Abstract
Discovery of epigenetic chemical probes is an important area of research with potential to deliver drugs for a multitude of diseases. However, commercially available libraries frequently used in drug discovery campaigns contain molecules that are focused on a narrow range of chemical space primarily driven by ease of synthesis and previously targeted enzyme classes (e.g., kinases) resulting in low hit rates for epigenetic targets. Here we describe the design and synthesis of a compound collection that augments current screening collections by the inclusion of privileged isosteres for epigenetic targets.
This work describes the photochemical synthesis of a tetrahydroquinoline library enriched with chemotypes privileged for epigenetic targets and exploring new regions of chemical space.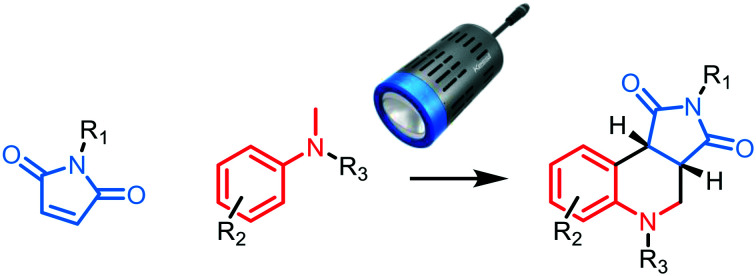
Introduction
The screening of libraries of small molecules underpins many drug discovery efforts.1 Most commonly, the chemistry used to construct these compound collections is selected to be easily expanded in throughput to provide hundreds or thousands of compounds.2 This allows for a large number of compounds to be tested but can mean compounds included in such libraries have simple structures or lack diversity. Certain motifs are overrepresented because of their facile incorporation into high-throughput synthesis workflows. An analysis of 66 submissions describing clinical candidates to the Journal of Medicinal Chemistry between 2016 and 2017 revealed that 30% targeted kinases, 17% GPCRs and 9% epigenetic targets,3 yet epigenetic bioisosteres are infrequently included in screening collections.4 This is compounded by the prediction that much of purchasable chemical space is targeted towards proteases, GPCRs, and kinases.5 This deficiency in diversity and lack of bioisostere inclusion means that hit rates are often low against epigenetic targets.6 Consequently, we4 and others,7 have called for bespoke screening collections to be designed and synthesized to address these deficiencies.
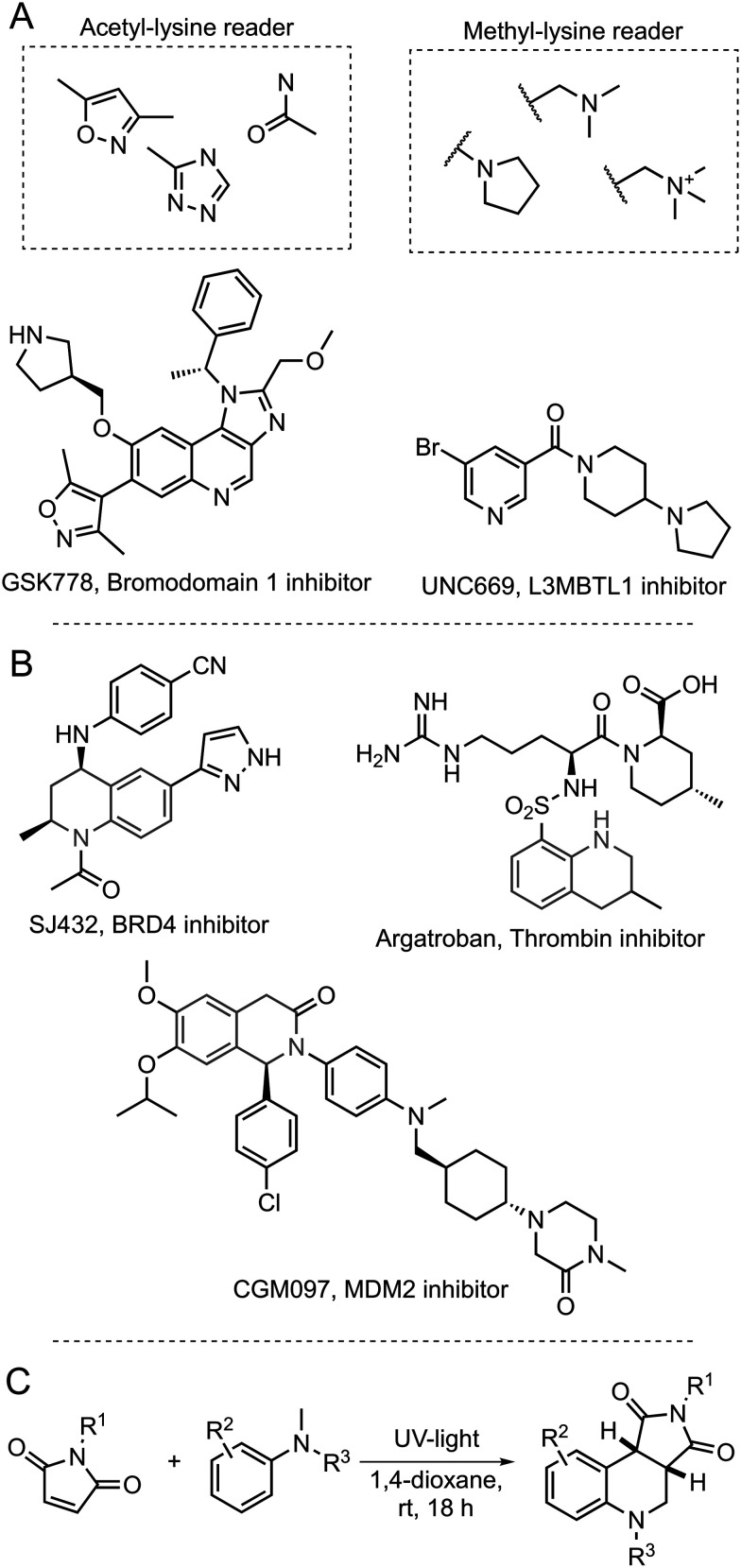
Drug discovery often utilizes privileged chemical motifs, or bioisosteres, as a starting point to identify chemical matter for a specific target class. Examples include hinge-binding motifs for kinases8 and acetyl lysine mimics for bromodomains.9 While effective chemotypes for kinases are frequently included in commercially available compound collections similarly effective chemotypes are not included for epigenetic targets. This is particularly true for privileged chemotypes that target acetyl- and methyl- lysine reader, writer, and eraser proteins which are of significant interest for the treatment of cancers, inflammation, and other diseases.4,10,11 Examples of highly effective chemotypes include 3,5-dimethylisoxazoles12 and 1,2,4-triazoles13 for bromodomains (acetyl lysine readers), and N-alkyl amines for methyl lysine readers14 (Fig. 1A).
Fig. 1. Exemplar epigenetic bioisosteres and ligands containing a tetrahydroquinoline pharmacophore highlighted in green. A: Examples of chemotypes employed as part of inhibitors of acetyl-lysine readers and methyl-lysine readers.23–25 B: Examples of bioactive molecules with tetrahydroquinoline or tetrahydroisoquinoline scaffolds.26–28 C: EDA-Mediated photochemical annulation of maleimides with N,N-dimethylaniline to yield a scaffold with a tetrahydroquinoline core.21.
One issue with screening libraries is the narrow toolkit of reactions that has typically been used to synthesize small molecules, limiting the types of scaffolds that can be accessed for biological testing.15 However, recently developed photochemical cyclizations have enabled the synthesis of new molecular entities.16–19 Specifically, the synthesis of tetrahydroquinolines is of particular interest as the scaffold is a highly effective and versatile pharmacophore, with over 10 000 bioactive molecules containing this pharmacophore reported in the ChEMBL database (Fig. 1B).20 An example of their synthesis has been reported via a photochemically induced radical annulation between maleimides and N-alkyl anilines.21 This reaction is enabled by the formation of an electron donor–acceptor (EDA) complex between the two reactants. EDA complexes are Lewis acid–base complexes that can absorb visible light and undergo single-electron transfer (SET) to generate radical intermediates.22 The main benefit of EDA-mediated reactions is that they do not require a catalyst, or anything beyond the two starting materials, and are therefore amenable for the rapid synthesis of small molecule screening collections. Whilst this EDA-mediated scaffold synthesis has been previously reported, its application to the synthesis of biologically relevant compound collections has not. We therefore sought to construct a bespoke collection of small molecules bearing chemotypes known to bind epigenetic targets based upon this tetrahydroquinoline scaffold (Fig. 1C).21
Results and discussion
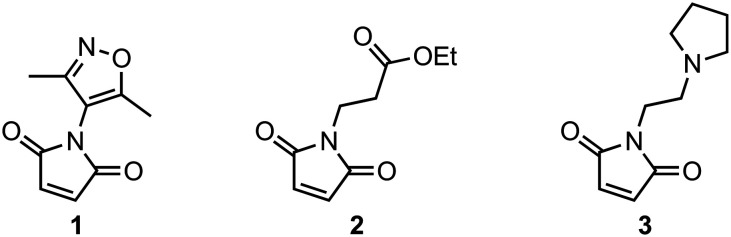
We envisaged that by introducing chemotypes from epigenetic ligands to the maleimide substrates it would be possible to prepare a library of compounds that target epigenetic reader, writer, or eraser protein domains. Therefore, three maleimides were synthesized from their respective primary amines via a condensation reaction with maleic anhydride or cis-5-norbornene-endo-2,3-dicarboxylic anhydride followed by a retro Diels-Alder reaction29 to function as substrates for the photochemical annulation. Maleimides bearing a 3,5-dimethylisoxazole (1) to target bromodomains, an ethyl ester (2) that could be used as a precursor for the synthesis of other functionalized products, and an N-alkyl pyrrolidine (3) to target methyl-lysine reader domains were obtained in moderate yields (Fig. 2).
Fig. 2. Maleimide building blocks used in the synthesis of the epigenetic focused tetrahydroquinoline library.
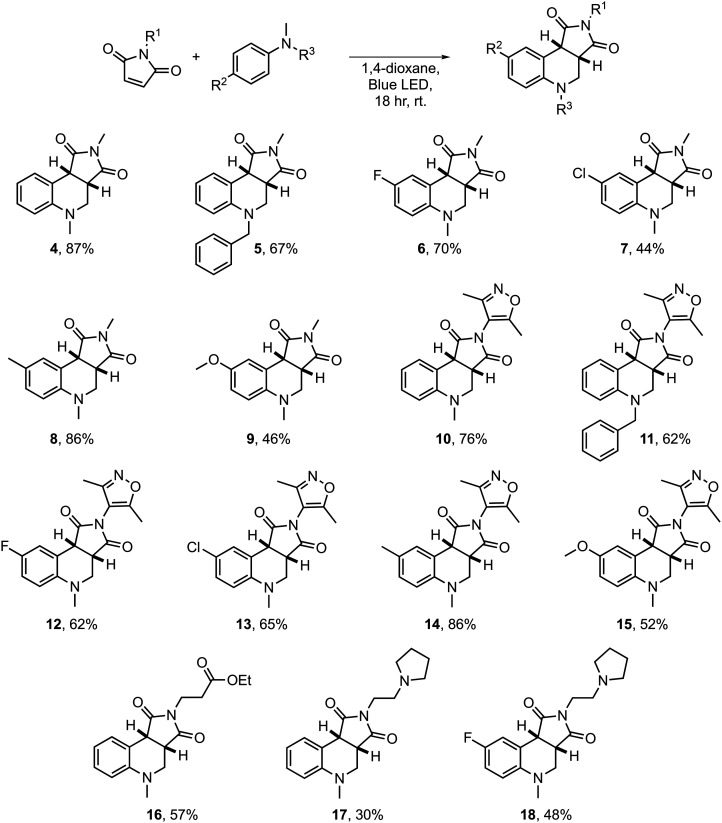
The maleimide substrates were then reacted with a range of N-methylanilines in the EDA-mediated α-alkylamino radical annulation to create the epigenetic focused compound collection (Scheme 1). A range of substituted anilines were employed to add extra utility to the compound collection. For example, 4-fluoro-N,N-dimethylaniline afforded the fluorinated products 6, 12 and 18 that could be tested in 19F ligand-observed NMR bioassays. Similarly, N-benzyl-N-methylaniline afforded products 5 and 11 that could be subsequently deprotected to reveal a secondary amine.
Scheme 1. Synthesis of the tetrahydroquinoline scaffold and epigenetic focused analogues.
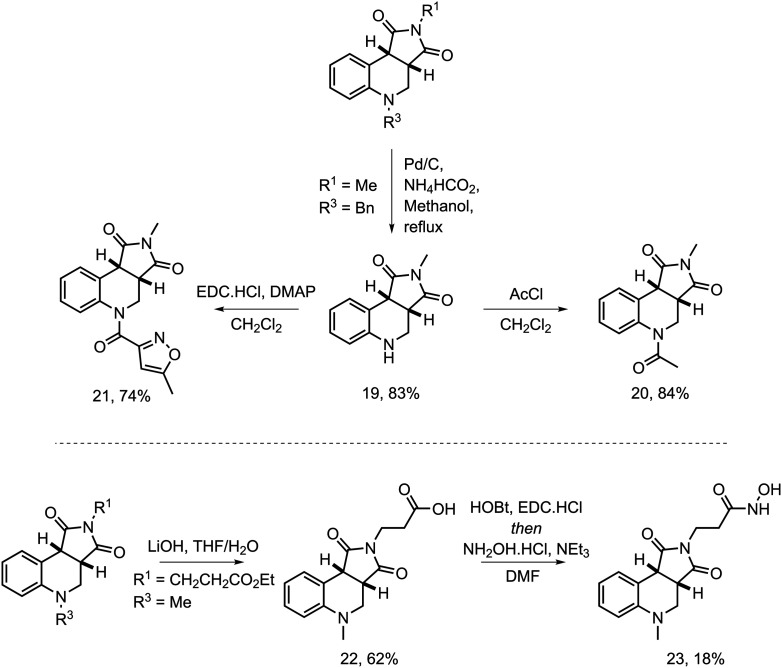
To further expand the range of epigenetic chemotypes found in the library and include other derivatizations, compound 5 was deprotected by hydrogenation to reveal the secondary aniline 19 in a good yield, which was decorated via two exemplar acylations (Scheme 2). An acetylation with acetyl chloride in dichloromethane afforded 20 bearing the N-acetyltetrahydroquinoline pharmacophore found in several bromodomain 4 (BRD4) ligands.30 An amide coupling with 5-methylisoxazole-3-carboxylic acid using N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC) and dimethylaminopyridine (DMAP) in dichloromethane afforded compound 21. Additionally, compound 16 was hydrolyzed using lithium hydroxide in tetrahydrofuran and water to afford the acid 22 and subsequently converted to the hydroxamic acid 23 yielding a putative HDAC inhibitor. Collectively, this yielded a small and focused collection of compounds bearing a range of privileged epigenetic chemotypes.
Scheme 2. Derivatizations of the tetrahydroquinoline scaffold.
Cheminformatics
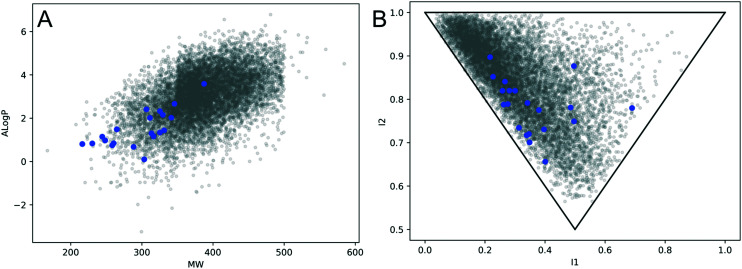
To demonstrate the relevance and value of our epigenetic focused library, the molecular properties of the compounds were calculated (Fig. 3). Most of the compounds (19 out of 20, 95%) possess lead like properties of A Log P <3 and molecular weight <350 Da, and all the molecules have A Log P between 0 and 4 and weigh under 400 Da making them ideal starting points for optimization via medicinal chemistry.31 Analysis of the principal moment of inertia (PMI) for the focused library shows that all the molecules are shifted from the rod-like region of the plot where many commercially available compounds reside, and some are shifted from the rod–disc axis, demonstrating promising 3D structures.
Fig. 3. Comparison of molecular properties for this compound collection (blue) and a commercially available epigenetic compound collection (grey). A: Molecular weight vs. A Log P. B: Principal moment of inertia plot. The commercially available epigenetic library is shown in grey and our focused library described is shown in blue. A Log P and principal moment of inertia (PMI) were calculated in RDKit. PMI represents the three-dimensional shape of a molecule where rod-like molecules are in the top left, disc-like molecules are at the lower middle, and sphere-like molecules are in the top right of the plot.
The tetrahydroquinolinedione core scaffold is also unique across several databases including ChEMBL, the NIH Molecular Libraries Small Molecule Repository (MLSMR), and a commercially available epigenetic library. The molecular fingerprint of compound 4 was compared by Tanimoto similarity to the same databases, where similar compounds typically have values ≥0.7.32 Searching the MLSMR database identified 19 molecules with similarity ≥0.6 (0.004%) and no molecules ≥0.7 (approx. 440 000 molecules). Conducting the same analysis with a commercially available epigenetic library found no molecules with similarity ≥0.6 (approx. 10 000 molecules). Finally, a comparison of compound 10 to BRD4 ligands deposited to ChEMBL found only four molecules with similarity ≥0.6 (0.1%) and no molecules ≥0.7 (3832 molecules) demonstrating that unique molecules can still be obtained despite enriching our focused library with privileged chemotypes for epigenetic targets.
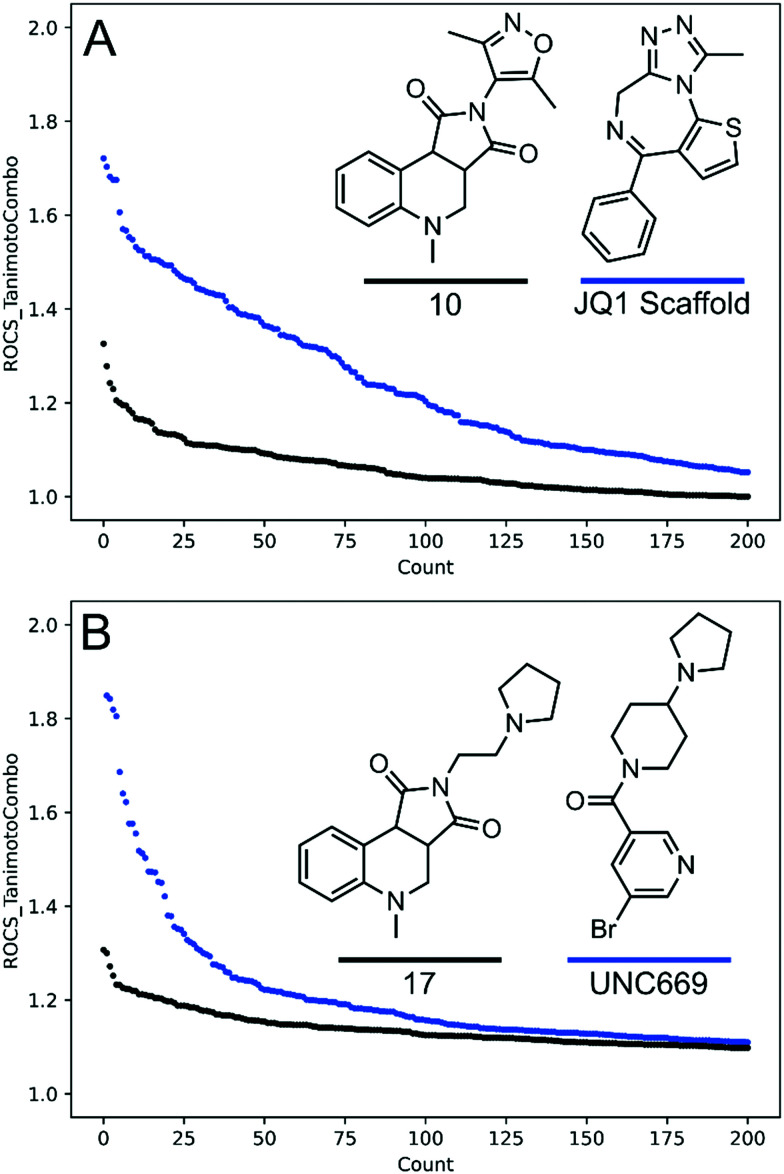
While substructure searches and Tanimoto similarity ranking are useful tools for identifying molecules with new chemical structures, they do not quantify similarities in 3-dimensional shape. We conducted a Rapid Overlay of Chemical Structures (ROCS) analysis of compound 10versus BRD4 ligands deposited in ChEMBL and compound 17versus lethal 3 malignant brain tumor-like protein 1 (L3MBTL1) ligands. An ensemble of conformers was generated in OMEGA for each ligand in ChEMBL and compared to the molecular shape of compound 10 or 17. Generally, ROCS scores greater than 1.4 indicate molecular shapes with significant similarity between the query molecule and the test compound.32,33 Analysis for compound 10 reveals that there are no shape similar compounds within the BRD4 ligand set, and only six ligands were identified with ROCS scores ≥1.2 (Fig. 4A). Identical analysis of compound 17 also showed that no ligands with scores ≥1.4 were contained within the L3MBTL1 set, and 19 ligands had scores ≥1.2 (Fig. 4B). This demonstrates that these new compounds are distinct in shape from previously utilized chemical matter for exemplar drug targets containing acetyl- or methyl- lysine reader domains and may form new interactions with proteins, thus augmenting current screening collections.
Fig. 4. Analysis of the top 200 ROCS ranked BRD4 or L3MBTL1 ligands in the ChEMBL database compared with exemplar compounds bearing privileged epigenetic chemotypes. Count refers to the ROCS ranking where 1 is the best ranked compound (most similar). A: Compound 10 (black) or the scaffold of JQ1 (blue) vs. BRD4 ligands deposited to ChEMBL (2168 molecules). B: Compound 17 (black) or UNC669 (blue) vs. L3MBTL1 ligands deposited to ChEMBL (10 847 molecules, UNC669 was removed from the test set).
To test whether shape similar compounds could be identified from these test sets we repeated the ROCS analysis using either the molecular scaffold of JQ1,34 a potent BRD4 ligand, or the L3MBTL1 ligand UNC66925 (Fig. 4). When using the JQ1 scaffold, 41 BRD4 ligands were identified with ROCS scores ≥1.4 and 101 ligands had scores ≥1.2. Similarly, 22 L3MBTL1 ligands were identified with scores ≥1.4 and 66 with scores ≥1.2. This analysis supports the finding that there are essentially no ligands that are similar to the compounds contained within our focused library despite employing privileged chemotypes that are enriched within the BRD4 or L3MBTL1 ChEMBL sets. This demonstrates that it is possible to construct screening collections that are enriched with bioisosteres or privileged chemotypes for epigenetic targets without compromising the uniqueness of the compounds contained within the library. This design strategy therefore enables access to new highly relevant chemical space that has not been otherwise explored.
Conclusions
In summary, the strategy for the design and synthesis of a focused compound collection described here has the potential to address the low hit rates frequently observed in drug discovery campaigns against epigenetic targets and can be applied to a range of chemistries. Here we utilized a convergent synthetic route to generate a small exemplar library of compounds with a unique combination of scaffold and anchoring moiety. The ideal molecular properties of these compounds, and their location in underpopulated regions of chemical space, may facilitate the identification and optimization of ligands for previously undrugged epigenetic targets. Future development of the library will focus on the inclusion of new scaffolds, additional isosteres and evaluation of the hit rate in various assays to further validate this design approach. It may also be possible to facilitate the discovery of dual inhibitors of epigenetic targets using this approach through the inclusion of multiple isosteres in a single bifunctional molecule further broadening the utility of hits from this library.35–37 We envisage that the chemo-informatically driven development of synthetic routes to novel scaffolds decorated with privileged isosteres will alleviate some of the issues with identifying epigenetic inhibitors.
Conflicts of interest
There is no conflict of interest to declare.
Supplementary Material
Acknowledgments
We thank the Penn Chemistry NMR Facility for access to instrumentation. The Bruker AVANCE NEO 400 MHz NMR spectrometer was supported by the NSF Major Research Instrumentation Program (NSF CHE-1827457) and Vagelos Institute for Energy Science and Technology. The Bruker AVANCE NEO 600 MHz NMR spectrometer was supported by the NIH supplement awards 3R01GM118510-03S1 and 3R01GM087605-06S1, and Vagelos Institute for Energy Science and Technology. We are grateful to OpenEye Scientific for an Academic License to support our research. We thank members of the Burslem lab for useful discussions.
Electronic supplementary information (ESI) available. See DOI: 10.1039/d1md00193k
References
- Hughes J. P. Rees S. Kalindjian S. B. Philpott K. L. Br. J. Pharmacol. 2011;162:1239–1249. doi: 10.1111/j.1476-5381.2010.01127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volochnyuk D. M. Ryabukhin S. V. Moroz Y. S. Savych O. Chuprina A. Horvath D. Zabolotna Y. Varnek A. Judd D. B. Drug Discovery Today. 2019;24:390–402. doi: 10.1016/j.drudis.2018.10.016. [DOI] [PubMed] [Google Scholar]
- Brown D. G. Boström J. J. Med. Chem. 2018;61:9442–9468. doi: 10.1021/acs.jmedchem.8b00675. [DOI] [PubMed] [Google Scholar]
- Green A. I. Burslem G. M. J. Med. Chem. 2021;64:7231–7240. doi: 10.1021/acs.jmedchem.1c00592. [DOI] [PubMed] [Google Scholar]
- Irwin J. J. Gaskins G. Sterling T. Mysinger M. M. Keiser M. J. J. Chem. Inf. Model. 2018;58:148–164. doi: 10.1021/acs.jcim.7b00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S. K. Tian X. LaFrance L. V. Duquenne C. Suarez D. P. Newlander K. A. Romeril S. P. Burgess J. L. Grant S. W. Brackley J. A. Graves A. P. Scherzer D. A. Shu A. Thompson C. Ott H. M. Aller G. S. V. Machutta C. A. Diaz E. Jiang Y. Johnson N. W. Knight S. D. Kruger R. G. McCabe M. T. Dhanak D. Tummino P. J. Creasy C. L. Miller W. H. ACS Med. Chem. Lett. 2012;3:1091–1096. doi: 10.1021/ml3003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions Z. Sánchez-Cruz N. Prieto-Martínez F. D. Alves V. M. Santos H. P. Muratov E. Tropsha A. Medina-Franco J. L. Drug Discovery Today. 2020;25:2268–2276. doi: 10.1016/j.drudis.2020.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czodrowski P. Hölzemann G. Barnickel G. Greiner H. Musil D. J. Med. Chem. 2015;58:457–465. doi: 10.1021/jm501597j. [DOI] [PubMed] [Google Scholar]
- Schiedel M. Moroglu M. Ascough D. M. H. Chamberlain A. E. R. Kamps J. J. A. G. Sekirnik A. R. Conway S. J. Angew. Chem., Int. Ed. 2019;58:17930–17952. doi: 10.1002/anie.201812164. [DOI] [PubMed] [Google Scholar]
- Ganesan A. Arimondo P. B. Rots M. G. Jeronimo C. Berdasco M. Clin. Epigenet. 2019;11:174. doi: 10.1186/s13148-019-0776-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer J. N. Raniszewski N. R. Burslem G. M. ChemBioChem. 2021;22:17–42. doi: 10.1002/cbic.202000459. [DOI] [PubMed] [Google Scholar]
- Hewings D. S. Wang M. Philpott M. Fedorov O. Uttarkar S. Filippakopoulos P. Picaud S. Vuppusetty C. Marsden B. Knapp S. Conway S. J. Heightman T. D. J. Med. Chem. 2011;54:6761–6770. doi: 10.1021/jm200640v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov O. Lingard H. Wells C. Monteiro O. P. Picaud S. Keates T. Yapp C. Philpott M. Martin S. J. Felletar I. Marsden B. D. Filippakopoulos P. Müller S. Knapp S. Brennan P. E. J. Med. Chem. 2014;57:462–476. doi: 10.1021/jm401568s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold J. M. James L. I. Korboukh V. K. Gao C. Coil K. E. Bua D. J. Norris J. L. Kireev D. B. Brown P. J. Jin J. Janzen W. P. Gozani O. Frye S. V. MedChemComm. 2012;3:45–51. doi: 10.1039/C1MD00195G. [DOI] [Google Scholar]
- Blakemore D. C. Castro L. Churcher I. Rees D. C. Thomas A. W. Wilson D. M. Wood A. Nat. Chem. 2018;10:383–394. doi: 10.1038/s41557-018-0021-z. [DOI] [PubMed] [Google Scholar]
- Perumal G. Kandasamy M. Ganesan B. Govindan K. Sathya H. Hung M.-Y. Chandru Senadi G. Wu Y.-C. Lin W.-Y. Tetrahedron. 2021;80:131891. doi: 10.1016/j.tet.2020.131891. [DOI] [Google Scholar]
- Itoh K. Nagao S. Tokunaga K. Hirayama S. Karaki F. Mizuguchi T. Nagai K. Sato N. Suzuki M. Hashimoto M. Fujii H. Chem. – Eur. J. 2021;27:5171–5179. doi: 10.1002/chem.202004186. [DOI] [PubMed] [Google Scholar]
- Nikitas N. F. Theodoropoulou M. A. Kokotos C. G. Eur. J. Org. Chem. 2021;2021:1168–1173. doi: 10.1002/ejoc.202001593. [DOI] [Google Scholar]
- Manley D. W. Mills A. O'Rourke C. Slawin A. M. Z. Walton J. C. Chem. – Eur. J. 2014;20:5492–5500. doi: 10.1002/chem.201304929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ChEMBL, https://www.ebi.ac.uk/chembl/, (accessed 30 March 2021)
- Hsu C.-W. Sundén H. Org. Lett. 2018;20:2051–2054. doi: 10.1021/acs.orglett.8b00597. [DOI] [PubMed] [Google Scholar]
- Crisenza G. E. M. Mazzarella D. Melchiorre P. J. Am. Chem. Soc. 2020;142:5461–5476. doi: 10.1021/jacs.0c01416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak J. Y. W. Wu K.-C. Gupta P. K. Barbero S. McLaughlin M. G. Lucke A. J. Tng J. Lim J. Loh Z. Sweet M. J. Reid R. C. Liu L. Fairlie D. P. J. Med. Chem. 2021;64:2186–2204. doi: 10.1021/acs.jmedchem.0c01967. [DOI] [PubMed] [Google Scholar]
- Gilan O. Rioja I. Knezevic K. Bell M. J. Yeung M. M. Harker N. R. Lam E. Y. N. Chung C. Bamborough P. Petretich M. Urh M. Atkinson S. J. Bassil A. K. Roberts E. J. Vassiliadis D. Burr M. L. Preston A. G. S. Wellaway C. Werner T. Gray J. R. Michon A.-M. Gobbetti T. Kumar V. Soden P. E. Haynes A. Vappiani J. Tough D. F. Taylor S. Dawson S.-J. Bantscheff M. Lindon M. Drewes G. Demont E. H. Daniels D. L. Grandi P. Prinjha R. K. Dawson M. A. Science. 2020;368:387–394. doi: 10.1126/science.aaz8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold J. M. Wigle T. J. Norris J. L. Lam R. Korboukh V. K. Gao C. Ingerman L. A. Kireev D. B. Senisterra G. Vedadi M. Tripathy A. Brown P. J. Arrowsmith C. H. Jin J. Janzen W. P. Frye S. V. J. Med. Chem. 2011;54:2504–2511. doi: 10.1021/jm200045v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavish P. J. Chi L. Yun M.-K. Tsurkan L. Martinez N. E. Jonchere B. Chai S. C. Connelly M. Waddell M. B. Das S. Neale G. Li Z. Shadrick W. R. Olsen R. R. Freeman K. W. Low J. A. Price J. E. Young B. M. Bharatham N. Boyd V. A. Yang J. Lee R. E. Morfouace M. Roussel M. F. Chen T. Savic D. Guy R. K. White S. W. Shelat A. A. Potter P. M. Cancer Res. 2020;80:3507–3518. doi: 10.1158/0008-5472.CAN-19-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikumoto R. Tamao Y. Tezuka T. Tonomura S. Hara H. Ninomiya K. Hijikata A. Okamoto S. Biochemistry. 1984;23:85–90. doi: 10.1021/bi00296a014. [DOI] [PubMed] [Google Scholar]
- Holzer P. Masuya K. Furet P. Kallen J. Valat-Stachyra T. Ferretti S. Berghausen J. Bouisset-Leonard M. Buschmann N. Pissot-Soldermann C. Rynn C. Ruetz S. Stutz S. Chène P. Jeay S. Gessier F. J. Med. Chem. 2015;58:6348–6358. doi: 10.1021/acs.jmedchem.5b00810. [DOI] [PubMed] [Google Scholar]
- Clevenger R. C. Turnbull K. D. Synth. Commun. 2000;30:1379–1388. doi: 10.1080/00397910008087165. [DOI] [Google Scholar]
- Brand M. Measures A. M. Wilson B. G. Cortopassi W. A. Alexander R. Höss M. Hewings D. S. Rooney T. P. C. Paton R. S. Conway S. J. ACS Chem. Biol. 2015;10:22–39. doi: 10.1021/cb500996u. [DOI] [PubMed] [Google Scholar]
- Nadin A. Hattotuwagama C. Churcher I. Angew. Chem., Int. Ed. 2012;51:1114–1122. doi: 10.1002/anie.201105840. [DOI] [PubMed] [Google Scholar]
- Maggiora G. Vogt M. Stumpfe D. Bajorath J. J. Med. Chem. 2014;57:3186–3204. doi: 10.1021/jm401411z. [DOI] [PubMed] [Google Scholar]
- Martin Y. C. Kofron J. L. Traphagen L. M. J. Med. Chem. 2002;45:4350–4358. doi: 10.1021/jm020155c. [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P. Qi J. Picaud S. Shen Y. Smith W. B. Fedorov O. Morse E. M. Keates T. Hickman T. T. Felletar I. Philpott M. Munro S. McKeown M. R. Wang Y. Christie A. L. West N. Cameron M. J. Schwartz B. Heightman T. D. La Thangue N. French C. A. Wiest O. Kung A. L. Knapp S. Bradner J. E. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalley J. P. Cowley S. M. Hodgkinson J. T. Molecules. 2020;25:4394. doi: 10.3390/molecules25194394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. Li Y. Zhang J. Zhang M. Wei A. Liu H. Xie Z. Ren W. Duan W. Zhang Z. Shen A. Hu Y. Eur. J. Med. Chem. 2021;209:112868. doi: 10.1016/j.ejmech.2020.112868. [DOI] [PubMed] [Google Scholar]
- Atkinson S. J. Soden P. E. Angell D. C. Bantscheff M. Chung C. Giblin K. A. Smithers N. Furze R. C. Gordon L. Drewes G. Rioja I. Witherington J. Parr N. J. Prinjha R. K. MedChemComm. 2014;5:342–351. doi: 10.1039/C3MD00285C. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.