Abstract
Myzus persicae is a globally important pest with the ability to adjust to a wide range of environmental situations, and many molecular technologies have been developed and applied to understand the biology and/or control this pest insect directly. Reverse-transcription quantitative real-time PCR (RT-qPCR) is a primary molecular technology that is used to quantify gene expression. Choosing a stable reference gene is significantly important for precisely clarifying the expression level of the target gene. Actin and 18S have been recommended as stable compounds for real-time RT-qPCR in M. persicae under the tested biotic and abiotic conditions. In this study, we checked the stability of Actin and 18S by analyzing the relative expression levels of the cytochrome 450 monooxygenase family member genes CYP6CY3 and CYP6-1, carboxylesterase gene E4 and vacuolar protein sorting gene VPS11 via RT-qPCR under various conditions. The expression levels of these four target genes were normalized using both Actin and 18S individually and the combination of these two genes. Our results confirmed that Actin and 18S can be used as reference genes to normalize the expression of target genes under insecticide treatment and starvation in M. persicae. However, at the developmental stages of M. persicae, the expression of the four tested target genes was normalized stably by Actin but not 18S, with the latter presenting a problematic change with the developmental stages. Thus, the stability of reference genes in response to diverse biotic and abiotic factors should be evaluated before each RT-qPCR experiment.
Introduction
Myzus persicae is commonly known as green peach aphid and represents the most well-known and dangerous agricultural pest worldwide. This pest induces injury to plants via direct feeding and spreading various plant viruses; thus, it leads to severe yield losses in economically important crop commodities [1, 2]. For the control of this insect, many molecular technologies have been developed and applied to understand the biology of M. persicae and/or control this pest insect directly [3–5].
Reverse-transcription quantitative real-time PCR (RT-qPCR) is a primary molecular technology used to quantify gene expression [6]. Typically, this technique requires the systematic normalization of the expression level of a target gene with one or more reference genes as internal controls to account for the different gene input quantities due to the different RNA samples [7]. Housekeeping genes are defined as genes expressed ubiquitously in all cell types and tissues regardless of the physiological status, development phase and treatment, and they should be easily detected and expressed at a constant level; consequently, they are used extensively as reference genes for RT-qPCR analysis [7, 8]. However, a single housekeeping gene that can satisfy all given experimental conditions has not been identified. Housekeeping genes are tissue-specific and condition-dependent, and these characteristics greatly affect the reliability and accuracy of RT-qPCR analysis [8, 9]. The use of unsuitable reference genes in RT-qPCR has been shown to lead to false results and then to erroneous interpretations [10–14]. Choosing the most stable reference gene in a specific tissue and treatment is significantly important to precisely clarify the expression level of the target gene [14, 15]. To date, more than one reference gene is required for accurate normalization in RT-qPCR analysis [8, 9].
In insect species, many experiments have been conducted in gene expression studies to select and evaluate reference genes for RT-qPCR analysis [16–19]. With the number of analysis tools, experimental factors and reference genes as parameters, the current trends in the selection of reference genes in insects were reviewed systematically based on a total of 90 representative papers, which were published mainly between 2013 and 2017 [12]. This review indicates that it is necessary to conduct reference gene screening experiments or validate the reasonability of reference genes used in each RT-qPCR. In addition, reference genes were selected and evaluated from a total of 78 insect species, including M. persicae, according to the 90 cited papers [12, 19]. In that publication, the stability of nine housekeeping genes, namely, 18S ribosomal RNA (18S), ribosomal protein gene (RPL27, RPL7 and RPL32), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), acetylcholinesterase (ACE), β-tubulins, Actin and elongation factor 1 alpha (EF1A), was evaluated comprehensively with one method and three software under various biotic and abiotic conditions [19]. Among those nine reference genes, Actin and 18S were confirmed as the most stable for RT-qPCR in M. persicae under all tested biotic and abiotic conditions (development stage, tissue, host post, wing dimorphism, insecticide, photoperiod and temperature) [19]. However, the suitability of these two reference genes may fluctuate according to the specific tissues and conditions.
In this case study, to validate the two reference genes Actin and 18S recommended by Kang et al. in M. persicae [19], we analyzed the relative expression levels of four target genes, including the two cytochrome 450 monooxygenase family member genes CYP6CY3 and CYP6-1, carboxylesterase gene E4 and vacuolar protein sorting gene VPS11, with RT-qPCR analysis under both biotic and abiotic conditions. CYP6CY3 and E4 are belong to metabolic enzymes, which have been studied widely and deeply and associated to insecticide resistance in M. persicae [1]. The over-expression of these two genes induced by insecticide treatment even as sublethal doses have been observed in many insects including M. persicae [20–22]. CYP6-1 is a member of cytochrome 450 monooxygenase family. VPS11 is involved in vesicle transport to vacuoles which plays an important role in segregation of intracellular molecules into distinct organelles [23]. However, both CYP6-1 and VPS11 are unfamiliar in M. persicae. The changes in the expression pattern of these two genes induced by the changes of both biotic and abiotic conditions would be unpredictable, which will give the relative comparison. Additionally, the expression levels of these four target genes were normalized using both Actin and 18S individually and the two-gene combination with the 2-ΔΔCt method.
Material and methods
Insects
A field population of M. persicae collected from Nanping, Fujiang Province in 2018 was used in this study. Aphids were grown on the leaves of Chinese cabbage (Brassica napus L var chinensis) in an incubator without the use of any pesticides at 22 ± 2°C, 65 ± 5% relative humidity and a 16 Light: 8 Dark photoperiod [1, 3].
Insecticidal stress
Two insecticides, flonicamid 97.1% and pymetrozine 98%, were used to treat M. persicae in this research, which are normally useful in the control of M. persicae. According to the bioassay results for these two insecticides (data not shown), three concentrations of flonicamid (17.2, 34.4 and 68.8 mg/L) and pymetrozine (10.0, 20.0 and 40.0 mg/L), which induced sublethal effects, were used to treat adults of M. persicae with the leaf dip method 01]. In the control group, Chinese cabbage leaves were treated with distilled water containing 1% EtOH. A total of 30 adults were collected to expose each dose of the two tested insecticides under standard conditions, 22 ± 2°C, 65 ± 5% relative humidity and a 16 Light: 8 Dark photoperiod. Three replicates were conducted for each treatment, and mortality was recorded after 24 h.
Starvation treatments
To create starvation stress, a group of 30 newly emerged adults of M. persicae as one treatment were put into glass petri dishes without food for a certain amount of time, including 6, 12, 24 or 48 hours, and placed in a climatic chamber under controlled conditions [1]. The aphids fed Chinese cabbage leaves corresponding to a certain time were used as the control group. All treatments were replicated three times, and the relevant mortality was recorded.
Aphid developing phases
Five different growing stages of M. persicae (green peach aphids), including 1st, 2nd, 3rd, and 4th instar nymphs and newly emerged adults, were collected as samples [24]. For each nymph sample, a total of 60 aphids were collected, and for each adult sample, a total of random 30 aphids were collected. All samples were collected three times.
RNA isolation and cDNA synthesis
A total of 20–30 adult or 50–60 nymph M. persicae from all treatments were assembled per biological replication to isolate RNA with RNeasy® Mini Kits (Qiagen, ON, Canada) according to the manufacturer’s protocol. A NanoDrop ND_1000 (NanoDrop Technologies, Wilmington, DE, USA) was used to conduct both qualitative and quantitative analyses. RNA integrity was confirmed by evaluating the samples by gel electrophoresis (1% gel). RNA samples with an absorbance ratio range (OD) of 260/280 between 1.90 and 2.20 were used in further studies [24, 25]. Primary stand cDNA was created from 1 μg of the total RNA in a 20 μl volume by using the Omniscript® Reverse Transcript Kit (Qiagen, ON, Canada) and then stored at -20°C for future analysis.
Gene expression with RT-qPCR
Primers used for the RT-qPCR analysis of the target genes CYP6CY3, CYP6-1, E4 and VPS11 were designed with online software (http://www.idtdna.com/scitools/applications/primerquest/) based on sequences in the National Center for Biotechnology Information (NCBI) database (Table 1). The primers used for the reference genes Actin and 18S are also shown in Table 1 and were cited from published previous studies [26, 27]. The amplification efficiency for each pair of specific primers was detected by the slope of the standard curve generated using a series of 10-fold dilutions of cDNAs [25].
Table 1. Primers used in real-time qRT-PCR.
| Genes | Primer sequences (5’-3’) | Length (bp) | PCR efficiency (%) | R 2 | Origin of primers |
|---|---|---|---|---|---|
| CYP6CY3 | F: CGGGGTGACGATCATCTATT | 128 | 94.2 | 0.983 | Accession number: HM009309 |
| R: GGGTGGTCTTTTGACAAAGC | |||||
| CYP6-1 | F: TCAACGAATGTGGCGACGAA | 120 | 102 | 0.982 | Accession number: AJ310561 |
| R: CGCAGGTGGCAATTACGTCT | |||||
| E4 | F: AAACTTTCCTTTTACACCGTT | 160 | 100 | 0.991 | Accession number: X74554 |
| R: TCTAAGCCAAGAAATGTTGAAA | |||||
| VPS11 | F: GGGTCAAGGAATAACTGGAATGGC | 180 | 105 | 0.991 | Accession number: XM_022313476 |
| R: TCAGGTCCATTCACTAACACCGA | |||||
| Actin | F: GGTGTCTCACACACAGTGCC | 142 | 100 | 0.996 | Bass et al., 2011 [22] |
| R: CGGCGGTGGTGGTGAAGCTG | |||||
| 18S | F: TCAACACGGGAAACCTCACCA | 80 | 96.6 | 0.992 | Coleman et al., 2015 [23] |
| R: CACCACCCACCGAATCAAGAA |
R2, regression coefficient.
RT-qPCR was evaluated using TransStart® Top Green qPCR SuperMix (Transgen, Beijing, China) with QuantStudio 3 Real-Time PCR Systems (Applied Biosystems, USA). According to the manufacturer’s instructions, the whole 20 μL reaction mixture was completed in a 50 μL tube containing 10 μL of 2 × TransStart® Top Green qPCR SuperMix, 0.4 μL of each primer, 8.2 μL of distilled ddH2O, and 1 μL of cDNA template. The RT-qPCR conditions were 30 seconds at 94°C, followed by 40 cycles at 94°C for 30 seconds and then annealing at 72°C for 30 seconds. The RT-qPCR study involved three self-determining biological replicates for each treatment. The relative expression of the four tested target genes was calculated with the Actin and 18S individually as reference genes according to Pfaffl [6] and with the two used together as reference genes according to the methods developed by Vandesompele et al. (2002) [8].
Single control normalization error E
According to a report by Vandesompele et al. (2002) [8], the possible errors of using only one reference gene to normalize the expression levels of the target gene, termed single control normalization errors (Es), were calculated.
Statistical analysis
Data were statistically evaluated using ANOVA and monitored by the LSD test in SPSS software version 22.0. The results were considered significant when the P value was < 0.05. For the expression of relative quantities of genes, Tukey’s comparison was used to test the mean differences between treatments across generations and separated by LSD tests.
Results
Amplification efficiency of PCR
As shown in Fig 1, a unique PCR product detected by 2% agarose gel electrophoresis analysis ensured that a single amplicon was produced for each target and reference gene. The PCR efficiency and regression coefficient (R2) were evaluated by the establishment of standard curves. As shown in Table 1, the efficiency values of all primers ranged from 94.2 to 105% with regression coefficient values from 0.98 to 1.00, which reached the threshold of 90.0 to 110% for the PCR efficiency and 0.98 to 1.00 for the regression coefficient (R2) [24, 28]. These results suggested that all the primer pairs were appropriate for the RT-qPCR analysis.
Fig 1. Gel electrophoresis analysis of PCR amplification for each target and reference gene.
M, 100 bp DNA Ladder (Takara Bio INC, Beijing, China); 1–6, Actin, 18S, CYP6CY3, CYP6-1, E4 and VPS11.
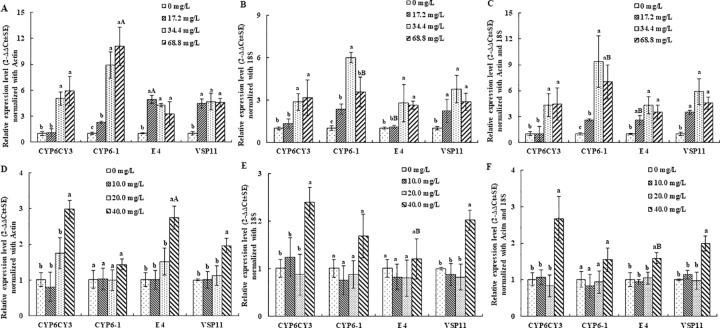
Expression of target genes normalized to Actin and 18S under the insecticide treatment
After exposure for 24 hours with flonicamid and pymetrozine by the leaf dip method, the mortality of M. persicae was less than 10.0% for the two tested insecticides, even at the highest doses of 68.8 mg/L for flonicamid and 40.0 mg/L for pymetrozine (Table 2). In the RT-qPCR assay, the fold expression of four target genes was normalized using Actin and 18S as the internal control. When M. persicae was treated with flonicamid, remarkable changes in the four target genes (CYP6CY3, CYP6-1, E4 and VPS11) induced by insecticide treatment were observed with both Actin and 18S as reference genes. A comparison of the expression data normalized to Actin and 18S showed that a similar tendency occurred for CYP6CY3 and VPS11 with different concentrations of flonicamid (Fig 2A and 2B) while significant differences occurred between CYP6-1 and E4 at flonicamid doses of 17.2 mg/L and 68.8 mg/L, respectively. For pymetrozine, the relative expression levels showed a similar expression pattern for CYP6CY3, CYP6-1 and VPS11 with either Actin or 18S used alone for normalization, and CYP6CY3 and VPS11 were obviously increased at the highest dose of 40.0 mg/L (Fig 2D and 2E). However, the relative expression level of E4 was increased significantly when normalized to Actin but not to 18S at the highest dose of 40.0 mg/L pymetrozine. In addition, the relative expression levels of target genes were also normalized using the combination of Actin and 18S (Fig 2C and 2F). The results showed that similar expression profiles were observed for all four tested genes compared with those normalized with 18S alone (Fig 2B, 2C, 2E and 2F).
Table 2. Mortality of M. persicae after exposure to different doses of flonicamid and pymetrozine for 24 hours.
| Flonicamid | Pymetrozine | ||
|---|---|---|---|
| Concentrations (mg/L) | Mortality (%) | Concentrations (mg/L) | Mortality (%) |
| 0 | 0 | 0 | 0 |
| 17.2 | 5.55 ± 3.84 | 10.0 | 4.44 ± 3.84 |
| 34.4 | 8.89 ± 1.92 | 20.0 | 6.67 ± 3.33 |
| 68.8 | 10.0 ± 3.33 | 40.0 | 8.89 ± 3.84 |
Fig 2. Fold expression of four target genes normalized to Actin and 18S under insecticide treatment.
A, B, C, Relative expression levels normalized to Actin and 18S alone and in combination under the flonicamid treatment; and D, E, F, relative expression levels normalized to Actin and 18S alone and in combination under the pymetrozine treatment. Values are shown as the mean ± SEM. Bars with different letters are significantly different from each other (P < 0.05). Lowercase letters indicate the comparison among different concentrations for one target gene normalized with the same housekeeping gene. Uppercase letters represent the comparison among different normalized genes for one target gene under the same treatment of one insecticide.
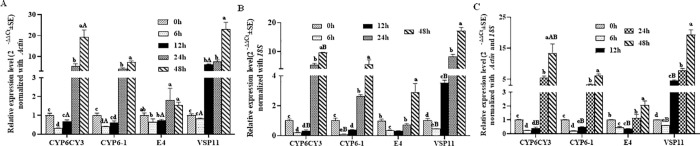
Expression of the target genes normalized to Actin and 18S in M. persicae after starvation
As shown in Fig 3, the tendency of expression changes of the four target genes normalized to Actin was generally consistent with those normalized to 18S and was initially inhibited and then promoted with changes in starvation time (Fig 3A and 3B). After starvation for 6 h, the relative expression levels of four target genes were all inhibited significantly compared to those without starvation treatment (starvation for 0 hours) normalized both with Actin and 18S as reference gene respectively, except for E4 and VSP11 normalized with Actin. The relative expression levels of four target genes were all increased significantly in M. persicae after starvation for 48 h normalized both with Actin and 18S as reference gene respectively. A comparison between the relative expression levels normalized with Actin and 18S showed that significant differences occurred for all tested target genes, with CYP6CY3 at 12 h and 48 h, CYP6-1 at 6 h, E4 at 12 h, and VPS11 at 6 h and 12 h. As shown in Fig 3C, the expression fold normalized to the combination of Actin and 18S showed no differences compared with those normalized to 18S individually (Fig 3B and 3C).
Fig 3.
Relative expression levels of target genes in M. persicae after starvation normalized to Actin (A) and 18S (B) alone and in combination (C). Values are shown as the mean ± SEM. Bars with different letters are significantly different from each other (P < 0.05). Lowercase letters indicate the comparison among different starvation time for one target gene normalized with the same housekeeping gene. Uppercase letters represent the comparison among different normalized genes for one target gene under the same starvation time.
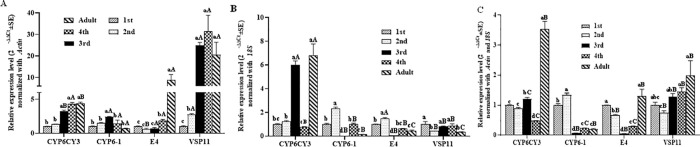
Expression of target genes normalized to Actin and 18S at different developmental stages of M. persicae
With the 1st instar nymphs of M. persicae as the control group, the expression levels of the four target genes (CYP6CY3, CYP6-1, E4 and VPS11) at different developmental stages of M. persicae are shown in Fig 4, which illustrates that the tendency of expression changes of the four target genes with the developmental stage were obviously different based on the normalization to Actin or 18S (Fig 4A and 4B). When normalized to Actin alone, the relative expression ratios of the four tested target genes changed in a certain regular pattern with the developmental stages of M. persicae. However, the fold change in expression of the four tested genes normalized to 18S alone fluctuated irregularly with the developmental stage in M. persicae. The mean threshold cycle (Ct) and standard error values of Actin and 18S in all samples tested for the developmental stages were 16.5 ± 1.5 and 14.4 ± 4.8, respectively. However, the mean Ct and standard error values of Actin and 18S in all samples tested for the insecticide treatment and starvation conditions were 19.6 ± 0.3 and 18.3 ± 2.0 and 12.9 ± 0.9 and 11.7 ± 1.3, respectively. Additionally, we calculated the stability of Actin and 18S with the software program NormFinder at the different developmental stages of M. persicae [29]. This result indicated that Actin (0.87) was more stable than 18S (2.06) in this subset. The relative expression levels of all four tested target genes normalized to Actin and 18S together are shown in Fig 4C, which indicated that correction could not be made due to the instability of 18S.
Fig 4.
Expression of target genes at different developmental stages of M. persicae normalized to Actin (A) and 18S (B) alone and in combination (C). Values are shown as the mean ± SEM. Bars with different letters are significantly different from each other (P < 0.05). Lowercase letters indicate the comparison among different developmental stages for one target gene normalized with the same housekeeping gene. Uppercase letters represent the comparison among different normalized genes for one target gene under the same developmental stage.
Single control normalization error E
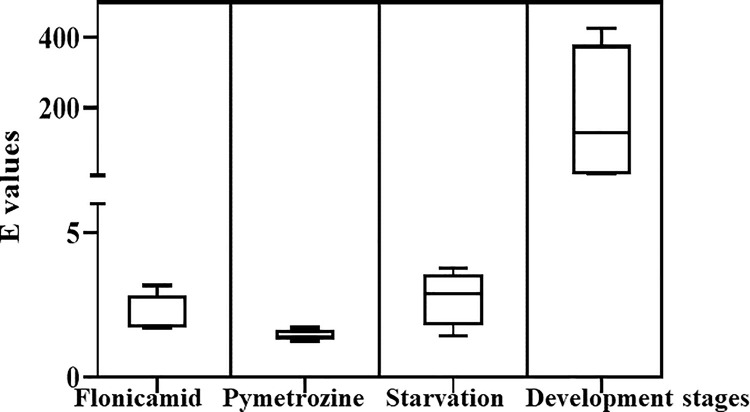
To evaluate the possible errors induced by using only Actin or 18S as a reference gene to normalize the relative expression levels of the four tested target genes, we calculated the single control normalization error E according to the method reported by Vandesompele et al. (2002) [8]. Under the insecticide treatment, the E values indicating the ratio of the fold expression between normalization to Actin and 18S ranged from 1.68 (for VSP11) to 3.17 (for CYP6-1) under flonicamid treatment and from 1.25 (for E4) to 1.71 (for CYP6-1) under pyrethrozine treatment (Fig 5). Under starvation conditions, the minimum E value was 1.42 for VSP11 and the maximum E value was 3.78 for E4. However, the E value during the developmental stages reached a maximum of 426 for CYP6-1 and a minimum of 6.58 for VSP11 (Fig 5).
Fig 5. Single control normalization error E for four target genes under different biotic and abiotic conditions in M. persicae.
The boxes indicate the 25th and 75th percentiles. Lines across the boxes represent the medians, and upper and lower dots represent the maximum and minimum values, respectively.
Discussion
RT-qPCR was developed and is frequently used to determine messenger RNA expression levels elaborated in many biological procedures [30, 31]. Different types of reference genes are used as internal controls to normalize the expression of target genes between different samples in many experiments [14, 24]. However, the expression of a specific reference gene changes under different environments and treatments, which can affect the real expression of a particular target gene. Therefore, a stable endogenous reference gene is essential for relative quantification in gene expression analyses. Currently, many reference genes have been recommended in different insect species, including sweet potato whitefly Bemisia tabaci (Gennadius) [32–34], brown planthopper Nilaparvata lugens (Stål) [35, 36], diamondback moth Plutella xylostella (Linnaeus) [15, 17], fruit fly Drosophila melanogaster (Meigen) [37], pea aphid Acyrthosiphon pisum (Harris) [38] and peach aphid M. persicae (Sülzer) [19]. According to the report of Kang et al. (2017) [19], 18S and Actin were recommended as the most stable reference genes in M. persicae for abiotic (photoperiod, temperature and insecticide susceptibility) and biotic (host plant, developmental stage, tissues and wing dimorphism) studies. In this paper, 18S and Actin were used to confirm their stability as reference genes for normalizing the selected four target genes CYP6CY3, CYP6-1, E4 and VPS11 under different treatments, including insecticide, starvation and development.
Actin encodes a major structural protein and is expressed at various levels in many cell types. It is regarded as an ideal internal control and widely used as a reference gene for RT-qPCR in many organisms. Recently, the stability of Actin as a housekeeping gene has been investigated frequently in insects [12, 39]. Actin is the most stable reference gene, including ribosomal protein L (RPL), tubulin, GAPDH, ribosomal protein S (RPS), 18S, EF1A, TATA box binding protein (TATA), heat shock protein (HSP) and succinate dehydrogenase complex subunit A (SDHA), across different developmental stages of many insects, including honeybee Apis mellifera (Linnaeus) [16], diamondback moth P. xylostella (Linnaeus) [15, 17], fruit fly D. melanogaster (Meigen) [37], striped stem borer Chilo suppressalis (Walker) [40], etc. [12, 39]. Previous studies have indicated that Actin is less stable in several members of the Coccinellidae family, including the seven-spotted lady beetle Coccinella septempunctata (Linnae) [41], ladybird beetle Coleomegilla maculata (DeGeer) [42] and convergent lady beetle Hippodamia convergens (Guérin-Méneville) [43]. 18S ribosomal RNA is a part of the ribosomal RNA (rRNA), which accounts for > 80% of the total RNA pool [12, 13], whereas mRNA accounts for only 3–5%. Several recent studies concluded that the use of ribosomal RNA as an endogenous control in a variety of cellular systems is consistently the best choice compared with other methods [39, 44]. However, it is generally acknowledged that the use of rRNA forms as reference genes to normalize the expression of target genes of mRNA species is problematic due to the potential to mask subtle changes in target genes [12, 45].
In this study, the values of single control normalization error E for Actin and 18S ranged from 1.25 to 3.78 under the two tested conditions. According to Vandesompele et al., the average 75th and 90th percentile E values were 3.0 (range 2.1–3.9) and 6.4 (range 3.0–10.9), respectively [8]. Additionally, the expression pattern of target genes with changes in condition was similar between the normalization of Actin and 18S. These data confirmed again that Actin and 18S are stable reference genes for the normalization of the expression levels of multiple genes under the starvation and insecticide treatment conditions. However, when comparing the detailed expression levels normalized with Actin and 18S individually, the relative expression ratios with Actin as a reference gene were higher than those normalized with 18S in some cases. The relative expression levels of the combination of Actin and 18S were fully consistent with 18S alone as a reference gene under both the insecticide and starvation treatments. According to Kang et al., the stability of nine reference genes in M. persicae under an insecticide treatment was ranked from the most stable to the least stable as follows: L27 (ribosomal protein L27) > 18S > RPL7 >EF1A> RPL32 > GAPDH > Actin > β‐tubulin >ACE (acetylcholinesterase) [19]. The stability of the nine selected candidate reference genes, including Actin and 18S, was not evaluated directly under starvation conditions [19]. According to our data, Actin and 18S were also stable in M. persicae under these application scenarios, which provides formative guidance for the application of Actin and 18S in M. persicae. Furthermore, the expression levels of the four tested target genes in M. persicae could be more accurately normalized with the combination of Actin and 18S. Therefore, the importance of carefully selecting more than one stable and suitable reference gene for RT-qPCR normalization was suggested, although target gene expression could be normalized with either Actin or 18S alone.
The developmental stage is an important experimental factor that has been widely investigated for its effect on the stability of reference genes in insects [12, 39]. In this study, the tendency of expression changes of four target genes with the development stage obviously differed when normalized to Actin or 18S individually and in combination. The mean Ct and standard error value value of Actin in all samples tested for developmental stages was 16.5 ± 1.5, and the stability calculated with the software program NormFinder was 0.87 [8]. In the same subset, the mean Ct and standard error and stability values for 18S were 14.4 ± 4.9 and 2.06, respectively. The E value during the developmental stages reached a maximum of 426 for CYP6-1 and a minimum of 6.58 for VSP11. These results indicated that Actin is stable in normalizing the expression of the four tested target genes in M. persicae at different developmental stages. However, 18S, which has been regarded as a more stable reference gene than Actin in M. persicae [19], showed a problematic change with the development stages. Unstable reference genes either alone or in combination cannot be used to normalize the expression of target genes in the analysis of RT‐qPCR result. It is recommended to evaluate the stability of reference genes in response to diverse biotic and abiotic factors before each RT-qPCR experiment.
Acknowledgments
The field population of M. persicae was kindly provided by Dr. Jinfeng Hu, Institute of Plant Protection, Fujian Academy of Agricultural Sciences, Fuzhou, Fujian, PR China.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was financially supported by the National Key Research and Development Program of China (2016YFD0200500). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Sial MU, Zhao Z, Zhang L, Zhang Y, Mao L, Jiang H. Evaluation of insecticides induced hormesis on the demographic parameters of Myzus persicae and expression changes of metabolic resistance detoxification genes. Sci. Rep. 2018; 8(1): 16601. doi: 10.1038/s41598-018-35076-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Charaabi K, Boukhris-Bouhachem S, Makni M, Denholm I. Occurrence of target-site resistance to neonicotinoids in the aphid Myzus persicae in Tunisia, and its status on different host plants. Pest Manag. Sci. 2018; 74: 1297–1301. doi: 10.1002/ps.4833 [DOI] [PubMed] [Google Scholar]
- 3.Sial MU, Zhao Z, Zhang L, Zhang Y, Mao L, Jiang H. Loop-mediated isothermal amplification for the detection of R81T mutation in nAChR with crude genomic DNA extracted from individual Myzus persicae. J. Pest Sci. 2020; 93: 531–541. [Google Scholar]
- 4.Rubio ‐ Meléndez ME, Sepúlveda DA, Ramírez CC. Temporal and spatial distribution of insecticide‐resistance mutations in the green peach aphid Myzus persicae (Hemiptera: Aphididae) on primary and secondary host plants in central Chile. Pest Manag. Sci. 2018; 74: 340–347. doi: 10.1002/ps.4708 [DOI] [PubMed] [Google Scholar]
- 5.Correy GJ, Zaidman D, Harmelin A, Carvalho S, Mabbitt PD, Calaora V, et al. Overcoming insecticide resistance through computational inhibitor design. Proc Natl Acad Sci USA. 2019; 116: 21012–21021. doi: 10.1073/pnas.1909130116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001; 29: e45. doi: 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huggett J, Dheda K, Bustin S, Zumla A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005; 6: 279–284. doi: 10.1038/sj.gene.6364190 [DOI] [PubMed] [Google Scholar]
- 8.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002; 3(7): RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chapman JR, Waldenström J. With reference to reference genes: A systematic review of endogenous controls in gene expression studies. PLoS One. 2015; 10(11): e0141853. doi: 10.1371/journal.pone.0141853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Panina Y, Germond A, Masui S, Watanabe TM. Validation of common housekeeping genes as reference for qPCR gene expression analysis during iPS reprogramming process. Sci. Rep. 2018; 8(1): 8716. doi: 10.1038/s41598-018-26707-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caracausi M, Piovesan A, Antonaros F, Strippoli P, Vitale L, Pelleri MC. Systematic identification of human housekeeping genes possibly useful as references in gene expression studies. Mol. Med. Rep. 2017; 6: 2397–2410. doi: 10.3892/mmr.2017.6944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lü J, Yang C, Zhang Y, Pan H. Selection of reference genes for the normalization of RT-qPCR data in gene expression studies in insects: A systematic review. Front Physiol. 2018; 9: 1560. doi: 10.3389/fphys.2018.01560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kozera B, Rapacz M. Reference genes in real-time PCR. Appl. Genet. 2013; 54: 391–406. doi: 10.1007/s13353-013-0173-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chervoneva I, Li Y, Schulz S, Croker S, Wilson C, Waldman SA, et al. Selection of optimal reference genes for normalization in quantitative RT-PCR. BMC Bioinformatics. 2010; 11: 253. doi: 10.1186/1471-2105-11-253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.You Y, Xie M, Vasseur L, You M. Selecting and validating reference genes for quantitative real-time PCR in Plutella xylostella (L.). Genome. 2018; 61: 349–358. doi: 10.1139/gen-2017-0176 [DOI] [PubMed] [Google Scholar]
- 16.Moon K, Lee SH, Kim YH. Validation of quantitative real-time PCR reference genes for the determination of seasonal and labor-specific gene expression profiles in the head of Western honey bee, Apis mellifera. PLoS One. 2018; 13(7): e0200369. doi: 10.1371/journal.pone.0200369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu W, Xie W, Zhang Z, Wang S, Wu Q, Liu Y, et al. Exploring valid reference genes for quantitative real-time PCR analysis in Plutella xylostella (Lepidoptera: Plutellidae). Int. J. Biol. Sci. 2013; 9: 792–802. doi: 10.7150/ijbs.5862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Majerowicz D, Alves-Bezerra M, Logullo R, Fonseca-de-Souza AL, Meyer-Fernandes JR, Braz GR, et al. Looking for reference genes for real-time quantitative PCR experiments in Rhodnius prolixus (Hemiptera: Reduviidae). Insect Mol. Biol. 2011; 20: 713–722. doi: 10.1111/j.1365-2583.2011.01101.x [DOI] [PubMed] [Google Scholar]
- 19.Kang ZW, Liu FH, Tian HG, Zhang M, Guo SS, Liu TX. Evaluation of the reference genes for expression analysis using quantitative real-time polymerase chain reaction in the green peach aphid, Myzus persicae. Insect Sci. 2017; 24: 222–234. doi: 10.1111/1744-7917.12310 [DOI] [PubMed] [Google Scholar]
- 20.Rix RR, Ayyanath MM, Cutler GC. Sublethal concentrations of imidacloprid increase reproduction, alter expression of detoxification genes, and prime Myzus persicae for subsequent stress. J Pest Sci. 2016; 89: 581–589. [Google Scholar]
- 21.Gerami S, Jahromi K, Ashouri A, Rasoulian G, Heidari A. Sublethal effects of imidacloprid and pymetrozine on the life table parameters of Aphis gossypii Glover (Homoptera: Aphididae). Comm Agric Appl Biol Sci. 2004; 70: 779–785. [PubMed] [Google Scholar]
- 22.Margaritopoulos JT, Kati AN, Voudouris CC, Skouras PJ, Tsitsipis JA. Long-term studies on the evolution of resistance of Myzus persicae (Hemiptera: Aphididae) to insecticides in Greece. Bull Entomol Res. 2021; 111:1–16. doi: 10.1017/S0007485320000334 [DOI] [PubMed] [Google Scholar]
- 23.Tichopad A. Standardized determination of real-time PCR efficiency from a single reaction set-up. Nucleic Acids Res. 2003; 31: 122e–122. doi: 10.1093/nar/gng122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004; 26: 509–515. doi: 10.1023/b:bile.0000019559.84305.47 [DOI] [PubMed] [Google Scholar]
- 25.Derveaux S, Vandesompele J, Hellemans J. How to do successful gene expression analysis using real-time PCR. Methods. 2010; 50: 227–230. doi: 10.1016/j.ymeth.2009.11.001 [DOI] [PubMed] [Google Scholar]
- 26.Bass C, Puinean AM, Andrews M, Cutler P, Daniels M, Elias J, Paul VL, Crossthwaite AJ, Denholm I, Field LM, Foster SP, Lind R, Williamson MS, Slater R. Mutation of a nicotinic acetylcholine receptor β subunit is associated with resistance to neonicotinoid insecticides in the aphid Myzus persicae. BMC Neurosci. 2011; 12: 51. doi: 10.1186/1471-2202-12-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coleman AD, Wouters RH, Mugford ST, Hogenhout SA. Persistence and transgenerational effect of plant-mediated RNAi in aphids. J. Exp. Bot. 2015; 66: 541–548. doi: 10.1093/jxb/eru450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hong F, Han HL, Pu P, Wei D, Wang J, Liu Y. Effects of five host plant species on the life history and population growth parameters of Myzus persicae (Hemiptera: Aphididae). J. Insect Sci. 2019; 19: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anstaett OL, Brownlie J., Collins ME, Thomas CJ. Validation of endogenous reference genes for RT-qPCR normalisation in bovine lymphoid cells (BL-3) infected with Bovine Viral Diarrhoea Virus (BVDV). Vet Immunol. Immunopathol. 2010; 137: 201–207. doi: 10.1016/j.vetimm.2010.05.006 [DOI] [PubMed] [Google Scholar]
- 30.Fleige S, Pfaffl MW. RNA integrity and the effect on the real-time qRT-PCR performance. Mol. Aspects Med. 2006; 27: 126–139. doi: 10.1016/j.mam.2005.12.003 [DOI] [PubMed] [Google Scholar]
- 31.Ahmed FE. Quantitative real-time RT-PCR: Application to carcinogenesis. Cancer Genomics Proteomics. 2005; 2: 317–332. [PubMed] [Google Scholar]
- 32.Li R, Xie W, Wang S, Wu Q, Yang N, Yang X, et al. Reference gene selection for qRT-PCR analysis in the sweetpotato whitefly, Bemisia tabaci (Hemiptera: Aleyrodidae). PLoS One. 2013; 8: e53006. doi: 10.1371/journal.pone.0053006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaur R, Gupta M, Singh S, Pandher S. Evaluation and validation of experimental condition-specific reference genes for normalization of gene expression in Asia II-I Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae). Gene Expr. Patterns. 2019; 34: 119058. doi: 10.1016/j.gep.2019.119058 [DOI] [PubMed] [Google Scholar]
- 34.Su Y, He WB, Wang J, Li JM, Liu SS, Wang XW. Selection of endogenous reference genes for gene expression analysis in the Mediterranean species of the Bemisia tabaci (Hemiptera: Aleyrodidae) complex. J. Econ. Entomol. 2013; 106: 1446–1455. doi: 10.1603/EC12459 [DOI] [PubMed] [Google Scholar]
- 35.Wan PJ, Tang YH, Yuan SY, He JC, Wang WX, Lai FX, et al. Reference genes for quantitative real-time PCR analysis in symbiont Entomomyces delphacidicola of Nilaparvata lugens (Stal). Sci. Rep. 2017; 7: 42206. doi: 10.1038/srep42206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ponton F, Chapuis MP, Pernice M, Sword GA, Simpson SJ. Evaluation of potential reference genes for reverse transcription-qPCR studies of physiological responses in Drosophila melanogaster. J. Insect Physiol. 2011; 57: 840–850. doi: 10.1016/j.jinsphys.2011.03.014 [DOI] [PubMed] [Google Scholar]
- 37.Yuan M, Lu Y, Zhu X, Wan H, Shakeel M, Zhan S, et al. Selection and evaluation of potential reference genes for gene expression analysis in the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae) using reverse-transcription quantitative PCR. PLoS One. 2014; 9(1): e86503. doi: 10.1371/journal.pone.0086503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang C, Pan H, Liu Y, Zhou X. Selection of reference genes for expression analysis using quantitative real-time PCR in the pea aphid, Acyrthosiphon pisum (Harris) (Hemiptera, Aphidiae). PLoS One. 2014; 9(11): e110454. doi: 10.1371/journal.pone.0110454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shakeel M, Rodriguez A, Tahir UB, Jin F. Gene expression studies of reference genes for quantitative real-time PCR: an overview in insects. Biotechnol. Lett. 2018; 40: 227–236. doi: 10.1007/s10529-017-2465-4 [DOI] [PubMed] [Google Scholar]
- 40.Xu J, Lu MX, Cui YD, Du YZ. Selection and evaluation of reference genes for expression analysis using qRT-PCR in Chilo suppressalis (Lepidoptera: Pyralidae). J. Econ. Entomol. 2017; 110: 683–691. doi: 10.1093/jee/tow297 [DOI] [PubMed] [Google Scholar]
- 41.Yang C, Preisser EL, Zhang H, Liu Y, Dai L, Pan H, et al. Selection of reference genes for RT-qPCR analysis in Coccinella septempunctata to assess un-intended effects of RNAi transgenic plants. Front Plant Sci. 2016; 7: 1672. doi: 10.3389/fpls.2016.01672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang C, Pan H, Noland JE, Zhang D, Zhang Z, Liu Y, et al. Selection of reference genes for RT-qPCR analysis in a predatory biological control agent, Coleomegilla maculata (Coleoptera: Coccinellidae). Sci. Rep. 2015; 5: 18201. doi: 10.1038/srep18201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pan H, Yang X, Siegfried BD, Zhou XA. Comprehensive selection of reference genes for RT-qPCR analysis in a predatory lady beetle, Hippodamia convergens (Coleoptera: Coccinellidae). PLoS One. 2015; 10(4): e0125868. doi: 10.1371/journal.pone.0125868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuchipudi SV, Tellabati M, Nelli RK, White GA, Perez BB, Sebastian S, et al. 18S rRNA is a reliable normalisation gene for real time PCR based on influenza virus infected cells. Virol. J. 2012; 9: 230. doi: 10.1186/1743-422X-9-230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang S, An S, Li Z, Wu F, Yang Q, Liu Y, et al. Identification and validation of reference genes for normalization of gene expression analysis using qRT-PCR in Helicoverpa armigera (Lepidoptera: Noctuidae). Gene. 2015; 555: 393–402. doi: 10.1016/j.gene.2014.11.038 [DOI] [PubMed] [Google Scholar]