Abstract
Serological determination of hepatitis C virus (HCV) subtypes has been hampered by the lack of suitable assays. Therefore, a recombinant immunoblot assay has been established for serological differentiation of HCV subtypes 1a, 1b, 2a, 2b, 3a, and 4a. It consists of recombinant HCV proteins from the NS-4 region propagated in Escherichia coli. To confirm the serotyping assay results, the results were compared with those obtained by nucleotide sequencing of the NS-5 region. Sera from 157 patients with chronic HCV infection were examined by this assay, and specific antibodies could be detected in 86% (n = 135) of them. The HCV genotype was determined correctly in all but one sample, and the subtypes determined by the serotyping assay corresponded to the HCV subtypes detected by nucleotide sequencing for 95% (n = 128) of the samples. These data indicate that HCV subtypes can be distinguished serologically. The assay that is described provides an easier means of identification of infection with different HCV subtypes for wider clinical and epidemiological applications.
Hepatitis C virus (HCV) is a single-stranded RNA virus of about 9,500 bp which is assumed to cause chronic hepatitis in more than 90% of HCV-infected individuals (13). HCV has been classified into at least six major genotypes and a number of more closely related subtypes (15, 24, 33, 41). Although some studies fail to find a correlation between HCV genotype and clinical outcome (29, 46), others demonstrate that the subtype of the infecting HCV strain seems to influence the clinical course of the infection as well as the outcome of therapy with alpha interferon (19, 27, 40). Therefore, several different tests like PCR with genotype-specific primers, restriction fragment length polymorphism assay, or hybridization techniques have been developed for determination of HCV genotypes (1, 18, 21, 23, 25). An advantage of serological tests such as the immunoblot assay or the enzyme-linked immunosorbent assay (ELISA) is their easy performance. The main problem when developing serological tests for the purpose of genotyping arises from the need not only for protein sequences with antigenic properties but also for protein sequences with type-specific properties.
Several different parts of the HCV genome, namely, the 5′ noncoding region (3, 7, 8, 37), the core region (5, 7, 8), the envelope region (7, 34, 38), the NS-3 region (4, 8), the NS-4 region (35, 42), and the NS-5 region (10, 20, 30, 33), have been shown to be appropriate for use in the grouping of HCV isolates into different types by nucleotide sequencing. Epitope mapping of the NS-4 region has revealed the existence of antigenic determinants in regions with considerable variability between the different genotypes of HCV (35, 43). ELISAs with peptides derived from these regions have been used to detect antibodies specific for different HCV genotypes (2, 35, 42, 43).
Until now HCV subtypes could not be distinguished by commercially available serological tests. Therefore, we have established an immunoblot assay based on subtype-specific recombinant proteins derived from the NS-4 region (NS-4 IBA). This assay allows determination of HCV subtypes 1a, 1b, 2a, 2b, 3a, and 4a, which are the most common in Western Europe and the United States (19). The subtypes determined from the results obtained by the NS-4 IBA were compared to the subtypes determined by sequencing of a part of the NS-5 region by following the classification proposed by Simmonds et al. (33).
MATERIALS AND METHODS
Patients.
Serum samples were collected from 147 patients who had chronic HCV infections and who lived around the city of Hamburg, Germany. Ten of them were born in Egypt and had acquired HCV infection prior to immigration into Germany. All of them tested positive by a second-generation ELISA (Abbott Laboratories, North Chicago, Ill.) for HCV, and antibody reactivity was confirmed by an in-house recombinant immunoblot assay (12). HCV viremia was proven by PCR as described previously (11). None of the patients was treated with alpha interferon at the time of this investigation. A group of 30 individuals who had no clinical or biochemical signs of liver disease and who tested negative by the second-generation ELISA for HCV, HCV immunoblot assay for HCV, and PCR for HCV served as negative controls. Serum samples from patients infected with HCV subtypes 2a and 2b were, in part, provided by courtesy of the National Reference Laboratory for HCV in Essen, Germany.
Nucleotide sequencing of a region within the NS-5 gene for genotyping.
HCV RNA extraction was performed by a modified guanidinium-thiocyanate-phenol-chloroform method as described previously (11). The isolated RNA was resuspended in 50 μl of diethyl pyrocarbonate-treated H2O. For cDNA synthesis 50 pmol of primer 51 (5′-AGTCATAGCCTCCGTGAA-3′; nucleotide positions 8290 to 8273, as described previously [9]) and 200 U of Moloney murine leukemia virus reverse transcriptase (Superscript; BRL-Life Technologies, Gaithersburg, Md.) were added to 11 μl of RNA. After reverse transcription, amplification of the HCV cDNA was performed with 5 μl of cDNA in a buffer containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 2 mM MgCl2, 160 μM (each) deoxynucleotide triphosphates, 30 pmol of each sense or antisense primer, and 2 U of Pfu thermostabile DNA polymerase (Stratagene, La Jolla, Calif.). For the first round of the nested PCR, primers 50 (5′-ATGGGGCAAAGGACGTCCG-3′; positions 7567 to 7585) and 51 were used. Five microliters of the first-round product was used in a second amplification step with primers 52 (5′-ACTGAATTCTCGTATGATACCCGC-3′; positions 7911 to 7925) and 53 (5′-GTCAAGCTTCACAGATAACG-3′; positions 8233 to 8222). The amplification products were purified by agarose gel electrophoresis and were cloned into pBluescript II (Stratagene). After transformation into Escherichia coli DH5α (BRL-Life Technologies), the nucleotide sequences were determined by the dideoxy chain termination method with modified T7 DNA polymerase (Sequenase version 2.0 kit; United States Biochemical Corp., Cleveland, Ohio). The sequences were analyzed and the percentages of similarity were calculated by using the BESTFIT program of the GCG program package developed at the University of Wisconsin and provided by the German Cancer Research Center (DKFZ), Heidelberg, Germany. Genotypes were determined by following the classification proposed by Simmonds et al. (33).
Establishment of the NS-4 IBA. (i) PCR amplification of NS-4 sequences.
Serum specimens containing different HCV genotypes were selected, and the sequences of the NS-4 of HCV RNA region were isolated. Therefore, HCV RNA extraction was performed by a modified guanidinium-thiocyanate-phenol-chloroform method as described previously (11). Eleven microliters of the RNA was reverse transcribed with primer 48 (5′-GTCAAGCTTTTATTCCACATGTGCTT-3′; positions 5642 to 5629), and 5 μl of the cDNA was amplified by nested PCR with primers 48 and 41 (5′-ACCGAATTCACGAAATACATC-3′; positions 5269 to 5280) in the first round. A second amplification step was performed with 5 μl of the first-round product with inner primers 43 (5′-CTTCGGATCCTACAACAGGCAGCGTG-3′; positions 5368 to 5382) and 44 (5′-GAGACTGCAGTTACTGTTTGAACTGCT-3′; positions 5513 to 5498). The nucleotide sequences of the amplification products were determined as described above to check for correctness.
(ii) Expression and purification of recombinant NS-4 proteins.
For expression of purified NS-4, DNA fragments were cloned into the pTrxFus expression vector (Invitrogen, San Diego, Calif.). After transformation into E. coli GI724 (Invitrogen), further steps were performed as recommended by the manufacturer. Briefly, cells were cultured in tryptophan-free medium (2% Casamino Acids, 1% glycerol, 1 mM MgCl2, 1× M9 salts, 100 μg of ampicillin per ml) overnight at 30°C. A total of 500 μl of each culture was inoculated into 10 ml of fresh medium and was allowed to grow at 30°C to an optical density at 550 nm of 0.5. Tryptophan was added to a final concentration of 100 μg/ml, and the induced cultures were propagated at 37°C for another 4 h. For protein extraction, the cells were lysed by ultrasonification at 100 W in eight cycles of 20 s each on ice. The samples were centrifuged at 12,000 × g for 5 min at 4°C to pellet the cell debris, and the supernatants were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. HCV fusion proteins were separated from other bacterial proteins by affinity chromatography with Thiobond Resin columns (Invitrogen). Four milliliters of the supernatants was mixed with 2 ml of Thiobond Resin, and the mixture was rocked for 30 min at room temperature. The settled resins were washed twice with a buffer consisting of 50 mM Tris-HCl (pH 7.4), 1 mM EDTA, 200 mM NaCl, and 1 mM 2-mercaptoethanol. Elution of the HCV fusion proteins was performed with washing buffer containing 100 mM 2-mercaptoethanol. The purities of the fusion proteins were confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Immunoblots.
Five nanograms of each soluble type-specific HCV fusion protein was transferred onto a polyvinylidene difluoride membrane (Millipore, Eschborn, Germany). The immunoblot assay was performed as described previously (12). Forty nanograms and 15 ng of immunoglobulin G from a standard HCV-negative serum sample (Behring, Marburg, Germany) was applied to each strip and served as an internal control.
In the first step, sera were tested in their native state to determine the HCV genotype. Most of the sera from subtype 1a- or subtype 1b-infected patients contained antibodies that reacted with epitopes of both the subtype 1a and subtype 1b recombinant proteins. The sera from subtype 2a- and subtype 2b-infected patients contained antibodies which also reacted with the subtype 2a and the subtype 2b recombinant proteins. To detect those antibodies which are directed against the subtype-specific epitopes of the respective recombinant proteins, serum samples needed to be preabsorbed with a solution that contained a surplus of subtype 1a and subtype 2a recombinant proteins. All sera were tested by the NS-4 IBA before and after preabsorption, and the patterns of reactivity were compared as described recently (32). After preabsorption, HCV subtypes could definitely be determined by the detection of the reactivity against a single subtype-specific recombinant protein that remained.
Identical results were always obtained when different batches were used for immunoblotting.
Nucleotide sequence accession numbers.
All new sequences described in this report have been submitted to the EMBL gene bank and have been given accession nos. Z35502 to Z35594 and X88564 to X88607.
RESULTS
The amino acid sequences of the type-specific NS-4 recombinant proteins that we had chosen for our NS-4 IBA were compared to those published for the HCV prototype strains (6, 9, 15, 22, 23, 45). Each of the proteins used in the NS-4 IBA shared more than 90% identity at the amino acid level with the corresponding reference isolate (data not shown).
A comparison between the peptides of the isolates used in this assay revealed differences that ranged from 18% between genotypes 1 and 4 to 68% between genotypes 2 and 3. The amino acid sequences of subtypes of genotypes 1 and 2 differed by 18 and 20%, respectively (Fig. 1). Of 157 serum samples from patients with chronic HCV infection, serological typing was possible for 86% (n = 135) (Table 1). The remaining samples which could not be classified by the NS-4 IBA also exhibited either low antibody titers in the second-generation ELISA or no reactivity against the NS-4 region at all. The latter was confirmed by a recently described antibody test containing four recombinant proteins from different regions of HCV (12).
FIG. 1.
Comparison of amino acid sequences of recombinant proteins used for NS-4 IBA. Bars indicate identical amino acids between subtypes 1a and 1b and between subtypes 2a and 2b. Asterisks indicate amino acids that are conserved among all isolates. Dots within the amino acid sequences indicate deletions.
TABLE 1.
Comparison of HCV subtypes determined by NS-4 IBA and nucleotide sequencinga
| Subtype by nucleotide sequencing | No. of isolates of the following subtype by serotyping:
|
|||||
|---|---|---|---|---|---|---|
| 1a | 1b | 2a | 2b | 3a | 4a | |
| 1a | 37 | 4 | ||||
| 1b | 2 | 63 | 1 | |||
| 2a | 5 | |||||
| 2b | 5 | |||||
| 3a | 9 | |||||
| 4a | 9 | |||||
Of 135 isolates, 128 (95%) had identical results by both methods.
A positive result by the NS-4 IBA could not be observed for any of the HCV-negative control samples.
All sera were tested in parallel in their native state and after absorption, and in all 135 serum samples that reacted in the NS-4 IBA, the HCV subtype could definitely be determined (Fig. 2). Serotype 1, with subtypes 1a and 1b, was detected in 78.5% (n = 106) of the serum samples tested. HCV serotype 2, with subtypes 2a and 2b, was detected by the immunoblot assay in 7.4% (n = 10) of the serum samples tested. Serotype 3a could be detected in 6.7% (n = 9) of the serum samples. All of serotype 3a-positive samples were derived from patients who were intravenous drug users. HCV serotype 4a was detected in 7.4% (n = 10) of the serum samples tested. The serotype 4a-positive samples were derived from Egyptian patients who had acquired HCV infection prior to their immigration into Germany. A comparison of the subtypes obtained by nucleotide sequencing and the immunoblot assay revealed identical results by either method for 95% (n = 135) of the samples. Only six samples were serologically assigned to a different subtype by the immunoblot assay compared to that to which it was assigned by nucleotide sequencing. Of the serum specimens containing HCV subtype 1a or 1b, two were classified as subtype 1b by sequencing but as subtype 1a by NS-4 IBA, whereas four were classified as subtype 1a by sequencing but as subtype 1b by immunoblot assay. The serum specimens containing either HCV subtype 2a or 2b were all correctly typed by the serological assay.
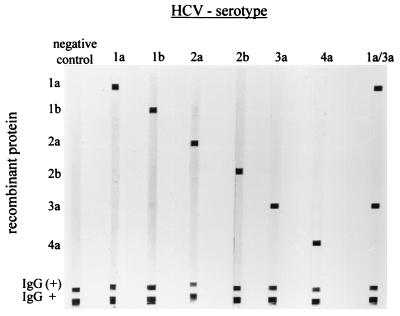
FIG. 2.
Pattern of reactivity by NS-4 IBA with serum samples from patients infected with different HCV subtypes, as described in the Materials and Methods section. HCV subtypes are determined by the antibody reactivities against the respective subtype-specific recombinant protein. Lane 1a/3a, a serum specimen derived from a patient who was sequentially infected with two strains with different HCV subtypes (subtypes 1a and 3a). Antibodies against both subtypes can clearly be detected. IgG, immunoglobulin G.
Regarding the HCV genotypes, total agreement between the results obtained by the serological assay and those obtained by nucleotide sequencing could be observed for all but one sample (Table 1). The sample with discrepant results originated from an Egyptian patient and was type 4a by the serological assay but was type 1b by nucleotide sequencing.
DISCUSSION
Although different methods for determination of HCV subtypes have been established, determination of HCV subtypes is still very laborious (1, 18, 21, 23, 25) and hitherto has required PCR. For the first time we present an immunoblot assay for the serological differentiation of HCV genotypes and subtypes on the basis of the use of recombinant proteins derived from the NS-4 region of HCV. The amino acid sequence of this region has been shown to contain considerable differences in isolates of different HCV genotypes and subtypes (2, 32). By following the classification proposed by Simmonds et al. (33), distinct HCV genotypes were found to share sequence identities of no more than 70%. The more closely related subtypes of a certain genotype shared identities of 74 to 81%, and different isolates of the same subtype had identities of 87 to 98%. Although the percent homology may vary depending on which region of the HCV genome is examined, the classification into genotypes and subtypes can readily be obtained by use of sequences from throughout large parts of the entire genome (36). However, the amino acid sequence of the region used for the assay described here is very similar in subtypes 1b and 4a. This might be the reason for the cross-reactivity that can occasionally be observed between these subtypes when serum specimens are tested in their native state. However, preabsorption of sera, which was necessary to distinguish subtypes, allowed differentiation of subtypes 1b and 4a. It has been described earlier that a high percentage of viruses which are serologically type 4 can be classified as type 1b by PCR-based genotyping assays (26). In our study only one sample determined by serological assay to contain genotype 4 virus contained a virus of a different type by nucleotide sequencing. This sample was derived from one of the Egyptian immigrants who had acquired HCV infection before moving to Germany. Since HCV subtype 4a is the most predominant subtype in Egypt and the Middle East, we cannot exclude the possibility of sequential infection with different HCV strains in this patient. Serological assays like the NS-4 IBA have the potential to detect double or sequential infections. It has been shown that in patients who were known to be infected with two HCV strains of different subtypes, only the prevailing strain can be detected by PCR (44). However, even when one subtype is suppressed, antibodies against both strains may remain detectable for months. Further studies must be performed to highlight the value of serological subtyping assays for the detection of sequential infections with different HCV strains.
One problem with serotyping assays may arise from missing reactivity due to low levels of antibody production in patients with immunosuppressing conditions (31). We found no reactivity of the NS-4 IBA with 14% of the samples tested. On the other hand, the serological assay described here has the advantage of allowing HCV subtyping of virus in samples with levels of HCV viremia below the detection limit of PCR. It has been demonstrated earlier that various PCR-based genotyping assays fail to determine HCV subtypes in up to 16.5% of samples due to low-level viremia (16).
Genomic sequence analysis of HCV isolates derived from 447 German patients revealed that more than 90% were infected with HCV subtype 1a or subtype 1b (14). By the immunoblot assay described here, distinct patterns of reactivity between HCV subtypes 1a and 1b were obtained. Discordant results at the subtype level between the NS-4 IBA and nucleotide sequencing occurred for only six (5%) samples. One reason for the discrepancies might be that different genomic regions share high degrees of homology with different HCV subtypes. While serotyping is performed with proteins from the NS-4 region, proteins from the NS-5 region are used for nucleotide sequencing. However, a consistent feature of studies on this topic is that the sequence relationships between subgenomic regions always reflect those of the complete genome (36, 39). Examination of samples with different results by genotyping and serotyping assays revealed only a few amino acid substitutions in the NS-4 region that might have accounted for the discrepancies (28).
It has been shown earlier that differences in genotypes between serological and PCR-based methods are predominantly found for isolates in samples from individuals with multiple exposures to different HCV types. It remains unclear whether the detection of antibodies in such samples corresponds to double infection or to previous expression of a genotype different from that detected by PCR (28). Double infections with different HCV genotypes have been found in 1 to 20% of HCV-infected patients. A higher degree of double infections is often found by PCR with genotype-specific primers (16, 17), restriction fragment length polymorphism analysis of PCR products (21, 22), or PCR followed by hybridization with specific oligonucleotides (1, 37). However, mutations in the initial infecting HCV strain or incorrect incorporation of nucleotides during PCR or reverse transcription could lead to an overestimation of the prevalence of coinfection with different HCV strains when genotyping is performed by the methods mentioned above. The results of these tests would more likely be influenced by point mutations than would the results of genotyping by nucleotide sequencing or serological assays. In one serum sample, antibody reactivity against the subtype 3a and additionally against the subtype 1a recombinant protein was detected by NS-4 IBA. The genotype was determined to be 3a by nucleotide sequencing. This sample was derived from a patient who was an intravenous drug user. It is well known that both subtypes 1a and 3a are found in a high percentage of intravenous drug users (14). This patient had a high risk of multiple exposures to HCV, and sequential infection with HCV strains of different subtypes had to be assumed. Therefore, serum samples which had been drawn up to 28 months prior to retrieval of the sample used in the present study were examined. It could be demonstrated by both sequencing and serological testing that the patient was initially infected with HCV subtype 1a. This demonstrates that the NS-4 IBA has the potential to detect double or sequential infections with different HCV strains.
The proteins of genotype 2 share homologies with the other genotypes of only about 40%. This type is common in Asia, and a certain percentage of strains in Italy, Spain, and other European countries are genotype 2 (19). In our collection of serum samples, genotype 2 seems to occur very seldom, as we have previously demonstrated (14). Therefore, some sera which were obtained from the National Reference Center for HCV had to be tested to examine the reliabilities of the subtype 2a and 2b recombinant proteins. All sera could be correctly typed as either HCV subtype 2a or HCV subtype 2b; however, only a small panel of serum specimens containing genotype 2 was available. The assay described here should be tested in an area with a high prevalence of HCV subtypes 2a and 2b to assess its value for determination of these subtypes in the daily routine.
In conclusion, a reliable assay for serological determination of HCV subtypes is described. The frequency of discrepant results between nucleotide sequencing and the immunoblot assay was very low, and the simplicity and rapidity of this serotyping assay suggest that it may be a suitable alternative for subtyping of HCV.
REFERENCES
- 1.Andonov A, Chaudhary R K. Subtyping of hepatitis C virus isolates by a line probe assay using hybridization. J Clin Microbiol. 1995;33:254–256. doi: 10.1128/jcm.33.1.254-256.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhattacherjee V, Prescott L E, Pike I, Rodgers B, Bell H, El-Zayadi A R, Kew M C, Conradie J, Lin C K, Marsden H, Saeed A A, Parker D, Yap P-L, Simmonds P. Use of NS-4 peptides to identify type-specific antibody to hepatitis C virus genotypes 1, 2, 3, 4, 5 and 6. J Gen Virol. 1995;76:1737–1748. doi: 10.1099/0022-1317-76-7-1737. [DOI] [PubMed] [Google Scholar]
- 3.Bukh J, Purcell R H, Miller R H. Sequence analysis of the 5′ non coding region of hepatitis C virus. Proc Natl Acad Sci USA. 1992;89:4942–4946. doi: 10.1073/pnas.89.11.4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bukh J, Purcell R H, Miller R H. At least 12 genotypes of hepatitis C virus predicted by sequence analysis of the putative E1 gene of isolates collected worldwide. Proc Natl Acad Sci USA. 1993;90:8234–8238. doi: 10.1073/pnas.90.17.8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cha T A, Beall E, Irvine B, Kolberg J, Chien D, Kuo G, Urdea M S. At least five related, but distinct, hepatitis C viral genotypes exist. Proc Natl Acad Sci USA. 1992;89:7144–7148. doi: 10.1073/pnas.89.15.7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chamberlain R W, Adams N, Saeed A A, Simmonds P, Elliott R M. Complete nucleotide sequence of a type 4 hepatitis C virus variant, the predominant genotype in the Middle East. J Gen Virol. 1997;78:1341–1347. doi: 10.1099/0022-1317-78-6-1341. [DOI] [PubMed] [Google Scholar]
- 7.Chan S W, McOmish F, Holmes E C, Dow B, Peutherer J F, Follet E, Yap P L, Simmonds P. Analysis of a new hepatitis C virus type and its phylogenetic relationship to existing variants. J Gen Virol. 1992;73:1131–1141. doi: 10.1099/0022-1317-73-5-1131. [DOI] [PubMed] [Google Scholar]
- 8.Chayama K, Tsubota A, Arase Y, Saitoh S, Koida I, Ikeda K, Matsumoto T, Kobayashi M, Iwasaki S, Koyama S, Morinaga T, Kumada H. Genotypic subtyping of hepatitis C virus. J Gastroenterol Hepatol. 1993;8:150–156. doi: 10.1111/j.1440-1746.1993.tb01507.x. [DOI] [PubMed] [Google Scholar]
- 9.Choo Q L, Richman K H, Han J H, Berger K, Lee C, Dong C, Gallegos C, Coit D, Medina Selby R, Barr P J, Weiner A J, Bradley D W, Kuo G, Houghton M. Genetic organization and diversity of hepatitis C virus. Proc Natl Acad Sci USA. 1991;88:2451–2455. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enomoto N, Takada A, Nakao T, Date T. There are two major types of hepatitis C virus in Japan. Biochem Biophys Res Commun. 1990;170:1021–1025. doi: 10.1016/0006-291x(90)90494-8. [DOI] [PubMed] [Google Scholar]
- 11.Feucht H H, Zöllner B, Laufs R. Comparison of conventional autoradiography with a new DNA enzyme immunoassay for the detection of hepatitis C virus-polymerase chain reaction amplification products. J Virol Methods. 1995;55:105–110. doi: 10.1016/0166-0934(95)00049-z. [DOI] [PubMed] [Google Scholar]
- 12.Feucht H H, Zöllner B, Polywka S, Laufs R. Study on the reliability of commercially available hepatitis C virus antibody tests. J Clin Microbiol. 1995;33:620–624. doi: 10.1128/jcm.33.3.620-624.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feucht H H, Zöllner B, Laufs R. Correlation of hepatitis C virus antibodies with viremia. Ann Intern Med. 1997;126:666–667. doi: 10.7326/0003-4819-126-8-199704150-00031. [DOI] [PubMed] [Google Scholar]
- 14.Feucht H H, Schröter M, Zöllner B, Polywka S, Nolte H, Laufs R. The influence of age on the prevalence of hepatitis C virus subtypes 1a and 1b. J Infect Dis. 1997;175:685–688. doi: 10.1093/infdis/175.3.685. [DOI] [PubMed] [Google Scholar]
- 15.Kato N, Hijikata M, Ootsuyama Y, Nakagawa M, Ohkoshi S, Sugimura T, Shimotohno K. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc Natl Acad Sci USA. 1990;87:9524–9528. doi: 10.1073/pnas.87.24.9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lau J Y N, Mizokami M, Kolberg J A, Davis G L, Prescott L E, Ohno T, Perillo R P, Lindsay K L, Gish R G, Qian K-P, Kohara M, Simmonds P, Urdea M S. Application of six hepatitis C virus genotyping systems to sera from chronic hepatitis C patients in the United States. J Infect Dis. 1995;171:181–189. doi: 10.1093/infdis/171.2.281. [DOI] [PubMed] [Google Scholar]
- 17.Matsubara T, Sumazaki R, Shin K, Nagai Y, Takita H. Genotyping of hepatitis C virus: coinfection by multiple genotypes detected in children with chronic posttransfusion hepatitis C. J Pediatr Gastroenterol Nutr. 1996;22:79–84. doi: 10.1097/00005176-199601000-00013. [DOI] [PubMed] [Google Scholar]
- 18.McOmish F, Chan S-W, Dow B C, Gillon J, Frame W D, Crawford R J, Yap P L, Follet E A C, Simmonds P. Detection of three types of hepatitis C virus in blood donors: investigation of type-specific differences in serologic reactivity and rate of alanine aminotransferase abnormalities. Transfusion. 1993;33:7–13. doi: 10.1046/j.1537-2995.1993.33193142314.x. [DOI] [PubMed] [Google Scholar]
- 19.McOmish F, Yap P L, Bow B C, Follet E A C, Seed C, Keller A J, Cobain T J, Krusius T, Kolho E, Naukkarinen R, Lin C, Lai C, Leong S, Medgyesi G A, Hejjas M, Kiyokawa H, Fukuda K, Cuypers T, Saeed A A, Al-Rasheed A M, Lin M, Simmonds P. Geographical distribution of hepatitis C virus genotypes in blood donors: an international collaborative survey. J Clin Microbiol. 1994;32:884–892. doi: 10.1128/jcm.32.4.884-892.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mori S, Kato N, Yagyu A, Tanaka T, Ikeda Y, Petchclai B, Chiewsilp P, Kurimura T, Shimotohno K. A new type of hepatitis C virus in patients in Thailand. Biochem Biophys Res Commun. 1992;183:334–342. doi: 10.1016/0006-291x(92)91648-a. [DOI] [PubMed] [Google Scholar]
- 21.Nakao T, Enomoto N, Takada N, Takada A, Date T. Typing of hepatitis C virus genomes by restriction fragment length polymorphism. J Gen Virol. 1991;72:2105–2112. doi: 10.1099/0022-1317-72-9-2105. [DOI] [PubMed] [Google Scholar]
- 22.Okamoto H, Okada S, Sugiyama Y, Kurai K, Iizuka H, Machida A, Miyakawa Y, Mayumi M. Nucleotide sequence of the genomic RNA of hepatitis C virus isolated from a human carrier: comparison with reported isolates for conserved and divergent regions. J Gen Virol. 1991;72:2697–2704. doi: 10.1099/0022-1317-72-11-2697. [DOI] [PubMed] [Google Scholar]
- 23.Okamoto H, Kurai K, Okada S, Yamamoto K, Lizuka H, Tanaka T, Fukuda S, Tsuda F, Mishiro S. Full-length sequence of a hepatitis C virus genome having poor homology to reported isolates: comparative study of four distinct genotypes. Virology. 1992;188:331–341. doi: 10.1016/0042-6822(92)90762-e. [DOI] [PubMed] [Google Scholar]
- 24.Okamoto H, Sugiyama Y, Okada S, Kurai K, Akahane Y, Sugai Y, Tanaka T, Sata K, Tsuda F, Miyakawa M. Typing hepatitis C virus by polymerase chain reaction with type specific primers: application to clinical surveys and tracing infectious sources. J Gen Virol. 1992;73:673–679. doi: 10.1099/0022-1317-73-3-673. [DOI] [PubMed] [Google Scholar]
- 25.Okamoto H, Tokita H, Sakamoto M, Horikita M, Kojima M, Iizuka H, Mishiro S. Characterization of the genomic sequence of type V (or 3a) hepatitis C virus isolates and PCR primers for specific detection. J Gen Virol. 1993;74:2385–2390. doi: 10.1099/0022-1317-74-11-2385. [DOI] [PubMed] [Google Scholar]
- 26.Pawlotsky J M, Prescott L, Simmonds P, Pellet C, Laurent-Puig P, Labonne C, Darthuy F, Remire J, Duval J, Buffet C, Etienne J P, Dhumeaux D, Dussaix E. Serological determination of hepatitis C virus genotype: comparison with a standardized genotyping assay. J Clin Microbiol. 1997;35:1734–1739. doi: 10.1128/jcm.35.7.1734-1739.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prati D, Capelli C, Zanella A, Mozzi F, Bosoni P, Pappalettera M, Zanuso F, Vianello L, Locatelli E, De Fazio C, Ronchi G, Del Ninno E, Colombo M, Sirchia G. Influence of different hepatitis C virus genotypes on the course of asymptomatic hepatitis C virus infection. Gastroenterology. 1996;110:178–183. doi: 10.1053/gast.1996.v110.pm8536854. [DOI] [PubMed] [Google Scholar]
- 28.Prescott L E, Berger A, Pawlotsky J M, Conjeevaram P, Pike I, Simmonds P. Sequence analysis of hepatitis C virus variants producing discrepant results with two different genotyping assays. J Med Virol. 1997;53:237–244. [PubMed] [Google Scholar]
- 29.Romeo R, Colombo M, Rumi M, Soffredini R, Del Ninno E, Donato M F, Russo A, Simmonds P. Lack of association between type of hepatitis C virus, serum load and severity of liver disease. J Viral Hepat. 1996;3:183–190. doi: 10.1111/j.1365-2893.1996.tb00093.x. [DOI] [PubMed] [Google Scholar]
- 30.Sakamoto M, Akahane Y, Tsuda F, Tanaka T, Woodfield D G, Okamoto H. Entire nucleotide sequence and characterization of a hepatitis C virus of genotype V/3a. J Gen Virol. 1994;75:1761–1768. [Google Scholar]
- 31.Schröter M, Feucht H H, Schäfer P, Zöllner B, Laufs R. High percentage of seronegative HCV-infections in hemodialysis patients: the need for PCR. Intervirology. 1997;40:277–278. doi: 10.1159/000150558. [DOI] [PubMed] [Google Scholar]
- 32.Schröter M, Feucht H H, Zöllner B, Schäfer P, Laufs R. Serological differentiation between HCV subtypes 1a and 1b by a recombinant immunoblot assay. Microbiol Immunol. 1998;42:387–391. doi: 10.1111/j.1348-0421.1998.tb02299.x. [DOI] [PubMed] [Google Scholar]
- 33.Simmonds P, Holmes E C, Cha T A, Chan S-W, McOmish F, Irvine B, Beall E, Yap P L, Kolberg J, Urdea M S. Classification of hepatitis C virus into six major genotypes and a series of subtypes by phylogenetic analysis of the NS-5 region. J Gen Virol. 1993;74:2391–2399. doi: 10.1099/0022-1317-74-11-2391. [DOI] [PubMed] [Google Scholar]
- 34.Simmonds P, McOmish F, Yap P L, Chan S W, Lin C K, Dusheiko G, Saeed A A, Holmes E C. Sequence variability of the 5′ non coding region of hepatitis C virus: identification of a new virus type and restriction on sequence diversity. J Gen Virol. 1993;74:661–668. doi: 10.1099/0022-1317-74-4-661. [DOI] [PubMed] [Google Scholar]
- 35.Simmonds P, Rose K A, Graham S, Chan S W, McOmish F, Dow B C, Follet E A C, Yap P L, Marsden H. Mapping of serotype-specific immunodominant epitopes in the NS-4 region of hepatitis C virus (HCV): use of type-specific peptides to serologically differentiate infections with HCV types 1, 2, and 3. J Clin Microbiol. 1993;31:1493–1503. doi: 10.1128/jcm.31.6.1493-1503.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simmonds P, Smith D B, McOmish F, Yap P L, Kolberg J, Urdea M S, Holmes E C. Identification of genotypes of hepatitis C virus by sequence comparisons in the core, E1 and NS-5 regions. J Gen Virol. 1994;75:1053–1061. doi: 10.1099/0022-1317-75-5-1053. [DOI] [PubMed] [Google Scholar]
- 37.Stuyver L, Vanarnhem W, Wyseur A, Deleys R, Maertens G. Analysis of the putative e1 envelope and NS-4a epitope regions of HCV type-3. Biochem Biophys Res Commun. 1993;192:635–641. doi: 10.1006/bbrc.1993.1462. [DOI] [PubMed] [Google Scholar]
- 38.Stuyver L, Rossau R, Wyseur A, Duhamel M, Vanderborght B, Van Heuverswyn H, Maertens G. Typing of hepatitis C virus isolates and characterization of new subtypes using a line probe assay. J Gen Virol. 1993;74:1093–1102. doi: 10.1099/0022-1317-74-6-1093. [DOI] [PubMed] [Google Scholar]
- 39.Stuyver L, Vanarnhem W, Wyseur A, Hernandez F, Delaporte E, Maertens G. Classification of hepatitis C viruses based on phylogenetic analysis of the envelope 1 and nonstructural 5b regions and identification of five additional subtypes. Proc Natl Acad Sci USA. 1994;91:10134–10138. doi: 10.1073/pnas.91.21.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takada A, Tsutsumi M, Zhang S C, Okanoue T, Matsushima T, Fujiyama S, Komatsu M. Relationship between hepatocellular carcinoma and subtypes of hepatitis C virus: a nationwide analysis. J Gastroenterol Hepatol. 1996;11:166–169. doi: 10.1111/j.1440-1746.1996.tb00055.x. [DOI] [PubMed] [Google Scholar]
- 41.Takamizawa A, Mori C, Fuke I, Manabe S, Murakami S, Fujita J, Onishi E, Andoh T, Yoshida I, Okayama H. Structure and organization of the hepatitis C virus genome isolated from human carriers. J Virol. 1991;65:1105–1113. doi: 10.1128/jvi.65.3.1105-1113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanaka T, Tsukiyama Kohara K, Yamaguchi K, Yagi S, Tanaka S, Hasegawa A, Ohta Y, Hattori N, Kohara M. Significance of specific antibody assay for genotyping of hepatitis C virus. Hepatology. 1994;19:1347–1353. [PubMed] [Google Scholar]
- 43.Tsukiyama Kohara K, Yamaguchi K, Maki N, Ohta Y, Miki K, Mizakami M, Ohba K, Tanaka S, Hattori V, Nomoto A, Kohara M. Antigenicities of group I and II hepatitis C virus polypeptides—molecular basis of diagnosis. Virology. 1993;192:430–437. doi: 10.1006/viro.1993.1058. [DOI] [PubMed] [Google Scholar]
- 44.Widell A, Mansson S, Persson N H, Thysell H, Hermodsson S, Blohme I. Hepatitis C superinfection in hepatitis C virus (HCV)-infected patients transplanted with an HCV-infected kidney. Transplantation. 1995;60:642–647. doi: 10.1097/00007890-199510150-00004. [DOI] [PubMed] [Google Scholar]
- 45.Yamada N, Tanihara K, Mizokami M, Ohba K, Takada A, Tsutsumi M, Date T. Full-length sequence of the genome of hepatitis C virus type 3a: comparative study with different genotypes. J Gen Virol. 1994;75:3279–3284. doi: 10.1099/0022-1317-75-11-3279. [DOI] [PubMed] [Google Scholar]
- 46.Zhou S, Terrault N A, Ferrell L, Hahn J A, Lau J Y, Simmonds P, Roberts J P, Lake J R, Ascher N L, Wright T L. Severity of liver disease in liver transplantation recipients with hepatitis C virus infection: relationship to genotype and level of viremia. Hepatology. 1996;24:1041–1046. doi: 10.1002/hep.510240510. [DOI] [PubMed] [Google Scholar]