Abstract
Protein biochemistry protocols typically include disulfide bond reducing agents to guard against unwanted thiol oxidation and protein aggregation. Commonly used disulfide bond reducing agents include dithiothreitol, β-mercaptoethanol, glutathione, and the tris(alkyl)phosphine compounds tris(2-carboxyethyl)phosphine (TCEP) and tris(3-hydroxypropyl)phosphine (THPP). While studying the catalytic activity of the NAD(P)H-dependent enzyme Δ1-pyrroline-5-carboxylate reductase, we unexpectedly observed a rapid non-enzymatic chemical reaction between NAD+ and the reducing agents TCEP and THPP. The product of the reaction exhibits a maximum ultraviolet absorbance peak at 334 nm and forms with an apparent association rate constant of 231–491 M−1 s−1. The reaction is reversible, and nuclear magnetic resonance characterization (1H, 13C, and 31P) of the product revealed a covalent adduct between the phosphorus of the tris(alkyl)phosphine reducing agent and the C4 atom of the nicotinamide ring of NAD+. We also report a 1.45 Å resolution crystal structure of short-chain dehydrogenase/reductase with the NADP+–TCEP reaction product bound in the cofactor binding site, which shows that the adduct can potentially inhibit enzymes. These findings serve to caution researchers when using TCEP or THPP in experimental protocols with NAD(P)+. Because NAD(P)+-dependent oxidoreductases are widespread in nature, our results may be broadly relevant.
Tris(alkyl)phosphine compounds such as tris(2-carboxyethyl)phosphine (TCEP) and tris(3-hydroxypropyl)phosphine (THPP) have become widely used as disulfide reducing agents in protein biochemistry research.1–5 TCEP and THPP have gained in popularity because of their several advantages over the classical reducing agent dithiothreitol (DTT, Cleland’s reagent), including irreversible reduction of disulfides, higher water solubility, lower susceptibility to oxidation, and a greater ability to reduce a wide range of disulfide substrates to their corresponding thiols.6,7 TCEP and THPP are typically used at ≤1.0 mM during protein purification and long-term storage of proteins, whereas in other applications such as enzyme assays, protein crystallography, protein refolding, and cysteine proteomic studies, the concentrations can be considerably higher (5–50 mM).4,8–12 We suspect that most biochemistry researchers, like us, have assumed that TCEP and THPP are innocuous.
We routinely use TCEP and THPP in the purification of human l-Δ1-pyrroline-5-carboxylate (l-P5C) reductase (PYCR), an enzyme that reduces l-P5C to l-proline using NAD(P)H. PYCR catalyzes the last step of l-proline biosynthesis and is important in the plant stress response13 and bacterial pathogenesis14 and, in humans, is of significant interest in bioenergetics and cancer research.15–19 Because l-P5C is not commercially available or amenable for long-term storage, the activity of PYCR in the reverse direction is sometimes studied using l-proline or the analogue l-thiazolidine-4-carboxylic acid (L-T4C) as the substrate, and NAD(P)+ as the hydride-accepting cofactor.20–23 Assaying PYCR in the reverse direction is attractive for high-throughput screening of compound libraries seeking to identify small molecule inhibitors of PYCR.24 We therefore sought to test the reverse activity of human PYCR1 and PYCR2 isoforms. In our assays, however, we did not find reverse activity with NAD(P)+ and l-proline. Instead, we unexpectedly discovered a chemical reaction between NAD+ and tris(alkyl)phosphine compounds.
PYCR1 and PYCR2 reverse activity with l-proline and NAD+ was tested, but no evidence for NADH formation was observed (SI Figure 1). Learning that some protocols for PYCR reverse activity include TCEP,24 we added THPP or TCEP to the assay and observed a rapid increase in absorbance around 340 nm (SI Figure 1). Suspecting a non-enzymatic reaction, we repeated the assay without PYCR, and again a sharp increase in absorbance at 340 nm was observed with THPP or TCEP (SI Figure 2).
Because the absorbance increase occurred shortly after the addition of TCEP and THPP, we examined the reaction by stopped-flow ultraviolet–visible spectrophotometry. Upon rapidly mixing THPP or TCEP with NAD+, we detected an absorbance peak at 334 nm within 200 ms (Figure 1a,b). Rate constants for the observed increase at 334 nm were then determined for TCEP and THPP. NAD+ was rapidly mixed with increasing concentrations of THPP and TCEP. The observed rate constant (kobs) versus THPP (Figure 1c) and TCEP (Figure 1d) concentration was then plotted to determine the apparent forward association (k+app) and reverse dissociation (k−app) rate constants. SI Table 1 summarizes the rate constants and shows that both the forward (491 M−1 s−1) and reverse (24 s−1) rate constants with TCEP are faster than with THPP. The equilibrium association constant (Kaapp), however, is slightly higher with THPP (35 M−1) than with TCEP (20 M−1). The reaction between NAD+ and TCEP (25 mM) was repeated in different buffers, and similar kobs values in the range of 30–35 s−1 for the increase at 334 nm were obtained, indicating product formation is independent of the buffer composition (SI Figure 3).
Figure 1.
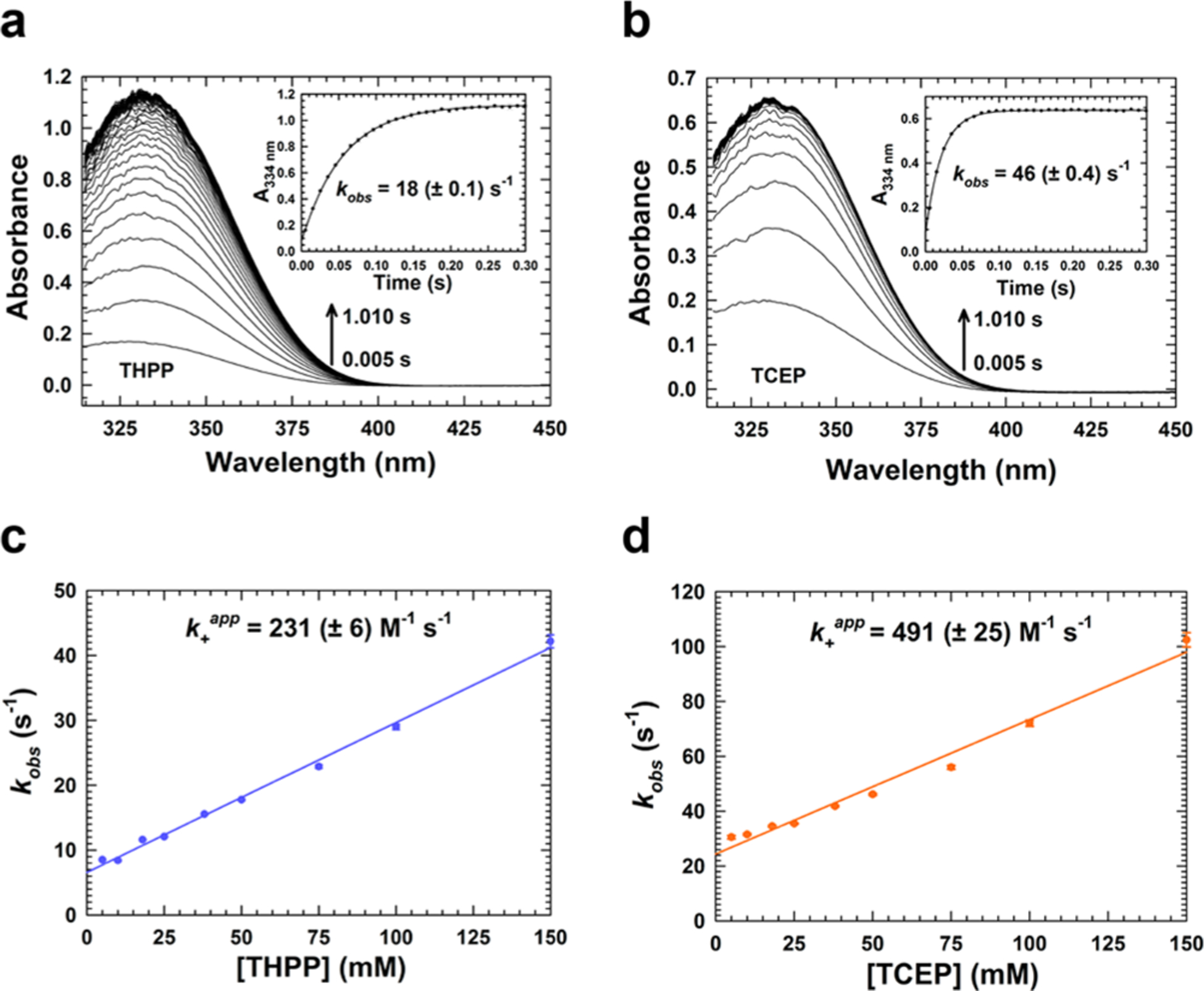
Stopped-flow kinetics of the NAD+–tris(alkyl)phosphine reaction. NAD+ (0.5 mM) was rapidly mixed with 50 mM (a) THPP and (b) TCEP in 100 mM HEPES (pH 7.5) buffer. Plots of absorbance at 334 nm fit to a single-exponential equation are shown in the insets along with the observed rate constant (kobs). The dependence of kobs on THPP and TCEP concentration was determined by holding the NAD+ concentration at a fixed value (0.5 mM) and varying the THPP and TCEP concentration (5–150 mM). kobs values were determined from single-exponential fits of single-wavelength data collected at 334 nm. The apparent forward association (k+app) and reverse dissociation (k−app) rate constants were determined from the plot of kobs vs (c) THPP and (d) TCEP concentration. Data are plotted as the mean ± the standard deviation of three technical replicates and fit to a linear equation. All concentrations are after mixing.
We next considered the possibility that TCEP and THPP had reduced NAD+ to NADH. l-Lactate dehydrogenase (LDH) and pyruvate were added to the TCEP/THPP–NAD+ reaction product, but no decrease in absorbance (330–340 nm) was observed, indicating that the product was not used by l-lactate dehydrogenase to catalyze the reduction of pyruvate. We also did not find evidence for NADH as the reaction product by mass spectrometry analysis.
Nuclear magnetic resonance (NMR) was then used to characterize and identify the product of the reaction between TCEP or THPP and NAD+. Initial NMR experiments included one-dimensional (1H and 31P) and two-dimensional (1H–31P HSQC and 1H–31P HSQC-TOCSY) analyses (Figure 2 and SI Figure 5; corresponding spectral data summarized in SI Table 4). The resulting data showed through-bond coupling of the phosphorus nucleus of THPP to protons of the nicotinamide ring of NAD+. Additionally, the 1H–31P HSQC-TOCSY data in SI Figure 5 indicate the reaction was reversible as there were 31P correlations for both the covalent adduct nicotinamide ring protons and the lone NAD+ nicotinamide ring protons. 1H–31P HSQC-TOCSY correlations to NAD+ nicotinamide ring protons meant the tris(alkyl)phosphine compound dissociated from the nicotinamide ring during the TOCSY spin lock.
Figure 2.
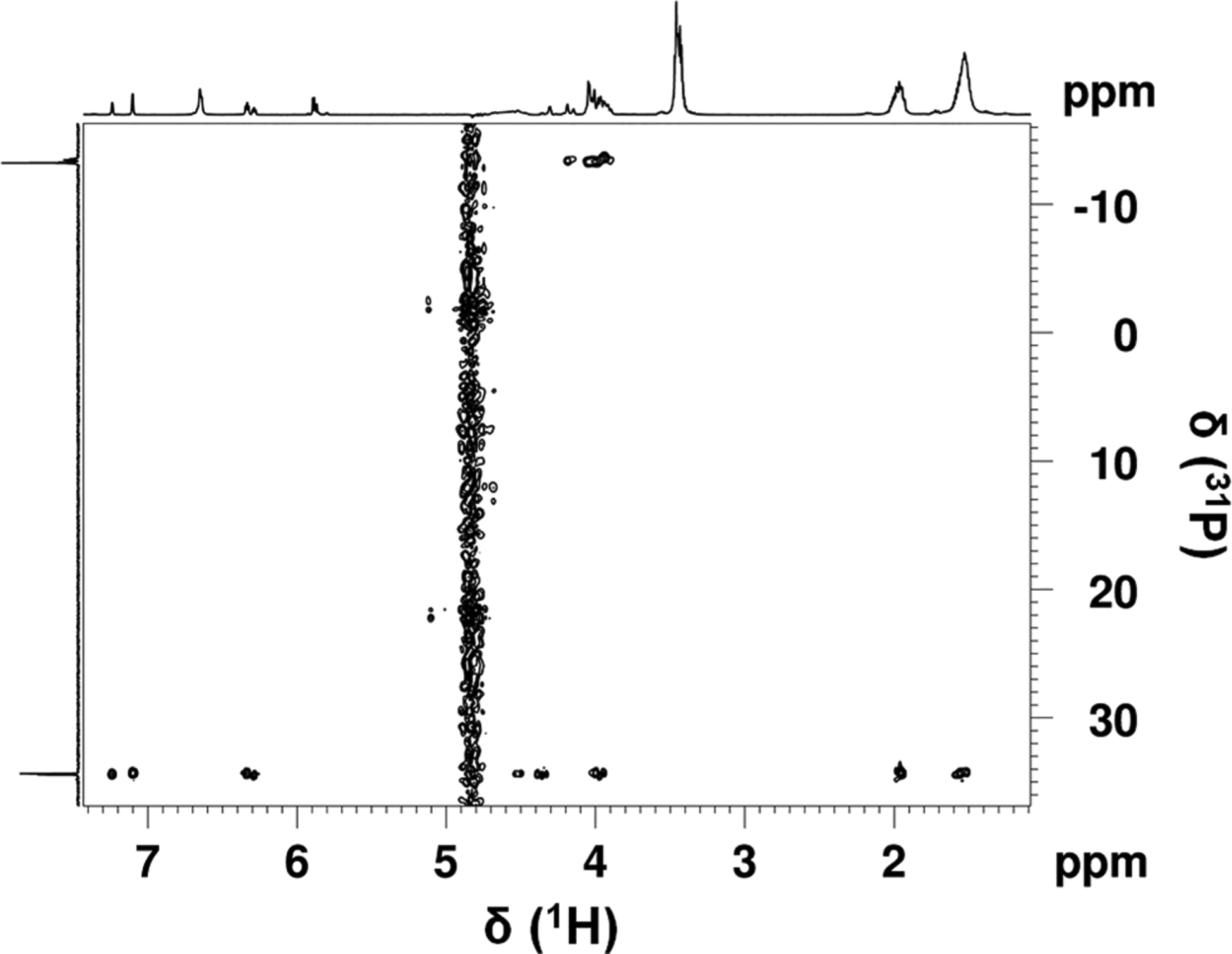
Two-dimensional 1H–31P HSQC spectrum of the NAD+ and THPP reaction mixture. The spectrum was acquired with a J(HP) coupling constant of 11 Hz.
Both one-dimensional (31P) and two-dimensional (1H–31P HSQC and 1H–31P HSQC-TOCSY) NMR analyses showed phosphorus signals at 34 and −13 ppm for the NAD+ and THPP reaction mixture. The phosphorus nuclei at 34 ppm show through-bond coupling to the nicotinamide ring protons of NAD+, as previously described. The phosphorus signals at −13 ppm are attributed to the phosphates in ADP; as expected, these phosphorus nuclei show through-bond coupling to ribose ring protons.
To further confirm the structural assignment, additional NMR experiments were conducted on the NAD+ and THPP reaction mixture. The sample had been stored at 4 °C for 3 weeks and showed minor decomposition; however, the reaction product was still the main component. These additional experiments included one-dimensional (13C) and two-dimensional (1H–1H COSY, 1H–1H TOCSY, 1H–13C HSQC, and 1H–13C HMBC) analyses (SI Figures 6 and 7 and SI Table 5). The additional data indicated that THPP attacked C4 of the nicotinamide ring of NAD+, the same position at which hydride transfer occurs, resulting in two diastereomers in an ~1:0.6 ratio (Figure 3a). NMR analyses of NAD+ alone and NADH alone (spectral data summarized in SI Tables 2 and 3, respectively) also support these structural assignments. Lastly, the one-dimensional proton spectra of the reaction product of NAD+ with THPP and TCEP were very similar (SI Figure 4), indicating a similar covalent adduct is formed with both tris(alkyl)phosphine compounds (Figure 3a,b).
Figure 3.
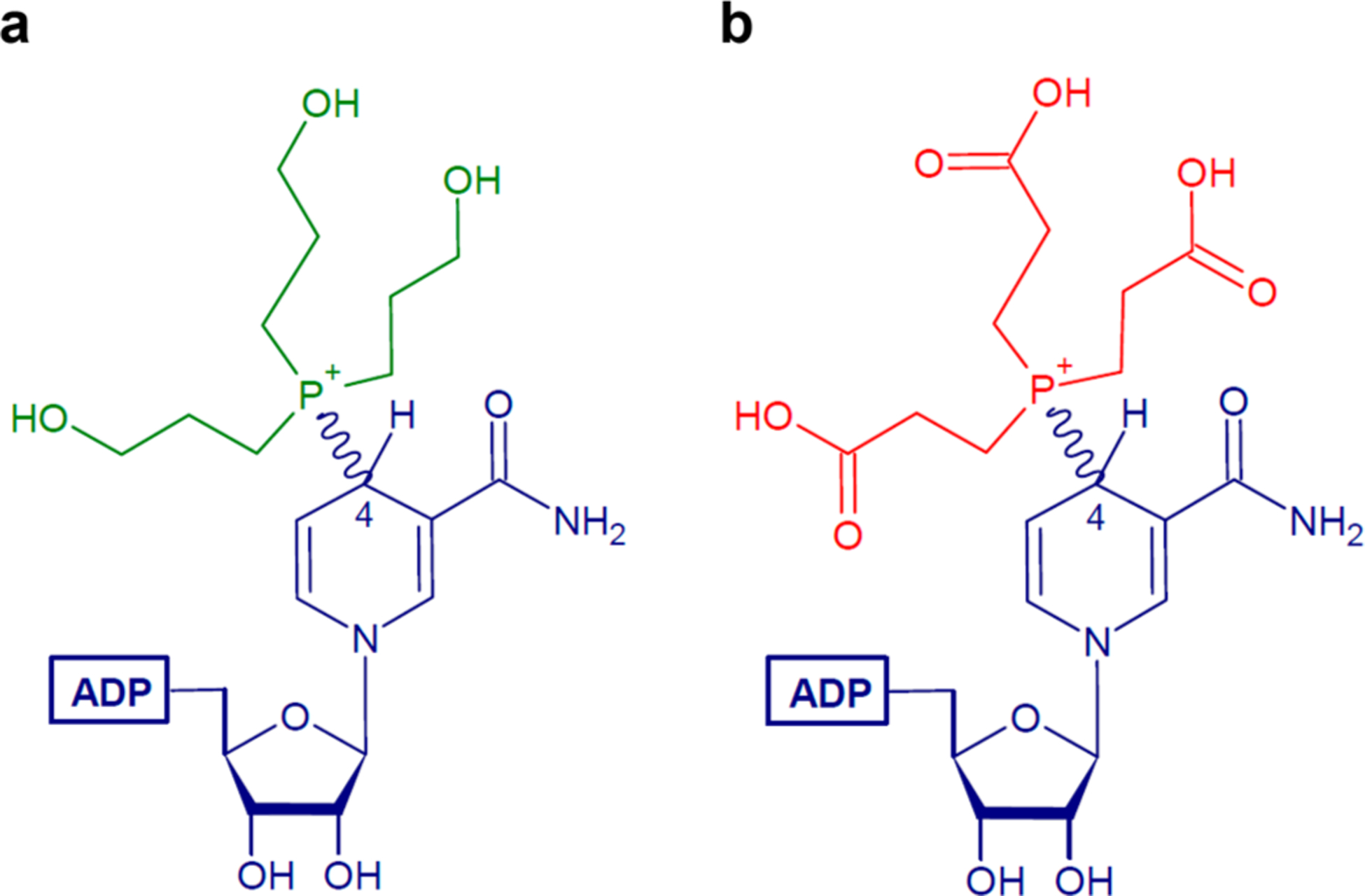
Chemical structures of the reaction products. Resolved covalent adduct products from the reaction of (a) NAD+ with THPP and (b) NAD+ with TCEP.
The covalent adduct between NADP+ and TCEP was also characterized in the context of an oxidoreductase active site using X-ray crystallography. The 1.45 Å resolution structure of a short-chain dehydrogenase/reductase (SDR) from Burkholderia ambifaria was determined from a crystal grown in the presence of 5 mM NADP+ and 1 mM TCEP (Protein Data Bank entry 5VPS). Whereas NADP+ was explicitly added to the crystallization experiment to obtain a structure of the functional enzyme, TCEP was included as an innocuous component of the enzyme stock solution buffer. The electron density maps clearly showed NADP+ bound to the Rossmann fold in the expected conformation (Figure 4a). Unexpectedly, strong electron density indicated that the NADP+ cofactor had been covalently modified at the C4 atom of the nicotinamide ring (Figure 4b). The electron density is consistent with the structure of the adduct determined from solution NMR. The structure shows that several active site residues form hydrogen bonds and ion pairs with the carboxyethyl groups of the TCEP (Figure 4b). These results suggest that the reaction of tris(alkyl)phosphine reducing agents with NAD(P)+ inactivates the biological cofactor and that the species generated can potentially inhibit enzymes.
Figure 4.
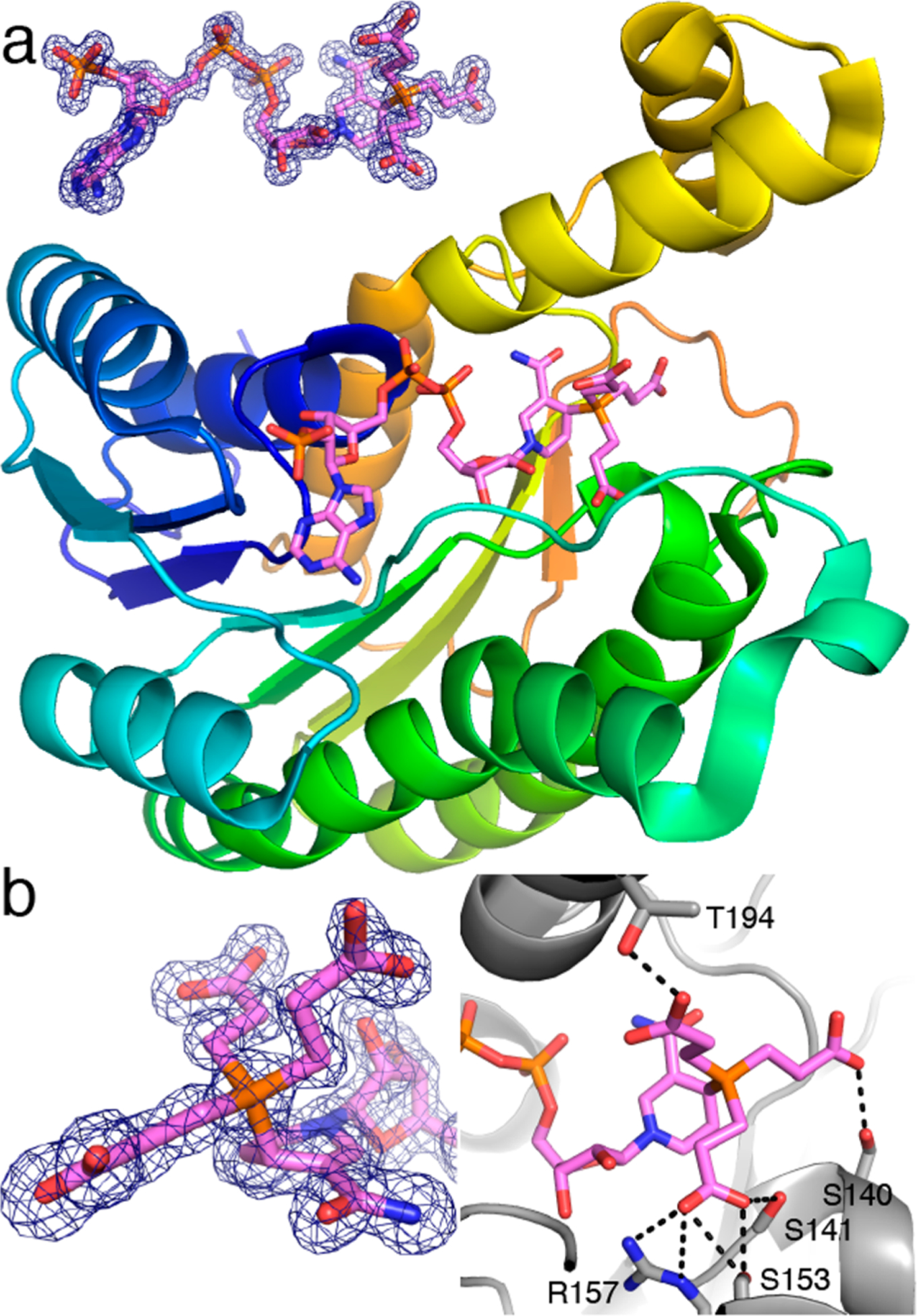
Structure of B. ambifaria SDR with a NADP+–TCEP adduct in the active site. (a) Cartoon drawing of the protein with the adduct colored pink. The inset shows electron density for the adduct (Polder omit, 3.0σ). (b) Close-ups of the adduct showing electron density for the P–C4 bond (left) and interactions with the carboxyethyl groups (right).
To the best of our knowledge, this is the first characterization of a covalent adduct between NAD(P)+ and TCEP or THPP. The reaction occurs at physiological pH within a range of concentrations of TCEP and THPP (0.5–50 mM) typically used in protein biochemistry and structural biology experiments. The reaction is reversible and occurs on a time scale comparable to that of TCEP reduction of disulfide bonds in DTT (43 M−1 s−1), peptide substrates containing a CXXC motif (650 M−1 s−1), and oxidized cysteines in proteins (1.5–813 M−1 s−1).4,9 The reaction between NAD+ and TCEP or THPP most likely proceeds through a nucleophilic attack by the phosphine at the C4 atom of the nicotinamide ring similar to that of a tertiary phosphine attacking an alkyl halide to form a phosphonium ion. The nicotinamide ring of NAD+ has electrophilic character and is susceptible to modifications by nucleophiles. Biologically relevant is the fact that sulfhydryl compounds such as cysteine and glutathione can form adducts at the C4 atom of the nicotinamide with absorbance maxima at 330–335 nm.25,26 In urocanase, a covalent intermediate between imidazolepropionate and NAD+ exhibited an absorbance peak at 335 nm.27 In that case, NMR showed that the imidazole nitrogen formed a covalent adduct at the C4 atom of the nicotinamide ring. The absorbance peak of 334 nm observed here for the covalent NAD–phosphine adduct is similar to that previously reported for various covalent NAD species. NAD addition reactions are also known for cyanide, bisulfite, and dihydroxyacetone.28 LDH has been shown to catalyze NAD adduct formation with cyanide and pyruvate.29 The NAD–pyruvate adduct inhibits LDH at high concentrations of pyruvate.29 Other examples include isoniazid–NAD/NADP adducts found in Mycobacterium tuberculosis that inhibit different enzymes such as dihydrofolate reductase.30
We showed that tris(alkyl)phosphine reducing agents inactivate the biological cofactor NAD(P)+, and the inactivated cofactor can, at least in one case, bind the active site of an enzyme. These results suggest that including TCEP or THPP in crystallization trials of NAD(P)+-dependent oxidoreductases could have unintended consequences. In the SDR structure, the active site is large enough to accommodate the bulky TCEP group, and several residues serendipitously stabilize the carboxyethyl groups (Figure 4). In other enzymes, the adduct may be too large to fit in the active site, which could result in weak or no electron density for NAD(P)+ despite the inclusion of the cofactor in the crystallization setup. One can also imagine cases in which the covalent modification causes the cofactor to bind in an atypical pose, making it difficult to infer the biochemical significance of the structure. Finally, even if the adduct does bind to the enzyme in crystallo, if the active site cannot stabilize the conformation of the tris(alkyl)-phosphine molecule, the nicotinamide may appear disordered, which would complicate the interpretation of the structure.
TCEP and THPP could also cause problems with enzyme assays and binding studies. To illustrate how TCEP could interfere with the assay of a dehydrogenase, we measured the activity of glucose-6-phosphate dehydrogenase (G6PDH) in the presence of 1–50 mM TCEP. With the glucose 6-phosphate concentration fixed at 5 mM and the NAD+ concentration varying from 0.25 to 0.4 mM, the apparent reaction velocity steadily decreased with an increase in TCEP concentration at all NAD+ concentrations tested (SI Figure 8). The effect of TCEP was more pronounced at the lower NAD+ concentrations and was significant even at the lowest TCEP concentration tested. For example, the inclusion of 1 mM TCEP decreased the apparent rate by ~20% when NAD+ was used in the range of 0.025–0.2 mM. The artifactual underestimation of the catalytic activity is presumably due to the reaction of TCEP with NAD+ decreasing the effective concentration of the latter. Another potential problem is that adding TCEP or THPP to NAD(P)+-dependent assays can generate a rapid absorbance increase around 340 nm that, without proper controls, could be misinterpreted as enzyme activity. This particularly could be a problem in enzyme-coupled reactions using NAD+/NADP+ and high-throughput assays that use only end point measurements. Also, the adduct may inhibit the enzyme, as implied by the SDR structure (Figure 4). With regard to binding studies, the addition of tris(alkyl)phosphine compounds would potentially lead to lower concentrations of free NAD(P)+, as in the glucose-6-phosphate dehydrogenase example (SI Figure 8), resulting in anomalous errors in the determined binding constant. In addition, data analysis could be complicated by unknowingly having two species present, NAD(P)+ and the NAD(P)+–tris(alkyl)phosphine covalent adduct. Methods that measure changes in heat to quantify binding events, such as isothermal titration calorimetry, would have complications due to the heat of the reaction between NAD(P)+ and TCEP or THPP.31
In summary, we characterized the chemical reaction kinetics of NAD+ and tris(alkyl)phosphines. Evidence from stopped-flow spectrophotometry, multidimensional NMR, and X-ray crystallography indicates that the product is a reversible covalent adduct between the phosphorus of the tris(alkyl)-phosphine and the C4 atom of the NAD+ nicotinamide. Altogether, our findings here serve as a cautionary note when using TCEP and THPP in biological assays and structural studies of NAD(P)+-dependent oxidoreductases.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank X. Liang for generating the PYCR2 expression clone.
Funding
This research was supported in part by the National Institute of General Medical Sciences, National Institutes of Health Grant R01GM065546, and by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Federal Contract HHSN272201700059C.
ABBREVIATIONS
- l-P5C
l-Δ1-pyrroline-5-carboxylate
- PYCR
l-Δ1-pyrroline-5-carboxylate reductase
- PYCR1/2
human l-Δ1-pyrroline-5-carboxylate reductase 1/2
- SDR
short-chain dehydrogenase/reductase
- TCEP
tris(2-carboxyethyl)phosphine
- THPP
tris(3-hydroxypropyl)phosphine
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.biochem.0c00490.
Methods, SI Tables 1–6, and SI Figures 1–8 (PDF)
Accession Codes
UniProt Accession IDs: PYCR1, P32322; PYCR2, Q96C36; G6PDH, P11411; SDR, B1YU47. Protein Data Bank entry 5VPS.
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.biochem.0c00490
The authors declare no competing financial interest.
Contributor Information
Sagar M. Patel, Department of Biochemistry and Redox Biology Center, University of Nebraska—Lincoln, Lincoln, Nebraska 68588, United States
Thomas G. Smith, Department of Chemistry, University of Nebraska—Lincoln, Lincoln, Nebraska 68588, United States
Martha Morton, Department of Chemistry, University of Nebraska—Lincoln, Lincoln, Nebraska 68588, United States;.
Kyle M. Stiers, Department of Biochemistry, University of Missouri, Columbia, Missouri 65211, United States
Javier Seravalli, Department of Biochemistry and Redox Biology Center, University of Nebraska—Lincoln, Lincoln, Nebraska 68588, United States.
Stephen J. Mayclin, Seattle Structural Genomics Center for Infectious Disease, UCB Pharma, Bainbridge Island, Washington 98110, United States
Thomas E. Edwards, Seattle Structural Genomics Center for Infectious Disease, UCB Pharma, Bainbridge Island, Washington 98110, United States
John J. Tanner, Department of Biochemistry, University of Missouri, Columbia, Missouri 65211, United States.
Donald F. Becker, Department of Biochemistry and Redox Biology Center, University of Nebraska—Lincoln, Lincoln, Nebraska 68588, United States.
REFERENCES
- (1).Levison ME, Josephson AS, and Kirschenbaum DM (1969) Reduction of biological substances by water-soluble phosphines: gamma-globulin (IgG). Experientia 25, 126–127. [DOI] [PubMed] [Google Scholar]
- (2).Ruegg UT, and Rudinger J (1977) Reductive cleavage of cystine disulfides with tributylphosphine. Methods Enzymol. 47, 111–116. [DOI] [PubMed] [Google Scholar]
- (3).Burns JA, Butler JC, Moran J, and Whitesides GM (1991) Selective reduction of disulfides by tris(2-carboxyethyl)-phosphine. J. Org. Chem 56, 2648–2650. [Google Scholar]
- (4).Cline DJ, Redding SE, Brohawn SG, Psathas JN, Schneider JP, and Thorpe C (2004) New water-soluble phosphines as reductants of peptide and protein disulfide bonds: reactivity and membrane permeability. Biochemistry 43, 15195–15203. [DOI] [PubMed] [Google Scholar]
- (5).Abo M, Li C, and Weerapana E (2018) Isotopically-labeled iodoacetamide-alkyne probes for quantitative cysteine-reactivity profiling. Mol. Pharmaceutics 15, 743–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Getz EB, Xiao M, Chakrabarty T, Cooke R, and Selvin PR (1999) A comparison between the sulfhydryl reductants tris(2-carboxyethyl)phosphine and dithiothreitol for use in protein biochemistry. Anal. Biochem 273, 73–80. [DOI] [PubMed] [Google Scholar]
- (7).McNulty J, Krishnamoorthy V, Amoroso D, and Moser M (2015) Tris(3-hydroxypropyl)phosphine (THPP): A mild, air-stable reagent for the rapid, reductive cleavage of small-molecule disulfides. Bioorg. Med. Chem. Lett 25, 4114–4117. [DOI] [PubMed] [Google Scholar]
- (8).Domkin V, and Chabes A (2015) Phosphines are ribonucleotide reductase reductants that act via C-terminal cysteines similar to thioredoxins and glutaredoxins. Sci. Rep 4, 5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Parsons ZD, and Gates KS (2013) Thiol-dependent recovery of catalytic activity from oxidized protein tyrosine phosphatases. Biochemistry 52, 6412–6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Wojdyla K, and Rogowska-Wrzesinska A (2015) Differential alkylation-based redox proteomics-lessons learnt. Redox Biol. 6, 240–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Seto A, Ikushima H, Suzuki T, Sato Y, Fukai S, Yuki K, Miyazawa K, Miyazono K, Ishitani R, and Nureki O (2009) Crystallization and preliminary X-ray diffraction analysis of GCIP/HHM transcriptional regulator. Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun 65, 21–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Willis MS, Hogan JK, Prabhakar P, Liu X, Tsai K, Wei Y, and Fox T (2005) Investigation of protein refolding using a fractional factorial screen: a study of reagent effects and interactions. Protein Sci. 14, 1818–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Fichman Y, Gerdes SY, Kovacs H, Szabados L, Zilberstein A, and Csonka LN (2015) Evolution of proline biosynthesis: enzymology, bioinformatics, genetics, and transcriptional regulation. Biol. Rev. Camb. Philos. Soc 90, 1065–1099. [DOI] [PubMed] [Google Scholar]
- (14).Christgen SL, and Becker DF (2019) Role of proline in pathogen and host interactions. Antioxid. Redox Signaling 30, 683–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).De Ingeniis J, Ratnikov B, Richardson AD, Scott DA, Aza-Blanc P, De SK, Kazanov M, Pellecchia M, Ronai Z. e., Osterman AL, and Smith JW (2012) Functional specialization in proline biosynthesis of melanoma. PLoS One 7, No. e45190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Phang JM (2019) Proline metabolism in cell regulation and cancer biology: recent advances and hypotheses. Antioxid. Redox Signaling 30, 635–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Liu W, Hancock CN, Fischer JW, Harman M, and Phang JM (2015) Proline biosynthesis augments tumor cell growth and aerobic glycolysis: involvement of pyridine nucleotides. Sci. Rep 5, 17206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Tanner JJ, Fendt SM, and Becker DF (2018) The proline cycle as a potential cancer therapy target. Biochemistry 57, 3433–3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Schworer S, Berisa M, Violante S, Qin W, Zhu J, Hendrickson RC, Cross JR, and Thompson CB (2020) Proline biosynthesis is a vent for TGFbeta-induced mitochondrial redox stress. EMBO J. 39, No. e103334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Meng Z, Lou Z, Liu Z, Li M, Zhao X, Bartlam M, and Rao Z (2006) Crystal structure of human pyrroline-5-carboxylate reductase. J. Mol. Biol 359, 1364–1377. [DOI] [PubMed] [Google Scholar]
- (21).Kenklies J, Ziehn R, Fritsche K, Pich A, and Andreesen JR (1999) Proline biosynthesis from L-ornithine in Clostridium sticklandii: purification of delta1-pyrroline-5-carboxylate reductase, and sequence and expression of the encoding gene, proC. Microbiology 145 (Part 4), 819–826. [DOI] [PubMed] [Google Scholar]
- (22).Deutch CE, Klarstrom JL, Link CL, and Ricciardi DL (2001) Oxidation of L-thiazolidine-4-carboxylate by delta1-pyrroline-5-carboxylate reductase in Escherichia coli. Curr. Microbiol 42, 442–446. [DOI] [PubMed] [Google Scholar]
- (23).Nocek B, Chang C, Li H, Lezondra L, Holzle D, Collart F, and Joachimiak A (2005) Crystal structures of delta1-pyrroline-5-carboxylate reductase from human pathogens Neisseria meningitides and Streptococcus pyogenes. J. Mol. Biol 354, 91–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Grünewald S, Steckel M, Husemann M, Meyer H, Han W, and Ding Z (2018) Inhibitors and antagonists of human PYCR1. EP3406253A1. [Google Scholar]
- (25).Dittmer DC, and Kolyer JM (1963) Addition compounds of thiols and 1-substituted nicotinamides. J. Org. Chem 28, 1720–1722. [Google Scholar]
- (26).Van Eys J, and Kaplan NO (1957) The addition of sulfhydryl compounds to diphosphopyridine nucleotide and its analogues. J. Biol. Chem 228, 305–314. [PubMed] [Google Scholar]
- (27).Matherly LH, DeBrosse CW, and Phillips AT (1982) A covalent nicotinamide adenine dinucleotide intermediate in the urocanase reaction. Biochemistry 21, 2789–2794. [DOI] [PubMed] [Google Scholar]
- (28).Ciotti MM, and Kaplan NO (1956) Chemistry and properties of the 3-acetylpyridine analogue of diphosphopyridine nucleotide. J. Biol. Chem 221, 823–832. [PubMed] [Google Scholar]
- (29).Holbrook JJ, Liljas A, Steindel SJ, and Rossmann MG (1975) Lactate dehydrogenase. In The Enzymes (Boyer PD, Ed.) pp 191–293, Academic Press, London. [Google Scholar]
- (30).Timmins GS, and Deretic V (2006) Mechanisms of action of isoniazid. Mol. Microbiol 62, 1220–1227. [DOI] [PubMed] [Google Scholar]
- (31).Di Trani JM, Moitessier N, and Mittermaier AK (2018) Complete kinetic characterization of enzyme inhibition in a single isothermal ttration calorimetric experiment. Anal. Chem 90, 8430–8435. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


