Supplemental Digital Content is available in the text.
Keywords: critical care, data science, machine learning, mortality, Pediatric Index of Mortality, pediatrics
OBJECTIVES:
Pediatric Index of Mortality 3 is a validated tool including 11 variables for the assessment of mortality risk in PICU patients. With the recent advances in explainable machine learning algorithms, we aimed to assess feasibility of application of these machine learning models to simplify the Pediatric Index of Mortality 3 scoring system in order to decrease time and labor required for data collection and entry for Pediatric Index of Mortality 3.
DESIGN:
Single-center, retrospective cohort study. Data from the Virtual Pediatric Systems for patients admitted to Cleveland Clinic Children`s PICU between January 2008 and December 2019 was obtained. Light Gradient Boosting Machine Regressor (a gradient boosting decision tree algorithm) was used for building the machine learning models. Variable importance was analyzed by SHapley Additive exPlanations. All of the 11 Pediatric Index of Mortality 3 variables were used as input variables in the machine learning models to predict Pediatric Index of Mortality 3 risk of mortality as the outcome variable. Mean absolute error, root mean squared error, and R-squared were calculated for each of the 11 machine learning models as model performance parameters.
SETTING:
Quaternary children’s hospital.
PATIENTS:
PICU patients.
INTERVENTIONS:
None.
MEASUREMENTS AND MAIN RESULTS:
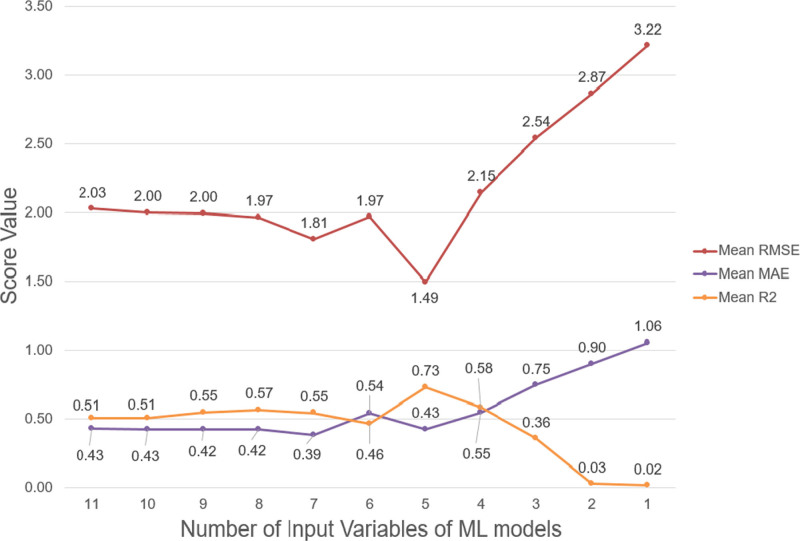
Five-thousand sixty-eight patients were analyzed. The machine learning models were able to maintain similar predictive error until the number of input variables decreased to four. The machine learning model with five input variables (mechanical ventilation in the first hour of PICU admission, very-high-risk diagnosis, surgical recovery from a noncardiac procedure, low-risk diagnosis, and base excess) produced lowest mean root mean squared error of 1.49 (95% CI, 1.05–1.93) and highest R-squared of 0.73 (95% CI, 0.6–0.86) with mean absolute error of 0.43 (95% CI, 0.35–0.5) among all the 11 machine learning models.
CONCLUSIONS:
Explainable machine learning methods were feasible in simplifying the Pediatric Index of Mortality 3 scoring system with similar risk of mortality predictions compared to the original Pediatric Index of Mortality 3 model tested in a single-center dataset.
Pediatric Index of Mortality 3 (PIM 3) is a mortality risk assessment scoring system for PICU patients (1). It was validated in various PICU settings from different countries and is widely used in clinical practice (2–5). PIM 3 is also used to calculate standardized mortality ratio (observed/expected mortality) to assess and compare outcomes between different PICUs. The PIM 3 score is calculated by using 11 variables collected from the time of initial patient contact to 1 hour after arrival in the PICU. Unfortunately, some of the variables (e.g., Pao2) may not be available before or within the 1 hour of PICU admission. Therefore, PIM 3 was designed to replace those missing input variables with normal values.
With recent advances in the predictive capabilities of machine learning (ML) algorithms and new methods explaining how input variables contribute to the ML models` outputs, we aimed to explore the feasibility of applying these ML methods to create a simpler version of PIM 3 with reduced number of input variables to decrease data collection time and workload for PICUs utilizing PIM 3.
MATERIALS AND METHODS
In addition to the patient demographics, PIM 3 variables (pupillary examination findings, type of PICU admission [elective or not], mechanical ventilation in the first hour of PICU admission [yes/no], base excess [mmol/L], systolic blood pressure [SBP] [mm Hg], [SBP]2/1,000, 100 × [Fio2/Pao2], surgical recovery [yes/no], and weighted diagnostic category [very-high-risk (VHR), high-risk (HR), and low-risk (LR)]) and already calculated individual risk of mortality (ROM) were extracted from the Virtual Pediatric Systems (VPS) database for noncardiac patients admitted to the Cleveland Clinic Children`s PICU between January 2008 and December 2019. Individual ROM calculated by the original PIM 3 algorithm that was already present in the VPS dataset was selected as the output variable for all ML models. Light Gradient Boosting Machine Regressor, a gradient boosting decision tree ML algorithm, was used to build the ML models (6). The dataset was divided into training, validation and test datasets with 2:1:1 ratio, respectively. Hyperparameter tuning was performed in validation dataset (Supplemental Digital Content 1, http://links.lww.com/CCX/A822). No imputations were performed to replace missing data. Contribution of each input variable to the ML model outcome was analyzed by SHapley Additive exPlanations (SHAP) values (7, 8). The first ML model was built using all 11 of the original PIM 3 variables as input variables. The least contributing input variable identified by SHAP analyses was eliminated and a new ML model was built with the remaining input variables. Following this methodology, also known as recursive feature elimination (9), a total of 11 ML models were built sequentially. To assess performance of the ML models in predicting the original PIM 3 ROM value, mean absolute error (MAE), root mean squared error (RMSE), and R-squared (R2) were calculated for each of the 11 ML models. In order to follow Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis guidelines (10), performance of all the ML models was only evaluated in the separate test dataset. Supplemental Digital Content 2 (http://links.lww.com/CCX/A823) includes the “Jupyter” Notebook file of data processing and analyses code. The study was approved by the Institutional Review Board (IRB) of the Cleveland Clinic (IRB Number 19-897).
RESULTS
Data from 5,068 PICU admissions from 3,665 unique patients were analyzed. Table 1 summarizes the patient characteristics and PIM 3 variables in the training, validation, and test datasets. The PIM 3 variables reliant on an arterial blood gas being done within the appropriate timeframe for scoring were missing for 97.2% of base excess and 97.3% of 100 × (Fio2/Pao2) values in the entire dataset. Figure 1 shows the summary of change in performance parameters of the ML models evaluated in the testing dataset as the number of variables decrease. The ML model with original 11 PIM 3 variables resulted in RMSE of 2.03 (95% CI, 1.42–2.63), MAE of 0.43 (95% CI, 0.35–0.5), and R2 of 0.51 (95% CI, 0.2–0.82) while the ML model with five input variables (mechanical ventilation in the first hour of PICU admission, VHR diagnosis, surgical recovery from a noncardiac procedure, LR diagnosis, and base excess) produced RMSE of 1.49 (95% CI, 1.05–1.93), MAE of 0.43 (95% CI, 0.35–0.5), and R2 of 0.73 (95% CI, 0.6–0.86). Supplemental Digital Content 3 (http://links.lww.com/CCX/A824) shows SHAP analyses, calibration graphs, and performance analyses results of all the ML models.
TABLE 1.
Summary of Patient Characteristics in Training, Validation, and Test Datasets and Pediatric Index of Mortality 3 Variables
| Patient Characteristics | Training Dataset | No. Missing Values in Training Dataset (%) | Validation Dataset | No. Missing Values in Validation Dataset (%) | Test Dataset | No. Missing Values in Test Dataset (%) |
|---|---|---|---|---|---|---|
| Total number of patients | 2,534 | 1,267 | 1,267 | |||
| Age, yr, mean (sd) | 8.2 (6.7) | 0 | 8.1 (6.7) | 0 | 8.7 (6.9) | 0 |
| Gender, n (%) | 0 | 0 | 0 | |||
| Female | 1,212 (47.8) | 626 (49.4) | 597 (47.1) | |||
| Male | 1,322 (52.2) | 641 (50.6) | 670 (52.9) | |||
| Race, n (%) | 333 (13.1) | 169 (13.3) | 147 (11.6) | |||
| Asian/Pacific Islander | 20 (0.9) | 6 (0.5) | 6 (0.5) | |||
| Black | 502 (22.8) | 272 (24.8) | 254 (22.7) | |||
| Non-White Hispanic | 7 (0.3) | 4 (0.4) | 6 (0.5) | |||
| White | 1,672 (76.0) | 813 (74.0) | 854 (76.2) | |||
| Patient type, n (%) | 0 | 0 | 0 | |||
| Scheduled (≥ 12 hr in advance) | 798 (31.5) | 407 (32.1) | 406 (32.0) | |||
| Unscheduled | 1,736 (68.5) | 860 (67.9) | 861 (68.0) | |||
| Patient origin, n (%) | 785 (30.9) | 397 (31.3) | 365 (28.8) | |||
| Emergency department | 860 (49.2) | 431 (49.5) | 446 (49.4) | |||
| General care floor | 96 (5.5) | 45 (5.2) | 47 (5.2) | |||
| Operating room | 619 (35.4) | 316 (36.3) | 329 (36.5) | |||
| Postanesthesia care unit | 169 (9.7) | 77 (8.9) | 76 (8.4) | |||
| Step-down unit | 4 (0.2) | 0 (0) | 2 (0.2) | |||
| Other | 1 (0.1) | 1 (0.1) | 2 (0.2) | |||
| Primary diagnosis category, n (%) | 34 (1.3) | 13 (1) | 13 (1) | |||
| Respiratory | 854 (34.2) | 394 (31.4) | 390 (31.1) | |||
| Cardiovascular | 45 (1.8) | 18 (1.4) | 24 (1.9) | |||
| Neurologic | 573 (22.9) | 319 (25.4) | 277 (22.1) | |||
| Endocrine | 129 (5.2) | 72 (5.7) | 81 (6.5) | |||
| Gastrointestinal | 116 (4.6) | 43 (3.4) | 57 (4.5) | |||
| Infectious | 67 (2.7) | 41 (3.3) | 42 (3.3) | |||
| Injury/poisoning/adverse effects | 154 (6.2) | 83 (6.6) | 86 (6.9) | |||
| Other | 562 (22.4) | 284 (22.6) | 297 (23.6) | |||
| Trauma, n (%) | 0 | 0 | 0 | |||
| No | 2,531 (99.9) | 1,266 (99.9) | 1,265 (99.8) | |||
| Yes | 3 (0.1) | 1 (0.1) | 2 (0.2) | |||
| PIM 3 variables | ||||||
| Pupillary reaction, > 3 mm and both fixed, n (%) | 3 (0.1) | 2 (<0.1) | 3 (0.2) | 2 (0.1) | 1 (0.1) | 2 (0.1) |
| Elective admission, n (% yes) | 778 (30.7) | 0 | 401 (31.6) | 0 | 397 (31.3) | 0 |
| Mechanical ventilation in first hour, yes, n (%) | 383 (15.1) | 0 | 157 (12.4) | 0 | 190 (15.0) | 0 |
| Base excess, mmol/L, mean (sd) | –5.5 (4.9) | 2,462 (97.1) | –6.8 (6.6) | 1,243 (98.1) | –6.6 (5.9) | 1,223 (96.5) |
| SBP, mm Hg, mean (sd) | 113.3 (19.1) | 24 (0.9) | 113.9 (18.8) | 9 (0.7) | 113.8 (19.6) | 9 (0.7) |
| (SBP)2/1,000, mean (sd) | 13.2 (4.4) | 24 (0.9) | 13.3 (4.5) | 9 (0.7) | 13.3 (4.7) | 9 (0.7) |
| 100 × (Fio2/Pao2), mean (sd) | 0.5 (0.5) | 2,462 (97.1) | 0.4 (0.3) | 1,244 (98.1) | 0.4 (0.4) | 1,225 (96.6) |
| Surgical recovery, yes, n (%) | 804 (31.7) | 0 | 396 (31.3) | 0 | 411 (32.5) | 0 |
| Very-high-risk disease, yes, n (%) | 96 (3.8) | 0 | 41 (3.2) | 0 | 52 (4.1) | 0 |
| High-risk disease, yes, n (%) | 124 (4.9) | 0 | 51 (4.0) | 0 | 64 (5.1) | 0 |
| Low-risk disease, yes, n (%) | 867 (34.2) | 0 | 419 (33.1) | 0 | 424 (33.5) | 0 |
| PIM 3 risk of mortality, %, mean (sd) | 1.2 (4.4) | 0 | 1.1 (4.8) | 0 | 1.2 (4.1) | 0 |
| PICU medical length of stay, d, mean (sd) | 2.6 (6.4) | 22 (0.8) | 2.7 (6.2) | 7 (0.5) | 2.8 (5.6) | 21 (1.6) |
| Mortality, n (%) | 26 (1.0) | 0 | 14 (1.1) | 0 | 13 (1.0) | 0 |
PIM 3 = Pediatric Index of Mortality 3, SBP = systolic blood pressure.
Figure 1.
Change in mean absolute error (MAE), root mean squared error (RMSE), and R-squared (R2) values of the machine learning (ML) models as the number of variables decrease.
DISCUSSION
This study suggests that explainable ML models can achieve similar ROM predictions with fewer input variables compared with the original PIM 3 model. The ML models were able to maintain comparable performance metrics until the number of input variables decreased to four. In fact, the ML model with five variables achieved the highest R2 and lowest RMSE among all other ML models. Utilizing fewer input variables (5 vs 11) of the PIM 3 model may decrease the labor and time required for data collection and entry for PICUs or databases using PIM 3 for trending observed to expected mortality ratio. Consequently, resources can be diverted into other areas such as quality improvement or direct clinical care, particularly in resource-limited countries, for example, where data extraction is performed by clinical team members (5). From this perspective, it can be argued that this proof of concept study has potential to indirectly improve care of critically ill children by making it easier to monitor the quality of care being provided.
Mechanical ventilation in the first hour of PICU admission, VHR diagnosis, surgical recovery from a noncardiac procedure, LR diagnosis, and base excess were the most important contributing input variables in PICU mortality prediction of our model. From a clinical standpoint, with the exception of base excess, these are variables that are immediately available upon admission to the PICU without any laboratory measurements required. This would allow the risk stratification of these patients to occur immediately upon their arrival. Not unexpectedly, children with HR medical conditions admitted to the PICU (e.g., cardiac arrest prior to PICU admission) faced the highest ROM, along with those intubated, which is consistent with previous research (11). Furthermore, a PICU stay in recovery from a noncardiac surgery or admission with a LR diagnosis was associated with lower mortality risk, which is consistent with previous findings (1).
Some of the PIM 3 variables were mostly missing (97.2% of base excess and 97.3% of 100 × (Fio2/Pao2) in this study`s dataset. Despite this, SHAP analyses showed that the variable of 100 × (Fio2/Pao2) was not a significant contributor to the model performance. In contrast, base excess was ranked higher among other input variables according to its importance for the ML models. More interestingly, pupillary examination finding was reported to have the highest odds ratio of 45.7 (95% CI, 31.71–65.9) in the original PIM 3 article (1) but was found to have the least contribution to the ML models in the present study. These differences in variable importance highlight how different predictive algorithms can vary widely in processing and assigning importance to the input variables.
To our knowledge, this is the first study assessing performance of ML methods in simplifying widely accepted ROM scoring tools. Therefore, this study may be considered as a proof of concept in exploring the role of ML to simplify commonly used predictive scoring systems without diminishing predictive power. Nonetheless, these results require further validation on external datasets including patients from multiple centers before they are regarded as valid and generalizable.
CONCLUSIONS
Explainable ML methods effectively simplified the 11 variable PIM 3 scoring system down to five variables with similar ROM predictions in a single-center dataset. Despite the promising preliminary findings, further external validation with multicenter data is necessary.
Supplementary Material
Footnotes
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s website (http://journals.lww.com/ccejournal).
The authors have disclosed that they do not have any potential conflicts of interest.
REFERENCES
- 1.Straney L, Clements A, Parslow RC, et al. ; ANZICS Paediatric Study Group and the Paediatric Intensive Care Audit Network. Paediatric index of mortality 3: An updated model for predicting mortality in pediatric intensive care*. Pediatr Crit Care Med. 2013; 14:673–681 [DOI] [PubMed] [Google Scholar]
- 2.Wolfler A, Osello R, Gualino J, et al. ; Pediatric Intensive Therapy Network (TIPNet) Study Group. The importance of mortality risk assessment: Validation of the pediatric index of mortality 3 score. Pediatr Crit Care Med. 2016; 17:251–256 [DOI] [PubMed] [Google Scholar]
- 3.Jung JH, Sol IS, Kim MJ, et al. Validation of pediatric index of mortality 3 for predicting mortality among patients admitted to a pediatric intensive care unit. Acute Crit Care. 2018; 33:170–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arias López MDP, Boada N, Fernández A, et al. ; Members of VALIDARPIM3 Argentine Group. Performance of the pediatric index of mortality 3 score in PICUs in Argentina: A prospective, national multicenter study. Pediatr Crit Care Med. 2018; 19:e653–e661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Solomon LJ, Naidoo KD, Appel I, et al. Pediatric index of mortality 3-an evaluation of function among ICUs in South Africa. Pediatr Crit Care Med. 2021; 22:813–821 [DOI] [PubMed] [Google Scholar]
- 6.Ke G, Meng Q, Finley T, et al. LightGBM: A highly efficient gradient boosting decision tree. 31st Conference on Neural Information Processing Systems (NIPS). Long Beach, CA, December 4–9, 2017 [Google Scholar]
- 7.Lundberg SM, Lee SI. A unified approach to interpreting model predictions. Advances in Neural Information Processing Systems. Long Beach, CA, December 4–9, 2017, pp 4765–4774 [Google Scholar]
- 8.Rodríguez-Pérez R, Bajorath J. Interpretation of machine learning models using shapley values: Application to compound potency and multi-target activity predictions. J Comput Aided Mol Des. 2020; 34:1013–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Degenhardt F, Seifert S, Szymczak S. Evaluation of variable selection methods for random forests and omics data sets. Brief Bioinform. 2019; 20:492–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins GS, Reitsma JB, Altman DG, et al. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): The TRIPOD statement. Ann Intern Med. 2015; 162:55–63 [DOI] [PubMed] [Google Scholar]
- 11.Verlaat CW, Wubben N, Visser IH, et al. ; SKIC (Dutch collaborative PICU research network). Retrospective cohort study on factors associated with mortality in high-risk pediatric critical care patients in the Netherlands. BMC Pediatr. 2019; 19:274. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.