Abstract
Global plant sulphur (S) deficiency is increasing because of a reduction in sulphate-based fertiliser application combined with continuous S withdrawal during harvest. Here, we applied 13C, 15N, 14C, and 35S quad labelling of the S-containing amino acids cysteine (Cys) and methionine (Met) to understand S cycling and microbial S transformations in the soil. The soil microorganisms absorbed the applied Cys and Met within minutes and released SO42− within hours. The SO42− was reutilised by the MB within days. The initial microbial utilisation and SO42− release were determined by amino acid structure. Met released 2.5-fold less SO42− than Cys. The microbial biomass retained comparatively more C and S from Met than Cys. The microorganisms decomposed Cys to pyruvate and H2S whereas they converted Met to α-ketobutyrate and S-CH3. The microbial stoichiometries of C, N, and S derived from Cys and Met were balanced after 4 d by Cys-derived SO42− uptake and Met-derived CO2 release. The microbial C:N:S ratio dynamics showed rapid C utilisation and loss, stable N levels, and S accumulation. Thus, short-term organic S utilisation by soil microorganisms is determined by amino acid structure whilst long-term organic S utilisation by soil microorganisms is determined by microbially controlled stoichiometry.
Subject terms: Biogeochemistry, Biogeochemistry
Introduction
Sulphur (S) is a vital plant macronutrient. It plays crucial roles in coenzyme A, chlorophyll, biotin, thiamine and glutathione (GSH) biosynthesis [1, 2]. Plant S deficiency has been reported worldwide and is attributed to reductions in atmospheric sulphur dioxide emissions over the last 30–40 years, low-S/high-N/high-P fertiliser application, a decrease in soil S return from manure or straw, and large soil S demand and removal by high-yield crops [1, 3]. Organic S accounts for >90% of the total soil S except in Gypsisols and other soils with gypsic qualifiers. Organic S must first be converted to inorganic SO42− before it can be absorbed by the roots [4]. Trace amounts of organic S mainly in the form of amino acids are directly absorbed by the roots [5]. Despite increasing incidences of plant S deficiency, belowground biogeochemical S cycling has not been fully elucidated. Hence, a strategy for predicting and controlling soil organic S mineralisation and recycling is urgently needed [6].
Cysteine (Cys) is the central metabolite coordinating S, C, and N assimilation in all photoautotrophic and chemoautotrophic organisms. It is the terminal metabolite in S assimilation and the starting point for the biosynthesis of methionine (Met), GSH, and other S metabolites [1]. Met is the precursor of microbially generated volatile organic S compounds such as methanethiol, dimethyl sulphide, and dimethyl disulphide. SO42− generation from Met is usually negligible [7]. It is energetically more efficient for microorganisms to use Met rather than Cys to build proteins because the former releases minimal SO42− [8]. Nevertheless, the microbial taxa utilising S-containing amino acids and their associated regulatory mechanisms have not been identified.
Much of the S in soil organic matter (SOM) exists in the form of the S-containing amino acids Cys and Met and originates from manure, animal residues, dead plants and microorganisms. Proteins are extracellularly decomposed by proteases into short peptides and individual amino acids. These substances are highly bioavailable N and S sources for plant roots and microorganisms [9, 10]. Microorganisms entirely remove Cys and Met from the soil solution and mineralise them within minutes to hours [11, 12]. Portions of the C, N, and S are immobilised as microbial biomass (MB) [5]. However, little is known about short-term (minutes to hours) C, N, and S cycling or the mechanisms by which microorganisms decompose small organic molecules.
Microorganisms absorb elements from the ambient environment and maintain relatively stable concentrations in their biomass. This phenomenon is known as stoichiometric homeostasis [13, 14]. Organic matter decomposition and nutrient cycling rates may be functions of stoichiometric imbalances between the substrates and the MB [15–17]. Element cycling comprises the restoration of C:N:S homeostasis by soil microbes in response to stoichiometric imbalances and the impact of stoichiometric homeostasis on organic matter decomposition [18, 19]. The crop residue C:N:S ratio is in the range of ~550:8–13:1–2. For bacteria and fungi, the C:N:S ratios are 38:9.5:1 and 105:11:1, respectively [20]. When the C:N or C:S ratios decline below certain thresholds, net N and S mineralisation occurs and supplementary N and S are released to maintain MB stoichiometry [21, 22]. Stoichiometric imbalances between substrates and decomposers strongly affect microbial physiology and microbe-mediated biogeochemistry [23].
The C, N, and P cycling processes are decoupled in response to N and P addition [24] and extreme temperatures [25] at the scales of landscapes, plant-soil ecosystems, soil profiles and microbial hotspots [26]. However, little is known about micro-scale decoupling mechanisms based on the presence of specific S-containing substrates such as amino acids. Certain amounts of Met and Cys are necessary for anabolic microbial biosynthesis. They are decomposed by mineralisation to SO42−. If Met and Cys are mainly immobilised in MB protein, then C, N, and S will be coupled. Otherwise, C-N-S decoupling occurs as there is an imbalance in microbial nutrient demand. Cys and Met constitute the bulk of low-molecular-weight organic S. Their C:N:S mass ratios are 38:9.5:1 and 105:11:1, respectively. Their large N and S proportions make them useful for investigations into C, N, and S utilisation during microbial mineralisation. They are also suitable for the examination of the regulatory mechanisms maintaining microbial C:N:S stoichiometry. It is necessary to clarify the nexus between organic S decomposition and soil element cycling because it will help explain the biochemistry, biology, and evolution of soil S cycling.
Farmyard manure (FYM) increases organic C content in soil and biological activity. In contrast, mineral fertiliser provides sufficient N, P, and S but no C [27, 28]. Manure input directly affects belowground biogeochemical processes by modifying organic C and adding nutrients. It also indirectly affects belowground biogeochemical processes by adjusting biotic activity [27, 29]. Topsoil and subsoil differ in terms of nutrient and element content, MB and community structure, soil C age, bioavailability, accessibility, and SOM decomposition rates [30]. Soils receiving long-term FYM or mineral fertiliser treatment were selected to elucidate the mechanisms of organic S decomposition. All soils were the same type and were exposed to the same climate conditions. However, they substantially differed in terms of organic matter content and MB. Our objectives were to (1) identify the key processes involved in soil Cys and Met decomposition, (2) determine whether Cys and Met decomposition couples or decouples C, N, and S utilisation and induces stoichiometrically regulated decomposition, (3) establish whether Cys and Met are rapidly decomposed in the soil solution in the presence of few microorganisms, and (4) verify whether long-term manure application affects this process. We hypothesised that microorganisms balance their C:N:S ratios by releasing NH4+ and SO42− after Cys and Met uptake. In this manner, the external C and S content in the soil could affect this release. To test this hypothesis, we subjected Cys and Met to quad labelling (13C, 15N, 14C, and 35S).
Materials and methods
Experimental site and treatments
Soil classified as udipsamment (US Soil Taxonomy) or brown sand with a sandy-loam texture was collected from selected treatments in a long-term organic manuring experiment at Woburn Experimental Farm, Bedfordshire, UK, on 1 June 2018. The experiment was initiated in 1964 to evaluate the influences of mineral fertilisers and organic manures on crop production and soil fertility. Soils were collected from the following treatments: (1) mineral fertilisers only (N, P, and K) equivalent to FYM at 25–50 t ha−1 y−1 (NM, 28 y), (2) FYM applied at 10 t ha−1 y−1 for 16 y (MM), and (3) FYM applied at 25–50 t ha−1 y−1 for 28 y (HM). All treatments were performed in quadruplicate. All plots were under a five-course rotation of winter rye, spring barley, winter bean, winter wheat, and forage maize since 2003. NM and MM received mineral P, K, and S fertilisers at 20, 83, and 36 kg ha−1, respectively. For NM, the total N, P, and S (organic and inorganic) inputs during 1964–2018 were 2.46, 1.77, and 0.96 t ha−1, respectively. For HM, the total C, N, P, and S inputs were 112.5, 5.80, 1.26, and 1.22 t ha−1, respectively. For MM, the total C, N, P, and S inputs were 14.1, 2.63, 1.69, and 1.00 t ha−1, respectively. Detailed information about the treatments was previously reported [31, 32]. Topsoil (0–23 cm plough layer) and subsoil (23–38 cm depth) samples were collected in quadruplicate, passed through a 5 mm mesh sieve to remove stones, roots, and other organic matter, and stored at 4 °C until the subsequent analyses. Soil physicochemical properties are listed in Table S1.
Plant-derived protein decomposition
To obtain 35S-labelled protein, maize plants were hydroponically cultivated with Na235SO4 and their roots and shoots were harvested after 30 d. Tissues were cut into small pieces, frozen at −80 °C, and pulverised at a 1:2 tissue:water ratio. To avoid interference from protein decomposition, no extraction solution (such as Tris-HCl) was used. Tissue suspensions were centrifuged at 4000 × g and 20 °C for 15 min. The supernatants were recovered and precooled (−20 °C) acetone was added to form a final 1:0.5 supernatant:acetone ratio. The mixtures were incubated at 4 °C for 12 h and centrifuged at 10,000 × g and 4 °C for 15 min. The sediments were washed several times with acetone and the residual acetone was removed by vacuuming for 15 min. The sediments comprised the extracted plant protein. The S content was determined by inductively coupled plasma-optical emission spectrometry (ICP-OES; Varian 710 ES, Agilent Technologies, Santa Clara, CA, USA).
To measure 35S-protein mineralisation at 20 °C, 0.5 mL of 50 µM S-protein (6 kBq mL−l) was dropped onto the surfaces of each 5 g field-moist soil sample collected from each NM, MM, and HM plot at 0–23 and 23–38 cm depth. Each treatment was prepared in quadruplicate. Each sample consisted of six analytical units so that measurements could be made at 0.25, 1, 2, 4, 6, and 12 d after protein addition. At each time point, the 35S in one analytical unit per replicate was extracted with 25 mL of 0.01 M CaCl2. The difference between the amount of 35S in the total protein and the CaCl2-extracted 35S from the soil samples at each time point was taken as the quantity of 35S immobilised in the MB (35SMB = 35Stotal − 35SCaCl2) because sandy soil has a relatively lower absorption rate. For the CaCl2 extractions, 0.5 mL of purified water or 1 M BaCl2 was added to every millilitre of extract to precipitate the 35SO42−. The suspensions were then centrifuged at 18,000 × g and 20 °C for 5 min. Differences in 35S activity between the water and BaCl2 treatments corresponded to the quantities of 35SO42− derived from labelled protein (35SSO42- = 35SCaCl2+H2O − 35SCaCl2+BaCl2). Total 35S in the soil extract less the 35S-SO42− was assumed to be the amount of 35S-protein remaining in the soil solution (35Sleft = 35SCaCl2 − 35SSO42-) [5].
S-containing amino acid decomposition
To trace temporal C, N, and S transformation in the various soil treatments, intact Cys or Met remaining in the soil solution, CO2 release, C, N, and S assimilated into the MB, and NH4+, NO3−, and SO42− generated were detected with 14C-, 35S-, or 15N-labelled Cys or Met. A 13C-PLFA (phospholipid-derived fatty acid) analysis was conducted to identify the microbial community assimilating Cys and Met.
To trace C and S transformation, 0.5 mL of 50 µM 14C- or 35S-labelled Cys (14C: 20.6 kBq mL−l; 35S: 50.4 kBq mL−l) or Met (14C: 21.8 kBq mL−l; 35S: 51.8 kBq mL−l) was added to each 5 g field-moist soil sample from each NM, MM, and HM plot at 0–23 cm and 23–38 cm depth. Forty parallel 5 g soil samples were prepared for NM, MM, and HM. There were four replicates and ten time points. The 14CO2 released from the soil was captured in a trap containing 1 mL of 1 M NaOH set above the soil [5, 12]. The 14C or 35S remaining in the soil was extracted with 25 mL of 0.5 M K2SO4 or 0.01 M CaCl2 at 2 min, 5 min, 15 min, 0.5 h, 1 h, 3 h, 9 h, 24 h, 48 h, and 96 h after labelled Cys or Met was added to the soil samples [33]. Incubation times <3 h were considered short-term (h) whereas incubation times in the range of 24–96 h were regarded as medium-term (d). The 35S-SO42− was extracted with 0.01 M CaCl2 and detected as previously described. The difference in 35S activity corresponded to the quantity of SO42− generated from labelled Cys and Met (35SSO42- = 35SCaCl2+H2O − 35SCaCl2+BaCl2). The difference between the total 35S added and the 35S-SO42− in the soil solution extracted with 0.01 M CaCl2 was taken as the residual soil 35S-Cys or 35S-Met (35Sleft = 35SCaCl2 − 35SSO42-). The difference between the total 35S and the 35S extracted from the soil solution was taken as the amount of 35S immobilised in the MB (35SMB = 35Stotal - 35SCaCl2) [5]. Total 14C activity minus the residual soil 14C plus the quantity of 14CO2 released was taken as the amount of 14C immobilised in the MB (14CMB = 14Ctotal − 14CK2SO4 − 14CCO2) [34]. Each sample was mixed with 4 mL Scintisafe 3 scintillation cocktail (Fisher Scientific, Loughborough, UK) [34] and 14C or 35S activity was determined with a Wallac 1404 liquid scintillation counter fitted with automated quench correction (Wallac EG&G, Milton Keynes, UK).
N transport from Cys and Met was detected by 15N labelling. In brief, 2 mL of 50 µM 99.8% atom 13C,15N dual-labelled Cys and Met was added to each 20 g field-moist sample collected from NM, MM, and HM at 0–23 and 23–38 cm depth. Each soil sample was extracted with 80 mL of K2SO4 after 0.25, 1, 9, 24, and 96 h and centrifuged at 18,000 × g and 20 °C for 5 min. NO3− and NH4+ were evaluated by microdetection [35]. The NH4+ and NO3− 15N isotopic ratios in the soil extracts were detected by the diffusion method [36]. The soil solutions were freeze-dried (Labconco, Kansas City, MO, USA) and their 15N abundances were detected with an elemental analysis-isotope ratio mass spectrometer (IsoPrime100; Isoprime Ltd., Cheadle Hulme, UK). The differences between the total 15N and the 15N extracted from the soil solutions were taken as the amounts of 15N immobilised in the MB (15NMB = 15Ntotal − 15NK2SO4).
Three hours after 13C,15N dual-labelled Cys or Met was added to the soil samples and the PLFAs were extracted, fractionated, and purified as previously described [27]. After 3 h, the C from Cys and Met was partly immobilised into PLFA. The δ13C of each PLFA was determined by gas chromatography combustion isotope ratio mass spectrometry (GC-C-IRMS; Thermo Fisher Scientific, Waltham, MA, USA). Cyclopropyl and monounsaturated fatty acids were indicators of Gram-negative (G−) bacteria. The 16:1 w7c, 17:1 w8c, 17:0 cyclo w7c, 18:2 w6c, 18:1 w9c, 18:1 w7c, 18:1 w5c, and 19:0 cyclo w7c show the 13C-labelled indicators. Iso- and anteiso-branched chain fatty acids (14:0 iso, 15:0 iso, 15:0 anteiso, 16:0 iso, 17:0 iso, and 17:0 anteiso) were indicators of Gram-positive (G+) bacteria. The 10Me-branched PLFAs (16:0 10-methyl, 17:0 10-methyl, 18:1 w7c 10-methyl, and 18:0 10-methyl) were used as actinobacterial biomarkers. The saturated straight-chain fatty acids (15:0, 16:0, and 18:0) occurring in a wide variety of microorganisms were used as universal PLFAs. The 15:0 DMA was an anaerobe indicator. The 18:1 w9c was an indicator of G− bacteria in agricultural soils and fungi in forest soils [27] and served here as a G− indicator.
Organic S decomposition in the soil solution
To assess organic S decomposition in the soil solution, 5 g fresh soil sample collected from NM, MM, and HM at 0–23 cm and 23–38 cm depth was extracted with 20 mL purified sterile water, oscillated at 180 × g for 5 min, and centrifuged at 18,000 × g and 20 °C for 5 min. The supernatants were removed and the soil samples were extracted twice with 10 mL purified sterile water each time. The supernatants were combined and centrifuged at 18,000 × g and 20 °C for 5 min. Then 0.5 mL of 50 µM 14C- or 35S-labelled Cys or Met was added to each supernatant and 0.5 mL aliquots were collected at 0.25, 0.5, 1, 3, 9, 24, 48, and 96 h. The 14C or 35S remaining in the solution and the 35SO42− generated (35SSO42- = 35SH2O − 35SH2O+BaCl2) were measured as previously described. After centrifugation at 18,000 × g, 0.1–0.2% microbial DNA remained in the soil solution. The MB was indicated by the DNA content in the soil samples and solutions and extracted with a FastDNA SPIN kit (MP Biomedicals, Irvine, CA, USA) according to the manufacturer’s instructions. DNA concentrations were determined in a NanoDrop ND-1000 UV-Vis spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA).
Determination of microbial C or S requirement from Cys and Met decomposition
To estimate S-containing amino acid mineralisation after the C, N, and S amendments, 5 g field-moist soil was placed in a 50 mL centrifuge tube and 0.5 mL of 50 µM 14C- or 35S-labelled Cys or Met was added along with supplementary C, N, or S. Only the HM topsoil was used because the Cys and Met decomposition rates were similar for all treatments and independent of soil depth or manure application rate. The soil samples were treated with 2 mg g−1 C as glucose, 0.1 mg g−1 N as NH4NO3, and 0.1 mg g−1 S as Na2SO4 dissolved in 50 µM 14C- or 35S-labelled Cys or Met [37]. The controls were 50 µM 14C- or 35S-labelled Cys or Met alone. The initial soil C:N:S ratio was 13.1:2.2:1.0. After C, N, and S addition, the ratios were 57.2:2.2:1.0, 13.1:4.4:1.0, and 4.1:0.7:1.0, respectively. The amounts of 14CO2, 14C-MB, 35S-MB, 35S-SO42−, 14C- and 35S-Cys, and 14C- and 35S-Met in the solutions were measured at 0.5 h, 1 h, 3 h, 6 h, 9 h, 24 h, 48 h, and 96 h as previously described. After amendment, MB carbon (MB-C) was detected by the fumigation method [38].
Statistical and data analysis
Substrate mineralisation was generally biphasic and described by a two-process, double first-order kinetic decay model [39]:
| 1 |
where f is the 14C-Cys or Met remaining in the soil (% of total addition); a1 and a2 are the quantities of 14C partitioned into primary (pool 1; catabolic process; microbial release as 14CO2) and slower secondary (pool 2; biomass production) mineralisation, respectively (%); k1 and k2 are the exponential coefficients of pools 1 and 2, respectively; and t is the time (h).
The half-life (t½, h) of pool 1 or pool 2 was calculated as:
| 2 |
One-way ANOVA followed by Tukey’s post-hoc test at P < 0.05 were performed in SAS v. 8.2 (SAS Institute Inc., Cary, NC, USA) after testing the ANOVA residuals for normality and homogeneity. Exponential decay equations were calculated with SigmaPlot v. 10.0 (SPSS Inc., Chicago, IL, USA). The 13C-labelled PLFA indicator data were evaluated by principal component analysis with CANOCO v. 5.0 (Microcomputer Power, Ithaca, NY, USA) to determine the influences of manure application and the soil active microbial communities. Images were plotted with Origin v. 8.1 (OriginLab, Northampton, MA, USA).
Results
Decomposition of S-containing amino acids and protein
Cys and Met decomposed rapidly in the soil (t½ < 1 min; Figs. 1 and 2) and were quickly immobilised into the MB (35S-MB: 31% for Cys and 67% for Met at 2 min). 3 h after microbial uptake, 37% of the Cys and 15% of the Met were released as SO42−. The released SO42− was reincorporated into the MB. The 35S-MB activity levels from Cys and Met were 61% and 70%, respectively. Within 15 min, Cys and Met released 28% and 34% NH4+, respectively. NH4+ production decreased over time and some of the NH4+ was oxidised to NO3−. The protein rapidly decomposed (t½ < 8 min) at low concentrations whereas nearly half the S-protein was transformed to SO42− within 48 h (Fig. 2).
Fig. 1. Transformation of C, N, and S from cysteine and methionine in soil.
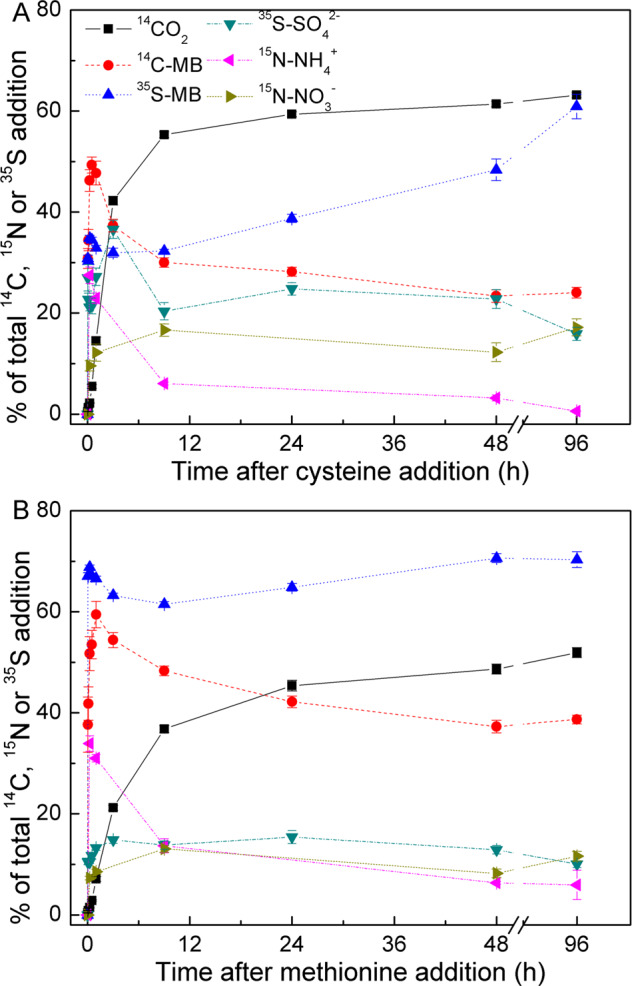
CO2 release, C and S assimilation into microbial biomass (MB), and NH4+, NO3−, and SO42− production were detected with 14C-, 35S-, or 15N-labelled cysteine (A) and methionine (B) at the indicated times after amino acid addition. There were three treatments and two soil depths (n = 6). Means ± SE for four replicates per treatment per soil depth are shown.
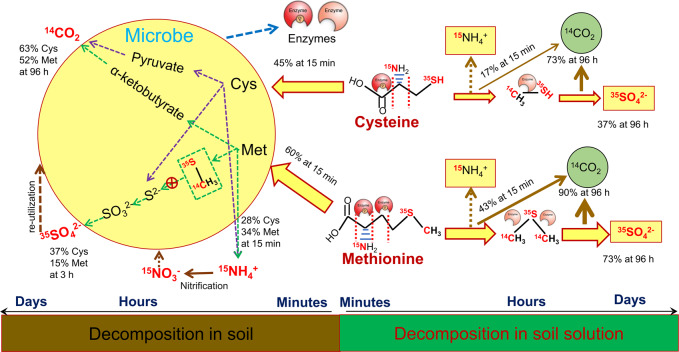
Fig. 2. Simplified cycling mechanisms of sulphur in proteins (Prot), cysteine (Cys), and methionine (Met) in soil.
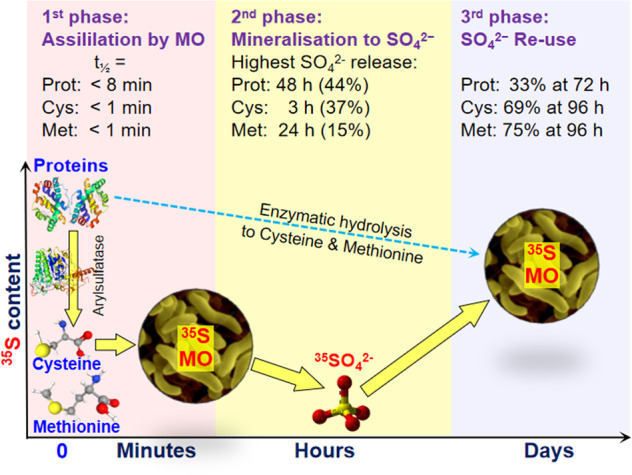
Intact Cys and Met are almost entirely assimilated within minutes by microorganisms (MO) that can also utilise protein-S. Some of the S immobilised in MO is released as SO42−, which is then reutilised by MO for growth. Proteins were decomposed to Cys and Met by arylsulfatase, and then utilized by MO.
After immobilisation in the MB, microbial C, N, and S stoichiometry differed between Cys and Met for ≤9 h after addition (P < 0.05). In contrast, they were similar after 96 h. In the short term, CO2 was released more rapidly from Cys than Met. As incubation progressed, however, the total amount of C released from Met in the form of CO2 was similar to that released from Cys. In the short term, more SO42−-S was released from Cys than Met. Hence, relatively little S was retained in the MB. Nevertheless, the SO42− released was reutilised and the 35S-MB content was similar for both Cys and Met at 96 h (Fig. 3). Microbial communities utilising Cys and Met were indistinguishable according to the 13C-PLFA analysis (Figs. S1 and S2). However, there was threefold more 13C-labelled PLFA derived from Met than Cys (Fig. S1).
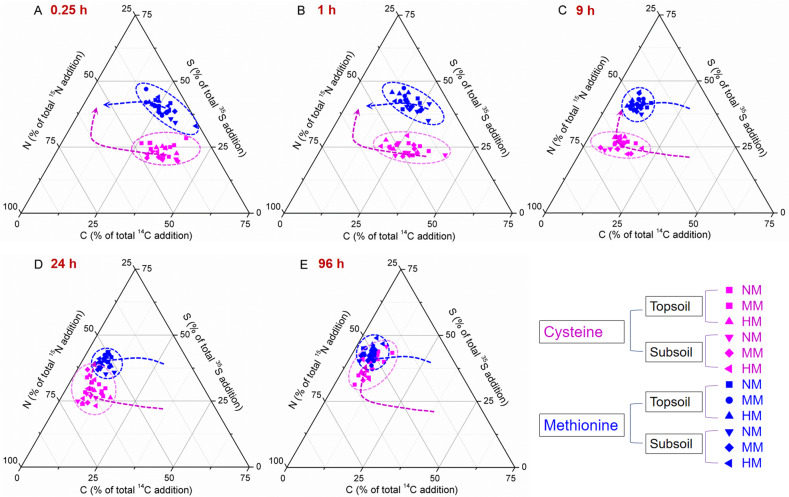
Fig. 3. Response of soil microbial C, N, and S stoichiometry to cysteine (Cys) and methionine (Met) addition.
The microbial C, N, and S stoichiometry (% of total addition to microbial biomass) after Cys and Met addition for 0.25 h (A), 1 h (B), 9 h (C), 24 h (D), and 96 h (E). NM, MM, and HM are zero, medium, and high application rates of farmyard manure (FYM), respectively. Data are normalised for C + N + S = 100%. Blue and pink arrows show convergence of the Cys and Met microbial transformation pathways in the soil. Despite their initial differences, the C:N:S stoichiometries of Cys and Met converged after 96 h.
Organic S decomposition in the soil solution
Cys and Met were rapidly mineralised in the soil solutions. There were lower microbial densities in the extracted soil solutions than the soil itself. Thus, 17.3% of the 14C-Cys and 43.3% of the 14C-Met decomposed after 15 min (Fig. 4). For 35S, 5.5% of the Cys and 16.0% of the Met were transformed into SO42−. These rates were substantially lower than those for 14C-Cys, 14C-Met, 35S-Cys, and 35S-Met decomposition in the soil (Fig. S3). The Cys and Met decomposition rates in the soil solution were high at ≤15 min but markedly lower between 15 min and 24 h after substrate addition. The Cys and Met decomposition rates were comparatively faster in the topsoil and at high manure application rates (Fig. S3).
Fig. 4. Transformation of cysteine (Cys) and methionine (Met) 14C and 35S in the soil solution.
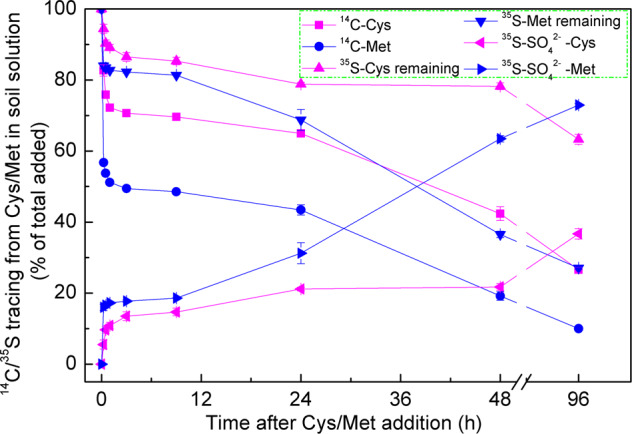
14C or 35S remaining in the soil solution and 35SO42− produced from Cys and Met are shown. There were three treatments and two soil depths (n = 6). Means ± SE for four replicates per treatment per soil depth are shown.
Requirement of microbial C or S derived from Cys and Met decomposition
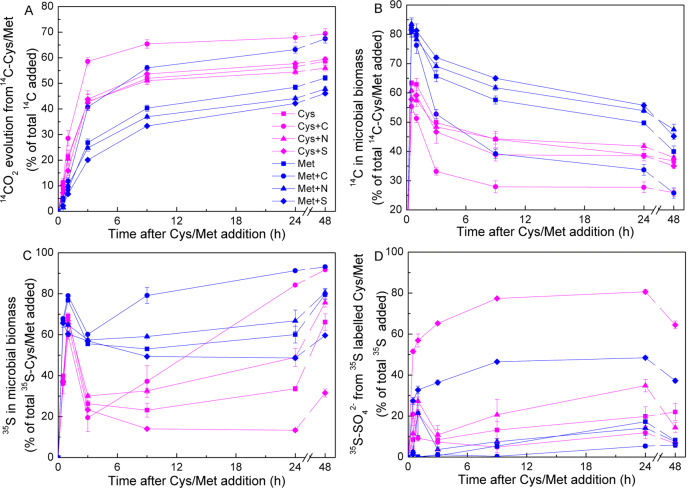
The addition of C and S dramatically influenced organic S decomposition but had a limited effect on C and S uptake in the MB (Fig. 5). C addition increased 14CO2 release from Cys and Met (Cys: from 52% to 67%; Met: from 59% to 69%; 48 h after addition) but simultaneously decreased the 14C content in the MB. N and S addition had minimal impact on C metabolism or 35S uptake in the MB. S addition increased SO42− release from Cys and Met by 193% and 363%, respectively.
Fig. 5. Effects of C, N, and S addition on 14C- or 35S-labelled cysteine (Cys) and methionine (Met) decomposition as functions of incubation time.
Soil samples were treated with 2 mg g−1 C as glucose, 0.1 mg g−1 N as NH4NO3, and 0.1 mg g−1 S as Na2SO4. 14CO2 evolution (A), 14C in microbial biomass (B), 35S in microbial biomass (C), and 35SO42− generated from labelled Cys and Met (D) are shown. Data are means ± SE; n = 4. Topsoil subjected to high FYM application rate was used.
Discussion
S-containing protein and amino acid decomposition
The half-lives of plant-derived 35S-protein and amino acid decomposition in the soil were <8 min and <1 min, respectively. Therefore, rapid decomposition and high bioavailability of proteins and amino acids are vital to organic S mineralisation [40, 41].
Protein and amino acid mineralisation and SO42− release and reutilisation occur in three steps (Fig. 2). (1) Proteins and amino acids are rapidly absorbed by the MB. Proteins are hydrolysed and assimilated into the MB in the form of amino acids and peptides. Here, 34.8% of the 35S-Cys and 68.8% of the 35S-Met were retained in the MB whereas 20% of the S-protein was retained in the MB after 15 min (Fig. 2). (2) S in the MB is released in the form of SO42−. The highest S release rates were observed 2 d after protein addition (50%), 3 h after Cys addition (37%), and 24 h after Met addition (15%). Soil N cycling is regulated by C:N imbalances between the substrates and the MB [22]. The results of the present study suggest that microbial SO42− release after the uptake of substrates with high S content and disequilibrium between substrates and microorganisms play critical roles in soil S cycling. The intensity of SO42− release might depend on substrate properties because a relatively small amount of S was released from Met. (3) The released SO42− is resorbed and reutilised by microorganisms. S-MB reached its highest concentration 4 d after protein addition. The 35S-MB content derived from Cys and Met was highest at the end of incubation (Figs. 1 and 2). Thus, SO42− is highly bioavailable to microorganisms.
When C demand drives microbial Cys and Met decomposition, readily accessible C sources such as glucose decrease 14C uptake and 14CO2 release from these amino acids. Reduced 14C uptake has been demonstrated by applying organic N (L-trialanine) and P (glucose-6-phosphate) after glucose addition. Therefore, C demand drives organic N and P decomposition [37, 42]. However, high C or S addition levels had negligible effects on the uptake of 14C derived from Cys or Met (Fig. 5). Hence, microorganisms may rapidly acquire these amino acids regardless of soil element status. In addition, S addition did not markedly alter Cys or Met C mineralisation. Therefore, the soil S supply met microbial demand [43]. At 0.5 h, S addition increased SO42− release from Cys and Met by 6- and 12-fold, respectively, compared with no S addition. High S addition reduced microbial S demand and most of the S was released in the form of SO4. C addition enhanced SO42− reutilisation possibly by increasing microbial growth and S demand (Fig. S4) [44]. Relatively large quantities of 35SO42− were released from Cys and Met in response to S addition because high amounts of unlabelled Na2SO4 diminished microbial S demand. N supply had no apparent effect on any of the foregoing processes possibly because the existing soil N stocks were adequate. The microbial Cys and Met uptake rates were not influenced by C or S status but were substantially affected by C and S metabolism and SO42− reutilisation. Therefore, S-containing amino acids are rapidly decomposed in the soil and SO42− release is affected by inorganic S bioavailability.
Mechanisms of C, N, and S decoupling from Cys and Met via decomposition and stoichiometric balance
Decoupling of decomposition was assessed for C, N, and S derived from Cys and Met (Fig. 3). Stoichiometric theory dictates that Met should release more CO2 and less SO42− than Cys because Met has a relatively higher C:S ratio [45]. Soil microorganisms efficiently utilised the C and S originating from Met. We observed that 46% and 52% of the 14C and 35% and 69% of the 35S were retained in the MB whilst Cys and Met released 21% and 10% of their 35SO42−, respectively, after 15 min. Thus, Met slowly released SO42− rather than rapidly releasing CO2. Moreover, Cys and Met were comparable as N sources because 38% and 40% of the inorganic N (NH4+ and NO3−) were produced by Cys and Met, respectively. In contrast, 40% of the N derived from alanine was utilised by soil microorganisms and converted to NH4+ within 5 min [12]. Similar values were obtained for Cys and Met.
According to C, N, and S stoichiometry, microorganisms should have higher N than S demand and release a higher proportion of SO42− than NH4+ after Cys and Met uptake (Fig. S5). However, our results did not corroborate these expectations. Organic S decomposition partially upheld the stoichiometric theory based on the SO42− and NH4+ production levels. At <9 h after Cys and Met addition, however, the relative N and S quantities actually released did not match the expected stoichiometry. Therefore, this model is not perfectly suited for the short-term decomposition of certain substrates. SOM quality is more influential to microbial element utilisation efficiency than stoichiometry [45].
Soil microorganisms use specific mechanisms to maintain stoichiometric balance after the uptake of various substrates. Following immobilisation in the MB, the C, N, and S from Cys and Met differed in terms of microbial stoichiometry at <9 h after addition because of variations in microbial C and S use efficiency (P < 0.05). At 0.25 h, the C:N:S ratio derived from Cys was 52:63:35 whereas that derived from Met was 47:59:69. At 9 h, the C:N:S ratio derived from Cys was 15:77:32 whereas that derived from Met was 15:73:62. These discrepancies resulted in short-term differences in CO2 and SO42− production. Nevertheless, there were no stoichiometric differences at 96 h. By that time, the C:N:S ratio derived from Cys was 13:82:61 whereas that derived from Met was 9:82:70. Cys initially released CO2 more rapidly than Met but as the incubation progressed, the cumulative CO2-C release rates were similar for both Met and Cys. In the short term, more SO42−-S was released from Cys than Met. Consequently, the S retention ratio in the MB was very low. The released SO42− was reutilised and the 35S-MB content was similar for both Cys and Met at 96 h. Microorganisms balance stoichiometry via their elemental demands [46]. This mechanism explains substrate-level elemental imbalances in soils and the MB. The relative elemental content of the MB follows a homeostatic regulatory curve with C, N, and S substrate content. Thus, microorganisms have a unifying element assimilation mechanism [13, 14, 47].
Amino acid decomposition mechanisms in soil and soil solution
The microbial communities and metabolic pathways involved might explain the relative differences in the Cys and Met decomposition rates. Shifts in microbial element stoichiometry are associated with changes in the soil microbial community. Modifications to agricultural management practices are responsible for the latter [23, 48, 49]. Microbial community dynamics can alleviate stoichiometric constraints and cause the system to behave in a manner that cannot be predicted by stoichiometric theory alone [50]. However, our 13C-PLFA analysis disclosed no such differences between the microbial communities utilising Cys and those assimilating Met (Fig. S1). Nevertheless, about threefold more Met-C than Cys-C was immobilised in the PLFA. The 13C-PLFA content was consistent with the 14C-MB and indicated that much of the Met-C was integrated into the MB. Therefore, Cys and Met metabolism rather than Cys and Met assimilation by various microbial communities influences microbial utilisation of the elements derived from them.
When a substrate is assimilated into microbial cells, its original molecular structure determines the biomolecules that will be derived from it [51, 52]. Substrate quality is the main factor controlling C mineralisation and assimilation into the MB [51, 53]. The glucose C use efficiency is nearly 80% whereas that for oxalic acid is only 20%. An earlier continuous sampling study showed that mineralisation is key for Cys but not Met [54]. Only a small quantity of SO42− was released even at relatively high Met uptake rates. The mechanisms of Cys and Met biosynthesis from 3-phosphoglycerate or pyruvate/Cys/oxaloacetate in E. coli consume 24.7 and 34.3 high-energy phosphate bonds, respectively [55]. Met decomposition releases more energy than Cys decomposition. The former induces microbial C and S assimilation. The reduced sulphur moiety (thiol, –SH) in Cys is strongly nucleophilic. Therefore, Cys is readily transformed to SO42− [56]. Our data suggest that L-Cys desulphydrase decomposed Cys to pyruvate, NH4+, and H2S, the pyruvate was decomposed to CO2, and the H2S was oxidised to SO42− [57]. The Met was decomposed to α-ketobutyrate, methanethiol (HS-CH3), and NH4+ [57] and might have been metabolised to β-thiomethylpropylamine, S-adenosylmethionine, or α-keto-γ-thiomethylbutyrate [56, 58]. However, the activity levels of the latter processes must have been low as Met released high NH4+ levels. Nevertheless, this conclusion was based solely on our current analyses. Therefore, position-specific isotopic labelling combined with specific metabolite detection is required [52]. This analysis would furnish validation data for Cys and Met transformation during microbial metabolism and substrate-controlled morphological changes in soil element cycling.
There was relatively greater SO42− release from Cys than Met in the soil (Fig. 6). Hence, microorganisms trapped within aggregates and attached to mineral surfaces prevail during decomposition. In the soil solution, Cys decomposition released SO42− more slowly than Met decomposition. The higher C content in Met might promote relatively greater enzyme activity, C cycling, and microbial growth in the soil solution. Thus, Met decomposition releases comparatively larger amounts of CO2-C and is, by extension, associated with higher SO42− release rates (Fig. 6). Rapid decomposition in response to high manure application rates may have been stimulated by increases in MB and enzyme activity and a decrease in S content [59]. Therefore, S-containing amino acids may be rapidly decomposed both in the soil and in the soil solution. In general, microorganisms are highly effective at Met uptake and metabolism.
Fig. 6. Mechanism of cysteine (Cys) and methionine (Met) decomposition in soil and soil solution.
Cys and Met are rapidly decomposed in the soil and soil solution but the underlying metabolic processes differ between them. In the soil, Cys and Met were quickly immobilised into microbial biomass (MB). More C was released as CO2 and more S was released as SO42− from Cys than Met. At 3 h after microbial uptake, 37% of the Cys but only 15% of the Met was decomposed to SO42−, which was then absorbed into the MB. 35S-MB derived from Cys and Met were 61% and 70%, respectively. Within 15 min, Cys and Met released 28% and 34% NH4+, respectively. NH4+ production decreased with time and some of it was oxidised to NO3−. The reduced sulphur moiety (thiol, –SH) in Cys was strongly nucleophilic. Hence, Cys was easily transformed to SO42−. L-Cysteine desulphydrase decomposed Cys to pyruvate, NH4+, and H2S, pyruvate was decomposed to CO2, and H2S was oxidised to SO42−. Met was decomposed to α-ketobutyrate, methanethiol (HS-CH3), and NH4+. Met decomposition released relatively more CO2-C and, by extension, more SO42− in the soil solution than Cys.
Conclusions
Sulphur (S) cycling involves Cys and Met uptake by microorganisms from the soil solution within minutes, intracellular mineralisation of these amino acids, SO42− release within hours, and SO42− reuptake and reutilisation within days. Cys releases more CO2 and SO42− than Met because of the differences in the metabolic pathways between these amino acids rather than the compositions of the microbial communities utilising them. The molecular structures of Cys and Met determine their short-term utilisation. Cys initially decomposes to pyruvate whereas Met is initially converted to α-ketobutyrate and -S-CH3. These differences account for the divergence in microbial stoichiometry between Cys and Met utilisation. As C is continuously lost by respiration, the microbial C:N:S ratios derived from Cys and Met based on 13C/14C, 15N, and 35S were 52:63:35 and 47:59:69, respectively, after 0.25 h, but had changed to 13:82:61 and 9:82:70, respectively, after 4 d. Hence, short-term organic S utilisation by soil microorganisms is determined by amino acid structure whilst medium-term organic S utilisation by soil microorganisms is determined by microbially controlled stoichiometry.
Supplementary information
Acknowledgements
This work was supported by the National Key Research and Development Program of China (SQ2020YFD10006-02), the National Natural Science Foundation of China (Nos. 31872180 and 31801936), the UK-China Virtual Joint Centre for Agricultural Nitrogen (CINAg. No. BB/N013468/1), and the Newton Fund via UK BBSRC, NERC, and the Chinese Ministry of Science and Technology. The Rothamsted Long-Term Experiments National Capability (LTE-NCG) was supported by the UK Biotechnology and Biological Science Research Council (No. BBS/E/C/000J0300) and the Lawes Agricultural Trust. The authors thank the curators of the Electronic Rothamsted Archive (e-RA) for access to data from the Rothamsted Long-Term Experiments. The authors also thank Mr Steve Freeman for his assistance with sample collection. YK recognises the Government Program of Competitive Growth of Kazan Federal University and the “RUDN University Program 5-100” for their support.
Compliance with ethical standards
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-021-00999-7.
References
- 1.Dong Y, Silbermann M, Speiser A, Forieri I, Linster E, Poschet G, et al. Sulfur availability regulates plant growth via glucose-TOR signaling. Nat Commun. 2017;8:1174. doi: 10.1038/s41467-017-01224-w. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 2.Freney JR, Melville GE, Williams CH. Soil organic matter fractions as sources of plant-available sulphur. Soil Biol Biochem. 1975;7:217–21. doi: 10.1016/0038-0717(75)90041-3. [DOI] [Google Scholar]
- 3.Kopittke PM, Dalal RC, Finn D, Menzies NW. Global changes in soil stocks of carbon, nitrogen, phosphorus, and sulphur as influenced by long‐term agricultural production. Global Change Biol. 2017;23:2509–19. doi: 10.1111/gcb.13513. [DOI] [PubMed] [Google Scholar]
- 4.Ciaffi M, Paolacci AR, Celletti S, Catarcione G, Kopriva S, Astolfi S. Transcriptional and physiological changes in the S assimilation pathway due to single or combined S and Fe deprivation in durum wheat (Triticum durum L.) seedlings. J Exp Bot. 2013;64:1663. doi: 10.1093/jxb/ert027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma Q, Luo Y, Wen Y, Hill PW, Chadwick DR, Wu L, et al. Carbon and sulphur tracing from soil organic sulphur in plants and soil microorganisms. Soil Biol Biochem. 2020;150:107971. doi: 10.1016/j.soilbio.2020.107971. [DOI] [Google Scholar]
- 6.Piotrowska-Długosz A, Siwik-Ziomek A, Długosz J, Gozdowski D. Spatio-temporal variability of soil sulfur content and arylsulfatase activity at a conventionally managed arable field. Geoderma. 2017;295:107–18. doi: 10.1016/j.geoderma.2017.02.009. [DOI] [Google Scholar]
- 7.Fitzgerald JW, Watwood ME. Amino-acid metabolism in forest soil—Isolation and turnover of organic matter covalently labelled with 35S-methionine. Soil Biol Biochem. 1988;20:833–8. doi: 10.1016/0038-0717(88)90089-2. [DOI] [Google Scholar]
- 8.Ma Q, Wen Y, Pan W, Macdonald A, Hill PW, Chadwick DR, et al. Soil carbon, nitrogen, and sulphur status affects the metabolism of organic S but not its uptake by microorganisms. Soil Biol Biochem. 2020;149:107943. doi: 10.1016/j.soilbio.2020.107943. [DOI] [Google Scholar]
- 9.Jan MT, Roberts P, Tonheim SK, Jones DL. Protein breakdown represents a major bottleneck in nitrogen cycling in grassland soils. Soil Biol Biochem. 2009;41:2272–82. doi: 10.1016/j.soilbio.2009.08.013. [DOI] [Google Scholar]
- 10.Farrell M, Macdonald LM, Hill PW, Wanniarachchi SD, Farrar J, Bardgett RD, et al. Amino acid dynamics across a grassland altitudinal gradient. Soil Biol Biochem. 2014;76:179–82. doi: 10.1016/j.soilbio.2014.05.015. [DOI] [Google Scholar]
- 11.Wilkinson A, Hill PW, Farrar JF, Jones DL, Bardgett RD. Rapid microbial uptake and mineralization of amino acids and peptides along a grassland productivity gradient. Soil Biol Biochem. 2014;72:75–83. doi: 10.1016/j.soilbio.2014.01.026. [DOI] [Google Scholar]
- 12.Hill PW, Jones DL. Plant–microbe competition: does injection of isotopes of C and N into the rhizosphere effectively characterise plant use of soil N? N Phytol. 2018;221:796–806. doi: 10.1111/nph.15433. [DOI] [PubMed] [Google Scholar]
- 13.Godwin CM, Cotner JB. Aquatic heterotrophic bacteria have highly flexible phosphorus content and biomass stoichiometry. ISME J. 2015;9:2324–27. doi: 10.1038/ismej.2015.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartman WH, Ye R, Horwath WR, Tringe SG. A genomic perspective on stoichiometric regulation of soil carbon cycling. ISME J. 2017;11:2652–65. doi: 10.1038/ismej.2017.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manzoni S, Jackson RB, Trofymow JA, Porporato A. The global stoichiometry of litter nitrogen mineralization. Science. 2008;321:684–6. doi: 10.1126/science.1159792. [DOI] [PubMed] [Google Scholar]
- 16.Cui J, Zhu Z, Xu X, Liu S, Jones DL, Kuzyakov Y, et al. Carbon and nitrogen recycling from microbial necromass to cope with C:N stoichiometric imbalance by priming. Soil Biol Biochem. 2020;142:107720. doi: 10.1016/j.soilbio.2020.107720. [DOI] [Google Scholar]
- 17.Wei X, Zhu Z, Liu Y, Luo Y, Deng Y, Xu X, et al. C:N:P stoichiometry regulates soil organic carbon mineralization and concomitant shifts in microbial community composition in paddy soil. Biol Fert Soils. 2020;56:1093–107. doi: 10.1007/s00374-020-01468-7. [DOI] [Google Scholar]
- 18.Maria M, Wolfgang W, Sophie ZB, Andreas R. Stoichiometric imbalances between terrestrial decomposer communities and their resources: mechanisms and implications of microbial adaptations to their resources. Front Microbiol. 2014;5:22. doi: 10.3389/fmicb.2014.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiao N, Xu XL, Hu YH, et al. Carbon and nitrogen additions induce distinct priming effects along an organic-matter decay continuum. Sci Rep-UK. 2016;6:19865. doi: 10.1038/srep19865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirkby CA, Kirkegaard JA, Richardson AE, Wade LJ, Blanchard C, Batten G. Stable soil organic matter: a comparison of C:N:P:S ratios in Australian and other world soils. Geoderma. 2011;163:197–208. doi: 10.1016/j.geoderma.2011.04.010. [DOI] [Google Scholar]
- 21.Manzoni S, Čapek P, Mooshammer M, Lindahl BD, Richter A, Šantrůčková H. Optimal metabolic regulation along resource stoichiometry gradients. Ecol Lett. 2017;20:1182–91. doi: 10.1111/ele.12815. [DOI] [PubMed] [Google Scholar]
- 22.Mooshammer M, Wanek W, Hämmerle I, Fuchslueger L, Hofhansl F, Knoltsch A, et al. Adjustment of microbial nitrogen use efficiency to carbon:nitrogen imbalances regulates soil nitrogen cycling. Nat Commun. 2014;5:3694. doi: 10.1038/ncomms4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delgado-Baquerizo M, Reich PB, Khachane AN, Campbell CD, Thomas N, Freitag TE, et al. It is elemental: soil nutrient stoichiometry drives bacterial diversity. Environ Microbiol. 2017;19:1176–88. doi: 10.1111/1462-2920.13642. [DOI] [PubMed] [Google Scholar]
- 24.Mayor JR, Mack MC, Schuur EAG. Decoupled stoichiometric, isotopic, and fungal responses of an ectomycorrhizal black spruce forest to nitrogen and phosphorus additions. Soil Biol Biochem. 2015;88:247–56. doi: 10.1016/j.soilbio.2015.05.028. [DOI] [Google Scholar]
- 25.Mooshammer M, Hofhansl F, Frank AH, Wanek W, Hämmerle I, Leitner S, et al. Decoupling of microbial carbon, nitrogen, and phosphorus cycling in response to extreme temperature events. Sci Adv. 2017;3:e1602781. doi: 10.1126/sciadv.1602781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen J, Seven J, Zilla T, Dippold MA, Blagodatskaya E, Kuzyakov Y. Microbial C:N:P stoichiometry and turnover depend on nutrients availability in soil: A 14C, 15N and 33P triple labelling study. Soil Biol Biochem. 2019;131:206–16. doi: 10.1016/j.soilbio.2019.01.017. [DOI] [Google Scholar]
- 27.Ma Q, Wu L, Wang J, Ma J, Zheng N, Hill PW, et al. Fertilizer regime changes the competitive uptake of organic nitrogen by wheat and soil microorganisms: An in-situ uptake test using 13C, 15N labelling, and 13C-PLFA analysis. Soil Biol Biochem. 2018;125:319–27. doi: 10.1016/j.soilbio.2018.08.009. [DOI] [Google Scholar]
- 28.Jiang G, Zhang W, Xu M, Kuzyakov Y, Zhang X, Wang J, et al. Manure and Mineral Fertilizer Effects on Crop Yield and Soil Carbon Sequestration: a Meta‐Analysis and Modeling Across China. Glob Biogeochem Cy. 2018;32:1659–72. doi: 10.1029/2018GB005960. [DOI] [Google Scholar]
- 29.Liu S, Wang J, Pu S, Blagodatskaya E, Kuzyakov Y, Razavi BS. Impact of manure on soil biochemical properties: a global synthesis. Sci Total Environ. 2020;745:141003. doi: 10.1016/j.scitotenv.2020.141003. [DOI] [PubMed] [Google Scholar]
- 30.Cheng L, Zhang N, Yuan M, Xiao J, Qin Y, Deng Y, et al. Warming enhances old organic carbon decomposition through altering functional microbial communities. ISME J. 2017;11:1825–35. doi: 10.1038/ismej.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma Q, Wen Y, Wang D, Sun X, Hill PW, Macdonald A, et al. Farmyard manure applications stimulate soil carbon and nitrogen cycling by boosting microbial biomass rather than changing its community composition. Soil Biol Biochem. 2020;144:107760. doi: 10.1016/j.soilbio.2020.107760. [DOI] [Google Scholar]
- 32.Ma Q, Wen Y, Ma J, Macdonald A, Hill PW, Chadwick DR, et al. Long-term farmyard manure application affects soil organic phosphorus cycling: a combined metagenomic and 33P/14C labelling study. Soil Biol Biochem. 2020;149:107959. doi: 10.1016/j.soilbio.2020.107959. [DOI] [Google Scholar]
- 33.Jones DL, Hill PW, Smith AR, Farrell M, Ge T, Banning NC, et al. Role of substrate supply on microbial carbon use efficiency and its role in interpreting soil microbial community-level physiological profiles (CLPP) Soil Biol Biochem. 2018;123:1–6. doi: 10.1016/j.soilbio.2018.04.014. [DOI] [Google Scholar]
- 34.Jones DL, Magthab EA, Gleeson DB, Hill PW, Sánchez-Rodríguez AR, Roberts P, et al. Microbial competition for nitrogen and carbon is as intense in the subsoil as in the topsoil. Soil Biol Biochem. 2018;117:72–82. doi: 10.1016/j.soilbio.2017.10.024. [DOI] [Google Scholar]
- 35.Mariano E, Jones DL, Hill PW, Trivelin PCO. Mineralisation and sorption of dissolved organic nitrogen compounds in litter and soil from sugarcane fields. Soil Biol Biochem. 2016;103:522–32. doi: 10.1016/j.soilbio.2016.10.004. [DOI] [Google Scholar]
- 36.Corre M, Brumme R, Veldkamp EF. Changes in nitrogen cycling and retention processes in soils under spruce forests along a nitrogen enrichment gradient in Germany. Glob Change Biol. 2010;13:1509–27. doi: 10.1111/j.1365-2486.2007.01371.x. [DOI] [Google Scholar]
- 37.Spohn M, Kuzyakov Y. Phosphorus mineralization can be driven by microbial need for carbon. Soil Biol Biochem. 2013;61:69–75. doi: 10.1016/j.soilbio.2013.02.013. [DOI] [Google Scholar]
- 38.Wu J, Joergensen RG, Pommerening B, Chaussod R, Brookes PC. Measurement of soil microbial biomass C by fumigation-extraction—an automated procedure. Soil Biol Biochem. 1990;22:1167–9. doi: 10.1016/0038-0717(90)90046-3. [DOI] [Google Scholar]
- 39.Glanville H, Hill PW, Schnepf A, Oburger E, Jones DL. Combined use of empirical data and mathematical modelling to better estimate the microbial turnover of isotopically labelled carbon substrates in soil. Soil Biol Biochem. 2015;94:154–68. doi: 10.1016/j.soilbio.2015.11.016. [DOI] [Google Scholar]
- 40.Greenfield LM, Hill PW, Paterson E, Baggs EM, Jones DL. Methodological bias associated with soluble protein recovery from soil. Sci Rep-UK. 2018;8:11186. doi: 10.1038/s41598-018-29559-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Näsholm T, Kielland K, Ganeteg U. Uptake of organic nitrogen by plants. N Phytol. 2010;182:31–48. doi: 10.1111/j.1469-8137.2008.02751.x. [DOI] [PubMed] [Google Scholar]
- 42.Farrell M, Prendergast-Miller M, Jones DL, Hill PW, Condron LM. Soil microbial organic nitrogen uptake is regulated by carbon availability. Soil Biol Biochem. 2014;77:261–7. doi: 10.1016/j.soilbio.2014.07.003. [DOI] [Google Scholar]
- 43.Niknahad-Gharmakher H, Piutti S, Machet JM, Benizri E, Recous S. Mineralization-immobilization of sulphur in a soil during decomposition of plant residues of varied chemical composition and S content. Plant Soil. 2012;360:391–404. doi: 10.1007/s11104-012-1230-7. [DOI] [Google Scholar]
- 44.Vong PC, Piutti S, Slezackdeschaumes S, Benizri E, Guckert A. Effects of low-molecular-weight organic compounds on sulphur immobilization and re-mineralization and extraction of immobilized sulphur by hot-water and acid hydrolysis. Eur J Soil Sci. 2010;61:287–97. doi: 10.1111/j.1365-2389.2010.01228.x. [DOI] [Google Scholar]
- 45.Takriti M, Wild B, Schnecker J, Mooshammer M, Knoltsch A, Lashchinskiy N, et al. Soil organic matter quality exerts a stronger control than stoichiometry on microbial substrate use efficiency along a latitudinal transect. Soil Biol Biochem. 2018;121:212–20. doi: 10.1016/j.soilbio.2018.02.022. [DOI] [Google Scholar]
- 46.Zhu Z, Ge T, Luo Y, Liu S, Xu X, Tong C, et al. Microbial stoichiometric flexibility regulates rice straw mineralization and its priming effect in paddy soil. Soil Biol Biochem. 2018;121:67–76. doi: 10.1016/j.soilbio.2018.03.003. [DOI] [Google Scholar]
- 47.Xu X, Hui D, King AW, Song X, Thornton PE, Zhang L. Convergence of microbial assimilations of soil carbon, nitrogen, phosphorus and sulfur in terrestrial ecosystems. Sci Rep-UK. 2015;5:17445. doi: 10.1038/srep17445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fanin N, Fromin N, Buatois B, Ttenschwiler SH. An experimental test of the hypothesis of non-homeostatic consumer stoichiometry in a plant litter-microbe system. Ecol Lett. 2013;16:764–72. doi: 10.1111/ele.12108. [DOI] [PubMed] [Google Scholar]
- 49.Peters MK, Hemp A, Appelhans T, Becker JN, Behler C, Classen A, et al. Climate–land-use interactions shape tropical mountain biodiversity and ecosystem functions. Nature. 2019;568:88–92. doi: 10.1038/s41586-019-1048-z. [DOI] [PubMed] [Google Scholar]
- 50.Kaiser C, Franklin O, Dieckmann U, Richter A. Microbial community dynamics alleviate stoichiometric constraints during litter decay. Ecol Lett. 2014;17:680–90. doi: 10.1111/ele.12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu X, Schimel JP, Thornton PE, Song X, Yuan F, Goswami S. Substrate and environmental controls on microbial assimilation of soil organic carbon: a framework for Earth system models. Ecol Lett. 2014;17:547–55. doi: 10.1111/ele.12254. [DOI] [PubMed] [Google Scholar]
- 52.Apostel C, Dippold M, Kuzyakov Y. Biochemistry of hexose and pentose transformations in soil analyzed by position-specific labeling and 13C-PLFA. Soil Biol Biochem. 2015;80:199–208. doi: 10.1016/j.soilbio.2014.09.005. [DOI] [Google Scholar]
- 53.Manzoni S, Taylor P, Richter A, Porporato A, Ågren GI. Environmental and stoichiometric controls on microbial carbon‐use efficiency in soils. N Phytol. 2012;196:79–91. doi: 10.1111/j.1469-8137.2012.04225.x. [DOI] [PubMed] [Google Scholar]
- 54.Fitzgerald JW, Hale DD, Swank WT. Sulphur-containing amino acid metabolism in surface horizons of a hardwood forest. Soil Biol Biochem. 1988;20:825–31. doi: 10.1016/0038-0717(88)90088-0. [DOI] [Google Scholar]
- 55.Akashi H, Gojobori T. Metabolic efficiency and amino acid composition in the proteomes of Escherichia coli and Bacillus subtilis. P Natl Acad Sci USA. 2002;99:3695–700. doi: 10.1073/pnas.062526999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nozaki T, Ali V, Tokoro M. Sulfur-Containing Amino Acid Metabolism in Parasitic Protozoa. Adv Parasit. 2005;60:1–99. doi: 10.1016/S0065-308X(05)60001-2. [DOI] [PubMed] [Google Scholar]
- 57.Takagi H, Ohtsu I. l-Cysteine Metabolism and Fermentation in Microorganisms. Adv Biochem Eng Biotechnol. 2016;159:129–51. doi: 10.1007/10_2016_29. [DOI] [PubMed] [Google Scholar]
- 58.Bustos I, Miguel AM, Fouad A, Carmen P, Teresa R, CM M. Volatile sulphur compounds-forming abilities of lactic acid bacteria: C-S lyase activities. Int J Food Microbiol. 2011;148:121–7. doi: 10.1016/j.ijfoodmicro.2011.05.011. [DOI] [PubMed] [Google Scholar]
- 59.Assefa MK, Tucher SV, Schmidhalter U. Soil sulfur availability due to mineralization: soil amended with biogas residues. J Soil Sci Enviro Manag. 2014;5:13–9. doi: 10.5897/JSSEM13.0408. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.