Abstract
Phylogeographical studies of Philippine vertebrates have demonstrated that genetic variation is broadly partitioned by Pleistocene island aggregation. Contemporary island discontinuity is expected to influence genetic differentiation but remains relatively undocumented, perhaps because the current episode of island isolation started in relatively recent times. We investigated inter- and intra-island population structure in a Philippine endemic bird genus (Sarcophanops) to determine whether genetic differentiation has evolved during the recent period of isolation. We sequenced thousands of genome-wide restriction site associated DNA (RAD) markers from throughout the Mindanao group to assess fine-scale genetic structure across islands. Specifically, we investigated patterns of gene flow and connectivity within and between taxonomic and geographical bounds. A previous assessment of mitochondrial DNA detected deep structure between Sarcophanops samarensis and a sister species, Sarcophanops steerii, but was insufficient to detect differentiation within either species. Analysis of RAD markers, however, revealed structure within S. samarensis between the islands of Samar/Leyte and Bohol. This genetic differentiation probably demonstrates an effect of recent geographical isolation (after the Last Glacial Maximum) on the genetic structure of Philippine avifauna. We suggest that the general lack of evidence for differentiation between recently isolated populations is a failure to detect subtle population structure owing to past genetic sampling constraints, rather than the absence of such structure.
Keywords: allopatric, Last Glacial Maximum, Pleistocene, RADseq, wattled broadbill
INTRODUCTION
The Philippine Archipelago is recognized as one of the most biologically diverse hotspots in the world (Myers et al., 2000), largely as a result of a complex geological and climatic history that has catalysed the evolution of endemic biodiversity (Brown et al., 2013). Owing to cyclic changes in sea level, the extent of land above water in the Philippine Archipelago has varied dramatically throughout its geological history. Specifically, changing climate regimens during the Last Glacial Maximum (LGM; 19–25 kyr BP) resulted in lower global sea levels, consequently uncovering shallow land bridges between islands. This network of shallow land bridges dramatically increased connectivity across the archipelago (Heaney, 1985), forming clustered groups of interconnected islands, or Pleistocene aggregate island complexes (PAICs; Diesmos et al., 2002; Brown et al., 2013). Of the > 7000 islands found in the present-day Philippine Archipelago (Kennedy et al., 2000), nearly all were reduced to six large PAICs (Luzon, Palawan, Mindoro, Negros-Panay, Mindanao and Sulu; Heaney, 1985).
The endemic Philippine avifauna generally adhere to the patterns of geographical and phylogenetic structure predicted under the PAIC model, at least when additional complexities, such as topography, palaeoclimatic factors and colonization history, are acknowledged (Hosner et al., 2013, 2014; Sánchez-González et al., 2015). This is to say, populations present on a particular PAIC (e.g. Mindanao PAIC) are likely to be closely related to one another but genetically distinct from populations confined to different PAICs during the LGM (e.g. Luzon PAIC; Sánchez-González & Moyle, 2011). As a presumed consequence of this complex geological history, the total diversity of Philippine avifauna includes a remarkably high proportion (~45%) of endemic species (BirdLife International, 2017). Although broad attempts at understanding Plio-Pleistocene diversification across the archipelago have been possible for some time, the power to detect fine-scale differentiation has been limited by DNA sequencing depth. Furthermore, much of the work on Philippine biodiversity has focused on the patterns and processes shaping diversity throughout the archipelago, despite the fact that not all lineages have distributions spanning its entirety. Hence, the generation of recent population genetic structure attributable to Holocene isolation on individual islands within the same PAIC remains largely theoretical (but see Hosner et al. 2018).
Here, we investigate the effect of individual islands on the generation of genetic differentiation in the endemic Philippine broadbills (Eurylaimidae: Sarcophanops), in which all extant lineages occur on one previously connected landmass (the Greater Mindanao PAIC), which now comprises many islands (Fig. 1). We used restriction site associated DNA sequencing (RADseq) to produce a genome-wide panel of thousands of single nucleotide polymorphisms (SNPs), which permits the assessment of subtle population genomic structure across islands that were part of the same PAIC as recently as the LGM. The two species of Philippine broadbill, Sarcophanops steerii Sharpe, 1876 and Sarcophanops samarensis Steere, 1890, occur in non-overlapping ranges within multiple subregions of the Mindanao PAIC (as described by Hosner et al. 2018). Specifically, S. steerii is found in Dinagat/Siargao, Eastern Mindanao and the Zamboanga Peninsula (referred to collectively as Mindanao), whereas S. samarensis is found on Samar/Leyte and Bohol (referred to collectively as Visayan). Inferring differentiation at this evolutionary time scale has not, to our knowledge, been documented in Philippine avifauna. Focusing on a genus (Sarcophanops) endemic to a single PAIC enables us to: (1) examine inter- or intra-island population structure within Sarcophanops species to obtain a glimpse into genetic connectivity of avifauna endemic to the Mindanao PAIC; and (2) expand our understanding of the population history of these enigmatic taxa.
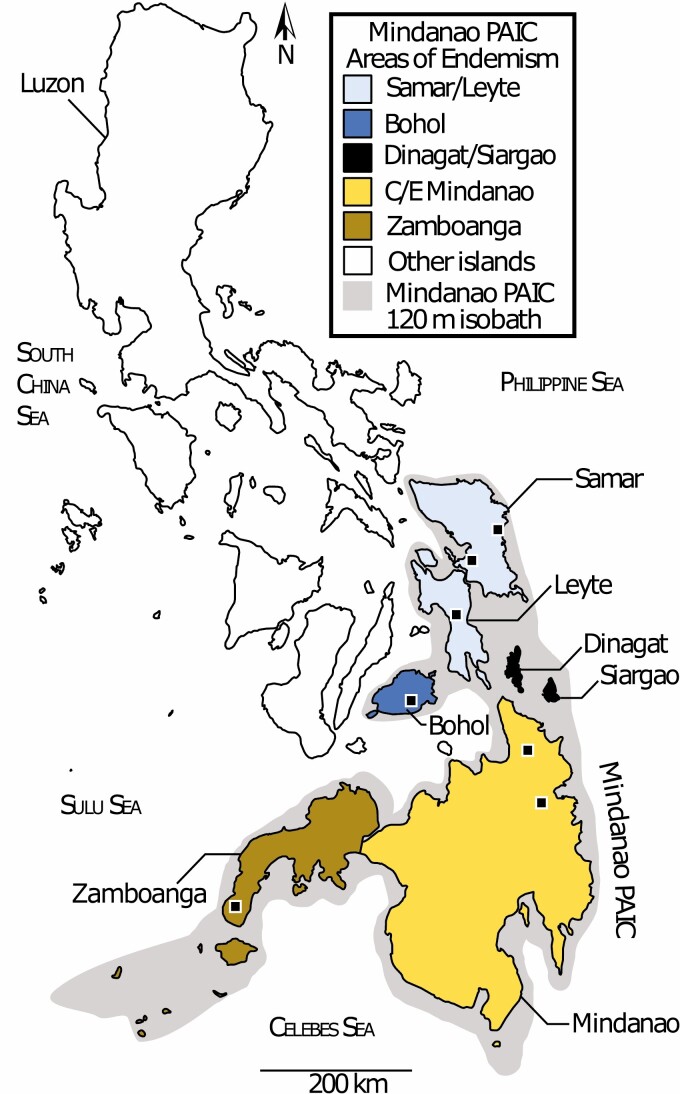
Figure 1.
Map of Philippine Archipelago, adapted from Hosner et al. (2018), with subregions of the Mindanao Pleistocene aggregate island complex (PAIC) shown (Mindanao in yellow and Visayas in blue). Approximate sampling localities are shown as black squares. Note the shallow (120 m) isobath surrounding the entire PAIC.
MATERIAL AND METHODS
Sampling and DNA extraction
We obtained tissue samples (N = 22) of Sarcophanops from across their distribution range in the Philippines and used two individuals of Serilophus lunatus Swainson, 1837 as the outgroup (Table 1; Fig. 1; for information on outgroup selection, see Moyle et al., 2006). Tissue samples (frozen and/or ethanol-preserved muscle tissue) and associated voucher specimens were collected under the auspices of Gratuitous Permits to Collect Biological Specimens issued by the Biodiversity Monitoring Bureau (formerly Parks and Wildlife Bureau) of the Philippine central government. Museum specimens and additional genetic material are housed in the Biodiversity Institute at the University of Kansas.
Table 1.
List of samples used in this study and their associated sequencing statistics
| Species | Museum number | Locality | Number of reads | RAD-tags | Coverage median | Coverage SD | Percentage missing from 50% CM/70% CM dataset |
|---|---|---|---|---|---|---|---|
| Sarcophanops steerii | KU 19047 | Mindanao | 874 532 | 20 185 | 30 | 33.29 | 15.31/3.75 |
| Sarcophanops steerii | KU 19050 | Mindanao | 991 797 | 20 902 | 35 | 37.70 | 13.46/3.52 |
| Sarcophanops steerii | KU 19061 | Mindanao | 2 408 639 | 31 817 | 72 | 83.53 | 3.46/0.48 |
| Sarcophanops steerii | KU 28295 | Mindanao | 2 600 919 | 39 135 | 74 | 80.55 | 14.20/13.43 |
| Sarcophanops steerii | KU 19186 | Zamboanga | 1 782 408 | 26 521 | 53 | 67.01 | 0.00/0.00 |
| Sarcophanops samarensis | KU 20929 | Bohol | 2 180 969 | 32 211 | 65 | 72.52 | 5.99/1.45 |
| Sarcophanops samarensis | KU 20930 | Bohol | 1 638 944 | 23 861 | 53 | 62.67 | 6.38/0.70 |
| Sarcophanops samarensis | KU 20932 | Bohol | 1 371 645 | 26 271 | 43.5 | 49.98 | 8.98/1.14 |
| Sarcophanops samarensis | KU 28181 | Bohol | 712 002 | 17 718 | 26 | 29.03 | 15.85/5.02 |
| Sarcophanops samarensis | KU 28182 | Bohol | 2 285 682 | 28 961 | 65 | 78.28 | 3.25/0.84 |
| Sarcophanops samarensis | KU 28213 | Bohol | 1 197 631 | 31 888 | 37 | 38.54 | 13.53/3.57 |
| Sarcophanops samarensis | KU 28231 | Bohol | 1 981 016 | 32 462 | 60 | 68.49 | 4.69/0.97 |
| Sarcophanops samarensis | KU 28247 | Bohol | 1 465 002 | 31 170 | 44 | 51.05 | 6.26/0.48 |
| Sarcophanops samarensis | KU 27374 | Leyte | 1 722 020 | 23 775 | 56 | 65.06 | 3.62/0.00 |
| Sarcophanops samarensis | KU 27376 | Leyte | 2 833 494 | 30 055 | 88 | 98.85 | 3.36/0.00 |
| Sarcophanops samarensis | KU 27448 | Leyte | 1 525 375 | 25 388 | 46 | 58.30 | 3.81/0.00 |
| Sarcophanops samarensis | KU 31598 | Samar | 450 539 | 22 184 | 15 | 14.55 | 23.32/2.03 |
| Sarcophanops samarensis | KU 31601 | Samar | 808 201 | 25 697 | 23 | 28.23 | 14.32/0.31 |
| Sarcophanops samarensis | KU 31612 | Samar | 1 793 799 | 39 214 | 54 | 55.63 | 5.87/1.23 |
| Sarcophanops samarensis | KU 31616 | Samar | 3 347 326 | 44 974 | 87 | 100.42 | 6.17/1.50 |
| Sarcophanops samarensis | KU 31618 | Samar | 889 378 | 24 240 | 26 | 31.44 | 9.35/0.35 |
| Sarcophanops samarensis | KU 31619 | Samar | 986 355 | 25 079 | 32 | 35.91 | 7.87/0.13 |
| Serilophus lunatus | KU 23405 | Vietnam | 3 515 364 | 55 899 | 63 | 91.42 | 4.52/0.00 |
| Serilophus lunatus | KU 23552 | Vietnam | 1 302 599 | 27 169 | 35 | 43.52 | 12.44/0.00 |
RADseq methods
We used a modified RADseq (Miller et al., 2007) protocol to prepare genomic libraries of putatively neutral loci from across the genome. Initially, we digested genomic DNA with a single restriction enzyme (NdeI). We chose this enzyme and protocol because of success in sequencing other Passeriformes species in the laboratory from unrelated projects (e.g. Manthey & Moyle, 2015). Next, we ligated custom adapters with attached barcodes (Andolfatto et al., 2011) to all samples (Supporting Information, Table S1). All individuals were pooled and subsequently purified with AMPure magnetic beads (Agencourt). To reduce genomic coverage in the library further, we used a Pippin Prep electrophoresis cassette (Sage Science) to size select fragments between 500 and 600 bp. We purified the library again with magnetic beads, performed a brief polymerase chain reaction (PCR) in duplicate (14 cycles) and performed a final purification. The final PCR step dual-indexed the samples (with standard Illumina indices) for multiplexing. We tested the library for DNA quality and quantity using quantitative PCR and an Agilent Tapestation at the University of Kansas Genome Sequencing Core Facility. The multiplexed library was then pooled with libraries from unrelated projects and sequenced on three lanes of an Illumina HiSeq2500 flow cell.
Single nucleotide polymorphism dataset construction
To assemble loci de novo and create SNP datasets from our sequencing data, we used the STACKS (Catchen et al., 2013) pipeline. Initially, we assigned sequences to individuals and removed reads with poor quality using the process_RADtags python script included in STACKS. In order to be included in downstream analyses, sequences were required to have an average phred score of ten in sliding windows of 15 bp, not to contain the adapter sequence and to contain the restriction cut site. Next, we used the ustacks, cstacks and sstacks modules of STACKS to assemble reads into loci and compare loci across individuals. We used ustacks with the default settings. In cstacks, we tested various numbers of mismatches allowed between stacks when assembling loci (N = 1–7). Here, genetic diversity within sampling localities generally increased with greater numbers of mismatches allowed, whereas genetic differentiation between populations was generally constant between N = 3 and N = 7 (Supporting Information, Table S2). Based on these initial patterns, we chose a value of N = 4 for subsequent analyses. We then used the sstacks module with default settings. Finally, we used the populations module of STACKS to filter all loci and create two SNP datasets: (1) a 50% coverage matrix (requiring an SNP to be represented in ≥ 50% of individuals; 50CM); and (2) a 75% coverage matrix (75CM). In addition to individual coverage, we required all loci to have a minimum read depth of five and maximum observed heterozygosity < 50% to reduce the inclusion of paralogues. We assessed how changing the minimum read depth of loci (m = 1, 5, 10, 15, 20) would affect population genetic estimates. Although the number of loci decreased with increasing minimum read depth, estimates of genetic diversity within and genetic differentiation between localities did not change substantially (Supporting Information, Table S2). Lastly, we assessed coverage across the genome by matching loci identified in STACKS to chromosomes in the zebra finch [Taeniopygia guttata Vieillot, 1817)] using the BLAST+ utility (Camacho et al., 2009). Here, we required a minimum of 70% sequence identity across ≥ 25 bp, and a maximum e-value of 0.001 to limit the number of expected matches by chance in order to define a match.
Phylogenetic analysis
We used two methods to identify phylogenetic relationships among individuals using a concatenated matrix of all full-length sequences: RAxML v.8 (Stamatakis, 2014) and MrBayes v.3.2.6 (Ronquist & Huelsenbeck, 2003). Initially, we estimated an appropriate model of sequence evolution (GTR+I+G in this case) based on the Bayesian information criterion (BIC) using PAUP v.4.0.151 (Swofford, 2011). In RAxML, we estimated a maximum likelihood tree and assessed support using 1000 rapid bootstrap replicates. In MrBayes, we ran four chains for five million generations, excluding the first 50% of trees as burn-in, and sampling every 5000 generations.
We also used the programs STRUCTURE (Pritchard et al., 2000) and Discriminant Analysis of Principal Components (DAPC; Jombart et al., 2010) to investigate population genetic structure for the stricter 75CM dataset. For both analyses, we subset our datasets to include only one SNP per locus (two replicates each) to minimize potential linkage effects. We ran STRUCTURE initially with the number of populations (K) limited to one to infer lambda. Next, we used a constant lambda (set to the inferred value) to run the admixture model with correlated allele frequencies for a number of likely values of K (K = 1–5, with five runs for each value of K). We defined the burn-in period as the first 100 000 Markov chain Monte Carlo generations, with the subsequent 100 000 iterations sampled. To determine the most likely number of genetic clusters, we implemented the ΔK method of Evanno et al. (2005) using the program STRUCTURE HARVESTER (Earl & vonHoldt, 2012). The DAPC analyses were performed in R (R Core Team, 2013), using the package ‘adegenet’ (Jombart, 2008; Jombart & Ahmed, 2011). For DAPC, the most likely number of populations was determined based on BIC values.
RESULTS
Sequencing coverage across individuals was variable (Table 1), with a median of ~1.6 million reads per individual (SD = 822 669 reads). From these reads, we recovered ~25 000 RAD-tags per individual (SD = 8533). The 50 and 75% coverage matrices had 1737 and 885 loci, respectively, corresponding to 4310 and 2271 SNPs (Supporting Information, Table S1). All raw sequence data from RADseq are available at the NCBI Sequence Read Archive (BioProject ID: 522809) https://www.ncbi.nlm.nih.gov/bioproject/522809. The number of loci per chromosome, based on BLAST results to the zebra finch, was positively related to chromosome size (R2 > 0.95; Supporting Information, Table S3), suggesting that we obtained relatively even sequencing coverage across the genome. Nearly 4% of ingroup variant SNPs were fixed between Visayan (S. samarensis) and Mindanao (S. steerii) species, with a large proportion being private within each group (~85%; Supporting Information, Table S4).
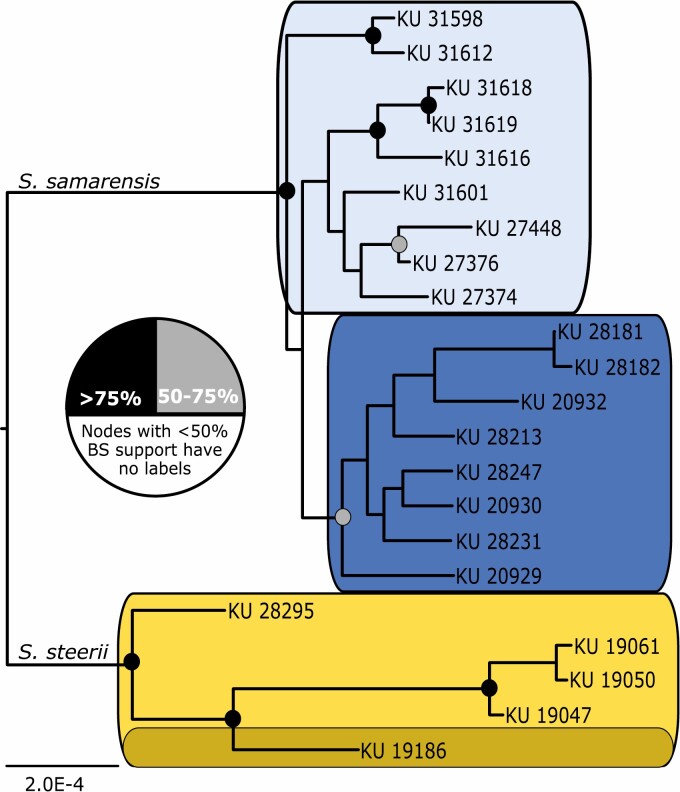
Genetic differentiation, measured by the fixation index FST, between sampling localities within a given species was generally low (FST < 0.15), but high between species (FST > 0.30; Table 2). The number of bi-allelic markers (>1000 SNPs) in our study permitted confident estimates of FST, despite the small sample sizes within populations (Willing et al., 2012). Likewise, population structure was most apparent across species (i.e. between S. samarensis and S. steeri) in phylogenetic and population genetic analyses (Fig. 2). Phylogenetic analysis in RAxML and MrBayes recovered very similar topologies for both coverage matrices, with all trees supporting a deep split between species. We present only the maximum likelihood tree for the 75CM herein, but all remaining trees can be found in the Supporting Information (Figs S1–S3). We did not recover significant differentiation between the individual from Zamboanga Peninsula and the rest of the Mindanao samples, despite the potential inflation of FST values attributable to the inclusion of only one individual from Zamboanga (Campagna et al., 2015). Relationships within S. samarensis were more concordant with our expectations (i.e. subspecific genetic structuring aligned with subregion geographical range). Specifically, we found the the Bohol population was identified as monophyletic, separate from Samar/Leyte.
Table 2.
Pairwise estimates of FST for the 75 and 50% coverage matrices above and below the diagonal, respectively
| Bohol | Leyte | Samar | Mindanao | Zamboanga | |
|---|---|---|---|---|---|
| Bohol | 0.114 | 0.122 | 0.323 | 0.321 | |
| Leyte | 0.116 | 0.106 | 0.369 | 0.452 | |
| Samar | 0.114 | 0.116 | 0.340 | 0.355 | |
| Mindanao | 0.310 | 0.346 | 0.333 | 0.204 | |
| Zamboanga | 0.309 | 0.447 | 0.357 | 0.214 |
Figure 2.
Maximum likelihood phylogeny for the 75CM dataset. Colours around clades correspond to Figure 1, with Visayan Islands in shades of blue and Mindanao in shades of yellow. Node support was drawn from 1000 rapid bootstrap replicates. Abbreviation: BS, bootstrap. Additional trees can be found in the Supporting Information (Figs S1–S3).
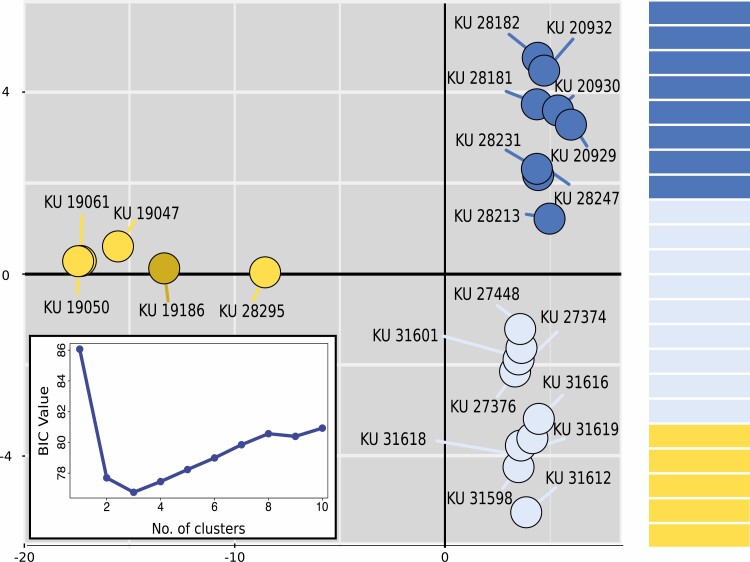
Population genetic analyses recovered a similar overall pattern. In STRUCTURE, the ΔK method most strongly supported two genetic clusters, separating populations along species boundaries with no subregion separation. We also inspected STRUCTURE results for K = 3, which recovered an additional break between Bohol and Samar/Leyte (Supporting Information, Tables S5–S7). Higher values of K did not clearly show finer-scale genetic partitioning between geographical subregions. When running DAPC on all individuals, we observed three distinct clusters corresponding to individuals from Mindanao, Bohol and Samar/Leyte (Fig. 3).
Figure 3.
Discriminant Analysis of Principal Components (DAPC) results. The Bayesian information criterion (BIC) model selection supported three genetic clusters (inset, bottom left). Coloration of points follows Figure 1. The bar chart on the right provides a STRUCTURE-like plot of the DAPC cluster assignment for all individuals.
Discussion
When comparing diversification in Sarcophanops with other endemic fauna from the Mindanao PAIC, we observe that many taxa show a similar pattern of differentiation. For example, in Cyrtodactylus geckos (Welton et al., 2010) and Crocidura shrews (Esselstyn et al., 2009) the Visayan and Mindanao populations form independent genetic clusters, which is consistent with our phylogenomic and population genetic analyses, which recover a deep split between the Mindanao (S. steerii) and Visayan (S. samarensis) species. Recently published findings based on Bayesian species delimitation of mitochondrial DNA sequence data also revealed the same deep split between Mindanao and Visayan species (Hosner et al., 2018) but failed to identify a signature of divergence within S. samarensis that we found here. The well-supported phylogenetic split between the Mindanao and Visayan species in both the mitochondrial DNA (mtDNA) and nuclear DNA suggests that they remained isolated during the LGM, despite the fact all these islands formed a single contiguous island, the Mindanao PAIC (Fig. 1). It is possible that Pleistocene isolation of this nature relates to the role of environmental suitability. Based on palaeoclimate projections, Hosner et al. (2014) found that the shallow Leyte Gulf (the land bridge uniting the northern and southern islands of the Mindanao PAIC) was unsuitable for most species in their study and still acted as a barrier to gene flow despite increased land connectivity. Although we did not perform niche modelling in the present study, the Leyte Gulf could also have been unsuitable habitat for Sarcophanops, thus facilitating the divergence of Mindanao and Visayan populations.
In our study, RADseq data revealed fine-scale inter-island diversification within the Visayan broadbills, which was not evident in mtDNA alone (Hosner et al., 2018). This suggests that the shallow split between Bohol and Samar/Leyte is rather recent, possibly post-LGM. Single nucleotide polymorphism-based genetic structure (Fig. 3) revealed a high probability of two distinct populations within S. samarensis: Samar/Leyte and Bohol. Although the identification of genetically distinct populations in different subregions of the Mindanao PAIC is not unprecedented (Hosner et al. 2018), this is interesting given that there are no current subspecific taxa from the Visayan Islands. In contrast, Mindanao contains two described subspecies (S. steerii sterrii and S. steerii mayri), but we recovered only one S. steerii population in the RADseq dataset, with no evidence to support separation of the Zamboanga population, as seen in the mtDNA dataset. The estimate of FST between Mindanao and Zamboanga populations could be inflated owing to the small (N = 1) sample size for the Zamboanga population (Campagna et al., 2015), suggesting that there could be even less population structure than reported herein. There are not, however, any modern genetic samples available from Dinagat/Siargao Islands, part of the described range of S. s. mayri; therefore, we cannot confidently claim that there are not any distinct S. steerii populations missed owing to our sampling.
Although all present-day islands in the Visayas were connected at one point during the LGM, the narrow (0.8–1.6 km) and shallow (maximum 20 m) San Juanico Strait separating Samar and Leyte probably extended terrestrial connectivity between these two islands longer relative to other neighbouring islands in the Mindanao PAIC. Rising sea levels at the end of the Pleistocene would have isolated Bohol first, whereas prolonged connectivity between Samar and Leyte could have promoted gene flow, thus obscuring population genetic effects of inter-island diversification. This pattern is not unique to broadbills and can be observed in many birds endemic to this area (Hosner et al., 2018). Given that little is known about the current population status of these birds, and because little appropriate forested habitat remains on Bohol in particular, understanding the genetic connectivity across the Visayan Islands is an important contribution towards addressing the conservation needs of this enigmatic genus properly.
Conclusions
Numerous studies have investigated the effect of PAICs in generating endemism in the Philippines (for review, see Brown et al., 2013). Nevertheless, the nature of those studies has provided a limited understanding of recent, between-island differentiation. Focusing on an endemic lineage restricted to a single and well-established island group, we were able to recover both deep and subtle genetic differentiation between islands. Although this differentiation was not well supported in the ‘fast evolving’ mtDNA (Hosner et al., 2018), we suggest that the two, previously undocumented, Visayan lineages probably arose after the LGM and are therefore detectable only in a deep, genome-wide scan of thousands of loci using a method such as RADseq. This study represents a step forward in understanding subtle genetic differentiation between recently isolated populations, not limited to island populations, that has been undocumented owing to past genomic sampling constraints.
SUPPORTING INFORMATION
Additional Supporting Information may be found in the online version of this article at the publisher’s web-site:
Figure S1. MrBayes tree for 50CM dataset.
Figure S2. RAxML tree for 50CM dataset.
Figure S3. MrBayes tree for 75CM dataset.
Table S1. Summary statistics of all samples.
Table S2. Testing different parameters in STACKS and their effects on number of loci, genetic diversity within populations and genetic differentiation between populations.
Table S3. BLAST results to zebra finch genome.
Table S4. Number of private, shared and fixed polymorphisms for each population (above) or for the two main clades (i.e. each species; below).
Table S5. STRUCTURE results for K = 3
Table S6. STRUCTURE results for K = 4
Table S7. STRUCTURE results for K = 5
ACKNOWLEDGEMENTS
We would like to thank Mark Robbins from the University of Kansas Natural History Museum. We thank the KU Genome Sequencing Core (supported by US National Institutes of Health grant 5P20GM103638 to E. A. Lundquist) and the KU Advanced Computing Facility (partially funded by National Science Foundation grant CNS 1337899 to A. T. Peterson). The National Science Foundation (DEB-0743491; DEB-1418895), American Ornithologists’ Society, American Museum of Natural History Chapman Fund and the University of Kansas Panorama Fund supported fieldwork; the National Science Foundation (DEB-1110619; DEB-1557053) and the University of Kansas Graduate Student Research Fund supported laboratory work. We would like to thank John A. Allen and two anonymous reviewers for their comments that helped to improved our manuscript. L.C.C. and J.D.M. contributed equally to this work.
References
- Andolfatto P, Davison D, Erezyilmaz D, Hu TT, Mast J, Sunayama-Morita T, Stern DL. 2011. Multiplexed shotgun genotyping for rapid and efficient genetic mapping. Genome Research 21: 610–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BirdLife International . 2017. IUCN red list for birds. Available at: http://www.birdlife.org [Google Scholar]
- Brown RM, Siler CD, Oliveros CH, Esselstyn JA, Diesmos AC, Hosner PA, Linkem CW, Barley AJ, Oaks JR, Sanguila MB, Welton LJ, Blackburn DC, Moyle RG, Townsend Peterson A, Alcala AC. 2013. Evolutionary processes of diversification in a model island archipelago. Annual Review of Ecology, Evolution, and Systematics 44: 411–435. [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campagna L, Gronau I, Silveira LF, Siepel A, Lovette IJ. 2015. Distinguishing noise from signal in patterns of genomic divergence in a highly polymorphic avian radiation. Molecular Ecology 24: 4238–4251. [DOI] [PubMed] [Google Scholar]
- Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA. 2013. Stacks: an analysis tool set for population genomics. Molecular Ecology 22: 3124–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diesmos AC, Brown RM, Alcala AC, Sison RV, Afuang LE, Gee GV. 2002. Philippine amphibians and reptiles: an overview of species diversity, biogeography, and conservation. Philippine biodiversity conservation priorities: a second iteration of the National Biodiversity Strategy and Action Plan, 26–44. [Google Scholar]
- Earl DA, vonHoldt BM. 2012. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources 4: 359–361. [Google Scholar]
- Esselstyn JA, Timm RM, Brown RM. 2009. Do geological or climatic processes drive speciation in dynamic archipelagos? The tempo and mode of diversification in Southeast Asian shrews. Evolution 63: 2595–2610. [DOI] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology 14: 2611–2620. [DOI] [PubMed] [Google Scholar]
- Heaney L. 1985. Zoogeographic evidence for middle and late Pleistocene landbridges to the Philippine Islands. Modern Quaternary Research in Southeast Asia 9: 127–143. [Google Scholar]
- Hosner PA, Campillo LC, Andersen MJ, Sánchez-González LA, Oliveros CH, Urriza RC, Moyle RG. 2018. An integrative species delimitation approach reveals fine-scale endemism and substantial unrecognized avian diversity in the Philippine Archipelago. Conservation Genetics 19: 1153–1168. [Google Scholar]
- Hosner PA, Nyári ÁS, Moyle RG. 2013. Water barriers and intra-island isolation contribute to diversification in the insular Aethopyga sunbirds (Aves: Nectariniidae). Journal of Biogeography 40: 1094–1106 [Google Scholar]
- Hosner PA, Sánchez-González LA, Peterson AT, Moyle RG. 2014. Climate-driven diversification and Pleistocene refugia in Philippine birds: evidence from phylogeographic structure and paleoenvironmental niche modeling: Philippine avian phylogeography. Evolution 68: 2658–2674. [DOI] [PubMed] [Google Scholar]
- Jombart T. 2008. Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24: 1403–1405. [DOI] [PubMed] [Google Scholar]
- Jombart T, Devillard S, Balloux F. 2010. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genetics 11: 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jombart T, Ahmed I. 2011. Adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27: 3070–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy RS, Gonzales PC, Dickinson EC, Miranda HC, Fisher TH. 2000. A guide to the birds of the Philippines. Oxford; New York: Oxford University Press. [Google Scholar]
- Manthey JD, Moyle RG. 2015. Isolation by environment in white-breasted nuthatches (Sitta carolinensis) of the Madrean Archipelago sky islands: a landscape genomics approach. Molecular Ecology 24: 3628–3638. [DOI] [PubMed] [Google Scholar]
- Miller MR, Dunham JP, Amores A, Cresko WA, Johnson EA. 2007. Rapid and cost-effective polymorphism identification and genotyping using restriction site associated DNA (RAD) markers. Genome Research 17: 240–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyle RG, Chesser RT, Prum RO, Schikler P, Cracraft J. 2006. Phylogeny and evolutionary history of Old World suboscine birds (Aves: Eurylaimides). American Museum Novitates 3544: 1–22. [Google Scholar]
- Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GA, Kent J. 2000. Biodiversity hotspots for conservation priorities. Nature 403: 853–858. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . 2013. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Sánchez-González LA, Hosner PA, Moyle RG. 2015. Genetic differentiation in insular lowland rainforests: insights from historical demographic patterns in Philippine birds. PLoS ONE 10: e0134284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-González LA, Moyle RG. 2011. Molecular systematics and species limits in the Philippine fantails (Aves: Rhipidura). Molecular Phylogenetics and Evolution 61: 290–299. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. 2011. PAUP*: phylogenetic analysis using parsimony, version 4.0b10. Sunderland: Sinauer Associates. [Google Scholar]
- Welton LJ, Siler CD, Linkem CW, Diesmos AC, Brown RM. 2010. Philippine bent-toed geckos of the Cyrtodactylus agusanensis complex: multilocus phylogeny, morphological diversity, and descriptions of three new species. Herpetological Monographs 24: 55–85. [Google Scholar]
- Willing EM, Dreyer C, van Oosterhout C. 2012. Estimates of genetic differentiation measured by FST do not necessarily require large sample sizes when using many SNP markers. PLoS ONE 7: e42649. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.