Abstract
Video capsule endoscope (VCE) is an emerging technology that allows examination of the entire gastrointestinal (GI) tract with minimal invasion. While traditional endoscopy with biopsy procedures are the gold standard for diagnosis of most GI diseases, they are limited by how far the scope can be advanced in the tract and are also invasive. VCE allows gastroenterologists to investigate GI tract abnormalities in detail with visualization of all parts of the GI tract. It captures continuous real time images as it is propelled in the GI tract by gut motility. Even though VCE allows for thorough examination, reviewing and analyzing up to eight hours of images (compiled as videos) is tedious and not cost effective. In order to pave way for automation of VCE-based GI disease diagnosis, detecting the location of the capsule would allow for a more focused analysis as well as abnormality detection in each region of the GI tract. In this paper, we compared four deep Convolutional Neural Network models for feature extraction and detection of the anatomical part within the GI tract captured by VCE images. Our results showed that VGG-Net has superior performance with the highest average accuracy, precision, recall and, F1-score compared to other state of the art architectures: GoogLeNet, AlexNet and, ResNet.
Keywords: Video capsule endoscopy, Convolutional neural network, Gradient-weighted class activation mapping (Grad-CAM), Gastrointestinal tract, VGG-net, ResNet, AlexNet, GoogLeNet
1. Introduction
Gastrointestinal (GI) endoscopy with biopsy is essential for detecting different diseases and abnormalities within the GI tract [14]. Numerous conditions with significant comorbidities, such as inflammatory bowel disease, celiac disease, barrett’s esophagus, diverticulitis, and GI malignancy can present with signs like ulcers, occult GI bleeding, erosions, among others within different GI tract regions [14]. For many of these conditions, some of the early symptoms are nonspecific, including abdominal pain, nausea, vomiting, diarrhea, constipation, and blood in stool. Due to this, diagnosis of the diseases early on is key to effective treatment and management. The GI tract consists of different anatomical landmarks; mouth, esophagus, stomach, small intestine (comprised of three parts: duodenum, jejunum, and ileum), large intestine, and anus. Identification of the anatomical components of the GI tract facilitates the diagnostic process as it enables gastroenterologists to focus on the abnormalities particular to an area of the GI tract. Even though traditional endoscopy with biopsy has been widely used, and remains the gold standard for various GI diseases diagnosis, it is limited by how far it can be advanced in to the bowel such as reaching parts of the small intestine (upper endoscopy) and terminal ileum (colonoscopy) along with being invasive, time-consuming, and costly [8,10]. These limitations are addressed by video capsule endoscopy (VCE) which is comparatively pain-free, noninvasive, and more economical alternative to traditional endoscopy [10].
Video capsule is an emerging endoscopic device in the shape of a capsule used to collect images from the GI tract for disease diagnosis. The capsule is ingested by the patient which moves within the GI tract continuously capturing real time images that are wirelessly sent to the data recorder or storage device [15]. While VCE is an innovative breakthrough [8], the captured images still need to be analyzed by gastroenterologists for detection of pathological signs of diseases such as bleeding, polyps, ulcers, among others [5,6]. An entire VCE procedure can last up to 8 h producing between 50,000 to 100,000 images per patient. The assessment and analysis of the images, compiled as a video, by the gastroenterologist can take up to 1 to 2 h on average. This time-consuming process adds a considerable cost to the endoscopy procedure, thereby limiting the general application of VCE. Since traditional upper GI endocsopy has the limited ability to advance into all three parts of the small intestine, VCE is done more often for diseases of the small bowel [9,10]. Developing deep learning models for efficient identification of different anatomical parts within the entire VCE image sequence will not only save time by allowing directed and focused assessment of different regions of the GI tract, it will also pave way for eventual automation of VCE based disease diagnosis [14,15].
Detecting anatomical landmarks in VCE images intuitively means recognizing higher semantic image features that discriminate one area of the GI tract from another. In this paper, we investigated multiple state of the art convolutional netural network (CNN) architectures for feature extraction and identification of anatomical landmarks in VCE images. We also compared performance of these state of the art models and visualized their decision making process using gradient weighted-class activation mapping (Grad-CAM).
2. Related Work
While significant amount of efforts have been put into minimizing time spent by gastroenterologists analyzing VCE images, less attention has been paid to developing models that automatically discriminate the different region of the GI tract [3,13,21]. Initial approaches have focused on lower level feature extraction techniques combined with dimensionality reduction [2] to solve the challenge of segmenting anatomical parts within the GI tract. Specifically, [13] applied color change pattern analysis to segment video into anatomic parts. This approach used intestinal contraction as a discriminatory feature of the digestive organs to extract energy-based feature in frequency domain to detect event boundaries by using high frequency content (HFC) function. The author in [2] proposed locality preserving projections which performed dimensionality reduction on visual feature vectors describing the capsule images. In [3], the author proposed using Discrete Cosine Transform (DCT) and Principal Component Analysis (PCA) as classifiers to disciminate stomach, intestine, and colon. Texture-based and other lower level feature extraction methods struggle with images containing occlusions from bubbles that are often found in VCE images. Despite this, CNNs have demonstrated superior performance in medical image analysis due to which they have recently gained significant attention. CNNs have shown state of the art results in high level feature extraction from images including VCE images [4]. CNN-based models create feature maps that are invariant to translation, scale, luminance, and rotation. Video capsules capture images at different angles, distances and lighting conditions within the GI tract. Due to these peculiar characteristics of VCE images, CNN-based models are more suitable for feature extraction and subsequent analysis than other lower level feature extraction techniques. The models use layers of repetitive computation, generally including convolution and pooling [12,16] with the aim of learning the input image mapping to an output class. With the use of CNNs, higher order image features can be extracted and further classified to relevant classes [1].
With each layer of a neural network focusing on abstraction of different features such as edges, textures, patterns, parts, and whole object within a dataset, the network (a composition of multiple layers) is able to detect complex discriminating properties between the different classes. Zou et al. proposed a deep CNN model for classifying digestive organs in VCE images [21] with 95.5% accuracy. In our work, we investigated the performance of four (4) state of the art CNN architectures - (Google-Net, VGG-Net, Alex-Net and Res-Net) on VCE images to identify anatomical GI landmarks. Our work complements and extends earlier work through comparison of different CNN-based models in addition to applying Grad-CAMs for interpretation and visualization of the classification done by the models.
The rest of the paper is structured as follows: (1) the structure of the deep learning architectures investigated in this work is described in Sect. 3 below; (2) experiment and results are presented in Sect. 4; and (3) conclusions along with future directions in Sect. 5.
3. Methods
3.1. CNN-Based Deep Learning Models
There are various CNN-based deep learning models that have shown excellent performance for image classification tasks. We investigated four (4) different models on our VCE dataset. The architecture of the models are described as follows:
VGG-Net.
The VGG-Network was proposed in the 2014 paper titled Very Deep Convolutional Networks for Large-Scale Image Recognition [18] by the Visual Geometry Group at Oxford. The model outperformed other models in the 2014 ImageNet Large Scale Visual Recognition Challenge (ILSVRC). The significant characteristics of the model are the use of a large number of small filters, specifically, filters with size 3 × 3 and 1 × 1 with stride of one. The max pooling layers of 2 × 2 are used after most, but not all, convolutional layers. Specifically, the network uses examples of two, three, and even four convolutional layers stacked together before a max pooling layer is used. With the number of filters increasing with depth of the model, many variants of the model have been developed and evaluated, named for the number of layers: VGG-16 and VGG-19 for 16 and 19 learned layers, respectively. For our study, we used VGG-19.
GoogLeNet.
The GoogLeNet model was proposed in the 2015 paper titled Going Deeper with Convolutions [19]. The model achieved top results in the 2014 version of ILSVRC challenge with a novel inception module. The inception module is a block of parallel convolutional layers with different sized filters and a max pooling layer. The results of this module are then concatenated. The main features of the model include: heavy use of the 1 × 1 convolution to reduce the number of channels; use of error feedback at multiple points in the network; development of very deep (22 layers) models; and, use of global average pooling for the output of the model.
ResNet.
The residual network (ResNet) was proposed in a 2016 paper titled Deep Residual Learning for Image Recognition [7]. The model with 152 layers proposed the use of residual blocks that uses shortcut connections. These are simple connections in the network architecture where the input is kept as is (not weighted) and passed on to a deeper layer. A residual block is a pattern of two convolutional layers with ReLU activation where the output of the block is combined with the input of the block.
The network takes the input as a 224 × 224 pixel image. The ResNet-50 architecture performs the initial convolution and max-pooling using 7 × 7 and 3 × 3 kernel sizes, respectively. The convolution operation in the residual blocks is performed with stride 2; thus, the size of input is reduced to half in terms of height and width even though the channel width is doubled. As we progress from one stage to another, the channel width is doubled and the size of the input is reduced to half.
AlexNet.
The AlexNet architecture was proposed in the paper ImageNet Classification with Deep Convolutional Neural Networks [11] and competed in the ImageNet LSVRC-2012 achieving a top-5 test error rate of 15.3%. The network has eight layers; the first five are convolutional layers, some of them followed by max-pooling layers, and the last three were fully connected layers. It used the non-saturating ReLU activation function, which showed improved training performance over tanh and sigmoid. To reduce over-fitting in the fully-connected layers, AlexNet employs the dropout regularization method.
4. Experiment and Results
4.1. Dataset Preparation, Annotation and Augmentation
Our dataset consisted of nine patient videos or approximately, 200,000 capsule endoscopy frames. We divided our dataset based on the four main anatomical regions: esophagus, stomach, small-bowel, and the colon. Each video was processed into frames and annotated by two medical research experts to identify the different anatomical regions of the GI tract. The capsule spends varying amounts of time in the different parts of the GI tract, resulting in a significant difference in the number of frames captured in each region. This results in significant class imbalance with more than 80% of the video capturing images of only the small-bowel. We addressed this class imbalance problem by up-sampling the images from other regions such as the esophagus, stomach and colon to balance the class distribution. We randomly rotated and cropped the images to generate another set of examples for these regions. Since the focus of our experiment is to compare the performance in each region across the four (4) architectures and not the performance of a single model across the four (4) regions of the GI tract. Therefore, the augmentation process was performed only on the training set to balance the classes while the test set was left intact.
4.2. Implementation and Results
The models were implemented using the Pytorch framework and trained for 50 epochs on NVIDIA GPU 4 Core, 160 GB Machine. Hyper-parameters were set as follows: batch size of 64 and learning rate of 0.001 with 70–30 train-test split. We used stochastic gradient descent for training the system. We evaluated the models based on the accuracy, precision, recall and, F1-scores. Table 1 shows comparative performance of the models. The VGG network, on average, demonstrated superior performance over other architectures on our VCE image dataset with the highest accuracy of 99.1%, closely followed by the AlexNet architecture with 97.3% accuracy.
Table 1.
Comparison of models performance
| Model | Metrics | |||
|---|---|---|---|---|
| Accuracy | Precision | Recall | F1 score | |
| VGGNet | 0.991 | 0.935 | 0.973 | 0.953 |
| ResNet | 0.878 | 0.619 | 0.935 | 0.704 |
| GoogleNet | 0.854 | 0.570 | 0.931 | 0.642 |
| AlexNet | 0.973 | 0.887 | 0.950 | 0.916 |
4.3. Visualization
Grad-CAMs [17] provide visual explanations for decisions from a large class of CNN-based models, making them more transparent and explicable. They use the gradients of any target concept flowing into a network layer to produce a coarse localization map highlighting the important image regions for predicting the concept. For each of our models, we combined the Grad-CAMs with high resolution class-discriminative visualizations to highlight the regions driving the decision-making process of the models (for each anatomical part of the GI tract). For the purpose of accessibility, Grad-CAMs also help gain a better understanding of why a good model is performing well versus why other models are not localizing the discriminative features for each class [20]. Identifying which anatomical part certain VCE images belong to varies widely in terms of difficulty when viewed independently. We selected a difficult example for each region to visualize the Grad-CAMs in order to explain the discriminative features that each model learned. These features were correlated with the performance of the model as shown in Table 2.
Table 2.
Normalized confusion Matrix
| True-Label | Model | Predicted Label |
|||
|---|---|---|---|---|---|
| Esophagus | Stomach | Small bowel | Colon | ||
|
| |||||
| Esophagus | VGGNet | 0.990 | 0.000 | 0.009 | 0.001 |
| ResNet | 0.995 | 0.000 | 0.003 | 0.002 | |
| GoogleNet | 0.957 | 0.001 | 0.012 | 0.031 | |
| AlexNet | 0.979 | 0.000 | 0.019 | 0.002 | |
|
| |||||
| Stomach | VGGNet | 0.000 | 0.926 | 0.000 | 0.074 |
| ResNet | 0.000 | 0.907 | 0.000 | 0.093 | |
| GoogleNet | 0.000 | 0.963 | 0.000 | 0.037 | |
| AlexNet | 0.000 | 0.870 | 0.000 | 0.130 | |
|
| |||||
| Small bowel | VGGNet | 0.004 | 0.000 | 0.991 | 0.005 |
| ResNet | 0.070 | 0.003 | 0.848 | 0.079 | |
| GoogleNet | 0.072 | 0.006 | 0.825 | 0.097 | |
| AlexNet | 0.019 | 0.000 | 0.972 | 0.009 | |
|
| |||||
| Colon | VGGNet | 0.001 | 0.002 | 0.013 | 0.984 |
| ResNet | 0.004 | 0.004 | 0.001 | 0.991 | |
| GoogleNet | 0.008 | 0.008 | 0.003 | 0.981 | |
| AlexNet | 0.003 | 0.004 | 0.017 | 0.977 | |
Figure 1, 2, 3, and 4 compares the GradCAM for all four models for the different parts of the GI tract. Discriminating features for the VCE images lie within the tissue areas. For each of the anatomical components, the best performing model should only focus on the tissue area of the image against occluding bubbles or liquid in determining the class of the image. For the esophagus (Fig. 1), small bowel (Fig. 3), and colon (Fig. 4), the GoogLeNet model intersectred the tissue areas with the bubbles as features that are not related to the anatomical part of the GI tract. However, the model is able to focus only on the tissue areas for the images from the stomach (Fig. 2). For all the regions, the VGG-Net did not transcend the tissue area in extracting discriminating features for its classification. AlexNet also demonstrably focused only on the tissue areas for all the four regions.
Fig. 1.
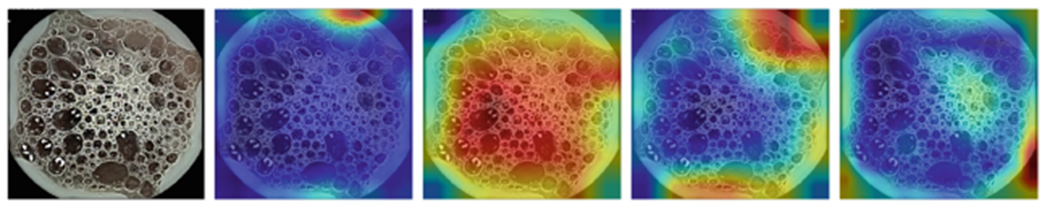
GradCAM comparison for Esophagus (Alexnet, Googlenet, Resnet-50, VGG)
Fig. 2.
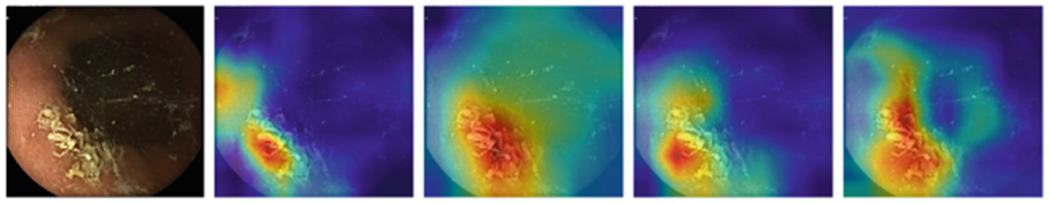
GradCAM comparison for Stomach (Alexnet, Googlenet, Resnet-50, VGG)
Fig. 3.
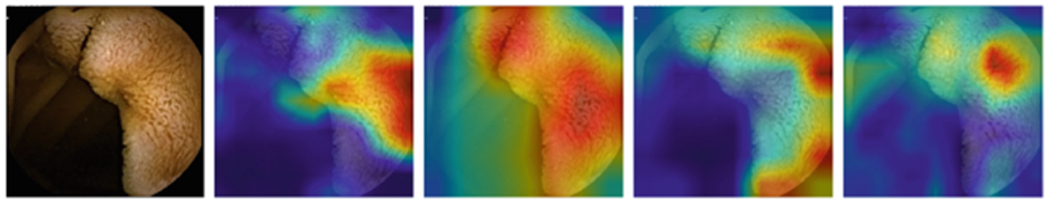
GradCAM comparison for Small Bowel (Alexnet, Googlenet, Resnet-50, VGG)
Fig. 4.
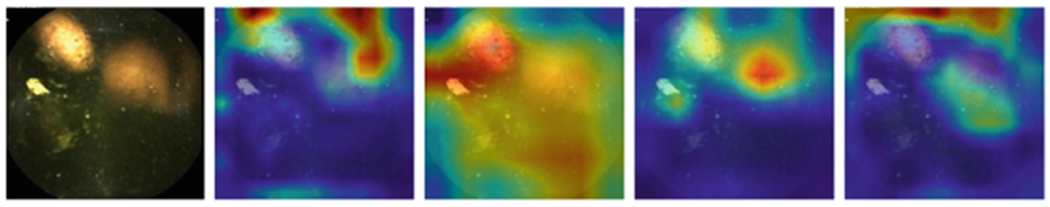
GradCAM comparison for Colon (Alexnet, Googlenet, Resnet-50, VGG)
5. Conclusion
In this work, we demonstrated the performance of different deep learning architectures for the recognition of anatomical components within the GI tract using VCE images. Experimental results shows that the VGG-network was able to learn more discriminative features for detecting different parts of the GI tract compared to other state of the art architectures. The VGG network had average classification accuracy of 99.1% while the least performing network (GoogLeNet) had an average classification accuracy of 85.4%. For future work, developing model to distinguish abnormal from normal frames for each region of the GI tract will allow gastroenterologists minimize time spent reviewing VCE videos by allowing them focus on images that are flagged as abnormal for any region of interest.
Acknowledgment.
This work was supported by Engineering in Medicine seed grant by the University of Virginia (Porter, Syed).
References
- 1.Anwar SM, Majid M, Qayyum A, Awais M, Alnowami M, Khan MK: Medical image analysis using convolutional neural networks: a review. J. Med. Syst 42(11), 226 (2018) [DOI] [PubMed] [Google Scholar]
- 2.Azzopardi C, Hicks YA, Camilleri KP: Exploiting gastrointestinal anatomy for organ classification in capsule endoscopy using locality preserving projections. In: 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), pp. 3654–3657. IEEE; (2013) [DOI] [PubMed] [Google Scholar]
- 3.Berens J, Mackiewicz M, Bell D: Stomach, intestine, and colon tissue discriminators for wireless capsule endoscopy images. In: Medical Imaging 2005: Image Processing, vol. 5747, pp. 283–290. International Society for Optics and Photonics; (2005) [Google Scholar]
- 4.Ding Z, Shi H, Zhang H, Meng L, Fan M, Han C, Zhang K, Ming F, Xie X, Liu H, et al. : Gastroenterologist-level identification of small-bowel diseases and normal variants by capsule endoscopy using a deep-learning model. Gastroenterology 157(4), 1044–1054 (2019) [DOI] [PubMed] [Google Scholar]
- 5.Eliakim R, Fireman Z, Gralnek IM, Yassin K, Waterman M, Kopelman Y, Lachter J, Koslowsky B, Adler SN: Evaluation of the pillcam colon capsule in the detection of colonic pathology: results of the first multicenter, prospective, comparative study. Endoscopy 38(10), 963–970 (2006) [DOI] [PubMed] [Google Scholar]
- 6.Eliakim R: The pillcam colon capsule-a promising new tool for the detection of colonic pathologies. Curr. Colorectal Cancer Rep 4(1), 5–9 (2008) [Google Scholar]
- 7.He K, Zhang X, Ren S, Sun J: Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778 (2016) [Google Scholar]
- 8.Iddan G, Meron G, Glukhovsky A, Swain P: Wireless capsule endoscopy. Nature 405(6785), 417–417 (2000) [DOI] [PubMed] [Google Scholar]
- 9.Klang E, Barash Y, Margalit RY, Soffer S, Shimon O, Albshesh A, Ben-Horin S, Amitai MM, Eliakim R, Kopylov U: Deep learning algorithms for automated detection of crohn’s disease ulcers by video capsule endoscopy. Gastrointest. Endosc 91(3), 606–613 (2020) [DOI] [PubMed] [Google Scholar]
- 10.Koh JEW, Hagiwara Y, Oh SL, Tan JH, Ciaccio EJ, Green PH, Lewis SK, Acharya UR: Automated diagnosis of celiac disease using DWT and non-linear features with video capsule endoscopy images. Future Gener. Comput. Syst 90, 86–93 (2019) [Google Scholar]
- 11.Krizhevsky A, Sutskever I, Hinton GE: Imagenet classification with deep convolutional neural networks. In: Advances in Neural Information Processing Systems, pp. 1097–1105 (2012) [Google Scholar]
- 12.LeCun Y, Bengio Y, Hinton G: Deep learning. Nature 521(7553), 436–444 (2015) [DOI] [PubMed] [Google Scholar]
- 13.Lee J, Oh J, Shah SK, Yuan X, Tang SJ: Automatic classification of digestive organs in wireless capsule endoscopy videos. In: Proceedings of the 2007 ACM Symposium on Applied Computing, pp. 1041–1045 (2007) [Google Scholar]
- 14.Li H, Hou X, Lin R, Fan M, Pang S, Jiang L, Liu Q, Ling F: Advanced endoscopic methods in gastrointestinal diseases: a systematic review. Quantitative Imaging Med. Surg 9(5), 905 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paul BD, Babu C: Robust image compression algorithm for video capsule endoscopy: a review. In: 2019 International Conference on Intelligent Sustainable Systems (ICISS), pp. 372–377. IEEE; (2019) [Google Scholar]
- 16.Razzak MI, Naz S, Zaib A: Deep learning for medical image processing: overview, challenges and the future. In: Classification in BioApps, pp. 323–350. Springer; (2018) [Google Scholar]
- 17.Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D: Gradcam: visual explanations from deep networks via gradient-based localization. In: Proceedings of the IEEE International Conference on Computer Vision, pp. 618–626 (2017) [Google Scholar]
- 18.Simonyan K, Zisserman A: Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556 (2014) [Google Scholar]
- 19.Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A: Going deeper with convolutions. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 1–9 (2015) [Google Scholar]
- 20.Zhou B, Khosla A, Lapedriza A, Oliva A, Torralba A: Learning deep features for discriminative localization. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2921–2929 (2016) [Google Scholar]
- 21.Zou Y, Li L, Wang Y, Yu J, Li Y, Deng WJ: Classifying digestive organs in wireless capsule endoscopy images based on deep convolutional neural network. In: 2015 IEEE International Conference on Digital Signal Processing (DSP), pp. 1274–1278. IEEE; (2015) [Google Scholar]


