Abstract
Background
To reduce the high incidence and mortality of gastric cancer (GC), we aimed to develop deep learning-based models to assist in predicting the diagnosis and overall survival (OS) of GC patients using pathological images.
Methods
2333 hematoxylin and eosin-stained pathological pictures of 1037 GC patients were collected from two cohorts to develop our algorithms, Renmin Hospital of Wuhan University (RHWU) and the Cancer Genome Atlas (TCGA). Additionally, we gained 175 digital pictures of 91 GC patients from National Human Genetic Resources Sharing Service Platform (NHGRP), served as the independent external validation set. Two models were developed using artificial intelligence (AI), one named GastroMIL for diagnosing GC, and the other named MIL-GC for predicting outcome of GC.
Findings
The discriminatory power of GastroMIL achieved accuracy 0.920 in the external validation set, superior to that of the junior pathologist and comparable to that of expert pathologists. In the prognostic model, C-indices for survival prediction of internal and external validation sets were 0.671 and 0.657, respectively. Moreover, the risk score output by MIL-GC in the external validation set was proved to be a strong predictor of OS both in the univariate (HR = 2.414, P < 0.0001) and multivariable (HR = 1.803, P = 0.043) analyses. The predicting process is available at an online website (https://baigao.github.io/Pathologic-Prognostic-Analysis/).
Interpretation
Our study developed AI models and contributed to predicting precise diagnosis and prognosis of GC patients, which will offer assistance to choose appropriate treatment to improve the survival status of GC patients.
Funding
Not applicable.
Keywords: Gastric cancer, Diagnosis, Overall survival, Deep learning, Multiple instance learning
Research in context.
Evidence before this study
We first searched PubMed and learned about relevant researches, and then carried out our project (Dec. 5, 2019). We searched for the keywords “gastric cancer” AND “deep learning” OR "artificial intelligence", with no restrictions on language or publication date. We learned that gastric cancer (GC) was the fifth most common type of malignant disease, and it ranks as the third leading cause of cancer-related deaths worldwide. Pathological evaluation remains the gold standard for the diagnosis of GC. When deciding on the necessity for further expensive and painful adjuvant treatments after surgery, clinicians tend to make decisions according to evidence-based information about the risk of death. Convolutional neural network (CNN) is a high-efficient deep learning method for image recognition and has excelled in quite a few image interpretation tasks and might be utilized to abstract additional characteristics from pathological images of GC patients. There had been a number of AI studies focusing on GC, most of which relied on endoscopy and medical radiologic technology. A few articles had focused on the application of deep learning algorithm to pathology of GC. There was still much room for improvement in developing AI models to diagnose GC and predict survival outcome through pathological pictures, especially to improve the performance of AI models.
Added value of this study
In this study, we designed a CNN-based model, GastroMIL, for the accurate diagnosis of GC directly from digital H&E-stained pictures. Encouragingly, this diagnosis model achieved excellent performance in differentiating GC from normal tissues in the training and internal validation sets (AUC nearly 1.000). Moreover, we have successfully developed a deep learning-based model, MIL-GC, to automatically predict OS in patients with GC with C-index of 0.728 and 0.671 in the training and internal validation sets. And we also used an independent external validation set, and the two models showed good diagnostic (AUC = 0.978) and prognostic (C-index = 0.657) prediction performance, indicating good robustness of the two designed models. And the risk score computed by MIL-GC was proved to be of independent prognostic value of GC by univariate and multivariable Cox analyze. In the comparison with human pathologists, the diagnostic model (GastroMIL) achieved accuracy better than that of the junior pathologist and comparable to that of expert pathologists. More importantly, we further constructed the first webpage (https://baigao.github.io/Pathologic-Prognostic-Analysis/) for the automatic diagnosis of GC and survival prediction.
Implications of all the available evidence
Our models can be adopted to make diagnosis with high accuracy and help clinicians select the appropriate adjuvant therapy following surgery, by identifying patients at high risk who would benefit from intensive regimens as well as patients at low risk who might be cured through surgery alone. It will help improve the survival status of GC patients and reduce the high mortality.
Alt-text: Unlabelled box
Introduction
Gastric cancer (GC) is the fifth most common type of malignant disease, and it ranks as the third leading cause of cancer-related deaths worldwide [1]. For patients with early GC, the 5-year survival rate can exceed 90% [2]. However, approximately half of patients with GC already proceed the advanced stage at the time of diagnosis, with the 5-year survival rate dropping below 30% [3,4]. To reduce the mortality of GC, early detection and appropriate treatment are crucial, and precise and efficient pathology services are indispensable to realize this goal.
Pathological evaluation remains the gold standard for the diagnosis of GC. Conventionally carried out by pathologists, this method is labor-intensive, tedious, and time-consuming. A severe shortage of pathologists and a heavy workload of diagnosis are widespread problems globally, which negatively affect the diagnostic accuracy [5]. Accordingly, it is necessary to design a new method to conveniently and accurately diagnose GC using pathological pictures.
Surgery is the main treatment for GC, followed by adjuvant treatments including chemoradiotherapy and molecular targeted therapy [6], [7], [8]. When deciding on the necessity for further expensive and painful adjuvant treatments, clinicians tend to make decisions according to evidence-based information about the risk of death. Clinical practice has confirmed that prognoses of almost all human cancers, including GC, are closely related to pathological criteria [9], especially the tumour-node-metastasis (TNM) staging system specified and revised by the American Joint Committee on Cancer (AJCC) [10]. However, manual histological analysis of tumour tissues is still not accurate enough to stratify and identify those who may benefit from adjuvant treatment. Hence, there is an urgent need to develop succinct and reliable methods to predict overall survival (OS) of patients with GC, which could assist in developing individualized therapeutic strategies and maximizing the benefits.
In recent years, deep learning has gradually attracted the attention of oncologists. Deep learning belongs to the class of machine learning that can successively identify more abstract information from the input data [11], [12], [13]. Deep learning has progressed remarkably in the field of oncology, and has been demonstrated to be superior to conventional machine learning techniques [14,15]. Convolutional neural network (CNN) is a high-efficient deep learning method for image recognition and has excelled in quite a few images interpretation tasks [16,17].
Many studies have reported that artificial intelligence (AI) trained with endoscopic images could detect GC precisely [18], [19], [20], [21]. When it comes to the field of tumour detection and prediction of prognosis of GC using AI through pathological images, some progress has been made. Song et al. [22] reported a histopathological diagnosis system for GC detection using deep learning with the sensitivity near 100% and average specificity of 80.6%. Another research developed recalibrated multi-instance deep learning for whole slide gastric image classification with 86% accuracy [23]. Wang et al. [24] successfully predicted GC outcome from resected lymph node histopathology images using deep learning.
Before proposing our models, we had established a number of challenges that needed to be overcome in order to make the developed AI models better applicable to clinical practice. First of all, a large sample size from multiple centres should be available for training and validating the proposed model to ensure the robustness. While ensuring the effectiveness of model training, it is preferable not to rely on extensive manual pixel-level annotation, which would be laborious and time consuming and might hinder the development of AI in the field of pathology. The developed model should be able to be applied in clinical practice, and it should be simple, affordable and accessible enough to be easily used by people without an AI background or in places where the economy is not particularly developed. We hoped to accomplish these challenges better than previous studies.
In this study, we developed deep learning-based models, named GastroMIL and MIL-GC, for precisely and conveniently detecting tumour and predicting outcome of GC by analyzing pathological pictures, respectively. GastroMIL and MIL-GC were proved to be novel and strong predictors for diagnosis and outcome of GC patients on both internal and independent external validation sets. In the comparison with human pathologists, our GastroMIL model outperformed the junior pathologist and achieved a great agreement with expert pathologists. Furthermore, we designed an online website (https://baigao.github.io/Pathologic-Prognostic-Analysis/) based on our analysis to make this prediction more available to users who have no knowledge of AI.
Methods
Patient Population
Three different cohorts were retrospectively collected to achieve a broad patient representation and thereby improve the ability to generalize results to other cohorts. In the Renmin Hospital of Wuhan University (RHWU; Wuhan, Hubei, China), we continuously collected 871 candidate patients with GC from 2012 to 2017, together with corresponding 588 tumour tissue blocks (made from surgically removed tumour tissue, which was formalin-fixed and paraffin-embedded) and 1276 pathological images. 1057 digital H&E-stained pictures of 449 GC patients from The Cancer Genome Atlas (TCGA) public dataset were collected, 934 of which were malignant and 123 were normal. In addition, 91 GC patients with 175 digital pictures were acquired from National Human Genetic Resources Sharing Service Platform (NHGRP; Shanghai, China) and served as the independent validation set to evaluate the robustness of our models.
We adopted the following inclusion criteria for developing the diagnostic model: (a) patients unequivocal diagnosed with GC by preoperative biopsy or postoperative pathological examination; (b) patients older than 18 years old and assentient to participate in this study; and (c) pathological images available and clear rather than loss, destruction or mildew.
Pictures used in the diagnostic model were excluded from the prognostic model when they met the following conditions: (a) identified as normal; (b) lack of follow-up information; and (c) no critical clinicopathologic information available.
Sample collection
Digital images of H&E-stained pathological images were used to construct the computer frameworks. For each GC patient, we selected two representative images in principle, which included tissues from not only GC tumour but also surrounding normal gastric tissues. For candidate patients from RHWU, the corresponding formalin-fixed, paraffin-embedded tumour tissue blocks, made from surgically removed tumour tissue, and their H&E-stained slides were obtained. Next, two expert pathologists A and B selected preferred blocks and marked areas that were cancerous or normal independently. When it came to a disagreement between them, the diagnostic opinion of another expert pathologist C was final adopted. Expert pathologists A and B were associate chief pathologists, while expert pathologists C was chief pathologist. The marked areas were utilized to construct tissue microarrays (TMAs), which were then photographed to obtain 1276 digital H&E-stained images, of which 640 were malignant and 636 were normal. For the TCGA cohort, 1057 pathological pictures (malignant 936, normal 123) were downloaded from the website (https://www.cbioportal.org/study/summary?id=lihc_tcga). In view of the uneven number of cancerous and normal pictures in the TCGA cohort, the data augmentation technique was used to equalize the distribution of images. In the external validation cohort from NHGRP, there were 91 malignant digital pathological images and 84 normal.
Clinical and pathological information was additionally needed for survival analysis, including survival state, OS time, age, sex, tumour size, neoplasm histologic grade, and pathologic T,N, M and TNM stages (according to the American Joint Committee on Cancer (AJCC) Cancer Staging Manual, Eighth Edition, 2017) [25]. Clinicopathological data of patients from the RHWU cohort were collected through electronic medical records, and those of the TCGA and NHGRP cohorts were downloaded directly from the official website.
This retrospective study was checked and approved by the clinical ethics committees of RHWU (No. WDRY2021-K002). And informed consents were gained from patients.
Diagnostic Model
Firstly, we designed a diagnostic model, named GastroMIL, to distinguish GC images from normal gastric tissue images. In order to avoid complex manual annotation, we applied weak supervised learning to our algorithm framework, specifically multiple instance learning (MIL) [26], [27], [28], [29]. Based on the assumption of MIL, each input image was a bag, and the tiles it contained were the example instances. To develop the model, we only needed coarse-grained labels of bags, that is, pathological diagnosis of each image. When the target picture was marked positive, at least one tile was positive; if the target picture was marked negative, all tiles should be negative.
Given that the images used to train the model had different magnifications, specifical the original magnification of images from TCGA was 20 × (without fixed size, could larger than 30000*30000 pixels), whereas that of RHWU was 30 × (3200*2400 pixels), we uniformly processed these images into 5 ×, 10 ×, and 20 × magnification and use them to develop the algorithm separately.
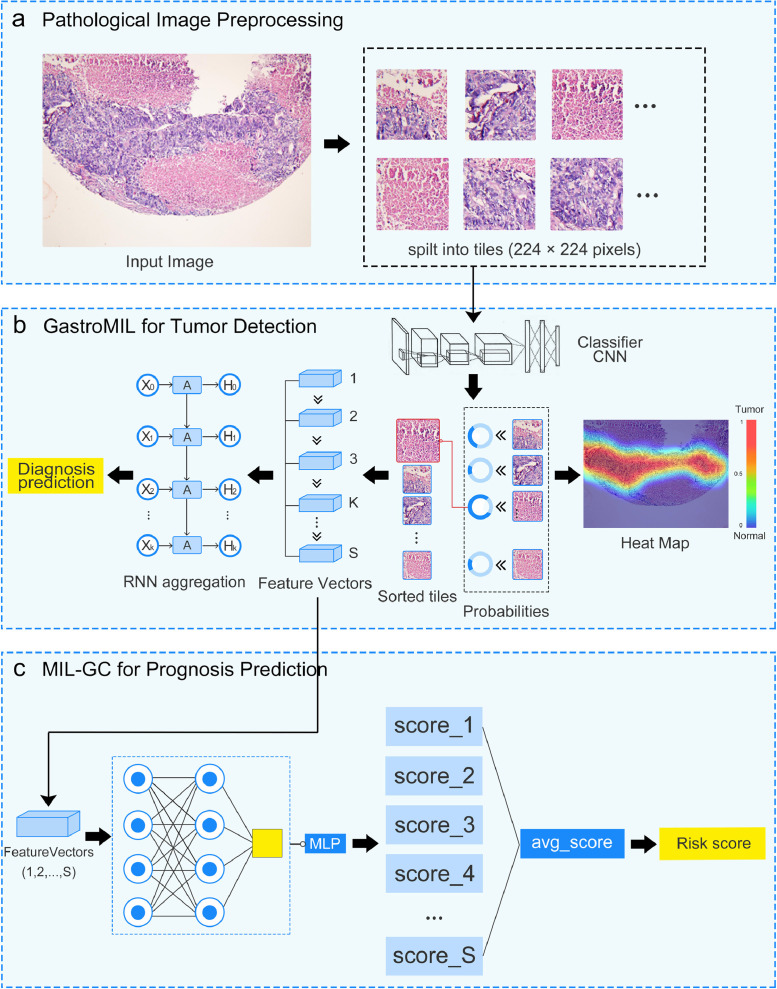
Our GastroMIL model comprised two-step algorithms (Fig. 1a-b). First of all, each input image was split into fixed-size tiles with 224 × 224 pixels, the labels of which were the same as the pathological diagnoses of the image itself. These tiles were used as training data for the first step algorithm, the MIL classifier. There were 10548460 tiles used for training and 4523755 tiles used for internal validation. Considering accuracy and efficiency, we chose RegNet developed by Facebook to constitute the backbone of MIL. The output of RegNet was the probability of these tiles being malignant. To obtain the inference results of the complete pictures, we introduced a recurrent neural network (RNN) as the second step classifier. Feature vectors with dimension 608 of the 32 most suspicious tiles gained from each picture by the first step were sequentially passed on to the RNN classifier to predict the probability of malignancy of the entire picture.
Fig. 1.
Flow chart of the developed models. The framework of GastroMIL is shown in a-b, and that of MIL-GC is shown in a-c. Pathological images are input and tiles with 224 × 224 pixels of each image are generated (a). Through CNN classifier of the MIL model, the probability of these tiles being malignant is output. Heat map visualizes ROIs identified by the model. Feature vectors with dimension 608 of the most suspicious tiles are extracted. Feature vectors of the K most suspicious tiles are input to the second layer of MIL and aggregated by RNN, and then the final diagnosis prediction of the input image is generated. In this study we took K as 32 (b). Feature vectors of the most S suspicious tiles are input to the prognosis model (in this study S = 128). In the MIL-GC model, each feature vector yields a probability value through a MLP algorithm. Probability values of the 128 most suspicious tiles of the input picture were merged to generate an average value as the output risk score (c). CNN, convolutional neural network; RNN, recurrent neural network; MIL, multiple instance learning; MLP, multilayer perceptron; ROI, region of interest.
The GastroMIL model could thus not only distinguish malignant images from normal, but also identify regions of interest (ROIs) by the segmentation and analysis of tiles. ROI indicated the area recognized as malignant by GastroMIL, which could be visualized in the form of heat map (Fig. 1b) and provide additional guiding information for clinicians.
Prognostic Model
After identifying the malignant images, we developed another model, MIL-GC, to predict the prognosis of GC patients. As shown in Fig. 1a-c, the first step of MIL-GC was similar to that of the GastroMIL model, and the 128 most suspicious tiles were selected and output as feature vectors with dimension 608. In the second step, each feature vector would finally yield a probability value between 0 and 1, through a multilayer perceptron (MLP) algorithm. Probability values of the 128 most suspicious tiles of the input picture were merged to generate an average value as the output risk score.
Statistical analysis
Receiver operating characteristic (ROC) curves and areas under the curve (AUCs), analysed with scikit-learn, a Python software package for machine learning, were used to quantify the performance of the diagnostic classifier as well as accuracy, sensitivity, and specificity. The cut-off value of ROC curves was set as 0.5. Cohen's kappa coefficient was used to assess the inter-observer agreement of the diagnostic model (GastroMIL) and human pathologists. To assess the predictive performance of the prognostic classifier, we adopted Harrell's concordance index (C-index) as a metric. Kaplan-Meier survival curve plotted with GraphPad Prism_9 was used to evaluate the correlation between risk score generated by prognostic models and OS of GC patients and Log-Rank test was performed. Prognostic factors were identified using univariate and multivariate Cox proportional hazards models implemented in SPSS26.0. The statistical significance level was set at 0.05 (two-tailed). Statistical significance threshold was adjusted for multiple comparisons using the Bonferroni correction. Python and Pytorch were employed to build the algorithm.
Role of the funding source
Not applicable.
Results
Patient characteristics
A total of 871 GC patients were initially screened from the RHWU cohort, and 588 with tumour tissue blocks were eligible for the study. There were 449 GC patients with digital H&E-stained pathological images were eligible for this study in the TCGA cohort and 91 in the NHGRP cohort. A total of 1276 images from the RHWU cohort and 1057 images from the TCGA cohort were obtained for the development of the GastroMIL model. Through data augmentation, 3221 pictures (malignant: normal = 1574: 1647) were finally enrolled in the GastroMIL model and 70% (N = 2261) were randomly assigned to the training set while the remaining 30% (N = 960) were included in the internal validation set. 175 pictures from the independent NHGRP cohort were used as the external validation set. The detailed data distribution was shown in Supplementary Table 1.
A total of 199 malignant pathological pictures with intact follow-up and clinicopathological information from the RHWU cohort and 440 from the TCGA cohort participated in the construction of the prognostic model and then randomly spilt into training set (N = 443) and internal validation set (N = 196) at a ratio of 70: 30. 91 GC digital pathological pictures with the required information from NHGRP were included in the external validation set.
Table 1 exhibits the baseline characteristics of the pictures used in MIL-GC. It is worth noting that the OS time of the TCGA cohort was significantly lower than that of the RHWU cohort (median of 13.8 months vs. 43 months, P < 0.0001, Kruskal-Wallis nonparametric test) (Bonferroni-adjusted significance threshold P’ < 0.017) and NHGRP cohort (median of 13.8 months vs. 44 months, P < 0.0001, Kruskal-Wallis nonparametric test) (Bonferroni-adjusted significance threshold P’ < 0.017) (Supplementary Fig. 1a). Differences between the RHWU cohort (median 43 months) and NHGRP cohort (median 44 months) were not statistically significant (P = 0.075, Kruskal-Wallis nonparametric test) (Bonferroni-adjusted significance threshold P’ < 0.017). Since pictures in the TCGA cohort were collected from different medical centres, the distribution of their OS time was much more heterogeneous. To better adapt our models to the heterogeneity caused by patients from different sources and to improve the generalizability of the developed models, we used a mixture of images from the RHWU and TCGA cohort together as the training and internal validation sets. The independent NHGRP cohort of images was used as the external validation set. The OS time of the external validation set (median 44 months) was significantly higher compared with the training set (median 20.2 months, P < 0.0001, Kruskal-Wallis nonparametric test) (Bonferroni-adjusted significance threshold P’ < 0.017) and the internal validation set (median 22.4 months, P < 0.0001, Kruskal-Wallis nonparametric test) (Bonferroni-adjusted significance threshold P’ < 0.017) (Supplementary Fig. 1b).
Table 1.
Baseline characteristics in the prognostic model (MIL-GC).
| RHWU (N = 199) | TCGA (N = 440) | NHGRP (N = 91) | |
|---|---|---|---|
| Age (years) | 60 (54, 66) | 67 (58, 73) | 63.5±10.8 |
| Sex | |||
| Male | 146 (73.4%) | 284 (64.5%) | 31 (34.1%) |
| Female | 51(25.6%) | 156 (35.5%) | 60 (65.9%) |
| Missing | 2 (1.0%) | 0 | 0 |
| Tumor size (cm) | 4.0 (2.8, 6.0) | 1.8 (1.1, 2,4) | 5.0 (4.0, 7.0) |
| Histologic grade | |||
| 1 | 12 (6.0%) | 11 (2.5%) | 0 |
| 2 | 57 (28.6%) | 159 (36.1%) | 16 (17.6%) |
| 3 | 125 (62.8%) | 261 (59.3%) | 69 (75.8%) |
| 4 | 0 | 0 | 6 (6.6%) |
| Missing | 5 (2.5%) | 9 (2.0%) | 0 |
| pT stage | |||
| pT1 | 39 (19.6%) | 22 (5.0%) | 2 (2.2%) |
| pT2 | 22 (11.1%) | 93 (21.1%) | 10 (11%) |
| pT3 | 2 (1.0%) | 197 (44.8%) | 47 (51.6%) |
| pT4 | 134 (67.3%) | 118 (26.8%) | 32 (35.2%) |
| Missing | 2 (1.0%) | 10 (2.3%) | 0 |
| pN stage | |||
| pN0 | 71 (35.7%) | 130 (29.5%) | 19 (20.9%) |
| pN1 | 33 (16.6%) | 119 (27.0%) | 19 (20.9%) |
| pN2 | 45 (22.6%) | 85 (19.3%) | 22 (24.2%) |
| pN3 | 50 (25.1%) | 88 (20.0%) | 31 (34.1%) |
| Missing | 0 | 18 (4.1%) | 0 |
| pM stage | |||
| pM0 | 168 (84.4%) | 388 (88.2%) | 89 (97.8%) |
| pM1 | 31 (15.6%) | 30 (6.8%) | 2 (2.2%) |
| Missing | 0 | 22 (5.0%) | 0 |
| pTNM stage | |||
| Stage I | 40 (20.1%) | 59 (13.4%) | 7 (7.7%) |
| Stage II | 34 (17.1%) | 132 (30.0%) | 27 (29.7%) |
| Stage III | 92 (46.3%) | 187 (42.6%) | 55 (60.5%) |
| Stage IV | 31 (15.6%) | 44 (10.0%) | 2 (2.2%) |
| Missing | 2 (1.0%) | 18 (4.1%) | 0 |
| Survival status | |||
| Alive | 146 (73.4%) | 268 (60.9%) | 37 (40.7%) |
| Dead | 53 (26.6%) | 172 (39.1%) | 54 (59.3%) |
| OS time (months) | 43 (33, 54) | 13.8 (7.1, 24.5) | 44 (15, 71) |
Performance of the Diagnostic Model
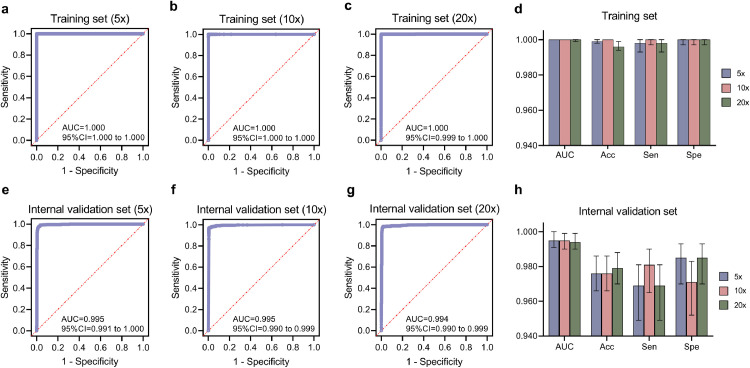
In the diagnostic model, GastroMIL, all 3221 pictures after data augmentation were mixed and then randomly split into the training set and the internal validation set at a ratio of 7: 3. As shown in Fig. 2, ROC curves and AUCs represented the ability to discriminate malignant pathological images of the GastroMIL model when pictures were at 5 ×, 10 × and 20 × magnification. The accuracy (Acc), sensitivity (Sen), and specificity (Spe) of each magnification in the training (Fig. 2d) and internal validation (Fig. 2h) sets are shown in Table 2 a.
Fig. 2.
Diagnostic abilities of GastroMIL at different magnification in the training and internal validation sets. a-c, ROC curves in the training set when images at 5 ×, 10 × and 20 × magnification, respectively; e-g, ROC curves in the internal validation set when images at 5 ×, 10 × and 20 × magnification, respectively. The AUC, Acc, Sen and Spe of the training and internal validation sets were exhibited in d and h, respectively. ROC, receiver operating characteristic; AUC, area under the curve; Acc, accuracy; Sen, sensitivity; Spe, specificity.
Table 2.
Accuracy, sensitivity and specificity of the diagnostic model (GastroMIL) and human pathologists.
| a. Accuracy, sensitivity and specificity in the diagnostic model (GastroMIL) when images at different magnification. | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Accuracy (95%) | Sensitivity (95%) | Specificity (95%) | ||||||||||
| Training set | 5 × | 0.999 (0.998, 1.000) | 0.998 (0.993, 1.000) | 1.000 (0.997, 1.000) | ||||||||
| 10 × | 1.000 (1.000, 1.000) | 1.000 (0.997, 1.000) | 1.000 (0.997, 1.000) | |||||||||
| 20 × | 0.996 (0.994, 0.999) | 0.998 (0.993, 1.000) | 1.000 (0.997, 1.000) | |||||||||
| Internal validation set | 5 × | 0.976 (0.966, 0.986) | 0.969 (0.949, 0.981) | 0.985 (0.970, 0.993) | ||||||||
| 10 × | 0.976 (0.966, 0.986) | 0.981 (0.965, 0.990) | 0.971 (0.952, 0.983) | |||||||||
| 20 × | 0.979 (0.970, 0.988) | 0.969 (0.949, 0.981) | 0.985 (0.970, 0.993) | |||||||||
| b. Accuracy, sensitivity and specificity in the diagnostic model (GastroMIL) with 10 × magnified images | ||||||||||||
| Accuracy (95%) | Sensitivity (95%) | Specificity (95%) | ||||||||||
| Training set | 1.000 (1.000, 1.000) | 1.000 (0.997, 1.000) | 1.000 (0.997, 1.000) | |||||||||
| Internal validation set | 0.976 (0.966, 0.986) | 0.981 (0.965, 0.990) | 0.971 (0.952, 0.983) | |||||||||
| External validation set | 0.920 (0.879, 0.961) | 0.934 (0.864, 0.969) | 0.905 (0.823, 0.951) | |||||||||
| c. Diagnostic performance of the GastroMIL model and human pathologists in the external validation set with images at 10 × magnification | ||||||||||||
| Accuracy (95%) | Sensitivity (95%) | Specificity (95%) | P-value* | Kappa# | ||||||||
| GastroMIL Model | 0.920 (0.879, 0.961) | 0.934 (0.864, 0.969) | 0.905 (0.823, 0.951) | - | - | |||||||
| Expert Pathologist D | 0.971 (0.947, 0.996) | 1.000 (0.960, 1.000) | 0.952 (0.884, 0.981) | 1.000 | 0.805 | |||||||
| Expert Pathologist E | 0.983 (0.963, 1.002) | 0.967 (0.908, 0.991) | 1.000 (0.956, 1.000) | 0.332 | 0.806 | |||||||
| Expert Pathologist F | 0.983 (0.963, 1.002) | 0.967 (0.908, 0.991) | 1.000 (0.956, 1.000) | 0.332 | 0.806 | |||||||
| Junior Pathologist G | 0.874 (0.825, 0.924) | 0.758 (0.661, 0.835) | 1.000 (0.956, 1.000) | <0.0001 | 0.617 | |||||||
*Difference of Accuracy between the GastroMIL Model and each human pathologist, tested by paired chi‐square test (McNemars test).
#Inter-observer agreement of the GastroMIL Model and each human pathologist, evaluated by Cohen's kappa coefficient.
In the training set, the AUCs achieved 1.000 at three different magnifications. The differences of Acc between the three groups was statistically significant (P = 0.003, Chi-square test) (significance threshold P < 0.05). The group of 10 × magnification (Acc = 1.000) outperformed that of 20 × (Acc = 0.996) (P = 0.004, Chi-square test) (Bonferroni-adjusted significance threshold P’ < 0.017). There was no statistically significant difference in Acc between the groups of 5 × and 10 × (0.999 vs. 1.000, P = 0.250, Chi-square test) (Bonferroni-adjusted significance threshold P’ < 0.017) and between the groups of 5 × and 20 × (0.999 vs. 0.996, P = 0.109, Chi-square test) (Bonferroni-adjusted significance threshold P’ < 0.017).
In the internal validation set, the AUC achieved 0.995 when pictures were magnified 10 times. The AUCs were also very close to it when images were magnified 5 times (AUC = 0.995) and 20 times (AUC = 0.994). The differences in Acc among the three groups of different magnifications (0.976, 0.976, and 0.979) were not statistically significant (P = 0.870, Chi-square test) (Bonferroni-adjusted significance threshold P’ < 0.017). It can be seen that our GastroMIL model achieved excellent diagnostic ability for the differentiation of malignant and normal gastric pathological pictures, and the generalization performance was excellent for images at these three magnifications.
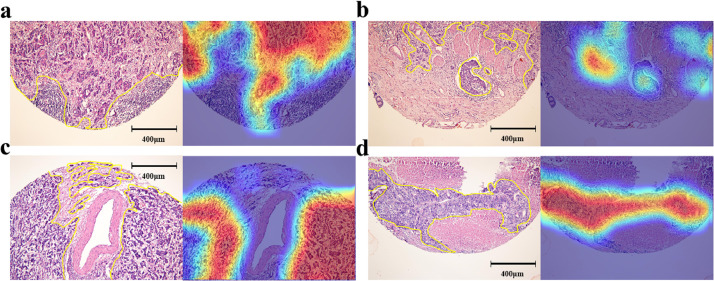
The diagnosis prediction by the GastroMIL model was based on the classification probability output of all tiles, which can be used to visualize the localization of highly suspected lesions on positive sections. That is, tiles predicted positive could show chromatic aberration, and through appropriate strides, suspected areas would overlap many times, colour of which thus became warmer and darker than other areas. The warmer the colour, the higher the probability that GastroMIL predicted malignant in this area. Fig. 1b showed how heat maps generated by GastroMIL. Heat maps of the RHWU (Fig. 3) and TCGA (Supplementary Fig. 2) cohorts could almost accurately outline the area where the tumour was located, regardless of different cohorts or pathological TNM stage, indicating excellent generalization performance of GastroMIL.
Fig. 3.
Heat maps of the RHWU cohort. a-d, pathological images and corresponding heat maps with pathological TNM stage I, II, III, and IV from the RHWU cohort, respectively. The actual tumor regions annotated by expert pathologists were shown with yellow lines.
Performance of the prognostic models
In the process of outcome prediction, 639 malignant images from RHWU and TCGA cohorts were mixed and then randomly divided into the training set and the internal validation set at a 7:3 ratio. Considering that the discriminatory power of the GastroMIL model for different magnification images was basically identical to each other, we chose 10 × images to be applied in the prognosis model. The MIL-GC model performed well in both the training set and the internal validation set, with C-index of 0.728 and 0.671, respectively.
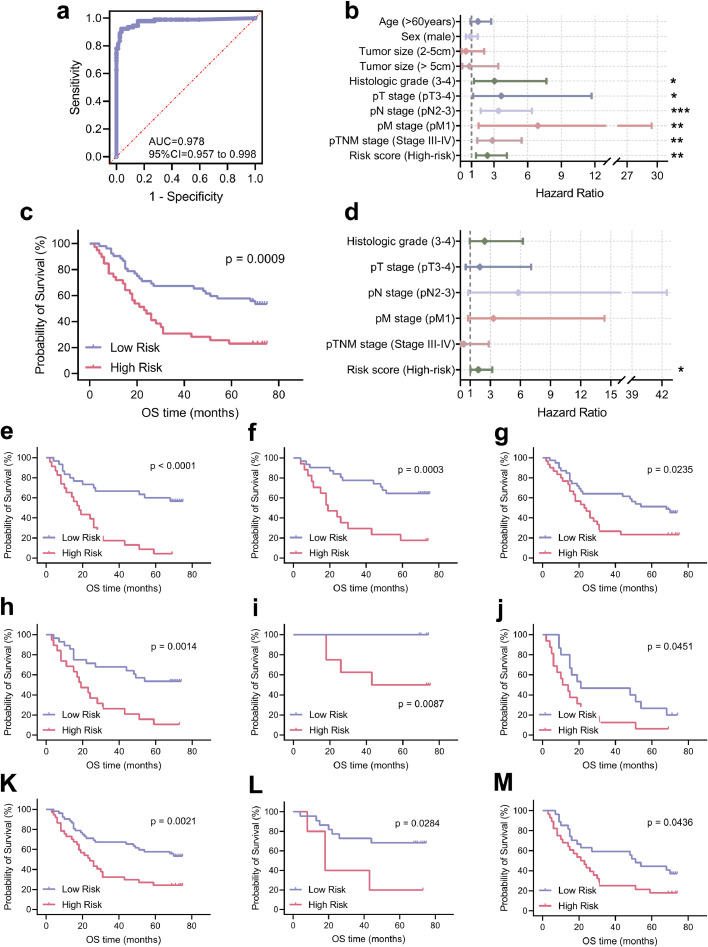
Prognostic model assigned risk score to each picture, and we divided the GC patients into high-risk and low-risk score groups. We used the median value of risk score in the training set as the threshold for stratifying patients. Then we adopted Kaplan-Meier plots and univariate and multivariable Cox models to assess the association between risk score and prognosis among patients with GC. In the training set, the MIL-GC classifier was a strong predictor of survival in the univariate analysis (HR = 4.209, P < 0.0001, Cox analyse; Supplementary Table 2 and Supplementary Fig. 3). The classifier remained strong in multivariable analysis (HR = 3.549, P < 0.0001, Cox analyse; Supplementary Table 2) after adjusting for significant prognostic indexes in univariable analyses: age, pT stage, pN stage, pM stage and pTNM stage.
In the internal validation set, our model stratified the population accurately based on univariate analysis (HR = 3.249, P < 0.0001, Cox analyse; Fig. 4a and c). The MIL-GC classifier predicted survival even after stratification for other features (such as age, histologic grade, pT grade, pN grade and pTNM grade; Fig. 4). The risk score computed by MIL-GC was of independent prognostic value (HR = 2.976, P < 0.0001, Cox analyse; Fig. 4b). The results showed that the prognostic model based on CNN was equipped to predict OS of GC and might provide a basis for the choice of treatment.
Fig. 4.
Prognostic significance of the risk score generated by MIL-GC in the internal validation set. HRs for prediction of survival by the MIL-GC model and other clinicopathological indexes based on univariate (a) and multivariate (b) analyses. The output score was converted into a binary score (high or low risk), using the median value of the training set as a threshold. KM survival curves for the internal validation set (c) and some other subgroups: age ≤60 (d); age > 60 (e); histologic grade 1-2 (f); histologic grade 3-4 (g); pT stage 3-4 (h); pN stage 0-1 (i); pN stage 2-3 (j); pTNM stage 1-2 (k) and pTNM stage 3-4 (l). ****, P < 0.0001; **, P < 0.01; *, P < 0.05. The P-value of Kaplan-Meier survival curve was evaluated by Log-Rank test. The P-value of HR was calculated by Cox analyse.
Independent external validation of developed models
Figures from the independent NHGRP cohort were employed as the external validation set for the diagnosis prediction model, GastroMIL and outcome prediction model, MIL-GC. The magnification of the original images is 20 × (3900*3900 pixels), and we pre-processed them into 10 × magnification. The GastroMIL model showed good performance in identifying malignant pathological images on the external validation set (AUC = 0.978, Fig. 5a). Heat maps of the independent external validation set were displayed in Supplementary Fig. 4.
Fig. 5.
Predicting diagnosis and prognostic performance in the external validation set. ROC curve (a) and HRs based on univariate (b) and multivariate (d) analyses are exhibited. KM survival curves for the external validation set (c) and some other subgroups: age > 60 (e); tumour size ≤5 (f); histologic grade 3 (g); pT stage 3 (h); pN stage 0 (i); pN stage 3 (j); pM stage 0 (k); pTNM stage 2 (l) and pTNM stage 3 (m). ***, P < 0.001; **, P < 0.01; *, P < 0.05. ROC, receiver operating characteristic; AUC, area under the curve. The P-value of Kaplan-Meier survival curve was evaluated by Log-Rank test. The P-value of HR was calculated by Cox analyse.
The C-index of MIL-GC in the external validation set was 0.657. The MIL-GC classifier was a strong predictor of OS in the univariate analysis (HR = 2.414, P < 0.0001, Cox analyse; Fig. 5b and c). The MIL-GC classifier predicted survival among the various subgroups (such as age > 60, tumour size ≤ 5, histologic grade 3, pT stage 3, pN stage 0 and 3, pM stage 0, pTNM stage II and pTNM stage III; Fig. 5). After adjusting for significant prognostic indexes in univariable analyses: histologic grade, pT stage, pN stage, pM stage and pTNM stage, the risk score output by MIL-GC remained strong in multivariable analysis (HR = 1.803, P = 0.043, Cox analyse; Fig. 5d). Good diagnostic and prognostic prediction performance demonstrated on the external validation set, indicating good robustness of the two designed models.
Comparing diagnostic performance with human pathologists
To explore how the diagnostic performance of our model compared to that of human pathologists, we employed three expert pathologists D, E, and F who were chief or associate chief pathologists and one junior pathologist G who was under training to diagnose images in the external validation set. Human pathologists were blind for patients’ information before examination. The accuracy, sensitivity, and specificity of manual diagnosis were exhibited in Table 2c. The performance of our GastroMIL model (Accuracy = 0.920) was significantly better than the junior pathologist G (Accuracy = 0.874) (P < 0.0001, paired chi‐square test). There was no significantly difference when our model compared to expert pathologist D (Accuracy = 0.971) (P > 0.05, paired chi‐square test), expert pathologist E (Accuracy = 0.983) (P > 0.05, paired chi‐square test), and expert pathologist F (Accuracy = 0.983) (P > 0.05, paired chi‐square test), respectively. And the diagnostic model achieved substantial interobserver agreement with the expert pathologists (kappa = 0.805, 0.806, and 0.806, respectively). Moreover, we designed an online website (https://baigao.github.io/Pathologic-Prognostic-Analysis/) to make the process of prediction more available and much easier for users without AI knowledge. The detail process of prediction is seen in Supplementary Fig. 5.
Analysis of representative predictive tiles
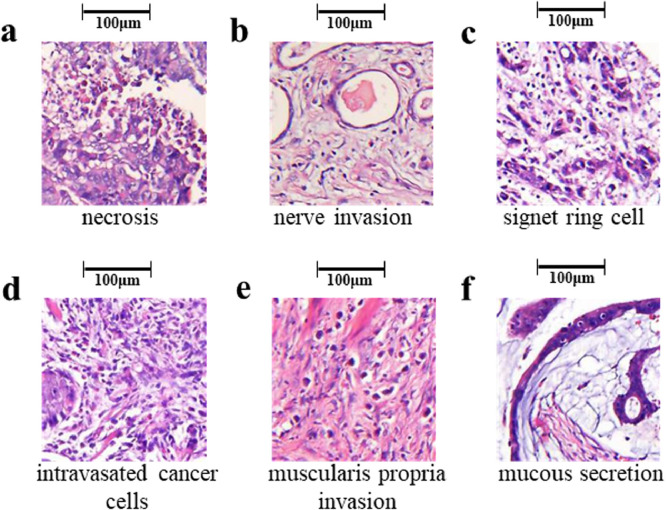
Our models predicted diagnosis and outcome of GC patients from the 32 and 128 most suspicious tiles automatically gained from the input HE-stained pathological pictures, respectively. We extracted these suspicious tiles and had them reviewed by expert pathologists A and B. Here we displayed some representative predictive tiles with interpretation by pathological experts (Fig. 6). These tiles were of obvious tumour heterogeneity, including necrosis, nerve invasion, signet ring cell, intravasated cancer cells, muscularis propria invasion, and mucous secretion, hiding significant diagnostic and prognostic information. These suspicious tiles provide a preliminary indication that our model could automatically identify regions of pathological significance and classify GC pathology images based on these regions. Further details on the analysis of deep learning, which has been traditionally treated as a black box, deserve our further study in the future.
Fig. 6.
Representative predictive tiles produced by our model. These tiles were of obvious tumour heterogeneity, including necrosis (a), nerve invasion (b), signet ring cell (c), intravasated cancer cells (d), muscularis propria invasion (e), and mucous secretion (f).
Discussion
In this study, we designed a CNN-based model, Gastro-MIL, for the accurate diagnosis of GC directly from digital H&E-stained pictures. Encouragingly, this diagnosis model achieved excellent performance in differentiating GC from normal tissues in the training and internal validation sets (AUC nearly 1.000). Moreover, we have successfully employed deep learning to automatically predict OS in patients with GC with C-index of 0.728 and 0.671 in the training and internal validation sets. The predictive models can be adopted to help clinicians select the appropriate adjuvant therapy following surgery, by identifying patients at high risk who would benefit from intensive regimens as well as patients at low risk who might be cured through surgery alone. And we also used an independent external validation set, and the two models showed good diagnostic (AUC = 0.978) and prognostic (C-index = 0.657) prediction performance, indicating good robustness of the two designed models. Moreover, the risk score computed by MIL-GC was proved to be of independent prognostic value of GC by univariate and multivariable Cox analyse. In the comparison with human pathologists, our diagnostic model GastroMIL outperformed the junior pathologists and demonstrated a high degree of consistence with expert pathologists (kappa > 0.8). More importantly, we further constructed the first webpage (https://baigao.github.io/Pathologic-Prognostic-Analysis/) for the automatic diagnosis of GC and survival prediction.
In recent years, deep learning, such as CNN, has attracted much attention and has achieved particular success in computer vision tasks. In our previous study, we developed an AI model to distinguish abnormal images from normal images in small bowel capsule endoscopy, and it was validated to exceed human performance [30]. In this study, we adopted CNN to analyse digital H&E-stained GC pathological images. Song et al. [9] reported deep learning model for GC detection by analysing histopathological images with validation in multicentre sample. They reached good performance and developed the system for pathologists to use the proposed model. However, they applied a large number of pixel-level manual annotations to train the model, which consumed a lot of time and effort of pathologists. The need for extensive manual annotation was also seen in the studies using AI for GC [23] and bladder [31] pathological diagnosis, different from the models proposed in our study. Due to the adoption of a weakly supervised model (specifical MIL) in our study, the only label we needed for training was the reported diagnoses made by pathologists in the course of their daily work, eliminating large manually annotated tasks that used to hinder the development and clinical practice of AI in pathology. And the system Song et al. [9] have developed may be too expensive (Small hospital: $84, 000-$87, 000; Large hospital: $161, 000-$164, 000) for economically underdeveloped areas with a shortage of pathologists, limiting its promotion in primary hospitals to a certain extent. We designed a website based on our analysis, simple and easy to use, and all the users need is a computer with an Internet connection or even an Internet-connected phone or tablet. When a histological image is uploaded without any professional annotation, the webpage will show a brief result of the primary type and survival prediction. The website identifies suspicious areas, thus improving diagnostic accuracy in a limited amount time, which will prove particularly useful in areas with a shortage of pathologists and in improving the diagnosis performance of junior pathologists who are under training.
There have been a number of AI studies focusing on GC [19,20,[32], [33], [34], [35]]. Most of these previous studies relied on endoscopy, and a small amount of selected medical radiologic technology, such as computed tomography. Moreover, previous studies generally employed a small sample size from a single centre, lacking effective proof to validate the robustness of models. We constructed models on much larger datasets from two different cohorts, and validated on another independent external validation set, greatly enhancing the universality of the diagnostic and survival models. In the meantime, images in the external validation were diagnosed by human pathologists. By comparing the diagnosis performance of our model with that of junior and expert pathologists, the accuracy and reliability of our model was further confirmed.
For survival prediction, we designed MIL-GC algorithm in this study. The risk score generated by MIL-GC exhibited a distinguished performance in predicting OS among patients with GC, as reflected by the C-index of 0.728, 0.671, and 0.657 in the training, internal validation and external validation sets, respectively. Furthermore, we applied Cox regression analysis to determine whether the risk score generated by MIL-GC was an independent biomarker for the prediction of OS in GC patients. Fortunately, the risk score generated by MIL-GC remained strong in multivariate regression (HR = 2.976, P < 0.0001 in the internal validation set, and HR = 1.803, P = 0.043 in the external validation set) after the adjustment for established prognostic features, indicating that the risk score generated by MIL-GC will be a promising supplement to the established markers and help refine risk stratification among GC patients.
A recent study [24] predicting the outcome of GC from resected lymph node histopathology images also yielded meaningful results. Our study focused on the pathological histological features from the stomach tissue, while their study concentrated on the lymph node metastasis of GC. Both of these two studies able to make predictions about the prognosis of patients with GC. If the key points of the two researches could be combined in the future, we may achieve a more satisfactory performance in predicting GC patients’ prognosis. Furthermore, our webpage could conveniently provide predictions of patient prognosis, serving as an important reference for selection of adjuvant therapy after surgery in patients with GC.
There are some limitations to our study. First, the survival time of GC individuals from the TGCA cohort was different from that in the RHWU cohort due to the progress of treatment. Therefore, it is not appropriate for us to use the TCGA or RHWU cohort as the training set and the other as the validation set. Hence, we mixed them together and randomly split them into the training or internal validation set. Furthermore, datasets we collected in the RHWU, TCGA and NHGRP cohorts were retrospective and thus suffered from inherent biases. In the future, we plan to conduct a prospective, randomized, multicentre clinical trial to validate the performance of precisely diagnosing GC and stratifying patients into high-risk and low-risk score groups to assist in selecting the suitable individualized adjuvant treatment regiments.
In conclusion, we developed deep learning models to diagnose GC and predict the survival outcomes of GC patients by analyzing H&E-stained pathological images. To make our models more intuitive and easier to use, an online website (https://baigao.github.io/Pathologic-Prognostic-Analysis/) based on developed algorithms was designed. Our models assist oncologists in the identification of GC and selection of appropriate treatment, thus reducing the physical and economic burdens of patients.
Contributors
W.D. was associated with Conceptualization, Funding acquisition, Resources, and Supervision of the study. B.H., S.T., N.Z., J.M., Y.L., P.H., B.D., and J.H. conducted Data curation. Z.H., C.Z., H.Z., and F.M. took charge of Formal analysis, Methodology and Software. B.H., S.T., N.Z., and J.M. performed the Investigation and Project administration. S.T., B.H., Z.H., C.Z., H.Z., and F.M. completed the Validation and Visualization. B.H. and S.T. completed Writing – original draft. W.D., B.H., S.T., F.L., M.J., and J.Z. conducted Writing – review & editing. All authors verified the underlying data and reviewed and approved the final manuscript.
Declaration of Competing Interest
All authors have no conflicts of interest to disclose.
Acknowledgments
The authors are grateful to all colleagues involved in collecting samples and developing models.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103631.
Appendix. Supplementary materials
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551. 2019. [DOI] [PubMed] [Google Scholar]
- 2.Nishida T, Sugimoto A, Tomita R. Impact of time from diagnosis to chemotherapy in advanced gastric cancer: a propensity score matching study to balance prognostic factors. World J Gastrointest Oncol. 2019;11(1):28–38. doi: 10.4251/wjgo.v11.i1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Satolli MA, Buffoni L, Spadi R, Roato I. Gastric cancer: the times they are a-changin'. World J Gastrointest Oncol. 2015;7(11):303–316. doi: 10.4251/wjgo.v7.i11.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suzuki R, Yamamoto E, Nojima M. Aberrant methylation of microRNA-34b/c is a predictive marker of metachronous gastric cancer risk. J Gastroenterol. 2014;49(7):1135–1144. doi: 10.1007/s00535-013-0861-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Metter DM, Colgan TJ, Leung ST, Timmons CF, Park JY. Trends in the US and Canadian Pathologist Workforces From 2007 to 2017. JAMA Netw Open. 2019;2(5) doi: 10.1001/jamanetworkopen.2019.4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang L, Li W, Liu Y, Zhang C, Gao W, Gao L. Clinical study on the safety, efficacy, and prognosis of molecular targeted drug therapy for advanced gastric cancer. Am J Transl Res. 2021;13(5):4704–4711. [PMC free article] [PubMed] [Google Scholar]
- 7.Nakamura Y, Kawazoe A, Lordick F, Janjigian YY, Shitara K. Biomarker-targeted therapies for advanced-stage gastric and gastro-oesophageal junction cancers: an emerging paradigm. Nat Rev Clin Oncol. 2021;18(8):473–487. doi: 10.1038/s41571-021-00492-2. [DOI] [PubMed] [Google Scholar]
- 8.Jácome AA, Sankarankutty AK, dos Santos JS. Adjuvant therapy for gastric cancer: what have we learned since INT0116? World J Gastroenterol. 2015;21(13):3850–3859. doi: 10.3748/wjg.v21.i13.3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song Z, Zou S, Zhou W. Clinically applicable histopathological diagnosis system for gastric cancer detection using deep learning. Nat Commun. 2020;11(1):4294. doi: 10.1038/s41467-020-18147-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao L-L, Lu J, Li P. Evaluation of the Eighth Edition of the American Joint Committee on Cancer TNM staging system for gastric cancer: an analysis of 7371 patients in the SEER database. Gastroenterol Res Pract. 2019;2019 doi: 10.1155/2019/6294382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang S, Yang J, Fong S, Zhao Q. Artificial intelligence in cancer diagnosis and prognosis: opportunities and challenges. Cancer Lett. 2020;471:61–71. doi: 10.1016/j.canlet.2019.12.007. [DOI] [PubMed] [Google Scholar]
- 12.Deo RC. Machine learning in medicine. Circulation. 2015;132(20):1920–1930. doi: 10.1161/CIRCULATIONAHA.115.001593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong D, Yip S. Machine learning classifies cancer. Nature. 2018;555(7697):446–447. doi: 10.1038/d41586-018-02881-7. [DOI] [PubMed] [Google Scholar]
- 14.Li Y, Tian S, Huang Y, Dong W. Driverless artificial intelligence framework for the identification of malignant pleural effusion. Transl Oncol. 2020;14(1) doi: 10.1016/j.tranon.2020.100896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alakwaa FM, Chaudhary K, Garmire LX. Deep Learning Accurately Predicts Estrogen Receptor Status in Breast Cancer Metabolomics Data. J Proteome Res. 2018;17(1):337–347. doi: 10.1021/acs.jproteome.7b00595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skrede O-J, De Raedt S, Kleppe A. Deep learning for prediction of colorectal cancer outcome: a discovery and validation study. Lancet. 2020;395(10221):350–360. doi: 10.1016/S0140-6736(19)32998-8. [DOI] [PubMed] [Google Scholar]
- 17.Coudray N, Ocampo PS, Sakellaropoulos T. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med. 2018;24(10):1559–1567. doi: 10.1038/s41591-018-0177-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao K, Uedo N, Muto M. Development of an e-learning system for the endoscopic diagnosis of early gastric cancer: an international multicenter randomized controlled trial. EBioMedicine. 2016;9:140–147. doi: 10.1016/j.ebiom.2016.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagao S, Tsuji Y, Sakaguchi Y. Highly accurate artificial intelligence systems to predict the invasion depth of gastric cancer: efficacy of conventional white-light imaging, nonmagnifying narrow-band imaging, and indigo-carmine dye contrast imaging. Gastrointest Endosc. 2020 doi: 10.1016/j.gie.2020.06.047. [DOI] [PubMed] [Google Scholar]
- 20.Horiuchi Y, Hirasawa T, Ishizuka N. Performance of a computer-aided diagnosis system in diagnosing early gastric cancer using magnifying endoscopy videos with narrow-band imaging (with videos) Gastrointest Endosc. 2020 doi: 10.1016/j.gie.2020.04.079. [DOI] [PubMed] [Google Scholar]
- 21.Luo H, Xu G, Li C. Real-time artificial intelligence for detection of upper gastrointestinal cancer by endoscopy: a multicentre, case-control, diagnostic study. Lancet Oncol. 2019;20(12):1645–1654. doi: 10.1016/S1470-2045(19)30637-0. [DOI] [PubMed] [Google Scholar]
- 22.Song Z, Zou S, Zhou W. Clinically applicable histopathological diagnosis system for gastric cancer detection using deep learning. Nat Commun. 2020;11(1):4294. doi: 10.1038/s41467-020-18147-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang S, Zhu Y, Yu L. RMDL: Recalibrated multi-instance deep learning for whole slide gastric image classification. Med Image Anal. 2019;58 doi: 10.1016/j.media.2019.101549. [DOI] [PubMed] [Google Scholar]
- 24.Wang X, Chen Y, Gao Y. Predicting gastric cancer outcome from resected lymph node histopathology images using deep learning. Nat Commun. 2021;12(1):1637. doi: 10.1038/s41467-021-21674-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buyyounouski MK CP, Kattan M, Prostate Amin MB, editors. AJCC Cancer staging manual. 8th ed. Springer; Chicago: 2017. pp. 715–726. [Google Scholar]
- 26.Xu Y, Zhu JY, Chang EI, Lai M, Tu Z. Weakly supervised histopathology cancer image segmentation and classification. Med Image Anal. 2014;18(3):591–604. doi: 10.1016/j.media.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 27.Verma N. 2012. Learning from data with low intrinsic dimension: University of California at San Diego. [Google Scholar]
- 28.Viola PA, Platt JC, Zhang C. Advances in Neural Information Processing Systems 18 [Neural Information Processing Systems, NIPS 2005, December 5-8, 2005. British Columbia; Vancouver: 2005. Multiple instance boosting for object detection. Canada]; 2005. [Google Scholar]
- 29.Wang X, Tang F, Chen H. UD-MIL: Uncertainty-driven deep multiple instance learning for OCT image classification. IEEE J Biomed Health Inform. 2020;24(12):3431–3442. doi: 10.1109/JBHI.2020.2983730. [DOI] [PubMed] [Google Scholar]
- 30.Ding Z, Shi H, Zhang H. Gastroenterologist-level identification of small-bowel diseases and normal variants by capsule endoscopy using a deep-learning model. Gastroenterology. 2019;157(4) doi: 10.1053/j.gastro.2019.06.025. 1044-54 e5. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Z, Chen P, McGough M. Pathologist-level interpretable whole-slide cancer diagnosis with deep learning. Nat Mach Intell. 2019;1(5):236–245. [Google Scholar]
- 32.Zhang L, Dong D, Zhang W. A deep learning risk prediction model for overall survival in patients with gastric cancer: a multicenter study. Radiother Oncol. 2020;150:73–80. doi: 10.1016/j.radonc.2020.06.010. [DOI] [PubMed] [Google Scholar]
- 33.Kuroki K, Oka S, Tanaka S. Clinical significance of endoscopic ultrasonography in diagnosing invasion depth of early gastric cancer prior to endoscopic submucosal dissection. Gastric Cancer. 2020 doi: 10.1007/s10120-020-01100-5. [DOI] [PubMed] [Google Scholar]
- 34.Dong D, Fang MJ, Tang L. Deep learning radiomic nomogram can predict the number of lymph node metastasis in locally advanced gastric cancer: an international multicenter study. Ann Oncol. 2020;31(7):912–920. doi: 10.1016/j.annonc.2020.04.003. [DOI] [PubMed] [Google Scholar]
- 35.Challoner BR, von Loga K, Woolston A. Computational image analysis of T-cell infiltrates in resectable gastric cancer: association with survival and molecular subtypes. J Natl Cancer Inst. 2020 doi: 10.1093/jnci/djaa051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.