Summary
Upon viral infection, natural killer (NK) cells expressing certain germline-encoded receptors are selected, expanded, and maintained in an adaptive-like manner. Currently, these are thought to differentiate along a common pathway. However, by fate mapping of single NK cells upon murine cytomegalovirus (MCMV) infection, we identified two distinct NK cell lineages that contributed to adaptive-like responses. One was equivalent to conventional NK (cNK) cells while the other was transcriptionally similar to type 1 innate lymphoid cells (ILC1s). ILC1-like NK cells showed splenic residency and strong cytokine production but also recognized and killed MCMV-infected cells, guided by activating receptor Ly49H. Moreover, they induced clustering of conventional type 1 dendritic cells and facilitated antigen-specific T cell priming early during MCMV infection, which depended on Ly49H and the NK cell-intrinsic expression of transcription factor Batf3. Thereby, ILC1-like NK cells bridge innate and adaptive viral recognition and unite critical features of cNK cells and ILC1s.
Keywords: single-cell fate mapping, NK cells, MCMV, adaptive-like NK cell responses, ILC1-like NK cells, ILC1
Graphical abstract

Highlights
-
•
Adaptive-like NK cell responses to MCMV encompass conventional and ILC1-like lineages
-
•
ILC1-like NK cells show enhanced cytokine production and splenic residency
-
•
ILC1-like NK cells show EOMES expression, target-specific cytotoxicity, and clonal expansion
-
•
ILC1-like NK cells drive cDC1 clustering and CD8+ T cell priming dependent on Ly49H and Batf3
Adaptive-like responses to MCMV are thought to originate from circulating conventional NK cells. Here, Flommersfeld et al. use single-cell fate mapping to show that a separate lineage of spleen-resident ILC1-like NK cells critically contributes to such responses. These ILC1-like NK cells induce cDC1 clustering and optimal CD8+ T cell priming, which depends on the transcription factor Batf3.
Introduction
Natural killer (NK) cells are of critical importance for the early control of virus infections. They are capable of directly recognizing virus-infected cells through an array of inhibitory and activating NK cell receptors that are triggered by the absence of major histocompatibility complex (MHC) molecules (Babić et al., 2010) and the increased expression of specific viral ligands (Arase et al., 2002; Smith et al., 2002). The combinatorial expression of these NK cell receptors and differences in their expression level outfit certain NK cells with an enhanced responsivity to defined pathogens (Adams et al., 2019; Brown et al., 2001; Hammer et al., 2018; Lee et al., 2001). Upon infection, NK cells harboring these receptors or receptor combinations have been shown to be selected, expanded, and maintained in an “adaptive-like” manner (Sun et al., 2009). Recently, single NK cells expressing the activating receptor Ly49H, which recognizes the glycoprotein m157 encoded by murine cytomegalovirus (MCMV), were found to clonally expand up to 10,000-fold when adoptively transferred to immunodeficient hosts and exposed to MCMV infection (Grassmann et al., 2019).
Adaptive-like NK cell responses are thought to originate from conventional NK (cNK) cells that arise in the bone marrow and then recirculate between blood and secondary lymphoid organs. During the steady state, these cNK cells differentiate from an immature (CD11b–CD27+, KLRG-1–), to a mature (CD11b+CD27+, KLRG-1+/–), to a terminally differentiated (CD11b+CD27–, KLRG-1+) state. Upon maturation their cytolytic capacity increases while their proliferative competence declines (Jeevan-Raj et al., 2017). This linear differentiation pathway is thought to be recapitulated during adaptive-like responses, with activated immature NK cells giving rise to mature and terminally differentiated offspring (Kamimura and Lanier, 2015).
Complementary to circulating cNK cells, type 1 innate lymphoid cells (ILC1s) contribute to host defense against viral infections. Rather than providing direct target recognition and cytotoxicity, ILC1s receive and amplify host-derived cytokine signals during the early phase of an immune response. Moreover, ILC1s are tissue resident (Gasteiger et al., 2015) and rapidly respond to localized viral infection while circulating NK cells have to first migrate to the infected tissue or respond only upon systemic spread of the virus (Weizman et al., 2017).
Recently, the borders between cNK cells and tissue-resident ILC1s have somewhat blurred. Tissue-resident NK cells have been identified in the liver and salivary glands (Erick et al., 2016; Sojka et al., 2014), and some of these are phenotypically highly similar to ILC1s (Cortez et al., 2014). In tumors, NK cells have even been suggested to convert into ILC1s, leading to a reduction of anti-tumoral activity (Cortez et al., 2017; Gao et al., 2017). On the other hand, adaptive-like immune responses to viral infection, which are initiated in the secondary lymphoid organs, are still considered to originate exclusively from cNK cells.
Here, we set out to test this hypothesis by studying adaptive-like NK cell responses at single-cell resolution; by mapping the fate of individual Ly49H+ NK cells and their offspring during MCMV infection and dissecting their heterogeneity at the steady state via single-cell RNA sequencing (RNA-seq), we found that responding NK cells followed two distinct paths of differentiation. These originated from two distinct NK cell lineages present already during the steady state and stably maintained throughout infection. One of these consisted of CD62L+CD160– circulating cNK cells and the other of CD62L–CD160+ NK cells that retained an “immature” phenotype, showed splenic residence, and shared transcriptional and functional characteristics with ILC1s; however, expression of transcription factor Eomesodermin (Eomes), direct recognition of MHC class I-deficient or virus-infected cells, vigorous clonal expansion upon MCMV infection, and generation of cytolytic progeny allocated these “ILC1-like NK cells” firmly within the NK cell lineage.
ILC1-like NK cells were characterized by selective expression of Basic leucin zipper transcriptional factor ATF-like 3 (Batf3) and harbored a distinct molecular signature indicative of NK-dendritic cell (DC) interactions that was absent from cNK cells and ILC1s. Indeed, ILC1-like NK cells localized close to conventional type 1 DCs (cDC1s) during the steady state and were strongly overrepresented within cDC1 clusters that formed during the first 24 h of MCMV infection. Cluster formation of ILC1-like NK cells and cDC1s as well as co-localization of cDC1s with MCMV-infected cells were largely abrogated in Ly49H-deficient hosts, leading to a subsequent reduction in antigen-specific priming of CD8+ T cells. This deficiency in cDC1 clustering and CD8+ T cell priming was virtually phenocopied in mixed chimeric mice lacking Batf3 expression in Ly49H+ NK cells. Taken together, these data identify Batf3+ ILC1-like NK cells as a distinct contributor to adaptive-like immune responses and an early sentinel of viral infection that bridges innate and adaptive immunity.
Results
Individual Ly49H+ NK cells follow two distinct paths of differentiation upon MCMV infection
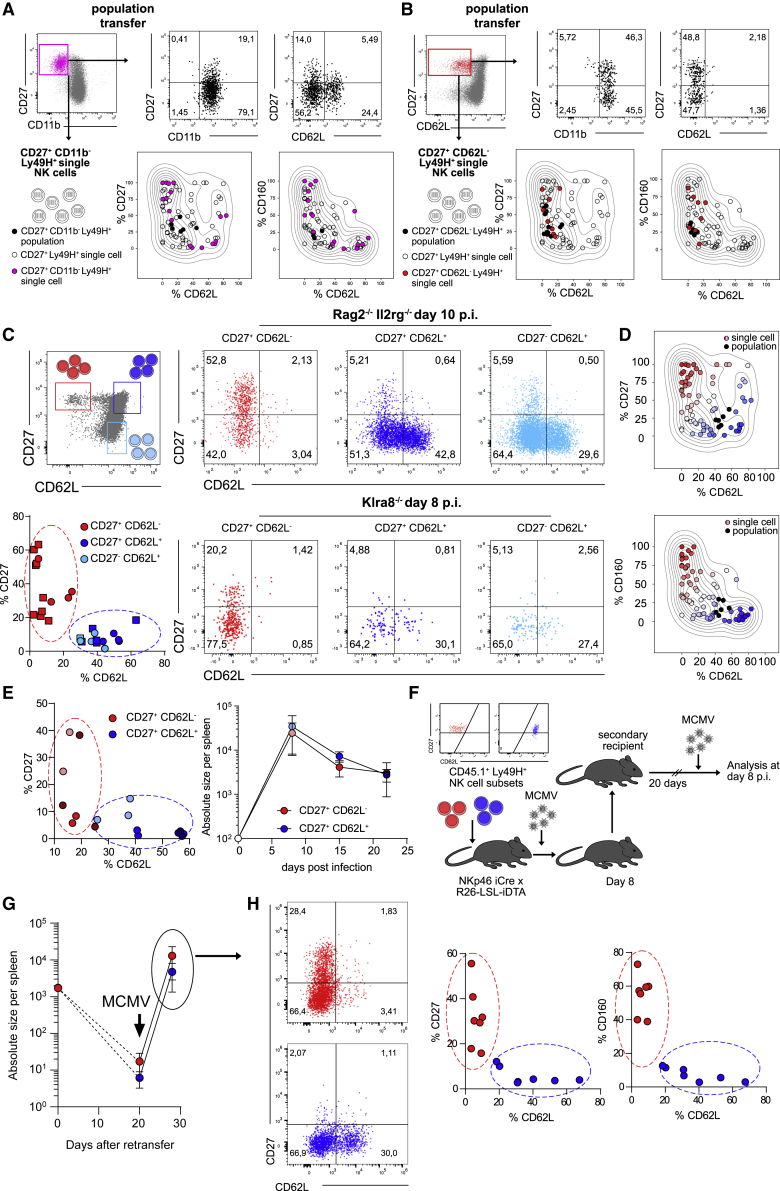
To establish the overall differentiation pattern of Ly49H+ NK cells at the peak of an adaptive-like immune response against MCMV, we adoptively transferred small populations of Ly49H+ NK cells into Klra8–/– (Ly49H-deficient) or Rag2–/– Il2rg–/– recipients and analyzed these at day 7 or 8 post infection (p.i.) (Figure 1A). While the resulting NK cell populations were almost uniformly positive for CD11b, other markers such as CD27, CD160, or CD62L were detectable in only a fraction of activated NK cells (Figures 1B and 1C)—a phenotypic heterogeneity that was also conserved when transferring only Ly49H+ NK cells that expressed the immaturity marker CD27 (Figure 1D). This could be due to phenotypic diversification during infection-driven expansion, allowing every NK cell to generate a diverse progeny (Figure S1A, left). Alternatively, single-cell-derived NK cell responses may be phenotypically restricted, and only by combining multiple NK cell clones would one achieve phenotypic heterogeneity (Figure S1A, right). We reasoned that single-cell fate mapping should allow us to discriminate between these two modes of differentiation. As described previously (Grassmann et al., 2019, 2020), we used retrogenic color barcoding to track single-cell-derived NK cell responses against MCMV. We transduced hematopoietic stem cells (HSCs) with retroviral constructs encoding various fluorescent proteins and generated retrogenic mice by transplanting these HSCs into irradiated hosts (Figure S1B). After several weeks, single color-barcoded CD27+ Ly49H+ NK cells were sorted via flow cytometry from the spleens of these retrogenic donors and adoptively transferred to Rag2–/– Il2rg–/– recipients in a multiplexed fashion (Figure 1E; Figures S1B and S1C). After infection with MCMV, we analyzed the resulting single-cell-derived clones at day 8 p.i. As previously observed (Grassmann et al., 2019), individual Ly49H+ NK cells generated a highly variable output, ranging from 10 to almost 100,000 daughter cells (Figure 1F). All single-cell-derived clones were uniformly positive for CD11b and Ly49H (Figure 1G, black; Figure S1D) but otherwise segregated into two distinct response patterns: a CD62L– pattern that contained a relatively large fraction of CD160+ and CD27+ cells (Figure 1G, red) and a CD62L+ pattern that lacked CD160 expression and showed relatively low percentages of CD27+ cells (Figure 1G, blue). Population-derived NK cell responses appeared as a mix of these two patterns (Figures 1B–1D). Thus, progeny derived from single Ly49H+ NK cells showed limited diversification and instead adopted one of two distinct fates (Figure S1A, right).
Figure 1.
Individual Ly49H+ NK cells follow two distinct paths of differentiation upon MCMV infection
(A) 3 × 104 or 1 × 103 CD45.1+ Ly49H+ NK cells were adoptively transferred into Klra8–/– or Rag2–/– Il2rg–/– mice followed by infection of recipients with MCMV and analysis of expanded NK cell populations in spleen at day 7 or 8 post infection (p.i.), respectively.
(B) Representative dot plots show marker expression by transferred Ly49H+ NK cells in Klra8–/– recipients at day 7 p.i.
(C) As in (B) but in Rag2–/– Il2rg–/– recipients at day 8 p.i.
(D) Representative dot plots show marker expression by transferred CD27+ Ly49H+ NK cells in Rag2–/– Il2rg–/– recipients at day 8 p.i.
(E–G) Multiplexed adoptive transfer of single color-barcoded CD27+ Ly49H+ NK cells into Rag2–/– Il2rg–/– mice, followed by infection of recipients with MCMV and analysis of expanded NK cell clones in spleen at day 8 p.i. (E) Schematic depiction. (F) Absolute size of NK cell responses derived from single or 100 transferred cells. (G) Representative dot plots and histograms depicting marker expression of NK cell clones that either lacked (red) or contained (blue) CD62L expressing cells. Endogenous Ly49H+ NK cell population is shown in the gray histogram.
Data in (B), (C), and (D) are representative of at least two independent, similar experiments. Data in (F) are pooled from seven independent, similar experiments. Lines indicate mean, and error bars represent SD. Data in (G) are representative of four independent experiments. In (B): n = 2–4, in (C), (D): n = 3 mice per group per experiment, in (F): n = 82 NK cell clones from 6–12 recipient mice per experiment, (G): n = 54 NK cell clones from 6–12 recipient mice per experiment.
See also Figure S1.
CD62L– NK cell clones originate from a defined NK cell subset
The separation into two main fates could be the result of (1) an early fate decision during expansion or (2) pre-existing functional diversity among steady-state NK cells (Figure S2A). Since expression of CD27 is lost upon steady-state maturation of NK cells, we first hypothesized that CD62L– clones, which contained a relatively large fraction of CD27+ cells, arose from more immature NK cells than CD62L+ clones, which showed lower percentages of CD27+ cells. However, single Ly49H+ NK cells, sorted from the immature CD27+CD11b– compartment, generated both types of response patterns after MCMV infection (Figure 2A). We found that during steady-state maturation, immature Ly49H+ NK cells showed a further subdivision into CD62L+ and CD62L– subsets (Figure S2B), and when sorting single Ly49H+ NK cells from the CD27+CD62L– compartment, these exclusively generated CD62L– clones in response to MCMV infection (Figure 2B). This observation was further corroborated when transferring populations of NK cells sorted according to their expression of CD27 and CD62L (Figure 2C). Upon infection, only CD27+CD62L– NK cell populations generated CD62L– response patterns, while CD62L+ NK cells generated patterns encompassing CD62L– and CD62L+ cells (Figure 2C), fitting well to the expression patterns observed in NK cell clones (Figure 2D). Furthermore, adaptive-like NK cell responses derived from CD27+CD62L– or CD27+CD62L+ NK cells showed similar kinetics while remaining phenotypically distinct with respect to expression of CD27 and CD62L until 3 weeks p.i. (Figure 2E). To assess the potential of the two subsets to mount memory-like responses, we transferred CD45.1+ CD27+CD62L– or CD45.1+ CD27+CD62L+ NK cells into NK cell-deficient NKp46iCre (Narni-Mancinelli et al., 2011) × R26-LSL-iDTA mice. In these mice, conditional Cre-mediated excision of a lox-stop-lox site leads to constitutive expression of the diphtheria toxin alpha chain (DTA) and depletion of NKp46-expressing cells. On day 8 p.i. with MCMV, we harvested CD45.1+ NK cells derived from each subset and re-transferred them into uninfected secondary hosts. After a resting period of 20 days, secondary hosts were re-infected and analyzed on day 8 p.i. (Figure 2F). Both subsets robustly re-expanded (Figure 2G) while maintaining their specific phenotype with regard to expression of CD27, CD62L, and CD160 (Figure 2H). These data indicated that adaptive-like NK cell responses were indeed composed of two distinct NK cell lineages that maintained their distinct phenotype throughout the memory phase and upon reinfection.
Figure 2.
CD62L– NK cell clones originate from a defined NK cell subset
(A) Adoptive transfer of Ly49H+CD27+CD11b– NK cell populations or multiplexed adoptive transfer of single color-barcoded Ly49H+CD27+CD11b– NK cells into Klra8–/– or Rag2–/– Il2rg–/– recipients followed by MCMV infection and analysis in spleen at day 7 or 8 p.i., respectively. Representative dot plots (upper panels) show phenotype of population-derived responses. Kernel density estimate (KDE) plots (lower panels) show percentage of CD27, CD62L, and CD160 expressing cells within population-derived responses (black dots) or within NK cell clones derived from all Ly49H+CD27+ single cells (circles) or specifically from Ly49H+CD27+CD11b– single cells (filled pink circles).
(B) As in (A) but adoptive transfer of Ly49H+CD27+CD62L– NK cells. Of note: NK cell clones derived from Ly49H+CD27+CD62L– single cells (filled red circles) uniformly lack CD62L expressing cells.
(C) CD27+CD62L–, CD27+CD62L+,and CD27–CD62L+ populations of Ly49H+ NK cells were adoptively transferred into Klra8–/– or Rag2–/– Il2rg–/– recipients followed by MCMV infection and analysis in spleen at day 8 and 10 p.i., respectively. Schematic dot plot depicts sort gating (upper left panel). Dot plots depict marker expression within expanded NK cell populations (right panels). Scatterplot depicts frequency of CD27 and CD62L expressing cells within expanded populations at day 8/10 p.i. in Klra8–/– (squares) and Rag2–/– Il2rg–/– recipients (circles) (lower left panel).
(D) KDE plots show expression of CD27, CD62L, and CD160 within NK cell responses derived from Ly49H+CD27+ populations (black dots) or from single cells sorted as Ly49H+CD27+, Ly49H+CD27+CD11b–,or Ly49H+CD27+CD62L– (circles). Red versus blue color coding delineates clones containing high versus low percentages of CD62L and low versus high percentages of CD27 and CD160 expressing cells.
(E) CD27+CD62L– and CD27+CD62L+ populations of Ly49H+ NK cells were sorted and adoptively transferred into NKp46 iCre x R26-LSL-iDTA recipients, followed by MCMV infection and analysis of expanded populations in spleen at day 8, 15, and 22 p.i. Scatterplot depicts frequency of CD27 and CD62L expressing cells within expanded populations at day 8 (light red/blue), 15 (bright red/blue), and 22 (dark red/blue) (left panel). Absolute size of expanded NK cell populations are depicted in the right panel.
(F–H) CD27+CD62L– and CD27+CD62L+ populations of Ly49H+ NK cells were sorted and adoptively transferred into NKp46 iCre x R26-LSL-iDTA recipients. At day 8 p.i. with MCMV 1,5–2 × 103 CD45.1+ Ly49H+ NK cells derived from either subset were sorted from spleens of primary recipients, re-transferred into uninfected Rag2–/–Il2rg–/– mice, rested for 20 days, and analyzed before or 8 days after secondary exposure to MCMV. (F) Initial sort purity and schematic depiction. (G) Absolute size of re-transferred NK cell populations at indicated time points. (H) Representative flow cytometry plots (left) and scatterplots depicting frequency of marker expressing cells within expanded populations at day 8 after recall infection (right).
Population data in (A) and (B) are representative of at least two independent experiments. Single cell data in (A) and (B) are pooled from two to three independent experiments. Data in (C) are pooled from five, and data in (D) from seven independent experiments. Data in (E) are representative of three independent experiments. Data in (G) and (H) are pooled from two independent experiments. Dots represent mean, and error bars indicate SD. In (A–C), (E), (G),and (H) n = 3, 2–4, 2–4, 2–3, and 3–4 mice per population-based experiment. In (A), (B), and (D) n = 22, 8, and 76 NK cell clones from 6–12 recipient mice per experiment.
See also Figure S2.
Steady-state CD27+CD62L– NK cells show a distinct transcriptional profile
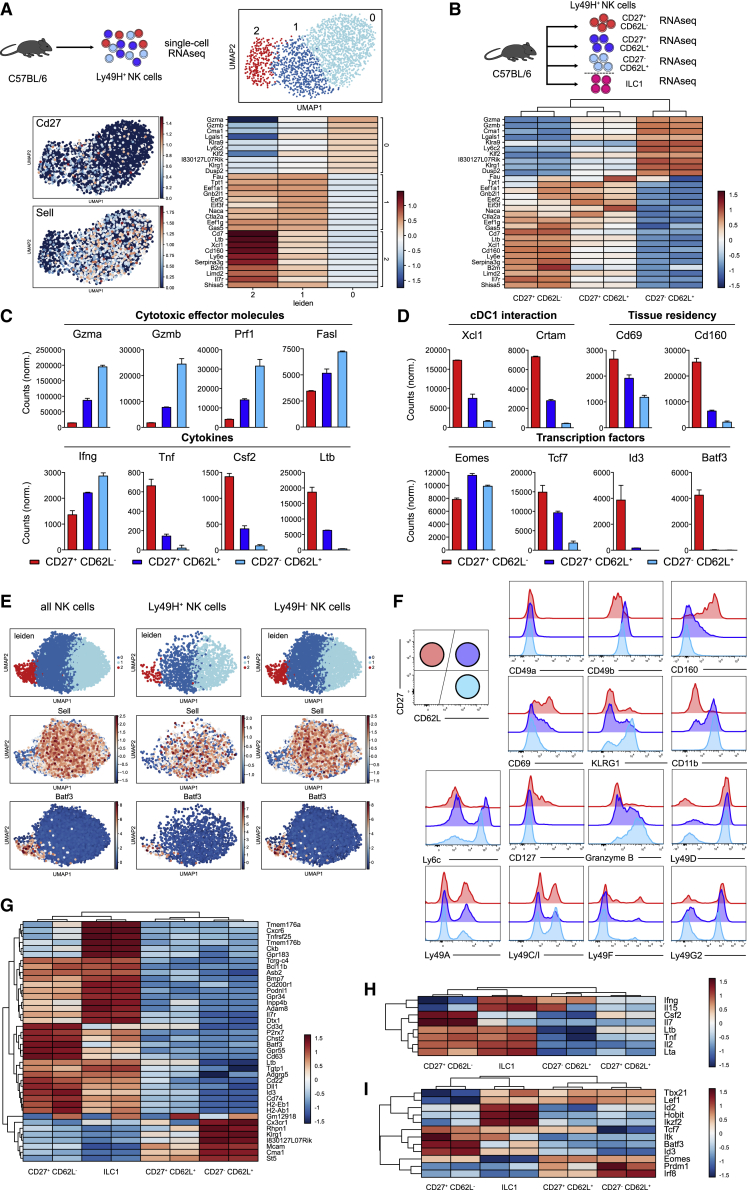
To investigate whether the functional subdivision, suggested by fate mapping of single NK cells, aligned with the transcriptional composition of the steady-state NK cell compartment, we performed single-cell RNA-seq (scRNA-seq) of Ly49H+ NK cells (Figure 3A) and bulk RNA-seq of Ly49H+ populations defined by CD27 and CD62L expression (Figure 3B; Figure S3A). Leiden clustering of scRNA-seq data identified three NK cell clusters (Figure 3A) whose borders almost perfectly aligned with expression of CD62L (negative in cluster 2, but positive in clusters 1 and 0) and CD27 (negative in cluster 0, but positive in clusters 2 and 1) (Figure 3A). Comparing the top 10 differentially expressed genes found in scRNA-seq profiles to RNA-seq data from sorted NK cell populations, we found that cluster 2 clearly mapped onto the profile of CD27+CD62L– NK cells while clusters 1 and 0 shared the expression patterns of CD27+CD62L+ and CD27–CD62L+ NK cell populations, respectively (Figures 3A and 3B). Among the defining genes for cluster 2, we found Cd160, Xcl1, Cd7, Ltb, and Il7r. In contrast, transcripts encoding Gzma, Gzmb, Klrg1, and Klf2, the transcription factor controlling CD62L expression (Bai et al., 2007), were underrepresented in this cluster and among sorted CD27+CD62L– NK cells (Figures 3A and 3B). Furthermore, CD27+CD62L– NK cells showed lower expression of cytotoxic effector molecules (Figure 3C, first row) and interferon γ (IFN-γ) but higher expression of other cytokines such as tumor necrosis factor alpha (TNF-α), granulocyte-macrophage colony-stimulating factor (GM-CSF), and Lt-b (Figure 3C, second row). Moreover, transcripts encoding the chemokine XCL-1, which attracts XCR1+ cDC1s and CRTAM, a receptor binding to Necl2 expressed on cDC1s (Galibert et al., 2005), suggested potential interactions between CD27+CD62L– NK cells and DCs. Furthermore, CD27+CD62L– NK cells expressed more transcripts encoding for CD69 and CD160, which have been described as markers for tissue-resident NK cells and intraepithelial ILC1s (Figure 3D, first row; Fuchs et al., 2013). Among selected transcription factors, Eomes was similarly expressed in all NK cell subsets while Id3 and Batf3 were strongly overrepresented in CD27+CD62L– NK cells (Figure 3D, second row). Analysis of our scRNA-seq dataset further confirmed the allocation of these genes to the respective NK cell subsets (Figure S3B and data not shown). Next, we asked whether CD27+CD62L– NK cells were restricted to the Ly49H+ compartment. To this end, we made use of an Eomes-GFP-reporter mouse and subjected splenic NK1.1+ Eomes-GFP+ NK cells to scRNA-seq. Among these, both Ly49H+ and Ly49H– NK cells showed a transcriptional cluster that closely matched the expression profile of CD27+CD62L– NK cells, showing e.g., high Batf3 and low Sell expression (Figure 3E). Last, we validated surface marker expression predicted by our transcriptional analysis via flow cytometry. We confirmed that CD27+CD62L– NK cells showed high surface expression of CD160 already before infection, expressed CD49b (albeit less than CD62L+ subsets), and, like these, lacked CD49a. Maturation markers KLRG1, CD11b, and Ly6c were negative among CD27+CD62L– NK cells. They expressed CD69 and were the only NK cell subset that contained a fraction of CD127+ cells but lacked granzyme B (GzmB) under steady-state conditions. In terms of their Ly49 receptor profile, no significant differences were found between CD27+CD62L– NK cells and the other NK cell subsets (Figure 3F).
Figure 3.
Steady-state CD27+CD62L– NK cells show a distinct transcriptional profile that partially overlaps with that of ILC1s
(A) During the steady state, splenic Ly49H+ NK cells were sorted for transcriptome analysis by single-cell RNA sequencing (scRNA-seq). UMAP-based Leiden clustering of scRNA-seq data is represented in the upper right panel. Relative expression of CD27 and CD62L in UMAP projection (lower left panel) and heatmap of top 10 genes defining each cluster are represented in the lower right panel.
(B) CD27+CD62L–, CD27+CD62L+, and CD27–CD62L+ populations of Ly49H+ NK cells as well as ILC1s (defined as CD19/CD3/TCRα/β– NK1.1+ CD27+ CD62L–, Ly49A/CI/G2 negative) were sorted from spleens of C57BL/6 mice for bulk RNA-seq (upper panel). Expression of top 10 cluster defining genes from scRNA-seq in bulk RNA-seq data of sorted populations are represented in the lower panel.
(C and D) Bar graphs depict mRNA normalized counts of indicated genes in sorted NK cell populations.
(E) UMAP-based Leiden clustering of scRNA-seq transcriptomes and relative expression of Sell and Batf3 in UMAP projection for all, Ly49H+ and Ly49H– NK cells sorted from Eomes-GFP reporter as CD19/CD3/TCRg/d– NK1.1+ Eomes-GFP+ and identified via hashtag antibody staining.
(F) Histograms show flow-cytometric detection of various markers in CD27+CD62L–, CD27+CD62L+, and CD27–CD62L+ subsets of Ly49H+ splenic NK cells.
(G) Bulk RNA expression profile of NK cell subsets compared to ILC1s. Depicted are the 40 most variable genes.
(H and I) As in (G) but comparing expression of selected (H) cytokines and (I) transcription factors.
Data in (A)–(D) and (F)–(I) are representative of two independent experiments.
See also Figures S3 and S4.
Transcriptional profiles of CD27+CD62L– NK cells and ILC1s partially overlap
Transcriptional analysis suggested that CD27+CD62L– NK cells were weakly cytolytic and harbored markers of tissue residency. Since these features also characterize ILC1s, we next compared the transcriptional profile of Ly49H+ NK cell subsets with that of ILC1s, enriched to high purity (Figure S4A). Considering the 40 most variable genes that defined the three NK cell subsets and ILC1s, we found substantial overlap in the expression profile of CD27+CD62L– NK cells and ILC1s but also identified many transcripts that were selectively enriched in either cell type (Figure 3G). This was confirmed by the accompanying principal component analysis, which showed high similarity of CD27+CD62L– NK cells and ILC1s in PC1 but clear segregation in PC2 (Figure S4B; Table S1). We found that CD27+CD62L– NK cells and ILC1s expressed similar amounts of Tnf, Lta, and Ltb, while only CD27+CD62L– NK cells expressed GM-CSF (Figure 3H). Both ILC1s and CD27+CD62L– NK cells expressed the chemokine receptor Cxcr6, which has been shown to mark memory-like NK cells in hapten-induced models of NK cell memory (O’Leary et al., 2006; Paust et al., 2010). Furthermore, Cd200r and the gene encoding the integrin Cd61 were expressed predominantly in both ILC1s and CD27+CD62L– NK cells, suggesting similar homing profiles (Figure S4C). While expression of CD61 could be confirmed using flow cytometry, CD200r and CXCR6 did not show a clear staining, especially when compared to highly positive liver ILC1s (Figure S4D). Concerning transcription factors, both ILC1 and CD27+CD62L– NK cells expressed Tcf7. In contrast, Id3, Batf3, and Eomes were enriched in CD27+CD62L– NK cells, while Id2 and Hobit characterized ILC1s. (Figure 3I). Expression of Eomes was further confirmed by flow cytometry, and Eomes deficiency in NKp46iCre Eomesflox/flox mice led to a similar reduction in numbers of CD27+CD62L– and CD62L+ NK cells (Figure S4E). Finally, the expression profile of sorted ILC1s perfectly matched to an ILC1 gene signature established previously (Weizman et al., 2017), while that of CD27+CD62L– NK cells only partly overlapped (Figure S4F). In conclusion, CD27+CD62L– NK cells were transcriptionally similar to ILC1s but also showed many transcriptional features of NK cells, which prompted us to term them “ILC1-like NK cells.”
ILC1-like NK cells show enhanced cytokine production but also target-specific cytotoxicity
Our transcriptional analysis had suggested that CD27+CD62L– ILC1-like NK cells produced a broader spectrum of cytokines than CD62L+ cNK cells. Indeed, when stimulated with phorbol myristate acetate (PMA)/ionomycin ex vivo, ILC1-like NK cells produced IFN-γ, GM-CSF, and TNF-α, while other NK cell subsets only produced IFN-γ (Figures 4A and 4B). Moreover, after stimulation with anti-NK1.1 and upon MCMV infection, ILC1-like NK cells showed stronger IFN-γ production than cNK cells (Figures 4C and 4D). Direct target recognition via activating Ly49 receptors or absence of MHC class I is an important feature that sets apart NK cells and ILC1s functionally. To test whether ILC1-like NK cells actually receive signaling via Ly49H, we performed co-culture experiments with Ba/F3-m157 cells, a pro-B cell line expressing the ligand for Ly49H. ILC1-like NK cells showed ligand-specific downregulation of Ly49H upon co-culture with Ba/F3-m157 cells (Figures 4E and 4F). Furthermore, although ILC1s can show some expression of granzymes, strong GzmB production and pronounced cytotoxicity are hallmarks of NK cells. Importantly, NK cell clones derived from single ILC1-like NK cells became uniformly positive for GzmB at the peak of their MCMV-driven expansion (Figures 4G and 4H). Moreover, ILC1-like NK cells were capable of missing self-recognition, killing RMA-S target cells that lack MHC class I expression, as efficient as CD27+CD62L+ NK cells (Figure 4I). Together, these functional analyses indicated that ILC1-like NK cells, despite broad cytokine production, belong to the NK cell family.
Figure 4.
ILC1-like NK cells show enhanced cytokine production but also target-specific cytotoxicity
(A) Ly49H+ NK cells were sorted into CD27+CD62L– (ILC1-like), CD27+CD62L+, and CD27–CD62L+ subsets and stimulated with PMA and ionomycin. Representative histograms show IFN-γ, TNF-α, and GM-CSF production of sorted NK cell subsets and unstimulated Ly49H+ NK cells as negative control.
(B) Bar graph depicts subset-specific cytokine production normalized to that of stimulated bulk Ly49H+ NK cells within the same experiment.
(C and D) C57BL/6 mice were infected with MCMV or left uninfected. Splenocytes were harvested 24 h later and stimulated with plate-bound aNK1.1 antibodies or left unstimulated. (C) Representative dot plots and (D) bar graph show IFN-ɣ production of Ly49H+ NK cell subsets.
(E and F) Ly49H+ NK cells sorted into ILC1-like and CD27+CD62L+ populations and co-incubated for 24 h with Ba/F3 (WT) and Ba/F3-m157 cells. (E) Dot plots show sort purity, and histograms show Ly49H expression profile. (F) Bar graph depicts Ly49H geometric mean fluorescence intensity (MFI).
(G) Color-barcode (left panel) and representative anti-Granzyme B staining (right panel) of Ly49H+ NK cell clones derived from a single ILC1-like (red) or a single CD27+CD62L+ NK cell (blue) detected in spleens of Rag2–/– Il2rg–/– recipients at day 8 p.i. with MCMV.
(H) Bar graph depicts average geometric MFI of anti-granzyme B staining in NK cell clones derived from ILC1-like versus CD27+CD62L+ NK cells.
(I) Ly49H+ ILC1-like (red) and Ly49H+ CD27+CD62L+ NK cells (blue) were sorted from the spleen of C57BL/6 mice 24 h p.i. with MCMV and incubated with equal numbers of GFP+ RMA and BFP+ RMA-S cells for 42 h. Bar graph depicts normalized counts of RMA and RMA-S cells in the absence of NK cells (gray) and after co-incubation with the respective subsets.
Data in (A), (C)–(G), and (I) are representative of two to three independent experiments. Data in (B) and (H) are pooled from three independent experiments. Bars indicate mean. Error bars represent SD. Significances in (B), (F), and (I) are calculated using two-way ANOVA, followed by Tukey’s multiple comparisons test. Significances in (D) are calculated using multiple t tests. Significances in (H) are calculated using Mann-Whitney test. In (C) and (D): n = 2 mice per group per experiment, in (H): n = 18 NK cell clones (4 derived from single ILC1-like NK cells, 14 derived from single CD27+CD62L+ NK cells) from 10–12 recipient mice per experiment.
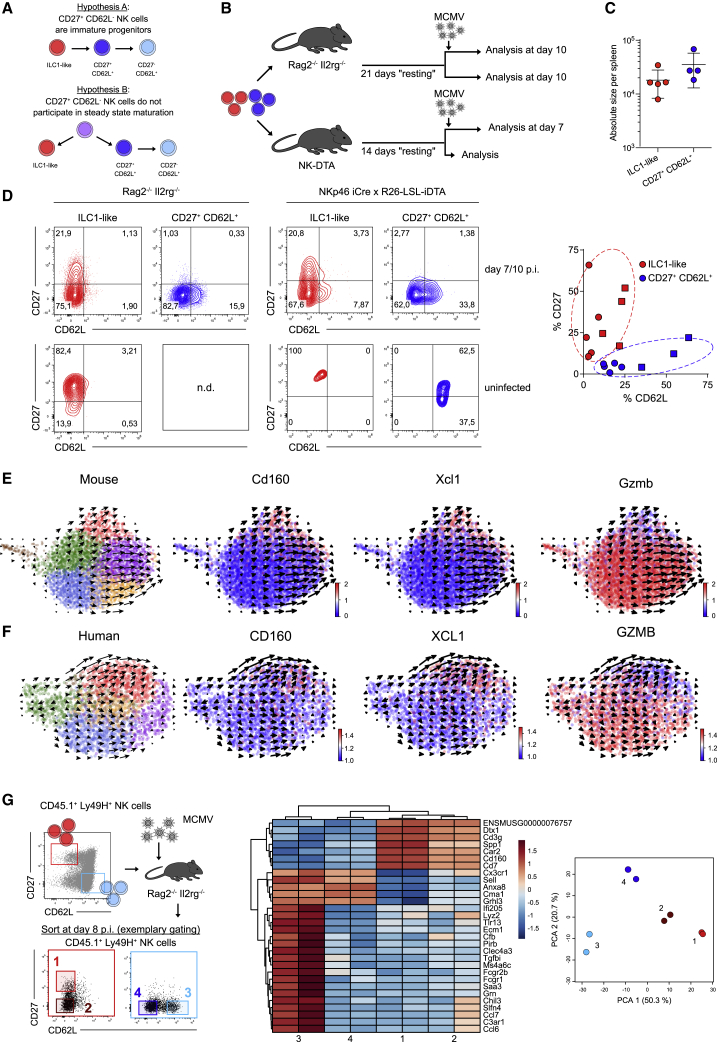
ILC1-like NK cells do not serve as precursors of CD62L+ NK cells
Since NK cells continuously mature under homeostatic conditions, we first hypothesized that ILC1-like NK cells could represent a transient, immature state, from which cells regularly differentiate into the cNK cell compartment. Alternatively, ILC1-like NK cells could represent a separate lineage that does not participate in steady-state maturation (Figure 5A). To test these hypotheses, we transferred low numbers of Ly49H+ ILC1-like or cNK cells into NKp46iCre × R26-LSL-iDTA or Rag2–/– Il2rg–/– mice and rested the transferred cells under homeostatic conditions for 2 or 3 weeks, respectively. After this resting period, we either infected mice with MCMV and analyzed subset composition 7–10 days later or left them uninfected (Figure 5B). Upon MCMV infection, ILC1-like and cNK cells mounted responses of similar size (Figure 5C). However, the two subsets retained their distinct response behaviors, generating CD62L– or CD62L+ response patterns, respectively (Figure 5D). Importantly, even without MCMV infection, no evidence for maturation of ILC1-like NK cells into the CD62L+ subsets was found, while CD27+CD62L+ NK cells further matured into CD27–CD62L+ cells (Figure 5D). Since adoptive transfer may have biased the developmental capacity of the indicated NK cell subsets, we further investigated their developmental potential under steady-state conditions in situ. Therefore, we performed scRNA-seq of splenic Ly49H+ NK cells within recently established retrogenic chimeras (Figure S5A). We reasoned that this setting was ideal to study the relationship of ILC1-like and CD62L+ NK cell subsets, due to their recent development starting out from HSCs. The newly generated NK cells again showed a distinct ILC1-like cluster characterized by transcripts such as Cd160, Xcl1, Cd7, and Ltb and the absence of Gzmb, Gzma, and Klf2 (Figure 5E; Figure S5B). Moreover, RNA velocity analysis of single-cell differentiation trajectories in situ (La Manno et al., 2018) indicated that this ILC1-like cluster showed almost no differentiation activity toward cNK cells (Figure 5E). These data provide further evidence that ILC1-like NK cells do not serve as an immature progenitor of cNK cells but instead constitute a separate lineage. RNA velocity analysis of a previously reported scRNA-seq dataset of human NK cells (Crinier et al., 2018) suggested a similar bifurcation into two lineages segregated from each other by the mutually exclusive expression of CD160 and XCL1 versus GZMB (Figure 5F). Lastly, we asked whether infection leads to a blurring of differentiation boundaries between ILC1-like and cNK cells. Indeed, both subsets generated CD27–CD62L– progeny upon MCMV infection. However, when comparing the transcriptional signature of these double-negative cells derived from ILC1-like or cNK cells, they remained clearly distinct from one another and retained key lineage defining features (Figure 5G; Table S2).
Figure 5.
ILC1-like NK cells do not serve as precursors of CD62L+ NK cells
(A) Schemes depict two possible scenarios of NK cell differentiation. ILC1-like NK cells represent a transient, immature differentiation state (hypothesis A) or a separate lineage that does not participate in steady-state maturation (hypothesis B).
(B–D) Ly49H+ NK cells were sorted into ILC1-like and CD27+CD62L+ populations and adoptively transferred into NKp46 iCre x R26-LSL-iDTA or Rag2–/– Il2rg–/– mice that were infected with MCMV 14 or 21 days later and analyzed at 7 or 10 days p.i., respectively. Uninfected controls were analyzed 14 (14+0) or 31 (21+10) days after initial adoptive transfer. (B) Schematic representation. (C) Absolute size of expanded ILC1-like and CD27+CD62L+ NK cell populations in Rag2–/– Il2rg–/– mice at day 10 p.i. (D) Representative contour plots show CD27 and CD62L expression profiles of NK cell populations at day 7 or 10 p.i. or in uninfected controls. Scatterplot depicts frequency of CD27 and CD62L expressing cells within expanded NK cell populations in NKp46 iCre x R26-LSL-iDTA (squares) and Rag2–/– Il2rg–/– mice (circles).
(E) RNA velocities in scRNA-seq data of splenic Ly49H+ NK cells derived from retrogenic chimeras 4 weeks after HSC transfer. Colors indicate Leiden clustering (left panel) or relative expression of indicated genes (right panels).
(F) As in (E) but scRNA-seq data of one human donor (blood and spleen) from (Crinier et al., 2018).
(G) Ly49H+ NK cells were sorted into ILC1-like (red) and CD27–CD62L+ (blue) populations and adoptively transferred into Rag2–/– Il2rg–/– recipients, followed by MCMV infection. Sub-compartments 1–4 with indicated expression of CD27 and CD62L were sorted from the expanded populations (red or blue) detected in spleen at day 8 p.i. Representative dot plots show sorting strategy. Heatmap depicts 30 most variable genes. Principal component analysis of transcriptome data from sorted sub-compartments 1–4.
Data in (C) and (D) are pooled from two and four independent experiments, respectively. Data in (E) are representative of two independent experiments. In (C), (D), and (G): n = 2–3, 1–3, and 2 mice per group per experiment, respectively.
See also Figure S5.
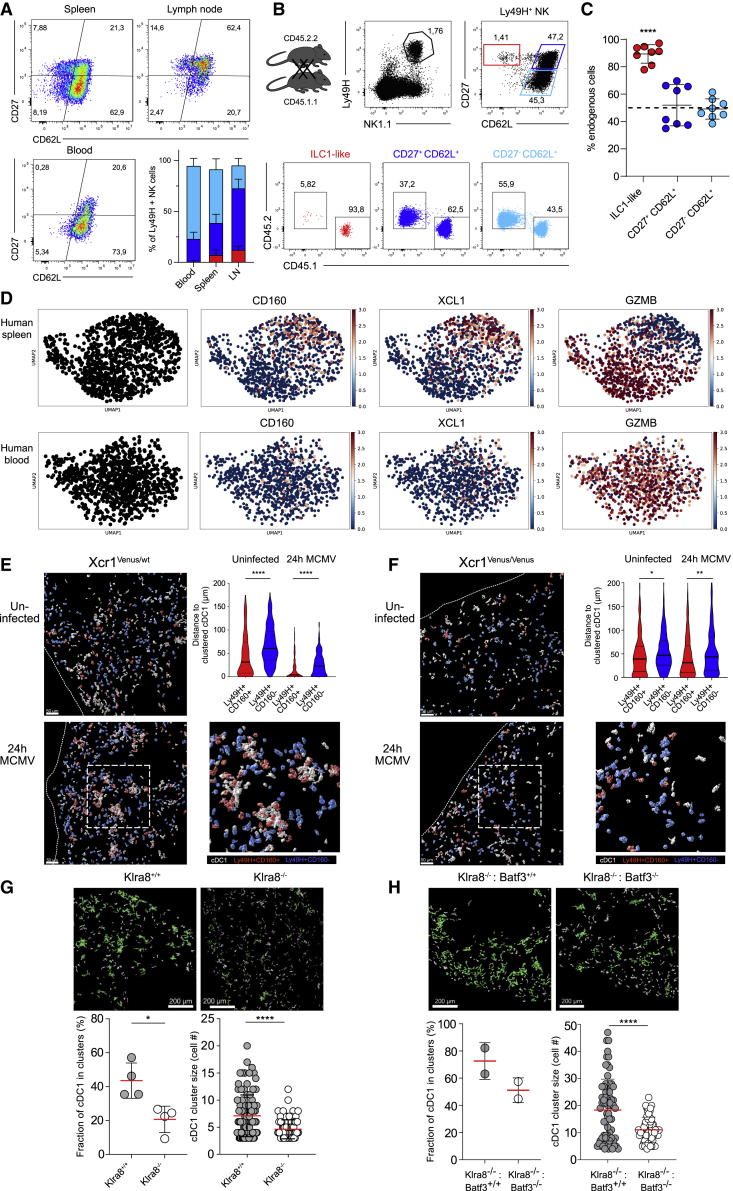
ILC1-like NK cells are overrepresented in lymphoid tissues and tissue-resident in the spleen
While largely absent from peripheral blood, we found CD27+CD62L– ILC1-like NK cells in spleen and lymph nodes (Figure 6A). Moreover, when subjecting lymph node NK cells to scRNA-seq, we found that a subset score learned from splenic ILC1-like and cNK cells (Figure S6A) nicely overlayed with the two main clusters found in lymph nodes (Figures S6B and S6C). In addition, RNA velocities indicated a developmental separation of these clusters similar to that found in the spleen (Figure S6D). Due to additional expression of tissue-residency markers like CD69 and CD160, we hypothesized that in contrast to cNK cells, ILC1-like NK cells may not recirculate but rather stably localize within lymphoid organs. Indeed, when we performed parabiosis of mice expressing distinct congenic markers, we found that ILC1-like NK cells remained almost entirely spleen resident, while cNK cells equilibrated between the two parabionts (Figures 6B and 6C). We wondered whether this tissue residency meant that transferred ILC1-like NK cells would home exclusively to the spleen even during infection. To investigate this, we transferred ILC1-like NK cells or cNK cells into NKp46 iCre x R26-LSL-iDTA mice, infected them with MCMV, and analyzed NK cell homing into different organs. Both ILC1-like NK cells and cNK cells were not only found in the spleen but also in the liver and lymph nodes. Importantly, in all of these locations, they retained their distinct lineage-specific phenotypes (Figure S6E). Similarly, when comparing previously reported scRNA-seq datasets of human NK cells from spleen and blood (Crinier et al., 2018), we found that CD160 and XCL1 expressing NK cells were strongly overrepresented in spleen, while GZMB expressing NK cells were similarly distributed between spleen and blood (Figure 6D).
Figure 6.
ILC1-like NK cells are spleen-resident and share a functional niche with cDC1s
(A) Representative pseudocolor plots and stacked bar graphs show frequency of ILC1-like, CD27+CD62L+, and CD27–CD62L+ NK cells among Ly49H+ NK cells in spleen, lymph nodes, and blood.
(B) Spleens of CD45.1 and CD45.2 parabionts were analyzed 4 weeks after surgery by flow cytometry. Representative dot plots show gating of ILC1-like, CD27+CD62L+, and CD27+CD62L− Ly49H+ NK cells and frequency of CD45.1 and CD45.2 cells within each subset.
(C) Frequency of endogenous cells among Ly49H+ NK cell subsets in each parabiont.
(D) scRNA-seq data from one human donor from (Crinier et al., 2018) showing expression of key marker genes CD160, XCL1, and GZMB.
(E) Visualization of cDC1s, Ly49H+CD160+ NK cells, and Ly49H+CD160– NK cells in spleens of MCMV-infected or uninfected Xcr1Venus/wt mice by confocal microscopy. Depicted are three-dimensional objects corresponding to individual cells identified via histo-cytometry. Staining for NK1.1, Ly49H, and CD160 was used to identify NK cell subsets, and anti-GFP (cross-reactive with Venus) was used to identify cDC1s. Violin plot shows quantification of distances between CD160+ or CD160– Ly49H+ NK cells and clustered cDC1 with and without MCMV infection. A cDC1 cluster was defined as at least three cDC1s within a distance of ≤5μm (see STAR Methods and Figure S7A).
(F) As in (E) but using XCR1-deficient Xcr1Venus/Venus mice.
(G) Visualization of cDC1s (identified by staining for XCR1) in spleens of Klra8+/+ and Klra8–/– littermates 24 h p.i. (number of cDC1 per mm2: 249.6 in Klra8+/+, 325.0 in Klra8–/–). Clustered cDC1s are indicated in green. Fraction of cDC1s in clusters (each dot represents data for one spleen section) and cDC1 cluster sizes in Klra8+/+ and Klra8–/– mice are shown.
(H) As in (G) but for Klra8–/–: Batf3+/+ and Klra8–/–: Batf3–/– mixed bone marrow chimeras (number of cDC1 per mm2: 241.5 in Klra8–/–: Batf3+/+, 258.2 in Klra8–/–: Batf3–/–).
Data in (B), (E), and (G) are representative of two independent experiments. Data in (C) are pooled from two independent experiments. Lines represent mean, and error bars indicate SD. Significances in (C) were calculated using one-sample t test (hypothetical value = 50). Significances in (E) and (F) were calculated using Kruskal-Wallis test, followed by Dunn’s multiple comparisons analysis. Significances in (G) and (H) are calculated using Mann-Whitney test. In (A): n = 8 mice, in (B) and (C): n = 4 mice per experiment, in (E)–(G): n = 2 mice per group per experiment, and in (H): n = 1–2 mice per group.
See also Figures S6 and S7.
ILC1-like NK cells preferentially cluster with cDC1s during early MCMV infection
RNA-seq analyses had revealed that ILC1-like NK cells express molecules such as XCL-1 or CRTAM that facilitate interactions with XCR1+ cDC1s. To investigate the microanatomical localization of ILC1-like Ly49H+ NK cells within the spleen, we employed confocal microscopy during the steady state and 24 h after MCMV infection. Since absence of CD62L expression is difficult to determine in dense tissue sections (due to cellular overlap), we used Ly49H together with CD160 as an alternative marker to discriminate ILC1-like NK cells from cNK cells and ILC1s (Figure S7A). In Xcr1-reporter mice (Xcr1Venus/wt), we observed that already during the steady state a fraction of cDC1s formed clusters with CD160+ ILC1-like but not CD160– cNK cells (Figure 6E; Figure S7B). Upon infection, selective co-clustering of ILC1-like NK cells and cDC1s was further enhanced (Figure 6E). Moreover, analysis of XCR1-deficient (Xcr1Venus/Venus) mice showed that this clustering was, at least in part, mediated via the XCL-1/XCR1 axis (Figure 6F). While positioned at similar distance as Ly49H– NK cells during the steady state, Ly49H+ NK cells located significantly closer to clustered cDC1s upon MCMV infection (Figure S7C), suggesting a functional relevance of Ly49H in this process. Indeed, we found that early clustering of cDC1s was virtually abrogated in Klra8–/– hosts (Figure 6G), and their co-localization with CD160+ ILC1-like NK cells was substantially reduced (Figure S7D). Furthermore, by using a GFP-expressing MCMV strain, we found that cDC1s were located significantly closer to MCMV-infected cells in Klra8+/+ versus Klra8–/– hosts (Figures S7E and S7F).
To next answer whether cDC1 clustering is specifically mediated by ILC1-like NK cells within the Ly49H+ compartment, we decided to investigate the functional relevance of the selective Batf3 expression that characterizes this NK cell lineage. Since Batf3 deficiency leads to a block of cDC1 development (Hildner et al., 2008), we generated mixed bone marrow chimeras consisting of Klra8–/– recipients reconstituted with one half of Klra8–/– and one half of Batf3–/– or Batf3+/+ bone marrow. Although numbers of ILC1-like NK cells are normal in Batf3–/– mice and in Klra8–/–: Batf3–/– chimeras (Figure S7G), we hypothesized that function of ILC1-like NK cells may be impaired in such mice. Indeed, we found that both the fraction of cDC1s in clusters and the size of these clusters were significantly reduced in Klra8–/–: Batf3–/– compared to Klra8–/–: Batf3+/+ chimeras (Figure 6H). Importantly, with 258.2 versus 241.5 of cDC1s observed in microscopic sections of Klra8–/–: Batf3–/– versus Klra8–/–: Batf3+/+ chimeras, absolute numbers of cDC1 were virtually identical.
Optimal CD8+ T cell priming upon MCMV infection depends on Ly49H and expression of Batf3 in ILC1-like NK cells
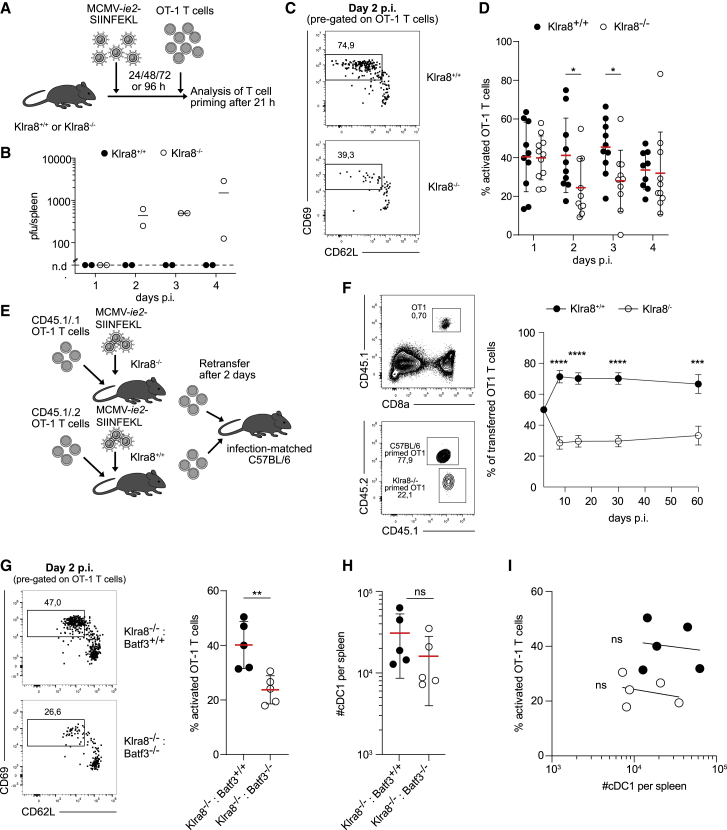
Uptake and cross-presentation of exogenous antigens on MHC class I by cDC1s is particularly important for efficient priming of CD8+ T cells upon MCMV infection (Busche et al., 2013). In absence of Ly49H, cDC1s showed diminished clustering and reduced co-localization with MCMV-infected cells. In absence of NK-cell intrinsic Batf3 expression, cDC1 clustering was similarly diminished. Thus, we hypothesized that delivery of MCMV-associated antigens to cDC1s and subsequent CD8+ T cell priming may also be deficient under these conditions. To test this hypothesis, we infected Klra8+/+ or Klra8–/– mice with recombinant MCMV, encoding the experimental antigen SIINFEKL under control of the viral ie2 promoter (MCMV-ie2-SIINFEKL). 24, 48, 72, or 96 h later, we then transferred naive OT-1 T cells, which harbor a transgenic T cell receptor (TCR) specific for SIINFEKL presented on MHC class I, and measured their activation 21 h later (Figure 7A). Klra8–/– mice are known to show delayed control of MCMV infection. Indeed, when measuring virus titers, we found that replication of MCMV-ie2-SIINFEKL was detectable at least until 96 h p.i. in Klra8–/– hosts while no sizeable virus load was found in wild-type mice (Figure 7B). However, despite reduced viral load, OT-1 T cells were primed more efficiently in wild type than in Klra8–/– mice, as measured by shedding of CD62L and upregulation of CD69 (Figures 7C and 7D). In the long run, reduced NK cell-mediated control of MCMV infection leads to stronger T cell activation (Mitrović et al., 2012), likely due to an increased number of latently infected cells and a parallel increase of available antigen (Redeker et al., 2014). We aimed to isolate the effects of reduced early T cell priming from these later-acting compensatory mechanisms. To achieve this, we co-transferred equal numbers of OT-1 T cells, primed in wild-type or Klra8–/– mice, into “infection-matched” wild-type hosts, all of which had been infected with MCMV-ie2-SIINFEKL 2 days prior (Figure 7E). Under these conditions, OT-1 T cells primed in wild-type versus Klra8–/– mice expanded significantly better and maintained this increased expansion throughout the chronic latent phase of MCMV infection (Figure 7F). Last, we asked whether optimal CD8+ T cell priming specifically required the activity of Batf3 within the Ly49H+ NK cell compartment. We found that priming of OT-1 T cells was significantly reduced in Klra8–/–: Batf3–/– versus Klra8–/–: Batf3+/+ chimeras and virtually phenocopied the priming deficit in Klra8–/– mice (Figure 7G). Importantly, the absolute numbers of cDC1s did not significantly differ between both groups of chimeras (Figure 7H), and T cell priming efficacy per chimera did not correlate with cDC1 numbers (Figure 7I). Together these data strongly argue that Batf3-expressing ILC1-like NK cells play a critical role for optimal CD8+ T cell priming upon MCMV infection.
Figure 7.
Optimal CD8+ T cell priming upon MCMV infection depends on Ly49H and expression of Batf3 in ILC1-like NK cells
(A) Klra8+/+ and Klra8–/– mice were infected with MCMV-ie2-SIINFEKL. 1 × 106 OT-1 T cells were adoptively transferred at 24, 48, 72, and 96 h p.i. and priming determined in spleen 21 h later.
(B) MCMV titers in Klra8+/+ and Klra8–/– mice at indicated time points p.i.
(C and D) Frequency of activated OT-1 T cells (defined as CD69+ CD62L–) in spleens of infected Klra8+/+ and Klra8–/– mice. (C) Representative flow cytometry data. (D) Summary statistics.
(E) 1 × 106 CD45.1/.1 and CD45.1/.2 OT-1 T cells were adoptively transferred into Klra8–/– and Klra8+/+ littermates, respectively, and subsequently infected with MCMV-ie2-SIINFEKL. At day 2 p.i., 100 CD45.1/.1 and 100 CD45.1/.2 OT-1 T cells were co-transferred into secondary infection-matched C57BL/6 recipients.
(F) Representative contour plots show frequency of all CD45.1+ and fraction of CD45.1/.1 and CD45.1/.2 OT-1 T cells in peripheral blood of secondary recipients at day 30 p.i. Dot plot shows frequency of CD45.1/.1 and CD45.1/.2 cells among transferred OT-1 T cells over time.
(G) As in (A), (C), and (D) on day 2 but performed in Klra8–/–: Batf3+/+ or Klra8–/–: Batf3–/– mixed chimeras.
(H) Quantification of cDC1s in spleens of Klra8–/–: Batf3+/+ and Klra8–/–: Batf3–/– mixed chimeras.
(I) Scatterplot shows no correlation between percentage of primed OT-1 T cells and number of cDC1s per host.
Data in (C) are representative of five independent experiments. Data in (D) are pooled from five independent experiments. Lines represent mean, and error bars indicate SD. Data in (F) are pooled from four independent experiments. Dots represent mean, and error bars indicate SEM. Data in (G)–(I) are pooled from two independent experiments. Lines represent mean, and error bars indicate SD. Significances in (D) were calculated by multiple Mann-Whitney tests for each individual time point. Significances in (F) were calculated using multiple t tests, and statistical significance was determined using the Holm-Sidak method, with alpha = 5.0%. Significances in (G) and (H) were calculated by Mann-Whitney test. Correlation in (I) was measured as Spearman correlation. Line indicates non-linear fit. In (B): n = 2 mice per group, in (C), (D), and (G–(I): n = 2–3, and in (F): n = 3–6 mice per group per experiment.
Discussion
By mapping the fate of individual NK cells during their adaptive-like response to MCMV infection, we found evidence that these responses are composed of two distinct NK cell lineages. Individual NK cells either generated CD62L+ clones, further characterized by low expression of CD27, or CD62L– clones, with high expression of CD27. These distinct response patterns of individual NK cells originated from CD62L+ cNK cells or CD27+CD62L– NK cells, respectively. Transcriptionally, CD27+CD62L– NK cells were similar to ILC1s, prompting us to term these cells ILC1-like NK cells.
Further phenotypic and functional analyses defined ILC1-like NK cells as a subset of innate lymphocytes that unites critical aspects of both ILC1s and cNK cells. According to recent classifications, CD127, CD160, and CD69 are exclusively or predominantly expressed on ILC1s while expression of activating Ly49 receptors, and Eomes is considered to be characteristic of cNK cells (Vivier et al., 2018). ILC1-like NK cells expressed all of the aforementioned markers. Reminiscent of ILC1s, ILC1-like NK cells were found to be tissue resident in the spleen and showed a broader cytokine profile than cNK cells, encompassing TNF-α, GM-CSF, and LT-b. On the other hand, ILC1-like NK cells were capable of direct recognition of MHC class I-deficient or virus-infected cells via missing-self recognition or the activating receptor Ly49H and mounted cytotoxic, adaptive-like NK cell responses as efficiently as cNK cells.
Previously, ILC1-like cells have been described as innate lymphoid cells arising via conversion from ILC3s in tissues (Cella et al., 2019) or via transdifferentiation from tumor-infiltrating NK cells (Cortez et al., 2017; Gao et al., 2017). In both cases, this term described innate lymphoid cells and not NK cells. Instead, the ILC1-like NK cells described showed key features of bona fide NK cells and were maintained as a stable NK cell lineage—at least during homeostatic turnover as well as during MCMV-driven expansion.
Thus, we argue that ILC1-like NK cells form a missing link between NK cells and ILC1s, uniting the target recognition, target killing, and adaptive-like expansion capacity of NK cells with the optimal tissue positioning and cytokine secretion capacity of ILC1s.
Their expression of genes that are described to mediate interactions with cDC1s, such as XCL-1 (Böttcher et al., 2018; Dorner et al., 2009), CRTAM (Galibert et al., 2005; Takeuchi et al., 2009), and GM-CSF (Sallusto and Lanzavecchia, 1994), prompted us to investigate whether ILC1-like NK cells shared a functional niche with cDC1s in vivo. Indeed, we found CD160+ ILC1-like NK cells to be in close vicinity to cDC1s already during the steady state. Ly49H+ NK cells have been described to interact with cDC1s during MCMV infection (Andrews et al., 2003). Here, however, we could ascribe this interaction primarily to ILC1-like NK cells. Following infection with MCMV, we found that ILC1-like NK cells, but not cNK cells, formed clusters with infected cells and cDC1s as early as 24 h p.i. On the side of the cDC1s, this interaction was XCR1-dependent. With respect to NK cells, cluster formation was dependent on both specific target recognition via Ly49H and activity of Batf3—a transcription factor specifically expressed in ILC1-like but not cNK cells. Moreover, both genes were also found to be important for enabling optimal CD8+ T cell priming. Taken together, these data indicate that ILC1-like NK cells not only fulfill functions previously ascribed to ILC1s or cNK cells separately but assume a unique role in bridging innate and adaptive recognition of viral infection.
Many of the characteristics identified for ILC1-like NK cells show strong parallels to human NK cell biology. Following established classifications, ILC1-like NK cells form part of the most “immature” (CD27+CD11b–KLRG-1–) NK cell subset. This fits well to human studies, which found tissue-resident NK cell subsets to predominantly share markers with “immature” CD56bright NK cells (Harmon et al., 2016; Melsen et al., 2016). Analogous to CD27+CD11b– NK cells, CD56bright cells in humans are thought to be progenitors of CD56dim NK cells (Chan et al., 2007). Recent elegant studies using hematopoietic stem cell barcoding in rhesus macaques found, however, that barcodes from CD56+ NK cells, which are overrepresented in lymph nodes, did not fully overlap with barcodes in CD56– NK cells that dominate in blood (Wu et al., 2018), suggesting that lymphoid tissue-resident lineages may be distinct from cNK cells also in humans and non-human primates. Moreover, comparing published scRNA-seq datasets from human spleen (Crinier et al., 2018) with our murine data, we observed similar RNA velocity trajectories suggesting that human NK cells differentiate along two paths characterized by the mutually exclusive expression of CD160/XCL1 and GZMB. Transcriptional comparison of ILC1-like NK cells and CD56bright tissue-resident NK cells reveals further parallels between the two subsets; in a recent report, CD56bright tissue-resident NK cells in the liver showed strong expression of Eomes and low expression of T-bet (Harmon et al., 2016), a pattern also found in ILC1-like NK cells in this study. Moreover, TCF7 expression is found both in human intra-epithelial (ie) ILC1s and CD56bright NK cells, but Hobit expression is limited to ieILC1s (Collins et al., 2019), mirroring the expression profiles of bona fide ILC1s and ILC1-like NK cells detected in our murine study. Despite all these parallels, it should be mentioned that ILC1-like NK cells and CD56bright NK cells differ considerably in the expression of other key markers. For example, CD62L is expressed in CD56bright NK cells and CD160 in CD56dim NK cells (Poli et al., 2009).
The existence of two separate paths of NK cell differentiation has major implications for our understanding of NK cell biology. In solid tumors, ILC1s have previously been described to arise via conversion from NK cells (Cortez et al., 2017; Gao et al., 2017). The existence of ILC1-like NK cells in the spleen and lymph nodes of mice under steady-state conditions, as well as their capacity to infiltrate extralymphatic organs such as the liver upon infection, raises the question whether converted ILC1s in tumors may originate selectively from this distinct NK cell lineage.
Our findings underscore the importance of single-cell fate mapping for the study of differentiation processes such as conversion or selection emerging from a heterogenous population of NK cells. With the technological advances presented here, the dynamics of this heterogeneity can now be studied in much greater detail, enabling us to better understand how innate lymphocytes help govern a host’s response against viral infection and cancer (Adams et al., 2020).
Limitations of the study
While our study provides various lines of evidence that ILC1-like NK cells are a transcriptionally, phenotypically, and functionally distinct NK cell lineage, it does not resolve when and where this lineage diverges from the common path of cNK cell maturation. Our data show that ILC1-like NK cells do not serve as progenitors for cNK cells under steady-state conditions or upon MCMV infection, but this may differ in other infection models or during development. While Batf3 was relevant for ILC1-like NK cell function, it was not required for their development. Further research may provide deeper insight into the transcriptional requirements for ILC1-like NK cell formation and thereby enable a more direct investigation of this lineage’s functional relevance and its developmental ties to tissue- or tumor-resident NK cell subsets. Furthermore, while resolving some of the interaction partners required for clustering of cDC1s and ILC1-like NK cells, our study does not answer how the splenic retention of the latter is achieved. It also does not resolve in mechanistic detail how ILC1-like NK cells orchestrate optimized antigen accessibility to cDC1s and optimized CD8+ T cell priming. Finally, we did not provide an in-depth evaluation of the potential human equivalent of this NK cell lineage. Given its important role in optimization of CD8+ T cell priming upon MCMV infection, it will be exciting to investigate whether a human equivalent of ILC1-like NK cells plays a similar role upon human CMV infection specifically and for anti-viral immunity in general.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-mouse NK1.1 | BioLegend | RRID: AB_2564304, Clone: PK136 |
| Anti-mouse NK1.1 | Life Technologies | RRID: AB_469479, Clone: PK136 |
| Anti-mouse Ly49H | Life Technologies | RRID: AB_2573489, Clone: 3D10 |
| Anti-mouse CD45.1 | BioLegend | RRID: AB_313495, Clone: A20 |
| Anti-mouse CD45.2 | BioLegend | RRID: AB_492872, Clone: 104 |
| Anti-mouse CD27 | BioLegend | RRID: AB_2565547, Clone: LG.3A10 |
| Anti-mouse CD27 | Life Technologies | RRID: AB_469370, Clone: LG7.F9 |
| Anti-mouse CD62L | BioLegend | RRID: AB_493380, Clone: MEL-14 |
| Anti-mouse CD62L | Becton Dickinson | RRID: AB_394666, Clone: MEL-14 |
| Anti-mouse CD160 | BioLegend | RRID: AB_10960743, Clone: 7H1 |
| Anti-mouse CD160 | R&D Systems | Cat# AF3899 |
| Anti-mouse CD11b | Becton Dickinson | RRID: AB_396679, Clone: M1/70 |
| Anti-mouse CD11b | Biolegend | RRID: AB_312795, Clone: M1/70 |
| Anti-mouse KLRG1 | Life Technologies | RRID: AB_469469, Clone: 2F1 |
| Anti-human Granzyme B (crossreactive) | Life Technologies | RRID: AB_2536538, Clone: GB11 |
| Anti-mouse CD3e | Becton Dickinson | RRID: AB_394595, Clone: 145-2C11 |
| Anti-mouse CD3e | BioLegend | RRID: AB_312673, Clone: 145-2C11 |
| Anti-mouse TCR beta | Becton Dickinson | RRID: AB_394683, Clone: H57-597 |
| Anti-mouse CD4 | Life Technologies | RRID: AB_10373812, Clone: RM4-5 |
| Anti-mouse CD8a | BioLegend | RRID: AB_2563057, Clone: 53-6.7 |
| Anti-mouse CD19 | Becton Dickinson | RRID: AB_11154223, Clone: 1D3 |
| Anti-mouse Ly6A/E (Sca-1) | BioLegend | RRID: AB_313345, Clone: D7 |
| Anti-mouse CD16/32 | BioLegend | RRID: AB_312801, Clone: 93 |
| Anti-mouse CD49a | BioLegend | RRID: AB_10945160, Clone: HMa1 |
| Anti-mouse CD49b | BioLegend | RRID: AB_2265144, Clone: DX5 |
| Anti-mouse Ly-6C | Becton Dickinson | RRID: AB_394628, Clone: AL-21 |
| Anti-mouse CD69 | BioLegend | RRID: AB_493564, Clone: H1.2F3 |
| Anti-mouse CD127 | BioLegend | RRID: AB_1937252, Clone: A7R34 |
| Anti-mouse Ly49A | Life Technologies | RRID: AB_1311270, Clone: A1(Ly49A) |
| Anti-mouse Ly49I | Life Technologies | RRID: AB_465301, Clone: YLI-90 |
| Anti-mouse Ly49F | Miltenyi Biotec | RRID: AB_2652744, Clone: HBF-719 |
| Anti-mouse Ly49G2 | Miltenyi Biotec | RRID: AB_2652759, Clone: 4D11 |
| Anti-mouse Ly49D | Life Technologies | RRID: AB_10717290, Clone: eBio4E5 |
| Anti-mouse IFN-gamma | Life Technologies | RRID: AB_1257211, Clone: XMG1.2 |
| Anti-mouse TNF-alpha | BioLegend | RRID: AB_315424, Clone: MP6-XT22 |
| Anti-mouse GM-CSF | BioLegend | RRID: AB_315381, Clone: MP1-22E9 |
| Anti-mouse XCR1 | BioLegend | RRID: AB_2565230, Clone: ZET |
| Anti-mouse CD200r | eBioscience | RRID: AB_2074081, Clone: 0X110 |
| Anti-mouse CD61 | BioLegend | RRID: AB_2561734, Clone: 2c9.g2 |
| Anti-mouse CXCR6 | BioLegend | RRID: AB_2566545, Clone: SA051D1 |
| Anti-sheep IgG | Jackson Immuno Research | RRID: AB_2340747 |
| Donkey anti-rabbit IgG | Biolegend | RRID: AB_1575130 |
| Donkey anti-rabbit IgG | Biolegend | RRID: AB_2563306 |
| Anti-GFP | Life Technologies | RRID: AB_221477 |
| Anti-Collagen IV | Abcam | Cat# ab6586 |
| TotalSeq™-B0301 anti-mouse Hashtag 1 Antibody | Biolegend | RRID: AB_2814067 |
| TotalSeq™-B0302 anti-mouse Hashtag 2 Antibody | Biolegend | RRID: AB_2814068 |
| TotalSeq™-B0304 anti-mouse Hashtag 4 Antibody | Biolegend | RRID: AB_2814070 |
| TotalSeq™-B0305 anti-mouse Hashtag 5 Antibody | Biolegend | RRID: AB_2814071 |
| TotalSeq™-B0306 anti-mouse Hashtag 6 Antibody | Biolegend | RRID: AB_2814072 |
| TotalSeq™-B0307 anti-mouse Hashtag 7 Antibody | Biolegend | RRID: AB_2814073 |
| Bacterial and Virus Strains | ||
| MCMV-wt (Smith strain) | HZI Braunschweig | N/A |
| MCMV-ie2-SIINFEKL | HZI Braunschweig | N/A |
| MCMV-ie2-SIINFEKL-GFP | HZI Braunschweig | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Propidium Iodide (PI) | Life Technologies | Cat# P1304MP |
| Ethidium Monoazide Bromide (EMA) | Life Technologies | Cat# E1374 |
| Recombinant murine IL-3 | PeproTech | Cat# 213-13 |
| Recombinant murine IL-6 | PeproTech | Cat# 216-16 |
| Recombinant murine SCF | PeproTech | Cat# 250-03 |
| RetroNectin® | Takara Bio Europe | Cat# T100B |
| Phorbol-12-myristat-13-acetat | Sigma Aldrich | Cat# P1585 |
| Ionomycin | Sigma Aldrich | Cat# I9657 |
| Golgi Plug™ | Becton Dickinson | Cat# 555029 |
| Percoll | Sigma-Aldrich | Cat# GE17-0891-01 |
| Antigenfix | Diapath | Cat# P0016 |
| TissueTek OCT freezing medium | Sakura Finetek | Cat# 4583 |
| Tris | AppliChem | Cat# A1379 |
| Triton X-100 | AppliChem | Cat# A4975 |
| Mouse serum | Sigma Aldrich | Cat# NS03L |
| Mowiol 4-88 | Sigma Aldrich | Cat# 81381 |
| Phophate buffered saline (PBS) | Life Technologies | Cat# 14190144 |
| Bovine serum albumin (BSA) | Sigma Aldrich | Cat# A3294 |
| Ethylenediaminetetraacetic acid (EDTA) | Carl Roth | Cat# 8040.3 |
| Skim milk powder | Sigma Aldrich | Cat# 70166 |
| Cytofix/Cytoperm™ Fixation/ Permeabilization Solution Kit | Becton Dickinson | RRID: AB_2869008 |
| eBioscience™ Foxp3 Transcription Factor Staining Buffer Set | Life Technologies | Cat# 00-5523-00 |
| True-Nuclear™ Transcription Factor Buffer Set | Biolegend | Cat# 424401 |
| Experimental Models: Cell Lines | ||
| Ba/F3 | UCSF | N/A |
| Ba/F3-m157 | UCSF | N/A |
| RMA | MSKCC | N/A |
| RMA-S | MSKCC | N/A |
| Platinum-E | Cell Biolabs | Cat# RV-101 |
| M2-10B4 | ATCC | RRID:CVCL_5794 |
| Experimental Models: Organisms/Strains | ||
| C57BL/6-Tg(TcraTcrb)1100Mjb/J | The Jackson Laboratory | RRID: IMSR_JAX:003831 |
| B6.SJL-Ptprca Pepcb/BoyJ | The Jackson Laboratory | RRID: IMSR_JAX:002014 |
| C;129S4-Rag2tm1.1Flv Il2rgtm1.1Flv/J | The Jackson Laboratory | RRID: IMSR_JAX:014593 |
| B6.BXD8-Klra8Cmv1-del/WumJ | The Jackson Laboratory | RRID: IMSR_JAX:008633 |
| B6.Cg-Ncr1 tm1.1(icre)Viv | E. Vivier, Centre d’Immunologie de Marseille-Luminy, France | RRID: IMSR_EM:05625 |
| Gt(ROSA)26Sortm1(DTA)Jpmb/J | The Jackson Laboratory | RRID: IMSR_JAX:006331 |
| B6.Cg-Xcr1tm2(HBEGF/Venus)Ksho | T. Kaisho, RIKEN Research Center for Allergy and Immunology, Japan | RRID: IMSR_RBRC09485 |
| B6.129S(C)-Batf3tm1Kmm/J | The Jackson Laboratory | RRID: IMSR_JAX:013755 |
| Eomestm2.1Rob/Eomes+ | Y. Tanriver, UK Freiburg, Germany | RRID: MGI:5646120 |
| Eomes < tm1Srnr > | S. Reiner, University of Pennsylvania | RRID: MGI:3802572 |
| C57BL/6JOlaHsd | Envigo | RRID: MGI:5657800 |
| BALB/cOlaHsd | Envigo | RRID: MGI:5659069 |
| Oligonucleotides | ||
| GFP family Not1 fwd5′ ATTAGCGGCCG CGCCACCATGGTGAGCAAGGGCG 3′ |
Sigma Aldrich | N/A |
| GFP family EcoR1 rev5′ TAATGAATTCTT ACTTGTACAGCTCG 3′ |
Sigma Aldrich | N/A |
| Recombinant DNA | ||
| EGFP | MIH Munich | N/A |
| EYFP | MIH Munich | N/A |
| CFP | MIH Munich | N/A |
| Ametrine | MIH Munich | N/A |
| EBFP2 | MIH Munich | N/A |
| Software and Algorithms | ||
| FlowJo V10 | FlowJo LLC | https://www.flowjo.com |
| Prism 9 | Graphpad | https://www.graphpad.com |
| Imaris 9.5 | Bitplane | https://imaris.oxinst.com/ |
| SCANPY (version 1.4.4) | Theis lab (GitHub) | https://github.com/theislab/scanpy |
| Velocyto (version 0.17.17) | Velocyto-team (GitHub) | https://velocyto.org |
| Cellranger (5.0.1) | 10x Genomics | https://support.10xgenomics.com |
| Deposited data | ||
| Single cell RNA sequencing data | This paper | GEO: GSE180978 |
| Bulk-RNA sequencing data | This paper | GEO: GSE180978 |
| Human scRNaseq data | Crinier et al., 2018 | GEO: GSE119562 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Veit R. Buchholz (veit.buchholz@tum.de).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Mice
C57BL/6JOlaHsd (females, 6-12 weeks) and BALB/cOlaHsd (males, 3 weeks) were purchased from Envigo. 6-14 weeks old female and male Rag2–/– Il2rg–/– (C;129S4-Rag2tm1.1Flv Il2rgtm1.1Flv/J), R26-LSL-iDTA (Gt(ROSA)26Sortm1(DTA)Jpmb/J), Klra8–/– (B6.BXD8-Klra8Cmv1-del/WumJ), CD45.1 (B6.SJL-Ptprca Pepcb/BoyJ), Batf3–/– (B6.129S(C)-Batf3tm1Kmm/J) and SIINFEKL peptide-specific TCR transgenic OT-1 (C57BL/6-Tg(TcraTcrb)1100Mjb/J) mice were originally obtained from The Jackson Laboratory and bred under specific pathogen-free conditions at our mouse facility at the Technical University of Munich. NKp46iCre (B6.Cg-Ncr1 tm1.1(icre)Viv) mice were generously provided by Eric Vivier. Xcr1Venus (B6.Cg-Xcr1tm2(HBEGF/Venus)Ksho) mice were a kind gift from Tsuneyasu Kaisho. Eomes-GFP (Eomestm2.1Rob/Eomes+) mice were generously provided by Yakup Tanriver. Eomesflox/flox (Eomes < tm1Srnr > ) were a kind gift from Steven Reiner. All animal experiments were approved by local authorities and performed in accordance with national guidelines.
Viruses
MCMV-wt (Smith strain) was prepared from the BAC-derived mouse cytomegalovirus clone pSM3fr 3.3. The MCMV-ie2-SIINFEKL was generated by fusing the SIINFEKL epitope at the C terminus of the ie2 sequence using en passant mutagenesis. MCMV was recovered from BACs by transfecting mouse embryo fibroblasts (MEFs). After reconstitution, virus was propagated on M2-10B4 cells and virus stocks were prepared as described previously (Chaudhry et al., 2020). In brief, virus was pelleted from supernatant of infected cells (26000 x g for 3.5 h). Subsequently, the pellet was re-suspended in VSB buffer (0.05 M Tris-HCl, 0.012 M KCl, and 0.005 M EDTA, adjusted to pH 7.8) and then purified by centrifugation through a 15% sucrose cushion in VSB buffer (53000 x g), and a subsequent slow centrifugation step (3000xg, 5 min) to remove cellular debris. The virus stocks were titrated on MEFs to measure virus stock titers.
Tissue culture
The Platinum-E packaging cell line, Ba/F3 cell lines, M2-10B4 and MEFs were grown in cDMEM (DMEM (Life Technologies), supplemented with 10% FCS, 0.025% L-Glutamine, 0.1% HEPES, 0.1% gentamycin and 1% penicillin/streptomycin). MEFs were made in house from C57BL/6. RMA cell lines were grown in cRPMI (RPMI (Life Technologies), supplemented with 10% FCS, 0.025% L-Glutamine, 0.1% HEPES, 0.1% gentamycin and 1% penicillin/streptomycin). All cell lines were maintained in a standard incubator at 37°C with 5% CO2 in humidified atmosphere. The sex of Platinum-E cells is female. The sex of M2-10B4, Ba/F3 and RMA cell lines is unknown.
Method details
Generation of retroviral fluorescent color-barcodes
For retrovirus production, Platinum-E packaging cells were transfected via calcium phosphate precipitation with retroviral vectors (mp71, a kind gift from Wolfgang Uckert) encoding for the five fluorescent proteins GFP, YFP, CFP, BFP and Ametrine. Virus-containing supernatants were collected 48 h after transfection and purified from remaining cells by centrifugation (1500 rpm, 4°C, 7 min). Supernatants were stored at 4°C and used within 4 weeks after harvest.
Generation of retrogenic mice
Bone marrow cells were isolated from tibia and femur of 8-15 weeks old CD45.1 or C57BL/6 mice and stained with anti-mouse Ly6A/E (Sca-1) and anti-mouse CD3/19 antibodies. Propidium iodide was added for life/dead discrimination. Sorted CD3– CD19– Sca-1+ stem cells were cultured at 37°C in cDMEM, supplemented with 20 ng/mL murine IL-3, 50 ng/mL murine IL-6 and 50 ng/mL murine SCF, for 3-4 days in a tissue-culture treated 48-well plate (250.000-300.000 cells/400 μl). Expanded stem cells were retrovirally transduced by spinoculation. Therefore, a tissue-culture untreated 48-well plate was coated with RetroNectin according to the manufacturer’s instructions. 400 μl of the pooled Platinum-E supernatants were added per well and centrifuged at 3000 g and 32°C for 2 h. Then, 200 μl of the supernatant was discarded and 200 μl of stem cells were added in cDMEM supplemented with 40 ng/mL mIL-3, 100 ng/mL mIL-6 and 100 ng/mL mSCF (final cell density: 300.000 cells/400 μl). Cells were centrifuged at 800 g and 32°C for 1.5 h. After 2 days in culture the transduced stem cells were suspended in FCS at 500.000 – 1.000.000 cells/100 μl and injected i.v. into irradiated C57BL/6 recipient mice (two times 4.5 Gy, with a resting period of 4 h).
Cell sorting and adoptive transfer of Ly49H+ NK cells
Spleens of CD45.1 or retrogenic donor mice were harvested and brought into single-cell suspension by mashing through a 40 μm cell strainer, followed by red blood cell lysis. For adoptive transfer of single Ly49H+ NK cells or rare NK cell populations, NK1.1+ cells were enriched by flow cytometric sorting (MoFlo XDP, Beckman Coulter). After enrichment, cells were stained with respective antibodies and propidium iodide (PI) was used for life/dead discrimination. For single-cell adoptive transfers CD3– CD19– NK1.1+ Ly49H+ CD27+ cells harboring distinct color-barcodes were sorted (expression of CD62L and CD11b was indexed or considered for sort decisions where indicated). 15-20 single cells with distinct barcodes were sorted into the same well and transferred into the same host. Because color-barcodes are distinguishable via flow cytometry, these multiplexed NK cells could nevertheless be analyzed as individual NK cell clones. For population transfers CD3– CD19– NK1.1+ Ly49H+ cells with distinct expression of CD27, CD62L and CD11b, respectively, were sorted. Sorting of Ly49H+ NK cells was performed on a BD FACSAria III (Becton Dickinson), MoFlo XDP or MoFlo Astrios cell sorter (Beckman Coulter). NK cells were sorted into a 96-well v-bottom plate containing 400.000 feeder splenocytes in 200 μl FCS. NK cells were injected i.v. into Rag2–/– Il2rg–/–, Klra8–/– or NKp46iCre x R26-LSL-iDTA recipient mice.
Retransfer of Ly49H+ NK cells
Ly49H+ NK cell subsets were sorted and adoptively transferred into NKp46iCre x R26-LSL-iDTA recipients as described above, followed by infection with MCMV-wt. Spleens of primary recipients were harvested at day 8 p.i. and CD45.1+ cells were enriched by flow cytometric sorting (BD FACSAria III, Becton Dickinson or MoFlo Astrios, Beckman Coulter). Afterward, enriched cells were stained with respective antibodies and CD19– CD3– CD45.1+ NK1.1+ Ly49H+ cells were sorted (BD FACSAria III, Becton Dickinson or MoFlo Astrios, Beckman Coulter) into a 96-well v-bottom plate containing 400.000 Rag2–/– Il2rg–/– splenocytes in 200 μl FCS. NK cells were injected i.v. into Rag2–/– Il2rg–/– as secondary recipients. Secondary recipients were analyzed after 20 days by flow cytometry as described or infected with MCMV-wt. Infected mice were analyzed at day 8 p.i.
Generation of virus stocks from salivary glands
3 weeks old male BALB/c mice were infected intraperitoneally (i.p.) with 2x105 plaque-forming units (pfu) of “wildtype” Smith strain MCMV, MCMV-ie2-SIINFEKL or MCMV-ie2-SIINFEKL-GFP. After 21 days, salivary glands were homogenized in 5% skim milk/DMEM using a dounce tissue grinder (loose pestle). Cells were pelleted (800 g, 4°C, 5 min) and supernatants were transferred into a fresh dounce tissue grinder. After a second round of homogenization (tight pestle) and centrifugation, supernatants were aliquoted and stored at −80°C. Concentration of virus stocks was determined by plaque-assay. For passaging MCMV, lower virus doses (500-2.500 pfu) were used. For infection experiments, 3 times passaged virus stocks were used.
Infections
Upon adoptive transfer experiments, Rag2–/– Il2rg–/–, Klra8–/– and NKp46iCre x R26-LSL-iDTA mice were infected i.p. with 100 pfu MCMV-wt (3rd passage). C57BL/6 and Xcr1Venus mice were infected with 2.000 pfu MCMV-wt (3rd passage) by intraperitoneal injection. Klra8+/+ and Klra8–/– mice were infected i.p. with 2.000 pfu MCMV-wt or MCMV-ie2-SIINFEKL-GFP (3rd passage) for confocal immunofluorescence imaging. Klra8–/–: Batf3+/+ or Klra8–/–: Batf3–/– mixed bone marrow chimeras, Klra8+/+ and Klra8–/– mice were infected with MCMV-ie2-SIINFEKL (500 pfu, 3rd passage) by intraperitoneal injection.
Flow cytometry
Spleens and lymph nodes were harvested and mashed through a 40 μm cell strainer to generate a single-cell suspension. Livers were homogenized by using a dounce tissue grinder. The liver cell suspension was pelleted and resuspended in 40% Percoll (Sigma-Aldrich). Separation of the lymphocyte fraction was performed by centrifugation at 2600 rpm, RT for 20 min (accel = 5, decel = 0). For blood screenings, 100 μl blood were collected in heparin tubes. Following red blood cell lysis, cells were incubated with EMA and anti-mouse CD16/CD32 for 20 min at 4°C exposed to light. Afterward, cells were washed with FACS buffer (PBS with 0,5% BSA and 2 mM EDTA) and stained with respective antibodies for 30 min at 4°C in the dark. After washing with FACS buffer, optional transcription factor staining was performed according to the manufacturer’s instructions using eBioscience Foxp3 Transcription Factor Staining Buffer Set (Thermo Fisher) or True-Nuclear Transcription Factor Buffer Set (Biolegend). After final washing, cells were resuspended in FACS buffer containing 1% paraformaldehyde and analyzed by flow cytometry (Cytoflex S or LX, Beckman Coulter). FlowJo software (FlowJo LLC) was used for analysis.
Stimulation with PMA/ionomycin
Ly49H+ NK cells were sorted from CD45.1 splenocytes as described into a 96-well U-bottom plate containing 400.000 C57BL/6 splenocytes per well in cRPMI. After sorting, Phorbol-12-myristat-13-acetat (PMA) and ionomycin (Sigma-Aldrich) were added at a final concentration of 20 ng/mL and 1.25 μg/mL, respectively. An equal volume of DMSO, diluted 1/10 in cRPMI, was added to the negative control. After one h, Golgi Plug™ (Beckton Dickinson) was added 1/1000 to the culture. 4 h later, cells were washed with FACS buffer and life/dead discrimination as well as staining of surface molecules was performed as described. Intracellular cytokine staining was done according to the manufacturer’s instructions using Cytofix/Cytoperm™ Fixation/ Permeabilization Solution Kit (Beckton Dickinson).
Ex vivo cytokine staining
For restimulation with plate-bound antibodies, a 96-well ELISA plate (Nunc) was coated with 1 μg/mL purified anti-mouse NK1.1 overnight at 4°C. Afterward, 5x106 freshly isolated splenocytes in cRPMI were added per well. Identical numbers of splenocytes were added to uncoated wells without restimulation. After 1 h, Golgi Plug™ was added and intracellular cytokine staining was performed as described.
Ba/F3 co-culture
Ba/F3-neo (WT) and Ba/F3-m157 were a kind gift from Hisashi Arase and Lewis Lanier. 2.000 CD27+ CD62L+ or CD27+ CD62L- Ly49H+ NK cells were sorted from spleens of CD45.1 mice and co-incubated with 50.000 Ba/F3 cells for 24 h in cRPMI. After co-culture, cells were stained and analyzed by flow cytometry.
Killing
RMA and RMA-S cells (a kind gift from Joseph Sun) were retrovirally transduced with fluorescent proteins GFP and BFP, respectively, as described. 3.000 CD27+ CD62L+ or CD27+ CD62L- Ly49H+ NK cells were sorted from spleens of C57BL/6 mice 24 h p.i. with MCMV and co-incubated with 1.500 GFP+ RMA and 1.500 BFP+ RMA-S cells mice for 42 h in cRPMI. After co-culture, PI was added for life/dead discrimination and the number of living GFP+ and BFP+ cells was analyzed by flow cytometry.
Mixed bone marrow chimeras
Bone marrow cells were isolated from femur and tibia of 10-15 weeks old Klra8–/–, Batf3–/– and Batf3+/+ mice. A total number of 5 × 106 bone marrow cells (as 1:1 mixture of the respective bone marrows) in 200 μl FCS were injected i.v. into irradiated Klra8–/– recipient mice (two times 4.5 Gy, with a resting period of 4 h). Reconstitution of the Ly49H+ NK cell compartment was analyzed after 6 weeks by blood sampling.
Priming of OT-1 T cells
Klra8–/–: Batf3+/+ and Klra8–/–: Batf3–/– mixed bone marrow chimeras, Klra8+/+ and Klra8–/– mice were infected with MCMV-ie2-SIINFEKL. CD45.1+ Rag1–/– OT-1 splenocytes were harvested and stained with antibodies directed against CD8 and CD44 and PI for life/dead discrimination. Absolute number of naive OT-1 T cells (CD8+ CD44low) in the sample was determined by flow cytometry and concentration was adjusted to 1x106 naive OT-1 T cells per 200 μl FCS. 1x106 naive OT-1 T cells were injected i.p. at 24, 48, 72 or 96 h p.i. After 21 h, spleens were harvested and analyzed by flow cytometry as described.
Retransfer of OT-1 T cells
1x106 CD45.1+/+ and CD45.1+/– Rag1–/– OT-1 splenocytes were adoptively transferred into Klra8–/– and Klra8+/+ mice, respectively, as described above. Mice were subsequently infected with MCMV-ie2-SIINFEKL. 2 days p.i., splenocytes were harvested and CD45.1+ cells were enriched by flow cytometric sorting (MoFlo Astrios, Beckman Coulter). Afterward, enriched cells were stained with respective antibodies and CD19– CD4– CD45.1+ CD8+ OT-1 T cells were sorted (MoFlo Astrios, Beckman Coulter) into a 96-well v-bottom plate containing 400.000 C57BL/6 splenocytes in 200 μl FCS. Identical numbers of CD45.1+/+ OT-1 cells from Klra8–/– and CD45.1+/– OT-1 cells from Klra8+/+ mice were sorted into the same well and co-transferred into infection-matched C57BL/6 recipient mice. Expansion of OT-1 cells was monitored by flow cytometric analysis of peripheral blood samples.
Bulk RNA sequencing
Samples were sorted and immediately frozen in liquid N2 and stored at −80°C until sequencing. RNA was isolated using the Arcturus Pico Pure Isolation Kit according to the manufacturer’s recommendations. Subsequently, cDNA was generated using the SMART Seq V4 Ultra low input RNA Kit by TaKaRa followed by library preparation with the Nextera XT DNA Kit by Illumina according to the manufacturer’s recommendations. Samples were then sequenced on a HiSeq 4000 (Illumina).
Generation of gene expression count matrix
The gene expression count matrix was obtained using Salmon (version 0.9.1; Genome Reference Consortium Mouse Build 38). The R package DESeq2 (version 1.26.0) was used to perform size factor normalization.
scRNaseq
NK cells were sorted and applied to droplet-based scRNaseq using the Chromium Single Cell 3′ Library & Gel Bead Kit (10x genomics). Sequencing was performed after library preparation with a Chromium Single Cell 3′ Library Construction Kit on a HiSeq 4000, HiSeq 2500 or NovaSeq 6000 (Illumina). For selected experiments hashtag antibodies were used (TotalSeq B 301, 302, 304, 305, 306 and 307). Preparation of antibody feature barcode libraries was done using Chromium Single Cell 3′ Feature Barcode Library Kit (10x genomics). Human scRNaseq data were derived from (Crinier et al., 2018) (donor 1, blood and spleen).
Generation of gene expression count matrix
The software Cell Ranger (version 5.0.1; 10x Genomics) was used for demultiplexing and alignment (mouse reference genome GRCm38 release 84) using default parameters. Velocyto (version 0.17.17) performed counting of spliced and un-spliced RNA molecules.
Pre-processing and quality control
Pre-processing and quality control of the data was carried out using the Python software package SCANPY. Cells that expressed low gene numbers, reading depth (counts) or high mitochondrial or ribosomal gene percentage were removed, the borders for these parameters were adapted according to the experiment. Cell cycle, mitochondrial genes and facultatively counts and gene numbers were regressed out. For Figure 3E counts were regressed out due to low sequencing depth. More detail can be found in the provided MetaData sheets.
Dimensionality reduction, clustering and visualization
SCANPY was also used to perform dimensionality reduction and clustering. The neighborhood graphs were based on n = 12-40 principal components and 30-80 neighbors. Clustering was performed using the Leiden algorithm with resolution r = 0.4 - 0.9 depending on the specific biological question. UMAP dimensionality reduction was computed using SCANPY’s default parameters. To answer the question whether Ly49 genes influence differentiation of NK cells in Figure 3E, all Klra genes were excluded before dimensionality reduction.
Trajectory Inference
The software tool scvelo (version 0.1.25) was used to compute RNA velocities within a deterministic model.
Confocal Immunofluorescence imaging and image analysis
Immunofluorescence microscopy was performed as described previously with minor modifications (Böttcher et al., 2018). In brief, spleens were fixed in Antigenfix solution (Diapath), dehydrated in 30% sucrose prior to embedding in TissueTek OCT freezing medium (Sakura Finetek) and stored at −80 °C. 30μm sections were permeabilized, blocked, and stained in 0.1M Tris (AppliChem) supplemented with 1% BSA, 0.3% Triton X-100 (AppliChem) and normal mouse serum (Sigma). Serial sections were prepared and visually inspected by epifluorescence light microscopy before acquisition of representative areas by confocal microscopy. The following antibodies were used for staining: anti-CD3 (clone 17A2, Biolegend), anti-CD160 (sheep polyclonal, R&D Systems), anti-Collagen IV (rabbit polyclonal, Abcam), anti-GFP (cross-reactive with Venus, rabbit polyclonal, Life Technologies), anti-Ly49H (clone 3D10, Life Technologies), anti-NK1.1 (clone PK136, Biolegend), anti-XCR1 (clone ZET, Biolegend). Anti-Ly49H, anti-NK1.1 and anti-XCR1 antibodies were injected i.v. prior to organ fixation. Therefore, 3 μg of the respective antibodies were diluted in 200 μl PBS and mice were sacrificed 5 min after injection.
Stained sections were mounted in Mowiol (Sigma) and analyzed on an inverted TCS SP8 confocal microscope (Leica) for tiled imaging using a HC PL APO CS2 40x/1.30NA or HC PL APO CORR CS2 63x/1.30NA objective. Tiled xy stacks were acquired with 1μm or 1.5μm z spacing to provide three-dimensional image volumes of 20μm or 30μm depth, depending on the experiment. 3D Image analysis was performed using Imaris 9.5 software (Bitplane) following adaptive image deconvolution using the Leica TCS SP8 LIGHTNING tool. Automated analyses using the Imaris surface generation tool was used to reconstruct 3D surfaces for individual cDC1 and NK cells. For histocytometric analysis of NK cell subsets, statistics for mean voxel intensities for fluorescence channels and positional information for surface objects were exported into Excel (Microsoft) and plotted in Flowjo software (TreeStar Inc) after conversion into CSV files. Following gating in Flowjo, annotation of Imaris surface IDs was used for visualization of NK cell subsets in microscopic images. Clustered cDC1 were identified using the spot-to-spot closest distance function on cDC1 surfaces (Distance ≤5μm). Clusters of less than three cells were removed manually.
Parabiosis
C57BL6 mice (CD45.2) were parabiotically joined to congenic B6.SJL (CD45.1) mice. Parabiosis was performed in accordance to previously published protocols (Coleman and Hummel, 1969; Harris, 1997). Briefly, mice were placed under isoflurane anesthesia (1.5%–2%), and a longitudinal incision was made along one flank of each mouse, around 2 inches from the elbow to the knee. Mice were joined by the femurs and humeri by suturing, and skin was joined using sutures and wound clips. Meloxicam (2 mg/kg) was administered subcutaneously immediately prior to surgery and every 24 h after surgery for three days for analgesia.
Measurement of MCMV titers
Spleens were harvested from MCMV infected Klra8–/– and Klra8+/+ mice at indicated time points and virus titration was done as described before by (Chaudhry et al., 2020). In brief, MEFs were used to titrate the organ homogenate diluted in DMEM. The infection was enhanced by centrifugation for 30 min at 1000 x g. The plates were incubated for another 30 min at 37°C, 5% CO2. The organ homogenates were then removed and cells layered with methylcellulose. The plates were incubated for 4 days at 37°C, 5% CO2 and the plaques were quantified by visual inspection under inverse microscope.
Quantification and statistical analysis
Prism software (v9.1.0, GraphPad) was used for quantification and statistical analysis. Statistical details of each experiment can be found at the end of the respective figure legend. Significance is defined as ∗p value < 0.05, ∗∗p value < 0.01,∗∗∗p value < 0.001, ∗∗∗∗p value < 0.0001. Normality tests were performed to decide whether to use parametric or nonparametric tests, where applicable.
Acknowledgements
This work was supported by the European Research Council starting grant (949719 – SCIMAP) to V.R.B.; the Else Kröner-Fresenius-Stiftung (EKFS 2019_A91) to V.R.B.; the German Research Foundation (DFG) – SFB 1054 (210592381) to V.R.B. and D.H.B.; an International Junior Research Group grant (N-LW-2016-370) from the Elite Network of Bavaria, Bavarian State Ministry of Science and the Arts to J.P.B.; the DFG – (424926990) to J.P.B.; the BMBF project Quan-T cell (e:Med initiative on Systems Medicine, FKZ 01ZX1505) to M.F.; the BMBF project TIDY (Computational Life Sciences, FKZ 031L0170A) to T.H.; the Helmholtz association project (PIE-008) to L.C.S.; and the DFG – TRR 179 (031L0170A) to M.S. In memoriam of J.E. who was a Cancer Research Institute/Hearst Foundation postdoctoral fellow and died in October 2020.
Author contributions
V.R.B. and S.G. designed and supervised the study. V.R.B., S.G., and S.F. wrote the manuscript. S.G., S.F., J.P.B., P.M., L.O.P, J.L. I.H., Q.Z., and M.Z.C. conducted experiments. I.A. and M.S. performed cell sorting. M.F. and S.J. performed data analysis. L.C.-S. and M.Z.C. provided virus preparations. Q.Z and T.H. performed sequencing experiments. J.E. and G.D.V. contributed parabiosis experiments. D.H.B., J.C.S., and G.G. contributed to the study design and provided critical discussion of the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: August 25, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.immuni.2021.08.002.
Contributor Information
Veit R. Buchholz, Email: veit.buchholz@tum.de.
Simon Grassmann, Email: simon.grassmann@tum.de.
Supplemental information
Data and code availability
All sequencing data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. This paper analyzes existing, publicly available data. The accession numbers for the datasets are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Adams N.M., Geary C.D., Santosa E.K., Lumaquin D., Le Luduec J.B., Sottile R., van der Ploeg K., Hsu J., Whitlock B.M., Jackson B.T. Cytomegalovirus Infection Drives Avidity Selection of Natural Killer Cells. Immunity. 2019;50:1381–1390. doi: 10.1016/j.immuni.2019.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams N.M., Grassmann S., Sun J.C. Clonal expansion of innate and adaptive lymphocytes. Nat. Rev. Immunol. 2020;20:694–707. doi: 10.1038/s41577-020-0307-4. [DOI] [PubMed] [Google Scholar]
- Andrews D.M., Scalzo A.A., Yokoyama W.M., Smyth M.J., Degli-Esposti M.A. Functional interactions between dendritic cells and NK cells during viral infection. Nat. Immunol. 2003;4:175–181. doi: 10.1038/ni880. [DOI] [PubMed] [Google Scholar]
- Arase H., Mocarski E.S., Campbell A.E., Hill A.B., Lanier L.L. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. 2002;296:1323–1326. doi: 10.1126/science.1070884. [DOI] [PubMed] [Google Scholar]
- Babić M., Pyzik M., Zafirova B., Mitrović M., Butorac V., Lanier L.L., Krmpotić A., Vidal S.M., Jonjić S. Cytomegalovirus immunoevasin reveals the physiological role of “missing self” recognition in natural killer cell dependent virus control in vivo. J. Exp. Med. 2010;207:2663–2673. doi: 10.1084/jem.20100921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai A., Hu H., Yeung M., Chen J. Kruppel-like factor 2 controls T cell trafficking by activating L-selectin (CD62L) and sphingosine-1-phosphate receptor 1 transcription. J. Immunol. 2007;178:7632–7639. doi: 10.4049/jimmunol.178.12.7632. [DOI] [PubMed] [Google Scholar]
- Böttcher J.P., Bonavita E., Chakravarty P., Blees H., Cabeza-Cabrerizo M., Sammicheli S., Rogers N.C., Sahai E., Zelenay S., Reis e Sousa C. NK Cells Stimulate Recruitment of cDC1 into the Tumor Microenvironment Promoting Cancer Immune Control. Cell. 2018;172:1022–1037. doi: 10.1016/j.cell.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M.G., Dokun A.O., Heusel J.W., Smith H.R., Beckman D.L., Blattenberger E.A., Dubbelde C.E., Stone L.R., Scalzo A.A., Yokoyama W.M. Vital involvement of a natural killer cell activation receptor in resistance to viral infection. Science. 2001;292:934–937. doi: 10.1126/science.1060042. [DOI] [PubMed] [Google Scholar]
- Busche A., Jirmo A.C., Welten S.P., Zischke J., Noack J., Constabel H., Gatzke A.K., Keyser K.A., Arens R., Behrens G.M., Messerle M. Priming of CD8+ T cells against cytomegalovirus-encoded antigens is dominated by cross-presentation. J. Immunol. 2013;190:2767–2777. doi: 10.4049/jimmunol.1200966. [DOI] [PubMed] [Google Scholar]
- Cella M., Gamini R., Sécca C., Collins P.L., Zhao S., Peng V., Robinette M.L., Schettini J., Zaitsev K., Gordon W. Subsets of ILC3-ILC1-like cells generate a diversity spectrum of innate lymphoid cells in human mucosal tissues. Nat. Immunol. 2019;20:980–991. doi: 10.1038/s41590-019-0425-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A., Hong D.-L., Atzberger A., Kollnberger S., Filer A.D., Buckley C.D., McMichael A., Enver T., Bowness P. CD56bright human NK cells differentiate into CD56dim cells: role of contact with peripheral fibroblasts. J. Immunol. 2007;179:89–94. doi: 10.4049/jimmunol.179.1.89. [DOI] [PubMed] [Google Scholar]
- Chaudhry M.Z., Casalegno-Garduno R., Sitnik K.M., Kasmapour B., Pulm A.K., Brizic I., Eiz-Vesper B., Moosmann A., Jonjic S., Mocarski E.S., Cicin-Sain L. Cytomegalovirus inhibition of extrinsic apoptosis determines fitness and resistance to cytotoxic CD8 T cells. Proc. Natl. Acad. Sci. USA. 2020;117:12961–12968. doi: 10.1073/pnas.1914667117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman D.L., Hummel K.P. Effects of parabiosis of normal with genetically diabetic mice. Am. J. Physiol. 1969;217:1298–1304. doi: 10.1152/ajplegacy.1969.217.5.1298. [DOI] [PubMed] [Google Scholar]
- Collins P.L., Cella M., Porter S.I., Li S., Gurewitz G.L., Hong H.S., Johnson R.P., Oltz E.M., Colonna M. Gene Regulatory Programs Conferring Phenotypic Identities to Human NK Cells. Cell. 2019;176:348–360. doi: 10.1016/j.cell.2018.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez V.S., Fuchs A., Cella M., Gilfillan S., Colonna M. Cutting edge: Salivary gland NK cells develop independently of Nfil3 in steady-state. J. Immunol. 2014;192:4487–4491. doi: 10.4049/jimmunol.1303469. [DOI] [PubMed] [Google Scholar]
- Cortez V.S., Ulland T.K., Cervantes-Barragan L., Bando J.K., Robinette M.L., Wang Q., White A.J., Gilfillan S., Cella M., Colonna M. SMAD4 impedes the conversion of NK cells into ILC1-like cells by curtailing non-canonical TGF-β signaling. Nat. Immunol. 2017;18:995–1003. doi: 10.1038/ni.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crinier A., Milpied P., Escalière B., Piperoglou C., Galluso J., Balsamo A., Spinelli L., Cervera-Marzal I., Ebbo M., Girard-Madoux M. High-Dimensional Single-Cell Analysis Identifies Organ-Specific Signatures and Conserved NK Cell Subsets in Humans and Mice. Immunity. 2018;49:971–986. doi: 10.1016/j.immuni.2018.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner B.G., Dorner M.B., Zhou X., Opitz C., Mora A., Güttler S., Hutloff A., Mages H.W., Ranke K., Schaefer M. Selective expression of the chemokine receptor XCR1 on cross-presenting dendritic cells determines cooperation with CD8+ T cells. Immunity. 2009;31:823–833. doi: 10.1016/j.immuni.2009.08.027. [DOI] [PubMed] [Google Scholar]
- Erick T.K., Anderson C.K., Reilly E.C., Wands J.R., Brossay L. NFIL3 Expression Distinguishes Tissue-Resident NK Cells and Conventional NK-like Cells in the Mouse Submandibular Glands. J. Immunol. 2016;197:2485–2491. doi: 10.4049/jimmunol.1601099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs A., Vermi W., Lee J.S., Lonardi S., Gilfillan S., Newberry R.D., Cella M., Colonna M. Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-γ-producing cells. Immunity. 2013;38:769–781. doi: 10.1016/j.immuni.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galibert L., Diemer G.S., Liu Z., Johnson R.S., Smith J.L., Walzer T., Comeau M.R., Rauch C.T., Wolfson M.F., Sorensen R.A. Nectin-like protein 2 defines a subset of T-cell zone dendritic cells and is a ligand for class-I-restricted T-cell-associated molecule. J. Biol. Chem. 2005;280:21955–21964. doi: 10.1074/jbc.M502095200. [DOI] [PubMed] [Google Scholar]
- Gao Y., Souza-Fonseca-Guimaraes F., Bald T., Ng S.S., Young A., Ngiow S.F., Rautela J., Straube J., Waddell N., Blake S.J. Tumor immunoevasion by the conversion of effector NK cells into type 1 innate lymphoid cells. Nat. Immunol. 2017;18:1004–1015. doi: 10.1038/ni.3800. [DOI] [PubMed] [Google Scholar]
- Gasteiger G., Fan X., Dikiy S., Lee S.Y., Rudensky A.Y. Tissue residency of innate lymphoid cells in lymphoid and nonlymphoid organs. Science. 2015;350:981–985. doi: 10.1126/science.aac9593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassmann S., Pachmayr L.O., Leube J., Mihatsch L., Andrae I., Flommersfeld S., Oduro J., Cicin-Sain L., Schiemann M., Flossdorf M., Buchholz V.R. Distinct Surface Expression of Activating Receptor Ly49H Drives Differential Expansion of NK Cell Clones upon Murine Cytomegalovirus Infection. Immunity. 2019;50:1391–1400. doi: 10.1016/j.immuni.2019.04.015. [DOI] [PubMed] [Google Scholar]
- Grassmann S., Mihatsch L., Mir J., Kazeroonian A., Rahimi R., Flommersfeld S., Schober K., Hensel I., Leube J., Pachmayr L.O. Early emergence of T central memory precursors programs clonal dominance during chronic viral infection. Nat. Immunol. 2020;21:1563–1573. doi: 10.1038/s41590-020-00807-y. [DOI] [PubMed] [Google Scholar]
- Hammer Q., Rückert T., Borst E.M., Dunst J., Haubner A., Durek P., Heinrich F., Gasparoni G., Babic M., Tomic A. Peptide-specific recognition of human cytomegalovirus strains controls adaptive natural killer cells. Nat. Immunol. 2018;19:453–463. doi: 10.1038/s41590-018-0082-6. [DOI] [PubMed] [Google Scholar]
- Harmon C., Robinson M.W., Fahey R., Whelan S., Houlihan D.D., Geoghegan J., O’Farrelly C. Tissue-resident Eomes(hi) T-bet(lo) CD56(bright) NK cells with reduced proinflammatory potential are enriched in the adult human liver. Eur. J. Immunol. 2016;46:2111–2120. doi: 10.1002/eji.201646559. [DOI] [PubMed] [Google Scholar]
- Harris R.B. Loss of body fat in lean parabiotic partners of ob/ob mice. Am. J. Physiol. 1997;272:R1809–R1815. doi: 10.1152/ajpregu.1997.272.6.R1809. [DOI] [PubMed] [Google Scholar]
- Hildner K., Edelson B.T., Purtha W.E., Diamond M., Matsushita H., Kohyama M., Calderon B., Schraml B.U., Unanue E.R., Diamond M.S. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 2008;322:1097–1100. doi: 10.1126/science.1164206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeevan-Raj B., Gehrig J., Charmoy M., Chennupati V., Grandclément C., Angelino P., Delorenzi M., Held W. The Transcription Factor Tcf1 Contributes to Normal NK Cell Development and Function by Limiting the Expression of Granzymes. Cell Rep. 2017;20:613–626. doi: 10.1016/j.celrep.2017.06.071. [DOI] [PubMed] [Google Scholar]
- Kamimura Y., Lanier L.L. Homeostatic control of memory cell progenitors in the natural killer cell lineage. Cell Rep. 2015;10:280–291. doi: 10.1016/j.celrep.2014.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Manno G., Soldatov R., Zeisel A., Braun E., Hochgerner H., Petukhov V., Lidschreiber K., Kastriti M.E., Lönnerberg P., Furlan A. RNA velocity of single cells. Nature. 2018;560:494–498. doi: 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.-H., Girard S., Macina D., Busà M., Zafer A., Belouchi A., Gros P., Vidal S.M. Susceptibility to mouse cytomegalovirus is associated with deletion of an activating natural killer cell receptor of the C-type lectin superfamily. Nat. Genet. 2001;28:42–45. doi: 10.1038/ng0501-42. [DOI] [PubMed] [Google Scholar]
- Melsen J.E., Lugthart G., Lankester A.C., Schilham M.W. Human Circulating and Tissue-Resident CD56(bright) Natural Killer Cell Populations. Front. Immunol. 2016;7:262. doi: 10.3389/fimmu.2016.00262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrović M., Arapović J., Jordan S., Fodil-Cornu N., Ebert S., Vidal S.M., Krmpotić A., Reddehase M.J., Jonjić S. The NK cell response to mouse cytomegalovirus infection affects the level and kinetics of the early CD8(+) T-cell response. J. Virol. 2012;86:2165–2175. doi: 10.1128/JVI.06042-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narni-Mancinelli E., Chaix J., Fenis A., Kerdiles Y.M., Yessaad N., Reynders A., Gregoire C., Luche H., Ugolini S., Tomasello E. Fate mapping analysis of lymphoid cells expressing the NKp46 cell surface receptor. Proc. Natl. Acad. Sci. USA. 2011;108:18324–18329. doi: 10.1073/pnas.1112064108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary J.G., Goodarzi M., Drayton D.L., von Andrian U.H. T cell- and B cell-independent adaptive immunity mediated by natural killer cells. Nat. Immunol. 2006;7:507–516. doi: 10.1038/ni1332. [DOI] [PubMed] [Google Scholar]
- Paust S., Gill H.S., Wang B.-Z., Flynn M.P., Moseman E.A., Senman B., Szczepanik M., Telenti A., Askenase P.W., Compans R.W., von Andrian U.H. Critical role for the chemokine receptor CXCR6 in NK cell-mediated antigen-specific memory of haptens and viruses. Nat. Immunol. 2010;11:1127–1135. doi: 10.1038/ni.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli A., Michel T., Thérésine M., Andrès E., Hentges F., Zimmer J. CD56bright natural killer (NK) cells: an important NK cell subset. Immunology. 2009;126:458–465. doi: 10.1111/j.1365-2567.2008.03027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redeker A., Welten S.P.M., Arens R. Viral inoculum dose impacts memory T-cell inflation. Eur. J. Immunol. 2014;44:1046–1057. doi: 10.1002/eji.201343946. [DOI] [PubMed] [Google Scholar]
- Sallusto F., Lanzavecchia A. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor alpha. J. Exp. Med. 1994;179:1109–1118. doi: 10.1084/jem.179.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H.R., Heusel J.W., Mehta I.K., Kim S., Dorner B.G., Naidenko O.V., Iizuka K., Furukawa H., Beckman D.L., Pingel J.T. Recognition of a virus-encoded ligand by a natural killer cell activation receptor. Proc. Natl. Acad. Sci. USA. 2002;99:8826–8831. doi: 10.1073/pnas.092258599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sojka D.K., Plougastel-Douglas B., Yang L., Pak-Wittel M.A., Artyomov M.N., Ivanova Y., Zhong C., Chase J.M., Rothman P.B., Yu J. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. eLife. 2014;3:e01659. doi: 10.7554/eLife.01659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J.C., Beilke J.N., Lanier L.L. Adaptive immune features of natural killer cells. Nature. 2009;457:557–561. doi: 10.1038/nature07665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi A., Itoh Y., Takumi A., Ishihara C., Arase N., Yokosuka T., Koseki H., Yamasaki S., Takai Y., Miyoshi J. CRTAM confers late-stage activation of CD8+ T cells to regulate retention within lymph node. J. Immunol. 2009;183:4220–4228. doi: 10.4049/jimmunol.0901248. [DOI] [PubMed] [Google Scholar]
- Vivier E., Artis D., Colonna M., Diefenbach A., Di Santo J.P., Eberl G., Koyasu S., Locksley R.M., McKenzie A.N.J., Mebius R.E. Innate Lymphoid Cells: 10 Years On. Cell. 2018;174:1054–1066. doi: 10.1016/j.cell.2018.07.017. [DOI] [PubMed] [Google Scholar]
- Weizman O.-E., Adams N.M., Schuster I.S., Krishna C., Pritykin Y., Lau C., Degli-Esposti M.A., Leslie C.S., Sun J.C., O’Sullivan T.E. ILC1 Confer Early Host Protection at Initial Sites of Viral Infection. Cell. 2017;171:795–808. doi: 10.1016/j.cell.2017.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Espinoza D.A., Koelle S.J., Yang D., Truitt L., Schlums H., Lafont B.A., Davidson-Moncada J.K., Lu R., Kaur A. Clonal expansion and compartmentalized maintenance of rhesus macaque NK cell subsets. Science immunology. 2018;3:eaat9781. doi: 10.1126/sciimmunol.aat9781. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. This paper analyzes existing, publicly available data. The accession numbers for the datasets are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.