Abstract
Rationale: Suboptimal vaccine immunogenicity and antigenic mismatch, compounded by poor uptake, means that influenza remains a major global disease. T cells recognizing peptides derived from conserved viral proteins could enhance vaccine-induced cross-strain protection.
Objectives: To investigate the kinetics, phenotypes, and function of influenza virus–specific CD8+ resident memory T (Trm) cells in the lower airway and infer the molecular pathways associated with their response to infection in vivo.
Methods: Healthy volunteers, aged 18–55, were inoculated intranasally with influenza A/California/4/09(H1N1). Blood, upper airway, and (in a subgroup) lower airway samples were obtained throughout infection. Symptoms were assessed by using self-reported diaries, and the nasal viral load was assessed by using quantitative PCR. T-cell responses were analyzed by using a three-color FluoroSpot assay, flow cytometry with MHC I–peptide tetramers, and RNA sequencing, with candidate markers being confirmed by using the immunohistochemistry results for endobronchial biopsy specimens.
Measurements and Main Results: After challenge, 57% of participants became infected. Preexisting influenza-specific CD8+ T cells in blood correlated strongly with a reduced viral load, which peaked at Day 3. Influenza-specific CD8+ T cells in BAL fluid were highly enriched and predominantly expressed the Trm markers CD69 and CD103. Comparison between preinfection CD8+ T cells in BAL fluid and blood by using RNA sequencing revealed 3,928 differentially expressed genes, including all major Trm-cell markers. However, gene set enrichment analysis of BAL-fluid CD8+ T cells showed primarily innate cell–related pathways and, during infection, included upregulation of innate chemokines (Cxcl1, Cxcl10, and Cxcl16) that were also expressed by CD8+ cells in bronchial tissues.
Conclusions: CD8+ Trm cells in the human lung display innate-like gene and protein expression that demonstrates blurred divisions between innate and adaptive immunity.
Clinical study registered with www.clinicaltrials.gov (NCT 02755948).
Keywords: influenza, CD8+ T cell, resident memory, controlled human infection
At a Glance Commentary
Scientific Knowledge on the Subject
Resident memory T (Trm) cells constitute a primarily tissue-restricted T-cell subset that is specialized to protect sites of early pathogen encounter from reinfection. In mice, Trm cells preferentially protect against influenza compared with circulating counterparts of the same specificity and confer cross-strain immunity. Although it is known that Trm cells are abundant in human lung tissue, how they respond to influenza infection in vivo is poorly understood.
What This Study Adds to the Field
In this study, volunteers were experimentally infected with influenza virus, and T-cell responses were analyzed in blood and bronchoscopic lower airway samples. Circulating T-cell frequencies correlated strongly with reduced disease severity, but these cells were phenotypically distinct from their CD8+ Trm counterparts in BAL fluid. This was supported by transcriptomic analysis of blood and BAL-fluid CD8+ T cells, which showed thousands of differentially expressed genes that distinguished these cells. Surprisingly, during infection, gene expression in airway CD8+ T cells was enriched for pathways conventionally associated with innate rather than adaptive immune cells. Thus, we show the pathways underlying the functional role of CD8+ Trm cells in the human lung, which may represent targets to enhance vaccine-induced immunity.
Despite vaccines, influenza remains one of the most important causes of severe infection worldwide (1). Even with annual reengineering to cover circulating strain mutation (2), vaccine effectiveness is often poor because of suboptimal immunogenicity and antigenic mismatch (3, 4). Seasonal influenza therefore still affects 10–46% of the global population each year, resulting in a mortality rate of 290–650,000 per annum (1), which increases during pandemics.
Because T cells recognize peptides derived from conserved viral proteins, they can confer cross-strain protection (5). However, systemic T cell–inducing vaccines have shown limited efficacy. This may be due to the tissue-restricted nature of most respiratory viral disease and the need for antiviral effectors to act rapidly at the site of infection. In this context, circulating T cells may represent surrogates rather than correlates of protection (with surrogates being statistically associated with protection but not mechanistically responsible for it and with correlates being both) (6). Resident memory T (Trm) cells are a tissue-restricted memory subset, poised at sites of potential infection to prevent disease through rapid responses to antigen reexposure (7). In murine influenza models, they offer greater heterosubtypic protection than patrolling central memory CD8+ T cells of the same specificity (8). However, the essential determinants of Trm-cell generation and maintenance in humans remain unclear. Furthermore, unlike Trm cells at other anatomic sites, pulmonary Trm cells do not persist, which is due to attrition and relocation to draining lymph nodes (9). Maintaining them at the site of infection may therefore require specific local signals (10, 11). Recent studies have shown that local amplification can occur through in situ proliferation after secondary infection (12) or through recruitment of specialized memory cells that preferentially reestablish tissue residency (13). Mucosal vaccine delivery could therefore optimize these responses in the human respiratory tract (11, 14).
In humans, almost all published work on lung Trm cells has used postmortem or ex vivo resected tissue (15, 16). To investigate human virus–specific CD8+ T cells in vivo, we previously challenged volunteers with respiratory syncytial virus and revealed the correlation between lower airway Trm cells and reduced disease severity (17). Here, we have experimentally infected individuals with influenza A/California/4/09(H1N1) virus to further investigate the differentiation of pulmonary CD8+ Trm cells. Using MHC class I–peptide tetramers, RNA sequencing, and confocal microscopy, we identify influenza-specific CD8+ T-cell responses in the lower airways and demonstrate the molecular pathways associated with human Trm cells during in vivo infection. Some of the results of these studies have been previously reported in the form of abstracts (18, 19).
Methods
Study Design
Healthy adults (18–55 yr) with hemagglutination inhibition (HAI) titer ⩽1:10 against influenza A/California/4/09(H1N1) were enrolled. Clinical sampling and sample processing were undertaken as previously described (17). The study was approved by the UK Research Ethics Service (reference number 11/LO/1826).
Viral Load
Quantitative PCR was performed on nasal lavage samples as previously described (17) by using pan–influenza A virus M gene primers and probes (forward: GACCRATCCTGTCACCTCTGAC, reverse: AGGGCATTYTGGACAAAKCGTCTA, probe: TGCAGTCCTCGCTCACTGGGCACG) (17).
FluoroSpot Assay
Human IFN-γ, IL-2 and FluoroSpot kits (Mabtech AB) were used according to the manufacturer’s instructions to analyze cryopreserved peripheral blood mononuclear cells (PBMCs) with pooled nonamers representing conserved immunodominant MHC class I–peptide epitopes (BEI Resources). Spots were read by using AID EliSpot version 3.1.1 HR software (Autoimmune Diagnostika GmbH).
Immunohistochemistry and Confocal Microscopy
Endobronchial tissue biopsy specimens were processed as previously described (17). Four-micron-thick microtome sections were costained by using an anti-CD8, anti–influenza A NP (nucleoprotein), anti-CD14, anti-viperin, anti–MIP3-α, anti-CXCR1, anti-CXCL16, anti-IP10, anti-FABP4 (anti–fatty acid binding protein 4), anti-BAFF (anti–B-cell activation factor), anti-CD69, and anti-CD103 antibodies (all Abcam; see Table E2 in the online supplement). An HCF1-Zeiss LSM-780 inverted microscope was used for visualization, and Fiji software was used for analysis.
Flow Cytometry
Whole-blood and BAL-fluid cells were stained by using HLA class I (A*01:01-CTELKLSDY [#TS-M045–2] and A*02:01-GILGFVFTL [#TB-0012-M2]) tetramers (MBL International) as previously described (17). Antibodies used for phenotypic analysis are listed in Table E2, and analysis was conducted by using FlowJo software (FlowJo LLC). Cell sorting was achieved by using a BD FACSAria II sorter (BD Biosciences).
RNA Sequencing
RNA sequencing was performed on an Illumina HiSeq 1500 system, reads were aligned by using Spliced Transcripts Alignment to a Reference software, and quality control was ensured by using the FastQC Screen tool (20, 21). Further analysis was performed by using the edgeR, glmFit, and tmod R packages (R Foundation for Statistical Computing) (22–24). Data were deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (accession number GSE175551).
Statistical Analysis
Statistical analysis was performed by using GraphPad Prism and R software. Two-group comparisons were tested by using a Mann-Whitney test for unpaired groups, and a Wilcoxon matched-pair signed rank test was used for paired groups. Multiple group comparisons were made by using a Friedman test with post hoc Dunn tests.
Results
Influenza Viral Load Correlates with Symptom Severity in Experimentally Infected Adults
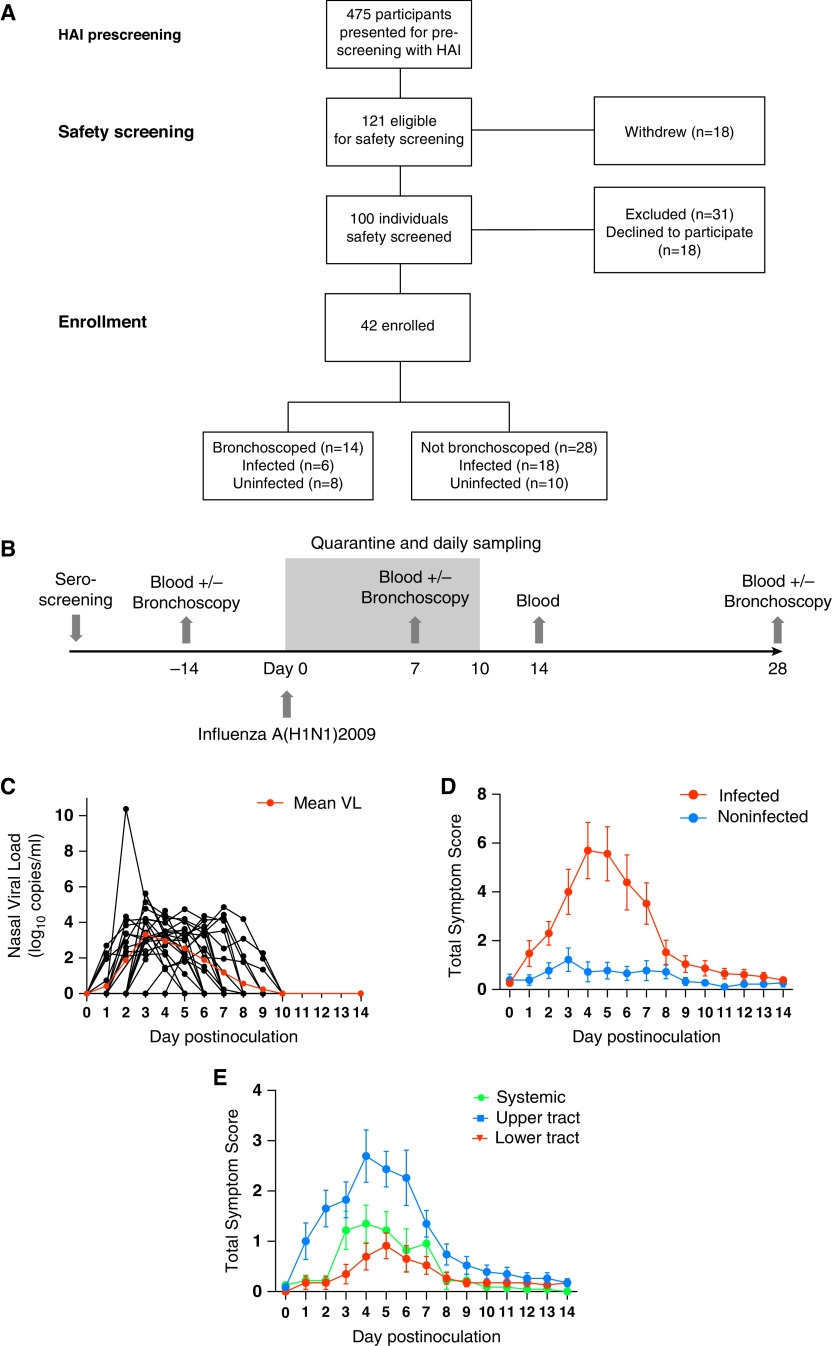
Forty-two participants with HAI titers ⩽1:10 were enrolled (Figures 1A and 1B). Fourteen of the 42 individuals additionally underwent up to three bronchoscopies (before inoculation and at Day 7 and Day 28 after inoculation). After inoculation, 24 participants (57.1%) developed symptomatic PCR-confirmed infection. There were no significant demographic differences between infected and uninfected individuals (Table 1). The median viral load peaked 3 days after infection at 3.33 (range, 0–5.63) log10 copies/ml. No virus was detected beyond Day 9 after inoculation (Figure 1C). Of the six infected individuals who underwent bronchoscopies, two had a detectable viral load at the Day 7 bronchoscopy. Symptoms (primarily upper respiratory) began 24 hours after infection and peaked around Day 5 (Figures 1D and 1E). Eleven infected individuals (45.8%) had a temperature of >37.9°C at least once. Thus, experimental influenza A/California/4/09(H1N1) infection caused a range of clinical outcomes, despite uniformly low preinoculation HAI antibody titers.
Figure 1.
Experimental human infection with influenza A/California/4/09(H1N1) virus induces a diversity of clinical outcomes. (A) Forty-two healthy adult volunteers were intranasally inoculated with influenza A/California/4/09(H1N1) and sampled periodically after inoculation. A Consolidated Standards of Reporting Trials diagram is shown. (B) A schematic showing the schedule of sampling. (C) The viral load (VL) of 42 individuals as determined by using M gene RT-PCR findings from nasal lavage. Individual data points and the mean (red) VL are shown. (D) Self-reported total daily symptom scores for infected (red, n = 23) and uninfected (blue, n = 18) individuals. Data are shown as the mean ± SEM. (E) Upper respiratory tract (blue), lower respiratory tract (red), and systemic (green) symptom scores in infected participants. Data are shown as the mean ± SEM. HAI = hemagglutination inhibition.
Table 1.
Study Participant Demographics
| Outcome | Bronchoscopy |
No Bronchoscopy |
Overall |
|||
|---|---|---|---|---|---|---|
| PCR-Positive | PCR-Negative | PCR-Positive | PCR-Negative | PCR-Positive | PCR-Negative | |
| Number | 5 | 9 | 19 | 9 | 24 | 18 |
| Age, yr, median (range) | 51 (43–53) | 53 (30–54) | 35 (18–55) | 40 (32–49) | 42.5(18–55) | 43 (30–54) |
| Sex, M:F | 2:3 | 8:1 | 14:5 | 6:3 | 16:8 | 14:4 |
| Ethnicity | ||||||
| White | 3 | 6 | 9 | 8 | 6 | 14 |
| Black British/Black other | 1 | 1 | 3 | 0 | 3 | 1 |
| Asian British/Indian/Asian other | 1 | 0 | 4 | 1 | 2 | 1 |
| Mixed | 0 | 0 | 2 | 0 | 2 | 0 |
| Other | 0 | 2 | 1 | 0 | 0 | 2 |
| Median height, cm | 1.63 | 1.71 | 1.76 | 1.73 | 1.77 | 1.72 |
| Median weight, kg | 66 | 82.6 | 73.5 | 75.5 | 73.2 | 76.75 |
| Median BMI, kg/m2 | 27.83 | 26.11 | 24.3 | 24.81 | 25.9 | 25.87 |
Definition of abbreviation: BMI = body mass index.
Demographic data summary of all subjects inoculated with influenza A/California/4/09(H1N1) virus. The data shown compare individuals who underwent bronchoscopies with those who did not undergo bronchoscopies and all included individuals.
Circulating Memory CD8+ T cells Correlate with Reduced Influenza Disease
The contribution of CD4+ and CD8+ T cells as correlates of protection against influenza disease has varied depending on the study design and virus strain (25, 26). To test the association of circulating CD8+ T cells with protection against influenza A/California/4/09(H1N1), PBMCs from 39 participants were assessed by using a FluoroSpot assay with pooled conserved CD8+ T-cell epitopes. Before infection, influenza-specific T cells could be quantified in almost all participants, albeit at modest frequencies (median, 28 spots/million PBMCs; range, 5–74 spots/million PBMCs) (Figure 2A). There was no difference in the influenza-specific T-cell frequencies between those who resisted infection and those who did not (P = 0.9373). However, the frequency of preexisting influenza-specific CD8+ T cells correlated strongly with a reduced viral load at the subsequent infection (Spearman rank correlation coefficient = −0.7918; P = 0.0007) (Figure 2B), and a similar trend was seen with symptom scores (Figure E1A).
Figure 2.
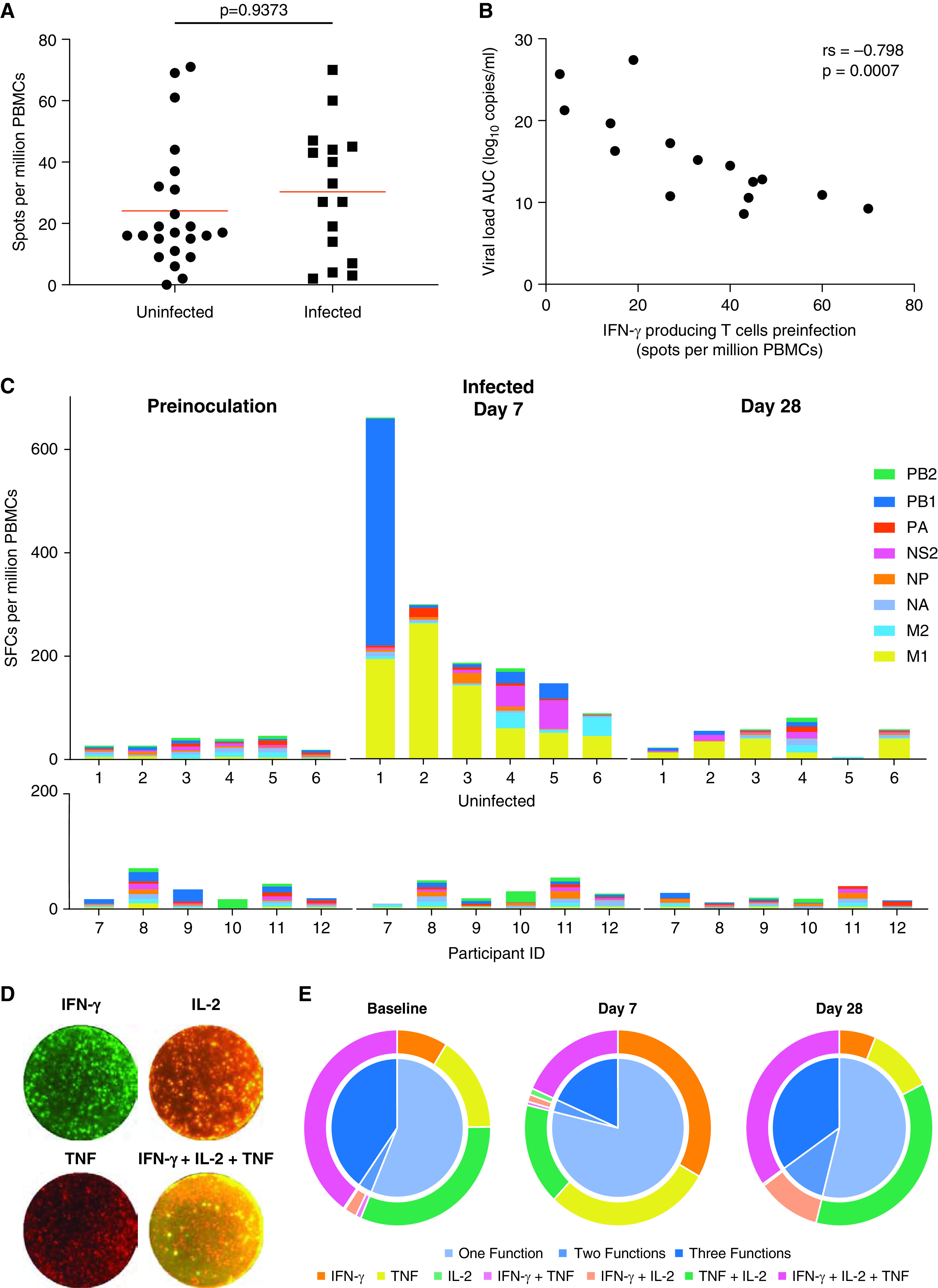
Circulating memory CD8+ T cells correlate strongly with reduced disease severity in influenza. Influenza-specific CD8+ T cells were quantified in blood by using a FluoroSpot assay after stimulation with conserved MHC class I epitopes. (A) When cell numbers allowed, IFN-γ–secreting influenza-specific T cells were measured in peripheral blood mononuclear cells (PBMCs) obtained before inoculation from participants who subsequently became infected (n = 16) or remained uninfected (n = 23). (B) Spearman correlation between preinoculation influenza-specific T cells and the viral load in individuals who subsequently had PCR-positive results (n = 16). (C) Frequency of IFN-γ–secreting T cells stimulated with influenza peptides derived from internal influenza proteins before infection and at the Day 7 and Day 28 time points. T-cell responses to conserved peptide derived from M1 and M2 (matrix), NA (neuraminidase), NP (nucleoprotein), NS2 (nonstructural protein 2), and the RNA polymerase complex (PA, PB1, and PB2) are shown as the median and interquartile range. (D) FluoroSpot images from one representative donor at Day 7 after infection are shown. (E) The proportion of T cells secreting IFN-γ, TNFα, IL-2, or combinations of multiple cytokines in peptide-stimulated PBMCs is shown. Pie chart sections represent the proportion of cells with one, two, or three functions. Individual cytokines and their combinations are denoted by arcs. AUC = area under the curve; ID = identifier; rs = Spearman’s rank correlation coefficient; SFC = spot-forming cell; TNF = tumor necrosis factor.
In 12 participants, cell numbers allowed analysis of complete time courses, and the breadth of CD8+ T-cell responses was analyzed. CD8+ T cells expanded significantly in all infected participants (P = 0.0475; mean fold-change, 7.68) (Figures 2C and E1B) but did not expand significantly in those who remained uninfected. By Day 28, these had contracted to leave a minimally enlarged memory pool (P = 0.4703). Both the size and the breadth of the responses were heterogeneous, but M1-specific T cells were the most consistently expanded. In some individuals PB1-, NS2 (nonstructural protein 2)–, and M2-specific T-cell populations also contributed substantially to the total response.
Polyfunctional T cells that express multiple cytokines or effector molecules are associated with greater antiviral efficacy (27). By using a three-color FluoroSpot assay, the capacity of individual cells to coexpress IFN-γ, TNF, and IL-2 was enumerated in infected individuals (Figures 2D, 2E and E2). Before infection, most influenza-specific CD8+ T cells expressed a single cytokine, but 40% were polyfunctional and expressed all three. At Day 7, single cytokine–producing T cells preferentially increased. However, by the convalescent time point, these proportions had returned to baseline. Together, these data suggest that during acute infection, the expansion of M1-, PB1-, M2-, and NS2-specific T cells is mainly made up of single cytokine–secreting cells, which disappear once infection resolves.
Influenza-Specific CD8+ T Cells Are Enriched in the Respiratory Tract throughout Infection
Circulating influenza-specific memory T cells may represent surrogates rather than correlates of protection, with direct effector mechanisms being more likely to act in the respiratory tract. Seven days after inoculation, the bronchial mucosa of the five infected individuals showed macroscopic inflammation with erythema and microhemorrhages that resolved by Day 28 (Figure 3A). Concurrently, CD8+ T cells analyzed by using flow cytometry of the blood of nine participants and the BAL fluid of four participants showed transient upregulation of CD38 and Ki-67 (markers of activation and proliferation, respectively) in both the blood (P = 0.0002) and the BAL fluid (P = 0.0286) at Day 7 (Figures 3B and 3C). At this time point, the frequency of activated CD8+ T cells was also significantly higher in BAL fluid than in blood (P = 0.0101).
Figure 3.
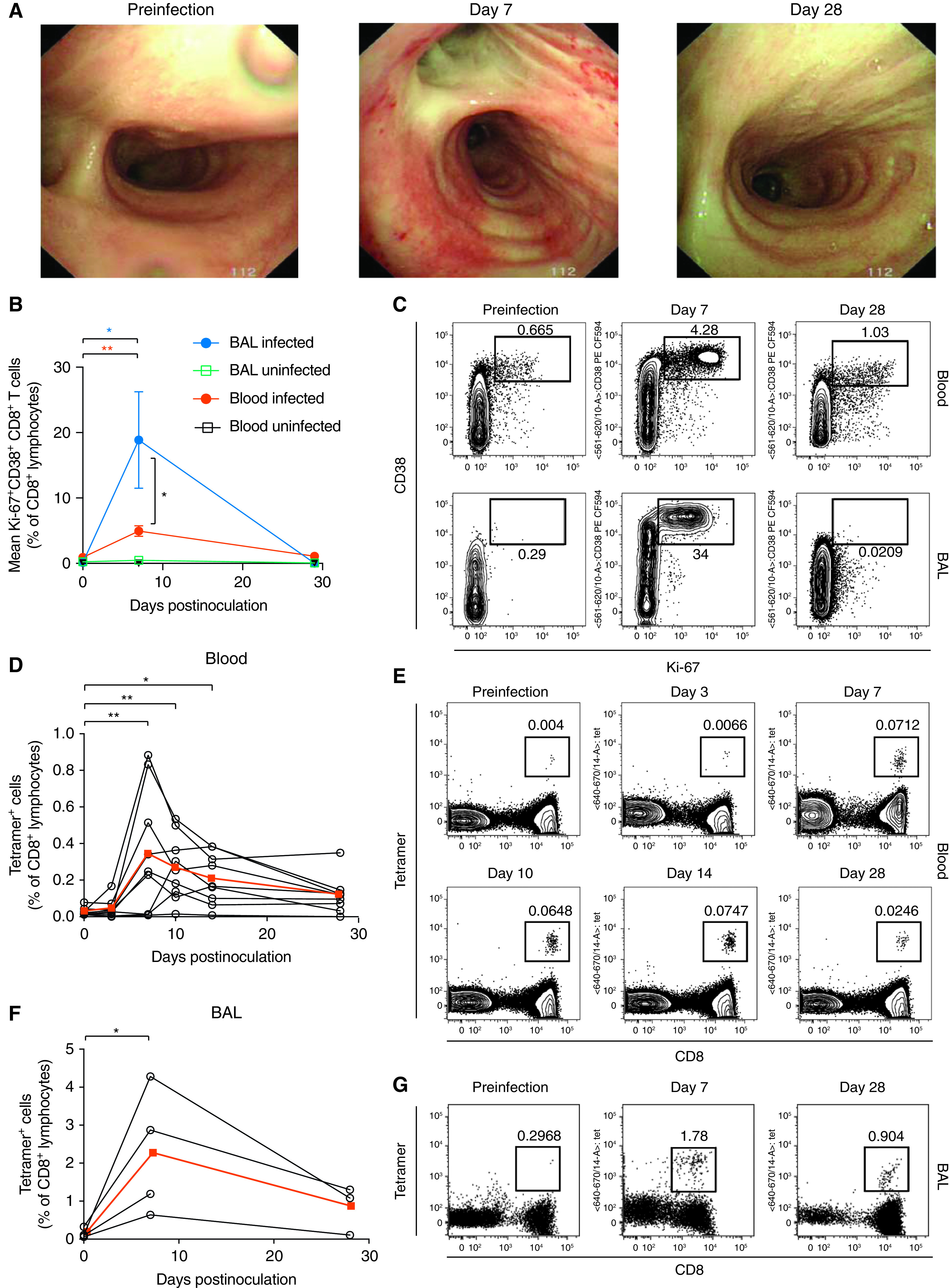
Influenza-specific CD8+ T cells are preferentially expanded in the human airway during experimental infection. Healthy adult volunteers were experimentally infected with influenza A virus and serially sampled for blood and BAL fluid. (A) The macroscopic appearances of the lower airway were assessed by using bronchoscopy before infection and at Day 7 and Day 28. One representative individual is shown. (B) CD8+ T cells in the blood and BAL fluid were costained with anti–Ki-67 and anti-CD38 for analysis by flow cytometry. P values from Wilcoxon matched-pair tests for pair-wise comparisons and from Mann-Whitney tests for unpaired comparisons are shown. (C) Flow cytometry plots are shown for one representative individual. (D) CD8+ T cells labeled with the A*01:01-CTELKLSDY tetramer or the A*02:01-GILGFVFTL tetramer were quantified in the blood. Individual data and the medians (red) are shown. P values from Friedman tests with Dunn’s multiple comparison tests are shown. (E) CD8+ T cells in blood were labeled with the A*02:01-GILGFVFTL tetramer. Flow cytometry plots are shown for one representative individual. (F) CD8+ T cells labeled with the A*01:01-CTELKLSDY tetramer or the A*02:01-GILGFVFTL tetramer were quantified in BAL fluid. Individual data and the medians (red) are shown. (G) CD8+ T cells in BAL were labeled with the A*02:01-GILGFVFTL tetramer. Flow cytometry plots are shown for one representative individual. P values from Wilcoxon matched-pair tests are shown.*P < 0.05 and **P < 0.01.
MHC–peptide tetramers were then used to track epitope-specific cells in participants with suitable HLA types. In the blood, tetramer-positive CD8+ T cells underwent classical proliferation followed by contraction (Figures 3D and 3E). This was paralleled by BAL-fluid tetramer-positive populations that showed a similar pattern of proliferation between Day 0 and Day 7 (P = 0.0286), although this proliferation pattern occurred more frequently by fivefold (P = 0.0112) (Figures 3F and 3G).
Trafficking of Activated Influenza-Specific CD8+ T Cells Alters Subset Composition in BAL Fluid
Epitope-specific CD8+ T cells also upregulated Ki-67 and CD38 at Day 7 after infection (P = 0.0039) (Figures 4A and E3A), with a similar pattern being shown in BAL fluid. CD8+ Trm cells are recognized by expression of CD69 and CD103, but in the blood, few influenza-specific CD8+ T cells expressed these markers, despite their recognized roles as activation markers (Figure 4B). In contrast, although tetramer-positive CD8+ T cells were only present at low frequencies in BAL fluid before infection, almost all coexpressed CD69 and CD103. After infection, the expanded tetramer-positive population became composed of both CD69+CD103+ and CD69+ subpopulations. A CD69+ cell population was not, however, seen in the blood, implying that it was upregulated only within the lung or airway. During convalescence, a trend toward increased CD103 coexpression then suggested progressive differentiation to the full Trm phenotype.
Figure 4.
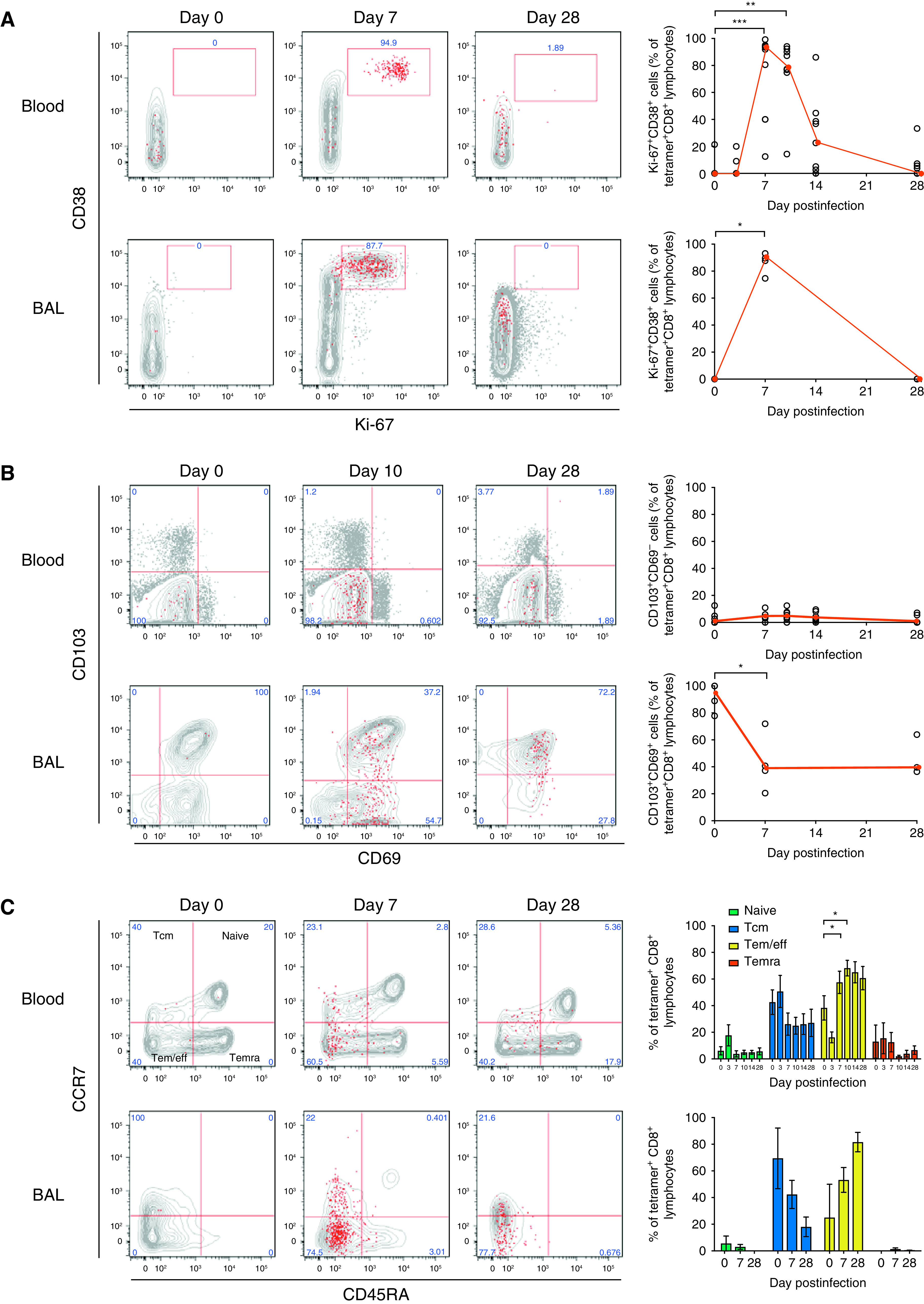
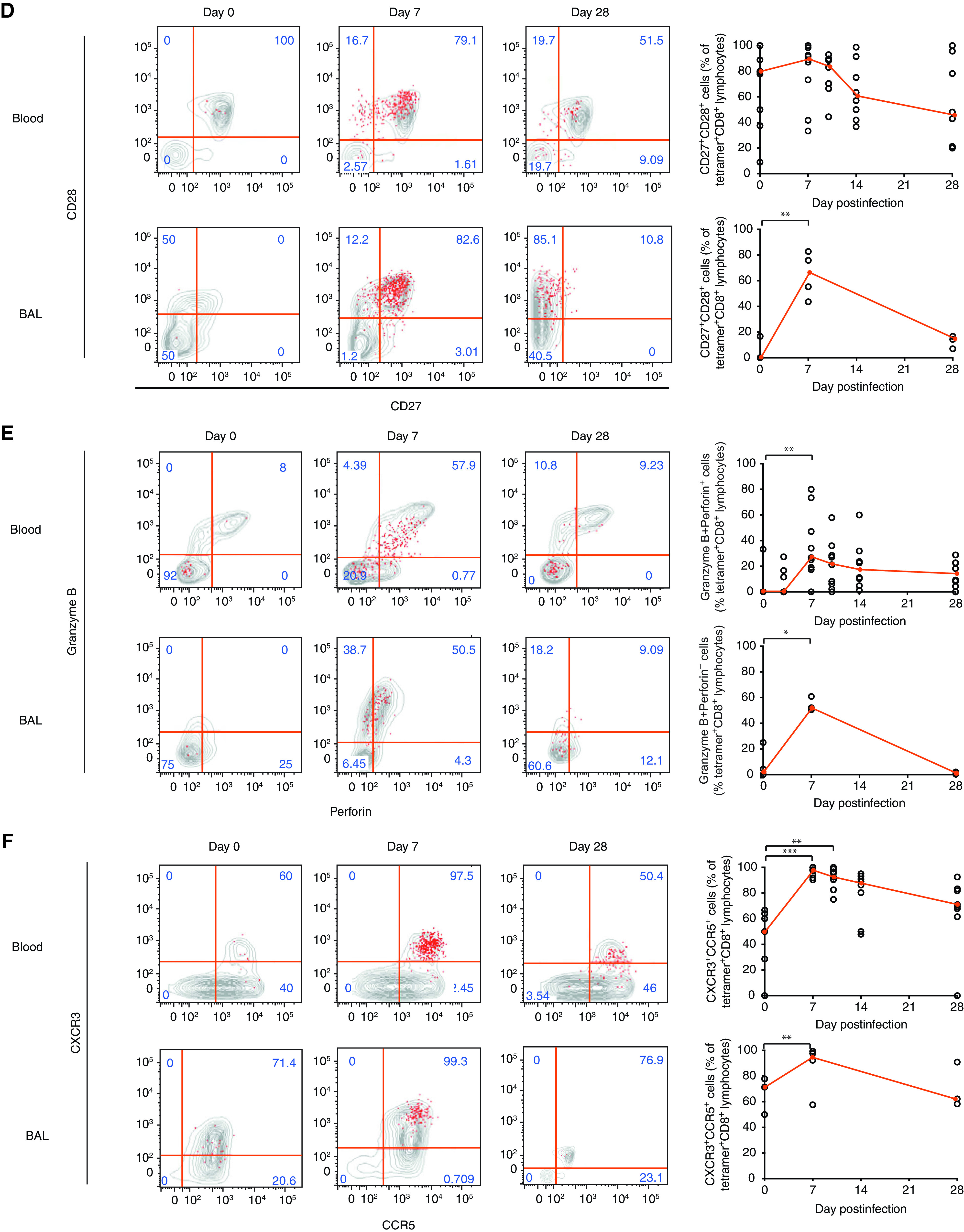
Influenza infection induces divergent phenotypic changes in the blood and BAL fluid. Whole-blood/peripheral blood mononuclear cells (n = 8) and BAL fluid (n = 4) from influenza-infected individuals were stained with a tetramer, anti-CD3, and CD4 and were costained with (A) Ki-67 and CD38; (B) CD69 and CD103; (C) CD45RA and CCR7 (the median and interquartile range are shown); (D) CD27 and CD28; (E) granzyme B and perforin; and (F) CCR5 and CXCR3 for analysis by flow cytometry. Medians (red) and individual data are shown. P values from Wilcoxon matched-pair tests are shown for pair-wise comparisons of BAL-fluid cells, and P values from Friedman tests with Dunn’s multiple comparison tests are shown for multiple comparisons of blood cells. *P < 0.05 and **P < 0.01. eff = effector; Tcm = central memory T; Tem = eff memory T; Temra = terminally differentiated Tem.
Blood and BAL-fluid CD8+ T cells also differed markedly in terms of subset composition, with almost no naive T cells appearing in BAL fluid at any time point (Figure 4C). With infection, influenza-specific CD8+ T cells in both blood and BAL fluid primarily displayed an effector/effector memory phenotype. This altered balance of T-cell subsets persisted to Day 28, despite proliferation having ceased. In the blood, the change in subset composition was also reflected in the costimulatory molecules CD27 and CD28 (Figure 4D), with high expression of both markers at Day 7 followed by downregulation of CD27 by Day 28 being shown. In contrast, the majority of BAL-fluid CD8+ Trm cells did not express CD27 and CD28 at rest but became predominantly double positive during infection (median, 65.6%; interquartile range [IQR], 81.1–46.5%) before being rapidly downregulated. This was paralleled by the expression of the cytotoxicity markers perforin and granzyme B in BAL-fluid cells, with the majority (median, 52.0%; IQR, 58.7–50.8%) significantly upregulating these markers at Day 7 but demonstrating almost no expression at Day 28 (Figure 4E).
Finally, activation was associated with upregulation of the chemokine receptors CCR5 and CXCR3, promoting chemotaxis to inflamed tissues and the lung respectively. In blood, 50% (IQR, 62.9–7.15) of influenza-specific CD8+ T cells expressed both receptors at rest, which increased to 97.8% (IQR, 99.5–92.4%) during infection (Figure 4F). In BAL fluid, even resting influenza-specific Trm cells were mostly CCR5+CXCR3+ (median, 71.4%; IQR, 77.8–50%), which further increased to 94.5% (IQR, 98.88–66.28%) at Day 7. These data therefore show distinct phenotypic changes that occur between BAL fluid and blood, supporting preclinical data of acutely activated influenza-specific CD8+ T cells that home to the lung via chemokine receptors.
Transcriptomic Signatures of Compartmentalization in Human BAL-Fluid and Blood CD8+ T Cells
The molecular determinants of Trm-cell generation in humans are poorly understood, so CD8+ T cells were sorted from blood and BAL fluid of 12 individuals for analysis by using RNA sequencing (Figures E3B and E3C). Because of limited BAL-fluid cell numbers, bulk CD45+CD14−CD19−CD3+ CD8+ cells were analyzed. All sorts were at least 95% pure, with any remaining non-CD8+ cells being negative for the other leukocyte markers CD45, CD14, and CD19 (Figure E3B).
Differential gene expression comparing blood and BAL-fluid CD8+ T cells before infection revealed 4,699 differentially expressed genes (DEGs) (false discovery rate < 0.05; Table E3). However, because immunophenotyping had shown the markedly different composition of CD8+ T cells from these two compartments (Figure 4), some genes were likely to have represented only differences between naive and memory T cells. To enrich the DEG list for genes specifically associated with Trm cells in the lung, DEGs from two previous studies (28, 29) that distinguished naive from memory T cells were therefore excluded. This resulted in 3,928 DEGs for continued analysis (Figure 5A, Table E4). Among the top 20% most significant DEGs in this list, all major markers associated with Trm cells were present, including CD69, ITGAE (CD103), ITGA1 (CD49a), FABP4, and S1PR1 (downregulated in BAL-fluid CD8+ T cells) (Figures 5B and 5C). In addition in the full gene list, 29 of the 31 genes previously proposed (30) as a core signature of Trm cells across different tissues were concordant (CA10, ITGA1, ITGAE, IL2, IL10, CXCR6, CXCL13, KCNK5, RGS1, DUSP6, PDCD1, and IL23R [upregulated in Trm cells] and STK38, TTC16, SELL, KLF2, KLF3, SBK1, TTYH2, NPDC1, KRT72, S1PR1, SOX13, KRT73, TSPAN18, PTGDS, RAP1GAP2, and CX3CR1 [downregulated in Trm cells]) (Figure 5B, Table E3).
Figure 5.
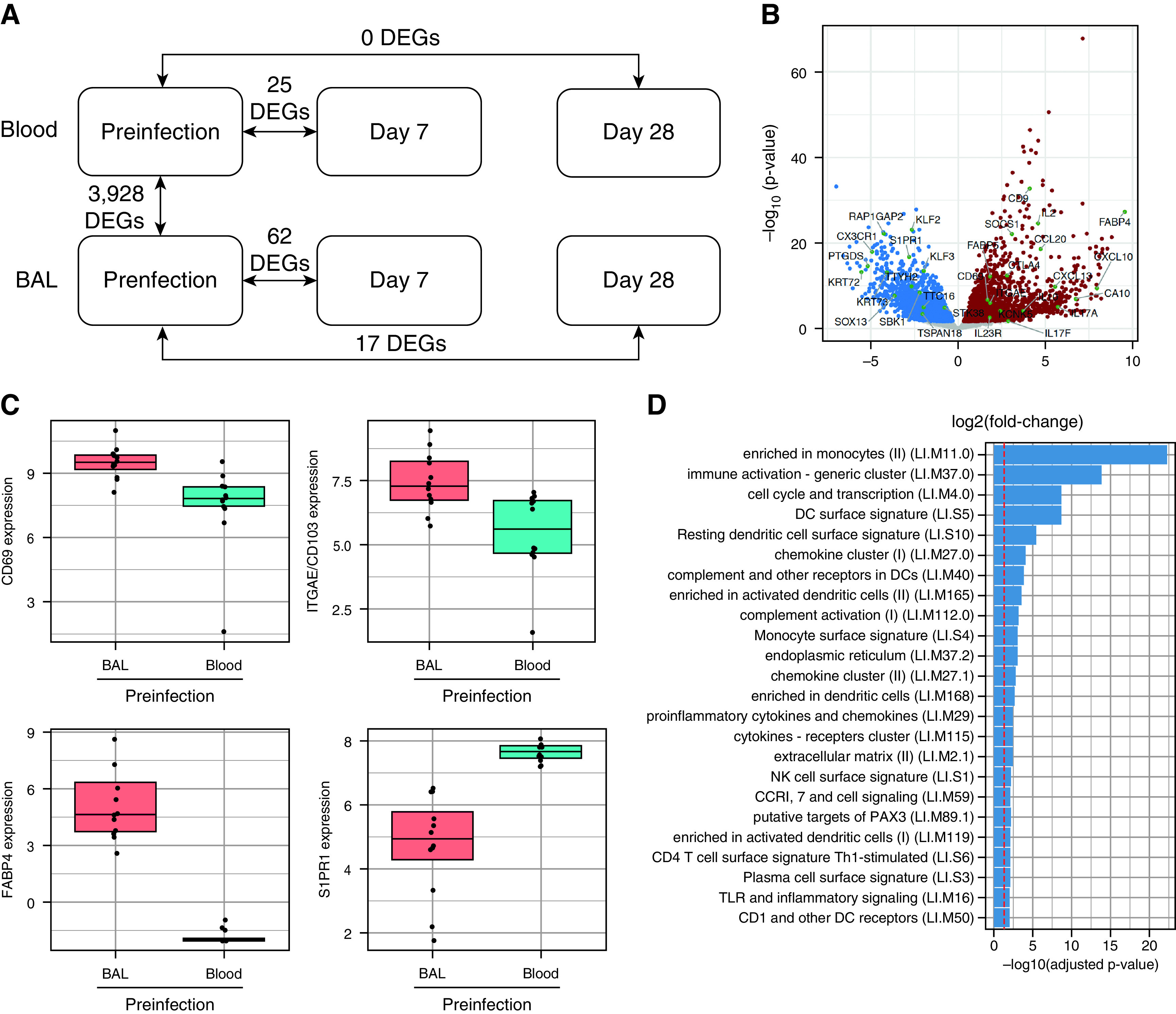
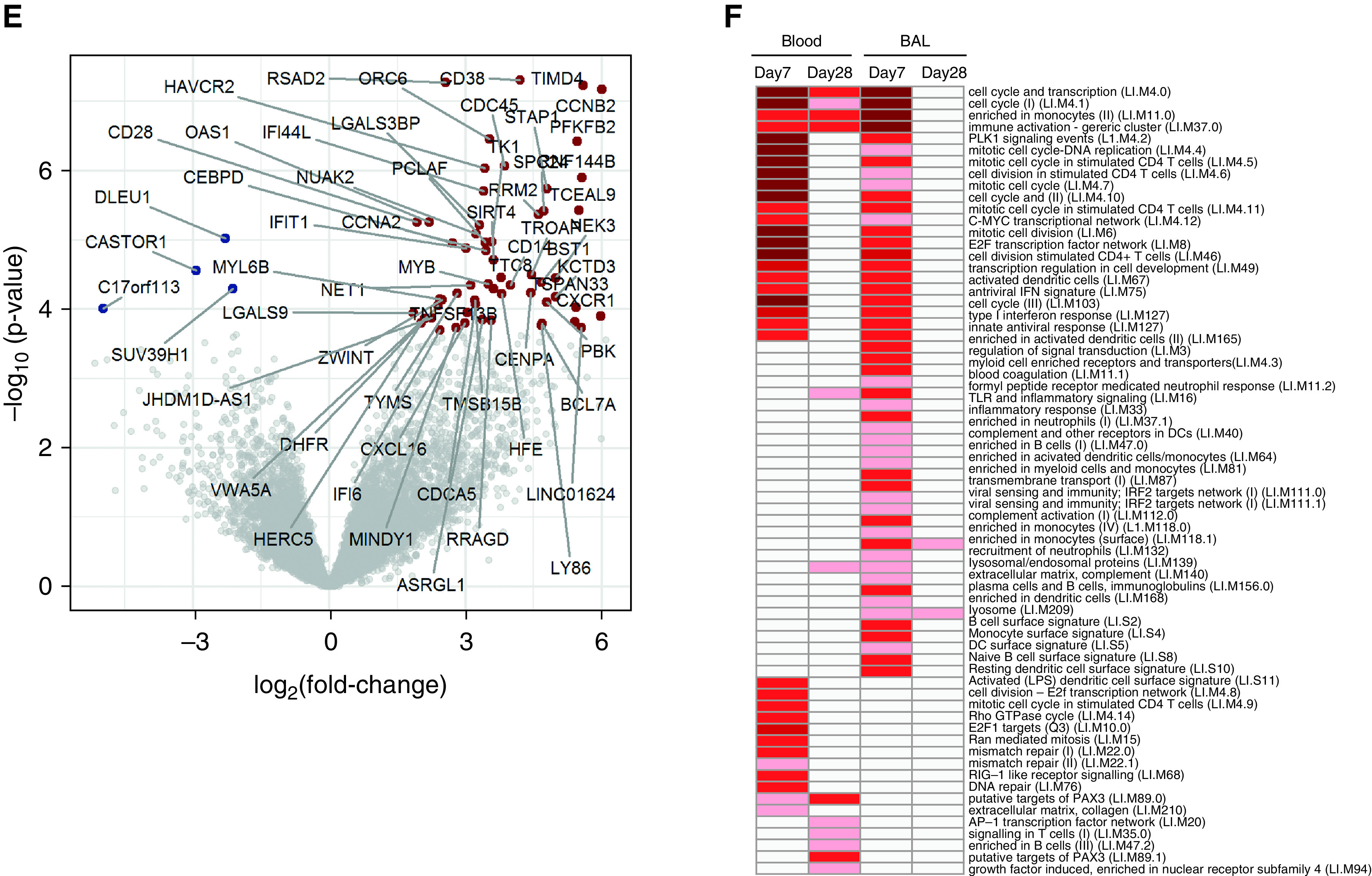
Resident memory CD8+ T cells in the lower airway responding to influenza infection display innate-like signatures. Live CD3+CD4−CD14−CD19−CD8+ T cells from the blood and BAL fluid of 12 individuals inoculated with influenza A virus at Day 7 before infection and Day 28 after infection were sorted by using a fluorescence-activated cell sorter. (A) Diagrammatic representation of differential gene expression (DEG) comparing compartments and time points. (B) Volcano plot showing DEGs in BAL fluid versus blood CD8+ T cells before infection. Red indicates genes upregulated in BAL fluid, and blue indicates genes downregulated in BAL fluid. (C) Preinfection expression of the resident memory T-cell markers CD69, CD103/ITGAE (integrin subunit alpha E), FABP4 (fatty acid binding protein 4), and S1PR1 were compared between BAL fluid and blood CD8+ T cells. (D) Functionally annotated gene sets significantly enriched (log10-adjusted P value) with DEGs comparing BAL fluid and blood CD8+ T cells were identified by using the tmod R package. The red line denotes the cutoff for significance (false discovery rate < 0.05). (E) Volcano plot demonstrating significant DEGs in BAL-fluid CD8+ T cells from five infected individuals at Day 7 after infection compared with before infection. Significantly upregulated DEGs are highlighted in red, significantly downregulated DEGS are highlighted in blue, and significant DEGs are labeled with their gene symbols. (F) Functionally annotated gene sets significantly enriched with DEGs comparing CD8+ T cells at each time point with the corresponding preinfection time point in blood and BAL fluid were identified by using the tmod R package. All statistically significant DEGs without Bonferroni-Hochberg correction are included. The darker the red coloration, the more significant the pathway enrichment. DC = dendritic cell; NK = natural killer; Th1 = T-helper cell type 1; TLR = Toll-like receptor.
Next, to identify functional pathways, gene set enrichment analysis was performed by using the Coincident Extreme Ranks in Numerical Observations algorithm (22). Surprisingly, the most significantly enriched gene sets were not associated with known T-cell functions. Instead, the top gene set was annotated as “enriched in monocytes,” with 10 of 24 of the most significantly enriched gene sets being labeled as monocyte pathways, dendritic-cell pathways, and other innate-cell pathways (Figure 5D, Table E5). Surprisingly, these included the myeloid-cell marker and LPS receptor CD14, despite this having been used to dump non-T cells for fluorescence-activated cell sorter sorting. The remaining pathways comprised generic cellular activities (cell cycle and transcription, endoplasmic reticulum, extracellular matrix) and common immune response pathways (including immune activation, TLR [Toll-like receptor] and inflammatory signaling, and cytokine- and chemokine-related pathways). Only one gene set (CD4 T-cell surface signature T-helper cell type 1–stimulated gene set) was specifically T cell–related.
Differential Gene Expression by CD8+ T Cells in BAL Fluid during Influenza Is Dominated by Innate-like Signatures
Comparing the transcriptomes of CD8+ cells between blood and BAL fluid is confounded by steady-state differences in the subset composition. However, longitudinal analysis of cells from the same anatomic compartment removes the contribution of cells that remain unchanged during infection. By comparing CD8+ T cells in BAL fluid at Day 7 after infection with preinfection samples, 62 DEGs (false discovery rate < 0.05) were identified (Figure 5E). These included the activation marker CD38 and the costimulatory marker CD28, which were also upregulated at Day 7 in our cytometric data (Figure 4). In contrast, only 25 DEGs were identified in the blood at the same time points (Figure E5, Tables E5 and E6).
By using the tmod R package, three major patterns of functional enrichment were revealed. Around half of the significantly enriched pathways upregulated at Day 7 after infection were shared by both blood and BAL-fluid CD8+ cells (Figure 5F). These were primarily related to cell division or pattern recognition receptor and innate IFN signaling. The second pattern involved upregulation only in BAL-fluid CD8+ cells at Day 7 after infection. These partly recapitulated the findings in resting BAL-fluid CD8+ T cells, with upregulated pathways predominantly relating to monocytes and dendritic cells, as well as cell-cycle and effector functions such as the lysosomal/endosomal pathways (Table E7). Finally, a smaller set of pathways was uniquely enriched only in blood CD8+ cells, which was again related to cellular proliferation as well as DNA repair and RIG-I signaling. By Day 28, these changes had largely resolved in both the blood and the BAL fluid (Tables E8 and E9). No mucosal-associated invariant T (MAIT) or natural killer T (NKT) cell modules were found to be enriched.
CD8+ T Cells Infiltrating the Bronchial Mucosa during Influenza Infection Express Innate-like Markers
To identify CD8+ Trm cells infiltrating the bronchial mucosa, endobronchial biopsies were costained with anti-CD8, anti-CD69, and anti-CD103 (Figures 6A–6D). At the acute time point, CD8+ cells were abundant in both the epithelium and the subepithelium, despite no detectable staining for influenza virus antigen (Figures 6A, 6B, and E6A). CD69 was coexpressed in 39% of CD8+ cells with strong colocalization (correlation coefficient = 0.79), whereas CD103 was coexpressed in 78% (correlation coefficient = 0.58) (Figures 6E and 6F). Coexpression of CD69 and CD103 was observed in 36% of CD8+ cells, which was similar to what was seen in BAL-fluid CD8+ Trm cells quantified by using flow cytometry (Figure 4B). In addition, FABP4 was coexpressed on 30% of CD8+ cells (correlation coefficient = 0.64; Figure E6B).
Figure 6.
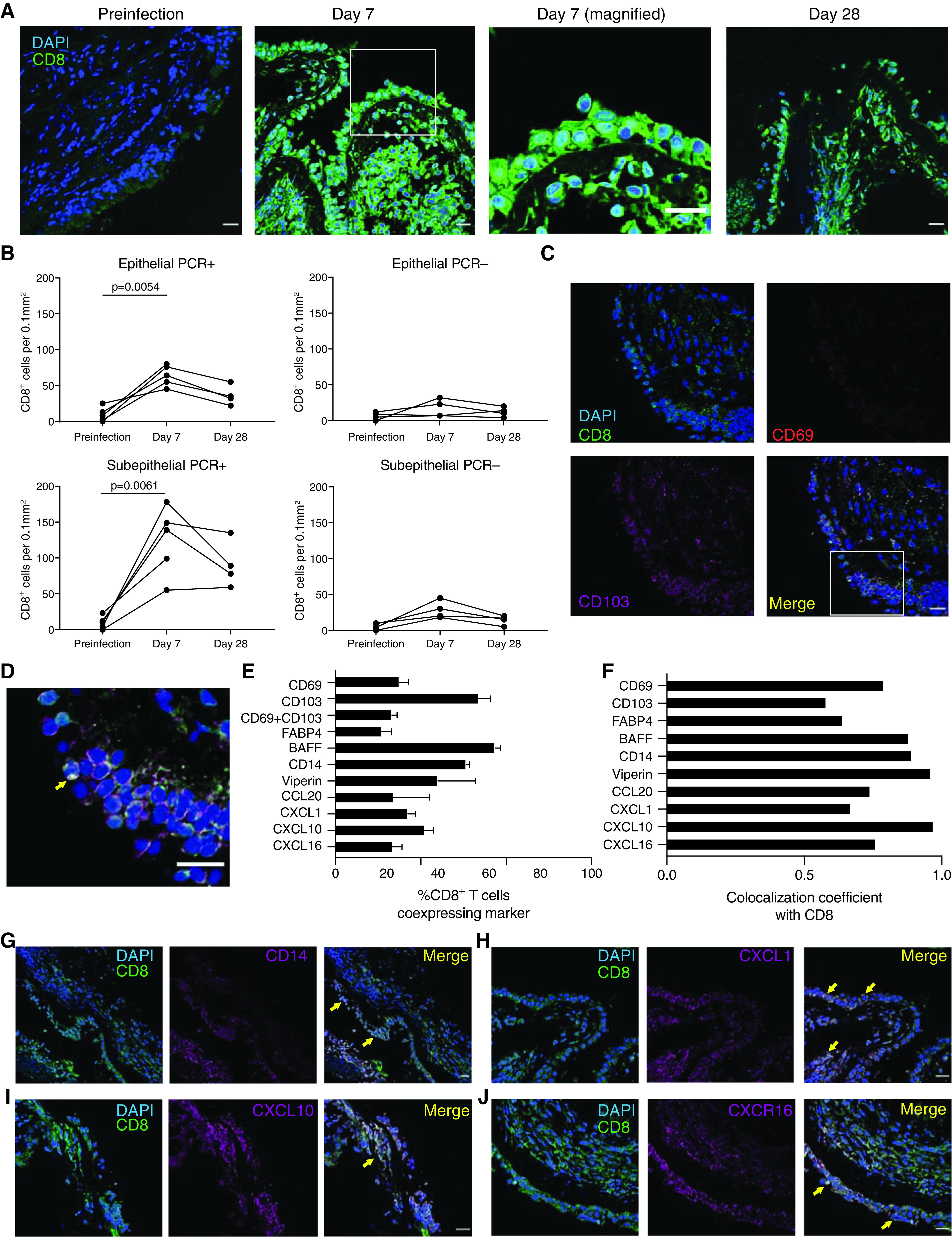
CD8+ cells in the bronchial mucosa during influenza coexpress resident memory T and innate-like markers. Endobronchial biopsy specimens were obtained through bronchoscopy of healthy volunteers (n = 9) inoculated with influenza A virus. (A) Sections obtained from bronchial biopsy specimens of participants with PCR-positive results were stained with anti-CD8 and DAPI. Cells expressing CD8 (green) in the epithelial and subepithelial layers at baseline (left), Day 7 (with a magnified image at Day 7), and Day 28 (right) from one representative individual are shown. (B) Cells expressing CD8 were quantified in the mucosal epithelium and subepithelium in participants with PCR-positive (n = 5) and PCR-negative (n = 4) results before infection and at Day 7 and Day 28 after inoculation. (C) Sections were costained with anti-CD8 (fluorescein isothiocyanate, green), anti-CD103 (cyanine 5 [Cy5], magenta), anti-CD69 (Cy3, red), and DAPI (blue) at Day 7 after infection. Representative images from one individual are shown. (D) Colocalization (yellow) of resident memory T markers (CD69 and CD103) with CD8 observed in the bronchial mucosa is represented in the magnified image. (E) The frequency of CD8+ cells expressing markers of tissue residence (CD69, CD103, and FABP4 [fatty acid binding protein 4]), stimulatory molecules (BAFF [B-cell activation factor] and CD14), IFN-stimulated genes (viperin), and chemokines (CCL20, CXCL1, CXCL10, and CXCL16) was quantified in the mucosal epithelium. (F) Colocalization correlation coefficients of each marker with CD8 in the mucosal epithelium were calculated. Biopsy sections (Day 7) were costained with anti-CD8 (fluorescein isothiocyanate, green) and (G) anti-CD14 (Cy5, magenta), (H) anti-CXCL1 (Cy5, magenta), (I) anti-CXCL10 (Cy5, magenta), and (J) anti-CXCL16 (Cy5, magenta) with colocalization shown in yellow. One representative individual is shown. Data are representative of three independent experiments performed on biopsy specimens obtained from five infected individuals (i.e., five biological replicates). Yellow arrows indicate cells with coexpression of CD8+ cells with other markers. Scale bars, 20 μm.
To confirm the unexpected finding of innate-like gene expression in BAL-fluid CD8+ T cells, bronchial tissue was stained for selected markers representative of the most statistically significant enriched pathways. The topmost significant DEGs contributing to the myeloid-cell and viral sensing pathways were selected to validate genes not typically associated with CD8+ T cells. CD14 was the most significant DEG in “myeloid cell enriched receptors and transporters” (Figure 6G); TNFSF13B (or BAFF) was the most significant DEG in “viral sensing and immunity: IRF2 targets network” (Figure E6C); RSAD2 (viperin) was the most significant DEG in “enriched in activated dendritic cells” (Figure E6D); CXCL1 was the most significant DEG in “enriched in activated dendritic cells” (Figure 6H); CXCL10 had the fourth highest significance in “viral sensing and immunity: IRF2 targets network (II)” (Figure 6I); CXCL16 had the second highest significance in “DC [dendritic-cell] surface signature” (Figure 6J); and CCL20 (MIP-3b) had the second highest significance in “enriched in activated dendritic cells” (Figure E6E). For these markers, coexpression was observed in 32–94% of CD8+ cells at Day 7 after infection with correlation coefficients of 0.74–0.97, indicating that all markers were strongly correlated with CD8 in coexpressed cells (Figures 6E and 6F).
Thus, we identified distinctive innate-like patterns of gene expression by human BAL-fluid CD8+ T cells responding to influenza infection in vivo, consistent with the rapid innate-like effector functions that Trm cells possess.
Discussion
In this study, we tracked CD8+ Trm cells in adults challenged with influenza A/California/4/09(H1N1) in vivo. Preexisting circulating memory CD8+ T cells correlated strongly with a reduced viral load (in support of previous studies [25, 31, 26]) and were temporally associated with virus-specific counterparts in the lung. However, antigen-specific CD8+ T cells in the blood and airway displayed markedly divergent kinetics, phenotypes, and functional characteristics, with airway CD8+ cells unexpectedly being dominated by transcriptomic signatures usually associated with innate immune cells. This suggests that although peripheral blood memory T cells may be strong surrogates of protection, they poorly represent Trm cells that act most directly to eliminate virus.
Despite correlations between circulating memory T cells and reduced disease, systemic T cell–inducing vaccines have yet to demonstrate substantial efficacy. We propose that this is due to differences in the tissue localization of T cells generated after infection and systemic vaccination (32). In murine influenza, Trm cells prevent lethal infection better than their systemic counterparts and are sufficient to confer cross-strain immunity (8). Pulmonary Trm-cell generation requires not only antigen stimulation but also costimulation and signals from the microenvironment for differentiation and the tuning of responses (33). Importantly, these signals are tissue restricted and require local monocytes and dendritic cells (34). This is supported by our data, which show influenza-specific CD69+CD103-CD8+ T cells appearing in the bronchial mucosa and BAL fluid but not in the blood during acute infection, together with increased acquisition of the CD69+CD103+ phenotype into convalescence, implying local upregulation of CD69 and later upregulation of CD103. However, even with these signals, murine lung-resident Trm cells wane over time, which is caused by redistribution and an imbalance between apoptosis and replenishment in the amino acid–poor lung environment (11, 35). In humans, lung transplant data suggest that both in situ Trm-cell maintenance and de novo recruitment can occur (36). Furthermore, that some Trm cells may reenter the circulation after resolution but preferentially return at repeat infection, enabling more rapid recall on local restimulation, suggests a role for mucosal vaccination to enhance these responses (13).
One previous study using postmortem tissues identified a core transcriptional signature common to Trm cells from multiple sites (30). Although that study sorted T cells on the basis of CD69 expression, the limited cell numbers obtained from in vivo BAL fluid meant that this was not possible here. Therefore, a proportion of the DEGs we identified between blood and BAL-fluid CD8+ cells are likely to have represented differences in the subset composition. Nevertheless, 94% of the genes in the earlier core profile were identified. However, the most significantly enriched pathways were functionally annotated as being monocyte- and dendritic cell–related, despite no evidence of contamination by myeloid cells.
Classical T cells make up by far the largest proportion of CD3+CD8+ cells in the lung, although nonclassical CD8-expressing T cells are also present. These include MAIT cells, invariant NKT cells and γδ T cells, all of which have subpopulations expressing CD8. MAIT cells make up around 2% of CD3+ cells in both the blood and the BAL fluid of healthy adults, so in numerical terms, these cells are unlikely to contribute substantially to differential gene expression. Similarly, invariant NKT cells are rare in humans, ranging in frequency between 0.1% and 1% in peripheral blood, and are mostly CD8−. In mice, they are mainly localized in the pulmonary vasculature and interstitium (37). Although γδ T cells make up 8–20% of pulmonary lymphocytes in mice, the majority are CD4−CD8− cells (38). Nevertheless, to mitigate the contribution of unconventional T cells, we undertook longitudinal analysis, controlling for systematic differences in the CD8+ subset composition. Furthermore, these signatures were unlikely to have been due to innate cells at the Day 7 time point, as most (including MAIT cells) normally respond more immediately and to lower peak abundance, although this remains to be definitively shown through using single-cell techniques (39).
We therefore conclude that CD8+ Trm cells in the airway express innate-like features that underlie their sentinel function. These pathways included pathogen-associated molecular pattern recognition by TLRs, which is implied by the upregulation of CD14 detected in CD8+ cells in bronchial biopsy specimens. We hypothesize that CD14 here is intracellular and would therefore only be revealed after detergent antigen retrieval for confocal microscopy. TLR4 and CD14 have previously been implicated in respiratory syncytial virus and influenza A virus recognition by alveolar macrophages, with CD14 being observed on CD8+ T cells during HIV infection, in severe asthma, and after in vitro stimulation allowing direct recognition of LPS (40–42). Downstream signaling regulates expression of proinflammatory and IFN-stimulated genes, including viperin, which has antiviral activity and is involved in the regulation of CD4+ T-cell responses (43, 44). In addition, our data suggest that CD8+ Trm-cell activation is associated with the production of innate mediators, including BAFF, which has a role in T-cell costimulation (45), and the chemokines CXCL1, CXCL10, CXCL16, and CCL20 (which are all commonly expressed by myeloid cells, but, in the case of CXCL10 and CXCL16, have also been reported in T cells). Together, these findings lead us to hypothesize that these mechanisms function to enhance chemoattraction, rapidly reinforcing the antiviral inflammatory environment (46, 47).
The major limitation of this study is the small number of infected participants who underwent lower airway sampling, which led to further restrictions on sample size once HLA matching to enable tetramer-labeled lower airway T-cell analysis was undertaken. Nevertheless, differences in these cells were sufficiently consistent for us to show that human pulmonary CD8+ memory T cells share phenotypic and transcriptional characteristics with other immune cells that act immediately when pathogens are encountered. These findings parallel those of recent transcriptomic studies of pulmonary CD8+ Trm cells in mice, which also upregulate cell-cycle genes, macrophage-associated chemokines (such as CCL3, CCL9, and CXCL10), and IFN-stimulated genes (48). We therefore propose that these are essential features of the CD8+ Trm-cell response that may be targeted through adjuvantation (e.g., by direct signaling through the TLR4 pathway [40]) and indicate the blurred distinction between innate and adaptive immunity in this setting.
Footnotes
Supported by the European Union 7th Framework PREPARE (Preparedeness Against (Re-) emerging Epidemics Platform foR European Preparedness) (grant HEALTH-F3-2013-602525) and Innovative Medicines Initiative BioVacSafe (Biomarkers for Enhanced Vaccines Immunosafety) grants; the Kwok Foundation; and the United States Department of Defense DARPA Prometheus program. P.J.O. and C.C. are supported by the Biomedical Research Centre award to the Imperial College Healthcare National Health Service Trust. Infrastructure support was provided by the National Institute for Health Research (NIHR) Imperial Biomedical Research Centre and the National Institute for Health Research Imperial Clinical Research Facility. The views expressed are those of the author(s) and not necessarily those of the National Health Service, the NIHR, or the Department of Health and Social Care, United Kingdom.
Author Contributions: C.C., P.J.O., S.H.E.K., C.W.W., and T.V. designed the work. S.P., S.K., S.K.U., Z.G., E.B., S.A., M.K., A. Jha, H.J., A. Jozwik, B.P.N., P.M., O.M.K., and C.C. acquired data. S.P., S.K., J.Z., J.M., H.-J.M., J.W., A.J., P.J.O., and C.C. analyzed and interpreted the data. S.P., S.K., and C.C. drafted the work. All authors reviewed, revised, and approved the final version to be published.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.202103-0620OC on July 13, 2021
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.World Health Organization. Geneva, Switzerland: World Health Organization; 2018. https://www.who.int/news-room/fact-sheets/detail/influenza-(seasonal) [Google Scholar]
- 2. Krammer F, Palese P. Advances in the development of influenza virus vaccines. Nat Rev Drug Discov. 2015;14:167–182. doi: 10.1038/nrd4529. [DOI] [PubMed] [Google Scholar]
- 3. Cobey S, Gouma S, Parkhouse K, Chambers BS, Ertl HC, Schmader KE, et al. Poor immunogenicity, not vaccine strain egg adaptation, may explain the low H3N2 influenza vaccine effectiveness in 2012-2013. Clin Infect Dis. 2018;67:327–333. doi: 10.1093/cid/ciy097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pebody R, Warburton F, Ellis J, Andrews N, Thompson C, von Wissmann B, et al. Low effectiveness of seasonal influenza vaccine in preventing laboratory-confirmed influenza in primary care in the United Kingdom: 2014/15 mid-season results. Euro Surveill. 2015;20:21025. [PubMed] [Google Scholar]
- 5. Koutsakos M, Illing PT, Nguyen THO, Mifsud NA, Crawford JC, Rizzetto S, et al. Human CD8+ T cell cross-reactivity across influenza A, B and C viruses. Nat Immunol. 2019;20:613–625. doi: 10.1038/s41590-019-0320-6. [DOI] [PubMed] [Google Scholar]
- 6. Sadoff JC, Wittes J. Correlates, surrogates, and vaccines. J Infect Dis. 2007;196:1279–1281. doi: 10.1086/522432. [DOI] [PubMed] [Google Scholar]
- 7. Schenkel JM, Masopust D. Tissue-resident memory T cells. Immunity. 2014;41:886–897. doi: 10.1016/j.immuni.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu T, Hu Y, Lee YT, Bouchard KR, Benechet A, Khanna K, et al. Lung-resident memory CD8 T cells (TRM) are indispensable for optimal cross-protection against pulmonary virus infection. J Leukoc Biol. 2014;95:215–224. doi: 10.1189/jlb.0313180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stolley JM, Johnston TS, Soerens AG, Beura LK, Rosato PC, Joag V, et al. Retrograde migration supplies resident memory T cells to lung-draining LN after influenza infection. J Exp Med. 2020;217:e20192197. doi: 10.1084/jem.20192197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Van Braeckel-Budimir N, Varga SM, Badovinac VP, Harty JT. Repeated antigen exposure extends the durability of influenza-specific lung-resident memory CD8+ T cells and heterosubtypic immunity. Cell Rep. 2018;24:3374–3382, e3. doi: 10.1016/j.celrep.2018.08.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Slütter B, Van Braeckel-Budimir N, Abboud G, Varga SM, Salek-Ardakani S, Harty JT. Dynamics of influenza-induced lung-resident memory T cells underlie waning heterosubtypic immunity. Sci Immunol. 2017;2:1–11. doi: 10.1126/sciimmunol.aag2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Park SL, Zaid A, Hor JL, Christo SN, Prier JE, Davies B, et al. Local proliferation maintains a stable pool of tissue-resident memory T cells after antiviral recall responses. Nat Immunol. 2018;19:183–191. doi: 10.1038/s41590-017-0027-5. [DOI] [PubMed] [Google Scholar]
- 13. Fonseca R, Beura LK, Quarnstrom CF, Ghoneim HE, Fan Y, Zebley CC, et al. Developmental plasticity allows outside-in immune responses by resident memory T cells. Nat Immunol. 2020;21:412–421. doi: 10.1038/s41590-020-0607-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mackay LK, Stock AT, Ma JZ, Jones CM, Kent SJ, Mueller SN, et al. Long-lived epithelial immunity by tissue-resident memory T (TRM) cells in the absence of persisting local antigen presentation. Proc Natl Acad Sci USA. 2012;109:7037–7042. doi: 10.1073/pnas.1202288109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sathaliyawala T, Kubota M, Yudanin N, Turner D, Camp P, Thome JJ, et al. Distribution and compartmentalization of human circulating and tissue-resident memory T cell subsets. Immunity. 2013;38:187–197. doi: 10.1016/j.immuni.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. de Bree GJ, van Leeuwen EM, Out TA, Jansen HM, Jonkers RE, van Lier RA. Selective accumulation of differentiated CD8+ T cells specific for respiratory viruses in the human lung. J Exp Med. 2005;202:1433–1442. doi: 10.1084/jem.20051365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jozwik A, Habibi MS, Paras A, Zhu J, Guvenel A, Dhariwal J, et al. RSV-specific airway resident memory CD8+ T cells and differential disease severity after experimental human infection. Nat Commun. 2015;6:10224. doi: 10.1038/ncomms10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paterson S.24.
- 19.Kar S.Fifth European Congress of Immunology.
- 20.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner Bioinformatics 20132915–21.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wingett SW, Andrews S. FastQ screen: a tool for multi-genome mapping and quality control. F1000 Res. 2018;7:1338. doi: 10.12688/f1000research.15931.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zyla J, Marczyk M, Domaszewska T, Kaufmann SHE, Polanska J, Weiner J. Gene set enrichment for reproducible science: comparison of CERNO and eight other algorithms. Bioinformatics. 2019;35:5146–5154. doi: 10.1093/bioinformatics/btz447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li S, Rouphael N, Duraisingham S, Romero-Steiner S, Presnell S, Davis C, et al. Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nat Immunol. 2014;15:195–204. doi: 10.1038/ni.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chaussabel D, Quinn C, Shen J, Patel P, Glaser C, Baldwin N, et al. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity. 2008;29:150–164. doi: 10.1016/j.immuni.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wilkinson TM, Li CK, Chui CS, Huang AK, Perkins M, Liebner JC, et al. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat Med. 2012;18:274–280. doi: 10.1038/nm.2612. [DOI] [PubMed] [Google Scholar]
- 26. Sridhar S, Begom S, Bermingham A, Hoschler K, Adamson W, Carman W, et al. Cellular immune correlates of protection against symptomatic pandemic influenza. Nat Med. 2013;19:1305–1312. doi: 10.1038/nm.3350. [DOI] [PubMed] [Google Scholar]
- 27. Lin L, Finak G, Ushey K, Seshadri C, Hawn TR, Frahm N, et al. COMPASS identifies T-cell subsets correlated with clinical outcomes. Nat Biotechnol. 2015;33:610–616. doi: 10.1038/nbt.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Akondy RS, Fitch M, Edupuganti S, Yang S, Kissick HT, Li KW, et al. Origin and differentiation of human memory CD8 T cells after vaccination. Nature. 2017;552:362–367. doi: 10.1038/nature24633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kaech SM, Hemby S, Kersh E, Ahmed R. Molecular and functional profiling of memory CD8 T cell differentiation. Cell. 2002;111:837–851. doi: 10.1016/s0092-8674(02)01139-x. [DOI] [PubMed] [Google Scholar]
- 30. Kumar BV, Ma W, Miron M, Granot T, Guyer RS, Carpenter DJ, et al. Human tissue-resident memory T cells are defined by core transcriptional and functional signatures in lymphoid and mucosal sites. Cell Rep. 2017;20:2921–2934. doi: 10.1016/j.celrep.2017.08.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McMichael AJ, Gotch FM, Noble GR, Beare PA. Cytotoxic T-cell immunity to influenza. N Engl J Med. 1983;309:13–17. doi: 10.1056/NEJM198307073090103. [DOI] [PubMed] [Google Scholar]
- 32. Liu Y, Ma C, Zhang N. Tissue-specific control of tissue-resident memory T cells. Crit Rev Immunol. 2018;38:79–103. doi: 10.1615/CritRevImmunol.2018025653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Szabo PA, Miron M, Farber DL. Location, location, location: tissue resident memory T cells in mice and humans. Sci Immunol. 2019;4:1–11. doi: 10.1126/sciimmunol.aas9673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dunbar PR, Cartwright EK, Wein AN, Tsukamoto T, Tiger Li ZR, Kumar N, et al. Pulmonary monocytes interact with effector T cells in the lung tissue to drive TRM differentiation following viral infection. Mucosal Immunol. 2020;13:161–171. doi: 10.1038/s41385-019-0224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McMaster SR, Wein AN, Dunbar PR, Hayward SL, Cartwright EK, Denning TL, et al. Pulmonary antigen encounter regulates the establishment of tissue-resident CD8 memory T cells in the lung airways and parenchyma. Mucosal Immunol. 2018;11:1071–1078. doi: 10.1038/s41385-018-0003-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Snyder ME, Finlayson MO, Connors TJ, Dogra P, Senda T, Bush E, et al. Generation and persistence of human tissue-resident memory T cells in lung transplantation. Sci Immunol. 2019;4:1–16. doi: 10.1126/sciimmunol.aav5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hinks TSC, Zhou X, Staples KJ, Dimitrov BD, Manta A, Petrossian T, et al. Innate and adaptive T cells in asthmatic patients: Relationship to severity and disease mechanisms. J Allergy Clin Immunol. 2015;136:323–333. doi: 10.1016/j.jaci.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kalyan S, Kabelitz D. Defining the nature of human γδ T cells: a biographical sketch of the highly empathetic. Cell Mol Immunol. 2013;10:21–29. doi: 10.1038/cmi.2012.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. van Wilgenburg B, Loh L, Chen Z, Pediongco TJ, Wang H, Shi M, et al. MAIT cells contribute to protection against lethal influenza infection in vivo. Nat Commun. 2018;9:4706. doi: 10.1038/s41467-018-07207-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Komai-Koma M, Gilchrist DS, Xu D. Direct recognition of LPS by human but not murine CD8+ T cells via TLR4 complex. Eur J Immunol. 2009;39:1564–1572. doi: 10.1002/eji.200838866. [DOI] [PubMed] [Google Scholar]
- 41. Turner D, Hoffman M, Yust I, Fried M, Bleiberg M, Tartakovsky B. Overexpression of a novel lymphocyte population, positive for an intracellular CD14-like antigen, in patients positive for human immunodeficiency virus type 1. Clin Diagn Lab Immunol. 2004;11:1040–1044. doi: 10.1128/CDLI.11.6.1040-1044.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Singhania A, Wallington JC, Smith CG, Horowitz D, Staples KJ, Howarth PH, et al. Multitissue transcriptomics delineates the diversity of airway T cell functions in asthma. Am J Respir Cell Mol Biol. 2018;58:261–270. doi: 10.1165/rcmb.2017-0162OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang X, Hinson ER, Cresswell P. The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe. 2007;2:96–105. doi: 10.1016/j.chom.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 44. Qiu L-Q, Cresswell P, Chin K-C. Viperin is required for optimal Th2 responses and T-cell receptor-mediated activation of NF-κB and AP-1. Blood. 2009;113:3520–3529. doi: 10.1182/blood-2008-07-171942. [DOI] [PubMed] [Google Scholar]
- 45. Hu S, Wang R, Zhang M, Liu K, Tao J, Tai Y, et al. BAFF promotes T cell activation through the BAFF-BAFF-R-PI3K-Akt signaling pathway. Biomed Pharmacother. 2019;114:108796. doi: 10.1016/j.biopha.2019.108796. [DOI] [PubMed] [Google Scholar]
- 46. Peperzak V, Veraar EA, Xiao Y, Babala N, Thiadens K, Brugmans M, et al. CD8+ T cells produce the chemokine CXCL10 in response to CD27/CD70 costimulation to promote generation of the CD8+ effector T cell pool. J Immunol. 2013;191:3025–3036. doi: 10.4049/jimmunol.1202222. [DOI] [PubMed] [Google Scholar]
- 47. Shashkin P, Simpson D, Mishin V, Chesnutt B, Ley K. Expression of CXCL16 in human T cells. Arterioscler Thromb Vasc Biol. 2003;23:148–149. doi: 10.1161/01.atv.0000043906.61088.4b. [DOI] [PubMed] [Google Scholar]
- 48. Low JS, Farsakoglu Y, Amezcua Vesely MC, Sefik E, Kelly JB, Harman CCD, et al. Tissue-resident memory T cell reactivation by diverse antigen-presenting cells imparts distinct functional responses. J Exp Med. 2020;217:1–13. doi: 10.1084/jem.20192291. [DOI] [PMC free article] [PubMed] [Google Scholar]