Abstract
Background
Carbapenem-resistant Enterobacterales (CRE) harboring blaKPC have been endemic in Chicago-area healthcare networks for more than a decade. During 2016–2019, a series of regional point-prevalence surveys identified increasing prevalence of blaNDM-containing CRE in multiple long-term acute care hospitals (LTACHs) and ventilator-capable skilled nursing facilities (vSNFs). We performed a genomic epidemiology investigation of blaNDM-producing CRE to understand their regional emergence and spread.
Methods
We performed whole-genome sequencing on New Delhi metallo-beta-lactamase (NDM)+ CRE isolates from 4 point-prevalence surveys across 35 facilities (LTACHs, vSNFs, and acute care hospital medical intensive care units) in the Chicago area and investigated the genomic relatedness and transmission dynamics of these isolates over time.
Results
Genomic analyses revealed that the rise of NDM+ CRE was due to the clonal dissemination of an sequence type (ST) 147 Klebsiella pneumoniae strain harboring blaNDM-1 on an IncF plasmid. Dated phylogenetic reconstructions indicated that ST147 was introduced into the region around 2013 and likely acquired NDM around 2015. Analyzing the relatedness of strains within and between facilities supported initial increases in prevalence due to intrafacility transmission in certain vSNFs, with evidence of subsequent interfacility spread among LTACHs and vSNFs connected by patient transfer.
Conclusions
We identified a regional outbreak of blaNDM-1 ST147 that began in and disseminated across Chicago area post-acute care facilities. Our findings highlight the importance of performing genomic surveillance at post-acute care facilities to identify emerging threats.
Keywords: NDM, Klebsiella pneumonia, ST147, genomic epidemiology, carbapenem resistance
Whole-genome sequencing of carbapenem-resistant Enterobacterales in Chicago area healthcare facilities revealed clonal dissemination of blaNDM-1Klebsiella pneumoniae sequence type 147 within and between post-acute care facilities.
Carbapenem-resistant Enterobacterales (CRE) represent an urgent antibiotic-resistance threat due to their resistance to first-line antibiotics and transmissibility in healthcare settings [1, 2]. The emergence of epidemic lineages of CRE that are resistant to nearly all antibiotics and that cause infections with high mortality rates, such as Klebsiella pneumoniae carbapenemase (KPC) containing Klebsiella pneumoniae (KPC-Kp) sequence type (ST) 258 [3], has further escalated the need for more effective strategies to interrupt CRE transmission. Most interventions to prevent the spread of CRE and other healthcare-associated antibiotic-resistance threats have been implemented at the level of individual healthcare facilities. However, there is now a multitude of evidence that the frequent movement of colonized and infected patients among regional healthcare facilities necessitates regional surveillance and infection prevention strategies [4].
Long-term acute care hospitals (LTACHs) and ventilator-capable skilled nursing facilities (vSNFs) are potentially high-impact settings for implementation of regional CRE surveillance and infection prevention interventions [5, 6]. Patients in these facilities have been shown to be colonized with CRE at high rates, likely due to a combination of their chronic severe illness, long lengths of stay, and high rates of prior or ongoing antibiotic exposure. Modeling and epidemiologic studies have suggested that the high CRE prevalence in LTACHs in particular has a significant impact on connected healthcare facilities with which they share patients [7, 8]. Currently, less is known about the regional influence of vSNFs, although the even longer lengths of patient stay and more limited resources for infection prevention indicate that they might also be important settings in regional amplification of antibiotic resistance.
A bundled infection prevention intervention [9] (Chicago PROTECT [10]) was initiated in July 2017 to control CRE in Chicago-area healthcare facilities, including in vSNFs and LTACHs. Serial point-prevalence surveys conducted to monitor the impact of the intervention demonstrated that KPC-Kp levels remained stable across regional facilities. However, during the intervention period, New Delhi metallo-beta-lactamase (NDM) containing CRE prevalence unexpectedly increased in a subset of surveyed vSNFs. Here, we applied whole-genome sequencing to investigate the underlying transmission dynamics related to the increase in NDM prevalence in the region.
METHODS
Study Isolates and Metadata
From October 2016 to July 2019, 20 medical intensive care units (ICUs) in 20 short-term acute care hospitals, 7 LTACHs, and 8 vSNFs in the Chicago region were invited to participate in serial 1-day point-prevalence surveys of residents. In vSNFs, surveys were performed in ventilator wards. Medical ICUs were surveyed once in 2016–2017; vSNFs and LTACHs were surveyed every 6–12 months. Patients who were present in their room at the time of the survey were considered eligible for participation. Written informed consent was waived for this project, and patients who were competent were provided a standardized verbal explanation of the rationale for surveillance and were asked for verbal assent. Local staff obtained a rectal swab sample from each participating patient and collected deidentified patient information assessed at the time of the survey (age up to 90 years, sex, respiratory support status, length of stay, contact precautions status, facility awareness of resident CRE status). Swabs were processed at a central laboratory within 6 hours of collection. Overnight growth from MacConkey agar plates was screened for 5 carbapenemase gene families (KPC, NDM, VIM, IMP, and OXA-48) using multiplex polymerase chain reaction assays (Acuitas MDRO gene test and Acuitas Resistome test, OpGen, Gaithersburg, MD, during 2016–2017; Xpert Carba-R, Cepheid, Sunnyvale, CA, during 2018–2019).
For all genomic analyses, only the first isolate of a given ST (for K. pneumoniae isolates) or species (for all other species) was used for each patient. We used a Fisher exact test to test for the statistical significance of the difference in NDM or KPC prevalence between the first and last surveys. Fisher exact P values were corrected using the Benjamini-Hochberg method.
Whole-Genome Sequencing
Genomic DNA was extracted from cultures derived from single subcultured colonies. Genomic libraries were prepared with the NEBNext Ultra DNA library prep kit and sequenced at the University of Michigan Advanced Genomics Core on an Illumina NovaSeq 6000. All sequenced isolates have been deposited under BioProject PRJNA686897.
Genomic Analysis
We processed whole-genome sequences [11, 12] and identified in silico multilocus sequence types [13, 14], generated and annotated assemblies [12, 15–20], called single-nucleotide variants (SNVs) [21–28], identified phylogenetic clustering of facilities [8], and calculated pairwise SNV distances between isolates [29]. Reference-based whole-genome alignments of study and public ST147 isolates [30–40] were used to generate a phylogenetic tree using IQ-TREE v1.6.12 [41, 42]. We inferred ancestral dates of the phylogeny with the R package BactDating v1.0.12 [43–45]. NDM-containing plasmids were identified from publicly available complete plasmids [21, 29, 46–50]. See Supplementary Methods for details of the genomic analysis.
Determining Patient Flow Between Facilities
We constructed a patient transfer matrix of the Chicago metropolitan region using the Centers for Medicare and Medicaid Services’ minimum dataset, Medicare Provider Analysis and Review limited dataset, Medicaid Analytic eXtract Data from 2010–2012. Using the patient transfer matrix, we constructed a directed weighted patient transfer network of healthcare facilities in the Chicago area using R igraph v1.2.6 [51], including all healthcare facilities in the study, and patient flow was determined as in [8]. See Supplementary Methods for details about calculating patient flow. Comparison of patient flow for interfacility isolate pairs ≤12 SNVs vs >12 SNVs was performed using Wilcox tests.
Data Analysis and Visualization
Data analysis and visualization was performed in R v4.0.2 [52]. Data visualization used the following packages: tidyverse v1.3.0 [53], pheatmap v1.0.12, lubridate v1.7.9.2, tidytree v0.3.3, treeio v1.12.0, ggtree v2.2.4 [54, 55], ggplotify v0.0.5, ggnewscale v0.4.4, and cowplot v1.1.0. Code for analysis and visualization can be found here: https://github.com/Snitkin-Lab-Umich/ndm-st147-chicago-ms.
Ethical Review
Bacterial isolates and deidentified clinical metadata were collected under a prior surveillance project that underwent ethical review at the Centers for Disease Control and Prevention and was determined to be a nonresearch activity (public health surveillance). The project was also evaluated independently at each participating healthcare facility and deemed either a public health assessment or human subjects research and approved by local review boards where applicable.
RESULTS
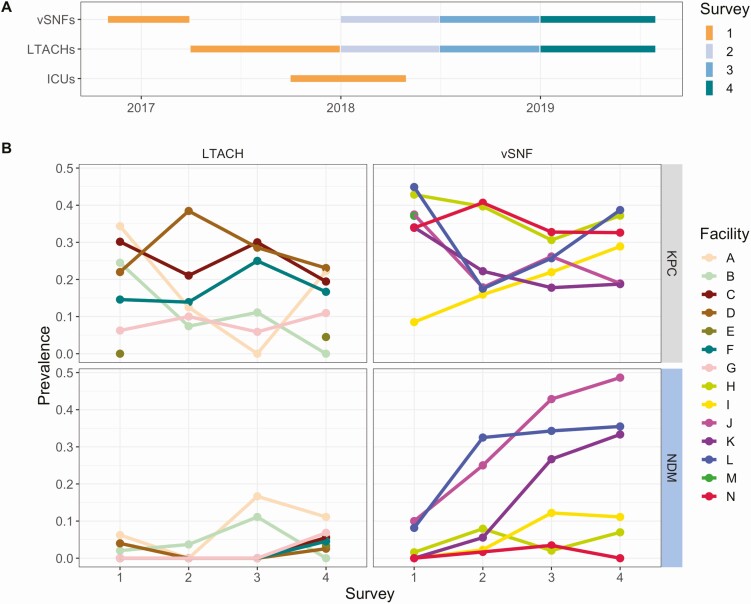
Prevalence of NDM, but not KPC, increased over time in certain vSNFs that were not closely connected by patient transfer. We first detected the presence of NDM+ CRE isolates in vSNFs and LTACHs during a regional point-prevalence survey conducted in 2017 and subsequently performed 3 follow-up surveys (Figure 1A). A summary of the patient population for each facility type across the 4 surveys can be found in Table 1. We found that while the prevalence of KPC+ CRE generally remained stable over time, the prevalence of NDM+ CRE increased in 3 vSNFs not closely connected by patient transfer (Fisher exact P < .05 for vSNFs J, K, and L; Figure 1B, Supplementary Figures 1, 2; Supplementary Table 1). The majority of NDM+ isolates were K. pneumoniae ST147 and carried blaNDM-1 on an IncF plasmid.
Figure 1.
Prevalence of New Delhi metallo-beta-lactamase (NDM) increased over time in certain vSNFs. A, Time window of when facilities were tested for each survey. Observance of NDM+ carbapenem-resistant Enterobacterales in vSNFs and LTACHs in survey 1 led to targeted follow-up surveys (2, 3, and 4) in these facilities. B, Proportion of NDM and Klebsiella pneumoniae carbapenemase–positive samples across surveys and facilities. vSNF O was not included as there was uneven sampling across surveys. ICUs are not shown in panel B because of very low prevalence (see Table 1). Abbreviations: ICU, intensive care unit; LTACH, long-term acute care hospital; vSNF, ventilator-capable skilled nursing facility.
Table 1.
Summary of Point-Prevalence Survey Results from Intensive Care Units, Long-Term Acute Care Hospitals, and Ventilator-Capable Skilled Nursing Facilities for Surveys 1, 2, 3, and 4
| Variable | Intensive Care Unit | Long-Term Acute Care Hospital | Ventilator-Capable Skilled Nursing Facility |
|---|---|---|---|
| Number of surveys | 1 | 4 | 4 |
| Number of facilities | 20 | 7 | 8 |
| Number of patients eligible | 238 | 1338 | 1325 |
| Number of patients surveyed, n (% of eligible) | 212 (89) | 1188 (89) | 1154 (87) |
| Age, mean (standard deviation), y | 62 (17) | 62 (15) | 60 (15) |
| Male, n (%) | 119 (56) | 644 (54) | 627 (54) |
| Length of stay, median (interquartile range), d | 5 (3–10) | 21 (11–37) | 126 (33–410) |
| Mechanical ventilation, n (%) | 102 (48) | 405 (21) | 477 (33) |
| Tracheostomy collar, n (%) | 0 (0) | 250 (21) | 384 (33) |
| Contact precautions, n (%) | 58 (27) | 736 (62) | 679 (59) |
| Carbapenemase gene | |||
| Klebsiella pneumoniae carbapenemase, n (%) | 11 (5) | 182 (15) | 360 (31) |
| New Delhi metallo-beta-lactamase, n (%) | 2 (1) | 30 (3) | 146 (13) |
| OXA-48, n (%) | 1 (0) | 0 (0) | 1 (0) |
| IMP, n (%) | 0 (0) | 1 (0) | 0 (0) |
| VIM, n (%) | 0 (0) | 4 (0) | 66 (6) |
| Any carbapenemase gene, n (%) | 14 (7) | 201 (17) | 479 (42) |
| Of carbapenemase-positive | |||
| With contact precautions, n/N (%) | 9/14 (64) | 169/201 (84) | 364/479 (76) |
| Known to facility, n/N (%) | 6/14 (43) | 107/201 (53) | 303/479 (63) |
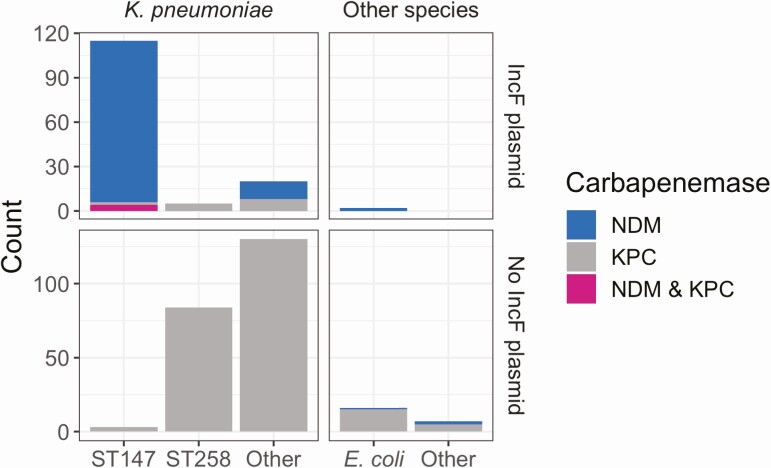
To understand the molecular basis for the increase in NDM+ CRE, we performed whole-genome sequencing on all CRE isolates from survey 1 and NDM+ isolates from the subsequent 3 follow-up surveys. We found that the presence of NDM was highly correlated with the presence of a suite of genes present on an NDM+ IncF plasmid isolated from K. pneumoniae [56] (Supplementary Figure 3). Most isolates that contained the IncF plasmid were blaNDM-1K. pneumoniae ST147; however, 1 blaNDM-1Escherichia coli ST354 isolate also contained the plasmid (Figure 2). While short-read sequencing data alone are insufficient to provide structural data to associate NDM with the plasmid backbone, the high degree of correlation between NDM and the IncF-associated plasmid genes in concert with the phylogenetic relationships between isolates strongly suggests that these genes are co-inherited (Supplementary Figure 3). In addition to NDM, the IncF plasmid harbored a number of antibiotic-resistance genes from several different resistance classes and the qacE gene, which may confer reduced susceptibility to common biocides [57] (Supplementary Figure 3).
Figure 2.
The majority of NDM+ isolates are Klebsiella pneumoniae sequence type 147 and carry NDM on an IncF plasmid. Number of sequenced isolates of various species and sequence types, what carbapenemase(s) they contain, and whether they have the IncF plasmid. Abbreviations: KPC, Klebsiella pneumoniae carbapenemase; NDM, New Delhi metallo-beta-lactamase.
Regional ST147 Isolates Are Phylogenetically Distinct from All Public Isolates
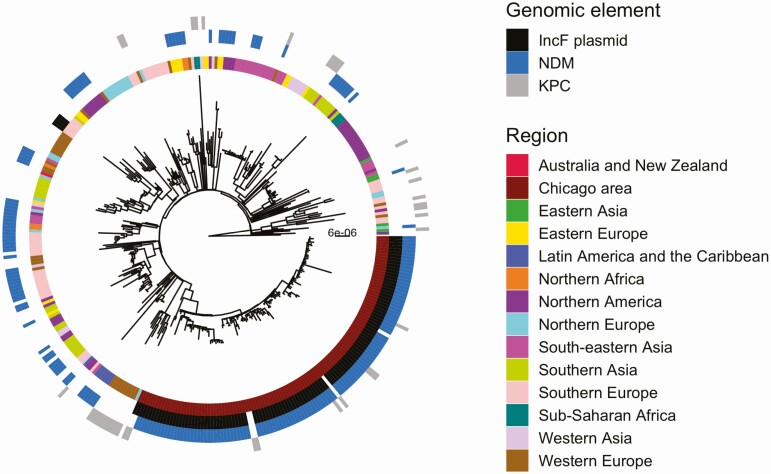
We investigated the phylogeography of ST147 in the Chicago area to determine whether circulating ST147 could be attributed to 1 or multiple importation events into the region. To this end, we constructed a whole-genome phylogeny that included publicly available ST147 genomes from across the globe. Examination of the phylogenetic reconstruction revealed that all of the ST147 isolates from the study form a monophyletic cluster, consistent with a single regional introduction (Figure 3). We also noted that while none of the NDM+ ST147 public isolates harbored the IncF plasmid, the majority of ST147s in the current study contained the plasmid. Moreover, 1 of the ST147 isolates in the KPC+/NDM– outgroup (from survey 3) contained the IncF plasmid but lacked NDM (Supplementary Figure 4), suggesting that the plasmid may have been acquired in a locally circulating ST147 strain, followed by integration of a mobile element harboring NDM. A dated phylogenetic analysis of circulating ST147 yielded an estimate of August 2015 for when the NDM+ clade of ST147 first arose in the region (95% credible interval [CI], February 2015–March 2016; Figure 4, Supplementary Figure 5, Supplementary Table 2) compared with an estimate of July 2013 for when ST147 first entered the region (CI, October 2011–October 2014; Supplementary Table 2).
Figure 3.
Study isolates are clonally separated from all publicly available isolates outside the Chicago region. Maximum likelihood phylogeny of study and public isolates annotated by geographic region and genomic element. Abbreviations: KPC, Klebsiella pneumoniae carbapenemase; NDM, New Delhi metallo-beta-lactamase.
Figure 4.
New Delhi metallo-beta-lactamase–positive sequence type 147 Klebsiella pneumoniae was introduced into the region around 2015. Dated phylogeny generated by BactDating. Gray bar on the root is the lower and upper bounds of the confidence interval (2015.09 to 2016.17). Abbreviations: ICU, intensive care unit; LTACH, long-term acute care hospital; vSNF, ventilator-capable skilled nursing facility.
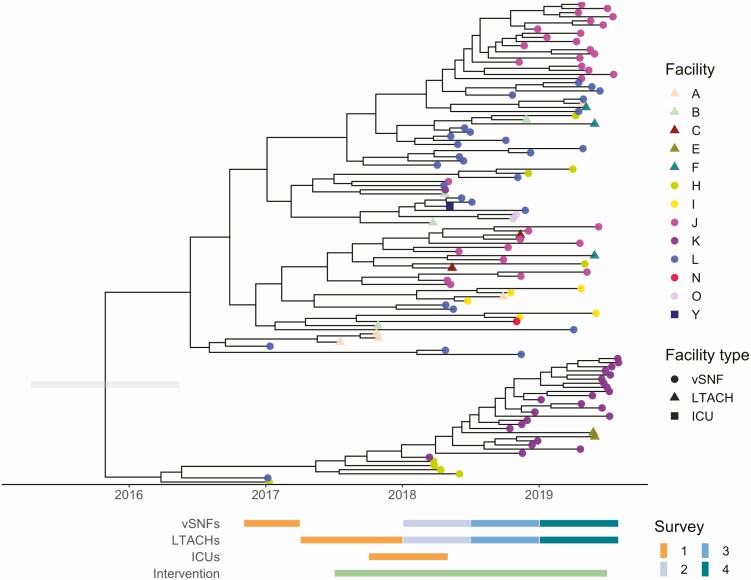
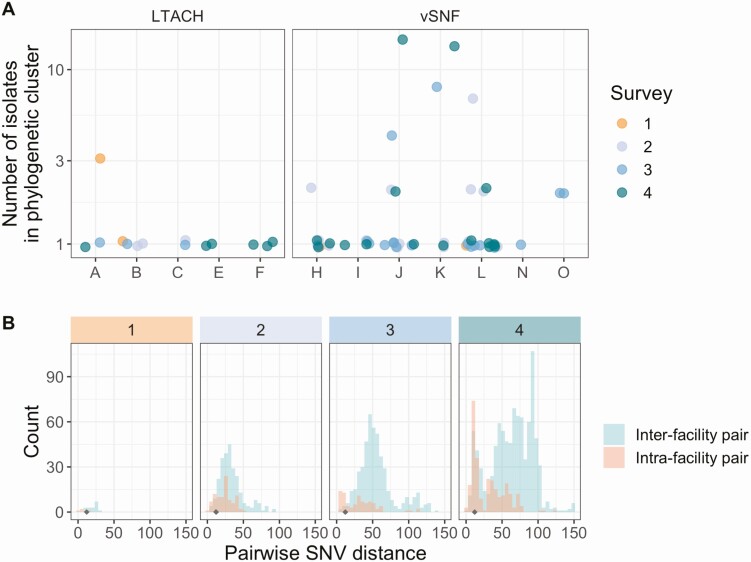
Genomic Evidence Indicates That Intrafacility Transmission is Driving Prevalence at High-Prevalence vSNFs
After determining that the increase in NDM prevalence corresponds to a clonal outbreak of blaNDM-1 ST147, we investigated the potential transmission dynamics of this clone within and between healthcare facilities. We observed a substantial clustering of isolates from certain vSNFs on the phylogeny (Figure 5A, Supplementary Figure 6), which suggests potential intrafacility transmission. To further investigate whether these clusters may represent within-facility transmission, we calculated pairwise SNV distances among all pairs of isolates and compared these distances for isolates from the same facility (intrafacility pairs) to isolates from different facilities (interfacility pairs) across surveys (Figure 5B). Indeed, starting in survey 3, we observed a disproportionate representation of small SNV distances (≤12 SNVs; see Methods section for threshold selection), which is consistent with intrafacility transmission in vSNFs. Of note, in survey 4, we observed spikes in small SNV distances for both intra- and interfacility pairs, with closely related intrafacility pairs being primarily from vSNFs and closely related interfacility pairs being from both vSNF–LTACH and vSNF–vSNF pairs (Supplementary Figure 7). Putting these closely related interfacility pairs in the context of the regional patient transfer network supports a potential role of patient transfer in regional blaNDM-1 ST147 spread in survey 4, with vSNF–LTACH and vSNF–vSNF isolate pairs less than 12 SNVs apart being from facilities with higher patient flow between them than isolate pairs with 12 or more SNVs (Wilcox P < .001; Supplementary Figure 8; higher patient flow indicates more movement of patients from source to destination facility, see Supplementary Methods for details).
Figure 5.
Intrafacility transmission is driving prevalence at high-prevalence vSNFs. A, Number of isolates in the largest subclade of the maximum likelihood phylogeny containing ≥90% of isolates from the given facility (see Methods section for more details). Note that the y-axis is log10-scaled. B, Pairwise SNV distances of isolates from the same and different facilities across surveys. The gray diamond at a pairwise SNV distance of 12 indicates the threshold for closely related isolates (see Methods section for details). Abbreviations: LTACH, long-term acute care hospital; SNV, single-nucleotide variant; vSNF, ventilator-capable skilled nursing facility.
DISCUSSION
We performed genomic analyses of CRE isolates collected through serial point-prevalence surveys in the Chicago area to investigate an increase in NDM+ CRE prevalence across a regional healthcare network. Our analysis supports the increase in NDM+ CRE being due to the clonal dissemination of a single blaNDM-1 ST147 strain of K. pneumoniae that emerged in 2015. Putting genomic analysis in the context of the regional healthcare network supports this strain first reaching high prevalence in a small number of vSNFs due to intrafacility transmission, followed by interfacility spread to connected healthcare facilities.
Whole-genome sequencing showed that the majority of blaNDM-1 ST147 harbored an IncF multidrug-resistance plasmid. Incorporating public data into the analysis revealed that these isolates formed a monophyletic clade, suggesting a single introduction into the region, either through importation of a preexisting NDM+ ST147 strain or acquisition of blaNDM by a locally circulating ST147 strain. Furthermore, examination of the global phylogeny indicates that while NDM+ ST147 has evolved multiple times in different locations and sometimes resulted in clonal outbreaks, none of the global NDM+ ST147 isolates we included in our analysis harbor the IncF plasmid found in our study isolates. The rapid and widespread dissemination of this strain in the region indicates that the NDM-carrying IncF plasmid we identified here can stably associate with an ST147 strain with epidemic potential. Given the potential negative impact of epidemic NDM-carrying K. pneumoniae, this possibility warrants close monitoring.
By combining regional surveillance with genomic analysis, we were able to discern that NDM initially spread in 3 vSNFs, likely via intrafacility transmission, with evidence of subsequent spread to healthcare facilities connected by patient transfer. There are several factors that likely contributed to the spread of this NDM+ ST147 clone. First, vSNF patients are a high-risk population for carriage of CRE as they are chronically ill, are usually admitted from ICUs or LTACHs, and are often exposed to antibiotics [58]. Second, patients in vSNFs generally have long lengths of stay, often much longer than patient stays at LTACHs [59], meaning that they have a longer period of time to acquire a multidrug-resistant organism. Furthermore, multibed rooms are common and the facilities themselves are often underresourced from a staffing and infection control perspective [6], both of which could facilitate intrafacility spread. Our findings paired with these observations indicate that vSNFs may be important healthcare facilities to detect emerging threats and potentially contain them before widespread dissemination. In the current study, we note that NDM+ isolates were uncommon in ICUs, and the outbreak of ST147 might not have been detectable until much later if sampling were restricted to ICUs.
Our study has several strengths. Active surveillance of diverse types of healthcare facilities in the region allowed us to identify and investigate a potential multidrug-resistant organism threat earlier than would have been possible if serial point-prevalence surveys across several facility types were not ongoing. Furthermore, cross-sectional patient sampling within each survey allowed us to obtain a complete snapshot of CRE prevalence at a given facility at a given point in time and to detect the increase in NDM+ isolates over time. Finally, we were able to leverage information from whole-genome sequencing to investigate the relatedness of isolates, as well as the intra- and interfacility transmission dynamics of NDM across the healthcare network.
Our study also has several important limitations. First, the cross-sectional study design could have led to potential biases in the number of NDM+ isolates sequenced at facilities, given that the patients at these facilities had different average lengths of stay, and precluded a more nuanced examination of NDM-1 intrafacility transmission dynamics and associated patient risk factors. Second, we lacked data from short-term acute care or community settings, particularly in the last 3 surveys, which limited our ability to examine the relative importance of other regional reservoirs for NDM+ ST147. However, the short-term acute care data that were available did not support their role in blaNDM-1 ST147 expansion. Third, we used facility-level aggregate patient transfer data to infer the likelihood of patient-level exposure to facilities; lack of patient-level facility exposure data precluded us from performing a more detailed exposure network analysis [60]. Last, we used short-read sequencing data, which limited our ability to investigate more complex plasmid dynamics. While we plan to perform long-read sequencing on a subset of these isolates in the future, we find it notable that we were able to leverage publicly available complete plasmid sequences to determine that NDM was carried on the same plasmid in the majority of isolates.
In conclusion, we identified an emerging blaNDM-1 ST147 clone of K. pneumoniae with epidemic potential. The identification of this clone and characterization of its ability to disseminate within and between healthcare facilities were made possible through whole-genome sequencing of NDM+ isolates from serial point-prevalence surveys at vSNFs, LTACHs, and ICUs. Our findings highlight the importance of performing surveillance of multidrug-resistant organisms not only in acute care hospital ICUs but also in post-acute care facilities such as LTACHs and vSNFs. vSNFs in particular appear to be especially important as sentinel sites of active surveillance for rare and emerging resistant pathogens.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Author Contributions. All authors developed the methodology and reviewed and edited the manuscript. M. L., E. S., M. H., A. M. J., and Z. L. conceptualized the research goals and aims. Z. L., R. C., A. P., and E. S. developed and implemented the software and curated the data. Z. L., R. C., and A. P. performed formal analysis. M. L., M. H., and E. S. provided resources. Z. L. and E. S. wrote the original draft. Z. L. and R. C. visualized the results. M. L., M. H., A. M. J., and E. S. provided supervision. M. L., M. H., and E. S. managed the project. M. L., M. H., and E. S. acquired funding.
Acknowledgments. The authors gratefully acknowledge the patients and staff of the facilities for their participation in this study. They thank officials from the Chicago Department of Public Health, Cook County Department of Public Health, Illinois Department of Public Health, and Centers for Disease Control and Prevention (CDC) for their direct involvement in and support of Chicago PROTECT. They also thank Louis Fogg and Vincent Young for fruitful discussion of the analyses presented here. They thank Ellen Benson, Mary Carl Froilan, Claire Heshmat, Jinal Makhija, and Mitali Shah for their role in specimen and data collection at participating healthcare facilities and Pamela Bell and Karen Lolans for their role in laboratory analysis. OpGen, Inc. (Gaithersburg, MD) provided Acuitas multidrug-resistant organism gene test and Acuitas Resistome test in kind during 2016–2017.
Disclaimer. Any opinions, findings, conclusions, or recommendations expressed here are those of the authors and do not necessarily reflect the views of the CDC, National Science Foundation, or the National Institutes of Health.
Financial support. This work was supported by CDC Cooperative Agreement (grantU54 CK000481) and SHEPheRD (task order 200-2011-42037), the National Science Foundation Graduate Research Fellowship Program (grant DGE 1256260 to Z. L.), the National Institutes of Health via the Molecular Mechanisms of Microbial Pathogenesis Training Grant (T32AI007528, A.M-J.), and the National Institutes of Health (1R01AI148259-01 to E. S. S.).
Potential conflicts of interest. M. L. and M. H. have received research support in the form of contributed product from OpGen, LLC, and from Sage Products (now part of Stryker Corporation). M. L. has also received an investigator-initiated grant from CareFusion Foundation (now part of BD). M. H. and R. W. have participated in clinical studies where participating healthcare facilities received contributed product from Sage Products Inc, Molnlycke, Clorox, or Medline. Neither M. H., R. W., nor their hospitals received product, funding, payments, or any other form of compensation. M. L. reports honorarium from antibiotic resistance symposium (Medical College of Wisconsin) and participates on CDC’s Healthcare Infection Control Practices Advisory Committee. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Tacconelli E, Carrara E, Savoldi A, et al. ; WHO Pathogens Priority List Working Group . Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 2018; 18:318–27. [DOI] [PubMed] [Google Scholar]
- 2. Ansari U, Lawsin A, Campbell D, et al. Molecular characterization of carbapenem-resistant Enterobacteriaceae in the USA, 2011–2015. Open Forum Infect Dis 2017; 4:S179. [Google Scholar]
- 3. Logan LK, Weinstein RA. The epidemiology of carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. J Infect Dis 2017; 215:28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ciccolini M, Donker T, Köck R, et al. Infection prevention in a connected world: the case for a regional approach. Int J Med Microbiol 2013; 303:380–7. [DOI] [PubMed] [Google Scholar]
- 5. Lin MY, Lyles-Banks RD, Lolans K, et al. ; Centers for Disease Control and Prevention Epicenters Program . The importance of long-term acute care hospitals in the regional epidemiology of Klebsiella pneumoniae carbapenemase-producing Enterobacteriaceae. Clin Infect Dis 2013; 57:1246–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pacilli M, Kerins JL, Clegg WJ, et al. Regional emergence of Candida auris in Chicago and lessons learned from intensive follow-up at 1 ventilator-capable skilled nursing facility. Clin Infect Dis 2020; 71:e718–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee BY, Bartsch SM, Lin MY, et al. How long-term acute care hospitals can play an important role in controlling carbapenem-resistant Enterobacteriaceae in a region: a simulation modeling study. Am J Epidemiol 2020. Available at: 10.1093/aje/kwaa247. Accessed 20 February 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Han JH, Lapp Z, Bushman F, et al. Whole-genome sequencing to identify drivers of carbapenem-resistant Klebsiella pneumoniae transmission within and between regional long-term acute-care hospitals. Antimicrob Agents Chemother 2019; 63. Available at: http://aac.asm.org/content/63/11/e01622-19. Accessed 20 February 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hayden MK, Lin MY, Lolans K, et al. Prevention of colonization and infection by Klebsiella pneumoniae carbapenemase–producing Enterobacteriaceae in long-term acute-care hospitals. Clin Infect Dis 2015; 60:1153–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lin M, Froilan MC, Makhija J, et al. Regional impact of a CRE intervention targeting high risk postacute care facilities (Chicago PROTECT). Infect Control Hosp Epidemiol 2020; 41:s48–9. [Google Scholar]
- 11. Andrews S. s-andrews/FastQC.2021. Available at: https://github.com/s-andrews/FastQC. Accessed 20 February 2021.
- 12. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014; 30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Crawford R. rdcrawford/MLSTyper.2020. Available at: https://github.com/rdcrawford/MLSTyper. Accessed 20 February 2021.
- 14. Hunt M, Mather AE, Sánchez-Busó L, et al. ARIBA: rapid antimicrobial resistance genotyping directly from sequencing reads. Microb Genom 2017; 3. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5695208/. Accessed 20 February 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Snitkin-Lab-Umich/assemblage. Snitkin-lab-umich, 2021. Available at: https://github.com/Snitkin-Lab-Umich/assemblage. Accessed 20 February 2021.
- 16. Li H. lh3/seqtk.2021. Available at: https://github.com/lh3/seqtk. Accessed 20 February 2021.
- 17. Ondov BD, Treangen TJ, Melsted P, et al. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol 2016; 17:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bankevich A, Nurk S, Antipov D, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012; 19:455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Walker BJ, Abeel T, Shea T, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 2014; 9:e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Aziz RK, Bartels D, Best AA, et al. The RAST server: rapid annotations using subsystems technology. BMC Genom 2008; 9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009; 25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. broadinstitute/picard. Broad Institute, 2021. Available at: https://github.com/broadinstitute/picard. Accessed 20 February 2021.
- 23. Li H, Handsaker B, Wysoker A, et al. ; 1000 Genome Project Data Processing Subgroup . The sequence alignment/map format and SAMtools. Bioinformatics 2009; 25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McKenna A, Hanna M, Banks E, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010; 20:1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Poplin R, Ruano-Rubio V, DePristo MA, et al. Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv 2018:201178. [Google Scholar]
- 26. Croucher NJ, Page AJ, Connor TR, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res 2015; 43:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Arndt D, Grant JR, Marcu A, et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res 2016; 44:W16–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kurtz S, Phillippy A, Delcher AL, et al. Versatile and open software for comparing large genomes. Genome Biol 2004; 5:R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Paradis E, Schliep K. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 2019; 35:526–8. [DOI] [PubMed] [Google Scholar]
- 30. Davis JJ, Wattam AR, Aziz RK, et al. The PATRIC bioinformatics resource center: expanding data and analysis capabilities. Nucleic Acids Res 2020; 48:D606–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Greninger AL, Chorny I, Knowles S, Ng VL, Chaturvedi V. Draft genome sequences of four NDM-1-producing Klebsiella pneumoniae strains from a health care facility in northern California. Genome Announc 2015; 3:e00421–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Khong WX, Marimuthu K, Teo J, et al. Tracking inter-institutional spread of NDM and identification of a novel NDM-positive plasmid, pSg1-NDM, using next-generation sequencing approaches. J Antimicrob Chemother 2016; 71:3081–9. [DOI] [PubMed] [Google Scholar]
- 33. Wyres KL, Wick RR, Judd LM, et al. Distinct evolutionary dynamics of horizontal gene transfer in drug resistant and virulent clones of Klebsiella pneumoniae. PLoS Genet 2019; 15:e1008114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Becker L, Kaase M, Pfeifer Y, et al. Genome-based analysis of carbapenemase-producing Klebsiella pneumoniae isolates from German hospital patients, 2008–2014. Antimicrob Resist Infect Control 2018; 7. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5930415/. Accessed 21 February 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pérez-Vázquez M, Sola Campoy PJ, Ortega A, et al. Emergence of NDM-producing Klebsiella pneumoniae and Escherichia coli in Spain: phylogeny, resistome, virulence and plasmids encoding blaNDM-like genes as determined by WGS. J Antimicrob Chemother 2019; 74:3489–96. [DOI] [PubMed] [Google Scholar]
- 36. Mansour W, Grami R, Jaidane N, et al. Epidemiology and whole-genome analysis of NDM-1-producing Klebsiella pneumoniae KP3771 from Tunisia. Microbial Drug Resistance 2019; 25:644–51. [DOI] [PubMed] [Google Scholar]
- 37. Roach D, Waalkes A, Abanto J, et al. Whole genome sequencing of Peruvian Klebsiella pneumoniae identifies novel plasmid vectors bearing carbapenem resistance gene NDM-1. Open Forum Infect Dis 2020; 7. doi: 10.1093/ofid/ofaa266. Accessed 21 February 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Marí-Almirall M, Cosgaya C, Pitart C, et al. ; MERCyCAT Study Group . Dissemination of NDM-producing Klebsiella pneumoniae and Escherichia coli high-risk clones in Catalan healthcare institutions. J Antimicrob Chemother 2021; 76:345–54. [DOI] [PubMed] [Google Scholar]
- 39. Falcone M, Giordano C, Barnini S, et al. Extremely drug-resistant NDM-9-producing ST147 Klebsiella pneumoniae causing infections in Italy, May 2020. Eurosurveillance 2020; 25:2001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li H. lh3/wgsim.2021. Available at: https://github.com/lh3/wgsim. Accessed 20 February 2021.
- 41. Minh BQ, Schmidt HA, Chernomor O, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol 2020; 37:1530–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Minh BQ, Nguyen MA, von Haeseler A. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol 2013; 30:1188–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Didelot X, Croucher NJ, Bentley SD, Harris SR, Wilson DJ. Bayesian inference of ancestral dates on bacterial phylogenetic trees. Nucleic Acids Res 2018; 46:e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Didelot X, Siveroni I, Volz EM. Additive uncorrelated relaxed clock models for the dating of genomic epidemiology phylogenies. Mol Biol Evol 2021; 38:307–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Plummer M, Best N, Cowles K, et al. coda: output analysis and diagnostics for MCMC. 2020. Available at: https://CRAN.R-project.org/package=coda. Accessed 20 February 2021.
- 46. Crawford RD, Snitkin ES. cognac: rapid generation of concatenated gene alignments for phylogenetic inference from large, bacterial whole genome sequencing datasets. BMC Bioinformatics 2021; 22:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 2012; 28:3150–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Revell LJ. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol Evol 2012; 3:217–23. [Google Scholar]
- 49. Alcock BP, Raphenya AR, Lau TTY, et al. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res 2020; 48:D517–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 1990; 215:403–10. [DOI] [PubMed] [Google Scholar]
- 51. Csardi G, Nepusz T. The igraph software package for complex network research. InterJournal 2005; Complex Systems:1695. Available at: http://interjournal.org/manuscript_abstract.php?361100992. [Google Scholar]
- 52. R Core Team. R: the R project for statistical computing. Available at: https://www.r-project.org/. Accessed 20 February 2021.
- 53. Wickham H, Averick M, Bryan J, et al. Welcome to the Tidyverse. J Open Source Softw 2019; 4:1686. [Google Scholar]
- 54. Yu G, Smith DK, Zhu H, Guan Y, Lam TT-Y. ggtree: an r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol 2017; 8:28–36. [Google Scholar]
- 55. Yu G, Lam TT, Zhu H, Guan Y. Two methods for mapping and visualizing associated data on phylogeny using ggtree. Mol Biol Evol 2018; 35:3041–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kwon T, Yang JW, Lee S, et al. Complete genome sequence of Klebsiella pneumoniae subsp. pneumoniae KP617, coproducing OXA-232 and NDM-1 carbapenemases, isolated in South Korea. Genome Announc 2016; 4. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4714118/. Accessed 20 February 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Abuzaid A, Hamouda A, Amyes SG. Klebsiella pneumoniae susceptibility to biocides and its association with cepA, qacΔE and qacE efflux pump genes and antibiotic resistance. J Hosp Infect 2012; 81:87–91. [DOI] [PubMed] [Google Scholar]
- 58. Rossow J, Ostrowsky B, Adams E, et al. Factors associated with Candida auris colonization and transmission in skilled nursing facilities with ventilator units, New York, 2016–2018. Clin Infect Dis 2021; 72:e753–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Fisch J, Lansing B, Wang L, et al. New acquisition of antibiotic-resistant organisms in skilled nursing facilities. J Clin Microbiol 2012; 50:1698–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Won SY, Munoz-Price LS, Lolans K, Hota B, Weinstein RA, Hayden MK; Centers for Disease Control and Prevention Epicenter Program . Emergence and rapid regional spread of Klebsiella pneumoniae carbapenemase-producing Enterobacteriaceae. Clin Infect Dis 2011; 53:532–40. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.