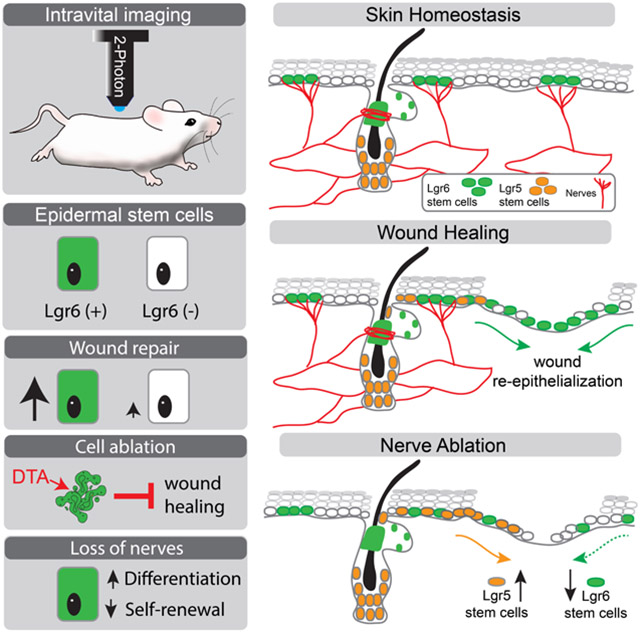
Abstract
Stem cells support the lifelong maintenance of adult organs but their specific roles during injury are poorly understood. Here, we demonstrate that Lgr6 marks a regionally restricted population of epidermal stem cells that interact with nerves and specialize in wound re-epithelialization. Diphtheria toxin-mediated ablation of Lgr6 stem cells delays wound healing, and skin denervation phenocopies this effect. Using intravital imaging to capture stem cell dynamics after injury, we show that wound re-epithelialization by Lgr6 stem cells is diminished following the loss of nerves. This induces the recruitment of other stem cell populations, including hair follicle stem cells, which partially compensate to mediate the wound closure. Single-cell lineage tracing and gene expression analysis reveal that the fate of Lgr6 stem cells is shifted towards differentiation following the loss of their niche. We conclude that Lgr6 epidermal stem cells are primed for injury response and interact with nerves to regulate their fate.
Graphical abstract
eTOC blurb
Huang et al. use intravital imaging to analyze the dynamics of discrete populations of stem cells during skin injury. This study shows that the epidermis contains a population of stem cells that specialize in wound healing and depend on nerves for regulating their fate.
Introduction
Long-term maintenance of adult tissues and their regeneration after injury are driven by resident stem cells. In the skin, the continuous replenishment of the stratified epidermis and the periodic regeneration of hair follicles and other appendages are fueled by populations of stem cells, which are organized in compartmentalized niches within the tissue (Rompolas and Greco, 2014; Gonzales and Fuchs, 2017; Belokhvostova et al., 2018). Despite their stereotypic and regionally restricted behaviors during skin homeostasis, stem cells can exhibit high plasticity and contribute to the regeneration of other epithelial compartments after injury (Schepeler, Page and Jensen, 2014; Dekoninck and Blanpain, 2019). In this study, we investigate the requirement, contribution, and regulation of distinct populations of epidermal stem cells to wound re-epithelialization.
Members of the leucine-rich repeat-containing family of G-protein coupled receptors (LGRs) are known to be preferentially expressed in stem cells of various adult organs and tissues, including simple and stratified epithelia (Barker et al., 2007; Jaks et al., 2008; Snippert et al., 2010; Barker, Tan and Clevers, 2013; Lehoczky and Tabin, 2015). In the skin, Lgr5 and Lgr6 mark specific stem cell niches in the hair follicle (Jaks et al., 2008; Snippert et al., 2010). Notably, Lgr6 also marks a sub-population of basally located stem cells in the interfollicular epidermis, whose function has not been resolved (Füllgrabe et al., 2015). Lineage tracing studies demonstrate that during homeostasis, Lgr5 and Lgr6 stem cells exhibit local clonal dynamics and contribute to self-renewing and differentiated lineages exclusively within their respective compartments (Rompolas and Greco, 2013; Füllgrabe et al., 2015). However, following injury, stem cells from hair follicles are known to exit their niche and contribute to the re-epithelialization of the epidermis (Taylor et al., 2000; Ito et al., 2005; Levy et al., 2007; Nowak et al., 2008; Schepeler, Page and Jensen, 2014; Vagnozzi, Reiter and Wong, 2015; Donati et al., 2017; Joost et al., 2018). While hair follicle stem cells are largely dispensable for the eventual healing of incisional wounds, the absence of hair appendages greatly affects the dynamics of wound re-epithelialization (Langton, Herrick and Headon, 2008; Garcin et al., 2016). This suggests that the tissue regulates the recruitment of stem cells from diverse niches by modulating their fate to coordinate the wound healing process. In this study, we set out to test this hypothesis by investigating the role of the epidermal pool of stem cells marked by Lgr6, in skin regeneration and wound healing.
Functionally discrete compartments in the skin are characterized by specific niche microenvironments that establish the identity of their resident stem cells and regulate their distinct dynamics (Gonzales and Fuchs, 2017). In homeostasis, these niches are crucial to supporting the efficient replenishment of epidermal structures and stereotypic growth of appendages (Rompolas and Greco, 2014). The collapse of boundaries between compartments and the radical changes in stem cell activity are phenomena observed after injury. These are in great part brought about by the drastic alterations in the tissue microenvironment, highlighting the importance of the niche as a crucial regulator of stem cell fate (Schepeler, Page and Jensen, 2014; Vagnozzi, Reiter and Wong, 2015; Donati et al., 2017; Joost et al., 2018). The skin harbors a wide assortment of cell types that are key constituents of the stem cell niche microenvironment. The cues and interactions that these niche factors provide are critical for the regulation of stem cell function in homeostasis but their role during wound healing remains largely unexplored.
Most adult organs are richly innervated and peripheral nerves constitute integral components of tissue microenvironments. In the skin, nerves directly interact with keratinocytes to relay sensory information to the central nervous system (Ansel et al., 1997; Lumpkin and Caterina, 2007; Talagas et al., 2018). However, this crosstalk is required for the expression of stem cell-specific genes, including Gli1 (Brownell et al., 2011). Importantly, a study by Nguyen and colleagues, demonstrated that Lgr6 expression by keratinocytes depends on skin innervation (Liao and Nguyen, 2014). The nature of the interaction between nerves and Lgr6-expressing stem cells in the skin, and their requirement for tissue regeneration and wound healing have yet to be determined.
Stem cells are characterized by their dynamic activity and nerves by their complex, 3-dimensional organization within tissues. These are major challenges in elucidating the role of epidermal Lgr6 stem cells and their association with cutaneous nerves. Moreover, the interactions between nerves and stem cells are not sufficiently resolved using routine histological approaches. To address these challenges, we developed state-of-the-art imaging tools and single-cell lineage tracing approaches to investigate the contribution of distinct stem cell populations to wound healing in real-time using intravital two-photon microscopy.
Results
Distinct dynamics of Lgr6 stem cells during wound re-epithelialization
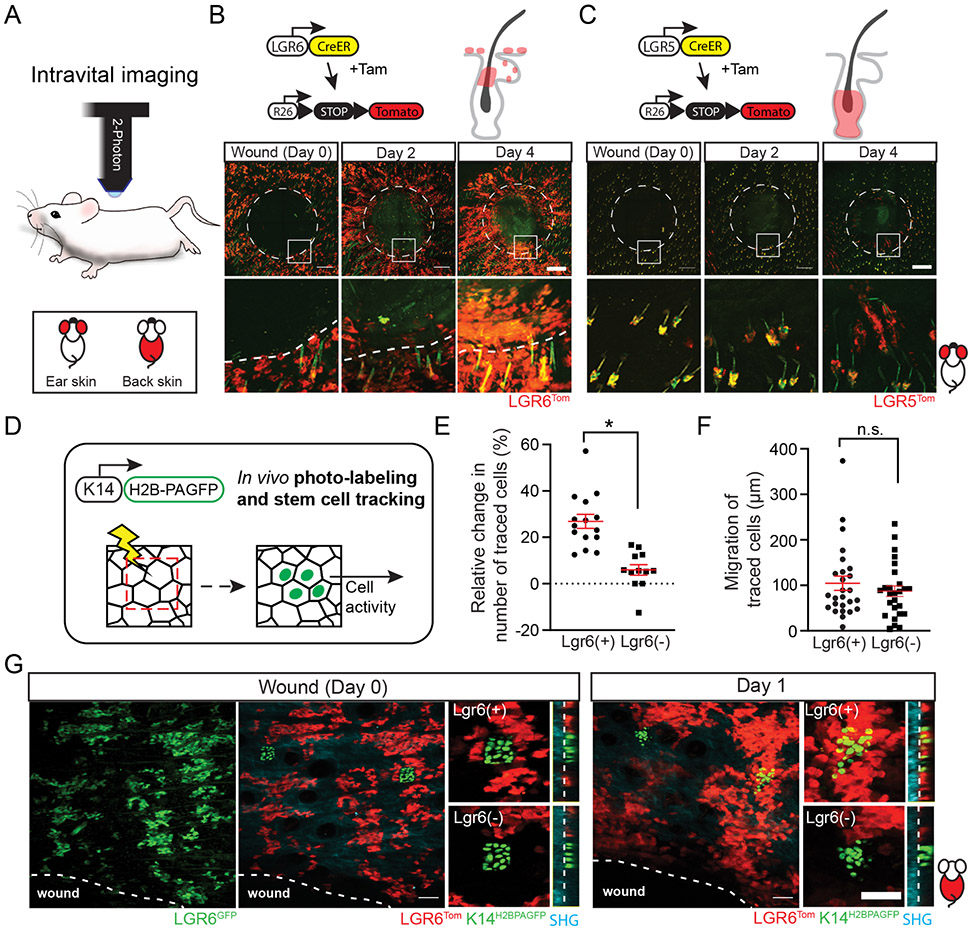
Stem cell populations with distinct localization and function during homeostasis can display plasticity and regenerate all skin structures in response to injury. In the skin, a typical wound healing response involves well defined stages of inflammation, clot formation, and re-epithelialization (Gurtner et al., 2008; Sun, Siprashvili and Khavari, 2014). Re-epithelialization of the wound bed is a critical step that allows the injured skin to be fully repaired; however, the timing and extent by which each stem cell population contributes to wound re-epithelialization has not been fully resolved. To capture the dynamics of discrete stem cell populations in the skin of live mice, we utilized previously established in vivo reporters, combined with two-photon intravital imaging (Barker et al., 2007; Snippert et al., 2010; Rompolas et al., 2012; Pineda et al., 2015; Huang and Rompolas, 2017). We confirmed that during homeostasis, Lgr6GFP and Lgr5GFP stem cells were found in mutually exclusive niches within the pilosebaceous unit (PSU) (Fig. S1A-B). Regardless of the phase of the hair cycle, Lgr5GFP stem cells were confined in the bulge and lower compartments of the hair follicle (Fig. S1A). Conversely, within the PSU, Lgr6GFP marked stem cells in the isthmus, as well as the basal layer of the sebaceous glands (Fig. S1B).
The interfollicular epidermis (IFE) was completely devoid of any Lgr5GFP stem cells, while a significant portion of basal cells in the IFE was positively marked by Lgr6GFP (Fig. S1A-B). In contrast to the hair follicle, which comprises of anatomically well-defined compartments, the IFE in the back and ear skin is characteristically uniform, seemingly lacking any structurally distinct niches (Doupé and Jones, 2012). Previous studies showed that Lgr6GFP stem cells comprised a minor fraction of the total population within the IFE (Füllgrabe et al., 2015). Using two-photon microscopy, we obtained a “bird’s eye” live view of mouse skin. We observed distinct territories of Lgr6GFP(+) and Lgr6GFP(−) stem cells within the IFE, consistent with the prior data. To test whether these territories represent defined niches of discrete stem cell populations, rather than the manifestation of transient Lgr6 expression within the IFE, we re-imaged the same areas of the live skin over the course of several weeks. We found that the topology of Lgr6GFP stem cells was relatively stable and their territories were easily identified in the epidermis across the different timepoints (Fig. S1C).
During homeostasis, Lgr6 stem cells were found to compete neutrally within the IFE (Füllgrabe et al., 2015). We hypothesized that Lgr6 epidermal stem cells may have a specific role in wound re-epithelialization. To test this, we devised a Cre-recombinase based, in vivo lineage tracing strategy to visualize distinct stem cell populations and track them in real time as they re-epithelialize the wound. After labeling stem cells with a Cre reporter (Lgr6Tom), full-thickness wounds were created; the entire area of the skin that encompasses the wound was imaged by two-photon microscopy and reconstructed at single-cell resolution using digital image tiling (Fig. 1A-C). By re-imaging the same wounds over time, we observed that Lgr6Tom stem cells were found within the wound bed two days after injury, indicating that they commenced re-epithelializing the wound soon after injury (Fig. 1B). In the same timeframe, Lgr5Tom stem cells from hair follicles around the wound had not yet exited their niches. By day four, a small number of Lgr5Tom cells were seen contributing to re-epithelializing the wound (Fig. 1C).
Figure 1. Live imaging of stem cell dynamics during wound healing.
(A) Stem cell activity during wound re-epithelialization was visualized by intravital two-photon microscopy. The legend indicates the area of the mouse skin where imaging was performed. (B - C) Genetic alleles used for lineage tracing by longitudinal live imaging. Mice were induced three days before wounding to permanently label the respective stem cell populations. (B) Lgr6Tom stem cells were found inside the re-epithelialized wound bed two days post-wounding. (C) During the same period, Lgr5Tom stem cells were still confined within their respective hair follicle niches and only started to contribute to the wound re-epithelialization 4 days after wounding. Wound diameter: 2.5 mm. (D) Genetic alleles and strategy to lineage trace epidermal stem cells by in vivo photo-labeling. Immediately after wounding, equivalent areas within the Lgr6(+) and Lgr6(−) niches were scanned to activate the H2B-PAGFP reporter. The same areas were re-imaged to track the changes in the respective stem cell population. (E, F) Quantification of the relative growth and mobility of tracked epidermal stem cells., n = 27, 3 mice, p = 0.0104 (E); n= 52, 5 mice, p = 0.4255 (F). (G) Representative example of the tracking of Lgr6(+) and Lgr6(−) epidermal stem cells with the use of photo-activatable reporter. Panels B and C (top rows) show tiled images of the mouse skin constructed from multiple fields-of-view. Scale bars: 500 μm (B, C), 100 μm (G).
Next, we investigated whether Lgr6(+) stem cells display distinct dynamics during wound re-epithelialization compared to the remaining stem cell population within the IFE. To overcome the lack of mutually exclusive genetic drivers to label Lgr6(+) and Lgr6(−) epidermal stem cells, we engineered a mouse that expresses a photo-activatable variant of GFP (K14H2BPAGFP) to specifically mark each respective population on-demand (Fig. 1D). We first treated the mice with Tamoxifen to permanently mark the Lgr6 stem cells with the Tomato Cre reporter, and subsequently photo-labeled equivalent groups of stem cells in the Lgr6(+) and Lgr6(−) areas of the epidermis, immediately after wounding. By tracking cells located at equal distance from the wound edge, we found that Lgr6(+) epidermal stem cells displayed more robust growth compared to their Lgr6(−) neighbors during the early stages of wound re-epithelialization (Fig. 1E-G). This was confirmed by a measured increase in the Lgr6Tom population, and their higher proliferation rate compared to the rest of the cells in the IFE, although the outflow of the highly proliferative Lgr6 stem cells from the hair follicle isthmus likely also contributes to this growth (Fig. S2A-D). Taken together, our data show that Lgr6 expression demarcates a distinct population of epidermal stem cells, which exhibit a pronounced response during the initial stages of wound re-epithelialization.
Ablation of Lgr6 stem cells impairs wound healing
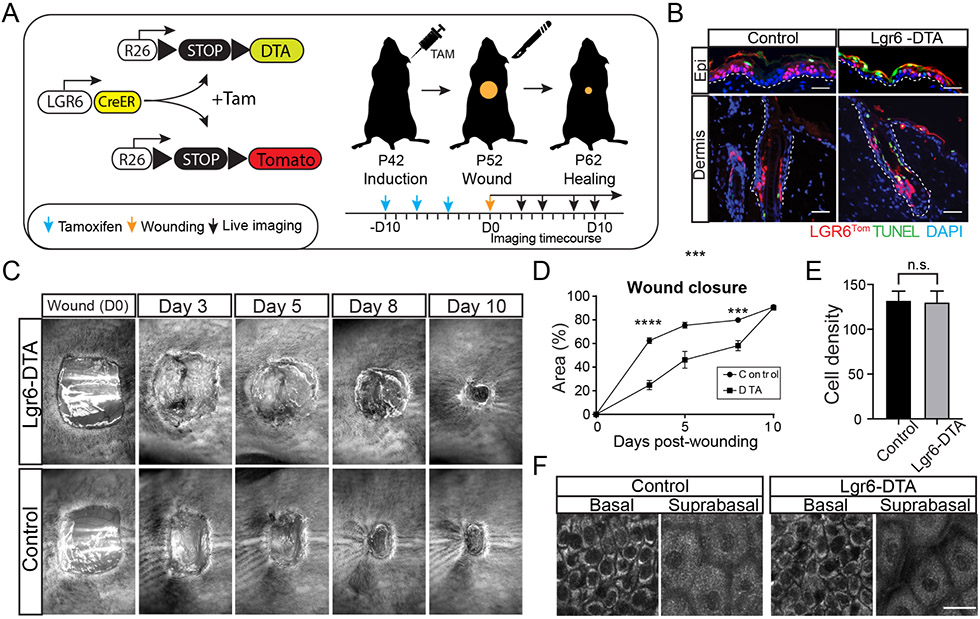
Our initial experiments suggested that epidermal Lgr6 stem cells may have a specialized role during injury. To test if this population was required for a wound healing response, we designed a genetic approach to specifically ablate Lgr6 stem cells using an inducible diphtheria toxin allele (DTA). Tamoxifen-induced activation of the Cre-recombinase resulted in DTA expression exclusively in the Lgr6 population (Fig. 2A). Terminal deoxynucleotidyl transferase nick end labeling (TUNEL) analysis of skin harvested from tamoxifen-treated mice one week after induction showed widespread cell death among the Lgr6 stem cell population (Fig. 2B). We next performed large incisional wounds on the back skin of mice with ablated (Lgr6-DTA) or intact (Control) Lgr6 stem cells and tracked the rate of wound closure. Mice with ablated Lgr6 stem cells experienced a significant delay in wound closure, especially during the first four days following injury (Fig. 2C, D).
Figure 2. Ablation of Lgr6 stem cells impairs wound healing.
(A) Schematic of the experimental design to ablate Lgr6 stem cells using an inducible diphtheria toxin-expressing allele (DTA). (B) TUNEL signal co-localizes with Lgr6Tom stem cells in Lgr6-DTA but not control mice, confirming the efficacy and specificity of the cell ablation process. The dashed line demarcates the border between the epidermis and dermis. (C) Representative images from a wound healing time course show a delay in wound closure in the skin of Lgr6-DTA mice compared to control. Wound diameter: 1 cm2. (D) Quantification of the rate of wound closure in control and Lgr6-DTA skin wounds. Statistical significance determined using an unpaired t-test method. Each timepoint was analyzed individually, without assuming a consistent S.D.; n = 14 mice, (Day 3) p < 0.0001; (Day 5)p = 0.0004, (Day 8) p = 0.0004, P = 0.5604 (Day 10); (E) Quantification of cell population density in the basal layer of the epidermis at the time prior to wounding; n = 29 from 6 mice, p = 0.8474. (F) Representative top-down views of the basal and suprabasal layers of the live epidermis, imaged using 750nm excitation wavelength to visualize all cells by endogenous autofluorescence immediately prior to wounding. Scale bars: 50 μm (B), 20 μm (F).
Previously, it was estimated that Lgr6 stem cells accounted for up to 20% of the total population in the epidermis (Füllgrabe et al., 2015). To test whether the delay in wound healing was due to the specific elimination of Lgr6 stem cells, rather than a general reduction in the number of basal progenitors, we quantified cell density in the IFE at the time of wounding. We found no differences in number of cells present in the basal or differentiated layers between the Lgr6-DTA and control mice (Fig. 2E, F). These data suggested that neighboring stem cells within the epidermis quickly compensated for the loss of Lgr6 stem cells following their DTA ablation. We hypothesized that the newly generated basal progenitors lacked an Lgr6 stem cell identity, and therefore, were unable to efficiently contribute to wound re-epithelialization. To test this, we re-imaged the same area of the skin of Lgr6-DTA mice over four weeks to monitor the elimination and possible recovery of the Lgr6GFP population (Fig. S3A, B). We found that 10 days after induction, a time equivalent to the wounding in the previous experiment, the epidermis was mostly devoid of Lgr6GFP stem cells. They however, began to gradually appear several days later (Fig. S3A). Interestingly, the re-emerging Lgr6GFP stem cells occupied similar topological niches to those present in the epidermis before the DTA ablation, suggesting that Lgr6 stem cells may be specified by the tissue microenvironment (Fig. S3A). Previous studies that analyzed the transcriptome of Lgr6(+) and Lgr6(−) stem cells during homeostasis also found no unique gene expression profiles between the two populations (Füllgrabe et al., 2015). This is consistent with our hypothesis for the regulation of Lgr6 stem cells by an extrinsic niche.
Epidermal Lgr6 stem cells physically interact with sensory nerves
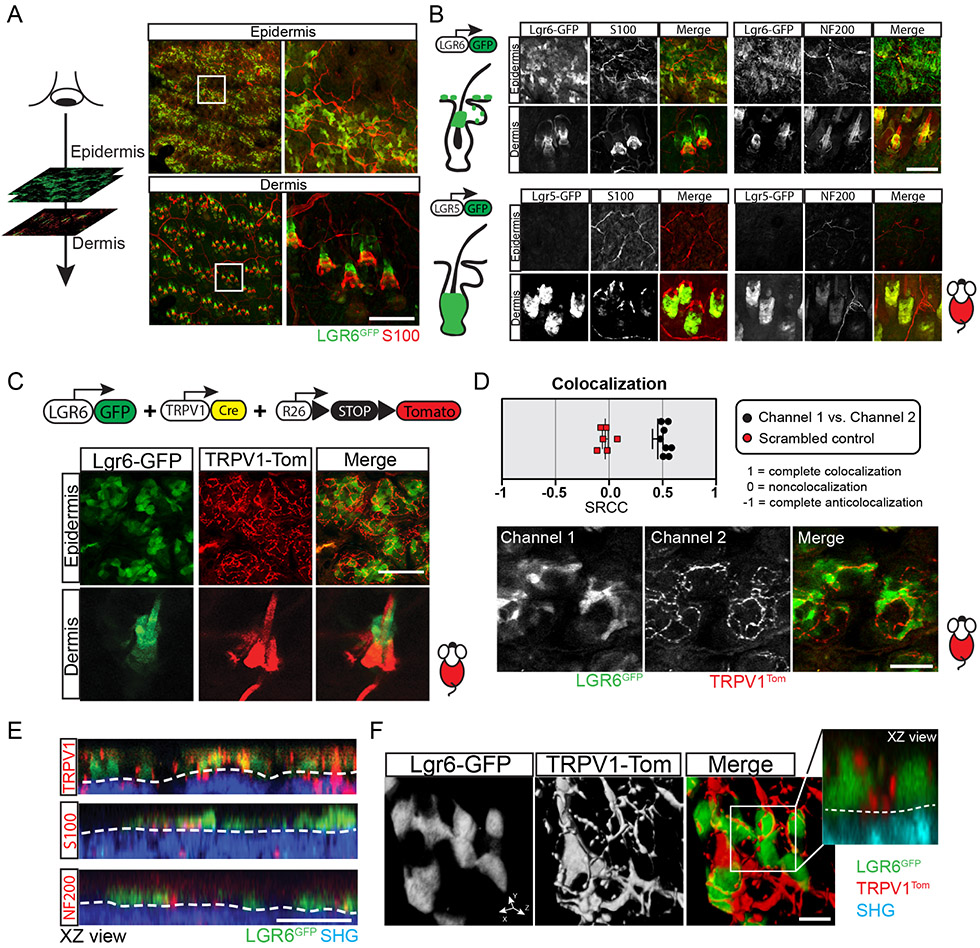
In the isthmus, which represents the specific anatomical site of hair follicle innervation, the expression of the stem cell marker Gli1, as well as expression of Lgr6, is abolished after surgical denervation of the skin (Brownell et al., 2011; Liao and Nguyen, 2014). This evidence prompted us to hypothesize that the possible interaction between Lgr6 stem cells in the IFE and cutaneous nerves may be important for their role in wound re-epithelialization. To characterize the precise localization of nerves with respect to Lgr6 stem cells, we performed high-resolution imaging and 3-dimensional analysis of whole-mounted, immuno-stained skin samples (Fig. 3A, B). Large nerve bundles that stained positive for NF200 (myelinated Aβ and Aδ axons) and S100 (Schwann cells) formed an extensive honeycomb-like network in the deeper layers of the dermis (Fig. 3A). Smaller diameter fibers emanating from these nerves were directed toward individual skin appendages and specifically wrapped around the Lgr6GFP stem cell population located in the isthmus (Fig. 3B; Movie S1). In contrast, Lgr5GFP hair follicle stem cells that occupy the bulge and secondary hair germ did not co-localize with nerves (Fig. 3B; Movie S2).
Figure 3. Colocalization of Lgr6 stem cells with sensory nerves in the epidermis.
(A) Schematic of serial optical sectioning, performed by two-photon microscopy, to provide a bird’s-eye view of the epidermis and dermis. Right panels show representative low and high magnification views of the corresponding skin compartments. (B) Examples of Lgr6GFP stem cells in the epidermis and hair follicles (dermis) and their locations are relative to the cutaneous nerves (top panels; see also Movie S1). Lgr5GFP stem cells are absent from the epidermis and do not co-localize with nerves within the hair follicles (bottom panels; see also Movie S2). (C) Genetic scheme to visualize interactions between nerves (TRPV1Tom) and Lgr6GFP stem cells by live imaging. Bottom panels show representative high magnification images of Lgr6GFP stem cells co-localizing with sensory nerves (TRPV1Tom) in the epidermis of live mice (see also Movie S3). (D) Colocalization analysis of Lgr6GFP stem cells and TRPV1Tom sensory nerve terminals within the epidermis. Spearman's rank correlation coefficient = 0.4552 (experimental), - 0.035 (control); n = 16 image samples from 3 mice. Bottom panel shows a representative example of the analyzed images. (E) Representative side views (XZ) of the mouse skin, reconstructed from serial optical sections showing the co-localization of Lgr6GFP stem cells with sensory nerves in the epidermis. Second harmonic generation (SHG), originating from the extracellular matrix, is used to demarcate the border between epidermis and dermis (white dashed line). (F) High-magnification 3D surface rendering demonstrate the physical proximity of epidermal Lgr6GFP stem cells with sensory nerves terminals (TRPV1Tom). Panel A (left column) shows tiled images of the mouse skin constructed from multiple fields-of-view. Scale bars: 100 μm (B-C), 50 μm (D-E), 10 μm (F)
To further analyze the neuro-epithelial interactions in the skin in vivo, we combined a genetic driver that labels TrpV1-Cre positive sensory afferents (TRPV1Tom) with the Lgr6GFP reporter allele to visualize both cell types in the live tissue by two-photon microscopy (Fig. 3C). We found that TRPV1Tom nerve fibers not only co-localized with the Lgr6GFP stem cells in the hair follicle but were also visible above the dermis, in physical contact with stem cells in the basal layer of the IFE (Fig. 2C). High-resolution, 3-dimensional analysis of the serial optical sections showed that TRPV1Tom nerve terminals co-localized preferentially with Lgr6GFP stem cells within the IFE (Fig. 3D-F; Movie S3). Myelinated Aβ and Aδ nerve fibers and Schwann cells were hardly detected in the interfollicular epidermis (Fig. 3E). Altogether, these data suggest that cutaneous nerves may represent a possible niche for Lgr6 stem cells, confirming previous findings (Liao and Nguyen, 2014). Consistent with this hypothesis, we observed that ablation of Lgr6 stem cells did not affect the innervation state of the skin (Fig. S3C).
Skin denervation impairs Lgr6 stem cell contribution to wound re-epithelialization
Our data so far support our hypothesis that Lgr6 stem cells may be specialized for wound response and they interact with nerves to maintain their identity. Prior work demonstrated that skin denervation alters the dynamics of wound healing, yet the mechanism remains poorly understood (Smith and Liu, 2002; Brownell et al., 2011; Buckley et al., 2012; Rinkevich et al., 2014; Alapure et al., 2018). To explore how sensory nerves affect stem cell dynamics during wound healing, we implemented a previously described surgical denervation method (Peterson et al., 2015; Peterson, Brownell and Wong, 2016). To denervate the back skin unilaterally and leave the left side intact for internal control, only the nerves emanating from the right side of spinal cord (segments T3-12) were transected (Fig. 4A; S4A, B). The mice were then allowed to recover for one month, for any transient inflammatory responses from the surgery to fully resolve. In the absence of any injury, no major functional or morphological differences were detected between the intact and denervated areas of the skin, and we found no evidence that denervation directly impeded hair growth or affected the morphology of the pilosebaceous unit (Fig. S4C-G).
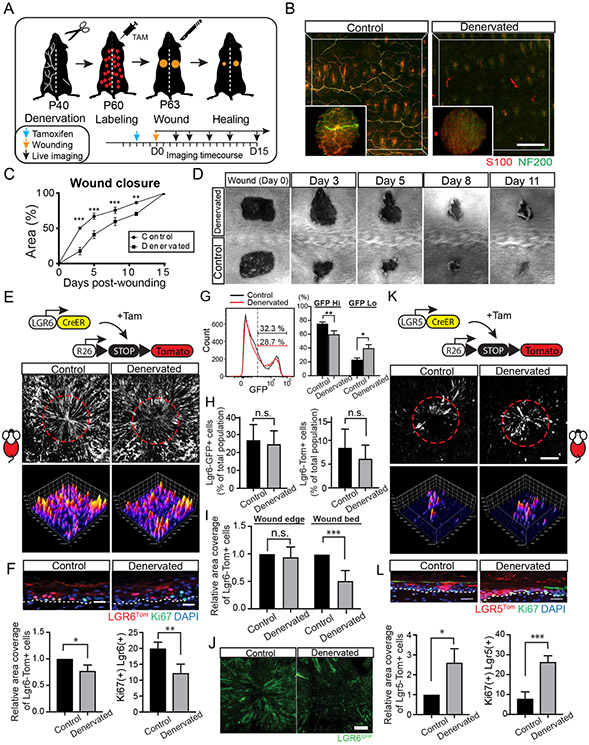
Figure 4. Denervation impairs wound re-epithelialization by Lgr6 stem cells.
(A) Schematic of the experimental design to test the effect of skin denervation to wound healing. (B) Representative low and high magnification maximum projection images of biopsies collected from the intact or denervated side of the back skin one month after denervation surgery. Whole-mount immunostaining for S100 and NF200 show the absence of nerves in the denervated side of the back skin. (C) Quantification of the wound closure rate in control and denervated skin wounds. Statistical significance was determined using a paired t-test method. Each time point was analyzed individually, without assuming a consistent S.D.; n = 7 wound pairs, (Day 3) p = 0.0004; (Day 5) p = 0.0003; (Day 8) p = 0.0006, (Day 11) p = 0.0015. (D) Representative time course of wound healing in intact and denervated back skin. Wound diameter: 0.5 cm2; two symmetrical wounds per mouse. (E) Lineage tracing of Lgr6Tom stem cells in wound healing. Full-thickness image projections (top panels) and surface plot analysis (bottom panels) of wounds taken at the end of the healing process. Wound diameter: 2.5 mm; two symmetrical wounds per mouse. (F) Top panel shows representative side views of the re-epithelialized wounds. Left graph shows quantification of Lgr6Tom signal in the re-epithelialized wounds. A paired t-test was used to measure statistical significance; n = 5 wound pairs, p = 0.0104. Right graph shows quantification of double-positive Ki67/Lgr6Tom cells in healed wounds; n = 880 (control), 618 (denervated) cells from 4 mice, p = 0.0031. (G) Histograms show the relative abundance and signal distribution of Lgr6-GFP expressing cells, analyzed by flow cytometry. Graph shows the fraction of Lgr6 stem cells with high or low GFP expression levels; n = 7 wound biopsy pairs, (GFP High) p = 0.0096; (GFP low) p = 0.0129. (H) Graphs show quantification by flow cytometry of the fraction of Lgr6GFP and Lgr6Tom labeled cells in the epidermis at the time of wounding; n = 7 wound biopsy pairs, (left graph; p = 0.148) (right graph, p = 0.23). (I) Quantification of Lgr6Tom signal at the edge of the wound (left graph; n = 5 wound pairs, p = 0.5043) and within the re-epithelialized wound bed (right graph; n = 6 wound pairs, p = 0.0001) (J) Representative examples of healed wounds show the presence of Lgr6GFP stem cells in the re-epithelialized wound bed. (K-L) Lineage tracing of Lgr5 stem cells in wound healing and equivalent quantifications as shown in panels E-F. Bottom left graph; n = 4 wound pairs, p = 0.019. Bottom right graph; n = 218 (control), 857 (denervated) cells from 4 mice, p = 0.0003. Panels B, E, J and K show tiled images of the mouse skin constructed from multiple fields-of-view. Scale bars: 200μm (B, E and K), 100μm (J), 20μm (F-L).
To analyze the effect of sensory nerve ablation on wound healing, we created full-thickness wounds of equal size in both the denervated and intact side of the back skin. Skin biopsies were concurrently collected from the wounds and evaluated by histology which validated effective nerve removal prior to wounding (Fig. 4B). Quantification of wound closure rate revealed a significant delay in the denervated side of the back skin. (Fig. 4C, D). More importantly, the rates of wound closure between the two areas showed the greatest difference during the first three days following injury, which phenocopied the observed kinetics of Lgr6 stem cell-deficient wound healing (Fig. 4C & 2D).
To this end, our data led us to hypothesize that the defect in wound healing of denervated skin may be due to an impairment in the ability of Lgr6 stem cells to re-epithelialize the wound. To test this, we tracked Lgr6Tom stem cells by live imaging and quantified their contribution to wound healing of denervated and intact skin (Fig. 4E). In intact skin, epidermal stem cells from the area surrounding the site of injury contributed to the wound re-epithelialization, generating radially oriented stripes after lineage tracing, consistent with previous work (Aragona et al., 2017; Park et al., 2017). In contrast, Lgr6Tom signal from wounds created on the denervated side of the back skin was largely diminished, and the re-epithelialized wound bed was devoid of Lgr6Tom clonal stripes (Fig. 4E, F). It was previously reported that denervation leads to a decrease in Lgr6 expression in the skin, which could directly affect the extent of Cre-induced recombination and therefore the number of labeled Lgr6Tom stem cells (Liao et al. 2014). To test this, we quantified the number of Lgr6GFP labeled cells in the denervated and control areas of the skin. While there was no significant difference in the percentage of the total basal population that expressed Lgr6, the level of GFP signal was overall decreased in denervated skin (Fig. 4G-H). This shift toward lower Lgr6-GFP expression levels did not affect the rate of Cre-recombination nor the labeling of Lgr6Tom cells before wounding (Fig. 4H). In spite of consistent labeling prior to wound healing, Lgr6Tom cell numbers were reduced in the wound bed after re-epithelialization in denervated skin (Fig. 4I). We therefore conclude that the observed phenotype was due to decreased contribution of Lgr6 stem cells during wound re-epithelialization (Fig. 4E, F, I). Furthermore, we observed that the Lgr6 cells that re-epithelialized the wound in normal skin retained GFP expression, but fewer Lgr6GFP cells were detected in the healed wound bed of denervated skin (Fig 4J). These data indicate that epidermal Lgr6 stem cells require interaction with cutaneous sensory nerves to manifest their role in wound re-epithelialization.
Compensatory recruitment of hair follicle stem cells in denervated skin during wound healing
Our experiments thus far showed that the ablation of Lgr6 stem cells impairs early stages of wound healing, and this phenomenon is phenocopied by the removal of nerves from the skin prior to injury. However, despite the initial delay, these wounds eventually heal. Remaining stem cells within the epidermis likely play a major role in compensating for the loss of their Lgr6 neighbors, and our observation that Lgr6 stem cell activity was overall reduced in absence of nerves support this (Fig. S5A-D). For example, we found that Lgr6Tom(−) cells proliferated more when compared to either Lgr6 Tom(+) cells from the same areas of the denervated skin or Lgr6Tom(−) cells from the contralateral intact skin (Fig. S5B). We then asked whether skin denervation and the subsequent impaired activity of Lgr6 stem cell have far reaching effects, affecting other stem cell populations outside the epidermis. Our earlier experiments showed that during homeostasis, Lgr5Tom stem cells from hair follicles contributed to wound re-epithelialization much later and in fewer numbers compared to Lgr6Tom cells (Fig 1C). Therefore, we hypothesized that the diminished contribution of Lgr6 stem cells to wound re-epithelialization after skin denervation might be also partially compensated by an increase in the activation of hair follicle stem cells.
First, we tested whether skin denervation affects hair follicle stem cells in the absence of any injury. By performing single cell lineage tracing and histological analysis, we found no difference in the activity or identity of hair follicle stem cells between control and denervated skin (Fig. S5E, F). Furthermore, Lgr5+ stem cells that were labeled before denervation did not exit the niche, nor moved beyond the upper bulge (Fig. S5G). We then performed lineage tracing of Lgr5Tom stem cells during wound healing to test their activity in control and denervated skin (Fig. 4K). We found that the contribution of Lgr5Tom cells to the re-epithelialized wounds increased by at least two-fold in denervated skin (Fig. 4K, L). This was mediated by an increase in the proliferation of IFE-bound progeny of Lgr5 stem cells rather than the growth of the Lgr5 stem cell pool within the hair follicle niche (Fig. 4L; Fig. S5H). Taken together, these experiments provide an insight in the intricate balancing of stem cell activity across the tissue that is influenced by the state of the niche.
The fate of epidermal Lgr6 stem cells is altered upon loss of nerve interaction
To this end, our findings indicate that sensory nerves modulate the activity of Lgr6 stem cells by regulating their stem cell identity and fate. To further explore whether Lgr6 cells maintain their identity and stem cell potential after losing their interaction with the niche, we performed in vitro experiments by isolating basal keratinocytes from Lgr6Tom mice that were previously induced for lineage tracing. Immediately after plating, all the cells that expressed the Tomato Cre reporter were also Lgr6GFP(+) (Fig. 5A). Although both Lgr6Tom(+) and Lgr6Tom(−) cells grew in culture, the former began to lose the GFP expression, supporting the idea that this was dependent on the interaction with nerves (Fig. 5A). To test whether Lgr6Tom(+) cells isolated from denervated skin have reduced growth potential, we performed colony-forming assays (Fig. 5B). We found that Lgr6 stem cells were less efficient in forming colonies when isolated from skin in which they previously lost their interaction with the neuronal niche (Fig. 5B).
Figure 5. Effect of skin denervation on the fate of Lgr6 stem cells.
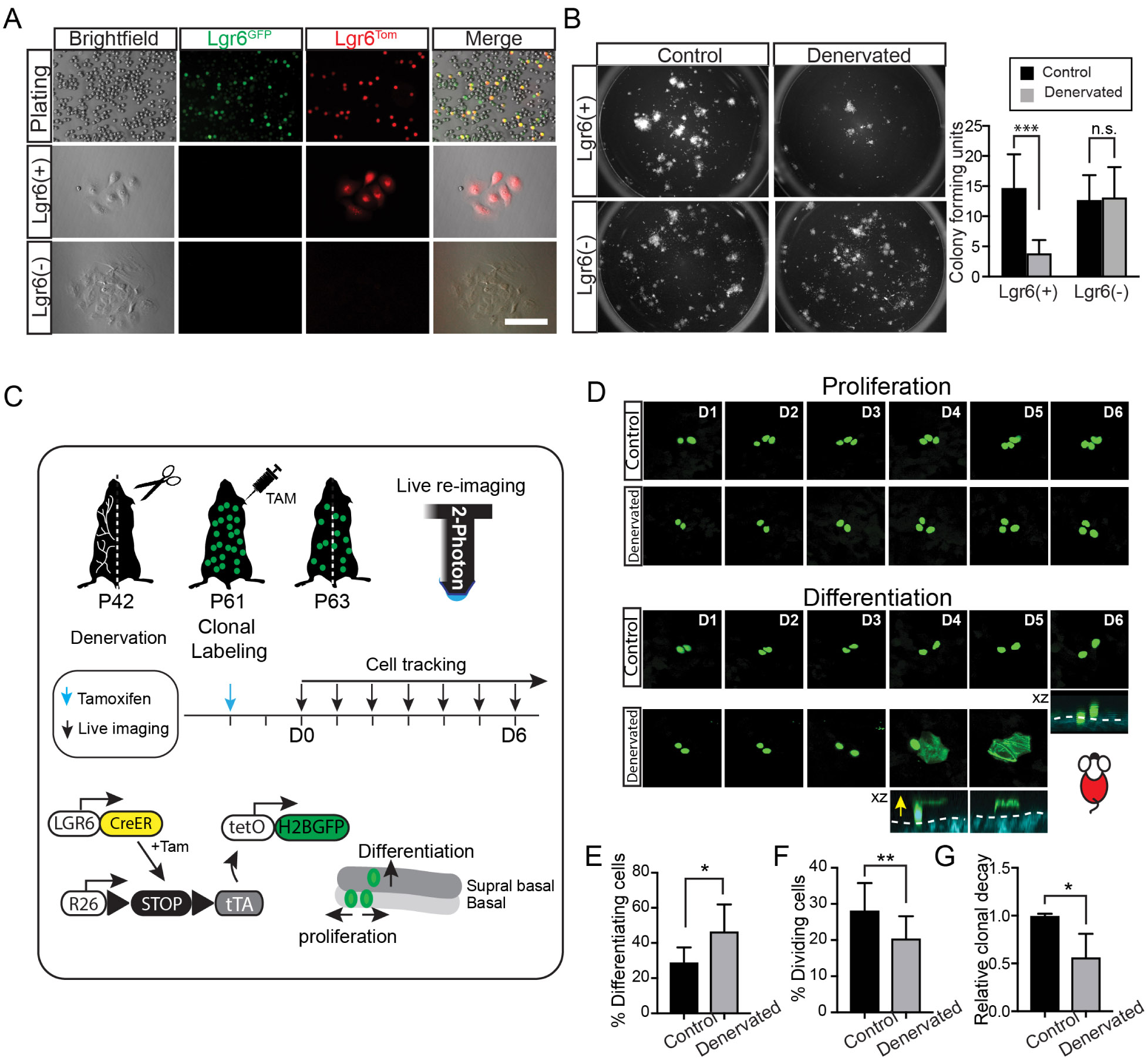
(A) Representative example of epidermal keratinocytes isolated from the mouse skin after labeling of Lgr6Tom stem cells. Immediately after sorting virtually all the Lgr6Tomare Lgr6GFP(+) (top panel). After 3 weeks in culture, Lgr6Tom cells no longer express GFP, indicating a loss of Lgr6 stem cell identity (lower panels). (B) Colony forming efficiency assay. Left panel shows representative plates containing colonies that originate from either Lgr6(+) or Lgr6(−) stem cells, isolated from normal or denervated skin. Lgr6(+); n = 7, p = 0.0004. Lgr6(−); n = 7, p = 0.8641. (C) Schematic of experimental design to quantify changes in the fate of Lgr6 stem cells in the presence or absence of nerves by single cell lineage tracing and live imaging. (D) Representative time series acquired by live imaging at the indicated time points, capturing the fate decisions of single-labeled Lgr6 stem cells. Stem cell progeny that commits to a terminal differentiation fate is seen leaving the basal layer and progressively moving upward until the nucleus of the cell is dissolved and the GFP signal is diffused inside the cornified envelop, indicating the final step of terminal differentiation. Yellow arrow shows the direction of cell movement when cells differentiate from basal layer to suprabasal layer of epidermis. (E-F) Quantification of differentiation and proliferation rates of single-traced Lgr6H2BGFP cells; n = 316 (control), 271 (denervated) cells from 4 mice, (E) p = 0.0194, (F) p = 0.0064. (G) Quantification of clonal decay measured as the fraction of remaining Lgr6H2BGFP cells at the end of the 6-day tracing period; n = 284 (control), 252 (denervated) traced cells from 4 mice , p = 0.0174. Scale bars: 10 μm.
To further confirm these findings, we performed in vivo lineage tracing at the single stem cell level to directly quantify the fates of epidermal Lgr6 stem cells and determine how these are altered after skin denervation. We marked individual Lgr6 stem cells with a bright nuclear reporter (Lgr6H2BGFP) using our previously established genetic scheme (Rompolas et al., 2016; Farrelly, Kuri and Rompolas, 2019). Since this nuclear label can distinguish individual stem cells from their neighbors, their fate can be directly quantified by longitudinal live imaging (Fig. 5C). After induction, the fate of single-marked Lgr6H2BGFP stem cells was directly captured and quantified based on whether the cells underwent cell division (self-renewal) or they left the basal layer and moved upwards, to be eventually shed from the skin (differentiation) (Fig. 5D). This analysis confirmed that in the absence of nerves, epidermal Lgr6H2BGFP stem cells have a higher propensity to leave the basal layer and undergo terminal differentiation (Fig. 5E-G).
Ablation of nerves alters the gene expression program of Lgr6 stem cells
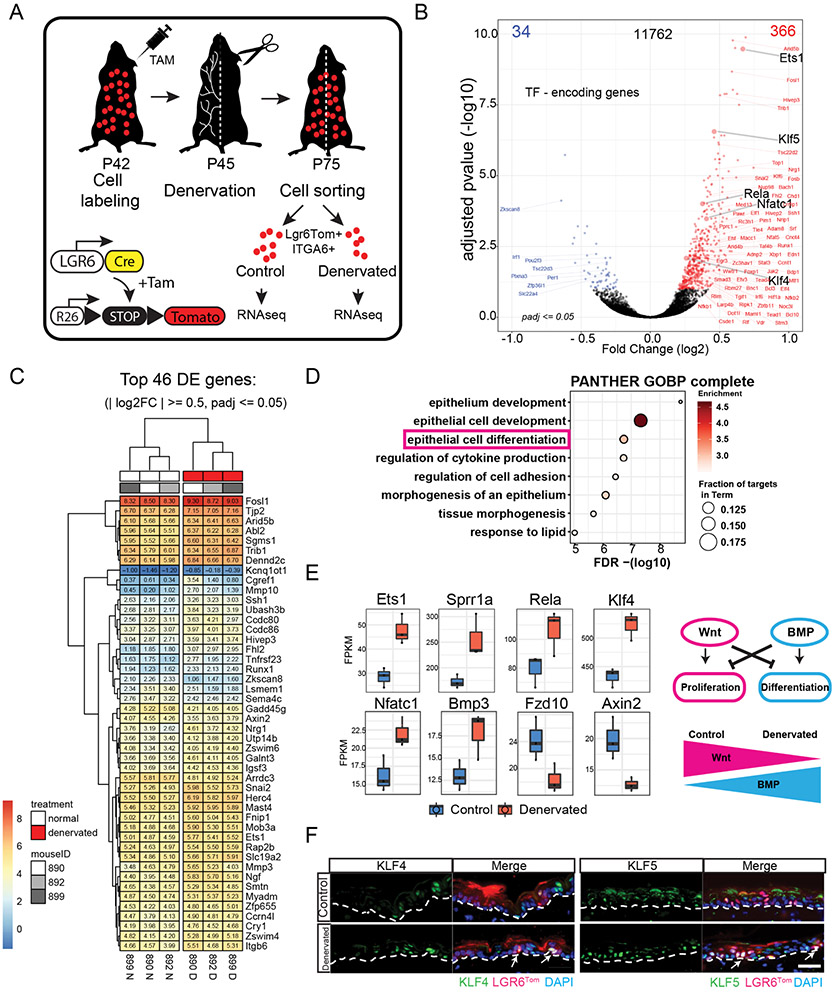
Our results thus far support a role for nerves in the regulation of Lgr6 stem cell dynamics during wound re-epithelialization. However, the genes that control the behavior of epidermal stem cells in a nerve-dependent manner are unknown. To resolve the differences in gene expression between Lgr6 stem cells in the presence or absence of nerves, we performed RNA sequencing analysis of Lgr6Tom stem cells isolated from intact or denervated skin (Fig. 6A). Comparative expression analysis revealed ~400 genes that were differentially expressed (±1.5-fold, adjusted p < 0.05) (Fig. 6B, C). These observed differences in gene expression were exhibited within a single stem cell population that was isolated from the same mouse and should therefore be attributed solely to the presence or absence of nerves. Interestingly, most differentially expressed genes (366 vs. 34) were upregulated in the absence of sensory nerves, suggesting that nerves may regulate stem cell activity by suppressing the expression of genes that control cell fate (Fig. 6B).
Figure 6. Gene expression changes in Lgr6 stem cells induced by skin denervation.
(A) Schematic of the experiment design to evaluate gene expression changes in Lgr6Tom stem cells isolated from denervated or intact (control) areas of the back skin. Replicates of paired samples were generated from 3 different mice and sorted at different times. (B) Volcano plot of RNA-seq data comparing control and denervated basal Lgr6Tom cell transcripts. Red dots represent upregulated genes, blue dots represent downregulated genes; adjusted p < = 0.05. (C) Heatmap of top differentially expressed genes from the RNA-seq analysis. (D) Gene ontology analysis of genes upregulated in denervated basal Lgr6Tom cells. (E) Differential expression analysis of transcription factor genes commonly related to keratinocyte differentiation, WNT and BMP signaling pathways. (F) Immunofluorescence analysis of KLF4 and KLF5 transcription factors in control and denervated skin. White arrows denote Lgr6Tom cells with high expression of KLF4 and KLF5. White dashed line demarcates the border between epidermis and dermis. Scale bars: 20μm.
To explore this possibility, we performed gene ontology analysis of the genes upregulated in Lgr6Tom cells isolated from denervated skin. The analysis revealed enrichment in genes with established roles in epithelial development, cell differentiation, regulation of cell adhesion, and morphogenesis (Fig. 6D). For example, genes critical for keratinocyte differentiation, including Ets1, Sprr1a, Rela, Klf4 and Klf5 showed higher expression in Lgr6Tom stem cells harvested from skin with ablated nerves (Fig. 6E, F). The gene signature for developmental pathways primarily included Wnt and BMP signaling, which are antagonizing pathways that promote proliferation and differentiation, respectively. Examples of genes upregulated in Lgr6Tom stem cells that lacked nerve interaction included BMP member Bmp3 and its target gene Nfatc1(Fig. 6E). Conversely, we found that the expression of Wnt ligand Frz10 and Wnt target gene Axin2 were downregulated in the same group of cells (Fig. 6E).
These data indicate that nerves in the epidermis suppress the expression of genes that promote stem cell differentiation. To confirm that this effect is specific to the Lgr6 population, we compared gene expression changes between Lgr6Tom cells and the remaining unlabeled epidermal population harvested from denervated skin using qRT-PCR. The pro-differentiation gene expression signature was indeed unique to Lgr6Tom stem cells, underscoring the functional interaction between this stem cell pool and cutaneous nerves (Fig. S6A, B). Intriguingly, the list of differentially expressed genes also included members of the circadian clock as well as genes involved in calcium signaling, suggesting that these pathways may also be critical for mediating the interaction between nerves and Lgr6 stem cells (Fig. S6C-G). This type of crosstalk may be important for maintaining Lgr6 epidermal stem cells in a primed state that allows them to respond quickly when fast growth is required, such as during wound re-epithelialization.
Discussion
Our study demonstrates that a distinct pool of epidermal stem cells, marked by Lgr6, is required for a robust wound re-epithelialization. The heterogeneity of stem cells in the epidermis has been the subject of intense research for many decades. The current consensus, based on many studies, including recent ones that employ cutting edge lineage tracing strategies and live imaging analyses, support the hypothesis that the mouse interfollicular epidermis is maintained by stem cells that divide once every 2-3 days to produce two daughter cells (Clayton et al., 2007; Rompolas et al., 2016). These progeny cells can independently decide to either continue to divide and replenish their pool or stop their cell cycle and initiate a deterministic program of terminal differentiation. The latter begins with their departure from the basal layer and ends with their eventual shedding from the surface of the skin a few days later. Tissue-wide, these decisions appear to be balanced and therefore, deemed to be stochastic during homeostasis. Regional adaptations due to the anatomical and functional idiosyncrasies of the ear, tail and back skin of the mouse as well as the differential expression of recently identified markers among basal cells raise the question of whether distinct pools of stem cells in fact exist in the epidermis (Mascré et al., 2012; Sada et al., 2016). More importantly, it is unknown if such stem cell pools have diverging roles in homeostatic maintenance versus wound healing. Our observations and those from prior studies suggest that ~20 % of basal epidermal keratinocytes express Lgr6 (Füllgrabe et al., 2015). Consistent with other reports, we found no significant differences in the activity or fate of epidermal Lgr6 stem cells under homeostasis. Despite representing a minor fraction of the total population, Lgr6 epidermal stem cells respond quickly during the initial stages of wound re-epithelialization and are required to ensure a robust wound healing process.
Homeostatic maintenance of the skin epidermis is highly stereotypical and has evolved to expend the least possible energy and resources while supporting the functionality and replenishment of the tissue. In contrast, wounding evokes a wide range of profound changes to the tissue that are required to ensure a successful tissue repair and in many cases the survival of the individual that bears the injuries. The inflammatory and immunological aspects of wound healing have been studied extensively, but the complexity of stem cell behaviors during wound re-epithelialization is only recently beginning to be appreciated. Here, we show that epidermal Lgr6 stem cells constitute a distinct epidermal stem cell population that is “primed” to respond to wound healing. Lgr6 epidermal stem cells quickly respond to injury and are among the first to begin the re-epithelialization process. Lgr5 stem cells, however, become activated and begin migrating out of their niches into the wound much later. A key question is how does the tissue regulate the activation and recruitment of distinct stem cell populations to ensure robustness in different physiological contexts? We found that when the contribution of Lgr6 stem cells was impaired, the number of Lgr5 stem cells that re-epithelialized the wound significantly increased, allowing a complete wound closure to be ultimately achieved. Nonetheless, the loss of Lgr6 stem cells still caused a significant delay in the wound healing. Taken together, these observations indicate a mechanism by which the skin regulates the fate of distinct stem cell pools to optimize the wound healing process. Even when one stem cell population is compromised, the tissue can still ensure a successful outcome, albeit with sub-optimal dynamics, by modulating the activity of the remaining stem cell populations.
Our wound healing experiments support the hypothesis of a distinct population of epidermal stem cells with a specific role in early injury response and wound re-epithelialization. Is this “primed” state an intrinsic property of these stem cells or subject to external regulation? The skin is a complex tissue, in which a wide range of cell types with diverse developmental origins and function cohabitate and interact. A wealth of experimental evidence supports the notion that these interactions often serve as key regulatory components of stem cell activity and are collectively referred to as the stem cell niche (Scadden, 2014; Mesa, Rompolas and Greco, 2015). Peripheral nerves are pervasive in virtually every tissue, including the skin, but their role as stem cell niches is less understood (Ansel et al., 1997; Lumpkin and Caterina, 2007; Talagas et al., 2018). The state-of-the-art imaging tools and genetic models established for this study allowed us to visualize and modulate the interactions between cutaneous nerves and stem cells in the intact skin of live mice. Thus, we provide evidence to support the distinct physical interactions of cutaneous nerves with Lgr6 stem cells in the hair follicle isthmus as well as similar interactions between nerves and a previously uncharacterized Lgr6 stem cell population in the interfollicular epidermis. By surgically denervating the skin, we demonstrate that the fate of epidermal Lgr6 stem cells and their ability to re-epithelialize wounds are linked to the presence of nerves. This indicates that the crosstalk between nerves and Lgr6 stem cells is functionally important.
Nerve-dependent niches define stem cell populations that are marked by expression of Lgr6 in both the hair follicle as well as the IFE (Liao and Nguyen, 2014). This raises the question of whether the stem cell populations in these two compartments are subject to common regulatory influences by nerves and if they have a common function in skin regeneration. Sensory nerves were shown to signal Gli1-expressing stem cells in the hair follicle via Sonic hedgehog ligands (Brownell et al., 2011). However, no equivalent Gli1+ stem cells exist in the IFE. Furthermore, tracking of hair follicle Gli1-positive stem cells, which were genetically marked before skin denervation, showed no significant changes in their contribution to wound healing in the short term (Brownell et al., 2011). Our results indicate that denervation specifically affects the contribution of epidermal Lgr6 stem cells during the early stages of wound re-epithelialization. Sympathetic nerves have also been proposed to regulate hair growth; a recent study demonstrated that the stimulation of photosensitive retinal ganglion cells can activate stem cells and promote hair growth via sympathetic innervation (Fan et al., 2018). In our experiments, we transected the dorsal roots, aiming to only remove the somatic sensory fibers from the skin. Although we cannot exclude that sympathetic nerves could also be affected in some cases, tyrosine hydroxylase staining confirmed their presence in skin that was devoid of sensory nerve fibers after surgery. Taken together, these data support our hypothesis that nerve-dependent Lgr6 epidermal stem cells are functionally distinct from other stem cell pools found in the IFE and hair follicles, and their mode of regulation may be adapted to a specific role in wound re-epithelialization.
The family of leucine-rich-repeat-containing G protein coupled receptors (Lgr4, 5 and 6) is preferentially expressed in unique epithelial stem cell populations throughout the body (Snippert et al., 2010). Lgr receptors bind to R-spondins to augment Wnt signaling responses (de Lau, Snel and Clevers, 2012; Raslan and Yoon, 2019). Canonical Wnt signaling plays a critical role in skin regeneration and wound healing (Cheon et al., 2006; Ito et al., 2007; Choi et al., 2013; Lim et al., 2013; Whyte et al., 2013). While Lgr4 and 5 are thought to be the main mediators of canonical Wnt signaling, the role and requirement of Lgr6 are less understood (Clevers, Loh and Nusse, 2014). Moreover, it is unclear why epidermal stem cells are induced by their interaction with nerves to express Lgr6. Our data show that Lgr6 expression in epidermal stem cells is augmented by their interaction with the nerves. However, the persistence of a fraction of epidermal stem cells that still express Lgr6 after denervation – albeit at lower levels – suggests that additional factors may play a role in the regulation of this gene. The specific function of the Lgr6 receptor in stem cell regulation is currently not well understood. The wide expression of Lgr4 in the skin and the well supported functional redundancy of Lgr genes further cloud our understanding of their specific requirement for skin regeneration (de Lau et al., 2011). Interestingly, while Lgr6−/− mice have morphologically normal digits during homeostatic growth, amputation results in dysmorphic nail regeneration (Lehoczky and Tabin, 2015). Their wild type counterparts, however, fully regenerate their lost digit tip. This observation indicates that Lgr6-expressing cells may have distinct roles in homeostasis versus wound response, further supporting our current findings.
The sensory role of cutaneous nerves is well established but there is also significant clinical evidence to suggest that innervation is critical for skin physiology (Paus et al., 1994; Botchkarev et al., 1997, 2004; Brain, 1997; Laverdet et al., 2015). One of the most common complications in patients with diabetes is peripheral neuropathy, which is characterized by progressive loss of pain perception and the degeneration of cutaneous nerve fibers (Vinik et al., 2000). Diabetes-induced nerve loss leads to insufficient epidermal maintenance, hair loss, and impaired healing of diabetic skin wounds (Cheng et al., 2013). In psoriasis, another major skin disease, the role of the nervous system in disease pathogenesis is supported by a significant increase in nerve fibers in affected areas and spontaneous remissions of psoriatic plaques after nerve injury (Ostrowski et al., 2011; Zhu et al., 2016). While the etiology of such skin diseases is multi-factorial, they are all characterized by abnormal stem cell proliferation and differentiation. It is therefore plausible that defective cutaneous nerves may contribute to their manifestation by influencing the activity and fate of certain stem cell populations. Experiments in mouse and rat models show that denervated skin exhibits delayed wound healing response (Engin et al., 1996; Smith and Liu, 2002; Brownell et al., 2011; Buckley et al., 2012; Rinkevich et al., 2014; Alapure et al., 2018). The data from these studies indicate that impairments in both wound contraction and re-epithelialization are responsible for the observed phenotype. Our identification of an epidermal stem cell population whose response to injury is specifically modulated by nerves sheds critical light in the regulation of the re-epithelialization phase of wound healing.
Limitations of the Study
As most similar studies, this relies heavily on the use of knock-in and transgenic fluorescent reporters which serve as surrogates to evaluate stem cell identity and function by lineage tracing analysis. Specifically, we use Lgr6 to mark and interrogate the role of a subpopulation of stem cells in the epidermis. Previous reports have demonstrated that Lgr6-expressing epidermal cells have stem cell properties, but whether this gene is involved in the regulation of stemness has not yet been fully elucidated. Therefore, it is unclear how faithfully expression levels of Lgr6 reflect the specific stem cell properties of this population. Leakiness of inducible Cre mouse lines has been described in the literature and can have confounding effects in lineage tracing analysis. We evaluated the extent of potential unspecific cell labeling by live imaging mice carrying Lgr6 and Lgr5 inducible Cre drivers prior to induction of recombination and we found that neither line exhibits any leakiness when combined with the fluorescent Cre reporters used in this study.
STAR Methods Text
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Panteleimon Rompolas (rompolas@pennmedicine.upenn.edu).
Materials Availability
Mouse lines generated in this study are available upon request to the Lead Contact.
Data and Code Availability
The RNA-sequencing data are available through the Gene Expression Omnibus of the National Center for Biotechnology Information, accession number GSE171662. No previously unpublished custom code was used to analyze the data.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
All procedures involving animal subjects were performed with the approval of the Institutional Animal Care and Use Committee (IACUC) of the University of Pennsylvania. The following strains were obtained from Jackson laboratories: Lgr6-EGFP-IRES-CreERT2 (Lgr6GFP in text, Stock No. #016934), Lgr5-EGFP-IRES-CreERT2 (or Lgr5GFP in text, Stock No. #008875), TrpV1Cre mice (Stock No. #017769), R26loxP-stop-loxP-tTA (R26−tTA in text, Stock No. #008600), TetO-H2BGFP (Stock No. #005104), R26loxP-stop-loxP-tdTom (Stock No #007908), R26loxP-nTomato-stop-loxP-nGFP (Stock No #023035) and R26loxP-stop-loxP-DTA (Stock No #009669). K14-H2B-PAGFP (K14H2BPAGFP in text) mice were generated by the Center for Animal Transgenesis and Germ Cell Research, at the School of Veterinary Medicine of the University of Pennsylvania. All mice that were used in this study were bred for multiple generations into a Crl:CD1(ICR) mixed background. Mice between 2-6 months of age were used for experiments, with equal male/female representation. There was no apparent difference in phenotype between genders. Mice were housed under standard laboratory conditions and received food and water ad libitum.
METHOD DETAILS
Tamoxifen induction of mice
To induce Cre-recombinase activation, mice were injected intraperitoneally with Tamoxifen (Sigma) dissolved in corn oil (Sigma). To induce ubiquitous recombination, mice were administered a single dose of 2 mg Tamoxifen between postnatal day 42 and 45. Lgr6-EGFP-Ires-CreERT2; R26loxP-stop-loxP-tdTom; R26loxP-stop-loxP-DTA were induced 3 days prior to injury. To induce single stem cell labelling, Lgr6-EGFP-Ires-CreERT2; R26loxP-stop-loxP-tTA; TetO-H2BGFP and Lgr5-EGFP-Ires-CreERT2; R26loxP-stop-loxP-tTA; TetO-H2BGFP mice were administered a single dose of 20 μg tamoxifen. Single cell tracing experiments began at day 3 post induction (Day 0).
Denervation surgery
Dorsal skin peripheral nerves were severed using microsurgery as previously described with minor adaptations (Ostrowski et al., 2011; Peterson et al., 2015). In brief, mice were anesthetized with an intraperitoneal injection of ketamine/xylazine cocktail in PBS (0.1 ml / 20 g body weight: 87.5 mg / kg Ketamine, 12.5 mg / kg Xylazine). Once a surgical plane of anesthesia was verified by absence of pedal reflex responses following physical stimulation, a 4.5-5 cm incision was made along the dorsal midline using surgical scissors to expose cutaneous nerves on the left side of the animal (T3-T12). The nerves on the left side were then removed via blunt dissection at a point close to the anatomical entry site into the skin. All nerves on the right side were left intact. The initial incision was closed with sutures. Surgical denervation of the ear was adapted from previously described procedures (Buckley et al., 2012). After reaching a surgical plane of anesthesia, an area of approximately 2 cm2 at the base of the left ear was shaved and swabbed with 70% alcohol. 200 μl of sterile phosphate-buffered saline (Gibco) was injected intradermally in the center of the shaved area to form a raised bleb. A 1 cm incision was then made across the bleb using a size 11 scalpel, to access the rostral auricular nerve innervating the ear from its base. Under direct vision using an operating microscope, microsurgical scissors were used to transect the rostral auricular nerve and several other nerve bundles at the ear base with high precision without inflicting any damage to the surrounding tissues or blood vessels. A mock surgery was performed in the right ear to serve as a contralateral control, in which, after the 1 cm incision was made on the raised saline bleb, all nerves were left intact. On both sides, the incision was closed with sutures. Mice were always housed individually post-surgery and administered Meloxicam (5 mg/kg) as analgesic every 12 hours for the 2 days following surgery. Mice were closely monitored until full recovery and the resumption of normal behavior.
Wounding assays
Mice that were at least eight weeks old were anaesthetized by intraperitoneal injection of ketamine/xylazine cocktail in PBS (0.1 ml / 20 g body weight: 87.5 mg / kg Ketamine, 12.5 mg / kg Xylazine). A surgical plane of anesthesia was verified by absence of pedal reflex responses following physical stimulation. In the assays that required only one central injury site, a full thickness 1 cm2 square wound was performed on the back skin using a scalpel on both experimental and control mice. In assays that used a contralateral site as control, a square wound of 0.5 cm2 squares was performed on each side. In cases where high resolution live imaging of the injury site was required, wounds were performed using a circular punch biopsy tool of 1.5 or 2.5 mm in diameter for ear and dorsal skin, respectively. The ear is a more convenient region to image and was therefore chosen for some experiments that required two-photon microscopy. Wounds to the ear were made from the dorsal side and did not penetrate the cartilage. Although dermal components are different in ear and back skin, we obtained similar results in both areas.
Intravital imaging of the mouse skin
Imaging preparation and procedures followed those previously described (Pineda et al., 2015; Huang & Rompolas, 2017). Mice were anaesthetized by intraperitoneal injection of ketamine/xylazine cocktail in PBS (0.1 ml / 20 g body weight; 87.5 mg / kg Ketamine, 12.5 mg / kg Xylazine). A surgical plane of anesthesia was verified by absence of pedal reflex responses following physical stimulation and was maintained during the imaging period with 1 % vaporized isoflurane in oxygen and air delivered through a nose cone. The skin was mounted on a custom-made stage with a glass coverslip placed directly against it. To maintain body temperature, mice were placed on a heating pad throughout the experiment. Image acquisition was performed with an upright Olympus FV1200MPE microscope, equipped with a Chameleon Vision II Ti: Sapphire laser. The laser beam was focused through 10X, 20X or 25X objective lenses (Olympus UPLSAPO10X2, N.A. 0.40; UPLSAPO20X, N.A. 0.75; XLPLN25XWMP2, N.A. 1.05). Emitted fluorescence was collected by two multi-alkali and two gallium arsenide phosphide (GaAsP) non-descanned detectors (NDD). The following wavelengths were collected by each detector: NDD1 419-458 nm; NDD1 458–495 nm; GaAsP-NDD1 495–540 nm; GaAsP-NDD2 575–630 nm. GFP and Tomato reporters were excited at 930 nm and their signal was collected by GaAsP-NDD1 and GaAsP-NDD2, respectively. Second harmonic generation signal was generated using 930nm excitation wavelength and detected in NDD2. Serial optical sections were acquired in 2.5-3 mm steps, starting from the surface of the epidermis and capturing the entire thickness of the epidermis and a partial section of the dermis (epidermis ~30 μm, dermis ~60-80 μm). To image all the cells in skin epidermis and measure the cell density in a defined area, the live skin was scanned using 750nm excitation wavelength. To image larger areas, a square matrix was defined with 10% overlap between individual tiles, which the microscope acquires automatically and sequentially using a motorized platform. Stitching is performed by the microscope software (Fluoview, Olympus USA) to produce a single final image. Multi-day tracing experiments were performed by re-imaging the same field of view at the indicated times after the initial acquisition, with vasculature and micro-tattoos used as landmarks to identify the imaging region at low magnification and clusters of hair follicles used as landmarks higher magnification. After each imaging session, the mice were monitored and allowed to recover in a warm chamber before being returned to the housing facility.
In vivo photo-labeling
Photo-labeling experiments with the K14H2BPAGFP; Lgr6-EGFP-Ires-CreERT2; R26loxP-stop-loxP-tdTom mice were carried out with the same equipment and imaging setup 3 days after Tamoxifen induction and right after wounding the skin tissue. The pre-activated form of the H2B-PAGFP and PAGFP fluorescent proteins was visualized by exciting with 850 nm wavelength and emission signal was collected in GaAsP-NDD1. Excitation with 930 nm verified that no signal is emitted by the reporters before activation. Photo-labeling was achieved by scanning a defined region-of-interest (ROI) at the plane of the basal layer of the epidermis, with the laser tuned to 750nm wavelength, for 5-10 sec, using 5-10% laser power. Immediately after photo-activation, a series of optical sections, with a range that includes the entire thickness of the skin, were acquired using the same acquisition settings as for GFP. Visualizing the signal of the activated form of PAGFP only within the ROI confirmed the successful photo-labeling of basal layer keratinocytes. Following the initial image acquisition immediately after photo-labeling, the same area of skin was re-imaged at the indicated times to evaluate the changes of the labeled basal layer Lgr6(+) cell population and their movements compared to Lgr6(−) cell group.
Immunostaining
For whole-mount staining, the dorsal skin was dissected, laid flat on the epidermal side to allow for removal of subcutaneous fat through scratching and fixed in 4% paraformaldehyde in PBS overnight at 4°C. The fixed skin was punched using a 2 mm diameter punch biopsy tool to obtain multiple samples that were incubated with primary antibodies 72 hours at room temperature. Samples were washed in 0.3% Triton X-100 PBS for 6-8 hours prior to secondary antibody incubation diluted for 48 hours at room temperature. After secondary antibody staining, samples were washed by PBS with 0.3% Triton-X 100 for 6-8 hours and then dehydrated in 100% methanol. Tissues were cleared in BABB (Benzyl Alcohol/ Benzyl Benzoate, 1:2 ratio) solution for 30 min. All antibodies were diluted in blocking solution (5% normal goat or horse serum, 20% DMSO in 0.3% Triton X-100 PBS). For section staining, dorsal skins were dissected, laid flat and directly fixed with 4% paraformaldehyde in PBS for overnight at 4°C. Fixed tissues were paraffin embedded and sectioned. Slides were deparaffinized using xylene substitute and then rehydrated using a graded ethanol series. For antigen retrieval, slides were places in a 1:50 dilution of Antigen Unmasking Solution (Vector Laboratories) and heated in a boiling bath for 10 min. Sections were washed in 0.2 % Tween 20 PBS and incubated in blocking solution (10 % normal goat or horse serum in 0.5 % Tween 20 PBS for 2 hours, then incubated with primary antibody diluted in staining solution (2% normal goat or horse serum 0.2% Tween 20 PBS) overnight at 4°C. After 2 hours of washing in 0.2% Tween 20 PBS, tissue sections were incubated with secondary antibody and DAPI diluted in staining solution for 1 hour at room temperature. To increase signal strength in the Klf5 immunostaining, a TSA amplification procedure was incorporated to the protocol, which involved the following steps: quenching of endogenous peroxidase by incubating in 3% Hydrogen peroxide in ddH2O for 15 min after antigen retrieval, a secondary antibody incubation with ImmPRESS Polymer Reagent (Vector Laboratories) for 1 hour at room temperature followed by washes and an incubation with a Fluorescein Tyramide Solution (Perkin Elmer) according to manufacturer’s instructions. Secondary antibody incubation was followed by washing in 0.2 % Tween 20 PBS and slide mounting using Fluor-Gel mounting medium (Electron Microscopy Sciences). TUNEL assay was performed using the In Situ Cell Death Detection Kit (Roche) according to manufacturer’s instructions. In brief, following deparaffinization and slide rehydration, slides were treated with a 20 μg/ml Proteinase K (Denville Scientific) solution in PBS for 15 min at room temperature, washed in 0.2 % Tween 20 PBS and incubated in TUNEL reaction mixture for 1 hour at 37°C, then washed prior to mounting. As a positive control for the reaction, we included a slide treated with DNase I for 15 min at room temperature prior to TUNEL reaction mixture incubation. For Hematoxylin and eosin (H&E) staining, sections were fixed in 4% paraformaldehyde for 10 min, washed in PBS and stained in hematoxylin for 8 min and eosin for 1 min. For BrdU labeling experiments, we administered mice with BrdU (50 μg/g) (Sigma-Aldrich) 2 hours before the tissue collection at indicated time points. The following primary antibodies were used: Anti-Ki67 (1:100, 14-5698, eBioscience), anti-RFP (1:500, 600-401-379, Rockland), anti-Keratin 14 (1:1000, 905301, BioLegend), anti-Keratin 10 (1:500, 905401, BioLegend), anti-Loricrin (1:500, 905101, BioLegend), anti-CD34 (1:50, 553731, BD Pharmingen), anti-S100 (1:400, Z031129-2, Dako), anti-NF200 (1:1000, ab2313552, Aves Labs), anti-TH (1:100, MAB7566, R&D Systems), anti-KLF4 (1:50, AF3158, R&D Systems), anti-KLF5 (1:50, AF3758, R&D Systems), anti-LRIG1 (1:50, AF3688, R&D Systems) and anti-BrdU (1:100, B-35128, Invitrogen). The following secondary antibodies were used: Alexa Fluor 488 Goat anti-Mouse (1:500, A-11001, Invitrogen), Alexa Fluor 594 Goat anti-Rabbit (1:500, A-11012, Invitrogen), Alexa Fluor 488 Goat anti-chicken (1:500, A-110039, Invitrogen), Alexa Fluor 594 Donkey anti-goat (1:500, A-110058, Invitrogen) and Alexa Fluor 647 Donkey anti-goat (1:500, A-21447, Invitrogen). Images were acquired using an Olympus BX51 equipped a Hamamatsu Orca CCD camera or a Leica DM6 B equipped with a Leica DFC9000 GT fluorescent camera and a Leica DMC2900 brightfield camera.
Cell preparation for flow cytometry and sorting
The dorsal skin was dissected and laid flat for subcutaneous fat scratching and then floated epidermis side up in 0.25 % Trypsin (Gibco) in PBS for 1 hour at 37°C to separate epidermal and dermal layers. Following the incubation, the epidermis was carefully scraped off and transferred to 10% FBS (fetal bovine serum) in DMEM (Gibco), in which it was manually gently dissociated using a serological pipette. The entire volume was filtered through a 70 μm cell strainer (VWR) to obtain a single cell suspension that was then centrifuged and resuspended in 2 % FBS in PBS. For cell staining, antibodies were directly added to the cell suspension and incubated for 10 min on ice. Afterwards, cells were washed using 2 % FBS PBS and resuspended for sorting in 5 mM EDTA PBS. The following antibodies were used: anti-CD49f-APC (1:300, 313615, BioLegend), anti-CD49f-PercP/Cy5.5 (1:300, 313618, BioLegend), anti-CD49f-FITC (1:300, 313606, BioLegend), anti-CD34-PE (1:50, 551387, BD Pharmingen), anti-CD34-Alexa Fluor 647 (1:50, 560230, BD Pharmingen), anti-Sca1-Ly-6A/E-Violet 605 (1:200, 108133, BioLegend) and DAPI (1:1000, Biotium). Flow cytometry was performed on a BD LSRII cytometer (BD Biosciences), sortings on a BD FACS Aria II sorter (BD Biosciences). Flow cytometry data was collected and exported using BD FACs Diva software (BD Biosciences) and analyzed and plotted using FlowJo software.
Colony forming efficiency assay
Lgr6GFP(+)RFP(+) and Lgr6GFP(−)RFP(−) cells were FACs isolated from control and denervated back skin, equal numbers (3x103) of live cells were plated, in triplicate, onto 24-well plates in CnT-07 medium (CELL N TEC, CnT-BM.1) supplemented with Rock Inhibitor (Y27632, EMD Millipore, SCM075). After 2-3 weeks in culture, cells were fixed and stained with DAPI (1:1000). Colony diameter was measured from scanned images of plates using image J and colony numbers were counted.
RNA-sequencing and data analysis
Lgr6-EGFP-Ires-CreERT2; R26loxP-stop-loxP-tdTom mice were administered a single dose of 20 μg tamoxifen at P42 to activate Cre-recombination. Four weeks after denervation surgery, Lgr6Tom were stained with anti-CD49f-APC (1:300, Biolegend) and isolated from both denervated and control sides of the same mouse. RNA was extracted using the RNeasy Mini Kit (Qiagen) following manufacturer’s instructions. The libraries for sequencing were prepared using NEBNext poly(A) mRNA magnetic isolation module followed by NEBNext Ultra Directional RNA library preparation kit for Illumina (both from New England Biolabs). Library quality was confirmed using an Agilent BioAnalyzer 2100 (Agilent), and library concentration was obtained using library quantification kits (New England Biolabs). Libraries were then sequenced in a NextSeq500 platform (75-base-pair [bp] single-end reads) (Illumina). Replicates were generated from 3 different mice, sorted at different times. RNA-seq reads were analyzed through Illumina’s Base Space RNA Express app (v1.1.0). FPKM (fragments per kilo-base per million mapped fragments) generation and differential expression analysis between denervated and control samples were performed using DESeq2 (Love et al., 2014). In brief, reads were aligned with STAR aligner (v2.3.1.s) to Mus musculus UCSC mm10 gene annotation (representing 24,421 genes). Genes were considered as statistically significantly differentially expressed using a relaxed threshold (i.e., DESeq2’s adjusted p-value <= 0.1), which yielded 478 up-regulated genes and 60 down-regulated genes. Gene ontology analysis of the 478 up-regulated genes in denervated samples was performed using PANTHER overrepresentation test (released 2019-04-29) and GO Ontology complete database (Thomas et al., 2003) (released 2019-02-02), using the set of genes designated as “expressed” based on the RNA-seq analysis (12,300 expressed genes) and applying a Fisher test with False Discovery Rate (FDR) correction. Ontology terms with a fold enrichment <= 2.5 and −log10(FDR) <=5 were removed. All figures related to RNA-seq analysis were generated with R (3.5.0) and R Studio (Version 1.1.383) using custom script, available upon request, on a x86_64-apple-darwin15.6.0 (64-bit) platform running under: mac OS Sierra 10.12.6.
QUANTIFICATION AND STASTICAL ANALYSIS
Image analysis
Raw digital files from two-photon imaging were acquired and saved in the OIB format using the microscope manufacturer’s software (FLUOVIEW, Olympus USA). These raw image stacks were then imported into ImageJ/Fiji (NIH Image) using Bio-Formats or to Imaris (Bitplane) for further analysis. Individual optical planes and maximum projections or three-dimensional renderings of sequential optical sections were used to assemble figures. Supplementary movies of 3D renderings were created using Imaris. To quantify population clonal dynamics, following induction, high-resolution optical sections were obtained sequentially and used to construct 3-dimensional tiled views of the epidermis. The same mice were then re-imaged using identical acquisition parameters. From each time point, the equivalent areas of the epidermis were sampled and processed by supervised segmentation to quantify the area of labeled clones within. For single-cell lineage tracing, individual high-magnification serial optical sections were obtained for each traced clone and 3-dimensional analysis was performed once the entire imaging time course was competed to analyze the state of basal and suprabasal cells in each time point. Departure of a cell from the basal layer and subsequent upward transit was scored as differentiation while continuous increase in the basal cell number was scored as self-renewal. Clone measurements were performed manually. Images shown in figures typically represent maximum projections or single optical sections selected from the z-stacks unless otherwise specified. Co-localization analysis was performed using EzColocalization (Stauffer et al., 2018).
qRT-PCR
Total RNA was isolated from Lgr6Tom cells from both denervated and control sides of the same mouse using RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. cDNA was synthesized using the ProtoScript First-Strand cDNA Synthesis Kit (NEB). The cDNA was used as a template in quantitative real time PCR reactions, performed in triplicate with SYBR green Master Mix reagents (Applied Biosystems) on the ABI 7500 Real-Time PCR System. The data was analyzed using GAPDH as an internal gene for normalization. Primers used in this study are listed in Table S1.
Statistics and reproducibility
Sample sizes were not pre-determined but are similar with what was reported previously (Rompolas et al. 2016). Data were collected and quantified randomly, and their distribution was assumed normal, but this was not formally tested. Lineage tracing experiments were successfully reproduced under similar conditions using at least three different mouse cohorts. The values of “n” (sample size) refer to data points obtained from all mice within the cohort, unless otherwise indicated, and are provided in the figure legends. Statistical calculations and graphical representation of the data were performed using the Prism 8 software package (GraphPad). Data are expressed as percentages or mean ± S.E.M and unpaired Student's t-test was used to analyze data sets with two groups, unless otherwise stated in the figure legends. For all analyses, P-values < 0.05 were designated as significant and symbolized in figure plots as *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, with precise values supplied in figure legends. No data were excluded from the analysis. Statistical calculations were performed using Prism 8 (GraphPad).
Data and materials availability:
Data sets and reagents presented in this study are available from the corresponding author upon request.
Supplementary Material
Movie S1. Co-localization of nerves with Lgr6 stem cells. Refers to Figure 3B.
Movie S2. Co-localization of nerves with Lgr5 hair follicle stem cells. Refers to Figure 3B.
Movie S3. Co-localization of Lgr6 stem cells with sensory nerves in the skin epidermis. Refers to Figure 3C.
Key resource table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Ki-67 Monoclonal Antibody (SolA15) | eBioscience | Cat# 14-5698; RRID:AB_10853185 |
| Anti-RFP (Rabbit) Antibody | Rockland | Cat# 600-401-379; RRID:AB_2209751 |
| Keratin 14 Polyclonal Antibody (Clone: Poly19053) | BioLegend | Cat# 905301; RRID:AB_2565048 |
| Keratin 10 Polyclonal Antibody (Clone: Poly19054) | BioLegend | Cat# 905401; RRID:AB_2565049 |
| Loricrin Polyclonal Antibody (Clone: Poly19051) | BioLegend | Cat# 905101; RRID:AB_2565046 |
| S100 beta Antibody | Agilent | Cat# Z031129-2; RRID:AB_2315306 |
| Neurofilament, Heavy Chain (NF-H) Antibody (NF200) | Aves Labs | Cat# ab2313552; RRID:AB_2313552 |
| Tyrosine Hydroxylase (TH) Antibody | Novus | Cat# MAB7566; RRID:AB_2885201 |
| Anti-KRT20/CK20/Cytokeratin 20 Antibody | LSBio | Cat# LS-B5677; RRID:AB_10915572 |
| Mouse KLF4 Antibody | R&D Systems | Cat# AF3158; RRID:AB_2130245 |
| HumanKLF5 Antibody | R&D Systems | Cat# AF3758; RRID:AB_2130246 |
| BrdU Monoclonal Antibody (MoBU-1) | Thermo Fisher Scientific | Cat# B35128; RRID:AB_2536432 |
| Mouse LRIG1 Polyclonal Antibody | R&D Systems | Cat# AF3688; RRID:AB_2138836 |
| Rat Anti-Mouse CD34-PE (Clone RAM34) | BD Biosciences | Cat# 553731; RRID:AB_395015 |
| Alexa Fluor-647 Rat Anti-Mouse CD34 (Clone RAM34) | BD Biosciences | Cat# 560230; RRID:AB_1645200 |
| FITC Anti-Human/Mouse CD49f Antibody (Clone GoH3) | BioLegend | Cat# 313606; RRID:AB_345300 |
| APC Anti-Human/Mouse CD49f Antibody (Clone GoH3) | BioLegend | Cat# 313615; RRID:AB_2734290 |
| PerCP/Cyanine5.5 Anti-Human/Mouse CD49f Antibody (Clone GoH3) | BioLegend | Cat# 313618; RRID:AB_2249260 |
| Brilliant Violet 605 Anti-Mouse Ly-6A/E(Sca-1) Antibody | BioLegend | Cat# 108133; RRID:AB_2562275 |
| Goat Anti-Chicken IgY (H+L) Secondary Antibody, Alex Fluor 488 | Invitrogen | Cat# A-11039; RRID:AB_142924 |
| Donkey Anti-Goat IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 594 | Invitrogen | Cat# A-11058; RRID:AB_2534105 |
| Donkey Anti-Goat IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Invitrogen | Cat# A-21447; RRID:AB_141844 |
| Goat Anti-Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Invitrogen | Cat# A-11001; RRID:AB_2534069 |
| Goat Anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 594 | Invitrogen | Cat# A-11012; RRID:AB_141359 |
| Chemicals, peptides, and recombinant proteins | ||
| Tamoxifen | Sigma | Cat# T5648 |
| Ketamine | Midwest Veterinary Supply | Cat# 17033-100-10 |
| Xylazine | Midwest Veterinary Supply | Cat# 310-01150-3 |
| Isoflurane, USP | Piramal | Cat# NDC6679401325 |
| 5-Bromo-2’-deoxyuridine (BrdU) | Sigma | Cat# HMBH0685V |
| CnT-07 medium | CELL N Tec | Cat# CnT-BM.1 |
| ROCK Inhibitor Y-27632 | EMD Millipore | Cat# SCM075 |
| Critical commercial assays | ||
| In Situ Cell Death Detection Kit, Fluorescein | Roche | Cat# 11684795910 |
| ProtoScript First Strand cDNA Synthesis Kit | New England BioLabs | Cat# E6300S |
| TRIzol reagent | Life Technologies | Cat# 252606 |
| VECTASHIELD Mounting Medium with DAPI | Vector Laboratories | Cat# H-1200 |
| ImmPRESSTM HRP REAGENT KIT anti-Goat IgG | VECTOR | Cat# MP-7405 |
| Power SYBR Green PCR Master Mix | Applied Biosystems | Cat# 4367659 |
| RNeasy mini Kit | QIAGEN | Cat# 74104 |
| NEBNext Poly(A) mRNA magnetic isolation module | New England Biolabs | Cat# E7490S |
| NEBNext Ultra Directional RNA Library Prep Kit for Illumina | New England Biolabs | Cat# E7420L |
| NEBNext Oligos for Illumina (Index Primer Set 1) | New England Biolabs | Cat# E7335L |
| NEBNext Oligos for Illumina (Index Primer Set 1) | Cat#E7500S | |
| Agilent BioAnalyzer 2100 | Agilent | N/A |
| NEBNext Library Quantification Kit for Illumina | New England Biolabs | Cat# E7630L |
| Deposited data | ||
| RNA-seq of Lgr6 stem cells from control and denervated skin | This study | GEO: GSE171662 |
| Experimental models: organisms/strains | ||
| Mouse: Crl:CD1(ICR) | Charles River | RRID:IMSR_CRL:022 |
| Mouse: Lgr6-EGFP-IRES-CreERT2 | Snippert et al., 2010; PMID 20223988 | RRID:IMSR_JAX:016934 |
| Mouse: Lgr5-EGFP-IRES-CreERT2 | Barker et al., 2007; PMID 17934449 | RRID:IMSR_JAX:008875 |
| Mouse: Rosa26-stop-tdTomato | Madisen et al., 2010; PMID 20023653 | RRID:IMSR_JAX:007908 |
| Mouse: Rosa26-nTomato-stop-nGFP | The Jackson Laboratory | RRID:IMSR_JAX:023035 |
| Mouse: Rosa26-stop-tTA | Wang et al., 2008; PMID 18054242 | RRID:IMSR_JAX:008600 |
| Mouse: TetO-H2BGFP | Tumbar et al., 2004; PMID 14671312 | RRID:IMSR_JAX:005104 |
| Mouse: Rosa26-stop-DTA | Voehringer et al., 2008; PMID 18354198 | RRID:IMSR_JAX:009669 |
| Mouse: TrpV1-Cre | Cavanaugh et al., 2011; PMID 21451044 | RRID:IMSR_JAX:017769 |
| Mouse: Advillin-CreERT2 | Lau et al., 2011; PMID 22188729 | RRID:IMSR_JAX:032027 |
| Mouse: Keratin14-H2BPAGFP | Farrelly et al., 2021; PMID 33984283 | N/A |
| Oligonucleotides | ||
| qRT-PCR primers 25 nmole DNA Oligo bases | Integrated DNA Technologies | N/A |
| Oligonucleotides for qRT-PCR see Table S1 | This study | N/A |
| Software and algorithms | ||
| GraphPad Prism 8 | https://www.graphpad.com/scientific-software/prism/ | N/A |
| Olympus Fluoview | https://www.olympus-lifescience.com/en/support/downloads/ | N/A |
| ImageJ | Schneider et al., 2012; PMID 22930834 | https://imagej.nih.gov/ij/ |
| Adobe Photoshop and Illustrator | https://www.adobe.com | N/A |
| Imaris | https://imaris.oxinst.com/ | N/A |
| FACS Diva software | https://www.bdbiosciences.com/en-us/instruments/research-instruments/research-software/flow-cytometry-acquisition/facsdiva-softwar | N/A |
| FlowJo software | https://www.flowjo.com/ | N/A |
| NextSeq500 | https://www.illumina.com | N/A |
| Illumina BaseSpace | https://www.illumina.com/products/by-type/informatics-products/basespace-sequence-hub.html | N/A |
| STAR | Dobin et al., 2013; PMID 23104886 | N/A |
| DESeq2 | Love et al., 2014; PMID 25516281 | N/A |
| PANTHER | http://www.pantherdb.org | N/A |
| R version 3.5.0 | https://www.r-project.org/ | N/A |
| R-Studio version 1.1.383 | https://www.rstudio.com/ | N/A |
| Other | ||
| Sterile 1mm, 1.5mm, 2mm, 2.5mm 4mm and 6mm Biopsy punch | Integra Miltex | Cat# 33-31AA, 33-31A, 33-31, 33-31B, 33-34, 33-6 |
Highlights.
Lgr6 stem cells in the skin epidermis show distinct response to injury
Ablation of Lgr6 stem cells impairs wound healing
Skin denervation alters the contribution of stem cells during wound healing
Loss of sensory nerves changes the fate of Lgr6 stem cells
Acknowledgments:
We thank Michael Rendl, Christopher Lengner, and Kenneth Zaret for our invaluable discussions and their critical comments. We are especially grateful to George Cotsarelis, John Stanley, John Seykora and Yana Kamberov for their advice that guided many aspects of this study. We thank Yoko Suzuki-Horiuchi for her contribution with the K14-H2BPAGFP mouse line. The Penn Skin Biology and Diseases Resource-based Center (SBDRC) provided critical services through Core A and Core B. We also acknowledge the support of the Institute for Regenerative Medicine and the entire stem cell community at Penn. S.H. was supported by a Penn IRM postdoctoral fellowship. O.F. was supported by training grant T32HD083185 from NIH/NICHD. G.R. was supported by training grant T32GM007229 from NIH/NIGMS. P.K. was supported by the American Association for Cancer Research, with the John and Elizabeth Leonard Family Foundation Basic Cancer Research Fellowship. P.R. was supported by grants from NIH/NEI (R01EY030599) and from the American Cancer Society (RSG1803101DCC). Penn SBDRC is supported by center core grant P30AR069589 from NIH/NIAMS.
Footnotes
Competing interests: The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alapure B. v. et al. (2018) “Surgical Denervation of Specific Cutaneous Nerves Impedes Excisional Wound Healing of Small Animal Ear Pinnae,” Molecular Neurobiology, 55(2), pp. 1236–1243. doi: 10.1007/s12035-017-0390-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansel JC et al. (1997) “Interactions of the skin and nervous system,” in Journal of Investigative Dermatology Symposium Proceedings. Nature Publishing Group, pp. 23–26. doi: 10.1038/jidsymp.1997.6. [DOI] [PubMed] [Google Scholar]
- Aragona M et al. (2017) “Defining stem cell dynamics and migration during wound healing in mouse skin epidermis,” Nature Communications, 8. doi: 10.1038/ncomms14684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N et al. (2007) “Identification of stem cells in small intestine and colon by marker gene Lgr5,” Nature, 449(7165), pp. 1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Barker N, Tan S and Clevers H (2013) “Lgr proteins in epithelial stem cell biology,” Development (Cambridge), 140(12), pp. 2484–2494. doi: 10.1242/dev.083113. [DOI] [PubMed] [Google Scholar]
- Belokhvostova D et al. (2018) “Homeostasis, regeneration and tumour formation in the mammalian epidermis,” International Journal of Developmental Biology, 62(6-8), pp. 571–582. doi: 10.1387/ijdb.170341fw. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchkarev VA et al. (1997) “Hair cycle-dependent plasticity of skin and hair follicle innervation in normal murine skin,” Journal of Comparative Neurology, 386(3), pp. 379–395. doi: . [DOI] [PubMed] [Google Scholar]
- Botchkarev VA et al. (2004) “Epithelial growth control by neurotrophins: Leads and lessons from the hair follicle,” in Progress in Brain Research. Elsevier, pp. 493–513. doi: 10.1016/S0079-6123(03)46031-7. [DOI] [PubMed] [Google Scholar]
- Brain SD (1997) “Sensory neuropeptides: Their role in inflammation and wound healing,” Immunopharmacology Immunopharmacology, pp. 133–152. doi: 10.1016/S0162-3109(97)00055-6. [DOI] [PubMed] [Google Scholar]
- Brownell I et al. (2011) “Nerve-derived sonic hedgehog defines a niche for hair follicle stem cells capable of becoming epidermal stem cells,” Cell Stem Cell, 8(5), pp. 552–565. doi: 10.1016/j.stem.2011.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley G et al. (2012) “Denervation affects regenerative responses in MRL/MpJ and repair in C57BL/6 ear wounds,” Journal of Anatomy, 220(1), pp. 3–12. doi: 10.1111/j.1469-7580.2011.01452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanaugh DJ, Chesler AT, Jackson AC, Sigal YM, Yamanaka H, Grant R, O'Donnell D, Nicoll RA, Shah NM, Julius D, & Basbaum AI (2011). Trpv1 reporter mice reveal highly restricted brain distribution and functional expression in arteriolar smooth muscle cells. The Journal of neuroscience : the official journal of the Society for Neuroscience, 31(13), 5067–5077. 10.1523/JNEUROSCI.6451-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C et al. (2013) “Loss of Innervation and Axon Plasticity Accompanies Impaired Diabetic Wound Healing,” PLoS ONE, 8(9). doi: 10.1371/journal.pone.0075877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheon SS et al. (2006) “Beta-catenin regulates wound size and mediates the effect of TGF-beta in cutaneous healing,” The FASEB Journal, 20(6), pp. 692–701. doi: 10.1096/fj.05-4759com. [DOI] [PubMed] [Google Scholar]
- Choi YS et al. (2013). “Distinct functions for Wnt/β-catenin in hair follicle stem cell proliferation and survival and interfollicular epidermal homeostasis,” Cell Stem Cell, 13(6), pp. 720–733. 10.1016/j.stem.2013.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton E et al. (2007) “A single type of progenitor cell maintains normal epidermis,” Nature, 446(7132), pp. 185–189. doi: 10.1038/nature05574. [DOI] [PubMed] [Google Scholar]
- Clevers H, Loh KM and Nusse R (2014) “An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control,” Science. American Association for the Advancement of Science. doi: 10.1126/science.1248012. [DOI] [PubMed] [Google Scholar]
- Dekoninck S and Blanpain C (2019) “Stem cell dynamics, migration and plasticity during wound healing,” Nature Cell Biology. Nature Publishing Group, pp. 18–24. doi: 10.1038/s41556-018-0237-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati G et al. (2017) “Wounding induces dedifferentiation of epidermal Gata6 + cells and acquisition of stem cell properties,” Nature Cell Biology, 19(6), pp. 603–613. doi: 10.1038/ncb3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, & Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England), 29(1), 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doupé DP and Jones PH (2012) “Interfollicular epidermal homeostasis: Dicing with differentiation,” Experimental Dermatology. Exp Dermatol, pp. 249–253. doi: 10.1111/j.1600-0625.2012.01447.x. [DOI] [PubMed] [Google Scholar]
- Engin C et al. (1996) “Delayed effect of denervation on wound contraction in rat skin,” Plastic and Reconstructive Surgery, 98(6), pp. 1063–1067. doi: 10.1097/00006534-199611000-00021. [DOI] [PubMed] [Google Scholar]
- Fan SMY et al. (2018) “External light activates hair follicle stem cells through eyes via an ipRGC–SCN–sympathetic neural pathway,” Proceedings of the National Academy of Sciences of the United States of America, 115(29), pp. E6880–E6889. doi: 10.1073/pnas.1719548115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrelly O, Kuri P, & Rompolas P (2019). “In Vivo Genetic Alteration and Lineage Tracing of Single Stem Cells by Live Imaging.” Methods in molecular biology (Clifton, N.J.), 1879, pp. 1–14. 10.1007/7651_2018_172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Füllgrabe A et al. (2015) “Dynamics of Lgr6+ progenitor cells in the hair follicle, sebaceous gland, and interfollicular epidermis,” Stem Cell Reports, 5(5), pp. 843–855. doi: 10.1016/j.stemcr.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrelly O, Suzuki-Horiuchi Y, Brewster M, Kuri P, Huang S, Rice G, Bae H, Xu J, Dentchev T, Lee V, & Rompolas P (2021). Two-photon live imaging of single corneal stem cells reveals compartmentalized organization of the limbal niche. Cell stem cell, 28(7), 1233–1247.e4. 10.1016/j.stem.2021.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcin CL et al. (2016) “Hair Follicle Bulge Stem Cells Appear Dispensable for the Acute Phase of Wound Re-epithelialization,” Stem Cells, 34(5), pp. 1377–1385. doi: 10.1002/stem.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales KAU and Fuchs E (2017) “Skin and Its Regenerative Powers: An Alliance between Stem Cells and Their Niche,” Developmental Cell. Cell Press, pp. 387–401. doi: 10.1016/j.devcel.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurtner GC et al. (2008) “Wound repair and regeneration,” Nature. Nature Publishing Group, pp. 314–321. doi: 10.1038/nature07039. [DOI] [PubMed] [Google Scholar]
- Huang S and Rompolas P (2017) “Two-photon microscopy for intracutaneous imaging of stem cell activity in mice,” Experimental Dermatology. Blackwell Publishing Ltd, pp. 379–383. doi: 10.1111/exd.13221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M et al. (2005) “Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis,” Nature Medicine, 11(12), pp. 1351–1354. doi: 10.1038/nm1328. [DOI] [PubMed] [Google Scholar]
- Ito M et al. (2007) “Wnt-dependent de novo hair follicle regeneration in adult mouse skin after wounding,” Nature, 447(7142), pp. 316–320. doi: 10.1038/nature05766. [DOI] [PubMed] [Google Scholar]
- Jaks V et al. (2008) “Lgr5 marks cycling, yet long-lived, hair follicle stem cells,” Nature Genetics, 40(11), pp. 1291–1299. doi: 10.1038/ng.239. [DOI] [PubMed] [Google Scholar]
- Joost S et al. (2018) “Single-Cell Transcriptomics of Traced Epidermal and Hair Follicle Stem Cells Reveals Rapid Adaptations during Wound Healing,” Cell Reports, 25(3), pp. 585–597.e7. doi: 10.1016/j.celrep.2018.09.059. [DOI] [PubMed] [Google Scholar]
- Langton AK, Herrick SE and Headon DJ (2008) “An extended epidermal response heals cutaneous wounds in the absence of a hair follicle stem cell contribution,” Journal of Investigative Dermatology, 128(5), pp. 1311–1318. doi: 10.1038/sj.jid.5701178. [DOI] [PubMed] [Google Scholar]
- de Lau WBM, Snel B and Clevers HC (2012) “The R-spondin protein family,” Genome Biology. Genome Biol. doi: 10.1186/gb-2012-13-3-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lau W et al. (2011) “Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling,” Nature, 476(7360), pp. 293–297. doi: 10.1038/nature10337. [DOI] [PubMed] [Google Scholar]
- Lau J, Minett MS, Zhao J, Dennehy U, Wang F, Wood JN, & Bogdanov YD (2011). Temporal control of gene deletion in sensory ganglia using a tamoxifen-inducible Advillin-Cre-ERT2 recombinase mouse. Molecular pain, 7, 100. 10.1186/1744-8069-7-100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laverdet B et al. (2015) “Skin innervation: Important roles during normal and pathological cutaneous repair,” Histology and Histopathology. Histology and Histopathology, pp. 875–892. doi: 10.14670/HH-11-610. [DOI] [PubMed] [Google Scholar]
- Lehoczky JA and Tabin CJ (2015) “Lgr6 marks nail stem cells and is required for digit tip regeneration,” Proceedings of the National Academy of Sciences of the United States of America, 112(43), pp. 13249–13254. doi: 10.1073/pnas.1518874112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy V et al. (2007) “Epidermal stem cells arise from the hair follicle after wounding,” The FASEB Journal, 21(7), pp. 1358–1366. doi: 10.1096/fj.06-6926com. [DOI] [PubMed] [Google Scholar]
- Liao XH and Nguyen H (2014) “Epidermal expression of Lgr6 is dependent on nerve endings and Schwann cells,” Experimental Dermatology, 23(3), pp. 195–198. doi: 10.1111/exd.12340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim X et al. (2013) “Interfollicular epidermal stem cells self-renew via autocrine Wnt signaling,” Science, 342(6163), pp. 1226–1230. doi: 10.1126/science.1239730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W and Anders S (2014) “Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2,” Genome Biology, 15(12). doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumpkin EA and Caterina MJ (2007) “Mechanisms of sensory transduction in the skin,” Nature. Nature Publishing Group, pp. 858–865. doi: 10.1038/nature05662. [DOI] [PubMed] [Google Scholar]
- Mascré G et al. (2012) “Distinct contribution of stem and progenitor cells to epidermal maintenance,” Nature, 489(7415), pp. 257–262. doi: 10.1038/nature11393. [DOI] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, & Zeng H (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nature neuroscience, 13(1), 133–140. 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesa KR, Rompolas P and Greco V (2015) “The Dynamic Duo: Niche/Stem Cell Interdependency,” Stem Cell Reports. Cell Press, pp. 961–966. doi: 10.1016/j.stemcr.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak JA et al. (2008) “Hair follicle stem cells are specified and function in early skin morphogenesis,” Cell Stem Cell, 3(1), pp. 33–43. doi: 10.1016/j.stem.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski SM et al. (2011) “Cutaneous denervation of psoriasiform mouse skin improves acanthosis and inflammation in a sensory neuropeptide-dependent manner,” Journal of Investigative Dermatology, 131(7), pp. 1530–1538. doi: 10.1038/jid.2011.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S et al. (2017) “Tissue-scale coordination of cellular behaviour promotes epidermal wound repair in live mice,” Nature Cell Biology, 19(2), pp. 155–163. doi: 10.1038/ncb3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paus R et al. (1994) “Hair growth induction by substance P,” Laboratory Investigation, 71(1), pp. 134–140. Available at: http://europepmc.org/article/med/7518880 (Accessed: February 28, 2021). [PubMed] [Google Scholar]
- Peterson SC et al. (2015) “Basal cell carcinoma preferentially arises from stem cells within hair follicle and mechanosensory niches,” Cell Stem Cell, 16(4), pp. 400–412. doi: 10.1016/j.stem.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson SC, Brownell I and Wong SY (2016) “Cutaneous surgical denervation: A method for testing the requirement for nerves in mouse models of skin disease,” Journal of Visualized Experiments, 2016(112). doi: 10.3791/54050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda CM et al. (2015) “Intravital imaging of hair follicle regeneration in the mouse,” Nature Protocols, 10(7), pp. 1116–1130. doi: 10.1038/nprot.2015.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raslan AA and Yoon JK (2019) “R-spondins: Multi-mode WNT signaling regulators in adult stem cells,” International Journal of Biochemistry and Cell Biology. Elsevier Ltd, pp. 26–34. doi: 10.1016/j.biocel.2018.11.005. [DOI] [PubMed] [Google Scholar]
- Rinkevich Y et al. (2014) “Clonal analysis reveals nerve-dependent and independent roles on mammalian hind limb tissue maintenance and regeneration,” Proceedings of the National Academy of Sciences of the United States of America, 111(27), pp. 9846–9851. doi: 10.1073/pnas.1410097111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompolas P et al. (2012) “Live imaging of stem cell and progeny behaviour in physiological hair-follicle regeneration,” Nature, 487(7408), pp. 496–499. doi: 10.1038/nature11218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompolas P et al. (2016) “Spatiotemporal coordination of stem cell commitment during epidermal homeostasis,” Science, 352(6292), pp. 1471–1474. doi: 10.1126/science.aaf7012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompolas P and Greco V (2014) “Stem cell dynamics in the hair follicle niche,” Seminars in Cell and Developmental Biology. Elsevier Ltd, pp. 34–42. doi: 10.1016/j.semcdb.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompolas P, Mesa KR and Greco V (2013) “Spatial organization within a niche as a determinant of stem-cell fate,” Nature, 502(7472), pp. 513–518. doi: 10.1038/nature12602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sada A et al. (2016) “Defining the cellular lineage hierarchy in the interfollicular epidermis of adult skin,” Nature Cell Biology, 18(6), pp. 619–631. doi: 10.1038/ncb3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scadden DT (2014) “Nice neighborhood: Emerging concepts of the stem cell niche,” Cell. Cell Press, pp. 41–50. doi: 10.1016/j.cell.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepeler T, Page ME and Jensen KB (2014) “Heterogeneity and plasticity of epidermal stem cells,” Development (Cambridge). Company of Biologists Ltd, pp. 2559–2567. doi: 10.1242/dev.104588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, & Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nature methods, 9(7), 671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PG and Liu M (2002) “Impaired cutaneous wound healing after sensory denervation in developing rats: Effects on cell proliferation and apoptosis,” Cell and Tissue Research, 307(3), pp. 281–291. doi: 10.1007/s00441-001-0477-8. [DOI] [PubMed] [Google Scholar]
- Snippert HJ et al. (2010) “Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin,” Science, 327(5971), pp. 1385–1389. doi: 10.1126/science.1184733. [DOI] [PubMed] [Google Scholar]
- Stauffer W, Sheng H, & Lim HN (2018). EzColocalization: An ImageJ plugin for visualizing and measuring colocalization in cells and organisms. Scientific reports, 8(1), 15764. 10.1038/s41598-018-33592-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun BK, Siprashvili Z and Khavari PA (2014) “Advances in skin grafting and treatment of cutaneous wounds,” Science. American Association for the Advancement of Science, pp. 941–945. doi: 10.1126/science.1253836. [DOI] [PubMed] [Google Scholar]
- Talagas M et al. (2018) “What about physical contacts between epidermal keratinocytes and sensory neurons?,” Experimental Dermatology, 27(1), pp. 9–13. doi: 10.1111/exd.13411. [DOI] [PubMed] [Google Scholar]
- Taylor G et al. (2000) “Involvement of follicular stem cells in forming not only the follicle but also the epidermis,” Cell, 102(4), pp. 451–461. doi: 10.1016/S0092-8674(00)00050-7. [DOI] [PubMed] [Google Scholar]
- Thomas PD et al. (2003) “PANTHER: A library of protein families and subfamilies indexed by function,” Genome Research, 13(9), pp. 2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, & Fuchs E (2004). Defining the epithelial stem cell niche in skin. Science (New York, N.Y.), 303(5656), 359–363. 10.1126/science.1092436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagnozzi AN, Reiter JF and Wong SY (2015) “Hair follicle and interfollicular epidermal stem cells make varying contributions to wound regeneration,” Cell Cycle, 14(21), pp. 3408–3417. doi: 10.1080/15384101.2015.1090062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinik AI et al. (2000) “Diabetic neuropathies,” Diabetologia. Diabetologia, pp. 957–973. doi: 10.1007/s001250051477. [DOI] [PubMed] [Google Scholar]
- Voehringer D, Liang HE, & Locksley RM (2008). Homeostasis and effector function of lymphopenia-induced "memory-like" T cells in constitutively T cell-depleted mice. Journal of immunology (Baltimore, Md. : 1950), 180(7), 4742–4753. 10.4049/jimmunol.180.7.4742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Sharma K, Deng HX, Siddique T, Grisotti G, Liu E, & Roos RP (2008). Restricted expression of mutant SOD1 in spinal motor neurons and interneurons induces motor neuron pathology. Neurobiology of disease, 29(3), 400–408. 10.1016/j.nbd.2007.10.004 [DOI] [PubMed] [Google Scholar]
- Whyte JL et al. (2013) “Augmenting Endogenous Wnt Signaling Improves Skin Wound Healing,” PLoS ONE, 8(10). doi: 10.1371/journal.pone.0076883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu TH et al. (2016) “The Role of the Nervous System in the Pathophysiology of Psoriasis: A Review of Cases of Psoriasis Remission or Improvement Following Denervation Injury,” American Journal of Clinical Dermatology. Springer International Publishing, pp. 257–263. doi: 10.1007/s40257-016-0183-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1. Co-localization of nerves with Lgr6 stem cells. Refers to Figure 3B.
Movie S2. Co-localization of nerves with Lgr5 hair follicle stem cells. Refers to Figure 3B.
Movie S3. Co-localization of Lgr6 stem cells with sensory nerves in the skin epidermis. Refers to Figure 3C.
Data Availability Statement
The RNA-sequencing data are available through the Gene Expression Omnibus of the National Center for Biotechnology Information, accession number GSE171662. No previously unpublished custom code was used to analyze the data.