Abstract
Background
The SARS-CoV-2 virus is responsible for the COVID-19 pandemic. Researchers have been studying the pathogenesis of the virus with the aim to improve our current diagnosis and management strategies. The microbiota have been proposed to play a key role in the pathogenesis of the disease.
Purpose
To investigate and report on the current available evidence on any associations between the gut and/or airway microbiota and the pathogenesis of COVID-19.
Methods
Using a predefined protocol in compliance with the PRISMA guidelines, a search was conducted on MEDLINE, Science Direct, DOAJ and Cochrane databases on primary research studies assessing the association between COVID-19 infection and the gut and/or airway microbiota.
Results
Twenty-two studies were included in the current review; nineteen studies concluded an association between the gut and/or airway dysbiosis and SARS-CoV-2, while 3 studies failed to observe a significant association between the airway microbiome and SARS-CoV-2 infection. Specifically, most studies reported a decrease in microbial diversity and therefore development of intestinal dysbiosis in COVID-19-positive patients compared to healthy controls as well as a possible association between increased intestinal dysbiosis and disease severity.
Conclusion
During infection with SARS-CoV-2, there are significant changes in the composition of the gut and airway microbiota. Furthermore, the gut microbiota may have a more important role than the airway microbiota in COVID-19 infection. In the future, studies should be more carefully designed to derive more conclusive evidence on the role of the gut and airway microbiota following infection with SARS-CoV-2 which will lead to the formulation of better management strategies in combating COVID-19.
Supplementary Information
The online version contains supplementary material available at 10.1007/s15010-021-01715-5.
Keywords: COVID-19, SARS-CoV-2, Gut microbiota, Airway microbiota, Dysbiosis
Introduction
SARS-CoV-2 is a novel beta coronavirus, responsible for the 2019 novel coronavirus disease (COVID-19) [1]. SARS-CoV-2 can cause Acute Respiratory Distress Syndrome (ARDS) which may develop into Systemic Inflammatory Response Syndrome (SIRS), subsequently leading to multi-organ failure [2] and death [3–5]. COVID-19 has had a severe impact on health [6]; from the beginning of the pandemic until October 1, 2021, COVID-19 has affected 220 countries accounting for 233,503,524 cases and causing 4,777,503 deaths [7].
The respiratory tract is believed to be the main mode of entry of the virus [8]. Angiotensin-Converting Enzyme 2 (ACE2) which is expressed heavily on both the respiratory and the gastrointestinal epithelium is the receptor to which the viral spike binds [9]. Respiratory droplets are believed to be the major source of transmission; yet faecal transmission may also play a role in transmission [10]. Therefore, understanding the association between the gut and airway microbiota and COVID-19 pathogenesis provides an important foundation to formulate better diagnostic and management strategies on combating the pandemic [11].
The microbiota in our body consist of bacteria, archaea, fungi and viruses that can affect the host health and help prevent diseases [12]. The microbiota play a pivotal role in the gut by modulating immune homeostasis, thereby providing an overall protection from pathogens [13, 14]. On the other hand, the airway microbiome is believed to be part of the first barrier against respiratory viral infections and disease progression [15]. In fact, evidence suggests that gut and airway microbiota may also play a role in the pathogenesis of other respiratory viruses such as influenza [16]. It has been proposed that changes in the composition of the intestinal microbiota may negatively impact lung function through systemic immunological effects, whereas lung inflammation in the context of respiratory viral infections may lead to gut dysbiosis [17]. The interrelation and bidirectional effect between the microbiota of the aforementioned anatomical compartments is referred to as the gut–lung microbiota axis. Previous studies have shown that alteration in the composition of the intestinal microbiota, through probiotic or antibiotic administration, affects the outcome of respiratory viral infections such as influenza [18], whereas commensal bacteria in the respiratory tract may be able to strengthen the mucosal immunity of the respiratory tract [19]. Similar mechanisms have been proposed with the pathogenesis of COVID-19 infection [20]. In particular, the gut microbiota may play a role in regulating the ACE2 receptor which binds to SARS-CoV-2 [20, 21], while the respiratory microbiota may protect against infection with SARS-CoV-2 by enhancing respiratory tract immune responses [22]. This scoping review aims to identify the latest evidence derived from primary research studies investigating the possible alteration of gut and/or airway microbiota during infection with SARS-CoV-2. Understanding of the mechanisms involved in this association will support the development of effective strategies to diagnose, manage and prevent COVID-19 disease.
Methods
This scoping review was conducted based on the methodological framework developed by key authors in the field. All the steps involved comply with the most recent relevant guidance [23, 24].
Review questions
- Is there an association between infection with SARS-CoV-2 and changes in the gut and/or airway microbiota?How does the gut and/or airway microbiota of COVID-19 patients compare with the microbiota of patients infected with other viruses and healthy participants?
What is the association between the gut and/or airway microbiota, the immune system and COVID-19 pathogenesis and disease severity?
Is there any evidence for a potential benefit of using faecal microbiota transplantation (FMT) on COVID-19 patients during their recovery?
Search strategy
A search on Pubmed, Science Direct, DOAJ and Cochrane databases until 14 May 2021 was conducted by researcher DL and validated by researcher KR. Examples of search terms used on Pubmed are listed in Supplementary Material 1, while search terms were adapted in other databases as appropriate.
Inclusion criteria
Included studies were observational studies or clinical trials assessing the association between gut and/or airway microbiota and COVID-19 infection in human participants (Table 1).
Table 1.
Inclusion criteria of studies included in scoping review
| Population | COVID-19 patients |
|---|---|
| Interest | Patients’ changes of airway and/or gut microbiota composition |
| Comparison | Healthy individuals or patients with other viruses |
| Outcome | Type of microbiome detected after COVID-19 infection |
| Study type | Quantitative method |
Exclusion criteria
Non-human, non-English studies and in-progress clinical trials were excluded.
Data extraction
Data extraction was carried out by researcher DL and validated by researcher KR. The following data were extracted: author name, type of study, country in which the study was conducted, sample size, median age of the participants in the study, study objective, whether and which serum inflammatory markers were evaluated, whether any antimicrobials or probiotics were administered to the participants of the study, sample collection and evaluation methods and key findings (including microbiota affected).
Data synthesis
The process of data synthesis involved summarising the key findings of the included studies and exploring the relationship between studies in a narrative form. In addition, the mean and the SEM of any numeric figures were calculated. The studies were grouped into studies investigating: (a) gut microbiota (Table 2) or (b) airway microbiota (Table 3) and (c) both gut and airway microbiota (Table 4).
Table 2.
Studies investigating the association between gut microbiota and COVID-19 infection
| Study | Study type | Country | Sample size | COVID-19 Patients’ median age in yearsa | Study objective | Addressing co-relationship between microbiota and serum inflammatory markers | Antimicrobial and/or probiotic administration in participants | Sample collection and evaluation methods | Key findings |
|---|---|---|---|---|---|---|---|---|---|
| Gu et al. [26] | Cross- sectional study | China |
Total: 84 (30 hospitalised COVID-19 patients, 24 hospitalised H1N1 patients, 30 healthy individuals) |
55 | To investigate the alteration of gut microbiota during COVID-19 infection | Yes |
Participants excluded from study if received antibiotics and/or probiotics within 4 weeks before enrollment No reference in study if participants received antibiotics/probiotics after infection |
Samples: Faecal samples collected after COVID-19 infection Assessment: 16S ribosomal RNA gene at the V3-V4 region of extracted DNA from fecal samples |
Microbial profiles among COVID-19 and H1N1 patients were significantly less diversified than the control group Lower serum concentration of lymphocytes and higher concentration of IL-6 and TNF-α in COVID-19 patients compared to healthy control H1N1 group Butyrate producing bacteria Lachnospiraceae, Ruminococcaceae, Blautia, Agathobacter, Anaerostipes, Fusicatenibacter, Eubacterium halii, Dorea, Faecalibacterium significantly decreased compared to the healthy control group COVID-19 group Depletion of Fusicatenibacter, Romboutsia, Anaerostipes, Eubacterium hallii, Ruminococcus torques and Blautia in COVID-19 patients compared to the healthy control group Opportunistic pathogens Streptococcus, Rothia, Veillonella, Erysipelatoclostridium and Actinomyces significantly increased compared to the healthy control group Butyrate producing bacteria (BPB) Ruminococcaceae, Lachnospiraceae, Fusicatenibacter, Anaerostipes, Agathobacter, Eubacterium hallii significantly decreased compared to the healthy control group Higher coverage of Fusicatenibacter, Romboutsia, Intestinibacter, Actinomyces, Erysipelatoclostridium compared to control group Main difference between H1N1 and COVID19 groups Higher abundance of Streptococcus, Fusicatenibacter, Collinsella, Dorea, Agathobacter, Eubacterium hallii, Ruminococcus torques in COVID-19 subjects compared to H1N1 group |
| Tang et al. [27] | Cohort study | China |
Total: 57 (20 mild hospitalised COVID-19 patients, 19 severe hospitalised COVID-19 patients, 18 critical hospitalised COVID-19 patients) |
59 | To investigate the difference in dysbiosis associated with different COVID-19 severity | Yes |
Participants included in study if treated with antibiotics 50.9% of total participants received antibiotics 5.3% of total participants received antifungals 12.3% of total subjects received probiotics |
Samples: Faecal samples collected after COVID-19 infection No report if samples were collected before or after antibiotics/probiotics were given Assessment: qPCR |
Intestinal dysbiosis progressed according to the severity of the disease 55.6% of critical patients presented with intestinal microecological failure All COVID-19 patients Significant reduction of probiotic bacteria Lactobacillus and Bifidobacterium compared to healthy individuals Significant reduction of anti-inflammatory bacteria (Butyrate producing bacteria) F. prausnitzii, C. butyricum, C. leptum, E. rectale compared to healthy individuals Mild group C. butyriucm negatively correlated with CRP concentrations Lactobacillus negatively correlated with prothrombin time Severe group F. prausnitzii and C. leptum positively correlated with neutrophil concentration E. rectale positively correlated with IL-6 concentrations Critical group C. butyriucm negatively correlated with CRP concentrations Bifidobacterium negatively correlated with prothrombin time and LDH Atopobium negatively correlated with D-dimer Bacteroides negatively correlated with LDH and CK concentrations |
| Zuo et al. [29] | Cohort study | Hong Kong |
Total: 69 (30 hospitalised COVID-19 patients, 9 hospitalised community-acquired pneumonia (CAP) patients, 30 healthy individuals) |
46 | To examine the changes in intestinal microbiota among COVID-19 patients during hospitalisation and recovery | No |
16 patients received antibiotics 19 patients received antiviral agents |
Samples: Faecal samples collected after COVID-19 infection No report if antibiotics/probiotics/antivirals were used before infection and if before or after sample collection No report on when samples were collected in relation to antibiotic treatment Assessment: Shotgun metagenomics sequencing technique |
Both COVID-19 and CAP patients were presented with more heterogeneous mycobiome 53% of COVID-19 patients showed instability with their fecal mycobiome and this continued after their hospitalisation Significant increase of opportunistic fungal pathogens C. albicans, Candida auris, Aspergillus flavus and Aspergillus niger in COVID-19 group Similar heterogeneous mycobiome composition found in the community-acquired pneumonia group |
| Zuo et al. [28] | Cohort study | Hong Kong |
Total: 15 hospitalised COVID-19 patients |
55 | To investigate the relationship between COVID-19 viral activity in the stool and intestinal microbiota | No | No report regarding use of antibiotics/probiotics by participants of the study |
Samples: Faecal samples were collected after COVID-19 infection Serial samples were collected until nasopharyngeal or throat swabs were negative on two consecutive samples Assessment: Shotgun metagenomics sequencing technique |
Confirmed faecal-oral transmission of COVID-19 even without any GI manifestation Dysbiosis correlated to infectivity of COVID-19 Stool samples with high COVID-19 infectivity Significant increase of Collinsella aerofaciens, Collinsella tanakaei, Streptococcus infantis and Morganella morganii compared to samples with low COVID-19 infectivity Significant increase of inflammatory and pathogenic bacteria R. gnavus, Clostridium hathewayi and Enterococcus avium upon clearance of COVID-19 virus from the stool Stool samples with low COVID-19 infectivity Higher abundance of bacteria that can boost immunity Parabacteroides merdae, Bacteroides stercoris, Alistipes onderdonkii and Lachnspiracea bacterium 1_1_57FAA compared to samples with high COVID-19 infectivity |
| Chen et al. [33] | Cohort study | China |
Total: 30 hospitalised COVID-19 patients |
53.5 | To examine the relationship between gut microbiota richness and COVID-19 from illness onset to 6 months post hospitalisation | Yes | No report regarding use of antibiotics/probiotics by participants of the study |
Samples: Faecal samples collected after COVID-19 infection Assessment: 16S rDNA sequencing technique |
Microbiota richness lowest during beginning of disease, and remained unchanged throughout the course of the disease Microbiota richness failed to return to normal concentration even after 6 months post hospitalisation Patients with lowest microbiota richness presented with highest concentration of CRP and illness severity |
| Lv et al. [31] | Cohort study | China |
Total: 150 (67 hospitalised COVID-19 patients, 35 hospitalised H1N1 patients, 48 healthy individuals) |
52 | To study the relationship between intestinal mycobiota alteration and clinical features of COVID-19 | Yes |
All subjects who received antifungals, probiotic treatment or both within 4 weeks before enrolment were excluded All patients with H1N1 infection or COVID-19 were treated with antiviral drugs No report if patients were placed on antibiotics and probiotics after sample collection or infection |
Samples: Fecal samples collected after COVID-19 infection No report if antiviral agents were used before or after sample collection No report when samples were collected in relation to antiviral treatment Assessment: Quantitative PCR with ITS1f and ITS2r primers |
More inflammation in COVID-19 patients compared to healthy individuals according to serum results Aspergillus niger, abundant in COVID-19 patients and was positively related to diarrhoea symptoms COVID-19 group Significant decrease of Ascomycota, Basidiomycota, Chromista and Mucoromycota compared to healthy individuals Ascomycota (especially Aspergillus niger and Aspergillus rugulosus) was negatively correlated with BPB Lachnospiraceae and Ruminococcaceae Mucoromycota was positively correlated with opportunistic pathogens Peptostreptococcaceae, Fusicatenibacter, Intestinibater, Aspergillus and Agathobacter Aspergillus was positively correlated with the incidence of diarrhea Penicillium citrinum was negatively correlated with CRP concentration Rhodotorula mucilaginosa was negatively correlated with angiotensin-converting enzyme concentration H1N1 group Significant increase of Ascomycota, Basidiomycota, Chlorophyta and Saccharomyces cerevisiae compared to healthy individuals Significant decrease of Cladosporium, Aspergillus, Penicillium, Aspergillus niger, Penicillium, Trechispora sp., Rhodotorula mucilaginosa, Moesziomyces aphidis and Wallemia sebi compared to healthy individuals Aspergilluspositively correlated with BPB Lachnospiraceae, Ruminococcaceae, Erysipelotrichaceae Penicillium and Penicillium polonicum positively correlated with Akkermansia Ascomycota negatively correlated with Roseburia and Marvinbryantia Aspergillus positively correlated with CRP concentration Mucoromycota negatively correlated with procalcitonin concentration Aspergillus penicillioides positively correlated with TNF-α, IL-2 and IL-10 concentrations |
| Mazzarelli et al. [35] | Cohort study | Italy |
Total: 23 (6 COVID-19 patients in the ICU (i-COVID19); 9 COVID-19 patients in the infectious disease wards (w-COVID-19), 3 non-COVID19 hospitalised patients in the ICU, 5 non-COVID-19 patients in general ward) |
67 | To explore the changes of intestinal microbiota among COVID-19 patients in the intensive care unit | Yes |
11 patients received antibiotic therapy (5 w-COVID19, 3 i-COVID19 and 3 non-COVID-19 patients) |
Samples: Rectal swabs collected after COVID-19 infection 11 patients received antibiotics (8 COVID-19 and 3 non-COVID-19 patients) 1 day or two at most before rectal swabs Assessment: PCR amplicons targeting the hypervariable regions V2, V4, V8 and V3-6, 7–9 of the 16S gene |
Microbial richness was reduced in i-COVID19 group compared to the w-COVID19 group w-COVID19 Significant increase of opportunistic pathogens Proteobacteria, Peptostreptococcaceae, Enterobacteriaceae, Staphylococcaceae, Vibrionaceae, Aerococcaceae, Dermabacteraceae, Actinobacteria compared to non-COVID-19 patients Significant decrease of Spirochaetes and Fusobacteria compared to non-COVID-19 patients i-COVID19 Compared to the w-COVID19, further increase of opportunistic pathogens Staphylococcaceae, Microbacteriaceae, Micrococcaceae, Pseudonocardiaceae and Erysipelotrichales and other bacteria: Erysipelotrichaceae, Microbacteriaceae, Mycobacteriaceae, Pseudonocardiaceae and Brevibacteriaceae Significant decrease of Carnobacteriaceae, Coriobacteriaceae, Mycoplasmataceae, Pectobacteriaceae, Moritellaceae, Selenomonadaceae and Micromonosporaceae compared to the w-COVID19 group |
| Yeoh et al. [30] | Cohort study | Hong Kong |
Total: 178 (100 hospitalised COVID-19 patients, 78 healthy individuals) |
36.4 | To examine the relationship between intestinal mycobiota alteration, COVID-19 severity and immune response | Yes |
34 hospitalized COVID-19 patients received antibiotics during hospitalization 73 hospitalized COVID- 19 patients received antiviral agents during hospitalization No report if patients received probiotics |
Samples: Fecal samples collected after COVID-19 infection No report on whether samples were collected in relation to antibiotic/antiviral treatments Assessment: Shotgun metagenomics sequencing technique |
Intestinal dysbiosis is corelated with disease severity followed by the use of antibiotics Higher inflammatory markers (CRP, TNFα and IL-10) among more severe COVID-19 patients Intestinal microbiota composition may be associated with the severity of immune response COVID-19 patients: Significant increase of Bacteroidetes, Ruminococcus gnavus, Rminococcus torques, Bacteroides dorei compared to healthy individuals Significant decrease of Actinobacteria, Bifidobacterium adolescentis, Faecalibacterium prausnitzii, Eubacterium rectale compared to healthy individuals Further decrease of BPB Faecalibacterium prausnitzii, Eubacterium rectale, Bifidobacterium bifidum and Bifidobacterium adolescentis in more severe COVID19 patients compared to healthy individuals Bacteroides dorei and Akkermansia muciniphila were positively correlated with IL-1B, IL-6 and CXCL8 compared to healthy individuals Significant increase of Lactobacillus ruminis and decrease of Eubacterium rectale, R. bromii, Faecalibacterium prausnitzii and Bifidobacterium longum in recovered COVID-19 patients regardless of their antibiotic treatment compared to healthy individuals |
| Yu et al. [32] | Cohort study | China | Total: 3 hospitalised COVID-19 patients | 78 | To investigate the changes of the immune system and gut dysbiosis among COVID19 patients with severe refractory hypoxaemia | Yes | All patients received antibiotics and antiviral treatments |
Samples: Rectal Swabs were collected after COVID-19 infection No report if samples were collected before or after management with antibiotics/probiotics Assessment: Nanopore Targeted Sequencing |
Level of hypoxemia closely related to the immune system markers CD3, CD4, CD8 T cells count, CD19 B cells and NK cells markedly below the normal range in COVID-19 patients Intestinal dysbiosis may be a key factor leading to severe COVID-19 infection COVID-19 patients Proportion of probiotics, such as Bifidobacterium, Lactobacillus and Eubacterium, was markedly reduced compared to healthy individuals Significant increase of Actinobacteria, Corynebacterium, Kluyneromyces (fungus), Firmicutes and Aspergillus (Fungus) in COVID-19 patients compared to healthy individuals |
| Liu et al. [34] | Randomised Clinical trial | China |
Total: 11 COVID-19 patients discharged from hospital more than 1 month ago prior to recruitment |
49 | To study the effect of faecal microbiota transplantation (FMT) on discharged resolved COVID-19 patients | Yes |
Patients were excluded if they received antibiotics or anti-inflammatory treatments 2 weeks prior to sample collection 11 patients received FMT after sample collection |
Samples: Faecal samples were collected after resolved COVID-19 infection Samples were collected before and after FMT Samples were collected after 4 days of FMT and then 1 week further after FMT Assessment: 16S rDNA sequencing technique |
FMT improves dysbiosis post COVID-19 infection FMT improves 100% of COVID-19 patients’ GI symptoms post discharge FMT decrease the naive B cells and increase memory B cells and non-switched B cells, therefore improve immune system during recovery from COVID-19 Significant decrease of Firmicutes and Actinobacteria in discharged COVID19 patients compared to the general population higher proportion of Bacteroidetes and Proteobacteria in discharged COVID19 patients compared to the general population FMT restored dysbiosis by increasing the abundance of Actinobacteria and decrease the abundance of Proteobacteria in post COVID-19 patients |
|
Cohort studies = 8 Cross sectional study = 1 Randomised Clinical trial = 1 |
China = 6 Hong Kong = 3 Italy = 1 |
Mean = 62.18 SEM = 16.41 |
Mean = 55.09 SEM = 3.41 |
Studies which reported Dysbiosis during COVID-19 = 8 Studies which reported association between Microbiota composition and COVID-19 severity = 4 Studies which reported alteration of microbiota composition during post COVID-19 infection = 2 Studies which reported the effectiveness of FMT on COVID19 treatment = 1 |
Yes = 9 No = 1 |
Studies which Included patients on antibiotics/probiotics = 5 Studies which excluded patients on antibiotics/probiotics = 3 Studies which did not report if participants received antibiotics/probiotics = 2 |
Studies which collected faecal samples = 8 Studies which collected rectal swabs = 1 Studies which collected samples after infection = 10 Studies which did not report if samples were collected before or after antibiotic/probiotic/antiviral treatments = 9 Studies which reported sample collection time with regards to antibiotic/probiotic use = 1 |
Studies that reported increase in opportunistic pathogens = 5 Studies that reported decrease in beneficial bacteria = 6 |
aIn case of multiple groups of COVID-19 patients, the median age of the largest group is used in the analysis
Table 3.
Studies investigating the association between airway microbiota and COVID-19 infection
| Study | Study type | Country | Sample size | COVID-19 Patients’ median age in yearsa | Study objective | Addressing co-relationship between microbiota and serum inflammatory markers | Antimicrobial and/or probiotic administration in participants | Sample collection and evaluation methods | Key findings |
|---|---|---|---|---|---|---|---|---|---|
| De Maio et al. [36] | Cohort study | Italy |
Total: 40 (18 mild COVID-19 patients, 22 healthy individuals) |
Not reported | To compare the nasopharyngeal microbiota composition among COVID-19 and non-COVID-19 patients | No | No report of antibiotic/probiotic use in the study |
Samples: Nasopharyngeal swabs collected after COVID-19 infection No report of antibiotic/probiotic use before or after swab collection in the study Assessment method: 16 s rRNA sequencing |
No significant differences in microbiota richness, diversity and composition between mild COVID-19 and control groups |
| Rueca et al. [37] | Cohort study | Italy |
Total: 39 (10 COVID-19 ICU patients, 11 mild to moderate COVID-19 patients, 8 other coronaviruses patients, 10 healthy individuals |
50 | To investigate the difference of nasopharyngeal microbiota composition among COVID-19 patients, other coronavirus patients and healthy individuals | No | No report of antibiotics/probiotic use in the study |
Samples: Nasopharyngeal swabs obtained after COVID-19 infection No report of antibiotics/probiotic use before or after sample collection in the study Assessment method: 16 s rRNA sequencing |
Altered nasopharyngeal microbiota richness among COVID-19 patients, particularly the ICU patients Absence of Deinococcus Thermus, Alicyclobacillaceae, Chromobacteriaceae, Deinococcacaee, Hydrogenophilaceae, Thermoanaerobacteraceae, Sporomusaceae and Thermoanaerobacterales FamilyIII. Incertae Sedis, Johnsonella, Tepidiphilus, Thermoanaerobacter, Ther- moanaerobacterium, Thermosinus and Variovorax in COVID-19 patients and other coronavirus patients Complete depletion of BPB Bifidobacterium and Clostridium in COVID-19 ICU patients Significant decrease in Candidatus Saccharibacteria in COVID-19 ICU patients and other coronavirus patients compared to healthy individuals Opportunistic pathogens Salmonella, Scardovia, Serratia and Pseudomonadaceae were only found in COVID-19 ICU patients Bulleidia, Halanaerobium, Streptobacillus, Epsilonproteobacteria Moraxellaceae, Mycoplasmataceae and Tenericutes were only found in paucisymptomatic COVID-19 patients Pectobacteriaceae were found exclusively to SARS-CoV-2 ICU patients |
| Shen et al. [43] | Cohort study | China |
Total: 53 (8 COVID-19 patients, 25 community-acquired pneumonia (CAP) patients, 20 healthy individuals) |
Not reported | To examine the mutation rate of COVID-19 and also the variances of nasopharyngeal microbiota between COVID-19 and community-acquired pneumonia (CAP) patients | No |
Pneumonia patients included in study received antibiotic Study did not describe the period of antibiotic use |
Samples: Bronchoalveolar lavage fluid through bronchoscope Sample collection was performed after COVID-19 infection Study did not describe if antibiotics were used before or after the sample collection Assessment method: Metatranscriptome sequencing |
No significant differences on the microbiota profile of COVID-19 and CAP patients 25% of COVID-19 sample had more than 5% variation(SARS-CoV-2 mutation rate comparable to Ebola virus) |
| Nardelli et al. [39] | Cohort study | Italy |
Total: 38 (18 COVID-19 patients, 8 recovered COVID19 patients, 12 healthy individuals) |
Not reported | To examine any differences in nasopharyngeal microbiota composition among recovered COVID-19 patients, current COVID-19 patients and healthy individuals | No | No report of antibiotics/probiotic use in the study |
Samples: Nasopharyngeal swabs were obtained after COVID-19 infection No report of antibiotics/probiotic use before or after sample collection in the study Assessment method: 16S rRNA sequencing |
Microbiome significantly different in COVID-19 group compared to the control group Difference in microbiota composition remained after patients’ recovery Fusobacterium periodonticum may increase the susceptibility to COVID-19 infection Significant increase of Firmicutes, Bacteroidetes, Actinobacteria in COVID-19 group compared to control group Significant decrease of Proteobacteria, Fusobacteria, Leptotrichia and Haemophil us compared to A further reduction of Fusobacterium was reported in more severe patients compared to less severe COVID-19 patients |
| Ventero et al. [40] | Cohort study | Italy |
Total: 74 (19 mild COVID-19 patients without hospitalisation, 18 severe COVID-19 patients with hospitalisation, 19 critical COVID19 patents admitted to intensive care units, 18 COVID-19 negative individuals with comorbidities) |
66 | Relationship of nasopharyngeal microbiota composition and COVID-19 severity | No | No report of antibiotics/probiotic use in the study |
Samples: Nasopharyngeal swabs collected after COVID-19 infection No report on whether the antibiotics used were or after the sample collection Assessment method: 16 s rDNA sequencing |
Marked alteration of nasopharyngeal microbiota composition between COVID-19 and control groups Decreased Network complexity of the microbiota was associated with with more severe disease Significant increase of Firmicutes, Bacteroidota, Proteobacteria, Actinobacteria in covid-19 patients compared to non-COVID-19 patients Significant increase of opportunistic pathogens Streptococcus, Prevotella, Veillonella, Haemophilus, Moraxella and Leptotrichia in covid-19 patients compared to non-COVID-19 patients Higher abundance of Prevotella was found in more severe COVID-19 patients compared to less severe COVID-19 patients |
| Rosas-Salazar et al. [38] | Cohort study | USA |
Total: 59 (38 mild-moderate symptomatic COVID-19 patients, 21 uninfected healthy control) |
30 | To examine the difference in composition of airway microbiome between COVID-19 and non-COVID-19 subjects | No |
Participants included in study did not receive any antibiotics for the previous 2 weeks or use any current intranasal medications No report of antibiotics/probiotic use in the study |
Samples: Nasopharyngeal swabs collected after COVID-19 infection Study did not describe whether antibiotics were used after sample collection Assessment method: 16 s rRNA sequencing |
Higher species index of upper respiratory tract microbiota in COVID-19 group compared to healthy control group Marked alteration of airway microbiota composition in COVID-19 patients compared to healthy control group Viral load proportional to the level of alteration of airway micromiome Increased abundance of Corynebacterium, Lawsonella,Staphylococcus, Dolosigranulum and Peptoniphulus in COVID-19 patients compared to healthy control group Increased abundance of Corynebacterium_1,, Moraxella, Dolosigranulum Staphylococcus, and Neisseria in non-COVID-19 subjects compared to COVID-19 subjects More abundance of Neisseriacea, Anaerococcus, Peptoniphulus, Campylobacter, and Enterococcus in COVID-19 patients wih higher viral load compared to healthy control group More abundance of Corynebacterium_1, Staphylococcus, Granilucatella, Neisseria, and Prevotella in COVID-19 patients with lower viral load |
| Miao et al. [41] | Cohort study | China |
Total: 397 (229 mild covid-19 patients, 78 severe covid-10 patients, 16 critical COVID-19 patients, 20 intubated non-COVID19 patients, 31 non-intubated non-COVID viral pneumonia patients, 23 non-intubated healthy subjects) |
70.5 |
To investigate co-infection rate and rate of antimicrobial usage among COVID-19 patients across disease severity To examine any differences in airway microbiota composition between critical COVID-19 patients and other non-COVID-19 patients |
No |
48 mild COVID-19 cases received antibiotics, none received carbapenems 60 severe COVID-19 cases received antibiotics and 3 received carbapenems 16 of critically severe COVID-19 cases received antibiotics and 13 received carbapenems Study did not describe whether antibiotics were given before, after or during COVID19 infection |
Samples: Nasopharyngeal swabs (Bronchoalveolar fluid lavage, Endotracheal aspiration) Sample collection performed after COVID-19 infection Study did not describe whether the antibiotics used were used before or after the sample collection Assessment method: Metagenomic Next-generation Sequencing |
Significantly higher co-infection rate among critical COVID-19 patients (81.3%) compared to severe patients (5.1%) and mild patients (0%) Klebsiella, Enterococcus, Coagulase-negative Staphylcocci, S. wiggsiad and M. hominis were the most common bacterial causes of co-infection in COVID-19 patients Candida, Aspergillus and Cryptococcus were the most common fungal causes of co-infection in COVID-19 patients Cytomegalovirus, Herpes Simplex Virus, Epstein-Barr Virus, Torque Teno Virus, Human Parvovirus B19 and JC Polyomavirus were the most common viral causes of co-infection in COVID-19 patients Anti-microbials commonly used in COVID-19 patients (21% mild patients, 76.9% severe patients, 100% critical patients) Alteration of airway microbiome profile in critical COVID-19 patients was likely due to intubation, rather than COVID-19 infection |
| Braun et al. [44] | Case control study | Israeli |
Total: 33 (21 COVID-19 positive subjects, 12 COVID-19 negative subjects) |
52 | To examine any differences in airway microbiota composition in COVID-19 positive and negative samples | No | No report of antibiotics/probiotic use in the study |
Samples: Nasopharyngeal swabs were obtained after COVID-19 infection Study did not report antibiotics/probiotic use before or after swab collection Assessment method: 16 s rRNA sequencing |
COVID-19 did not insert any significant effect on the composition of airway microbiota No significant difference between COVID-19 positive and negative groups |
| Zhang et al. [42] | Cohort study | China |
Total: 187 (62 COVID-19 patients, 125 non-COVID pneumonia patients) |
Not reported |
To investigate changes of diversity of airway microbiome among COVID-19 patients Gene markers to better diagnose the disease |
Yes | No report of antibiotics/probiotic use in the study |
Samples: Nasopharyngeal swabs from sputum samples Swabs collected after COVID-19 infection Study did not report antibiotic/probiotic usage before or after swab collection Assessment method: RT-PCR and Metatranscriptomic NGS Sequencing |
Airway microbiome in COVID-19 samples were less diversified Certain microbiota were associated with CRP concentration 47.4% of COVID-19 samples revealed an increase of presence in opportunistic pathogens compared to 52% of non-COVID-19 samples 36 differentially expressed genes related to immune pathway such as cytokine signalling were found in COVID-19 samples, suggesting a possible diagnostic marker for COVID-19 Increased abundance of Human influenza virus, Respiratory syncytial viruses, Human alphaherpesvirus 1 and Candida albicans in COVID-19 patients compared to non-COVID-19 patients |
| Mostafa et al. [45] | Cohort study | USA |
Total: 50 (40 COVID-19 patients, 10 patients suspected with COVID-19 infection but with negative results) |
50.5 |
To examine the accuracy of metagenomic next-generation sequencing (mNGS) on COVID-19 diagnosis, the Coinfection in COVID-19 patients and any Changes in the composition of the airway microbiome |
No | No report of antibiotic/probiotic use in the study |
Samples: Nasopharyngeal swabs collected after infection or suspected with COVID-19 No report of antibiotic/probiotic usage before or after swab collection in the study Assessment method: metagenomic sequencing |
mNGS achieved 77.5% accuracy compared to traditional method RT-PCR 12.5% of COVID-19 positive samples contained other opportunistic pathogens Significant decrease in their diversity of the airway microbiota composition in COVID-19 patients, especially in more severe infection compared to negative COVID19 patients High abundance of opportunistic pathogens, including Haemophilus influenzae, Moraxella catarrhalis, human metapneumovirus and human alphaherpesvius in COVID-19 positive samples compared to the negative samples |
| Merenstein et al. [46] | Cohort study | USA |
Total: 113 (83 hospitalised COVID-19 patients < 30 healthy control) |
64 | To examine any alteration in the composition of airway microbiota in COVID-19 patents and its association with disease severity | Yes |
72 COVID- 19 subjects received antibacterial agents 20 COVID- 19 subjects received antifungals agents 18 COVID-19 subjects received antiviral agents 13 Non-COVID-19 subjects received antibacterial Study did not mention the timing of antimicrobial agents used |
Samples: Nasopharyngeal swabs from Endotracheal aspiration- Oropharyngeal sampling Samples obtained after COVID-19 infection Study did not report antibiotic/probiotic usage before or after swab collection Assessment method: 16S rRNA gene sequencing |
The airway microbiome communities of COVID-19 were markedly different from that of the healthy control group The diversity of the microbiome was significant decreased in COVID-19 cases, especially in more severe patients compared to healthy control Level of dysbiosis was associated with COVID-19 severity The microbiome composition as associated with lymphocyte to neutrophil ratio and a specific peripheral blood mononuclear cell profile Unclear whether the systemic immune response was directed by the airway microbiota or other factors such as disease severity Lower abundance of Proteobacteria, Actinobacteria, Haemophilus, Actinomyces and Nisseria in severe COVID-19 samples compared to samples from healthy individuals Higher presence of Bacteroidtes, Anelloviridae, Redondoviridaee in COVID-19 samples compared to samples from healthy individuals Significant increase of opportunistic pathogens including Staphylococcus, Enterococcus, Stenotrophomonas, Enterobacteriaceae and Enterobacterales in COVID-19 patients compared to healthy control Anelloviridae and Redondoviridae showed more frequent colonization and higher titers in severe disease |
|
Cohort studies = 10 Case control study = 1 |
Studies conducted in Italy = 4 Studies conducted in China = 3 Studies conducted in USA = 3 Studies conducted in Israel = 1 |
Mean = 98.45 SEM = 31.31 |
Mean = 54.71 SEM = 4.78 |
Airway microbiota composition during COVID-19 = 10 Coinfection in COVID-19 = 2 Association between airway microbiota and COVID-19 severity = 2 Airway microbiota composition and susceptibility of COVID-19 = 1 |
No = 9 Yes = 2 |
Studies in which antibiotic/probiotic use was not reported = 7 Studies in which antibiotic/probiotic use were included = 3 Studies in which antibiotic/probiotic use by partitcipants was excluded = 1 |
Studies which used samples from Nasopharyngeal swabs = 10 Studies which used samples from Endotracheal aspiration = 2 Studies which used samples from Bronchoalveolar fluid lavage = 2 Studies which used samples from Sputum samples = 1 Studies which used samples from Swabs are collected after COVID-19 infection = 11 Studies with no reports of antibiotics/probiotic use before or after swab collection in the study = 11 Study with no report of sample collection after treatment initiation = 4 |
Studies that reported increase in opportunistic pathogens = 6 Studies that reported decrease in beneficial bacteria = 2 Studies that reported no significant difference in microbiological composition = 3 |
aIn cases of multiple groups of COVID-19 patients, the median age of the largest group is used in the analysis
Table 4.
Studies investigating the association between gut and airway microbiota and COVID-19 infection
| Study | Study type | Country | Sample size | COVID-19 Patients’ median age in years | Addressing co-relationship between microbiota and serum inflammatory markers | Study objective | Antimicrobial and/or probiotic administration in participants | Sample collection and evaluation methods | Key findings |
|---|---|---|---|---|---|---|---|---|---|
| Xu et al. [47] | Cohort study | China |
Total: 64 (35 COVID-19 patients, 10 non-COVID-19 patients with other diseases, 19 healthy subjects) |
Not mentioned | Yes |
To investigate changes of airway and gut microbiota composition during COVID-19 infection and to examine the temporal association between airway and gut microbiota composition during the course of COVID-19 infection |
16 COVID-19 patients were reported that have received antibiotics and/or antiviral agents Probiotic use in COVID-19 patients was not reported Duration of antibiotic use was not reported |
Samples: Throat swabs and anal swabs collected after COVID-19 infection No report on when samples was collected i.e. before or after antibiotic treatment Assessment method: 16 s rRNA seequencing |
The diversity, richness and evenness of both airway and gut microbiome was significantly lower in COVID-19 patients compared to healthy control group Synchronous restoration of microbiota in both the airway and the gut in mild COVID-19 patients Functional bacteria such as Bifidobacterium, Lactobacillus and Faecalibacterium were found to be negatively corelated with the abundance of opportunistic pathogens In the gut: Significant increase of Peudomonas and opportunistic pathogens Neisseria and Actinomyces in COVID-19 patients compared to healthy control In the airway: Significant increase of Alloprevotella, Bacteroidales, Pseudomonas, Actinomycetales in the airway of COVID-19 patients compared to healthy control Significant increase of opportunistic pathogens Rothia, Porphyromonas, Fusobacterium, Neisseria and Saccharibacteria incertae sedis in the airway of COVID-19 patients compared to healthy control |
Results
Overview of included studies
Study selection
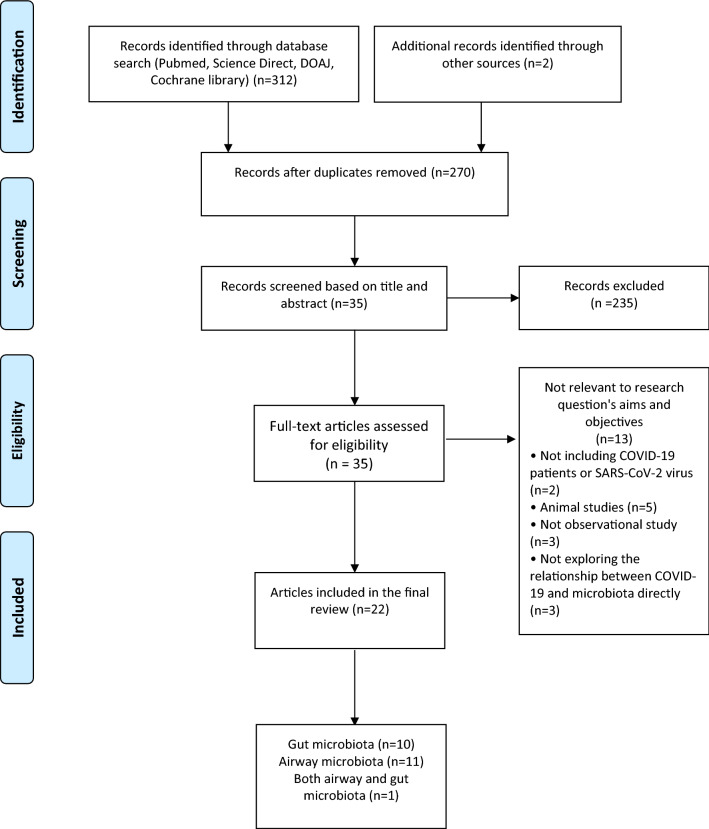
The PRISMA flow diagram [25] shows the number of studies identified, studies excluded and final studies included in the current review paper (Fig. 1). The initial search retrieved 312 articles through database search and two extra papers were found through manual checking of other literature reviews. 44 articles were eliminated due to duplication, giving rise to 270 articles. After screening the papers by reviewing the abstracts and titles and applying the inclusion and exclusion criteria, 35 papers were eligible to be included in this review. During the full manuscript review process, 13 papers were deemed ineligible since they were not relevant to our current research question (2 of them did not include any COVID-19 patients or investigate SARS-CoV-2 virus, 5 of them were animal studies, 3 of them were not observational studies or clinical trials and 3 of them did not investigate the direct relationship between COVID-19 and airway or gut microbiota). In total, 22 studies were included in the review.
Fig. 1.
PRISMA flow diagram on the selection of studies included in the current scoping review
Study characteristics
Ten of the 22 studies included in the review investigated the association between COVID-19 infection and the composition of the gut microbiota, 11 studies examined the changes in the composition of the airway microbiota during infection with COVID-19 and one study focused on the association between COVID-19 and both the airway and gut microbiota. The main characteristics of the studies are shown in Tables 2, 3, 4. Ten studies investigated the association between gut microbiota and COVID-19 [26–35], 11 studies investigated the association between airway microbiota and COVID-19 [36–46] and one study investigated the association between both gut and airway microbiota and COVID-19 [47]. All studies collected their samples after onset of infection with SARS-CoV-2.
Studies investigating the association between gut microbiota and COVID-19
Among the 10 studies investigating the association between gut microbiota and COVID-19 [26–35], eight studies were cohort studies [27–33, 35], one study was a cross-sectional study [26], and one study was a clinical trial [34] (Table 2).
Nine studies focused on the intestinal dysbiosis during COVID-19 infection [26–33, 35], 4 studies examined the relationship between COVID-19 severity with gut microbiota composition [27, 30, 32, 35]. 2 studies focused on intestinal dysbiosis post COVID-19 infection [28, 33] and one study assessed the efficacy of faecal microbiota transplantation post COVID-19 infection [34]. Eight studies collected the microbiota DNA from stool samples [26–31, 34, 43] while 2 studies collected their samples from rectal swabs [32, 35]. Five studies included participants who were administered antimicrobials and/or probiotics prior to sample collection [27, 28, 30, 32, 35], 3 studies excluded these participants [26, 31, 32] while 2 studies did not report this information [29, 33]. Six studies had recruited their participants from China [26, 27, 31–34], three studies from Hong Kong [28–30] and one from Italy [35]. The mean sample size of all studies was 62 (SEM = 16) and the median age of COVID-19 subjects was 55.09 (SEM = 3.41). Only one study [35] mentioned the sample collection time with regards to antibiotic/probiotic/antiviral use; all other studies did not disclose this information.
Studies investigating the association between airway microbiota and COVID-19
The search identified 10 cohort studies [36–43, 45, 46] and one case–control study [44] investigating the association between airway microbiota and COVID-19 (Table 3).
Eleven studies investigated the composition of the airway microbiota during COVID-19 infection [36–46], 2 studies examined the association between the composition of airway microbiota and COVID-19 severity [40, 46] and 2 studies assessed the prevalence of co-infection during COVID-19 [41, 45]. Ten studies examined the DNA of the microbiota collected from nasopharyngeal swabs [36–42, 44–46], 2 studies collected DNA samples from endotracheal aspiration [41, 46] and 2 studies collected samples using bronchoalveolar lavage [41, 43]. Three studies included participants who were administered with antimicrobials and/or probiotics prior to sample collection [41, 43, 46], one study excluded these participants [37] and 7 studies did not report this information [36, 37, 39, 40, 42, 44, 45]. None of the studies reported if antibiotics or probiotics were used before or after the sample collection.
Subjects were recruited from Italy in four studies [36, 37, 39, 40], from China in three studies [41–43], from the US in three studies [38, 45, 46] and from Israel in one study [44]. The mean sample size of all studies was 98 (SEM = 31). The average median age of the COVID-19 subjects in the studies was 54.71 (SEM = 4.78).
Studies investigating the association between both gut and airway microbiota and COVID-19
Only one study investigated the association between both the gut and airway microbiota and COVID-19 [47]. Sixty-four participants were recruited to study the temporal changes of airway and gut microbiota composition during COVID-19 infection. Both anal and throat swabs were used to collect the samples, while subjects had received antimicrobial treatment prior to sample collection (Table 4).
Study outcomes
Studies investigating the association between gut microbiota and COVID-19
Among the 10 studies investigating the role of the gut microbiota on COVID-19, 5 studies concluded that COVID-19 infection increased the amount of opportunistic pathogens in the gut [26–29, 31] and 6 studies suggested that COVID-19 infection reduced the number of beneficial gut microbiota [26, 27, 29–32] (Table 2).
Comparison of gut microbiota in COVID-19 patients vs. patients infected with other viruses and healthy participants
Gu et al. [26] examined the difference in composition of gut microbiota of COVID-19 patients compared to patients infected with H1N1 and healthy individuals due to the similarities of clinical presentation and transmission routes of H1N1 and SARS-CoV-2. The microbial profiles of the COVID-19 and H1N1 patients were significantly less diversified than those of the healthy control group. Moreover, even though the clinical presentations of COVID-19 group and H1N1 group were similar, a lower plasma concentration of lymphocytes and a higher concentration of IL-6 and TNF-α were found in the COVID-19 patients compared to the healthy control group [26] (Table 2).
Lv et al. [31] investigated the association between COVID-19 clinical features and intestinal microbiota, and they found an association between Aspergillus niger and diarrhea symptoms, which was present in 20.89% of COVID-19 patients in this study [31] (Table 2).
Zuo et al. [29] investigated the changes in intestinal fungal microbiota among patients with COVID-19 during their hospitalisation and their recovery. The results of the study showed that the COVID-19 group and healthy control group demonstrated a similar faecal mycobiome profile at baseline; however, patients with COVID-19 and community-acquired pneumonia (CAP) presented with high mycobiome heterogeneity during the course of the disease. Fifty-three percent of COVID-19 patients showed instability with their fecal mycobiome and this continued after their discharge [29].
Gut microbiota, the immune system and disease severity
Yu et al. [32] described the changes of the immune system and gut dysbiosis among 3 COVID-19 patients with severe refractory hypoxaemia and reported a different gut microbiota composition in COVID-19 patients compared to that of healthy individuals. The researchers concluded that the intestinal dysbiosis may be a key factor influencing COVID-19 severity [32] (Table 2).
Yeoh et al. [30] compared the stool samples from hospitalised COVID-19 patients with non-COVID-19 healthy individuals to investigate if intestinal microbiota ecology can reflect COVID-19 clinical severity and immunological profile. It was reported that intestinal dysbiosis being the most important contributing factor for disease severity, while the use of antibiotics was the second most important factor. More importantly, as higher inflammation markers, including CRP, TNF-α and IL-10, were found among COVID-19 patients with severe disease, it was proposed that the alteration of intestinal microbiota composition may be associated with hyper-inflammatory responses [30].
Zuo et al. [28] detected a high level of COVID-19 genome in 7 out of 15 patients’ stool samples, while none of these patients presented with any GI symptoms. A higher abundance of opportunistic bacterial species were found among the stool samples with high COVID-19 infectivity. On the other hand, bacteria associated with immune priming, such as butyrate-producing bacteria, were found in the faecal samples of patients with low SARS-CoV-2 infectivity. These results highlight the importance of the potential faecal–oral transmission route even in patients without GI manifestations [28].
Tang et al. [27] recruited COVID-19 hospitalised patients and categorised them by disease severity. Analysis of stool samples showed a decreased abundance of beneficial bacteria and an increased abundance in opportunistic pathogens in all COVID-19 patients. The intestinal dysbiosis progressed according to the severity of the disease and 55.6% of critical patients presented with intestinal micro-ecological failure. Based on these findings, it was suggested that the intestinal microbial profile could be used as a diagnostic biomarker and a prognostic factor of the severity of COVID-19 [27].
Chen et al. [33] conducted a 6-month follow-up study to determine the diversity of the intestinal microbiota among COVID-19 patients during different timepoints of the disease. It was concluded that the microbiota richness was reduced after disease onset; microbiota diversity decreased during the beginning of the disease and remained unchanged throughout the course of the disease. Interestingly, microbiota richness failed to return to normal even after 6 months post hospitalisation. Patients with the lowest diversity presented with the highest CRP concentration and disease severity, therefore indicating a potential association between the extension of intestinal dysbiosis, inflammatory response and clinical severity [33].
By comparing COVID-19 patients in the intensive care units (ICU) and the infectious diseases wards (ID), Mazzarelli et al. [35] found that microbial richness was reduced in the ICU group compared to the ID group. Furthermore, contrary to non-COVID-19 patients, Proteobacteria were found to be abundant while Spirochaetes and Fusobacteria were depleted in COVID-19 patients.
Fecal microbiota transplantation in COVID-19 patients
Liu et al. [34] were interested in the potential benefit of using faecal microbiota transplantation (FMT) on COVID-19 patients during their recovery. Eleven COVID-19 patients who had been discharged 1 month before the start of the study were recruited and received FMT for 4 days in the form of an oral capsule. Blood and stool samples were collected before and after FMT to observe any immunological profile alterations or changes in the composition of the gut microbiota. Upon receiving FMT, there was a decrease in the naive B cells and an increase in memory B cells and non-switched B cells, suggesting the treatment may have positive effects on the subjects’ immune system. FMT also resulted in an improvement of dysbiosis post COVID-19 infection. At the same time, 5 out of 11 subjects reported GI symptoms after the recovery from COVID-19, and upon receiving FMT, all 5 patients reported a relief of their symptoms [34].
Studies investigating the association between airway microbiota and COVID-19
Among the 11 studies that investigated the association between airway microbiota and COVID-19, 8 studies concluded that there was an association between COVID-19 infection and alteration of airway microbiota composition [37–40, 42, 43, 45, 46], while three studies suggested that there was no significant difference [34, 40, 45]. Six studies concluded that COVID-19 infection increased the coverage of opportunistic pathogens in the airway [37, 39, 40, 42, 45, 46], while 2 studies reported a reduction in commensal bacteria in the airway due to COVID-19 [35, 44] (Table 3).
Airway Microbiota in COVID-19 patients vs. patients infected with other viruses vs healthy participants
Rueca et al. [37] examined the differences in the nasopharyngeal microbiota population among COVID-19 patients, other coronavirus patients and healthy individuals. They reported a decrease in nasopharyngeal microbiota richness among the COVID-19 patients, particularly in those admitted to the ICU. Opportunistic pathogens such as Pseudomonaceae presented exclusively in the COVID-19 ICU group [37].
Shen et al. [43] investigated the mutation rate of SARS-CoV-2 and also the variances in terms of nasopharyngeal microbiota composition between COVID-19 and community-acquired pneumonia (CAP) patients. The results of the study suggested that the mutation speed of SARS-CoV-2 allele may be comparable to that of the Ebola virus. On the other hand, comparison of airway microbiota of COVID-19 and CAP patients to that of healthy controls suggests lung dysbiosis, even though no significant changes were found in microbiome composition between COVID-19 and CAP patients [43].
Nardelli et al. [39] analysed the difference in the composition of the nasopharyngeal microbiota among COVID-19 patients compared to recovered COVID-19 patients and healthy individuals. Compared to the healthy control group, it was found that the microbiome was significantly different in the COVID-19 group in comparison to the non-COVID-19 group. This difference remained unchanged even after the patients’ recovery. Interestingly, Fusobacterium periodonticum (FP), had a negative correlation with symptom severity. Previous studies have demonstrated the role of FP in the metabolism of sialic acid, which may be one of the key receptors of SARS-CoV-2 [48]. Therefore, the reduction of FP may increase patients’ susceptibility to COVID-19 infection [39].
Rosas-Salazar et al. [38] examined if there were any differences in the composition of upper respiratory tract microbiota between mild COVID-19 patients and an uninfected control group. They reported a significant difference in the airway microbiome of nasal swab samples between the two groups. The difference was more prominent in COVID-19 patients with higher viral loads compared to those with lower viral loads [38].
Zhang et al. [42] examined possible changes in the diversity of the airway microbiome among COVID-19 patients compared to non-COVID pneumonia patients. They reported that the airway microbiome in samples from COVID-19 patients was less diversified and more abundant with opportunistic pathogens. The research team also identified various differentially expressed host genes in samples from COVID-19 patients, most of which were related to cytokine signalling deregulation, suggesting an important role in the immunopathogenesis of COVID-19 [42].
Mostafa et al. [45] applied meta-genomic next-generation sequencing (mNGS) on COVID-19-positive and -negative nasopharyngeal swab specimens, and found that mNGS achieved 77.5% accuracy compared to regular diagnostic RT-PCR test. In addition, 12.5% of the samples showed a high abundance of opportunistic pathogens. COVID-19 samples also showed a significant decrease in their airway bacterial diversity which was directly proportionated to disease severity [45].
Merenstein et al. [46] reported a marked difference of airway microbiome communities between the COVID-19 patients and the healthy control group. Microbiota composition was significantly less diverse in patients presenting with severe disease. In addition to the above, the loss of microbiota diversity was also associated with a lower lymphocyte to neutrophil ratio, a well described biomarker linked to disease severity. However, the researchers acknowledged that it remains unclear whether the systemic immune response was directed by the airway microbiota or other factors [46].
Airway microbiota, the immune system and COVID-19 disease severity
Ventero et al. [40] collected microbiota samples from nasopharyngeal swabs of COVID-19 patients and compared them to negative control subjects. The COVID-19 patients were further categorised according to disease severity. A significant difference in the nasopharynx microbiota composition between the two groups was reported. The network complexity of the microbiota was decreased in patients with more severe disease, suggesting a potential biomarker for COVID-19 severity [40].
Studies showing no association between COVID-19 infection and airway microbiota composition
Contrary to the reports of other studies, Braun et al. [44] found no differences in the composition of the airway microbiome between samples from confirmed COVID-19 patients compared to negative COVID-19 patients [44]. Consistently, when De Maio et al. [36] compared the nasopharyngeal microbiota composition among COVID-19 and healthy controls they found that there were no significant differences in microbiota richness, diversity and composition between the SARS-CoV-2-infected patients and the healthy control group [36].
Miao et al. [42] examined if there were any differences in the composition of the airway microbiota between critically ill COVID-19 patients compared to a non-COVID-19 control group. They reported that critically ill COVID-19 patients had a significantly higher co-infection rate (81.3%) compared to those with severe (5.1%) or mild disease (0%). Anti-microbials were also commonly used in all patients (100% of critically severe patients, 76.9% of severe patients and 21% of mild patients). In terms of differences in the airway microbiome, it was concluded that critically ill and intubated COVID-19 patients had a distinct airway microbiome compared to the non-intubated and non-COVID-19 patients. However, the microbiota profile in the intubated COVID-19 patients was similar to that of the intubated non-COVID-19 groups. Therefore, the research team suggested that the differences in airway microbiome in critically COVID-19 patients may be caused by intubation and mechanical ventilation, rather than COVID-19 infection per se [42].
Studies investigating the association between both airway and gut microbiota and COVID-19
Only one cohort study in China was identified in our search to have examined the association between both the gut and airway microbiota and COVID-19 [47] (Table 4). Xu et al. [47] conducted both throat and anal swabs on COVID-19 patients, non-COVID-19 patients and healthy adults to assess any changes between the airway and gut microbiota during the course of SARS-CoV-2 infection. The team reported that the diversity, richness and evenness of both the airway and the gut microbiome were significantly lower in COVID-19 patients, in comparison to both non-COVID-19 and the healthy control group. This alteration was gradually restored back to normal towards the end of their infection in both the upper respiratory tract and the intestine among COVID-19 patients with mild disease. Furthermore, the richness of the microbiome in both organ systems seemed to be negatively associated with the serum level of lipopolysaccharides. In terms of the microbiota composition, an elevated coverage of opportunistic pathogens was found in both the airway and the gut. Interestingly, the absence of functional bacteria, such as Bifidobacterium, Lactobacillus and Faecalibacterium, in the gut was found to be negatively correlated with the abundance of opportunistic pathogens [47].
Discussion
The aim of this review was to report the current evidence from research studies investigating the association between COVID-19 infection and the composition of the gut and airway microbiota. Nineteen studies concluded a correlation between airway and/or gut dysbiosis and SARS-CoV2 [26–35, 37–40, 42, 43, 45–47], while 3 studies failed to observe any significant association between the airway microbiome and SARS-CoV2 infection [36, 41, 44]. The findings of the current review suggest that the gut microbiota may have a more important role than the airway microbiota in COVID-19 infection. Specifically, the studies reported a higher colonization with opportunistic pathogens in both the gut and airway samples of SARS-CoV-2-infected patients. These opportunistic pathogens included Streptococcus, Clostridium, Enterococcus, Peptostreptococcaceae in the gut and Salmonella, Pseudomonadaceae, Bacteroidetes, Streptococcus, Staphylococcus Haemophilus, parainfluenzae, Neisseria, Rothia, Porphyromonas, Sarccharibacteria incertae sedis, Human influenza virus, Respiratory syncytial viruses and Human alphaherpesvirus 1 in the airway. In addition, there was a reported downregulation of commensal bacteria, such as butyrate-producing bacteria (BPB) Ruminococcaceae, Lachnospiraceae, Faecalibacterium and Bifidobacterium in the gut and Bacteroides and Bifidobacterium in the airway.
Interestingly, one of the studies addressed the potential linkage between the respiratory and the gut microbiome [47]. Compared to the gut, the respiratory tract is more susceptible to pathogenic invasions [49, 50], and thus dysbiosis, if any, in the respiratory tract is likely to occur earlier in comparison to intestinal dysbiosis [47]. Through microbe-induced inflammation and swallowing, bacteria may translocate from the oropharynx to the gastrointestinal tract, which potentially explains the association between respiratory and intestinal dysbiosis [50, 51].
Most of the studies included in the current review acknowledged that it may be difficult to identify whether the imbalance in the microbiota composition is the cause or consequence of SARS-CoV-2 infection. In fact, the samples from all studies were collected after the onset of SARS-CoV-2 infection; therefore, there were no baseline microbiota compositions for researchers to compare microbiome diversity during and after SARS-CoV-2 infection. Animal studies may be able to provide some answers, as samples can be collected before, during and after the infection with SARS-CoV-2 to observe any changes in microbiota. By infecting macaques with the SARS-CoV virus, Sokol et al. [52] observed a clear cause and effect relationship between SARS-CoV-2 and intestinal microbiota composition. In particular, faecal samples from infected subjects were dominated by Bacteroidetes, Acinetobacter and Firmicutes. When comparing to SARS-CoV-2 viral load both nasopharyngeally and rectally, it was reported that the coverage of Acinetobacter was proportionated with the nasopharyngeal viral load, while the coverage of Peptostreptococcaceae was proportionated to the rectal viral load. These changes of microbiota ecology were also accompanied with a marked reduction of short-chain fatty acids (SCFAs) produced by the microbiome which may induce local immunomodulatory changes in the subjects [52]. The aforementioned immunomodulatory and taxonomic diversity changes in the microbiota composition, may account for the loss of microbiome functional redundancy and increased susceptibility to colonization from pathogenic bacteria in patients with COVID-19 infection.
Butyrate (one of the key SCFAs)-producing bacterium is significantly reduced during COVID-19 infection [27, 29, 31, 35, 53]. The resulting reduction of butyrate production in the gut may be linked to a pro-inflammatory state [54], thus increasing susceptibility to pulmonary viral infections, including COVID-19 [55–57]. Although evidence suggests that SCFA does not interfere with COVID-19 infection in the intestine [53], a reduction in SCFA may promote a systemic pro-inflammatory state in both macaques [52] and humans [58]. In support of this, dietary interventions may alter the composition of airway and intestinal microbiota, which may potentially affect the clinical course of COVID-19 infection by modulating systemic immune responses [59].
A decrease in SCFA can also downregulate the ACE2 [60], one of the key receptors of SARS-CoV-2 virus [61]. The depletion of ACE2 has been shown to promote epithelial damage in the intestine and thus increase susceptibility to inflammation in animal studies [62]. The abundance of SARS-CoV-2 virus in the gastrointestinal tract was proven to provide a gastrointestinal route for COVID-19 infection [63] which may worsen intestinal dysbiosis. Therefore, there is a pathogenic feedback loop between the reduced production of SCFA and SARS-CoV-2 [31]. A similar phenomenon can be observed in the airway through the downregulation of pharyngeal mucosal ACE2 by the imbalance of the airway microbiome [62, 64, 65].
The increase in opportunistic pathogens in the gut and in the airway during the course of COVID-19 also plays a significant role in worsening disease prognosis. Co-infection has been shown to be a key prognostic factor in COVID-19 [66]; for example, pulmonary aspergillosis was estimated to occur in 19–33% of COVID-19 patients [67–69], and was reported in two of the studies included in this review [29, 31]; Pseudomonas is one of the most commonly reported pathogens isolated from COVID-19 patients with bacterial co-infection [70], which was reported in one study included in this review as well [37].
Several limitations have been identified in the studies included in the current review. All papers studied COVID-19 cases retrospectively and there was lack of longitudinal data to determine microbiota composition at baseline since this is not routinely assessed in the management of COVID-19 patients. More animal studies in the future may give us further insight on the progression of the microbiome alteration during the whole course of disease [71]. Another limitation is that 9 out of 10 gut microbiota studies came from either China or Hong Kong, while subjects from the airway microbiota studies were recruited in 6 different countries. This suggests that the literature on intestinal microbiome could contain a higher level of selection bias due to lack of geographic heterogeneity. Moreover, the high average median ages in our analysis may represent a relatively homogeneous demographic exposed to similar environmental factors. This could affect the objectivity when studying the patients’ microbiota profiles [72]. The small average sample size of all studies is also an important limitation of the studies presented in the current paper. However, it is important to note that most studies were pilot studies due to the recent occurrence of COVID-19 pandemic. In addition, only one study included non-hospitalised COVID-19 patients [40], while 5 studies recruited patients from the Intensive Care Unit for comparison [27, 35, 37, 40, 41]. The composition of the microbiota could have been affected by the hospital [73] and ICU [74] environment, and may have therefore affected the objectivity of the studies. Furthermore, only 4 out of 22 studies excluded subjects who received antimicrobial and/or probiotic treatment prior to the sample collection [26, 31, 34, 38], while 9 studies chose to include these participants [27, 28, 30, 32, 35, 41, 43, 46, 47] and 9 studies did not report this information [29, 33, 36, 37, 39, 40, 42, 44, 45]. In addition, with the exception of one study [35], the remaining studies did not report the time of sample collection in regards to the administration of antibiotic or probiotic. In fact, the use of antimicrobials have been shown to affect intestinal [75] and respiratory tract microbiota composition [49], therefore introducing bias in the findings. However, the difficulty to exclude these subjects is understandable, especially when antibiotics are commonly prescribed among critically ill COVID-19 patients [30]. In terms of sample collection, although certain studies suggest that rectal swabs are inferior to faecal sample collection [76], other studies suggested that they may be interchangeable [77, 78]. However, in airway microbiota studies, bronchoalveolar fluid lavage has been found to be a better sample collection method compared to nasopharyngeal swabs or throat swabs [79, 80], suggesting a better alternative for future study design. On the other hand, the composition of the microbiota may be easily affected during the process of sample collection by the personnel involved in sample collection as well as the hospital environment [81].
Conclusion
Our current review supports that there is overwhelming evidence that significant changes of the gut/airway microbiota composition are associated with SARS-CoV-2 infection and influence disease progression and prognosis. In particular, the gut microbiota may have a more important role than the airway microbiota in COVID-19 infection. However, studies with larger cohorts, longer duration, broader age group, more diverse subject demographics from different geographical locations, as well as studies that take into consideration provision of antimicrobials and probiotic administration in COVID-19 patients are needed to derive more conclusive results on identifying individualized changes of microbiota composition alterations of the gut-lung axis during SARS-CoV-2 infection. The completion of such studies will derive more conclusive evidence of the role of the microbiota in COVID-19 disease and will allow the development of better management strategies to be applied in combating COVID-19. As the pandemic progresses, a novel group of chronic patients with long-COVID is expected to pose a new challenge for healthcare settings worldwide. Sustained loss of intestinal microbiota diversity could be implicated in prolonged immunological changes that, in theory, could account for chronic symptoms associated with SARS-CoV-2 infection. Unravelling the dynamics of microbiome changes in the context of COVID-19 may offer a novel target for therapeutic interventions that extend beyond the acute phase of the disease.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
Search terms were designed with the support of Mrs Carrie Rodomar, Head Librarian at the University of Nicosia Medical School.
Abbreviations
- SARS-CoV-2
Severe Acute Respiratory Syndrome Coronavirus 2
- COVID-19
2019 Novel coronavirus disease
- ARDS
Acute Respiratory Distress Syndrome
- SIRS
Systemic Inflammatory Response Syndrome
- ACE2
Angiotensin-converting enzyme 2
- CAP
Community-Acquired Pneumonia
- ICU
Intensive Care Units
- FMT
Fecal Microbiota Transplantation
- F. periodonticum
Fusobacterium periodonticum
- mNGS
Metagenomic Next-Generation Sequencing
- BPB
Butyrate-Producing Bacteria
- SCFAs
Short-Chain Fatty Acids
Funding
No external funding was received for this work.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Lipsitch M, Swerdlow DL, Finelli L. Defining the epidemiology of Covid-19: studies needed. N Engl J Med. 2020;382:1194–1196. doi: 10.1056/NEJMp2002125. [DOI] [PubMed] [Google Scholar]
- 2.Chan JF-W, Yuan S, Kok K-H, To KK-W, Chu H, Yang J, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qiu H, Wu J, Hong L, Luo Y, Song Q, Chen D. Clinical and epidemiological features of 36 children with coronavirus disease 2019 (COVID-19) in Zhejiang, China: an observational cohort study. Lancet Infect Dis. 2020;20:689–696. doi: 10.1016/S1473-3099(20)30198-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu J, Ji P, Pang J, Zhong Z, Li H, He C, et al. Clinical characteristics of 3062 COVID-19 patients: a meta-analysis. J Med Virol. 2020;92:1902–1914. doi: 10.1002/jmv.25884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.WHO. Coronavirus disease (COVID-2019) situation reports 2021
- 7.WHO. WHO Coronavirus (COVID-19) Dashboard 2021
- 8.van Doremalen N, Bushmaker T, Morris DH, Holbrook MG, Gamble A, Williamson BN, et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N Engl J Med. 2020;382:1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jia HP, Look DC, Shi L, Hickey M, Pewe L, Netland J, et al. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J Virol. 2005;79:14614–14621. doi: 10.1128/JVI.79.23.14614-14621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020 doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mutua MP, Muya S, Muita GM. A general perspective of microbiota in human health and disease. Arch Clin Microbiol. 2020 doi: 10.36648/1989-8436.11.2.106. [DOI] [Google Scholar]
- 13.Vemuri R, Gundamaraju R, Shastri MD, Shukla SD, Kalpurath K, Ball M, et al. Gut microbial changes, interactions, and their implications on human lifecycle: an ageing perspective. BioMed Res Int. 2018;2018:1–13. doi: 10.1155/2018/4178607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vemuri R, Shankar EM, Chieppa M, Eri R, Kavanagh K. Beyond just bacteria: functional biomes in the gut ecosystem including virome, mycobiome archaeome and helminths. Microorganisms. 2020;8:483. doi: 10.3390/microorganisms8040483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuki K, Fujiogi M, Koutsogiannaki S. COVID-19 pathophysiology: a review. Clin Immunol. 2020;215:108427. doi: 10.1016/j.clim.2020.108427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi HY, Zhu X, Li WL, Mak JWY, Wong SH, Zhu ST, et al. Modulation of gut microbiota protects against viral respiratory tract infections: a systematic review of animal and clinical studies. Eur J Nutr. 2021 doi: 10.1007/s00394-021-02519-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen C-J, Wu G-H, Kuo R-L, Shih S-R. Role of the intestinal microbiota in the immunomodulation of influenza virus infection. Microbes Infect. 2017;19:570–579. doi: 10.1016/j.micinf.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 18.Sencio V, Machado MG, Trottein F. The lung–gut axis during viral respiratory infections: the impact of gut dysbiosis on secondary disease outcomes. Mucosal Immunol. 2021;14:296–304. doi: 10.1038/s41385-020-00361-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci. 2011;108:5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chattopadhyay I, Shankar EM. SARS-CoV-2-indigenous microbiota nexus: Does gut microbiota contribute to inflammation and disease severity in COVID-19? Front Cell Infect Microbiol. 2021;11:590874. doi: 10.3389/fcimb.2021.590874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chhibber-Goel J, Gopinathan S, Sharma A. Interplay between severities of COVID-19 and the gut microbiome: implications of bacterial co-infections? Gut Pathog. 2021;13:14. doi: 10.1186/s13099-021-00407-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Di Stadio A, Costantini C, Renga G, Pariano M, Ricci G, Romani L. The microbiota/host immune system interaction in the nose to protect from COVID-19. Life. 2020;10:345. doi: 10.3390/life10120345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peters MDJ, Godfrey CM, Khalil H, McInerney P, Parker D, Soares CB. Guidance for conducting systematic scoping reviews. Int J Evid Based Healthc. 2015;13:141–146. doi: 10.1097/XEB.0000000000000050. [DOI] [PubMed] [Google Scholar]
- 24.Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. PLOS Med. 2021;18:e1003583. doi: 10.1371/journal.pmed.1003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021;372:n71. doi: 10.1136/bmj.n71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu S, Chen Y, Wu Z, Chen Y, Gao H, Lv L, et al. Alterations of the gut microbiota in patients with Coronavirus Disease 2019 or H1N1 influenza. Clin Infect Dis. 2020;71:2669–2678. doi: 10.1093/cid/ciaa709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang L, Gu S, Gong Y, Li B, Lu H, Li Q, et al. Clinical significance of the correlation between changes in the major intestinal bacteria species and COVID-19 severity. Engineering. 2020;6:1178–1184. doi: 10.1016/j.eng.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zuo T, Liu Q, Zhang F, Lui GC-Y, Tso EY, Yeoh YK, et al. Depicting SARS-CoV-2 faecal viral activity in association with gut microbiota composition in patients with COVID-19. Gut. 2020 doi: 10.1136/gutjnl-2020-322294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zuo T, Zhan H, Zhang F, Liu Q, Tso EYK, Lui GCY, et al. Alterations in fecal fungal microbiome of patients with COVID-19 during time of hospitalization until discharge. Gastroenterology. 2020;159:1302–1310.e5. doi: 10.1053/j.gastro.2020.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeoh YK, Zuo T, Lui GC-Y, Zhang F, Liu Q, Li AY, et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut. 2021;70:698–706. doi: 10.1136/gutjnl-2020-323020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lv L, Gu S, Jiang H, Yan R, Chen Y, Chen Y, et al. Gut mycobiota alterations in patients with COVID-19 and H1N1 infections and their associations with clinical features. Commun Biol. 2021;4:480. doi: 10.1038/s42003-021-02036-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu L, Tong Y, Shen G, Fu A, Lai Y, Zhou X, et al. Immunodepletion with hypoxemia: a potential high risk subtype of Coronavirus Disease 2019. Infect Dis (Except HIV/AIDS) 2020 doi: 10.1101/2020.03.03.20030650. [DOI] [Google Scholar]
- 33.Chen Y, Gu S, Chen Y, Lu H, Shi D, Guo J, et al. Six-month follow-up of gut microbiota richness in patients with COVID-19. Gut. 2021 doi: 10.1136/gutjnl-2021-324090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu F, Ye S, Zhu X, He X, Wang S, Li Y, et al. Gastrointestinal disturbance and effect of fecal microbiota transplantation in discharged COVID-19 patients. J Med Case Rep. 2021;15:60. doi: 10.1186/s13256-020-02583-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mazzarelli A, Giancola ML, Farina A, Marchioni L, Rueca M, Gruber CEM, et al. 16S rRNA gene sequencing of rectal swab in patients affected by COVID-19. PLoS ONE. 2021;16:e0247041. doi: 10.1371/journal.pone.0247041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Maio F, Posteraro B, Ponziani FR, Cattani P, Gasbarrini A, Sanguinetti M. Nasopharyngeal microbiota profiling of SARS-CoV-2 infected patients. Biol Proced Online. 2020;22:18. doi: 10.1186/s12575-020-00131-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rueca M, Fontana A, Bartolini B, Piselli P, Mazzarelli A, Copetti M, et al. Investigation of nasal/oropharyngeal microbial community of COVID-19 patients by 16S rDNA sequencing. Int J Environ Res Public Health. 2021;18:2174. doi: 10.3390/ijerph18042174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosas-Salazar C, Kimura KS, Shilts MH, Strickland BA, Freeman MH, Wessinger BC, et al. SARS-CoV-2 infection and viral load are associated with the upper respiratory tract microbiome. J Allergy Clin Immunol. 2021;147:1226–1233.e2. doi: 10.1016/j.jaci.2021.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nardelli C, Gentile I, Setaro M, Di Domenico C, Pinchera B, Buonomo AR, et al. Nasopharyngeal microbiome signature in COVID-19 positive patients: Can we definitively get a role to fusobacterium periodonticum? Front Cell Infect Microbiol. 2021;11:625581. doi: 10.3389/fcimb.2021.625581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ventero MP, Cuadrat RRC, Vidal I, Andrade BGN, Molina-Pardines C, Haro-Moreno JM, et al. Nasopharyngeal microbial communities of patients infected with SARS-CoV-2 that developed COVID-19. Front Microbiol. 2021;12:637430. doi: 10.3389/fmicb.2021.637430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miao Q, Ma Y, Ling Y, Jin W, Su Y, Wang Q, et al. Evaluation of superinfection, antimicrobial usage, and airway microbiome with metagenomic sequencing in COVID-19 patients: a cohort study in Shanghai. J Microbiol Immunol Infect. 2021 doi: 10.1016/j.jmii.2021.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang H, Ai J-W, Yang W, Zhou X, He F, Xie S, et al. Metatranscriptomic characterization of COVID-19 identified a host transcriptional classifier associated with immune signaling. Clin Infect Dis. 2021;73:376. doi: 10.1093/cid/ciaa663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen Z, Xiao Y, Kang L, Ma W, Shi L, Zhang L, et al. Genomic diversity of severe acute respiratory Syndrome-Coronavirus 2 in patients with Coronavirus Disease 2019. Clin Infect Dis. 2020;71:713–720. doi: 10.1093/cid/ciaa203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Braun T, Halevi S, Hadar R, Efroni G, Glick Saar E, Keller N, et al. SARS-CoV-2 does not have a strong effect on the nasopharyngeal microbial composition. Sci Rep. 2021;11:8922. doi: 10.1038/s41598-021-88536-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mostafa HH, Fissel JA, Fanelli B, Bergman Y, Gniazdowski V, Dadlani M, et al. Metagenomic next-generation sequencing of nasopharyngeal specimens collected from confirmed and suspect COVID-19 patients. MBio. 2020;11:e01969–e2020. doi: 10.1128/mBio.01969-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Merenstein C, Liang G, Whiteside SA, Cobián-Güemes AG, Merlino MS, Taylor LJ, et al. Signatures of COVID-19 severity and immune response in the respiratory tract microbiome. Infect Dis (Except HIV/AIDS) 2021 doi: 10.1101/2021.04.02.21254514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu R, Lu R, Zhang T, Wu Q, Cai W, Han X, et al. Temporal association between human upper respiratory and gut bacterial microbiomes during the course of COVID-19 in adults. Commun Biol. 2021;4:240. doi: 10.1038/s42003-021-01796-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morniroli D, Giannì ML, Consales A, Pietrasanta C, Mosca F. Human sialome and Coronavirus Disease-2019 (COVID-19) pandemic: An understated correlation? Front Immunol. 2020;11:1480. doi: 10.3389/fimmu.2020.01480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Man WH, de Steenhuijsen Piters WAA, Bogaert D. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat Rev Microbiol. 2017;15:259–270. doi: 10.1038/nrmicro.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dang AT, Marsland BJ. Microbes, metabolites, and the gut–lung axis. Mucosal Immunol. 2019;12:843–850. doi: 10.1038/s41385-019-0160-6. [DOI] [PubMed] [Google Scholar]
- 51.Wypych TP, Wickramasinghe LC, Marsland BJ. The influence of the microbiome on respiratory health. Nat Immunol. 2019;20:1279–1290. doi: 10.1038/s41590-019-0451-9. [DOI] [PubMed] [Google Scholar]
- 52.Sokol H, Contreras V, Maisonnasse P, Desmons A, Delache B, Sencio V, et al. SARS-CoV-2 infection in nonhuman primates alters the composition and functional activity of the gut microbiota. Gut Microbes. 2021;13:1–19. doi: 10.1080/19490976.2021.1893113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pascoal LB, Rodrigues PB, Genaro LM, dos Gomes ABSP, Toledo-Teixeira DA, Parise PL, et al. Microbiota-derived short-chain fatty acids do not interfere with SARS-CoV-2 infection of human colonic samples. Gut Microbes. 2021;13:1–9. doi: 10.1080/19490976.2021.1874740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qin N, Zheng B, Yao J, Guo L, Zuo J, Wu L, et al. Influence of H7N9 virus infection and associated treatment on human gut microbiota. Sci Rep. 2015;5:14771. doi: 10.1038/srep14771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Friedland RP, Haribabu B. The role for the metagenome in the pathogenesis of COVID-19. EBioMedicine. 2020;61:103019. doi: 10.1016/j.ebiom.2020.103019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tang Y, Liu J, Zhang D, Xu Z, Ji J, Wen C. Cytokine storm in COVID-19: the current evidence and treatment strategies. Front Immunol. 2020;11:1708. doi: 10.3389/fimmu.2020.01708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chemudupati M, Kenney AD, Smith AC, Fillinger RJ, Zhang L, Zani A, et al. Butyrate reprograms expression of specific interferon-stimulated genes. J Virol. 2020;94:e00326–e420. doi: 10.1128/JVI.00326-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Esquivel-Elizondo S, Ilhan ZE, Garcia-Peña EI, Krajmalnik-Brown R. Insights into butyrate production in a controlled fermentation system via gene predictions. mSystems. 2017 doi: 10.1128/mSystems.00051-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gasmi A, Tippairote T, Mujawdiya PK, Peana M, Menzel A, Dadar M, et al. The microbiota-mediated dietary and nutritional interventions for COVID-19. Clin Immunol. 2021;226:108725. doi: 10.1016/j.clim.2021.108725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Robinson CM, Pfeiffer JK. Viruses and the microbiota. Annu Rev Virol. 2014;1:55–69. doi: 10.1146/annurev-virology-031413-085550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Melin AD, Janiak MC, Marrone F, Arora PS, Higham JP. Comparative ACE2 variation and primate COVID-19 risk. Commun Biol. 2020;3:641. doi: 10.1038/s42003-020-01370-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hashimoto T, Perlot T, Rehman A, Trichereau J, Ishiguro H, Paolino M, et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. 2012;487:477–481. doi: 10.1038/nature11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xiao F, Tang M, Zheng X, Liu Y, Li X, Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158:1831–1833.e3. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Koester ST, Li N, Lachance DM, Morella NM, Dey N. Variability in digestive and respiratory tract Ace2 expression is associated with the microbiome. PLoS ONE. 2021;16:e0248730. doi: 10.1371/journal.pone.0248730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Khatiwada S, Subedi A. Lung microbiome and coronavirus disease 2019 (COVID-19): possible link and implications. Hum Microbiome J. 2020;17:100073. doi: 10.1016/j.humic.2020.100073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cox MJ, Loman N, Bogaert D, O’Grady J. Co-infections: potentially lethal and unexplored in COVID-19. Lancet Microbe. 2020;1:e11. doi: 10.1016/S2666-5247(20)30009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alanio A, Dellière S, Fodil S, Bretagne S, Mégarbane B. Prevalence of putative invasive pulmonary aspergillosis in critically ill patients with COVID-19. Lancet Respir Med. 2020;8:e48–e49. doi: 10.1016/S2213-2600(20)30237-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Koehler P, Cornely OA, Böttiger BW, Dusse F, Eichenauer DA, Fuchs F, et al. COVID-19 associated pulmonary aspergillosis. Mycoses. 2020;63:528–534. doi: 10.1111/myc.13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van Arkel A, Rijpstra TA, Belderbos H, van Wijngaarden P, Verweij PE, Bentvelsen RG. COVID-19-associated pulmonary aspergillosis. Am J Resp Crit Care Med. 2020;202(1):132–135. doi: 10.1164/rccm.202004-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Qu J, Cai Z, Liu Y, Duan X, Han S, Liu J, et al. Persistent bacterial coinfection of a COVID-19 Patient caused by a genetically adapted pseudomonas aeruginosa chronic colonizer. Front Cell Infect Microbiol. 2021;11:641920. doi: 10.3389/fcimb.2021.641920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang T, Chakraborty S, Saha P, Mell B, Cheng X, Yeo J-Y, et al. Gnotobiotic rats reveal that gut microbiota regulates colonic mRNA of Ace2, the receptor for SARS-CoV-2 infectivity. Hypertension. 2020 doi: 10.1161/HYPERTENSIONAHA.120.15360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Scepanovic P, Hodel F, Mondot S, Partula V, Byrd A, The Milieu Intérieur Consortium et al. A comprehensive assessment of demographic, environmental, and host genetic associations with gut microbiome diversity in healthy individuals. Microbiome. 2019;7:130. doi: 10.1186/s40168-019-0747-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ticinesi A, Milani C, Lauretani F, Nouvenne A, Mancabelli L, Lugli GA, et al. Gut microbiota composition is associated with polypharmacy in elderly hospitalized patients. Sci Rep. 2017;7:11102. doi: 10.1038/s41598-017-10734-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Aardema H. Marked changes in gut microbiota in cardio-surgical intensive care patients: a longitudinal cohort study. Front Cell Infect Microbiol. 2020;9:10. doi: 10.3389/fcimb.2019.00467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ramirez J, Guarner F, Bustos Fernandez L, Maruy A, Sdepanian VL, Cohen H. Antibiotics as major disruptors of gut microbiota. Front Cell Infect Microbiol. 2020;10:572912. doi: 10.3389/fcimb.2020.572912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mottawea W, Butcher J, Li J, Abujamel T, Manoogian J, Mack D, et al. The mucosal–luminal interface: an ideal sample to study the mucosa-associated microbiota and the intestinal microbial biogeography. Pediatr Res. 2019;85:895–903. doi: 10.1038/s41390-019-0326-7. [DOI] [PubMed] [Google Scholar]
- 77.Bassis CM, Moore NM, Lolans K, Seekatz AM, Weinstein RA, For the CDC Prevention Epicenters Program et al. Comparison of stool versus rectal swab samples and storage conditions on bacterial community profiles. BMC Microbiol. 2017;17:78. doi: 10.1186/s12866-017-0983-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Reyman M, van Houten MA, Arp K, Sanders EAM, Bogaert D. Rectal swabs are a reliable proxy for faecal samples in infant gut microbiota research based on 16S-rRNA sequencing. Sci Rep. 2019;9:16072. doi: 10.1038/s41598-019-52549-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Marsh RL, Kaestli M, Chang AB, Binks MJ, Pope CE, Hoffman LR, et al. The microbiota in bronchoalveolar lavage from young children with chronic lung disease includes taxa present in both the oropharynx and nasopharynx. Microbiome. 2016;4:37. doi: 10.1186/s40168-016-0182-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Van Driessche L, Valgaeren BR, Gille L, Boyen F, Ducatelle R, Haesebrouck F, et al. A deep nasopharyngeal swab versus nonendoscopic bronchoalveolar lavage for isolation of bacterial pathogens from preweaned calves with respiratory disease. J Vet Intern Med. 2017;31:946–953. doi: 10.1111/jvim.14668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim D, Hofstaedter CE, Zhao C, Mattei L, Tanes C, Clarke E, et al. Optimizing methods and dodging pitfalls in microbiome research. Microbiome. 2017;5:52. doi: 10.1186/s40168-017-0267-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.