Abstract
Mycobacterium avium complex (MAC) is composed of environmental mycobacteria found widely in soil, water, and aerosols that can cause disease in animals and humans, especially disseminated infections in AIDS patients. MAC consists of two closely related species, M. avium and M. intracellulare, and may also include other, less-defined groups. The precise differentiation of MAC species is a fundamental step in epidemiological studies and for the evaluation of possible reservoirs for MAC infection in humans and animals. In this study, which included 111 pig and 26 clinical MAC isolates, two novel allelic M. avium PCR-restriction enzyme analysis (PRA) variants were identified, differing from the M. avium PRA prototype in the HaeIII digestion pattern. Mutations in HaeIII sites were confirmed by DNA sequencing. Identification of these isolates as M. avium was confirmed by PCR with DT1-DT6 and IS1245 primers, nucleic acid hybridization with the AccuProbe system, 16S ribosomal DNA sequencing, and biochemical tests. The characterization of M. avium PRA variants can be useful in the elucidation of factors involved in mycobacterial virulence and routes of infection and also has diagnostic significance, since they can be misidentified as M. simiae II and M. kansasii I if the PRA method is used in the clinical laboratory for identification of mycobacteria.
The Mycobacterium avium complex (MAC) consists of two closely related species, M. avium and M. intracellulare, and possibly other, less-well-defined organisms. MAC is a group of environmental mycobacteria found widely in soil, water, and aerosols and causes disease in animals and humans (14). There appear to be clinically as well as genetically significant differences between M. avium and M. intracellulare. More than 90% of MAC isolates from AIDS patients are M. avium, as are most of the pathogenic isolates from swine and cattle. The biochemical tests for the identification of the species of MAC do not accurately resolve the two species. Consequently, many laboratories report isolates simply as members of MAC.
Several alternative methods for identifying the species of MAC isolates have been described. MAC can be classified into 28 serovars; isolates belonging to serovars 1 to 6, 8 to 11, and 21 are M. avium, and those belonging to serovars 7, 12 to 20, and 25 are M. intracellulare (23). Serovars 22 to 24 and 26 to 28 may represent M. intracellulare, M. scrofulaceum, and other, nonclassifiable mycobacteria (30). However, reagents for serotyping are not commercially available, and up to 15% of isolates cannot be typed.
Recent studies have emphasized DNA-based methods for the identification of MAC species. The most widely used system is a commercial hybridization assay (Gen-Probe) in which species-specific labeled DNA probes bind to the rRNA sequences. Specific probes are available for the identification of M. avium and M. intracellulare (10). The second-generation format of the assay uses a chemiluminiscent, acridinium ester-labeled probe detected with a luminometer and has a sensitivity of 95% (17). This system is considered the current “gold standard” in spite of the fact that some MAC serovars do not hybridize with either of the species-specific probes (30).
Based on data indicating significant nucleotide sequence variation at the hsp65 locus among different species of mycobacteria, Telenti et al. (28) proposed a method for species identification based on analysis of polymorphisms of restriction digests of that gene. That approach, designated PCR-restriction enzyme analysis (PRA), which involves amplification of a 441-bp fragment of hsp65 followed by digestion with BstEII and HaeIII, enabled the differentiation of 29 mycobacterial species and subspecies, including both M. avium and M. intracellulare. New algorithms were proposed by Taylor et al. (27) and by Devalois et al. (5), with the last including 34 species and subspecies. We have recently applied this technique to 18 MAC strains, including reference strains as well as animal and human isolates, and confirmed the usefulness of this technique in the differentiation of M. avium, M. intracellulare, and M. scrofulaceum (24).
A PCR with primers derived from the DT1 and DT6 fragments, identified in a genomic library of M. avium serotype 2, was developed by Thierry et al. (29). DT6-positive strains correspond to M. avium, DT1-positive strains correspond to M. intracellulare, and strains positive with both pairs of primers can be identified as M. avium serotype 2 or 3. It was reported that DNA from none of the other mycobacterial species was amplified with these primers. A comparative evaluation of this identification system and the AccuProbe method showed that the two methods can be equally sensitive for species identification of M. avium and M. intracellulare (4).
Insertion sequences (IS) are species specific and can therefore be used for species identification. Elements identified in the M. avium complex include IS900 in M. avium subsp. paratuberculosis (11); IS902 in M. avium subsp. silvaticum (19); IS901, IS1110, IS1245, and IS1311 in M. avium (12, 13, 16, 22); and IS1141 in M. intracellulare (18). IS1245 and IS1311, which have 85% identity at the DNA level, were found to be consistently present in M. avium strains and have been used for analysis of strain relatedness (7). Guerrero et al. (12) reported that IS1245 is limited to the M. avium group (M. avium subsp. avium, M. avium subsp. paratuberculosis, and M. avium subsp. silvaticum), while M. intracellulare appears to be devoid of this element.
Here we identified two novel allelic variants of M. avium in 111 swine isolates and 26 clinical MAC isolates by PRA. A third allelic variant, which corresponds to the variant characterized previously by Telenti et al. (28), was also identified in these isolates. These M. avium isolates were identified by PCR with the DT1-DT6 and IS1245 primers, nucleic acid hybridization with the AccuProbe system, DNA sequencing, and biochemical tests. Our results are relevant because an increase in the occurrence of mycobacteriosis was observed in the last years in the southern region of Brazil, where the pig isolates were obtained. This is an important center of swine production, being responsible for approximately 60% of the entire country’s production (1,540,000 tons in 1997) (8). The producers, industry, local meat inspection services, official institutes, and universities have united to elaborate a control program that includes (i) the quantification of the economic losses, (ii) a case-control study to access the risk factors for the infection, and (iii) the identification and molecular characterization of the mycobacteria isolated from animals in that area. In addition, MAC is also an important opportunistic pathogen in Brazil, especially among AIDS patients. In São Paulo, located in the southeastern region, where the clinical isolates were obtained, 23 MAC cultures (18.4%) were obtained from 125 bone marrow aspirates from AIDS patients between 1990 and 1992. Between 1985 and 1990, only 11 MAC-positive cultures were isolated among 60,000 cultures from human immunodeficiency virus-negative patients in the same region (1).
MATERIALS AND METHODS
Mycobacterial isolates.
During meat inspection, farms having two or more swine carcasses with lymph node enlargement were sampled. The collected tissues were stored at −20°C until bacteriological examination. Samples were decontaminated by the Petroff method, inoculated onto Löwenstein-Jensen and Stonebrink-Lesslie media, and incubated for 60 days. After the examination of 64 sampled farms, 196 acid-fast bacillus isolates were obtained from 131 animals on 52 farms. One hundred eleven isolates from 90 animals on 45 farms were identified by molecular methods. Of these, 107 isolates were identified as M. avium.
Twenty-six clinical isolates from São Paulo, located in the southeastern region of Brazil, were obtained at Instituto Adolfo Lutz. These isolates were previously identified as MAC by biochemical tests and were submitted for molecular identification by PRA.
Sample preparation.
A loopful of mycobacteria grown on solid medium (Löwenstein-Jensen or Stonebrink-Lesslie medium) was suspended in 0.4 ml of 10 mM Tris–1 mM EDTA (pH 8.0) with 1% Triton X-100 and was subjected to three cycles of freezing and thawing. Five to 10 microliters was used for the PCRs. Liquid cultures in 7H9 medium supplemented with OADC (Difco Laboratories, Detroit, Mich.) were obtained and used for AccuProbe and biochemical identification.
PRA.
Primers Tb11 (5′-ACCAACGATGGTGTGTCCAT-3′) and Tb12 (5′-CTTGTCGAACCGCATACCCT-3′) were used to amplify a 441-bp fragment. Amplification reactions were performed in 50-μl reaction mixtures containing 20 mM Tris-Cl (pH 8.4), 50 mM KCl, 1.5 mM MgCl2, 200 μM (each) four deoxyribonucleoside triphosphates (dNTPs), 10% glycerol, 25 pmol of each primer, and 1 U of Taq polymerase (Gibco-BRL Life Technologies and CENBIOT-UFRGS). Samples were incubated at 94°C for 5 min to denature the DNA. Forty-five cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min were followed by a final extension at 72°C for 7 min. Fifteen microliters of each reaction mixture was digested with BstEII and HaeIII (Gibco-BRL). The digestion products were separated in 4% agarose gels (Gibco-BRL), and a 25-bp ladder (Gibco-BRL) was used as an external marker. After electrophoresis at 5 V/cm, the gels were photographed on a UV transilluminator. Restriction patterns were copied with an Epson ES 1000C scanner and Adobe Photoshop LE for Macintosh. Migration profiles were converted into molecular size data by using Molecular Analyst (Bio-Rad).
DT1-DT6 PCR.
Primers AV6 and AV7 (5′-ATGGCCGGGAGACGATCTATGCCGGCGTAC-3′ and 5′-CGTTCGATCGCAGTTTGTGCAGCGCGTACA-3′) and primers IN38 and IN41 (5′-GAACGCCCGTTGGCTGGCCATTCACGAAGGAG-3′ and 5′-GCGCAACACGGTCGGACAGGCCTTCCTCGA-3′) were used to amplify the 187- and 666-bp fragments, respectively. Amplifications were performed in 50 μl containing 20 mM Tris-Cl (pH 8.4), 50 mM KCl, 2 mM MgCl2, 200 μM dNTPs, 100 μg of bovine serum albumin per ml, 100 pmol of each primer, and 2 U of Taq polymerase. The amplification mixture was denatured at 95°C for 5 min, followed by 25 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min and a final extension at 72°C for 7 min. DNA from M. avium serotype 2 (ATCC 25291), which is amplified with both sets of primers, was used as a positive control (29).
IS1245 PCR.
Primers P1 and P2 (5′-GCCGCCGAAACGATCTAC-3′ and 5′-AGGTGGCGTCGAGGAAGAC-3′), amplifying a 427-bp fragment, were used in a reaction mixture consisting of 20 mM Tris (pH 8.4), 50 mM KCl, 1.5 mM MgCl2, 200 μM dNTPs, 10% glycerol, 25 pmol of each primer, and 1 U of Taq polymerase. After an initial denaturation at 94°C for 5 min, samples were incubated at 94°C for 1 min, 60°C for 1 min, and 72°C 1 min for 30 cycles, with a final extension at 72°C for 7 min.
Detection of amplified products.
Five to 10 microliters of each PCR mixture was separated by size in 1% agarose–Tris-borate-EDTA gels containing ethidium bromide at a final concentration of 5 μg/ml. A 1-kb DNA ladder (Gibco-BRL) was used as a DNA marker.
AccuProbe test.
Nucleic acid hybridization was performed with acridinium ester-labeled, single-stranded DNA probes complementary to the rRNAs of MAC, M. avium, and M. intracellulare. Bacteria from single colonies, grown in OADC-supplemented 7H9 medium (Difco), were lysed by sonication in a tube for 15 min and incubated with the lyophilized DNA probe at 60°C for 15 min. The selection reagent was added, and the mixture was incubated at 60°C for a further 5 min and kept at room temperature for 5 min. Results were expressed as relative light units after reading in a LEADER luminometer.
DNA sequencing.
Sequencing of the hsp65 fragment amplified with the TB11 and TB12 primers from the three PRA variants, M. avium ATCC 25291, and two M. intracellulare strains (one clinical isolate and ATCC 13950) was performed. The amplified products were cloned in the pCR2.1 vector (TA cloning kit; Invitrogen) and sequenced by using M13 direct and reverse primers in an automated ABI Prism 377 sequencer (Perkin-Elmer) according to the manufacturer’s instructions. Sequences were aligned by using the DNAsis program (Hitachi)
Biochemical identification.
Swine and human isolates were identified by the methods of Collins et al. (2) and Kent and Kubica (15) at Instituto Adolfo Lutz, São Paulo, Brazil.
Nucleotide sequence accession numbers.
The nucleotide sequences of the hsp65 genes determined in this study have been deposited in GenBank under the following accession numbers: M. avium subsp. avium ATCC 25291, AF126030; M. avium I, AF126031; M. avium II, AF126032; M. avium III, AF126033; M. intracellulare clinical isolate CC1400, AF126034; M. intracellulare ATCC 13950, AF126035.
RESULTS
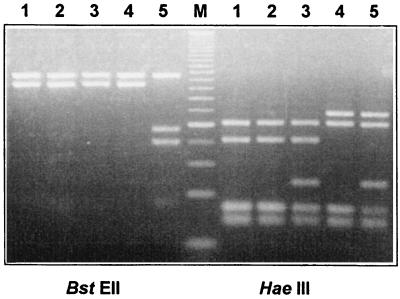
Analysis of mycobacterial isolates by PRA identified three distinct M. avium-like patterns in 107 of the 111 swine isolates (Fig. 1). The most common M. avium PRA pattern was observed in 76 isolates and corresponded to the M. avium pattern described by Telenti et al. (28) (M. avium I). Twenty-four isolates showed a similar pattern, with an additional band of approximately 60 bp after HaeIII digestion (M. avium II). Seven isolates showed a different M. avium-like pattern, with a fragment of approximately 145 bp instead of the 105-bp fragment after HaeIII digestion (M. avium III). These patterns were not described in the algorithms proposed by Telenti et al. (28), Taylor et al. (27), and Devalois et al. (5). When the HaeIII digestion patterns of hsp65 amplicons from M. avium ATCC 25291, M. intracellulare ATCC 13950, and the three variants were compared, it was observed that, coincidentally, both the additional fragment of variant II and the larger fragment of variant III were present in the HaeIII digest of M. intracellulare (Fig. 1). Sequencing of the 16S rRNA gene hypervariable fragment A (21) was performed for one isolate from each group and confirmed the identification of M. avium (data not shown).
FIG. 1.
PRA (4% agarose gel stained with ethidium bromide) of M. avium ATCC 25291 (lanes 1), M. avium I (lanes 2), M. avium II (lanes 3), M. avium III (lanes 4), and M. intracellulare ATCC 13950 (lanes 5). Lane M, 25-bp DNA ladder (Gibco-BRL).
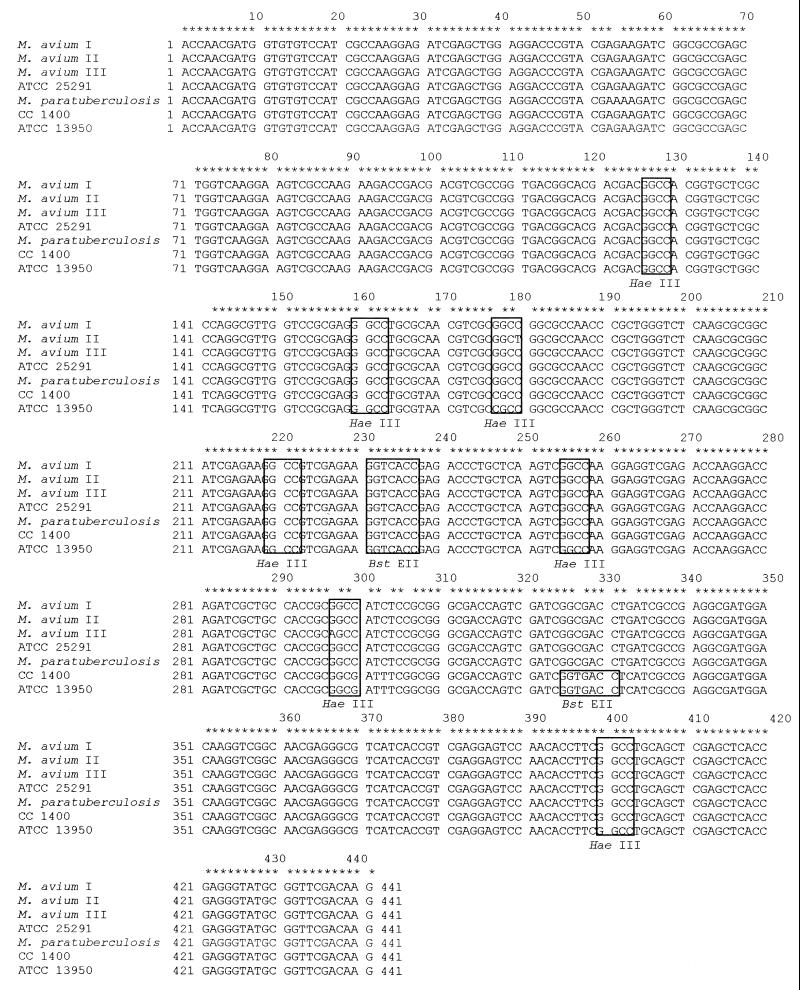
Sequencing of hsp65 amplified fragments of these variants showed that mutations in HaeIII sites which explain the observed restriction pattern polymorphism have occurred (Fig. 2). While the sequence of the variant I fragment was identical to that of the prototype M. avium ATCC 25291 fragment, the mutation in the third HaeIII site in variant II (GGCC→GGCT) resulted in the appearance of a 60-bp band, corresponding to the uncut 17-bp plus 42-bp fragments. This band is also evident in the M. intracellulare strains, but the mutation is located in a different position in the site (GGCC→CGCC). In the same manner, the mutation in the sixth HaeIII site in variant III (GGCC→AGCC) resulted in the appearance of the 145-bp band, corresponding to the uncut 42-bp plus 103-bp fragments. Again in M. intracellulare there is a mutation in the same restriction site, affecting a different position (GGCC→GGCG). The two M. intracellulare sequences also showed seven other mutations that have not been found in the M. avium sequences. The comparison of the obtained sequences with the hsp65 sequence of M. paratuberculosis deposited in GenBank under accession number X74518 revealed another mutation: an A at position 322 in the M. paratuberculosis sequence was substituted for a G in all other sequences (Fig. 2). All mutations observed occur in the third codon position and result in no amino acid substitution.
FIG. 2.
Alignment of hsp65 sequences (total length, 441 positions) from M. avium I, M. avium II, M. avium III, M. avium ATCC 25291, M. paratuberculosis (GenBank accession no. X74518), M. intracellulare (clinical isolate CC 1400), and M. intracellulare ATCC 13950. Nucleotides common to all mycobacterial isolates are indicated by asterisks. HaeIII and BstEII sites are boxed.
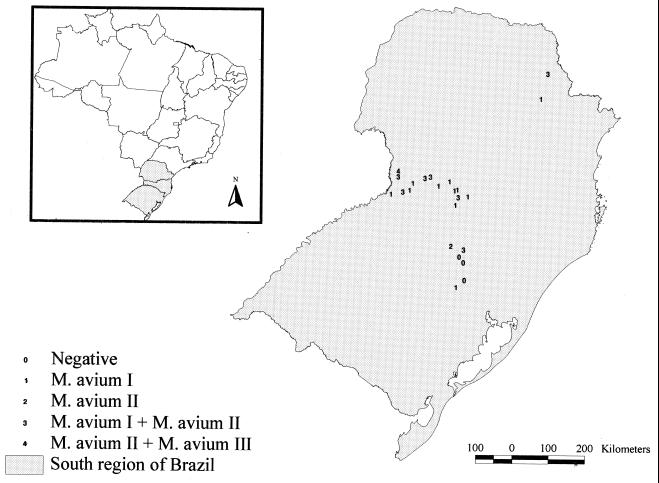
The map in Fig. 3 depicts the origins of the isolates showing the three PRA M. avium variant patterns. Variants I and II were isolated in different regions included in the study. Variant III was isolated in only one region.
FIG. 3.
Geographical distribution of swine M. avium isolates, showing the localization of PRA variants.
For further characterization of these variants, PCR with the DT1, DT6, and IS1245 primers was performed. Amplification with the DIG-derived primers gave positive results with DNAs from all isolates, and no amplification of the DT1 fragment was observed. DT1-negative and DT6-positive amplification is specific for M. avium, according to Thierry et al. (29) and Sola et al. (25). DNAs from all isolates except the seven isolates giving PRA M. avium variant III and one isolate from variant II were positive by amplification with the IS1245 primers. The negative results were confirmed after repeated tests, on different occasions, and with different DNA sample preparations.
The IS1245-negative isolates and seven other isolates representing variants I and II, chosen at random, were subjected to the AccuProbe test with M. avium- and M. intracellulare-specific probes. All isolates hybridized exclusively with the M. avium-specific probe. Taken together, these results confirmed that these isolates represented M. avium variants (Table 1).
TABLE 1.
Identification of pig isolates from M. avium PRA variants I, II, and III by PCR with DT1, DT6, and IS1245 primers, AccuProbe analysis, 16S ribosomal DNA sequencing, and biochemical tests
| PRA variant | Result from:
|
|||||
|---|---|---|---|---|---|---|
| DT1 | DT6 | IS1245 | AccuProbe | 16S ribosomal DNA sequence | Biochemical identification | |
| I | − | + | + | M. avium | M. avium | MAC |
| II | − | + | +a | M. avium | M. avium | MAC |
| III | − | + | − | M. avium | M. avium | MAC |
DNA from one strain of this group was not amplified with this primer.
To estimate the presence of these M. avium PRA variants in clinical specimens, 26 samples from São Paulo, previously identified as MAC by routine biochemical tests, were subjected to PRA. The molecular identification revealed the existence of 2 M. intracellulare, 16 M. avium variant I, 6 M. avium variant II, and 2 M. avium variant III isolates.
DISCUSSION
Among three distinct hsp65 gene PRA patterns observed in 107 of 111 isolates studied, one was characteristic of M. avium and the other two were mixed patterns showing bands common to both the M. avium and M. intracellulare patterns described by Telenti et al. (28). Amplification with DT1- and DT6-derived primers and results obtained by the other methods (biochemical identification, AccuProbe test, and 16S rRNA gene sequencing) were consistent with the identification of the 107 isolates as M. avium. The remaining four isolates were identified as M. bovis and Nocardia sp. by PRA and biochemical identification (data not shown).
It was initially suspected that the M. avium variants could represent intermediate subspecies of MAC between M. avium and M. intracellulare. Comparative sequence analysis of the hsp65 gene fragments was essential for the testing of this hypothesis. The two M. intracellulare sequences showed nine identical mutations compared to the M. avium prototype sequence. Each PRA variant showed only one mutation located in a HaeIII restriction site, in a position different from the mutations in M. intracellulare. Therefore, it appears that the 60- and 145-bp bands occurring in these variants and also in M. intracellulare have different origins and cannot be regarded as evidence of the relatedness of these strains.
Allelic variants were found in swine isolates from different geographic regions, suggesting that the occurrence of these mutations was not fortuitous and localized. Swine variant III isolates were found in a unique region, but conclusions about this variant cannot be drawn because only six isolates were obtained. It could be speculated that at least this variant could represent a single strain that had disseminated in that area. However, the finding of three variants in clinical isolates obtained from a different region of the country suggests that the occurrence of the three variants can be common. Sources of M. avium human infection and routes of transmission are not well known. Ingestion followed by invasion through the gastrointestinal tract could be the main route of infection in AIDS patients, because these organisms can be isolated from stool specimens from these patients (3). The proportions of each variant found in pig and human M. avium isolates were similar: M. avium I was found in 71% of the pig and 66.6% of the human isolates, M. avium II was found in 22.5% of pig and 25.9% of human isolates, and the proportions of M. avium III in pig and human isolates were 6.5 and 8.3%, respectively. In the light of the results shown here, the possibility of transmission of these M. avium variants through swine products or from common sources cannot be ruled out.
The presence of these three variants in clinical isolates with confirmed biochemical identification as MAC has important consequences. MAC bacteria cause chronic pulmonary infections in elderly people and lymphadenitis in children, and they are the most common cause of systemic bacterial infections in patients with AIDS in the developed world (14). Since the beginning of the AIDS epidemic, the incidence of infections with M. avium, especially disseminated infections, has increased dramatically. The success of the treatment of M. avium infections depends on quick identification and rapid institution of appropriate antibiotic therapy. Many laboratories are using molecular techniques for quicker identification of atypical mycobacteria, especially in AIDS patients, and the PRA method is a useful alternative for this purpose. By using the algorithm proposed by Devalois et al. (5), the M. avium variants can be misidentified if no other identification method is employed. For example, M. avium II can be misidentified as M. kansasii I (140-, 105-, and 80-bp fragments on HaeIII digestion; 245- and 220-bp fragments on BstEII digestion) (5), and M. avium III can be misidentified as M. simiae II (155- and 140-bp fragments on HaeIII digestion; 245- and 220-bp fragments on BstEII digestion) (5). This could have important consequences for the definitive diagnosis and treatment of these patients.
Although several works referring to PRA of MAC strains have been published recently, none described M. avium PRA variants. Devalois et al. (6) studied 14 MAC strains showing discordant results (DT1-positive amplification and negative reaction with the M. intracellulare AccuProbe probe). Five distinct profiles, different from the patterns found here, were observed, showing a marked heterogeneity of these strains, which were tentatively identified as M. intracellulare. Devalois et al. (5) also reported PRA variants of M. kansasii, M. chelonae, M. fortuitum, M. gordonae, M. flavescens, M. simiae, M. nonchromogenicum, M. abscessus, and M. peregrinum. Three strains of M. paratuberculosis and two of wood pigeon and Crohn’s disease mycobacteria gave the M. avium prototype profile, and no M. avium PRA variants were reported.
The 441-bp hsp65 gene segments amplified by PCR with PRA primers for 56 isolates, comprising M. avium, M. intracellulare, and M. scrofulaceum, were sequenced, and 360 bp was analyzed, by Swanson et al. (26). Those authors found 11 unique sequences showing different point mutations that were grouped in three clusters. Cluster A comprised 30 of the 56 isolates, identified as M. avium, and had two alleles with sequences identical to those of variants I and II described here. They did not report variant III. Cluster B comprised six alleles and 23 strains, identified as M. intracellulare. They did not report the M. intracellulare sequences obtained here. Cluster C corresponded to three isolates showing three different alleles. Although many mutations identified in that study occurred in HaeIII sites, creating new sites in some alleles and destroying it in others, those authors did not report the existence of the PRA variants.
The DNAs of all isolates identified by PRA as M. avium III and one identified as M. avium II were not amplified with IS1245 primers. This IS has been found exclusively in M. avium strains (12). Pestel-Caron and Arbeit (20) reported that this insertion sequence was present in 159 M. avium isolates representing 40 distinct restriction fragment length polymorphism strains studied. The results shown here suggest that this element can be missing in isolates identified by other phenotypic and genotypic methods as M. avium. Another possibility is that mutations in regions complementary to IS primers prevented amplification. The DNAs of all of the IS1245-negative isolates were positive by amplification with primers derived from another M. avium-specific IS, IS1311, which shows 85% sequence similarity with IS1245 (data not shown) (22). The negative amplification of the IS and the geographic distribution of M. avium variant III suggest that they may represent a unique M. avium cluster.
The significance of these variants in terms of virulence has to be determined. Despite its genetic diversity, it is accepted that the MAC forms a well-defined phylogenetic group, as has been demonstrated by internal transcribed spacer sequence comparison of different isolates (9). Characterization of the precise genetic changes that separate these M. avium variants will certainly help in the definition of epidemiological connections and will shed light on factors involved in mycobacterial virulence and routes of infection. This will have a marked influence in the elaboration of control programs.
ACKNOWLEDGMENTS
We acknowledge Robert Arbeit and Dick van Soolingen for fruitful discussions, Rosângela Siqueira de Oliveira for technical assistance with cultures, Suely Yoko Mizuka Ueki for biochemical identification, and Vânia lara Bortolotto and Deise Aparecida Mota for assistance with AccuProbe tests. We thank Petra de Haas for 16S rRNA sequencing and Maria Jesus Garcia for providing us with IS1311 primers.
M.P.S. is the recipient of a fellowship (97/06917-9) from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP). S.C.L. is a member of the Red de Latinoamérica y del Caribe de Tuberculosisis (RELACTB). M.R.S.B. received a grant from FAPESP. This work was supported by FAPESP.
REFERENCES
- 1.Barreto J A, Palaci M, Ferrazoli L, Martins M C, Suleiman J, Lourenço R, Ferreira O C, Jr, Riley L W, Johnson W D, Jr, Galvão P A. Isolation of Mycobacterium avium complex from bone marrow aspirates of AIDS patients in Brazil. J Infect Dis. 1993;168:777–779. doi: 10.1093/infdis/168.3.777. [DOI] [PubMed] [Google Scholar]
- 2.Collins C H, Grange J M, Yates M D. Tuberculosis bacteriology. Oxford, United Kingdom: Butterworth Heinemann; 1997. [Google Scholar]
- 3.Damsker B, Bottone E J. Mycobacterium avium-Mycobacterium intracellulare from the intestinal tracts of patients with the acquired immunodeficiency syndrome: concepts regarding acquisition and pathogenesis. J Infect Dis. 1985;151:179–181. doi: 10.1093/infdis/151.1.179. [DOI] [PubMed] [Google Scholar]
- 4.Devalois A, Picardeau M, Goh K S, Sola C, Vincent V, Rastogi N. Comparative evaluation of PCR and commercial DNA probes for detection and identification to species level of Mycobacterium avium and Mycobacterium intracellulare. J Clin Microbiol. 1996;34:2756–2759. doi: 10.1128/jcm.34.11.2756-2759.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Devalois A, Goh K S, Rastogi N. Rapid identification of mycobacteria to species level by PCR-restriction fragment length polymorphism analysis of the hsp65 gene and proposition of an algorithm to differentiate 34 mycobacterial species. J Clin Microbiol. 1997;35:2969–2973. doi: 10.1128/jcm.35.11.2969-2973.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Devalois A, Picardeau M, Paramasivan C N, Vincent V, Rastogi N. Molecular characterization of Mycobacterium avium complex isolates giving discordant results in AccuProbe tests by PCR-restriction enzyme analysis, 16S rRNA gene sequencing, and DT1-DT6 PCR. J Clin Microbiol. 1997;35:2767–2772. doi: 10.1128/jcm.35.11.2767-2772.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devalois A, Rastogi N. Computer-assisted analysis of Mycobacterium avium fingerprints using insertion elements IS1245 and IS1311 in a Caribbean setting. Res Microbiol. 1997;148:703–713. doi: 10.1016/S0923-2508(99)80069-2. [DOI] [PubMed] [Google Scholar]
- 8.Favero J A. Balanço positivo para a suinocultura eficiente. An Suin Ind. 1999;21:16–22. [Google Scholar]
- 9.Frothingham R, Wilson K H. Molecular phylogeny of the Mycobacterium avium complex demonstrates clinically meaningful divisions. J Infect Dis. 1994;169:305–312. doi: 10.1093/infdis/169.2.305. [DOI] [PubMed] [Google Scholar]
- 10.Gonzales R, Hanna B A. Evaluation of Gen-Probe DNA hybridization systems for the identification of Mycobacterium tuberculosis and Mycobacterium avium-intracellulare. Diagn Microbiol Infect Dis. 1987;8:69–78. doi: 10.1016/0732-8893(87)90152-0. [DOI] [PubMed] [Google Scholar]
- 11.Green E P, Tizard M L V, Moss M T, Thompson J, Winterbourne T J, McFadden J J, Hermon-Taylor J. Sequence and characteristics of IS900, an insertion element identified in a human Crohn’s disease isolate of Mycobacterium paratuberculosis. Nucleic Acids Res. 1989;17:9063–9073. doi: 10.1093/nar/17.22.9063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guerrero C, Bernasconi C, Burki D, Bodmer T, Telenti A. A novel insertion element from Mycobacterium avium, IS1245, is a specific target for analysis of strain relatedness. J Clin Microbiol. 1995;33:304–307. doi: 10.1128/jcm.33.2.304-307.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hernandez-Perez M, Fomukong N G, Hellyer T, Brown I N, Dale J W. Characterization of IS1110, a highly mobile element from Mycobacterium avium. Mol Microbiol, 1994;12:717–724. doi: 10.1111/j.1365-2958.1994.tb01059.x. [DOI] [PubMed] [Google Scholar]
- 14.Inderlied C B, Kemper C A, Bermudez L E M. The Mycobacterium avium complex. Clin Microbiol Rev. 1993;6:266–310. doi: 10.1128/cmr.6.3.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kent P T, Kubica G P. Public health mycobacteriology—a guide for the level III laboratory. Atlanta, Ga: Centers for Disease Control; 1985. [Google Scholar]
- 16.Kunze Z M, Wall S, Appelberg R, Silva M T, Portaels F, McFadden J J. IS901, a new member of a widespread class of atypical insertion sequences, is associated with pathogenicity in Mycobacterium avium. Mol Microbiol. 1991;5:2265–2272. doi: 10.1111/j.1365-2958.1991.tb02157.x. [DOI] [PubMed] [Google Scholar]
- 17.Lebrun L, Espinasse F, Poveda J, Vincent-Levy-Frebault V. Evaluation of nonradioactive DNA probes for identification of mycobacteria. J Clin Microbiol. 1992;30:2476–2478. doi: 10.1128/jcm.30.9.2476-2478.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McAdam R, Guilhot C, Gicquel B. Transposition in mycobacteria. In: Bloom B R, editor. Tuberculosis: pathogenesis, protection, and control. Washington, D.C: American Society for Microbiology; 1994. pp. 199–216. [Google Scholar]
- 19.Moss M T, Malik Z P, Tizard M L V, Green E P, Sanderson J D, Hermon-Taylor J. IS902, an insertion element of the chronic-enteritis-causing Mycobacterium avium subsp. silvaticum. J Gen Microbiol. 1992;138:139–145. doi: 10.1099/00221287-138-1-139. [DOI] [PubMed] [Google Scholar]
- 20.Pestel-Caron M, Arbeit R D. Characterization of IS1245 for strain typing of Mycobacterium avium. J Clin Microbiol. 1998;36:1859–1863. doi: 10.1128/jcm.36.7.1859-1863.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rogall T, Wolters J, Floher T, Böttger E C. Towards a phylogeny and definition of the species at the molecular level within the genus Mycobacterium. Int J Syst Bacteriol. 1990;40:323–330. doi: 10.1099/00207713-40-4-323. [DOI] [PubMed] [Google Scholar]
- 22.Roiz M P, Palenque E, Guerrero C, Garcia M J. Use of restriction fragment length polymorphism as a genetic marker for typing Mycobacterium avium strains. J Clin Microbiol. 1995;33:1389–1391. doi: 10.1128/jcm.33.5.1389-1391.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saito H, Tomioka H, Sato K, Tasaka H, Dawson D J. Identification of various serovar strains of Mycobacterium avium complex by using DNA probes specific for Mycobacterium avium and Mycobacterium intracellulare. J Clin Microbiol. 1990;28:1694–1697. doi: 10.1128/jcm.28.8.1694-1697.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sircili, M. P., E. Roxo, and S. C. Leão. Discrimination of members of the Mycobacterium avium complex by polymerase chain reaction. J. Brazilian Soc. Microbiol., in press.
- 25.Sola C, Devalois A, Goh K S, Legrand E, Rastogi N. Molecular characterization of Mycobacterium avium complex isolates from Caribbean patients by DT1/DT6-PCR, nonradioactive Southern hybridization, and the AccuProbe system. Cur Microbiol. 1996;33:352–358. doi: 10.1007/s002849900127. [DOI] [PubMed] [Google Scholar]
- 26.Swanson D S, Kapur V, Stockbauer K, Pan X, Frothingham R, Musser J M. Subspecific differentiation of Mycobacterium avium complex strains by automated sequencing of a region of the gene (hsp65) encoding a 65-kilodalton heat shock protein. Int J Syst Bacteriol. 1997;47:414–419. doi: 10.1099/00207713-47-2-414. [DOI] [PubMed] [Google Scholar]
- 27.Taylor T B, Patterson C, Hale Y, Safranek W W. Routine use of PCR-restriction fragment length polymorphism analysis for identification of mycobacteria growing in liquid media. J Clin Microbiol. 1997;35:79–85. doi: 10.1128/jcm.35.1.79-85.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Telenti A, Marchesi F, Balz M, Bally F, Böttger E C, Bodmer T. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction analysis. J Clin Microbiol. 1993;31:175–178. doi: 10.1128/jcm.31.2.175-178.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thierry D, Vincent V, Clement F, Guesdon J L. Isolation of specific DNA fragments of Mycobacterium avium and their possible use in diagnosis. J Clin Microbiol. 1993;31:1048–1054. doi: 10.1128/jcm.31.5.1048-1054.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomioka H, Saito H, Sato K, Tasaka H, Dawson D J. Identification of Mycobacterium avium complex strains belonging to serovars 21-28 by three commercial DNA probe tests. Tubercle Lung Dis. 1993;74:91–95. doi: 10.1016/0962-8479(93)90033-T. [DOI] [PubMed] [Google Scholar]