ABCG17 and ABCG18 ABA importers redundantly regulate ABA homeostasis and long-distance translocation.
Abstract
The effects of abscisic acid (ABA) on plant growth, development, and response to the environment depend on local ABA concentrations. Here, we show that in Arabidopsis, ABA homeostasis is regulated by two previously unknown ABA transporters. Adenosine triphosphate–binding cassette subfamily G member 17 (ABCG17) and ABCG18 are localized to the plasma membranes of leaf mesophyll and cortex cells to redundantly promote ABA import, leading to conjugated inactive ABA sinks, thus restricting stomatal closure. ABCG17 and ABCG18 double knockdown revealed that the transporters encoded by these genes not only limit stomatal aperture size, conductance, and transpiration while increasing water use efficiency but also control ABA translocation from the shoot to the root to regulate lateral root emergence. Under abiotic stress conditions, ABCG17 and ABCG18 are transcriptionally repressed, promoting active ABA movement and response. The transport mechanism mediated by ABCG17 and ABCG18 allows plants to maintain ABA homeostasis under normal growth conditions.
INTRODUCTION
Abscisic acid (ABA) is a plant hormone that regulates growth and responses to the changing environment. For example, seed dormancy, germination, drought tolerance, stomatal closure, and lateral root emergence are modulated by this important hormone under normal conditions and also in response to stimuli (1–7). The ABA response is regulated at multiple steps: biosynthesis, transport, catabolism, perception, and signal transduction. ABA travels long distances throughout the plant, a characteristic that was shown about 50 years ago to affect stomatal conductance and responses to drought (8–11). For decades, the concept stated in textbooks was that in response to drought, root-derived ABA travels to shoot guard cells via the xylem sap to prevent water loss (12, 13). Several studies support the idea that root-derived ABA synthesis is required for ABA-induced stomatal closure to elicit drought tolerance (14–16). Recent studies, however, provide evidence that stomatal closure is triggered by shoot-specific ABA synthesis (17–20). While the complete map of ABA biosynthesis is not entirely clear yet, studies showed that ABA could be synthesized directly in guard cells during abiotic stresses and in the leaf vasculature (21).
Multiple lines of evidence support the hypothesis that ABA transport is necessary for proper ABA responses (22–24). First, ABA, being a weak acid, exists primarily in its charged and membrane-impermeable form depending on the pH value in the cytoplasm environment, suggesting the need for transporter-facilitated movement across membranes (25–29). Second, using deuterium-labeled ABA and reciprocal grafting between wild-type (WT) and ABA-biosynthetic mutant tomatoes, it was found that foliage-derived ABA promotes primary root growth but inhibits the development of lateral roots (30). A recent study demonstrated that leaf-derived ABA participates in rice seed development in response to environmental temperature variations (31). These highlight the importance of long-distance ABA transport in governing plant physiological and morphological aspects, beyond stomata control. Third, phloem-specific ABA synthesis complements ABA activity in stomatal aperture, indicating that ABA movement within the leaf is important (32–34). Fourth, ABA transport is required for processes such as germination (23, 35, 36).
In recent years, several ABA transporters have been identified and characterized (15, 23, 37, 38). For example, in Arabidopsis seeds, adenosine triphosphate–binding cassette subfamily G member 25 (ABCG25) and ABCG31 export ABA out of endosperm, while ABCG40 and ABCG30 import ABA into the embryo to regulate seed germination. In leaves, ABCG25 is mainly expressed in the vasculature, promoting ABA transport to guard cells, while ABCG40 imports ABA into guard cells to control the stomatal aperture (23, 37, 39–41). MtABCG20 was recently characterized as an ABA importer that is involved in germination and lateral root formation in Medicago truncatula (42). Additional transporters include, a transporter of the multidrug and toxic compound extrusion transporter family (DTX50), and NITRATE TRANSPORT1/PEPTIDE TRANSPORTER FAMILY 4.6 (NPF4.6) (also known as NRT1.2 and AIT1) (15, 23, 25, 34, 43, 44).
In addition to transport, ABA concentrations are tightly controlled by ABA metabolism (36, 45). ABA is conjugated with glucose by uridine diphosphate (UDP)–glucosyltransferases (ABA-glucosyltransferase) to catalyze the formation of the stored form of ABA, ABA–glucose ester (ABA-GE) (46). Previous studies showed that there are 26 subfamilies of UDP-glucosyltransferases (UGTs) in Arabidopsis. Seven UGTs, which belong to different groups of UDP subfamily 1 (encoded by UGT84B1, UGT75B1, UGT84B2, UGT71B6, UGT75B2, UGT73B1, and UGT71C5), have ABA to ABA-GE catalysis activity (47, 48). However, our understanding on the spatiotemporal aspects of ABA homeostasis and the mechanisms underlying ABA translocation to control ABA responses under normal and stress conditions is limited.
Here, we exploited a genome-scale artificial microRNA (amiRNA) screen targeting the Arabidopsis transportome to identify novel ABCG ABA transporters. ABCG17 and ABCG18 are localized to the plasma membrane and import ABA. ABCG17- and ABCG18-knockdown lines displayed enhanced ABA accumulation, induced ABA response, and reduced stomatal aperture size, conductance, and reduced transpiration with increased water use efficiency compared to WT. In addition, seedlings with double loss of function of ABCG17 and ABCG18 showed enhanced long-distance ABA translocation from the shoot to the root, which led to lateral root outgrowth inhibition. ABCG17 and ABCG18 are primarily expressed in the shoot mesophyll and stem cortex cells, where they drive ABA import, allowing ABA-GE formation. In turn, these ABA-GE sinks prevent active ABA accumulation in guard cells and limit ABA long-distance travel to lateral root formation sites. Under abiotic stress conditions, ABCG17 and ABCG18 are transcriptionally repressed, promoting active ABA movement, accumulation, and response.
RESULTS
Transportome-scale amiRNA screen indicates involvement of class A ABCGs in ABA-mediated activity
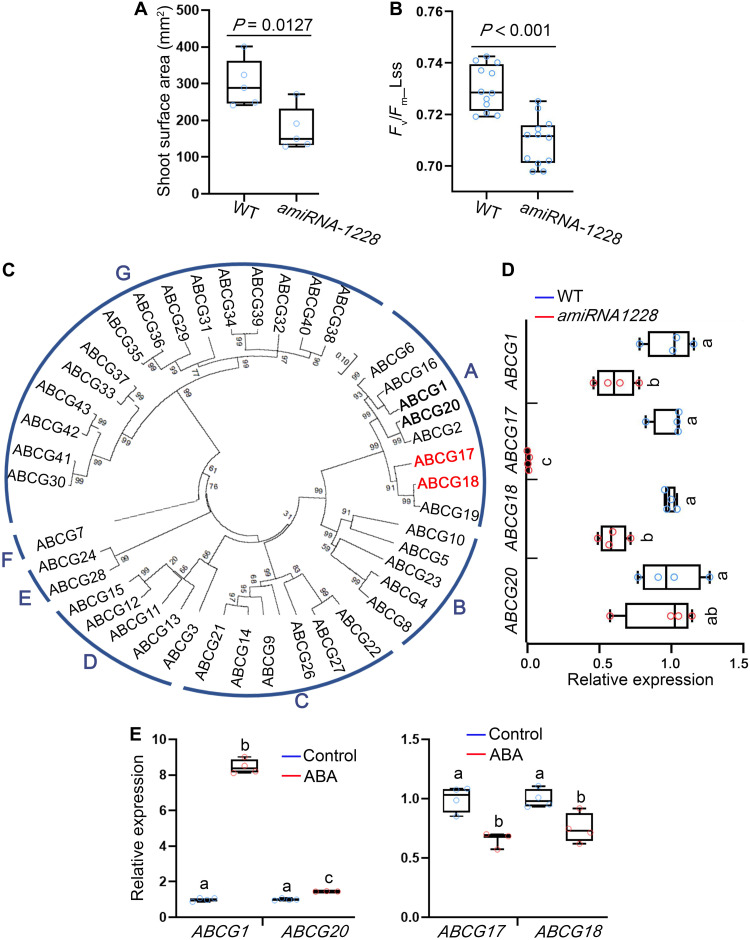
Over 75% of the genes in the Arabidopsis genome belong to gene families, complicating functional analyses (49). To partially overcome functional redundancy and reveal genetic factors involved in ABA homeostasis and translocation, we used a transportome-specific amiRNA collection of 3000 lines targeting multiple transporters from the same family (50). The transporter amiRNA sublibrary is part of the broad PHANTOM amiRNA tool (49). The seedlings in the amiRNA collection were screened for defects in shoot growth and photosynthesis-related parameters. The screen identified amiRNA-1228 line as an interesting candidate; this line had slight but significant shoot growth inhibition (Fig. 1A and fig. S1A). In addition, the amiRNA-1228 line showed a significant reduction in photosystem II quantum yield of light-adapted samples at steady state (Fig. 1B), indicating limited activity of photosystem II (51). We therefore hypothesized that the genes targeted by amiRNA-1228 might be related to ABA activity and homeostasis (52).
Fig. 1. Reduced expression of several class A ABCG members results in moderate shoot growth and decreased photosynthesis rate.
(A) Shoot surface area of 18-day-old WT and amiRNA-1228. Shown are averages (±SD), n = 5; P value indicates significant differences, Student’s t test. (B) Fv/Fm_Lss, indicating photosystem II quantum yield of light-adapted samples at steady state, measured for 18-day-old WT and amiRNA-1228. Shown are averages (±SD), n = 12; P < 0.001 (P = 1.15208 × 10−5) indicates significant differences, Student’s t test. (C) Phylogenetic tree of Arabidopsis ABCG family based on amino acid sequences. Red and bold fonts indicate proteins coded by putative amiRNA-1228 target genes. (D) Relative expression of the indicated ABCG amiRNA-1228–targeted genes in 12-day-old WT and amiRNA-1228 seedlings, quantified by qRT-PCR. Shown are averages (±SD), n = 4; different letters represent significant differences, one-way analysis of variance (ANOVA) with Student’s t test (P < 0.05). (E) Relative expression, quantified by qRT-PCR, of the indicated ABCG amiRNA-1228–targeted genes in response to ABA treatment (5 μM ABA for 3 hours) in 7-day-old seedlings. Shown are averages (±SD), n = 4; different letters represent significant differences, one-way ANOVA with Student’s t test (P < 0.05).
amiRNA-1228 putatively targets four class A ABCG subfamily members with different extents of complementarity in the potential recognition sites (fig. S1B). ABCG17 and ABCG18 are grouped in one branch, ABCG1 and ABCG20 are classified into another, of the Arabidopsis ABCG family phylogenetic tree (Fig. 1C). To test whether amiRNA-1228 indeed targets these genes, we examined their expression levels by quantitative reverse transcription polymerase chain reaction (qRT-PCR) in amiRNA-1228 plants compared to that in WT plants (Fig. 1D). ABCG1, ABCG17, and ABCG18 were significantly down-regulated in amiRNA-1228, but there was no difference in levels of ABCG20 (Fig. 1D). Next, we tested the transcriptional responses of these genes to ABA treatment. ABCG1 and ABCG20 were induced by 5 μM ABA treatment for 3 hours, whereas ABCG17 and ABCG18 were repressed (Fig. 1E). ABCG1 and ABCG16 were previously shown to regulate pollen and reproductive development (53, 54) and suberin formation in roots (55). ABCG20, ABCG2, and ABCG6 regulate suberin barriers in roots and seed coats (54). The functions of ABCG17 and ABCG18, located on separate phylogenetic branch (Fig. 1C), have not been characterized. Together, our results indicate that ABCG17 and ABCG18 genes are interesting candidates in the context of ABA-mediated activity.
ABCG17 and ABCG18 are redundantly required for ABA response and stomatal closure
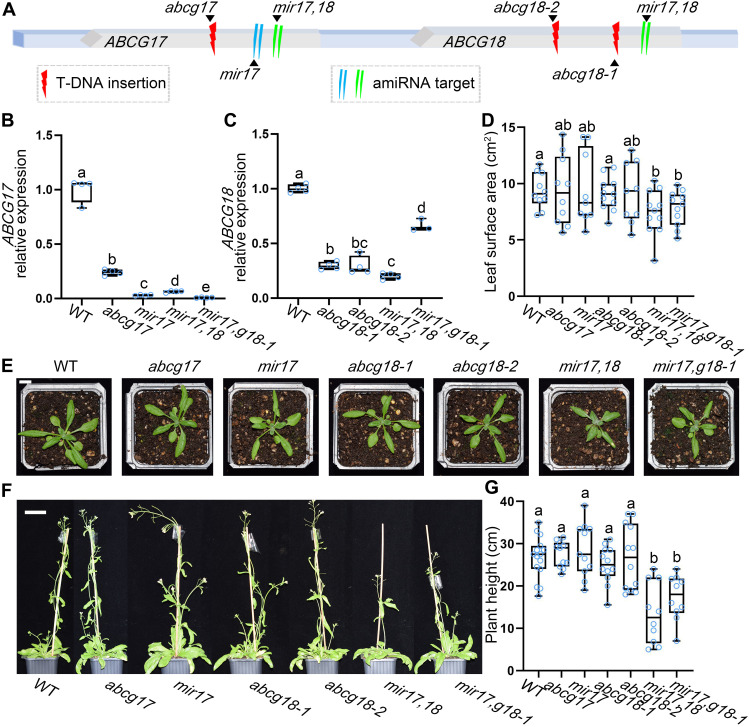
To determine whether ABCG17 and ABCG18 contribute to the amiRNA-1228 phenotype and to dissect their activity, we obtained transferred DNA (T-DNA) insertion lines for both genes (Fig. 2A). Because ABCG17 and ABCG18 are genetically linked on chromosome 3, we could not generate double mutants by crossing T-DNA insertion lines. Therefore, two independent amiRNA double-knockdown lines for ABCG17 and ABCG18 were generated. The first was an amiRNA targeting both ABCG17 and ABCG18 (mir17,18). The second was an amiRNA targeting ABCG17 (mir17) transformed into the abcg18-1 T-DNA insertion background (mir17,g18-1) (Fig. 2A). We examined the expression levels of ABCG17 and ABCG18 in the single- and double-knockdown lines compared to WT. Both ABCG17 and ABCG18 were significantly down-regulated in the respective backgrounds (Fig. 2, B and C). Next, we monitored the shoot phenotypes of abcg17 and abcg18 single-mutant and double-knockdown lines grown in soil under normal conditions. None of the single mutants showed shoot growth impairment, but both double-knockdown lines, mir17,18 and mir17,g18-1, displayed significant reductions in shoot surface area compared to WT and the single mutants (Fig. 2, D and E). Similarly, 50-day-old double-knockdown plants had shorter inflorescence stems compared to WT and single mutants (Fig. 2, F and G) but showed no differences in bolting time (except abcg18-2) (fig. S3). The results are in line with the amiRNA-1228 phenotype and indicate that ABCG17 and ABCG18 are redundantly required for plant shoot growth.
Fig. 2. ABCG17 and ABCG18 redundantly regulate plant growth.
(A) Illustration of the ABCG17 and ABCG18 genome region. T-DNA insertions and amiRNA knockdown sites are indicated. (B and C) Relative expression levels of ABCG17 (B) and ABCG18 (C) in the indicated genotypes. mir17,18 is the abcg17 and abcg18 double-knockdown amiRNA line; mir17,g18-1 is mir17 (amiRNA-ABCG17) transformed into the background of abcg18-1 T-DNA insertion line. Shown are averages (±SD), n = 4; different letters represent significant differences, one-way ANOVA with Student’s t test (P < 0.05). (D) Leaf surface area of the indicated lines. Shown are averages (±SD), n ≥ 9 plants; different letters represent significant differences, one-way ANOVA with Student’s t test (P < 0.05). (E) Shoot phenotypes of 25-day-old plants grown in soil under normal conditions. Scale bar, 1 cm. (F) Phenotypes of 50-day-old plants grown in soil under normal conditions. Scale bar, 2 cm. (G) Quantification of inflorescence stem height of the indicated genotypes. Shown are averages (±SD), n ≥ 10 plants; different letters represent significant differences, one-way ANOVA with Student’s t test (P < 0.05). Photo credit: Yuqin Zhang, Tel Aviv University.
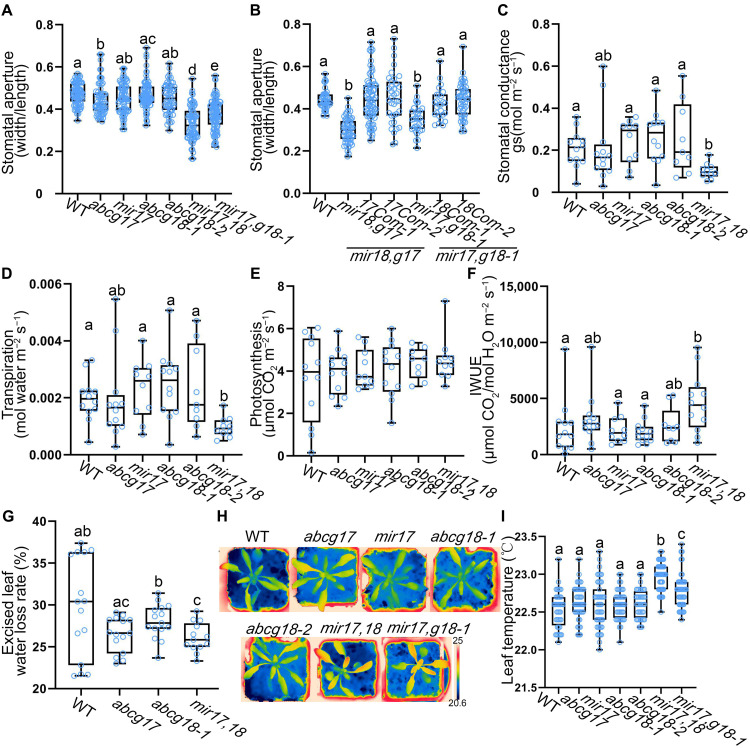
Since amiRNA-1228 showed photosynthesis-related phenotypes, we tested whether it resulted from ABCG17/ABCG18-dependent alteration of stomatal aperture size. Single abcg17 and abcg18 mutants did not differ from WT plants in stomatal aperture size. However, both mir17,18 and mir17,g18-1 had significantly smaller stomatal apertures than WT and single mutants (Fig. 3A).
Fig. 3. Stomatal aperture, conductance, and transpiration are regulated by ABCG17 and ABCG18.
(A) Stomatal aperture measurements of 25-day-old plants. Shown are averages (±SD), n ≥ 45. (B) Thirty-day-old plants stomatal aperture complementation analyses of two independent ABCG17 and ABCG18 double-knockdown lines. Com stands for complementation lines (pABCG17:ABCG17 or pABCG18:ABCG18). Shown are averages (±SD), n ≥ 26. (C) Plant stomatal conductance measurements of 25-day-old plants. (D) Leaf transpiration rates of the 25-day-old plants. For (C) and (D), shown are averages (±SD), n = 5 plants. (E) Leaf photosynthetic properties of 25-day-old plants, n = 5 plants. Results were not significant at P > 0.05 by one-way ANOVA with Student’s t test (P < 0.05). (F) Instantaneous water use efficiency (IWUE) of plants under normal conditions. Shown are averages (±SD), n = 5 plants. (G) Water loss rates of leaves excised from 30-day-old plants (90 min, room temperature air). Shown are averages (±SD), n ≥ 13. (H) Thermal images of 25-day-old plants grown on soil under normal conditions. Color-coded scale bar indicates temperature. Black scale bar, 1 cm. (I) Leaf temperatures of the indicated genotypes in (H). Shown are averages (±SD), n ≥ 40. For all graphs, different letters represent significant differences, one-way ANOVA with Student’s t test (P < 0.05).
To further validate whether the phenotypes observed in mir17,18 and mir17,g18-1 are indeed an on-target effect driven by ABCG17 and ABCG18, we generated independent CRISPR and complementation lines. CRISPR17,18 targets both ABCG17 and ABCG18 using a single sgRNA. CRISPR17,18 plants were sequenced and found to be mutated in both ABCG17 and ABCG18 genes (T2 generation), showing delayed shoot phenotype and significantly smaller stomatal apertures (fig. S2, A to C). In addition, we generated ABCG17 and ABCG18 complementation lines, pABCG17:ABCG17 in the background of mir18,g17 double knockdown, and pABCG18:ABCG18 in the background of mir17,g18-1. In both cases, the respective genomic construct is complementing a T-DNA mutation (g17 or g18-1). The complementation lines showed complete rescue of stomatal apertures phenotype (Fig. 3B), together validating the activity of ABCG17 and ABCG18 in regulating stomatal aperture.
To further study ABCG17 and ABCG18 activity in guard cell regulation, we tested stomatal conductance and gas exchange (i.e., transpiration and photosynthesis) in the mutant lines. The double-knockdown line had significantly reduced stomatal conductance compared to WT and single mutants (Fig. 3C). In line with stomatal conductance and aperture defects, the double-knockdown line had significantly lower transpiration rates than WT and single mutants (Fig. 3D). However, photosynthesis rates were not changed within experimental error in both single mutants and the double-knockdown line mir17,18 compared to WT plants (Fig. 3E). In agreement with this finding, the double-knockdown line mir17,18 showed an increase in instantaneous water use efficiency under well-watered conditions (Fig. 3F). Analysis of water loss rates from excised leaves of single mutants and mir17,18 showed delayed water loss rates compared to WT when exposed to air (Fig. 3G and fig. S4). Evaluation of leaf temperature using thermal imaging showed higher leaf temperature in both the double-knockdown lines compared to WT and single mutants (Fig. 3, H and I, and fig. S5). These data support the hypothesis that ABCG17 and ABCG18 are required for stomatal activity under normal conditions.
To further characterize ABCG17 and ABCG18 activity, we generated transgenic plants overexpressing (35S promoter) each of the two genes. Two independent lines for each gene showed a 50- to 100-fold change in transcriptional activation (fig. S6, A and B). ABCG17- or ABCG18-overexpressing lines showed no differences in shoot surface area from that of WT plants, but ABCG18 overexpression resulted in a slight plant height increase (fig. S6, C to F). Neither ABCG17- nor ABCG18-overexpressing lines showed differences in bolting time (fig. S6G). ABCG17 or ABCG18 overexpression resulted in reduced stomatal aperture (fig. S7, A and B), elevated leaf temperature (fig. S7, C and D), and induced ABA response, reflected by the induction of pRAB18:GFP (56) and pMAPKKK18:LUC (57) ABA reporters (fig. S7, E to G) compared to the control.
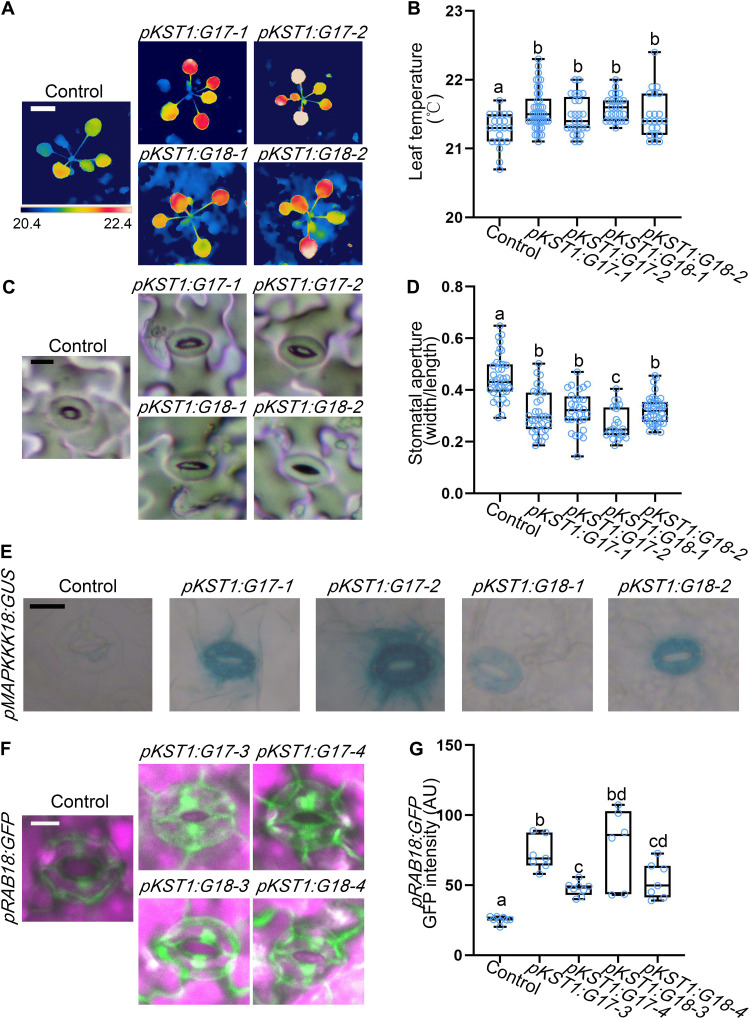
To further understand ABCG17 and ABCG18 function, we tested whether their ectopic activity in guard cells may promote ABA accumulation and responses. We therefore expressed ABCG17 and ABCG18 driven by a guard cell–specific (pKST1) promoter (58). pKST1:ABCG17 and pKST1:ABCG18 lines showed higher leaf surface temperature and closed stomatal aperture compared to the control (Fig. 4, A to D). In addition, both ABA reporters pMAPKKK18:GUS and pRAB18:GFP showed significantly stronger ABA responses in pKST1:ABCG17 or pKST1:ABCG18 background lines compared to the control (Fig. 4, E to G), implying that ABCG17 and ABCG18 may participate in ABA import. Together, these results suggest that ABCG17 and ABCG18 redundantly regulate stomatal aperture and might function as ABA transporters.
Fig. 4. Ectopic expression of ABCG17 or ABCG18 in guard cells promotes local ABA responses.
(A) Thermal images of 22-day-old plants of the indicated genotypes grown on soil under normal conditions. All lines are in the pMAPKKK18:GUS background. Color-coded scale bar indicates temperature. White scale bar, 1 cm. (B) Leaf temperatures of plants of the indicated genotypes measured in (A). Shown are averages (±SD), n ≥ 22; different letters represent significant differences, one-way ANOVA with Student’s t test (P < 0.05). (C) Stomatal impressions of the indicated genotypes. Scale bar, 10 μm. (D) Width/length ratio of stomatal apertures of 22-day-old plants of the indicated genotypes. All lines are in the pMAPKKK18:GUS background. Shown are averages (±SD), n ≥ 24; different letters represent significant differences, one-way ANOVA with Student’s t test (P < 0.05). (E) pMAPKKK18:GUS signal in guard cells of 12-day-old plants expressing pKST1:ABCG17 (pKST1:G17-1 and pKST1:G17-2) or pKST1:ABCG18 (pKST1:G18-1 and pKST1:G18-2). Scale bar, 10 μm. (F) pRAB18:GFP intensity in guard cells of 12-day-old plants expressing pKST1:ABCG17 (pKST1:G17-3 and pKST1:G17-4) or pKST1:ABCG18 (pKST1:G18-3 and pKST1:G18-4). Scale bar, 10 μm. Green stands for green fluorescent protein (GFP) fluorescent signal, and purple stands for chlorophyll. (G) Quantification of respective GFP signal intensity in (F). Shown are averages (±SD), n ≥ 6; different letters represent significant differences, one-way ANOVA with Student’s t test (P < 0.05). AU, arbitrary units.
ABCG17 and ABCG18 are plasma membrane–localized ABA importers
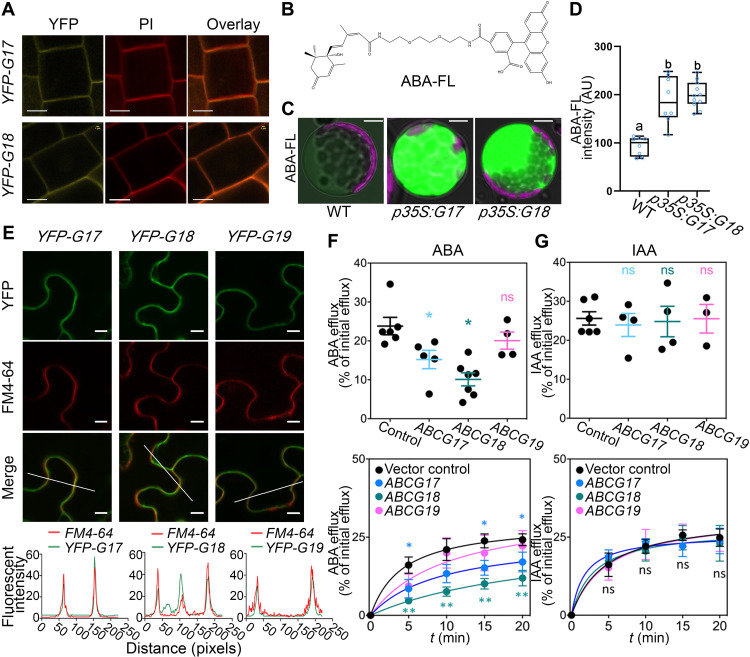
To determine the subcellular localizations of ABCG17 and ABCG18, we cloned the coding sequences of ABCG17 and ABCG18 and constructed vectors for expression of these proteins with N-terminal yellow fluorescent protein (YFP) tags under the control of the 35S promoter. Confocal microscopy of p35S:YFP-ABCG17 and p35S:YFP-ABCG18 transgenic lines in the root meristem revealed that ABCG17 and ABCG18 are localized to the plasma membrane (Fig. 5A).
Fig. 5. ABCG17 and ABCG18 are ABA importers localized to the plasma membrane.
(A) ABCG17 and ABCG18 subcellular localization shown by p35S:YFP-ABCG17 (YFP-G17) and p35S:YFP-ABCG18 (YFP-G18) stable transgenic Arabidopsis lines. Fluorescence was imaged in the root meristem epidermis layer. Yellow indicates YFP-ABCG17 or YFP-ABCG18 fluorescence, and red indicates propidium iodide (PI). Scale bars, 5 μm. (B) Molecular structure of ABA-FL: A fluorescein moiety is linked to the ABA carboxylic group. (C) ABA-FL fluorescent signal in Arabidopsis protoplasts overexpressing ABCG17 or ABCG18. p35S:G17 and p35S:G18 indicate p35S:ABCG17 and p35S:ABCG18, respectively. Protoplasts were treated with 5 μM ABA-FL for 12 hours. Green stands for ABA-FL fluorescent signal, and purple stands for chlorophyll. Scale bars, 10 μm. (D) Quantification of ABA-FL fluorescence intensity in protoplasts overexpressing ABCG17 or ABCG18. Shown are averages (±SD), n ≥ 8 protoplasts; different letters represent significant differences, one-way ANOVA with Student’s t test (P < 0.05). (E) Confocal imaging of tobacco leaves transfected with p35S:YFP-ABCG17 (YFP-G17), p35S:YFP-ABCG18 (YFP-G18), or p35S:YFP-ABCG19 (YFP-G19), stained with FM4-64 plasma membrane marker. Scale bars, 10 μm. Bottom graphs are quantified fluorescence intensities of pixels along the white line within the images. X axis represents distance along the line. (F) [3H]ABA efflux from tobacco protoplasts as percentage of initial efflux. Top graph is 15-min [3H]ABA, n ≥ 4, *P < 0.05, Welch’s t test. (G) [14C] indole acetic acid ([14C]IAA) efflux from tobacco protoplasts as percentage of initial efflux. Top graph is 15 min [14C]IAA, n ≥ 4; ns indicates not significant.
To examine whether ABCG17 and ABCG18 facilitate ABA transport, we synthesized fluorescently labeled ABA molecules by adding a fluorescein tag to the carboxylic group (Fig. 5B). Unlike the fluorescently labeled gibberellin molecules that we previously reported (59), the fluorescent ABA (ABA-FL) was only slightly bioactive in root growth assays, suggesting low binding affinity for the PYR/PYL/RCAR ABA receptor (fig. S8). We used the new ABA-FL molecule to test ABCG17 and ABCG18 transport activity. Isolated protoplasts from leaf mesophylls of transgenic Arabidopsis plants overexpressing ABCG17 or ABCG18 and WT as a control were treated with 5 μM ABA-FL for 12 hours and imaged. Protoplasts overexpressing ABCG17 or ABCG18 had enhanced ABA-FL fluorescent signal compared to WT protoplasts (Fig. 5, C and D), suggesting that both ABCG17 and ABCG18 promote ABA import.
To validate ABCG17 and ABCG18 ABA transport activity, we carried out a biochemical transport assay with radiolabeled ABA ([3H]ABA) in tobacco plants that overexpress each of these two transporters. ABCG17 or ABCG18 overexpression in tobacco leaves showed plasma membrane localization (Fig. 5E) and resulted in a significant reduction in measured protoplast export activity compared to the control (Fig. 5F). Reduced net export is in line with an import activity for both transporters that would function here as reimporters of [3H]ABA.
To test whether ABCG17 and ABCG18 transport ABA-GE, in addition to ABA, we quantified [3H]ABA uptake in protoplasts expressing ABCG18 in a competition experiment, in which unlabeled ABA-GE (or the solvent control) was offered in a 100-fold excess. ABA-GE inhibited [3H]ABA uptake into ABCG18-expressing protoplasts, indicating that ABA-GE might also serve as a substrate for ABCG18 (fig. S9).
In addition to the in planta transport assays, we examined the transport activity of ABCG17 and ABCG18 in yeast and Xenopus oocyte transport systems. Both genes were expressed individually and also in combination in these two systems, respectively. Significant ABA transport activity was observed for ABCG17, ABCG18, or combined ABCGs in yeast compared to the controls (fig. S10A). The influx of ABA into mock Xenopus oocytes is attributed to diffusion of protonated noncharged ABA when oocytes are incubated in acidic assay buffer containing ABA (60). The previously characterized ABCG25 (40) showed the expected export activity, reflected by reduced accumulation of ABA in oocytes (fig. S10B). Conversely, increased accumulation compared to mock indicated import activity. In agreement with the yeast system, ABCG17 and combined ABCGs showed significant import activity compared to the mock in the Xenopus oocyte system; however, although there is an import activity trend in comparison to the mock, no significant transport activity was observed for ACBG18 in oocytes (fig. S10B and table S1).
To determine the specificity of ABCG17 and ABCG18 in ABA transport activity, we tested the effects of overexpressing ABCG19, the closest gene in the cluster (Fig. 1C). Notably, despite a trend, overexpression of ABCG19 did not result in a significant ABA transport activity (Fig. 5F). We also evaluated transport of a radioactively labeled auxin ([14C]IAA), such as ABA, an organic acid and a known substrate of members of the ABCB subclass. ABCG17- or ABCG18-overexpressing lines did not reveal altered transport of auxin compared to the control (Fig. 5G). In summary, these experiments indicate that ABCG17 and ABCG18 are plasma membrane–localized ABA importers.
ABCG17 and ABCG18 limit long-distance ABA transport to regulate lateral root development
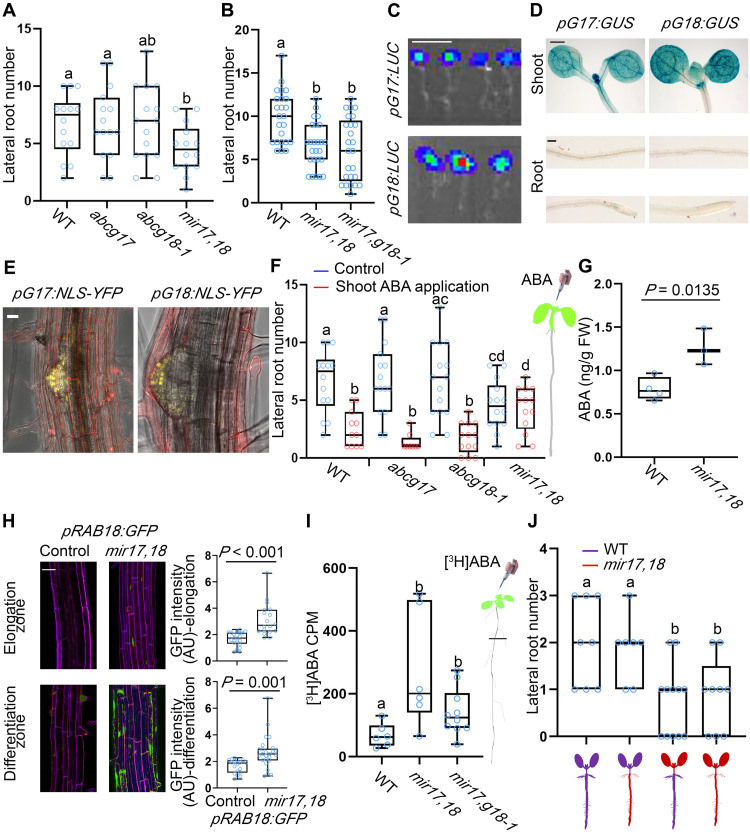
In addition to the stomatal conductance and aperture size phenotypes, double knockdown of ABCG17 and ABCG18 resulted in reduced lateral root number compared to WT and single mutants (Fig. 6A). More specifically, mir17,18 and mir17,g18-1 showed normal lateral root initiation (fig. S11, B and C, and tables S2 and S3) but reduced lateral root primordia outgrowth compared to WT under normal conditions (Fig. 6B and fig. S11A). This result is intriguing because the expression of ABCG17 and ABCG18 is largely restricted to the shoot: Luciferase reporter lines (pABCG17:LUC and pABCG18:LUC), β-glucuronidase (GUS) reporter lines (pABCG17:GUS and pABCG18:GUS), yellow fluorescent protein (YFP) reporter lines (pABCG17:NLS-YFP and pABCG18:NLS-YFP), and qRT-PCR measurements showed strong signals in shoots (Fig. 6, C to E, and fig. S12). A very weak signal was found in lateral root emerging primordia using the pABCG17:NLS-YFP pABCG18:NLS-YFP lines, only when applying maximum laser power and using a high-sensitivity gallium arsenide phosphide (GaAsP) detector (Fig. 6E and fig. S13, A and B). ABA regulates key stages of lateral root postemergence development (61). Specifically, ABA signaling plays a crucial role in salt stress–induced lateral root suppression (5, 62) and lateral root growth recovery (63), associated with an increase in miR165a and a reduction in PHB (PHABULOSA) levels (64). In addition to ABA, it might be possible that reduced photosynthetic assimilation affects lateral root number. Duan et al. (65) recently reported on the importance of CYP38-dependent photosynthetic activity in supporting root growth. Because the photosynthesis rates of the ABCG17,18 double and single mutants are not significantly different from WT (Fig. 3E), we speculated that ABCG17,18 and CYP38 function through distinct mechanisms to regulate lateral root formation.
Fig. 6. ABCG17 and ABCG18 are required for long-distance ABA transport and regulation of lateral root formation.
(A or B) Lateral root numbers of 10-day-old seedlings, n ≥ 14 (A) and n ≥ 18 (B). (C) Bioluminescence signal driven by pABCG17:LUC (pG17:LUC) or pABCG18:LUC (pG18:LUC). Scale bar, 1 cm. (D) GUS staining for pABCG17:GUS (pG17:GUS) and pABCG18:GUS (pG18:GUS) in shoot and root of 5-day-old seedlings. Scale bars, 0.5 mm for shoots and 0.1 mm for roots. (E) Representative images of pABCG17:NLS-YFP (pG17:NLS-YFP) and pABCG18:NLS-YFP (pG18:NLS-YFP) lateral root emergence sites. Scale bar, 20 μm. (F) Lateral root numbers for the indicated genotypes. Shoots of 4-day-old plants were treated with 5 μM ABA for 4 days, n ≥ 13. Right is shoot-specific ABA application illustration. (G) ABA quantification in roots of indicated genotypes (12-day-old plants). FW, fresh weight. n ≥ 3; P value indicates significant differences, Student’s t test. (H) pRAB18:GFP signal in the background of mir17,18 and control roots (left) and the quantification of respective GFP signal intensity (right). Green represents GFP fluorescent signal, and purple represents propidium iodide. Scale bar, 20 μm. n ≥ 7; P value indicates significant differences, Student’s t test. (I) [3H]ABA counts per minute (CPM) in roots only of the indicated phenotypes (roots were isolated as indicated by a black line in the illustration on the right). Background measurements from control samples (no [3H]ABA treatment) were deduced, n ≥ 6. (J) Lateral root number quantification of the indicated grafted lines, n ≥ 7. For all graphs, shown are averages (±SD); different letters represent significant differences, one-way ANOVA with Student’s t test (P < 0.05).
To test whether shoot-born ABA can affect lateral root development, we applied ABA specifically to shoots and quantified lateral root length and lateral root number. The results indicated that ABA application to WT shoots inhibited lateral root elongation and lateral root development (fig. S14, A and B). Moreover, ABA application to the shoots induced expression of the pMAPKKK18:GUS ABA reporter (57) in the root (fig. S14C). Shoot-specific ABA application inhibited lateral root emergence in the single mutants similarly to WT, but the mir17,18 double-knockdown mutant was insensitive to ABA application to shoots (Fig. 6F). We speculated that mir17,18 already accumulates high enough ABA levels in the root compared to WT under normal conditions.
To test this hypothesis, we quantified ABA levels in roots. The double-knockdown mir17,18 plants accumulated significantly higher ABA amounts in the root compared to WT (Fig. 6G). The data showed more than 85% accuracy, among different replicates and experiments, suggesting good recovery of the analytes after sample preparation steps, with minimal losses (table S4). In addition, we analyzed the ABA reporter pRAB18:GFP in the background of ABCG17 and ABCG18 loss-of-function lines. We found enhanced pRAB18:GFP signal in mir17,18 roots compared to control roots (Fig. 6H), supporting the hypothesis that ABCG17 and ABCG18 knockdown causes abnormally high levels of ABA accumulation in the root.
To further understand an apparent ABCG17 and ABCG18 activity in limiting ABA shoot-to-root translocation, we treated shoots with radioactively labeled ABA (0.1 μCi/ml [3H]ABA) and quantified counts in roots after 24 hours. Both of the two double-knockdown lines mir17,18 and mir17,g18-1 roots accumulated higher levels of [3H]ABA than the WT roots (Fig. 6I), supporting the hypothesis that ABCG17 and ABCG18 activities restrict long-distance transport of ABA to the root. Because auxin is a key mobile molecule required for lateral root formation, we carried similar experiments with shoot-applied radioactively labeled [14C]IAA. Root extractions did not reveal significant differences between mir17,18 and WT (fig. S15). To further confirm ABCG17 and ABCG18 activity in long-distance ABA translocation, we carried out reciprocal WT and mir17,18 grafting assays. The results showed that, within experimental error, WT scion/mir17,18 rootstock grafts developed the same number of lateral roots as WT self-grafted plants. However, mir17,18 scion/WT rootstock grafts developed significantly fewer lateral roots, with lateral root numbers similar to mir17,18 self-grafted plants (Fig. 6J). These results indicate that ABCG17 and ABCG18 redundantly restrict shoot-to-root ABA translocation, mediating lateral root development under normal conditions.
ABCG17 and ABCG18 are expressed in shoot mesophyll and cortex cells under nonabiotic stress conditions
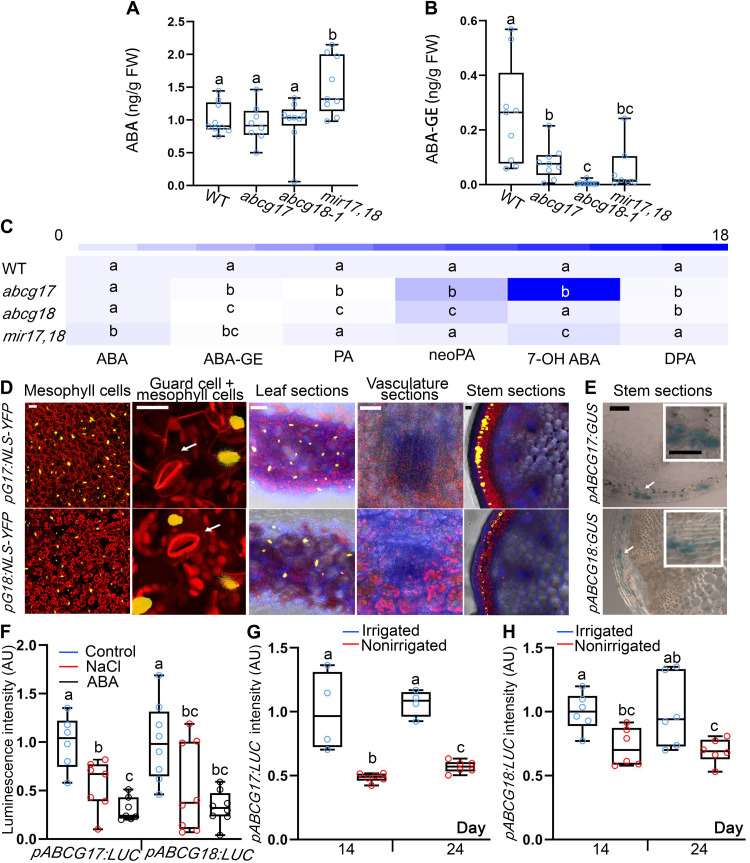
The obtained results suggest that ABCG17 and ABCG18 limit ABA levels in guard cells and roots, through a process that takes place in the shoot. Therefore, we speculated that ABCG17 and ABCG18 redundantly regulate ABA homeostasis, possibly by allowing ABA storage. To test this hypothesis, we evaluated ABA and ABA-GE levels in shoots. The double-knockdown mir17,18 plants accumulated higher ABA levels in the shoot than WT or single-mutant plants (Fig. 7A), consistent with results showing that double-knockdown lines have reduced stomatal aperture size, conductance, transpiration, and elevated leaf temperature (Fig. 3). The double-knockdown mir17,18 shoots accumulated less ABA-GE than WT shoots (Fig. 7B), suggesting that ABCG17 and ABCG18 might promote ABA-GE production catalyzed by UGTs. To better characterize ABCG17 and ABCG18 function in ABA metabolism, we quantified additional ABA metabolites. Several ABA-inactive catabolites and conjugated forms showed profound effects in ABCG17 and ABCG18 mutants (Fig. 7C and fig. S16). High accuracy and repeatability were measured for shoot ABA and ABA metabolite recovery method (table S4 and dataset S1).
Fig. 7. ABCG17 and ABCG18 are primarily expressed in mesophyll and cortex cells, allowing ABA-GE formation under normal conditions.
(A and B) Quantification of ABA (A) and ABA-GE (B) in shoots of 12-day-old plants of the indicated genotypes. Shown are averages (±SD), n ≥ 7; different letters represent significant differences, one-way ANOVA with Student’s t test (P < 0.05). (C) ABA metabolites profile heatmap in shoots of 12-day-old plants of the indicated genotypes. Absolute values and abbreviations are presented in fig. S16. Shown are averages (±SD), n ≥ 7, different letters represent significant differences, one-way ANOVA with Student’s t test (P < 0.05). (D) pABCG17:NLS-YFP (pG17:NLS-YFP) and pABCG18:NLS-YFP (pG18:NLS-YFP) signal (yellow) in leaves and stem. YFP signal is detected in mesophyll and cortex cells. Chlorophyll is in red. Fluorescent brightener is in blue. White arrows point to the guard cells. Scale bars, 20 μm. (E) GUS staining for pABCG17:GUS (pG17:GUS) and pABCG18:GUS (pG18:GUS) in inflorescence stem of 30-day-old plants. Scale bar, 100 μm. Insets are magnifications of areas indicated by white boxes. Scale bars, 50 μm. (F) Luminescence intensity of pABCG17:LUC (pG17:LUC) and pABCG18:LUC (pG18:LUC) with and without 5 μM ABA or 100 mM NaCl treatment. Shown are averages (±SD), n ≥ 6; different letters represent significant differences, one-way ANOVA with Student’s t test (P < 0.05). (G and H) Luminescence intensity of pABCG17:LUC (pG17:LUC) (G) and pABCG18:LUC (pG18:LUC) (H). Plants were irrigated for 15 days followed by water withhold for 3 weeks. Control plants (irrigated) were watered throughout the experiment. Shown are averages (±SD), n ≥ 4; different letters represent significant differences, one-way ANOVA with Student’s t test (P < 0.05).
To identify the exact cell types that accumulate ABA because of transport facilitated by ABCG17 and ABCG18, we characterized the expression patterns of ABCG17 and ABCG18 in leaf and stem sections. pABCG17:NLS-YFP and pABCG18:NLS-YFP lines showed that ABCG17 and ABCG18 are primarily expressed in parenchyma shoot cells, which are the leaf mesophyll cells (Fig. 7, D and E, and fig. S17, A and B). We did not detect expression in the leaf or stem veins or bundle sheath cells (Fig. 7, D and E). These results imply that under normal conditions, ABCG17 and ABCG18 promote ABA uptake into shoot parenchyma cells, leading to increased ABA-GE levels. Thus, under nonstress conditions, low levels of free shoot-born ABA are available to guard cells and for long-distance movement to roots.
Next, we wanted to elucidate how ABCG17 and ABCG18 function in ABA-mediated homeostasis under abiotic stress conditions, when high levels of ABA are needed. Both of the two double-knockdown lines exhibited stronger resistance by showing more and longer lateral roots in 100 mM NaCl treatment compared to WT and single mutants (fig. S11, A, D, and E). Characterizing lateral root number, lateral root length, and lateral root primordium of ABCG17-overexpressing lines with and without 100 mM NaCl treatment did not show significant difference with NaCl treatment compared to WT (fig. S18 and tables S5 and S6). However, one of the ABCG18-overexpressing lines presented more lateral root compared to WT with 100 mM NaCl treatment (fig. S19). To further study this, we expressed ABCG17 or ABCG18 driven by a phloem-specific (pSUC2:XVE) promoter (estradiol-induced system). pSUC2:XVE:ABCG17 and pSUC2:XVE:ABCG18 lines did not change the lateral root number compared to WT but showed less primordium with estradiol treatment (fig. S20 and tables S9 to S11). pSUC2:XVE:ABCG18 lines displayed an increase in lateral root number but with a similar response to WT in response to 100 mM NaCl treatment (fig. S21 and tables S12 to S14).
To study how ABCG17 and ABCG18 function in ABA-mediated homeostasis under stress conditions, we monitored the expression levels of ABCG17 and ABCG18 in response to abiotic stresses using pABCG17:NLS-YFP and pABCG18:NLS-YFP plants, as well as pABCG17:LUC and pABCG18:LUC plants. ABCG17 and ABCG18 were transcriptionally repressed following ABA or NaCl treatment (Fig. 7F and fig. S22, A and B), consistent with qRT-PCR experiments following ABA treatment (Fig. 1E). Similarly, long-period water-withhold experiments showed that ABCG17 and ABCG18 are repressed as luminescence intensity of promoter-driven luciferase reporters was reduced in plants not irrigated for several days (14 and 24 days) (Fig. 7, G and H, and fig. S22C). Simultaneously, we checked radioactively labeled ABA in the roots of double-mutant lines once applied only to shoot under NaCl or no NaCl treatment. The accumulated levels of [3H]ABA in WT roots under NaCl stress reached that in the double-mutant lines (fig. S22D), further demonstrating that under stress conditions, ABCG17 and ABCG18 are repressed and cannot maintain ABA levels in the shoot mesophyll cells.
In summary, our experiments imply that ABCG17 and ABCG18 are necessary for ABA homeostasis by creating ABA-GE sinks in mesophyll and cortex cells. These ABA-GE sinks limit ABA accessibility to guard cells and ABA availability for long-distance translocation to regulate lateral root formation. The ABA homeostasis shoot sink is released once the plant senses abiotic stress conditions.
DISCUSSION
Here, we used a transportome-scale amiRNA screen to overcome functional redundancy to identify two previously unstudied ABCG transporters, ABCG17 and ABCG18. Compared to WT and single abcg17 and abcg18 mutants, the double-knockdown lines had higher ABA content and reduced ABA-GE content in shoots. Several ABA catabolites, including Phaseic acid (PA) and neo Phaseic acid (neoPA) showed significant metabolic changes in the ABCG17 and ABCG18 single mutants without displaying pronounced physiological or developmental phenotypes. This may suggest that the two ABCG transporters might be partially redundant with differences that might reflect changes in substrate specificity toward ABA metabolites or slight differences in expression patterns. The dissimilarity between metabolite abundance and phenotypic differences can also be seen in the previous study (66).
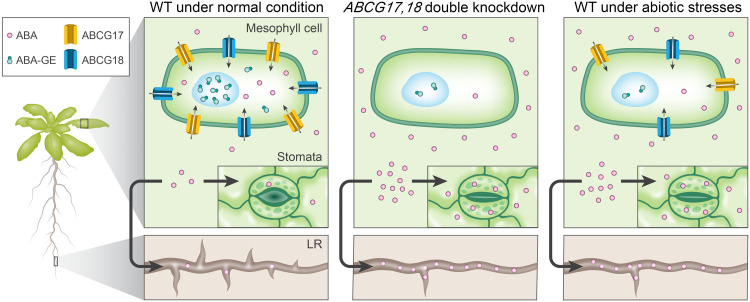
Given that the double-knockdown lines show high ABA content and reduced ABA-GE content in shoots compared to WT and that ABCG17 and ABCG18 are primarily expressed in the shoot cortex and mesophyll cells, localized to the plasma membrane, and promote ABA import, we hypothesized that the two proteins are redundantly required for ABA accumulation and storage in these cells (Fig. 8). The ABCG17- and ABCG18-dependent ABA sink in mesophyll cells limits the hormone translocation and activity to guard cells and the lateral root emergence sites, two distinct ABA-mediated processes. The two redundant transporters are required for ABA homeostasis under normal conditions, whereas the suggested conjugated–ABA sink mechanism is restricted in abiotic stress environments, enabling rapid ABA responses at distinct target sites (Fig. 8). It should be noted that the data presented in fig. S22D, showing that WT accumulates higher (but not statistically significant) levels of ABA in roots, following application in shoots, under nonstress and stress conditions, do not fit the proposed model.
Fig. 8. Proposed model illustrating ABCG17 and ABCG18 function in regulating ABA homeostasis under normal and abiotic stress conditions.
Under normal conditions, ABCG17 and ABCG18 promote ABA import into mesophyll cells. Under abiotic stress environments, ABCG17 and ABCG18 are transcriptionally repressed (similar to the double-knockdown lines), leading to free ABA availability to guard cells and long-distance translocation to the root. LR, lateral root.
Our model raises several questions. It is unknown how free ABA, which is not taken into the shoot cortex or mesophyll cells, finds its way to the guard cells or lateral root formation sites, which requires short- or long-distance transport, respectively. ABA might move through the apoplast, passively diffuse, or actively translocate over the plasma membrane. Several ABA transporters have been identified so far. In Arabidopsis, ABCG40 imports ABA into guard cells (15, 45), and in tomato, AIT1.1 was recently shown to play a role in ABA import into guard cells in an aspartic acid–glutamic acid–leucine–leucine–alanine (DELLA)–dependent manner (67). In addition, NPF4.6 regulates vasculature ABA import, and ABCG25 and DTX50 regulate vasculature ABA export (24). We speculate that it is unlikely that free ABA can travel from shoot to root by simple diffusion. Therefore, additional transporters remain to be identified to mediate shoot-to-root ABA transport. These transporters are expected to load ABA into the shoot phloem and unload ABA in the root to regulate lateral root development. Our proposed model suggests that ABCG17 and ABCG18 mediate the rate-limiting step for ABA accumulation in shoots, thus allowing ABA translocation to roots, while ABA-GE accumulates within the shoot cortex and mesophyll cells (Fig. 8). However, it is currently unclear whether ABA-GE itself can travel from root to shoot and vice versa (29), and further tools are required to characterize such mechanisms.
Previous studies have shown that ABA translocation from root to shoot via the xylem is required for stomatal closure, which restricts transpiration and water loss under drought stress (12–16). Little is known, however, about shoot-to-root transport via the phloem. McAdam et al. (30) presented evidence that most of the ABA found in the tomato root is synthesized in the shoot. It is therefore possible that ABCG17 and ABCG18 might serve a role in the mechanism regulating this process in Arabidopsis. Further studies in tomato and other species are needed to confirm whether the ABCG17- and ABCG18-mediated shoot-to-root ABA translocation mechanism is conserved. Manzi et al. showed that the accumulation of ABA in roots after long-term water stress largely relies on shoot-to-root ABA transport (68), a process that is likely conserved in Arabidopsis (17), citrus (69), and other species. We show that ABCG17 and ABCG18 are down-regulated under abiotic stress conditions, supporting the idea that their reduced activity promotes shoot-to-root ABA translocation under stress conditions.
The model proposed here suggests that ABCG17 and ABCG18 drive an inactive ABA-GE sink that restricts stomatal activity and lateral root formation. However, we currently have little information on the cell-type or subcellular distributions of ABA and ABA-GE or of the associated ABA-glucosyltransferase localization. Because ABA-GE competition assays significantly inhibited ABA import by ABCG18, it is possible that ABCG18 can also transport ABA-GE, a mechanism that would further enhance ABA-GE sink into shoot cortex and mesophyll cells. Future work, including direct transport assays, is needed to confirm this observation.
A detailed map of ABA-glucosyltransferase activity is required to plot ABA and ABA-GE movement in high resolution. Such a task remains challenging as this large family of enzymes is likely regulated at the protein level, and profiling the expression pattern will not be sufficient. High-resolution fluorescent probes specific for ABA-glucosyltransferase that can be used in live plants would be extremely useful tools in the field. There are some indications for ABA-GE accumulation in intracellular storage organelles such as vacuoles (70), but it remains unclear exactly how ABA and ABA-GE distribute within the mesophyll cells. We speculate that ABCG17 and ABCG18 activity leads to high levels of free ABA within the mesophyll and shoot cortex cells, which is then converted to ABA-GE by UDP-glycosyltransferases. Current models suggest that the ABA-GE is translocated to the endoplasmic reticulum and vacuole for storage (45, 71). Burla et al. (72) suggested that two distinct membrane transport mechanisms are active in Arabidopsis mesophyll cells: a proton gradient-driven and an adenosine triphosphate–binding cassette transporter–mediated. They demonstrated that ABCC1 and ABCC2 are localized in tonoplasts and exhibit ABA-GE transport activity in a yeast heterologous expression system (72, 73). Therefore, it is possible that ABCC and ABCG transporters work in concert: ABCG17 and ABCG18 import free ABA into mesophyll cells, and ABCC1 and ABCC2 mediate storage of ABA-GE in vacuoles. In addition to the need to identify subcellular ABA-GE transporters, there is no direct evidence of ABA and ABA-GE distribution to different organelles. New biochemical and genetic tools are required to generate a comprehensive ABA profile at subcellular resolution.
METHODS
Plant material and growth conditions
All Arabidopsis thaliana lines are in Col-0 background. For assays on plates, sterilized seeds were plated on 16 × 16 cm square petri dishes or 8.5-cm round petri dishes with growth media containing 0.5× Murashige-Skoog (MS) medium (pH 5.95 to pH 6.1, 1% sucrose, 0.8% plant agar). The sterilized seeds were transferred to growth chambers (Percival, CU41L5) at 21°C, light intensity (150 μEm−2 S−1) under long-day conditions (16-hour light/8-hour dark) after 3-day stratification at 4°C in the dark. For soil experiments, seedlings were transferred onto soil and grown in dedicated growth rooms under long-day conditions (16-hour light/8-hour dark) at 21°C. The following ABA reporters were previously described: pMAPKKK18:GUS, pMAPKKK18:LUC (57), and pRAB18:GFP (56).
Agrobacterium transformation
GV3101 electrocompetent Agrobacterium tumefaciens strain was incubated on ice with ~100 ng of plasmid for 5 min and electroporated using a MicroPulser (Bio-Rad Laboratories; 2.2 kV, 5.9 ms). Immediately after electroporation, 600 μl of LB medium was added, and the sample was shaken for 2 hours at 28°C. The Agrobacterium was then plated on selective LB agar plates containing the relevant antibiotics for 2 days at 28°C.
Arabidopsis transformation
Agrobacterium vectors were validated by colony PCR and sequencing before growing in 100 ml of LB medium containing gentamycin (25 μg/ml), rifampicin (50 μg/ml), and vector-specific antibiotic for ~36 hours at 28°C. Agrobacterium was harvested by centrifugation for 10 min at 4000 rpm, supernatant was discarded, and the bacteria pellet was resuspended in 40 ml of 5% sucrose + 0.05× MS + 0.02% Silwet L-77. Arabidopsis flowers were dipped into the agrobacterial solution for ~5 min. After dipping, plants were kept in the dark for ~18 hours and grown until seeds were harvested. T1 seeds were sown on 0.5× MS media containing the appropriate antibiotics for selecting transgenic plants or on soil with 0.1% basta spraying on ~10-day-old plants.
Genotyping
T-DNA insertion lines for single mutants ordered from Gabi Kat (www.gabi-kat.de) and The Arabidopsis Information Resource (www.arabidopsis.org/) are listed in table S15. Primers for the T-DNA insertion mutant genotyping were designed using the T-DNA Primer Design Tool powered by Genome Express Browser Server (http://signal.salk.edu/tdnaprimers.2.html). Homozygous mutants were characterized by PCR carried out with primers listed in table S16.
Cloning
ABCG17, ABCG18, and ABCG19 coding regions were amplified using Phusion High-fidelity Polymerase (New England Biolabs) from Col-0 complementary DNA (cDNA) using primers listed in table S17. Promoters of ABCG17 and ABCG18 were amplified from Col-0 DNA using Phusion High-fidelity Polymerase (New England Biolabs) using primers listed in table S17. Promoters of ABCG17 and ABCG18 are 1276 and 1580 base pairs long, respectively, including 5′ untranslated region. ABCG17, ABCG18, and ABCG19 coding regions as well as ABCG17 and ABCG18 promoter fragments were cloned into pENTR/D-TOPO (Invitrogen K2400), verified by sequencing, and subsequently cloned into binary destination vectors using LR Gateway reaction (Invitrogen 11791). p35S:YFP-ABCG17 and p35S:YFP-ABCG18 were generated using the pH7WGY2 vector and were selected using spectinomycin in Escherichia coli and hygromycin in plants. p35S:ABCG17 and p35S:ABCG18 were generated using pH2GW7 vector and selected using spectinomycin in E. coli and hygromycin in plants. p35S:XVE:ABCG17 and p35S:XVE:ABCG18 were generated using pMDC7 vector and selected using spectinomycin in E. coli and hygromycin in plants. pABCG17:LUC and pABCG18:LUC were generated using the pFlash vector and selected using spectinomycin in E. coli and gentamycin in plants. pABCG17:NLS-YFP and pABCG18:NLS-YFP were generated using R1-R2:NLS-YFP in pART27 vector and selected using spectinomycin in E. coli and kanamycin in plants. pABCG17:GUS and pABCG18:GUS were generated using pWGB3 vector, kanamycin and hygromycin in E. coli and hygromycin in plants. p35S:amiRNA lines were generated using the pH2GW7 vector, spectinomycin in E. coli and hygromycin in plants. Using primers with restriction enzyme Asc I, pABCG17 and pABCG18 were ligated into ABCG17 CDS and ABCG18 CDS using Asc I site. LR Gateway reaction was carried with the binary vector pGWB1. pABCG17:ABCG17 and pABCG18:ABCG18 were transformed into mir18,g17 and mir17,g18-1, respectively, for phenotype complementation assays. To generate pSUC2:XVE:ABCG17CDS:NosT, pSUC2:XVE:ABCG18CDS:NosT, pKST1:ABCG17CDS:NosT, and pKST1:ABCG18CDS:NosT, multisite Gateway reaction was carried (LR+): pSUC2:XVE or pKST1 (in p1p4r position) with ABCG17CDS or ABCG18CDS, respectively (in pENTR), and NosT terminator (in 2R3e position) and cloned into pB7m34GW.
The WMD3 website was used to design amiRNAs (http://wmd3.weigelworld.org/cgi-bin/webapp.cgi) (table S18). The amiRNA sequences were synthesized by Syntezza Bioscience Ltd. and were cloned into the pUC57 vector with Gateway system borders and then cloned the amiRNAs into the pH2GW7 destination vector using the Gateway system.
A single single-guide RNA was designed to target both ABCG17 and ABCG18 and cloned into Cas9/Chimera vector. Cloning primers and sequencing primers are listed in tables S19 and S20, respectively.
Plant genetics
All transgenic lines were generated from the destination vectors transformed into Col-0, with the following exceptions: pSUC2:XVE:ABCG17/18CDS:NosT and pKST1:ABCG17/18CDS:NosT were transformed into the ABA reporters pMAPKKK18:GUS or pRAB18:GFP, respectively. T1 seeds were collected and selected on 1/2 MS plates containing appropriate antibiotics or on soil with basta spraying. At least 10 independent lines for each construct were generated. Two representative homozygous lines were obtained for further characterization. To generate overexpression lines in the background of the ABA reporters, the homozygous ABCG17 or ABCG18 overexpression lines were crossed with the ABA reporter line pRAB18:GFP, pMAPKKK18:GUS, or pMAPKKK18:LUC. F1 heterozygous seeds for both constructs were used for analysis. As a control, the respective ABA reporters were crossed with Col-0 to generate F1 seeds. To generate double-knockdown line mir17,18 in the background of the ABA reporters, the homozygous mir17,18 line was crossed with pMAPKKK18:GUS to obtain F1 seeds. Heterozygous seeds were used for further analysis. As a control, the pMAPKKK18:GUS ABA reporter was crossed with Col-0 to generate F1 seeds.
Phylogenetic tree
A phylogenetic tree of Arabidopsis ABCG family members, based on protein sequences, was constructed using sequences alignment by Clustal W (www.clustal.org/clustal2/e). The one neighbor-joining phylogenetic tree was constructed using MEGA7.0 (Molecular Evolutionary Genetics Analysis) software with 1000 bootstrap replications.
Gas exchange measurements
Gas exchange was measured using a LI-COR LI-6800 portable gas exchange system. Plants were grown in short-day conditions (8-hour light/16-hour dark). Photosynthesis was induced under light (150 μmol m−2 s−1) with CO2 (400 μmol mol−1) surrounding the leaves. The amount of blue light was set to 10% of the photosynthetically active photon flux density. The flow rate was set to 200 μmol air s−1, and the leaf-to-air vapor pressure deficit was kept around 1 kPa during the measurement. Leaf temperature was ~22°C. Measurements were performed between 10:00 a.m. and 1:00 p.m. (3 hours after the light was switched on).
Fluorescently labeled ABA synthesis
The synthesis and characterization of ABA-FL are described in the Supplementary Materials (figs. S23 and S24).
Fluorescently labeled ABA protoplast experiments
ABA-FL was dissolved in dimethyl sulfoxide to 5 mM stock solution. Protoplasts isolation was carried as previously described (74) with the following adaptations: Protoplasts were treated with ABA-FL for 12 hours at a final concentration of 5 μM ABA-FL, washed with 500 μl of W5 solution [2 mM solution (pH 5.7) containing 154 mM NaCl, 125 mM CaCl2, and 5 mM KCl] three times, resuspended in 100 μl of W5 solution, and mounted on slides for confocal microscopy.
Stomatal aperture measurements
Stomatal aperture was determined using a rapid and almost permanent imprinting technique previously described (75). Leaves of similar size were cut from 25-day-old plants. The abaxial side of the leaves was attached to 0.5-ml silicone impression material (elite HD+, Zhermack Clinical). The leaves were removed after the silicone impression material dried. Subsequently, transparent nail varnish was placed on the epidermal side. The samples were placed on slides with coverslips and imaged with a light microscope (75). The stomatal aperture size was quantified using Fiji software.
RNA extraction, cDNA synthesis, and quantitative PCR
RNA was isolated using the SV Total RNA Isolation System (Promega). For cDNA synthesis, the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) was used according to manufacturer information. Fast SYBR Green Master Mix (Applied Biosystems) was used for quantitative PCR as described before (76). The primers used for quantitative PCR are listed in table S21.
Thermal imaging
Thermal images were obtained using an FLIR T6xx series instrument. The camera was placed vertically ~50 cm above the plants. Leaf temperature was quantified using a customized region-of-interest tool implemented according to the manufacturer’s instructions.
Histology, microscopy, and phenotypic characterization
Sections were made using the Vibratome technology as previously described (77). Sections were stained with 0.01% (grams per milliliter) Fluorescent Brightener 28 in 1× phosphate-buffered saline (PBS) for 20 min, washed with 1× PBS three times (10 min each wash), and subsequently imaged with a Zeiss LSM780 confocal microscope. Plant shoot images were taken using a Nikon camera (WEM ED IF Aspherical Micro 1:1 Φ 62). For root growth assays, seedlings were grown on plates and scanned using HP Scanjet G3110. Leaf size and root length were measured using Fiji software (https://fiji.sc/).
GUS staining and histology
Histochemical detection of GUS activity was carried out using 5-bromo-4-chloro-3-indolyl-β-d-glucuronide as a substrate as previously described (50). Samples were placed on slides with glass coverslips and imaged with a Zeiss binocular microscope. Histology of GUS-stained samples was performed using protocol as described previously (78).
Leaf water loss assay
Leaves of 30-day-old plants were cut and weighed immediately to determine the fresh weight. Leaves were then exposed to room temperature air and weighed at 30, 60, 90, 120, 180, and 480 min.
Luciferase assays
ABCG17 and ABCG18 promoters were cloned into the pFlash vector using the Gateway system to generate promoter:LUC reporter constructs. Constructs were then transformed into Col-0 plants, and homozygous transgenic plants were treated with luciferin (Promega; 10 mg/ml) and placed in the dark for 5 min. The luminescence (LUC) signal intensity was subsequently detected using the BioSpace system (camera: IS1643N7056).
Lateral root primordium and bolting time quantifications
Five-day-old seedlings were treated with 100 mM NaCl for five more days (control plants were grown on regular MS media), and lateral root primordium number of 10-day-old seedlings at different developmental stages was monitored. The seedlings were fixed in fresh 4% Paraformaldehyde (PFA) (4 g of PFA powder dissolved into 100 ml of 1× PBS solution), with 0.01% Triton in vacuum for 1 hour. Samples were washed three times with 1× PBS, each time 3 min. The solution was removed, and ClearSee solution was added (10% xylitol, 25% urea, and 15% sodium deoxycholate in water; sodium deoxycholate was added last) for at least 3 days at room temperature. Samples were placed on slides, and lateral root primordium at different developmental stages was monitored using a Zeiss fluorescent binocular Axio Zoom V16.
For plant bolting time quantification, a standard 5-mm stem inflorescence was set as a bolting point. Plants were monitored every day to mark the bolting point.
Grafting assays
Six-day-old plants were used for grafting assay according to procedures described previously (79). Both of the two cotyledons were cut from seedlings, media used in the grafting assay contained 0.5% sucrose, and grafted plants grew vertically in short-day conditions. Lateral roots were scored 8 days after grafting.
Radioactive ABA translocation and transport assays
ABA long-distance shoot-to-root translocation was measured using [3H]ABA. Shoots were treated with 2 μl of [3H]ABA (0.1 μCi/ml). After 24 hours, roots were excised and placed into 3 ml of scintillation liquid (Opti-Fluor) for at least 24 hours in darkness before monitoring in a liquid scintillation counter.
[3H]ABA (ART2192; 1 mCi/ml and 10 Ci/mmol) and [14C]IAA (ARC0160; 0.1 mCi/ml and 55 mCi/mmol) export from tobacco (Nicotiana benthamiana) mesophyll protoplasts were analyzed as described (80). Tobacco mesophyll protoplasts were prepared 4 days after agrobacterium-mediated transfection of p35S:YFP-ABCG17, p35S:YFP-ABCG18, p35S:YFP-ABCG19, or empty vector control. Relative export from protoplasts is calculated from exported radioactivity into the supernatant as follows: (radioactivity in the supernatant at time t = x min) − (radioactivity in the supernatant at time t = 0) × (100%)/(radioactivity in the supernatant at t = 0 min); presented are mean values from eight ([3H]ABA) and four ([14C]IAA) independent transfections.
For ABA-GE competition experiments, protoplasts from tobacco leaves transfected with p35S:YFP-ABCG18 or vector control were prepared and assayed as described previously (81). In short, unlabeled ABA-GE (or 50% methanol as solvent control) was included in the transport buffer at a 1000-fold excess compared to [3H]ABA (adjusted to 10 nM). Relative import into protoplasts is calculated as follows: (radioactivity in the protoplasts at time t = x min) − (radioactivity in the protoplasts at time t = 0) × (100%)/(radioactivity in the protoplasts at t = 0 min); presented are mean values from four independent transfections.
Xenopus oocyte transport assay
For complementary RNA (cRNA) synthesis, coding DNA sequences of the ABCG genes were cloned into Xenopus laevis oocyte expression vector pNB1u using USER cloning as described previously (82). DNA template for in vitro transcription was generated by PCR using Phusion High-Fidelity DNA Polymerase (NEB) using forward primer (5′-AATTAACCCTCACTAAAGGGTTGTAATACGACTCACTATAGGG-3′) and reverse primer (5′-TTTTTTTTTTTTTTTTTTTTTTTTTTTTTATACTCAAGCTAGCCTCGAG-3′). The PCR products were purified using the E.Z.N.A. Cycle Pure Kit (Omega Bio-tek). Capped cRNA was in vitro transcribed using the mMessage mMachine T7 Kit (Ambion) following the manufacturer’s instructions.
X. laevis oocytes, stage V or VI, were purchased from Ecocyte Bioscience (Germany). Oocytes were injected with 25 ng of cRNA for each transporter expressed, i.e., regardless of whether the transporters were expressed individually or in combination. cRNA was injected using a Drummond NANOJECT II (Drummond Scientific Company, Broomall Pennsylvania). The injected oocytes were incubated for 3 days at 16°C in Hepes-based kulori buffer [90 mM NaCl, 1 mM KCl, 1 mM MgCl2, 1 mM CaCl2, and 5 mM Hepes (pH 7.4)] supplemented with gentamycin (100 μg/ml).
Five days after cRNA injection, transport assay was performed as described previously (60), with some modifications. Oocytes were preincubated in Kulori (pH 5.8) for 5 min, and then the oocytes were incubated in 10 μM ABA containing Kulori buffer (pH 5.8) for 3 hours. Subsequently, oocytes were washed four times in Kulori buffer (pH 5.8) and then homogenized with 50% methanol and stored at −20°C overnight. Oocyte extracts were spun down at 15,000g for 10 min at 4°C, and supernatant was spun down at 10,000g for 10 min at 4°C. The supernatant was then diluted with water, filtered through a 0.22-μm polyvinylidene difluoride–based filter plate (MSGVN2250, Merck Millipore), and analyzed by analytical liquid chromatography coupled to mass spectrometry (LC-MS/MS) detailed in table S1.
Yeast transport assay
pENTR-ABCG17 and pENTR-ABCG18 cDNA were subcloned into pAG423-GPD-ccdB and pAG426-GPD-ccdB using LR Gateway reaction, respectively. Yeast strain YMM12 (39) was transformed with pAG423-GPD-ccdB, pAG426-GPD-ccdB, pAG423-GPD-ABCG17, and pAG426-GPD-ABCG18. For coexpression of both transporters, pAG423-GPD-ABCG17 and pAG426-GPD-ABCG18 were cotransformed into YMM12, and pAG423-GPD-ccdB and pAG426-GPD-ccdB were cotransformed for control. The yeast cells were grown in synthetic complete medium (SC)–His, SC-Ura, and SC-His-Ura for pAG423-GPD-ABCG17, pAG426-GPD-ABCG18, and cotransformed vectors, respectively. For transport assays, yeast cultures were grown overnight in a 50 ml of culture to an optical density at 600 nm (OD600) = 0.4 to 0.6 in corresponding SC media. Cells were collected by centrifugation and resuspended to OD600 = 5.0 in 0.1 M MES buffer (pH 4.6) supplemented with 2% dextrose. Twelve replicates of each sample with 100 μl of resuspended yeast cells were aliquoted into a 1.5-ml Eppendorf tube, and an equal volume of 30 nM [3H]ABA (American Radiolabeled Chemicals Inc., MO, USA) was added for a final concentration of 15 nM [3H]ABA. The yeast cells were incubated for 3 hours with occasional mixing and rinsed three times with 0.1 M MES buffer (pH 4.6). Last, the cells were suspended in 100 μl of 0.1 M MES buffer (pH 4.6) supplemented with 2% dextrose and then moved to scintillant.
Hormone quantification
Labeled and nonlabeled standards of ABA and related metabolites were procured from Sigma-Aldrich (St. Louis, USA), Olchemim Ltd. (Olomouc, Czech Republic), and National Research Council (NRC-CNRC, Canada). Standard-grade solvents were used for hormone extraction and sample preparation, including methanol, acetic acid (LiChrosolv, Sigma-Aldrich, USA), acetonitrile (J.T.Baker, Avantor, PA, USA), formic acid (Honeywell Fluka, Thermo Fisher Scientific, MA, USA), and deionized water (Milli-Q, Synergy-UV Millipore system, USA). Plant samples frozen in liquid nitrogen were grounded using motor and pestle. Around 200 mg of samples (shoot or root) was measured from ground powder and extracted with ice-cold methanol/water/formic acid (15/4/1 v/v/v) added with an isotope-labeled (deuterium) internal standards (ISs). Similar concentrations of ABA’s ISs and its metabolites were added into samples and calibration standards. The samples were purified using Oasis MCX SPE cartridges (Waters, USA) according to the manufacturer’s protocol. The samples were injected on Acquity ultraperformance liquid chromatography (UPLC) BEH C18 column (1.7 μm, 2.1 × 100 mm; Waters; with gradients of 0.1% acetic acid in water or acetonitrile), connected to an Acquity UPLC H class system [with Waters Acquity Quaternary Solvent Manager (QSM), Flow Through Needle (FTN) sample manager, and Photodiode Detector Array (PDA)] coupled with a UPLC–electrospray ionization (ESI)–MS/MS triple quadrupole mass spectrometer (Xevo TQ-S, Waters, equipped with ESI probe) for identification and quantification of hormones. The hormones were measured using an MS detector, both in positive and negative modes, with two multiple reaction monitoring (MRM) transitions for each compound. With the exception of ABA that was determined in positive mode, other metabolites, including PA, Dihydrophaseic acid (DPA), neoPA, 7-hydroxy ABA, and ABA-GE, were determined in negative mode. External calibration curves that were constructed with serial dilutions of hormone standards, added with ISs, were used for quantification and calculated through Target Lynx (v4.1; Waters) software by comparing the ratios of MRM peak areas of analyte to that of IS (endogenous levels). To evaluate reproducibility and linearity of the method, parameters such as accuracy and precision were measured from five replicates for each of A. thaliana (Col-0) shoot and root samples and were spiked with three different concentrations of nonlabeled hormone standards (low/medium/high), along with added ISs. Accuracy was measured after comparing the known concentration of standard with that of endogenous hormone levels and spiked samples. Limit of detection (LOD) and limit of quantification (LOQ) were given as signal-to-noise ratios of 3:1 and 10:1, respectively. The repeatability of the method, known as precision, was calculated in terms of percent relative SD (table S4). Heatmap was generated using the BAR HeatMapper Plus tool (http://bar.utoronto.ca/ntools/cgi-bin/ntools_heatmapper_plus.cgi).
Statistical analysis
Multiple comparisons were performed by using one-way analysis of variance (ANOVA) with the least significant difference post hoc test in SPSS 19.0. Two-tailed Student’s t test was used for graphs with only two groups. Statistical significance was determined at P < 0.05 unless otherwise claimed.
Acknowledgments
We thank J. Schroeder (UCSD) and S. Cutler (UCR) for sharing ABA reporter seeds and constructs, pRAB18:GFP (56), pMAPKKK18:GUS, and pMAPKKK18:LUC (57) and C. Crocoll and the DynaMo Metabolomics Facility for help with data acquisition (oocyte transport assays). We thank Y. Lee (Pohang University of Science and Technology) for sharing the yeast strain YMM12.
Funding: This work was supported by grants from the Israel Science Foundation (2378/19 and 3419/20 to E.S.), the Human Frontier Science Program (HFSP—RGY0075/2015 and HFSP—LIY000540/2020 to E.S. and H.H.N.-E.), Danmarks Grundforskningsfond (DNRF99 to H.H.N.-E.), the European Research Council (757683-RobustHormoneTrans to E.S. and 679189 GAtransport to R.W.), the PBC postdoctoral fellowship (to Y.Z.), and the ADAMA Center for Novel Delivery Systems in Crop Protection fellowship (to S.L.) and by the Swiss National Funds (31003A-165877/1 to M.G.).
Author contributions: Y.Z. and E.S. conceived and designed the study and wrote the manuscript. Y.Z. performed the research. H.V.K. carried out ABA measurements presented in Figs. 6 and 7 and fig. S16. J.L. and L.C. performed hormone transport assays presented in Fig. 5 and fig. S9. H.B. assisted in ABCG17,18 cloning and vector constructions. S.L. synthesized the chemical ABA-Fluorescein. A.E. carried out gas exchange measurements presented in Fig. 3. D.R. carried out GUS stem section in Fig. 7. Z.M.B. and N.W. performed hormone transport assays in the Xenopus oocyte system presented in fig. S10. S.D. performed hormone transport assay in the yeast system presented in fig. S10. L.C., H.H.N.-E., A.A., L.R., L.S., N.S., R.W., and M.G. designed and supervised the work and edited the manuscript. All authors discussed the results and commented on the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All the data supporting the findings of this study are available within the article and the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S24
Tables S1 to S21
General synthetic procedures
Synthesis of compound 2 (ABA-FL)
HPLC-MS analysis conditions
Preparative HPLC purification conditions
Quantitifying ABA by LC-MS/MS (oocytes transport assays)
Dataset S1
REFERENCES AND NOTES
- 1.Zhu J.-K., Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53, 247–273 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cutler S. R., Rodriguez P. L., Finkelstein R. R., Abrams S. R., Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 61, 651–679 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Zhu J.-K., Abiotic stress signaling and responses in plants. Cell 167, 313–324 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeevaart J., Creelman R., Metabolism and physiology of abscisic acid. Annu. Rev. Plant. Physiol. Plant. Mol. Biol. 39, 439–473 (1988). [Google Scholar]
- 5.Duan L., Dietrich D., Ng C. H., Chan P. M. Y., Bhalerao R., Bennett M. J., Dinneny J. R., Endodermal ABA signaling promotes lateral root quiescence during salt stress in Arabidopsis seedlings. Plant Cell 25, 324–341 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robbins N. E., Trontin C., Duan L., Dinneny J. R., Beyond the barrier: Communication in the root through the endodermis. Plant Physiol. 166, 551–559 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chater C. C., Oliver J., Casson S., Gray J. E., Putting the brakes on: Abscisic acid as a central environmental regulator of stomatal development. New Phytol. 202, 376–391 (2014). [DOI] [PubMed] [Google Scholar]
- 8.GOLDBACH H., GOLDBACH E., Abscisic acid translocation and influence of water stress on grain abscisic acid content. J. Exp. Bot. 28, 1342–1350 (1977). [Google Scholar]
- 9.Setter T. L., Brun W. A., Brenner M. L., Abscisic acid translocation and metabolism in soybeans following depodding and petiole girdling treatments. Plant Physiol. 67, 774–779 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Setter T. L., Brun W. A., Brenner M. L., Effect of obstructed translocation on leaf abscisic acid, and associated stomatal closure and photosynthesis decline. Plant Physiol. 65, 1111–1115 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Milthorpe F., Moorby J., Vascular transport and its significance in plant growth. Annu. Rev. Plant Physiol. 20, 117–138 (1969). [Google Scholar]
- 12.Wilkinson S., Davies W. J., ABA-based chemical signalling: The co-ordination of responses to stress in plants. Plant Cell Environ. 25, 195–210 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Sauter A., Davies W. J., Hartung W., The long-distance abscisic acid signal in the droughted plant: the fate of the hormone on its way from root to shoot. J. Exp. Bot. 52, 1991–1997 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Zhang H., Zhu H., Pan Y., Yu Y., Luan S., Li L., A DTX/MATE-type transporter facilitates abscisic acid efflux and modulates ABA sensitivity and drought tolerance in Arabidopsis. Mol. Plant 7, 1522–1532 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Kuromori T., Seo M., Shinozaki K., ABA transport and plant water stress responses. Trends Plant Sci. 23, 513–522 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Ikegami K., Okamoto M., Seo M., Koshiba T., Activation of abscisic acid biosynthesis in the leaves of Arabidopsis thaliana in response to water deficit. J. Plant Res. 122, 235–243 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Christmann A., Weiler E. W., Steudle E., Grill E., A hydraulic signal in root-to-shoot signalling of water shortage. Plant J. 52, 167–174 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Holbrook N. M., Shashidhar V., James R. A., Munns R., Stomatal control in tomato with ABA-deficient roots: Response of grafted plants to soil drying. J. Exp. Bot. 53, 1503–1514 (2002). [PubMed] [Google Scholar]
- 19.Christmann A., Hoffmann T., Teplova I., Grill E., Müller A., Generation of active pools of abscisic acid revealed by in vivo imaging of water-stressed Arabidopsis. Plant Physiol. 137, 209–219 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan B. C., Joseph L. M., Deng W. T., Liu L., Li Q. B., Cline K., McCarty D. R., Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 35, 44–56 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Bauer H., Ache P., Lautner S., Fromm J., Hartung W., al-Rasheid K. A. S., Sonnewald S., Sonnewald U., Kneitz S., Lachmann N., Mendel R. R., Bittner F., Hetherington A. M., Hedrich R., The stomatal response to reduced relative humidity requires guard cell-autonomous ABA synthesis. Curr. Biol. 23, 53–57 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Park J., Lee Y., Martinoia E., Geisler M., Plant hormone transporters: What we know and what we would like to know. BMC Biol. 15, 93 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang J., Yim S., Choi H., Kim A., Lee K. P., Lopez-Molina L., Martinoia E., Lee Y., Abscisic acid transporters cooperate to control seed germination. Nat. Commun. 6, 8113 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lacombe B., Achard P., Long-distance transport of phytohormones through the plant vascular system. Curr. Opin. Plant Biol. 34, 1–8 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Boursiac Y., Léran S., Corratgé-Faillie C., Gojon A., Krouk G., Lacombe B., ABA transport and transporters. Trends Plant Sci. 18, 325–333 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Slovik S., Daeter W., Hartung W., REVIEW ARTICLE: Compartmental redistribution and long-distance transport of abscisic acid (ABA) in plants as influenced by environmental changes in the rhizosphere—A biomathematical model. J. Exp. Bot. 46, 881–894 (1995). [Google Scholar]
- 27.Wilkinson S., Davies W. J., Xylem sap pH increase: A drought signal received at the apoplastic face of the guard cell that involves the suppression of saturable abscisic acid uptake by the epidermal symplast. Plant Physiol. 113, 559–573 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dodd I. C., Tan L. P., He J., Do increases in xylem sap pH and/or ABA concentration mediate stomatal closure following nitrate deprivation? J. Exp. Bot. 54, 1281–1288 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Jiang F., Hartung W., Long-distance signalling of abscisic acid (ABA): The factors regulating the intensity of the ABA signal. J. Exp. Bot. 59, 37–43 (2007). [DOI] [PubMed] [Google Scholar]
- 30.McAdam S. A., Brodribb T. J., Ross J. J., Shoot-derived abscisic acid promotes root growth. Plant Cell Environ. 39, 652–659 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Qin P., Zhang G., Hu B., Wu J., Chen W., Ren Z., Liu Y., Xie J., Yuan H., Tu B., Ma B., Wang Y., Ye L., Li L., Xiang C., Li S., Leaf-derived ABA regulates rice seed development via a transporter-mediated and temperature-sensitive mechanism. Sci. Adv. 7, eabc8873 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merilo E., Yarmolinsky D., Jalakas P., Parik H., Tulva I., Rasulov B., Kilk K., Kollist H., Stomatal VPD response: There is more to the story than ABA. Plant Physiol. 176, 851–864 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuromori T., Sugimoto E., Shinozaki K., Inter-tissue signal transfer of abscisic acid from vascular cells to guard cells. Plant Physiol. 164, 1587–1592 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merilo E., Jalakas P., Laanemets K., Mohammadi O., Hõrak H., Kollist H., Brosché M., Abscisic acid transport and homeostasis in the context of stomatal regulation. Mol. Plant 8, 1321–1333 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Lee K. P., Piskurewicz U., Turečková V., Strnad M., Lopez-Molina L., A seed coat bedding assay shows that RGL2-dependent release of abscisic acid by the endosperm controls embryo growth in Arabidopsis dormant seeds. Proc. Natl. Acad. Sci. U.S.A. 107, 19108–19113 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seo M., Koshiba T., Transport of ABA from the site of biosynthesis to the site of action. J. Plant Res. 124, 501–507 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Kang J., Park J., Choi H., Burla B., Kretzschmar T., Lee Y., Martinoia E., Plant ABC transporters. Arabidopsis Book 9, e0153 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Do T. H. T., Martinoia E., Lee Y., Functions of ABC transporters in plant growth and development. Curr. Opini. Plant Biol. 41, 32–38 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Kang J., Hwang J. U., Lee M., Kim Y. Y., Assmann S. M., Martinoia E., Lee Y., PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc. Natl. Acad. Sci. U.S.A. 107, 2355–2360 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuromori T., Miyaji T., Yabuuchi H., Shimizu H., Sugimoto E., Kamiya A., Moriyama Y., Shinozaki K., ABC transporter AtABCG25 is involved in abscisic acid transport and responses. Proc. Natl. Acad. Sci. U.S.A. 107, 2361–2366 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cho M., Lee S. H., Cho H.-T., P-Glycoprotein4 displays auxin efflux transporter-like action in arabidopsis root hair cells and tobacco cells. Plant Cell 19, 3930–3943 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pawela A., Banasiak J., Biała W., Martinoia E., Jasiński M., Mt ABCG20 is an ABA exporter influencing root morphology and seed germination of Medicago truncatula. Plant J. 98, 511–523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seo M., Hanada A., Kuwahara A., Endo A., Okamoto M., Yamauchi Y., North H., Marion-Poll A., Sun T. P., Koshiba T., Kamiya Y., Yamaguchi S., Nambara E., Regulation of hormone metabolism in Arabidopsis seeds: Phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J. 48, 354–366 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Kanno Y., Hanada A., Chiba Y., Ichikawa T., Nakazawa M., Matsui M., Koshiba T., Kamiya Y., Seo M., Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc. Natl. Acad. Sci. U.S.A. 109, 9653–9658 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma Y., Cao J., He J., Chen Q., Li X., Yang Y., Molecular mechanism for the regulation of ABA homeostasis during plant development and stress responses. Int. J. Mol. Sci. 19, 3643 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nambara E., Marion-Poll A., Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 56, 165–185 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Liu Z., Yan J. P., Li D. K., Luo Q., Yan Q., Liu Z. B., Ye L. M., Wang J. M., Li X. F., Yang Y., UDP-glucosyltransferase71c5, a major glucosyltransferase, mediates abscisic acid homeostasis in Arabidopsis. Plant Physiol. 167, 1659–1670 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen T.-T., Liu F. F., Xiao D. W., Jiang X. Y., Li P., Zhao S. M., Hou B. K., Li Y. J., The Arabidopsis UDP-glycosyltransferase75B1, conjugates abscisic acid and affects plant response to abiotic stresses. Plant Mol. Biol. 102, 389–401 (2020). [DOI] [PubMed] [Google Scholar]
- 49.Hauser F., Chen W., Deinlein U., Chang K., Ossowski S., Fitz J., Hannon G. J., Schroeder J. I., A genomic-scale artificial microRNA library as a tool to investigate the functionally redundant gene space in Arabidopsis. Plant Cell 25, 2848–2863 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y., Nasser V., Pisanty O., Omary M., Wulff N., di Donato M., Tal I., Hauser F., Hao P., Roth O., Fromm H., Schroeder J. I., Geisler M., Nour-Eldin H. H., Shani E., A transportome-scale amiRNA-based screen identifies redundant roles of Arabidopsis ABCB6 and ABCB20 in auxin transport. Nat. Commun. 9, 4204 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murchie E. H., Lawson T., Chlorophyll fluorescence analysis: A guide to good practice and understanding some new applications. J. Exp. Bot. 64, 3983–3998 (2013). [DOI] [PubMed] [Google Scholar]
- 52.Saradhi P. P., Suzuki I., Katoh A., Sakamoto A., Sharmila P., Shi D. J., Murata N., Protection against the photo-induced inactivation of the photosystem II complex by abscisic acid. Plant Cell Environ. 23, 711–718 (2000). [Google Scholar]
- 53.Liu L., Zhao L., Chen P., Cai H., Hou Z., Jin X., Aslam M., Chai M., Lai L., He Q., Liu Y., Huang X., Chen H., Chen Y., Qin Y., ATP binding cassette transporters ABCG1 and ABCG16 affect reproductive development via auxin signalling in Arabidopsis. Plant J. 102, 1172–1186 (2020). [DOI] [PubMed] [Google Scholar]
- 54.Yadav V., Molina I., Ranathunge K., Castillo I. Q., Rothstein S. J., Reed J. W., ABCG transporters are required for suberin and pollen wall extracellular barriers in Arabidopsis. Plant Cell 26, 3569–3588 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shanmugarajah K., Linka N., Gräfe K., Smits S. H. J., Weber A. P. M., Zeier J., Schmitt L., ABCG1 contributes to suberin formation in Arabidopsis thaliana roots. Sci. Rep. 9, 11381 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim T.-H., Hauser F., Ha T., Xue S., Böhmer M., Nishimura N., Munemasa S., Hubbard K., Peine N., Lee B. H., Lee S., Robert N., Parker J. E., Schroeder J. I., Chemical genetics reveals negative regulation of abscisic acid signaling by a plant immune response pathway. Curr. Biol. 21, 990–997 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Okamoto M., Peterson F. C., Defries A., Park S. Y., Endo A., Nambara E., Volkman B. F., Cutler S. R., Activation of dimeric ABA receptors elicits guard cell closure, ABA-regulated gene expression, and drought tolerance. Proc. Natl. Acad. Sci. U.S.A. 110, 12132–12137 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kelly G., Lugassi N., Belausov E., Wolf D., Khamaisi B., Brandsma D., Kottapalli J., Fidel L., Ben-Zvi B., Egbaria A., Acheampong A. K., Zheng C., Or E., Distelfeld A., David-Schwartz R., Carmi N., Granot D., The Solanum tuberosum KST1 partial promoter as a tool for guard cell expression in multiple plant species. J. Exp. Bot. 68, 2885–2897 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shani E., Weinstain R., Zhang Y., Castillejo C., Kaiserli E., Chory J., Tsien R. Y., Estelle M., Gibberellins accumulate in the elongating endodermal cells of Arabidopsis root. Proc. Natl. Acad. Sci. U.S.A. 110, 4834–4839 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu D., Veres D., Belew Z. M., Olsen C. E., Nour-Eldin H. H., Halkier B. A., Functional expression and characterization of plant ABC transporters in Xenopus laevis oocytes for transport engineering purposes. Meth. Enzymol. 576, 207–224 (2016). [DOI] [PubMed] [Google Scholar]
- 61.De Smet I., Signora L., Beeckman T., Inzé D., Foyer C. H., Zhang H., An abscisic acid-sensitive checkpoint in lateral root development of Arabidopsis. Plant J. 33, 543–555 (2003). [DOI] [PubMed] [Google Scholar]
- 62.Geng Y., Wu R., Wee C. W., Xie F., Wei X., Chan P. M. Y., Tham C., Duan L., Dinneny J. R., A spatio-temporal understanding of growth regulation during the salt stress response in Arabidopsis. Plant Cell 25, 2132–2154 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao Y., Xing L., Wang X., Hou Y.-J., Gao J., Wang P., Duan C.-G., Zhu X., Zhu J.-K., The ABA receptor PYL8 promotes lateral root growth by enhancing MYB77-dependent transcription of auxin-responsive genes. Sci. Signal. 7, ra53 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bloch D., Puli M. R., Mosquna A., Yalovsky S., Abiotic stress modulates root patterning via ABA-regulated microRNA expression in the endodermis initials. Development 146, dev177097 (2019). [DOI] [PubMed] [Google Scholar]
- 65.Duan L., Pérez-Ruiz J. M., Cejudo F. J., Dinneny J. R., Characterization of CYCLOPHILLIN38 shows that a photosynthesis-derived systemic signal controls lateral root emergence. Plant Physiol. 185, 503–518 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lashbrooke J., Cohen H., Levy-Samocha D., Tzfadia O., Panizel I., Zeisler V., Massalha H., Stern A., Trainotti L., Schreiber L., Costa F., Aharoni A., MYB107 and MYB9 homologs regulate suberin deposition in angiosperms. Plant Cell 28, 2097–2116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shohat H., Illouz-Eliaz N., Kanno Y., Seo M., Weiss D., The tomato DELLA protein PROCERA promotes Abscisic Acid responses in guard cells by upregulating an Abscisic Acid transporter. Plant Physiol. 184, 518–528 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Manzi M., Lado J., Rodrigo M. J., Zacarías L., Arbona V., Gómez-Cadenas A., Root ABA accumulation in long-term water-stressed plants is sustained by hormone transport from aerial organs. Plant Cell Physiol. 56, 2457–2466 (2015). [DOI] [PubMed] [Google Scholar]
- 69.Manzi Fraga M. J., Pitarch-Bielsa M., Arbona V., Gómez Cadenas A., Leaf dehydration is needed to induce abscisic acid accumulation in roots of citrus plants. Environ. Exp. Bot. 139, 116–126 (2017). [Google Scholar]
- 70.Dietz K. J., Sauter A., Wichert K., Messdaghi D., Hartung W., Extracellular β-glucosidase activity in barley involved in the hydrolysis of ABA glucose conjugate in leaves. J. Exp. Bot. 51, 937–944 (2000). [PubMed] [Google Scholar]
- 71.Schroeder J. I., Nambara E., A quick release mechanism for abscisic acid. Cell 126, 1023–1025 (2006). [DOI] [PubMed] [Google Scholar]
- 72.Burla B., Pfrunder S., Nagy R., Francisco R. M., Lee Y., Martinoia E., Vacuolar transport of abscisic acid glucosyl ester is mediated by ATP-binding cassette and proton-antiport mechanisms in Arabidopsis. Plant Physiol. 163, 1446–1458 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jarzyniak K. M., Jasiński M., Membrane transporters and drought resistance–a complex issue. Front. Plant Sci. 5, 687 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu F.-H., Shen S. C., Lee L. Y., Lee S. H., Chan M. T., Lin C. S., Tape-Arabidopsis Sandwich – a simpler Arabidopsis protoplast isolation method. Plant Methods 5, 16 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Geisler M., Nadeau J., Sack F. D., Oriented asymmetric divisions that generate the stomatal spacing pattern in Arabidopsis are disrupted by the too many mouths mutation. Plant Cell 12, 2075–2086 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tal I., Zhang Y., Jørgensen M. E., Pisanty O., Barbosa I. C. R., Zourelidou M., Regnault T., Crocoll C., Erik Olsen C., Weinstain R., Schwechheimer C., Halkier B. A., Nour-Eldin H. H., Estelle M., Shani E., The Arabidopsis NPF3 protein is a GA transporter. Nat. Commun. 7, 11486 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mitra P. P., Loqué D., Histochemical staining of Arabidopsis thaliana secondary cell wall elements. J. Vis. Exp. 2014, e51381 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.De Smet I., Chaerle P., Vanneste S., De Rycke R., Inzé D., Beeckman T., An easy and versatile embedding method for transverse sections. J. Microsc. 213, 76–80 (2004). [DOI] [PubMed] [Google Scholar]
- 79.Marsch-Martínez N., Franken J., Gonzalez-Aguilera K. L., de Folter S., Angenent G., Alvarez-Buylla E. R., An efficient flat-surface collar-free grafting method for Arabidopsis thaliana seedlings. Plant Methods 9, 14 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Henrichs S., Wang B., Fukao Y., Zhu J., Charrier L., Bailly A., Oehring S. C., Linnert M., Weiwad M., Endler A., Nanni P., Pollmann S., Mancuso S., Schulz A., Geisler M., Regulation of ABCB1/PGP1-catalysed auxin transport by linker phosphorylation. EMBO J. 31, 2965–2980 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zürcher E., Liu J., di Donato M., Geisler M., Müller B., Plant development regulated by cytokinin sinks. Science 353, 1027–1030 (2016). [DOI] [PubMed] [Google Scholar]
- 82.Nour-Eldin H. H., Hansen B. G., Nørholm M. H., Jensen J. K., Halkier B. A., Advancing uracil-excision based cloning towards an ideal technique for cloning PCR fragments. Nucleic Acids Res. 34, e122 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S24
Tables S1 to S21
General synthetic procedures
Synthesis of compound 2 (ABA-FL)
HPLC-MS analysis conditions
Preparative HPLC purification conditions
Quantitifying ABA by LC-MS/MS (oocytes transport assays)
Dataset S1