Abstract
Single-molecule fluorescence resonance energy transfer (smFRET) is a useful tool for observing the dynamics of protein-nucleic acid interactions. While the majority of smFRET measurements have used two fluorophores, multicolor smFRET measurements employing more than two fluorophores offer more information about how protein-nucleic acid complexes dynamically move, assemble, and disassemble. Multi-color smFRET experiments include three or more fluorophores and at least one donor-acceptor pair. This review highlights how multi-color smFRET is being used to probe the dynamics of three different classes of biochemical processes- protein-DNA interactions, chromatin remodeling, and protein translation.
Keywords: Multicolor single-molecule fluorescence microscopy, fluorescence resonance energy transfer, single-molecule biophysics, DNA-protein interaction, RNA-protein interaction
Introduction
The spatial resolution of traditional light microscopy is theoretically confined to 200–400 nm due to the diffraction limit [1]. Conformational rearrangements of individual biomolecules or the interactions between them often take place on a length scale of single nanometers, which makes it challenging to study with traditional microscopy, and even with super-resolution techniques that improve the resolution of light microscopy by more than an order of magnitude [2–4]. While Structural biology techniques such as X-ray crystallography and cryo-electron microscopy overcome this challenge, they come with a compromise on temporal resolution. Fluorescence resonance energy transfer (FRET) fills this gap [5,6]. FRET is the direct transfer of excitation energy from one fluorophore (donor) to another (acceptor) without an actual photo emission by the donor. FRET is distance dependent and occurs when the two fluorophores are brought close to one another, usually less than 10 nm apart. The FRET efficiency E is defined as the fraction of donor excitation events that result in energy transfer to the acceptor fluorophore. Practically, E is calculated by measuring the intensities of the acceptor (A) and donor (D) fluorophores when the donor is excited, and finding the fraction of total fluorescence intensity emitted by the acceptor, as shown in the equation below
The FRET efficiency is related to the distance R between the donor and acceptor fluorophores and is equal to 0.5 when R = R0 (the Forster radius, Figure 1a), which for most fluorophore pairs is 3–7 nm [7].
Figure 1.
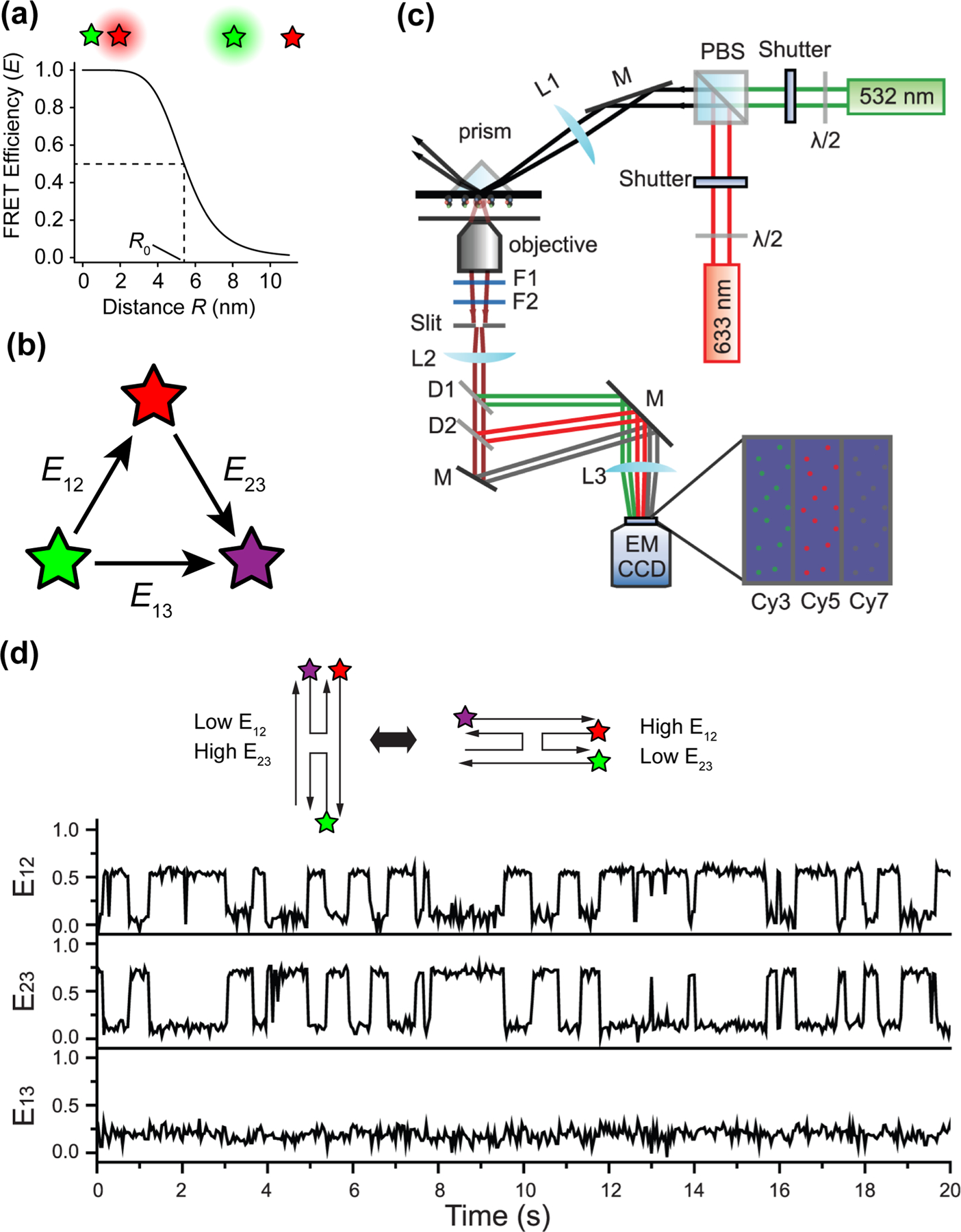
Multi-color single-molecule FRET. (a) Relationship between FRET efficiency and distance between FRET donor (green) and acceptor (red) fluorophores. R0 is the Forster radius, which is the distance between donor and acceptor where FRET efficiency equals 0.5. R0 = 5.4 nm for Cy3 and Cy5. (b) Directionality of energy transfer in a three-color FRET system. (c) Three-color FRET TIRF microscope setup. (d) Single-molecule traces of Holliday junction dynamics. E12, E23, and E13 refer to FRET efficiencies of the donor-acceptor pairs shown in b. (c) and (d) are adapted from Lee et al. [9].
While smFRET experiments measuring the distance between two fluorophores have been informative, the information content of these experiments is limited to determining the distance between two components in a single system. As we venture into more complex biological systems, the need to monitor distances between more than two components emerges and a single FRET pair becomes insufficient. The distance between three or more components can be measured using multi-color smFRET that utilize more than two colors of fluorophores attached to specific sites of interest on one or multiple molecules. In a 3-color smFRET experiment, for example using fluorophores Cy3, Cy5 and Cy7, the fluorophore excited by the shortest wavelength (Cy3) can transfer energy to Cy5 and Cy7, and Cy5 can also transfer energy to Cy7 (Figure 1b). Fluorescence signals at the single-molecule level are typically observed by a total internal reflection fluorescence (TIRF) or confocal microscope [8] equipped with multiple lasers and capable of recording multiple colors simultaneously. An example of a 3-color prism-based TIRF microscope is shown in Figure 1c [9].
The first 3-color smFRET study investigated Holliday junction dynamics [10] by simultaneously monitoring the correlated movements between three DNA strands on a single junction. The technique was later improved with the use of alternating excitation [11], followed by the increasing popularity of far-red dyes (Cy7, Alexa750, LD750 [9,12,13]) and the implementation of 4-color smFRET [14]. A set of FRET time traces visualizing Holliday junction dynamics with Cy3, Cy5 and Cy7 labeling three separate DNA strands is shown in Figure 1d [9]. The PacBio sequencing technology was more recently repurposed as a single-molecule microscope that can image four colors in a high-throughput manner [15].
In this review, we include studies utilizing a multi-color FRET scheme (Figure 1b), as well as studies that measure FRET between two fluorophores but utilize a third or fourth fluorophore for colocalization, focusing on protein-DNA and protein-RNA interactions studies from the past two years. Studies that only colocalize single molecules without measuring FRET are not reviewed here [16,17].
Multi-color FRET Studies on Protein-DNA Interactions
The interactions between proteins and DNA are fundamental to how genes are expressed and how the genome organization is maintained. smFRET is a useful tool to directly visualize and dissect these biological processes in vitro, including how proteins recognize their cognate DNA sequences [18–21]. Marklund et al. recently utilized a three-color smFRET assay to study the target search process of lac repressors (LacI) [20]. They utilized rhodamine-labeled LacI and dual-labeled DNA where Cy5 and Cy7 were placed at two separate LacI-binding motifs (Figure 2a). They observed that LacI rapidly switches from one site to another, at a rate that can be explained by one-dimensional diffusion (Figure 2a). They further tested this by inserting an unlabeled operator site in between the two labeled sites. In this construct, the rate of switching between the two labeled sites decreased (Figure 2b), consistent with the unlabeled site stalling LacI during its 1D sliding along the DNA. They subsequently used single-molecule confocal laser tracking combined with fluorescence correlation spectroscopy to demonstrate that LacI rotates along the DNA helical axis during 1D diffusion. They estimated LacI traverses ~40 bp per revolution [20], consistent with a greedy search approach where LacI does not scan every base pair along the DNA helix but instead occasionally hops out of the DNA grove, potentially missing a target sequence in exchange for a faster sliding rate.
Figure 2.
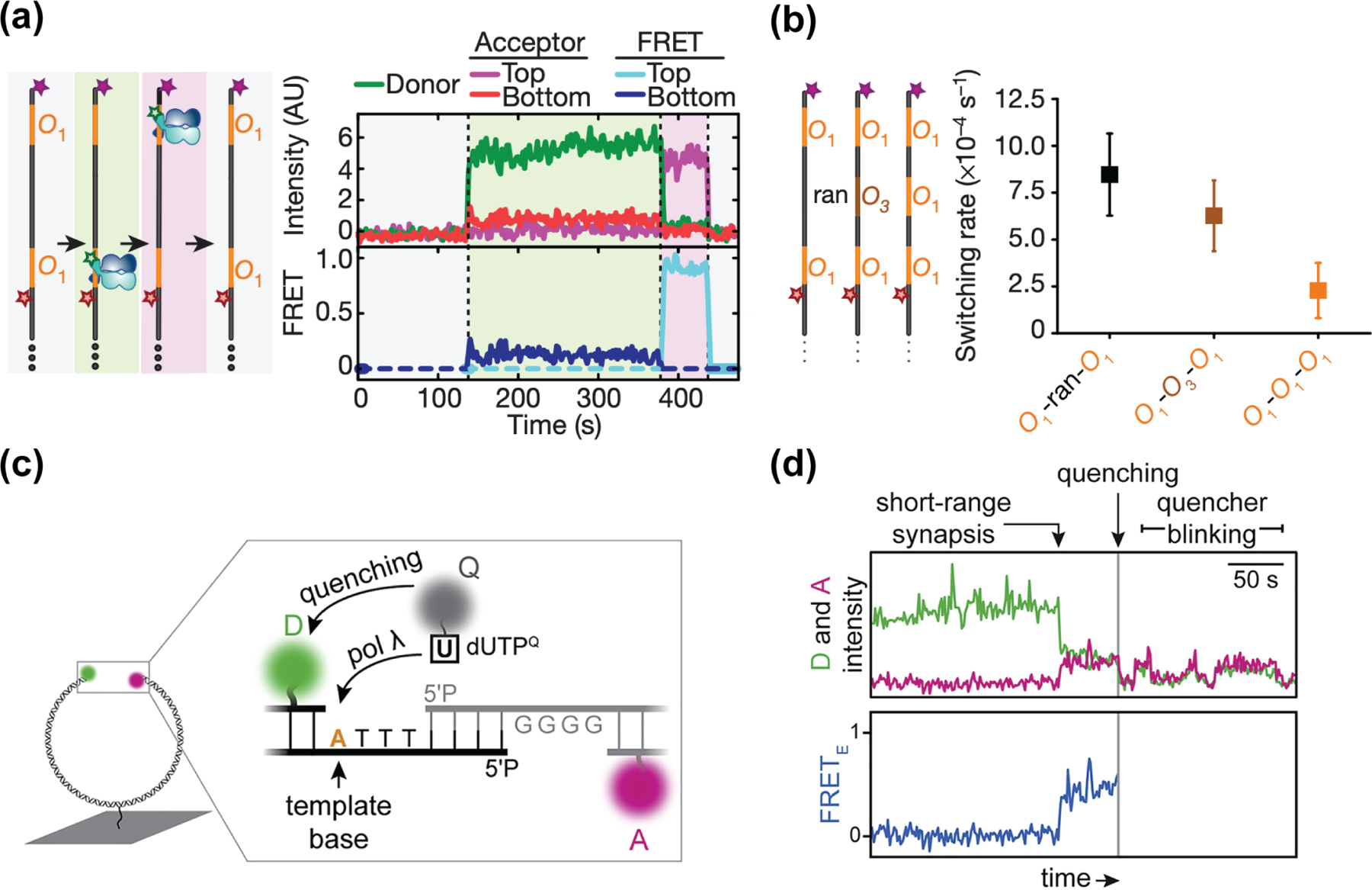
Multicolor FRET studies on protein-DNA interactions. (a) Left: Cartoons depicting possible scenarios of Cy3-labeled LacI bound to DNA. The DNA contains two LacI-binding sites (O1), each site distinctly labeled with Cy5 or Cy7. Right: A set of single-molecule time traces capturing the binding of a LacI dimer onto the bottom O1 site (light green region), followed by re-localization to the top O1 site due to LacI sliding along the DNA (light purple region). (b) Left: DNA constructs with various intervening sequences between the two high-affinity O1 sites. (ran: random DNA sequence; O3: low-affinity LacI-binding site) (c) Cartoons showing the placement of fluorophores in the smFRET assay to monitor DNA orientation during NHEJ. DNA molecules are dual-labeled with Cy3 and Cy5 on each end. dUTP nucleotides are labeled with BHQ10 quencher. (d) A set of single-molecule time traces capturing the sequential steps of NHEJ, where an increase in FRET efficiency (first arrow) showing the alignment of two DNA ends is followed by the incorporation of the quencher-labeled nucleotide (grey line). (a) (b) are adapted from Marklund et al. [20]; (c) (d) are adapted from Stinson et al. [22].
Many multi-component FRET experiments dedicate one fluorophore to report on enzyme activity, while using other fluorophores to monitor structural variations via FRET [22,23]. Stinson et al. used a three-component FRET system to study the mechanism of non-homologous end joining (NHEJ). Previously they used the Xenopus egg extract and a DNA construct with a FRET pair on both ends of the DNA to mimic the alignment of two DNA molecules during NHEJ [24]. The more recent study builds on the assay by utilizing a quencher-labeled dUTP to test whether DNA ends remain aligned during the initial ligation step and subsequent DNA polymerization (Figure 2c). The incorporation of BHQ-dUTP during DNA polymerization places the quencher in proximity to the Cy3 fluorophore and quenches the total fluorescence (Figure 2d, grey line). They found that DNA ends are processed while both ends are in an aligned high-FRET conformation.
Multi-color FRET Studies of Chromatin Remodeling
The base unit of chromatin is the nucleosome, a nucleoprotein complex consisting of 147 base pairs of DNA wrapped around an octamer of histone proteins [25]. A family of enzymes called chromatin remodelers actively establish and maintain chromatin architecture by manipulating nucleosome positions and the identity of the histones within the octamer [26]. Following up on pioneering two-color smFRET measurements of chromatin remodelers sliding nucleosomes [27–32], three-color smFRET measurements are providing more clarity as to how these enzymes manipulate DNA and histones [30,33–35].
The Deindl and Bowman labs utilized multi-color FRET to observe DNA propagate from one side of the nucleosome to the other during nucleosome sliding by the chromatin remodelers Chd1 and Snf2h [30]. Here, an asymmetric nucleosome construct was labeled with Cy3 and Cy7 on the short and long linker sides of DNA and Cy5 was on histone H2A (Figure 3a). Cy3-Cy5 FRET reports on DNA movement at the short-linker side (the DNA exit) and Cy5-Cy7 FRET reports on DNA movements on the opposite long-linker side (the DNA entry). During nucleosome sliding by using Chd1 and Snf2h the Cy5-Cy7 FRET signal increases first, followed by a decrease in Cy3-Cy5 FRET after a delay time τ (Figure 3b), suggesting that Chd1 and Snf2h first translocate nucleosomal DNA on the long-linker entry side, creating a 3 bp DNA bulge. After an average delay (τ) of 4.9 seconds (Figure 3c), the bulge then propagates to the short-linker exit side, causing a decrease in Cy3-Cy5 FRET as the histone octamer is repositioned on the DNA wrapped around it. The DNA sequence used to position the nucleosome in this study (the Widom 601 positioning sequence) is asymmetric in its flexibility with the more flexible side more stably bound to the histone core [36,37]. When the orientation of the long and short linkers on 601 were flipped (Figure 3d), the remodelers still manipulated DNA on the long-linker side before the short-linker, but the delay time τ was significantly longer (21 s) (Figure 3e and f). These measurements clearly show that the DNA bulge created travels from the long linker slide to the short linker, which may be a common mechanism of nucleosome sliding, and that propagation of DNA around the histone octamer is influenced by sequence-dependent DNA mechanics as was also shown for another remodeler INO80 [38].
Figure 3.
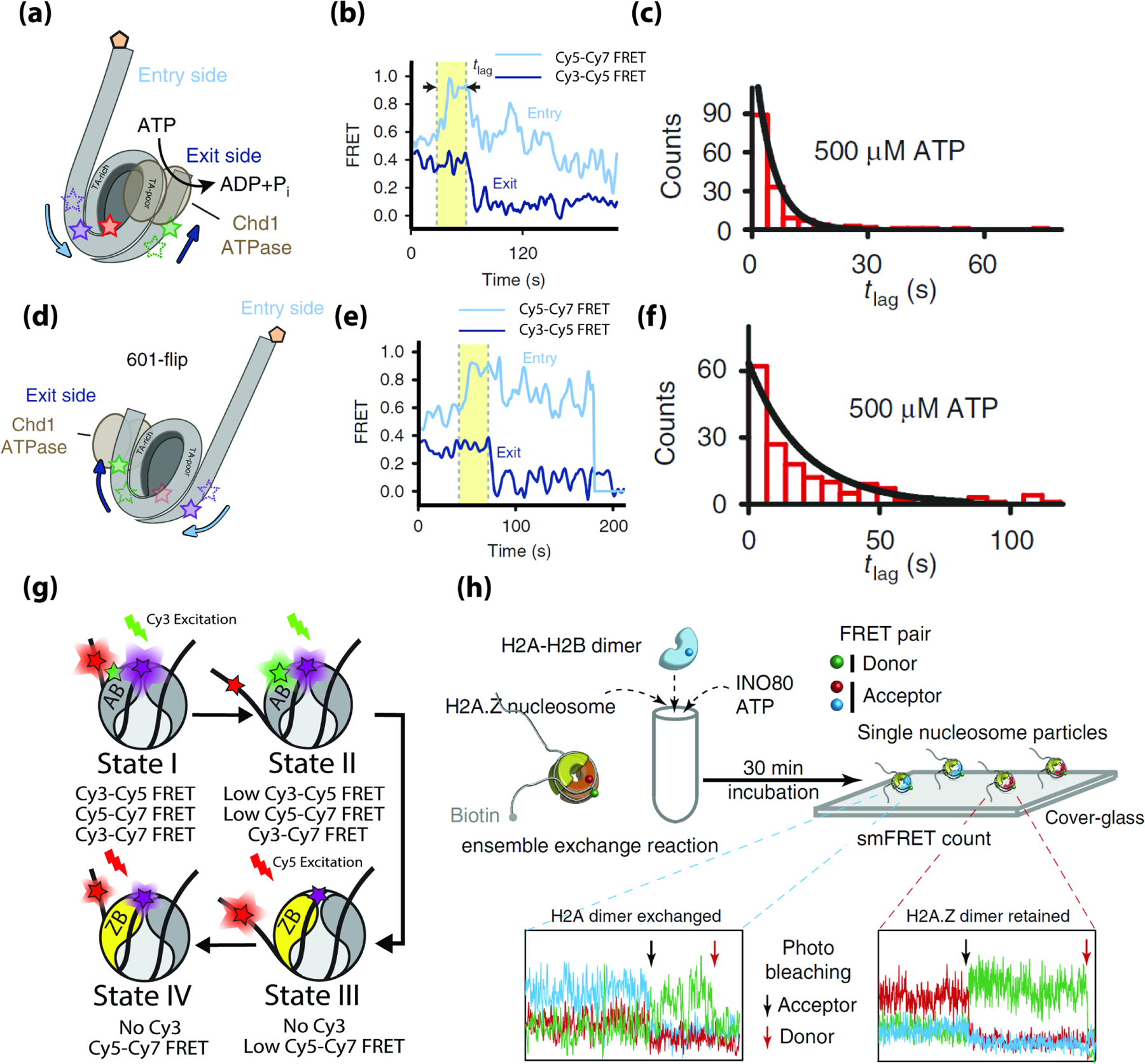
Multi-color FRET measurements of chromatin remodeling. (a) The 3-color nucleosome construct used by Sabantsev et al. [30] (b) Example FRET time traces showing the entry side movement precedes the exit side. (c) Histogram of the lag time (tlag) between entry DNA movement and exit side movement (d) The ‘601-flip’ nucleosome construct used by Sabantsev et al. (e) Example FRET time traces showing the entry DNA movement precedes the exit DNA for the ‘601-flip’ nucleosome, consistentwith the construct in (a). (f) Histogram of the lag time (tlag) between entry DNA movement and exit side movement for the ‘601-flip’ nucleosome. (g) Cartoons depicting the four intermediate states of a canonical nucleosome undergoing H2A.Z exchange as well as their expected pair-wise FRET efficiencies. (h) Experimental scheme and representative traces to examine histone exchange by INO80 in Brahma et al. [33] (a-f) are adapted from Sabantsev et al. [30] (h) is adapted from Brahma et al. [33].
The two remodelers in the INO80 subfamily, INO80 and SWR1, carry out histone exchange- the act of replacing nucleosomal histones with free histones. SWR1 exchanges nucleosomal H2A-H2B for free H2A.Z-H2B [39]. While previous measurements using two-color smFRET demonstrated that SWR1 can unwrap and rewrap nucleosomal DNA [40], the relationship and necessity of these DNA dynamics to the actual histone exchange reaction remained unclear. To address this, we have recently used multi-color FRET to observe SWR1 unwrap and rewrap nucleosomal DNA in real time during a histone exchange reaction. We utilized a three-color nucleosome construct with Cy5 and Cy7 on nucleosomal DNA and Cy3 on histone H2A, allowing DNA dynamics to be monitored via Cy5 and Cy7 FRET, while the eviction of H2A-H2B could be detected by the loss of Cy3 (Figure 3g). These measurements revealed several new intermediates in the exchange reaction, showing that SWR1 unwraps nucleosomal DNA transiently from the side of the nucleosome that it removes H2A-H2B from (MF Poyton et al., unpublished).
In contrast to SWR1, the chromatin remodeler INO80 has been suggested to replace nucleosomal H2A.Z-H2B with H2A-H2B, although its activity in vitro and in vivo is disputed [41–43]. Three-color smFRET measurements from the Lee and Bartholomew labs confirm that this exchange reaction can occur in vitro using multi-color FRET and single-molecule pulldown assays [33]. Here, the reaction was performed in bulk solution and the product was analyzed via FRET to Cy5.5 on the nucleosomal DNA after immobilizing it to the surface. Photobleaching traces unambiguously showed 10% of nucleosomes contain H2A-H2B, and that the reverse reaction did not take place (Figure 3h).
Ribosome Assembly and Translation Dynamics
Translation of mRNA into protein requires many different proteins and RNA to form a translating ribosomal complex [44]. Recently several studies utilized one FRET pair to monitor structural dynamics of the ribonucleoprotein complex and additional colors were used to report on an activity of interest (protein binding, nascent transcription etc.) through either FRET or colocalization. We studied ribosome assembly by colocalizing the arrival of Cy3-labeled ribosomal proteins to a previously characterized [45] dynamic complex of Cy5-labeled protein S4 and Cy7-labeled 16S rRNA [46]. By monitoring the shift in equilibrium between non-native and natively folded RNA states (distinguished by Cy5-Cy7 FRET), we were able to correlate the assembly of ribosomal proteins onto the 16S rRNA with the maturation of RNA structure.
More recently, the Puglisi lab studied the ribosome assembly process on nascent RNA [47,48]. They immobilized a stalled transcription complex, reinitiated transcription and monitored the progression of transcription via a gradual increase in Cy3.5 signal due to movement of template DNA towards the surface where the laser excitation is the strongest. When transcription is complete, the Cy3.5-labeled DNA template dissociates together with the RNA polymerase. The nascent RNA stays immobilized on the surface, available for a Cy3-oligo complementary to the 5’ end of nascent RNA and a Cy3.5-oligo complementary to the 3’ end to bind. Proper folding of the nascent RNA will bring the 5’ and 3’ ends close to one another, resulting in FRET increase (Figure 4a). Cy5.5-labeled ribosomal protein S7 was expected to bind to the natively folded RNA after the 3’−5’ helix is formed [47], consistent with the appearance of Cy5.5 signal after the FRET increase. They also utilized a similar scheme to measure the kinetics of the transition from translation initiation to elongation in a eukaryotic translation system [49]. Here, they formed a pre-initiation complex on the surface by binding the Cy3-labeled 40S subunit to an immobilized mRNA and detected the arrival of the Cy5-labeled 60S subunit (forming the initiation complex) and Cy5.5-labeled tRNA (forming the elongation complex) via FRET. A similar strategy was used more recently to study how a virulence factor SARS-CoV-2 NSP1 inhibits mRNA translation [50].
Figure 4.
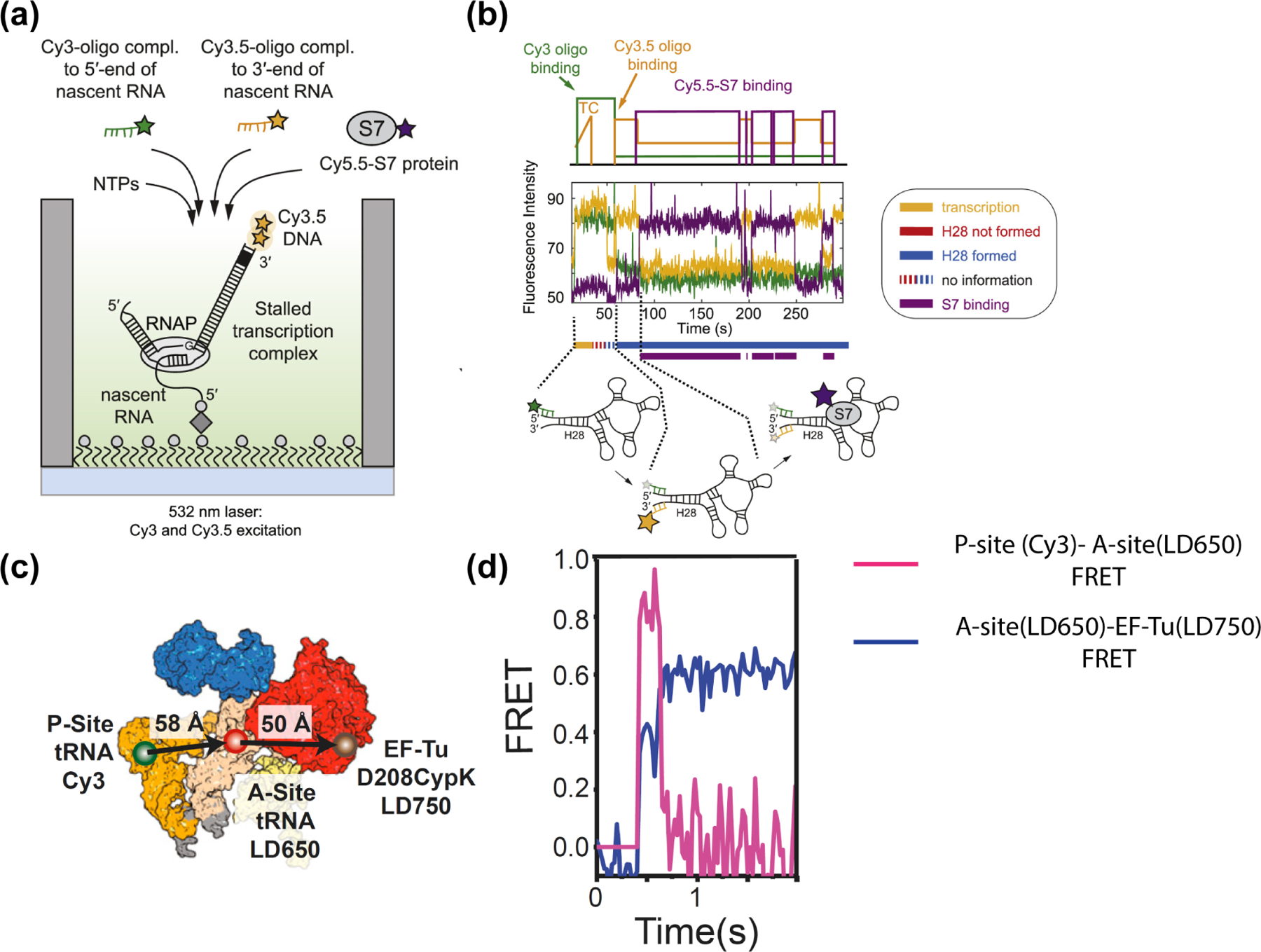
Multicolor smFRET studies on ribosome assembly and translation dynamics. (a) Experimental design to visualize nascent RNA folding and ribosomal protein binding on zero-mode waveguides. (b) Fluorescence time traces showing an initial gradual increase in Cy3.5 intensity due to transcription activity, presence of the Cy3-oligo complementary to the 5’ end of nascent RNA, followed by the dissociation of the RNAP-DNA complex (decrease of Cy3.5 intensity to background level), FRET between Cy3.5-oligo complementary to the 3’ end of nascent RNA and the Cy3-oligo indicates formation of the H28 helix, and subsequent gain and loss of FRET with Cy5.5-S7 protein due to S7 binding and disassociation. (c) The FRET construct used to observe EF-Tu-tRNA complex bind to a ribosomal protein complex with a tRNA in the P-site. (d) A real-time trajectory of EF-Tu-tRNA binding to the A-site of the (increase in both Cy3-LD650 and LD650-LD750 FRET), followed by subsequent unbinding of EF-Tu (LD650-LD750 FRET decrease) and the movement of the A-site tRNA into the accommodation corridor of the ribosome (Cy3-LD650 FRET decrease). (a-b) are adapted from Duss et al. [47] (c-d) are adapted from Morse et al. [51]
The selection of the correct tRNA during translation is facilitated by the G protein elongation factor Tu (EF-Tu) [51,52]. The Blanchard lab measured the kinetics of tRNA accommodation and subsequent unbinding of EF-Tu during tRNA selection using three-color smFRET [51]. Among many three-color constructs utilized, a key experiment employed a construct where an LD750-labeled EF-Tu was bound to an LD650-labeled tRNA (LD750 and LD650 are more photostable analogues of Cy7 and Cy5, respectively [13]). This dual-labeled tRNA-EF-Tu complex was introduced to a ribosomal complex containing a LD550-labeled tRNA (Figure 4c). Shortly after binding, the LD650-LD750 FRET decreased to zero due to unbinding of the EF-Tu accompanied by a dramatic increase in FRET between the two tRNAs, suggesting that the incoming tRNA enters the accommodation corridor of the ribosome only after the dissociation of EF-Tu.
Challenges and Outlook
The increased spatial information available in multicolor smFRET experiments has clear advantages over similar two-color FRET experiments as exemplified by the recent work discussed in this review. Nevertheless, multicolor smFRET measurements are far less common than their two-color counterparts. As the number of fluorophores increases, the statistical likelihood of observing single molecules containing all fluorophores decreases due to incomplete labeling and photobleaching. As a result, multi-color measurements typically require the observation of far more molecules than in two-color experiments. Advancements in imaging technology that increases the number of molecules imaged in a single experiment and improvements in fluorophore photostability, such as self-healing fluorophores [53,54] should make multicolor FRET measurements more accessible. Clearly, the increased information content makes multi-color FRET a powerful tool that will continue to illuminate how multiple molecules interact and rearrange during the most sophisticated biological processes.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interest
The authors declare no conflict of interest.
References
- 1.Abbe E: Beiträge zur Theorie des Mikroskops und der mikrosk opischen Wahrnehmung: I. Die Construction von Mikroskopen auf Grund der Theorie. Arch für mikroskopische Anat 1873, 9:413–418. [Google Scholar]
- 2.Hell SW, Wichmann J: Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt Lett 1994, 19:780. [DOI] [PubMed] [Google Scholar]
- 3.Rust MJ, Bates M, Zhuang X: Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat Methods 2006, 3:793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF: Imaging intracellular fluorescent proteins at nanometer resolution. Science (80- ) 2006, 313:1642–1645. [DOI] [PubMed] [Google Scholar]
- 5.Förster T: Zwischenmolekulare Energiewanderung und Fluoreszenz. Ann Phys 1948, 437:55–75. [Google Scholar]
- 6.Ha T, Enderle T, Ogletree DF, Chemla DS, Selvin PR, Weiss S: Probing the interaction between two single molecules: Fluorescence resonance energy transfer between a single donor and a single acceptor. Proc Natl Acad Sci U S A 1996, 93:6264–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lakowicz JR: Principles of fluorescence spectroscopy. Springer; 2006. [Google Scholar]
- 8.Chung HS, Gopich IV, McHale K, Cellmer T, Louis JM, Eaton WA: Extracting rate coefficients from single-molecule photon trajectories and FRET efficiency histograms for a fast-folding protein. J Phys Chem A 2011, 115:3642–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee S, Lee J, Hohng S: Single-Molecule Three-Color FRET with Both Negligible Spectral Overlap and Long Observation Time. PLoS One 2010, 5:e12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hohng S, Joo C, Ha T: Single-molecule three-color FRET. Biophys J 2004, 87:1328–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nam KL, Kapanidis AN, Hye RK, Korlann Y, Sam OH, Kim Y, Gassman N, Seong KK, Weiss S: Three-color alternating-laser excitation of single molecules: Monitoring multiple interactions and distances. Biophys J 2007, 92:303–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Mallon J, Wang H, Singh D, Jo MH, Hua B, Bailey S, Ha T: Real-time observation of cas9 postcatalytic domain motions. Proc Natl Acad Sci U S A 2021, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altman RB, Zheng Q, Zhou Z, Terry DS, Warren JD, Blanchard SC: Enhanced photostability of cyanine fluorophores across the visible spectrum. Nat Methods 2012, 9:428–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee J, Lee S, Ragunathan K, Joo C, Ha T, Hohng S: Single-Molecule Four-Color FRET. Angew Chemie Int Ed 2010, 49:9922–9925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen J, Dalal RV, Petrov AN, Tsai A, O’Leary SE, Chapin K, Cheng J, Ewan M, Hsiung PL, Lundquist P, et al. : High-throughput platform for real-time monitoring of biological processes by multicolor single-molecule fluorescence. Proc Natl Acad Sci U S A 2014, 111:664–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoskins AA, Friedman LJ, Gallagher SS, Crawford DJ, Anderson EG, Wombacher R, Ramirez N, Cornish VW, Gelles J, Moore MJ: Ordered and dynamic assembly of single spliceosomes. Science (80- ) 2011, 331:1289–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoskins AA, Rodgers ML, Friedman LJ, Gelles J, Moore MJ: Single molecule analysis reveals reversible and irreversible steps during spliceosome activation. Elife 2016, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li S, Zheng EB, Zhao L, Liu S: Nonreciprocal and Conditional Cooperativity Directs the Pioneer Activity of Pluripotency Transcription Factors. Cell Rep 2019, 28:2689–2703.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donovan BT, Chen H, Jipa C, Bai L, Poirier MG: Dissociation rate compensation mechanism for budding yeast pioneer transcription factors. Elife 2019, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.*. Marklund E, van Oosten B, Mao G, Amselem E, Kipper K, Sabantsev A, Emmerich A, Globisch D, Zheng X, Lehmann LC, et al. : DNA surface exploration and operator bypassing during target search. Nature 2020, 583:858–861. This study uses 3-color smFRET to specifically locate a DNA binding protein (LacI) onto one of two DNA binding sites on single DNA molecules. This assay allows them to conclude that LacI undergoes 1D diffusion along the DNA.
- 21.Mivelaz M, Cao AM, Kubik S, Zencir S, Hovius R, Boichenko I, Stachowicz AM, Kurat CF, Shore D, Fierz B: Chromatin Fiber Invasion and Nucleosome Displacement by the Rap1 Transcription Factor. Mol Cell 2020, 77:488–500.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stinson BM, Moreno AT, Walter JC, Loparo JJ: A Mechanism to Minimize Errors during Non-homologous End Joining. Mol Cell 2020, 77:1080–1091.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wortmann P, Götz M, Hugel T: Cooperative Nucleotide Binding in Hsp90 and Its Regulation by Aha1. Biophys J 2017, 113:1711–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graham TGW, Walter JC, Loparo JJ: Two-Stage Synapsis of DNA Ends during Non-homologous End Joining. Mol Cell 2016, 61:850–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ: Crystal structure of the nucleosome core particle at 2.8 Å resolution . Nature 1997, 389:251–260. [DOI] [PubMed] [Google Scholar]
- 26.Clapier CR, Iwasa J, Cairns BR, Peterson CL: Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat Rev Mol Cell Biol 2017, 18:407–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qiu Y, Levendosky RF, Chakravarthy S, Patel A, Bowman GD, Myong S: The Chd1 Chromatin Remodeler Shifts Nucleosomal DNA Bidirectionally as a Monomer. Mol Cell 2017, 68:76–88.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou CY, Johnson SL, Lee LJ, Longhurst AD, Beckwith SL, Johnson MJ, Morrison AJ, Narlikar GJ: The Yeast INO80 Complex Operates as a Tunable DNA Length-Sensitive Switch to Regulate Nucleosome Sliding. Mol Cell 2018, doi: 10.1016/j.molcel.2018.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deindl S, Hwang WL, Hota SK, Blosser TR, Prasad P, Bartholomew B, Zhuang X: ISWI remodelers slide nucleosomes with coordinated multi-base-pair entry steps and single-base-pair exit steps. Cell 2013, 152:442–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.**. Sabantsev A, Levendosky RF, Zhuang X, Bowman GD, Deindl S: Direct observation of coordinated DNA movements on the nucleosome during chromatin remodelling. Nat Commun 2019, 10:1–12. This study uses 3-color smFRET to monitor the timing of entry and exit DNA movement on single nucleosomes during nucleosome sliding by chromatin remodelers. They find that entry DNA movement precedes exit DNA movement by a few seconds. During the few seconds delay, a few extra base pairs are absorbed by the nucleosome and propagated towards the exit side.
- 31.Kirk J, Lee JY, Lee Y, Kang C, Shin S, Lee E, Song JJ, Hohng S: Yeast Chd1p Unwraps the Exit Side DNA upon ATP Binding to Facilitate the Nucleosome Translocation Occurring upon ATP Hydrolysis. Biochemistry 2020, 59:4481–4487. [DOI] [PubMed] [Google Scholar]
- 32.Armache JP, Gamarra N, Johnson SL, Leonard JD, Wu S, Narlikar GJ, Cheng Y: Cryo-EM structures of remodeler-nucleosome intermediates suggest allosteric control through the nucleosome. Elife 2019, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brahma S, Udugama MI, Kim J, Hada A, Bhardwaj SK, Hailu SG, Lee T-H, Bartholomew B: INO80 exchanges H2A.Z for H2A by translocating on DNA proximal to histone dimers. Nat Commun 2017, 8:15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huynh MT, Yadav SP, Reese JC, Lee TH: Nucleosome Dynamics during Transcription Elongation. ACS Chem Biol 2020, 15:3133–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee J, Lee TH: Single-Molecule Investigations on Histone H2A-H2B Dynamics in the Nucleosome. Biochemistry 2017, 56:977–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ngo TTM, Zhang Q, Zhou R, Yodh JG, Ha T: Asymmetric unwrapping of nucleosomes under tension directed by DNA local flexibility. Cell 2015, 160:1135–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Basu A, Bobrovnikov DG, Qureshi Z, Kayikcioglu T, Ngo TTM, Ranjan A, Eustermann S, Cieza B, Morgan MT, Hejna M, et al. : Measuring DNA mechanics on the genome scale. bioRxiv 2020, doi: 10.1101/2020.08.17.255042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Basu A, Bobrovnikov DG, Qureshi Z, Kayikcioglu T, Ngo TTM, Ranjan A, Eustermann S, Cieza B, Morgan MT, Hejna M, et al. : Measuring DNA mechanics on the genome scale. Nature 2021, 589:462–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C: ATP-Driven Exchange of Histone H2AZ Variant Catalyzed by SWR1 Chromatin Remodeling Complex. Science (80- ) 2004, 303:343–348. [DOI] [PubMed] [Google Scholar]
- 40.Willhoft O, Ghoneim M, Lin C, Chua YD, Wilkinson M, Chaban Y, Ayala R, Mccormack EA, Ocloo L, Rueda DS, et al. : Structure and dynamics of the yeast SWR1 : nucleosome complex. Science (80- ) 2018, [DOI] [PubMed] [Google Scholar]
- 41.Watanabe S, Radman-Livaja M, Rando OJ, Peterson CL: A histone acetylation switch regulates H2A.Z deposition by the SWR-C remodeling enzyme. Science (80- ) 2013, 340:195–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang F, Ranjan A, Wei D, Wu C: Comment on “A histone acetylation switch regulates H2A.Z deposition by the SWR-C remodeling enzyme.” Science (80- ) 2016, doi: 10.1126/science.aad5921. [DOI] [PubMed] [Google Scholar]
- 43.Watanabe S, Peterson CL: Response to comment on “A histone acetylation switc h regulates H2A.Z deposition by the SWR-C remodeling enzyme.” Science (80- ) 2016, 353:358b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramakrishnan V: Ribosome structure and the mechanism of translation. Cell 2002, 108:557–572. [DOI] [PubMed] [Google Scholar]
- 45.Kim H, Abeysirigunawarden SC, Chen K, Mayerle M, Ragunathan K, Luthey-Schulten Z, Ha T, Woodson SA: Protein-guided RNA dynamics during early ribosome assembly. Nature 2014, 506:334–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abeysirigunawardena SC, Kim H, Lai J, Ragunathan K, Rappé MC, Luthey-Schulten Z, Ha T, Woodson SA: Evolution of protein-coupled RNA dynamics during hierarchical assembly of ribosomal complexes. Nat Commun 2017, 8:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.**. Duss O, Stepanyuk GA, Puglisi JD, Williamson JR: Transient Protein-RNA Interactions Guide Nascent Ribosomal RNA Folding. Cell 2019, 179:1357–1369.e16. This study uses several multicolor FRET experiments to study the ribosome assembly process on pre-folded and nascent RNA. In a 4-color smFRET experiment, they monitored transcription of nascent RNA, folding states of nascent RNA and subsequent binding of ribosomal proteins on single RNA molecules.
- 48.Duss O, Stepanyuk GA, Grot A, O’Leary SE, Puglisi JD, Williamson JR: Real-time assembly of ribonucleoprotein complexes on nascent RNA transcripts. Nat Commun 2018, 9:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.**. Wang J, Johnson AG, Lapointe CP, Choi J, Prabhakar A, Chen DH, Petrov AN, Puglisi JD: eIF5B gates the transition from translation initiation to elongation. Nature 2019, 573:605–608. The authors utilize a 4-color smFRET experiment to visualize eukaryotic translation initiation in real time.
- 50.Lapointe CP, Grosely R, Johnson AG, Wang J, Fernández IS, Puglisi JD: Dynamic competition between SARS-CoV-2 NSP1 and mRNA on the human ribosome inhibits translation initiation. Proc Natl Acad Sci 2021, 118:e2017715118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.**. Morse JC, Girodat D, Burnett BJ, Holm M, Altman RB, Sanbonmatsu KY, Wieden HJ, Blanchard SC: Elongation factor-Tu can repetitively engage aminoacyl-tRNA within the ribosome during the proofreading stage of tRNA selection. Proc Natl Acad Sci U S A 2020, 117:3610–3620. This study uses 3-color smFRET experiments to visualize EF-Tu and tRNA selection during translation elongation.
- 52.Rodnina MV, Wintermeyer W: Ribosome fidelity: tRNA discrimination, proofreading and induced fit. Trends Biochem Sci 2001, 26:124–130. [DOI] [PubMed] [Google Scholar]
- 53.Zheng Q, Juette MF, Jockusch S, Wasserman MR, Zhou Z, Altman RB, Blanchard SC: Ultra-stable organic fluorophores for single-molecule research. Chem Soc Rev 2014, 43:1044–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pati AK, El Bakouri O, Jockusch S, Zhou Z, Altman RB, Fitzgerald GA, Asher WB, Terry DS, Borgia A, Holsey MD, et al. : Tuning the Baird aromatic triplet-state energy of cyclooctatetraene to maximize the self-healing mechanism in organic fluorophores. Proc Natl Acad Sci U S A 2020, 117:24305–24315. [DOI] [PMC free article] [PubMed] [Google Scholar]


