Abstract
We describe a purified ubiquitination system capable of rapidly catalyzing the covalent linkage of polyubiquitin chains onto a model substrate, phosphorylated IκBα. The initial ubiquitin transfer and subsequent polymerization steps of this reaction require the coordinated action of Cdc34 and the SCFHOS/β-TRCP-ROC1 E3 ligase complex, comprised of four subunits (Skp1, cullin 1 [CUL1], HOS/β-TRCP, and ROC1). Deletion analysis reveals that the N terminus of CUL1 is both necessary and sufficient for binding Skp1 but is devoid of ROC1-binding activity and, hence, is inactive in catalyzing ubiquitin ligation. Consistent with this, introduction of the N-terminal CUL1 polypeptide into cells blocks the tumor necrosis factor alpha-induced and SCF-mediated degradation of IκB by forming catalytically inactive complexes lacking ROC1. In contrast, the C terminus of CUL1 alone interacts with ROC1 through a region containing the cullin consensus domain, to form a complex fully active in supporting ubiquitin polymerization. These results suggest the mode of action of SCF-ROC1, where CUL1 serves as a dual-function molecule that recruits an F-box protein for substrate targeting through Skp1 at its N terminus, while the C terminus of CUL1 binds ROC1 to assemble a core ubiquitin ligase.
Regulation of protein stability by ubiquitin (Ub)-dependent proteolysis plays major roles in the control of multiple aspects of cell function, such as transcriptional control, cell cycle progression, and signal transduction (17). Ubiquitination involves three distinct enzymatic events that ultimately lead to the covalent linkage of Ub polymers to the lysine ɛ-amino groups of substrate proteins. Ub is initially charged in an ATP-dependent fashion by the Ub-activating enzyme, E1, to form a high-energy thiol-ester bond between the carboxyl group of its C-terminal glycine residue and E1. The thiol-ester-linked Ub is then transferred to an E2, which cooperates with an E3 ligase to catalyze the formation of an isopeptide bond between Ub and the substrate (16).
One well-characterized Ub-proteasome pathway is the degradation of IκBα required for the activation of the transcription factor NF-κB (26). IκBα, which sequesters NF-κB in the cell cytoplasm (3), is phosphorylated by the IKK kinase complex that is activated in response to proinflammatory cytokines (4, 9, 28, 36, 48, 53). Such phosphorylation triggers the rapid ubiquitination and subsequent degradation of IκBα, resulting in the release of NF-κB (5, 7, 37). Recent studies have demonstrated that the β-TRCP/HOS F-box protein family plays a direct role in targeting IκBα for ubiquitination and degradation (12, 15, 21, 22, 30, 42, 47, 50) by binding to this inhibitor at its N-terminally located DS(PO3)GΨXS(PO3) motif (1, 47, 49). More recently, the ubiquitination of IκBα has been reconstituted in vitro with purified components including SCFHOS/β-TRCP-ROC1 as the E3 ligase complex (44).
SCFHOS/β-TRCP-ROC1 contains Skp1, cullin 1 (CUL1), HOS/β-TRCP, and the newly identified ROC1 (also called Rbx1 or Hrt1). ROC1 is a novel RING-H2 finger protein that was initially isolated as a CUL4A-interacting protein by a yeast two-hybrid screen (30). It has also been biochemically purified as a common component of both the human (44) and yeast (38) SCF complexes, as well as the native human von Hippel-Lindau (VHL) tumor suppressor complex (19). In addition, the ROC1 homologue, ROC2 (also called SAG), was isolated as a redox-agent-induced gene product that protects cells from apoptosis (10). ROC1 plays an essential role in the Cdc34/SCF-mediated ubiquitination-degradation pathway. Yeast ROC1 encodes an essential gene whose reduced expression led to the accumulation of Sic1 and Clns (19, 30, 38, 40), both of which are previously identified substrates of the Cdc34-SCF ubiquitination apparatus (11, 35, 39). Yeast ROC1 is a component of the SCF complexes that mediates the in vitro ubiquitination of Sic1 by Cdc4 (19, 38) and Clns by Grr1 (40).
The in vitro reconstitution experiments (44) reveal that the human ROC1 protein is recruited by CUL1 to form the SCFHOS/β-TRCP-ROC1 complex (with Skp1 and HOS/β-TRCP). Like its yeast counterpart, human Skp1 links CUL1 to the F-box protein HOS/β-TRCP. In addition, Skp1 enhances the ability of HOS/β-TRCP to interact with phosphorylated IκBα. The purified recombinant SCFHOS/β-TRCP-ROC1 complex specifically binds IKKβ-phosphorylated IκBα and catalyzes its ubiquitination in the presence of Ub, E1, and Cdc34 as the E2-conjugating enzyme. Each of the four subunits within SCFHOS/β-TRCP-ROC1 is required for the substrate ubiquitination reaction. These studies suggest that SCFHOS/β-TRCP-ROC1 acts as an E3 holoenzyme that is both necessary and sufficient to initiate and catalyze ubiquitination. A recent study has identified Sgt1p as a novel protein that interacts with Skp1 and which is required for assembling the yeast kinetochore complex (20). However, the biochemical role of Sgt1p in the SCF-ROC1-mediated ubiquitination reaction remains to be determined.
In addition to its role in the SCF pathway, ROC1 may mediate other ubiquitin-dependent proteolysis events in the cell. It has been shown by cotransfection experiments that ROC1 binds five cullin family members (CUL1, CUL2, CUL3, CUL4A, and CUL4B), whereas ROC2 preferentially interacts with CUL5 (30). Consistent with this, ROC1 is found to be a component of the pVHL complex (19), whose additional subunits include pVHL, CUL2, and elongins C and B (24, 33, 43). Furthermore, ROC1 shares extensive homology with APC11 (19, 30, 38), a subunit of the anaphase-promoting complex (35), which interacts with the cullin-related protein APC2 (30). These findings suggest that ROC/APC11, through its combinatorial interaction with cullin/APC2, forms a dimeric core component common to a large family of multisubunit Ub ligases.
Using a sensitive 32P-Ub-incorporation assay, we have previously observed that the purified ROC1-CUL1 complex contains a ligase activity capable of catalyzing Ub polymerization in a substrate-independent manner (44). In addition, missense mutations in ROC1 significantly reduce Ub ligase activity without affecting its interaction with CUL1 (30). Consistent with these observations, a recently published study by Seol et al. (38) has shown that the Cdc53-Hrt1 subcomplex is capable of activating the autoubiquitination of Cdc34. These studies indicate that the ROC1-CUL1 complex constitutes a Ub ligase. Furthermore, yeast ROC1 has been shown to directly interact with Cdc34 (38, 40). Taken together, these findings raise the intriguing possibility that the ROC/APC11–cullin/APC2 subassembly within the E3 complex carries out a common Ub ligase function that facilitates the transfer of activated Ub from a cognate E2 to targeted substrates.
In this report, we show a rapid synthesis of polyubiquitin chains covalently linked to IκBα through a coordinated action between SCFHOS/β-TRCP-ROC1 and Cdc34. We further elucidate the mechanism of action of SCFHOS/β-TRCP-ROC1 in which it utilizes two distinct domains within CUL1 for substrate targeting and for Ub ligation.
MATERIALS AND METHODS
Plasmids.
N-terminally Flag-tagged CUL1 fragments were constructed by PCR by using the pcDNA-CUL1 plasmid (29) as the template. The following primers were used (boldfaced sequences indicate Flag tag): Flag-CUL1 (started at the initiation codon), 5′GCCACCATGGATTACAAGGATGACGACGATAAGATGTCGTCAACCCGGAGCC; Flag-CUL1 (amino acids 324 to 776), 5′GCCACCATGGAT TACAAGGATGACGACGATAAGAATC T TG TATC TAGAATCCAGGAT; Flag-CUL1 (terminated at amino acid 776), 5′GGACTAGTTAAGCCAAGTAACTGTAGGTG; Flag-CUL1 (1 to 452), 5′GACAACCATCACTTGATTGAG; Flag-CUL1 (1 to 544), 5′GGACCCGGAGCTCAGCACTTG; and Flag-CUL1 (1 to 645), 5′CTTTAATAAAATCTGTAAAACTTGCGCC. PCRs were performed with the Expand High Fidelity PCR System from Boehringer Mannheim per manufacturer's protocol, and the 3′ deoxyadenosine overhangs were added by using Taq DNA polymerase from Stratagene for a final 10-min extension step. The Flag-tagged CUL1 constructs were inserted into cytomegalovirus (CMV)-promoter-based expression vectors using either the Invitrogen Eukaryotic TA Cloning Kit (Flag-CUL1 fragments) or the Eukaryotic TOPO TA Cloning Kit (full-length Flag-CUL1) per manufacturer's instructions. All Flag-CUL1 truncations were verified by dideoxy sequencing.
The plasmids expressing HA-ROC1 and Skp1 have been described previously (29, 30).
Enzymes.
Human E1, mouse Cdc34 (mCdc34), SCFHOS-ROC1, and IKKβS177E,S181E were prepared as described previously (44). E2-25K was isolated from cytosolic extracts of HeLa cells by using a Ub-affinity column as previously described (16). E1 and other E2s present in the E2-25K preparation were removed by Q-Sepharose chromatography and glycerol gradient sedimentation. Ubc4 and Ubc5 were kindly provided by G. Fang (Harvard Medical School) and T. Ohta and Y. Xiong (University of North Carolina at Chapel Hill).
Transfection, metabolic labeling, and extract preparation.
Plates (150 by 25 mm) of 293T cells were grown on 20 ml of Dulbecco modified Eagle medium per plate (Gibco BRL), 10% heat-inactivated fetal bovine serum (Sigma), and 1% antibiotic-antimycotic agent (Gibco BRL). DNA(s) was transfected up to a concentration of 30 μg per plate by using the standard calcium phosphate precipitation method. For metabolic labeling, 45-h-posttransfected cells were washed with 10 ml of phosphate-buffered saline and were starved for 30 min with 6 ml of Dulbecco modified Eagle medium lacking l-methionine/l-cysteine per plate (Gibco BRL), 10% heat-inactivated and dialyzed fetal bovine serum (Gibco BRL), and 1% antibiotic-antimycotic agent. The media was then changed to include 100 μCi of Easy Tag Express-[35S] Protein labeling Mix (NEN) per ml. Labeling was allowed to occur for approximately 2 h.
To harvest the transfected cells, the plates were washed with 10 ml of phosphate-buffered saline, and cells were pelleted at 180 × g for 5 min with a Beckman CS-6KR centrifuge at 4°C. Cell pellets were resuspended in 0.4 ml of buffer A (10 mM Tris-HCl [pH 7.4], 10 mM NaCl, 0.5% NP-40, 1 mM phenylmethylsulfonyl fluoride, 2 μg of antipain per ml, and 2 μg of leupeptin per ml) per plate, and the resulting suspension was sonicated (seven repetitive 20-s treatments). Buffer B (20 mM Tris-HCl [pH 7.4], 1 M NaCl, 0.2% NP-40, 1 mM phenylmethylsulfonyl fluoride, 2 μg of antipain per ml, and 2 μg of leupeptin per ml) (0.6 ml per plate) was then added. The mixture was agitated for 60 min at 4°C followed by centrifugation (at 100,000 × g at 4°C for 60 min). Supernatants were saved.
In vitro ubiquitination of IκBα.
The in vitro ubiquitination of IκBα was carried out as previously described (44), with modifications. Glutathione S-transferase (GST)-IκBα (1 to 54) (3.3 pmol) was phosphorylated by purified IKKβS177E,S181E (0.1 pmol) in a reaction mixture (30 μl) that contained 50 mM Tris-HCl [pH 7.4], 0.6 mM dithiothreitol, 5 mM MgCl2, 2 mM NaF, 10 nM Okadaic Acid, and 50 μM [γ-32P]ATP. After incubation at 37°C for 20 min, SCFHOS-ROC1 (1 pmol) and ATP (to a final concentration of 4 mM) were added to the reaction mixture, and the second incubation was carried out at 0°C for 15 min. Ub (300 pmol), E1 (2 pmol), and mCdc34 (60 pmol or specified otherwise) were then added to the mixture, and the final incubation was at 37°C for times as indicated. The reaction was terminated by the addition of sodium dodecyl sulfate (SDS) loading buffer (15 μl), and the mixture was boiled for 3 min. Aliquots of the reaction products (20 μl) were separated by SDS–8.5% and 6% polyacrylamide gel electrophoresis (PAGE).
Immunoprecipitation.
Extracts, in the amounts indicated, were mixed with 10 μg of αHA (12CA5), αFlag (M2) antibody, or αSkp1 antibody (4 μl). Protein A-agarose beads (10 μl; Upstate Biotechnology) were added. The mixture was agitated for 1 h at 4°C. In experiments shown in Fig. 3B and 5B, protein extracts were mixed with M2-cross-linked antibody beads (15 μl; Sigma), and the mixture was agitated for 2 h at 4°C. Beads were washed three times with 0.5 ml of buffer C (buffer A and B mixed in equal volumes) then were washed two times with 0.5 ml of buffer D (25 mM Tris-HCl [pH 7.5], 1 mM EDTA, 0.01% NP-40, 10% glycerol, and 50 mM NaCl). Bound protein was released by boiling the beads for 3 min in the presence of 40 μl of SDS loading buffer. Twenty microliters of each eluate was used for SDS-PAGE followed by autoradiography.
FIG. 3.
The N terminus of CUL1 binds Skp1, but not ROC1. (A) Immunoprecipitation analysis of the CUL1 N-terminal polypeptide for its interaction with Skp1 and ROC1. 293T cells were transfected with various combinations of full-length Flag-CUL1, Flag-CUL1 (1 to 452), HA-ROC1, and Skp1. 35S-labeled extracts (approximately 0.2 mg of protein) were immunoprecipitated by using αFlag (lanes 1 to 6), αHA (lanes 7 to 12), or αSkp1 (lanes 13 to 14) antibodies, and immunoprecipitates were separated by SDS–12.5% PAGE followed by autoradiography. (B) Immunoprecipitation and immunoblot analyses of the CUL1 N-terminal protein for its interaction with endogenous SCF-ROC1 components. Approximately 1 mg of extract protein was immunoprecipitated by M2 antibody cross-linked beads, and the resulting precipitates were examined by immunoblot analysis for the presence of Flag-tagged CUL1 derivatives, Skp1, HA-tagged ROC1, endogenous ROC1, and β-TRCP.
FIG. 5.
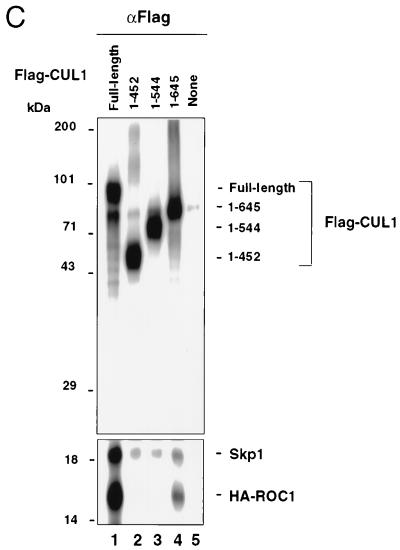
The C terminus of CUL1 interacts with ROC1 in a region that includes the cullin/APC2 consensus motif. (A) Immunoprecipitation analysis of Flag-CUL1 (324 to 776). 293T cells were transfected with HA-ROC1, Flag-CUL1, or Flag-CUL1 (324 to 776), alone or in combination. 35S-labeled extracts (approximately 0.2 mg of protein) were immunoprecipitated with αHA (lanes 1 to 5) or αFlag antibodies (lanes 6 to 10). Immunoprecipitates were separated by SDS–12.5% PAGE followed by autoradiography. (B) Immunoprecipitation and immunoblot analyses of Flag-CUL1 (324 to 776). Approximately 1 mg of extract protein was immunoprecipitated by M2 antibody cross-linked beads, and the resulting precipitates were examined by immunoblot analysis for the presence of Flag-tagged CUL1 derivatives, Skp1, HA-ROC1 and endogenous ROC1. (C) The ROC1 binding domain coincides with the cullin/APC2 homology region. 293T cells were transfected with CMV-based vectors expressing HA-ROC1, Flag-CUL1, Flag-CUL1 (1 to 452), Flag-CUL1 (1 to 544), or Flag-CUL1 (1 to 645), alone or in combination. 35S-labeled extracts (approximately 0.6 mg of protein) were immunoprecipitated with αFlag antibodies, and the immunoprecipitates were separated by SDS–12.5% PAGE. The portion of the gel containing both Skp1 and HA-ROC1 was exposed for four times as long as that containing CUL1.
IκBα degradation.
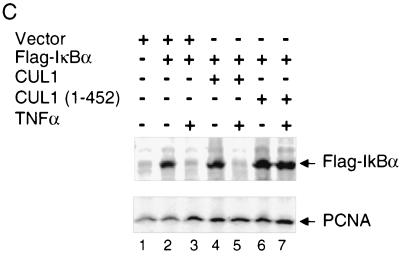
HeLa cells were transfected with 2 μg of CMV vectors expressing Flag-IκBα, CUL1, or CUL1 (1 to 452) and were treated with human tumor necrosis factor alpha (TNFα) (0.5 ng/ml) 24 h later. Cells were harvested 20 min after the addition of TNFα, and the level of Flag-IκBα in whole-cell extracts (100 μg) was analyzed by immunoblot analysis with the M2 monoclonal antibody.
Ub ligation assay.
For the reactions shown in Fig. 4A, the reaction mixture (30 μl) contained 50 mM Tris-HCl (pH 7.4), 5 mM MgCl2, 2 mM NaF, 10 nM Okadaic Acid, 2 mM ATP, 0.6 mM dithiothreitol, 5 μg of 32P-Ub, E1 (2 pmol), mCdc34 (10 pmol), and SCFHOS-ROC1 (0.1 pmol). For experiments shown in Fig. 4B and 6, the immunoprecipitated recombinant ROC1-CUL1 complexes, prepared as described above, were added to a Ub ligation mixture containing the same components as above except for the omission of SCFHOS-ROC1. The mixture was incubated at 37°C for 60 min unless otherwise specified. The reaction mixture was then treated with 20 μl of 4× concentrated Laemmli loading buffer and was boiled for 3 min prior to SDS–12.5% and/or 7.5% PAGE analysis.
FIG. 4.
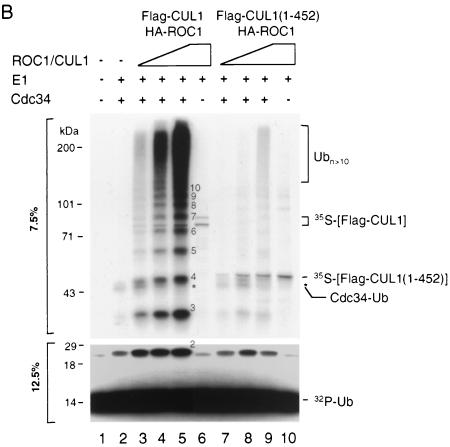
The N terminus of CUL1 is devoid of Ub ligase-activating activity and inhibits TNFα-induced degradation of IκBα. (A) Substrate-independent Ub self-polymerization catalyzed by Cdc34 and SCFHOS/β-TRCP-ROC1. The Ub ligation assay was carried out with indicated components as described in Materials and Methods. Aliquots of the reaction products were separated by SDS–7.5% and 12.5% PAGE. The autoradiogram of the 7.5% gel is shown at the top, while a region of the autoradiogram of the 12.5% gel is shown at the bottom. The numbers on the right indicate the polymerization status of the ligation products. (B) The N terminus of CUL1 is devoid of Ub ligase-activating activity. Three levels of 35S-labeled extracts containing HA-ROC1 and Flag-CUL1 (lanes 3, 4, and 5) or Flag-CUL1 (1 to 452) (lanes 7, 8, and 9), corresponding to 0.07, 0.21, and 0.7 mg of total proteins, respectively, were immunoprecipitated by αFlag antibodies. In each titration set, the molar amounts of full-length CUL1 and the truncated protein in the immunoprecipitates were approximately equal, based on quantitation of the 35S-labeled polypeptides by phosphorimager analysis. The immunoprecipitates were assayed for Ub ligase activity as described in Materials and Methods. Aliquots of the reaction products were separated by SDS–7.5% and 12.5% PAGE. The numbers in the middle indicate the polymerization status of the ligation products. The autoradiogram of the 7.5% gel is shown at the top, while a region of the autoradiogram of the 12.5% gel is shown at the bottom. The autoradiogram from the 7.5% gel was exposed for three times as long as that from the 12.5% gel. (C) Overexpression of CUL1 (1 to 452) inhibits the degradation of IκBα. TNFα-induced degradation of Flag-IκBα in HeLa cells was carried out as described in Materials and Methods. One hundred micrograms of extract proteins was separated by SDS–10% PAGE and was subjected to immunoblot analysis by using M2 antibody.
FIG. 6.
The C terminus of CUL1 fully supports Ub ligation. (A) Flag-CUL1 (324 to 776) complexes with ROC1 to catalyze Ub ligation. The αHA immunoprecipitates of 35S-labeled extracts (approximately 0.3 mg of total protein) containing HA-ROC1 and Flag-CUL1 (lanes 1 to 5) or Flag-CUL1 (324 to 776) (lanes 6 to 10) were assayed for Ub ligase activity (as shown on the left). The reaction was terminated at the time points indicated. The autoradiogram is shown. Equal molar mounts of Flag-CUL1 and Flag-CUL1 (324 to 776) were present in the immunoprecipitates, as determined by phosphorimager analysis of the 35S-labeled polypeptides. Quantitation of Ub ligation products (molecular masses greater than 70 kDa) is shown on the right. (B) Both CUL1 (324 to 776) and HA-ROC1 are required for Ub ligation. HA-ROC1, Flag-CUL1, and Flag-CUL1 (324 to 776), alone or in combination, were transfected into 293T cells. 35S-labeled extracts (approximately 0.3 mg of total protein) were immunoprecipitated with αHA antibodies and were assayed for ligase activity. (C) CUL1 (1 to 544) is inactive in supporting Ub ligation. 35S-labeled extracts containing HA-ROC1/Flag-CUL1 (lane 1) or HA-ROC1/Flag-CUL1 (1 to 544) (lanes 2 and 3) were immunoprecipitated with αFlag antibodies and were assayed for ligase activity. The immunoprecipitates used in lane 2 contained an equal molar amount of Flag-CUL1 (1 to 544) (derived from approximately 0.2 mg of total extract protein) compared to the Flag-CUL1 used in lane 1. A 0.6-mg sample of extract protein containing Flag-CUL1 (1 to 544) was used in lane 3.
RESULTS
Rapid ubiquitination of IκBα catalyzed by the Cdc34– SCFHOS/β-TRCP-ROC1 ligase system in vitro.
We have previously reconstituted the ubiquitination of IκBα in vitro with six purified components that include GST-IκBα (1 to 54) as the substrate, IKKβS177E,S181E, Ub, E1, Cdc34, and SCFHOS/β-TRCP- ROC1 (44). In this reaction, GST-IκBα (1 to 54) was phosphorylated with 32P by IKKβS177E,S181E and was subsequently bound to SCFHOS/β-TRCP-ROC1. Following the addition of Ub, E1, and Cdc34, covalent linkage of Ub to the substrate generated multiple high-molecular-weight substrate-Ub conjugates (Fig. 1A, lane 1) in a substrate-dependent manner (lane 2). The identity of these 32P-labeled, high-molecular-weight reaction products as GST-IκBα (1 to 54)-Ub conjugates was confirmed by anti-GST Western blot analysis (reference 44 and data not shown). To assess the kinetics of this reaction, we examined the efficiency of GST-IκBα (1 to 54) ubiquitination as a function of time. As shown in Fig. 1B, multiple high-molecular-weight substrate-Ub conjugates were readily formed after 3 min (lane 3), and the levels of polyubiquitinated substrates (containing six or more ubiquitin moieties) further increased over time (Fig. 1B, lanes 4 to 6 on the 6% gel, and Fig. 1C, middle and right panels). In contrast, time-dependent accumulation of mono-, di-, or triubiquitinated substrates was not observed (Fig. 1B, lanes 3 to 6), suggesting an efficient Ub chain polymerization by this reconstitution system. GST-IκBα (1 to 54) conjugated with four or five ubiquitin molecules comigrated with the autophosphorylated IKKβS177E,S181E species, precluding an assessment of their production. The reaction required Ub, E1, Cdc34, and SCFHOS/β-TRCP-ROC1 as their removal abolished ubiquitination (Fig. 1B, lane 1).
FIG. 1.
The SCFHOS/β-TRCP-ROC1/Cdc34 ligase system rapidly catalyzes the initial Ub transfer and subsequent polymerization onto phosphorylated IκBα. (A) Substrate-dependent ubiquitination of IκBα. The ubiquitination of GST-IκBα (1 to 54) was carried out as described in Materials and Methods in the presence (lane 1) or absence (lane 2) of GST-IκBα (1 to 54). The reaction was incubated at 37°C for 60 min. (B) Kinetics of the GST-IκBα (1 to 54) ubiquitination reaction. Ubiquitination reactions were carried out with wild-type Ub (lanes 2 to 6) or UbK48R (lanes 7 to 11). The reaction mixture was incubated for the times indicated, and aliquots of the reaction products were separated by both SDS/8.5% and 6% PAGE. The autoradiogram is shown, and the Ub polymerization status of the ubiquitinated GST-IκBα (1 to 54) is indicated. (C) Quantitation of the ubiquitinated GST-IκBα (1 to 54) products. Shown is the phosphorimager quantitation of the levels of each of the ubiquitinated species.
To analyze the initial catalysis leading to the formation of the first isopeptide bond between GST-IκBα (1 to 54) and Ub, we replaced Ub with UbK48R to inhibit Ub polymerization. A monoubiquitinated substrate was formed within 3 min of incubation at a level approximately sevenfold greater than that observed with wild-type Ub (Fig. 1B, compare lanes 3 and 8, also see Fig. 1C, left panel). This substrate-Ub conjugate peaked after 9 min (Fig. 1B, lanes 8 to 11, and Fig. 1C, left panel). Both di- and triubiquitinated GST-IκBα (1 to 54) appeared with slower kinetics compared to the monoubiquitinated product (Fig. 1B, lanes 8 to 11, and Fig. 1C, left panel). Significant levels of polyubiquitinated products (containing six or more ubiquitin moieties), formed through lysine residues other than K48, were only detected after 60 min of incubation (Fig. 1B, lanes 8 to 11 on the 6% gel). However, contrary to the distinct ladder of polyubiquitinated products synthesized with Ub (Fig. 1B, lanes 3 to 6 on the 6% gel), the high-molecular-weight products formed with UbK48R were indiscrete and heterogeneous (Fig. 1B, lanes 8 to 11 on the 6% gel).
These results demonstrate that (i) the initial isopeptide bond formed between GST-IκBα (1 to 54) and a thiol-ester-charged Ub is rapidly catalyzed upon mixing the substrate-E3 complex with Ub, E1, and Cdc34; (ii) once the first Ub is covalently attached, Ub chain polymerization proceeds promptly; and (iii) K48 is the preferred acceptor residue for Ub polymerization catalyzed by the Cdc34–SCFHOS/β-TRCP-ROC1 ligase system. The rapid reaction kinetics observed are in accord with in vivo observations that the proinflammatory cytokine-induced degradation of IκBα is an extremely rapid event (31).
Both Cdc34 and Ubc4 and -5 support ubiquitination of IκBα in vitro.
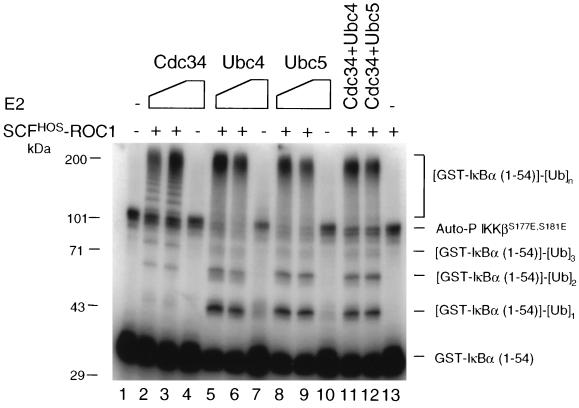
We compared Cdc34 and Ubc4 and -5 for their ability to support SCFHOS/β-TRCP-ROC1-catalyzed ubiquitination of IκBα by using the purified reconstitution system. Consistent with observations in Fig. 1, Cdc34 efficiently catalyzed the covalent linkage of polyubiquitin chains to the substrate in a concentration-dependent manner (Fig. 2, lanes 2, 3, and 13). Addition of Ubc4 (lanes 5 and 6) or Ubc5 (lanes 8 and 9) in place of Cdc34 also supported the ubiquitination of GST-IκBα (1 to 54). However, several differences were observed. First, incubation with Ubc4 or -5 resulted in more substrates converted into Ub conjugates than did incubation with Cdc34 (compare lanes 5 and 8 with lane 2), suggesting that Ubc4 and -5 promoted more-efficient substrate utilization. This is in keeping with previously published observations (30). Second, Ubc4 and -5 produced a greater abundance of both the mono- and diubiquitinated GST-IκBα (1 to 54) species than did Cdc34 (compare lanes 5, 6, 8, and 9 with lanes 2 and 3). This is consistent with the recent report that Cdc34 supported polyubiquitination of an IκBα peptide (residues 20 to 43) in the presence of the [β-TRCP]-containing SCF complex, whereas Ubc5 predominantly stimulated mono- and diubiquitination (46). These results suggest that Ubc4 or -5 generates mono- and diubiquitinated IκBα more efficiently than Cdc34. However, Cdc34 and Ubc4 or -5 appear to be equally efficient in producing polyubiquitinated species. Third, both Ubc4 and Ubc5, but not Cdc34, appeared to catalyze low levels of monoubiquitination of the substrate in the absence of SCFHOS/β-TRCP-ROC1 (compare lanes 7 and 10 with lane 4). This is consistent with the previous finding that yeast Ubc4 alone catalyzed the formation of low-molecular-mass ubiquitinated conjugates of IκBα (8). These results suggest that both Ubc4 and Ubc5 may bind nonspecifically to the phosphorylated GST-IκBα (1 to 54) and catalyze its monoubiquitination in an E3-independent manner. Fourth, while the high-molecular-weight Ub conjugates (containing six or more Ub moieties) (Fig. 1) formed with Cdc34 migrated as a protein ladder (lanes 2 and 3), those produced by Ubc4 (lanes 5 and 6) or Ubc5 (lanes 8 and 9) appeared as heterogeneous species. Fifth, a significant portion of autophosphorylated IKKβS177E,S181E was converted into high-molecular-weight species by Ubc4 and -5, but not by Cdc34 (compare lanes 5, 6, 8, and 9 with lanes 2 and 3). Thus, Ubc4 and -5 may also catalyze the ubiquitination of autophosphorylated IKKβS177E,S181E by unknown mechanisms. Lastly, mixing of Cdc34 with Ubc4 (lane 11) or Ubc5 (lane 12) produced conjugate patterns identical to those observed with Ubc4 or -5 alone, suggesting that Ubc4 and -5 acted in a dominant fashion under the conditions used.
FIG. 2.
Comparison of Cdc34 and Ubc4 and -5 in their ability to support the ubiquitination of IκBα. The reaction mixture contained 20 (lanes 2, 5, and 8) or 60 (lanes 3, 6, and 9) pmol of E2 as indicated, as well as other components as described in Materials and Methods. In reactions shown in lanes 11 and 12, 20 pmol of Cdc34 was mixed with 20 pmol of either Ubc4 or Ubc5. No ubiquitination agents (Ub, E1, E2, or SCFHOS/β-TRCP-ROC1) were added to the reaction shown in lane 1. SCFHOS/β-TRCP-ROC1 was omitted in lanes 4, 7, and 10, while E2 was omitted in lane 13. The reaction was incubated for 60 min, and the products were separated by SDS–8.5% PAGE. The autoradiogram is shown.
Both Cdc34 and Ubc4 and -5 have been reported to function as an E2 for the ubiquitination of IκBα (8, 13, 30, 42, 44, 46, 50). By expressing dominant negative mutants of E2s, Gonan et al. (13) showed that both Ubc5 and Cdc34 were involved in signal-induced ubiquitination and degradation of IκBα. While the above results are in accord with previous studies, they did demonstrate that Cdc34 and Ubc4 and -5 produce distinct conjugate patterns. Cdc34 appeared to predominantly promote synthesis of polyubiquitin chains, but not mono- and diubiquitinated substrate species. However, Ubc4 and -5 seemed to be more efficient than Cdc34 in engaging the ubiquitination of IκBα, albeit resulting in the accumulation of mono- and diubiquitin conjugates. It is presently not understood whether a mechanism exists in cells to coordinate these two different classes of E2s to catalyze the efficient and extensive polyubiquitination of IκBα.
The N terminus of CUL1 binds Skp1, but not ROC1, and is inactive in supporting Ub ligation.
The above results establish that SCFHOS/β-TRCP-ROC1 coordinates with Cdc34 to catalyze polyubiquitination reactions. The fact that the Ub ligase activity resides within the ROC1-CUL1 subcomplex of the E3 holoenzyme (30, 38, 44) implies that the core ligase module must interact with Cdc34 to catalyze efficient Ub transfer and chain polymerization. To determine structural domains required for the assembly of the ROC1-CUL1 core Ub ligase, we employed deletion analysis to define regions within CUL1 required for interacting with Skp1 and ROC1, as well as for the activation of the Ub ligase activity.
Treatment of extracts of 293T cells containing 35S-labeled Flag-CUL1, HA-ROC1, and Skp1 with either anti-Flag (Fig. 3A, lane 1) or anti-HA (lane 7) antibodies resulted in coprecipitation of all three proteins. This result is consistent with previous findings indicating that CUL1/Cdc53 is capable of simultaneously binding both Skp1 and ROC1 (19, 30, 38, 40, 44). However, anti-Flag immunoprecipitation of extracts containing overexpressed HA-ROC1, Skp1, and a Flag-tagged CUL1 N-terminal fragment (spanning amino acids 1 to 452) yielded Skp1, but not HA-ROC1 (lane 2). This finding was confirmed by a reciprocal immunoprecipitation experiment with anti-HA antibodies, which failed to detect Flag-CUL1 (1 to 452) or Skp1 (lane 8). Thus, ROC1 interacts with neither Skp1 nor the N terminus of CUL1.
Several additional points were revealed by this immunoprecipitation experiment. First, the full-length CUL1 protein interacted with HA-ROC1 in the absence of overexpressed recombinant Skp1 (lanes 3 and 9). Second, CUL1 bound Skp1 regardless of the presence of ROC1 (lane 4). Third, Flag-CUL1 (1 to 452) formed a complex with endogenous Skp1 (lane 5), consistent with the previous finding that the N terminus of Cdc53 is required for Skp1 binding (32). Detection of the interaction between CUL1 (1 to 452) or CUL1 and endogenous Skp1 (Fig. 3B) is due to the presence of high levels of Skp1 in 293T cells (54). Lastly, immunoprecipitation with anti-Skp1 antibodies confirmed a direct interaction between Skp1 and CUL1 (lane 14) or CUL1 (1 to 452) (lane 13).
Further immunoprecipitation and immunoblot analyses were carried out to better assess the ability of the recombinant CUL1 or CUL1 (1 to 452) to interact with the endogenous SCF-ROC1 components. As shown in Fig. 3B, both CUL1 and CUL1 (1 to 452) interacted with Skp1 independent of transfection of the Skp1-expressing plasmid. In addition, they were both associated with the endogenous β-TRCP, which appeared as a doublet of proteins at 58 to 60 kDa (47). In cells transiently expressing Flag-CUL1 and HA-ROC1, β-TRCP was complexed with Flag-CUL1 in levels significantly higher than those derived from other transfected cells (compare lane 3 with the rest). The reason for this discrepancy is not clear. Contrary to their comparable capacity to interact with both Skp1 and β-TRCP, the full-length CUL1, but not CUL1 (1 to 452), was able to bind both recombinant and endogenous ROC1.
Taken together, these results demonstrate that CUL1 interacts with Skp1 and ROC1 through distinct domains and that the N terminus of CUL1 is both necessary and sufficient for binding Skp1. Therefore, CUL1 recruits the F-box substrate-targeting protein through Skp1 at its N terminus.
We next examined the ability of the full-length and N-terminal CUL1s to support substrate-independent Ub ligation by using a 32P-Ub-incorporation assay as previously established (44). Incubation of purified E1, Cdc34, and SCFHOS/β-TRCP-ROC1 with [32P]-Ub resulted in the formation of a protein ladder of 32P-labeled Ub polymers in an E1-Cdc34-dependent manner (Fig. 4A, lanes 1 to 4), consistent with previously reported observations (44). As shown, the synthesized Ub polymers, ranging in size from dimers to octamers, migrated identically to those produced by E2-25K/E1 (compare lanes 4 and 5). E2-25K is known to be capable of polymerizing unanchored Ub chains (6). Of note, the addition of SCFHOS/β-TRCP-ROC1 did not further enhance the ability of E2-25K to catalyze Ub polymerization (data not shown), suggesting that these two enzymes do not cooperate. These results thus confirm that the human SCFHOS/β-TRCP-ROC1–Cdc34 ligase system catalyzes Ub self-polymerization. Significant cellular levels of unanchored polyubiquitin chains have been previously identified (14, 41, 45). The mechanism and biological significance of this self-ligation reaction is presently unclear. However, this reaction obviates the requirement for a specific substrate, thus providing a sensitive assay to measure the Ub ligase activity by Cdc34 and ROC1-CUL1.
It has been reported that in yeast, ROC1 complexed with either SCF or Cdc53 (CUL1 homologue) alone, can catalyze extensive autoubiquitination of Cdc34 (2, 38, 40). However, only low levels of autoubiquitination of Cdc34 by the human SCFHOS/β-TRCP-ROC1 complex were detected by immunoblot analysis (data not shown). Nevertheless, we cannot rule out that the high-molecular-weight Ub ligation products (with molecular weight greater than that of Ub9) contained autoubiquitinated Cdc34.
The addition of a reaction mixture containing 32P-Ub, E1, and Cdc34 to the 35S-labeled HA-ROC1/Flag-CUL1 complex, immobilized to anti-Flag-protein A beads, resulted in the synthesis of 32P-labeled Ub polymers (Fig. 4B, lanes 1 to 5). The synthesis of Ub polymers by ROC1-CUL1 required the presence of Cdc34 (lane 6). Substitution of the HA-ROC1/Flag-CUL1 complex with the Flag-CUL1 (1 to 452) immunoprecipitates abolished Ub polymerization reaction (Fig. 4B, lanes 7 to 9). These results demonstrate that the N terminus of CUL1, incapable of binding ROC1, is also catalytically inactive in supporting Ub ligation.
We have previously established that the SCF components HOS/β-TRCP and CUL1 are required for the degradation of IκB in HeLa cells induced by TNFα (12). The results shown in the present study indicate that CUL1 and CUL1 (1 to 452) interacted with endogenous SCF-ROC1 components to form SCFβ-TRCP-ROC1 or SCFβ-TRCP complexes lacking ROC1, respectively (Fig. 3B). Based on our previous finding that the SCFHOS/β-TRCP complex lacking ROC1 binds IκBα but does not support IκBα ubiquitination (44), we reasoned that expression of CUL1 (1 to 452) in cells would block the degradation of IκB. To test this possibility, we examined the levels of IκBα by immunoblot analysis in TNFα-induced cells that were transfected with CMV-based vectors expressing Flag-IκBα and either the full-length or the N-terminal fragment (amino acids 1 to 452) of CUL1. The results indicate that TNFα induced degradation of IκBα in the presence of vector alone (Fig. 4C, upper panel, lanes 2 and 3) or recombinant CUL1 (lanes 4 and 5). This result suggests that the assembled SCFβ-TRCP-ROC1 complex containing the recombinant full-length CUL1 detected in Fig. 3B is functional in promoting the IκBα degradation. In contrast, introduction of CUL1 (1 to 452) into cells significantly reduced Flag-IκBα degradation (lanes 6 and 7). Under these conditions, the levels of PCNA remained unchanged (Fig. 4C, lower panel), excluding the possibility of any nonspecific effects that might influence the levels of IκBα detected. Furthermore, overexpression of CUL1 (1 to 452) inhibited the degradation of endogenous IκBα as well (data not shown). These results establish that CUL1 (1 to 452) exerts a dominant negative effect by forming SCF complexes lacking ROC1 that bind phosphorylated IκB but which are unable to catalyze its ubiquitination.
These data collectively demonstrate that the N terminus of CUL1 provides an anchorage site for Skp1, which recruits a given F-box protein, capable of targeting its cognate substrates. This region alone, however, is devoid of ROC1-binding activity and, hence, is inactive in supporting Ub ligation and degradation of a physiological substrate.
ROC1 forms a complex with the C terminus of CUL1, that includes the cullin/APC2 consensus region, to catalyze Ub polymerization.
To determine the role of the C terminus of CUL1 in ubiquitination, we expressed a Flag-tagged CUL1 fragment spanning amino acids 324 to 776 in 293T cells. Immunoprecipitation with anti-Flag antibodies detected this C-terminal species, which migrated as a doublet at apparent molecular weights of ∼50 and 60 kDa (Fig. 5A, lane 7). Further experiments are required to determine whether this heterogeneity derives from protein modification or degradation. When Flag-CUL1 (324 to 776) and HA-ROC1 were coexpressed, the two formed a complex, as demonstrated by reciprocal immunoprecipitations with either anti-HA (lane 5) or anti-Flag (lane 10) antibodies. Phosphorimager analysis indicated that Flag-CUL1 (324 to 776) bound to ROC1 with an efficiency equal to that observed with the full-length CUL1 (lanes 4 and 9). Like the full-length protein, Flag-CUL1 (324 to 776) interacted with ROC1 in both recombinant and endogenous forms (Fig. 5B, lanes 2 and 3). Furthermore, this C-terminal polypeptide did not bind Skp1. In contrast, Flag-CUL1 (1 to 452) interacted with Skp1, but not ROC1 (Fig. 5B, lane 4), which is consistent with observations shown in Fig. 3B. As expected, anti-Flag immunoprecipitates derived from untransfected cells did not contain SCF-ROC1 components (Fig. 5B, lane 1).
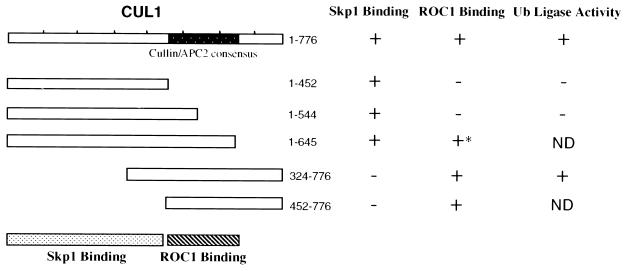
Previously described sequence analysis identified a cullin/Cdc53/APC2 region of homology at the C terminus spanning approximately 200 amino acids (the cullin box) (51, 52). In CUL1, the cullin box is located between amino acids 450 and 650. The apparent overlap between the cullin box and the ROC1-binding domain (Fig. 5A) prompted us to examine the intriguing possibility that this conserved cullin box is the region responsible for ROC1 binding. For this purpose, we expressed two additional Flag-tagged truncated CUL1 polypeptides spanning amino acids 1 to 544 and 1 to 645, respectively, in 293T cells. Immunoprecipitation experiments with anti-Flag antibodies indicate that both truncated proteins interacted specifically with endogenous Skp1, though each truncated CUL1 protein was bound to Skp1 with lower efficiency than that of the wild type (Fig. 5C, lanes 1 to 5). However, while full-length CUL1 and Flag-CUL1 (1 to 645) were found complexed with both Skp1 and HA-ROC1 (Fig. 5C, lanes 1 and 4, respectively), neither Flag-CUL1 (1 to 452) nor Flag-CUL1 (1 to 544) interacted with ROC1 (lanes 2 and 3, respectively). Of note, Flag-CUL1 (1 to 645) bound to HA-ROC1 with an efficiency 3.3-fold lower than that of the conjugate containing the full-length CUL1 (compare lanes 1 and 4; determined by phosphorimager analysis), suggesting that the C-terminal 130 amino acids of CUL1 may play a role in stabilizing the ROC1 binding. Furthermore, a truncated CUL1 protein (amino acids 452 to 776) was found to be able to bind ROC1 (data not shown), suggesting that a sequence stretch spanning amino acids 324 to 452 (upstream of the cullin box) is also dispensable for ROC1 binding. Taken together, these results suggest that the ROC1-binding domain coincides with the conserved cullin box (see Fig. 7).
FIG. 7.
Structural domains within the human CUL1 protein. A schematic representation of the truncated human CUL1 polypeptides analyzed is shown on the left. Structural and functional features of CUL1 are also shown. A summary of Skp1 and ROC1 binding as well as the Ub ligase activity of full-length and truncated CUL1 polypeptides is shown on the right. Note that CUL1 (1 to 645), marked by an asterisk, binds to ROC1 threefold less efficiently than the full-length protein (see Fig. 5C).
The rate of synthesis of Ub ligation products was examined and compared in the presence of immunopurified HA-ROC1/Flag-CUL1 (Fig. 6A, left panel, lanes 1 to 5) or HA-ROC1/Flag-CUL1 (324 to 776) (left panel, lanes 6 to 10) complex. With HA-ROC1/Flag-CUL1, multiple high-molecular-weight species were formed after 3 min of incubation and increased over time (left panel, lanes 2 to 5). An approximately 50%-higher rate of Ub ligation was observed with HA-ROC1/Flag-CUL1 (324 to 776) in comparison to HA-ROC1/Flag-CUL1 (left panel, lanes 7 to 10; see right panel for quantitation). Further experiments indicated that the presence of both ROC1 and CUL1 or CUL (324 to 776) is required for the assembly of the active Ub ligase complex (Fig. 6B). Low levels of Ub ligase activity observed with extracts from cells transfected with HA-ROC1 alone (Fig. 6B, lane 3) are due to the presence of a small amount of endogenous CUL1 in the immunoprecipitates (data not shown). Furthermore, CUL1 (1 to 544), which was incapable of binding ROC1, was also inactive in supporting Ub ligation (Fig. 6C), suggesting that the integrity of the cullin box is required for both ROC1 binding and Ub ligase activation. The low expression of CUL (1 to 645) and CUL1 (452 to 776) in transfected 293T cells (approximately 10-fold lower than the expression of the full-length CUL1) failed to yield levels of protein sufficient for measuring Ub ligase activity.
We conclude from these results that ROC1 binds the C terminus of CUL1 (amino acids 324 to 776) and that this complex is sufficient to catalyze Ub ligation. The interaction of these two proteins is most likely mediated by direct contact between ROC1 and the cullin box within CUL1. However, further experiments are required to determine whether the cullin box alone is sufficient to activate Ub ligase activity.
DISCUSSION
The kinetic analysis in the present study reveals that the SCFHOS/β-TRCP-ROC1–Cdc34 ligase system rapidly catalyzes the initial Ub transfer and subsequent polymerization onto a model substrate, phosphorylated IκBα. While both Cdc34 and Ubc4 and -5 support IκBα ubiquitination, they produce distinct conjugate patterns. Cdc34 appeared to predominantly promote the synthesis of polyubiquitin chains, but not mono- and diubiquitinated substrate species. However, Ubc4 and -5 seemed to be more efficient in engaging the ubiquitination of IκBα when compared to Cdc34, albeit resulting in the accumulation of mono- and diubiquitin conjugates.
Our previous reconstitution experiments have revealed that Skp1 is indispensable for tethering HOS/β-TRCP to CUL1 (44). The results of deletion analysis indicate that the N terminus of CUL1 (amino acids 1 to 452) is both necessary and sufficient for binding Skp1, and hence HOS/β-TRCP. Consistent with this, introduction of the N-terminal CUL1 polypeptide into cells blocks the TNFα-induced and SCF-mediated degradation of IκB by disrupting the endogenous SCF-ROC1 complexes. These observations confirm and extend the previous finding that deletion of Cdc53 (residues 9 to 280) completely disrupts Skp1–F-box protein binding in budding yeast cells (32). Our mapping analysis has localized the C terminus of CUL1 (amino acids 342 to 776) as the region responsible for interaction with ROC1, resulting in the assembly of a complex fully active in supporting ubiquitin polymerization. This interaction is most likely mediated by a direct contact between ROC1 and the cullin/APC2 consensus motif. Taken together, these results suggest the mode of action of SCF-ROC1 in ubiquitination (see Fig. 8A). In this model, CUL1 serves as a dual-function molecule: the N terminus binds Skp1, which then recruits the F-box protein for targeting a phosphorylated substrate, and the C-terminal region, including the cullin box, binds ROC1, leading to the assembly of a Ub ligase, which interacts with Cdc34 to catalyze Ub transfer and polymerization.
FIG. 8.

(A) Proposed structural organization of SCF-ROC1 and interactions between components of the E3 complex and the substrate as well as Cdc34. (B) A proposed molecular framework for the ROC-APC11-dependent Ub ligases. Note that the VCB-CUL2-ROC1 complex refers to the pVHL tumor suppressor complex containing pVHL, elongins C and B, CUL2, and ROC1. See the text for a description of the model presented.
Mutagenesis studies have shown that the conserved RING-H2 residues within ROC1 are critical for ubiquitination (19, 30, 40), implicating a role of the RING finger structure in contacting Cdc34. More-recent studies have uncovered a large number of RING-finger-containing proteins, including c-Cbl (18) and BRCA1 (25), that possess Ub ligase activity. As shown in Fig. 6C, ROC1 and CUL1 are both important for ubiquitin ligase activity. The identification of the core ligase formed between ROC1 and the C terminus of CUL1 should enable further detailed structural analysis, leading to the elucidation of the enzymatic action of the ubiquitin ligation reaction.
Previous studies have established that the human ROC1 binds five members of the cullin family (CUL1, CUL2, CUL3, CUL4A, and CUL4B), while ROC2 preferentially interacts with CUL5 (30). In addition, the ROC homologue APC11 binds APC2 (30). The localization of the ROC1-binding domain to the cullin box of CUL1 suggests a structural conservation in the assembly of a core ROC/APC11-cullin/APC2 complex through this conserved region. Furthermore, our data strongly suggests the interaction between ROC1 and the cullin box of CUL1 is required for the activation of the Ub ligase activity (Fig. 7), implying that other ROC-CUL complexes may contain Ub ligase activity as well. Indeed, both ROC1-CUL2 (unpublished data) and APC11-APC2 (30), assembled in transfected cells, are capable of catalyzing Ub ligation reactions in vitro.
We, therefore, propose a unified framework depicting the action of the ROC/APC11-dependent super-family of E3 ligases (Fig. 8B). Members of this E3 class each contain a common dimeric core Ub ligase element formed by combinatorial interactions between the ROC/APC11 and CUL/APC2 family proteins (Fig. 8, left). This core ligase module is recruited, via an adapter protein, to form a complex with a substrate-targeting molecule (Fig. 8B, middle). The resulting multisubunit E3 holoenzyme binds the substrate and catalyzes its ubiquitination in the presence of an E2 (Fig. 8B, right). The Skp1–F-box based SCF model is one such pathway that recruits the ROC1-CUL1 core Ub ligase to catalyze the ubiquitination of phosphorylated substrate proteins, such as IκBα. There is accumulating evidence that this mechanism may also apply to the pVHL complex. pVHL, which targets HIF α-subunits for oxygen-dependent degradation (27), forms a complex that structurally resembles the SCF-ROC1 complex (19, 43) and also contains Ub ligase activity (23). More recently, the N terminus of CUL2 has been shown to bind the elongin BC dimer, which interacts with pVHL (34). The underlying similarity between CUL2 and CUL1 (this study) suggests a general dual functional role for cullin molecules in substrate targeting and Ub ligation. Using the N terminus of these cullin molecules as bait may identify Skp1-like molecules that link respective substrate-targeting molecules, leading to the identification of novel ubiquitination and degradation pathways. Lastly, APC, comprised of 8 to 12 subunits (35), exhibits a greater degree of structural complexity than SCF-ROC1. Understanding the assembly of this E3 ligase requires a more detailed biochemical characterization of the APC subunits.
ACKNOWLEDGMENTS
We thank T. Ohta, Y. Xiong, and G. Fang for providing purified Ubc5 and Ubc4 proteins; J. W. Harper for providing β-TRCP antibodies; J. Hurwitz for helpful discussion; and S. Santagata for critical reading of the manuscript.
Z.-Q.P. was supported by the Life and Health Insurance Medical Research Fund, the New York Community Trust, and the Irma T. Hirschl Award. This study was supported by Public Health Service grants CA78419 to Z.R. and GM55059 to Z.-Q.P.
REFERENCES
- 1.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. β-Catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banerjee A, Gregori L, Xu Y, Chau V. The bacterially expressed yeast cdc34 gene product can undergo autoubiquitination to form a multiubiquitin chain-linked protein. J Biol Chem. 1993;268:5668–5675. [PubMed] [Google Scholar]
- 3.Beg A A, Ruben S M, Scheinman R I, Haskill S, Rosen C A, Baldwin A S., Jr IκB interacts with the nuclear localization sequences of the subunits of NF-κB: a mechanism for cytoplasmic retention. Genes Dev. 1992;6:1899–1913. doi: 10.1101/gad.6.10.1899. [DOI] [PubMed] [Google Scholar]
- 4.Beg A A, Finco T S, Nantermet P V, Baldwin A S J. Tumor necrosis factor and interleukin-1 lead to phosphorylation and loss of IκB: a mechanism for NF-κB activation. Mol Cell Biol. 1993;13:3301–3310. doi: 10.1128/mcb.13.6.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown K, Gerstberger S, Carlson L, Fransozo G, Siebenlist U. Control of IκB proteolysis by site-specific, signal-induced phosphorylation. Science. 1995;267:1485–1488. doi: 10.1126/science.7878466. [DOI] [PubMed] [Google Scholar]
- 6.Chen Z, Pickart C M. A 25-kilodalton ubiquitin carrier protein (E2) catalyzes multi-ubiquitin chain via K48 of ubiquitin. J Biol Chem. 1990;265:21835–21842. [PubMed] [Google Scholar]
- 7.Chen Z, Hagler J, Palombella V J, Melandri F, Scherer D, Ballard D, Maniatis T. Signal-induced site-specific phosphorylation targets IκB to the ubiquitin-proteasome pathway. Genes Dev. 1995;9:1586–1597. doi: 10.1101/gad.9.13.1586. [DOI] [PubMed] [Google Scholar]
- 8.Chen Z J, Parent L, Maniatis T. Site-specific phosphorylation of IkappaBalpha by a novel ubiquitination-dependent protein kinase activity. Cell. 1996;84:853–862. doi: 10.1016/s0092-8674(00)81064-8. [DOI] [PubMed] [Google Scholar]
- 9.DiDonato J A, Hayakawa M, Rothwarf D M, Zandi E, Karin M. A cytokine-responsive IκB kinase that activates the transcription factor NF-κB. Nature. 1997;388:548–554. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- 10.Duan H, Wang Y, Aviram M, Swaroop M, Loo J A, Bian J, Tian Y, Mueller T, Bisgaier C L, Sun Y. SAG, a novel zinc RING finger protein that protects cells from apoptosis induced by redox agents. Mol Cell Biol. 1999;19:3145–3155. doi: 10.1128/mcb.19.4.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feldman R M R, Correll C C, Kaplan K B, Deshaies R J. A complex of Cdc4p, Skp1p, and Cdc53p/Cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 12.Fuchs S Y, Chen A, Xiong Y, Pan Z-Q, Ronai Z. HOS, a human homologue of Slimb, forms an SCF complex with Skp1 and Cullin 1 and targets the phosphorylation-dependent degradation of IκB and β-catenin. Oncogene. 1999;18:2039–2046. doi: 10.1038/sj.onc.1202760. [DOI] [PubMed] [Google Scholar]
- 13.Gonen H, Bercovich B, Orian A, Carrano A, Takizawa C, Yamanaka K, Pagano M, Iwai K, Ciechanover A. Identification of the ubiquitin carrier proteins, E2s, involved in signal-induced conjugation and subsequent degradation of IkappaBalpha. J Biol Chem. 1999;274:14823–14830. doi: 10.1074/jbc.274.21.14823. [DOI] [PubMed] [Google Scholar]
- 14.Halderman M T, Finley D, Pickart C M. Dynamics of ubiquitin conjugation during erythroid differentiation in vitro. J Biol Chem. 1995;270:9507–9516. doi: 10.1074/jbc.270.16.9507. [DOI] [PubMed] [Google Scholar]
- 15.Hatakeyama S, Kitagawa M, Nakayama K, Shirane M, Matsumoto M, Hattori K, Higashi H, Nakano H, Okumura K, Onoe K, Good R A, Nakayama K. Ubiquitin-dependent degradation of IkappaBalpha is mediated by a ubiquitin ligase Skp1/Cul 1/F-box protein FWD1. Proc Natl Acad Sci USA. 1999;96:3859–3863. doi: 10.1073/pnas.96.7.3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hershko A, Heller H, Elias S, Ciechanover A. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J Biol Chem. 1983;258:8206–8214. [PubMed] [Google Scholar]
- 17.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 18.Joazeiro C A, Wing S S, Huang H, Leverson J D, Hunter T, Liu Y C. The Tyrosine kinase negative regulator c-Cbl as a RING-Type, E2-Dependent ubiquitin-protein ligase. Science. 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- 19.Kamura T, Koepp D M, Conrad M N, Skowyra D, Moreland R J, Iliopoulos O, Lane W S, Kaelin W G, Jr, Elledge S J, Conaway R C, Harper J W, Conaway J W. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science. 1999;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- 20.Kitagawa K, Skowyra D, Elledge S J, Harper J W, Hieter P. SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol Cell. 1999;4:21–33. doi: 10.1016/s1097-2765(00)80184-7. [DOI] [PubMed] [Google Scholar]
- 21.Laney J D, Hochstrasser M. Substrate targeting in the ubiquitin system. Cell. 1999;97:427–430. doi: 10.1016/s0092-8674(00)80752-7. [DOI] [PubMed] [Google Scholar]
- 22.Latres E, Chiaur D S, Pagano M. The human F box protein beta-Trcp associates with the Cul1/Skp1 complex and regulates the stability of β-catenin. Oncogene. 1999;18:849–854. doi: 10.1038/sj.onc.1202653. [DOI] [PubMed] [Google Scholar]
- 23.Lisztwan J, Imbert G, Wirbelauer C, Gstaiger M, Krek W. The von Hippel-Lindau tumor suppressor protein is a component of an E3 ubiquitin-protein ligase activity. Genes Dev. 1999;13:1822–1833. doi: 10.1101/gad.13.14.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lonergan K M, Iliopoulos O, Ohh M, Kamura T, Conaway R C, Conaway J W, Kaelin W G., Jr Regulation of hypoxia-inducible mRNAs by the von Hippel-Lindau tumor suppressor protein requires binding to complexes containing elongins B/C and Cul2. Mol Cell Biol. 1998;18:732–741. doi: 10.1128/mcb.18.2.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lorick K L, Jensen J P, Fang S, Ong A M, Hatakeyama S, Weissman A M. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maniatis T. A ubiquitin ligase complex essential for the NF-kappaB, Wnt/Wingless, and Hedgehog signaling pathways. Genes Dev. 1999;13:505–510. doi: 10.1101/gad.13.5.505. [DOI] [PubMed] [Google Scholar]
- 27.Maxwell P H, Wiesener M S, Chang G W, Clifford S C, Vaux E C, Cockman M E, Wykoff C C, Pugh C W, Maher E R, Ratcliffe P J. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 28.Mercurio F, Zhu H, Murray B W, Shevchenko A, Bennett B L, Li J, Young D B, Barbosa M, Mann M. IKK-1 and IKK-2: cytokine-activated IκB kinases essential for NF-κB activation. Science. 1997;278:860–866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- 29.Michel J, Xiong Y. Human CUL-1, but not other cullin family members, selectively interacts with SKP1 to form a complex with SKP2 and cyclin A. Cell Growth Differ. 1998;9:439–445. [PubMed] [Google Scholar]
- 30.Ohta T, Michel J J, Schottelius A J, Xiong Y. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol Cell. 1999;3:535–541. doi: 10.1016/s1097-2765(00)80482-7. [DOI] [PubMed] [Google Scholar]
- 31.Palombella V J, Rando O J, Goldberg A L, Maniatis T. The ubiquitin proteasome pathway is required for processing the NF-kappa B1 precursor protein and the activation of NF-kappa B. Cell. 1994;78:773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 32.Patton E E, Willems A, Sa D, Kuras L, Thomas D, Craig K L, Tyers M. Cdc53 is a scaffold protein for multiple Cdc34/Skp1/F-box protein complexes that regulate cell division and methionine biosynthesis in yeast. Genes Dev. 1998;12:692–705. doi: 10.1101/gad.12.5.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pause A, Lee S, Worrell R A, Chen D Y, Burgess W H, Linehan W M, Klausner R D. The von Hippel-Lindau tumor-suppressor gene product forms a stable complex with human CUL-2, a member of the Cdc53 family of proteins. Proc Natl Acad Sci USA. 1997;94:2156–2161. doi: 10.1073/pnas.94.6.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pause A, Peterson B, Schaffar G, Stearman R, Klausner R D. Studying interactions of four proteins in the yeast two-hybrid system: structural resemblance of the pVHL/elongin BC/hCUL-2 complex with the ubiquitin ligase complex SKP1/cullin/F-box protein. Proc Natl Acad Sci USA. 1999;96:9533–9538. doi: 10.1073/pnas.96.17.9533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peters J-M, King R W, Deshaies R. Cell cycle control by ubiquitin-dependent proteolysis. In: Peters J-M, Harris J R, Finley D, editors. Ubiquitin and the biology of the cell. New York, N.Y: Plenum Press; 1998. pp. 345–387. [Google Scholar]
- 36.Regnier C H, Song H Y, Gao X, Goeddel D V, Cao Z, Rothe M. Identification and characterization of an IκB kinase. Cell. 1997;90:373–383. doi: 10.1016/s0092-8674(00)80344-x. [DOI] [PubMed] [Google Scholar]
- 37.Scherer D C, Brockman J A, Chen Z, Maniatis T, Ballard D W. Signal-induced degradation of IκB requires site-specific ubiquitination. Proc Natl Acad Sci USA. 1995;92:11259–11263. doi: 10.1073/pnas.92.24.11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seol J H, Feldman R M, Zachariae W, Shevchenko A, Correll C C, Lyapina S, Chi Y, Galova M, Claypool J, Sandmeyer S, Nasmyth K, Deshaies R J. Cdc53/cullin and the essential hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module hat activates the E2 enzyme cdc34. Genes Dev. 1999;13:1614–1626. doi: 10.1101/gad.13.12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skowyra D, Craig K, Tyers M, Elledge S J, Harper J W. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 40.Skowyra D, Koepp D M, Kamura T, Conrad M N, Conaway R C, Conaway J W, Elledge S J, Harper J W. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and rbx1. Science. 1999;284:662–665. doi: 10.1126/science.284.5414.662. [DOI] [PubMed] [Google Scholar]
- 41.Spence J, Sadis S, Haas A L, Finley D. A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol Cell Biol. 1995;15:1265–1273. doi: 10.1128/mcb.15.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spencer E, Jiang J, Chen Z J. Signal-induced ubiquitination of IκBα by the F-box protein Slimb/β-TrCP. Genes Dev. 1999;13:284–294. doi: 10.1101/gad.13.3.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stebbins C E, Kaelin W G, Jr, Pavletich N P. Structure of the VHL-ElonginC-ElonginB complex: implications for VHL tumor suppressor function. Science. 1999;284:455–461. doi: 10.1126/science.284.5413.455. [DOI] [PubMed] [Google Scholar]
- 44.Tan P, Fuchs S Y, Chen A, Wu K, Gomez C, Ronai Z, Pan Z Q. Recruitment of a ROC1-CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of IκBα. Mol Cell. 1999;3:527–533. doi: 10.1016/s1097-2765(00)80481-5. [DOI] [PubMed] [Google Scholar]
- 45.Van Nocker S, Vierstra R D. Multiubiquitin chains linked through lysine 48 are abundant in vivo and are competent intermediates in the ubiquitin proteolytic pathway. J Biol Chem. 1993;268:24766–24773. [PubMed] [Google Scholar]
- 46.Vuillard L, Nicholson J, Hay R T. A complex containing betaTrCP recruits Cdc34 to catalyze ubiquitination of IkappaBalpha. FEBS Lett. 1999;455:311–314. doi: 10.1016/s0014-5793(99)00895-9. [DOI] [PubMed] [Google Scholar]
- 47.Winston J T, Strack P, Beer-Romero P, Chu C Y, Elledge S J, Harper J W. The SCFβ-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IκBα and β-catenin and stimulates IκBα ubiquitination in vitro. Genes Dev. 1999;13:270–283. doi: 10.1101/gad.13.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woronicz J D, Gao X, Cao Z, Rothe M, Goeddel D V. IκB kinase-β: NF-κB activation and complex formation with IκB kinase-α and NIK. Science. 1997;278:866–869. doi: 10.1126/science.278.5339.866. [DOI] [PubMed] [Google Scholar]
- 49.Yaron A, Gonen H, Alkalay I, Hatzubai A, Jung S, Beyth S, Mercurio F, Manning A M, Ciechanover A, Ben-Neriah Y. Inhibition of NF-κB cellular function via specific targeting of the IκB-ubiquitin ligase. EMBO J. 1997;16:6486–6494. doi: 10.1093/emboj/16.21.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yaron A, Hatzubal A, Davis M, Lavon I, Amit S, Manning A M, Andersen J S, Mann M, Mercurio F, Ben-Neriah Y. Identification of the receptor component of the IκBα-ubiquitin ligase. Nature. 1998;396:590–594. doi: 10.1038/25159. [DOI] [PubMed] [Google Scholar]
- 51.Yu H, Peters J-M, King R W, Page A M, Hieter P, Kirschner M W. Identification of a cullin homology region in a subunit of the anaphase-promoting complex. Science. 1998;279:1219–1222. doi: 10.1126/science.279.5354.1219. [DOI] [PubMed] [Google Scholar]
- 52.Zachariae W, Shevchenko A, Andrews P D, Ciosk R, Galova M, Stark M J R, Mann M, Nasmyth K. Mass spectrometric analysis of the anaphase-promoting complex from yeast: identification of a subunit related to cullins. Science. 1998;279:1216–1219. doi: 10.1126/science.279.5354.1216. [DOI] [PubMed] [Google Scholar]
- 53.Zandi E, Rothwarf D M, Delhase M, Hayakawa M, Karin M. The IκB kinase complex (IKK) contains two kinase subunits, IKKα and IKKβ, necessary for IκB phosphorylation and NF-κB activation. Cell. 1997;91:243–252. doi: 10.1016/s0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]
- 54.Zhang H, Kobayashi R, Galaktionov K, Beach D. p19/Skp1 and p45/Skp2 are essential elements of the Cyclin A-Cdk2 S phase kinase. Cell. 1995;82:915–925. doi: 10.1016/0092-8674(95)90271-6. [DOI] [PubMed] [Google Scholar]