≤1 and RHI ≤3 | 92% (69–100) | 56% (8–75) | 67.5% (36–82) | 82% (68–95.5) |
| Visibility of superficial microvessels ≤0 and RHI ≤3 | 92% (64–100) | 48% (15–67) | 62% (52.5–65) | 70% (57–83) |
| Crypt architecture ≤0 and NHI ≤1 | 45.5% (12–74) | 93% (52–100) | 80% (50–85) | 72% (54–91) |
| Infiltration of the cell ≤1 between the crypts and NHI ≤1 | 64% (29–91) | 76% (39–91) | 72.5% (46–83) | 70% (53–87) |
| Distance between the crypts ≤1 and NHI ≤1 | 91% (65–100) | 52% (8–72) | 62.5% (31–78) | 78% (62–94) |
| Visibility of superficial microvessels ≤0 and NHI ≤1 | 57% (49–59) | 90% (61–100) | 44% (15–63) | 67% (54–81) |
Serum Soluble Markers
By downstream modeling and logistic regression, we identified brain-derived neurotrophic factors (BDNF) and macrophage inflammatory proteins (MIP-1 α) that, when combined together, showed the highest predictive ability for RHI ≤3 (no neutrophils) and NHI ≤1 with an AUROC of 0.82 (95% CI, 0.69–0.97) (Fig. 2). When we investigated analytes separately for predicting HR, univariate logistic regression was significant for 2 other markers: leukemia inhibitory factor (LIF) and soluble vascular cell adhesion molecule 1 (sVCAM1), both showing higher values associated with active inflammation by histology. The AUROC was 0.74 for LIF (95% CI, 0.591–0.89) and 0.72 for sVCAM1 (95% CI, 0.53–0.90). Serum soluble markers did not correlate significantly at P < 0.05 with ECSS (LIF vs ECSS r = 0.31; sVCAM vs ECSS 0.37).
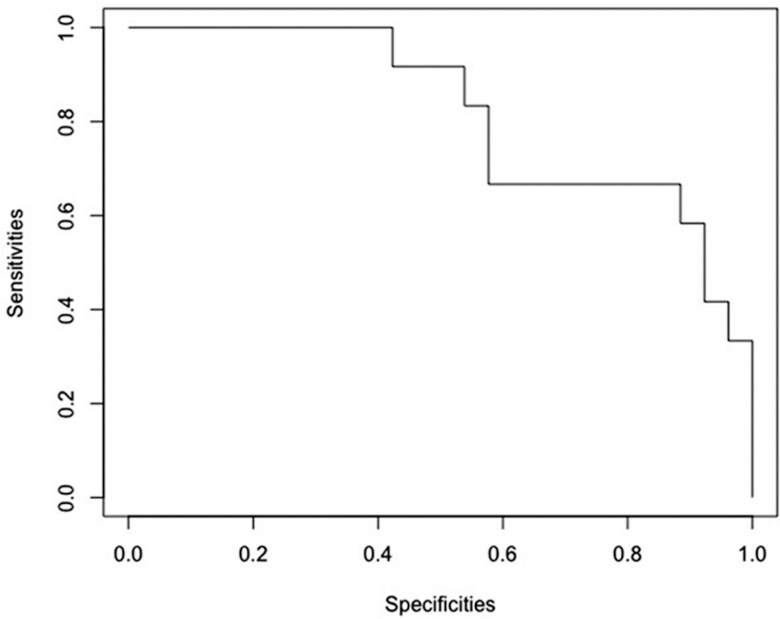
FIGURE 2.
ROC curves of serum BDNF and MIP-1 that, when combined together, were best at predicting histological healing (RHI ≤3 and NHI ≤1) with an AUROC of 0.82.
Culture Supernatant of UC Mucosal Biopsies
The supernatants from the mucosal biopsy cultures were analyzed for a panel of soluble markers. The following markers were identified as healed or nonhealed mucosa. (Table 7). Both RHI ≤3 and NHI ≤1 identified the same set of molecules—MMP9, TNFR2, IL-1RA, and hepatocyte growth factor (HGF)—as a panel associated with HR. Area under the receiver operating characteristic curve was 0.82 (95% CI, 0.5–1) for a panel of 4 analytes to predict HR by RHI or NHI. The ECSS score ≤3 identified sVCAM-1, TNFR2, and IL-1Ra as a panel—the latter 2 being the same identified by HR—for predicting healing with an AUROC of 0.75 (95% CI, 0.25–1). For prediction of ER assessed by MES = 0, the AUROC was 0.91 (95% CI, 0.5–1) for 4 analytes as a panel of sICAM-1, IL-6, IL-1Ra, and IL-15.
TABLE 7.
Biopsy Culture Derived Selected Makers From PLS-DA Modeling and Combined Marker AUC Values. Markers Were Selected Based on VIP >1.5
| Score used | Markers Selected From PLS-DA Model (VIP > 1.5) | AUC (95% CI) |
|---|---|---|
| RHI | MMP-9, TNFR2, IL-1ra, HGF | 0.82 (0.54–1) |
| NHI | MMP-9, TNFR2, IL-1ra, HGF | 0.82 (0.5–1) |
| Mayo | sICAM-1 IL-6 IL-1ra IL-15 | 0.91 (0.5–1) |
| ECCS | sVCAM-1, TNFR2, IL-1ra | 0.75 (0.25–1) |
RNA Sequence Analysis
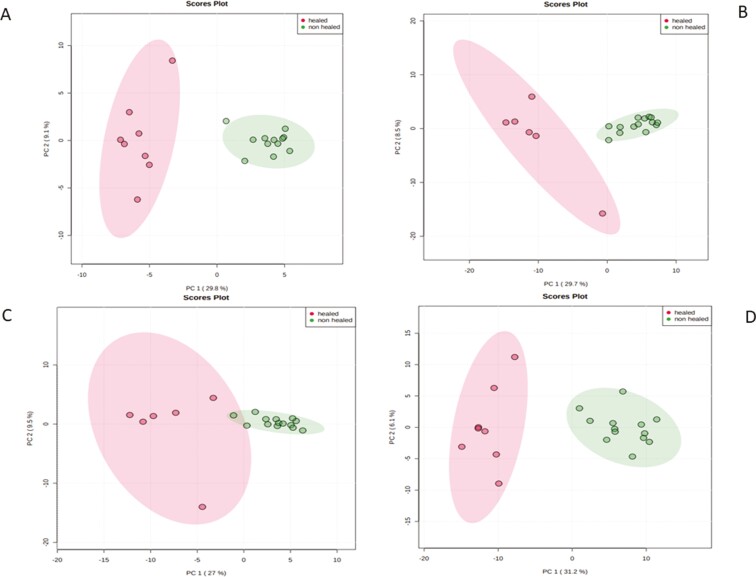
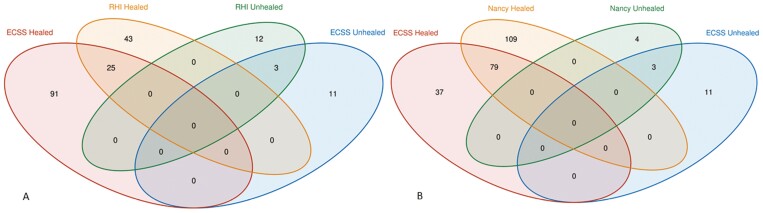
We performed RNA sequence analysis using each of the scores (MES, ECSS, RHI, NHI) and identified upregulated and downregulated genes in each of the outcome variables (Supplementary Table 1). Moreover, principal component analysis (PCA) shows clear separation of the healing vs nonhealing samples for all the scores, including ECSS, MES, RHI, and NHI index scores (Fig. 3). Comparing ECCS scores healing vs nonhealing, we found 116 genes upregulated and 14 genes downregulated. By RHI scores, 68 genes were upregulated and 15 genes were downregulated. Nancy Histological Index scores for healing vs nonhealing resulted in 188 upregulated and 7 downregulated genes. Those commonly identified by ECSS and RHI scores are summarized by the Venn diagram in Fig. 4. Twenty-five genes were overlapping between healed mucosa defined by ECSS and RHI score; 79 genes were overlapping between healed mucosa defined by ECSS and NHI index. (Supplementary Table 2)
FIGURE 3.
Principal component analysis score plots represented on the differentially expressed transcriptome datasets. Each of the plots demonstrating clustering of patients according to the healed vs nonhealed categories. Each of the dots (red or green) represent samples (or patients) and are colored according to the subject cohort (healed vs nonhealed). Ellipses represent 95% confidence healed or nonhealed patients. Results are plotted according to the top 2 principal components scores: principal component 1 (PC1) and principal component 2 (PC2). PC1 and PC2 scores, with the percentage variation explained by the x and y axis. Four different definitions of the healed vs nonhealed defined as (A) RHI scores, (B) NHI, (C) ECCS scores, and (D) Mayo score.
FIGURE 4.
Overlap of upregulated genes in healed and nonhealed mucosa as defined by (A) ECSS and RHI scores, and (B) ECSS and NHI scores.
We listed all the important genes prioritized by variable importance in projection of more than 1 in the PLS-DA model. We found that healing and nonhealing samples were separated by differentially expressed genes. For healing vs nonhealing comparison as defined by RHI, NHI, Mayo and ECCS, there were 32, 83, 127, and 60 differentially genes respectively (Supplementary Table 3). All these genes were highly predictive. For example, the AUC value using 60 genes together for healing vs nonhealing defined by ECCS was 1. The AUC value for each of the genes is listed in the Supplementary Table 4.
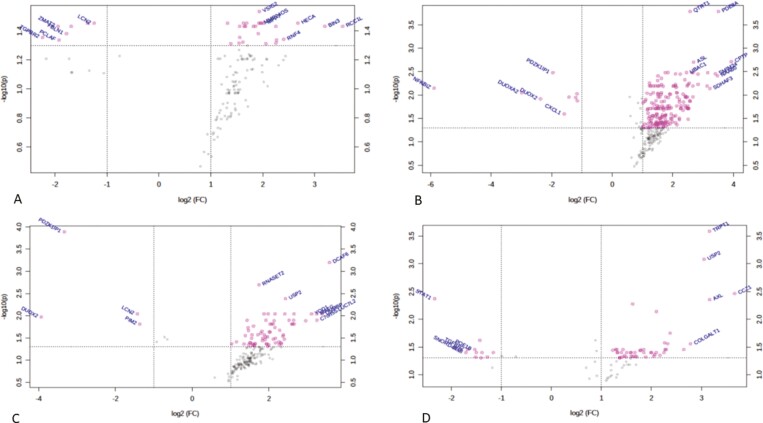
As shown by the volcano plots in Figure 5 these included genes relevant to TGF- β signaling such as TGFBR2, PDZK1IP1, USP2, and YOD1 and macrophage recruitment into tissues such as RNASET2, neutrophil and plasma cell function RNF4 and PIM2, and tumor suppressor genes HECA and BIN3. A number of those identified were shared by MES and ECSS defined healing, in addition to histological criteria defined healing (NHI and RHI).
FIGURE 5.
Volcano plot representations of the differentially expressed genes in healed vs nonhealed mucosa as defined by (A) ECSS, (B) Mayo, (C) NHI, and (D) RHI scores.
Gene Enrichment Analysis
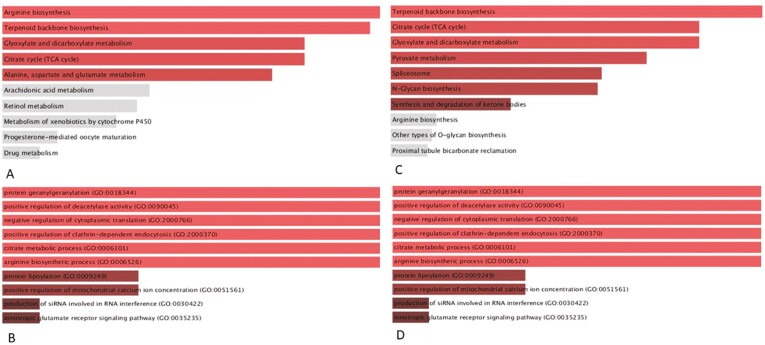
To investigate the roles of the gene pathways that might be involved in the healing process, gene enrichment analysis on the overlapping genes with KEGG and gene-ontology (GO) was used. Supplementary Tables 5 and 6 summarize the enriched pathways (P < 0.05) identified by KEGG and GO biological process analysis, respectively, for the differentially expressed genes as defined by each score. Those identified for the 25 differentially expressed genes that were commonly upregulated according to ECSS vs RHI scores and the 79 genes commonly upregulated between ECSS and NHI scores are illustrated in Figure 6 and summarized in Supplementary Table 7. In summary, the constitutive androstane receptor (CAR) and pregnane x receptor (PXR) pathways—in addition to the metabolic pathways related to aminoacids, terpinoid, and andarachidonic acid and the tricarboxylic acid (TCA) cycle were identified and considered mechanistically plausible. The biological relevance of these pathways to MH is presented in the discussion.
FIGURE 6.
GO and KEGG pathway enrichment analysis of commonly upregulated genes in healed mucosa defined by ECSS and RHI scores (A and B) and ECSS and NHI scores (C and D). Pathways for which P < 0.05 are shaded red.
DISCUSSION
Both ER and HR are considered therapeutic targets to prevent long-term complication in IBD. However, ER may not always translate accurately to HR, especially when previous generation of endoscopes is used. Although with the current generation of HD and VCE endoscopes, discrepancy between endoscopy and histology is smaller.17 Advances in endoscopy technologies have dramatically improved the way to assess the intestinal mucosa, allowing in vivo microstructural mucosal features to be visualized. The endocytoscope, although only available in limited number of centers, currently operates in a similar way to a standard endoscope and can be switched to electronic chromoendoscopy and ultra-high magnification in vivo microscopic mode at the press of a button. For ultra-high magnification, methylene blue spray is required, a technique familiar to gastroenterologists for dye spray endoscopy.
In this study, we explored whether the latest generation endocytoscope with ultra-high magnification can accurately assess subtle inflammatory changes in the colonic mucosa and better determine HR of patients in UC. We confirmed that mucosal ER defined by the latest-generation endocytoscope does correlate strongly with HR in UC. The best value of ECSS for predicting HR with RHI and NHI was ≤3. We further sought to detect the best predictors among each endocytoscope score items for their ability to differentiate between MH and mild inflammation. The best predictors were the distance between crypts and total ECSS (Table 4). Similarly, Natazako et al showed a good diagnostic accuracy of ECSS, with sensitivity of 77% (95% CI, 59–89), specificity of 97% (95% CI, 83–99) ,and accuracy of 86 % (95% CI, 75–93) to predict HR in patients with UC.22 Bessho et al developed the first ECSS score and showed good correlation of each item and histopathological grade.25 Therefore, this study also confirmed that endocytoscopy features such as crypt architecture, distance between crypts, cellular infiltration, and visibility of microvessels at endoscopy were strongly correlated with histological scores. Furthermore, each of these features could accurately predict histology defined using the validated scores such as RHI and NHI for UC. (Tables 4–6).
Of note, Maeda et al described a computer-aided diagnosis (CAD) endocytoscopy system to predict persistent histologic activity.46 It is likely that CAD diagnosis may also be related to long-term clinical prognoses. However, this requires a prospective longitudinal follow-up study with specific therapies. We have recently reported that, even with high definition and electronic chromoendoscopy, endoscopic remission and histologic remission equally predict clinical outcomes at 1 year.36 Thus endoscopy is getting closer to histology and AI might enable efficient use of endocytoscopy with minimal training and time.
We also investigated if the introduction of soluble markers in blood could provide a noninvasive method to predict HR in UC. We found that serum levels of BDNF + MIP-1α predicted HR defined by histological scores of RHI and NHI. Leukemia inhibitory factor and sVCAM1 showed higher values associated with active disease by histology. Interestingly, BDNF and MIP-1α have been associated with healing in different tissues and derived from specific subsets of macrophages and plasma cells and so could be mechanistically relevant.
In this study, sVCAM-1 concentration was related to HR in UC. A mechanistic explanation might be that mucosal VCAM-1 adheres to monocyte-expressed α4β7 integrin and directs in vivo gut homing.47 This facilitates recruitment of subtypes of macrophages that have been identified as important in restitution and repair in IBD, and hence, soluble markers such as sVCAM and MIP-1α are of interest.30 In the future, this may minimize and avoid invasive procedures to monitor response to therapy and assess MH. However, in this study soluble markers were not compared with fecal calprotectin. Recently, a new Monitr serological test was developed to assess mucosal inflammation by evaluating 13 biomarkers in CD patients.27, 48 There is no obvious overlap between this panel in CD and our panel in UC, except VCAM and further large studies are required to replicate and validate these preliminary findings.
A panel of cytokines and soluble proteins from mucosal biopsy cultures that could predict MH by endoscopic, endocytoscopic, and histologic criteria was also identified (Table 4). This was aimed at mechanistic assessment of MH and the process of mucosal restitution and repair. Several of these molecules (IL-1Ra, TNFR2) are associated with healing by antagonising inflammatory cytokines.49, 50 Matrix metalloproteinase (MMP) 9 has been demonstrated to be involved in wound healing and angiogenesis. Matrix metalloproteinase has been shown to be involved in intestinal healing in mice. It has also been implicated in inflammation, though blocking MMP9 did not improve active UC.51 Matrix metalloproteinases including MMP9 play a role in tissue remodeling, but further studies are required regarding MMP9 in MH.51, 52 Notably, MMP9 expression regulates epithelial barrier function, as evident by decreased paracellular permeability and reorganization of claudins; it also acts as a tumor suppressor in colitis-associated cancer by sustaining the epithelial mucosal integrity through/ the activation of EGFR-Sp1 signaling pathway.52
Hepatocyte growth factor (HGF) is a paracrine multifunctional protein involved in angiogenesis and regeneration of tissues.53
It is of note that sVCAM-1 was associated with HR in both the serum and biopsy culture supernatants.
RNA-seq analysis identified genes expressed in colonic biopsies that discriminated between healed and nonhealed mucosa and overlapped gene expression between ECSS defined healing and histology defined healing (Figs 4–5). These gene expression profiles relate to a number of metabolic pathways involved in vital tissue functions that may be damaged in inflammation related to tissue destruction (Fig. 6 and Supplementary Table 7) or involved in intestinal restitutive and repair processes triggered by damage.
Partial least squares discriminant analysis, volcano plot, gene enrichment, GO analysis, and KEGG analysis identified a number of genes and pathways that are biologically plausible to be involved in MH. It is interesting that a number of tumor suppressor genes are upregulated in healed mucosa (HECA, BIN3) compared with nonhealed inflamed mucosa. This may be relevant to dysplasia risk. In this study, the TGF-β pathway, neutrophil function, and macrophage recruitment were identified as important in defining healing by histological indices (RHI and NHI) and by ECSS. (Fig. 5). In addition, biological processes and pathway enrichment (GO, KEGG, and WIKI) identified a number of molecular functions and pathways that may be relevant for MH including the pregane X receptor (PXR)-JNK axis, which has been shown to be involved in healing in other organ such as skin.54 Constitutive androstane receptor (CAR) pathway regulates intestinal mucosal response to injury55 in mice. Activation of PXR, which is a close relative of CAR, may also enhance intestinal epithelial wound healing.56 Therefore, a small molecule inhibitor of the xenobiotic receptor CAR (eg, CINPA157) may be useful in MH and needs further translational studies. In addition to these pathways, this study identified several metabolic pathways including those involving arachidonic acid, amino acids, terpenoid biosynthesis, and the TCA cycle, whose involvement in healing is already recognized.51, 53, 54 Interestingly, the involvement of amino acids histidine and arginine in intestinal cell restitution may also involve TGF-β pathways.58 Transforming growth factor-β promotes protein translation at least in part by increasing the mitochondrial oxidation of glucose and glutamine carbons to support the energy demand of translation. In addition to stimulating the entry of glucose and glutamine carbon into the TCA cycle, TGFβ induced the biosynthesis of proline from glutamine in a Smad4-dependent fashion.59 Oxidoreductase activity is an important molecular function in intestinal healing due to intestinal damage resulting from neutrophil-derived reactive oxygen.60 Terpenoids are bioactive and can help anchor proteins to cell membranes and are shown to affect wound healing.61
The strengths of this study include the use of ultra-high magnification in in vivo microscopy to identify ultrastructural features of ER, the application of 2 validated histology scores (RHI and NHI), the study of molecular markers from mucosal biopsy culture, and the RNA sequencing analysis of mucosal biopsies and bioinformatics to identify potential molecules and molecular pathways that are relevant. In addition, this study has examined potential molecular pathways and soluble markers that might be relevant to the healing process and therefore indicators of healing or targets for therapy. The MES acted as a comparator of a routinely used endoscopy score in clinical practice, which poorly correlated with ECSS.
Limitations of the study include a relatively small number of patients giving large confidence intervals for results; thus validation in a larger independent cohort is needed. There were no fecal calprotectin assays from any patients, as this was not an aim for this study, but this biomarker is reported extensively in the context of advanced endoscopies recently.62 This study did not investigate CD patients, though mucosal biopsies may not represent molecular events in CD and UC. Of note, there was no focus on the mechanistic effect of the selected genes on the mucosal architecture because it was beyond the scope of this exploratory study; it will require minigut organoids, molecular imaging, and animal models using conditional gene knockdowns—a task for future studies.
In conclusion, ultra-high magnification endocytoscopy score strongly correlated with either RHI or NHI but not with MES. A number of soluble markers were identified, which could predict HR, including molecules such as sVCAM1, which was elevated in both peripheral blood and mucosal biopsy cultures. RNA transcriptomics analysis identified differentially expressed genes that were shared between ECSS and healing defined by histological score.
Supplementary Material
Glossary
Abbreviations
- IBD
inflammatory bowel disease
- UC
ulcerative colitis
- CD
Crohn’s disease
- MH
mucosal healing
- MES
Mayo endoscopic score
- HH
histological healing
- ER
endoscopic remission
- HR
histological remission
- HD
WLE, high definition white light endoscopy
- NBI
narrow banding imaging
- ECSS
endocytoscope scoring system
- UCEIS
ulcerative colitis endoscopic index score
- SES-CD
simple endoscopic score for CD
- PICaSSO
Paddington International virtual chromoendoscopy score
- CDAI
Crohn’s disease activity index
- HBI
Harvey–Bradshaw index
- RHI
Robarts Histological Index
- LP
lamina propria
- GO
gene ontogeny
- KEGG
kyoto encyclopedia of genes and genomes
- AUC
area under curve
- AUROC
area under receiver operating characteristic curve
- PLS-DA
partial least squares discriminant analysis
- PCA
principal component analysis
- VIP
variable importance projection
- VF
visual field
Author Contribution: MI and SG contributed to the study conception and design. MI, SS, OMN, US, DZ, and RC contributed to data acquisition. MI, AA, LJ, OMN, AB, JW, GK, and SG contributed to the analysis and interpretation of data. AA and AB contributed to the biostatistical analysis. MI, OMN, LJ, AA, and SG drafted the article. MI, AA, LJ, OMN, SS, US, JW DZ, RC, GK, and SG critically revised the manuscript. MI, AA, LJ, OMN, SS,.US, JW, DZ, RC, GK, and SG gave final approval of the manuscript.
Supported by: MI and SG are funded by the NIHR Birmingham Biomedical Research Centre at the University Hospitals Birmingham NHS Foundation Trust and the University of Birmingham. AA was supported by the National Institute for Health Research (NIHR) Surgical Reconstruction and Microbiology Research Centre (SRMRC), Birmingham, UK. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health. Olympus has kindly loaned an endocytoscope for this study.
Conflicts of Interest: None of the other authors have any conflict of interest to declare related to this manuscript.
REFERENCES
- 1. Agrawal M, Colombel JF. Treat-to-target in inflammatory bowel diseases, what is the target and how do we treat? Gastrointest Endosc Clin N Am. 2019;29:421–436. [DOI] [PubMed] [Google Scholar]
- 2. Peyrin-Biroulet L, Sandborn W, Sands BE, et al. Selecting therapeutic targets in inflammatory bowel disease (STRIDE): determining therapeutic goals for treat-to-target. Am J Gastroenterol. 2015;110:1324–1338. [DOI] [PubMed] [Google Scholar]
- 3. Pineton de Chambrun G, Blanc P, Peyrin-Biroulet L. Current evidence supporting mucosal healing and deep remission as important treatment goals for inflammatory bowel disease. Expert Rev Gastroenterol Hepatol. 2016;10:915–927. [DOI] [PubMed] [Google Scholar]
- 4. Baert F, Moortgat L, Van Assche G, et al. ; Belgian Inflammatory Bowel Disease Research Group; North-Holland Gut Club . Mucosal healing predicts sustained clinical remission in patients with early-stage Crohn’s disease. Gastroenterology. 2010;138:463–468; quiz e10. [DOI] [PubMed] [Google Scholar]
- 5. Shah SC, Colombel JF, Sands BE, et al. Mucosal healing is associated with improved long-term outcomes of patients with ulcerative colitis: a systematic review and meta-analysis. Clin Gastroenterol Hepatol. 2016;14:1245–1255 e1248. [DOI] [PubMed] [Google Scholar]
- 6. Shah SC, Colombel JF, Sands BE, et al. Systematic review with meta-analysis: mucosal healing is associated with improved long-term outcomes in Crohn’s disease. Aliment Pharmacol Ther. 2016;43:317–333. [DOI] [PubMed] [Google Scholar]
- 7. Bryant RV, Winer S, Travis SP, et al. Systematic review: histological remission in inflammatory bowel disease. Is ‘complete’ remission the new treatment paradigm? An IOIBD initiative. J Crohns Colitis. 2014;8:1582–1597. [DOI] [PubMed] [Google Scholar]
- 8. Bryant RV, Burger DC, Delo J, et al. Beyond endoscopic mucosal healing in UC: histological remission better predicts corticosteroid use and hospitalisation over 6 years of follow-up. Gut. 2016;65:408–414. [DOI] [PubMed] [Google Scholar]
- 9. Frøslie KF, Jahnsen J, Moum BA, et al. ; IBSEN Group . Mucosal healing in inflammatory bowel disease: results from a Norwegian population-based cohort. Gastroenterology. 2007;133:412–422. [DOI] [PubMed] [Google Scholar]
- 10. Iacucci M, Fort Gasia M, Hassan C, et al. Complete mucosal healing defined by endoscopic Mayo subscore still demonstrates abnormalities by novel high definition colonoscopy and refined histological gradings. Endoscopy. 2015;47:726–734. [DOI] [PubMed] [Google Scholar]
- 11. Narang V, Kaur R, Garg B, et al. Association of endoscopic and histological remission with clinical course in patients of ulcerative colitis. Intest Res. 2018;16:55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Park S, Abdi T, Gentry M, Laine L. Histological disease activity as a predictor of clinical relapse among patients with ulcerative colitis: systematic review and meta-analysis. Am J Gastroenterol. 2016;111:1692–1701. [DOI] [PubMed] [Google Scholar]
- 13. Ozaki R, Kobayashi T, Okabayashi S, et al. Histological risk factors to predict clinical relapse in ulcerative colitis with endoscopically normal mucosa. J Crohns Colitis. 2018;12:1288–1294. [DOI] [PubMed] [Google Scholar]
- 14. Christensen B, Erlich J, Gibson PR, et al. Histological healing is associated with decreased clinical relapse in patients with ileal Crohn’s disease. Gastroenterology. 2018;154(6):S-128–S-129. [Google Scholar]
- 15. Nardone OM, Cannatelli R, Zardo D, et al. Can advanced endoscopic techniques for assessment of mucosal inflammation and healing approximate histology in inflammatory bowel disease? Therap Adv Gastroenterol. 2019;12:1756284819863015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Iacucci M, Furfaro F, Matsumoto T, et al. Advanced endoscopic techniques in the assessment of inflammatory bowel disease: new technology, new era. Gut. 2019;68:562–572. [DOI] [PubMed] [Google Scholar]
- 17. Iacucci M, Cannatelli R, Gui X, et al. Assessment of endoscopic healing by using advanced technologies reflects histological healing in ulcerative colitis. J Crohns Colitis. 2020;14:1282–1289. [DOI] [PubMed] [Google Scholar]
- 18. Iacucci M, Smith SCL, Bazarova A, et al. OP26 The first real-life multicentre prospective validation study of the electronic chromoendoscopy score (Paddington International Virtual ChromoendoScopy ScOre) and its outcome in ulcerative colitis. J Crohns Colitis. 2020;14/S1. [Google Scholar]
- 19. Iacucci M, Smith SCL, Bazarova A, et al. The first real-life multicentre prospective validation study of the electronic chromoendoscopy score (Paddington International Virtual ChromoendoScopy ScOre) and its outcome in ulcerative colitis. Gastrointest Endosc. 2020;91:A1–A686. [DOI] [PubMed] [Google Scholar]
- 20. Takenaka K, Ohtsuka K, Fujii T, et al. Development and validation of a deep neural network for accurate evaluation of endoscopic images from patients with ulcerative colitis. Gastroenterology. 2020;158:2150–2157. [DOI] [PubMed] [Google Scholar]
- 21. Le Berre C, Sandborn WJ, Aridhi S, et al. Application of artificial intelligence to gastroenterology and hepatology. Gastroenterology. 2020;158:76–94.e2. [DOI] [PubMed] [Google Scholar]
- 22. Nakazato Y, Naganuma M, Sugimoto S, et al. Endocytoscopy can be used to assess histological healing in ulcerative colitis. Endoscopy. 2017;49:560–563. [DOI] [PubMed] [Google Scholar]
- 23. Nishiyama S, Oka S, Tanaka S, et al. Clinical usefulness of endocytoscopy in the remission stage of ulcerative colitis: a pilot study. J Gastroenterol. 2015;50:1087–1093. [DOI] [PubMed] [Google Scholar]
- 24. Ueda N, Isomoto H, Ikebuchi Y, et al. Endocytoscopic classification can be predictive for relapse in ulcerative colitis. Medicine (Baltimore). 2018;97:e0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bessho R, Kanai T, Hosoe N, et al. Correlation between endocytoscopy and conventional histopathology in microstructural features of ulcerative colitis. J Gastroenterol. 2011;46:1197–1202. [DOI] [PubMed] [Google Scholar]
- 26. Nardone OM, Shivaji UN, Ferruzza V, et al. Soluble blood markers of mucosal healing in inflammatory bowel disease: the future of noninvasive monitoring. Inflamm Bowel Dis. 2020;26:961–969. [DOI] [PubMed] [Google Scholar]
- 27. D’Haens G, Kelly O, Battat R, et al. Development and validation of a test to monitor endoscopic activity in patients with Crohn’s disease based on serum levels of proteins. Gastroenterology. 2020;158:515–526.e10. [DOI] [PubMed] [Google Scholar]
- 28. Verstockt B, Verstockt S, Veny M, et al. Expression levels of 4 genes in colon tissue might be used to predict which patients will enter endoscopic remission after vedolizumab therapy for inflammatory bowel diseases. Clin Gastroenterol Hepatol. 2020;18:1142–1151.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Arijs I, De Hertogh G, Lemmens B, et al. Effect of vedolizumab (anti-α4β7-integrin) therapy on histological healing and mucosal gene expression in patients with UC. Gut. 2018;67:43–52. [DOI] [PubMed] [Google Scholar]
- 30. Ho GT, Cartwright JA, Thompson EJ, et al. Resolution of inflammation and gut repair in IBD: translational steps towards complete mucosal healing. Inflamm Bowel Dis. 2020;26:1131–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dhanda AD, Creed TJ, Greenwood R, et al. Can endoscopy be avoided in the assessment of ulcerative colitis in clinical trials? Inflamm Bowel Dis. 2012;18:2056–2062. [DOI] [PubMed] [Google Scholar]
- 32. Schroeder KW, Tremaine WJ, Ilstrup DM. Coated oral 5-aminosalicylic acid therapy for mildly to moderately active ulcerative colitis. A randomized study. N Engl J Med. 1987;317:1625–1629. [DOI] [PubMed] [Google Scholar]
- 33. Travis SP, Schnell D, Krzeski P, et al. Developing an instrument to assess the endoscopic severity of ulcerative colitis: the Ulcerative Colitis Endoscopic Index of Severity (UCEIS). Gut. 2012;61:535–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Trivedi PJ, Kiesslich R, Hodson J, et al. The Paddington International Virtual Chromoendoscopy Score in ulcerative colitis exhibits very good inter-rater agreement after computerized module training: a multicenter study across academic and community practice (with video). Gastrointest Endosc. 2018;88:95–106.e2. [DOI] [PubMed] [Google Scholar]
- 35. Iacucci M, Daperno M, Lazarev M, et al. Development and reliability of the new endoscopic virtual chromoendoscopy score: the PICaSSO (Paddington International Virtual ChromoendoScopy ScOre) in ulcerative colitis. Gastrointest Endosc. 2017;86:1118–1127.e5. [DOI] [PubMed] [Google Scholar]
- 36. Iacucci M, Smith SC, Bazarova A, et al. An international multicenter real-life prospective study of electronic chromoendoscopy score PICaSSO in Ulcerative Colitis. Gastroenterology. 2021;S0016–5085(20)35563–3. Online ahead of print. [DOI] [PubMed] [Google Scholar]
- 37. Marchal-Bressenot A, Salleron J, Boulagnon-Rombi C, et al. Development and validation of the Nancy histological index for UC. Gut. 2017;66:43–49. [DOI] [PubMed] [Google Scholar]
- 38. Klenske E, Atreya R, Hartmann A, et al. Magnification endoscopy with optical chromoendoscopy shows strong correlation with histologic inflammation in patients with inflammatory bowel disease. Endosc Int Open. 2019;7:E1018–E1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pai RK, Khanna R, D’Haens GR, et al. Definitions of response and remission for the Robarts Histopathology Index. Gut. 2019;68:2101–2102. [DOI] [PubMed] [Google Scholar]
- 40. Garthwaite PH. An interpretation of partial least squares. J Am Stat Assoc. 1994;89:122–127. [Google Scholar]
- 41. Wold H. Estimation of Principal Components and Related Models by Iterative Least Squares. New York: Academic Press; 1966:391–420. [Google Scholar]
- 42. Li B, Ruotti V, Stewart RM, et al. RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics. 2010;26:493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen EY, Tan CM, Kou Y, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Maeda Y, Kudo SE, Mori Y, et al. Fully automated diagnostic system with artificial intelligence using endocytoscopy to identify the presence of histologic inflammation associated with ulcerative colitis (with video). Gastrointest Endosc. 2019;89:408–415. [DOI] [PubMed] [Google Scholar]
- 47. Schleier L, Wiendl M, Heidbreder K, et al. Non-classical monocyte homing to the gut via α4β7 integrin mediates macrophage-dependent intestinal wound healing. Gut. 2020;69:252–263. [DOI] [PubMed] [Google Scholar]
- 48. Sandborn WJ, Abreu MT, Dubinsky MC. A noninvasive method to assess mucosal healing in Patients* with Crohn’s disease. Gastroenterol Hepatol (N Y). 2018;14:1–12. [PMC free article] [PubMed] [Google Scholar]
- 49. Bersudsky M, Luski L, Fishman D, et al. Non-redundant properties of IL-1α and IL-1β during acute colon inflammation in mice. Gut. 2014;63:598–609. [DOI] [PubMed] [Google Scholar]
- 50. Bradford EM, Ryu SH, Singh AP, et al. Epithelial TNF receptor signaling promotes mucosal repair in inflammatory bowel disease. J Immunol. 2017;199:1886–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Neumann PA, Twardy V, Becker F, et al. Assessment of MMP-2/-9 expression by fluorescence endoscopy for evaluation of anastomotic healing in a murine model of anastomotic leakage. PLoS One. 2018;13:e0194249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pujada A, Walter L, Patel A, et al. Matrix metalloproteinase MMP9 maintains epithelial barrier function and preserves mucosal lining in colitis associated cancer. Oncotarget. 2017;8:94650–94665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fajardo-Puerta AB, Mato Prado M, Frampton AE, et al. Gene of the month: HGF. J Clin Pathol. 2016;69:575–579. [DOI] [PubMed] [Google Scholar]
- 54. Tanaka Y, Uchi H, Ito T, et al. Indirubin-pregnane X receptor-JNK axis accelerates skin wound healing. Sci Rep. 2019;9:18174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hudson GM, Flannigan KL, Erickson SL, et al. Constitutive androstane receptor regulates the intestinal mucosal response to injury. Br J Pharmacol. 2017;174:1857–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Terc J, Hansen A, Alston L, et al. Pregnane X receptor agonists enhance intestinal epithelial wound healing and repair of the intestinal barrier following the induction of experimental colitis. Eur J Pharm Sci. 2014;55:12–19. [DOI] [PubMed] [Google Scholar]
- 57. Cherian MT, Chai SC, Wright WC, et al. CINPA1 binds directly to constitutive androstane receptor and inhibits its activity. Biochem Pharmacol. 2018;152:211–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Matsui T, Ichikawa H, Fujita T, et al. Histidine and arginine modulate intestinal cell restitution via transforming growth factor-β1. Eur J Pharmacol. 2019;850:35–42. [DOI] [PubMed] [Google Scholar]
- 59. Schwörer S, Berisa M, Violante S, et al. Proline biosynthesis is a vent for TGFβ-induced mitochondrial redox stress. EMBO J. 2020;39:e103334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Matthews JD, Owens JA, Naudin CR, et al. Neutrophil-derived reactive oxygen orchestrates epithelial cell signaling events during intestinal repair. Am J Pathol. 2019;189:2221–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Barreto RS, Albuquerque-Júnior RL, Araújo AA, et al. A systematic review of the wound-healing effects of monoterpenes and iridoid derivatives. Molecules. 2014;19:846–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cannatelli R, Bazarova A, Zardo D, et al. Fecal calprotectin thresholds to predict endoscopic remission using advanced optical enhancement techniques and histological remission in IBD patients. Inflamm Bowel Dis. 2021;27:647–654. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.