Abstract
The impact of the extent of testing infectious individuals on suppression of COVID-19 is illustrated from the early stages of outbreaks in Germany, the Hubei province of China, Italy, Spain and the UK. The predicted percentage of untested infected individuals depends on the specific outbreak but we found that they typically represent 60–80% of all infected individuals during the early stages of the outbreaks. We propose that reducing the underlying transmission from untested cases is crucial to suppress the virus. This can be achieved through enhanced testing in combination with social distancing and other interventions that reduce transmission such as wearing face masks. Once transmission from silent carriers is kept under control by these means, the virus could have been fully suppressed through fast isolation and contact tracing of tested cases.
Subject terms: Applied mathematics, Epidemiology
Introduction
Daniel Defoe in “A Journal of the Plague Year” (1722) comments on the 1665 Great Plague of London that “. . . if all the infected persons were effectually shut in, no sound person could have been infected by them, because they could not have come near them. But the case was this (and I shall only touch it here): namely, that the infection was propagated insensibly, and by such persons as were not visibly infected, who neither knew whom they infected or who they were infected by.”1 COVID-19 is presenting similar problems today.
COVID-19 produced by the SARS-CoV-2 virus probably emerged in Wuhan, China in December 20192 and has subsequently spread. By 10 April 2020 there were 1,521,252 confirmed cases and 92,798 deaths distributed in 213 countries3. One year later, there were around 135 million confirmed cases and almost 3 million deaths from 220 countries4. The worldwide burden of the disease is still very high despite significant efforts in many countries to suppress the spread of the virus. Until the end of 2020, when vaccines became available, efforts to suppress the virus focused on non-pharmaceutical interventions which ranged from handwashing and social distancing to contact tracing and more stringent measures such as isolation of infected individuals, banning of large gatherings or strict lockdowns5–7.
Optimising interventions to mitigate or suppress the burden of COVID-19 during an epidemic is a pressing global challenge due to significant uncertainties regarding the transmissibility of SARS-CoV-2 and other factors such as potentially large numbers of undocumented infections as well as political, social and economic considerations8,9. Underreported cases may include asymptomatic infected individuals10–12 and some that present symptoms but are not reported or are diagnosed with an alternative disease13. There is no consensus on the proportion of unreported cases and their potential impact on the spread of SARS-CoV-2. For instance, a World Health Organization report in February 2020 suggested that “the proportion of truly asymptomatic infections is unclear but appears to be relatively rare and does not appear to be a major driver of transmission”14.
Studies testing for SARS-CoV-2 infection in both symptomatic and asymptomatic individuals10–12, however, suggest that reported asymptomatic carriers can represent 50% or more of the cases at the time of testing. These may include true asymptomatic that remain asymptomatic throughout their infection and pre-symptomatic individuals that become symptomatic after testing. A comprehensive review suggests that around 20% of infected individuals are true asymptomatic15. Most countries test individuals when they have symptoms, and unreported infections are likely to include most of the asymptomatic individuals and a fraction of those with symptoms. Such asymptomatic individuals can act as silent carriers for SARS-CoV-2 and have been suggested as a key factor promoting the rapid spread of the virus16, similar to what has previously been observed with other infectious diseases17. An important question is to what extent isolation and contact tracing that has been implemented by many countries are effective in preventing the spread of SARS-CoV-2 if there is a significant proportion of infectious individuals that are not tested for it. On the positive side, if recovery from infection leads to immunity, one could hope that untested positive individuals could significantly contribute to the build-up of herd immunity in the population18,19. However, population-based sero-epidemiological studies suggest that the levels of population immunity reached through natural infection are far from that needed for herd immunity even in countries that have experienced the largest epidemics20.
Overall, the levels of underreporting of cases and the importance of such silent carriers on interventions for mitigation and suppression21 of the virus are not clear. Mathematical modelling has been very successful in epidemiology22–24 to devise and simulate suppression strategies and there is an ongoing effort to describe the dynamics of COVID-19 epidemics with mathematical models5,16,19,21,25–37. The effect of undocumented cases on the spread of the virus has been studied previously16, but the influence of such cases on control strategies has not been analysed. Asymptomatic individuals have been included in some models5,28,30,33,34,37,38 but the role of silent carriers in terms of untested cases is not fully understood.
Here we use mathematical models to quantify the levels of underreporting in several COVID-19 outbreaks and understand how this influences the spread of the virus and the efficacy of non-pharmaceutical measures to suppress it.
Materials and methods
Data
Our analysis uses data on numbers of infected and deceased individuals by country or region obtained from the Wolfram Data Repository39. We focus on the outbreaks in Germany, Hubei (China), Italy, Spain and United Kingdom (UK). All these countries have implemented social distance measures to control the spread of the virus. These measures involved global interventions that aim for a reduction of transmission at the population level (e.g., a lockdown) and local interventions involving isolation of infectious individuals and their contacts (see details in Additional file 1). Nationwide lockdowns were ordered in all the studied countries/regions to suppress the virus at the beginning of the outbreaks (see Sect. 1 in Additional file 1). These lockdowns were relaxed after periods that ranged between 56 days in Spain and around 100 days in the UK. Once the initial nationwide lockdowns were relaxed, all study countries kept stay-at-home policies which were particularly strict in China (see Sect. 2 in Additional file 1). In addition, all countries increased testing and contact tracing efforts. Among the European countries, Germany performed the largest number of tests per thousand people and had the lowest positive rate at the early stages of the epidemic (March and April 2020; see Figs. 4.2 and 5.2 in Additional file 1). From June 2020, the UK experienced the most prominent increase in testing efforts and registered the lowest positive rate (see Figs. 4.1 and 5.1 in Additional file 1). This was associated with the launching of test and trace schemes in the four nations of the UK during May 2020 (13 May in Wales40, 20 May in Northern Ireland41 and 28 May in England42 and Scotland43). We did not find detailed data on daily tests performed in China but according to the COVID-19 Government Response Tracker44,45, testing and contact tracing have remained at high levels in China since the early stages of the epidemic (see Sects. 3.1 and 4 in Additional file 1). In fact, only the measures in China were able to avoid a resurgence of the virus so far. The rest of the regions considered here had further waves of infection and all of them except Spain ordered further lockdowns at the end of 2020 and/or beginning of 2021 (see Sect. 1 in Additional file 1).
Models
To describe the role of silent carriers in the spread of the virus, we used extensions of the SEIR model16,23,25,29 to include two types of infected individuals: tested (or reported) and untested (or unreported). The classification of infectious individuals into tested and untested classes intends to be in line with the available data which contains the number of tested cases. We remark that such classification does not make an explicit distinction between symptomatic and asymptomatic cases; in principle, both tested and untested classes may contain symptomatic and asymptomatic cases.
We propose two models: model 1 and model 2. Model 1 is a relatively simple extension of the SEIR model that we use to describe the early stages of epidemics in each of the regions. This model is fitted to data to estimate key epidemiological parameters and the proportion of tested and untested infectious individuals in each region (see parameter estimation methods below). Model 2 is a generalisation of model 1 to account for the isolation of infectious individuals and their contacts. The estimated parameters for model 1 are used to parameterise model 2. We now introduce both models, describe the main assumptions of the models and the methods for parameter estimation.
Model 1
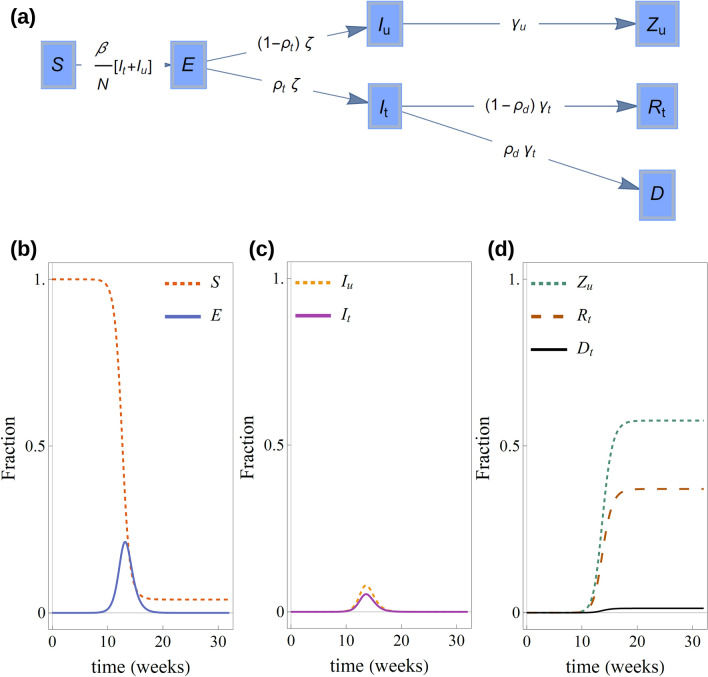
Model 1 assumes that individuals can be in any of five compartments: Susceptible (), exposed to the virus (), tested infectious (), untested infectious (), recovered tested cases (), dead after testing positive () and untested infectious that recover or die (). The flow between compartments is schematically shown in Fig. 1a. The change of the number of individuals in each compartment is described by deterministic, continuous-time dynamics given by the following differential equations:
| 1 |
Figure 1.
(a) Flow diagram for the compartment of model 1. Panels (b–d) show a typical time evolution of model variables. As the epidemic progresses, (b) the number of susceptible individuals decreases. The number of exposed individuals initially increases, reaches a peak and decreases at later stages of the epidemic. (c) The progression of the number of both tested and untested infected individuals also exhibits a peak. The decay of and after the peak induces a gradual weakening of the chain of transmission that leads to the end of the epidemic. (d) The number of tested and untested individuals that recover from infection or die increase monotonically during the epidemic.
Here, is the population size (see Table S1 in Additional file 2). According to these equations, susceptible individuals (compartment ) become exposed to the virus (compartment ) at a rate . Here, is the rate at which an infected individual transmits the infection to a susceptible individual and is the fraction of infectious individuals at time . Exposed individuals remain in this state during a latent period after which they become infectious. Of those that are infectious, a fraction are tested for infection and move to the tested infected compartment . The remaining fraction of infectious individuals, , are not tested for infection and move to the untested infected compartment, , after the incubation period. A fraction of infected individuals that were tested for infection, i.e. a fraction of , die at a rate and move to compartment, . The rest of the tested cases move to the recovered compartment . The fraction corresponds to the case fatality rate (CFR). The infection fatality rate (IFR) is the proportion of the total population that dies and can be determined from our models as . Infected individuals in the untested compartment, , move to the compartment . Assuming that recovered individuals are fully immune to infection, individuals in the compartment are effectively removed from the epidemic. See a typical time evolution of the model variables in Fig. 2b-d.
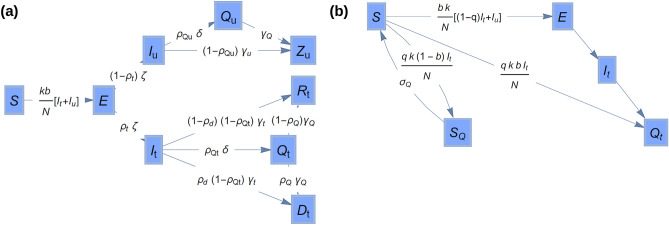
Figure 2.
Model 2. (a) Extension of the flow diagram of model 1 (see Fig. 1) to incorporate a compartment for isolation of tested infectious individuals and a compartment for isolation of untested infectious individuals. (b) Flow diagram showing the implementation of contact tracing in model 2.
The reproductive number corresponding to this model can be analytically calculated using the next generation method24 and conveniently expressed as the sum of two terms, , giving the contribution of tested and untested individuals, respectively. The dependence of these contributions on the parameters of the model are given by the following expressions:
| 2 |
Model 2—Isolation and contact tracing
Model 2 is an extension of model 1 to study the effect of isolation of infected individuals and their contacts. This is achieved by adding two more compartments to model 1, and , which contain isolated tested and untested infectious individuals, respectively (see Fig. 2a). The fraction of tested and untested infectious individuals who are isolated are denoted as and , respectively. Both types of infectious individuals are assumed to become isolated at the same rate, . Isolation of tested individuals is the most natural isolation strategy. Possible isolation of untested individuals, however, was incorporated to simulate scenarios in which some untested individuals may self-isolate without being tested if they exhibit symptoms or due to reasons that might not be linked to their infection (e.g. being advised to work from home or voluntarily trying to minimise contact). Contact tracing involves isolation of a fraction of susceptible individuals that were in contact with infected tested cases (see Fig. 2b). A fraction of these contacts would have acquired the virus through the contact (i.e. in the absence of contact tracing and isolation, they would have moved to the exposed compartment). These individuals are quarantined in the compartment before they become infections and will eventually recover or die. The remaining contacts, i.e. a fraction , remain susceptible after the interaction with the infected case and their isolation is represented by a compartment where they remain for a period . After this period, such individuals remain susceptible and return to the compartment. With these assumptions, the number of individuals in each compartment evolve according to the following differential equations:
| 3 |
Here, is the number of contacts per unit time. The transmission rate is given by . Splitting the transmission rate into the number of contacts per unit time, , and the fraction of successful transmissions in a contact, , is necessary to model contact tracing23. Note that model 1 can be formally recovered from model 2 when the rates related to isolation and contact tracing are zero, i.e. when in Eq. (3).
One can again use the next generation method24 to obtain an expression for the reproduction number of model 2 which can be expressed as the sum of two terms, , where
| 4 |
Control measures
Both model 1 and model 2 can be used to simulate measures such as a lockdown that achieve a reduction of transmission at the population level. This is simply implemented by reducing the intrinsic transmission rate by a factor (i.e. the reduced transmission is ).
The effect of the initial lockdown ordered in all studied countries and subsequent relaxation was simulated with a piece-wise dependence of on time (see details in Sect. 1 of Additional file 3). The gradual increase in testing efforts, isolation of cases and contact tracing was simulated with model 2 assuming sigmoid growth functions for , and (see details in Sect. 1 of Additional file 3).
Main assumptions of the models
Several simplifying assumptions were made to make the models operational and applicable to settings with limited data which are typical at early stages of epidemics. Here, we describe the main assumptions.
Homogeneous populations
The total number of individuals in each compartment of our models represent average values for the whole country. More precise descriptions at smaller scales such as geographical regions within countries or at the level of individuals would require accounting for spatial heterogeneity within the populations31,46 (e.g. cities and rural areas) as well as differences in both susceptibility and mortality across different age and vulnerability groups47,48 or topological heterogeneity of the network of contacts between individuals49.
Ignoring heterogeneities limits the ability of our models to identify specific ways to make interventions operational. For instance, reductions in transmission are treated at a generic level without specifying if they could be achieved by enhanced social distancing, school closure, etc. Accounting for such details would require using individual-based simulations21.
Deterministic dynamics
We focused on stages of epidemics in which the number of infectious individuals is large enough as for stochastic effects to be relatively unimportant on average27. Our models can be extended to incorporate stochasticity50,51. This would give a more accurate description of epidemics when SARS-CoV-2 has just invaded or at later stages when the number of infected individuals becomes very low.
All of the population at the start of the epidemic are susceptible
However, it may be that a proportion of the population are not susceptible for genetic reasons52 or due to cross immunity53,54.
Imported cases
We focused on epidemics that are at a stage in which imported cases are expected to play a secondary role relative to internal transmission. Accounting for imported cases is crucial, however, to prevent re-emergence of infection once an area has reached low numbers of infected individuals55,56. Imported cases could be included in our models in terms of an influx of infected individuals. In scenarios in which imported cases represent an important fraction of new infections, however, a stochastic version of the model would be more appropriate than the deterministic dynamics used here.
The transition times between compartments are exponentially distributed
This memory-less assumption is usual for classical compartmental models23. For COVID-19, however, transitions between compartments are better described in terms of gamma distributions5,47 and using models with memory would provide a more precise description of the dynamics5,23,57.
Tested and untested cases transmit infection at the same rate
The populations of tested and untested cases will typically consist of different proportions of symptomatic and asymptomatic cases. In spite of that, no statistical difference has been observed between the viral load of symptomatic and asymptomatic individuals58,59. It is therefore reasonable to assume that transmission is similar for symptomatic and asymptomatic individuals and therefore similar for tested and untested cases. Differences in overall transmission, however, are incorporated in the model through the assumption that recovery rates for tested and untested cases can be different.
Immunity follows after recovery from infection
Whether or not this is the case, and even if it is, the duration of the immunity is still unclear60. Our model could easily be extended to account for re-infections, should such data become available, and predictions might significantly change.
The latent and incubation periods coincide
We adopted a parsimonious approach which assumes that the latent period (i.e. the time between exposure to communicability) coincides with the incubation period (time between exposure and the appearance of symptoms)25. There is, however, a growing body of evidence for pre-symptomatic transmission61–65. There is the potential to incorporate this in our models by varying the incubation rate parameter, , or including a new compartment for pre-symptomatic infectious individuals31. We expect our predictions to be qualitatively independent of these details.
Reported deaths
We assume that the number of deaths in the datasets originates from individuals that were tested for infection.
Transmission in different regions
It is assumed that transmission of SARS-CoV-2 and the fraction of infectious cases are similar in different regions. This is reflected in the informative prior distributions used for and in the parameter estimation procedure (see Table 1).
Table 1.
Assumptions for the prior probability distribution of the estimated parameters.
| Parameter | Prior | Support |
|---|---|---|
| Fraction tested infected, | Uninformative for | |
| Fraction of tested infected that die, | Mean set to the global estimate of WHO85 | |
| Recovery rate for untested infected, | Range assumed from manual fit exploration (contains typical values for recovery rate25,47) | |
| Recovery rate for untested infected, | Similar assumptions as for | |
| Reproduction number, | Estimates from Ref.5 | |
| Transmission rate, | Derived from other parameters using Eq. (2) | |
| Number of exposed at the first data point, | Refs.16,25 and manual fit exploration |
denotes a normal distribution with mean and variance . denotes a uniform distribution in the interval .
Parameter estimation
We fitted the Model 1 to data. The value for the incubation rate was set to48 1/5.2 days-1. The free parameters in our fits were the rate of transmission, , proportion of infectious that were tested, , proportion of tested infectious that die, , rate to recovery of tested infectious individuals, rate of recovery of untested infectious individuals, , and initial number of exposed individuals, . We denote the free parameters by a vector The model was fit to the time series for the number of daily reported infected individuals and cumulative deaths, , in a period of days in the early stages of epidemics (here, is used to denote discrete time in days). In particular, we used days since the first data point with a positive number of deaths (see Table S1 in Additional file 2). We used data at early stage of each outbreak to minimise the influence of suppression strategies on our parameter estimates.
Using data on deaths is important to obtain reliable descriptions of COVID-19 epidemics since data on deaths is likely to be more accurately recorded than data on infected and recovered individuals5,19,38,66. In addition to deaths, we can use data on infected individuals which is represented by the tested infectious compartment, , in our models.
Our fitting procedure aims at calculating the posterior probability density function for the parameters given the data, . To this end, we use the procedure proposed in51 which can be regarded as an approximate Bayesian algorithm67. The posterior is approximated by the empirical distribution of a set of 500 point estimates of the model parameters. A point estimate is obtained by simulating epidemics with parameters sampled from a prior probability density . In each realization, a simulation of Model 1 produces deterministic evolution functions and for the number of tested cases and cumulative deaths. The functions and are used to build a random daily time series , where and are, respectively, the number of tested infected and deaths predicted at day . The point estimate is defined as the parameter vector corresponding to the realization that gives the closest prediction,, to the observations, . More explicitly, the point estimate for the model parameters is given by
| 5 |
where is a distance function. In particular, we used a weighted quadratic form for the distance:
| 6 |
Weighting by the observed values was used to account for the fact that the values taken by and differ by orders of magnitude. We checked, however, that fits with an unweighted distance give results that are statistically compatible with those reported in the main text. In addition, in the previous version of this work we obtained similar results by an approximate maximisation of a log-likelihood function in which and , i.e. the predicted number of tested infected and deaths were described as random variables obeying a Poisson distribution with mean and , respectively68.
The prior probability density is defined as the product of priors for each parameter:
| 7 |
The priors used in our fits are summarised in Table 1.
Results
Calibration of Model 1 at early stages of epidemics
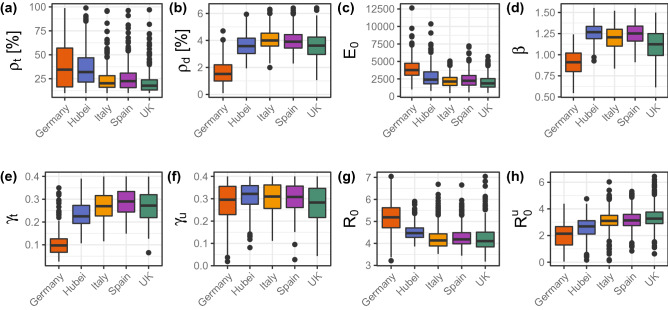
Figure 3 shows the estimates for the parameters of model 1 applied to the early stages of each outbreak (see numerical summary statistics in Additional file 2, Table S2). A comparison of the predicted trends and the data is shown in Additional file 2, Fig. S1.
Figure 3.
Boxplot statistics of the estimated parameters for model 1.
The values for the testing coverage, , reveal that during the early stage of outbreaks, Germany scored the highest in terms of testing for infection (34% [95% CI 10–96%]). Hubei follows Germany in terms of testing coverage, followed by Spain, Italy and the UK. Our prediction for Hubei (32% [95% CI 11–85%]) is statistically consistent with the 65% reporting estimated by Li et al.16 for China (not only Hubei) in the period 24 Jan–8 Feb 2020 which overlaps with the period 22 Jan–6 Feb 2020 used here. We note, however, that our estimate has a wide 95% CI and the comparison is not highly informative. The high testing percentage predicted for Germany agrees with the higher testing coverage at early stages of epidemics in this country (see Fig. 4.2 in Additional file 1). Taking the confidence intervals into account, we estimate that for each infected individual tested in the UK, there could have been between 1 and 9 untested infected individuals. At the other end of the testing spectrum, we estimate that for each infected individual tested in Germany, between 0.05 and 9 individuals might have not been tested at the beginning of the epidemic. A higher testing coverage for Germany is in qualitative agreement with estimates given elsewhere28,38. Our estimates for the reporting percentage, however, tend to be higher than those obtained by Jagodnik et al.38 and the differences we found between countries are not as extreme as those given by Chicchi et al.28.
From our estimates of , we predict that 66% [95% CI 4–90%] of infected individuals were not tested for infection in Germany. Bearing in mind that 50% of infected individuals might be reported as asymptomatic at testing10,12, we conclude that untested cases in Germany would mostly correspond to asymptomatic cases at testing. In contrast, untested cases in other countries might include a significant number of symptomatic individuals in addition to asymptomatic cases. In particular, these may include infections of care home residents that are known to be underreported in many countries at early stages of epidemics69.
The proportion of tested infected individuals that die, , is smaller for the outbreak in Germany than for the other outbreaks. This might be a combined effect of the fact that infected individuals in this country were relatively young at the beginning of the outbreak70 and the high testing rate. Indeed, the COVID-19 fatality rate is lower for the younger than for the elderly47 and, the higher the testing rate, the more individuals with mild or no symptoms will be included in the tested infected compartment of our model. The lower death rate of individuals with mild symptoms will lead to an effectively lower death rate for the whole set of infected individuals in this compartment. Accordingly, a lower value of does not necessarily mean a lower overall infection fatality rate. In fact, our estimates for does not vary much among countries (between 0.5% [95% CI 0.1–1.1%] for Germany and 1.1% [95% CI 0.4–2.9%] for Hubei). These values are compatible with an IFR of 0.68% [95% CI 0.53–0.82%] reported for several studies in a systematic review71. Related to this, we found that the predicted fraction of deaths by the end of unmitigated epidemics is not too different for different countries either: 0.2% [95% CI 0.03–0.8%] for Germany, 0.8% [95% CI 0.3–3.6%] for Hubei, 0.7% [95% CI 0.3–2.1%] for Italy, 0.8% [95% CI 0.3–3.6%] for Spain and 0.6% [95% CI 0.2–2.3%] for the UK.
Our estimates for the time from reporting of infection to recovery or death are 3.4 [95% CI 2.4–6.0] days for Spain, 3.7 [95% CI 2.6–6.3] days for Italy, 3.7 [95% CI 2.6–6.5] days for UK, 4.4 [95% CI 2.7–7.1] days for Hubei and 10 [95% CI 3–26] days for Germany. The time for Hubei is consistent with estimates for China in other studies16,35. In general, the values we obtained are smaller than the infectious period (time from infection to death or recovery) reported elsewhere for COVID-195,47,72. Our estimates thus probably reflect a reporting delay in all the studied outbreaks, in agreement with data on the onset of symptoms and reporting70,73,74. Under this hypothesis, our model predicts the smallest reporting delay for Germany, in agreement with a higher testing effort in this country at early stages of the epidemic.
The removal period for untested infected individuals, , ranges between 3.1 [95% CI 2.5–6.1] days for Hubei and 3.5 [95% CI 2.5–8.0] days for the UK. Comparing with the reporting-to-recovery period and bearing in mind the reporting delays in all outbreaks, our estimates of suggest that untested individuals remain infectious for a shorter time than tested individuals.
We predict that the number of exposed individuals at the beginning of our simulations, , is of the order of several thousand for all the countries, in qualitative agreement with estimates of a previous study for China16.
The estimated reproduction number is statistically similar in all the studied outbreaks. This reflects our prior assumption that transmission of SARS-CoV-2 is intrinsically similar in different regions. The transmission rate, , was derived from the estimates of and and it takes values that are around 1 for all countries.
Suppression strategies
Suppression of the virus is achieved when the reproductive number is smaller than 1. In terms of model 2, the condition can be achieved in several different ways, i.e. there are many combinations of the parameters of the model that can lead to suppression of the virus (see Eq. 4). A simple suppression strategy consists in reducing the transmission rate . We studied this strategy in April 2020 to make predictions related to the lockdowns that were active in Germany, Italy, Spain and the UK; the lockdown had just being lifted in Hubei68. We predicted that early reduction of transmission would delay the outbreaks but resurgence of infection was likely after relaxing the lockdowns unless transmission was kept reduced by a factor larger than 70% of its intrinsic values (see an update to these results in Sect. 2 of the Additional file 3). These results were prescient for Germany, Italy, Spain and the UK where further waves of infection occurred later in 2020.
In addition to nationwide lockdowns, all studied regions have implemented isolation and contact tracing programmes to suppress the virus. Following this, the reductions of mentioned above for scenarios without isolation and contact tracing should be interpreted as an effective measure of the reduction of transmission. Indeed, when isolation and contact tracing are taken into account, one needs a smaller reduction in to suppress the virus. Below we use model 2 to account for the combined effect of a reduced and enhanced testing, isolation and contact tracing in the design of suppression strategies bearing in mind the presence of silent carriers of the virus.
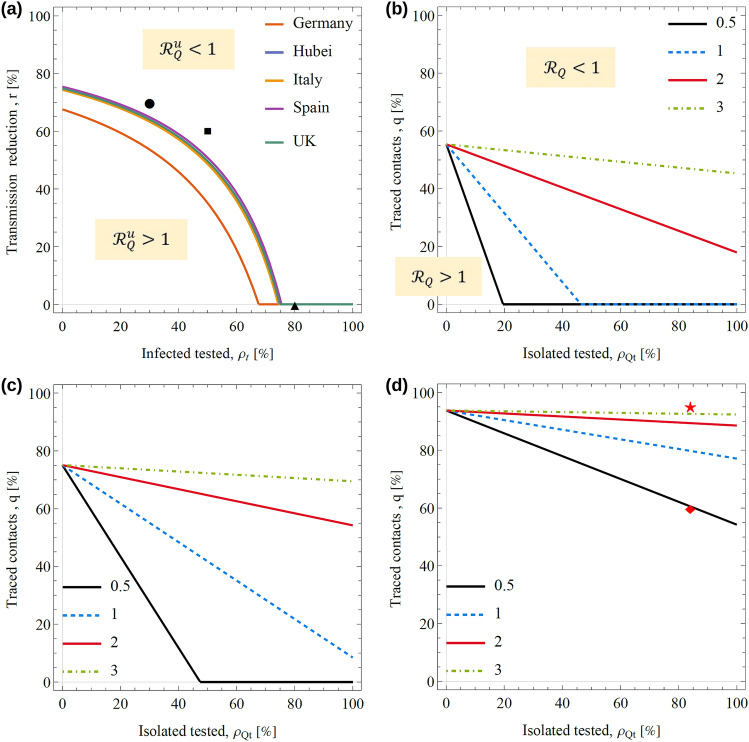
From Eq. (4) one can see that interventions that only isolate tested cases and their contacts can reduce the reproductive number associated with tested infected individuals. However, such strategy cannot suppress the virus since the reproductive number of untested cases is for all the studied epidemics (Fig. 3h). Ensuring that is then a crucial step for suppression of the virus. The reproduction number decreases with the testing rate, physical distancing and isolation of untested cases. Even in optimistic scenarios in which all tested cases would isolate in 0.5 days, we estimate that more than 25% of untested cases should isolate to suppress the virus. This is unlikely to occur by spontaneous isolation of untested cases (see Sect. 3 of the Additional file 3).
We now focus on more feasible suppression approaches that combine enhanced testing and physical distancing with isolation and contact tracing of tested cases (i.e. no isolation is assumed for untested infectious individuals). The condition can be achieved through physical distancing and/or enhanced testing that are described by a reduction in and an increase in , respectively. Figure 4a shows the threshold line corresponding to in the space , where is the reduction factor of . In particular, can be achieved without the need of physical distancing () if more than ~ of infected individuals are tested. The condition could also be achieved without any testing (i.e. with ) if a severe reduction of transmission with 80% is imposed. The later corresponds to a lockdown scenario studied above. Between these two extremes, the condition requires a combination of physical distancing and enhanced testing.
Figure 4.
Suppression of the virus with physicals distance, enhanced testing, isolation and contact tracing. (a) Threshold lines for the reproduction number of untested cases as a function of the testing percentage and reduction of transmission, . For a given country, above the line and below the line. The threshold lines for Italy and Hubei, Spain and UK are close to each other and hardly distinguishable in the plot. Taking the UK as an example, panels (b–d) show the threshold lines for suppression in terms of the isolation and contact tracing percentages of tested cases for different combinations of the testing percentage and reduction of transmission marked by the circle, square and triangle in panel (a). More explicitly, the parameters are in panel (b), in panel (c), and in panel (d). For a given panel, different lines indicate the threshold for different values of the time to isolation, , as marked by the legend. In panel (d), the diamond and star correspond to situations with and , respectively, which were used to simulate the effect of enhanced isolation and contact tracing on the daily deaths in the UK (see Sect. 4 in Additional file 3). All lines show estimates based on the median of the model parameters.
Once the condition is satisfied for untested cases, suppression can be achieved by reducing through isolation and contact tracing of tested cases. To illustrate this, we take the UK as an example and consider three scenarios with different levels of testing and social distancing.
Testing 30% of cases and reducing transmission by 70% through physical distancing leads to (circle in Fig. 4a) and suppression is possible if 20% of infected cases were isolated in = 0.5 days (see black line in Fig. 4b). In this case, contact tracing would not be needed. For slower isolation of tested cases, however, contact tracing would be needed even if 100% of tested cases were isolated. This is illustrated in Fig. 4b by the lines corresponding to isolation in = 2 or 3 days. Our results echo other studies in terms of the need to quickly isolate infected individuals75. In order to allow for a reasonable time between reporting of infection and isolation, carriers of the virus should be identified as early as possible, i.e. reporting delays should be minimised.
Suppression is also possible if 50% of cases are tested and strong physical distancing is imposed which keeps transmission reduced by a 60% (square in Fig. 4a). In this case, suppression without contact tracing is only possible for prompt isolation of tested cases (see black line in Fig. 4c). For slower isolation rates, a high coverage of contacts is needed even if all tested cases were isolated. For instance, if 100% of tested cases were isolated after 2 days, more than 60% of contacts should be traced to suppress the virus (see the red line in Fig. 4c).
In principle, mass testing and high compliance with isolation would make suppression possible without the need for physical distancing. For instance, suppression would be possible if > 80% of the cases were tested (triangle in Fig. 4a). As in the scenarios described above, high isolation and contact tracing would be needed to suppress the virus even if isolation is very fast (see Fig. 4d).
Detecting 80% of cases as assumed in the last scenario could be possible if, for instance, only truly asymptomatic cases escaped testing (around 20% of infected individuals are expected to be truly asymptomatic15). We tested the feasibility of this scenario for the second wave of infection observed in the UK after the relaxation of the initial lockdown. Stay-at-home and physical distancing policies remained active once the lockdown was relaxed in the UK. It is interesting, however, that the second wave of infection can be explained by model 2 in terms of a hypothetical scenario consisting of complete relaxation of the lockdown combined with enhanced testing and launching of test and trace schemes throughout the country. More explicitly, the scenario is as follows: Testing only misses asymptomatic cases (i.e.15 = 80%), 84% of those that tested positive self-isolate76 after 2 days of being tested77 and 60% of contacts are traced and isolated78 (this corresponds to the diamond in Fig. 4d that is below the red line, i.e. it is in the region with ). Full details of this analysis and a comparison of the predictions of model 2 with data are given in Sect. 4 of the Additional file 3. We checked that a similar scenario in which 95% of contacts are traced instead of 60% leads to suppression of the virus (see the star symbol in Fig. 4d which is above the red line where ).
Even though a scenario that assumes a complete relaxation of the lockdown can reproduce the data in the UK, a scenario with a partial relaxation of the lockdown is more realistic since stay-at-home and physical distancing policies remained active after the relaxation of the lockdown. This would indicate that the levels of testing, isolation and contact tracing were probably less effective than those used above for complete relaxation of the lockdown. For example, we checked that a scenario with post-lockdown transmission reduced by 20% and an isolation rate of 20% also captures the observed trend for the number of dealy deaths after the lockdown (see Fig. S4(c) in Sect. 4 of the Additional file 3). In general, there can be many combinations of the parameters of model 2 leading to a reasonable description of the data. In spite of that, our results suggest that high levels of testing, isolation and contact tracing could suppress the virus even if 20% of cases were asymptomatic and not tested for infection.
Conclusions
The main aim of our modelling work is to contribute to the understanding of the epidemiological patterns of SARS-Cov-2. The models should be viewed as a general guide of how the outbreak and interventions may play out rather than as an exact representation of COVID-19 epidemics. In spite of our simplifying assumptions, there are two main implications from the models which are relevant for health policy in dealing with the outbreak.
The first, involves the existence of a significant proportion of cases that are not tested and act as silent carriers of the infection. We found that the predicted percentage of untested infected individuals at the early stages of epidemics may have represented 60–80% of the cases. The specific percentage depends on the country and we found the lowest proportion of unreported cases in Germany. The levels of underreporting are expected to have gradually decreased during the course of the studied epidemics since testing capacity increased in all the studied regions. It is likely, however, that underreporting remains significant due to asymptomatic individuals or cases with mild symptoms.
Our model predicts that a resurgence of the virus was likely after relaxing the initial lockdown in 2020 in all the studied regions. This would be similar to second waves of infection observed in the 1918 influenza epidemics79. In fact, we made this prediction in April 2020 when lockdowns were still active in the studied European countries and it had just been relaxed in Hubei68. This prediction was confirmed by the resurgence of cases observed in European countries in August 202080.
The second implication involves the finding that unreported cases play an important role in the control of COVID-19 epidemics. In particular, unreported cases act as silent carriers and control strategies need to account for them or be prone to the risk of re-emergence or ineffective suppression of spread. For instance, we predict that isolation and contact tracing of tested cases can have a limited impact on the suppression of spread unless the underlying transmission of silent carriers is suppressed. The latter can be achieved by combining physical distancing and thorough testing of case contacts. Related to this, we found that physical distance might not be essential in an ideal scenario in which testing only misses 20% of cases (e.g. asymptomatic), there is a high adherence to self-isolation policies and contact tracing is highly effective. In the absence of pharmaceutical interventions, however, a certain level of physical distance is likely to be necessary for suppression. To summarise, in line with previous work19,81 and our predictions in April 2020, we suggest that widespread testing combined with contact tracing26,27, isolation of infected individuals and social distancing are necessary to suppress SARS-CoV-2 using non-pharmaceutical interventions without severe lockdowns.
Vaccinations are now available and our models could be extended to study the combined effect of vaccination and non-pharmaceutical interventions on epidemics while accounting for underreporting. Information on the effects of vaccines is still limited but it is expected that they will reduce the risk of individuals becoming infected and will protect against COVID-19 symptoms and severe illness82–84. Gradual reductions of the risk of infection and risk of death can be readily simulated by assuming gradually decreasing the transmission rate () and proportion of infected individuals that die (). A reduction in leads to a reduction of the reproduction number of both tested and untested infectious individuals and this aids suppression of the virus. Enhanced protection against symptoms plays a less clear role in the suppression and would require a more detailed analysis. Indeed, protection against symptoms might lead to an increased proportion of silent carriers of the virus. This might lead to an increase in the reproduction number that might ultimately reduce some of the benefits of the vaccine. Based on early evidence that vaccines reduce the risk of serious illness82, prioritising the vaccination of the most vulnerable individuals is crucial to make sure that a potential resurge of transmission associated with silent carriers occurs when those individuals have been already vaccinated. In addition to that, keeping high levels of testing and isolation of cases will be crucial to prevent potential negative effects associated with unreported cases.
Supplementary Information
Acknowledgements
We acknowledge fruitful discussions with Oliver Carrillo, Bruno Lopes, Christopher McGuigan, Veronica Morales, Stefano Polizzi, Ovidiu Rotariu, John Strachan and Eduard Vives.
Author contributions
F.P.R., K.F. and N.S. planned the project and designed the research. F.P.R. retrieved the data and run the numerical simulations. F.P.R. N.S. and K.F wrote the manuscript. All authors reviewed the manuscript and approved the final version of it.
Data availability
All data used in this work are available from the cited sources. The models were analysed implemented in Mathematica. A notebook that retrieves the data and runs the calculations can be downloaded from https://doi.org/10.6084/m9.figshare.14636199. Point estimates of the parameters of model 1 and an R script to generate Fig. 3 can also be downloaded from this link.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-00056-5.
References
- 1.Defoe, D. A Journal of the Plague Year. (E. Nutt, 1722).
- 2.Zhu N, et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization. Coronavirus disease 2019 (COVID-19) Situation Report – 81. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200410-sitrep-81-covid-19.pdf?sfvrsn=ca96eb84_2 (2020).
- 4.World Health Organization. Coronavirus disease 2019 (COVID-19) Situation Report – 203. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200810-covid-19-sitrep-203.pdf?sfvrsn=aa050308_4 (2020).
- 5.Flaxman, S., Mishra, S., Gandy, A. & al, E. Estimating the number of infections and the impact of non-pharmaceutical interventions on COVID-19 in 11 European countries. (2020) 10.25561/77731.
- 6.Ainslie, K. E. C., Han Fu, C. W. & al, E. Evidence of initial success for China exiting COVID-19 social distancing policy after achieving containment. (2020) 10.25561/77646. [DOI] [PMC free article] [PubMed]
- 7.European Centre for Disease Prevention and Control. Contact tracing for COVID-19: current evidence, options for scale-up and an assessment of resources needed. https://www.ecdc.europa.eu/en/publications-data/contact-tracing-covid-19-evidence-scale-up-assessment-resources (2020).
- 8.Anderson RM, Heesterbeek H, Klinkenberg D, Hollingsworth TD. How will country-based mitigation measures influence the course of the COVID-19 epidemic? Lancet. 2020;395:931–934. doi: 10.1016/S0140-6736(20)30567-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collinson, S. Contagion: The Economic and Social Impacts of Covid-19 on our Region. https://www.birmingham.ac.uk/news/thebirminghambrief/items/2020/03/contagion-the-economic-and-social-Impacts-of-covid-19-on-our-region.aspx.
- 10.Mizumoto K, Kagaya K, Zarebski A, Chowell G. Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Eurosurveillance. 2020;25:2000180. doi: 10.2807/1560-7917.ES.2020.25.10.2000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Day M. Covid-19: identifying and isolating asymptomatic people helped eliminate virus in Italian village. BMJ. 2020;368:m1165. doi: 10.1136/bmj.m1165. [DOI] [PubMed] [Google Scholar]
- 12.Tara John. Iceland lab’s testing suggests 50% of coronavirus cases have no symptoms. https://edition.cnn.com/2020/03/14/health/coronavirus-asymptomatic-spread/index.html.
- 13.Chavarria-Miró, G. et al. Sentinel surveillance of SARS-CoV-2 in wastewater anticipates the occurrence of COVID-19 cases. medRxiv 2020.06.13.20129627 (2020) 10.1101/2020.06.13.20129627.
- 14.World Health Organization. Report of the WHO-China Joint Mission on coronavirus disease 2019 (COVID-19). https://www.who.int/docs/default-source/coronaviruse/who-china-joint-mission-on-covid-19-final-report.pdf (2020).
- 15.Buitrago-Garcia D, et al. Occurrence and transmission potential of asymptomatic and presymptomatic SARS-CoV-2 infections: A living systematic review and meta-analysis. PLOS Med. 2020;17:e1003346. doi: 10.1371/journal.pmed.1003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li R, et al. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV2) Science. 2020 doi: 10.1126/science.abb3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stein RA. Super-spreaders in infectious diseases. Int. J. Infect. Dis. 2011;15:e510–e513. doi: 10.1016/j.ijid.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clive Cookson. Coronavirus may have infected half of UK population—Oxford study. https://www.ft.com/content/5ff6469a-6dd8-11ea-89df-41bea055720b.
- 19.Lourenço, J. et al. Fundamental principles of epidemic spread highlight the immediate need for large-scale serological surveys to assess the stage of the SARS-CoV-2 epidemic. medRxiv 2020.03.24.20042291 (2020). 10.1101/2020.03.24.20042291.
- 20.European Centre for Disease Prevention and Control. Immune responses and immunity to SARS-CoV-2. https://www.ecdc.europa.eu/en/covid-19/latest-evidence/immune-responses.
- 21.Ferguson, N. M., Laydon, D., Nedjati-Gilani, G. & et al. Impact of non-pharmaceutical interventions (NPIs) to reduce COVID-19 mortality and healthcare demand. 10.25561/77482 (2020). [DOI] [PMC free article] [PubMed]
- 22.Anderson, R. M. & May, R. M. Infectious diseases of humans: dynamics and control. (Oxford University Press, 1991).
- 23.Keeling, M. J. & Rohani, P. Modeling infectious diseases in humans and animals. (Princeton UP, 2007).
- 24.Diekmann, O., Heesterbeek, H. & Britton, T. Mathematical Tools for Understanding Infectious Disease Dynamics. (Princeton University Press, 2013).
- 25.Wang H, et al. Phase-adjusted estimation of the number of coronavirus disease 2019 cases in Wuhan China. Cell Discov. 2020;6:10. doi: 10.1038/s41421-020-0148-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hellewell J, et al. Feasibility of controlling COVID-19 outbreaks by isolation of cases and contacts. Lancet Glob. Health. 2020;8:e488–e496. doi: 10.1016/S2214-109X(20)30074-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keeling, M. J., Hollingsworth, T. D. & Read, J. M. The Efficacy of Contact Tracing for the Containment of the 2019 Novel Coronavirus (COVID-19). medRxiv 2020.02.14.20023036 (2020). 10.1101/2020.02.14.20023036. [DOI] [PMC free article] [PubMed]
- 28.Chicchi, L., Di Patti, F., Fanelli, D., Piazza, F. & Ginelli, F. Analysis and forecast of COVID-19 spreading. https://www.researchgate.net/profile/Francesco_Piazza3/project/Analysis-and-forecast-of-COVID-19-spreading/attachment/5e766abfcfe4a7809f8a0ff0/AS:871607706861568@1584818879039/download/COVID19_update21.03.2020.pdf?context=ProjectUpdatesLog (2020). [DOI] [PMC free article] [PubMed]
- 29.Read, J. M., Bridgen, J. R. E., Cummings, D. A. T., Ho, A. & Jewell, C. P. Novel coronavirus 2019-nCoV: early estimation of epidemiological parameters and epidemic predictions. medRxiv 2020.01.23.20018549 (2020). 10.1101/2020.01.23.20018549. [DOI] [PMC free article] [PubMed]
- 30.Kucharski, A. J. et al. Early dynamics of transmission and control of COVID-19: a mathematical modelling study. Lancet Infect. Dis.0, (2020). [DOI] [PMC free article] [PubMed]
- 31.Arenas, A. et al. A mathematical model for the spatiotemporal epidemic spreading of COVID19. medRxiv 2020.03.21.20040022 (2020). 10.1101/2020.03.21.20040022.
- 32.López L, Rodó X. The end of social confinement and COVID-19 re-emergence risk. Nat. Hum. Behav. 2020 doi: 10.1038/s41562-020-0908-8. [DOI] [PubMed] [Google Scholar]
- 33.Giordano G, et al. Modelling the COVID-19 epidemic and implementation of population-wide interventions in Italy. Nat. Med. 2020;26:855–860. doi: 10.1038/s41591-020-0883-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gatto M, et al. Spread and dynamics of the COVID-19 epidemic in Italy: Effects of emergency containment measures. Proc. Natl. Acad. Sci. 2020 doi: 10.1073/pnas.2004978117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang B, et al. Estimation of the transmission risk of the 2019-nCoV and its implication for public health interventions. J. Clin. Med. 2020;9:462. doi: 10.3390/jcm9020462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maier BF, Brockmann D. Effective containment explains subexponential growth in recent confirmed COVID-19 cases in China. Science. 2020;368:742–746. doi: 10.1126/science.abb4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stutt ROJH, Retkute R, Bradley M, Gilligan CA, Colvin J. A modelling framework to assess the likely effectiveness of facemasks in combination with ‘lock-down’ in managing the COVID-19 pandemic. Proc. R. Soc. Math. Phys. Eng. Sci. 2020;476:20200376. doi: 10.1098/rspa.2020.0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lachmann, A., Jagodnik, K. M., Giorgi, F. M. & Ray, F. Correcting under-reported COVID-19 case numbers: estimating the true scale of the pandemic. medRxiv 2020.03.14.20036178 (2020). 10.1101/2020.03.14.20036178.
- 39.Wolfram Research, Inc. Epidemic Data for Novel Coronavirus COVID-19" from the Wolfram Data Repository (2020). https://www.wolframcloud.com/obj/resourcesystem/published/DataRepository/resources/Epidemic-Data-for-Novel-Coronavirus-COVID-19.
- 40.Test, Trace, Protect (contact tracing for coronavirus (COVID-19)). GOV.WALEShttps://gov.wales/test-trace-protect-contact-tracing-coronavirus-covid-19.
- 41.Contact Tracing Service - Management Information Update | HSC Public Health Agency. https://www.publichealth.hscni.net/covid-19-coronavirus/testing-and-tracing-covid-19/contact-tracing-service-management-information.
- 42.NHS Test and Trace statistics (England): methodology. GOV.UKhttps://www.gov.uk/government/publications/nhs-test-and-trace-statistics-england-methodology/nhs-test-and-trace-statistics-england-methodology.
- 43.Test and Protect rolled out nationally - gov.scot. https://www.gov.scot/news/test-and-protect-rolled-out-nationally/.
- 44.COVID-19 Government Response Tracker. https://www.bsg.ox.ac.uk/research/research-projects/covid-19-government-response-tracker.
- 45.COVID-19: Testing and Contact Tracing. Our World in Datahttps://ourworldindata.org/covid-testing-contact-tracing.
- 46.How many confirmed cases are there in your area?https://www.bbc.com/news/uk-51768274 (2020).
- 47.Verity, R. et al. Estimates of the severity of coronavirus disease 2019: a model-based analysis. Lancet Infect. Dis.0, (2020). [DOI] [PMC free article] [PubMed]
- 48.Li Q, et al. Early transmission dynamics in Wuhan, China, of Novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Newman, M. E. J. Networks: an introduction. (Oxford University Press, 2010).
- 50.Pérez-Reche, F. J., Ludlam, J. J., Taraskin, S. N. & Gilligan, C. A. Synergy in spreading processes: From exploitative to explorative foraging strategies. Phys. Rev. Lett.106, (2011). [DOI] [PubMed]
- 51.Pérez-Reche, F. J., Neri, F. M., Taraskin, S. N. & Gilligan, C. A. Prediction of invasion from the early stage of an epidemic. J. R. Soc. Interface9, (2012). [DOI] [PMC free article] [PubMed]
- 52.Tanigawa, Y. & Rivas, M. Initial Review and analysis of COVID-19 host genetics and associated phenotypes. Preprints 2020030356. 10.20944/preprints202003.0356.v1.
- 53.King, A. Possible Biological Explanations for Kids’ Escape from COVID-19. https://www.the-scientist.com/news-opinion/possible-biological-explanations-for-kids-escape-from-covid-19-67273 (2020).
- 54.Grifoni A, et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davidson, H. China coronavirus infections rise as some Spanish companies prepare to restart work. https://www.theguardian.com/world/2020/apr/13/china-infections-rise-as-some-spanish-companies-prepare-to-restart-work (2020).
- 56.Wells CR, et al. Impact of international travel and border control measures on the global spread of the novel 2019 coronavirus outbreak. Proc. Natl. Acad. Sci. 2020;117:7504–7509. doi: 10.1073/pnas.2002616117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wearing HJ, Rohani P, Keeling MJ. Appropriate models for the management of infectious diseases. PLOS Med. 2005;2:e174. doi: 10.1371/journal.pmed.0020174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arons MM, et al. Presymptomatic SARS-CoV-2 Infections and transmission in a skilled nursing facility. N. Engl. J. Med. 2020;382:2081–2090. doi: 10.1056/NEJMoa2008457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zou L, et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lawton, G. Can you catch the coronavirus twice? We don’t know yet. New Scientisthttps://institutions.newscientist.com/article/mg24532754-600-can-you-catch-the-coronavirus-twice-we-dont-know-yet/.
- 61.Rothe C, et al. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N. Engl. J. Med. 2020;382:970–971. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pan X, et al. Asymptomatic cases in a family cluster with SARS-CoV-2 infection. Lancet Infect. Dis. 2020;20:410–411. doi: 10.1016/S1473-3099(20)30114-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kimball, A. Asymptomatic and presymptomatic SARS-CoV-2 infections in residents of a long-term care skilled nursing facility—king County, Washington, March 2020. MMWR Morb. Mortal. Wkly. Rep.69, (2020). [DOI] [PMC free article] [PubMed]
- 64.Bai Y, et al. Presumed asymptomatic carrier transmission of COVID-19. JAMA. 2020 doi: 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wei, W. E. Presymptomatic Transmission of SARS-CoV-2—Singapore, January 23–March 16, 2020. MMWR Morb. Mortal. Wkly. Rep.69, (2020). [DOI] [PMC free article] [PubMed]
- 66.Jombart, T. et al. Inferring the number of COVID-19 cases from recently reported deaths. medRxiv (2020) doi:10.1101/2020.03.10.20033761. [DOI] [PMC free article] [PubMed]
- 67.Marjoram P, Molitor J, Plagnol V, Tavaré S. Markov chain Monte Carlo without likelihoods. Proc. Natl. Acad. Sci. USA. 2003;100:15324–15328. doi: 10.1073/pnas.0306899100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Perez-Reche, F. J., Forbes, K. J. & Strachan, N. J. C. Importance of untested infectious individuals for the suppression of COVID-19 epidemics. medRxiv0, 2020.04.13.20064022 (2020).
- 69.Comas-Herrera, A., Zalakain, J., Litwin, C., Hsu, A. T. & Fernandez-Plotka, J.-L. Mortality associated with COVID-19 outbreaks in care homes: early international evidence. https://ltccovid.org/2020/04/12/mortality-associated-with-covid-19-outbreaks-in-care-homes-early-international-evidence/amp/?__twitter_impression=true.
- 70.Robert Koch Institut. Coronavirus Disease 2019 (COVID-19) Daily Situation Report of the Robert Koch Institute - 11/04/2020. https://www.rki.de/DE/Content/InfAZ/N/Neuartiges_Coronavirus/Situationsberichte/2020-04-11-en.pdf?__blob=publicationFile.
- 71.Meyerowitz-Katz G, Merone L. A systematic review and meta-analysis of published research data on COVID-19 infection fatality rates. Int. J. Infect. Dis. 2020;101:138–148. doi: 10.1016/j.ijid.2020.09.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Linton, N. M. et al. Incubation period and other epidemiological characteristics of 2019 novel coronavirus infections with right truncation: A statistical analysis of publicly available case data. 10.1101/2020.01.26.20018754. [DOI] [PMC free article] [PubMed]
- 73.Instituto de Salud Carlos III. Informe sobre la situación de COVID-19 en España. Informe COVID-19 no 21. 6 de abril de 2020. https://www.isciii.es/QueHacemos/Servicios/VigilanciaSaludPublicaRENAVE/EnfermedadesTransmisibles/Documents/INFORMES/Informes%20COVID-19/Informe%20n%C2%BA%2021.%20Situaci%C3%B3n%20de%20COVID-19%20en%20Espa%C3%B1a%20a%206%20de%20abril%20de%202020.pdf (2020).
- 74.Istituto Superiore di Sanita. Integrated surveillance of COVID-19 in Italy. https://www.epicentro.iss.it/en/coronavirus/bollettino/Infografica_11aprile%20ENG.pdf (2020).
- 75.Kretzschmar ME, et al. Impact of delays on effectiveness of contact tracing strategies for COVID-19: A modelling study. Lancet Public Health. 2020;5:e452–e459. doi: 10.1016/S2468-2667(20)30157-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Coronavirus and self-isolation after testing positive in England - Office for National Statistics. https://www.ons.gov.uk/peoplepopulationandcommunity/healthandsocialcare/healthandwellbeing/bulletins/coronavirusandselfisolationaftertestingpositiveinengland/1februaryto13february2021.
- 77.Weekly statistics for NHS Test and Trace (England): 3 December to 9 December. GOV.UKhttps://www.gov.uk/government/publications/nhs-test-and-trace-england-statistics-3-december-to-9-december/weekly-statistics-for-nhs-test-and-trace-england-3-december-to-9-december.
- 78.Keeling MJ, Hollingsworth TD, Read JM. Efficacy of contact tracing for the containment of the 2019 novel coronavirus (COVID-19) J. Epidemiol. Commun. Health. 2020 doi: 10.1136/jech-2020-214051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hatchett RJ, Mecher CE, Lipsitch M. Public health interventions and epidemic intensity during the 1918 influenza pandemic. Proc. Natl. Acad. Sci. 2007;104:7582. doi: 10.1073/pnas.0610941104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.European Centre for Disease Prevention and Control. Rapid Risk Assessment: Coronavirus disease 2019 (COVID-19) in the EU/EEA and the UK – eleventh update: resurgence of cases. https://www.ecdc.europa.eu/en/publications-data/rapid-risk-assessment-coronavirus-disease-2019-covid-19-eueea-and-uk-eleventh (2020).
- 81.World Health Organization. WHO Director-General’s opening remarks at the media briefing on COVID-19. https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19—16-march-2020 (2020).
- 82.Bernal JL, et al. Effectiveness of the Pfizer-BioNTech and Oxford-AstraZeneca vaccines on covid-19 related symptoms, hospital admissions, and mortality in older adults in England: Test negative case-control study. BMJ. 2021;373:n1088. doi: 10.1136/bmj.n1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.CDC. Coronavirus Disease 2019 (COVID-19). Centers for Disease Control and Preventionhttps://www.cdc.gov/coronavirus/2019-ncov/science/science-briefs/fully-vaccinated-people.html (2020).
- 84.Moore S, Hill EM, Tildesley MJ, Dyson L, Keeling MJ. Vaccination and non-pharmaceutical interventions for COVID-19: A mathematical modelling study. Lancet Infect. Dis. 2021;21:793–802. doi: 10.1016/S1473-3099(21)00143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.World Health Organization. WHO Director-General’s opening remarks at the media briefing on COVID-19 - 3 March 2020. https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---3-march-2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used in this work are available from the cited sources. The models were analysed implemented in Mathematica. A notebook that retrieves the data and runs the calculations can be downloaded from https://doi.org/10.6084/m9.figshare.14636199. Point estimates of the parameters of model 1 and an R script to generate Fig. 3 can also be downloaded from this link.