Abstract
The high degree of genetic diversity within human immunodeficiency virus type 1 (HIV-1), which includes two major groups, M (major) and O (outlier), and various env subtypes within group M (subtypes A to J), has made designing assays that will detect all known HIV-1 strains difficult. We have developed a generic primer set based on the conserved immunodominant region of transmembrane protein gp41 that can reliably amplify as few as 10 copies/PCR of viral DNA from near-full-length clones representing group M subtypes A to H (subtypes I and J were not available). The assay is highly sensitive in detecting plasma viral RNA from HIV-1 strains of diverse geographic origins representing different subtypes of HIV-1 group M as well as HIV-1 group O. Of the 253 group M plasma specimens (subtypes A, 68 specimens; B, 71; C, 19; D, 27; E, 23; F, 33; and G, 12), 250 (98.8%) were amplified by using the gp41 M/O primer set. More importantly, all 32 (100%) group O plasma samples were also amplified with these primers. In vitro spiking experiments further revealed that the assay could reliably detect as few as 25 copies/ml of viral RNA and gave positive signals in HIV-1-seropositive specimens with plasma copy numbers below the limits of detection by all commercially available viral load assays. In addition, analysis of five seroconversion panels indicated that the assay is highly sensitive for early detection of plasma viremia during the “window period.” Thus, the highly sensitive assay will be useful for early detection of HIV-1 in clinical specimens from all known HIV-1 infections, regardless of their genotypes and geographic origins.
Human immunodeficiency virus type 1 (HIV-1) is characterized by an unusually high degree of genetic variability in vivo. Analysis of the HIV-1 env gene of virus isolates from different geographic origins has revealed that HIV-1 can be divided into two major groups: M (major) and O (outlier). Phylogenetic analysis of the env gene has shown that HIV-1 group M can be subdivided into genetically equidistant subtypes, comprising subtypes A to J. Although most subtypes are common in Central Africa, the worldwide distribution of various HIV-1 subtypes is quite different, with subtype B being the most prevalent in North America and Europe; subtype A the most prevalent in Africa; subtype E the most prevalent in Thailand; subtype C the most prevalent in India and South Africa; and subtype F the most prevalent in Romania, Brazil, and Argentina (11). In addition, the highly divergent HIV-1 group O viruses are centered in Cameroon and its neighboring countries, such as Equatorial Guinea and Gabon (10, 19, 25). Although additional HIV-1 group O infections have been iden tified in Europe and the United States, most patients have hadlinks to West Central Africa. Recently, a new variant of HIV-1, designated group N, has been identified with its epicenter in Cameroon (24).
Current detection of HIV-1 infection in blood donations is based on serologic testing for antivirus antibodies and viral antigens. Detection of plasma viremia, however, would enable earlier detection of HIV-1, thus reducing the blood transfusion infections associated with “window period” cases (14, 23). In addition, several reports have shown that antibodies against some variants of HIV-1 group O are not reliably detected by all commercially available diagnostic assays (4, 17), primarily because of diversity in the immunodominant regions of HIV-1. Currently available nucleic acid-based diagnostic testing (22) and viral load assays also have reduced sensitivities for non-subtype B isolates (1). Since therapeutic decisions require accurate viral load measurements, the inability of current viral load assays to accurately detect all subtypes presents a dilemma for clinicians (3). Thus, a highly sensitive assay that will detect both HIV-1 groups M and O and that can be used for both qualitative and quantitative detection of HIV-1 is needed. In the present investigation, we have developed a highly sensitive molecular detection procedure that can detect viral RNA from plasma, can be used as an early diagnostic tool, and might potentially be used for viral load determination for both group M and group O HIV-1 infections.
MATERIALS AND METHODS
Study subjects.
The HIV-1-positive samples tested in the present study were obtained from various ongoing HIV-1 studies throughout the world. Samples included serum and/or plasma specimens from Uganda (n = 54), Thailand (n = 43), Cameroon (n = 40), Ivory Coast (n = 29), Argentina (n = 27), Brazil (n = 23), the United States (n = 16), Ghana (n = 8), Mexico (n = 8), China (n = 6), Lebanon (n = 5), Zimbabwe (n = 4), South Africa (n = 4), Spain (n = 4), and India (n = 3), as well as those from miscellaneous sources (n = 7). The specificity of the assay was tested against specimens from HIV-2-infected individuals from Ivory Coast (n = 16) and Ghana (n = 2), as well as HIV-seronegative donors from the United States (n = 41). HIV-1 early seroconversion panels (five members: no. 946 and 948 to 951) were obtained from Boston Biomedica, Inc., Boston, Mass.
Viral stocks.
Viral isolates representing various HIV-1 subtypes, HIV-2, and simian immunodeficiency virus (SIV) were expanded as described previously (20, 27). The SIV isolates either were obtained from the AIDS Reagent and Reference Program or were primary isolates obtained from naturally infected monkeys (unpublished observations). Viral RNA was extracted from the culture supernatant and used to test the initial sensitivity and specificity of the primers. In addition, near-full-length clones of HIV-1 representing subtypes A to H were also used. The detailed information about these clones was published elsewhere (8, 9, 16).
HIV-1 subtype analysis.
The C2V3 or gp41 region of the env gene was amplified from all specimens by using DNA lysates of uncultured peripheral blood lymphocytes or RNA extracts from infected plasmas, as described previously (26). PCR products were used for automated sequencing reactions with dye terminator labeling chemistry. Sequencing reactions were run in an automated DNA sequencer 373 (Applied Biosystems, Foster City, Calif.). The sequences were then aligned by using the CLUSTAL W (1.74) multiple sequence alignment program. The phylogenetic tree was constructed by the neighbor-joining method included in the PHYLIP 3.5c package (5). The tentative subtype of each isolate was assigned based on the clade pattern. Accession numbers for the sequences have been described elsewhere (21).
Development of generic primers for detection of group M and O viruses.
To define the regions with considerable homology for primer design, the DNA sequences of HIV-1 groups M and O from the Los Alamos database (15) and our own sequence database were aligned and carefully examined. Primer sets were designed based on the consensus sequences, and primer sequences were then compared with all known sequences in databases for assessment of specificity. Initially, we designed primer sets within the protease, integrase, and env genes. Oligonucleotides from all three regions were synthesized at the Biotechnology Core Facility, Centers for Disease Control and Prevention, Atlanta, Ga. Only the consensus primers for the gp41 region are described here. For reverse transcription (RT) and primary PCR, the primers were GP40F1 (forward; 5′TCTTAGGAGCAGCAGGAAGCACTATGGG; nucleotides 7789 to 7816 based on HXB2 [GenBank accession no. K03455]) (21a) and GP41R1 (reverse; 5′AACGACAAAGGTGAGTATCCCTGCCTAA; nucleotides 8347 to 8374). For the nested PCR, the primers were GP46F2 (forward; 5′ACAATTATTGTCTGGTATAGTGCAACAGCA; nucleotides 7850 to 7879) and GP47R2 (reverse; 5′TTAAACCTATCAAGCCTCCTACTATCATTA; nucleotides 8281 to 8310).
RNA extraction, RT, and PCR.
Viral RNA was extracted from plasma by using the QIAamp viral RNA kit according to the manufacturer’s protocol (Qiagen, Valencia, Calif.). Briefly, 200 μl of plasma was mixed with 800 μl of lysis buffer. After a 10-min incubation, 800 μl of 100% ethanol was added to the lysate. The mixture was filtered through a column by centrifugation. After being washed with buffer, the RNA was eluted from the column by adding 50 μl of RNase-free water. For negative controls, RNA from normal human plasma was also extracted. Three to 10 μl of the RNA extract was used to synthesize cDNA with primer GP41R1 (20 μM) and the GeneAmp RNA PCR kit following the manufacturer’s protocol (Perkin-Elmer Cetus, Norwalk, Conn.). The 20-μl cDNA reaction mixture was then added to a PCR mixture containing 50 μM GP40F1 and 30 μM GP41R1, 1× GeneAmp PCR buffer II, 1.25 mM MgCl2, and 2.5 U of AmpliTaq DNA polymerase (Perkin-Elmer Cetus, Foster City, Calif.) and was brought to a final volume of 100 μl with sterile distilled water. After initial denaturation at 94°C for 2 min, 35 cycles of PCR were performed in the GeneAmp 9600 thermocycler (Perkin-Elmer Cetus). Each cycle consisted of denaturation at 94°C for 30 s, annealing at 50°C for 30 s, and extension at 72°C for 60 s, with a final extension at 72°C for 5 min. For nested PCR, 5 μl of the primary PCR product was added to a 100-μl PCR mixture containing reagents similar to those in the primary PCR, except that the primers were replaced by 25 μM each GP46F2 and GP47R2. The PCR mixtures were subjected to 35 cycles under the same conditions as the primary PCR. After PCR, the nested PCR products were electrophoresed in 1.5% agarose gels along with a 100-bp ladder (Gibco, Grand Island, N.Y.) and visualized under UV light by ethidium bromide staining.
Sensitivity and specificity of the gp41 M/O primer pair.
The sensitivity of the gp41 M/O primers was tested in duplicate by using spiked plasma samples with known copy numbers of HIV-1. An HIV-1 subtype B stock with known copy numbers was obtained from Abbott Laboratories (North Chicago, Ill.) and used to spike normal human plasma at 1,000, 100, 50, 25, 10, and 1 copy/ml. Viral RNA was then extracted by using 200 μl of the spiked plasma as described above. Ten microliters of the RNA extract from each dilution was used to synthesize cDNA. The total cDNA reaction mixture from each dilution was then used for primary PCR. Following primary PCR, 5 μl of the PCR product was used for the nested PCR. In addition, the sensitivity of the assay for HIV-1 group M subtypes A to H was also tested with near-full-length virus clones (8, 9, 16) at 100, 10, 5, 1, and 0.1 copy per 100-μl PCR mixture. The amplification was confirmed by agarose gel electrophoresis and ethidium bromide staining. To prevent carryover contaminations, RNA extraction, RT-PCR master mixture preparation, and RNA or DNA additions were carried out in physically separated UV light-treated chambers and with interspersed negative controls in each assay. The sensitivity for the near-full-length clones of HIV-1 group M subtypes was also confirmed independently in the laboratory of one of the collaborators. To determine the specificity of the gp41 M/O primers, RNAs from 41 normal human plasma specimens and 18 HIV-2-infected human plasma specimens were also tested with the gp41 M/O primers.
RESULTS
Sensitivity and specificity of gp41 M/O primers.
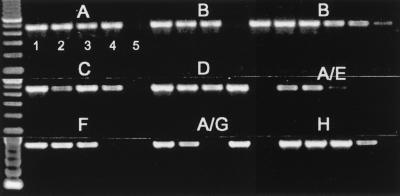
For pilot experiments, we evaluated several different primer sets from the protease, integrase, and gp41 regions for detection of viral RNA from culture supernatants as well as viral DNA from cultured cells of HIV-1 group M (subtypes A to H), HIV-1 group O, and HIV-2 viral stocks (20, 27). From these studies, we established that the primer set in the gp41 region, named gp41 M/O, was highly sensitive for RNA and DNA detection of all available group M (subtypes A to H [viral isolates for subtypes I and J were not available]) and group O isolates. The sensitivity of the gp41 M/O primer set was examined by using near-full-length clones of HIV-1 subtypes A to H (8, 9, 16). Testing of serial dilutions of known copy numbers of near-full-length clones representing subtypes A (92UG037.1), B (YU2 and SG3), C (92BR025.8), D (94UG114.1), A/E (90CF402.8), F (93BR020.1), A/G (92NG003.1), and H (90CF056.1) resulted in consistent amplification of 10 or more copies/PCR; however, in some cases, 1 to 5 copies/PCR were also detectable (Fig. 1). Thus, the gp41 M/O primer set was highly sensitive for detection of viral DNA regardless of viral genotypes.
FIG. 1.
Sensitivity of gp41 M/O primers in detecting HIV-1 group M subtypes A to H near-full-length molecular reference clones. The known numbers of copies per PCR of the clones A (92UG037.1), B (YU2 [left set of lanes] and SG3 [right set of lanes]), C (92BR025.8), D (94UG114.1), A/E (90CF402.8), F (93BR020.1), A/G (92NG003.1), and H (90CF056.1) were amplified with gp41 M/O primers. For each subtype, lane 1 is 100 copies/PCR, lane 2 is 10 copies/PCR, lane 3 is 5 copies/PCR, lane 4 is 1 copy/PCR, and lane 5 is 0.1 copy/PCR.
Further analysis also revealed that the gp41 M/O primer set was specific for HIV-1 only, since none of the culture supernatants from primary HIV-2 isolates (GB87, IC77618, IC310319, IC310072, and SLRHC) (20), the SIV isolates (MAC239, SMM 1, SIV-ST, M156, SM55, and SM74), or the SIV chimpanzee isolate (CPZ) could be amplified with these primers (data not shown).
Detection of HIV-1 RNA by RT-PCR assay, sensitivity, and specificity.
We next examined the sensitivity of viral RNA detection in plasma by using a panel of plasma for which the viral subtype was established by phylogenetic analysis of the env region (Table 1). RT-PCR analysis of 68 specimens with subtype A obtained from diverse geographic locations revealed that 67 (98.5%) could be amplified by the gp41 M/O primers (Table 1). Likewise, 69 of 71 (97%) subtype B, 19 of 19 (100%) subtype C, 27 of 27 (100%) subtype D, 23 of 23 (100%) subtype E, 33 of 33 (100%) subtype F, and 12 of 12 (100%) subtype G specimens gave positive signals with the gp41 M/O primers. The overall sensitivity of the gp41 M/O primer set for detection of HIV-1 group M subtypes in plasma was 98.8% (250 of 253). The three specimens that did not amplify with the gp41 M/O primers did give positive signals with the protease region primers (data not shown). More importantly, the gp41 M/O primers were also able to amplify viral RNA from plasma from individuals infected with HIV-1 group O viruses. Of 32 plasma samples containing group O virus representing specimens from Cameroon, Spain, and the United States, all (100%) were amplified with the gp41 M/O primers.
TABLE 1.
Detection of viral RNA in plasma from phylogenetically divergent HIV-1 group M and group O specimens with gp41 M/O primers
| HIV type | Country of origin | No. of specimens tested | gp41 M/O primer set
|
|
|---|---|---|---|---|
| No. of specimens positive | % Positivity | |||
| HIV-1 group M | ||||
| Subtype A | ||||
| Ivory Coast | 20 | 19 | 95 | |
| Uganda | 24a | 24 | 100 | |
| Cameroon | 15 | 15 | 100 | |
| Ghana/Lebanon | 6/3 | 6/3 | 100 | |
| Total | 68 | 67 | 98.5 | |
| Subtype B | ||||
| United States | 11 | 11 | 100 | |
| Mexico | 8 | 8 | 100 | |
| Brazil | 19 | 17 | 90 | |
| China | 6 | 6 | 100 | |
| Thailandb | 20 | 20 | 100 | |
| Miscellaneous | 7 | 7 | 100 | |
| Total | 71 | 69 | 97 | |
| Subtype C | ||||
| Zimbabwe | 4 | 4 | 100 | |
| S. Africa | 4a | 4 | 100 | |
| India | 3 | 3 | 100 | |
| Uganda | 4 | 4 | 100 | |
| United States | 3 | 3 | 100 | |
| Lebanon | 1 | 1 | 100 | |
| Total | 19 | 19 | 100 | |
| Subtype D | ||||
| Uganda | 26 | 26 | 100 | |
| Lebanon | 1 | 1 | 100 | |
| Total | 27 | 27 | 100 | |
| Subtype E | ||||
| Thailand | 23 | 23 | 100 | |
| Subtype F | ||||
| Argentina | 27a | 27 | 100 | |
| Brazil | 4 | 4 | 100 | |
| Japan | 2 | 2 | 100 | |
| Total | 33 | 33 | 100 | |
| Subtype G | ||||
| Ghana/Lebanon | 2/1 | 3 | 100 | |
| Ivory Coast | 9 | 9 | 100 | |
| Total | 12 | 12 | 100 | |
| HIV-1 group O | ||||
| Cameroon | 25 | 25 | 100 | |
| Spain | 5 | 5 | 100 | |
| United States | 2 | 2 | 100 | |
| Total | 32 | 32 | 100 | |
| HIV-2 | Ivory Coast | 16 | 0 | 0 |
| Ghana | 2 | 0 | 0 | |
| HIV negative | United States | 41 | 0 | 0 |
One specimen each of the potential recombinant with discordance in the env and prt subtype. The specimens from Uganda represent A env and B to D prt, the specimens from South Africa represent C env and B prt, and the specimens from Argentina represent F env and B prt.
Subtype B viruses from Thailand are referred to as B′.
The gp41 M/O primer set is highly specific for plasma detection of HIV-1 only, since neither plasma samples from HIV-2-infected individuals (n = 18) nor those from uninfected controls (n = 41) were positive (Table 1).
Assessment of gp41 M/O primers for viremia detection during the window period.
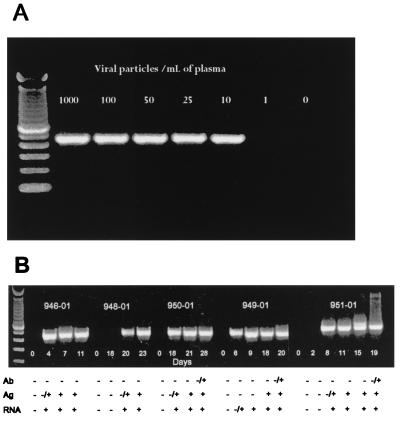
In vitro spiking of plasma with known copy numbers of HIV-1 subtype B viruses prior to RNA extraction and RT-PCR established that the gp41 M/O primers could reliably detect as few as 25 copies of HIV-1 RNA per ml (Fig. 2A). We next examined the sensitivity of the gp41 M/O primers for early detection of plasma viremia in cases of seroconversion. Analysis of five seroconverters from the United States (all subtype B) revealed that RT-PCR detection preceded both HIV-1 antibody and p24 antigen detections. The early detection levels were comparable to those of the three commercially available RNA-based detection assays, although in one case (panel 949), the gp41 M/O primer set was more sensitive for early detection of plasma viremia.
FIG. 2.
Sensitivity of gp41 M/O primers in detecting HIV-1 RNA in plasma with known copy numbers (A) and early seroconverters (B). (A) Normal plasma was spiked with known copy numbers of subtype B viruses prior to RNA extraction and amplification. (B) RNA extracts from longitudinal plasma specimens from five persons (panel no. 946 and 948 to 951) during the window period were amplified with gp41 M/O primers. The results with HIV-1 antibody (Ab), p24 antigen (Ag), and three commercial RNA tests (RNA) are shown as negative (−) and positive (+) at the bottom of each panel.
Assessment of gp41 M/O primers for potential viral load detection.
Quantification of HIV-1 RNA has become an important tool for monitoring antiretroviral therapy, as well as predicting HIV-1 disease progression. Currently, there are three commercially available assays for quantitation of HIV-1 viral RNA in plasma: the AMPLICOR HIV-1 Monitor Test, which is based on PCR (Roche); the Nuclisense HIV-1 QT assay, which is based on an isothermal nucleic acid sequence-based amplification (OTC; Organon Technika); and the Quantiplex HIV RNA assay, which uses branched-DNA signal amplification techniques (Chiron) (see reference 3 for review). While the sensitivity of all three assays is fairly high, they have been shown to miss specimens with extremely low viral loads (<50 copies/ml). In addition, some of these assays are less sensitive for certain non-B subtypes, and none are capable of detecting group O specimens (2, 22). Since the gp41 M/O primer set had shown broad sensitivities for plasma HIV-1 RNA detection, we next examined the utility of this assay for detecting plasma viremia in a subset of specimens shown in Table 1. Of the 50 specimens representing group M (subtypes A to F), 29 had detectable viral load in all three tests, 11 were negative by one of the three tests (details about these specimens are given in Table 2), 7 were negative by all three tests, and 3 were also negative by the ultrasensitive versions of two tests (Roche and OTC) (Table 2). All 50 specimens, including 21 specimens which had previously been shown to have virus at low or undetectable levels by commercial viral load assays were positive by gp41 M/O primers. These results suggest that inclusion of these newly identified primers in commercial viral load assays would provide better sensitivities for plasma viral RNA detection for all known subtypes of HIV-1 group M and group O viruses.
TABLE 2.
Sensitivity of detection of HIV-1 by the gp41 M/O primer set compared with those of commercial viral load assays
| Specimen | Country of origin | Subtype (env/prt)a | Sensitivity of viral load assay (no. of copies/ml)b
|
Result with gp41 M/O primers | ||
|---|---|---|---|---|---|---|
| Chiron | OTC | Roche | ||||
| 301-06 | Ghana | A | 4 × 103 | 5 × 103 | BLD | Positive |
| 301-07 | Ghana | A | 5 × 103 | 2 × 103 | BLD | Positive |
| 301-12 | Ghana | A | 6 × 104 | 3 × 104 | BLD | Positive |
| 301-21 | Uganda | A/A | 7 × 102 | BLD | BLD | Positive |
| 301-24 | Uganda | A | BLD | 9 × 102 | 6 × 102 | Positive |
| 301-27 | Uganda | A/BD | BLD | BLD | BLD | Positive |
| TIA175 | Ivory Coast | A | ND | BLDc | BLDc | Positive |
| 301-14 | South Africa | C/B | 4 × 103 | BLD | 7 × 102 | Positive |
| 301-16 | South Africa | C | 3 × 103 | 9 × 102 | BLD | Positive |
| 301-29 | Zimbabwe | C | >8 × 105 | 1 × 103 | BLD | Positive |
| 301-44 | India | C | 4 × 103 | BLD | 3 × 103 | Positive |
| 301-23 | Uganda | D | BLD | BLD | BLD | Positive |
| 301-26 | Uganda | D/D | 2 × 103 | BLD | 5 × 102 | Positive |
| 301-39 | Zimbabwe | E/A | 4 × 103 | BLD | BLD | Positive |
| 301-40 | Thailand | E/A | 2 × 103 | BLD | BLD | Positive |
| 301-47 | Argentina | F/B | BLD | BLD | BLD | Positive |
| 301-03 | Ghana | G | 103 | BLD | BLD | Positive |
| 301-08 | Ghana | G | 1 × 103 | 7 × 102 | BLD | Positive |
| DAA3671 | Ivory Coast | G | ND | BLDc | BLDc | Positive |
| DAA3760 | Ivory Coast | G | ND | BLDc | BLDc | Positive |
| 301-01 | Cameroon | O | BLD | BLD | BLD | Positive |
Subtype was ascribed based on phylogenetic analysis of the env region. In some cases, the protease region (prt) was also sequenced to identify potential dual or recombinant infections. Specimen no. 310-14, 301-27, 301-39, and 301-47 represent dual infection with multiple subtypes or potential recombinants of two different genotypes.
BLD, below limit of detection. The limits of detection of each assay were 500 copies/ml for the Chiron assay and 400 copies/ml for the OTC and Roche assays. ND, not done.
Specimen still negative by the ultrasensitive versions of the OTC and Roche assays (both with a detection limit of 50 copies/ml).
DISCUSSION
In the present investigation, we have identified a highly conserved region in the transmembrane protein gp41 of HIV-1 and devised a primer set that allows molecular detection of available group M (subtypes A to H) and group O viruses. Thus, the assay is a highly sensitive and specific diagnostic tool for HIV-1 DNA and RNA detection of both group M and O viruses, and the primer set might be adapted for quantitation of HIV-1 viral loads in infected patients. While several molecular detection assays have been described for detection of proviral DNA from peripheral blood (7, 18, 22), to the best of our knowledge, this is the only assay that can amplify both group M viruses with equal sensitivities for different subtypes and group O viruses from plasma. Of the 253 plasma specimens tested containing group M virus, the primers were able to amplify 250 specimens, yielding a test sensitivity of 98.8%. The three specimens negative with the gp41 M/O primers were amplified with pol primers, suggesting that the sample integrity and the RNA extraction procedure were not the likely reasons for the lack of gp41 amplification. Whether the lack of amplification is due to highly divergent strains remains to be determined.
More importantly, our results showed that all 32 (100%) specimens with group O HIV-1 were also amplified. In general, the sequence variability among group O sequences is similar to that observed among the various group M subtypes (12, 13), and therefore more specific and sensitive primers are needed for diagnostic detection of group O strains (18). In fact, nucleotide sequence alignment of 28 group O sequences in the gp41 region has revealed remarkable sequence conservation (data not shown), further supporting our findings that these primers can be used for both qualitative and quantitative detection of all known HIV-1 strains. Additional sequencing of approximately 200 selected group M strains, together with a compilation of the existing database sequences, has further confirmed the sequence conservation of all group M sequences in this region (13). The high conservation in this region of gp41 suggests that this site may represent a crucial functional domain of the HIV-1 envelope protein. Thus, we believe that the gp41 region identified here provides a reliable and highly conserved region that can be exploited for diagnostic assays. While the nucleotide sequences selected for the gp41 M/O primers are highly conserved, the gp41 region amplified by these primers shows considerable divergence at nucleotide and amino acid levels between the immunodominant epitopes of groups M and O; thus, most serologic assays based on the detection of antibodies to the gp41 region have to add a group O-specific peptide to enhance antibody detection (4, 17).
In addition to being highly sensitive for detection of both group M and group O HIV-1 viruses, the assay was highly specific for HIV-1. Neither DNA from any of the HIV-2 and SIV isolates nor plasma from HIV-2-infected patients could be amplified. Furthermore, none of the plasma from HIV-seronegative individuals resulted in any false-positive signal.
In the United States, the rate of HIV-1 transmission by blood transfusion with blood screened with the current antibody and antigen tests is estimated to be two donations per million units of blood donated (23). To further reduce transfusion-related transmission of HIV-1, efforts have focused on implementing direct detection of HIV-1 nucleic acids as markers for viral infection by using individual or pooled plasma testing (to be implemented by early 1999 in the United States). Because of the high sensitivity achievable with the gp41 M/O primers, they may have an impact on the safety of the blood supply by further reducing HIV infections by blood transfusion associated with donors during the window period. As demonstrated in the present study, the assay is capable of detecting the presence of viral RNA from seroconversion panels of subtype B earlier than antibody and p24 antigen detection. However, sensitivities for other subtypes of group M and for group O still remain to be determined. In addition, because gp41 M/O primers detect all subtypes (A to H) tested of group M (with the exception of three specimens) and all group O strains, the accuracy of testing of donated blood will not be dependent on the geographic origin of the donors. Thus, the high sensitivity of the assay for any group M or O infections makes it an ideal candidate to be included as a molecular screening tool in developed countries where pooled plasma donations are currently being considered for genetic screening.
The other important aspect of our study is the fact that we were able to detect viral RNA of all HIV-1 variants in plasma with high efficiency by using the gp41 M/O primer set. The measurement of plasma HIV-1 RNA has become an important tool in the clinical management of HIV-1-infected patients, because RNA levels in plasma change dynamically in response to successful therapy. However, detection of certain non-B subtypes and group O infections continues to be a problem with current commercial viral load assays (1, 6). The consensus gp41 M/O primers not only allowed amplification of all subtypes of group M and group O, but also amplified RNA from specimens with which even ultrasensitive commercial viral load assays had failed. Thus, it appears that the current assay can potentially be adapted to accurately quantify virtually any HIV-1 RNA or DNA targets from any specimens, regardless of their geographic origins or viral genotypes. However, the efficiency of quantitative detection of non-subtype B viruses for viral load determination still remains to be tested.
In summary, our assay provides a highly sensitive and reliable assay for diagnostic detection of HIV-1 group M and group O viruses. The assay may have a broad range of applications, from utilization as a diagnostic tool for early detection of HIV-1 infection in blood donor settings, thus further reducing the risk of transfusion-related HIV-1 transmission, to potential adaptation for quantitative measurement of viral load for clinical management of the infected patient.
ACKNOWLEDGMENTS
We thank Richard George, Nancy Young, Nathan Shaffer, Steve Alexander, Artur Ramos, Charles Schable, and Kanchit Limpakarnjanarat for providing some of the specimens tested and Donna Rudolph and Silvina Masciotra for sample preparation.
This work was supported in part by grant G.0134.97 of the Fonds voor Wetenschappelijk Onderzoek, Brussels, Belgium, and grant IC18-CT97.0246 of the EC project to G. van der Groen.
ADDENDUM IN PROOF
The gp41 M/O assay can also amplify the recently identified HIV-1 group N virus (http://hiv-weblanl.gov/HTML/reviews/reviews.html).
REFERENCES
- 1.Alaeus A, Lidman K, Sönnerborg A, Albert J. Subtype-specific problems with quantification of plasma HIV-1 RNA. AIDS. 1997;11:859–865. doi: 10.1097/00002030-199707000-00004. [DOI] [PubMed] [Google Scholar]
- 2.Apetrei C, Loussert-Ajaka I, Descamps D, Damond F, Saragosti S, Brun-Vezinet F, Simon F. Lack of screening test sensitivity during HIV-1 non-subtype B seroconversions. AIDS. 1996;10:F57–F60. doi: 10.1097/00002030-199612000-00002. [DOI] [PubMed] [Google Scholar]
- 3.Cavert W. In vitro detection and quantitation of HIV in blood and tissues. AIDS. 1998;12(Suppl. A):S27–S34. [PubMed] [Google Scholar]
- 4.Eberle J, Loussert-Ajaka I, Brust S, Zekeng L, Hauser P H, Kaptue L, Knapp S, Damond F, Saragosti S, Simon F, Gürtler L G. Diversity of the immunodominant epitope of gp41 of HIV-1 subtype O and its validity for antibody detection. J Virol Methods. 1997;67:85–91. doi: 10.1016/s0166-0934(97)00079-7. [DOI] [PubMed] [Google Scholar]
- 5.Felsenstein J. PHYLIP—phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 6.Fransen K, Colebunders B, Heyndrickx L, Janssens W, van de Groen G. How many different HIV-1 viral load test kits do we need? AIDS. 1998;12:230–231. [PubMed] [Google Scholar]
- 7.Fransen K, Zhong P, De Beenhouwer H, Carpels G, Peeters M, Louwagie J, Janssens W, Piot P, van der Groen G. Design and evaluation of new, highly sensitive and specific primers for polymerase chain reaction detection of HIV-1-infected primary lymphocytes. Mol Cell Probes. 1994;8:317–322. doi: 10.1006/mcpr.1994.1043. [DOI] [PubMed] [Google Scholar]
- 8.Gao F, Robertson D L, Carruthers C D, Morrison S G, Jian B, Chen Y, Barré-Sinoussi F, Girard M, Srinivasan A, Abimiku A G, Shaw G M, Sharp P M, Hahn B H. A comprehensive panel of near-full-length clones and reference sequences for non-subtype B isolates of human immunodeficiency virus type 1. J Virol. 1998;72:5680–5698. doi: 10.1128/jvi.72.7.5680-5698.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghosh S K, Fultz P N, Keddie E, Saag M S, Sharp P M, Hahn B H, Shaw G M. A molecular clone of HIV-1 tropic and cytopathic for human and chimpanzee lymphocytes. Virology. 1993;194:858–864. doi: 10.1006/viro.1993.1331. [DOI] [PubMed] [Google Scholar]
- 10.Gürtler L G, Zekeng L, Tsague J M, von Brunn A, Ze E A, Eberle J, Kaptue L. HIV-1 subtype O: epidemiology, pathogenesis, diagnosis, and perspectives of the evolution of HIV. Arch Virol. 1996;11(Suppl.):195–202. doi: 10.1007/978-3-7091-7482-1_17. [DOI] [PubMed] [Google Scholar]
- 11.Hu D, Dondero T J, Mastro T D, Gayle H D. Global and molecular epidemiology of HIV. In: Wormser G P, editor. AIDS and other manifestations of HIV infection—1998. Philadelphia, Pa: Lippincott-Raven Publishers; 1998. pp. 27–40. [Google Scholar]
- 12.Hunt J C, Golden A M, Lund J K, Gürtler L G, Zekeng L, Obiang J, Kaptue L, Hampl H, Vallari A, Devare S G. Envelope sequence variability and serologic characterization of HIV type 1 group O isolates from Equatorial Guinea. AIDS Res Hum Retroviruses. 1997;13:995–1005. doi: 10.1089/aid.1997.13.995. [DOI] [PubMed] [Google Scholar]
- 13.Korber B, Loussert-Ajaka I, Blouin J, Saragosti S. A comparison of HIV-1 group M and group O functional and immunogenic domains in the gag p24 protein and the C2V3 region of the envelope protein. In: Myers G, Korber B, Foley B M, Jeang K T, Mellows J, Wain-Hobson S, editors. Human retrovirus and AIDS—1996, molecular immunology database. Los Alamos, N.Mex: Los Alamos National Laboratory; 1996. pp. IV63–IV79. [Google Scholar]
- 14.Laperche S, Moreau P, Lair J, Couroucé A M. Two successive HIV contaminations from subjects in the window period. AIDS. 1998;12:1397–1398. doi: 10.1097/00002030-199811000-00027. [DOI] [PubMed] [Google Scholar]
- 15.Leitner T. Genetic subtypes of HIV-1. In: Myers G, Korber B, Foley B M, Jeang K T, Mellows J, Wain-Hobson S, editors. Human retrovirus and AIDS—1996, a compilation of nucleic acids and amino acid sequences. Los Alamos, N.Mex: Los Alamos National Laboratory; 1996. pp. III28–III40. [Google Scholar]
- 16.Li Y, Hui H, Burgess C J, Price R W, Sharp P M, Hahn B H, Shaw G M. Complete nucleotide sequence, genome organization, and biological properties of human immunodeficiency virus type 1 in vivo: evidence for limited defectiveness and complementation. J Virol. 1992;66:6587–6600. doi: 10.1128/jvi.66.11.6587-6600.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loussert-Ajaka I, Chaix M-L, Korber B, Letourneur F, Gomas E, Allen E, Ly T-D, Brun-Vézinet F, Simon F, Saragosti S. Variability of human immunodeficiency virus type 1 group O strains isolated from Cameroonian patients living in France. J Virol. 1995;69:5640–5649. doi: 10.1128/jvi.69.9.5640-5649.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loussert-Ajaka I, Descamps D, Simon F, Brun-Vézinet F, Ekwalanga M, Saragosti S. Genetic diversity and HIV detection by polymerase chain reaction. Lancet. 1995;346:912–913. doi: 10.1016/s0140-6736(95)92762-x. [DOI] [PubMed] [Google Scholar]
- 19.Mauclère P, Loussert-Ajaka I, Damon F, Fagot P, Souquieres S, Lobe M M, Keou F M, Barré-Sinoussi F, Saragosti S, Brun-Vézinet F, Simon F. Serological and virological characterization of HIV-1 group O infection in Cameroon. AIDS. 1997;11:445–453. doi: 10.1097/00002030-199704000-00007. [DOI] [PubMed] [Google Scholar]
- 20.Owen S M, Ellenberger D, Rayfield M, Wiktor S, Michel P, Grieco M H, Gao F, Hahn B H, Lal R B. Genetically divergent strains of human immunodeficiency virus type 2 use multiple coreceptors for viral entry. J Virol. 1998;72:5425–5432. doi: 10.1128/jvi.72.7.5425-5432.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pieniazek D, Yang C, Lal R B. Phylogenetic analysis of gp41 envelope of HIV-1 groups M, N, and O strains provides an alternate region for subtype determination. In: Korber B, Foley B, McCutchan F, Mellors J, Hahn B H, Sodroski J, Kuiken C, editors. Human retrovirus and AIDS—1998. Los Alamos, N.Mex: Los Alamos National Laboratory; 1998. pp. 112–117. [Google Scholar]
- 21a.Pillai, S. (HIV Sequence Database). 13 November 1998, posting date. [Online.] http://hiv-web.lanl.gov/NUM-HXB2/HXB2.MAIN/html. [10 June 1999, last date accessed.]
- 22.Respess R A, Butcher A, Wang H, Chaowanachan T, Young N, Shaffer N, Mastro T D, Biryahwaho B, Downing R, Tanuri A, Schechter M, Pascu R, Zekeng L, Kaptue L, Gürtler L, Eberle J, Ellenberger D, Fridlund C, Rayfield M, Kwok S. Detection of genetically diverse human immunodeficiency virus type 1 group M and O isolates by PCR. J Clin Microbiol. 1997;35:1284–1286. doi: 10.1128/jcm.35.5.1284-1286.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schreiber G B, Busch M P, Kleinman S H, Korelitz J J The Retrovirus Epidemiology Donor Study. The risk of transfusion-transmitted viral infections. N Engl J Med. 1996;334:1685–1690. doi: 10.1056/NEJM199606273342601. [DOI] [PubMed] [Google Scholar]
- 24.Simon F, Mauclere P, Roques P, Loussert-Ajaka I, Muller-Trutwin M, Saragosti S, Georges-Courbot M C, Barré-Sinoussi F, Brun-Vézinet F. Identification of a new human immunodeficiency virus type 1 distinct from group M and group O. Nat Med. 1998;4:1032–1037. doi: 10.1038/2017. [DOI] [PubMed] [Google Scholar]
- 25.Takehisa J, Zekeng L, Ido E, Mboudjeka I, Moriyama H, Miura T, Yamashita M, Gürtler L G, Hayami M, Kaptue L. Various types of HIV mixed infections in Cameroon. Virology. 1998;245:1–10. doi: 10.1006/viro.1998.9141. [DOI] [PubMed] [Google Scholar]
- 26.Tanuri A, Swanson T, Devare S, Berro O J, Savedra A, Costa L J, Telles J G, Brindeire R, Schable C, Pieniazek D, Rayfield M. HIV-1 subtypes among blood donors from Rio de Janeiro, Brazil. J Acquired Immune Defic Syndr Hum Retrovirol. 1999;20:60–66. doi: 10.1097/00042560-199901010-00009. [DOI] [PubMed] [Google Scholar]
- 27.Xiao L, Owen S M, Goldman I, Lal A A, deJong J J, Goudsmit J, Lal R B. CCR-5 coreceptor usage of NSI HIV-1 isolates is independent of phylogenetically distinct isolates. Virology. 1998;240:83–92. doi: 10.1006/viro.1997.8924. [DOI] [PubMed] [Google Scholar]