Abstract
Amyloid fibril formation occurs in restricted environment, such as the interface between intercellular fluids and bio-membranes. Conformational interconversion from α-helix to β-structure does not progress in fluids; however, it can occur after sedimentary aggregation during amyloid fibril formation induced by heat treatment of hen egg white lysozyme (HEWL). Secondary structures of various proteins and denatured proteins titrated with 2,2,2-trifluoroethanol (TFE) were examined using their CD spectra. Gaussian peak/trough and singular value decompositions (SVD) showed that the spectral pattern of the α-helix comprised a sharp trough at wavelength 207 nm and a broad trough at 220 nm. Conversely, we distinguished two patterns for β-sheet—a spread barrel type, corresponding to ConA, and a tightly weaved type, corresponding to the soybean trypsin inhibitor. Herein, we confirmed that the spectral/conformational interconversion of the heat-treated HEWL was not observed in the dissolved fluid.
Keywords: Secondary structure, Singular value decomposition, Amyloid fibrils
Highlights
-
•
The application of singular value decomposition (SVD) of the CD spectrum to secondary protein structure analysis is a useful method.
-
•
We distinguished two patterns for β-sheet—a spread barrel type and a tightly weaved type.
-
•
We confirmed that the spectral/conformational interconversion of the heat-treated HEWL was not observed in the dissolved fluid.
1. Introduction
Alzheimer's dementia [1], Parkinson's extrapyramidal disorders [2], Huntington's chorea [3], type II diabetes [4], amyotrophic lateral sclerosis (ALS) [5], and other national-designated intractable cases are some of the examples of amyloid diseases; 40 types of human proteins associated with such diseases have been identified until 2018 [6], the occurrence of which is caused by irregular aggregation and localization induced by steric changes [7]. The corresponding rearrangements of such proteins is disturbed by any pathological fragmentation under conditions that deviate from environments suitable for metabolic pathways and physiological homeostasis [8]. Therefore, the mechanism underlaying the three-dimensional structural changes should be elucidated to better understand the onset processes in vivo, and develop treatment strategies against these diseases [9].
Amyloid fibrils and precursors are prepared via heat treatment and vigorous stirring, of which the yield is regulated by medium pH, concentration of electrolytes, and reductive condition [10]. The pH equivalent to the isoelectric point and ionic strength, sufficient to achieve a shielding effect, is predominantly responsible for protein denaturation and precipitation, thereby inducing the formation of amyloid fibrils of hen egg white lysozyme (HEWL) [11]. Furthermore, a preceding aggregation of this protein occurs and the secondary structure is rearranged.
Amyloid fibrils are formed due to the rewinding of α-helical or other coil structures to β-strand/sheet [12]. However, another study disagreed with the scheme where the most stable conformation in the aqueous solution was spontaneously reformed to an isolated β-strand [11]. If the protein misfolding is achieved, the segment will be antecedently affected by the environmental conversion to enforce the conformational change [13]. This scenario is essential for the formation of β-strand/sheet structures [14]. Furthermore, the association with other similarly changed fragments would likely lead to an amyloid fibril [15].
In a previous study, using a polarized attenuated total reflection Fourier transform infrared (ATR-FTIR) spectroscopy, the quantities of secondary structures from absorbance peaks for the amyloid fibril precipitate were assigned as α-helix at wavenumber 1654 cm−1, β-strand/sheet at 1633 and 1684 cm−1, and disordered segment at 1654 cm−1 [16,17]; it remains unclear as to whether aggregation or β-strand formation occurred first. Therefore, further investigations on the conformational changes that occur in solution/dispersion or in precipitation are warranted.
Based on the analysis of the secondary structure of a protein in solution, the far UV circular dichroism (CD) spectropolarimetry is invoked [18]. Woody et al. proposed the existence of CD spectral patterns specific to their artificial α-helix and β-strand, and those observed for their deemed “random coils” (namely, a disordered segment), using their poly lysyl chain [19]. We have preliminarily analyzed our CD spectra of proteins to be approximated as the linear combination of Woody's primitive patterns, and obtained general proportions of the artificial α-helical, β-strand/sheet, and “random coil” segments [20].
The CD spectrum is predominantly induced by the electronic transition on the amide π-system in the polypeptide backbone [21]. Each amide moiety has one particular space orientation depend on the formation of standard conformations by hydrogen bonds between main chain atoms, such as α-helices, β-strands/sheets, various turns, and disordered segments [22]. Because the molecular geometry of amide moieties in α-helices, β-strands/sheets, and several turn segments are constrained by hydrogen bonding with proton donners and/or acceptors in their adjacent residues, their electronic transitions track their routine. Therefore, the individual components of the CD spectra may comprise their characteristic patterns. In contrast, the three-dimensional transformation of amide moieties in a disordered segment is irregular and flexible [23], and hence the spectral components specific to amide moieties contained by the disordered segment should be ignored. In contrast, the pattern of the CD spectrum derived from the “random coil” deemed by Woody et al.'s proposal should only appear in structural consequence by chance, without causality toward the molecular geometries of amide moieties in a disordered segment.
In this study, we explore a predictive correspondence between the X-ray crystallographic structures and the apparent CD spectra of proteins. HEWL forms amyloid [11] comprises a hybrid sequence containing α-helical segments and a tiny β-strand [24]. Before inspecting the CD spectrum for HEWL, we identified models for available proteins under their peculiar folding, suitable for analyses. Bovine and human serum albumins (BSA and HSA, respectively) were examined as typical models predominantly comprising α-helices [25,26], whereas a legume lectin concanavalin A (ConA) was manipulated as the opposite example, comprising 14 β-strand components of antiparallel sheets in the diverse ligand-accessible barrel structure [27,28]. Trypsin inhibitors (TIs) and other enzyme inhibitors are products of legume seed protected from herbivore digestion for sowing through animal excretion. The protease tolerant protein TI comprises a robust and dense antiparallel β-sheet structure minutely woven with a three-fold symmetry [29].
Various protein structures are transformed into α-helices depending on the content of the proton donating solvent, 2,2,2-trifluoroethanol (TFE), believed to be a stabilizing reagent for α-helical structures in proteins, such as chemically modified lysozyme [30], β-lactoglobulin [31], sperm whale apomyoglobin [32], human acylphosphatase [33], β-barrel protein fibroblast growth factor [34], and thermophilic bacterial acylphosphatase [35]. The typical pattern of the CD spectrum for α-helix in HEWL was expanded by TFE (50% v/v) [36]. The titration of TFE could induce changes in ConA unfolding [37]. Furthermore, acetonitrile has been reported as a β-structure stabilizer of BSA [38] or viral protein [39], the protocol of which could be used for various proteins.
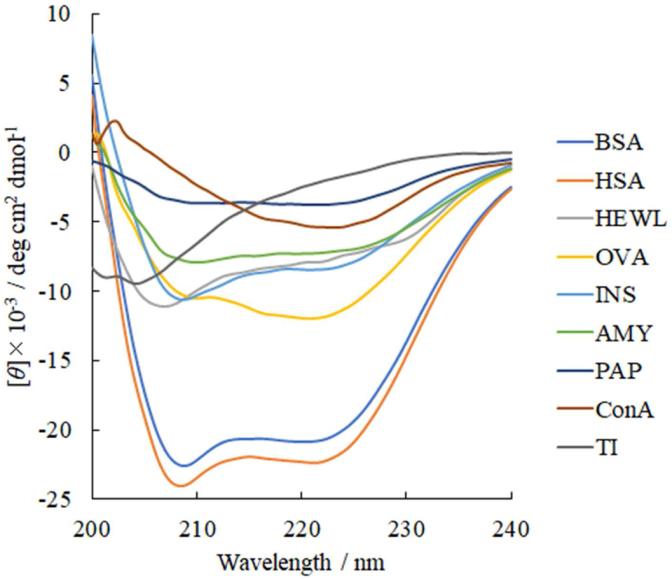
For the hybrid examples possessing α-helical and β-strand/sheet segments, we used insulin (INS), amylase (AMY), papain (PAP), and ovalbumin (OVA). Their amino acid residual proportions, with which the α-helical, β-strand/sheet, and disordered segments of HEWL and other proteins, retrieved from Protein Data Bank (PDB) entries, appear in the caption of Fig. 1.
Fig. 1.
The observed CD spectra of HEWL are 55%, 7%, and 39% (residual proportions constituting the α-helical, β-strands/sheets, and disordered segments, respectively); BSA (84%, 0%, 16%); HSA (82%, 0%, 18%); INS (66%, 3%, 31%); AMY (34%, 18%, 48%); OVA (34%, 33%, 33%); PAP (30%, 18%, 52%); ConA (6%, 42%, 52%); and TI (5%, 30%, 65%). Their residual numbers described were retrieved from PDB entries of 6QWW, 4F5S, 1BJ5, 1GUJ, 1AQM, 1OVA, 2CIO, 1BXH, and 1AVU, respectively.
2. Experimental
Materials HEWL (UniProtKB P00698), BSA (UniProtKB P02769), HSA (UniProtKB P02768), INS (UniProtKB P01308), AMY (UniProtKB P2768), OVA (UniProtKB P01308), and TI (UniProtKB P01070) were purchased from Wako Pure Chemical Industries (Osaka, Japan), and PAP (UniProtKB P00784) was obtained from Merck (Darmstadt, Germany). ConA (UniProtKB P02866) was obtained as a lyophilized powder containing approximately 15% protein, primarily balanced with NaCl from Sigma-Aldrich (St. Lewis, MO, USA). TFE, acetonitrile, and other reagents were of the highest grade commercially available.
Residual proportions of secondary structures and Ramachandran geometrical coordinates of the investigated proteins were retrieved from their corresponding entries of PDB of the Cambridge Crystallographic Data Centre/Brookhaven National Laboratory published on the website https://www.rcsb.org/, of which domestic mirror site PDBj at the Institute for Protein Research (IPR), Osaka University, was accessed in 2019–2020.
CD spectropolarimetry and ATR-FTIR spectroscopy for proteins Samples of HEWL, BSA, I, INS, AMY, OVA, and PAP were dissolved in 10 mmol/L phosphate/NaOH buffer (pH 6.0), TI was dissolved in 10 mmol/L phosphate/NaOH buffer (pH 7.4), and ConA was dissolved in 150 mmol/L NaCl, 10 mmol/L phosphate/NaOH buffer (pH 7.4). The measured concentration of each sample was 50 μg/mL except for the measured concentration of ConA, which was 45 μg/mL. The far-ultraviolet CD spectra of HEWL and the other proteins were recorded on a J-820 spectropolarimeter (JASCO, Tokyo, Japan). The spectra were collected at 25 °C using a rectangular quartz cell with a path length of 1 cm at wavelengths ranging from 190 to 250 nm with an interval of 0.1 nm, a response time of 1 s, and a scanning speed of 100 nm/min under a nitrogen atmosphere. The CD data were expressed in terms of mean residue ellipticity in deg∙cm2∙dmol−1.
HEWL samples were dissolved in a vial bottle with a tiny protrusion at the center bottom. The salt concentration was adjusted by adding the required concentration of the NaCl solution. Final HEWL concentrations were adjusted to 0.45 mg/mL in 10 mmol/L phosphate/NaOH buffer (pH 6.0). The final heat-treated HEWL were obtained by incubation at 65 °C while stirring with a 15 mm magnetic stirrer bar at 100 rpm for 5 h, following previously reported procedures [[40], [41]]. The non-heated control was incubated at 25 °C for 5 h. The prepared HEWL samples were centrifuged at 15,000 rpm for 10 min. Then, their supernatants and precipitates were obtained. The lower layer was analyzed by an attenuated total reflection-Fourier transform infrared ATR-FTIR spectrometer. The IR spectra were recorded with a Fourier UATR instrument (PerkinElmer, Norwalk, CT, USA) at 25 °C, and represented as averages of 64 or 128 scans recorded from 1700 to 1600 cm−1 at an interval of 4 cm−1 [11]. Curve-fitting approximations of the observed spectra in the range from 1700 to 1600 cm−1 were performed using the SOLVER module of Microsoft Excel 2016. The CD spectra were recorded with the equipment mentioned above at 25 °C.
The protein concentration was determined using spectrometry with the bicinchoninic acid (BCA) copper (I) complex method [42]. For the supernatants of the heat-treated HEWL sample, its protein concentration was obtained at an absorbance 562 nm, after the sample kit solution (200 μL) and upper layer protein solution (25 μL) were mixed.
Curve fittings of a linear combination of Woody's patterns and peak/trough decomposition with that of Gaussian functions The spectral pattern of each secondary structure reported by Woody was created by printing a figure on graph paper and acquiring [θ] at 0.5 nm intervals. The spectral pattern was named as Woody's patterns and defined as basis sets of secondary structures. Woody's patterns for his artificial α-helical, β-strand/sheet, and “random coil” segments were used for reconstructing the corresponding CD spectrum by their liner combination. The coefficient optimizing procedure of this liner combination were performed using the SOLVER module of Microsoft Excel 2016.
A Gaussian bell-shaped function was expressed with parameters given to the componential peak height and/or trough depth A, the averaged position of wavelength (for CD)/wavenumber (for ATR-FTIR) (μ), and the standard deviation (σ). The number of functions required for the Gaussian peak/trough decomposition was determined by the practical trials. The coefficients in the linear combination were tentatively optimized until the minimal solution set of the given parameters was obtained.
Procedure of singular value decomposition (SVD) The -th observed CD spectrum {|} of the sample protein is represented as the m-dimensional vertical vector measured at the wavelength {|}. The matrix M comprised a horizontal sequence of vectors from the first spectral vector through the -th spectral vectors, with an m × n rectangular matrix defined as follows, where m ≥ n:
and are real and transposed matrices, respectively. Their products and become orthogonal matrices and eigenvectors are the rows of U and V. The matrices describing M are transformed into the formulas:
The diagonal matrix Σ constitutes the diagonal elements {|} of the positive real values in descending order. These elements are singular values, indicating dispersion [43]. The -th column of the orthogonal matrix V is the coefficient vector corresponding to the singular value , and the vector is called a singular vector. The -th column of the orthogonal matrix U is called a basic vector . The principal component vector was the coefficient vector multiplied by the corresponding singular value .
The matrix U has rows that are the basis function vectors [44]. Here, it is different from the ω used in our past papers cited and ω worked as the coefficient of ψ.
From diagram for logarithm of the singular value in descending order versus the index corresponding to the documental spectra, we practically decided the dimensionality, namely the minimum dimensionality of the basis functions required to reproduce the vector space of the documental spectra. It might be practically negligible with singular value less than a several hundredth of the highest singular value of the first principal components. Due to the dimensionality decided under this criterion instead of the mathematical rank , the yielded principal components approximately reproduces the vector space including documental spectrum as the ..-th feature vector composed with the -th elements :
3. Results and discussion
Peak/trough decomposition of α-helical spectra with Gaussian functionsFig. 1 shows the far-ultraviolet CD spectra of HEWL, BSA, HSA, jack bean (Canavalia ensiformis) concanavalin A (ConA), human pancreatic insulin (INS), bacterial (Bacillus subtilis) amylase (AMY), papaya (Carica papaya L.) papain (PAP), and hen egg ovalbumin (OVA). The CD spectra were decomposed to the spectral components by the curve-fitting approximation using Woody's straightforward basis set containing the artificial α-helix, β-sheet, and “random coil”. The fitted parameters Woody's stripped basis set in Table S1 indicate the α-helix:β-sheet proportion in ConA is 1:3 (0.0628:0.161), which agrees with the ratio obtained from the PDB entry record (1:7 (0.13:0.87)). The observed CD spectrum of ConA showed poor coincidence, yet the observed spectrum would graphically agree to a linearly combined spectrum for α-helical and β-sheet segments [[45], [46]]. However, the account of the disordered segment was 47% with no difference due to this curve-fitting. Except for ConA, the proportions of α-helical and β-sheet segments of selected proteins disagreed with the results of the curve-fitting with Woody's straightforward basis set and the residual proportion retrieved from PDB entry records.
To improve the approximation of the curve-fitting procedure, we neglected the component of the CD spectrum assigned to the “random coil” from Woody's straightforward basis set since it assumes no spectral pattern due to a particular electronic transition attributed to the chaotic structure. Table S1 lists the approximations of the observed CD spectra via a linear combination of the Woody's stripped patterns for the artificial α-helical and β-strand/sheet segments. Contrary to the expectation that root mean square (RMS) values decrease since there is no straightforward basis set of a “random coil” segment, those of the examined proteins resulted in RMS values larger than the approximations conducted with Woody's straightforward basis set.
Figure S2 A presents the spectral differences of the approximated curves of the observed CD spectra. The spectral difference curves of BSA, HSA, INS, and AMY shared features of troughs at approximately 208 and 227 nm with a peak at approximately 222 nm. Although the curve of OVA had an individual peak at 205 nm, it also featured similar features of 208 and 227 nm troughs and 222 nm peaks. We found partially common features on the curves of ConA and HEWL.
Such systematic errors on the approximations indicated a misinterpretation in the curve-fitting procedure or in Woody's basis set [[47], [48]]. Moreover, in the curve-fitting approximation limited to using the stripped basis set containing α-helical and β-sheet segments, the spectral difference curves are illustrated in Fig. S2B. Because both spectral differences showed in Figs. S2A and S1 have several same peaks in each protein, we considered that Woody's “random coil” pattern was not directly involved with the systematic errors on the approximations. Therefore, the results indicate that components in both Woody's straightforward and stripped basis sets, which might give the pattern of an α-helix, do not apply to a practical pattern obtained from a native protein.
Woody's primitive spectra of the artificial α-helical and β-strand/sheet segments were disposed by the peak/trough decomposition using minimal numbers of Gaussian functions. This resulted in the compositions of three Gaussian functions with troughs at wavelengths μ of 204, 209, and 221 nm for Woody's artificial α-helix and those of two Gaussian functions with a peak at μ of 196 nm and a trough at 217 nm for Woody's artificial β-strand/sheet (Table 1).
Table 1.
The functional parameters computed by the peak/trough decompositions for the spectrum of Woody's artificial α-helix, β-strand, and our observed CD spectra of the model proteins. The functional parameters A, σ, and μ indicate a peak height/trough depth, a standard deviation as half width of the peak/trough, and a central wavelength of the peak/trough, respectively. G1, G2, and G3 represent Gaussian functions to compose α-helix and are described by these parameters, ascending order in their wavelengths of which values of parameters μ were initiated at 204, 209, and 221 nm, respectively. G4 and G5 represent Gaussian functions to compose β-strand/sheet, and their values of μ were initiated to 196 and 217 nm, respectively.
| Parameters of Gaussian fitting | Gaussian components of α-helix G1, G2, G3 |
Gaussian components of β-sheet G4, G5 |
RMS | ||||
|---|---|---|---|---|---|---|---|
| G1 | G2 | G3 | G4 | G5 | |||
| Woody's artificial α-helix | A | −23.4 | −22.3 | −42.8 | – | – | 0.967 |
| σ | 2.53 | 3.78 | 8.06 | – | – | ||
| μ | 204.4 | 209.1 | 221.2 | – | – | ||
| Woody's artificial β-strand /sheet |
A | – | – | – | 41.6 | −12.6 | 0.300 |
| σ | – | – | – | 5.42 | 9.34 | ||
| μ | – | – | – | 196.3 | 216.7 | ||
| BSA | A | −11.7 | −9.02 | −20.2 | – | – | 0.0756 |
| σ | 3.25 | 4.66 | 8.58 | – | – | ||
| μ | 206.4 | 211.1 | 222.4 | – | – | ||
| HSA | A | −12 | −10.3 | −21.6 | – | – | 0.0625 |
| σ | 3.40 | 4.90 | 8.49 | – | – | ||
| μ | 206.1 | 210.8 | 222.6 | – | – | ||
| HEWL | A | −9.48 | −6.85 | −6.63 | 0.0221 | −1.19 | 0.0660 |
| σ | 3.98 | 4.84 | 6.52 | 4.05 | 2.70 | ||
| μ | 205.6 | 214.3 | 227.3 | 196.5 | 221.2 | ||
| INS | A | −8.40 | −4.56 | −6.45 | 9.79 | −2.00 | 0.0261 |
| σ | 3.45 | 3.92 | 8.21 | 5.58 | 5.40 | ||
| μ | 206.6 | 212.6 | 224.0 | 194.9 | 220.7 | ||
| AMY | A | −4.01 | −3.47 | −6.12 | 4.59 | −1.54 | 0.0355 |
| σ | 3.67 | 5.18 | 7.89 | 6.10 | 7.08 | ||
| μ | 206.7 | 212.3 | 225.5 | 188.3 | 215.6 | ||
| OVA | A | −6.34 | −1.45 | −9.24 | 0.100 | −2.58 | 0.0266 |
| σ | 3.60 | 2.80 | 8.73 | 2.00 | 8.85 | ||
| μ | 207.7 | 214.9 | 221.6 | 198.0 | 221.0 | ||
| PAP | A | −1.30 | −1.89 | −1.62 | 2.00 | −1.59 | 0.0242 |
| σ | 3.36 | 6.42 | 4.95 | 1.99 | 12.7 | ||
| μ | 207.2 | 214.2 | 225.4 | 191.8 | 220.0 | ||
| ConA) | A | 0.00 | −3.56 | −0.65 | 4.22 | −4.47 | 0.0333 |
| σ | 3.66 | 5.65 | 4.58 | 4.93 | 5.63 | ||
| μ | 205.0 | 215.2 | 238.8 | 196.5 | 225.5 | ||
| TI | A | −9.94 | −0.505 | −2.83 | 0.200 | −0.235 | 0.0340 |
| σ | 6.57 | 0.875 | 7.29 | 1.98 | 7.57 | ||
| μ | 203.4 | 205.7 | 218.9 | 181.9 | 210.0 | ||
The observed CD spectrum of BSA, an α-helix rich protein, was decomposed into three Gaussian functions (Fig. S1B). The reproducibility of the reconstructed spectrum increased compared to the spectrum approximated using Woody's straightforward basis set (Fig. S1A). The RMS value for the peak decomposition using Gaussian function degraded less than 0.01 (Table 1) while the spectral difference curve appeared as a plateau (Fig. S1B). The trough wavelengths μ of the decomposed Gaussian functions were determined as 206, 211, and 222 nm (Table 1), nominated as G1, G2, and G3, respectively.
HSA showed similar values (Table 1). Compared to the wavelengths of decomposed troughs of Woody's α-helix, G1, G2, and G3 shifted higher by approximately 2 nm. The wavelengths of the peaks and troughs observed in Fig. S2 are localized near those of G1, G2, and G3. Therefore, Woody's primitive pattern of the artificial α-helix had slightly misalignment from the α-helical patterns, which occupied over 70% of the CD spectra of BSA and HSA. Consequently, we provisionally determined the combination of G1, G2, and G3 acting as a spectral contribution for the α-helix, in which the proportion of peak/trough area under the curve () was 1:1:4 for these albumins. The combination of Gaussian functions G1 and G2 was regarded as a latent single trough (i.e., the excitation) while the usage of two Gaussian functions is favorable for various appearances, induced by thermal fluctuations or electronic state diversities or quantum uncertainty that is delocalization of π electrons or transition state uncertainty derived induced by excited unbonded electrons.
Peak/trough decomposition of β-strand/sheet spectra with Gaussian functions Irregular left-handed helical residues of 3–5 were observed in BSA and HSA with their 85 or 80 α-helical residues on their Ramachandran maps (Figs. S8B and S8C); most α-helical residues take a right-hand helix, stabilized by formation of hydrogen bond network parallel to the axis of the helix, and are resulted to take the constrained structure with a repeat length of 3.6 residues per turn [49]. Thus, the peak/trough intensity in the CD spectrum caused by the α-helix portion might be filled with a much larger preference for β-sheet, and the observed CD spectrum of ConA showed exactly that feature [50]. In contrast, amino acid residues belonging to β-strand/sheet structures may have a steric structure with large diversity, since various arrangements can lead to parallel or antiparallel orientations and straight or curved backbones. Furthermore, the residues in the β-structures extruding from its zigzag plane provide a wide margin of movement. The multiple variety at their backbone level and the amino acid residue level are manifested in the CD spectral diversity, which has been an inherent limitation of the secondary structure estimation. Therefore, the helix content could be predicted more or less accurately, without predicting the β-structural content [20,51]
According to the decomposition of Woody's primitive pattern for artificial β-structures, we prepared a basis set containing two Gaussian functions for peak/trough decomposition for these proteins besides three Gaussian functions for the α-helical pattern, as described above. These were denoted as G4 and G5, in which the initial values of wavelengths μ were 196 and 217 nm, respectively. Table 1 shows the two serum albumins contain fewer β-structures than their CD spectrum. After the peak/trough decompositions for HEWL, OVA, INS, AMY, and PAP, the respective CD spectra were reproduced with coefficients of linear combination of parameters from G1 to G5 functions (Table 1). All RMS values computed were less than 0.006. This indicated that the peak/trough decomposition was accomplished using the basis set containing five Gaussian functions, three reproduced the observed spectra of α-helix and two of β-structures. The proportions of the area under the peak/trough curve (AUC = Aσ√2π) of functions G1:G2:G3 were approximately 2:2:3 for HEWL, 3:1:4 for INS, and 1:1:3 for AMY (Table 1).
The trough at 222 nm in the observed CD spectral shapes of PAP and OVA were deeper and wider than the trough at 210 nm. This peculiarity is observed in the shape of Woody's pattern for the artificial α-helix compared to the observed spectra of BSA and HSA. If the 222 nm troughs of the observed CD spectra of PAP and OVA was n → π* excitation signal identical to the 217 nm component of the β-structures, then:
-
1)
More right-hand twisted antiparallel β-structures (trypsin inhibitor) transformed to the left-hand twisted antiparallel β-structures (lectin ConA), and more wavelengths of the n → π* excitation signal shifts from 200 nm to 221 nm, as described by Micsonai et al. [20].
-
2)
Although PAP contains less β-structures relative to OVA, the proportion (35%) of AUC for the G5 function (0.837) to the sum of AUCs for G1, G2, and G3 (2.423) was greater than OVA (0.3%). Comparing the Ramachandran maps of OVA and PAP (Figs. S8F and S8G), the plots in the second quadrant (especially the rectangular region of −180 < φ < −45 and + 45 < ψ < +180) contain several residues comprising non-β-structured (i.e., disordered) portions for PAP. AMY showed a similar tendency. These results suggest that the AUC for G5 function was not proportional to the number of β-structures. Thus, hypothetical estimations of common patterns to distinguish α-helices and β-structures may not be realistic.
-
3)
The trough at the wavelength range of 217–222 nm depends on the steric constraint or stability of the residual portion. For OVA, the n → π* excitation signal (G3 and G5) may grow due to the reduction of structural disorder, regardless of α-helix and/or β-structures. Thus, the regions comprising β-structured fragments and disordered segments of AMY and PAP could fluctuate. In our subjective sights, the strand and disordered segment tangles were in the AMY and PAP crystallographic structures, whereas the β-sheet and barrel structures of OVA were separated from other portions.
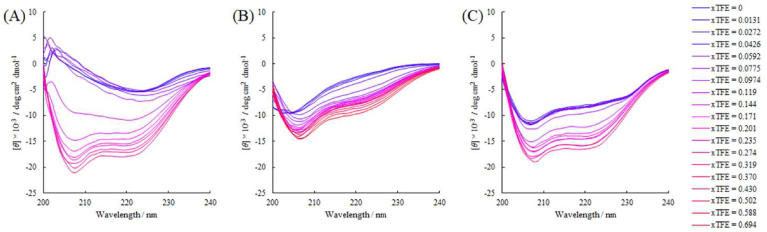
2,2,2-Trifluoroethanol titration of ConA can trace the interconversion pathway from β-structures to α-helix TFE stabilizes the α-helical structure for various proteins, including β-structure-rich proteins [52]. In this study, we used TFE to trace the interconversion pathway of the α-helix structure from native ConA. The α-helical pattern obtained in the TFE molar fraction was at least 0.14 (50%), agreeing with Xu and Keiderling [37]. The observed CD spectra of ConA regarding the molar fraction of TFE are shown in Fig. 2A.
Fig. 2.
Obtained CD spectra of ConA (A), TI (B), and HEWL (C) in the absence and presences of TFE at molar fractions (xTFE) of 0, 0.0131, 0.0272, 0.0426, 0.0592, 0.0775, 0.0974, 0.119, 0.143, 0.171, 0.201, 0235, 0274, 0.319, and 0.370. Those of TI at the xTFE of 0.430, 0.502, 0.588, and 0.694 were superimposed onto the diagram B.
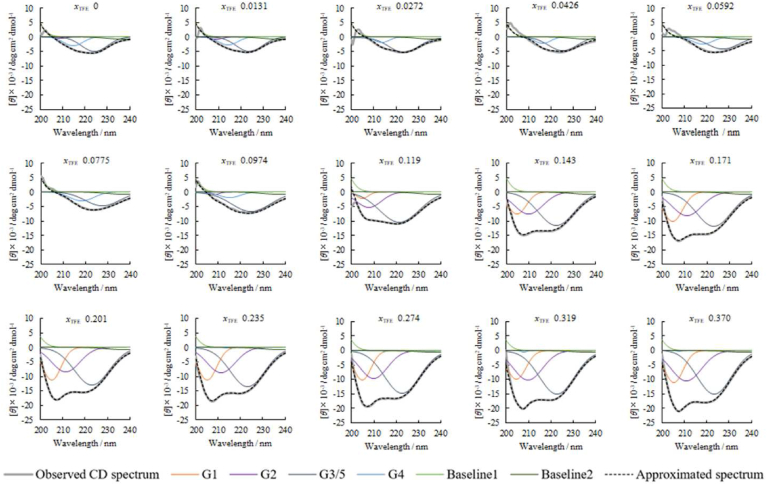
During the lower molar fraction of TFE, the trough at 221 nm shifted to a higher wavelength. At fraction 0.0974, the broad trough at 220 nm corresponding to the β-structure of ConA, gradually shifted to a higher wavelength (225 nm). Between 0.0974 and 0.110, the spectral shape was transformed into twin peaks. A fraction over 0.0974 showed a deep increasing trough at 207 nm. Considering the continued spectral change, the peak/trough decompositions were applied to these CD spectra (Fig. 3). Provided that the G5 function prepared for β-structure was integrated into the G3 function prepared for α-helix, both G3 and G5 represent the n → π* excitation signal. Baseline1 and Baseline2, determined using Gaussian function optimization at the TFE molar fraction of 0, were used to correct the baseline of the observed spectra.
Fig. 3.
Approximated spectrum (black solid curve) to the observed CD spectrum (grey solid curve), provided by the peak/trough decomposition (dashed curves) using Gaussian functions, G1, G2, G3/G5, and G4 for ConA in the absence and presences of TFE at molar fractions (xTFE) of 0, 0.0131, 0.0272, 0.0426, 0.0592, 0.0775, 0.0974, 0.119, 0.143, 0.171, 0.201, 0235, 0274, 0.319, and 0.370.
The obtained peak height A, center wavelength (μ), and standard deviations (σ) of G1, G2, G3/G5, G4, Baseline1, and Baseline2 functions are summarized in Figs. S3A, S3B, and S3C, respectively. Furthermore, the AUC of these functions was calculated using the integral of the Gaussian function Aσ√2π (Fig. S3D). The AUC, for G4 function assigned to the π → π* transition in the β-structure, vanished with increasing TFE molar fraction, whereas those for G1 and G2 functions assigned to the π → π* transition in the α-helix. For G3/G5 function, the AUC corresponding to this n → π* transition increased against the AUC decrement of G4 function. In contrast, the AUC of G3/G5 function decreased from 0.05 to 0.1, and then the AUC increased to over the TFE molar fraction of 0.1. This discontinuous turning point was synchronized with the starting point of the AUCs increment of G1 and G2 functions. The AUC of the G3/G5 function may create confusion between the observed spectral decrement of β-structure and the observed spectral increment of α-helix. Analysis of the AUCs of such peak/trough decomposed Gaussian functions showed that the composites of the spectral patterns for α-helix and β-structure could not re-enact the conformational interconversion pathway.
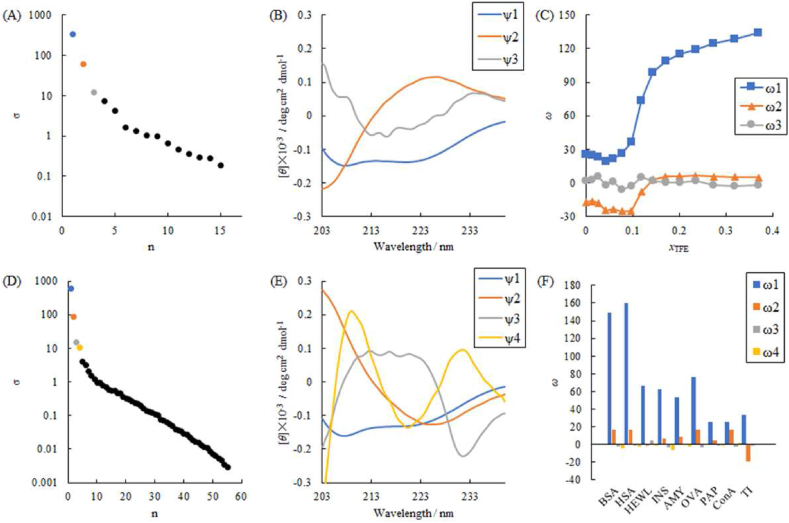
Singular value decomposition (SVD) approach for spectral transition The conformational interconversion from the antiparallel β-sheet to the α-helical structure in ConA by adding TFE, observed in the CD spectra (Fig. 2, Fig. 3), should include characteristic CD spectral changes. Principal component analysis and the SVD approach was used to extract the spectral differences dependent on TFE molar fraction [43]. For the observed CD spectrum at the i-th molar ratio of TFE, the intensity at the j-th wavelength λj (from 205 to 240 nm at an interval of 1 nm) was expressed as Φi(λj). The m × n rectangular matrix M comprises an n-dimensional horizontal sequence of m-dimensional vertical vectors Φi, where the SVD procedure decomposed to the basis matrix U, the singular value matrix Σ, and the singular vector matrix V. The contributions of the i-th basis set in the matrix U are represented by the i-th diagonal elements σi of the matrix Σ as a dispersion sorted in descending order. The logarithm of σ values was plotted as an index of this order.
Fig. 4A shows that the first and second components were significant, whereas the third component, namely noise, with a dispersion less than one five-hundredths, is insignificant. Fig. 4B shows the basis functions of the first, second, and insignificant third components, in which the first and second basis functions appear similar to the α-helical spectrum (with troughs at 207.5 and 220 nm) and the up-down inversion of the β-structural spectrum (trough at 226.5 nm), respectively. Fig. 4C shows the principal components, ω, corresponding to the first and second basis functions dependent on the molar fraction of TFE (xTFE). Compared to Fig. S3D, the results of the SVD analysis suggests that the ω values of the first and second basis functions consistently represent the proportions of α-helical and β-structural contributions, respectively. Therefore, consecutive changes were observed. Here, second basis functions shown in Fig. 4B is inverted from the spectrum pattern of β-sheet while they had specifical peaks of β-sheet. That is, the larger the negative value in the ω value, the larger the β-sheet content. It is the reason that negative values of ω2 values have been calculated.
Fig. 4.
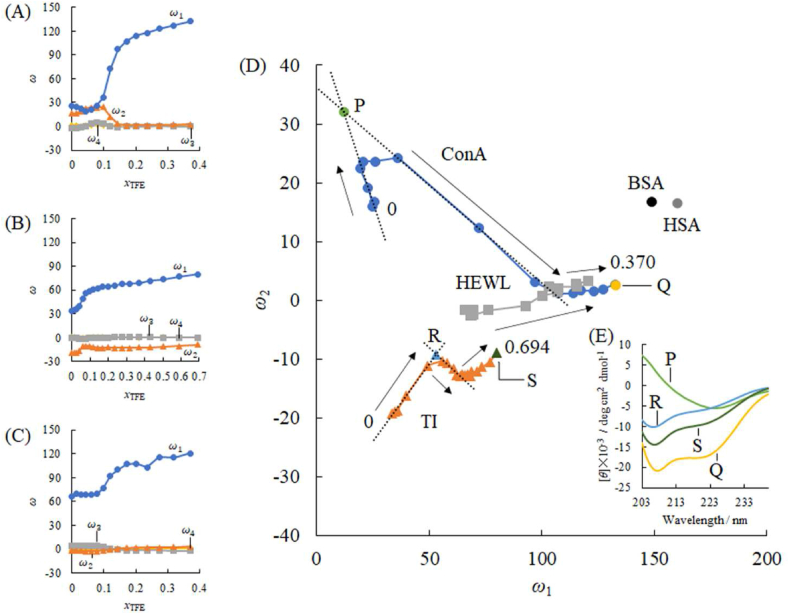
(A) The singular value decomposition (SVD) for 15 observed CD spectral transition of ConA dependent on the TFE molar fraction (xTFE): the singular values (σ) indicated the relative variances corresponding to the compositions, shown by their logarithm values as a function of the composition index n. (B) The spectra of basis functions for three compositions with the exceptionally high σ values in Fig. 4A. (C) The coefficients corresponding to three basis functions, which were given as the principal component vectors (ω) to the molar fraction of TFE (xTFE). (D) The singular values (σ) computed using the SVD analysis for 55 CD spectra of the investigated proteins, BSA, HSA, INS, AMY, OVA, PAP, 15 series of ConA, 19 series of TI, and 15 series of HEWL. (E) The spectra of basis functions for four compositions with the exceptionally high (σ) values in Fig. 4D. (F) The coefficients corresponding to four basis functions for the investigated proteins.
From the successful application of SVD, we supposed this basis set divided the observed CD spectrum into the contributions of α-helical and β-structural spectra. Therefore, SVD analysis was expected to show the spectra of examined protein samples integrated for all obtained spectra. First, the CD spectral changes of TI and HEWL were measured depending on xTFE (Fig. 2B–C). As the spectral transition of TI was seldom susceptible to the xTFE, further spectra at xTFE values of 0.430, 0.502, 0.588, and 0.694 were examined (Fig. 2B). Based on the SVD analysis integrated for 55 spectra (BSA, HSA, INS, AMY, OVA, PAP, 15 for ConA, 19 for TI, and 15 for HEWL), Fig. 4D shows the logarithm of singular values (σ) to the descending order index of components; four compounds were analyzed, while Fig. 4E shows the basis functions corresponding to the four singular values. The first and second basis functions also resembled the α-helical and β-structural spectra, respectively.
Fig. 4F shows that the principal components (ω) of the basis functions were depended on xTFE. BSA (containing 84% of α-helix) and HSA (containing 73% of α-helix) largely contributed to the first basis function, corresponding to the α-helical segments. ConA (containing 8% α-helix and 45% β-structure) showed a higher proportion of the second component (i.e., β-structure) than the first component (i.e., α-helix). The third and fourth components in AMY would be higher than those of other proteins, suggesting that these components correspond to the population of the anti-clockwise α-helix segments or turns found in the Ramachandran map of AMY (Fig. S8E). For TI, the negative coefficient for the second component corresponded to the spectral pattern of the β-structure. According to Schönburrner [53], TFE enhances the β-structure of TI, which indicates that the antiparallel β-sheet structure is too robust and dense to unravel the transformation to the α-helix. Finally, the integrated SVD procedure decomposed to proportions of the secondary structures, α-helix, β-structure, and minor characteristic fragments.
During the spectral/conformational interconversions induced by TFE, the coefficients for the first component corresponding to the α-helix increased based on xTFE for ConA, TI, and HEWL (Fig. 5). In contrast, those for the second component, corresponding to β-structure, also increased based on a xTFE to <0.1 for ConA and TI. This indicates that low xTFE can stabilize the β-structure. As HEWL contains a small number of β-structural segments, its coefficient for the α-helical component simply increased without an increase in the β-structural component. The difference of coefficients ω1 and ω2 for ConA in Fig. 4, Fig. 5A is occurred by their second basis functions that have similar shape, just flipped over each other.
Fig. 5.
The diagrams of the coefficients corresponding to four basis functions in Fig. 4E as a function of the molar fraction of TFE (xTFE) for ConA (A), TI (B), and HEWL (C). The diagrams of the coefficient ω1 for the first basis function (corresponding to the population of α-helix) versus the coefficient ω2 for the second basis function (corresponding to the population of β-structure), for ConA, TI, and HEWL. (D) Arrows represent directions of time series on the trajectory which indicated the spectral/conformational interconversion pathway. Along the trajectory of ConA, the tangential line initially heads forward to upper left (slope of −1.17 and intercept of 46.1), whereas the tangential line turns at the singularity point P and then turns to lower right (slope of −0.332 and intercept of 36.1). This singularity point is defined as the intersection of these lines. The trajectory arrived at the point Q. The re-enacted CD spectra at P and Q were shown in the inset. Along the TI trajectory, the tangential line initially heads forward to upper right (slope of +5.29 and intercept of −37.2), whereas the tangential line turns at the singularity point R and then turns forward to lower right (slope of −0.355 and intercept of 9.72). The trajectory arrived at the point S. The re-enacted CD spectrum at R was shown in the inset (E). Along the trajectory of HEWL, its tangential line simply followed toward upper right with a loose slope.
Fig. 5D shows the diagrams of the coefficients for the first and second components over time. Abscissa (ω1) and ordinate (ω2) indicate degrees of the conformational interconversion for α-helix and β-structure, respectively. The trajectory (interconversion pathway) of ConA indicated transient interconversion to higher β-structural properties under the low molar fraction of TFE. Beyond a certain established singularity point (P), the α-helical property increases, while the β-structural property decreases. This singularity point indicates the most stabilized β-structure of ConA; the reconstructed CD spectrum is shown in Fig. 5E (ConA at P).
The CD spectrum of ConA converged to the extreme point Q, of which the reconstructed CD spectrum is shown in Fig. 5E (ConA at Q). Point Q asymptotically approached the coordinates corresponding to BSA and HSA. The gradual approach of the plots of each protein in the direction of the BSA and HSA plots whose main constituent structure is α-helix suggests that the α-helix structure progresses. However, the trajectory of TI also went through an established singularity point R, and then decreased the degree of β-structural transformation. Furthermore, the intensity of ω2 had a negative sign and its trajectory arrived at point S, approaching the stationary point Q of ConA, at higher xTFE (0.694 corresponding to 90 vol%). At point R, the re-enacted CD spectrum is shown in Fig. 5E. According to the quantitative computations of Micsonai [20], the right-twisted antiparallel β-strand like those of TI provides a characteristic CD spectral pattern different from the flat antiparallel β-sheet like those of ConA, and its spectrum resembled the α-helix. The trajectory of HEWL drew a simple line parallel to the tail of that of ConA. This indicated that the terminal β-segment of HEWL readily inter was converted to α-helix.
Therefore, the proportions of the secondary structures, α-helix, β-structure, and minor fragments could be distinguished by the SVD analysis, except for proteins with squeezed structures such as TI.
Amyloid formation of HEWL induced by thermal stimulation According to Cannon and Donald [54], incubation of HEWL at 65 °C for 120 h induced amyloid fibril formation. A lower pH or higher salt concentration enhances amyloid formation [10,[55], [56], [57]]. The conformational interconversion from α-helix to β-sheet of HEWL could not occur in dissolved fluids [11]. Thus, confirming that amyloid formation depended quantitatively on the closeness of the pH to the isoelectric point and on the square root of ionic strength (J). Since the electrostatic repulsion between protein particles is decayed by electrolytes according to the reciprocal square law (Debye's shielding effect), the effects of pH and ionic strength indicate that the native protein aggregates initially precipitated and promoted an irregular approach to each other. We believe that the conformational rearrangement consecutively progressed under ATR-FTIR observations, inducing the interconversion from α-helices or disordered segments to β-strands. These physicochemical properties may influence the aggregation of colloidal protein molecules, although they may not directly regulate the conformational interconversion.
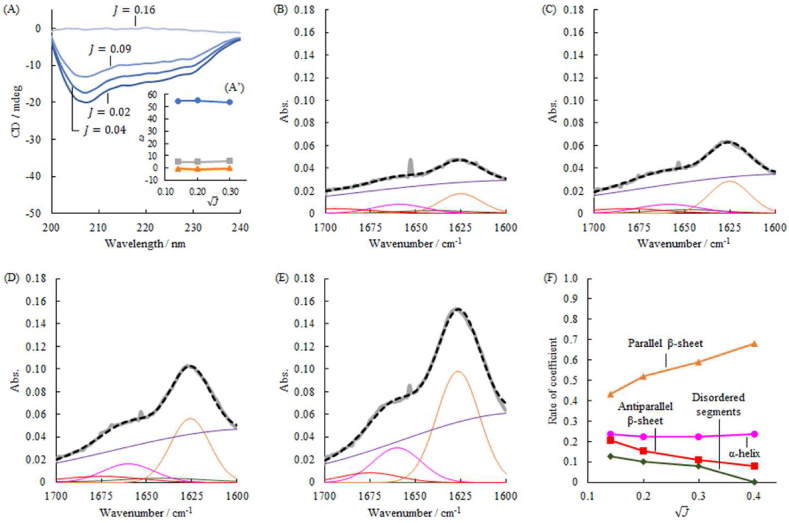
Fig. 6A shows the CD ellipticity [θ] observed for the centrifuged supernatant of the heat-treated HEWL at J of 0.02, 0.04, 0.09, and 0.16. The intensity of their ellipticity attenuated depending on the ionic strength, in which the specific ellipticity was determined by dividing the protein concentration measured using a BCA assay. The CD spectra for the supernatants were analyzed using the SVD procedure and integrated with the previous 55 spectra, except for the HEWL sample at J 0.16. The coefficients of the 1st, 2nd, 3rd, and 4th components are shown in the inset diagram (Fig. 6A). The heat treatments of HEWL at J of 0.02, 0.04, and 0.09 showed insignificant differences in these coefficients, which indicates that the aggregation process in solution did not affect their secondary structures.
Fig. 6.
The amyloid formation of the heat-treated HEWL: the CD spectra observed in the supernatants of the centrifuged samples (A). Except for the spectrum at the ionic strength of 0.16, these spectra were appended to the previous 55 spectra of protein samples, and then SVD analysis were performed. The obtained basis functions resembled the results in Fig. 4D–F. The coefficients of the 1st, 2nd, 3rd, and 4th basis functions were invariable to the square root of ionic strength as shown in the inset of Fig. 6A (A′). The ATR-FTIR spectra observed in the precipitates of the centrifuged sample at the ionic strengths 0f 0.02 (B), 0.04 (C), 0.09 (D), and 0.16 (E). The ATR-FTIR spectra were analyzed using the Gaussian peak decomposition method. Assignments of Gaussian functions to the secondary structures were described in the text, and the populations of secondary structures as a function of the square root of ionic strength (F).
ATR-FTIR spectra were measured for the precipitates of the heat-treated HEWL solution at J of 0.02, 0.04, and 0.09, as shown in Fig. 6B–E. According to Seo et al. [16], the decomposed peaks with Gaussian functions were assigned to α-helix (1648–1660 cm−1), parallel β-sheet (1610–1640 cm−1), antiparallel β-sheet (1675–1695 cm−1), and disordered segments (1640–1648 cm−1). Fig. 6F summarizes the proportions of secondary structures as a function of the square root of J. Moreover, we confirmed the progress of fibrosis with increased J by SEM shown in Fig. S6. Therefore, the conformational interconversion progressed only in the precipitates.
4. Conclusions
This comparative study showed that the (i) curve-fitting procedure with the linear combination of Woody's straightforward basis set for their artificial α-helix, β-strand/sheet, and "random coil" segments, (ii) Gaussian peak/trough decomposition procedure, and (iii) SVD procedure were verified to distinguish the contributions of the secondary structures on the observed far-ultraviolet CD spectra of proteins. However, there were systematic errors; in procedure (i), there was a deviation of the spectral patterns of Woody's artificial second structures from the practical CD spectra of the examined protein samples. Procedure (ii) appeared cumbersome and did not give a continuous smooth curve. These limitations were derived from the low intrinsic distinguishability for differences among spectral patterns of the secondary structures. Procedure (iii) gave relatively good results in our prepared okiselections. It recognized an irregular pattern in the CD spectrum of the trypsin inhibitor from those of ordinary α-helical and β-structural segments. This study clarifies that the common component by the SVD method could detailly trace secondary structural transformation of different proteins. Furthermore, the current comprehensive analysis suggests that the SVD approach plays a key role in regulating the solubility of such mixtures.
This study contributes to the simultaneous monitoring and its use of the conformational and spectral behavior of biologically functional proteins and other biopolymers in solution, such as secondary structural transformation.
Declaration of competing interest
The authors declare that they have no conflict of interest.
Acknowledgments
The authors are grateful to Professor Kenichi Sakai and Dr. Masaaki Akamatsu, Tokyo University of Science, for their productive advice and technical support. This work was partially supported by JSPS KAKENHI Grant Number 17K05366.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2021.101153.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Storey E., Cappai R. The amyloid precursor protein of Alzheimer's disease and the Abeta peptide. Neuropathol. Appl. Neurobiol. 1999;25:81–97. doi: 10.1046/j.1365-2990.1999.00164.x. [DOI] [PubMed] [Google Scholar]
- 2.Winslow A.R., Rubinsztein D.C. The Parkinson disease protein α-synuclein inhibits autophagy. Autophagy. 2011;7:429–431. doi: 10.4161/auto.7.4.14393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Landles C., Bates G.P. Huntingtin and the molecular pathogenesis of Huntington's disease. EMPO. Rep. 2004;5:958–963. doi: 10.1038/sj.embor.7400250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Westermark P., Wernstedt C., O’brien T.D., Hayden D.W., Johnson K.H. Islet amyloid in Type 2 human diabetes mellitus and adult diabetic cats contains a novel putative polypeptide hormone. Am. J. Pathol. 1987;127:414–417. [PMC free article] [PubMed] [Google Scholar]
- 5.Neumann M., Kwong L.K., Sampathu D.M., Trojanowski J.Q., Lee V.M.-Y. TDP-43 Proteinopathy in frontotemporal lobar degeneration and amyotrophic lateral sclerosis: protein misfolding diseases without amyloidosis. Arch. Neurol. 2007;64:1388–1394. doi: 10.1001/archneur.64.10.1388. [DOI] [PubMed] [Google Scholar]
- 6.Benson M.D., Buxbaum J.N., Eisenberg D.S., Merlini G., Saraiva M.J.M., Sekijima Y., Sipe J.D., Westermark P. Amyloid nomenclature 2020: update and recommendations by the international society of amyloidosis (ISA) nomenclature committee. Amyloid. 2020;27:217–222. doi: 10.1080/13506129.2020.1835263. [DOI] [PubMed] [Google Scholar]
- 7.Cuanalo-Contreras K., Mukherjee A., Soto C. Role of protein misfolding and proteostasis deficiency in protein misfolding diseases and aging. Int. J. Biochem. Cell Biol. 2013;2013:1–10. doi: 10.1155/2013/638083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hetz C., Glimcher L.H. Protein homeostasis networks in physiology and disease. Curr. Opin. Cell Biol. 2011;23:123–125. doi: 10.1016/j.ceb.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carrell R.W., Lomas D.A. Conformational disease. Lancet. 1997;350:134–138. doi: 10.1016/S0140-6736(97)02073-4. [DOI] [PubMed] [Google Scholar]
- 10.Chatani E., Imamura H., Yamamoto N., Kato M. Stepwise organization of the β-structure identifies key regions essential for the propagation and cytotoxicity of insulin amyloid fibrils. J. Biol. Chem. 2014;289:10399–10410. doi: 10.1074/jbc.M113.520874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sakaguchi T., Wada T., Kasai T., Shiratori T., Minami Y., Shimada Y., Otsuka Y., Komatsu K., Goto S. Effects of ionic and reductive atmosphere on the conformational rearrangement in hen egg white lysozyme prior to amyloid formation. Colloids Surf. B Biointerfaces. 2020;190:110845. doi: 10.1016/j.colsurfb.2020.110845. [DOI] [PubMed] [Google Scholar]
- 12.Serpell L.C. Alzheimer's amyloid fibrils: structure and assembly. Biochim. Biophys. Acta. 2000;1502:16–30. doi: 10.1016/s0925-4439(00)00029-6. [DOI] [PubMed] [Google Scholar]
- 13.Almeida Z.L., Brito R.M.M. Structure and aggregation mechanisms in amyloids. Molecules. 2020;25:1195. doi: 10.3390/molecules25051195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J., Du X., Hashim S., Shy A., Xu B. Aromatic-aromatic interactions enable α-helix to β-sheet transition of peptides to form supramolecular hydrogels. J. Am. Chem. Soc. 2017;139:71–74. doi: 10.1021/jacs.6b11512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soto C., Castaño E.M., Frangione B., Inestrosa N.C. The alpha-helical to beta-strand transition in the amino-terminal fragment of the amyloid beta-peptide modulates amyloid formation. J. Biol. Chem. 1995;270:3063–3067. doi: 10.1074/jbc.270.7.3063. [DOI] [PubMed] [Google Scholar]
- 16.Seo J., Hoffmann W., Wamke S., Huang X., Gewinner S., Schollkopf W., Bowers M.T., von Helden G., Pagel K. An infrared spectroscopy approach to follow β-sheet formation in peptide amyloid assemblies. Nat. Chem. 2017;9:39–44. doi: 10.1038/nchem.2615. [DOI] [PubMed] [Google Scholar]
- 17.Belbachir K., Noreen R., Gouspillou G., Petibois C. Collagen types analysis and differentiation by FTIR spectroscopy. Anal. Bioanal. Chem. 2009;395:829–837. doi: 10.1007/s00216-009-3019-y. [DOI] [PubMed] [Google Scholar]
- 18.Perez-Iratxeta C., Andrade-Navarro M.A. K2D2: estimation of protein secondary structure from circular dichroism spectra. BMC Struct. Biol. 2008;8:25. doi: 10.1186/1472-6807-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woody R.W. Circular dichroism. Methods Enzymol. 1995;246:34–71. doi: 10.1016/0076-6879(95)46006-3. [DOI] [PubMed] [Google Scholar]
- 20.Micsonai A., Wien F., Kernya L., Lee Y.-H., Goto Y., Réfrégiers M., Kardos J. Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc. Natl. Acad. Soc. U.S.A. 2015;112:E3095–E3103. doi: 10.1073/pnas.1500851112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woody R.W. Circular dichroism spectrum of peptides in the poly(pro)II conformation. J. Am. Chem. Soc. 2009;131:8234–8245. doi: 10.1021/ja901218m. [DOI] [PubMed] [Google Scholar]
- 22.Wu Y., Huang H.W., Olah G.A. Method of oriented circular dichroism. Biophys. J. 1990;57:797–806. doi: 10.1016/S0006-3495(90)82599-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kessler J., Andrushchenko V., Kapitán J., Bouř P. Insight into vibrational circular dichroism of proteins by density functional modeling. Phys. Chem. Chem. Phys. 2018;20:4926–4935. doi: 10.1039/c7cp08016f. [DOI] [PubMed] [Google Scholar]
- 24.Frare E., Leureto P.P.de, Zurdo J., Dobson C.M., Fontana A. A highly amyloidogenic region of hen lysozyme. J. Mol. Biol. 2004;340:1153–1165. doi: 10.1016/j.jmb.2004.05.056. [DOI] [PubMed] [Google Scholar]
- 25.Gülseren I., Güzey D., Bruce B.D., Weiss J. Structural and functional changes in ultrasonicated bovine serum albumin solutions. Ultrason. Sonochem. 2007;14:173–183. doi: 10.1016/j.ultsonch.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 26.Purcell M., Neault J.F., Tajmir-Riahi H.A. Interaction of Taxol with human serum albumin. Biochim. Biophys. Acta. 2000;1478:61–68. doi: 10.1016/s0167-4838(99)00251-4. [DOI] [PubMed] [Google Scholar]
- 27.Goto S., Masuda K., Miura M., Kanazawa K., Asaki M., Masui M., Shiramizu M., Terada H., Chuman H. Quantitative estimation of interaction between carbohydrates and concanavalin A by surface plasmon resonance biosensor. Chem. Pharm. Bull. Jpn. 2002;50:445–449. doi: 10.1248/cpb.50.445. [DOI] [PubMed] [Google Scholar]
- 28.Goto S., Terada H. Analysis of binding affinity of sugars to concanavalin A by surface plasmon resonance sensor Biacore. Spectroscopy. 2002;16:285–288. [Google Scholar]
- 29.Avilés-Gaxiola S., Chuck-Hernández C., Saldívar S.O.S. Inactivation methods of trypsin inhibitor in legumes: a review. J. Food Sci. 2018;83:17–29. doi: 10.1111/1750-3841.13985. [DOI] [PubMed] [Google Scholar]
- 30.Segawa S., Fukuno T., Fujiwara K., Noda Y. Local structures in unfolded lysozyme and correlation with secondary structures in the native conformation: helix-forming or -breaking propensity of peptide segments. Biopolymers. 1991;31:497–509. doi: 10.1002/bip.360310505. [DOI] [PubMed] [Google Scholar]
- 31.Shiraki K., Nishikawa K., Goto Y. Trifluoroethanol-induced stabilization of the α-helical structure of β-lactoglobulin: implication for non-hierarchical protein folding. J. Mol. Biol. 1995;245:180–194. doi: 10.1006/jmbi.1994.0015. [DOI] [PubMed] [Google Scholar]
- 32.Luo Y., Baldwin R.L. Trifluoroethanol stabilizes the pH 4 folding intermediate of sperm whale apomyoglobin. J. Mol. Biol. 1998;279:49–57. doi: 10.1006/jmbi.1998.1774. [DOI] [PubMed] [Google Scholar]
- 33.Taddei N., Chiti F., Fiaschi T., Bucciantini M., Capanni C., Stefani M., Serrano L., Dobson C.M., Ramponi G. Stabilisation of alpha-helices by site-directed mutagenesis reveals the importance of secondary structure in the transition state for acylphosphatase folding. J. Mol. Biol. 2000;300:633–647. doi: 10.1006/jmbi.2000.3870. [DOI] [PubMed] [Google Scholar]
- 34.Srisailam S., Kumar T.K.S., Rajalingam D., Kathir K.M., Sheu H.-S., Jan F.-J., Chao P.-C., Yu C. Amyloid-like fibril formation in an all β-barrel protein. Partially structured intermediate state(s) is a precursor for fibril formation. J. Biol. Chem. 2003;278:17701–17709. doi: 10.1074/jbc.M300336200. [DOI] [PubMed] [Google Scholar]
- 35.Plakoutsi G., Taddei N., Stefani M., Chiti F. Aggregation of the Acylphosphatase from Sulfolobus solfataricus: the folded and partially unfolded states can both be precursors for amyloid formation. J. Biol. Chem. 2004;279:14111–14119. doi: 10.1074/jbc.M312961200. [DOI] [PubMed] [Google Scholar]
- 36.Buck M., Radford S.E., Dobson C.M. A partially folded state of hen egg white lysozyme in trifluoroethanol: structural characterization and implications for protein folding. Biochemistry. 1993;32:669–678. doi: 10.1021/bi00053a036. [DOI] [PubMed] [Google Scholar]
- 37.Xu Q., Keiderling T.A. Trifluoroethanol-induced unfolding of concanavalin A: equilibrium and time-resolved optical spectroscopic studies. Biochemistry. 2005;44:7976–7987. doi: 10.1021/bi050003u. [DOI] [PubMed] [Google Scholar]
- 38.Kandori K., Uoya Y., Ishikawa T. Effects of acetonitrile on adsorption behavior of bovine serum albumin onto synthetic calcium hydroxyapatite particles. J. Colloid Interface Sci. 2002;252:269–275. doi: 10.1006/jcis.2002.8495. [DOI] [PubMed] [Google Scholar]
- 39.Chevalier C., Bazzal A.A., Vidic J., Février V., Bourdieu C., Bouguyon E., Le Goffic R., Vautherot J.-F., Bernard J., Moudjou M., Noinville S., Chich J.F., Da Costa B., Rezaei H., Delmas B. PB1-F2 influenza A virus protein adopts a β-sheet conformation and forms amyloid fibers in membrane environments. J. Biol. Chem. 2010;285:13233–13243. doi: 10.1074/jbc.M109.067710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang M., Dutta C., Tiwari A. Disulfide-bond scrambling promotes amorphous aggregates in lysozyme and bovine serum albumin. J. Phys. Chem. B. 2015;119:3969–3981. doi: 10.1021/acs.jpcb.5b00144. [DOI] [PubMed] [Google Scholar]
- 41.Wang S.S.S., Liu K.N., Wang B.W. Effects of dithiothreitol on the amyloid fibrillogenesis of Hen egg-white lysozyme. Eur. Biophys. 2010;39:1229–1242. doi: 10.1007/s00249-010-0576-0. [DOI] [PubMed] [Google Scholar]
- 42.Tangpasuthadol V., Pongchaisirikul N., Hoven V.P. Surface modification of chitosan films. Effects of hydrophobicity on protein adsorption. Carbohydr. Res. 2003;338:937–942. doi: 10.1016/s0008-6215(03)00038-7. [DOI] [PubMed] [Google Scholar]
- 43.Goto S., Komatsu K., Terada H. Topology of the interconversion pathway networks of cycloheptane conformations and those of related n-membered rings. Bull. Chem. Soc. Jpn. 2013;86:230–242. [Google Scholar]
- 44.Otsuka Y., Kuwashima W., Tanaka Y., Yamaki Y., Shimada Y., Goto S. Effects of heat treatment on indomethacin-cimetidine mixture; investigation of drug-drug interaction using singular value decomposition in FTIR spectroscopy. J. Pharm. Sci. 2021;110:1142–1147. doi: 10.1016/j.xphs.2020.09.049. [DOI] [PubMed] [Google Scholar]
- 45.Woody R.W. Improved calculation of the nπ* rotational strength in polypeptides. J. Chem. Phys. 1968;49:4797. doi: 10.1063/1.1669962. [DOI] [PubMed] [Google Scholar]
- 46.Becker J.W., Reeke G.N., Wang J.L., Cunningham B.A., Edelman G.M. The covalent and three-dimensional structure of concanavalin A. III. Structure of the monomer and its interactions with metals and saccharides. J. Biol. Chem. 1975;250:1513–1524. [PubMed] [Google Scholar]
- 47.Chen Y.H., Yang J.T., Chau K.H. Determination of the helix and β form of proteins in aqueous solution by circular dichroism. Biochemistry. 1974;13:3350–3359. doi: 10.1021/bi00713a027. [DOI] [PubMed] [Google Scholar]
- 48.Chang C.T., Wu C.S., Yang J.T. Circular dichroic analysis of protein conformation: inclusion of the β-turns1. Anal. Biochem. 1978;91:13–31. doi: 10.1016/0003-2697(78)90812-6. [DOI] [PubMed] [Google Scholar]
- 49.Crisma M., Formaggio F., Moretto A., Toniolo C. Peptide helices based on a-amino acids. Biopolymers. 2006;84:3–12. doi: 10.1002/bip.20357. [DOI] [PubMed] [Google Scholar]
- 50.Corrêa D.H.A., Ramos C.H.I. The use of circular dichroism spectroscopy to study protein folding, form and function. Afr. J. Biochem. Res. 2009;3:164–173. [Google Scholar]
- 51.Welch W.R.W., Kubelka J., Keiderling T.A. Infrared, vibrational circular dichroism, and Raman spectral simulations for β-sheet structures with various isotopic labels, interstrand, and stacking arrangements using density functional theory. J. Phys. Chem. B. 2013;117:10343–10358. doi: 10.1021/jp4056126. [DOI] [PubMed] [Google Scholar]
- 52.Nelson J.W., Kallenbach N.R. Stabilization of the ribonuclease S‐peptide α‐helix by trifluoroethanol. Proteins. 1986;1:211–217. doi: 10.1002/prot.340010303. [DOI] [PubMed] [Google Scholar]
- 53.Schönbrunner N., Wey J., Engels J., Georg H., Kiefhaber T. Native-like β-structure in a trifluoroethanol-induced partially folded state of the all-β-sheet protein tendamistat. J. Mol. Biol. 1996;260:432–445. doi: 10.1006/jmbi.1996.0412. [DOI] [PubMed] [Google Scholar]
- 54.Cannon D., Donald A.M. Control of liquid crystallinity of amyloid-forming systems. Soft Matter. 2013;9:2852–2857. [Google Scholar]
- 55.Diaz-Espinoza R., Nova E., Monasterio O. Overcoming electrostatic repulsions during amyloid assembly: effect of pH and interaction with divalent metals using model peptides. Arch. Biochem. Biophys. 2017;621:46–53. doi: 10.1016/j.abb.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 56.Kumar E.K., Prabhu N.P. Differential effects of ionic and non-ionic surfactants on lysozyme fibrillation. Phys. Chem. Chem. Phys. 2014;16:24076–24088. doi: 10.1039/c4cp02423k. [DOI] [PubMed] [Google Scholar]
- 57.Zeng H.J., Miao M., Yang R., Qu L.B. Effect of silybin on the fibrillation of hen egg-white lysozyme. J. Mol. Recogn. 2017;30 doi: 10.1002/jmr.2566. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.