Abstract
We present here the characterization of SPB1, an essential yeast gene that is required for ribosome synthesis. A cold-sensitive allele for that gene (referred to here as spb1-1) had been previously isolated as a suppressor of a mutation affecting the poly(A)-binding protein gene (PAB1) and a thermosensitive allele (referred to here as spb1-2) was isolated in a search for essential genes required for gene silencing in Saccharomyces cerevisiae. The two mutants are able to suppress the deletion of PAB1, and they both present a strong reduction in their 60S ribosomal subunit content. In an spb1-2 strain grown at the restrictive temperature, processing of the 27S pre-rRNA into mature 25S rRNA and 5.8S is completely abolished and production of mature 18S is reduced, while the abnormal 23S species is accumulated. Spb1p is a 96.5-kDa protein that is localized to the nucleolus. Coimmunoprecipitation experiments show that Spb1p is associated in vivo with the nucleolar proteins Nop1p and Nop5/58p. Protein sequence analysis reveals that Spb1p possesses a putative S-adenosyl-l-methionine (AdoMet)-binding domain, which is common to the AdoMet-dependent methyltransferases. We show here that Spb1p is able to bind [3H]AdoMet in vitro, suggesting that it is a novel methylase, whose possible substrates will be discussed.
Ribosomes from bacteria have been successfully assembled in vitro from their purified components (59). In contrast, this is not the case in eukaryotes in which ribosome biogenesis requires the completion of a series of events that must occur sequentially, each step being potentially the target of various regulatory mechanisms (64, 78). This complex process occurs in the nucleolus of the cell and starts with the transcription by RNA polymerase I of a precursor rRNA molecule (pre-rRNA). This pre-rRNA is assembled with ribosomal proteins and matured to yield the ribosomal particles of 40S and 60S that are exported to the cytoplasm. Maturation of the pre-rRNA includes several endo- and exonucleolytic cleavages and the modification of many of its nucleotides, a process that requires numerous small nucleolar RNAs (snoRNAs) and nucleolar proteins (69). The nucleolus of Saccharomyces cerevisiae contains ca. 100 snoRNAs that can be divided, based on their structure, into three groups: the C+D class, the H+ACA class, and the RNA component of the RNase MRP (71). The C+D class can base pair with the pre-rRNA and targets the 2′-O-ribose methylation, and the H+ACA class targets the pseudoridylation of the pre-rRNA (4, 5, 44, 71). In addition, two C+D snoRNAs, U3 and U14, and two H+ACA snoRNAs, snR10 and snR30, are required for the early cleavages of the pre-rRNA that lead to 18S formation (33, 51, 56, 70). The snoRNAs are associated with proteins to form the snoRNP particles. In yeast, the H+ACA snoRNAs associate with Gar1p (5, 18, 25), Cbf5p (46, 79), Nhp2p (79), and Nop10p (29). The C+D snoRNAs are associated with Nop1p (71, 72, 77), Nop5/58p (45, 81) and Nop56p (D. Lafontaine and D. Tollervey, unpublished data). Sof1p (36), Lcp5p (80), Mpp10p (6), and Imp3p and Imp4p (49) are exclusively associated with U3. Nop1p is required for several steps of pre-ribosome maturation. Several mutant alleles of NOP1 have been isolated in yeast that alter either synthesis of 18S and 25S (nop1-2 and nop1-5), assembly of 60S (nop1-4 and nop1-7) or methylation of the pre-rRNA (nop1-3) (73). Recently, it has been proposed that Nop1p may actually possess a domain able to bind S-adenosyl-l-methionine (AdoMet), a methyltransferase cofactor (57). Interestingly, the nop1-3 mutation which prevents pre-rRNA methylation falls into the predicted AdoMet-binding domain of Nop1p. Taken together, these observations suggest that Nop1p could be the methylase responsible for pre-rRNA methylation. NOP77 (also called NOP4) was isolated due to its genetic interaction with NOP1 (8). A small subset of Nop1p was found associated with Nop77p. It was proposed that Nop77p could be involved in pre-rRNA methylation (8), but this result was not confirmed by other authors (68). Nop4/77p is likely not a methyltransferase by itself but could assist Nop1p, for instance, in binding to the RNA through its multiple RNA binding elements. Finally, Nop2p is another putative methylase that has been implicated in a late methylation event of the 27S molecule. However, Nop2p failed to bind in vitro to [3H]AdoMet (32).
In addition to the pre-rRNA, several nucleolar and ribosomal proteins involved in ribosome synthesis are methylated. Gar1p (25), Nsr1p/gar2/nucleolin (50), Nop1p/fibrillarin (30, 48, 65), and Sbp1p (formerly Ssb1p) (37) possess a particular domain rich in glycine, arginine, and phenylalanine residues (i.e., the GAR domain), whose arginine residues have been shown, in some instances, to be dimethylated (12, 47, 52, 53). It is striking that Nop1p, which is now regarded as a putative methylase (57), is itself methylated on its arginine residues. None of the known proteins associated with Nop1p, Nop56p (21), Nop5/58p (21, 45, 81), Nop4/77p (8, 68), and Sof1p (36) are obvious candidates for being the methylase of nucleolar proteins.
Several years ago, a genetic screen was designed to isolate suppressors of a mutation of the poly(A)-binding protein (Pab1p) in yeast (spb). This screen led to the isolation of seven suppressors that all turned out to be impaired in their ability to produce normal 60S ribosomal subunits (61). One of these suppressors, spb2, encodes a ribosomal protein from the large subunit, while another, spb4, codes for a putative RNA helicase of the DEAD-box protein family that is required for normal 25S production (15, 62). Several other spb mutants affecting 60S particle synthesis have now been isolated. Some of them encode ribosomal proteins of the large subunit, while others correspond to proteins with yet unknown function (82; C. Bonnerot and B. Lapeyre, unpublished data). We report here an analysis of one of these spb mutants, spb1. Cloning and sequencing of the wild-type gene corresponding to this mutant has revealed that it had been independently identified as complementing a mutation that affects gene silencing in S. cerevisiae. This gene, for which no clear function had been assigned was simply designated by its open reading frame (ORF) name, YCL054w (54). Spb1p possesses an AdoMet-binding domain, suggesting that it could be a methyltransferase. We have analyzed rRNA processing in the spb1-2 strain and found that 27S rRNA maturation is blocked and an aberrant 23S species is accumulated while synthesis of 18S rRNA is reduced. We did not detect any variation in the global rRNA methylation pattern in the spb1-2 strain. Spb1p is localized to the nucleolus and is associated in vivo with the two nucleolar proteins Nop1p and Nop5/58p. We report here that Spb1p is able to bind [3H]AdoMet in vitro, suggesting that it is likely a novel methylase.
MATERIALS AND METHODS
Strains media and microbiological methods.
The strains used in this study are listed in Table 1. They were obtained from several laboratories as indicated or were constructed by using classical yeast techniques, using a derivative of the W303 strain with a deletion of the TRP1 gene (BMA64) that was kindly provided by F. Lacroute. The URA3 strain YBL4164 was obtained by transforming the W303 strain with a linear fragment containing the URA3 gene. YBL4365 was obtained by plasmid shuffling as follows: plasmid pAS16 was first transformed into the spb1Δ strain YDK14-1A/pDK423 HA-SPB1 CEN LEU2. Then, cells that became Leu− since they had lost the pDK423 plasmid were isolated by replica plating on media plus or minus leucine. Strains were grown either in complete yeast media or in synthetic media from Difco, as described elsewhere (26). 5-Fluoro-orotic acid (5-FOA) was bought from Toronto Research Chemicals, Toronto, Ontario, Canada. Yeast transformation was performed by using the lithium acetate method (24). Tetrad dissection was performed with a micromanipulator from Micro Video Instruments (Avon, Mass.) mounted on a Nikon microscope.
TABLE 1.
Strains used in this study
| Strain | Relevant genotypea | Plasmid | Origin |
|---|---|---|---|
| BMA64 | MATa/α trp1Δ | F. Lacroute | |
| YAS1667 | MATa pab1::HIS3 | pAS77 | A. Sachs |
| YBL4226 | MATa pab1::HIS3 trp1Δ | pBL471 | C. Bonnerot |
| YAS151 | MATα spb1-1 ADE2 | A. Sachs | |
| YBL4163 | MATα spb1-1 ADE2 URA3 | This work | |
| JR4710 | MATa spb1-2 ADE2 lys2Δ | J. Rine | |
| JR4712 | MATα spb1-2 lys2Δ | J. Rine | |
| YBL4164 | MATa URA3 | This work | |
| YBL4166 | MATa spb1-2 URA3 | This work | |
| YBL4383 | MATa spb1-1 | pAS16 | This work |
| YBL4384 | MATa spb1-2 ADE2 lys2Δ | pAS16 | This work |
| YBL4459 | MATa spb1-1 pab1::HIS3 | pBL471 | This work |
| YBL4258 | MATa spb1-2 pab1::HIS3 | pBL471 | This work |
| YBL4460 | MATa/α spb1::TRP1/SPB1 | This work | |
| YDK14-1A/403 | MATa spb1::HIS3MX6 | pDK403 | This work |
| YDK14-1A/423 | MATa spb1::HIS3MX6 | pDK423 | This work |
| YDK14-1A/376 | MATa spb1::HIS3MX6 | pDK376 | This work |
| YBL4365 | MATa spb1::HIS3MX6 | pAS16 | This work |
All strains are ade2-1 his3-11 leu2-3,112 trp1-1 ura3-1 can1-100 unless otherwise stated.
Recombinant DNA work.
Standard procedures were used (63). PCR was performed with genomic yeast DNA or plasmid DNA, as indicated, with oligonucleotide primers (Eurogentec) (Table 2) and PFU-Turbo DNA polymerase (Stratagene). PCR products were purified on spin columns (Boehringer Mannheim) prior to use. Gene disruption was done as described earlier (7) with the two oligonucleotides OBL139 and OBL140 for the PCR. Plasmids obtained from different laboratories or constructed for this study are listed on Table 3. The construction of the three plasmids pDK403, pDK376, and pDK423 that code either for Spb1p or for the N-terminally hemagglutinin (HA)-tagged Spb1p will be detailed elsewhere (D. Kressler, P. Linder, and J. de la Cruz, unpublished data). Briefly, two HA-tag motifs have been introduced after the initiator methionine to yield the sequence: M/asr(ypydvpdyag)2ssrvd/GKTQKKN. The uppercase letters are the natural residues from Spb1p; the lower case are the residues that have been inserted, and the underlined residues form the HA motif. The sequences for the three hybridization primers used to detect the pre-rRNA intermediate species have been previously described (72).
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Description | Sequence |
|---|---|---|
| OBL139 | Del SPB1 5′ | CTGCTTCAGTTTGTAGTTAGATTTAACTCAATAGAGGTGATTGGCAAAACTGATGCGGTATTTTCTCCT |
| OBL140 | Del SPB1 3′ | CCTTTATTTTCAATATTATACAAGGGAATGGAAAAATAATGCTCTTTGTTAGGGTGTTGGCGGGTGTC |
| 1 | ITS1 | CGGTTTTAATTGTCCTA |
| 2 | ITS1 | ATGAAAACTCCACAGTG |
| 3 | ITS2 | AAAGGCCAGCAATTTCAAGTTA |
TABLE 3.
Plasmids used in this study
| Plasmid | Description | Source or reference |
|---|---|---|
| pAS77 | PAB1 URA3 CEN | A. Sachs |
| pAS16 | SPB1 URA3 CEN | A. Sachs |
| pBL471 | PAB1 URA3 TRP1 CEN | This work |
| pDK403 | SPB1 LEU2 CEN | Kressler et al.a |
| pDK423 | HA-SPB1 LEU2 CEN | Kressler et al.a |
| pDK376 | GAL-HA-SPB1 LEU2 CEN | Kressler et al.a |
Kressler et al., unpublished data.
RNA work.
For pulse-chase analysis, cells were labeled essentially as previously described (72). One A600 unit of cells was labeled with either 0.1 mCi of l-[methyl-3H]methionine (85 Ci/mmol; Amersham) for 2 min or 0.1 mCi of [5,6-3H]uracil (45 Ci/mmol; Amersham) for 1 min. The chase was done with either cold methionine (50 mM) or uracil (20 mM) for the indicated times. At each time point, samples were taken and quickly centrifuged, and the cell pellets were frozen in liquid nitrogen. To label the spb1-2 strain, the amount of cells and radioactivity had to be increased, due to the low rRNA content of this mutant. Six A600 units were labeled with 0.5 mCi of either tritiated uracil or methionine. RNA was isolated from frozen cell pellets essentially as described earlier (14) with the following modifications. Each cell pellet was resuspended in 0.35 ml of buffer (0.3 M NaCl, 10 mM Tris-HCl [pH 7.5], 1 mM EDTA, 0.2% sodium dodecyl sulfate [SDS]), 0.35 ml of PCI (50% phenol, 48% CHCl3, 2% isoamyl alcohol), and 0.2 ml of zirconium beads (Biospec Products, Bartlesville, Okla.). The mixture was vortexed for 10 min at maximum speed in a cold room on a multimixer (VWR, South Plainfield, N.J.). The aqueous phase was reextracted once with an equal volume of PCI, and RNA was recovered by precipitation with cold ethanol. RNA samples were separated by electrophoresis on 1.2% agarose slab gels (Quantum) containing 20 mM MOPS (morpholinepropanesulfonic acid; pH 7.0), 5 mM sodium acetate, 0.5 mM EDTA, and 370 mM formaldehyde (11). Capillary transfer to positively charged Nylon (Schleicher and Schuell) was performed by using 40 mM NaOH as a transfer solution. To reveal tritiated molecules, membranes were exposed at −70°C for autoradiography by using Biomax MS film and a Transcreen LE (Kodak) as an intensifying screen. To prepare the radioactive probes, oligonucleotides were end-labeled with [γ-32P]ATP (Amersham) and T4 polynucleotide kinase (Appligene), and double-stranded DNA was labeled by the random priming method with [α-32P]dCTP (Amersham) and a Neblot kit (Biolabs).
Polysome analysis.
Polysomes were prepared according to the Sachs' laboratory protocol that was adapted from a previously published method (34). In brief, 20 ml of cells were grown to late mid-log phase (A600 ∼1 unit), and then cycloheximide was added to a final concentration of 0.1 mg/ml, and the cells were rapidly cooled in an ice-cold water bath. Cells were washed once in 20 ml of breaking buffer (100 mM KCl, 20 mM HEPES [pH 7.4], 2 mM Mg acetate, 14.8 mM β-mercaptoethanol, 0.1 mg of cycloheximide per ml) once in 1 ml of breaking buffer and then were resuspended to obtain a total volume of 0.4 ml (cells plus medium). Cold zirconium beads were added, and the tubes were frozen in liquid nitrogen. After thawing of the suspension, the cells were broken by vortexing on a multimixer for 45 s. Then, the extracts were clarified two times by centrifugation for 10 min at 16,000 × g. Approximately 10 A260 units were loaded onto a 15 to 50% sucrose gradient prepared as described elsewhere (55) in breaking buffer minus cycloheximide. The gradients were centrifuged in a Beckman SW41 rotor at 4°C for 2 h at 275,000 × g and collected at the bottom of the tube, while the A260 value was monitored by using a continuous flow cell UV detector (Pharmacia).
Immunofluorescence microscopy.
Cells were fixed directly in the culture medium with 4% paraformaldehyde (Electron Microscopy Science) for 30 min, washed three times in 0.1 M KPO4 (pH 6.4)–1.2 M sorbitol, and digested with zymolyase 20T (0.1 μg/μl) for 45 min at room temperature on a wheel. After one wash in the same buffer, cells were applied on a poly-l-lysine (Sigma)-coated slide and allowed to sediment for 10 min. Cells were dehydrated 3 min in cold methanol, rapidly washed in cold acetone, and dried at room temperature. After rehydration of the cells in phosphate-buffered saline (PBS) (27) containing 1% bovine serum albumin (BSA) (Fraction V; Sigma), immunodetection was achieved as follows. Primary antibodies were added in 1% BSA-PBS in a total volume of 4 μl at the following dilution: anti-Nop1p (3), 1/1,000; and anti-HA (3F10, Boehringer Mannheim), 1/200. The mixture was then incubated in the dark for 1 h. After three washes with 10 μl of 1% BSA-PBS, secondary antibodies were added at the following dilution: anti-mouse-fluorescein isothiocyanate (FITC), 1/60; and anti-rat-rhodamine, 1/160 (Sigma). The mixture was then incubated for another hour at room temperature. After three more washes to remove the fluorescently labeled secondary antibodies, the slides were mounted in 90% glycerol–10% PBS containing 50 ng of DAPI (4′,6′-diamidino-2-phenylindole; Sigma) and 1 mg of p-phenylenediamine per ml as anti-fading agent. Slides were viewed on a Leitz Microscope, and images were acquired with a high-resolution camera (Hamamatsu).
Protein extraction.
Native protein extracts were prepared from mid-log-phase cells that were harvested by centrifugation, washed in ice-cold buffer (0.9% NaCl, 1 mM NaN3, 10 mM EDTA, 50 mM NaF), and frozen in liquid nitrogen. Yeast pellets were weighed, thawed on ice, and resuspended in 5 volumes of ice-cold breaking buffer (50 mM Tris-Cl [pH 7.5], 15 mM MgCl2, 150 mM NaCl, 1% NP-40, 1 mM EDTA, 1 mM dithiothreitol [DTT]) supplemented with a complete protease inhibitor cocktail (Boehringer Mannheim). Zirconium beads were added to the mixture, and cells were lysed by vortexing at 4°C for 4 min on a multimixer. Extracts were clarified three times by centrifugation at 4°C for 10 min at 16,000 × g. Protein concentrations were determined by the Bradford protein assay (Sigma).
SDS-PAGE and Western blotting.
Routinely, 50 to 100 μg of proteins were analyzed by SDS–% polyacrylamide gel electrophoresis (PAGE) (63) and then transferred onto a nitrocellulose membrane (Protran; Schleicher and Schuell) by using a semidry blotter (OWL) for 3 h at 1 mA/cm2 in 39 mM glycine–48 mM Tris-base–0.037% SDS–20% methanol (pH 8.3). After transfer, the proteins were revealed by staining the membrane in 0.2% amido black (Sigma). Western blotting was done essentially as described elsewhere (27). Membranes were saturated in TBS (27), 0.05% Tween, and 3% nonfat milk. Antibodies were incubated in the same medium for 1 h at room temperature. Primary antibodies were added at the following dilution: anti-HA (3F10; Boehringer Mannheim), 1/1,000; anti-Nop1p (3), 1/10,000; anti-Qsr1p, 1/1,000 (75). Anti-rat, -mouse, and -rabbit secondary antibodies coupled to horseradish peroxidase were obtained from Sigma.
Preparation of an anti-HA affinity column.
The anti-HA affinity column was prepared essentially as previously described (27). First, 2 mg of anti-HA 12CA5 (58) was mixed with 1 ml of protein A-Sepharose (Pharmacia) in a total volume of 10 ml adjusted to 100 mM Tris-Cl (pH 8.0) and incubated for 1 h at room temperature. Beads were washed three times with 10 ml of 0.2 M Na2B4O7 (anhydrous) (pH 9.0) (Sigma) and resuspended in 10 ml of the same solution. Immunoglobulin G antibodies were cross-linked to protein A-Sepharose by the addition of dimethyl pimelimidate (Sigma) as follows: 52 mg of powder was added to the 10-ml suspension of beads (20 mM, final), and the mixture was incubated for 30 min at room temperature with gentle mixing. The reaction was stopped by washing the beads once in 0.2 M ethanolamine and then incubated for 2 h at room temperature in 0.2 M ethanolamine (pH 8.0) with gentle mixing. The beads were then washed five times in 10 ml of PBS and stored in 1.5 ml of 0.01% merthiolate in PBS.
Immunopurification and coimmunoprecipitation.
Coimmunoprecipitations were performed with 0.5 mg of native protein extract and 2 μl of monoclonal antibodies raised against Nop1p (A66) (3) or Nop5/58p (B47) (81) in a final volume of 30 μl of breaking buffer (see above, in the protein extraction section) containing 5 mg of BSA per ml and incubated for 2 h at 4°C. Antigen-antibody complexes were precipitated with 16 μl of a 50% slurry of protein A-Sepharose beads (Pharmacia) incubated for 2 h at 4°C. HA-Spb1p was immunoprecipitated with protein A-Sepharose beads covalently coupled to anti-HA antibodies as described above by using 20 μl of beads. The beads were recovered by centrifugation for 1 min at 400 × g and washed five times with 0.5 ml of breaking buffer. Samples were denatured for 5 min at 90°C in Laemmli loading buffer and analyzed by SDS-PAGE (43).
Gel filtration and cation-exchange chromatography.
A native protein extract (100 mg) was loaded onto a 120-ml bed gel filtration column (Superdex-200; Pharmacia) that was preequilibrated at 1 ml/min with 25 mM Tris-Cl (pH 7.5), 150 mM NaCl, 1 mM EDTA, and 2 mM DTT. Then, 60 fractions of 2 ml were collected, of which 20 μl was analyzed by Western blotting to detect the HA-Spb1 protein. A fraction containing HA-Spb1p was diluted five times with 25 mM MES (pH 6.1)–1 mM EDTA–2 mM DTT and loaded onto a 1-ml cation-exchange column (Mono-S; Pharmacia) equilibrated with the same buffer. The column was washed at 1 ml/min until the optical density of the flowthrough nearly reached the baseline. Elution was performed by a two step NaCl gradient (0 to 0.5 M and 0.5 to 1 M). Fractions were taken and subjected to Western blot analysis with anti-HA (3F10; Boehringer Mannheim), anti-Nop1p (A66), and anti-Qsr1p (75) antibodies.
In vitro [3H]AdoMet-binding assay.
This procedure was adapted from a previously published method (31) and from a protocol developed by John Aris (personal communication). Proteins were immunoprecipitated as described above, and then the beads were washed twice in 0.5 ml of AdoMet-binding buffer (50 mM Tris-Cl [pH 8.0], 100 mM NaCl, 2 mM DTT, plus complete protease inhibitor cocktail [Boehringer Mannheim]) and resuspended in 40 μl of the same buffer in a 0.5-ml Eppendorf tube to which 5.5 mCi of [3H]AdoMet (75 Ci/mmol; NEN) was added. Tubes containing the suspensions of beads were placed on ice, caps open, under a transilluminator table (six tubes of 15 W) that was held upside down at 4 cm from the top of the tubes. The suspension was UV irradiated for 30 min at 254 nm. Beads were then resuspended in Laemmli loading buffer, and the proteins were released by incubating them for 5 min at 90°C. Radioactive incorporation was measured by determining the trichloroacetic acid-precipitable counts on 1/10 of the volume. The remaining 9/10 was analyzed as described above by SDS-PAGE, transferred onto membrane, and exposed at −70°C to Biomax MS films with intensifying screens (Transcreen LE; Kodak).
RESULTS
spb1-1 and spb1-2 are both able to suppress the deletion of PAB1.
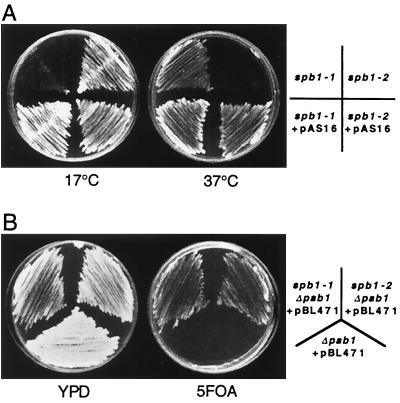
The wild-type SPB1 gene was cloned by transforming a strain harboring the cold-sensitive spb1-1 allele with a yeast genomic library prepared in a centromeric plasmid (A. B. Sachs, personal communication). Two clones were obtained that were able to complement the growth defect of the spb1-1 strain at 17°C, the shortest clone being called pAS16. Sequence analysis of the cloned fragment revealed the presence of an ORF termed YCL054w (A. B. Sachs, personal communication). Independently, a thermosensitive allele for that gene had been isolated in a genetic screen designed to identify essential genes involved in silencing (54). We will refer to this thermosensitive allele as spb1-2. Plasmid pAS16 or a derivative containing only the wild-type SPB1 gene was able to rescue the cold-sensitive phenotype of spb1-1 as well as the thermosensitive phenotype of spb1-2 (Fig. 1A). We then tested whether spb1-2 was able, like spb1-1, to bypass the deletion of PAB1 by assaying the growth on 5-FOA-containing plates of a pab1Δ strain complemented by a pPAB1 URA3 plasmid. An spb1-1 strain can grow in the absence of PAB1 and therefore is able to lose the plasmid and to grow on 5-FOA plates. As shown on Fig. 1B, the strain pab1Δ spb1-2 is able to grow at 25°C on 5-FOA plates, indicating that spb1-2 as well as spb1-1 is able to bypass the deletion of PAB1. Thus, the spb phenotype of spb1-1 is not specific for that allele.
FIG. 1.
Strains spb1-1 and spb1-2 are complemented by the wild-type SPB1 gene, and both behave as suppressors of pab1Δ. (A) The wild-type SPB1 gene can restore normal growth of both spb1-1 and spb1-2. Strains spb1-1 (YAS 151), spb1-2 (JR4710), or these strains transformed by the plasmid pAS16 that contains the wild-type SPB1 gene (YBL4383 and YBL4384, respectively) were streaked onto yeast-peptone-dextrose (YPD) plates and grown at 17°C (for 7 days) or at 37°C (for 3 days). (B) Double mutants spb1-1 pab1Δ (YBL4459) and spb1-2 pab1Δ (YBL4258) were streaked onto a YPD plate, along with a single pab1Δ strain as a control. These three strains harbor the plasmid pBL471 that bears a wild-type PAB1 gene that complements the absence of PAB1 on their chromosome and a URA3 gene as a marker. When streaked onto 5-FOA-containing plates, the pab1Δ strain is unable to grow due to its inability to lose the PAB1 gene (plasmid pBL471 containing the URA3 gene), while both spb1-1 and spb1-2 are able to lose this plasmid and thus to grow on these plates.
SPB1 is an essential gene that complements spb1-1 and spb1-2.
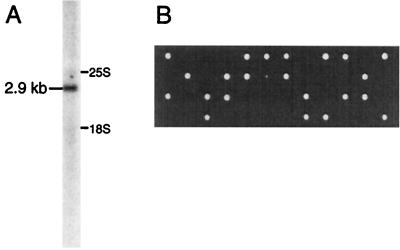
The YCL054w ORF was originally believed to be 2,175 nucleotides (nt) long and to encode a 725-amino-acid-long protein with a calculated molecular size of 83 kDa (60). However, recent sequence corrections have proposed extending the size of this ORF to 2,526 nt, encoding a protein with a molecular size of 96.5 kDa. This ORF is located very close to the PNB1 gene, leaving only 168 nt between the two ORFs. The SPB1 mRNA was visualized in a Northern blot experiment with a fragment of the coding region as a probe (Fig. 2A). The mRNA detected in wild-type cells has a size of 2.9 kb, as determined by using mature 25S and 18S rRNAs as molecular size markers. This is compatible with the ORF size of 2.5 kb, plus the 5′ untranslated region (5′UTR) the 3′UTR and the poly(A) tail. A PCR-based strategy was used to disrupt the SPB1 gene (7). A DNA fragment containing the TRP1 gene surrounded by the flanking sequences of the SPB1 gene was prepared and used to transform a wild-type diploid strain (BMA64). Transformants were tested by Southern blotting for the correct integration of the DNA at the SPB1 locus, then sporulated. Dissection of the tetrads thus obtained (Fig. 2B) shows that only two spores grew per tetrad, which were all Trp−, indicating that the SPB1 gene is essential for spore development. This result was confirmed by using an spb1Δ::HIS3MX6 strain containing a plasmid carrying the wild-type SPB1 gene and a URA3 marker (YBL4365). This strain was unable to lose the SPB1-containing plasmid and thus to grow on a 5-FOA-containing plate, demonstrating that SPB1 is essential for cell viability (data not shown). Finally, it has been shown that spb1-2 is allelic to YCL054 (SPB1) (54). A similar result has now been obtained for the spb1-1 allele (D. Kressler and J. de la Cruz, unpublished data).
FIG. 2.
Characterization of the SPB1 gene. (A) Total RNA from a wild-type W303 strain was probed in a Northern blot experiment with a fragment of the SPB1 gene. The positions of the 25S and 18S rRNA are indicated on the right of the lane. A single band, one corresponding to an mRNA of 2.9 kb, was detected by this probe. (B) One of the two copies of the SPB1 gene was deleted in a diploid strain by insertion of a TRP1 marker (YBL4460). These cells were then sporulated and the four daughter spores were dissected by micromanipulation and grown on YPD plates. In each tetrad, only two spores were able to grow, all of which were Trp−, indicating that the gene is essential for spore development.
SPB1 codes for a protein with a putative AdoMet-binding motif that is conserved throughout evolution.
Methylases that utilize AdoMet as a donor of methyl groups supposedly share a common domain structure able to bind this cofactor (66). However, there is still no precise definition of a consensus sequence for this domain. Recently, a search of the S. cerevisiae genome has led to the proposal that 33 ORFs could possess an AdoMet-binding motif, seven of which are known methyltransferases (57). Spb1p was not identified as a putative methyltransferase in this analysis, while in a previous study it was suggested to possess an AdoMet-binding domain due to its homology with YCR047c, another putative methyltransferase (9, 35, 40, 41). Interestingly, Nop1p is now considered as a candidate methyltransferase (57), a possibility that had escaped previous searches. We screened several databases for sequences having homologies with Spb1p from S. cerevisiae. Eight putative proteins from different organisms exhibit significant similarity with Spb1p, in particular with its N-terminal moiety which contains the putative AdoMet-binding motif. An alignment of the N-terminal 221 residues of Spb1p with proteins from various species is presented here (Fig. 3), with the four motifs that are proposed to form the AdoMet-binding domain indicated as motifs I, post-I, II, and III (57). However, there is still no evidence that any of these proteins might be able to bind AdoMet in vitro or in vivo. Interestingly, the strongest similarity between the various proteins do not always correspond to the four AdoMet-binding motifs. Motifs II and III are the least conserved, while there are blocks of perfect matches on both sides of these motifs.
FIG. 3.
Sequence alignment of the predicted AdoMet-binding region of Spb1p from S. cerevisiae with potential homologues from different species. Eight predicted proteins from different organisms possess significant similarities with the N-terminal moiety of Spb1p from S. cerevisiae (Sc; accession number, CAA42391) as follows: Bc, Botryotinia fuckeliana (AL111693); Sp, Schizosaccharomyces pombe pmt2 (CAA22605); Mm, Mus musculus (AI574417); Cp, Cryptosporidium parvum (AQ450296); At, Arabidopsis thaliana (CAB39594); Hs, Homo sapiens JM23 (HSA005892); Ce, Caenorhabditis elegans (CAA85279); and Ec, E. coli (74) (AAC76211). Residues identical between the different species are on a black background, and the ones that are similar are on a gray background. The four motifs (I, post I, II, and III) that form the putative AdoMet-binding domain are boxed.
Spb1p is required for normal rRNA synthesis.
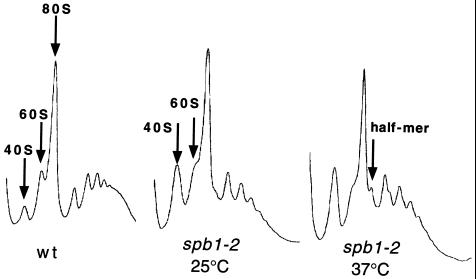
It has been previously shown that the spb1-1 strain had a reduced content in 60S ribosomal particles (61), while spb1-2 had a very low total rRNA content (54), suggesting that Spb1p could be involved in ribosome biogenesis. Polysomes were prepared from the spb1-2 strain, grown either at the permissive or at the nonpermissive temperature, and analyzed on a 15 to 50% sucrose gradient. As for the spb1-1 allele, the level of 60S particles in an spb1-2 strain is strongly reduced compared to the wild type, with a relative excess of 40S particles, even when the cells were grown at the permissive temperature. The defect was more pronounced at the restrictive temperature, with an accumulation of half-mers (28), a structure attributed to a stoichiometric imbalance of subunits due to the reduced level of 60S (Fig. 4). This phenotype is likely due to a defect in 60S synthesis, as shown for other spb mutants that correspond to ribosomal proteins of the 60S subunits or to proteins involved in the synthesis of these particles (62, 82; Bonnerot and Lapeyre, unpublished data).
FIG. 4.
Polysome profile analysis. Cellular extracts were loaded onto 15 to 50% sucrose gradients and centrifuged for 2 h at 275,0000 × g. A continuous A260 record for each gradient is presented with the top of the tube on the left. Extracts were prepared from the following: wt, wild-type strain (YBL4164); spb1-2 25°C, the spb1-2 strain (JR4710) grown at the permissive temperature of 25°C; and spb1-2 37°C, the spb1-2 strain grown for 1 h at the nonpermissive temperature of 37°C. The arrows indicate the peaks of 40S, 60S, and 80S subunits and of the half-mer.
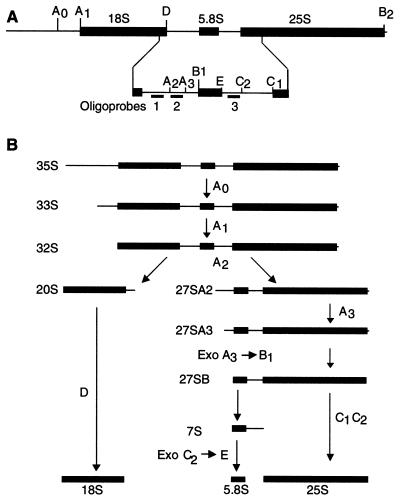
To analyze in greater detail the defect in ribosome synthesis in an spb1-2 strain, pulse-chase labeling experiments were performed in the presence of either l-[methyl-3H]methionine or [3H]uracil. Labeled uracil is incorporated into the pre-rRNA during transcription by RNA polymerase I, while the methyl group of methionine is transferred to the pre-rRNA posttranscriptionally. Due to the rapid metabolism of methionine, the half-lives of the pre-rRNA intermediates appear much shorter when using tritiated methionine to label the pre-rRNA than when using tritiated uracil that is only slowly chased by the addition of cold uracil. In the wild-type strain, the 35S pre-rRNA is cleaved sequentially at sites A0, A1, and A2 to generate successively the 33S, 32S, and then 20S and 27SA2 intermediates (Fig. 5). The 20S pre-rRNA is then processed to mature 18S, whereas the 27SA2 is processed to 27SA3, 27SB, 25S, and 7S, the 7S being then converted into 5.8S (69). Due to the very low rRNA synthesis in an spb1-2 strain, both the amount of cells labelled and the amount of the radioactive precursor added had to be increased to visualize pre-rRNA intermediates (see Materials and Methods). In the spb1-2 mutant strain, processing of the rRNA precursors is slowed down with an accumulation of 35S, even when the cells were grown at the permissive temperature (Fig. 6, lanes 4 to 6). However, the most striking defect is the accumulation of the 27S pre-rRNA in the mutant compared to the wild-type strain. In the wild-type strain grown at 37°C, labeling with tritiated methionine shows that processing of the 27S is complete after only 4.5 min of chase, while in the spb1-2 strain no 25S is formed after 6.5 min of chase (Fig. 6, lanes 10 to 12). In addition to the poor synthesis of rRNA molecules and the accumulation of 27S pre-rRNA in the mutant, there is also an accumulation of the abnormal 23S species. The 23S form is believed to result from the inhibition of cleavage at sites A0, A1, and A2 and therefore to extend from the 5′ end of the pre-rRNA to the A3 site. In contrast to mutations that affect mostly the 18S formation in which no 20S is formed, we observed here that both 20S and 23S are made, suggesting that in the mutant there is only a partial inhibition of the cleavage at sites A0, A1, and A2.
FIG. 5.
Scheme of pre-rRNA processing in yeast. (A) Structure of the rRNA transcript with the cleavage sites and the location of the hybridization probes. The primary transcript of 35S contains the 5′ external transcribed spacer, followed by the 18S rRNA, the ITS1, the 5.8S rRNA, the ITS2, the 25S rRNA, and the 3′ external transcribed spacer. The mature species of 18S, 5.8S, and 25S are represented by thick lines, and the spacers are represented by thin lines. The location of the three hybridization primers is also shown (Oligo 1, Oligo 2, and Oligo 3). (B) Simplified pre-rRNA processing scheme. The 35S pre-rRNA is cleaved successively at sites A0 and A1 to yield the 33S and 32S pre-rRNA species. Then, cleavage at A2 frees the 20S form that can further be processed into 18S rRNA and the 27SA2 that undergoes a more complex pathway. Cleavage at A3 leads to 27SA3 that can be processed into two different forms. In this simplified scheme, only 27SB is represented that will lead to 7S and 25S after cleavage at C1 and C2. 7S will be further processed into 5.8S.
FIG. 6.
Pulse-chase labeling of the pre-rRNA. Cells were grown either at 25°C or heat-shocked for 1 h at 37°C; they were then labeled for 2 min with tritiated methionine and chased with cold methionine for the indicated times. Total RNA was extracted and separated on a denaturing 1.2% agarose gel that was transferred to a nylon membrane and exposed with an intensifying screen for three days. Lanes 1 to 3 and 7 to 9, wild-type strain (YBL4164); lanes 4 to 6 and 10 to 12, spb1-2 (YBL4166); lanes 1 to 6, cells grown at 25°C; lanes 7 to 12, cells grown at 37°C.
Since Spb1p possesses a putative AdoMet-binding motif, suggesting that it could be a novel methylase, we investigated whether the spb1 mutant strain presents a defect in pre-rRNA methylation. Our data show clearly that global rRNA methylation is not affected by a mutation in the SPB1 gene, even when the cells were grown at the restrictive temperature (Fig. 6, lanes 10 to 12). We measured the incorporation of tritium when the cells were labeled either with l-[methyl-3H]methionine or with [3H]uracil and found that the ratio of methyl groups versus uracil was essentially the same in the wild-type and the mutant strains (data not shown). However, we cannot exclude the possibility that only a subset of the numerous methylated sites would be under the dependence of this methylase and that their absence would be masked by the bulk of the methyl groups.
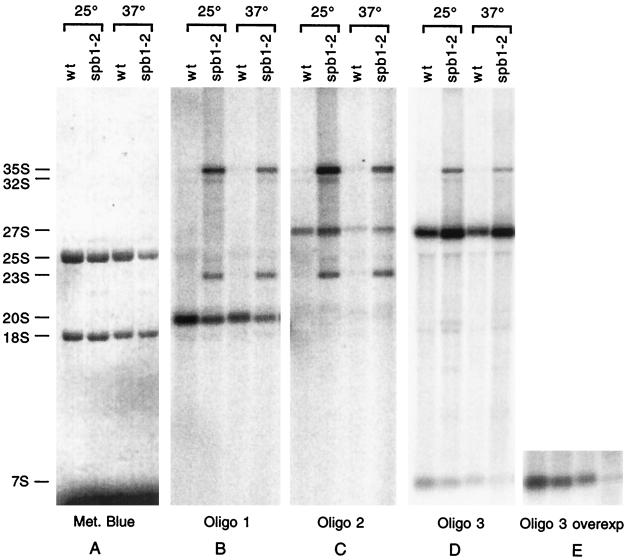
Northern analysis with oligonucleotides hybridizing to different parts of internal transcribed spacer 1 (ITS1) and -2 as probes reveals the identity of the various intermediates that are accumulated or are missing in the spb1-2 strain grown at permissive and restrictive temperatures (Fig. 7). The spb1-2 strain contains approximately six times less rRNA than the wild-type strain, making difficult to detect the various pre-rRNA intermediates. Therefore, for the mutant strain the amount of cells from which the RNA was prepared was increased in order to load similar amounts of rRNA for the wild-type and the mutant strains, as demonstrated by methylene blue staining (Fig. 7A). In the spb1-2 mutant, there is an accumulation of the 35S precursor that is not detected in the wild-type (Fig. 7B to D). Hybridization with oligonucleotide 3 reveals an accumulation of 27S in the mutant that is not converted into 25S and 7S (Fig. 7D), a phenotype that is more pronounced at the restrictive temperature, as evidenced by the absence of 7S at this temperature (Fig. 7E). Hybridization with oligonucleotide 2 detects the presence of 27SA2 among the bulk of the other 27S species, a result that is expected since there is some 20S pre-rRNA that is still made in the mutant (Fig. 7C). Another defect exhibited by the spb1-2 strain is the accumulation of the 23S species, which are detected by oligonucleotide 1 and 2 (Fig. 7B and C). The accumulation of the aberrant 23S species, which is most likely a dead-end product that cannot be converted into mature 18S rRNA, correlates with a decrease of the 20S precursor. Taken together, these data suggest that the strongest defect in the spb1-2 mutant is a complete inhibition of the processing of the 27S into 25S and 7S, which is accompanied by a partial inhibition of the cleavage at sites A0, A1, and A2.
FIG. 7.
Northern blot hybridization of the various pre-rRNA intermediate species. Pre-rRNA intermediates can be detected by Northern blotting by using probes complementary to various regions of the different spacers. Total RNA was extracted from wild-type (YBL4164) or spb1-2 (YBL4166) strains grown either at a permissive temperature (25°C) or heat-shocked for 1 h at a nonpermissive temperature (37°C) and tested by hybridization with oligonucleotide probes 1, 2, and 3 as indicated (see Fig. 5 for the positions of the probes). Equivalent amounts of total RNA extracted from the wild-type and the spb1-2 strains were loaded in each lane and visualized by methylene blue staining (left panel). Positions of the major bands are indicated on the left. Panel E corresponds to an overexposure of panel D in order to clearly detect the 7S species that is no longer accumulated in the spb1-2 strain grown at the nonpermissive temperature.
Spb1p is a nucleolar protein.
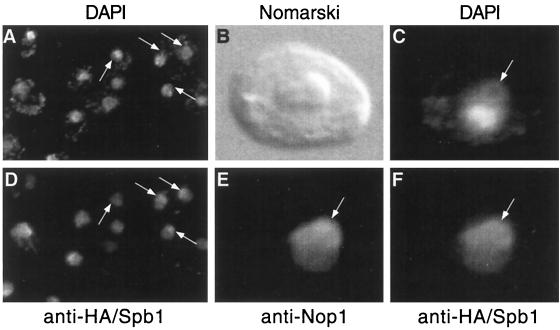
Since Spb1p is required for ribosome synthesis, we expected to find the protein associated with the nucleolus. A construction expressing a tagged version of Spb1p fused to the HA peptide (HA-Spb1p) was transformed into an spb1Δ strain. The fusion protein was detected by indirect immunofluorescence with an anti-HA antibody raised in rats. The bulk of the nuclear DNA is revealed by DAPI staining, which also shows a lightly stained crescent known to correspond to the nucleolus (67). The anti-HA antibodies decorate precisely this crescent with a light background on the remainder of the nucleus (Fig. 8A and D). A mouse monoclonal antibody directed against Nop1p (3) was used as another way to localize the nucleolus. The result of a triple-labeling experiment (DAPI, α-Nop1, and α-HA) indicates clearly that Spb1p is localized to the nucleolus, overlapping the distribution of Nop1p (Fig. 8C, E, and F). Double labeling (DAPI and α-HA) with a strain that does not express the HA-tagged Spb1p gave no signal (data not shown).
FIG. 8.
Indirect immunofluorescence detection of Spb1p within the nucleolus. Cells expressing HA-Spb1p (YDK14-1A/pDK423) were fixed with paraformaldehyde and processed for immunofluorescence as described in Materials and Methods. DAPI was added to the mounting medium to visualize the DNA within the mitochondria and the nucleus (DAPI). Anti-HA rat antibodies detect the HA peptide fused to Spb1p within the nucleolus (secondary anti-rat antibody coupled to Texas red). (A) A large field showing several cells labelled with DAPI; panel (D) the same cells observed through a Texas red filter show that HA-Spb1p is mostly localized to the nucleolus. Triple-labeling experiments have been performed by also using a mouse monoclonal antibody raised against Nop1p as a nucleolar marker. (B) Higher magnification of a cell photographed by using interferential contrast (Nomarski); (C) the same cell stained with DAPI reveals the crescent shape zone that corresponds to the nucleolus; (E) anti-Nop1p decorates the nucleolus (secondary anti-mouse antibody coupled to fluorescein); (F) anti-HA-Spb1p gives a signal that overlaps mostly with the signal obtained with the anti-Nop1p antibodies. Arrows show the positions of the nucleoli.
Spb1p is associated with the nucleolar proteins Nop1 and Nop5/58.
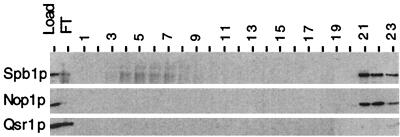
Several nucleolar components involved in ribosome biogenesis are known to be present within large ribonucleoprotein complexes. We investigated whether Spb1p was also associated with such large complexes. Native cell extracts were prepared from cells expressing the HA-Spb1 fusion protein and fractionated according to their size on a Sephadex column. Spb1p was recovered only in high-molecular-weight fractions, in the size range of 1 to 4 MDa, along with Nop1p and Qsr1p (16), a ribosomal protein associated in the cytoplasm with the 60S subunits (data not shown). A fraction containing Spb1p was then subjected to ion-exchange chromatography. Spb1p was not retained on an anion-exchange column (Mono-Q) (data not shown), while it bound to a cation-exchange column (Mono-S). Spb1p was eluted mostly at a high salt concentration, in the same fractions as had Nop1p, while Qsr1p did not bind to that column under these conditions (Fig. 9). It is noteworthy that the predicted pIs for Nop1p and Spb1p are, respectively, 10.3 and 8.0, and this suggests that the two proteins did not come out in the same fractions simply because they have the same biochemical characteristics.
FIG. 9.
Spb1p is eluted with Nop1p on a Mono-S column. Cellular extracts containing HA-Spb1p (YDK14-1A/pDK376) were fractionated by size chromatography on a Sephadex column. A high-molecular-weight fraction containing HA-Spb1p was then loaded onto a cation-exchange column (Mono-S). Elution was then performed with a gradient of NaCl that was applied in two steps. Fractions of 1 ml were collected, and a sample of each fraction was subjected to an SDS–10% PAGE, transferred to nitrocellulose, and probed with antibodies revealing HA-Spb1p (upper panel), Nop1p (middle), or the ribosomal protein Qsr1 (lower panel). Load, the fraction that was applied onto the Mono-S column; FT, flowthrough fraction that was not retained by the column; lanes 1 to 15, first slope of the gradient from 0 to 0.5 M NaCl; lanes 16 to 23, second slope of the gradient from 0.5 to 1 M NaCl.
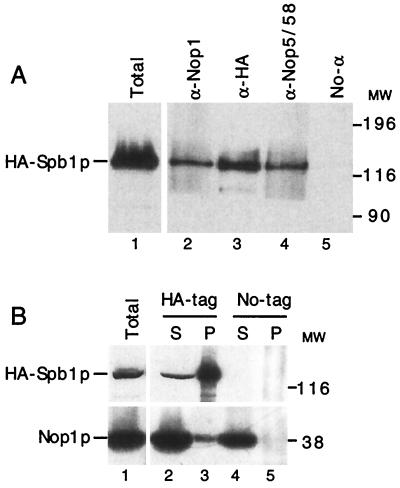
These findings led us to further characterize potential interactions between Spb1p and Nop1p by coimmunoprecipitation. We also searched for interactions with Nop5/58p, a protein associated with Nop1p and the C+D snoRNAs (21, 44, 81). Antibodies raised against Nop1p (Fig. 10A, lane 2) or against Nop5/58p (lane 4) immunoprecipitated HA-tagged Spb1p as revealed by Western blot analysis with an anti-HA antibody. The specificity of the reaction is given by the absence of HA-Spb1p when the antibodies have been omitted (Fig. 10A, lane 5) or when using a monoclonal antibody raised against Pab1p (1) (data not shown). When immunoprecipitation was first performed with anti-HA antibody to precipitate HA-Spb1p, then Nop1p was detected in the pellet (Fig. 10B, lane 3). Nop1p was not detected in the pellet when immunoprecipitation was done with a strain in which Spb1p was not HA tagged (Fig. 10B, lane 5) or when we were using an HA-tagged strain incubated without antibodies (data not shown). We can conclude from these experiments that Spb1p, Nop1p, and Nop5/58p are physically associated in vivo.
FIG. 10.
Spb1p is associated with Nop1p and Nop5/58p as shown by coimmunoprecipitation. (A) Cellular extracts prepared from a strain expressing HA-Spb1p (YDK14-1A/pDK376) were incubated with various antibodies, and the complexes were precipitated with protein A-Sepharose beads. After several extensive washes, proteins were eluted from the beads, separated by SDS–10% PAGE, and analyzed by Western blotting with anti-HA antibodies that detect HA-Spb1p. Lane 1, total cell extract corresponding to one-tenth the amount of protein used for the immunoprecipitation; lanes 2 to 5, pellets from the immunoprecipitation. Lane 2, anti-Nop1p; lane 3, anti-HA; lane 4, anti-Nop5/58; lane 5, no antibody added. MW, molecular weight markers in thousands. (B) Cellular extracts prepared from strains expressing either wild-type Spb1p or HA-tagged Spb1p were immunoprecipitated with the anti-HA antibodies. Supernatants (S) and pellets (P) were then analyzed by Western blotting with the anti-HA antibodies (upper panel) or with the anti-Nop1p antibodies (lower panel). Lane 1, total cell extract corresponding to one-tenth the amount of protein used for the immunoprecipitation; lanes 2 and 3, strain YDK14-1A/pDK423 that expresses HA-Spb1p; lanes 4 and 5, strain YDK14-1A plus pDK403 that expresses wild-type Spb1p. Lanes 2 and 4, one-tenth the supernatants; lanes 3 and 5, pellets. MW, molecular weight markers in thousands.
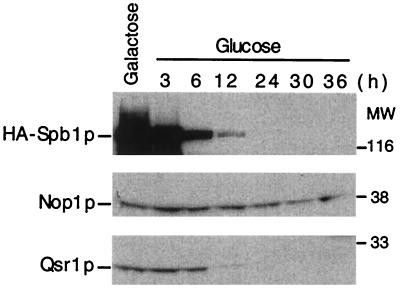
Finally, to test the possibility that the defect in ribosome synthesis observed in an spb1-2 mutant could be due to a destabilization of Nop1p, we examined the stability of Nop1p in a strain depleted of Spb1p. A strain harboring a conditional allele of SPB1, placed under the control of the GAL promoter, was shifted from galactose- to glucose-containing medium, and samples were taken after 3, 6, 12, 24, 30, and 36 h. Western blot analysis reveals that Spb1p is weakly detected after 12 h in glucose and has disappeared after 24 h (Fig. 11). At that time the 60S ribosomal protein Qsr1p has also disappeared, a result that supports the view that Spb1p is required for the synthesis or the stability of 60S ribosomal subunits. In contrast, Nop1p is still detected 36 h after the shift, indicating that depletion of Spb1p does not destabilize Nop1p. This may reflect the fact that only a small fraction of Nop1p may be associated with Spb1p, as suggested by coimmunoprecipitation experiments (Fig. 10B, lanes 2 and 3). However, indirect immunofluorescence reveals that in most cells Nop1p is no longer visible in the nucleolus. Some cells present a strong nuclear signal, while others are essentially not labelled (data not shown).
FIG. 11.
Steady-state level of Nop1p and Qsr1p upon Spb1p depletion. A strain expressing HA-Spb1p under the control of a GAL promoter (YDK14-1A/pDK376) was grown in galactose until early log phase and then transferred to glucose-containing medium to turn off the expression of Spb1p. Samples were taken at different times after the shift, and proteins were extracted. Equal amounts of proteins for each time point were separated by SDS–10% PAGE. Western blotting with anti-HA antibodies detects HA-Spb1p migrating at 120 kDa (upper panel). The anti-Nop1p revealed the protein at 38 kDa (middle panel), and Qsr1p was detected at 20 kDa (lower panel). MW, molecular weight markers in thousands.
Spb1p is able to bind in vitro to [3H]AdoMet.
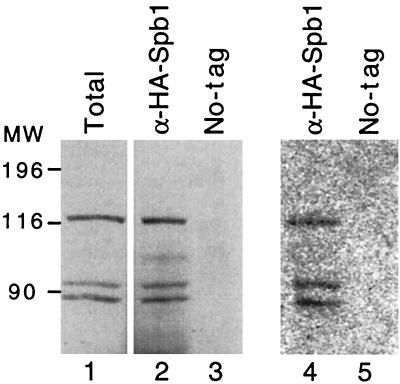
The only remarkable feature of the Spb1p sequence is the presence of a consensus AdoMet-binding motif. Due to the poor definition of this consensus, Spb1p has been regarded as a putative AdoMet-binding protein by some authors (41) and has not been retained by others (57). In order to get some insights into Spb1p's biochemical functions, we undertook to establish an in vitro assay to test for AdoMet-binding activity of Spb1p. Our assay was based on previous experiments performed with large quantities of recombinant proteins that were incubated with [3H]AdoMet and then cross-linked by using UV light irradiation before being analyzed by SDS-PAGE and autoradiography (31). The goal of our assay was to bind the protein to be tested to Sepharose beads and then to incubate these beads with [3H]AdoMet, followed by UV light cross-linking. Proteins labeled by [3H]AdoMet were separated by SDS-PAGE and visualized by autoradiography. We prepared a column of anti-HA antibodies by cross-linking purified anti-HA immunoglobulin G antibodies to protein A-Sepharose beads. These beads were then incubated with a native cellular extract prepared from cells expressing HA-tagged Spb1p. After extensive washes, HA-Spb1p bound to the beads was incubated with [3H]AdoMet and UV cross-linked. Complexes were then separated by SDS-PAGE, transferred to a nitrocellulose membrane, and revealed by autoradiography for the presence of tritiated molecules. The same membrane was then tested by Western blotting to identify unambiguously the proteins of interest. Spb1p is very sensitive to proteolysis and often gives raise to several peptides, most of which have the same N terminus, as evidenced by their detection with the anti-HA antibody. In the present experiment, three bands of 116, 93, and 87 kDa were detected by the antibody (Fig. 12, lane 1). The AdoMet-binding motif being located on the N-terminal moiety of the protein, they were all expected to be able to bind to [3H]AdoMet. This is precisely what was observed, since the three peptides were able to bind [3H]AdoMet with the same efficiency (Fig. 12, lane 4). Several controls demonstrate the specificity of the reaction. First, an extract prepared from a wild-type strain that does not express HA-Spb1p gave no signal (Fig. 12, lane 5), showing that the three bands of 116, 93, and 87 kDa are truly related to Spb1p. Second, large amounts of BSA or recombinant Gar1p were UV irradiated in the presence of [3H]AdoMet and gave no signal (data not shown). This result demonstrates the specificity of the treatment with the UV light that cross-linked only interacting molecules and not just molecules present in the solution. Third, when the samples were not treated with the UV light, no signal was visible with the HA-Spb1p related peptides, showing that a covalent bond was not formed in the absence of the cross-linking agent (data not shown). We conclude from these data that Spb1p is able to bind specifically in vitro to [3H]AdoMet, the cofactor for most methyltransferases.
FIG. 12.
Spb1p is radiolabelled after UV cross-linking with [3H]AdoMet. Western blotting with anti-HA antibodies revealed the presence of HA-Spb1p in a total cellular extract prepared from the strain (YDK14-1A/pDK376), along with two degradation products (lane 1). The anti-HA antibodies cross-linked to the protein A-Sepharose beads were used to immunoprecipitate HA-Spb1p from the same extract. The beads were then incubated with [3H]AdoMet before being UV cross-linked for 30 min. Proteins were recovered in a denaturing buffer and analyzed by Western blotting (lane 2). The same experiment was performed with a strain in which Spb1p is not tagged (lane 3). The membrane was exposed for autoradiography for 3 weeks at −70°C with an intensifying screen to reveal the tritiated molecules. Lanes 4 and 5 are identical to lanes 2 and 3. MW, molecular weight markers in thousands.
DISCUSSION
We report here the characterization of Spb1p, a novel nucleolar protein of S. cerevisiae, that is required for normal pre-rRNA processing. Spb1p copurifies with Nop1p through size chromatography and cation-exchange chromatography. Coimmunoprecipitation experiments demonstrate the association of Spb1p with Nop1p and Nop5/58p, another nucleolar protein previously shown to be associated with Nop1p (21, 81). A protein that migrates at 120 kDa was previously found to be associated with Nop1p and Nop5/58p (81). This size is in good agreement with the observation that Spb1p migrates on SDS-PAGE as a protein with a size of ∼120 kDa.
We present here evidence that the low level of 60S particles in the spb1-2 mutant is due to the inhibition of the processing of the 27S pre-rRNA into mature 25S and 5.8S rRNAs. We also detected a partial inhibition of the cleavage at sites A0, A1, and A2 that is observed in other yeast mutants that affect primarily 60S synthesis (42). However, we cannot conclude whether Spb1p is directly involved in this process, particularly since we have not been able to detect a specific association of Spb1p with the C+D snoRNAs. Spb1p could exert its effect on rRNA synthesis through the proteins with which it interacts. This view is supported by the observation that depletion of Spb1p is accompanied by a delocalization of Nop1p in most cells. However, one cannot conclude at this point whether this delocalization is a direct consequence of the depletion of Spb1p or if it is a consequence of a general disorganization of the nucleolus due to an arrest of ribosome synthesis. It is striking that the spb1-2 strain has a very low 18S and 25S rRNA content. This could result from the very slow synthesis of the 60S subunits in the mutant, with the 40S being then adjusted by a feedback mechanism. Otherwise, in the light of its isolation as an essential gene involved in silencing (54), Spb1p could have an additional function such as regulating rDNA transcription or recombination.
Comparing the sequence of Spb1p with the available databases reveals that the most striking feature is the presence of a putative AdoMet-binding domain, the cofactor of many methyltransferase enzymes. The presence of this motif in Spb1p was first reported (41) due to similarities to another putative AdoMet-binding protein encoded by the ORF YCR47c (9), both proteins being related to Escherichia coli FtsJ (74). We set up an in vitro assay to assess the AdoMet-binding activity of immunoprecipitated proteins. We found that Spb1p is able to bind in vitro to [3H]AdoMet, supporting the view that it could be a novel methylase. Native extracts prepared from a strain expressing HA-Spb1p contained three major peptides that derived from Spb1p by proteolysis and contained the same N-terminal domain, since they were all detected by the anti-HA antibodies (Fig. 12). These peptides were all able to bind [3H]AdoMet, thus demonstrating that they all contain the AdoMet-binding domain. This finding is in agreement with the observation that the putative AdoMet-binding domain lies within the first 160 residues of the protein.
Considering that Spb1p is a putative methylase, we searched for its potential substrates. By comparing the incorporation of [5,6-3H]uracil with l-[methyl-3H]methionine, we found that global methylation of the pre-rRNA was not decreased in the mutant cells, in contrast to the defect observed in the nop1-3 mutant (73). This result indicates that Spb1p is not the major methylase of the pre-rRNA, even though we cannot exclude that Spb1p could be required to methylate only a subset of the methyl groups, as has been proposed for Nop2p (32).
Another possibility would be that Spb1p is a protein methylase. Protein methylation is one of the numerous protein modifications that can modulate protein activity in vivo. Some modifications that appear to be reversible, such as the formation of methyl esters on the carboxyl groups, may participate in signal transduction (2). In contrast, methylation of the amino group of the side chain of lysine or arginine residues seems to be irreversible but leads to the formation of modified amino acid groups that extend the repertoire of biochemical reactions that a protein can perform (13). It has been known for a long time that proteins involved in RNA metabolism contain asymmetric dimethylarginines (19). Among these, there is a subset of proteins that are located within the nucleolus and that share a common domain known as the GAR domain, in which arginine residues are dimethylated (25). It has been suggested that the GAR domain could be involved in RNA binding (22, 23, 38) or in protein-protein interactions (10). A recent report indicates that methylation of the arginines could reduce their affinity for RNA (39). Other workers have found that methylation of the nuclear protein Hrp1, which is involved in mRNA 3′-end cleavage and export, is inhibited by its cognate RNA (17, 76).
The major activity for arginine methylation in yeast has been characterized as the product of the gene RMT1 (20). In an rmt1Δ strain, the amount of dimethylarginine is reduced to less than 15% compared to the wild type. However, an rmt1Δ strain is viable, indicating that the function of this methylase is not essential or that it is partially redundant with another enzyme. Since there is some arginine dimethylation activity left in an rmt1Δ strain, it is conceivable that Spb1p could be responsible for that activity, allowing the cell to survive. If Spb1p is truly a protein methylase, one obvious substrate would be the GAR protein Nop1p to which it is associated. Methylating the GAR domain of Nop1p could possibly control the activity of the protein, providing the cell with a tool for regulating ribosome synthesis.
AdoMet is used as a cofactor in a very broad spectrum of biochemical reactions. Its methyl group can be transferred to more than 40 different molecules. In addition to the AdoMet-binding domain, the broad variety of enzymes that utilize AdoMet as a cofactor must contain other motifs that should be characteristic of the classes to which they belong, as DNA-methylase, rRNA-methylase, protein-methylase, decarboxylase, etc. It is striking that the best similarity between the potential homologues of Spb1p is observed outside the four motifs that are proposed to form the AdoMet-binding domain. This might reveal that the AdoMet-binding domain has evolved to acquire a more specialized function. So far, sequence analyses do not readily detect the additional motifs that would allow us to sort these proteins and to predict their activity. However, data are now accumulating rapidly that may allow us in the near future to make this prediction. It is also conceivable that there is a second domain, interlaced with the AdoMet-binding one, that is specific for the Spb1p function and not for a class of methylases. We also are considering the possibility that Spb1p may not be a methyltransferase. Its ability to bind AdoMet could serve to regulate the availability and/or accessibility of AdoMet to other methylases such as Nop1p in order to regulate their activity. Experiments are in progress to assess the activity of a recombinant Spb1p protein in vitro and to identify its substrates in vivo.
ACKNOWLEDGMENTS
We are indebted to A. Sachs who initiated this work and who gave us generously the cloned SPB1 gene (pAS16), as well as many other reagents. We thank J. Aris who kindly provided several antibodies, including large amounts of anti-Nop1p and anti-Nop5/58p, and B. Trumpower for the anti-Qsr1p antibody. We thank J. de la Cruz and P. Linder for sharing materials and results prior to publication and for stimulating discussion. We are grateful to F. Martin for his help with the chromatography experiments. We thank members of our laboratory for fruitful discussions and J. Morrissey for careful reading of the manuscript.
This work was supported by the Centre National de la Recherche Scientifique and by grants from the Ligue contre le Cancer, from the Fondation pour la Recherche Médicale, and from the Philippe Foundation. L.P. had a fellowship from the MENESR. D.K. was supported by a grant from the Swiss National Science Foundation to P. Linder.
REFERENCES
- 1.Adam S A, Nakagawa T, Swanson M S, Woodruff T K, Dreyfuss G. mRNA polyadenylate-binding protein: gene isolation and sequencing and identification of a ribonucleoprotein consensus sequence. Mol Cell Biol. 1986;6:2932–2943. doi: 10.1128/mcb.6.8.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aletta J M, Cimato T R, Ettinger M J. Protein methylation: a signal event in post-translational modification. Trends Biochem Sci. 1998;23:89–91. doi: 10.1016/s0968-0004(98)01185-2. [DOI] [PubMed] [Google Scholar]
- 3.Aris J P, Blobel G. Identification and characterization of a yeast nucleolar protein that is similar to a rat liver nucleolar protein. J Cell Biol. 1988;107:17–31. doi: 10.1083/jcb.107.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bachellerie J P, Cavaille J. Guiding ribose methylation of rRNA. Trends Biochem Sci. 1997;22:257–261. doi: 10.1016/s0968-0004(97)01057-8. [DOI] [PubMed] [Google Scholar]
- 5.Balakin A G, Smith L, Fournier M J. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell. 1996;86:823–834. doi: 10.1016/s0092-8674(00)80156-7. [DOI] [PubMed] [Google Scholar]
- 6.Baserga S J, Agentis T M, Wormsley S, Dunbar D A, Lee S. Mpp10p, a new protein component of the U3 snoRNP required for processing of 18S rRNA precursors. Nucleic Acids Symp Ser. 1997;36:64–67. [PubMed] [Google Scholar]
- 7.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bergès T, Petfalski E, Tollervey D, Hurt E C. Synthetic lethality with fibrillarin identifies NOP77p, a nucleolar protein required for pre-rRNA processing and modification. EMBO J. 1994;13:3136–3148. doi: 10.1002/j.1460-2075.1994.tb06612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bork P, Ouzounis C, Sander C, Scharf M, Schneider R, Sonnhammer E. Comprehensive sequence analysis of the 182 predicted open reading frames of yeast chromosome III. Protein Sci. 1992;1:1677–1690. doi: 10.1002/pro.5560011216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bouvet P, Diaz J J, Kindbeiter K, Madjar J J, Amalric F. Nucleolin interacts with several ribosomal proteins through its RGG domain. J Biol Chem. 1998;273:19025–19029. doi: 10.1074/jbc.273.30.19025. [DOI] [PubMed] [Google Scholar]
- 11.Brown T, Mackey K. Analysis of RNA by Northern and slot blot hybridization. In: Ausubel F M, editor. Current protocols in molecular biology. Vol. 1. New York, N.Y: John Wiley & Sons; 1997. pp. 4.9.1–4.9.16. [DOI] [PubMed] [Google Scholar]
- 12.Christensen M E, Fuxa K P. The nucleolar protein, B-36, contains a glycine and dimethylarginine-rich sequence conserved in several other nuclear RNA-binding proteins. Biochem Biophys Res Commun. 1988;155:1278–1283. doi: 10.1016/s0006-291x(88)81279-8. [DOI] [PubMed] [Google Scholar]
- 13.Clarke S. Protein methylation. Curr Opin Cell Biol. 1993;5:977–983. doi: 10.1016/0955-0674(93)90080-a. [DOI] [PubMed] [Google Scholar]
- 14.Cross F R, Tinkelenberg A H. A potential positive feedback loop controlling CLN1 and CLN2 gene expression at the start of the yeast cell cycle. Cell. 1991;65:875–883. doi: 10.1016/0092-8674(91)90394-e. [DOI] [PubMed] [Google Scholar]
- 15.de la Cruz J, Kressler D, Rojo M, Tollervey D, Linder P. Spb4p, an essential putative RNA helicase, is required for a late step in the assembly of 60S ribosomal subunits in Saccharomyces cerevisiae RNA. Vol. 4. 1998. pp. 1268–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eisinger D P, Dick F A, Trumpower B L. Qsr1p, a 60S ribosomal subunit protein, is required for joining of 40S and 60S subunits. Mol Cell Biol. 1997;17:5136–5145. doi: 10.1128/mcb.17.9.5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frankel A, Clarke S. RNase treatment of yeast and mammalian cell extracts affects in vitro substrate methylation by type I protein arginine N-methyltransferases. Biochem Biophys Res Commun. 1999;259:391–400. doi: 10.1006/bbrc.1999.0779. [DOI] [PubMed] [Google Scholar]
- 18.Ganot P, Caizergues-Ferrer M, Kiss T. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev. 1997;11:941–956. doi: 10.1101/gad.11.7.941. [DOI] [PubMed] [Google Scholar]
- 19.Gary J D, Clarke S. RNA and protein interactions modulated by protein arginine methylation. Prog Nucleic Acid Res Mol Biol. 1998;61:65–131. doi: 10.1016/s0079-6603(08)60825-9. [DOI] [PubMed] [Google Scholar]
- 20.Gary J D, Lin W J, Yang M C, Herschman H R, Clarke S. The predominant protein-arginine methyltransferase from Saccharomyces cerevisiae. J Biol Chem. 1996;271:12585–12594. doi: 10.1074/jbc.271.21.12585. [DOI] [PubMed] [Google Scholar]
- 21.Gautier T, Bergès T, Tollervey D, Hurt E. Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol Cell Biol. 1997;17:7088–7098. doi: 10.1128/mcb.17.12.7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghisolfi L, Joseph G, Amalric F, Erard M. The glycine-rich domain of nucleolin has an unusual supersecondary structure responsible for its RNA-helix-destabilizing properties. J Biol Chem. 1992;267:2955–2959. [PubMed] [Google Scholar]
- 23.Ghisolfi L, Kharrat A, Joseph G, Amalric F, Erard M. Concerted activities of the RNA recognition and the glycine-rich C-terminal domains of nucleolin are required for efficient complex formation with pre-ribosomal RNA. Eur J Biochem. 1992;209:541–548. doi: 10.1111/j.1432-1033.1992.tb17318.x. [DOI] [PubMed] [Google Scholar]
- 24.Gietz D, St. Jean A, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Girard J-P, Lehtonen H, Caizergues-Ferrer M, Amalric F, Tollervey D, Lapeyre B. GAR1 is an essential small nucleolar RNP protein required for pre-rRNA processing in yeast. EMBO J. 1992;11:673–682. doi: 10.1002/j.1460-2075.1992.tb05099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guthrie C, Fink R G. Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:1–863. [PubMed] [Google Scholar]
- 27.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 28.Helser T L, Baan R A, Dahlberg A E. Characterization of a 40S ribosomal subunit complex in polyribosomes of Saccharomyces cerevisiae treated with cycloheximide. Mol Cell Biol. 1981;1:51–57. doi: 10.1128/mcb.1.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henras A, Henry Y, Bousquet-Antonelli C, Noaillac-Depeyre J, Gelugne J P, Caizergues-Ferrer M. Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO J. 1998;17:7078–7090. doi: 10.1093/emboj/17.23.7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henriquez R, Blobel G, Aris J P. Isolation and sequencing of NOP1. A yeast gene encoding a nucleolar protein homologous to a human autoimmune antigen. J Biol Chem. 1990;265:2209–2215. [PubMed] [Google Scholar]
- 31.Higman M A, Niles E G. Location of the S-adenosyl-l-methionine binding region of the vaccinia virus mRNA (guanine-7-)methyltransferase. J Biol Chem. 1994;269:14982–14987. [PubMed] [Google Scholar]
- 32.Hong B, Brockenbrough J S, Wu P, Aris J P. Nop2p is required for pre-rRNA processing and 60S ribosome subunit synthesis in yeast. Mol Cell Biol. 1997;17:378–388. doi: 10.1128/mcb.17.1.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hughes J M, Ares M J. Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991;10:4231–4239. doi: 10.1002/j.1460-2075.1991.tb05001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hutchison H T, Hartwell L H, McLaughlin C S. Temperature-sensitive yeast mutant defective in ribonucleic acid production. J Bacteriol. 1969;99:807–814. doi: 10.1128/jb.99.3.807-814.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ingrosso D, Fowler A V, Bleibaum J, Clarke S. Sequence of the d-aspartyl/l-isoaspartyl protein methyltransferase from human erythrocytes. Common sequence motifs for protein, DNA, RNA, and small molecule S-adenosylmethionine-dependent methyltransferases. J Biol Chem. 1989;264:20131–20139. [PubMed] [Google Scholar]
- 36.Jansen R, Tollervey D, Hurt E C. A U3 snoRNP protein with homology to splicing factor PRP4 and G beta domains is required for ribosomal RNA processing. EMBO J. 1993;12:2549–2558. doi: 10.1002/j.1460-2075.1993.tb05910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jong A Y, Clark M W, Gilbert M, Oehm A, Campbell J L. Saccharomyces cerevisiae SSB1 protein and its relationship to nucleolar RNA-binding proteins. Mol Cell Biol. 1987;7:2947–2955. doi: 10.1128/mcb.7.8.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kiledjian M, Dreyfuss G. Primary structure and binding activity of the hnRNP U protein: binding RNA through RGG box. EMBO J. 1992;11:2655–2664. doi: 10.1002/j.1460-2075.1992.tb05331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim S, Park G H, Paik W K. Recent advances in protein methylation: enzymatic methylation of nucleic acid binding proteins. Amino Acids. 1998;15:291–306. doi: 10.1007/BF01320895. [DOI] [PubMed] [Google Scholar]
- 40.Klimasauskas S, Timinskas A, Menkevicius S, Butkiene D, Butkus V, Janulaitis A. Sequence motifs characteristic of DNA[cytosine-N4]methyltransferases: similarity to adenine and cytosine-C5 DNA-methylases. Nucleic Acids Res. 1989;17:9823–9832. doi: 10.1093/nar/17.23.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koonin E V, Bork P, Sander C. Yeast chromosome III: new gene functions. EMBO J. 1994;13:493–503. doi: 10.1002/j.1460-2075.1994.tb06287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kressler D, de la Cruz J, Rojo M, Linder P. Dbp6p is an essential putative ATP-dependent RNA helicase required for 60S-ribosomal-subunit assembly in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:1855–1865. doi: 10.1128/mcb.18.4.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 44.Lafontaine D L, Tollervey D. Birth of the snoRNPs: the evolution of the modification-guide snoRNAs. Trends Biochem Sci. 1998;23:383–388. doi: 10.1016/s0968-0004(98)01260-2. [DOI] [PubMed] [Google Scholar]
- 45.Lafontaine D L, Tollervey D. Nop58p is a common component of the box C+D snoRNPs that is required for snoRNA stability. RNA. 1999;5:455–467. doi: 10.1017/s135583829998192x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lafontaine D L J, Bousquet-Antonelli C, Henry Y, Caizergues-Ferrer M, Tollervey D. The box H+ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 1998;12:527–537. doi: 10.1101/gad.12.4.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lapeyre B, Amalric F, Ghaffari S H, Rao S V, Dumbar T S, Olson M O. Protein and cDNA sequence of a glycine-rich, dimethylarginine-containing region located near the carboxyl-terminal end of nucleolin (C23 and 100 kDa) J Biol Chem. 1986;261:9167–9173. [PubMed] [Google Scholar]
- 48.Lapeyre B, Mariottini P, Mathieu C, Ferrer P, Amaldi F, Amalric F, Caizergues-Ferrer M. Molecular cloning of Xenopus fibrillarin, a conserved U3 small nuclear ribonucleoprotein recognized by antisera from humans with autoimmune disease. Mol Cell Biol. 1990;10:430–434. doi: 10.1128/mcb.10.1.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee S, Baserga S J. Imp3p and Imp4p: two specific components of the U3 small nucleolar ribonucleoprotein that are essential for pre-18S rRNA processing. Mol Cell Biol. 1999;19:5441–5452. doi: 10.1128/mcb.19.8.5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee W C, Xue Z X, Mélèse T. The NSR1 gene encodes a protein that specifically binds nuclear localization sequences and has two RNA recognition motifs. J Cell Biol. 1991;113:1–12. doi: 10.1083/jcb.113.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H D, Zagorski J, Fournier M J. Depletion of U14 small nuclear RNA (snR128) disrupts production of 18S rRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:1145–1152. doi: 10.1128/mcb.10.3.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lischwe M A, Cook R G, Ahn Y S, Yeoman L C, Busch H. Clustering of glycine and NG,NG-dimethylarginine in nucleolar protein C23. Biochemistry. 1985;24:6025–6028. doi: 10.1021/bi00343a001. [DOI] [PubMed] [Google Scholar]
- 53.Lischwe M A, Ochs R L, Reddy R, Cook R G, Yeoman L C, Tan E M, Reichlin M, Busch H. Purification and partial characterization of a nucleolar scleroderma antigen (Mr = 34,000; pI, 8.5) rich in NG,NG-dimethylarginine. J Biol Chem. 1985;260:14304–14310. [PubMed] [Google Scholar]
- 54.Loo S, Laurenson P, Foss M, Dillin A, Rine J. Roles of ABF1, NPL3, and YCL54 in silencing in Saccharomyces cerevisiae. Genetics. 1995;141:889–902. doi: 10.1093/genetics/141.3.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luthe D S. A simple technique for the preparation and storage of sucrose gradients. Anal Biochem. 1983;135:230–232. doi: 10.1016/0003-2697(83)90755-8. [DOI] [PubMed] [Google Scholar]
- 56.Morrissey J P, Tollervey D. Yeast snR30 is a small nucleolar RNA required for 18S rRNA synthesis. Mol Cell Biol. 1993;13:2469–2477. doi: 10.1128/mcb.13.4.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Niewmierzycka A, Clarke S. S-Adenosylmethionine-dependent methylation in Saccharomyces cerevisiae. Identification of a novel protein arginine methyltransferase. J Biol Chem. 1999;274:814–824. doi: 10.1074/jbc.274.2.814. [DOI] [PubMed] [Google Scholar]
- 58.Niman H L, Houghten R A, Walker L E, Reisfeld R A, Wilson I A, Hogle J M, Lerner R A. Generation of protein-reactive antibodies by short peptides is an event of high frequency: implications for the structural basis of immune recognition. Proc Natl Acad Sci USA. 1983;80:4949–4953. doi: 10.1073/pnas.80.16.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nomura M. Assembly of bacterial ribosomes. Science. 1973;179:864–873. doi: 10.1126/science.179.4076.864. [DOI] [PubMed] [Google Scholar]
- 60.Oliver S G, van der Aart Q J, Agostoni-Carbone M L, Aigle M, Alberghina L, Alexandraki D, Antoine G, Anwar R, Ballesta J P, Benit P, et al. The complete DNA sequence of yeast chromosome III. Nature. 1992;357:38–46. doi: 10.1038/357038a0. [DOI] [PubMed] [Google Scholar]
- 61.Sachs A B, Davis R W. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989;58:857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- 62.Sachs A B, Davis R W. Translation initiation and ribosomal biogenesis: involvement of a putative rRNA helicase and RPL46. Science. 1990;247:1077–1079. doi: 10.1126/science.2408148. [DOI] [PubMed] [Google Scholar]
- 63.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 64.Scheer U, Weisenberger D. The nucleolus. Curr Opin Cell Biol. 1994;6:354–359. doi: 10.1016/0955-0674(94)90026-4. [DOI] [PubMed] [Google Scholar]
- 65.Schimmang T, Tollervey D, Kern H, Frank R, Hurt E C. A yeast nucleolar protein related to mammalian fibrillarin is associated with small nucleolar RNA and is essential for viability. EMBO J. 1989;8:4015–4024. doi: 10.1002/j.1460-2075.1989.tb08584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schluckebier G, O'Gara M, Saenger W, Cheng X. Universal catalytic domain structure of AdoMet-dependent methyltransferases. J Mol Biol. 1995;247:16–20. doi: 10.1006/jmbi.1994.0117. [DOI] [PubMed] [Google Scholar]
- 67.Sillevis-Smitt W W, Vlak J M, Molenaar I, Rozijn T H. Nucleolar function of the dense crescent in the yeast nucleus. A biochemical and ultrastructural study. Exp Cell Res. 1973;80:313–321. doi: 10.1016/0014-4827(73)90302-9. [DOI] [PubMed] [Google Scholar]
- 68.Sun C, Woolford J L., Jr The yeast NOP4 gene product is an essential nucleolar protein required for pre-rRNA processing and accumulation of 60S ribosomal subunits. EMBO J. 1994;13:3127–3135. doi: 10.1002/j.1460-2075.1994.tb06611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tollervey D. Trans-acting factors in ribosome synthesis. Exp Cell Res. 1996;229:226–232. doi: 10.1006/excr.1996.0364. [DOI] [PubMed] [Google Scholar]
- 70.Tollervey D. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 1987;6:4169–4175. doi: 10.1002/j.1460-2075.1987.tb02763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tollervey D, Kiss T. Function and synthesis of small nucleolar RNAs. Curr Opin Cell Biol. 1997;9:337–342. doi: 10.1016/s0955-0674(97)80005-1. [DOI] [PubMed] [Google Scholar]
- 72.Tollervey D, Lehtonen H, Carmo-Fonseca M, Hurt E C. The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre-rRNA processing in yeast. EMBO J. 1991;10:573–583. doi: 10.1002/j.1460-2075.1991.tb07984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tollervey D, Lehtonen H, Jansen R, Kern H, Hurt E C. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell. 1993;72:443–457. doi: 10.1016/0092-8674(93)90120-f. [DOI] [PubMed] [Google Scholar]
- 74.Tomoyasu T, Yuki T, Morimura S, Mori H, Yamanaka K, Niki H, Hiraga S, Ogura T. The Escherichia coli FtsH protein is a prokaryotic member of a protein family of putative ATPases involved in membrane functions, cell cycle control, and gene expression. J Bacteriol. 1993;175:1344–1351. doi: 10.1128/jb.175.5.1344-1351.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tron T, Yang M, Dick F A, Schmitt M E, Trumpower B L. QSR1, an essential yeast gene with a genetic relationship to a subunit of the mitochondrial cytochrome bc1 complex, is homologous to a gene implicated in eukaryotic cell differentiation. J Biol Chem. 1995;270:9961–9970. doi: 10.1074/jbc.270.17.9961. [DOI] [PubMed] [Google Scholar]
- 76.Valentini S R, Weiss V H, Silver P A. Arginine methylation and binding of Hrp1p to the efficiency element for mRNA 3′-end formation. RNA. 1999;5:272–280. doi: 10.1017/s1355838299981633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Venema J, Tollervey D. Processing of pre-ribosomal RNA in Saccharomyces cerevisiae. Yeast. 1995;11:1629–1650. doi: 10.1002/yea.320111607. [DOI] [PubMed] [Google Scholar]
- 78.Warner J R. The nucleolus and ribosome formation. Curr Opin Cell Biol. 1990;2:521–527. doi: 10.1016/0955-0674(90)90137-4. [DOI] [PubMed] [Google Scholar]
- 79.Watkins N J, Gottschalk A, Neubauer G, Kastner B, Fabrizio P, Mann M, Luhrmann R. Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA-binding protein, are present together with Gar1p in all H BOX/ACA-motif snoRNPs and constitute a common bipartite structure. RNA. 1998;4:1549–1568. doi: 10.1017/s1355838298980761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wiederkehr T, Pretot R F, Minvielle-Sebastia L. Synthetic lethal interactions with conditional poly(A) polymerase alleles identify LCP5, a gene involved in 18S rRNA maturation. RNA. 1998;4:1357–1372. doi: 10.1017/s1355838298980955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu P, Brockenbrough J S, Metcalfe A C, Chen S, Aris J P. Nop5p is a small nucleolar ribonucleoprotein component required for pre-18S rRNA processing in yeast. J Biol Chem. 1998;273:16453–16463. doi: 10.1074/jbc.273.26.16453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhong T, Arndt K T. The yeast SIS1 protein, a DnaJ homolog, is required for the initiation of translation. Cell. 1993;73:1175–1186. doi: 10.1016/0092-8674(93)90646-8. [DOI] [PubMed] [Google Scholar]