FleN uses allostery and structural remodeling to fine-tune transcription activation by FleQ for monotrichous phenotype.
Abstract
Diverse sigma factors associate with the RNA polymerase (RNAP) core enzyme to initiate transcription of specific target genes in bacteria. σ54-Mediated transcription uses AAA+ activators that utilize their ATPase activity for transcription initiation. FleQ is a σ54-dependent master transcriptional regulator of flagellar genes in Pseudomonas aeruginosa. The ATPase activity of FleQ is regulated via a P-loop ATPase, FleN, through protein-protein interaction. We report a high-resolution crystal structure of the AAA+ domain of FleQ in complex with antiactivator FleN. The data reveal that FleN allosterically prevents ATP binding to FleQ. Furthermore, FleN remodels the region of FleQ essential for engagement with σ54 for transcription initiation. Disruption of the conserved protein-protein interface, by mutation, shows motility and transcription defects in vivo and multiflagellate phenotype. Our study provides a detailed mechanism used by monoflagellate bacteria to fine-tune the expression of flagellar genes to form and maintain a single flagellum.
INTRODUCTION
Transcription initiation is the pivotal regulatory checkpoint of gene expression in bacteria. One of the modes of regulation is through the widely distributed yet unique RNA polymerase (RNAP) subunit σ54 associated with the expression of genes required for a diverse range of physiological processes. The σ54 RNAP recognizes −12 and −24 elements in the promoter and forms a closed complex, where the DNA is still double stranded (1, 2). The adenosine triphosphatase (ATPase) activity of an upstream bound activator is essential for isomerization of the closed complex into transcriptionally competent open complex, where the two strands of the DNA separate to form the transcription bubble. The activators or the enhancer-binding proteins (EBPs) assemble into active hexamers that hydrolyze adenosine 5′-triphosphate (ATP), contact the σ54 RNAP holoenzyme, and drive conformational changes within the polymerase-promoter complex that are crucial for transcription initiation (3, 4).
FleQ is one such AAA+ (ATPase associated with various cellular activities) EBP that regulates the expression of about 50 genes responsible for flagellar biosynthesis, assembly, and regulation in conjunction with σ54 in Pseudomonas aeruginosa. Deletion of fleQ results in down-regulation of expression of flagellar genes, leading to nonmotile phenotype (5–8). The domain architecture of FleQ resembles the other σ54 transcription activators and includes N-terminal regulatory domain (REC), central AAA+ ATPase domain, and C-terminal DNA binding domain harboring the helix-turn-helix motif. The central domain is the most conserved of all the three domains and contains conserved motifs such as Walker A, involved in ATP binding, and Walker B, involved in hydrolysis (2, 9–11). The central domain is further composed of two subdomains, the α/β subdomain at the N terminus followed by the α-helical subdomain toward the C terminus. The nucleotide binds at the cleft between the two subdomains (9). In addition, the central domain contains two surface-exposed loops L1 and L2. The L1 loop includes the highly conserved GAFTGA sequence motif. It has been demonstrated for other bacterial EBPs (bEBPs) that the L1/L2 loops are vital for direct engagement with σ54 as well as the promoter. This interaction is critical for DNA strand separation, consequently leading to transcription initiation (12–14).
Despite pronounced sequence similarities, FleQ has distinct features that differentiate it from other σ54 activators. While other activators exist in inactive dimeric forms and, upon phosphorylation of the REC domain, assemble into active hexamers capable of ATP hydrolysis, by contrast, FleQ is not regulated through phosphorylation. It lacks the conserved aspartic acid residue that is known to be phosphorylated in the REC domain of other EBPs (12, 15, 16). On the contrary, the ATPase activity of FleQ is constitutively present and is negatively regulated by direct interaction with another P-loop ATPase, FleN. FleN is known to regulate the number of flagella and is key for establishing monotrichous phenotype in P. aeruginosa. Disruption of fleN leads to multiflagellate phenotype with motility and chemotactic defects (17). It has been shown that ATP-mediated dimerization in FleN is essential for its antagonist effect over FleQ. It exerts this effect without affecting the DNA binding ability of FleQ (18, 19). However, the lack of structural data on the protein-protein complex impedes a detailed understanding of the mechanism of transcription inhibition by antiactivators of σ54 regulators in general and by FleN in particular.
To decipher the detailed structural mechanism of transcription regulation, we determined the crystal structure of AAA+ domain of FleQ bound to FleN at 1.98 Å. The complex crystallized as a heterotetramer. Comparison of the complex structure with the earlier determined structures of FleN and ATPase domain of FleQ reinforces that the dimer of FleN is necessary for its antagonistic activity against FleQ and it allosterically inhibits the ATPase activity of the latter. There are drastic structural changes in FleQ upon binding of FleN such that the ATP binding site is contorted. Furthermore, the structure reveals that the C terminus of FleN is essential for binding with the activator and interacts specifically with the polymerase interacting loop of FleQ, conceivably blocking its association with σ54 RNAP and arresting transcription initiation. Perturbations in the conserved interaction interface through site-directed mutagenesis and in vivo complementation assays show transcription and motility defects with multiflagellate phenotype. Overall, we demonstrate that the antiactivator FleN uses a combination of allostery and structural remodeling to fine-tune transcription activation by master regulator FleQ for monotrichous phenotype in P. aeruginosa.
RESULTS
AAA+ ATPase domain of FleQ is necessary for interaction with FleN
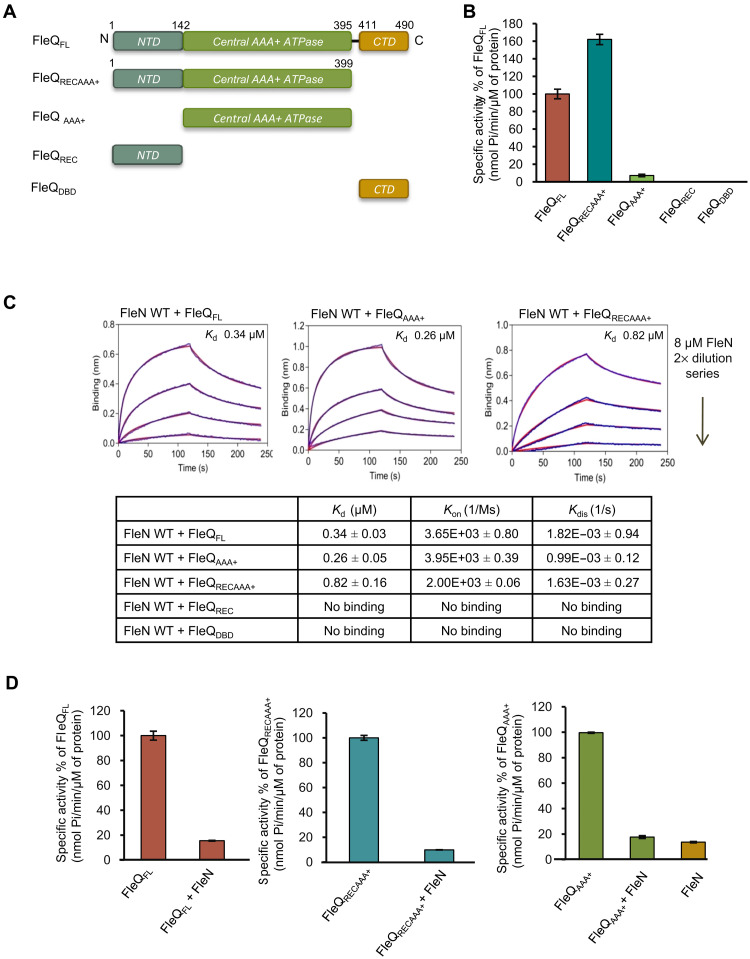
To dissect the structural and functional role of different domains of FleQ, we engineered a series of constructs of FleQ containing either the N-terminal domain (NTD) (FleQREC, residues 1 to 142) or the central domain (FleQAAA+, residues 142 to 395) or the C-terminal domain (CTD) (FleQDBD, residues 411 to 490) or the Central domain with NTD (FleQRECAAA+, residues 1 to 399) (Fig. 1A). FleQAAA+ shows a weak ATPase activity compared to wild-type (WT) protein. As expected, FleQREC and FleQDBD do not exhibit any ATPase activity (Fig. 1B). FleQRECAAA+ shows around 1.5-fold higher ATPase activity compared to full-length protein.
Fig. 1. Interaction of different domains of FleQ with FleN.
(A) Schematic representation of series of constructs of FleQ used in the manuscript. The domains FleQREC, FleQAAA+, and FleQDBD are shown in gray, green, and pale orange colors, respectively. (B) The ATPase activity of FleQFL and various domain constructs (1 μM) was measured in the presence of 2 mM ATP. The specific activities of ATP hydrolysis (nmol Pi/min per μM protein) are plotted in percentage of FleQFL on the y axis for each construct. (C) BLI curves showing interaction of FleN with either FleQFL or FleQAAA+ or FleQRECAAA+. The x and y axes represent time (in seconds) and nanometer shift on binding, respectively. The first part of the curve (0 to 120 s) represents association, and the second part of the curve (from 120 to 240 s) represents dissociation. The curves in blue and red colors indicate experimental and fitting data, respectively. The dissociation constant (KD, μM) of interaction between FleN and different constructs of FleQ is shown in the table. Rate of association and dissociation is shown as Kon (1/Ms) and Kdis (1/s), respectively. No binding was recorded in case of FleQREC and FleQDBD constructs, as no significant shift in nanometers on the y axis was observed. (D) Inhibition of the ATPase activity of FleQFL, FleQRECAAA+, and FleQAAA+ in the presence of FleN WT is shown as a specific activity (nmol Pi/min per μM protein) on the y axis. For the experiments, 1 μM FleQ or its constructs, 5 μM FleN WT, and 2 mM ATP were used.
Next, we characterized the binding affinity between FleQ and FleN using biolayer interferometry (BLI) assay in the presence of a nonhydrolyzable ATP analog, ATPγS (adenosine 5′-[γ-thio] triphosphate). FleQ interacts with FleN with a KD (dissociation equilibrium constant) of 0.34 μM, confirming a direct interaction between the two proteins. We further mapped the FleN binding domain of FleQ using the designed constructs through BLI. The assay revealed that the binding affinity of FleQAAA+ and FleQRECAAA+ with FleN is similar to FleQFL with a KD of 0.26 and 0.82 μM, respectively (Fig. 1C). However, FleQREC and FleQDBD did not interact with FleN (Fig. 1C and fig. S1). The data confirm that FleN directly interacts with FleQ through its central domain.
As FleN is a known inhibitor of the ATPase activity of FleQ (19), we further examined the ability of FleN to inhibit the ATPase activity of different domain constructs of FleQ. FleN reduces the ATPase activity of FleQFL, FleQAAA+, and FleQRECAAA+ by 80%, and the residual activity is similar to the ATPase activity of FleN (Fig. 1D). The above findings confirm that the central domain of FleQ is necessary and sufficient for interaction with FleN.
FleN-ATPγS-FleQAAA+ structure reveals a heterotetramer
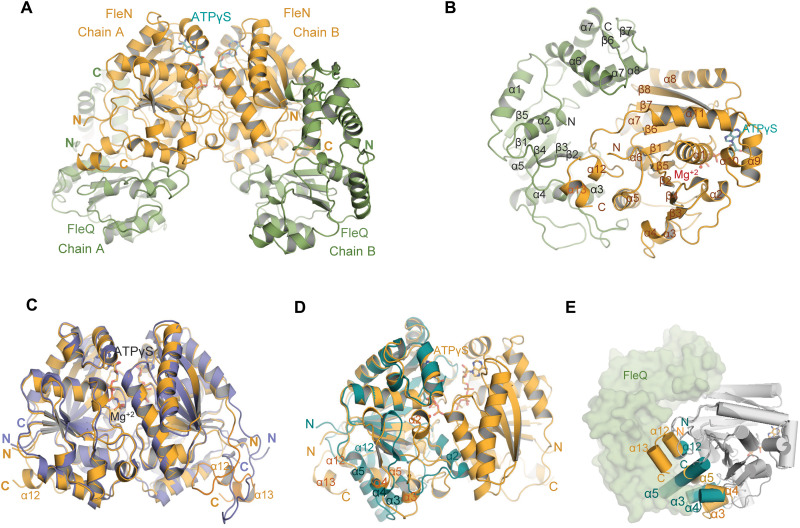
To gain detailed structural insights into the mechanism of transcription inhibition by FleN, we determined the crystal structure of FleQAAA+ in complex with full-length FleN (residues 1 to 280) in the presence of ATPγS and Mg2+.The complex crystallized in 10% PEG 8000 (polyethylene glycol, molecular weight 8000), 0.1 M Tris (pH 6.5), and 0.2 M calcium acetate. Phases were obtained using single-wavelength anomalous dispersion (SAD) with SeMet-labeled FleN crystal. Data collection and refinement statistics are displayed in Table 1. The FleN-ATPγS-FleQAAA+ structure revealed a complex in the asymmetric unit where a dimeric FleN is present in the complex with two molecules of FleQAAA+ (Fig. 2, A and B). Each monomer of FleN was also bound to one ATPγS and one Mg2+ ion.
Table 1. Data collection and refinement statistics.
| FleN-ATPγS-FleQAAA+ | SeFleN-ATPγS-FleQAAA+ | |
| Wavelength | 0.97951 | 0.97856 |
| Space group | P21 | P21 |
| Cell (Å) |
a = 64.22, b = 153.93, c = 64.57, α = 90.0, β = 95.03, γ = 90.0 |
a = 63.96, b = 156.51, c = 65.36, α = 90, β = 95.08, γ = 90 |
| Resolution (Å)* | 77–1.98 (2.02–1.98) | 156.51–2.8 (2.95–2.8) |
| Total no. of reflections | 267,897 | 183,159 |
| Unique reflections | 86,189 | 29,858 |
| Rsym or Rmerge | 9.2 (56.4) | 12.1 (45.3) |
| R meas | 11.1 (68.6) | 14.5 (58.5) |
| R pim | 6.2 (38.6) | 7.9 (36.4) |
| I/σI | 7.6 (1.9) | 9.5 (2.5) |
| Completeness (%) | 99.5 (99.5) | 94.7 (72.3) |
| Correlation CC (1/2) | 0.984 (0.556) | 0.978 (0.821) |
| Multiplicity | 3.1 (3.0) | 6.1 (4.2) |
| Refinement | ||
| Resolution (Å) | 64.319–1.98 | |
| Rwork/Rfree† | 17.88/21.8 | |
| Average B factors (Å2) | ||
| Chain A | 23.8 | |
| Chain B | 23.6 | |
| Chain C | 39.0 | |
| Chain D | 34.2 | |
| ATPγS-A | 19.7 | |
| ATPγS-B | 18.8 | |
| Mg2+ ions | 13.1 | |
| Water | 40.4 | |
| RMSD | ||
| Bond lengths (Å) | 0.008 | |
| Bond angles (°) | 1.034 | |
*Parentheses show the highest-resolution shell.
†To calculate Rfree, 5% of the data are excluded from refinement.
Fig. 2. Structure of FleN-ATPγS-FleQAAA+ complex.
(A) FleN-ATPγS-FleQAAA+ structure showing the FleN dimer in bright orange in complex with ATPγS in cyan, interacting with FleQAAA+ domains colored green forming a heterotetrameric complex. (B) Half of the complex containing one monomer of FleN and one molecule of FleQAAA+. All the secondary structure elements are labeled. ATPγS and Mg+2 are shown in cyan stick and red sphere, respectively. (C) Superimposition of dimeric FleN from AMP-PNP–bound structure [slate, Protein Data Bank (PDB): 5J1J] and FleQ-bound structure (bright orange) from the FleN-ATPγS-FleQAAA+ complex depicts structural changes at the C-terminal end. (D) Superimposition of the FleN dimer from the FleN-ATPγS-FleQAAA+ complex structure (bright orange) onto the Apo-FleN monomer (deep teal, PDB: 5JVF). The conformational changes in the helices are depicted. (E) Superimposition of FleN-Apo monomer (light gray with α3, α4, α5 and α12 in deep teal, PDB: 5JVF) onto FleN bound to ATPγS-FleQAAA+ (dark gray with α3, α4, α5, α12, and α13 in orange) showing steric clash between the monomeric FleN (deep teal) and FleQ (green) shown in surface representation. Helices are depicted as cylinders.
FleN is a P-loop ATPase with a deviant Walker A motif that has a signature lysine (K19). It exists as a dimer in the FleN-ATPγS-FleQAAA+ complex structure (Fig. 2A). The two ATPγS molecules are present at the interface of the FleN dimer such that each nucleotide is bound to one monomer and the binding is stabilized by residues K19, E153, and T155 of the adjacent monomer (fig. S2, A and B). The bound nucleotide interacts with the residues of Walker A, switch I, and switch II motifs and is also stabilized by Mg2+, which is coordinated through classical octahedral geometry (fig. S2A). The ion also forms polar interactions with the β- and γ-phosphate. As previously described for the FleN–AMP-PNP (adenylyl-imidodiphosphate) crystal structure, the dimeric contacts of FleN and the contacts with nucleotide analogs are all maintained in the current structure (fig. S2A) (19). The position and conformation of nucleotide concur well with the one observed in the FleN–AMP-PNP complex structure, and root mean square deviation (RMSD) of superimposition of FleN from FleN-ATPγS-FleQAAA+ onto FleN–AMP-PNP [Protein Data Bank (PDB): 5J1J] is 0.56 Å for 2953 atoms (Fig. 2C).
We also aligned FleN from FleN-ATPγS-FleQAAA+ onto the previously determined monomer structure of FleN (Fig. 2D). The superimposition yields RMSD of 1.3 (for 1312 atoms) and reveals substantial changes similar to the monomer to dimer transition observed earlier upon ATP binding (19). These nucleotide-driven changes are essential for dimerization. The FleN monomer forms severe clashes with the FleQAAA+ domain in the complex structure and confirms the inability of the FleN monomer to interact with FleQ (Fig. 2E). Thus, the structural data support our prior observation that ATP binding is essential for dimerization and subsequently for interaction with FleQ.
We have shown earlier that the Walker A residue K19 and a conserved E153 stabilize the dimer form of FleN by interacting with the bound nucleotide in trans (fig. S2A). Substitution of these residues with Ala affects ATP binding to FleN and consequently prevents dimer formation (19). We performed a BLI experiment with K19A and E153A substitution in FleN to assess their binding with FleQFL. Upon K19A and E153A substitution, there was no detectable binding (fig. S3). On the other hand, the D48A substitution, although defective in ATP hydrolysis, binds to the nucleotide and forms stable dimers. D48A substitution exhibited similar binding affinity as WT FleN with FleQFL in the BLI experiment (fig. S3). Together, these data further endorse that the dimeric FleN is necessary for binding with FleQ.
The most notable structural change in FleN is in the conformation of the amphipathic helix (α12) at the C terminus in the monomer structure from residues 257 to 273. This region is disordered in the FleN dimer and has been implicated in membrane binding in homologous proteins (20). However, in the complex structure, it adopts two short helical turns (α12 and α13) that are engaged in interaction with FleQ (Fig. 2E). This part of the structure attains different conformation in the monomer-, dimer-, and FleQ-bound form.
FleN allosterically inhibits binding of ATP to FleQ
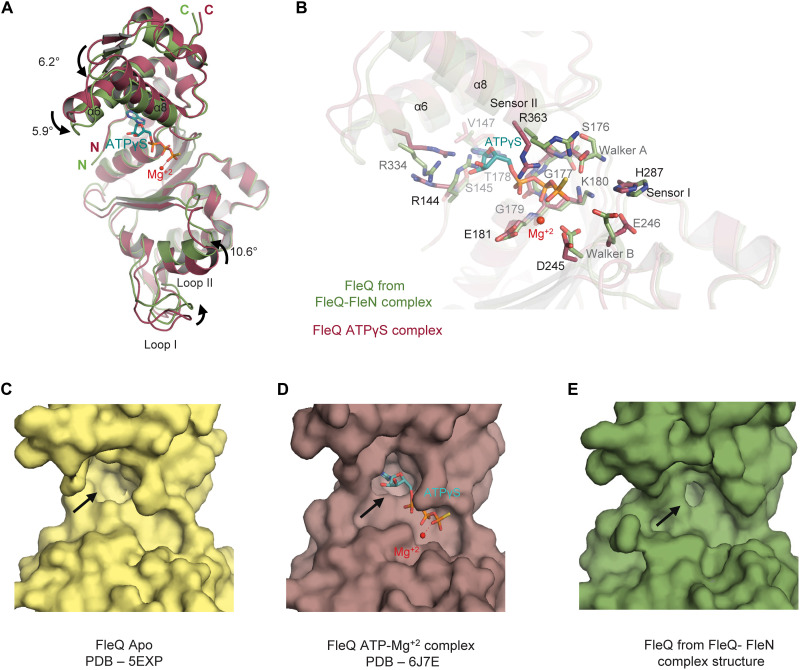
The AAA+ ATPase domain of FleQ is composed of two subdomains. The N-terminal α/β subdomain is composed of five stranded parallel β sheet and five α helices, whereas the C-terminal α-helical subdomain consists of four helices distinctive of AAA+ ATPases. Compared to the earlier determined structure of FleQAAA+-ATPγS (PDB: 6J7E), the conformation of FleQ in the FleN-bound structure reveals surprising features. Superimposition of the two structures yields an RMSD of 1.4 Å for 1769 atoms (Fig. 3A). There is about 2.5- and 3.5-Å rigid body movement of the α-helical and α/β subdomain, respectively, toward the bound FleN to form a more compact structure compared to the ATPγS-bound form. As a result, the ATP binding site of FleQ present at the interface of the two subdomains, becomes too constricted to accommodate the nucleotide triphosphate (Fig. 3, B to E, and fig. S4A). It was hence not surprising that there was no electron density for the nucleotide in the active site.
Fig. 3. Conformational changes in FleQ AAA+ domain.
(A) Superimposition of FleQ from ATPγS-Mg+2–bound structure (raspberry, PDB: 6J7E) onto FleQ (green) from the FleN-ATPγS-FleQAAA+ complex structure highlights the conformational changes on FleN binding. ATPγS and Mg+2 are shown in cyan stick and red sphere, respectively. The curved arrows show the movement of different helices and loop1. (B) Conformational changes of different residues in the ATP binding pocket of FleQAAA+ upon interaction with FleN are depicted. Color coding is the same as in (A). (C) Surface representation of FleQAAA+ in Apo structure (pale yellow, PDB: 5EXP). (D) Surface representation of FleQAAA+ in the ATPγS- and Mg+2-bound structure (raspberry, PDB: 6J7E). (E) Surface representation of FleQAAA+ in the FleN-ATPγS-FleQAAA+ structure (green) depicting the conformation of ATP binding pocket (shown with black arrows).
Besides the movement of the two subdomains, there are substantial conformational changes in the residues that are otherwise involved in interaction with the nucleotide in the ATPγS-bound structure of FleQAAA+ (9). K180 of the Walker A motif, which interacts with the phosphates of the nucleotide, exhibits altered conformation in the FleN-bound structure. R334 and R363 of sensor II that interact with the ATP analog and stabilize its binding to the protein in the ATPγS-bound structure have altered conformation in the FleN-bound structure. Similarly, R144 that forms stacking interaction with R334 in the nucleotide-bound structure occupies the ATP binding site in the FleN-bound structure. The side chains of the highly conserved residue that participate in catalysis, i.e., E246 and D245 of Walker B and sensor I H287, also display altered conformations (Fig. 3B). All these specific interactions essential for ATP binding and hydrolysis are disrupted in the FleN-bound structure.
FleN and FleQAAA+ interface is conserved
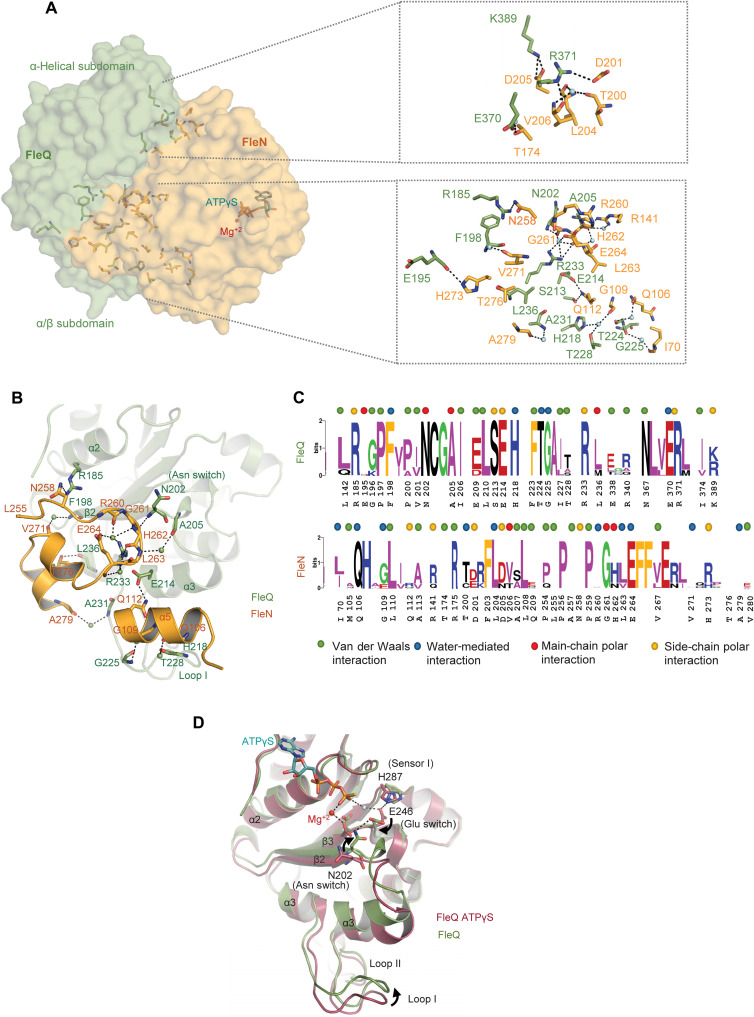
The complex structure shows that FleQ and FleN form extensive interactions. The interaction interface between the two proteins can be defined into two subcontact surfaces, one present on each subdomain. The α/β subdomain provides most of the interactions between FleQAAA+ and FleN with a buried surface area (BSA) of about 1084 Å2 (Fig. 4A). The orientation of the α-helical subdomain with respect to FleN is slightly different in the two FleQAAA+ monomers, as shown in fig. S4B, with BSA of 318 and 406 Å2, respectively. Consequently, the α-helical subdomain shows asymmetric interactions in the two monomers. Overall, there are fewer interactions with FleN at this interface. K389:NZ and R371:NE of FleQ form hydrogen bonds with D205 and main-chain oxygen of V206, respectively, of both the FleN monomers. Similarly, R371:NH1 of FleQ forms a salt bridge with D201:OD1 of FleN. E370:OE1 of FleQ monomer B interacts with T174:OG1 and R175:NH1 of monomer B of FleN. These interactions are, however, absent in monomer A (Fig. 4A).
Fig. 4. Interaction interface between FleQAAA+ and FleN and conformations of Glu switch.
(A) Surface representation of interaction interface between FleN (bright orange) and FleQ (green) in the FleN-ATPγS-FleQAAA+ structure. FleN interacts with both α/β and α-helical subdomains of FleQAAA+. The zoomed-in view illustrates the residues involved in the interaction at the interface. Waters are shown in aquamarine spheres. (B) Details of the interactions of the C terminus (L255 - V280) and α5 helix of FleN with α2 and α3 helices, β2 strand, and loop1 of FleQAAA+ are shown. The waters are depicted as spheres. (C) Sequence logos from alignment of FleQ and FleN from monoflagellated bacteria. Residues involved in main-chain polar, side-chain polar, water-mediated, and van der Waals interactions are denoted with red, orange, blue, and green dots on the top. (D) Conformational changes in α2 and α3 helices, β2 strand, and loop1 of FleQAAA+ on binding with FleN (FleN-ATPγS-FleQAAA+; green) are compared with ATPγS-bound FleQAAA+ structure (FleQAAA+-ATPγS-Mg+2; raspberry, PDB: 6J7E). Conformational changes of Glu switch (E246) and Asn switch (N202) are shown. The curved arrows show movement of Glu switch, Asn switch, and loop1.
The FleN-ATPγS-FleQAAA+ structure reveals that the interactions of FleN with the α/β subdomain are either through the C-terminal stretch (P254-V280) of the protein that forms a loop followed by two small helical fragments or through α5 that is inserted in the groove formed by the α3 and L1 loop of FleQ (Fig. 4, B and C). Most of the interactions between FleN and FleQAAA+ are identical in the two halves. Residue R185:NH2 of the α/β subdomain interacts with N258:ND2 of FleN. Similarly, the main chain of residues N202:N and A205:O of FleQAAA+ interacts with G261:O and R141:NH2 (present on α6) of FleN, respectively. In addition, R233:NH2 of FleQ interacts with the main-chain oxygen of H262. The main chain atoms of E195 and L236 of FleQ form interactions with side chains of H273:NE2 and T276:OG1, respectively. S213:OG of monomer A of FleQ forms a hydrogen bond with Q112:NE2 of monomer A of FleN. This interaction is water-mediated in monomer B. Q112:NE2 also interacts with E214:OE2 of FleQ in α5. Besides these, there are several water-mediated interactions between the two proteins (Fig. 4C).
Sequence comparison of FleN from different bacterial species harboring a single polar flagellum shows that the C-terminal stretch is conserved. G:261 of FleN present in the C-terminal loop is absolutely conserved in all the proteins (Fig. 4C). This residue provides flexibility to the loop that forms interaction with FleQ. Similarly, the sequence comparison of FleQ reveals that the residues involved in interaction with FleN are highly conserved (Fig. 4C).
The conformational changes linking FleN binding to transcription inhibition
A structural link exists between the nucleotide binding status in the active site of bEBPs to the conformation of the L1 and L2 loops that allow the highly conserved signature sequence, GAFTGA, present in the L1 loop to engage with σ54. This interaction between bEBP and σ54 is essential for transcription initiation. Structural and biochemical data have established that different nucleotide states trigger motions of the GAFTGA motif in the bEBPs (12, 21).
In the crystal structure of FleQ-ATPγS, the Walker B residue E246, present in the DEXX motif, interacts with sensor I H287 and a catalytic water molecule that makes a nucleophilic attack on the γ-phosphate of ATP for hydrolysis. This state is the catalytically competent state of the protein (9). However, in the FleN-bound form, due to the movement of the FleQ subdomains closer to each other causing the release of ATP, the side chain of E246 flips (90° rotation) and interacts with N202, also called the Asn switch (Fig. 4D). This conformation is incompatible with catalysis.
It is apparent from the complex structure that the residues of FleN from 254 to 280 form several interactions with the α/β subdomain of FleQ (Fig. 4C). Residues 260RGHLE264 of FleN are held together by a water molecule in the form of a rigid loop. The residue G261 present in the loop forms steric clash with the conformation of N202 in the ATPγS-bound form (fig. S4C). We propose that G261 causes flipping of N202 and favors the formation of N202-E246 interaction and consequently a catalytically nonproductive complex. These residues of FleN are conserved in the bacteria that harbor single polar flagella (Fig. 4C). In addition, the interaction also helps relocate α3 and simultaneously the GAFTGA L1 loop by about 3.6 Å. FleQ:R233 present in α3 interacts with the backbone atoms of H262 and E264 of FleN. Furthermore, FleN:A279 interacts with FleQ:A231, stabilizing the repositioned L1 loop (Fig. 4C). Compared to other EBPs, this conformation of the L1 loop is not compatible for interaction with the σ54 RNAP and hence transcriptionally inactive.
Thus, FleN exerts its effect by allosterically modulating the active site, switching it from an active configuration to an inactive one, simultaneously releasing the ATP. In addition, the interaction between the two proteins locks the L1 loop of FleQ in a conformation that may not be conducive for interaction with σ54.
C terminus of FleN is essential for inhibition of ATPase activity of FleQ
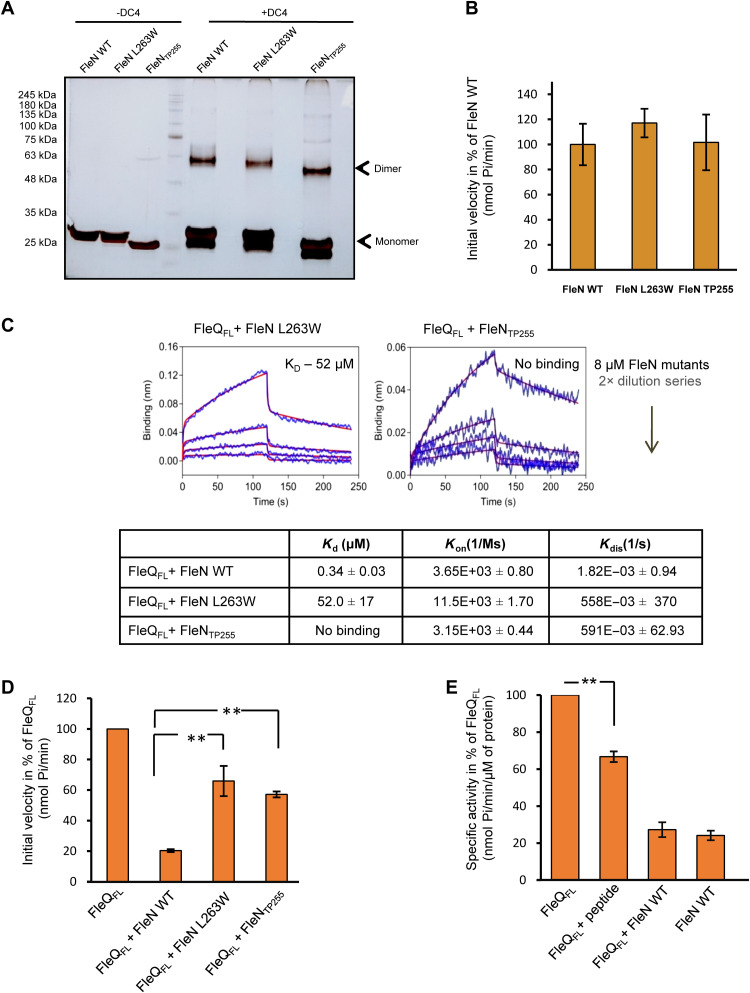
To corroborate that FleN drives the observed conformational changes in FleQ and to validate the crystallographic protein-protein interface, we next designed a set of mutations in FleN. We truncated 25 amino acids from the C terminus of FleN that were essential for triggering the structural changes in FleQ (FleNTP255, residues 1 to 255). In addition, a single substitution of L263W was also designed to perturb the formation of a stable complex. Previously, it was shown that dimerization of FleN is essential for inhibition of the ATPase activity of FleQ (19). To rule out any possibility of disruption of the overall structure and defect in dimerization, cross-linking of the FleN variants was performed using a DC4 cross-linker. As seen in Fig. 5A, the interface mutants (FleNTP255 and FleN L263W) in the presence of a cross-linker form additional bands near 60 and 56 kDa, respectively, which correspond to the dimer of these proteins. Furthermore, we also checked the ATPase activity of FleN mutants (L263W and FleNTP255). Mutants showed equivalent ATPase activity as the WT protein, which confirmed that the mutants were functional (Fig. 5B).
Fig. 5. Interaction interface between FleN and FleQAAA+ in solution.
(A) In-solution dimerization status of FleN WT and its variants was analyzed by chemical cross-linking using DC4 (1 mM). SDS-PAGE gel picture shows cross-linked products, with lanes indicating the presence and absence of cross-linker as +DC4 and −DC4, respectively. The bands corresponding to the monomeric and dimeric species are labeled in the gel picture. (B) The ATPase activity of FleN WT and its variants (5 μM) was measured in the presence of 2 mM ATP. The initial velocities of ATP hydrolysis are plotted in percentage of FleN WT on the y axis (nmol Pi/min). SD of three independent experiments is shown. (C) BLI curves showing interaction between FleQFL and FleN or its variants (FleN L263W and FleNTP255) at different concentrations. The curves in blue and red colors indicate experimental and fitting data, respectively. The dissociation constants (KD, μM), rate of association (Kon, 1/Ms), and dissociation (Kdis, 1/s) are shown in the table along with SDs. (D) The ATPase activity of FleQFL (1 μM) was measured at 2 mM ATP in the absence and presence of FleN WT and its variants (FleN L263W and FleNTP255) (5 μM). The initial velocities (nmol Pi/min) are plotted in percentage of FleQFL. Statistical significance (P value) of the defect in inhibition was calculated using unpaired t test (**P ≤ 0.01). (E) The ATPase activity of FleQFL (1 μM) was measured in the presence of FleN WT (5 μM) and C-terminal peptide of FleN (100 μM). The initial velocities (nmol Pi/min) are plotted in percentage of initial velocity of FleQFL. SD (±) is calculated from three independent experiments. The statistical significance (P value) was calculated by unpaired t test (**P ≤ 0.01; ***P ≤ 0.0001).
We then probed the ability of the mutant proteins to bind to full-length FleQ using BLI. No binding was observed between FleQ and FleNTP255 (Fig. 5C). Similarly, point mutation of L263W is also capable of disrupting this interaction, as the KD value was 100-fold higher than in the WT FleN (Fig. 5C). These data confirm that the C terminus of FleN is critical for the interaction between the two proteins.
We next measured the ATPase activity of FleQFL in the presence of C-terminally truncated FleNTP255 and L263W. FleN WT completely abolishes the ATPase activity of FleQFL; however, FleN L263W and FleNTP255 showed reduced ability to inhibit the activity of FleQFL with 65.9 and 57.1% residual activity, respectively (Fig. 5D). Next, we assayed the ATPase activity of FleQ in the presence of 100 μM of only the C-terminal peptide of FleN comprising the last 25 amino acids. In the presence of the peptide, FleQ exhibited around 66% of the ATPase activity (Fig. 5E). These results demonstrated that the observed defect in the interaction and inhibition by the interface mutants could be solely attributed to the disruption of the interaction interface in solution.
Destabilization of the interface between FleQ and FleN results in motility defects in P. aeruginosa
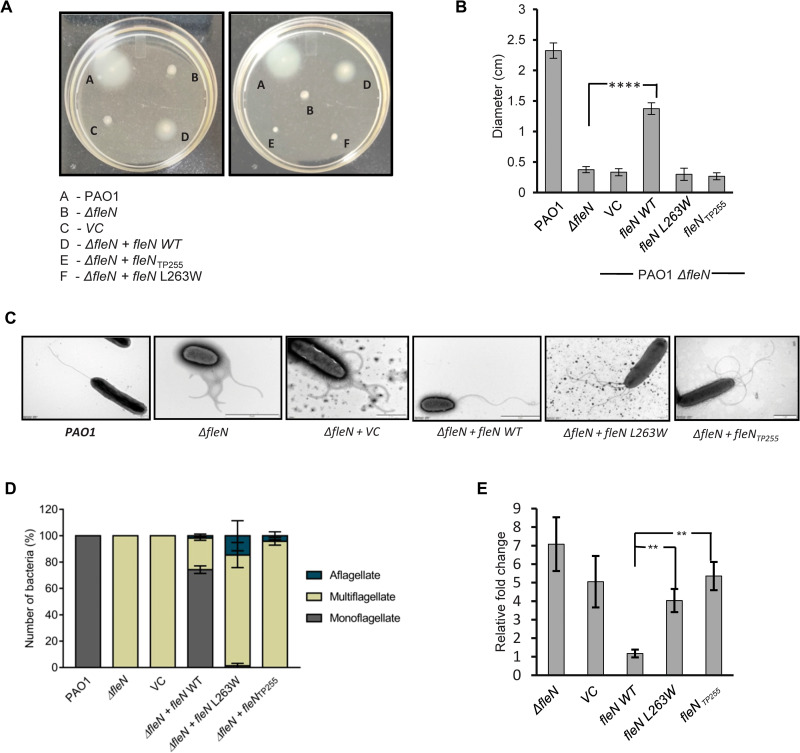
FleQ activates the transcription of flagellar genes, which is, in turn, essential for motility of the organism. The transcription activation by FleQ is further regulated by FleN. To investigate the effect of FleN C-terminal truncation on the motility of P. aeruginosa, the swimming motility assay was performed on soft agar plates and the diameter of the spread of the bacteria was measured. While the WT PAO1 displayed reasonable motility with a diameter of 2.3 cm, the PAO1 ΔfleN strain showed impaired motility and remained at the point of inoculation with a diameter of 0.37 cm only (Fig. 6, A and B). Complementation of fleNWT integrated on the plasmid pHERD20T restored motility with a diameter of 1.37 cm. However, complementation with fleN L263W and fleNTP255 failed to reinstate the motility and exhibited seven- and eightfold reduction in diameter, respectively. This suggests that the interaction between FleQ and FleN is essential for the swimming motility of P. aeruginosa (Fig. 6, A and B).
Fig. 6. Motility and flagellar phenotypes of WT and mutant P. aeruginosa.
(A) The swimming motility of P. aeruginosa WT strain PAO1, knockout strain (ΔfleN), vector control strain (VC), knockout strain complemented with WT fleN (ΔfleN + fleN WT), and its variants (ΔfleN + fleN L263W, ΔfleN + fleNTP255) is shown in soft agar plates. The swimming zone was calculated by measuring the diameters of the colonies. (B) Bar diagram represents the swimming zone for different strains of P. aeruginosa. VC (vector control) indicates knockout strain complemented with empty vector pHERD20T. The y axis shows the diameter of the swimming zone in centimeters. The statistical significance (P value) was calculated by unpaired t test (**P ≤ 0.01; ***P ≤ 0.001). (C) Electron micrographs showing flagellar phenotypes of different strains of P. aeruginosa—WT (PAO1), knockout (ΔfleN), knockout strain complemented with VC (ΔfleN + VC) and WT fleN (ΔfleN + fleN WT), or its variants (ΔfleN + fleN L263W and ΔfleN + fleNTP255). (D) The number of bacteria showing different phenotypes is shown on the y axis in percentage. Gray, brown, and blue bars denote monoflagellate, multiflagellate, and aflagellate phenotype, respectively. (E) Relative transcript levels of fleS in various strains of P. aeruginosa compared to the WT PAO1. Unpaired t test was performed to calculate the P value (**P ≤ 0.01; ***P ≤ 0.001). SD is calculated from three independent experiments in all cases.
Interaction between FleQ and FleN is essential for monoflagellate phenotype
The antagonistic activity of FleN against FleQ is essential for the monoflagellate phenotype of P. aeruginosa. A bacterial strain that is knockout of fleN displays multiple polar flagella as it fails to regulate the transcription activity of FleQ. To investigate the effect of C-terminal truncation and point mutation on the flagellar number, transmission electron microscopy was performed with the WT, ΔfleN, and various complemented strains of P. aeruginosa. Unsurprisingly, the PAO1 strain exhibits a single polar flagellum, whereas the ΔfleN strain was multiflagellate (Fig. 6, C and D). Complementation of the knockout strain with fleN restored the WT phenotype. However, the knockout complemented with fleN L263W substitution and fleNTP255 failed to restore monotrichous phenotype and displays multiple polar flagella, confirming that the FleN variants fail to interact with FleQ and are unable to regulate its activity in vivo (Fig. 6, C and D).
Interaction between FleQ and FleN is essential for transcription regulation
In P. aeruginosa, the expression of genes of the two-component system fleSR, which are part of the flagellar cascade, is directly activated by FleQ (7). We have further investigated the effect of disruption of the FleN and FleQ interface on the regulation of transcription by FleQ, which activates the expression of fleSR genes directly. We performed a real-time quantitative polymerase chain reaction (qRT-PCR) experiment to assess the effect of C-terminal deletion of FleN on the transcription regulation by FleQ. The fleN knockout (ΔfleN) strain showed higher levels of expression of fleS gene as compared to the PAO1 strain (Fig. 6E). Complementation of ΔfleN with the WT copy of fleN reduces the expression to a level comparable to the PAO1 strain. However, complementation of the ΔfleN bacterial strain with either fleN L263W or fleNTP255 could not reduce fleS expression. This suggests that FleN regulates flagellar gene expression. Furthermore, the interface mutants are unable to interact with FleQ and, therefore, are unable to regulate the transcription in vivo (Fig. 6E).
DISCUSSION
Most bEBPs are regulated through their diverse regulatory domains present at the N terminus that sense diverse environmental cues and regulate the activity of the AAA+ ATPase domain. The canonical bEBPs are regulated through either phosphorylation, ligand binding, or protein-protein interaction. The bEBPs that are part of two-component systems are regulated through phosphorylation of the conserved aspartate present in the N-terminal REC domain through the cognate histidine kinase (22–25). The EBPs regulated through ligand binding harbor effector binding domains such as GAF (cGMP-specific phosphodiesterases, adenylyl cyclases and FhlA) and PAS (Per-Arnt-Sim) at the N terminus (12, 25). Furthermore, the third group of bEBPs is regulated through protein-protein interactions using antiactivators. The ATPase activity of extensively studied bEBP, PspF, that lacks the NTD is regulated through interaction with phage shock protein A (PspA). The coiled-coil structure of the PspF interacting domain of PspA indicates similarity with the mechanism followed by unfoldase ClpB to regulate the ATPase activity (26). Similarly, NifA-NifL forms part of a two-component system and the activity of NifA is modulated through interaction of the central domain of NifA with the C-terminal part of NifL (27–29). It also regulates the DNA binding and interaction of NifA with RNAP and hence regulates the transcription of genes in adverse environmental conditions. However, the lack of structural data on the protein-protein complex impedes a detailed understanding of the mechanism of transcription inhibition by the antiactivators of σ54 regulators. Our complex structure being the first σ54 activator-antiactivator complex sheds light on the in-depth mechanism.
We have established that FleN and FleQAAA+ form a stable complex and determined a high-resolution crystal structure of the complex of two proteins in the presence of a nucleotide analog. In the structure, the primary interaction determinants on FleQ were the two interaction interfaces, one each present on the α-helical and α/β subdomain. FleN, on the other hand, interacts only in the dimeric form. The interaction interfaces are conserved within the monoflagellated bacteria. Furthermore, the interaction of FleN with the AAA+ domain of FleQ locks the L1 and L2 loops in a conformation that is not conducive for interaction with RNAP. We also demonstrate that perturbations in the protein-protein interface that support the crystallographic heterotetramer show transcription defects in vivo.
FleQ is a unique σ54 activator, as it contains the regulatory domain, but it is not required to be activated through phosphorylation. It forms constitutive hexamers and possibly other forms of oligomers that exist in equilibrium. Unlike in the case of negative regulators, the NTD of FleQ does not inhibit its ATPase activity; instead, it enables the formation of the functional hexameric form (15, 16). In addition, we have earlier demonstrated that FleQ sensor I is a histidine residue that is unique in place of highly conserved asparagine found in the majority of the other family members (9). This ensures that, although constitutive, the ATPase activity is depleted compared to the other AAA+ proteins that contain a conserved Asn in sensor I.
The ATPase activity of FleQ is further regulated through protein-protein interaction via FleN. FleN exists as a dimer and monomer in the presence and absence of the ATP, respectively. The binding of ATP triggers the dimeric assembly that is capable of inhibiting FleQ. After hydrolysis, the dimer breaks into monomers that are incompatible with the FleQ complex. Reversible dimerization allows FleN to fine-tune transcriptional activation of flagellar genes so as to prevent the formation of multiple flagella and enable the synthesis and maintenance of a single flagellum (19).
This viewpoint also explains the need to keep the Glu switch in the active conformation in FleQ, which is disabled on binding to FleN. Furthermore, FleN hydrolyzes ATP and dissociates from FleQ, which results in the formation of the active hexameric form and places the Glu switch in the catalytic competent state. This is in contrast to other σ54 activators as well as other AAA+ proteins that harbor Asn in sensor I and have an intrinsically high ATPase activity. Thus, the Glu switch is present in the inactive conformation even in the presence of ATP. The ATPase activity of these enzymes is triggered upon binding to the substrate. This is seen in case of NtrC1, PspF, ZraR, p97, RFC, etc. (30).
P. aeruginosa is an opportunistic pathogen that causes biofilm-mediated chronic infections in humans. FleQ regulates the expression of pel, psl, cdrA, and PA2440 genes involved in biofilm formation (8, 31, 32). FleQ represses the expression of exopolysaccharides by binding the pel and psl promoters, and this repression is released in the presence of bis-(3′-5′)-cyclic diguanosine monophosphate (c-di-GMP) (8). FleQ binds to sequences referred to as box1 and box2 of pel promoters as a hexamer and inhibits transcription initiation. Studies have shown a moderate effect of FleN regulation at these promoters (8, 15). It has been observed that FleN is required for complete derepression of pel, psl, and cdr operons in the presence of high levels of c-di-GMP (8). However, unlike the flagellar genes, the expression of the exopolysaccharide genes required for the formation of biofilms is σ70 dependent (7, 8, 33). We observed that the conformation of the c-di-GMP–bound AAA+ FleQ domain bears notable similarity with the FleN-bound AAA+ FleQ domain. C-di-GMP causes compaction of the active site of FleQ favoring N202-E246 interaction and locking of L1 loops in inactive conformation (figs. S4C and S5A) (15). This strongly suggests that the binding of FleN predisposes FleQ for interaction with c-di-GMP, thereby releasing the repression (fig. S5B).
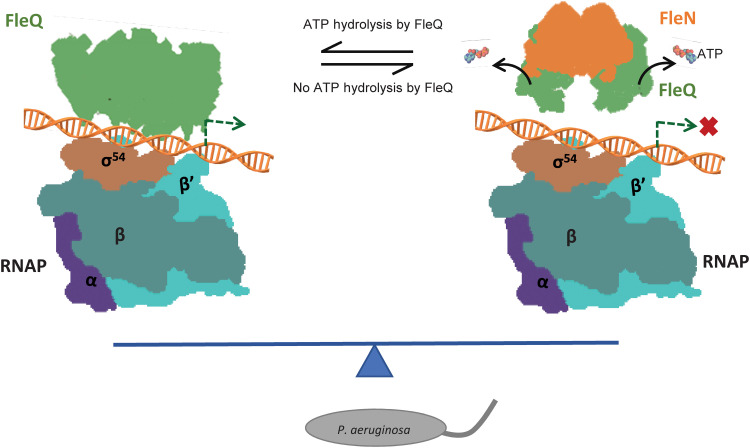
In light of current structural and previous biochemical data, we propose that the antiactivator FleN exerts its inhibitory effect on FleQ by a concerted mechanism involving (i) allosterically preventing ATP binding to FleQ and (ii) locking the L1 loop into a conformation that is unable to form a stable interaction with σ54 and (iii) by triggering the release of Glu switch residue E153 in an inactive conformation preventing catalysis. This allows FleN to fine-tune the transcription activation of flagellar genes by FleQ and ensures the assembly and maintenance of single flagella in monoflagellate bacteria (Fig. 7). Last, our structural data provide a robust platform for the development of therapeutic strategies against an important human pathogen, P. aeruginosa.
Fig. 7. Model for σ54-mediated transcription regulation by the antiactivator FleN.
The RNAP bound to DNA is depicted. The master regulator FleQ is in green. Antiactivator is shown in orange. The binding of antiactivator triggers the release of ATP by FleQ and prevents its interaction with RNAP. The fine-tuning of transcription allows formation of single flagella in bacteria (gray).
MATERIALS AND METHODS
Mutant construction and protein purification
Site-specific mutations in FleN were made using QuikChange II XL site-directed mutagenesis method described by Stratagene. pGEX-6p1-fleN WT was used as template backbone to prepare the mutants. Mutations were confirmed by sequencing. FleN WT, SeFleN, FleQFL, and different constructs were purified as described previously (9, 18, 19).
Crystallization of complex of FleN-ATPγS-FleQAAA+
Screening of the complex crystals was done with commercially available screen from Hampton Research. Hits were found at 4°C in a well containing 10% PEG 8000, 0.1 M imidazole (pH 8), and 0.2 M calcium acetate. Crystallization conditions were further optimized using hanging drop with equal volume of protein (0.8 μl) and reservoir solution. FleN and FleQAAA+ (0.2 mM/0.2 mM) were mixed in a 1:1 molar ratio. Both the proteins were preincubated with ATPγS in a 1:4 molar ratio. Crystals were cryoprotected by soaking into reservoir solution with 30% PEG 3350 and 5% glycerol and flash-frozen in liquid nitrogen.
Data collection and structure determination
X-ray diffraction datasets were collected at ID30B beamline of the European Synchrotron Radiation Facility (ESRF), France. Crystals of FleN-ATPγS-FleQAAA+ and SeFleN-ATPγS-FleQAAA+ diffracted to a resolution of 1.98 and 2.8 Å, respectively. The data were processed using DIALS package (34) in the CCP4i2 program suite. Scaling was performed using AIMLESS in the CCP4 program suite (35). The structure was determined by SAD phasing using selenium. Twelve of 16 selenium sites were located using AutoSol (36, 37). Model building was done using AutoBuild (37). High-resolution native datasets were then used for phase extension. Subsequently, model building and refinement were performed using COOT and PHENIX (38, 39).
Binding affinity measurements using BLI
Binding affinity between FleN and FleQFL or its constructs was measured through BLI using an Octet RED96 instrument (ForteBio). EZ-Link NHS-PEG4-Biotin (Thermo Fisher Scientific, catalog number 21330) was used for biotinylation of FleQFL or its constructs (0.2 mM), which were preincubated with ATPγS (0.8 mM) on ice for 30 min. For biotinylation and removal of excess biotin, a procedure described by manufacturers was used. The biotinylated FleQFL protein was then immobilized onto the streptavidin-coated biosensors in buffer containing 20 mM Hepes (pH 7), 250 mM NaCl, 1 mM dithiothreitol (DTT), 2 mM MgCl2, and 10 mM KCl. Binding studies were performed by dipping sensors into the varying concentrations of FleN and its mutants, which were preincubated with 0.5 mM ATPγS on ice for 30 min. For binding studies, immobilization buffer with ATPγS, bovine serum albumin, and 0.05% Tween 20 was used to reduce the nonspecific binding. An equal number of empty sensors were used to rule out nonspecific interactions. FleQFL or its construct immobilized sensors were dipped in a reference well with binding buffer. Thus, double referencing eliminated the nonspecific binding as well as baseline drift. Binding studies were performed at 25°C. The raw data were processed using ForteBio data analysis software with a global fit. A 2:1 model was used for fitting the data.
ATPase assay
The ATPase activity of WT and mutant proteins was measured using the EnzChek Phosphate Assay Kit (Invitrogen) as described previously (9, 19). Briefly, the ATPase activity of FleN WT and mutants (5 μM) was carried out in a reaction buffer containing 20 mM Hepes (pH 7), 2 mM MgCl2, 10 mM KCl, 1 mM DTT, and 250 mM NaCl. Absorbance at 360 nm was measured continuously at a 5-min interval immediately after adding 2 mM ATP. For inhibition assay, the ATPase activity of 1 μM FleQ alone as well as in the presence of FleN WT and mutants (5 μM) was carried out. C-terminal peptide was dissolved in the abovementioned ATPase reaction buffer. FleQ (1 μM) and peptide (100 μM) were used to perform the reaction.
In-solution chemical cross-linking assay
For detection of oligomeric state of proteins, in-solution chemical cross-linking with DC4 was performed. To carry out the reaction, 5 μM protein was preincubated with 100 μM ATPγS on ice for 30 min in a buffer containing 25 mM Hepes (pH 7), 300 mM NaCl, and 5 mM MgCl2. DC4 (0.5 mM) was then added to the reaction mixture and incubated at room temperature for 10 min. The reaction was stopped by adding SDS gel loading dye and boiled at 95°C for 5 min. Reaction products were analyzed using 10% SDS–polyacrylamide gel electrophoresis (PAGE). For visualization, silver staining method was used.
P. aeruginosa mutant strain preparation
To perform in vivo assays, fleN was cloned in the shuttle vector pHERD20T using Kpn I and Hind III restriction sites. Point mutations in fleN (L263W and a stop codon at 256th position) were prepared using QuikChange II XL site-directed mutagenesis method described by Stratagene. Mutations were confirmed by sequencing. pHERD20T-fleN WT, pHERD20T-vector, pHERD20T-fleN L263W, and pHERD20T-fleNTP255 were electroporated in the PAO1 ΔfleN strain obtained from the Manoil laboratory, University of Washington. For selection of complemented strains, bacteria were grown on carbenicillin (300 μg/ml) plates.
Motility assay
A soft agar plate assay was performed to assess the flagellar-based swimming motility of different strains of P. aeruginosa. From the glycerol stocks, bacterial strains were streaked on LB-carbenicillin plates and kept at 37°C for 16 hours (carbenicillin, 300 μg/ml). Single colonies of different strains (WT and mutants) were picked with the help of toothpick and poked into 0.3% agar plates. Plates were incubated at 37°C for 12 hours without disturbing. The diameter of each colony was measured.
Transmission electron microscopy
Bacteria were grown overnight and subcultured in a fresh LB media at 37°C until OD600 (optical density at 600 nm) reached 0.4. One milliliter of bacterial culture was centrifuged at 5000 rpm and washed two times with fresh LB and finally resuspended in 500 μl of fresh LB. Five microliters of culture was allowed to adhere to the glow-discharged carbon-coated copper grids for 5 min and drained off. The grids were washed two times with ultrapure water and stained with 2% aqueous solution of phosphotungstic acid for negative staining. Samples were examined with a JEOL JEM-1400 series 120 keV transmission electron microscope equipped with tungsten filament.
RNA isolation and qRT-PCR
WT P. aeruginosa (PAO1), ΔfleN, vector control, and strains complemented with fleNWT, fleN L263W, and fleNTP255 were grown overnight in LB medium. The overnight cultures were diluted 1:100 in LB medium to set up a secondary culture and grown to an OD600 of 0.4 to 0.6 for total RNA extraction. The cultures (3 ml) were mixed with 6 ml of a bacterial RNA protect solution (Qiagen), and the mixtures were centrifuged at 4000 rpm for 20 min to harvest the cells. One milliliter of TRIzol (Invitrogen) was added to resuspend the pellets, and the cells were lysed using sonication. The RNeasy Mini Kit (Qiagen) was used for RNA extraction and purification. The concentration (A260) and quality (A260/A280 > 2, A260/A230 > 2) of RNA were estimated using NanoDrop (Thermo Fisher Scientific). The SuperScript III Platinum Kit (Invitrogen) was used for one-step qRT-PCR. The rpoD gene was used as the reference gene. The relative levels of fleS gene expression of the cells were calculated by relative quantification using the reference gene. SDs were calculated from independent replicates.
Statistical analysis
To examine the statistically significant difference between two groups, unpaired t test was used. ±SD is represented for all the data. GraphPad Prism t test calculator was used to perform all statistical analyses. *P < 0.05, **P < 0.01, and ***P < 0.001 as indicated.
Acknowledgments
We acknowledge the help rendered by N. Foos (ID30B beamline, ESRF) during data collection and I. Barwal during transmission electron microscopy experiment. We also acknowledge D. N. Rao for providing plasmid pHERD20T for complementation assays. The knockout strains were procured from Pseudomonas aeruginosa mutant library maintained in the Manoil laboratory prepared using NIH grant no. NIH P30 DK089507. We thank D. T. Nair for critical reading of the manuscript.
Funding: D.J. acknowledges funding from the Department of Biotechnology, Ministry of Science and Technology ESRF Access Program of RCB grant BT/INF/22/SP22660/2017, Department of Biotechnology grant BT/PR23844/BRB/10/1598/2017, and Regional Centre for Biotechnology Intramural funds. C. thanks the Department of Biotechnology for research fellowship, and P.B. thanks the Department of Science and Technology for the INSPIRE fellowship.
Author contributions: C. cloned, purified protein, performed biochemical experiments, and collected x-ray diffraction data. P.B. purified protein, performed biochemical experiments, analyzed structural data, and prepared figures. S.R. performed in vivo experiments. H.N.G. solved the structure. D.J. conceptualized, acquired funding, supervised research, carried out model building and refinement, performed structural analysis, and wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Atomic coordinates have been deposited at the Protein Data Bank under accession no. PDB: 7EJW.
Supplementary Materials
This PDF file includes:
Figs. S1 to S5
REFERENCES AND NOTES
- 1.Saecker R. M., Record M. T., Dehaseth P. L., Mechanism of bacterial transcription initiation: RNA polymerase—Promoter binding, isomerization to initiation-competent open complexes, and initiation of RNA synthesis. J. Mol. Biol. 412, 754–771 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ghosh T., Bose D., Zhang X., Mechanisms for activating bacterial RNA polymerase. FEMS Microbiol. Rev. 34, 611–627 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Danson A. E., Jovanovic M., Buck M., Zhang X., Mechanisms of σ54-dependent transcription initiation and regulation. J. Mol. Biol. 431, 3960–3974 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang N., Buck M., A perspective on the enhancer dependent bacterial RNA polymerase. Biomolecules 5, 1012–1019 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arora S. K., Ritchings B. W., Almira E. C., Lory S., Ramphal R., A transcriptional activator, FleQ, regulates mucin adhesion and flagellar gene expression in Pseudomonas aeruginosa in a cascade manner. J. Bacteriol. 179, 5574–5581 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dasgupta N., Ferrell E. P., Kanack K. J., West S. E. H., Ramphal R., fleQ, the gene encoding the major flagellar regulator of Pseudomonas aeruginosa, is σ70 dependent and is downregulated by Vfr, a homolog of Escherichia coli cyclic AMP receptor protein. J. Bacteriol. 184, 5240–5250 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dasgupta N., Wolfgang M. C., Goodman A. L., Arora S. K., Jyot J., Lory S., Ramphal R., A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol. Microbiol. 50, 809–824 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Hickman J. W., Harwood C. S., Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol. Microbiol. 69, 376–389 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banerjee P., Chanchal, Jain D., Sensor I regulated ATPase activity of FleQ is essential for motility to biofilm transition in Pseudomonas aeruginosa. ACS Chem. Biol. 14, 1515–1527 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Dasgupta N., Ramphal R., Interaction of the antiactivator FleN with the transcriptional activator FleQ regulates flagellar number in Pseudomonas aeruginosa. J. Bacteriol. 183, 6636–6644 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jyot J., Dasgupta N., Ramphal R., FleQ, the major flagellar gene regulator in Pseudomonas aeruginosa, binds to enhancer sites located either upstream or atypically downstream of the RpoN binding site. J. Bacteriol. 184, 5251–5260 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bush M., Dixon R., The role of bacterial enhancer binding proteins as specialized activators of σ54-dependent transcription. Microbiol. Mol. Biol. Rev. 76, 497–529 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen B., Doucleff M., Wemmer D. E., De Carlo S., Huang H. H., Nogales E., Hoover T. R., Kondrashkina E., Guo L., Nixon B. T., ATP ground- and transition states of bacterial enhancer binding AAA+ ATPases support complex formation with their target protein, σ54. Structure 15, 429–440 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dey S., Biswas M., Sen U., Dasgupta J., Unique ATPase site architecture triggers cis-mediated synchronized ATP binding in heptameric AAA+-ATPase domain of flagellar regulatory protein FlrC. J. Biol. Chem. 290, 8734–8747 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsuyama B. Y., Krasteva P. V., Baraquet C., Harwood C. S., Sondermann H., Navarro M. V. A. S., Mechanistic insights into c-di-GMP–dependent control of the biofilm regulator FleQ from Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 113, E209–E218 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Su T., Liu S., Wang K., Chi K., Zhu D., Wei T., Huang Y., Guo L., Hu W., Xu S., Lin Z., Gu L., The REC domain mediated dimerization is critical for FleQ from Pseudomonas aeruginosa to function as a c-di-GMP receptor and flagella gene regulator. J. Struct. Biol. 192, 1–13 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Dasgupta N., Arora S. K., Ramphal R., fleN, a gene that regulates flagellar number in Pseudomonas aeruginosa. J. Bacteriol. 182, 357–364 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harshita, Chanchal, Jain D., Cloning, expression, purification, crystallization and initial crystallographic analysis of FleN from Pseudomonas aeruginosa. Acta Crystallogr. Struct. Biol. Commun. 72, 135–138 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chanchal, Banerjee P., Jain D., ATP-induced structural remodeling in the antiactivator FleN enables formation of the functional dimeric form. Structure 25, 243–252 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Schuhmacher J. S., Rossmann F., Dempwolff F., Knauer C., Altegoer F., Steinchen W., Dörrich A. K., Klingl A., Stephan M., Linne U., Thormann K. M., Bange G., MinD-like ATPase FlhG effects location and number of bacterial flagella during C-ring assembly. Proc. Natl. Acad. Sci. U.S.A. 112, 3092–3097 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joly N., Zhang N., Buck M., Zhang X., Coupling AAA protein function to regulated gene expression. Biochim. Biophys. Acta 1823, 108–116 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Moretfl E., Segovia L., The sigma 54 bacterial enhancer-binding protein family: Mechanism of action and phylogenetic relationship of their functional domains. J. Bacteriol. 175, 6067–6074 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doucleff M., Chen B., Maris A. E., Wemmer D. E., Kondrashkina E., Nixon B. T., Negative regulation of AAA+ ATPase assembly by two component receiver domains: A transcription activation mechanism that is conserved in mesophilic and extremely hyperthermophilic bacteria. J. Mol. Biol. 353, 242–255 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Bourret R. B., Borkovich K. A., Simon M. I., Signal transduction pathways involving protein phosphorylation in prokaryotes. Annu. Rev. Biochem. 60, 401–441 (1991). [DOI] [PubMed] [Google Scholar]
- 25.Gao R., Stock A. M., Molecular strategies for phosphorylation-mediated regulation of response regulator activity. Curr. Opin. Microbiol. 13, 160–167 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osadnik H., Schöpfel M., Heidrich E., Mehner D., Lilie H., Parthier C., Risselada H. J., Grubmüller H., Stubbs M. T., Brüser T., PspF-binding domain PspA1-144 and the PspA·F complex: New insights into the coiled-coil-dependent regulation of AAA+ proteins. Mol. Microbiol. 98, 743–759 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Little R., Colombo V., Leech A., Dixon R., Direct interaction of the NifL regulatory protein with the GlnK signal transducer enables the Azotobacter vinelandii NifL-NifA regulatory system to respond to conditions replete for nitrogen. J. Biol. Chem. 277, 15472–15481 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Narberhaus F., Lee H. S., Schmitz R. A., He L., Kustu S., The C-terminal domain of NifL is sufficient to inhibit NifA activity. J. Bacteriol. 177, 5078–5087 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barrett J., Ray P., Sobczyk A., Little R., Dixon R., Concerted inhibition of the transcriptional activation functions of the enhancer-binding protein NIFA by the anti-activator NIFL. Mol. Microbiol. 39, 480–493 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Zhang X., Wigley D. B., The “glutamate switch” provides a link between ATPase activity and ligand binding in AAA+ proteins. Nat. Struct. Mol. Biol. 15, 1223–1227 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baraquet C., Harwood C. S., FleQ DNA binding consensus sequence revealed by studies of FleQ-dependent regulation of biofilm gene expression in Pseudomonas aeruginosa. J. Bacteriol. 198, 178–186 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baraquet C., Murakami K., Parsek M. R., Harwood C. S., The FleQ protein from Pseudomonas aeruginosa functions as both a repressor and an activator to control gene expression from the Pel operon promoter in response to c-di-GMP. Nucleic Acids Res. 40, 7207–7218 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baraquet C., Harwood C. S., Cyclic diguanosine monophosphate represses bacterial flagella synthesis by interacting with the Walker A motif of the enhancer-binding protein FleQ. Proc. Natl. Acad. Sci. U.S.A. 110, 18478–18483 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winter G., Waterman D. G., Parkhurst J. M., Brewster A. S., Gildea R. J., Gerstel M., Fuentes-Montero L., Vollmar M., Michels-Clark T., Young I. D., Sauter N. K., Evans G., DIALS: Implementation and evaluation of a new integration package. Acta Crystallogr. D Struct. Biol. 74, 85–97 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Evans P., Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Terwilliger T. C., Adams P. D., Read R. J., McCoy A. J., Moriarty N. W., Grosse-Kunstleve R. W., Afonine P. V., Zwart P. H., Hung L. W., Decision-making in structure solution using Bayesian estimates of map quality: The PHENIX AutoSol wizard. Acta Crystallogr. D Biol. Crystallogr. 65, 582–601 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zwart P. H., Afonine P. V., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., McKee E., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Storoni L. C., Terwilliger T. C., Adams P. D., Automated structure solution with the PHENIX suite. Methods Mol. Biol. 426, 419–435 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., Adams P. D., Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S5