Abstract
From early life to adulthood, the microbiota play a crucial role in the health of the infant. The microbiota in early life are not only a key regulator of infant health but also associated with long-term health. Pregnancy to early life is the golden time for the establishment of the infant microbiota, which is affected by both environmental and genetic factors. Recently, there is an explosion of the studies on the role of microbiota in human diseases, but the application to disease or health is relatively limited because many aspects of human microbiota remain controversial, especially about the infant microbiota. Therefore, a critical and conclusive review is necessary to understand fully the relationship between the microbiota and the health of infant. In this article, we introduce in detail the role of microbiota in the infant from pregnancy to early life to long-term health. The main contents of this article include the relationship between the maternal microbiota and adverse pregnancy outcomes, the establishment of the neonatal microbiota during perinatal period and early life, the composition of the infant gut microbiota, the prediction of the microbiota for long-term health, and the future study directions of microbiota.
Keywords: microbiome, maternal, offspring, newborn, pregnancy, long-term health, brain development, immune system development
1 Introduction
The emergence of the concept of the microbiome and the development of molecular technology, especially 16S rRNA, have greatly increased the understanding of the microbiota in maternal-fetal interface and early life ( Table 1 for glossary of terms). This makes it possible to characterize the microbiota of pregnant women and their offspring without culturing. During pregnancy, the maternal microbiota affects the development of the fetus, especially the brain development, such as the uterine microbiota, the vagina microbiota, the gastrointestinal microbiota, the placental microbiota (controversial), and the oral microbiota. Notably, the disorder of the maternal microbiota can lead to adverse pregnancy outcomes (APOs), which seriously threaten the health of the offspring. After birth, the infant microbiota affected by both environmental and genetic factors are quickly established to ensure healthy growth. In this article, we describe the role of the microbiota (including the maternal microbiota during pregnancy and the infant microbiota during early life) in the health of offspring from pregnancy to long-term health. These provide a theoretical basis for improving infant health by adjusting the microbiota in pregnancy and early life and promote the understanding of maternal and fetal health.
Table 1.
Glossary of terms.
| Terms | Description |
|---|---|
| Microbiota | Microbial community in a particular environment. May be used interchangeably with “microbiome.” |
| Microbiome | Combined genetic material of microorganisms in a particular environment. May be used interchangeably with “microbiota.” |
| Microbe | A microorganism, especially a bacterium causing disease or fermentation. |
| Diversity | The range of different types of organisms and their relative abundance in a particular environment. |
| 16S rRNA | 16S ribosomal RNA is a subunit of the ribosomal RNA containing specific signature sequences useful for bacterial identification. |
| Abundance | Relative level of a microbe |
The search strategy of this article: We search literature and collect data through Web of Science and Pubmed. First, we use different keyword combinations to search, including infant or newborn or offspring, microbiota* or microbiome* or microbe*, maternal, pregnancy or pregnant or gestation*, etc. Secondly, we read the retrieved articles and eliminate irrelevant articles. Then, we read the rest of the literature carefully and developed a manuscript outline. Finally, we extract the target content from the literature through summary and induction.
2 The Maternal Microbiota and Offspring During Pregnancy
2.1 The Maternal Microbiota During Pregnancy
Previously, the culprit of intrauterine infections in the fetus was considered to be the microbes that rise from the vagina, such as bacteria, viruses, and fungi. However, with the development of science and technology, the oral and gut microbiota have been found to be also associated with the health of the fetus because the microbiota in these parts can spread through the blood. During pregnancy, the mother’s gut, oral cavity, and vagina microbiota all undergo changes. These changes are related to a variety of factors, including diet, antibiotic use, infections, stress, and host genes ( Figure 1 ) (1–5). The study of Romero et al. revealed that the stability of the vaginal microbiota of healthy pregnant women is higher than that of healthy nonpregnant women and Lactobacillus species were the main component of the vaginal microbiota of healthy pregnant women (6). Traditionally, the uterus is considered to be sterile, but this concept was broken until recent years. The endometrium has its own microbiota, and the biomass of these microbiota is very low (7). Regrettably, the current understanding of the endometrium microbiota is limited, and its role in fetal development and pregnancy outcome still needs to be fully elucidated.
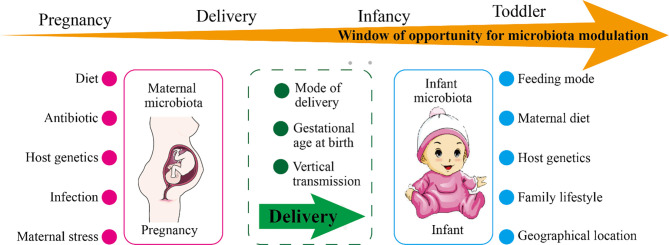
Figure 1.
The factors affecting maternal or infant microbiota and the windows of opportunity for microbiota modulation. As shown in the figure, the maternal microbiota and the infant microbiota are affected by environmental and genetic factors. During pregnancy, the main influencing factors of the maternal microbiota are diet, use of antibiotics, infection, stress, and host genetics. During delivery, the maternal microbiota will be transmitted vertically to the newborn, and the delivery mode and the gestational age of the newborn at birth affect the colonization of the newborn’s microbiota. The main influencing factors of infant microbiota are feeding mode, maternal diet, family lifestyle, geographic location, and host genetics. The window of opportunity for offspring microbiota modulation are pregnancy, delivery, infancy, and toddler.
Emerging evidence indicated that the richness and homogeneity of the gut microbiota of pregnant women are not significantly different from those of normal women, but the distribution and composition are changed obviously (8). In addition, pregnant women with pregnancy complications have a reduced diversity of their gut microbiota, which is detrimental to the health of both the mother and the fetus. For example, gut microbial remodeling during pregnancy in rats with chronic hypertension is impaired (9). The abundance of short-chain fatty acid-producing Coprococcus in the gut microbiota of pregnant women with preeclampsia is decreased (10). In 2014, Aagaard et al. found that the human placental microbiota are mainly composed of nonpathogenic symbiotic microbiota, including Fusobacteria phyla, Firmicutes, Bacteroidetes, Tenericutes, and Proteobacteria (11). The human placental microbiota are similar to the oral microbiota and limited diversity compared with the gut microbiota (11, 12). However, on the contrary, another study showed that the human placenta does not have microbiota but there may be potential pathogens (13). Also, the study concluded there is no correlation between bacterial infection of the placenta and common adverse pregnancy outcomes (APOs) (13).
The imbalance of the maternal microbiota leads to a variety of complications during pregnancy, such as obesity, hypertension, and preeclampsia. Here, we mainly introduce the impact of the maternal microbiota on the fetus, so we will not elaborate on the complications of pregnancy. The differential change of the maternal microbiota during pregnancy is an important cause of APOs. In the following paragraphs, we introduce the factors that affect the maternal microbiota and the impact of the maternal microbiota on APOs.
2.2 The Factors Affecting Maternal Microbiota During Pregnancy
At present, the factors that have been clearly proven to affect the microbiota of the vagina and other parts of pregnant women are diet, antibiotic use, infection, and maternal stress. Factors that need to be further studied are immune status, age, and genetic background. In addition to these factors, complications during pregnancy are also important influencing factors. For example, the changes in the composition of the gut microbiota, oral microbiota, and vaginal microbiota in patients with gestational diabetes are obvious (14). Pregnancy is one of the important windows for regulating the microbiota, and the influencing factors of the maternal microbiota have always been highly concerned. Although the abundance of many kinds of microbes has been proved to change significantly during pregnancy, the function of the specific microbes and their interaction with the host still need to be studied in depth. A precise understanding of the factors that affect the maternal microbiota is helpful to formulate an individualized plan, which can promote better management of the health of the mother and fetus during pregnancy.
2.3 The Role of the Maternal Gut Microbiota on Prenatal Development
The maternal nutritional status, immunity, and metabolism have always been the focus and difficulty in the study of the maternal-fetal interface. Maternal vital substances reach the placenta through blood circulation, and then spread or transport into the fetus’s body (15). During the first two-thirds of pregnancy, the mother exhibits hyperappetite and fat accumulation, at this time the development of the fetus is very limited (16). The fetus develops rapidly during the last third of gestation, when the maternal metabolism increases (16). The maternal microbiota are an important regulator of the metabolism and immunity during pregnancy, among which the gut microbiota are the most prominent (17–19). The gut microbiota can digest carbohydrates in the gut to produce vitamins, amino acids, and short-chain fatty acids (SCFAs) (20). As we all know, vitamins and amino acids are essential for fetal development, and SCFAs are also closely related to immunity and metabolism (21). Although the immune response and metabolic regulation mechanisms of adults are different from those of fetuses, it is also meaningful to explore the role of the maternal microbiota products in fetal development. The synthesis of SCFAs increases during pregnancy, which is necessary for the differentiation of regulatory T cells in the thymus of the fetus. In 2020, a study revealed that the changes in the diet of pregnant mice affect the growth and development of the fetus through the gut microbes (22). Specifically, pregnant mice fed undernourished (UN) or high-fat diet had lower plasma folic acid concentrations compared with the control group, and the relative abundance of the three Lactobacilli taxa in the gut of pregnant mice on a high-fat diet was higher compared with the control group. UN-fed or high-fat diet pregnant mice exhibit metabolic dysfunction and the fetal growth restriction. Furthermore, several studies suggested that the specific microbes in the maternal intestine are necessary for fetal immune system and neurodevelopment (3, 23, 24). The studies pointed out that segmented filamentous bacteria (SFB, common commensal bacteria in the cecum and ileum of mice) in the maternal intestine is the determinant of maintaining the level of IL-17A (IL-17A is related to the maternal immune activation). Consistently, the lack of IL-17A protects the fetus from behavioral disorders and neurodevelopmental disorders.
In summary, we conclude that the maternal gut microbiota and its metabolites are closely associated with the prenatal development of the fetus. However, we did not introduce the role of the maternal vagina and other parts of the microbiota in prenatal development because the current evidence in these areas is limited.
2.4 The Maternal Microbiota and APOs
Complications during pregnancy and APOs are the two most closely related to the maternal microbiota. We have previously introduced the former (14); here, we continue to introduce the latter. Currently, the APOs associated with the maternal microbiota include late abortion (LA), preterm rupture of membranes (PROM), hyperemesis gravidarum (HG), premature delivery (PTD), intrauterine growth restriction, and stillbirth (25). Worldwide, PTD is the most important factor leading to neonatal death (26). Also, intrauterine infection accounts for about 25%–40% of PTD (27). However, the cause of PTD caused by the maternal microbiota has not yet been revealed. The direct sources of the maternal microbiota that can cause fetal infection include placenta, umbilical cord, amniotic fluid, and fetal membranes; indirect sources include the gastrointestinal tract, vagina, skin and mucous membranes.
2.4.1 The Uterine Microbiota and APOs
The serious consequence of the bacterial invasion of the amniotic cavity is neonatal death (26). The uterine microbiota are minute and important (28). Bacterial culture experiments indicated that positive bacterial culture in amniotic fluid is positively correlated with gestational age, and the microbial composition of the first and third trimesters is different (29). The microbial load in the amniotic cavity of pregnant women with premature rupture of membranes is related to the severity of the inflammatory reaction in the amniotic membrane (30). The frequency of the microbiota in the amniotic cavity of PTD women is 40%, and 29% of them have sterile intraamniotic inflammation (31). The level of proinflammatory cytokines in the amniotic fluid of women who give birth within 34 weeks of gestation is upregulated (the full-age gestation period is approximately 280 days), which links PTD to intraamniotic infection (32, 33). Further research revealed that there are uncultivated microbes that may induce inflammatory response in the amniotic fluid of PTD women. In addition, culture-independent techniques showed 46% of women with PTD have intrauterine infections (34). The development of detection technology has increased the detection rate of microbes in amniotic fluid, which promotes the understanding of the relationship between the uterine microbiota and APOs. Currently, the clinical significance of PCR-positive amniotic fluid seems to be consistent with that of positive cultures (35). However, the mechanism by which the uterine microbes cause APOs, especially PTD, is still unclear.
2.4.2 The Vaginal Microbiota and APOs
The vaginal microbiota change not only during the female menstrual cycle but also during the reproductive process (36). The vaginal microbiota are closely related to women’s health, especially APOs. Malnourished women have an increased inflammatory response to the vaginal microbiota, which would lead to spontaneous preterm delivery (sPTD) (37, 38). Lactobacillus is the dominant species in the vagina, and its level may affect the APOs of pregnant women. Pregnant women with the low level of Lactobacilli in the vagina exhibit an increased risk of sPTD and diversity of Mycoplasma and Gardnerella vaginalis (38–40). Consistent with this, women with term deliveries have high levels of Lactobacillus and low bacterial diversity in the vagina (41, 42). However, not all Lactobacillus species in vagina are beneficial for pregnancy outcomes. For example, Lactobacillus iners is a risk factor leading to sPTD, but Lactobacillus crispatus is protective against sPTD (41, 42). A study in which participants were mainly African Americans showed that the diversity and abundance of vaginal microbes in term delivery women are significantly higher than that in PTD women (42). Concretely, Bifidobacterium longum/Bifidobacterium breve, Lactobacillus gasseri/Lactobacillus johnsonii, Lactobacillus iners/Ralstonia solanacearum, and Lactobacillus crispatus/Lactobacillus acidophilus of the vaginal microbiota might be related to decreased risk of PTD, and the vaginal community state type not dominated by Lactobacillus might be related to increased risk of PTD (43). In addition, the increased risk of PROM is associated with aerobic vaginitis (p < 0.05) (44). Consistently, Naomi Juliana et al. found that in Nigerian pregnant women, bacterial vaginosis is not associated with miscarriage and intrauterine growth retardation, but is associated with PROM, PTD, and low birth weight (45).
2.4.3 The Gut Microbiota and APOs
Among the maternal microbiota, the differences in the gut microbiota are the most striking and influential. However, the studies on the relationship between gut microbiota and APOs are limited, and most of the studies focus on gut microbiota and pregnancy complications. It seems that the gut microbiota is difficult to link directly to APOs because the intestine and the fetus are far apart in human anatomy. In recent years, the colonization of gut microbes in the vagina has linked the maternal gut microbiota with APOs. The migration of Listeria monocytogenes from the intestine to the vagina, leading to maternal-fetal infection, is a good example (46). A Danish report revealed that Listeria monocytogenes in pregnancy pose a serious threat to the fetus, and the rate of miscarriage or stillbirth among mothers infected with Listeria monocytogenes is as high as 32% (46). In addition, pregnant women have multiple identical microbiota in their vaginas and rectum, and 68% of them have the same genotype (47). From here, we can see that to a large extent there is a corresponding relationship between the vaginal microbiota and the rectal microbiota. The above evidence supports the link between the gut microbiota and APOs through the vaginal microbiota. Therefore, the gut microbiota can be used as a reservoir for vaginal probiotics, and regulating the gut microbiota is an effective strategy for improving pregnant women’s APOs.
Numerous microbial metabolites and components in the intestine have a powerful ability to regulate inflammatory response and immunity, which provides another link between maternal gut microbiota and APOs (21). Evidence indicated that the relative abundance of Prevotellaceae_UCG_003, Prevotella_1, and Selenomonas_1 in feces of aborted women is significantly reduced, and the level of proinflammatory cytokines in serum is increased (48). The study further pointed out that the metabolites of the gut microbiota are meaningful for recurrent miscarriage (48). Besides, the decrease in the alpha diversity of the gut microbiota is closely associated with the occurrence of sPTD, which may be regulated by the metabolites of the gut microbiota (49).
2.4.4 The Placental Microbiota and APOs
The existence of the placental microbiota is controversial. Some scholars believe that the human placental microbiota is unique and similar to the composition of oral microbiota (11, 12). Sweeney et al. observed that Ureaplasma species are the most common microbes in the placenta, and their relative abundance is related to PTD and term delivery (50, 51). In contrast, a study of 76 term delivery does not support the presence of placental microbiota (52). The study pointed out that the occasionally observed microbes may be caused by pollution, which does not represent the presence of placental microbes. Coincidentally, Kuperman et al. found that the fetus in the womb is sterile through the study of 28 human placenta and six mouse placenta (53). They believe even if the placental microbiota are present, their biomass is extremely low and the impact on the fetus may be small. There is no proof which view is right at present. The studies of placental microbiota should be carefully designed, analyzed, and conducted in an unbiased manner.
2.4.5 The Oral Microbiota and APOs
There is a Chinese proverb called “disease comes from the mouth”, which reflects the importance of oral microbes in human health from the side. There are many kinds of microbes that promote potentially APOs in the oral cavity, such as Campylobacter rectus, Porphyromonas gingivalis, Fusobacterium nucleatum, and Filifactor alocis. The pathways of the oral microbiota causing APOs may be (54): (1) Blood-borne spread of the oral microbes. (2) The immune response of the host and/or fetus to the pathogen causes inflammatory mediators to enter bloodstream. (3) Sexual behavior leads to the exchange of the oral and vaginal microbes. Porphyromonas gingivalis, the culprit of periodontitis, is a common anaerobic bacteria in the oral cavity and associated with many kinds of APOs, including fetal growth restriction (55), spontaneous abortion (56), and PTD (57). The mechanisms of Porphyromonas gingivalis causing APOs may be as follows: (1) Porphyromonas gingivalis directly invades the maternal tissue or placenta at the maternal-fetal interface (58). (2) Porphyromonas gingivalis evades the killing of the immune system by suppressing the immune response in the maternal tissues (59, 60). (3) Porphyromonas gingivalis leads to increased inflammatory response in maternal and fetal tissues via regulating the immune response (61). (4) Porphyromonas gingivalis promotes the increase of acute-phase proteins in maternal and fetal tissues, such as alpha-fetoprotein and C-reactive protein (62). (5) Porphyromonas gingivalis promotes the invasion and colonization of other maternal microbes. Therefore, the infection of Porphyromonas gingivalis could cause a systemic reaction to result in APOs, which is extremely detrimental to the health of the mother and fetus. It is very important for pregnant women to have good oral hygiene by controlling the oral microbiota.
3 The Microbiota and Offspring During Perinatal Period and Early Life
From birth to adulthood, environmental factors interact with genetic factors to maintain the healthy growth of the individual. During perinatal period and early life, the microbiota of offspring is closely related to the maternal microbiota, especially the neonatal and infant gut microbiota. Perinatal period and early life are the prime time for the colonization of the gut microbiota of offspring. The mode of delivery and gestational age at birth are important factors affecting the neonatal gut microbiota during the perinatal period. In early life, the determinants of the infant gut microbiota include feeding mode, maternal diet, environmental factors, and host genotype. In the following section, we introduce the establishment and development of neonatal gut microbiota and the driving forces of neonatal and infant gut microbiota.
3.1 Establishment and Development of Neonatal Microbiota
The establishment of neonatal gut microbes is the result of the interaction of environmental and host factors. Both beneficial and harmful microbes would colonize the neonatal intestines at birth and are profound for the health of infant (63) ( Figure 2 ). The microbes that newborns are exposed to at birth are very important, especially the maternal microbes (64). Bacteria from the maternal vagina and skin have been observed in the feces of newborns (65). The mouth, skin, and intestines of newborns delivered vaginally are rich in Lactobacillus which is the core of the maternal vaginal microbiota (66). After birth, newborns would continue to obtain microbes from the maternal microbiota in different parts. Also, the colonization of these microbes is durable and stable.
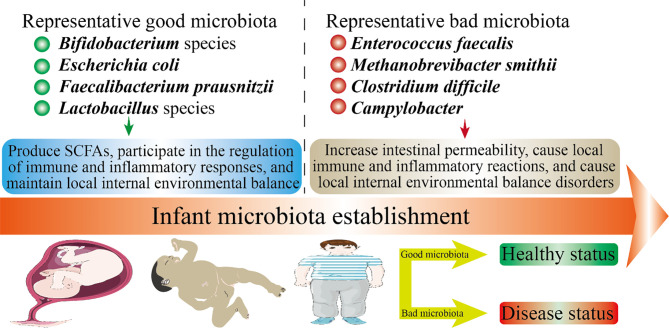
Figure 2.
Infant health status and microbiota establishment. The health of offspring is closely related to the microbiota. In this figure, we have listed several representative good microbiota and several representative bad microbiota. The microbiota will inhibit the developmental stages throughout the offspring. The mechanism by which the good microbiota is beneficial to health may be: produce SCFAs, participate in the regulation of immune and inflammatory responses, and maintain local internal environmental balance. The mechanism by which the bad microbiota is harmful to health may be: increase intestinal permeability, cause local immune and inflammatory reactions, and cause local internal environmental balance disorders.
The infant gut microbiota exhibits initially low diversity and then increases with early development. In early life, the microbes that colonize the intestine are usually aerobic bacteria because the intestine is aerobic. After adulthood, the microbes colonized in early life will be replaced by typical anaerobic bacteria in the intestine (67). Studies on the gut microbiota of infants indicated that Clostridia, Enterobacteria, and Streptococci were observed in the infant intestines in the first 2 days after birth (68). Clostridia, Bacteroides, and Bifidobacterium appear in the intestines of 40% of infant on the third day (68). On the 4th–7th days, Bifidobacterium became dominant, and its relative abundance is highest (68). Before weaning, the relative abundance of Bacteroides gradually increases to compete for Bifidobacterium in the infant intestine. After ingesting solid food, the composition of the infant gut microbiota is similar to that of adults (67). In 2–3 years after birth, the infant gut microbiota gradually develops towards the adult gut microbiota (66, 67).
3.2 The Driving Forces of the Infant Microbiota
The process from colonization to maturity of the gut microbiota is nonrandom and dynamic (69, 70). The process is affected by many factors during the perinatal period and early life, although it is difficult to quantify.
3.2.1 The Mode of Delivery
The mode of delivery is a crucial factor affecting vertical transmission (71). The composition of the gut microbiota of the infant delivered vaginally appears to be similar to that of the maternal vaginal microbiota: Lactobacillus dominates, followed by Senathia spp. and Prevotella. While the gut microbiota of infant delivered by cesarean section is similar to that of the maternal skin: Staphylococcus is dominant, followed by Propionibacterium and Corynebacterium (65, 72). Cesarean section leads to the imbalance of infant gut microbiota and the decrease of the diversity (73, 74). There was a study: researchers exposed infant delivered by cesarean section to vaginal fluid to promote the recovery of the infant gut microbiota. Interestingly, this approach effectively improves the oral, gut, and skin microbiota of infant delivered by cesarean section within 30 days after birth (75). This indicates that the vertical transmission of the maternal vaginal microbes can be reversed to some extent after birth.
It is worth noting that epidemiology has linked cesarean section to autoimmune diseases, asthma, obesity, and allergies (76). The difference in infant microbiota caused by vaginal delivery and cesarean section may be attributed to the “bacterial baptism” of vaginal fluid. Vaginal microbes spread vertically to the infant to promote the colonization of the infant microbes. However, there is another voice here: the differences in the infant microbiota caused by cesarean section may due to the use of antibiotics during delivery, the inactivity of the mother, and complications during pregnancy (77). The current studies may overestimate the effect of vaginal delivery on infant microbiota. Furthermore, the difference in infant microbiota caused by cesarean section is more moderate in rodents compared with humans. Of course, this may be caused by the spread of microbes in rodents more easily than in humans. Therefore, convincing evidence is necessary to clarify the differences in infant gut microbiota caused by the mode of delivery.
3.2.2 The Gestational Age at Birth
Gestational age at birth is also important for the colonization of infant microbiota. The immature microbiota poses a huge threat to premature because their immune and nervous systems are not yet fully developed. Usually, premature need to stay in the hospital for treatment. Factors such as the use of antibiotics and parenteral feeding can interfere with the natural establishment of infant microbiota. Compared with term infant, the intestines of premature cannot colonize symbiotic anaerobic bacteria such as Bacteroides and Bifidobacterium, and the relative abundance of Enterococcus and Enterobacteriaceae in their feces is increased (78–83). Studies showed that the gut microbiota of very low birth weight infant exhibits a transition from Bacilli to Gammaproteobacteria to Clostridia (84). The interaction between the immune system of premature infants and their immature microbiota may lead to inflammation and infectious diseases (85, 86). Compared with term infant, the SCFAs of the gut microbiota of preterm infant are significantly lower (81, 87). In short, the gestational age at birth affects the infant short-term health and even long-term health to a certain extent.
3.2.3 The Feeding Mode and Maternal Diet
The feeding mode is mainly associated with the infant gastrointestinal microbiota. The role of breastfeeding in regulating the gut microbiota of infant has been widely recognized (88, 89). In particular, the relative abundance of Bifidobacterium in the intestines of the breastfed infant is increased (90). On the one hand, breastfeeding provides infant with maternal microbes, nutrients, and antibacterial agents, which are important for infant health. On the other hand, the IgA in breast milk contributes to the “tolerability” and “regulation” of the infant immune system (91). It should be emphasized that human milk oligosaccharides (HMOs) in breast milk can selectively shape the formation of beneficial microbes in infant. The bioactive molecules and identified microbes in breast milk are shown in Table 2 (92–98). The gut microbiota of breast-fed infant is dominated by Bifidobacterium, while formula-fed is dominated by Bacteroides and Bifidobacterium (99). Besides, breastfeeding can effectively prevent infant from being overweight by improving the imbalance of the gut microbiota compared with formula-fed (100). Therefore, breast milk is the best food for babies. These also reflect the importance of infant microbiota in early health, and its impact on long-term health is also profound (101).
Table 2.
Bioactive molecules and identified microbes in breast milk.
| Category | Specific type |
|---|---|
| Identified microbes | Staphylococci, Streptococci, Corynebacteria, Propionibacteria, Lactobacillus spp., Bifidobacterium spp. |
| Anti-microbials | Immunoglobulins- Secretory IgA, IgM, IgG, lactoferrin, lactadherin/MFG E8, Lysozyme, complement C3, antiviral mucins—MUC1, MUC4 |
| Digestive enzymes | Amylase, Bile acid-stimulating esterase, bile acid-stimulating lipases, lipoprotein lipase |
| Growth factors | Epidermal growth factor (EGF), nerve growth factor (NGF), insulin-like growth factor (IGF), transforming growth factor (TGF), taurine, polyamines, heparin-binding EGF like growth factor (HB-EGF), vascular endothelial growth factor (VEGF), erythropoietin |
| Transporters | Lactoferrin, folate binder, cobalamin binder, IGF binder, thyroxine binder, corticosteroid binder |
| Hormones | Calcitonin, somatostatin, adiponectin, leptin, ghrelin |
| Oligosaccharides and glycans | Human milk oligosaccharides (HMOs), gangliosides, glycosaminoglycans |
| Cytokines, chemokines, and anti-inflammatory factors | Tumor necrosis factor-alpha (TNF-α), interferon-gamma (IFN-γ), Transforming growth factor-beta (TGF-β), prostaglandins, 1-antichymotrypsin, 1-antitrypsin, platelet-activating factor: acetyl hydrolase, interleukins-IL-6, IL-7, IL-8, IL-10 chemokines-granulocyte colony stimulating factor (G-CSF), macrophage migratory inhibitory factor (MIF) |
The maternal diet may affect the baby’s gut microbes through milk. Milk can be regarded as a kind of body fluid, and the maternal diet affects directly the microbial composition and diversity of breast milk (102, 103). During lactation, maternal fat consumption and fiber intake are important determinants of breast milk microbiota (104). Furthermore, maternal diet also affects the bioactive molecules in milk, such as HMOs (105). Therefore, breastfeeding mothers should diet carefully to give the best care to the baby.
3.2.4 The Environmental Factors
It is also meaningful to study the relationship between environmental factors and the colonization of microbiota in early life. Despite the lack of in-depth research, family members have been defined as potentially affecting the colonization of infant gut microbiota (63, 106). Interestingly, a Dutch study showed that the relative abundance of Bifidobacterium in the gut microbiota of infant with siblings (1 month old) is higher than that of those without siblings (107). In addition, the level of facultative anaerobes and anaerobes in the infant gut without big sisters is lower, while that of Clostridium and Escherichia coli is higher (108). A Danish study showed that the abundance and diversity of gut microbes of infant with siblings in early life are increased, while the influence of pets on them was not obvious (109). However, the concept of the “sibling effect” remains controversial because it is so extensive and difficult to quantify. Geographical location may also affect indirectly the microbiota in early life because geographic location determines eating habits and lifestyle (63, 110). For example, the diversity of infant microbiota living in rural Africa is significantly different from that of in cities (111).
3.2.5 Host Genetics
In recent years, the studies on host genetics and infant gut microbiota have gradually increased (112, 113). The most convincing evidence is that the microbial similarity of children with genetically identical twins (under 10 years of age) is higher than that of fraternal twins and unrelated controls (114). Additionally, the relative abundance of Bifidobacterium is associated with single nucleotide polymorphisms (SNPs) located at the LCT site of human genes (human lactase gene) (115). However, genetic factors are complex, and more evidence is needed to describe its association with the microbiota in early life.
4 Infant Gut Microbiota
In Table 3 , we summarize the changes in the microbial composition of some parts of the infant body from delivery to postpartum (116). In early life, the infant gut microbiota undergo a transition from being dominated by Lactobacillus and Bifidobacterium to being dominated by Firmicutes and bacteria (117), which represents the gut microbiota from infancy to maturity. Lactobacillus and Bifidobacterium can promote the development of infant-acquired immunity and innate immunity in early life. Importantly, the relative abundance of Bifidobacterium in feces in early life is related to the risk of developing noncommunicable diseases in later life, such as obesity and asthma (118, 119). The composition and diversity of infant gut microbiota change dramatically with the intake of solid foods. The dietary fiber fermented by the gut microbiota produces SCFAs, which are related to the host’s immunity and metabolism (21, 120). Below, we introduce in detail the gut microbiota of infant.
Table 3.
Changes in the microbial composition of various parts of the infant body from delivery to postpartum.
| Microbiota\sites | Gastrointestinal tract | Skin | Oral mucosa | Nares |
|---|---|---|---|---|
| Actinobacteria | ↓↓ | ↑ | ↓↓ | ↑ |
| Bacteroidetes | ↑↑ | ↓ | ↑↑ | ↑ |
| Firmicutes | ↑ | → | ↑↑ | ↓ |
| Fusobacteria | ↓ | ↓↓ | ↓ | ↑ |
| Proteobacteria | ↓ | ↓ | ↓↓ | ↑↑ |
| Others | ↓ | ↓↓ | ↓ | ↑ |
“↑“ represents an increase in the abundance of the microbe from delivery to postpartum. “↑↑“ represents a significant increase in the abundance of the microbe from delivery to postpartum. “↓“ represents a decrease in the abundance of the microbe from delivery to postpartum. “↓↓“ represents a significant decrease in the abundance of the microbe from delivery to postpartum. “→“ represents that the abundance of the microbe is basically unchanged from delivery to postpartum.
4.1 The Dominant Populations of Infant Gut Microbiota
The adult gut microbiota are relatively stable (121), while the infant (less than 1 year old) have a lower diversity of gut microbiota compared with other age groups (110). Infant gut microbiota are dominated by Bifidobacterium, but there is great variability among individuals. According to the dominant population and composition, the core of infant gut microbiota is mainly divided into six groups (122): group 1, Bifidobacteriales, Lactobacillales, Anaerostipes, Clostridiales, and Faecalibacterium; group 2, Verrucomicrobiales and Bacteroidales; group 3, Clostridiales; group 4, Enterobacteriales; group 5, Pasteurellales; and group 6, Selenomonadales. In Table 4 , we summarize the dominant populations and their functions of the infant gut microbiota.
Table 4.
The dominant populations of infant gut microbiota.
| Microbes | Key description | Refs. |
|---|---|---|
| Bifidobacteria | Bifidobacteria make a major metabolic contribution to their host through the degradation of diet-derived glycans and host-provided carbohydrates (known as host glycans and including mucins and HMOs). | (123) |
| Clostridia class | They are known as pathogenic microorganisms that may cause bacteremia and pseudomembranous colitis, and their presence at high densities is interpreted as an indicator of an unhealthy microbiota. | (124) |
| Genus Bacteroides | Members of this genus are classified as saccharoclastic bacteria that are able to metabolize host-produced glycans, such as HMOs and mucins, but also complex plant polysaccharides such as starch, cellulose, xylans, and pectins. | (125, 126) |
| Genera Veillonella | These bacteria are saccharolytic and utilize end products of carbohydrate fermentation (e.g., lactate) of other infant gut bacteria, such as Streptococcus spp. and Bifidobacterium spp., to produce propionate, forming an important trophic chain. | (127) |
| Genera Streptococcus | They are among the first established bacteria in the infant gut, where they can be identified within the first 24 h following birth. | (128) |
| Genus Collinsella | Members of the genus Collinsella have recently been shown to reach high numbers when they are associated with an infant gut microbiota dominated by bifidobacteria. | (129) |
| Genus Lactobacillus | Vertical transmission of Lactobacillus species presents the origin of infant Lactobacillus microbiota component. And they may be related to the HMO metabolism of infants. | (128) |
| Genus Akkermansia | They can provide a barrier for the baby’s intestines and may participate in the fermentation of HMO. | (130) |
4.1.1 Bifidobacteria
Bifidobacteria, belonging to the phylum Actinomycetes, anaerobic gram-positive bacilli, was isolated from the feces of the breastfed infant as early as 1899. At present, Bifidobacteria have been widely used in medicine and food. New strains are constantly being developed. At present, the ones that have been found to be closely related to infant health are Bifidobacterium breve and Bifidobacterium longum. The vertical transmission of the maternal Bifidobacterium to the offspring promotes its distribution in nature. The identification experiments of common Bifidobacterium in mothers and offspring confirmed this conclusion (69, 131).
HMOs in breast milk are natural prebiotics, and their absorption in the intestinal tract of infant mainly depends on Bifidobacteria (132). The evidence is that the level of HMOs in the feces of infant is negatively correlated with the level of Bifidobacteria. Interestingly, HMOs have no direct nutritional value for infant, and their important function is to shape the infant gut microbiota and benefit long-term health (133). After premature infant are supplemented with Bifidobacterium longum, the inflammatory response is weakened and the intestinal permeability is reduced (133). Premature infants supplemented with Bifidobacteria-based microbes can restore the gut microbiota to the level of term infant (134). Therefore, in early life, Bifidobacterium can effectively improve the health of infant, whether they are the premature infant or term infant.
4.1.2 The Genus Lactobacillus
The genus Lactobacillus are also known to be the dominant species in the infant gut microbiota and observed shortly after delivery (128). Various genus Lactobacillus are detected in meconium (the relative abundance: vaginal delivery is significantly greater than caesarean section), including Lactobacillus reuteri, Lactobacillus plantarum, Lactobacillus sakei, Lactobacillus brevis species, and Lactobacillus casei (135). Studies on the gut microbiota of infant in early life suggested that Lactobacillus gasseri and Lactobacillus rhamnosus are dominant in terms of the genus Lactobacillus (136). In addition to being abundantly present in the infant gut microbiota, the mother’s vagina and milk are also rich in the genus Lactobacillus. The former is passed vertically to the offspring during vaginal delivery and the latter is passed to the offspring during breastfeeding. The main types of the genus Lactobacillus in milk are L. plantarum and L. pentosus (137). As mentioned above, apart from Bifidobacteria, the genus Lactobacillus can also digest HMOs, and there are obvious differences in the ability of different strains to ferment HMOs (138). Furthermore, Lactobacillus reuteri DSM17938 has been shown to be effective in relieving colic of breast-fed infant (139).
4.1.3 The Genus Clostridium
The genus Clostridium can be divided into multiple genera, all of which belong to the Clostridia class. The genus Clostridium is the only genus of anaerobic bacillus, and their spores are round or ovoid, their diameter is wider than the bacteria. Most Clostridium species in the intestines of infants are pathogenic, especially Clostridium perfringens, Clostridium difficile, Clostridium tetani, and Clostridium botulinum (140). Clostridium species are common settler in infant intestine, but they are usually asymptomatic (141). The results of infant fecal microbial culture showed that the level of Clostridium perfringens and other clostridia may reach 107 CFU/g (142). A stool study of infant (<1 year old) in Jordan indicated that the colonization rate of Clostridium perfringens in the infant intestines is 27.2%, and the colonization rate of infant <6 months is higher (p < 0.004) (143). Interestingly, Rada et al. found that both the Clostridia and Bifidobacteria in the feces of infant (3–253Radadays old) grow vigorously on the prebiotic oligosaccharides, which may pose a challenge for supplementing prebiotics to infant lacking Bifidobacteria (144). In contrast, Enterococcus faecalis isolated from infant feces can inhibit the growth of Clostridium difficile, which may have potential applications for preventing Clostridium difficile colonization and infection (145). In short, in early life, we need to beware of Clostridium to prevent them from becoming the dominant species in infant gut microbiota.
However, Kim et al. found that neonatal acquisition of Clostridia species protects against colonization by bacterial pathogens (146). The article pointed out that the administration of Clostridiales enhances the colonization resistance of the infant intestine, which can prevent some pathogens from attacking the infant intestine. Consistent with this, commensal Clostridia is considered to play an important role in maintaining intestinal homeostasis (147, 148). These data improve our understanding of the function of infant microbes. The application of Clostridia species to regulate the homeostasis of the infant intestinal microbiota is still a huge challenge, because most Clostridia are pathogenic.
4.1.4 The Genus Bacteroides
From birth to adulthood, the human microbiota gradually mature, among which the infant microbiota are usually characterized by a low total level. The relative abundance of Bacteroides in the infant feces delivered vaginally is higher than that of cesarean section (p < 0.01) (149). The diversity of Bacteroides in the feces of formula-fed infant is higher than that of breast-fed infant (150). Furthermore, the delayed colonization of Bacteroides caused by cesarean section may be associated with Th1 response (151). Similar to Bifidobacteria and the genus Lactobacillus, Bacteroides, and Fragilis in infant intestine can also digest HMOs (125). Additionally, in the environment of carbohydrates and HMOs, Bacteroides thetaiotaomicron promotes the growth of bacteria that produce SCFAs (152). However, some Bacteroides are pathogenic. For example, Bacteroides fragilis can cause anaerobic meningitis in infant (153). The abundance of Bacteroides in the intestine of allergic infant (<2 years old) is higher than that of allergic infant (154).
4.1.5 The Genus Veillonella and Streptococcus
The genus Veillonella are strictly anaerobic and parasitizes in the mouth, intestine, and respiratory tract of humans and animals (155). They can produce endotoxins and therefore play a role in various mixed infections. They are often detected from soft tissue abscesses and blood and are usually normal microbiota (155). The genus Veillonella is also common in the intestine of infants. In recent years, the new species have been identified continuously. For example, in 2018, Mashima et al. isolated a new type of anaerobic Gram-negative cocci strain from the biofilm of children’s tongue (156). Later, they found that the strain is different from the previous Veillonella species through genetic sequencing, and named it Veillonella infantium. An interesting study showed that the relative abundance of Veillonella in the intestine of infant (2–6 months old) exposed to unsafe food caused by Hurricane Maria is reduced (157).
The genus Streptococcus is another major group of common Gram-positive cocci in pyogenic cocci. There are 69 species and subspecies, which are widely distributed in nature and the nasopharynx, gastrointestinal tract of the human body, and most of them belong to normal genus (158). Pathogenic Streptococci can cause a variety of purulent inflammation and hypersensitivity diseases in humans, among which group B Streptococcus (GBS) are the most threatening to infants (159). Although antibiotics can reduce GBS level during delivery, GBS is still an important cause of neonatal sepsis in some high-income contexts (160). Approximately 35% of pregnant women carry GBS, and the gut microbiota of the offspring of mothers who carry GBS are significantly different from those of mothers who do not carry GBS (161). Therefore, effective prevention of the colonization of the GBS is essential for infant health.
4.1.6 The Genus Collinsella and Akkermansia
In the infant gut microbiota dominated by Bifidobacteria, the level of the genus Collinsella can be very high (129). So far, the genus Collins includes five species, all isolated from the human gastrointestinal tract: Collinsella stercoris, Collinsella intestinalis, Collinsella aerofaciens, Collinsella tanakaei, and Collinsella ihuae (162–164). Akkermansia muciniphila is the only specie of the genus Akkermansia in the gut microbiota, although its relative abundance is very low (165, 166). The level of Akkermansia muciniphila in the intestine rises rapidly after weaning and increases with age (167). Animal experiments indicated that the abundance of Akkermansia muciniphila in the intestine of obese and type 2 diabetic mice is reduced, and administration of Akkermansia muciniphila can control the inflammation and reduce intestinal permeability (168). Notably, Akkermansia muciniphila has also been found to be involved in the fermentation of HMOs.
5 The Role of Infant Microbiota in Future Health Planning
From birth, the gut microbiota are responsible for the activation and development of the immune system, the development of the central nervous system (CNS), and the digestion and metabolism of food. Early life is a critical period for microbial colonization, which not only affects the health of infant, but also is profound to long-term health planning ( Figure 2 ). Below, we introduce in detail the relationship between infant gut microbiota and brain development and the shaping of the immune system. We also talk about the role of microbiota in early life in diseases and the direction of future study on human microbiota.
5.1 Infant Brain Development
Early life (less than 2 years old) is the period of rapid brain development. The brain volume of newborns is only about 1/3 the size of adults, but their brains will increase to 80%–90% of adults by the age of 2 (169). In early life, the infant brain undergoes successively the growth of axons and dendrites, the formation of synapses, the expansion of neurogliocyte, and the myelination of axons. Importantly, the first 2 years of life is also a golden period for the establishment of the gut microbiota. Therefore, the establishment of the gut microbiota in early life is essential for the development of the infant brain (169). In Figure 3 , we briefly describe the role of the gut microbiota in the development of the CNS early in life ( Figure 3 ).
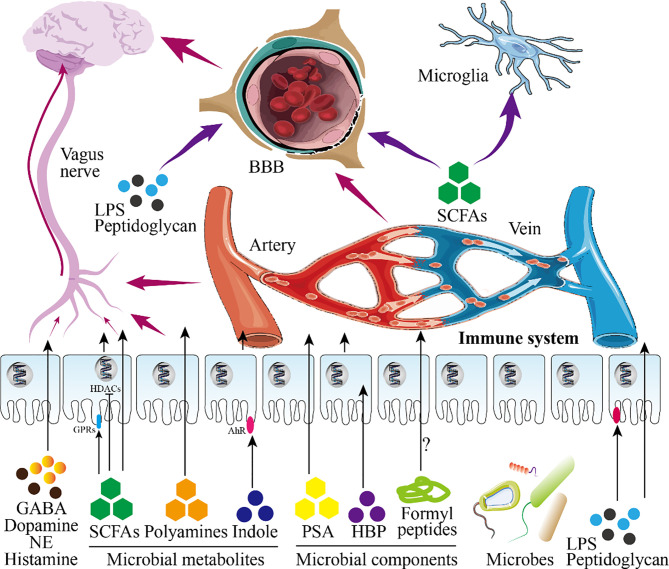
Figure 3.
Infant gut microbiota and central nervous system. This figure shows the role of infant microbiota in the central nervous system (CNS). Microbial metabolites and microbial components directly pass-through epithelial cells or act on receptors on epithelial cells to play a role (in this figure, intestinal epithelial cells are taken as an example). On the one hand, these substances will further reach the blood-brain barrier (BBB) after entering the blood circulation and enter the brain to participate in the regulation of the CNS after passing through the BBB. On the other hand, these substances continue to undergo biochemical reactions in the blood circulation to affect the CNS. Besides, SCFAs and LPS entering the blood circulation can also directly act on nerve cells. Shortly, these substances regulate the development of the brain and CNS.
There is a Chinese proverb: pull one hair and the whole body is affected. This sentence well reflects the role of the gut microbiota in brain development. What must be mentioned here is the ability of the metabolites or components of the microbiota to promote the development of the nervous system. These important molecules can enter the blood circulation through the intestinal barrier to affect brain development (170). For example, the indigenous spores produced by bacteria promote the synthesis of serotonin in enterochromaffin cells (ECs), thereby regulating the serotonin levels in the circulation, colon and ileum (171). Besides, SCFAs, the product of dietary fiber fermented by the gut microbiota, can directly regulate the sympathetic nervous system through free fatty acid receptors (belonging to G protein-coupled receptors) (21, 172). Kimura et al. found that SCFAs produced by the pregnant mother determine the development of the fetal intestine, pancreas, and nerves through GPR43 and GPR41 (173). SCFAs can also promote the maturation of microglia which differentiate retrogradely into other types of glial cells (174). Furthermore, the permeability of the blood-brain barrier (BBB) of germ-free mice is increased, and the administration of sodium butyrate (a type of SCFAs) improves the permeability defect of the BBB of germ-free mice by inhibiting histone deacetylases (HDACs) (175). In addition, the diversity of the gut microbiota is related to the release of intestinal peptides (such as oxytocin, ghrelin, glucagon-like peptide, cholecystokinin) (176). The disorder of the maternal gut microbiota causes the fetal brain to fail to develop normally, which is mediated by the metabolites of maternal gut microbes (177). The insufficiency of the gut microbiota is a great threat to premature babies because it weakens the ability of the intestinal wall to resist pathogens (170).
On the basis of its influence on brain development, the relationship between the gut microbiota and behavior in early life has also been established in recent years. For example, there are data showing that the diversity of the gut microbiota is associated with the cognitive ability of infant (170). In short, gut microbiota can shape the individual behaviors and social behaviors of infant (178).
5.2 Infant Immune System Development
Before birth, the maternal microbiota participate in regulating the innate immunity of the fetus. After birth, the infant microbiota continue to shape innate immunity and acquired immunity to promote the development of the immune system to a normal state.
5.2.1 Microbial Colonization Modulates Early Development of the Immune System in Mucosal Tissues
The microbiota on the mucosal surface in the early life changes dynamically until they reach an equilibrium point, then remain stable throughout long-term planning in the absence of external or internal insults (179). The colonization of microbes on the mucosal surface and the shaping of the immune system occur simultaneously in early life. Therefore, the microbiota on the mucosa may directly or indirectly shape the immune system until it matures. In Table 5 , we summarize the gut microbes that have been identified in recent years to regulate intestinal mucosal immunity. The immune system of newborns is very different from that of adults, because infant preferentially develop tolerance in response to pathogen invasion (195–197). The period after birth is the most critical period, during which exposure to the microbiota may have a profound impact on the structure and function of the infant immune system.
Table 5.
Identified gut microbes regulating intestinal mucosal immunity.
| Microbial species | Involved substances | Involved intermediates | Key description | Refs. |
|---|---|---|---|---|
| Lactobacilli, Clostridiales members | Tryptophan indole derivatives | AhR | IL-22 production; resistance to enteric pathogens; maintenance of intestinal homeostasis and barrier functions | (180–182) |
| Various microbes including Bacteroides spp. | SCFAs | Receptors GPR41, GPR43 and GPR109, HDAC inhibition, mTOR, STAT3, ERK and MAPK signaling | Protective inflammatory responses during pathogen infection; secretion of AMPs, chemokines and cytokines; controls IECs turnover and barrier functions; RALDH1 expression and vitamin A metabolism | (21) |
| Clostridium sporogenes | Indole 3-propionic acid | PXR | Regulation of intestinal permeability and intestinal inflammation, defense against intracellular pathogens | (183) |
| Clostridial species | Mechanosensors/mechanotransducer Piezo2 | Cellular forces | Serotonin release by enterochromaffin cells | (171, 184) |
| Bacteroides vulgatus, Enterococcus faecium | Nod2 | Peptidoglycan components; muramyl dipeptide | Restriction of bacterial growth or dissemination, expression of inflammatory genes, goblet cell function | (185) |
| C. rodentium, S. typhimurium, H. pylori | Unmethylated CpG bacterial DNA | NF-κB, TLR9 | Decreases intestinal inflammation and damage following bacterial challenge | (186) |
| Helicobacter hepaticus, S. typhimurium, Pasteurellaceae family | Autophagy | Cellular stresses | Control inflammation-induced apoptosis, necroptosis and maintains intestinal barrier, lysozyme secretion by Paneth cells, promotes bacterial clearance | (187) |
| Rotavirus EW | dsRNA | Nlrp9b inflammasome | Restricts rotavirus infection by IL-18 production and pyroptosis | (188) |
| Commensal gut microbes | Free fatty acids | TLR4, PPAR | Prevents development of metabolic syndrome; regulates expression of lysozyme and PPAR-controlled genes | (189) |
| C. rodentium | TLR ligands | MyD88 signalling | Secretion of AMPs, control of bacterial infiltration, enhanced barrier integrity | (190) |
| Toxoplasma gondii | ligands include extracellular ATP | P2X7R/NLRP3 inflammasome | IL-1β secretion and inhibition of parasitic proliferation | (191) |
| Lactobacillus rhamnosus, Ruminococcus gnavus | Pili | GPR and ERK/MAPK signaling | Expression of glycoroteins and mucus production by goblet cells; cytoprotective responses | (192) |
| Salmonella Typhimurium, C. rodentium | Flagellin | NAIP/NLRC4 inflammasome | Protects against enteric pathogen invasion; expulsion of pyroptotic IECs and release of eicosanoid and IL-18 | (193) |
| Clostridium difficile | Enterotoxins | Caspase-3/7-mediated apoptosis | Restricts C. difficile growth in vivo | (194) |
AhR, aryl hydrocarbon receptor; SCFAs, short-chain fatty acids; GPR, G protein-coupled receptor; PXR, pregnane X receptor; HDAC, histone deacetylase; RALDH1, aldehyde dehydrogenase 1; Nod2, nucleotidebinding oligomerization domain 2; TLR9, toll-like receptor 9; dsRNA, double-stranded RNA; Nlrp9b, NLR family, pyrin domain containing 9B; PPAR, peroxisome proliferator-activated receptor; P2X7R, P2X7 receptor; IECs, Intestinal epithelial cells.
5.2.2 The Mechanisms of the Microbiota Regulating Immunity
5.2.2.1 Microbial Metabolites
The metabolites of gut microbiota are diverse, and their sources include gut microbes digesting exogenous food components and fermenting endogenous compounds. These metabolites may act directly on the mucosa or enter the blood circulation through mucosal epithelial cells. The mechanism by which these metabolites modulate the immune response may be as follows (including but not limited to) (198): (1) Gut microbes digest dietary fiber to produce SCFAs which have wide-ranging effects on the immune system ( Figure 4 ); (2) metabolites of gut microbes bind to specific receptors on host cells, including aryl hydrocarbon receptor (AHR), G protein-coupled receptors 41 and 43, Toll-like receptors (TLRs), and PXR receptors (see Table 5 ); (3) polyamines, such as spermine, putrescine, and spermidine, are involved in gene transcription and translation and exist in most living cells.
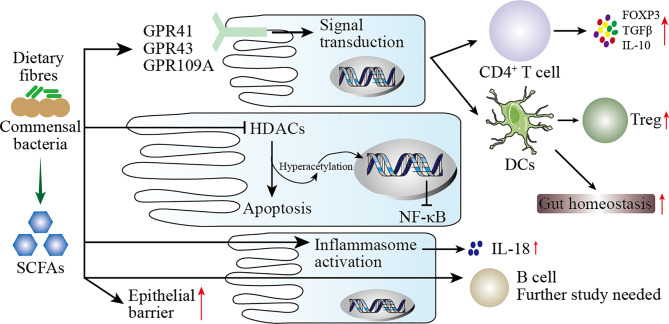
Figure 4.
Mechanisms of SCFAs regulating mucosal immunity. SCFAs are produced by the digestion of dietary fiber by the intestinal microbiota. On the one hand, SCFAs increase the barrier function of the intestinal mucosa to protect the body from harmful microbes. On the other hand, SCFAs participate in the mucosal immunity of the intestine, specifically: (1) SCFAs act on G protein-coupled receptors (GPR) on intestinal epithelial cells (IECs) to activate downstream cell signal transduction to regulate the immune response of T cells and dendritic cells (DCs). (2) After SCFAs enter IECs, they act as histone deacetylases (HDACs) inhibitors to affect the transcription of inflammatory genes. (3) SCFAs activate inflammasomes in IECs to promote the secretion of IL-18. (4) SCFAs directly pass through IECs to regulate the immune response of B cells.
5.2.2.2 Microbial Components
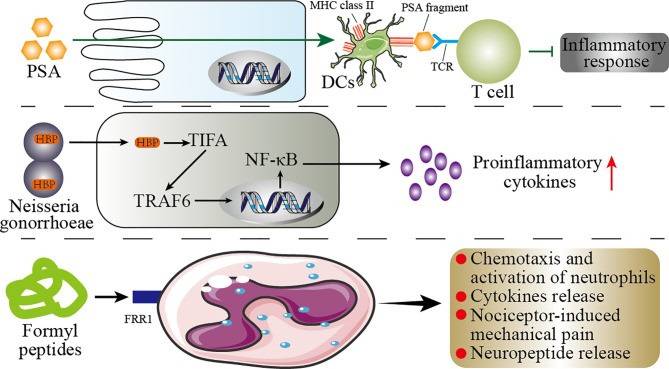
Pattern recognition receptors (PRRs) are important components of innate immunity, which are responsible for responding, detecting, and coordinating self and nonself-antigens (198). PRRs can respond to the components of a variety of microbes (such as fungi, viruses, and bacteria), including peptidoglycans, lipopolysaccharides, formyl peptides, unique nucleic acid structures, and flagellin. The activation of PRRs promotes the release of chemokines, apoptotic factors, and cytokines through a signal cascade to participate in disease occurrence (199). We summarize briefly the mechanism by which microbial components modulate the immune response in Figure 5 .
Figure 5.
Mechanisms of the microbial components regulating immunity. This figure shows the role of the microbial components polysaccharide A (PSA), formyl peptide, and HBP (d-glycero-β-d-mannoheptose-1,7-bisphosphate) in the host immunity. PSA can enter the circulatory system directly through the host’s intestinal epithelial cells (IECs). In the circulatory system, PSA connects dendritic cells (DCs) and T cells through MHC class II and T-cell receptors (TCR) to inhibit inflammation. In the host’s genitourinary tract, HBP promotes the secretion of proinflammatory cytokines by the host’s immune cells through the TIFA/TRAF6/NF-κB pathway. Formyl peptide binds to neutrophils by binding to formyl peptide receptors 1 (FRR1) to regulate the inflammatory response of neutrophils.
5.3 The Microbiota in Early Life as a Possible Predictor of the Human Health
From pregnancy to long-term planning, the microbiota plays an important role in the health of the baby. The normal microbiota of infant has a positive impact on the health in early life and long-term planning, while disturbance of the microbiota may be one of the causes of brain dysplasia, immune system defects, and metabolic disorders. In Table 6 , we summarize the microbial metabolites or components related to diseases that have been identified in recent years. Therefore, the formation factors of the infant microbiota are essential for establishing targeted strategies to minimize the early interference and long-term effects on the human health (213).
Table 6.
Microbial metabolites or components related to diseases.
| Microbial metabolites or components | Human disease or preclinical models | Refs. | |
|---|---|---|---|
| Cancer | Colorectal cancer | SCFAs, B vitamins, N1, N12-diacetylspermine | (200, 201) |
| Gynecological and reproductive disorders | Bacterial vaginosis and other sexually transmitted infections | Polyamines, HBP | (202) |
| Preterm labor | SCFAs | (203) | |
| Allergic and immune disorders | Asthma | SCFAs | (204, 205) |
| Inflammatory bowel disease | SCFAs, B vitamins | (206) | |
| Neurological disorders | Central nervous system dysfunction | SCFAs | (174) |
| Autism spectrum disorder | 4-EPS | (207) | |
| Metabolic disorders | Obesity and metabolic syndrome | TMAO | (208) |
| Kidney disease | p-Cresol, SCFAs | (209) | |
| Type 2 diabetes | TMAO | (210) | |
| Cardiovascular disease | TMAO | (209, 211) | |
| Other gastrointestinal disorders | Infectious colitis (Clostridium difficile) | Bile acids | (212) |
4-EPS, 4-ethyl phenol sulfate; HBP, d-glycero-β-d-manno-heptose-1,7-biphosphate; SCFAs, short-chain fatty acids; TMAO, trimethylamine N-oxide.
At present, the colonization of infant gut microbiota has been widely recognized for its role in brain development and shaping the immune system. The communication between the normal gut microbiota and the host contributes to the metabolism and immunity in early life (214). It is worth mentioning that the lack of colonization of specific microbiota in early life may induce inflammatory or allergic diseases later, such as asthma and inflammatory bowel disease (IBD) (215). Interestingly, the maternal agricultural exposure during pregnancy can reduce the risk of asthma in the offspring (216). This may be mediated by the microbiota, because there is evidence that agricultural exposure in early life increases the diversity of the microbiota (217). Consistently, the use of antibiotics within the first 6 months of life increases the risk of asthma and allergies at 6 years of age (218). Besides, antibiotics administered to infant within 1 year of age are associated with the development of type 2 diabetes, type 1 diabetes, overweight, and eczema later, which is associated with infant microbiota (219–221). Shortly, these data support the connection between disturbances in the microbiota in early life and long-term health. However, the specific influencing factors and mechanisms need to be further explored.
5.4 Future Study Directions of Microbiota
5.4.1 Characterizing Microbiome Composition
At present, it is possible to directly classify microbial populations without culturing due to the emergence of high-throughput DNA sequencing technology. Also, 16s RNA technology provides a reliable method for studying complex microbiota. Based on this, characterizing microbiome composition is the primary direction for future research on the role of microbiota in disease and health.
5.4.2 Investigating the Function of Microbiota
Hypertranscriptome sequencing and macrogenomics laid the foundation for exploring the functions of the microbiota. The elucidation of the functions of the microbiota or specific microbes helps to regulate clearly the health or disease of the host.
5.4.3 Causation or Correlation Between Microbiota and Host Disease
The treatment method based on micro-ecological failed to achieve the expected results, causing many conclusions in this field to be questioned. The article published by Nature explains that host variables can interfere with microbiome research, prompting researchers to fully consider host variables in the subsequent research process (222). Therefore, the causal relationship or correlation between the microbiota and the host diseases needs urgently to be studied in depth (223).
5.4.4 Analysis of the Structure and Biological Activity of Microbial Metabolites or Components
The application of mass spectrometry and chromatography is of great significance for the exploration of microbial metabolites or components (224–226). Both targeted and untargeted proteomics and metabolomics are feasible for revealing the diversity of microbial metabolites or components. Exploring and identifying potential microbial metabolites or components provides opportunities for diagnosing and treating associated diseases.
5.4.5 Isolation of Individual Strains
The isolation of individual strains or species in microbial communities has always been a challenge. Accurately capturing and analyzing the characteristics of individual microbial species is necessary for the implementation of research on microbial communities.
5.4.6 Microbial Trace
The complex interaction between the microbiota and the host is a difficult point in the study. The trace of microbes provides “visual proof” for the ecological specificity of them. This method is expected to have a thorough understanding of the mechanism of action of the microbiota in the host.
6 Conclusions
The maternal microbiota are closely associated with the health of the fetus, and their disorder can lead to adverse pregnancy outcomes, including late abortion (LA), preterm rupture of membranes (PROM), hyperemesis gravidarum (HG), premature delivery (PTD), intrauterine growth restriction, and stillbirth. The maternal microbiota would be transmitted vertically to the newborn. The birth method, gestational age at birth, feeding method, and maternal factors all determine the colonization of the infant microbiota. The gut microbiota is the most important in the infant microbiota, and their cores are Bifidobacteria, the genus Lactobacillus, the genus Clostridium, the genus Bacteroides, the genus Veillonella and Streptococcus, and the genus Collinsella and Akkermansia. The microbiota in early life is essential for the brain development of offspring and the shaping of the immune system, which affects both infant health and long-term health. Therefore, from pregnancy to early life, reasonable intervention is important to regulate the maternal or offspring microbiota for offspring’s health.
Author Contributions
YY and XC drafted the manuscript and assisted in reviewing literature. FW and YQY modified the manuscript. FC and CZ reviewed and edited the final manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (Grant Nos. 82003761 and 81873838) and Zhejiang Provincial Natural Science Foundation of China (Grant No. LY16H160025).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- 1. Goodrich JK, Davenport ER, Beaumont M, Jackson MA, Knight R, Ober C, et al. Genetic Determinants of the Gut Microbiome in UK Twins. Cell Host Microbe (2016) 19(5):731–43. doi: 10.1016/j.chom.2016.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhou P, Zhou Y, Liu B, Jin Z, Zhuang X, Dai W, et al. Perinatal Antibiotic Exposure Affects the Transmission Between Maternal and Neonatal Microbiota and Is Associated With Early-Onset Sepsis. mSphere (2020) 5(1):e00984–19. doi: 10.1128/mSphere.00984-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kim S, Kim H, Yim YS, Ha S, Atarashi K, Tan TG, et al. Maternal Gut Bacteria Promote Neurodevelopmental Abnormalities in Mouse Offspring. Nature (2017) 549(7673):528–32. doi: 10.1038/nature23910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jasarevic E, Howard CD, Misic AM, Beiting DP, Bale TL. Stress During Pregnancy Alters Temporal and Spatial Dynamics of the Maternal and Offspring Microbiome in a Sex-Specific Manner. Sci Rep (2017) 7:44182. doi: 10.1038/srep44182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Codagnone MG, Stanton C, O’Mahony SM, Dinan TG, Cryan JF. Microbiota and Neurodevelopmental Trajectories: Role of Maternal and Early-Life Nutrition. Ann Nutr Metab (2019) 74 Suppl 2:16–27. doi: 10.1159/000499144 [DOI] [PubMed] [Google Scholar]
- 6. Romero R, Hassan SS, Gajer P, Tarca AL, Fadrosh DW, Nikita L, et al. The Composition and Stability of the Vaginal Microbiota of Normal Pregnant Women Is Different From That of Non-Pregnant Women. Microbiome (2014) 2(1):4. doi: 10.1186/2049-2618-2-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Molina NM, Sola-Leyva A, Haahr T, Aghajanova L, Laudanski P, Castilla JA, et al. Analysing Endometrial Microbiome: Methodological Considerations and Recommendations for Good Practice. Hum Reprod (2021) 36(4):859–79. doi: 10.1093/humrep/deab009 [DOI] [PubMed] [Google Scholar]
- 8. Qin S, Liu Y, Wang S, Ma J, Yang H. Distribution Characteristics of Intestinal Microbiota During Pregnancy and Postpartum in Healthy Women. J Matern Fetal Neonatal Med (2021) 1-8. doi: 10.1080/14767058.2020.1812571 [DOI] [PubMed] [Google Scholar]
- 9. Ishimwe JA, Akinleye A, Johnson AC, Garrett MR, Sasser JM. Gestational Gut Microbial Remodeling Is Impaired in a Rat Model of Preeclampsia Superimposed on Chronic Hypertension. Physiol Genomics (2021) 53(3):125–36. doi: 10.1152/physiolgenomics.00121.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Altemani F, Barrett HL, Gomez-Arango L, Josh P, David McIntyre H, Callaway LK, et al. Pregnant Women Who Develop Preeclampsia Have Lower Abundance of the Butyrate-Producer Coprococcus in Their Gut Microbiota. Pregnancy Hypertens (2021) 23:211–9. doi: 10.1016/j.preghy.2021.01.002 [DOI] [PubMed] [Google Scholar]
- 11. Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The Placenta Harbors a Unique Microbiome. Sci Transl Med (2014) 6(237):237ra65. doi: 10.1126/scitranslmed.3008599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pelzer E, Gomez-Arango LF, Barrett HL, Nitert MD. Review: Maternal Health and the Placental Microbiome. Placenta (2017) 54:30–7. doi: 10.1016/j.placenta.2016.12.003 [DOI] [PubMed] [Google Scholar]
- 13. de Goffau MC, Lager S, Sovio U, Gaccioli F, Cook E, Peacock SJ, et al. Human Placenta has No Microbiome But can Contain Potential Pathogens. Nature (2019) 572(7769):329–34. doi: 10.1038/s41586-019-1451-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yao Y, Cai X, Chen C, Fang H, Zhao Y, Fei W, et al. The Role of Microbiomes in Pregnant Women and Offspring: Research Progress of Recent Years. Front Pharmacol (2020) 11:643. doi: 10.3389/fphar.2020.00643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nugent BM, Bale TL. The Omniscient Placenta: Metabolic and Epigenetic Regulation of Fetal Programming. Front Neuroendocrinol (2015) 39:28–37. doi: 10.1016/j.yfrne.2015.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lain KY, Catalano PM. Metabolic Changes in Pregnancy. Clin Obstet Gynecol (2007) 50(4):938–48. doi: 10.1097/GRF.0b013e31815a5494 [DOI] [PubMed] [Google Scholar]
- 17. Koren O, Goodrich JK, Cullender TC, Spor A, Laitinen K, Backhed HK, et al. Host Remodeling of the Gut Microbiome and Metabolic Changes During Pregnancy. Cell (2012) 150(3):470–80. doi: 10.1016/j.cell.2012.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Neuman H, Koren O. The Pregnancy Microbiome. Nestle Nutr Inst Workshop Ser (2017) 88:1–9. doi: 10.1159/000455207 [DOI] [PubMed] [Google Scholar]
- 19. Nuriel-Ohayon M, Neuman H, Koren O. Microbial Changes During Pregnancy, Birth, and Infancy. Front Microbiol (2016) 7:1031. doi: 10.3389/fmicb.2016.01031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. LeBlanc JG, Milani C, de Giori GS, Sesma F, van Sinderen D, Ventura M. Bacteria as Vitamin Suppliers to Their Host: A Gut Microbiota Perspective. Curr Opin Biotechnol (2013) 24(2):160–8. doi: 10.1016/j.copbio.2012.08.005 [DOI] [PubMed] [Google Scholar]
- 21. Yao Y, Cai X, Fei W, Ye Y, Zhao M, Zheng C. The Role of Short-Chain Fatty Acids in Immunity, Inflammation and Metabolism. Crit Rev Food Sci Nutr (2020) 1–12. doi: 10.1080/10408398.2020.1854675 [DOI] [PubMed] [Google Scholar]
- 22. Palladino E, Van Mieghem T, Connor KL. Diet Alters Micronutrient Pathways in the Gut and Placenta That Regulate Fetal Growth and Development in Pregnant Mice. Reprod Sci (2021) 28(2):447–61. doi: 10.1007/s43032-020-00297-1 [DOI] [PubMed] [Google Scholar]
- 23. Choi GB, Yim YS, Wong H, Kim S, Kim H, Kim SV, et al. The Maternal Interleukin-17a Pathway in Mice Promotes Autism-Like Phenotypes in Offspring. Science (2016) 351(6276):933–9. doi: 10.1126/science.aad0314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of Intestinal Th17 Cells by Segmented Filamentous Bacteria. Cell (2009) 139(3):485–98. doi: 10.1016/j.cell.2009.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Romero R. Prenatal Medicine: The Child Is the Father of the Man. 1996. J Matern Fetal Neonatal Med (2009) 22(8):636–9. doi: 10.1080/14767050902784171 [DOI] [PubMed] [Google Scholar]
- 26. Lawn JE, Cousens S, Zupan J, Lancet Neonatal Survival Steering T . 4 Million Neonatal Deaths: When? Where? Why? Lancet (2005) 365(9462):891–900. doi: 10.1016/S0140-6736(05)71048-5 [DOI] [PubMed] [Google Scholar]
- 27. Goldenberg RL, Hauth JC, Andrews WW. Intrauterine Infection and Preterm Delivery. N Engl J Med (2000) 342(20):1500–7. doi: 10.1056/NEJM200005183422007 [DOI] [PubMed] [Google Scholar]
- 28. Garcia-Grau I, Simon C, Moreno I. Uterine Microbiome-Low Biomass and High Expectationsdagger. Biol Reprod (2019) 101(6):1102–14. doi: 10.1093/biolre/ioy257 [DOI] [PubMed] [Google Scholar]
- 29. Watts DH, Krohn MA, Hillier SL, Eschenbach DA. The Association of Occult Amniotic Fluid Infection With Gestational Age and Neonatal Outcome Among Women in Preterm Labor. Obstet Gynecol (1992) 79(3):351–7. doi: 10.1097/00006250-199203000-00005 [DOI] [PubMed] [Google Scholar]
- 30. Theis KR, Romero R, Motomura K, Galaz J, Winters AD, Pacora P, et al. Microbial Burden and Inflammasome Activation in Amniotic Fluid of Patients With Preterm Prelabor Rupture of Membranes. J Perinat Med (2020) 48(2):115–31. doi: 10.1515/jpm-2019-0398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Romero R, Miranda J, Chaemsaithong P, Chaiworapongsa T, Kusanovic JP, Dong Z, et al. Sterile and Microbial-Associated Intra-Amniotic Inflammation in Preterm Prelabor Rupture of Membranes. J Matern Fetal Neonatal Med (2015) 28(12):1394–409. doi: 10.3109/14767058.2014.958463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Esplin MS, Romero R, Chaiworapongsa T, Kim YM, Edwin S, Gomez R, et al. Monocyte Chemotactic Protein-1 Is Increased in the Amniotic Fluid of Women Who Deliver Preterm in the Presence or Absence of Intra-Amniotic Infection. J Matern Fetal Neonatal Med (2005) 17(6):365–73. doi: 10.1080/14767050500141329 [DOI] [PubMed] [Google Scholar]
- 33. Hillier SL, Witkin SS, Krohn MA, Watts DH, Kiviat NB, Eschenbach DA. The Relationship of Amniotic Fluid Cytokines and Preterm Delivery, Amniotic Fluid Infection, Histologic Chorioamnionitis, and Chorioamnion Infection. Obstet Gynecol (1993) 81(6):941–8. [PubMed] [Google Scholar]
- 34. Mendz GL, Kaakoush NO, Quinlivan JA. Bacterial Aetiological Agents of Intra-Amniotic Infections and Preterm Birth in Pregnant Women. Front Cell Infect Microbiol (2013) 3:58. doi: 10.3389/fcimb.2013.00058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. DiGiulio DB. Diversity of Microbes in Amniotic Fluid. Semin Fetal Neonatal Med (2012) 17(1):2–11. doi: 10.1016/j.siny.2011.10.001 [DOI] [PubMed] [Google Scholar]
- 36. Mancabelli L, Tarracchini C, Milani C, Lugli GA, Fontana F, Turroni F, et al. Vaginotypes of the Human Vaginal Microbiome. Environ Microbiol (2021) 23(3):1780–92. doi: 10.1111/1462-2920.15441 [DOI] [PubMed] [Google Scholar]
- 37. Anahtar MN, Gootenberg DB, Mitchell CM, Kwon DS. Cervicovaginal Microbiota and Reproductive Health: The Virtue of Simplicity. Cell Host Microbe (2018) 23(2):159–68. doi: 10.1016/j.chom.2018.01.013 [DOI] [PubMed] [Google Scholar]
- 38. Fettweis JM, Serrano MG, Brooks JP, Edwards DJ, Girerd PH, Parikh HI, et al. The Vaginal Microbiome and Preterm Birth. Nat Med (2019) 25(6):1012–21. doi: 10.1038/s41591-019-0450-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brown RG, Al-Memar M, Marchesi JR, Lee YS, Smith A, Chan D, et al. Establishment of Vaginal Microbiota Composition in Early Pregnancy and Its Association With Subsequent Preterm Prelabor Rupture of the Fetal Membranes. Transl Res (2019) 207:30–43. doi: 10.1016/j.trsl.2018.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Elovitz MA, Gajer P, Riis V, Brown AG, Humphrys MS, Holm JB, et al. Cervicovaginal Microbiota and Local Immune Response Modulate the Risk of Spontaneous Preterm Delivery. Nat Commun (2019) 10(1):1305. doi: 10.1038/s41467-019-09285-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Abdelmaksoud AA, Koparde VN, Sheth NU, Serrano MG, Glascock AL, Fettweis JM, et al. Comparison of Lactobacillus Crispatus Isolates From Lactobacillus-Dominated Vaginal Microbiomes With Isolates From Microbiomes Containing Bacterial Vaginosis-Associated Bacteria. Microbiol (Reading) (2016) 162(3):466–75. doi: 10.1099/mic.0.000238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stout MJ, Zhou Y, Wylie KM, Tarr PI, Macones GA, Tuuli MG. Early Pregnancy Vaginal Microbiome Trends and Preterm Birth. Am J Obstet Gynecol (2017) 217(3):356 e1–356 e18. doi: 10.1016/j.ajog.2017.05.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tabatabaei N, Eren AM, Barreiro LB, Yotova V, Dumaine A, Allard C, et al. Vaginal Microbiome in Early Pregnancy and Subsequent Risk of Spontaneous Preterm Birth: A Case-Control Study. BJOG (2019) 126(3):349–58. doi: 10.1111/1471-0528.15299 [DOI] [PubMed] [Google Scholar]
- 44. Han C, Li H, Han L, Wang C, Yan Y, Qi W, et al. Aerobic Vaginitis in Late Pregnancy and Outcomes of Pregnancy. Eur J Clin Microbiol Infect Dis (2019) 38(2):233–9. doi: 10.1007/s10096-018-3416-2 [DOI] [PubMed] [Google Scholar]
- 45. Juliana NCA, Suiters MJM, Al-Nasiry S, Morre SA, Peters RPH, Ambrosino E. The Association Between Vaginal Microbiota Dysbiosis, Bacterial Vaginosis, and Aerobic Vaginitis, and Adverse Pregnancy Outcomes of Women Living in Sub-Saharan Africa: A Systematic Review. Front Public Health (2020) 8:567885. doi: 10.3389/fpubh.2020.567885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Smith B, Kemp M, Ethelberg S, Schiellerup P, Bruun BG, Gerner-Smidt P, et al. Listeria Monocytogenes: Maternal-Foetal Infections in Denmark 1994-2005. Scand J Infect Dis (2009) 41(1):21–5. doi: 10.1080/00365540802468094 [DOI] [PubMed] [Google Scholar]
- 47. El Aila NA, Tency I, Claeys G, Verstraelen H, Saerens B, Santiago GL, et al. Identification and Genotyping of Bacteria From Paired Vaginal and Rectal Samples From Pregnant Women Indicates Similarity Between Vaginal and Rectal Microflora. BMC Infect Dis (2009) 9:167. doi: 10.1186/1471-2334-9-167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu Y, Chen H, Feng L, Zhang J. Interactions Between Gut Microbiota and Metabolites Modulate Cytokine Network Imbalances in Women With Unexplained Miscarriage. NPJ Biofilms Microbiomes (2021) 7(1):24. doi: 10.1038/s41522-021-00199-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gershuni V, Li Y, Elovitz M, Li H, Wu GD, Compher CW. Maternal Gut Microbiota Reflecting Poor Diet Quality Is Associated With Spontaneous Preterm Birth in a Prospective Cohort Study. Am J Clin Nutr (2021) 113(3):602–11. doi: 10.1093/ajcn/nqaa361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sweeney EL, Kallapur SG, Gisslen T, Lambers DS, Chougnet CA, Stephenson SA, et al. Placental Infection With Ureaplasma Species Is Associated With Histologic Chorioamnionitis and Adverse Outcomes in Moderately Preterm and Late-Preterm Infants. J Infect Dis (2016) 213(8):1340–7. doi: 10.1093/infdis/jiv587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Prince AL, Ma J, Kannan PS, Alvarez M, Gisslen T, Harris RA, et al. The Placental Membrane Microbiome Is Altered Among Subjects With Spontaneous Preterm Birth With and Without Chorioamnionitis. Am J Obstet Gynecol (2016) 214(5):627.e1–627.e16. doi: 10.1016/j.ajog.2016.01.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sterpu I, Fransson E, Hugerth LW, Du J, Pereira M, Cheng L, et al. No Evidence for a Placental Microbiome in Human Pregnancies at Term. Am J Obstet Gynecol (2021) 224(3):296.e1–296.e23. doi: 10.1016/j.ajog.2020.08.103 [DOI] [PubMed] [Google Scholar]
- 53. Kuperman AA, Zimmerman A, Hamadia S, Ziv O, Gurevich V, Fichtman B, et al. Deep Microbial Analysis of Multiple Placentas Shows No Evidence for a Placental Microbiome. BJOG (2020) 127(2):159–69. doi: 10.1111/1471-0528.15896 [DOI] [PubMed] [Google Scholar]
- 54. Cobb CM, Kelly PJ, Williams KB, Babbar S, Angolkar M, Derman RJ. The Oral Microbiome and Adverse Pregnancy Outcomes. Int J Womens Health (2017) 9:551–9. doi: 10.2147/IJWH.S142730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tavarna T, Phillips PL, Wu XJ, Reyes L. Fetal Growth Restriction Is a Host Specific Response to Infection With an Impaired Spiral Artery Remodeling-Inducing Strain of Porphyromonas Gingivalis. Sci Rep (2020) 10(1):14606. doi: 10.1038/s41598-020-71762-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chanomethaporn A, Chayasadom A, Wara-Aswapati N, Kongwattanakul K, Suwannarong W, Tangwanichgapong K, et al. Association Between Periodontitis and Spontaneous Abortion: A Case-Control Study. J Periodontol (2019) 90(4):381–90. doi: 10.1002/JPER.18-0174 [DOI] [PubMed] [Google Scholar]
- 57. Ye C, Kobayashi H, Katagiri S, Miyasaka N, Takeuchi Y, Kuraji R, et al. The Relationship Between the Anti-Porphyromonas Gingivalis Immunoglobulin G Subclass Antibody and Small for Gestational Age Delivery: A Longitudinal Study in Pregnant Japanese Women. Int Dent J (2020) 70(4):296–302. doi: 10.1111/idj.12548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Daalderop LA, Wieland BV, Tomsin K, Reyes L, Kramer BW, Vanterpool SF, et al. Periodontal Disease and Pregnancy Outcomes: Overview of Systematic Reviews. JDR Clin Trans Res (2018) 3(1):10–27. doi: 10.1177/2380084417731097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Maekawa T, Krauss JL, Abe T, Jotwani R, Triantafilou M, Triantafilou K, et al. Porphyromonas Gingivalis Manipulates Complement and TLR Signaling to Uncouple Bacterial Clearance From Inflammation and Promote Dysbiosis. Cell Host Microbe (2014) 15(6):768–78. doi: 10.1016/j.chom.2014.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jia L, Han N, Du J, Guo L, Luo Z, Liu Y. Pathogenesis of Important Virulence Factors of Porphyromonas Gingivalis via Toll-Like Receptors. Front Cell Infect Microbiol (2019) 9:262. doi: 10.3389/fcimb.2019.00262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cheng WC, Hughes FJ, Taams LS. The Presence, Function and Regulation of IL-17 and Th17 Cells in Periodontitis. J Clin Periodontol (2014) 41(6):541–9. doi: 10.1111/jcpe.12238 [DOI] [PubMed] [Google Scholar]
- 62. Chopra A, Radhakrishnan R, Sharma M. Porphyromonas Gingivalis and Adverse Pregnancy Outcomes: A Review on its Intricate Pathogenic Mechanisms. Crit Rev Microbiol (2020) 46(2):213–36. doi: 10.1080/1040841X.2020.1747392 [DOI] [PubMed] [Google Scholar]
- 63. Rodriguez JM, Murphy K, Stanton C, Ross RP, Kober OI, Juge N, et al. The Composition of the Gut Microbiota Throughout Life, With an Emphasis on Early Life. Microb Ecol Health Dis (2015) 26:26050. doi: 10.3402/mehd.v26.26050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Korpela K, de Vos WM. Early Life Colonization of the Human Gut: Microbes Matter Everywhere. Curr Opin Microbiol (2018) 44:70–8. doi: 10.1016/j.mib.2018.06.003 [DOI] [PubMed] [Google Scholar]
- 65. Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery Mode Shapes the Acquisition and Structure of the Initial Microbiota Across Multiple Body Habitats in Newborns. Proc Natl Acad Sci USA (2010) 107(26):11971–5. doi: 10.1073/pnas.1002601107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tamburini S, Shen N, Wu HC, Clemente JC. The Microbiome in Early Life: Implications for Health Outcomes. Nat Med (2016) 22(7):713–22. doi: 10.1038/nm.4142 [DOI] [PubMed] [Google Scholar]
- 67. Clemente JC, Ursell LK, Parfrey LW, Knight R. The Impact of the Gut Microbiota on Human Health: An Integrative View. Cell (2012) 148(6):1258–70. doi: 10.1016/j.cell.2012.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Mitsuoka T. Intestinal Flora and Human Health. Asia Pac J Clin Nutr (1996) 5(1):2–9. [PubMed] [Google Scholar]
- 69. Avershina E, Lundgard K, Sekelja M, Dotterud C, Storro O, Oien T, et al. Transition From Infant- to Adult-Like Gut Microbiota. Environ Microbiol (2016) 18(7):2226–36. doi: 10.1111/1462-2920.13248 [DOI] [PubMed] [Google Scholar]
- 70. Avershina E, Storro O, Oien T, Johnsen R, Wilson R, Egeland T, et al. Bifidobacterial Succession and Correlation Networks in a Large Unselected Cohort of Mothers and Their Children. Appl Environ Microbiol (2013) 79(2):497–507. doi: 10.1128/AEM.02359-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yang R, Gao R, Cui S, Zhong H, Zhang X, Chen Y, et al. Dynamic Signatures of Gut Microbiota and Influences of Delivery and Feeding Modes During the First 6 Months of Life. Physiol Genomics (2019) 51(8):368–78. doi: 10.1152/physiolgenomics.00026.2019 [DOI] [PubMed] [Google Scholar]
- 72. Montoya-Williams D, Lemas DJ, Spiryda L, Patel K, Carney OO, Neu J, et al. The Neonatal Microbiome and Its Partial Role in Mediating the Association Between Birth by Cesarean Section and Adverse Pediatric Outcomes. Neonatology (2018) 114(2):103–11. doi: 10.1159/000487102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Akagawa S, Tsuji S, Onuma C, Akagawa Y, Yamaguchi T, Yamagishi M, et al. Effect of Delivery Mode and Nutrition on Gut Microbiota in Neonates. Ann Nutr Metab (2019) 74(2):132–9. doi: 10.1159/000496427 [DOI] [PubMed] [Google Scholar]
- 74. Reyman M, van Houten MA, van Baarle D, Bosch A, Man WH, Chu M, et al. Impact of Delivery Mode-Associated Gut Microbiota Dynamics on Health in the First Year of Life. Nat Commun (2019) 10(1):4997. doi: 10.1038/s41467-019-13014-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Dominguez-Bello MG, De Jesus-Laboy KM, Shen N, Cox LM, Amir A, Gonzalez A, et al. Partial Restoration of the Microbiota of Cesarean-Born Infants via Vaginal Microbial Transfer. Nat Med (2016) 22(3):250–3. doi: 10.1038/nm.4039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Stokholm J, Thorsen J, Blaser MJ, Rasmussen MA, Hjelmso M, Shah S, et al. Delivery Mode and Gut Microbial Changes Correlate With an Increased Risk of Childhood Asthma. Sci Transl Med (2020) 12(569):eaax9929. doi: 10.1126/scitranslmed.aax9929 [DOI] [PubMed] [Google Scholar]
- 77. Stinson LF, Payne MS, Keelan JA. A Critical Review of the Bacterial Baptism Hypothesis and the Impact of Cesarean Delivery on the Infant Microbiome. Front Med (Lausanne) (2018) 5:135. doi: 10.3389/fmed.2018.00135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hill CJ, Lynch DB, Murphy K, Ulaszewska M, Jeffery IB, O’Shea CA, et al. Evolution of Gut Microbiota Composition From Birth to 24 Weeks in the INFANTMET Cohort. Microbiome (2017) 5(1):4. doi: 10.1186/s40168-016-0213-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Rouge C, Goldenberg O, Ferraris L, Berger B, Rochat F, Legrand A, et al. Investigation of the Intestinal Microbiota in Preterm Infants Using Different Methods. Anaerobe (2010) 16(4):362–70. doi: 10.1016/j.anaerobe.2010.06.002 [DOI] [PubMed] [Google Scholar]
- 80. Jacquot A, Neveu D, Aujoulat F, Mercier G, Marchandin H, Jumas-Bilak E, et al. Dynamics and Clinical Evolution of Bacterial Gut Microflora in Extremely Premature Patients. J Pediatr (2011) 158(3):390–6. doi: 10.1016/j.jpeds.2010.09.007 [DOI] [PubMed] [Google Scholar]
- 81. Arboleya S, Binetti A, Salazar N, Fernandez N, Solis G, Hernandez-Barranco A, et al. Establishment and Development of Intestinal Microbiota in Preterm Neonates. FEMS Microbiol Ecol (2012) 79(3):763–72. doi: 10.1111/j.1574-6941.2011.01261.x [DOI] [PubMed] [Google Scholar]
- 82. Arboleya S, Sanchez B, Milani C, Duranti S, Solis G, Fernandez N, et al. Intestinal Microbiota Development in Preterm Neonates and Effect of Perinatal Antibiotics. J Pediatr (2015) 166(3):538–44. doi: 10.1016/j.jpeds.2014.09.041 [DOI] [PubMed] [Google Scholar]
- 83. Cong X, Xu W, Janton S, Henderson WA, Matson A, McGrath JM, et al. Gut Microbiome Developmental Patterns in Early Life of Preterm Infants: Impacts of Feeding and Gender. PloS One (2016) 11(4):e0152751. doi: 10.1371/journal.pone.0152751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. La Rosa PS, Warner BB, Zhou Y, Weinstock GM, Sodergren E, Hall-Moore CM, et al. Patterned Progression of Bacterial Populations in the Premature Infant Gut. Proc Natl Acad Sci USA (2014) 111(34):12522–7. doi: 10.1073/pnas.1409497111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Collado MC, Cernada M, Neu J, Perez-Martinez G, Gormaz M, Vento M. Factors Influencing Gastrointestinal Tract and Microbiota Immune Interaction in Preterm Infants. Pediatr Res (2015) 77(6):726–31. doi: 10.1038/pr.2015.54 [DOI] [PubMed] [Google Scholar]
- 86. Torrazza RM, Neu J. The Altered Gut Microbiome and Necrotizing Enterocolitis. Clin Perinatol (2013) 40(1):93–108. doi: 10.1016/j.clp.2012.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Arboleya S, Sanchez B, Solis G, Fernandez N, Suarez M, Hernandez-Barranco AM, et al. Impact of Prematurity and Perinatal Antibiotics on the Developing Intestinal Microbiota: A Functional Inference Study. Int J Mol Sci (2016) 17(5):649. doi: 10.3390/ijms17050649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. O’Sullivan A, Farver M, Smilowitz JT. The Influence of Early Infant-Feeding Practices on the Intestinal Microbiome and Body Composition in Infants. Nutr Metab Insights (2015) 8(Suppl 1):1–9. doi: 10.4137/NMI.S29530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Li N, Xie Q, Zhao L, Shi J, Evivie SE, Lv X, et al. Human Milk and Infant Formula Modulate the Intestinal Microbiota and Immune Systems of Human Microbiota-Associated Mice. Food Funct (2021) 12(6):2784–98. doi: 10.1039/d0fo03004j [DOI] [PubMed] [Google Scholar]
- 90. Bezirtzoglou E, Tsiotsias A, Welling GW. Microbiota Profile in Feces of Breast- and Formula-Fed Newborns by Using Fluorescence. Situ Hybridization (FISH) Anaerobe (2011) 17(6):478–82. doi: 10.1016/j.anaerobe.2011.03.009 [DOI] [PubMed] [Google Scholar]
- 91. Maynard CL, Elson CO, Hatton RD, Weaver CT. Reciprocal Interactions of the Intestinal Microbiota and Immune System. Nature (2012) 489(7415):231–41. doi: 10.1038/nature11551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Bode L, McGuire M, Rodriguez JM, Geddes DT, Hassiotou F, Hartmann PE, et al. It’s Alive: Microbes and Cells in Human Milk and Their Potential Benefits to Mother and Infant. Adv Nutr (2014) 5(5):571–3. doi: 10.3945/an.114.006643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Lonnerdal B. Bioactive Proteins in Human Milk: Mechanisms of Action. J Pediatr (2010) 156(2 Suppl):S26–30. doi: 10.1016/j.jpeds.2009.11.017 [DOI] [PubMed] [Google Scholar]
- 94. Walker A. Breast Milk as the Gold Standard for Protective Nutrients. J Pediatr (2010) 156(2 Suppl):S3–7. doi: 10.1016/j.jpeds.2009.11.021 [DOI] [PubMed] [Google Scholar]
- 95. Fernandez L, Ruiz L, Jara J, Orgaz B, Rodriguez JM. Strategies for the Preservation, Restoration and Modulation of the Human Milk Microbiota. Implications for Human Milk Banks and Neonatal Intensive Care Units. Front Microbiol (2018) 9:2676. doi: 10.3389/fmicb.2018.02676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Khodayar-Pardo P, Mira-Pascual L, Collado MC, Martinez-Costa C. Impact of Lactation Stage, Gestational Age and Mode of Delivery on Breast Milk Microbiota. J Perinatol (2014) 34(8):599–605. doi: 10.1038/jp.2014.47 [DOI] [PubMed] [Google Scholar]
- 97. Ballard O, Morrow AL. Human Milk Composition: Nutrients and Bioactive Factors. Pediatr Clin North Am (2013) 60(1):49–74. doi: 10.1016/j.pcl.2012.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Cacho NT, Lawrence RM. Innate Immunity and Breast Milk. Front Immunol (2017) 8:584. doi: 10.3389/fimmu.2017.00584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Harmsen HJ, Wildeboer-Veloo AC, Raangs GC, Wagendorp AA, Klijn N, Bindels JG, et al. Analysis of Intestinal Flora Development in Breast-Fed and Formula-Fed Infants by Using Molecular Identification and Detection Methods. J Pediatr Gastroenterol Nutr (2000) 30(1):61–7. doi: 10.1097/00005176-200001000-00019 [DOI] [PubMed] [Google Scholar]
- 100. Forbes JD, Azad MB, Vehling L, Tun HM, Konya TB, Guttman DS, et al. Association of Exposure to Formula in the Hospital and Subsequent Infant Feeding Practices With Gut Microbiota and Risk of Overweight in the First Year of Life. JAMA Pediatr (2018) 172(7):e181161. doi: 10.1001/jamapediatrics.2018.1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Alsharairi NA. The Infant Gut Microbiota and Risk of Asthma: The Effect of Maternal Nutrition During Pregnancy and Lactation. Microorganisms (2020) 8(8):1119. doi: 10.3390/microorganisms8081119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Maher SE, O’Brien EC, Moore RL, Byrne DF, Geraghty AA, Saldova R, et al. The Association Between the Maternal Diet and the Maternal and Infant Gut Microbiome: A Systematic Review. Br J Nutr (2020) 1–29. doi: 10.1017/S0007114520000847 [DOI] [PubMed] [Google Scholar]
- 103. Cortes-Macias E, Selma-Royo M, Garcia-Mantrana I, Calatayud M, Gonzalez S, Martinez-Costa C, et al. Maternal Diet Shapes the Breast Milk Microbiota Composition and Diversity: Impact of Mode of Delivery and Antibiotic Exposure. J Nutr (2021) 151(2):330–40. doi: 10.1093/jn/nxaa310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. LeMay-Nedjelski L, Asbury MR, Butcher J, Ley SH, Hanley AJ, Kiss A, et al. Maternal Diet and Infant Feeding Practices Are Associated With Variation in the Human Milk Microbiota at 3 Months Postpartum in a Cohort of Women With High Rates of Gestational Glucose Intolerance. J Nutr (2021) 151(2):320–9. doi: 10.1093/jn/nxaa248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Seferovic MD, Mohammad M, Pace RM, Engevik M, Versalovic J, Bode L, et al. Maternal Diet Alters Human Milk Oligosaccharide Composition With Implications for the Milk Metagenome. Sci Rep (2020) 10(1):22092. doi: 10.1038/s41598-020-79022-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Fouhy F, Ross RP, Fitzgerald GF, Stanton C, Cotter PD. Composition of the Early Intestinal Microbiota: Knowledge, Knowledge Gaps and the Use of High-Throughput Sequencing to Address These Gaps. Gut Microbes (2012) 3(3):203–20. doi: 10.4161/gmic.20169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, et al. Factors Influencing the Composition of the Intestinal Microbiota in Early Infancy. Pediatrics (2006) 118(2):511–21. doi: 10.1542/peds.2005-2824 [DOI] [PubMed] [Google Scholar]
- 108. Adlerberth I, Strachan DP, Matricardi PM, Ahrne S, Orfei L, Aberg N, et al. Gut Microbiota and Development of Atopic Eczema in 3 European Birth Cohorts. J Allergy Clin Immunol (2007) 120(2):343–50. doi: 10.1016/j.jaci.2007.05.018 [DOI] [PubMed] [Google Scholar]
- 109. Laursen MF, Zachariassen G, Bahl MI, Bergstrom A, Host A, Michaelsen KF, et al. Having Older Siblings Is Associated With Gut Microbiota Development During Early Childhood. BMC Microbiol (2015) 15:154. doi: 10.1186/s12866-015-0477-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Arrieta MC, Stiemsma LT, Amenyogbe N, Brown EM, Finlay B. The Intestinal Microbiome in Early Life: Health and Disease. Front Immunol (2014) 5:427(427). doi: 10.3389/fimmu.2014.00427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, et al. Impact of Diet in Shaping Gut Microbiota Revealed by a Comparative Study in Children From Europe and Rural Africa. Proc Natl Acad Sci USA (2010) 107(33):14691–6. doi: 10.1073/pnas.1005963107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J, et al. Individuality in Gut Microbiota Composition Is a Complex Polygenic Trait Shaped by Multiple Environmental and Host Genetic Factors. Proc Natl Acad Sci USA (2010) 107(44):18933–8. doi: 10.1073/pnas.1007028107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Turpin W, Espin-Garcia O, Xu W, Silverberg MS, Kevans D, Smith MI, et al. Association of Host Genome With Intestinal Microbial Composition in a Large Healthy Cohort. Nat Genet (2016) 48(11):1413–7. doi: 10.1038/ng.3693 [DOI] [PubMed] [Google Scholar]
- 114. Stewart JA, Chadwick VS, Murray A. Investigations Into the Influence of Host Genetics on the Predominant Eubacteria in the Faecal Microflora of Children. J Med Microbiol (2005) 54(Pt 12):1239–42. doi: 10.1099/jmm.0.46189-0 [DOI] [PubMed] [Google Scholar]
- 115. Ke X, Walker A, Haange SB, Lagkouvardos I, Liu Y, Schmitt-Kopplin P, et al. Synbiotic-Driven Improvement of Metabolic Disturbances Is Associated With Changes in the Gut Microbiome in Diet-Induced Obese Mice. Mol Metab (2019) 22:96–109. doi: 10.1016/j.molmet.2019.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Plovier H, Everard A, Druart C, Depommier C, Van Hul M, Geurts L, et al. A Purified Membrane Protein From Akkermansia Muciniphila or the Pasteurized Bacterium Improves Metabolism in Obese and Diabetic Mice. Nat Med (2017) 23(1):107–13. doi: 10.1038/nm.4236 [DOI] [PubMed] [Google Scholar]
- 117. Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the Human Infant Intestinal Microbiota. PloS Biol (2007) 5(7):e177. doi: 10.1371/journal.pbio.0050177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Bjorksten B, Sepp E, Julge K, Voor T, Mikelsaar M. Allergy Development and the Intestinal Microflora During the First Year of Life. J Allergy Clin Immunol (2001) 108(4):516–20. doi: 10.1067/mai.2001.118130 [DOI] [PubMed] [Google Scholar]
- 119. Kalliomaki M, Collado MC, Salminen S, Isolauri E. Early Differences in Fecal Microbiota Composition in Children may Predict Overweight. Am J Clin Nutr (2008) 87(3):534–8. doi: 10.1093/ajcn/87.3.534 [DOI] [PubMed] [Google Scholar]
- 120. Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, et al. Succession of Microbial Consortia in the Developing Infant Gut Microbiome. Proc Natl Acad Sci USA (2011) 108 Suppl 1:4578–85. doi: 10.1073/pnas.1000081107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Claesson MJ, Cusack S, O’Sullivan O, Greene-Diniz R, de Weerd H, Flannery E, et al. Composition, Variability, and Temporal Stability of the Intestinal Microbiota of the Elderly. Proc Natl Acad Sci U.S.A. (2011) 108 Suppl 1:4586–91. doi: 10.1073/pnas.1000097107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Valles Y, Artacho A, Pascual-Garcia A, Ferrus ML, Gosalbes MJ, Abellan JJ, et al. Microbial Succession in the Gut: Directional Trends of Taxonomic and Functional Change in a Birth Cohort of Spanish Infants. PloS Genet (2014) 10(6):e1004406. doi: 10.1371/journal.pgen.1004406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Yassour M, Vatanen T, Siljander H, Hamalainen AM, Harkonen T, Ryhanen SJ, et al. Natural History of the Infant Gut Microbiome and Impact of Antibiotic Treatment on Bacterial Strain Diversity and Stability. Sci Transl Med (2016) 8(343):343ra81. doi: 10.1126/scitranslmed.aad0917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J, et al. The First Microbial Colonizers of the Human Gut: Composition, Activities, and Health Implications of the Infant Gut Microbiota. Microbiol Mol Biol Rev (2017) 81(4):507-14. doi: 10.1128/MMBR.00036-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Marcobal A, Barboza M, Sonnenburg ED, Pudlo N, Martens EC, Desai P, et al. Bacteroides in the Infant Gut Consume Milk Oligosaccharides via Mucus-Utilization Pathways. Cell Host Microbe (2011) 10(5):507–14. doi: 10.1016/j.chom.2011.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the Human Gut Microbiome. Nature (2011) 473(7346):174–80. doi: 10.1038/nature09944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Vipperla K, O’Keefe SJ. The Microbiota and its Metabolites in Colonic Mucosal Health and Cancer Risk. Nutr Clin Pract (2012) 27(5):624–35. doi: 10.1177/0884533612452012 [DOI] [PubMed] [Google Scholar]
- 128. Solis G, de Los Reyes-Gavilan CG, Fernandez N, Margolles A, Gueimonde M. Establishment and Development of Lactic Acid Bacteria and Bifidobacteria Microbiota in Breast-Milk and the Infant Gut. Anaerobe (2010) 16(3):307–10. doi: 10.1016/j.anaerobe.2010.02.004 [DOI] [PubMed] [Google Scholar]
- 129. Dogra S, Sakwinska O, Soh SE, Ngom-Bru C, Bruck WM, Berger B, et al. Dynamics of Infant Gut Microbiota Are Influenced by Delivery Mode and Gestational Duration and Are Associated With Subsequent Adiposity. mBio (2015) 6(1):e02419-14. doi: 10.1128/mBio.02419-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Cha H. [Experiences of a Social Therapist. Endlessly, Limitlessly Addicted]. Krankenpfl Soins Infirm (1994) 87(8):50–1. [PubMed] [Google Scholar]
- 131. Milani C, Mancabelli L, Lugli GA, Duranti S, Turroni F, Ferrario C, et al. Exploring Vertical Transmission of Bifidobacteria From Mother to Child. Appl Environ Microbiol (2015) 81(20):7078–87. doi: 10.1128/AEM.02037-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Sakanaka M, Gotoh A, Yoshida K, Odamaki T, Koguchi H, Xiao JZ, et al. Varied Pathways of Infant Gut-Associated Bifidobacterium to Assimilate Human Milk Oligosaccharides: Prevalence of the Gene Set and Its Correlation With Bifidobacteria-Rich Microbiota Formation. Nutrients (2019) 12(1):71. doi: 10.3390/nu12010071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Underwood MA, German JB, Lebrilla CB, Mills DA. Bifidobacterium Longum Subspecies Infantis: Champion Colonizer of the Infant Gut. Pediatr Res (2015) 77(1-2):229–35. doi: 10.1038/pr.2014.156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Alcon-Giner C, Dalby MJ, Caim S, Ketskemety J, Shaw A, Sim K, et al. Microbiota Supplementation With Bifidobacterium and Lactobacillus Modifies the Preterm Infant Gut Microbiota and Metabolome: An Observational Study. Cell Rep Med (2020) 1(5):100077. doi: 10.1016/j.xcrm.2020.100077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Nagpal R, Tsuji H, Takahashi T, Kawashima K, Nagata S, Nomoto K, et al. Sensitive Quantitative Analysis of the Meconium Bacterial Microbiota in Healthy Term Infants Born Vaginally or by Cesarean Section. Front Microbiol (2016) 7:1997. doi: 10.3389/fmicb.2016.01997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Yang B, Chen Y, Stanton C, Ross RP, Lee YK, Zhao J, et al. Bifidobacterium and Lactobacillus Composition at Species Level and Gut Microbiota Diversity in Infants Before 6 Weeks. Int J Mol Sci (2019) 20(13):3306. doi: 10.3390/ijms20133306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Jamyuang C, Phoonlapdacha P, Chongviriyaphan N, Chanput W, Nitisinprasert S, Nakphaichit M. Characterization and Probiotic Properties of Lactobacilli From Human Breast Milk. 3 Biotech (2019) 9(11):398. doi: 10.1007/s13205-019-1926-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Thongaram T, Hoeflinger JL, Chow J, Miller MJ. Human Milk Oligosaccharide Consumption by Probiotic and Human-Associated Bifidobacteria and Lactobacilli. J Dairy Sci (2017) 100(10):7825–33. doi: 10.3168/jds.2017-12753 [DOI] [PubMed] [Google Scholar]
- 139. Sung V, D’Amico F, Cabana MD, Chau K, Koren G, Savino F, et al. Lactobacillus Reuteri to Treat Infant Colic: A Meta-Analysis. Pediatrics (2018) 141(1):e20171811. doi: 10.1542/peds.2017-1811 [DOI] [PubMed] [Google Scholar]
- 140. Kiu R, Caim S, Alcon-Giner C, Belteki G, Clarke P, Pickard D, et al. Preterm Infant-Associated Clostridium Tertium, Clostridium Cadaveris, and Clostridium Paraputrificum Strains: Genomic and Evolutionary Insights. Genome Biol Evol (2017) 9(10):2707–14. doi: 10.1093/gbe/evx210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Cui QQ, Yang J, Niu YN, Qiang CX, Li ZR, Xu KY, et al. Epidemiological Investigation of Clostridioides Difficile Colonization in Chinese Community Infants. Anaerobe (2019) 56:116–23. doi: 10.1016/j.anaerobe.2019.01.008 [DOI] [PubMed] [Google Scholar]
- 142. Mevissen-Verhage EA, Marcelis JH, de Vos MN, Harmsen-van Amerongen WC, Verhoef J. Bifidobacterium, Bacteroides, and Clostridium Spp. In Fecal Samples From Breast-Fed and Bottle-Fed Infants With and Without Iron Supplement. J Clin Microbiol (1987) 25(2):285–9. doi: 10.1128/JCM.25.2.285-289.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Al Radaideh AJ, Badran EF, Shehabi AA. Diversity of Toxin Genotypes and Antimicrobial Susceptibility of Clostridium Perfringens Isolates From Feces of Infants. Germs (2019) 9(1):28–34. doi: 10.18683/germs.2019.1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Rada V, Nevoral J, Trojanova I, Tomankova E, Smehilova M, Killer J. Growth of Infant Faecal Bifidobacteria and Clostridia on Prebiotic Oligosaccharides in In Vitro Conditions. Anaerobe (2008) 14(4):205–8. doi: 10.1016/j.anaerobe.2008.05.003 [DOI] [PubMed] [Google Scholar]
- 145. Romyasamit C, Thatrimontrichai A, Aroonkesorn A, Chanket W, Ingviya N, Saengsuwan P, et al. Enterococcus Faecalis Isolated From Infant Feces Inhibits Toxigenic Clostridioides (Clostridium) Difficile. Front Pediatr (2020) 8:572633. doi: 10.3389/fped.2020.572633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Kim YG, Sakamoto K, Seo SU, Pickard JM, Gillilland MG, 3rd, Pudlo NA, et al. Neonatal Acquisition of Clostridia Species Protects Against Colonization by Bacterial Pathogens. Science (2017) 356(6335):315–9. doi: 10.1126/science.aag2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Lopetuso LR, Scaldaferri F, Petito V, Gasbarrini A. Commensal Clostridia: Leading Players in the Maintenance of Gut Homeostasis. Gut Pathog (2013) 5(1):23. doi: 10.1186/1757-4749-5-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Maurice CF, Haiser HJ, Turnbaugh PJ. Xenobiotics Shape the Physiology and Gene Expression of the Active Human Gut Microbiome. Cell (2013) 152(1-2):39–50. doi: 10.1016/j.cell.2012.10.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Gregory KE, LaPlante RD, Shan G, Kumar DV, Gregas M. Mode of Birth Influences Preterm Infant Intestinal Colonization With Bacteroides Over the Early Neonatal Period. Adv Neonatal Care (2015) 15(6):386–93. doi: 10.1097/ANC.0000000000000237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Sanchez E, De Palma G, Capilla A, Nova E, Pozo T, Castillejo G, et al. Influence of Environmental and Genetic Factors Linked to Celiac Disease Risk on Infant Gut Colonization by Bacteroides Species. Appl Environ Microbiol (2011) 77(15):5316–23. doi: 10.1128/AEM.00365-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Jakobsson HE, Abrahamsson TR, Jenmalm MC, Harris K, Quince C, Jernberg C, et al. Decreased Gut Microbiota Diversity, Delayed Bacteroidetes Colonisation and Reduced Th1 Responses in Infants Delivered by Caesarean Section. Gut (2014) 63(4):559–66. doi: 10.1136/gutjnl-2012-303249 [DOI] [PubMed] [Google Scholar]
- 152. Chia LW, Mank M, Blijenberg B, Aalvink S, Bongers RS, Stahl B, et al. Bacteroides Thetaiotaomicron Fosters the Growth of Butyrate-Producing Anaerostipes Caccae in the Presence of Lactose and Total Human Milk Carbohydrates. Microorganisms (2020) 8(10):1513. doi: 10.3390/microorganisms8101513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Farah S, Alshehri MA, Alfawaz TS, Ahmad M, Alshahrani DA. Bacteroides Fragilis Meningitis in a Saudi Infant: Case Report and Literature Review. Int J Pediatr Adolesc Med (2018) 5(3):122–6. doi: 10.1016/j.ijpam.2018.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Suzuki S, Shimojo N, Tajiri Y, Kumemura M, Kohno Y. A Quantitative and Relative Increase in Intestinal Bacteroides in Allergic Infants in Rural Japan. Asian Pac J Allergy Immunol (2008) 26(2-3):113–9. [PubMed] [Google Scholar]
- 155. Feng D, Neuweiler I, Nogueira R, Nackenhorst U. Modeling of Symbiotic Bacterial Biofilm Growth With an Example of the Streptococcus-Veillonella Sp. System. Bull Math Biol (2021) 83(5):48. doi: 10.1007/s11538-021-00888-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Mashima I, Liao YC, Miyakawa H, Theodorea CF, Thawboon B, Thaweboon S, et al. Veillonella Infantium Sp. Nov., an Anaerobic, Gram-Stain-Negative Coccus Isolated From Tongue Biofilm of a Thai Child. Int J Syst Evol Microbiol (2018) 68(4):1101–6. doi: 10.1099/ijsem.0.002632 [DOI] [PubMed] [Google Scholar]
- 157. Wang L, de Angel Sola D, Acevedo Flores M, Schriefer A, Wang L, Geronimo Lopez K, et al. Prenatal Food Insecurity Post Hurricane Maria Is Associated With Decreased Veillonella in the Infant Gut. Pediatr Res (2020) 88(6):917–24. doi: 10.1038/s41390-020-0839-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Saad R, Tsoi K, Onac IA, Davies KA, Hajela VK, Tacu C. Streptococcus-Associated Vasculitis: A Role for Antibiotic Therapy? IDCases (2021) 24:e01071. doi: 10.1016/j.idcr.2021.e01071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Puopolo KM, Lynfield R, Cummings JJ, Committee On F. Newborn, and Committee On Infectious D. Management of Infants at Risk for Group B Streptococcal Disease. Pediatrics (2019) 144(2):e20191881. doi: 10.1542/peds.2019-1881 [DOI] [PubMed] [Google Scholar]
- 160. Madrid L, Seale AC, Kohli-Lynch M, Edmond KM, Lawn JE, Heath PT, et al. Infant Group B Streptococcal Disease Incidence and Serotypes Worldwide: Systematic Review and Meta-Analyses. Clin Infect Dis (2017) 65(suppl_2):S160–72. doi: 10.1093/cid/cix656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Cassidy-Bushrow AE, Sitarik A, Levin AM, Lynch SV, Havstad S, Ownby DR, et al. Maternal Group B Streptococcus and the Infant Gut Microbiota. J Dev Orig Health Dis (2016) 7(1):45–53. doi: 10.1017/S2040174415001361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Kageyama A, Benno Y, Nakase T. Phylogenetic and Phenotypic Evidence for the Transfer of Eubacterium Aerofaciens to the Genus Collinsella as Collinsella Aerofaciens Gen. Nov., Comb. Nov. Int J Syst Bacteriol (1999) 49 Pt 2:557–65. doi: 10.1099/00207713-49-2-557 [DOI] [PubMed] [Google Scholar]
- 163. Nagai F, Watanabe Y, Morotomi M. Slackia Piriformis Sp. Nov. And Collinsella Tanakaei Sp. Nov., New Members of the Family Coriobacteriaceae, Isolated From Human Faeces. Int J Syst Evol Microbiol (2010) 60(Pt 11):2639–46. doi: 10.1099/ijs.0.017533-0 [DOI] [PubMed] [Google Scholar]
- 164. Durand GA, Cadoret F, Lagier JC, Fournier PE, Raoult D. Description of ‘Gorbachella Massiliensis’ Gen. Nov., Sp. Nov., ‘Fenollaria Timonensis’ Sp. Nov., ‘Intestinimonas Timonensis’ Sp. Nov. And ‘Collinsella Ihuae’ Sp. Nov. Isolated From Healthy Fresh Stools With Culturomics. New Microbes New Infect (2017) 16:60–2. doi: 10.1016/j.nmni.2017.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Derrien M, Collado MC, Ben-Amor K, Salminen S, de Vos WM. The Mucin Degrader Akkermansia Muciniphila Is an Abundant Resident of the Human Intestinal Tract. Appl Environ Microbiol (2008) 74(5):1646–8. doi: 10.1128/AEM.01226-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia Muciniphila Gen. Nov., Sp. Nov., a Human Intestinal Mucin-Degrading Bacterium. Int J Syst Evol Microbiol (2004) 54(Pt 5):1469–76. doi: 10.1099/ijs.0.02873-0 [DOI] [PubMed] [Google Scholar]
- 167. Collado MC, Derrien M, Isolauri E, de Vos WM, Salminen S. Intestinal Integrity and Akkermansia Muciniphila, a Mucin-Degrading Member of the Intestinal Microbiota Present in Infants, Adults, and the Elderly. Appl Environ Microbiol (2007) 73(23):7767–70. doi: 10.1128/AEM.01477-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, et al. Cross-Talk Between Akkermansia Muciniphila and Intestinal Epithelium Controls Diet-Induced Obesity. Proc Natl Acad Sci USA (2013) 110(22):9066–71. doi: 10.1073/pnas.1219451110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Diaz Heijtz R. Fetal, Neonatal, and Infant Microbiome: Perturbations and Subsequent Effects on Brain Development and Behavior. Semin Fetal Neonatal Med (2016) 21(6):410–7. doi: 10.1016/j.siny.2016.04.012 [DOI] [PubMed] [Google Scholar]
- 170. Lu J, Claud EC. Connection Between Gut Microbiome and Brain Development in Preterm Infants. Dev Psychobiol (2019) 61(5):739–51. doi: 10.1002/dev.21806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Yano JM, Yu K, Donaldson GP, Shastri GG, Ann P, Ma L, et al. Indigenous Bacteria From the Gut Microbiota Regulate Host Serotonin Biosynthesis. Cell (2015) 161(2):264–76. doi: 10.1016/j.cell.2015.02.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Muller PA, Schneeberger M, Matheis F, Wang P, Kerner Z, Ilanges A, et al. Microbiota Modulate Sympathetic Neurons via a Gut-Brain Circuit. Nature (2020) 583(7816):441–6. doi: 10.1038/s41586-020-2474-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Kimura I, Miyamoto J, Ohue-Kitano R, Watanabe K, Yamada T, Onuki M, et al. Maternal Gut Microbiota in Pregnancy Influences Offspring Metabolic Phenotype in Mice. Science (2020) 367(6481):eaaw8429. doi: 10.1126/science.aaw8429 [DOI] [PubMed] [Google Scholar]
- 174. Erny D, Hrabe de Angelis AL, Jaitin D, Wieghofer P, Staszewski O, David E, et al. Host Microbiota Constantly Control Maturation and Function of Microglia in the CNS. Nat Neurosci (2015) 18(7):965–77. doi: 10.1038/nn.4030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Braniste V, Al-Asmakh M, Kowal C, Anuar F, Abbaspour A, Toth M, et al. The Gut Microbiota Influences Blood-Brain Barrier Permeability in Mice. Sci Transl Med (2014) 6(263):263ra158. doi: 10.1126/scitranslmed.3009759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Lach G, Schellekens H, Dinan TG, Cryan JF. Anxiety, Depression, and the Microbiome: A Role for Gut Peptides. Neurotherapeutics (2018) 15(1):36–59. doi: 10.1007/s13311-017-0585-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Vuong HE, Pronovost GN, Williams DW, Coley EJL, Siegler EL, Qiu A, et al. The Maternal Microbiome Modulates Fetal Neurodevelopment in Mice. Nature (2020) 586(7828):281–6. doi: 10.1038/s41586-020-2745-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Sampson TR, Mazmanian SK. Control of Brain Development, Function, and Behavior by the Microbiome. Cell Host Microbe (2015) 17(5):565–76. doi: 10.1016/j.chom.2015.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Spor A, Koren O, Ley R. Unravelling the Effects of the Environment and Host Genotype on the Gut Microbiome. Nat Rev Microbiol (2011) 9(4):279–90. doi: 10.1038/nrmicro2540 [DOI] [PubMed] [Google Scholar]
- 180. Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, et al. Tryptophan Catabolites From Microbiota Engage Aryl Hydrocarbon Receptor and Balance Mucosal Reactivity via Interleukin-22. Immunity (2013) 39(2):372–85. doi: 10.1016/j.immuni.2013.08.003 [DOI] [PubMed] [Google Scholar]
- 181. Metidji A, Omenetti S, Crotta S, Li Y, Nye E, Ross E, et al. The Environmental Sensor AHR Protects From Inflammatory Damage by Maintaining Intestinal Stem Cell Homeostasis and Barrier Integrity. Immunity (2019) 50(6):1542. doi: 10.1016/j.immuni.2019.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Wlodarska M, Luo C, Kolde R, d’Hennezel E, Annand JW, Heim CE, et al. Indoleacrylic Acid Produced by Commensal Peptostreptococcus Species Suppresses Inflammation. Cell Host Microbe (2017) 22(1):25–37.e6. doi: 10.1016/j.chom.2017.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Venkatesh M, Mukherjee S, Wang H, Li H, Sun K, Benechet AP, et al. Symbiotic Bacterial Metabolites Regulate Gastrointestinal Barrier Function via the Xenobiotic Sensor PXR and Toll-Like Receptor 4. Immunity (2014) 41(2):296–310. doi: 10.1016/j.immuni.2014.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Alcaino C, Knutson KR, Treichel AJ, Yildiz G, Strege PR, Linden DR, et al. A Population of Gut Epithelial Enterochromaffin Cells is Mechanosensitive and Requires Piezo2 to Convert Force Into Serotonin Release. Proc Natl Acad Sci USA (2018) 115(32):E7632–41. doi: 10.1073/pnas.1804938115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Pedicord VA, Lockhart AAK, Rangan KJ, Craig JW, Loschko J, Rogoz A, et al. Exploiting a Host-Commensal Interaction to Promote Intestinal Barrier Function and Enteric Pathogen Tolerance. Sci Immunol (2016) 1(3):eaai7732. doi: 10.1126/sciimmunol.aai7732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, et al. A Toll-Like Receptor Recognizes Bacterial DNA. Nature (2000) 408(6813):740–5. doi: 10.1038/35047123 [DOI] [PubMed] [Google Scholar]
- 187. Casanova JE. Bacterial Autophagy: Offense and Defense at the Host-Pathogen Interface. Cell Mol Gastroenterol Hepatol (2017) 4(2):237–43. doi: 10.1016/j.jcmgh.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Zhu S, Ding S, Wang P, Wei Z, Pan W, Palm NW, et al. Nlrp9b Inflammasome Restricts Rotavirus Infection in Intestinal Epithelial Cells. Nature (2017) 546(7660):667–70. doi: 10.1038/nature22967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Shi H, Kokoeva MV, Inouye K, Tzameli I, Yin H, Flier JS. TLR4 Links Innate Immunity and Fatty Acid-Induced Insulin Resistance. J Clin Invest (2006) 116(11):3015–25. doi: 10.1172/JCI28898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Friedrich C, Mamareli P, Thiemann S, Kruse F, Wang Z, Holzmann B, et al. MyD88 Signaling in Dendritic Cells and the Intestinal Epithelium Controls Immunity Against Intestinal Infection With C. Rodentium. PloS Pathog (2017) 13(5):e1006357. doi: 10.1371/journal.ppat.1006357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Murakami T, Ockinger J, Yu J, Byles V, McColl A, Hofer AM, et al. Critical Role for Calcium Mobilization in Activation of the NLRP3 Inflammasome. Proc Natl Acad Sci USA (2012) 109(28):11282–7. doi: 10.1073/pnas.1117765109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Graziani F, Pujol A, Nicoletti C, Dou S, Maresca M, Giardina T, et al. Ruminococcus Gnavus E1 Modulates Mucin Expression and Intestinal Glycosylation. J Appl Microbiol (2016) 120(5):1403–17. doi: 10.1111/jam.13095 [DOI] [PubMed] [Google Scholar]
- 193. Zhao Y, Yang J, Shi J, Gong YN, Lu Q, Xu H, et al. The NLRC4 Inflammasome Receptors for Bacterial Flagellin and Type III Secretion Apparatus. Nature (2011) 477(7366):596–600. doi: 10.1038/nature10510 [DOI] [PubMed] [Google Scholar]
- 194. Saavedra PHV, Huang L, Ghazavi F, Kourula S, Vanden Berghe T, Takahashi N, et al. Apoptosis of Intestinal Epithelial Cells Restricts Clostridium Difficile Infection in a Model of Pseudomembranous Colitis. Nat Commun (2018) 9(1):4846. doi: 10.1038/s41467-018-07386-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Ridge JP, Fuchs EJ, Matzinger P. Neonatal Tolerance Revisited: Turning on Newborn T Cells With Dendritic Cells. Science (1996) 271(5256):1723–6. doi: 10.1126/science.271.5256.1723 [DOI] [PubMed] [Google Scholar]
- 196. Forsthuber T, Yip HC, Lehmann PV. Induction of TH1 and TH2 Immunity in Neonatal Mice. Science (1996) 271(5256):1728–30. doi: 10.1126/science.271.5256.1728 [DOI] [PubMed] [Google Scholar]
- 197. Sarzotti M, Robbins DS, Hoffman PM. Induction of Protective CTL Responses in Newborn Mice by a Murine Retrovirus. Science (1996) 271(5256):1726–8. doi: 10.1126/science.271.5256.1726 [DOI] [PubMed] [Google Scholar]
- 198. Rooks MG, Garrett WS. Gut Microbiota, Metabolites and Host Immunity. Nat Rev Immunol (2016) 16(6):341–52. doi: 10.1038/nri.2016.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Yuan M, Jiang Z, Bi G, Nomura K, Liu M, Wang Y, et al. Pattern-Recognition Receptors Are Required for NLR-Mediated Plant Immunity. Nature (2021) 592(7852):105–9. doi: 10.1038/s41586-021-03316-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Donohoe DR, Holley D, Collins LB, Montgomery SA, Whitmore AC, Hillhouse A, et al. A Gnotobiotic Mouse Model Demonstrates That Dietary Fiber Protects Against Colorectal Tumorigenesis in a Microbiota- and Butyrate-Dependent Manner. Cancer Discov (2014) 4(12):1387–97. doi: 10.1158/2159-8290.CD-14-0501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Johnson CH, Dejea CM, Edler D, Hoang LT, Santidrian AF, Felding BH, et al. Metabolism Links Bacterial Biofilms and Colon Carcinogenesis. Cell Metab (2015) 21(6):891–7. doi: 10.1016/j.cmet.2015.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Srinivasan S, Morgan MT, Fiedler TL, Djukovic D, Hoffman NG, Raftery D, et al. Metabolic Signatures of Bacterial Vaginosis. mBio (2015) 6(2):e00204-15. doi: 10.1128/mBio.00204-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Voltolini C, Battersby S, Etherington SL, Petraglia F, Norman JE, Jabbour HN. A Novel Antiinflammatory Role for the Short-Chain Fatty Acids in Human Labor. Endocrinology (2012) 153(1):395–403. doi: 10.1210/en.2011-1457 [DOI] [PubMed] [Google Scholar]
- 204. Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, et al. Gut Microbiota Metabolism of Dietary Fiber Influences Allergic Airway Disease and Hematopoiesis. Nat Med (2014) 20(2):159–66. doi: 10.1038/nm.3444 [DOI] [PubMed] [Google Scholar]
- 205. Thorburn AN, McKenzie CI, Shen S, Stanley D, Macia L, Mason LJ, et al. Evidence That Asthma Is a Developmental Origin Disease Influenced by Maternal Diet and Bacterial Metabolites. Nat Commun (2015) 6:7320. doi: 10.1038/ncomms8320 [DOI] [PubMed] [Google Scholar]
- 206. Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, et al. Activation of Gpr109a, Receptor for Niacin and the Commensal Metabolite Butyrate, Suppresses Colonic Inflammation and Carcinogenesis. Immunity (2014) 40(1):128–39. doi: 10.1016/j.immuni.2013.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, et al. Microbiota Modulate Behavioral and Physiological Abnormalities Associated With Neurodevelopmental Disorders. Cell (2013) 155(7):1451–63. doi: 10.1016/j.cell.2013.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Schugar RC, Shih DM, Warrier M, Helsley RN, Burrows A, Ferguson D, et al. The TMAO-Producing Enzyme Flavin-Containing Monooxygenase 3 Regulates Obesity and the Beiging of White Adipose Tissue. Cell Rep (2017) 19(12):2451–61. doi: 10.1016/j.celrep.2017.05.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Meijers BK, Claes K, Bammens B, de Loor H, Viaene L, Verbeke K, et al. P-Cresol and Cardiovascular Risk in Mild-to-Moderate Kidney Disease. Clin J Am Soc Nephrol (2010) 5(7):1182–9. doi: 10.2215/CJN.07971109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Tanase DM, Gosav EM, Neculae E, Costea CF, Ciocoiu M, Hurjui LL, et al. Role of Gut Microbiota on Onset and Progression of Microvascular Complications of Type 2 Diabetes (T2DM). Nutrients (2020) 12(12):3719. doi: 10.3390/nu12123719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, et al. Intestinal Microbiota Metabolism of L-Carnitine, a Nutrient in Red Meat, Promotes Atherosclerosis. Nat Med (2013) 19(5):576–85. doi: 10.1038/nm.3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, et al. Precision Microbiome Reconstitution Restores Bile Acid Mediated Resistance to Clostridium Difficile. Nature (2015) 517(7533):205–8. doi: 10.1038/nature13828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Saavedra JM, Dattilo AM. Early Development of Intestinal Microbiota: Implications for Future Health. Gastroenterol Clin North Am (2012) 41(4):717–31. doi: 10.1016/j.gtc.2012.08.001 [DOI] [PubMed] [Google Scholar]
- 214. Houghteling PD, Walker WA. Why Is Initial Bacterial Colonization of the Intestine Important to Infants’ and Children’s Health? J Pediatr Gastroenterol Nutr (2015) 60(3):294–307. doi: 10.1097/MPG.0000000000000597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, et al. Microbial Exposure During Early Life has Persistent Effects on Natural Killer T Cell Function. Science (2012) 336(6080):489–93. doi: 10.1126/science.1219328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Twardziok M, Schroder PC, Krusche J, Casaca VI, Illi S, Bock A, et al. Asthmatic Farm Children Show Increased CD3(+)CD8(low) T-Cells Compared to Non-Asthmatic Farm Children. Clin Immunol (2017) 183:285–92. doi: 10.1016/j.clim.2017.09.009 [DOI] [PubMed] [Google Scholar]
- 217. Dicksved J, Floistrup H, Bergstrom A, Rosenquist M, Pershagen G, Scheynius A, et al. Molecular Fingerprinting of the Fecal Microbiota of Children Raised According to Different Lifestyles. Appl Environ Microbiol (2007) 73(7):2284–9. doi: 10.1128/AEM.02223-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Risnes KR, Belanger K, Murk W, Bracken MB. Antibiotic Exposure by 6 Months and Asthma and Allergy at 6 Years: Findings in a Cohort of 1,401 US Children. Am J Epidemiol (2011) 173(3):310–8. doi: 10.1093/aje/kwq400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Mai XM, Kull I, Wickman M, Bergstrom A. Antibiotic Use in Early Life and Development of Allergic Diseases: Respiratory Infection as the Explanation. Clin Exp Allergy (2010) 40(8):1230–7. doi: 10.1111/j.1365-2222.2010.03532.x [DOI] [PubMed] [Google Scholar]
- 220. Boursi B, Mamtani R, Haynes K, Yang YX. The Effect of Past Antibiotic Exposure on Diabetes Risk. Eur J Endocrinol (2015) 172(6):639–48. doi: 10.1530/EJE-14-1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Markle JG, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kampczyk U, et al. Sex Differences in the Gut Microbiome Drive Hormone-Dependent Regulation of Autoimmunity. Science (2013) 339(6123):1084–8. doi: 10.1126/science.1233521 [DOI] [PubMed] [Google Scholar]
- 222. Vujkovic-Cvijin I, Sklar J, Jiang L, Natarajan L, Knight R, Belkaid Y. Host Variables Confound Gut Microbiota Studies of Human Disease. Nature (2020) 587(7834):448–54. doi: 10.1038/s41586-020-2881-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Ni J, Wu GD, Albenberg L, Tomov VT. Gut Microbiota and IBD: Causation or Correlation? Nat Rev Gastroenterol Hepatol (2017) 14(10):573–84. doi: 10.1038/nrgastro.2017.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Lichtman JS, Marcobal A, Sonnenburg JL, Elias JE. Host-Centric Proteomics of Stool: A Novel Strategy Focused on Intestinal Responses to the Gut Microbiota. Mol Cell Proteomics (2013) 12(11):3310–8. doi: 10.1074/mcp.M113.029967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Marcobal A, Kashyap PC, Nelson TA, Aronov PA, Donia MS, Spormann A, et al. A Metabolomic View of How the Human Gut Microbiota Impacts the Host Metabolome Using Humanized and Gnotobiotic Mice. ISME J (2013) 7(10):1933–43. doi: 10.1038/ismej.2013.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Sridharan GV, Choi K, Klemashevich C, Wu C, Prabakaran D, Pan LB, et al. Prediction and Quantification of Bioactive Microbiota Metabolites in the Mouse Gut. Nat Commun (2014) 5:5492. doi: 10.1038/ncomms6492 [DOI] [PubMed] [Google Scholar]