Abstract
Design and synthesis of N-(trifluoromethyl)phenyl substituted pyrazole derivatives and their potency as antimicrobial agents are described. Several of these novel compounds are effective growth inhibitors of antibiotic-resistant Gram-positive bacteria and prevent the development of biofilms by methicillin-resistant Staphylococcus aureus (MRSA) and Enterococcus faecalis. These compounds eradicated the preformed biofilms effectively and were found to be more effective than the control antibiotic vancomycin. Potent compounds showed low toxicity to cultured human embryonic kidney cells with a selectivity factor of >20. The most promising compound is very potent against meropenem, oxacillin, and vancomycin-resistant clinical isolates of Enterococcus faecium. Investigations into the mode of action by performing macromolecular synthesis inhibition studies showed a broad range of inhibitory effects, suggesting targets that have a global effect on bacterial cell function.
Design and synthesis of N-(trifluoromethyl)phenyl substituted pyrazole derivatives and their potency as antimicrobial agents are described.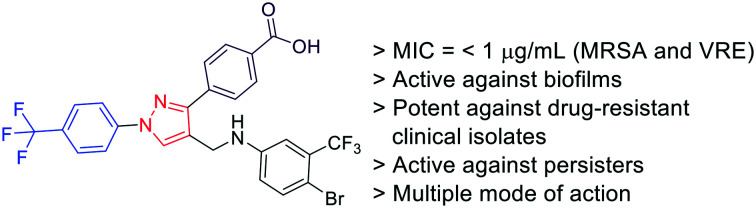
Introduction
Microbial resistance to antibiotics is an unresolved American and global concern.1 Drug-resistant infections cause more than 700 000 deaths worldwide annually, and the toll is expected to reach 10 million per year by 2050.2 At least 2.8 million people are infected with drug-resistant microbes and more than 35 000 people die each year as a result of these infections in the US alone.2,3 Without urgent and coordinated actions, we are moving toward an era in which normal infections or minor injuries may become fatal. The Centers for Disease Control and Prevention (CDC) recommends combatting antibiotic resistance by promoting the development of new antibiotics for drug-resistant bacteria.4,5
Almost 30% and 2% of people are carriers of Staphylococcus aureus and its drug-resistant variant, methicillin-resistant S. aureus (MRSA) respectively.6S. aureus is the leading cause of drug-resistant microbial infections and MRSA causes 10-fold more infections than all the multi-drug resistant (MDR) Gram-negative pathogens combined. Enterococcus faecalis and Enterococcus faecium have become the leading causes of nosocomial infections in recent years.7 These enterococci strains are intrinsically resistant to several commonly used antibiotics and rapidly acquire resistance to several potent antibiotics including vancomycin. Therefore, the choice of effective antibiotics to treat enterococcal infections is limited.8
Results and discussion
Synthesis of N-(trifluoromethyl)phenyl substituted pyrazole derivatives
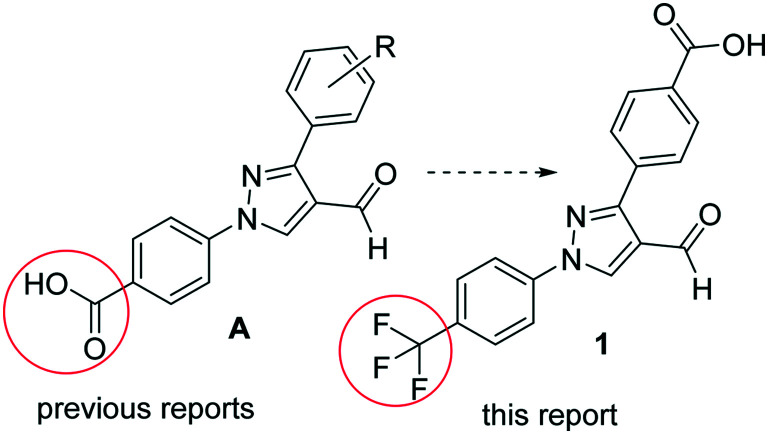
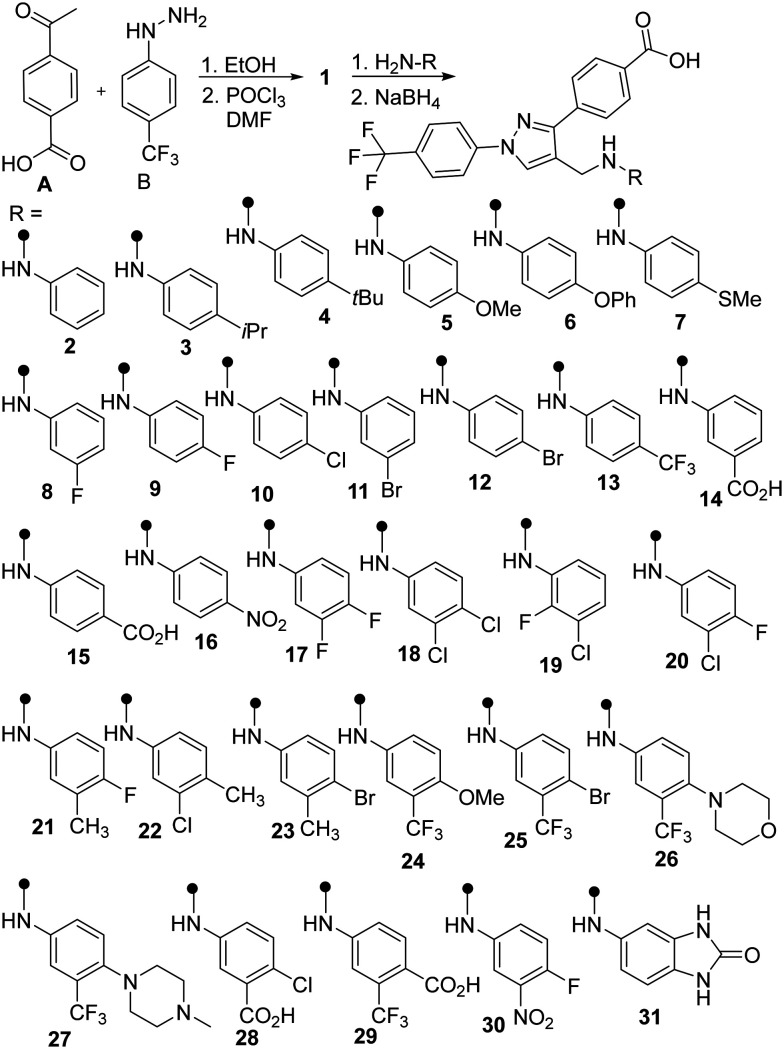
Pyrazole derivatives are known for their wide range of pharmacological activities including antibacterial properties.9–11 In a continuation of our effort to find potent antimicrobial agents, we have reported the synthesis and antimicrobial studies of several pyrazole-derived hydrazones12–16 and anilines.17 These compounds are 4-pyrazolobenzoic acid derivatives (Fig. 1A). In this report, we replaced the carboxylic acid with the trifluoromethyl (CF3) group. The CF3 group is found in many drugs and pharmacologically active molecules.18 It is a classic isostere of several non-polar groups.19 We have found that the presence of the CF3 group in the N-aryl moiety of pyrazole derivatives reduces the toxicity against human embryonic kidney (HEK293) cells while retaining their potency against MRSA.20 To reduce the lipophilicity of the molecule (1), we added a carboxylic acid group to the other phenyl ring (Fig. 1). The designed aldehyde derivative (1) was synthesized efficiently by using the methods of our previous reports (Scheme 1).17,21 The reaction of 4-acetylbenzoic acid (A) with 4-(trifluoromethyl)phenyl hydrazine (B) formed the intermediate hydrazone, which on reaction with in situ generated Vilsmeier reagent afforded pyrazole-derived aldehyde (1). This pure aldehyde derivative (1) was synthesized in multigram scale, which on reductive amination with different anilines afforded the target molecules (Scheme 1). We synthesized a series of 30 compounds containing mono- (1–16) and disubstituted (17–31) aniline derivatives (Table 1).
Fig. 1. Design of compounds based on the lead structures.
Scheme 1. Synthesis of pyrazole derived anilines.
Antimicrobial activity of synthesized compounds (2–31). Gram-positive antibiotic susceptible S. aureus ATCC 25923 (Sa23), antibiotic-resistant: S. aureus ATCC 33591 (Sa91), S. aureus ATCC 33592 (Sa92), S. aureus BAA-2312 (Sa12), and S. aureus ATCC 700699 (Sa99); S. epidermidis ATCC 700296 (Se), Bacillus subtilis ATCC 6623 (Bs), antibiotic susceptible Enterococcus faecalis ATCC 29212 (Efs12) and vancomycin-resistant Enterococcus faecium ATCC 700221 (Efm21). Gram-negative bacteria: A. baumannii ATCC 19606, A. baumannii ATCC BAA-1605, A. baumannii ATCC 747, Escherichia coli ATCC 25922, Enterobacter aerogenes ATCC 13048, P. aeruginosa ATCC 27833, K. pneumoniae ATCC 700603 (no activity against Gram-negative bacterial strains), and V = vancomycin.
| # | Sa23 | Sa91 | Sa92 | Sa12 | Sa99 | Se | Bs | Efs12 | Efm21 |
|---|---|---|---|---|---|---|---|---|---|
| 2 | 25 | 50 | 25 | 25 | 25 | 25 | 25 | NA | 25 |
| 3 | 3.12 | 12.5 | 3.12 | 6.25 | 3.12 | 12.5 | 3.12 | 6.25 | 3.12 |
| 4 | 1.56 | 3.12 | 1.56 | 3.12 | 1.56 | 3.12 | 1.56 | 3.12 | 1.56 |
| 5 | 50 | NA | 25 | NA | 25 | NA | 25 | NA | NA |
| 6 | 1.56 | 3.12 | 1.56 | 3.12 | 1.56 | 3.12 | 1.56 | 3.12 | 1.56 |
| 7 | 12.5 | 25 | 25 | 25 | 12.5 | 50 | 25 | 25 | 12.5 |
| 8 | 25 | 25 | 25 | 25 | 12.5 | 25 | 12.5 | 25 | 12.5 |
| 9 | 25 | 25 | 25 | 25 | 12.5 | 50 | 25 | NA | 25 |
| 10 | 6.25 | 6.25 | 6.25 | 12.5 | 6.25 | 12.5 | 6.25 | 12.5 | 6.25 |
| 11 | 3.12 | 3.12 | 6.25 | 6.25 | 3.12 | 12.5 | 6.25 | 12.5 | 6.25 |
| 12 | 6.25 | 6.25 | 3.12 | 6.25 | 3.12 | 12.5 | 3.12 | 6.25 | 6.25 |
| 13 | 12.5 | 12.5 | 6.25 | 12.5 | 3.12 | 12.5 | 6.25 | 25 | 12.5 |
| 14 | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| 15 | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| 16 | 25 | 25 | 12.5 | 25 | 12.5 | 25 | 12.5 | 25 | 25 |
| 17 | 12.5 | 25 | 6.25 | 25 | 6.25 | 25 | 12.5 | 25 | 12.5 |
| 18 | 1.56 | 1.56 | 1.56 | 1.56 | 0.78 | 3.12 | 0.39 | 3.12 | 1.56 |
| 19 | 3.12 | 6.25 | 3.12 | 6.25 | 3.12 | 12.5 | 1.56 | 12.5 | 6.25 |
| 20 | 3.12 | 12.5 | 3.12 | 6.25 | 3.12 | 12.5 | 3.12 | 12.5 | 6.25 |
| 21 | 12.5 | 25 | 6.25 | 25 | 6.25 | 25 | 12.5 | 25 | 12.5 |
| 22 | 3.12 | 6.25 | 3.12 | 3.12 | 1.56 | 6.25 | 1.56 | 6.25 | 3.12 |
| 23 | 3.12 | 6.25 | 3.12 | 3.12 | 1.56 | 12.5 | 1.56 | 6.25 | 3.12 |
| 24 | 12.5 | 25 | 12.5 | 12.5 | 6.25 | 25 | 6.25 | 25 | 12.5 |
| 25 | 0.78 | 1.56 | 0.78 | 1.56 | 0.78 | 1.56 | 0.39 | 3.125 | 0.78 |
| 26 | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| 27 | 25 | 50 | 12.5 | 25 | 12.5 | NA | 25 | 25 | 25 |
| 28 | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| 29 | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| 30 | 12.5 | 25 | 12.5 | 25 | 6.25 | 25 | 12.5 | NA | NA |
| 31 | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| V | 0.78 | 1.56 | 1.56 | 0.78 | 3.12 | 3.12 | 0.39 | 3.12 | >50 |
Antimicrobial studies novel compounds
These synthesized compounds were tested against different strains of Gram-positive and Gram-negative bacteria using our previously described methods.12,13 Several of these compounds showed potent activity against nine Gram-positive strains but they did not show any noticeable activity against Gram-negative strains. The phenyl aniline derivative (2) showed weak activity against the tested Gram-positive strains. Adding the hydrophobic alkyl substituents (3 and 4) increased the activity significantly and the tert-butyl substituted compound showed activity against the strains with minimum inhibitory concentration (MIC) values as low as 1.56 μg mL−1. The addition of the methoxy substituent (5) almost eliminated the activity. The phenoxy derivative (6) showed very good activity against the tested strains. This molecule (6) inhibited the growth of the staphylococcal strains efficiently with MIC values 1.56–3.12 μg mL−1. The growth of B. subtilis was also inhibited efficiently with MIC values as low as 1.56 μg mL−1. E. faecalis and E. faecium strains were inhibited by this compound (6) with the MIC values of 3.12 and 1.56 μg mL−1 respectively. The methyl sulfide derivative (7) showed weak activity against the tested strains. Fluoro-substitutions (8 and 9) also failed to show good activity against these strains. Chloro (10) and bromo derivatives (11 and 12) showed improved activity with an MIC value as low as 3.12 μg mL−1 against several tested strains. The trifluoromethyl-substituted derivative (13) showed good activity against the tested bacteria with the lowest MIC value of 3.12 μg mL−1 for one of the MRSA strains. Substitution with carboxylic acid eliminated the activity of the resultant compounds (14 and 15) whereas a nitro substituent (16) showed moderate activity against the tested strains. The difluoro aniline derivative (17) inhibited the growth of MRSA strains moderately with an MIC value as low as 6.25 μg mL−1. The dichloro substitution (18) resulted in one of the most potent compounds in the series. This molecule inhibited the growth of S. aureus strains with MIC values 0.78–1.56 μg mL−1. The growth of B. subtilis strain was inhibited efficiently at a sub μg mL−1 concentration and enterococcal strains were inhibited efficiently. Chlorofluoro substitution also resulted in potent molecules (19 and 20) with MIC values as low as 1.56 μg mL−1. The mixed substitution of methyl and halogens resulted in the formation of potent molecules (21, 22, and 23) as well. The methoxy and trifluoromethyl substituted compound (24) showed the moderate inhibition of bacterial growth. Bromo and trifluoromethyl substitutions gave the most potent compound (25) of the series, which inhibited the growth of three of the five S. aureus strains with an MIC value of 0.78 μg mL−1. This compound inhibited the growth of S. epidermidis and E. faecium with the MIC values of 1.56 and 0.78 μg mL−1 respectively and this compound (25) was the most potent against these two strains. Morpholine substitution eliminated the activity of the resultant compound (26) and the N-methyl piperazine derivative (27) showed moderate activity against the tested bacteria. Carboxylic acid substituted molecules (28 and 29) did not show any antimicrobial activity. Fluoro and nitro substitutions in the aniline moiety (30) showed moderate antibacterial activity with the MIC values as low as 6.25 μg mL−1. The last compound (31) with a protic substituent did not show any activity.
Based on these results, we can deduce a very good structure activity relationship (SAR) for these molecules. Compounds with a protic substituent (14, 15, 28, and 29) in the aniline moiety did not show any antimicrobial activity. Hydrophilic substituents (5, 26, and 27) resulted in weaker activity than that of nonpolar substituents (4 and 6). With increasing the size of halogen substituents (9, 10, 11, and 12) resulted in increased lipophilicity, there was an increase in the activity of the molecules. A similar pattern was observed for disubstituted compounds.
Cytotoxicity studies of potent antimicrobial agents
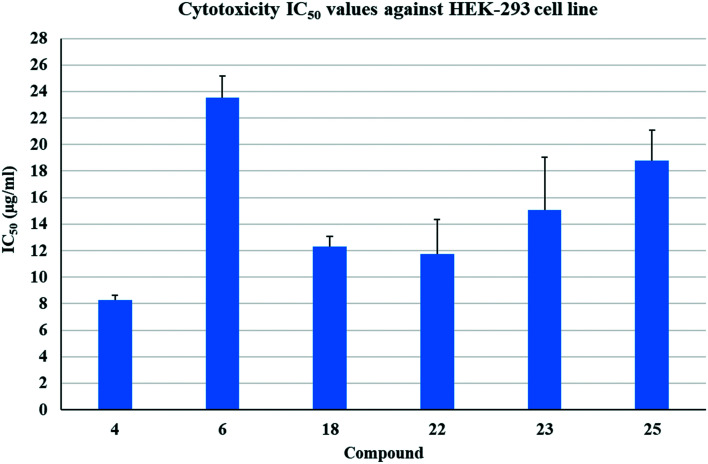
To narrow down the list of potent compounds for further studies, we evaluated the possible toxicity of potent compounds (MIC ≤1.56 μg mL−1) against human cells. Monosubstituted potent compounds (4 and 6) varied in their effect on human embryonic kidney cells (HEK293) with 50% inhibitory concentration (IC50) values of 8 and 23.5 μg mL−1 respectively (Fig. 2). Disubstituted potent compounds (18, 22, 23, and 25) showed toxicity for HEC293 cells with IC50 values >12 μg mL−1. Compounds (6, 23, and 25) showing the least toxicity for HEK293 cells were selected for further studies. Tamoxifen was used as a positive control, which showed 100% cell death at 74 μg mL−1 concentration.
Fig. 2. IC50 (μg mL) values for HEK293 cells treated with the most potent compounds.
Biofilm eradication studies
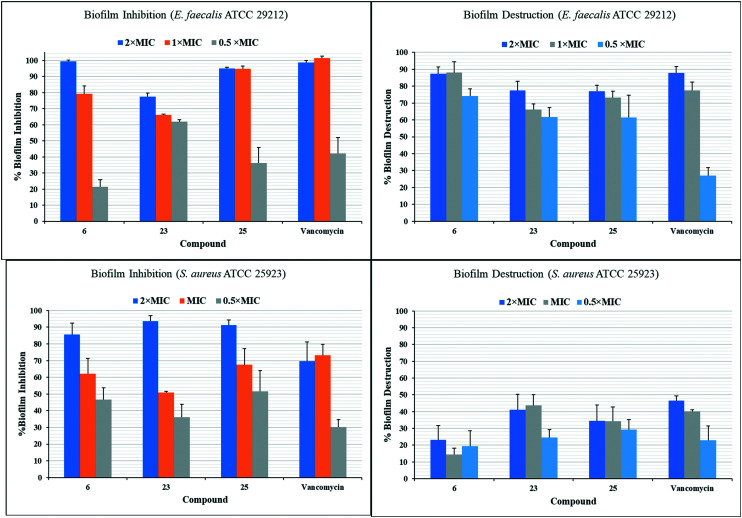
After finding molecules with both potent antibacterial activity as well as low toxicity to cultured human cells, we tested the three best compounds for their ability to inhibit the growth of bacteria in a biofilm context (Fig. 3). The phenoxyphenyl derivative (6) inhibited the growth of almost 100% and 80% of biofilms of E. faecalis at 2× and 1× MIC respectively and compound 25 showed similar activity, although, compound 23 was less effective. These compounds (6, 23, and 25) were also tested for their ability to eliminate the preformed biofilm of E. faecalis. The potency of these compounds appears to be comparable to the positive control, vancomycin. These compounds also inhibited the growth of S. aureus biofilm. Compound 23 inhibited the growth of S. aureus biofilm at 2× MIC by more than 90% that was significantly greater inhibition than the positive control vancomycin (paired T-Test, p < 0.05). Ability to prevent biofilm formation decreased with concentration, as inhibition generally parallels growth inhibition. However, compound 25 at 0.5× MIC was significantly better than vancomycin at inhibiting biofilm formation (p < 0.05), suggesting some direct effect. These compounds are weak eliminators of preformed S. aureus biofilm, but comparable to the positive control, vancomycin.
Fig. 3. Biofilm inhibition and eradication studies of three potent compounds (6, 23, and 25) against E. faecalis and S. aureus.
Calgary biofilm device (CBD) experiments
After finding molecules with both potent antibacterial activity as well as low toxicity to cultured human cells, we tested the three best compounds for their ability to inhibit the growth of bacteria in a biofilm context (Table 2). The Calgary biofilm device (CBD) is a well-established for determining biofilm susceptibilities to antibiotics in a quick and reproducible manner using a 96-well plate with a lid that contains pegs on which bacteria establish biofilm (Innovotech, product code: 19111).22 Biofilm eradication experiments utilizing CBD for selected potent compounds were carried out to determine the minimum biofilm eradication concentration (MBEC) using techniques reported in some publications with slight modifications.23–25
Summary of biofilm eradication studies against S. aureus ATCC 25923 and E. faecalis ATCC 29212. Each values in this table resulted from a minimum of three independent experiments. V = vancomycin.
| # |
S. aureus ATCC 25923 MBC/MBEC (μg mL−1) |
E. faecalis ATCC 29212 MBC/MBEC (μg mL−1) |
|---|---|---|
| 6 | 3.12/6.25 | 12.5/12.5 |
| 23 | 12.5/12.5 | 12.5/25 |
| 25 | 3.12/6.25 | 6.25/6.25 |
| V | 6.25/>50 | 25/>50 |
Although the presence and survival of planktonic cells in the CBD experiment depended on that fate of the biofilms in the challenge plate, determining MBC in this way allows us to compare MBC and MBEC in the same experiment. From this assay, we observed that our test compounds have MBC/MBEC ratios of 1 : 1 to 1 : 2 during experiments on various strains of bacteria (Table 2). The tested compounds demonstrated good biofilm killing capabilities against S. aureus biofilm: compound 6 (MBEC = 6.25 μg mL−1), 23 (MBEC = 12.5 μg mL−1) and 25 (MBEC = 6.25 μg mL−1) while conventional antibiotic, vancomycin, was not effective in killing biofilm even at highest test concentration (50 μg mL−1). Likewise, MBEC values for compounds 6, 23 and 25 are 12.5, 25 and 6.25 μg mL−1 respectively against E. faecalis biofilm in the CBD experiment. Compound 25 and vancomycin showed different MBEC values in Calgary plate method compared to similar experiments using the crystal violet stain method. This might be explained by the absence of growth medium (use of PBS instead) in the Calgary plate method, creating non-growing conditions in the challenge plates.
Time kill assay (TKA)
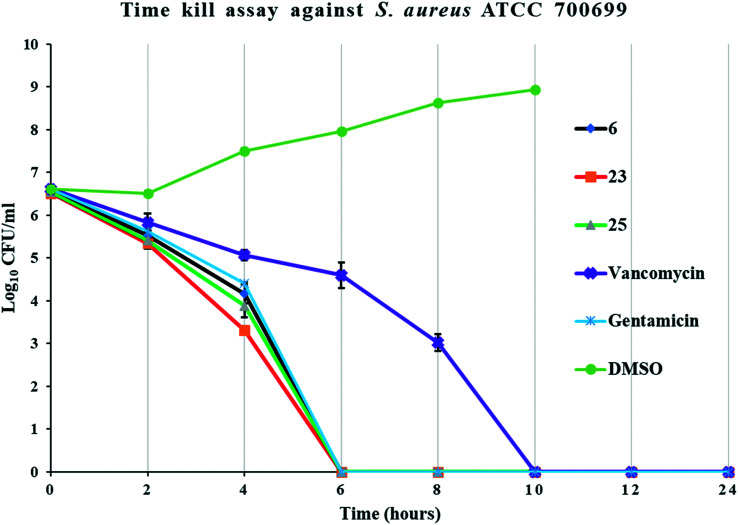
Time kill studies showed that our potent compounds (6, 23, and 25) are bactericidal. These compounds eliminated >99.9% bacteria by 6 h. One of the positive controls (gentamicin) also eliminated >99.9% bacteria by 6 h. The positive control vancomycin took 10 h to eliminate bacteria (Fig. 4). Thus, TKA studies show that our potent compounds rapidly kill bacteria like gentamicin.
Fig. 4. Time kill assay of potent compounds (6, 23, and 25). The error bars represent standard deviation of the plate count data performed in triplicates.
Activity against persisters
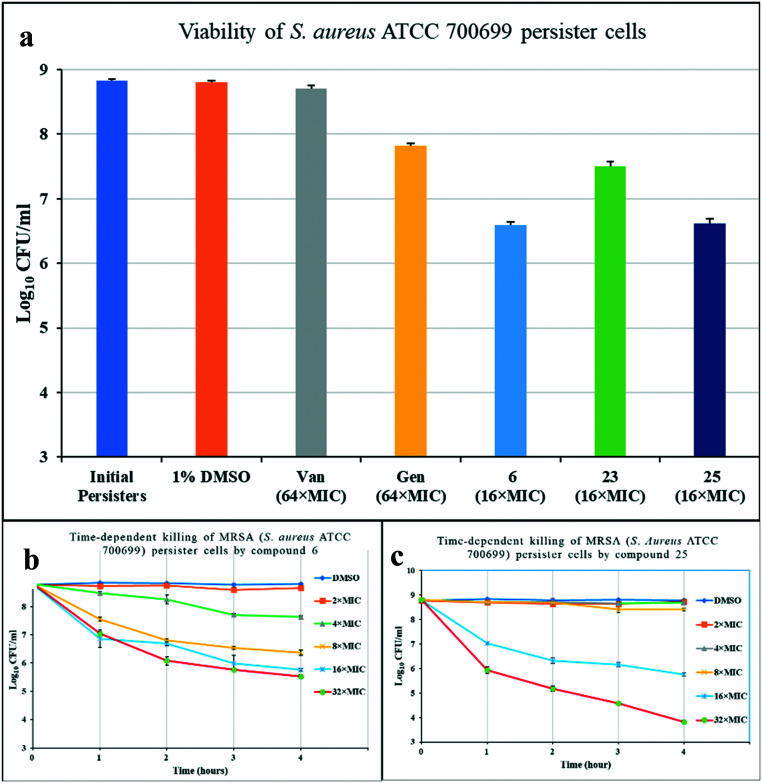
Persister cells, often associated with biofilms, are non-growing cells that show increased resistance to antibiotics.26 Stationary phase cells of S. aureus behave as persister cells and are tolerant to conventional antibiotics like vancomycin and gentamicin even at a very high MIC value.27S. aureus ATCC 700699 was grown to stationary phase and treated with control antibiotics and compounds 6, 23, and 25 to determine their activity (Fig. 5a). After 4 hours of exposure, vancomycin (64× MIC) showed no bactericidal activity whereas the other control gentamicin (64× MIC) reduced bacterial viability by one log. All three compounds tested (16× MIC) showed bactericidal activity against persister cells, with compounds 6 and 25 showing a greater than two log reduction in viable cells. The two compounds (6 and 25) showing the most potent activity against MRSA persister cells were further studied to observe their time-dependent killing activity using up to 32× MIC concentrations of compounds over a 4 h period (Fig. 5b and c). Both compounds showed steady, concentration-dependent rates of killing, although neither were significantly bactericidal at the lowest concentration of 2× MIC. For compound 6, 32× and 16× MIC were both observed to reduce persister cell concentration by almost three logs (Fig. 5b). Compound 25 showed no significant bactericidal activity over 4 h at concentrations up to 8× MIC (Fig. 5c). However, concentrations of 16× and 32× MIC decreased viability of persister cells by 3 and 5 logs respectively over the 4 h period. Vancomycin and gentamicin were used as positive controls. The steady decline in persister cell viability with treatment by compounds 6 and 25, suggests that bacterial death would continue beyond the 4 h period studied.
Fig. 5. (a) Viability of S. aureus ATCC 700699 persister cells after 4 h treatment at stated MIC of compounds. (b) Time-kill assay for compound 6, and (c) for compound 25 against S. aureus. The error bars represent standard deviation values of plate count performed in triplicates.
Activity against clinical isolates
The most promising compound (25) was tested against clinical isolates of E. faecalis, E. faecium, and S. aureus (Table 3). This molecule inhibited the growth of E. faecalis strains with MIC values as low as 2 μg mL−1. The potency of this compound is comparable to the potency of meropenem (M), and these E. faecalis strains are resistant to oxacillin (O) and vancomycin (V) antibiotics. This molecule was tested against five clinical isolates of E. faecium. One of the strains was inhibited at 4 μg mL−1 and the MIC value for the remaining strains were 8 μg mL−1. Our novel compound (25) was very potent against these strains of E. faecium compared to the positive controls, meropenem, oxacillin, and vancomycin which failed to show any activity against these strains. S. aureus clinical isolates were inhibited by this compound with an MIC value 4 μg mL−1. Thus, compared to the positive controls, the novel compound (25) showed very good activity against E. faecalis, potent activity against untreatable E. faecium strains, and comparable activity against the S. aureus clinical isolates.
Activity of compound 25 against clinical isolates. M = meropenem, O = oxacillin, and V = vancomycin.
| Collection number | Organism | 25 | M | O | V |
|---|---|---|---|---|---|
| 1088051 | E. faecalis | 4 | 2 | 64 | >32 |
| 1091563 | E. faecalis | 8 | 2 | 64 | >32 |
| 1091720 | E. faecalis | 8 | 8 | 64 | >32 |
| 1094112 | E. faecalis | 8 | 8 | >64 | >32 |
| 1100440 | E. faecalis | 8 | 4 | >64 | >32 |
| 1088168 | E. faecium | 4 | >128 | >64 | >32 |
| 1088228 | E. faecium | 8 | >128 | >64 | >32 |
| 1088925 | E. faecium | 8 | >128 | >64 | >32 |
| 1089544 | E. faecium | 8 | >128 | >64 | >32 |
| 1091282 | E. faecium | 8 | >128 | >64 | >32 |
| 1088060 | S. aureus | 4 | 0.06 | 0.5 | 0.5 |
| 1088105 | S. aureus | 4 | 0.12 | 0.25 | 1 |
| 1088626 | S. aureus | 4 | 0.06 | 0.25 | 0.5 |
| 1088064 | S. aureus | 4 | 4 | 64 | 0.5 |
| 1088610 | S. aureus | 4 | 4 | 64 | 0.5 |
| 1088888 | S. aureus | 8 | 8 | >64 | 1 |
| 1089106 | S. aureus | 4 | 2 | 64 | 1 |
Determination of the mode of action
After finding the potent activity of compound (25), we investigated the mode of action by using macromolecular synthesis assays (MMS) with S. aureus ATCC 29213, the model strain for these studies. The MIC value of this compound was 1–2 μg mL−1. The results of performing MMS inhibition assays at multiples of the MIC are shown in Fig. 6.
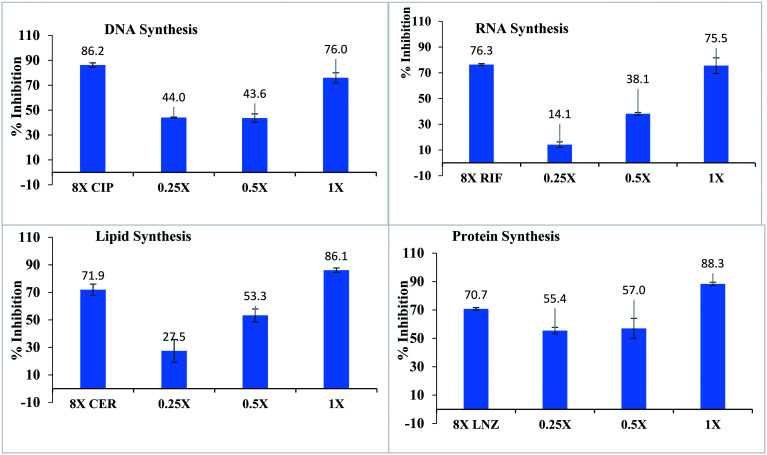
Fig. 6. Macromolecular synthesis (MMS) inhibition for the mode of action for compound 25. CIP (ciprofloxacin), RIF (rifampicin), CER (cerulenin) and LNZ (linezolid) are the positive controls for DNA synthesis, RNA synthesis, lipid synthesis, and protein synthesis respectively. All the positive controls were tested at 8xMIC and compound 25 was tested in the range of 0.25× to 1× MIC.
Nucleic acid and protein synthesis
The compound resulted in increasing the inhibition of both DNA and RNA synthesis from low concentration to high concentration. DNA synthesis was inhibited approximately 44% at 0.25× MIC to ∼75% inhibition at 1× MIC. The inhibition of RNA synthesis was 14.1% and > 75% at 0.25× MIC and 1× MIC respectively. Approved antibiotics, ciprofloxacin, and rifampicin showed comparatively very weak inhibition of DNA and RNA synthesis respectively. Similar the inhibition of protein synthesis was observed in the bacterial strain, 55.4% and 88.3% protein syntheses were inhibited at 0.25× and 1× MICs respectively. Linezolid, an approved antibiotic and a known protein synthesis inhibitor as a positive control, inhibited 70.7% protein synthesis at 8× MIC value. Thus, the potent compound showed a very good dose–response for the inhibition of nucleic acids and cellular protein.
Lipid synthesis
Lipid synthesis was inhibited significantly by the compound and a good dose–response was observed. At 0.25× MIC, 27.5% lipid synthesis was inhibited and the percent inhibition increased significantly with increasing the concentration of compound (25). A concentration of 1× MIC caused 86.1% inhibition of the lipid synthesis. The tested compound is very potent compared to the positive control, cerulenin, which inhibited 71.9% lipid synthesis at 8× MIC.
Interestingly, the same inhibition pattern occurred regardless of the macromolecular synthesis system being investigated, with substantial and nearly maximum inhibition occurring at a concentration of 2× MIC. One possible implication of these results is that the compound may be inhibiting some other aspect of bacterial cell function upon which all of these metabolic activities depend.
Conclusions
We have discovered potent antimicrobial agents based on our lead compounds with selective activity against drug-resistant Gram-positive bacteria. Multiple modes of the mechanism of action make these compounds very good candidates for antibiotics since bacteria will have difficulty developing resistance. These compounds are potent growth inhibitors of antibiotic sensitive and antibiotic-resistant bacteria both in planktonic and biofilm contexts. These compounds are very effective against persister cells compared to positive controls. Potent activity against E. faecium clinical isolates makes the lead molecules promising candidates for further antibiotic development.
Conflicts of interest
Authors declare no conflict of interest.
Supplementary Material
Acknowledgments
This publication was made possible by the Arkansas INBRE program, supported by a grant from the National Institute of General Medical Sciences, (NIGMS), P20 GM103429 from the National Institutes of Health to record the Mass Spectrometry data. This publication was made possible by the Arkansas INBRE Summer Research Grant, supported by a grant from NIGMS, P20 GM103429 from NIH. The Kays Foundation and ABI voucher support helped to complete this grant.
Electronic supplementary information (ESI) available. See DOI: 10.1039/d1md00230a
References
- Friedrich M. J. JAMA, J. Am. Med. Assoc. 2016;315:242–242. [Google Scholar]
- Mullard A. Nat. Rev. Drug Discovery. 2020;19:575–576. doi: 10.1038/d41573-020-00143-8. [DOI] [PubMed] [Google Scholar]
- CDC, Antibiotic/Antimicrobial Resistance (AR/AMR), https://www.cdc.gov/drugresistance/index.html, (accessed 12/23/2020, 2020)
- CDC, Antibiotic/Antimicrobial Resistance, https://www.cdc.gov/drugresistance/federal-engagement-in-ar/national-strategy/index.html, (accessed 10/19/2016, 2016)
- CDC, Antibiotic/Antimicrobial Resistance (AR/AMR), https://www.cdc.gov/drugresistance/biggest_threats.html, (accessed 9/17/2019, 2019)
- Chambers H. F. Deleo F. R. Nat. Rev. Microbiol. 2009;7:629–641. doi: 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahansepas A. Aghazadeh M. Rezaee M. A. Hasani A. Sharifi Y. Aghazadeh T. Mardaneh J. Microb. Drug Resist. 2018;24:76–82. doi: 10.1089/mdr.2017.0049. [DOI] [PubMed] [Google Scholar]
- Wang Y. Xiong Y. Wang Z. Zheng J. Xu G. Deng Q. Wen Z. Yu Z. J. Antibiot. 2021;74:143–151. doi: 10.1038/s41429-020-00374-2. [DOI] [PubMed] [Google Scholar]
- Verma R. Verma S. K. Rakesh K. P. Girish Y. R. Ashrafizadeh M. Sharath Kumar K. S. Rangappa K. S. Eur. J. Med. Chem. 2021;212:113134. doi: 10.1016/j.ejmech.2020.113134. [DOI] [PubMed] [Google Scholar]
- Mishra S. Patel S. Halpani C. G. Chem. Biodiversity. 2019;16:e1800366. doi: 10.1002/cbdv.201800366. [DOI] [PubMed] [Google Scholar]
- Hammad A. Abutaleb N. S. Elsebaei M. M. Norvil A. B. Alswah M. Ali A. O. Abdel-Aleem J. A. Alattar A. Bayoumi S. A. Gowher H. Seleem M. N. Mayhoub A. S. J. Med. Chem. 2019;62:7998–8010. doi: 10.1021/acs.jmedchem.9b00720. [DOI] [PubMed] [Google Scholar]
- Whitt J. Duke C. Sumlin A. Chambers S. A. Alnufaie R. Gilmore D. Fite T. Basnakian A. G. Alam M. A. Molecules. 2019;24:2051. doi: 10.3390/molecules24112051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitt J. Duke C. Ali M. A. Chambers S. A. Khan M. M. K. Gilmore D. Alam M. A. ACS Omega. 2019;4:14284–14293. doi: 10.1021/acsomega.9b01967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alnufaie R. Raj KC H. Alsup N. Whitt J. Chambers S. Andrew. Gilmore D. Alam M. A. Molecules. 2020;25:2758. doi: 10.3390/molecules25122758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alnufaie R. Alsup N. Kc H. R. Newman M. Whitt J. Chambers S. A. Gilmore D. Alam M. A. J. Antibiot. 2020;73:818–827. doi: 10.1038/s41429-020-0341-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delancey E. Allison D. KC H. R. Gilmore D. F. Fite T. Basnakian A. G. Alam M. A. Antibiotics. 2020;9:650. doi: 10.3390/antibiotics9100650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brider J. Rowe T. Gibler D. J. Gottsponer A. Delancey E. Branscum M. D. Ontko A. Gilmore D. Alam M. A. Med. Chem. Res. 2016;25:2691–2697. doi: 10.1007/s00044-016-1678-8. [DOI] [Google Scholar]
- Gillis E. P. Eastman K. J. Hill M. D. Donnelly D. J. Meanwell N. A. J. Med. Chem. 2015;58:8315–8359. doi: 10.1021/acs.jmedchem.5b00258. [DOI] [PubMed] [Google Scholar]
- Müller K., in Fluorine in Life Sciences: Pharmaceuticals, Medicinal Diagnostics, and Agrochemicals, ed. G. Haufe and F. R. Leroux, Academic Press, 2019, pp. 91–139, 10.1016/B978-0-12-812733-9.00002-7 [DOI] [Google Scholar]
- Allison D. Delancey E. Ramey H. Williams C. Alsharif Z. A. Al-khattabi H. Ontko A. Gilmore D. Alam M. A. Bioorg. Med. Chem. Lett. 2017;27:387–392. doi: 10.1016/j.bmcl.2016.12.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- US Pat., 10596153, 2020
- Ceri H. Olson M. E. Stremick C. Read R. R. Morck D. Buret A. J. Clin. Microbiol. 1999;37:1771–1776. doi: 10.1128/JCM.37.6.1771-1776.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison A. T. Abouelhassan Y. Norwood V. M. Kallifidas D. Bai F. Nguyen M. T. Rolfe M. Burch G. M. Jin S. Luesch H. Huigens R. W. J. Med. Chem. 2016;59:3808–3825. doi: 10.1021/acs.jmedchem.5b02004. [DOI] [PubMed] [Google Scholar]
- Yang H. Kundra S. Chojnacki M. Liu K. Fuse M. A. Abouelhassan Y. Kallifidas D. Zhang P. Huang G. Jin S. Ding Y. Luesch H. Rohde K. H. Dunman P. M. Lemos J. A. Huigens R. W. J. Med. Chem. 2021;64:7275–7295. doi: 10.1021/acs.jmedchem.1c00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison A. T. Abouelhassan Y. Kallifidas D. Tan H. Kim Y. S. Jin S. Luesch H. Huigens R. W. J. Med. Chem. 2018;61:3962–3983. doi: 10.1021/acs.jmedchem.7b01903. [DOI] [PubMed] [Google Scholar]
- Carvalho G. Balestrino D. Forestier C. Mathias J.-D. npj Biofilms Microbiomes. 2018;4:6. doi: 10.1038/s41522-018-0049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W. Zhu W. Hendricks G. L. Van Tyne D. Steele A. D. Keohane C. E. Fricke N. Conery A. L. Shen S. Pan W. Lee K. Rajamuthiah R. Fuchs B. B. Vlahovska P. M. Wuest W. M. Gilmore M. S. Gao H. Ausubel F. M. Mylonakis E. Nature. 2018;556:103–107. doi: 10.1038/nature26157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.