Abstract
Diagnosis of human monocytotropic ehrlichiosis (HME) generally depends on serology that detects the antibody response to immunodominant proteins of Ehrlichia chaffeensis. Protein immunoblotting was used to evaluate the reaction of the antibodies in patients’ sera with the recombinant E. chaffeensis 120- and 28-kDa proteins as well as the 106- and the 37-kDa proteins. The cloning of the genes encoding the latter two proteins is described in this report. Immunoelectron microscopy demonstrated that the 106-kDa protein is located at the surfaces of ehrlichiae and on the intramorular fibrillar structures associated with E. chaffeensis. The 37-kDa protein is homologous to the iron-binding protein of gram-negative bacteria. Forty-two serum samples from patients who were suspected to have HME were tested by immunofluorescence (IFA) using E. chaffeensis antigen and by protein immunoblotting using recombinant E. chaffeensis proteins expressed in Escherichia coli. Thirty-two serum samples contained IFA antibodies at a titer of 1:64 or greater. The correlation of IFA and recombinant protein immunoblotting was 100% for the 120-kDa protein, 41% for the 28-kDa protein, 9.4% for the 106-kDa protein, and 0% for the 37-kDa protein. None of the recombinant antigens yielded false-positive results. All the sera reactive with the recombinant 28- or the 106-kDa proteins also reacted with the recombinant 120-kDa protein.
Human monocytotropic ehrlichiosis (HME) is an emerging tick-borne infectious disease. The disease is characterized by nonspecific clinical findings which include fever (97.2%), malaise (84%), headache (81.3%), myalgia (68.1%), rigors (61.1%), and rash (36.2%) (20). Hematologic and clinical chemistry laboratory results typically include leukopenia, thrombocytopenia, and elevated levels of hepatic enzymes in serum (20). HME is a moderate-to-severe illness, even life-threatening in some cases (18, 26, 30). The disease was first reported in the United States in 1987 (19, 23). It has been documented serologically in 30 states of the United States (19). The etiologic agent, Ehrlichia chaffeensis, has been isolated from patients with ehrlichiosis in only a few cases since the first isolate, Arkansas strain, was cultivated in 1991 (3, 13, 15, 17, 27). E. chaffeensis is an obligately intracellular gram-negative bacterium that is closely related genetically and antigenically to Ehrlichia canis and Ehrlichia ewingii (canine pathogens), Ehrlichia muris (a Japanese rodent isolate), and Cowdria ruminantium (the etiologic agent of heartwater in cattle) (5, 31, 34). Amblyomma americanum ticks are the predominant vectors (4). The diagnosis of HME is difficult because the clinical findings are nonspecific and it is difficult to isolate the etiologic agent. Despite the availability of PCR amplification of nucleotide sequences of the E. chaffeensis genomic DNA, the diagnosis of HME usually depends on serology. Immunofluorescence assay (IFA) is the most commonly used method for diagnosis of HME, but IFA is difficult to standardize owing to the requirement for experienced persons to examine the slides and the subjective determination of the end point. IFA cannot distinguish whether the antibodies were stimulated by the homologous organism or by an antigenically related organism. Thus, false-positive results may result from infection with an organism with cross-reactive antigens. Moreover, IFA also requires cell-cultivated E. chaffeensis, which can be grown in only a few research laboratories.
Convalescent-phase human sera recognize several immunoreactive E. chaffeensis antigens, including the 120-, 66-, 55-, and 44-kDa proteins and the 28-kDa protein complex (8, 9, 11, 35). These immunoreactive proteins of E. chaffeensis are the molecular basis for the serologic diagnosis of HME. Molecular cloning techniques circumvent the fastidious growth characteristics of ehrlichiae and provide large amounts of pure ehrlichial proteins for serology. To determine the best candidate antigens for recombinant-protein-based serology or a vaccine, most of the genes encoding E. chaffeensis immunodominant proteins have been cloned and overexpressed, including the genes encoding the 120-, 58-, and 28-kDa proteins (25, 29, 36). The 120-kDa protein (27a) and the 28-kDa protein (25) have been demonstrated to be outer membrane proteins of E. chaffeensis. The 120-kDa protein has been shown to be a potential diagnostic serologic tool (37). The 28-kDa protein is a member of a family encoded by multiple homologous genes (25). The 28-kDa protein is highly varied genetically and antigenically among strains of E. chaffeensis (10, 38). The 58-kDa protein is an analog of the GroEL protein of Escherichia coli, which is a heat shock protein (29). In this study we have cloned and sequenced two additional E. chaffeensis genes, those encoding the 106- and 37-kDa proteins. We demonstrated that the 106-kDa protein is an outer membrane protein and the 37-kDa protein is an analog of an iron-binding protein of gram-negative bacteria. We expressed the E. chaffeensis 120-, 106-, 37-, and 28-kDa proteins in E. coli and evaluated the reactivities of these recombinant proteins with HME patients’ sera by using protein immunoblotting.
MATERIALS AND METHODS
Ehrlichiae.
E. chaffeensis (Arkansas strain) and E. canis (Oklahoma strain) were obtained from Jacqueline Dawson (Centers for Disease Control and Prevention, Atlanta, Ga.). Ehrlichiae were cultivated in DH82 cells, a canine macrophage-like cell line (16). DH82 cells were harvested with a cell scraper when 100% of the cells were infected with ehrlichiae. The cells were centrifuged at 17,400 × g for 20 min. The pellets were disrupted with a Braun-Sonic 2000 sonicator at a power setting of 40 W for 30 s twice on ice. The cell lysate was loaded onto discontinuous gradients of 42%–36%–30% Renografin and then centrifuged at 80,000 × g for 60 min. Ehrlichiae in the heavy and light bands were collected (32) and washed by centrifugation with sucrose-phosphate-glutamate buffer (218 mM sucrose, 3.8 mM KH2PO4, 7.2 mM K2HPO4, 4.9 mM glutamate [pH 7.0]).
Cloning the 106- and 37-kDa protein genes.
Ehrlichial DNA was extracted with phenol-chloroform according to a method described previously (28). Ehrlichial DNA was partially digested with the restriction endonuclease EcoRI, and DNA was separated by electrophoresis on a 1% agarose gel. The 1.0-to-10.0-kb DNA fraction was excised and purified from the agarose gel. Ehrlichial DNA was ligated into EcoRI-precut λgt11 phage cloning and expression vector (Promega Corporation, Madison, Wis.). The phage library was screened by using rabbit anti-E. chaffeensis serum as previously described (28). The DNA inserts in the positive clones were PCR amplified by using λgt11 forward and reverse primers (Promega Corporation) which are complementary to the λgt11 DNA sequences. PCR-amplified DNA was sequenced with an ABI Prism 377 DNA Sequencer (Perkin-Elmer Applied Biosystems, Foster City, Calif.). The DNA sequence and deduced amino acid sequences were analyzed by using the Genetics Computer Group (Madison, Wis.) software package and DNASTAR (Madison, Wis.) software. The deduced protein was analyzed by using the PSORT program (24a), which predicts the presence of signal sequences by the methods of McGeoch (24) and von Heijne (32) and detects potential transmembrane domains by the method of Klein et al. (22).
Adapter PCR.
Adapter PCR amplifies unknown DNA sequences that are adjacent to a known DNA sequence. Adapter PCR was used to find the unknown sequences up- and downstream of the cloned E. chaffeensis DNA fragment that contained the 106- and 37-kDa protein genes with the GenomeWalker Kit (Clontech Laboratories, Inc., Palo Alto, Calif.). E. chaffeensis Arkansas genomic DNA was digested completely with each of five restriction enzymes, DraI, EcoRV, PvuII, ScaI, and StuI. All five enzymes produced blunt-end DNA fragments. Each batch of digested genomic DNA fragments was ligated with an adapter to create genomic libraries. Restriction enzyme-digested E. chaffeensis DNA fragments were ligated with adapters. The adapter-ligated genomic DNA fragments were used as templates to amplify the unknown DNA sequences. Primers complementary to the known sequences near the 5′ end of the 106-kDa protein gene were used to amplify the missing sequences on the 5′ end of the 106-kDa protein gene. Primers derived from the sequences near the 3′ end of the 37-kDa protein gene were used to amplify the unknown DNA sequences downstream of the 37-kDa protein gene. The adapter has one blunt end and has the 5′ end overhanging. The adapter primer is complementary to the protruding strand. In the first cycle of PCR, DNA was amplified by the primer complementary to the E. chaffeensis DNA sequence. The overhanging adapter strand was amplified together with the E. chaffeensis DNA in the first cycle of PCR. The 5′ end of the PCR product from the first cycle was complementary to the overhanging adapter sequence. The products from the first cycle served as templates in the subsequent amplification with the adapter primer. Therefore, the unknown sequences of E. chaffeensis were specifically amplified by adapter PCR. To increase the sensitivity, nested PCR amplification was used.
Southern blotting.
The groEL analog heat shock protein gene of E. chaffeensis had been cloned by the same approaches as those used in this study (30). It was suspected that some of our clones would contain the heat shock protein gene. DNA hybridization was performed on all positive clones by plaque lift. The probe was labeled by using digoxigenin-11-dUTP with a DIG DNA Labeling and Detection Kit (Boehringer Mannheim Co., Indianapolis, Ind.) according to the manufacturer’s protocol. The probe was a 411-bp PCR product which was amplified from E. chaffeensis genomic DNA by using an E. chaffeensis heat shock protein gene primer pair. The forward primer (TAT CGT CAG TGG GCT GG) started at nucleotide 351 on the sense strand of the heat shock protein gene. The reverse primer (GCA AGA GCC AAT GGA TCC) started at nucleotide 726 on the antisense strand. Any clones hybridizing with the heat shock protein gene probe were excluded from further study.
Southern blotting was also used to determine the copy numbers of the E. chaffeensis 106- and 37-kDa protein genes. Ehrlichial DNA was digested completely with a single restriction enzyme or double digested with two restriction enzymes. DNA was separated in an agarose gel by electrophoresis. The DNA was vacuum transferred onto a nylon membrane and hybridized with a digoxigenin-labeled 106- or 37-kDa protein gene probe. The 106-kDa protein gene probe was a 2,195-bp DNA fragment which was PCR amplified from E. chaffeensis genomic DNA with primers 106f and 106r. The 106f primer (ATT TCA GAG TAC TTT GCA GCA) started from the DNA sequence corresponding to amino acid 252 of the 106-kDa protein. The 106r primer (TGT GTG CCT TTT TAC TGA GAT GT) was 46 nucleotides downstream of the TGA termination codon. The 37-kDa protein gene was amplified by PCR using a recombinant λgt11 clone (clone 9) as the template with 37f and λgt11 forward primer. The 37f primer (TCG CAA GGA AGA ATT ATT ACA) is 116 nucleotides downstream of the start codon of the gene. The reverse primer is λgt11 forward primer (Promega), which annealed to the vector sequence downstream of the TAG termination codon of the 37-kDa protein gene.
Expression of the ehrlichial genes.
The same DNA fragments used to prepare the 106- and 37-kDa protein gene probes were used to express the 106- and 37-kDa proteins, respectively. The PCR-amplified 106- and 37-kDa protein genes were first cloned into the pCRII vector (Invitrogen, Carlsbad, Calif.). Then the DNA inserts were cleaved from the recombinant pCRII clones by EcoRI and were cloned in-frame into the pGEX expression vector (Amersham Pharmacia Biotech, Piscataway, N.J.). The ehrlichial genes were expressed in E. coli BL21 with isopropyl-β-d-thiogalactopyranoside (IPTG) induction, and the recombinant proteins were purified by affinity purification with Glutathione Sepharose 4B (Amersham Pharmacia Biotech). The 120-kDa protein gene of E. chaffeensis has been cloned into the pGEX expression vectors previously (36). Plasmid pGEX120 contained the entire gene for the 120-kDa protein. Plasmid pGEX13 contained a DNA fragment encoding the first two repeat units of the 120-kDa protein. The 28-kDa protein gene was cloned previously (25). Plasmid p29/p28, containing the 28-kDa protein gene of E. chaffeensis, was provided by Y. Rikihisa (Department of Veterinary Biosciences, The Ohio State University, Columbus). The recombinant 28-kDa protein was purified by using the S-Tag Thrombin Purification Kit according to the instructions of the manufacturer (Novagen Inc., Madison, Wis.). The ehrlichial genes were expressed in E. coli as described above.
Production of antibodies to the recombinant ehrlichial proteins.
One rabbit each was immunized intradermally with 50 μg of recombinant 106- or 37-kDa protein four times. In the first injection, the antigens were mixed with Freund’s complete adjuvant. In the subsequent injections, the antigens were mixed with Freund’s incomplete adjuvant. Serum was collected from each rabbit prior to immunization as a negative control. Monospecific polyclonal antibodies to the recombinant 120-kDa protein were prepared in rabbits previously (37).
Immunoelectron microscopy.
Infected monolayers were fixed with a mixture of 0.5% glutaraldehyde, 2.5% formaldehyde, 0.03% trinitrophenol, and 0.03% CaCl2 in 0.05 M cacodylate buffer (pH 7.3) and embedded in LR White medium as described previously (12). The ultrathin-sectioned, LR White-embedded cells were reacted with rabbit antisera to the recombinant protein followed by colloidal gold-labeled anti-rabbit immunoglobulin G (IgG) (H+L) (AutoProbe EM GAR GIS; Amersham Life Science, Arlington Heights, Ill.).
Patients’ sera.
Forty-two human serum samples were provided by MRL Diagnostics (Cypress, Calif.) without the patients’ identification or clinical history. All these sera were submitted to MRL Diagnostics for the assay of antibodies to E. chaffeensis. Ten serum samples from nonfebrile disease patients with no history of ehrlichiosis were obtained from the University of Texas Medical Branch and used as negative controls.
IFA.
Antigen slides were prepared with E. chaffeensis Arkansas-infected DH82 cells. E. chaffeensis-infected DH82 cells from a 150-cm2 flask were collected when 100% of the cells were infected. The cells were centrifuged at 200 × g for 10 min. The pellet was resuspended in 10 ml of phosphate-buffered saline (PBS) with 0.1% bovine albumin. The ehrlichiae were inactivated by the addition of sodium azide to a final concentration of 0.01% at 4°C overnight. Ten microliters of antigen was applied to each well of 12-well slides. The slides were air dried and fixed in acetone for 5 min. Patients’ sera were diluted serially from an initial dilution of 1:64 in twofold increments. Ten microliters of each dilution of serum was applied to each well of the slides. The slides were incubated at room temperature for 30 min. Slides were washed with PBS twice for 5 min, rinsed with distilled water, and air dried. Fifteen microliters of 1:100 fluorescein isothiocyanate-labeled goat anti-human IgG, IgA, and IgM were applied onto each well of the slides. The slides were incubated and washed as above, then air dried and mounted with coverslips. The slides were examined in a UV microscope with barrier and exciter filters for fluorescein.
Protein immunoblotting.
Each recombinant E. chaffeensis protein was electrophoresed in a 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel with a preparative comb in order to separate the recombinant proteins from contaminating E. coli proteins. The proteins were electroblotted onto a nitrocellulose membrane by using a Trans-Blot SD Semi-Dry Transfer Cell (Bio-Rad, Hercules, Calif.). The membranes were stained by using 0.1% Ponceau S in 5% acetic acid (Sigma Chemical Co., St. Louis, Mo.). The position of the recombinant protein band on each membrane was marked with a pencil. An approximately 1-cm width of membrane around the recombinant protein band was cut out. The membranes were blocked with 5% nonfat dried milk. To screen all recombinant proteins with each patient serum sample simultaneously, the membrane strips containing each recombinant protein were placed one by one on a gasket of the Mini-Protein II Multiscreen Apparatus (Bio-Rad) with the antigen side face up. Five hundred microliters of a 1:100 dilution of each patient serum sample was added into each slot. The recombinant proteins on the membranes were incubated with the patients’ sera for 1 h with continuous rocking. The strips were removed from the Mini-Protein II Multiscreen Apparatus and washed three times with 0.1 M Tris-buffered saline (pH 7.4) for 10 min each time. The strips were incubated with peroxidase-labeled goat anti-human IgA, IgG, and IgM. After washing, the bound antibodies were detected by using 4-chloro-1-naphthol (Sigma Chemical Co.).
Nucleotide sequence accession number.
The GenBank accession no. for the nucleotide sequences of the 106- and 37-kDa protein genes is AF117273.
RESULTS
Cloning the 106- and 37-kDa protein genes.
Thirty clones reactive with anti-E. chaffeensis rabbit sera were obtained. Six recombinant λgt11 phage clones (clones 3, 6, 10, 11, 20, and 21) were selected for further analysis based on the fact that they did not hybridize with the heat shock protein gene. PCR amplification of the DNA inserts in these clones demonstrated DNA insert sizes of 4,252 bp in clone 3 and clone 10, 2,921 bp in clone 9, 2,791 bp in clone 19, 2,489 bp in clone 6, and 1,792 bp in clone 20. Except for clones 3 and 10, all the clones were at first thought to be different from each other because the sizes of the DNA inserts in these clones were different and none of them had an internal EcoRI cleavage site.
DNA sequence analysis demonstrated that all six clones overlapped. They started from the same position at an EcoRI restriction site at the 3′ end but stopped at different positions. No EcoRI cleavage site was found on the 5′ end of any clone. Therefore, we considered that all clones were generated from the same DNA fragment by EcoRI restriction enzyme star activity during the time of digestion of E. chaffeensis genomic DNA for construction of the genomic library.
The largest DNA insert in the clones was completely sequenced. DNA sequence analysis of 4,252 bp of nucleotides from the insert of clone 3 revealed two open reading frames (ORFs). The first was a truncated ORF of 2,818 bp on the 5′ end, and the second was a complete ORF of 1,041 bp on the 3′ end. The intergenic space between the two ORFs was 374 bp. The missing part of the first ORF and the DNA sequences adjacent to its 5′ end were amplified by adapter PCR. DNA sequencing of the PCR products revealed that the first ORF extended a further 50 nucleotides on its 5′ end. The first ORF was 2,868 bp, with the capacity of encoding a 108-kDa protein. The second ORF could encode a protein with a predicted molecular weight of 38 kDa. Signal sequences were predicted from the deduced amino acid sequences of both proteins. The 108-kDa protein was most likely cleaved between G28 and A29 and gave rise to a 106-kDa mature protein. The 38-kDa protein was predicted to be cleaved between S19 and F20, and the predicted molecular size of the mature protein was 37 kDa (Fig. 1). Hereafter, the protein encoded by the first ORF is designated the 106-kDa protein and the protein encoded by the second ORF is designated the 37-kDa protein.
FIG. 1.
DNA sequence of the 106- and 37-kDa protein genes of E. chaffeensis. The first ORF is the 106-kDa protein gene, and the second ORF is the 37-kDa protein gene. Arrows indicate the sequences and directions of primers that were used to amplify the DNA fragments to express the genes. The predicted signal sequences of amino acids at the beginning of each protein are underlined.
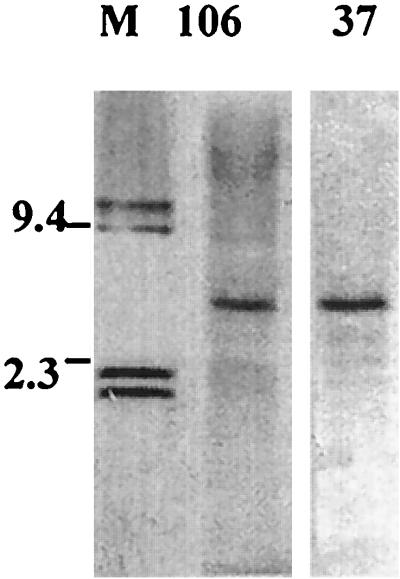
Restriction enzyme mapping analysis demonstrated that Alw26I and EcoRI each had a single restriction site in the 4,735-bp DNA fragment containing the 106- and 37-kDa protein genes. Alw26I cut the 4,735-bp DNA fragment at nucleotide 553, and EcoRI cut the fragment at nucleotide 4702. When Alw26I and EcoRI double-digested E. chaffeensis genomic DNA was hybridized with the 106- or the 37-kDa protein gene probe, Southern blotting demonstrated the presence of a single 4.1-kb band. Since both the 106- and 37-kDa protein gene probes were derived from portions of the Alw26I/EcoRI fragment, these results indicated that both the 106- and the 37-kDa protein gene have a single copy in the E. chaffeensis genome (Fig. 2).
FIG. 2.
Southern blotting revealed that the 106-kDa protein gene probe (lane 106) and the 37-kDa protein gene probe (lane 37) hybridized with Alw26I and EcoRI double-digested E. chaffeensis genomic DNA. Lane M, Digoxigenin-labeled DNA marker (in kilobases).
Homology of the 37-kDa protein with an iron-binding protein.
A search of the SwissProt database with a FASTA program revealed that the 37-kDa protein is homologous to the iron(III)-binding periplasmic protein precursor of bacteria including Serratia marcescens (28%), Haemophilus influenzae (26.6%), and Neisseria gonorrhoeae (27.4%) (Fig. 3). SwissProt database searching did not reveal any homolog for the 106-kDa protein.
FIG. 3.
DNA sequence homology of the E. chaffeensis 37-kDa protein gene (E.ch) with the S. marcescens iron-binding protein (S.ma).
Expression of the 106- and 37-kDa proteins.
The clone containing a fragment of the 106-kDa protein gene was designated pGEX106, and the clone containing the 37-kDa protein gene was designated pGEX37. Both pGEX106 and pGEX37 expressed proteins of the expected sizes. E. coli transformed with pGEX106 expressed a 75-kDa protein. E. coli transformed with pGEX37 expressed a 37-kDa protein. The pGEX106-expressed protein was smaller than 106 kDa because pGEX106 contained only a part of the 106-kDa protein gene.
Reaction of E. chaffeensis antigens with antisera to the recombinant proteins.
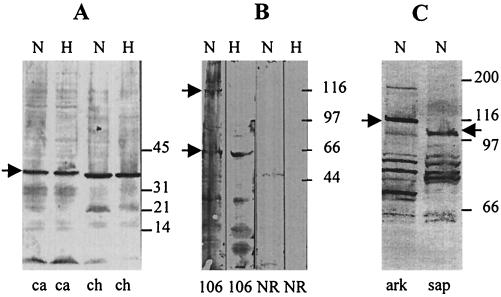
Rabbit antiserum to the recombinant 37-kDa protein reacted with a 37-kDa protein of E. chaffeensis and a 38-kDa protein of E. canis (Fig. 4A). The preimmunization serum did not react with the E. chaffeensis 37-kDa protein (data not shown). Rabbit antiserum to the recombinant 106-kDa protein reacted with a 106-kDa protein and a 60-kDa protein of the nonheated antigen of E. chaffeensis. When the antigen was heated, only the 60-kDa protein remained reactive with the antibodies to the 106-kDa protein (Fig. 4B). No reaction of the anti-106-kDa protein serum was observed with E. canis protein (data not shown). Rabbit antisera to the repeat units of the 120-kDa protein reacted with a 120-kDa protein of the Arkansas strain and a 97-kDa protein of the Sapulpa strain of E. chaffeensis (Fig. 4C).
FIG. 4.
Protein immunoblotting. (A) Rabbit antiserum to the recombinant 37-kDa protein reacted with E. canis (ca) and E. chaffeensis (ch) antigens. (B) Rabbit antiserum to the recombinant 106-kDa protein (106) and normal preimmunization rabbit serum (NR) reacted with E. chaffeensis antigens. (C) Rabbit antiserum to the recombinant 120-kDa protein reacted with E. chaffeensis Arkansas (ark) and E. chaffeensis Sapulpa (sap) antigens. Arrows indicate the ehrlichial proteins that reacted with rabbit antisera to each recombinant protein. H, heated antigen; N, non-heated antigen.
Immunoelectron microscopy.
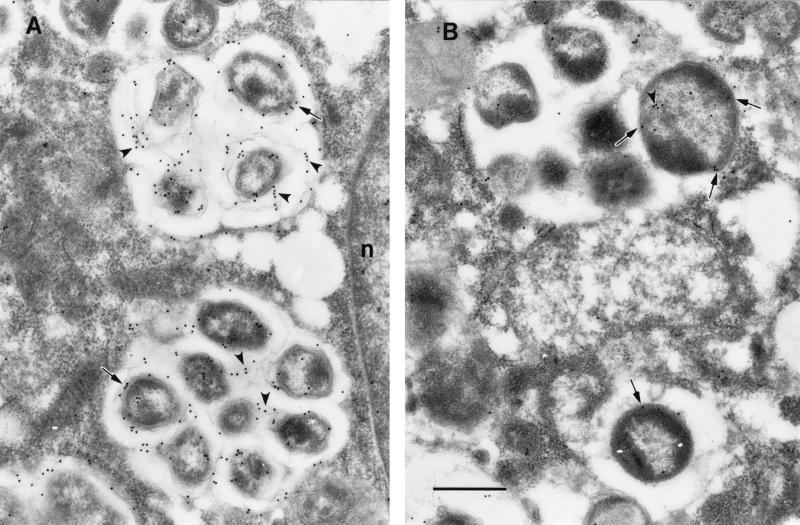
In ultrathin sections of the E. chaffeensis-infected cells, rabbit antiserum to the 106-kDa protein reacted with antigens located in the intramorular fibrillar matrices and at the surfaces of the ehrlichiae (Fig. 5A). This result indicated that the fibrillar matrices contained the 106-kDa protein that had been shed off the surfaces of the ehrlichiae. Gold label was also seen on the membrane limiting the ehrlichial morulae (Fig. 5A). In ultrathin sections, the antigen that reacted with the rabbit antiserum to the 37-kDa protein was located in the ehrlichial cytoplasm and periplasmic space (Fig. 5B).
FIG. 5.
Immunoelectron microscopic visualization of 106- and 37-kDa proteins in ultrathin sections of E. chaffeensis-infected DH82 cells. Bar, 0.5 μm. (A) The 106-kDa protein is located at the surfaces of ehrlichiae (arrows) and in intramorular fibrils originating from the ehrlichial cell surfaces (arrowheads). Gold particle label is also seen on the membrane limiting ehrlichial inclusions (morulae). n, host cell nucleus. (B) Gold particle label with antibodies to the 37-kDa protein is localized in the ehrlichial cell cytoplasm (arrowhead) and in the periplasmic space (arrows).
Reaction of recombinant proteins of E. chaffeensis with HME patients’ sera.
Preliminary testing with 16 serum samples demonstrated that the patients’ sera reacted equally with the entire recombinant 120-kDa glutathione S-transferase (GST) fusion protein and the recombinant GST fusion protein with two repeat units. None of the patients’ sera reacted with the thrombin-cleaved recombinant two-repeat-unit protein. The recombinant two-repeat-unit–GST fusion protein had a better yield than the entire 120-kDa protein in E. coli. Therefore, the GST fusion protein with two repeat units was used to represent the 120-kDa protein in subsequent protein immunoblotting reactions with patients’ sera. All other recombinant proteins used in the protein immunoblotting were also fusion proteins. The 106- and 37-kDa proteins were GST fusion proteins, and the 28-kDa protein was an S-Tag fusion protein.
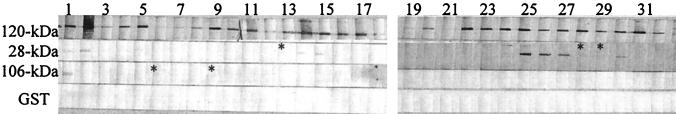
Protein immunoblotting demonstrated that among the 42 serum samples from MRL Diagnostics, 32 reacted with the recombinant 120-kDa protein, 13 reacted with the recombinant 28-kDa protein, 3 reacted with the recombinant 106-kDa protein, and none reacted with the recombinant 37-kDa protein. None of the serum samples reacted with the GST protein. All sera reactive with the recombinant 106-kDa protein and the recombinant 28-kDa protein were also reactive with the 120-kDa protein. By IFA, 32 of the 42 human serum samples from MRL Diagnostics reacted with E. chaffeensis at a titer of 1:64 or greater (Table 1; Fig. 6). The correlation of IFA and protein immunoblotting using the 120-kDa protein for diagnosis of HME was 100%. To confirm the specificity, 10 serum samples from subjects who did not have HME were tested for reactivity with the recombinant E. chaffeensis proteins. None of these sera reacted with any recombinant E. chaffeensis proteins (data not shown).
TABLE 1.
Patient sera reacting with E. chaffeensis antigen by IFA and with recombinant ehrlichial proteins by Western blotting
| Patient no. | IFA titer | Reaction with recombinant protein
|
||
|---|---|---|---|---|
| 120 kDa | 106 kDa | 28 kDa | ||
| 1 | 64 | + | + | + |
| 2 | 256 | + | − | + |
| 3 | 128 | + | − | − |
| 4 | 64 | + | − | − |
| 5 | 128 | + | − | − |
| 6 | 256 | + | + | − |
| 7 | 64 | + | − | − |
| 8 | 512 | + | − | − |
| 9 | 64 | + | + | − |
| 10 | 2,048 | + | − | − |
| 11 | 256 | + | − | − |
| 12 | 256 | + | − | + |
| 13 | 64 | + | − | + |
| 14 | 64 | + | − | + |
| 15 | 2,048 | + | − | − |
| 16 | 128 | + | − | − |
| 17 | 64 | + | − | − |
| 18 | 128 | + | − | − |
| 19 | 512 | + | − | − |
| 20 | 128 | + | − | − |
| 21 | 2,048 | + | − | − |
| 22 | 64 | + | − | − |
| 23 | 256 | + | − | + |
| 24 | 512 | + | − | + |
| 25 | 1,024 | + | − | + |
| 26 | 512 | + | − | + |
| 27 | 128 | + | − | + |
| 28 | 512 | + | − | + |
| 29 | 64 | + | − | + |
| 30 | 128 | + | − | + |
| 31 | 2,048 | + | − | − |
| 32 | 128 | + | − | − |
| 33–42 | − | − | − | |
FIG. 6.
Protein immunoblotting of HME patients’ sera with recombinant E. chaffeensis 120-, 28-, and 106-kDa proteins. Asterisks indicate the weak bands.
DISCUSSION
Our study and previous reports (25, 27a) demonstrated that the 120-, 106-, and 28-kDa proteins are all surface exposed. However, their immunogenicities are quite different. Based on the reactivities of the recombinant proteins with the HME patients’ sera, we concluded that the 120-kDa protein is the most immunodominant, the 28-kDa protein is less immunodominant, and the 106-kDa protein is the least immunodominant. These results are consistent with a previous report that demonstrated that more patients’ sera reacted with the 120-kDa protein than with the 28-kDa protein (10). The reason that many patients’ sera did not recognize the 28-kDa protein is either low antigenicity of the protein or antigenic diversity of the protein. Although the 28-kDa protein is conserved among species, it is relatively variable among the strains of E. chaffeensis (13, 38). In contrast, the 120-kDa protein is species specific and has very little variation among E. chaffeensis strains. The only difference in amino acid sequences of the 120-kDa protein among strains of E. chaffeensis is the deletion of one repeat unit in some strains. The Arkansas strain contains four repeat units, and the Sapulpa strain contains only three repeat units. The fact that the antisera of rabbits stimulated with the repeats of the 120-kDa protein of the Arkansas strain reacted with the 97-kDa antigen of the Sapulpa strain demonstrated that the 120-kDa proteins from various strains have similar immunogenicities regardless of the number of repeat units.
The reaction of patients’ sera with GST fusion protein, but not with the cleaved polypeptide of the recombinant 120-kDa protein that consists of only two repeat units, is not due to reaction with GST protein, because none of the sera reacted with GST. These results suggested that antigenic epitopes on the repeats of the 120-kDa protein are conformationally dependent. It is possible that the N-terminal GST peptide in the fusion protein helped the repeats of the 120-kDa protein to fold properly to form the conformational epitopes. Conformational antigenic epitopes have been found in repeats of the alpha C protein of group B streptococci (21).
The 37-kDa protein of E. chaffeensis is homologous to the iron-binding protein of gram-negative bacteria. The iron-binding proteins of gram-negative bacteria not only have amino acid sequence homology but also have similar genetic structures. In all bacteria investigated, the iron-binding protein is encoded by the first gene (gene A) of an operon of three genes. The remaining two genes (B and C) encode a permease and an ATP-binding protein, respectively. These genes are called the fbpABC operon in N. gonorrhoeae (1), hitABC in H. influenzae (2), sfuABC (6) in S. marcescens, and afuABC in Actinobacillus pleuropneumoniae (14). The three genes of the fbpABC, hitABC, and sfuABC operons are tandemly arranged and are located very close to each other in the chromosome. The distance between fbpA and fbpB is 58 nucleotides, and that between fbpB and fbpC is 20 nucleotides (1). Genes sfuA and sfuB are 33 bp apart, and sfuB and sfuC overlap one other (6). The distance between genes hitA and hitB is 118 nucleotides; hitbB and hitC overlap (2). The distances between genes afuA and afuB and between afuB and afuC are 105 and 47 bp, respectively (14). To characterize whether the 37-kDa protein gene of E. chaffeensis was followed by the genes analogous to the permease and ATP-binding protein in the genome, we amplified approximately 1 kb of DNA downstream of the 37-kDa protein gene by using the genome walking method. No analog of either the permease gene or the ATP-binding protein gene was found in the 1 kb of nucleotides downstream of the iron-binding protein gene (data not shown). A previous report demonstrated that desferroxamine, an intracellular iron chelator, completely prevents the survival of E. chaffeensis. Therefore E. chaffeensis is believed to be sensitive to intracytoplasmic iron depletion (7). We attempted to induce the iron-binding proteins of E. chaffeensis by treatment with desferroxamine and ethylenediamine dihydroxyphenylacetic acid (EDDA). However, no matter what the concentration of iron chelator and which iron chelator was used, in SDS-polyacrylamide gel electrophoresis no change was observed either in the proteins present or in the amounts of the corresponding proteins in iron-depleted and nondepleted E. chaffeensis. In protein immunoblotting the same density was observed when rabbit antiserum to the recombinant 37-kDa protein reacted with E. chaffeensis grown under iron-depleted and nondepleted conditions (data not shown). The iron-binding feature of the 37-kDa protein of E. chaffeensis remains to be determined.
ACKNOWLEDGMENTS
We thank Julie Wen and Violet Han for assistance in electron microscopy and Josie Ramirez-Kim for assistance in the preparation of the manuscript.
This study was supported by a grant from the National Institute of Allergy and Infectious Diseases (AI31431).
REFERENCES
- 1.Adhikari P, Berish S A, Nowalk A J, Veraldi K L, Morse S A, Mietzner T A. The fbpABC locus of Neisseria gonorrhoeae functions in the periplasm-to-cytosol transport of iron. J Bacteriol. 1996;178:2145–2149. doi: 10.1128/jb.178.7.2145-2149.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adhikari P, Kirby S D, Nowalk A J, Veraldi K L, Schryvers A B, Mietzner T A. Biochemical characterization of a Haemophilus influenzae periplasmic iron transport operon. J Biol Chem. 1995;270:25142–25149. doi: 10.1074/jbc.270.42.25142. [DOI] [PubMed] [Google Scholar]
- 3.Anderson B E, Dawson J E, Jones D C, Wilson K H. Ehrlichia chaffeensis, a new species associated with human ehrlichiosis. J Clin Microbiol. 1991;29:2838–2842. doi: 10.1128/jcm.29.12.2838-2842.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson B E, Sims K G, Olson J G, Childs J E, Piesman J F, Happ C M, Maupin G O, Johnson B J B. Amblyomma americanum: a potential vector of human ehrlichiosis. Am J Trop Med Hyg. 1993;49:239–244. doi: 10.4269/ajtmh.1993.49.239. [DOI] [PubMed] [Google Scholar]
- 5.Anderson B E, Greene C E, Jones D C, Dawson J E. Ehrlichia ewingii sp. nov., the etiologic agent of canine granulocytic ehrlichiosis. Int J Syst Bacteriol. 1992;42:299–302. doi: 10.1099/00207713-42-2-299. [DOI] [PubMed] [Google Scholar]
- 6.Angerer A, Gaisser S, Braun V. Nucleotide sequences of the sfuA, sfuB, and sfuC genes of Serratia marcescens suggest a periplasmic-binding-protein-dependent iron transport mechanism. J Bacteriol. 1990;172:572–578. doi: 10.1128/jb.172.2.572-578.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barnewall R E, Rikihisa Y. Abrogation of gamma interferon-induced inhibition of Ehrlichia chaffeensis infection in human monocytes with iron transferrin. Infect Immun. 1994;62:4804–4810. doi: 10.1128/iai.62.11.4804-4810.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brouqui P, Dumler J S, Raoult D, Walker D H. Antigenic characterization of ehrlichiae: protein immunoblotting of Ehrlichia canis, Ehrlichia sennetsu, and Ehrlichia risticii. J Clin Microbiol. 1992;30:1062–1066. doi: 10.1128/jcm.30.5.1062-1066.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brouqui P. Serologic diagnosis of human monocytic ehrlichiosis by immunoblot analysis. Clin Diagn Lab Immunol. 1994;1:645–649. doi: 10.1128/cdli.1.6.645-649.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen S M, Cullman L C, Walker D H. Western immunoblotting analysis of the antibody response of patients with human monocytotropic ehrlichiosis to different strains of Ehrlichia chaffeensis and Ehrlichia canis. Clin Diagn Lab Immunol. 1997;4:731–735. doi: 10.1128/cdli.4.6.731-735.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen S M, Dumler J S, Feng H M, Walker D H. Identification of the antigenic constituents of Ehrlichia chaffeensis. Am J Trop Med Hyg. 1994;50:52–58. [PubMed] [Google Scholar]
- 12.Chen S M, Popov V L, Feng H M, Walker D H. Analysis and ultrastructural localization of Ehrlichia chaffeensis proteins with monoclonal antibodies. Am J Trop Med Hyg. 1996;54:405–412. doi: 10.4269/ajtmh.1996.54.405. [DOI] [PubMed] [Google Scholar]
- 13.Chen S M, Yu X J, Popov V L, Westerman E L, Hamilton F G, Walker D H. Genetic and antigenic diversity of Ehrlichia chaffeensis: comparative analysis of a novel human strain from Oklahoma and previously isolated strains. J Infect Dis. 1997;175:856–863. doi: 10.1086/513982. [DOI] [PubMed] [Google Scholar]
- 14.Chin N, Frey J, Chang C F, Chang Y F. Identification of a locus involved in the utilization of iron by Actinobacillus pleuropneumoniae. FEMS Microbiol Lett. 1996;143:1–6. doi: 10.1111/j.1574-6968.1996.tb08452.x. [DOI] [PubMed] [Google Scholar]
- 15.Dawson J E, Anderson B E, Fishbein D B, Sanchez J L, Goldsmith C S, Wilson K H, Duntley C W. Isolation and characterization of an Ehrlichia sp. from a patient diagnosed with human ehrlichiosis. J Clin Microbiol. 1991;29:2741–2745. doi: 10.1128/jcm.29.12.2741-2745.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dawson J E, Rikihisa Y, Ewing S A, Fishbein D B. Serologic diagnosis of human ehrlichiosis using two Ehrlichia canis isolates. J Infect Dis. 1991;163:564–567. doi: 10.1093/infdis/163.3.564. [DOI] [PubMed] [Google Scholar]
- 17.Dumler J S, Chen S M, Asanovich K, Trigiani E, Popov V L, Walker D H. Isolation and characterization of a new strain of Ehrlichia chaffeensis from a patient with nearly fatal monocytic ehrlichiosis. J Clin Microbiol. 1995;33:1704–1711. doi: 10.1128/jcm.33.7.1704-1711.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fichtenbaum C J, Peterson L R, Weil G J. Ehrlichiosis presenting as a life-threatening illness with features of the toxic shock syndrome. Am J Med. 1993;95:351–357. doi: 10.1016/0002-9343(93)90302-6. [DOI] [PubMed] [Google Scholar]
- 19.Fishbein D B, Sawyer L A, Holland C J, Hayes E B, Okoroanyanwu W, Williams D, Sikes R K, Ristic M, McDade J E. Unexplained febrile illnesses after exposure to ticks: infection with an Ehrlichia? JAMA. 1987;257:3100–3104. [PubMed] [Google Scholar]
- 20.Fishbein D B, Dawson J E, Robinson L E. Human ehrlichiosis in the United States, 1985 to 1990. Ann Intern Med. 1994;120:736–743. doi: 10.7326/0003-4819-120-9-199405010-00003. [DOI] [PubMed] [Google Scholar]
- 21.Gravekamp C, Horensky D S, Michel J L, Madoff L C. Variation in repeat number within the alpha C protein of group B streptococci alters antigenicity and protective epitopes. Infect Immun. 1996;64:3576–3583. doi: 10.1128/iai.64.9.3576-3583.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klein P, Kanehisa M, DeLisi C. The detection and classification of membrane-spanning proteins. Biochim Biophys Acta. 1985;815:468–476. doi: 10.1016/0005-2736(85)90375-x. [DOI] [PubMed] [Google Scholar]
- 23.Maeda K, Markowitz N, Hawley R C, Ristic M, Cox D, McDade J E. Human infection with Ehrlichia canis, a leukocytic rickettsia. N Engl J Med. 1987;316:853–856. doi: 10.1056/NEJM198704023161406. [DOI] [PubMed] [Google Scholar]
- 24.McGeoch D J. On the predictive recognition of signal peptide sequences. Virus Res. 1985;3:271–286. doi: 10.1016/0168-1702(85)90051-6. [DOI] [PubMed] [Google Scholar]
- 24a.Nakai, Kenta. 18 February 1999, revision date. [Online.] PSORT program. Institute for Molecular and Cellular Biology, Osaka University, Osaka, Japan. http://psort.nibb.ac.jp.
- 25.Ohashi N, Zhi N, Zhang Y L, Rikihisa Y. Immunodominant major outer membrane proteins of Ehrlichia chaffeensis are encoded by a polymorphic multigene family. Infect Immun. 1998;66:132–139. doi: 10.1128/iai.66.1.132-139.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paddock C D, Suchard D P, Grumbach K L, Hadley W K, Kerschmann R L, Abbey N W, Dawson J E, Anderson B E, Sims K G, Dumler J S, Herndier B G. Fatal seronegative ehrlichiosis in a patient with HIV infection. N Engl J Med. 1993;329:1164–1167. doi: 10.1056/NEJM199310143291605. [DOI] [PubMed] [Google Scholar]
- 27.Paddock C D, Sumner J W, Shore G M, Bartley D C, Elie R C, McQuade J G, Martin C R, Goldsmith C S, Childs J E. Isolation and characterization of Ehrlichia chaffeensis strains from patients with fatal ehrlichiosis. J Clin Microbiol. 1997;35:2496–2502. doi: 10.1128/jcm.35.10.2496-2502.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27a.Popov, V. L., et al. Unpublished data.
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Sumner J W, Sims K G, Jones D C, Anderson B E. Ehrlichia chaffeensis expresses an immunoreactive protein homologous to the Escherichia coli GroEL protein. Infect Immun. 1993;61:3536–3539. doi: 10.1128/iai.61.8.3536-3539.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tal A, Shannahan D. Ehrlichiosis presenting as a life-threatening illness. Am J Med. 1995;98:318–319. doi: 10.1016/S0002-9343(99)80388-X. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 31.van Vliet A H M, Jongejan F, van Kleef M, van der Zeijst B A M. Molecular cloning, sequencing analysis, and expression of the gene encoding the immunodominant 32-kilodalton protein of Cowdria ruminantium. Infect Immun. 1994;62:1451–1456. doi: 10.1128/iai.62.4.1451-1456.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weiss E, Coolbaugh J C, Williams J C. Separation of viable Rickettsia typhi from yolk sac and L cell host components by Renografin density gradient centrifugation. Appl Microbiol. 1975;30:456–463. doi: 10.1128/am.30.3.456-463.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wen B H, Rikihisa Y, Mott J, Fuerst P A, Kawahara M, Suto C. Ehrlichia muris sp. nov., identification on the basis of 16S rRNA base sequences and serological, morphological, and biological characteristics. Int J Syst Bacteriol. 1995;45:250–254. doi: 10.1099/00207713-45-2-250. [DOI] [PubMed] [Google Scholar]
- 35.Yu X J, Brouqui P, Dumler J S, Raoult D. Detection of Ehrlichia chaffeensis in human tissue by using a species-specific monoclonal antibody. J Clin Microbiol. 1993;31:3284–3288. doi: 10.1128/jcm.31.12.3284-3288.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu X J, Crocquet-Valdes P A, Walker D H. Cloning and sequencing of the gene for a 120-kDa immunodominant protein of Ehrlichia chaffeensis. Gene. 1996;184:149–154. doi: 10.1016/s0378-1119(96)00586-0. [DOI] [PubMed] [Google Scholar]
- 37.Yu X J, Crocquet-Valdes P A, Cullman L C, Walker D H. The recombinant 120-kDa protein of Ehrlichia chaffeensis, a potential diagnostic tool. J Clin Microbiol. 1996;34:2853–2855. doi: 10.1128/jcm.34.11.2853-2855.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu X J, McBride J W, Walker D H. Genetic diversity of the 28-kilodalton outer membrane protein gene in human isolates of Ehrlichia chaffeensis. J Clin Microbiol. 1999;37:1137–1143. doi: 10.1128/jcm.37.4.1137-1143.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]