Abstract
Equids are chronically infected with parasitic strongyle nematodes. There is a rich literature on horse strongyles, but they are difficult to identify morphologically and genetic studies on strongyles infecting other equid species are few, hampering studies of host specificity. We sequenced expelled worms from two sympatric zebra species in central Kenya to expand the strongyle phylogeny and used DNA metabarcoding on faecal samples to genetically characterize zebra nemabiomes for the first time. We generated sequences for several species new to public genetic reference databases, all of which are typical strongyles in wild zebras (i.e., the three species of Cylindropharynx and Cyathostomum montgomeryi), and identified their closest relatives. We also discovered an apparent fungus infecting a quarter of the expelled Crossocephalus viviparus worms, a hyperabundant nematode species in the family Atractidae, hinting at the possibility that zebra host-parasite dynamics may involve a zebra-fungus mutualism. The two zebra species had similar nemabiomes; we found a complete overlap in the list of nematode species they carry and very similar prevalence (i.e., proportion of hosts infected) for the different nematode species. Our study suggests limited host-specificity in zebra strongyles and high potential for transmission between the plains zebra and the endangered Grevy's zebra.
Keywords: Equid parasitology, Zebra nematodes, DNA metabarcoding, Equid nemabiome, Strongyle phylogeny
Graphical abstract
Highlights
-
•
We genetically characterized plains and Grevy's zebra nemabiomes for the first time.
-
•
The zebra species hosted the same nematode taxa and may cross-infect each other.
-
•
Nematode community composition was similar, with weak clustering by host species.
-
•
We updated the strongyle phylogeny with 4 newly-sequenced species common in zebras.
-
•
Many expelled Crossocephalus viviparus worms appeared fungus-infected.
1. Introduction
Over a century of research has built a rich literature on the gastrointestinal parasites of equids (family Equidae, which comprises nine extant Equus species, five of which are globally threatened or near-threatened). The most prominent of these parasites are the strongyles (family Strongylidae), a diverse group of faecal-orally transmitted, monoxenous nematodes that chronically infect untreated equid hosts (Dvojnos and Kharchenko, 1994; Lichtenfels et al., 2008). Equid strongyles are thought to be monophyletic and to have coevolved with the host genus (Durette-Desset et al., 1994; Dvojnos, 1982), but most appear to be capable of infecting multiple equid species (Krecek et al., 1987b; Matthee et al., 2004; Ogbourne, 1976; Tolliver et al., 1985). Studies on these nematodes are hindered by the difficulty of identifying strongyles to the species level based on morphology – an impossible task for eggs shed in faeces and one requiring considerable experience when dealing with larvae or adult worms (Lichtenfels et al., 2008). Questions about host specificity would therefore be best addressed using molecular methods (Bredtmann et al., 2017), but most genetic work has thus far been restricted to the strongyles infecting one equid species – the domestic horse (E. caballus; Campbell et al., 1995; Gasser et al., 2004; Hung et al., 1999; Mitchell et al., 2019). A few studies have begun to expand our understanding of strongyle genetics in other equid hosts, and we build upon these efforts to genetically describe strongyle diversity in the present study (Bredtmann et al., 2019; Bu et al., 2013; Hung et al., 1996; Louro et al., 2021).
Equid strongyles are divided into two subfamilies: Strongylinae (comprising five genera: Strongylus, Triodontophorus, Bidentostomum, Oesophagodontus, and Craterostomum) and Cyathostominae (including 14 genera; Lichtenfels, 2008). Some species in the former subfamily have been found to be highly pathogenic (Duncan and Pirie, 1975). Potential variation in pathogenicity and widespread resistance to anthelmintics in the more speciose Cyathostominae (Nielsen et al., 2014) has spurred interest in the use of molecular methods to identify cyathostomin species and disentangle their effects (Gasser et al., 2004; Love et al., 1999). Research on strongyles infecting domestic horses has provided valuable information on their biology that can inform studies of closely related strongyles infecting other equid hosts, but such inferences require genetically sequencing strongyles from multiple equid species to resolve the strongyle phylogeny.
Understanding the extent of strongyle host specificity is of particular interest to conservationists working with endangered equids living sympatrically with other equid species. The Grevy's zebra (E. grevyi) is one such endangered species, and in recent decades its range overlap with the closely related plains zebra (E. quagga) in Kenya has increased (Rubenstein et al., 2016). Studies on zebra nematode communities have thus far relied on the morphological identification of nematodes extracted from faeces or culled animals, a difficult and labour-intensive task that limits the number of hosts examined and requires considerable expertise. To date, only two such studies have been conducted on wild Grevy's zebras (Muoria et al., 2005; Mwatenga, 2017), both using cultured larvae from faeces and identifying only a few strongyle genera (Oesophagostomum, Strongylus, and Trichonema, also known as Parapoteriostomum), potentially because larvae are difficult to identify (Lichtenfels et al., 2008). However, strongyle diversity in Grevy's zebras is likely to be much higher than indicated by these previous studies; culled plains and mountain zebras (E. zebra) in southern Africa were found to have rich strongyle communities in which many genera of cyathostomins and strongylins were highly prevalent (Krecek et al., 1987a, 1987b; Scialdo-Krecek et al., 1983). DNA metabarcoding of faecal samples is emerging as a powerful method to mitigate issues arising from the difficulty of morphological worm identification, allowing the characterization of entire nemabiomes (nematode communities) by capturing DNA from eggs and larvae shed in faeces (Avramenko et al., 2015). This method has been recently used to investigate horse nemabiomes (Mitchell et al., 2019; Poissant et al., 2021), but has not yet been used in other equids.
Equid strongyles appear to be relatively generalist parasites within Equus. Extensive but incomplete overlap was found in the strongyle species hosted by sympatric donkeys (E. asinus), horses, and plains and mountain zebras in southern Africa (Matthee et al., 2004). Similarly, in Ukraine, high overlap was found in the strongyle species infecting captive plains and Grevy's zebras, as well as the ponies with which they shared pasture (Kuzmina et al., 2013; Zvegintsova and Treus, 1999). However, these captive-born zebras were regularly dewormed and lacked most strongyle species that typically occur in wild African equids, indicating that they had lost much of their original strongyle fauna (Kuzmina et al., 2013). It remains unclear how many and which strongyle species might be transmitted between plains zebras and Grevy's zebras in the wild. In this study, our objectives were to (1) build upon genetic reference databases for equid nematodes and generate an updated strongyle phylogeny integrating the species that infect zebras; (2) genetically characterize the nemabiomes of plains and Grevy's zebras for the first time, using DNA metabarcoding; and (3) determine the extent to which wild plains and Grevy's zebras host the same strongyle species, and thus their potential for cross-infection.
2. Materials and methods
2.1. Field methods
We collected fresh zebra dung opportunistically whenever defecation was observed during zebra censuses (N = 186 and 223 samples from Grevy's and plains zebras, respectively) at three sites in central Kenya: Mpala Research Centre, Lewa Conservancy, and Samburu National Reserve and the contiguous Buffalo Springs National Reserve. Censuses at each site occurred across four years, with field seasons in July–September 2015, May–July 2016, March 2017, June–July 2017, and June–July 2018 to capture interseasonal and interannual variation (egg shedding tends to be seasonal but the amount and timing of rainfall in the region is highly variable across seasons and years). Within 20 min of observing a defecation, two or three faecal balls from the top of the pile were collected and stored on ice until our return to the research station later that day. All sampled zebras were infected with strongyle nematodes as evidenced by faecal egg counts that we conducted for other studies.
2.2. Worm collection, identification, and sequencing
We collected 144 nematodes expelled in zebra dung as reference samples for DNA analysis and morphological identification. Worms were collected from six Grevy's zebras, five plains zebras, and one Grevy's x plains hybrid zebra at Mpala in March 2018 and June–July 2018. Nematodes were picked out of faeces under a dissection microscope using soft metal forceps, washed three times with saline water, and viewed and photographed under a light microscope using both 40× and 100× total magnification. Each worm was cut into thirds and the head and tail were submerged in 70% EtOH while the mid-section was stored in DNA stabilizing lysis buffer (Zymo Xpedition Stabilization/Lysis Solution, Zymo Research) and heat-treated at 72 °C for 30 min as a precaution against foot-and-mouth disease. All samples were then frozen at −20 °C until transport (Kartzinel et al., 2015). The heads and tails were sent to HelmWest Laboratory, Missoula (MT, USA), where JMK clarified them with lactophenol and observed and photographed them under a light microscope with 100× magnification. JMK and KJT independently morphologically identified each reference worm using all observations and photographs as well as the species identification keys provided in Lichtenfels et al. (2008); results were then compared and any discrepancies discussed to finalize taxonomic identities. The mid-sections were processed for sequencing by KJT at Princeton University. DNA was extracted from worms by a standard proteinase-K based enzyme digestion protocol (SI1). We PCR amplified the internal transcribed spacer 2 (ITS2) locus of ribosomal DNA and the flanking 5.8S and 28S regions – a 292-461bp region known to exhibit much higher interspecific variation than intraspecific variation in equid strongyles (Hung et al., 2000) – using the primers NC1 and NC2 (Gasser et al., 1993), and following a PCR protocol modified from Sim et al. (2010) (SI1). Post-PCR DNA concentrations were measured using PicoGreen and samples were purified and Sanger sequenced in both directions by Macrogen USA. A total of 91 samples were successfully sequenced and were then used to reconstruct the strongyle phylogeny.
2.3. Phylogeny building
We used Geneious 11.0.4 (https://www.geneious.com) to align forward and reverse reads and generate the consensus sequences for each nematode reference sample. Each of these sequences was run through BLASTn to find the closest matches in GenBank and confirm morphological identification. Any worm for which morphological identification was uncertain was identified to species level if >98% similar to a GenBank reference and to genus level if >95% similar to a GenBank reference along the entire sequence. We chose these thresholds based on inter- and intraspecific sequence similarity (inferred using the European Bioinformatics Institute, or ‘EMBL-EBI’, MUSCLE algorithm; Madeira et al., 2019) between worms that we could confidently identify morphologically. We then conducted a multiple sequence alignment using MUSCLE (Edgar, 2004) and used MEGA-X (version 10.2.6; Kumar et al., 2018; Stecher et al., 2020) to test models for estimating evolutionary distance by maximum likelihood with default parameters. The model with the lowest Bayesian information criterion (BIC) score (the Tamura 3-parameter model with gamma-distributed substitution rates, γ = 0.45) was selected to build a phylogenetic tree with the neighbour-joining method (Saitou and Nei, 1987). The tree robustness was assessed using the bootstrap test (2000 replicates) and we plotted the consensus tree using the R package ggtree, labelling nodes that were highly supported with bootstrap values (i.e., the percentage of the replicate trees that had the same branch tips cluster together) (R version 4.0.0; R Core Team, 2019; Yu et al., 2017).
In addition to generating a phylogeny from our reference worms, we integrated the reference worms that we could identify to species level into a broader phylogenetic tree that included all sequenced strongyles with species-level identification extracted from the ‘Nemabiome’ nematode ITS2 database (version 1.2.0; www.nemabiome.ca), which draws data from GenBank and which included sequences contributed from the present study (Avramenko et al., 2015, 2017). In addition to the 329 Strongylidae sequences extracted from the database, we manually added the only available sequence for Strongylus asini (entered only as S. asini in GenBank) and our Caenorhabditis elegans positive control to serve as the outgroup. A sequence from one of the Coronocyclus labiatus references (MG738707) was excluded after it appeared as an outgroup in the phylogeny instead of C. elegans in an initial tree construction. We then processed the sequences as above (using the Tamura 3-parameter model with γ = 1.11). In addition, we pruned the tree by merging all immediate sister taxa with identical species assignments or synonymous assignments (i.e., Cylicodontophorus mettami and Parapoteriostomum mettami, Petrovinema poculatum and Cylicostephanus poculatus; Lichtenfels et al., 2008) in R with the ‘drop.tip’ function from the package ape (Paradis and Schliep, 2019) before visualizing the tree with ggtree.
2.4. Nemabiome characterization from faecal DNA
To DNA metabarcode nemabiomes, we first thoroughly homogenized the freshly-collected faecal samples by massaging sample bags. A pea-sized subsample (∼0.2g) was taken for DNA analysis, heat-treated at 72 °C for 30 min, and frozen in DNA lysis buffer until transport at −20 °C (Kartzinel et al., 2015). At Princeton University, we extracted DNA along with extraction controls with a Zymo Quick-DNA Faecal/Soil Microbe Miniprep kit. To multiplex amplicons before sequencing, we used NC1 and NC2 primers tagged with an 8-bp label to amplify the ITS2 region, following the protocol described above (and in SI1). DNA was purified using a Zymo DNA Clean and Concentrator kit and concentrations were quantified using a Qubit 4 fluorometer. We then standardized the concentration of PCR products to be multiplexed and submitted them for high-throughput paired-end sequencing on an Illumina MiSeq platform (2 × 250bp reads) at Princeton University's Lewis-Sigler Institute for Integrative Genomics.
Output sequences were run through the obitools workflow (Boyer et al., 2016) to (1) align paired-end reads, (2) demultiplex sequences for each sample, (3) dereplicate the sequences, (4) identify sequences that were likely to be derived from PCR/sequencing errors, and (5) assign sequences to a taxon against both a global database (derived from the EMBL database) and the local database we created from the nematode reference samples that we morphologically identified (see details in SI1) (Pansu et al., 2019). Preference was given to the local database in the case of a discrepancy and equal match scores. Additional filtering steps were conducted in R (SI1). All samples were rarefied to 1000 reads based on rarefaction curves constructed with the vegan package (Oksanen et al., 2019) to standardize library sizes and enable cross-sample comparison.
Prior to analyses, the 224 unique sequences that remained were grouped into clusters approximating species, called molecular operational taxonomic units (mOTUs), using the SUMATRA algorithm for sequence alignment (Mercier et al., 2013) and MCL for clustering (van Dongen, 2008). For our analyses, we used a 98% similarity threshold between sequences to define clusters. Interspecific differentiation in nemabiome communities was visualized using non-metric multidimensional scaling (NMDS) ordinations based on Jaccard distances (Jaccard, 1901) calculated with the package vegan (Oksanen et al., 2019) and using bipartite networks (constructed with the package bipartite; Dormann et al., 2008). Because of possible amplification biases between nematode taxa, the potentially misleading skew in egg shedding between taxa (Kuzmina et al., 2012), and our particular interest in host specificity, we compared zebra nemabiomes on a prevalence (presence/absence) basis. We focused on the most abundant taxa in nemabiome analyses (the 35 sequences that contribute >1% to the total pooled reads, clustered by 98% similarity into 20 mOTUs). Nemabiomes between the two zebra species were compared with an analysis of similarity (ANOSIM; using the ‘anosim’ function in vegan; Oksanen et al., 2019). This analysis produces a metric of dissimilarity, R, ranging from 0, indicating identical communities, to 1, indicating completely distinct nematode communities (Clarke, 1993).
To assess the sensitivity of the interspecific nemabiome overlap to the similarity threshold used in mOTU clustering, we reran these analyses using various similarity thresholds (90%, 95%, 97%, 98%, and 99%; SI3) and found that the results did not change. Similarly, we evaluated whether sampling location or season affected the level of nemabiome overlap between the two zebra species and found no evidence for it (SI4). Samples from different sites and seasons were therefore pooled for the analyses presented in this study. Finally, the correspondence in prevalence of nematode mOTUs in plains vs. Grevy's zebras was assessed with a linear regression, and the per-individual species richness was compared between host species using a Wilcoxon rank sum test.
3. Results
The expelled worms we found in zebra faeces that were successfully sequenced belonged to five genera and 18 species or mOTUs. Most of the collected worms were Parapoteriostomum species, followed by Crossocephalus viviparus (the only species collected from the family Atractidae), and only one was a strongylin (Craterostomum acuticaudatum). Four species were new to public genetic reference databases (Cylindropharynx brevicauda, C. intermedia, C. longicauda, and Cyathostomum montgomeryi). Our study also provided new information on within-species genetic variation in strongyles: out of the 91 worms sequenced, only three, all of which were identified as Cylicostephanus minutus, had species-level matches (>98% similarity) to existing GenBank entries, and these matches were only partial (85–90% of the sequence length).
The phylogeny of the nematodes we collected showed strong clustering by genus (i.e., most sequences from the same genus grouped together in one clade). There was high confidence in the nodes separating the three species of Cylindropharynx, showing that C. brevicauda and C. intermedia are more closely related to each other than they are to C. longicauda (SI2). There was also strong support for Cylicostephanus bidentatus and Cyathostomum montgomeryi being sister taxa. Apart from the node separating an unidentified Cylicostephanus species and Craterostomum acuticaudatum, much of the remaining tree structure was highly variable and most cross-genus relationships could not be resolved (i.e., most branch nodes were supported by fewer than 50% of the tree replicates in the bootstrap test; SI2).
In our broader phylogeny of strongyles integrating sequences from the Nemabiome database, the immediate relatives of the newly-sequenced strongyles could be confidently identified. The closest relatives of Cylindropharynx were Skrjabinodentus caragandicus, a cyathostomin, and Bidentosomum ivashkini, a strongylin (Fig. 1). The optimal tree further placed the three Triodontophorus species and Craterostomum acuticaudatum – which are all also strongylins (but see Hung et al., 2000) – as the next clade to branch from this cluster. The closest relative of Cyathostomum montgomeryi was identified with high confidence as Cyathostomum tetracanthum, and unlike the phylogeny of our reference worms, Cylicostephanus bidentatus was placed far from these species and in a basal position within the broader phylogeny, potentially because of the addition of many more taxa in this phylogeny. Parapoteriostomum and Poteriostomum were shown to be closely-related genera, and non-equid strongyles (Tziminema, Khalilia, Kiluluma, Quilonia, and Murshidia species – members of the Strongylidae family that infect tapirs, rhinoceroses, and elephants) formed a cluster with several well-supported internal nodes. Much uncertainty remains in the relationships between most cyathostomin genera, with Cylicostephanus, Cylicocyclus, Coronocyclus, and Cyathostomum species widely distributed throughout the phylogeny.
Fig. 1.
Phylogeny of all sequenced Strongylidae with species-level taxon assignments from GenBank and the present study, coloured by genus (the same colour code is used across all figures in this paper). Branch tips were pruned such that only one tip was kept of all immediate sister taxa with identical taxon assignments, and the number of samples merged in each tip is indicated in parentheses. Bootstrap percentages over 50% are displayed in bold to highlight nodes with high support. Branch lengths represent the number of base substitutions per site, estimated with the Tamura 3-parameter model assuming gamma-distributed substitution rate variation.
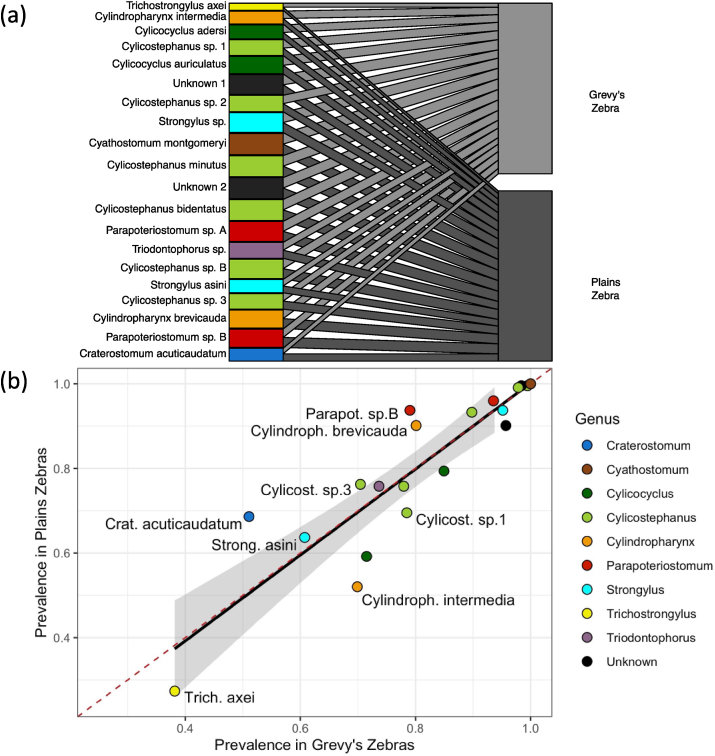
All nematode mOTUs were found in both host species. Nemabiome community composition differed slightly by host species as evidenced by some weak but statistically significant clustering in ordination plots (SI3, SI4). However, the ANOSIM revealed an overall R value of 0.013 (p < 0.05), a value much lower (indicating much more similar communities) than has been reported between plains and mountain zebras (R = 0.41; Matthee et al., 2004).
Prevalence of nematode taxa (the proportion of individual hosts infected) in plains and Grevy's zebras was correlated nearly one-to-one (linear model p < 0.001, r2 = 0.78, estimated slope = 1.02; Fig. 2b). The most prevalent nematodes – ‘Unknown 2’ (which had no reference matches >95% in public genetic databases), Cylicostephanus bidentatus, Cyathostomum montgomeryi, and Cylicostephanus sp. B – were equally prevalent in the two host zebra species (upper right corner of Fig. 2b). Potential weak host preferences as suggested by skew in prevalence were apparent in some nematode clades. For example, among Cylindropharynx, C. intermedia was more prevalent in Grevy's zebras (prevalence in Grevy's: 69.9%, in plains: 52.0%) and C. brevicauda in plains zebras (80.1% in Grevy's, 90.1% in plains; Fig. 2). While none of our zebra hosts carried the three principal strongylins of domestic horses (Strongylus vulgaris, S. edentatus, and S. equinus, each of which has ITS2 references in GenBank), an unknown mOTU likely to be in the Strongylus genus was prevalent in both zebra species, as were other strongylins such as S. asini, Triodontophorus sp., and Craterostomum acuticaudatum (51.1–95.2% prevalence). As a group, these relatives of the highly pathogenic S. vulgaris are highly prevalent in wild zebras – in fact, none of our zebra samples were wholly free of strongylin DNA. Finally, the two zebra species did not differ in per-individual species richness for the 20 nematode mOTUs analysed (Wilcoxon rank sum test p = 0.49; mean species richness per Grevy's zebra = 16.1 ± s.d. 2.8 and per plains zebra = 16.0 ± s.d. 2.4).
Fig. 2.
Nematode prevalence in plains vs. Grevy's zebras in (a) a bipartite graph, where edge widths indicate prevalence in each zebra species, and (b) a linear regression, shown in black with shading representing standard error and the one-to-one line indicated by the dashed red line. Only sequences comprising >1% of total reads were used and they were clustered into mOTUs by 98% similarity. Taxon labels followed by a letter signify species-level matches (>98% similarity) to reference worms identified only to genus (see SI2), while those followed by a number represent sequences that matched a reference only to the genus level (>95% similarity). Black boxes/points are taxa without a match of >95% to any identified sequences. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Forty of the 144 extracted individual worms were Crossocephalus viviparus, an atractid nematode that has yet to be sequenced and that would not amplify with our primers, even after a second round of DNA extraction. Our primers, designed for identifying strongyles, may be too divergent from atractid DNA to bind to the target site (atractids belong to a different superfamily). Moreover, we found ten C. viviparus worms (25% of the C. viviparus collected) to be infected with what appeared to be fungal hyphae sprouting from the mouth or anogenital orifices. The hyphae stained with lactophenol blue, indicating the presence of chitin, a major component in the cell walls of fungi, although cellulose also takes up this stain (Fig. 3). Our attempts to identify the fungus by amplifying the ITS region using the fungus-specific ITS1F and ITS2R primers (Gardes and Bruns, 1993), following Smith and Peay (2014), failed.
Fig. 3.
Crossocephalus viviparus extracted from fresh zebra faeces were often infected with an apparent fungus. Hyphae emerging from the head (left, right) and from the tail (centre) of infected worms and stained with lactophenol blue (right). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
Our sequenced reference samples provided many new contributions to public genetic databases. Four of the seven strongyle taxa that we could confidently identify to species-level had not previously been sequenced – these were all strongyles common in wild zebras (the three Cylindropharynx species and Cyathostomum montgomeryi; Krecek et al., 1987b; Matthee et al., 2004; Round, 1968; Scialdo-Krecek et al., 1983). We were further able to identify the closest relatives for each of these taxa, and the proximity of Cylindropharynx species with strongylins in our broad phylogeny indicates that their pathogenicity may be a worthwhile target for future research.
The high uncertainty of many phylogenetic nodes and the scattered distribution of many cyathostomin genera in the tree highlight the need to further resolve the strongyle phylogeny. The placement of Strongylus species among cyathostomins rather than with the other strongylins in our broad phylogeny was not based on nodes with high confidence, but it is mirrored by some previous strongyle phylogenies and appears to depend on the exact tree construction method used (Hung et al., 2000). It is possible that some of the uncertainty in the strongyle phylogeny arises from polymorphisms at the ITS2 locus. The amplification of the ITS2 locus from Coronocyclus coronatus and Cylicostephanus calicatus worms in a previous study produced one of two PCR products – a longer fragment and a shorter fragment – and sometimes both from a single individual. Combining information from ITS2 sequences and COI sequences appeared to provide better resolution (Bredtmann et al., 2019). However, despite demonstrated ITS2 haplotype diversity within Cylicocyclus nassatus and Coronocyclus labiatus (Louro et al., 2021), these sequences clustered well by species in our phylogeny using ITS2 (Fig. 1).
No nematode mOTU occurred in only one of the two host species in our study, suggesting high potential for cross-transmission, and the prevalence of nematode taxa in plains and Grevy's zebras was correlated almost one-to-one. The overlap in nemabiome community composition we found between plains and Grevy's zebras was much higher than the overlap previously found between plains and mountain zebras in southern Africa, some of which were sympatric (Matthee et al., 2004). It was noted that plains and mountain zebras shared more nematode species when living in sympatry, but an R value for interspecific dissimilarity within the same site was not provided, making a direct comparison of similarity difficult. The evolutionary divergence of plains and Grevy's zebras is more recent than that of plains and mountain zebras (Jónsson et al., 2014), so the nemabiomes of the former pair could be more similar than the latter.
In our broad strongyle phylogeny, strongyles found in elephants (Murshidia, Quilonia, and Khalilia species; McLean et al., 2012), rhinoceroses (Kiluluma and Khalilia species; Knapp et al., 1997), and Baird's tapirs (Tziminema unachi; Güiris et al., 2017) clustered separately from those found in equids, outlining potential limits to host generalism in strongyles (Fig. 1). The feeding habits, digestive morphology, and immune systems of different equid species may be close enough to enable many strongyle species to be successfully transmitted between them, but other perissodactyls or other large hindgut-fermenting herbivores appear to have coevolved with their own strongyle taxa. The fact that nematodes in the family Strongylidae infect perissodactyls and elephants may be linked to the convergent evolution in the gut morphology of these host species (Clauss, 2013).
We were unable to amplify Crossocephalus viviparus DNA, but the diversity and genetics of atractids deserves further study as they are likely the most abundant nematode in zebra nemabiomes. Atractids regularly reach infection intensities in the millions in zebras, black and white rhinoceroses, and occasionally in horses (Probstmayria vivipara is an atractid reported to infect horses and rhinoceroses but also zebras; Felippelli et al., 2015; Knapp et al., 1997; Scialdo-Krecek et al., 1983). Accordingly, atractids are very likely to be a key player in nematode competition dynamics within the host, but as viviparous nematodes they do not contribute to faecal egg counts and they are largely omitted from the literature on equid host-parasite dynamics. Crossocephalus viviparus has been reported in plains zebras in South Africa (Scialdo-Krecek et al., 1983), and our study confirms its presence in this species at the opposite end of its latitudinal range and documents it for the first time in Grevy's zebras.
Nematophagous fungi, such as Duddingtonia flagrans and Mucor circinelloides, are known to survive passage through the horse gut and to fight strongyle infections and improve horse body condition (Braga et al., 2009; Canhão-Dias et al., 2020). The use of D. flagrans as a biological control of cyathostomin nematodes has been successfully tested in plains zebras and African wild ass in a zoological park, and it is now commercially available in the form of inoculated nutritional pellets (Larsen, 2000; Palomero et al., 2018). If the hypha-like structures we found emerging from C. viviparus nematodes are confirmed to be a fungus, then a nematophagous fungus can also infect atractids and may potentially form a mutualistic relationship with its zebra hosts. Future research on the little-understood atractids and host-fungus-nematode dynamics is likely to make important contributions to our understanding of equid parasitology.
5. Conclusions
This study revealed an absence of strict host specificity among gastrointestinal nematodes infecting sympatric plains and Grevy's zebras in central Kenya, suggesting that the two species have high potential for cross-infecting one another. Cumulative zebra dung density on the landscape is thus likely a determinant of the exposure risk of both zebra species to these faecal-orally transmitted nematodes. We also found their nemabiomes to be highly similar in terms of the prevalence of the various nematode taxa. Several prevalent sequences from the metabarcoding analyses matched publicly available reference sequences only to genus level or broader. Given the absence of reference sequences in public databases for many nematodes infecting wildlife, and the wealth of information such databases provide for phylogenetic and DNA metabarcoding studies, we suggest that the development of extensive taxonomically verified reference libraries is a high priority for future research (Pringle and Hutchinson, 2020). We anticipate that further investigations of the diversity of zebra nematodes will likely result in the discovery of new species, some of which may be of considerable consequence to host health and conservation.
Declaration of competing interest
None.
Acknowledgments
This work was funded by the National Science Foundation (IBN-0309233, CNS-025214, IOB-9874523, IIS-0705822, IIS-0747369, IOS-1656527) and the High Meadows Environmental Institute and Department of Ecology and Evolutionary Biology at Princeton University. Sequence alignment, filtering, and taxonomic assignment were performed on computational resources managed and supported by Princeton Research Computing, a consortium of groups including the Princeton Institute for Computational Science and Engineering (PICSciE) and the Office of Information Technology's High Performance Computing Centre and Visualization Laboratory at Princeton University. We thank Mpala Research Centre, Kenya Wildlife Services (permit number KWS/BRM/5001), the National Environment Management Authority (NEMA/AGR/107/2018), and the National Commission for Science and Technology (A10450) for permission to conduct our research and for logistical support, as well as the United States Department of Agriculture for permission to transport the samples into and within the United States (USDA APHIS permits 130123, 137202, and 137555). We thank the wonderful team of research staff at Mpala Research Centre, including Rosemary Warungu, Margaret Mwangi, Anthony Mwangi, Timothy Sisanya, and Josphat Mwangi. We thank Emily Nonnamaker, Lily Reisinger, Laurel Easterling, Monica Seng, Madison Spinelli, and Lindsay Martinez for their generous assistance in the field. We thank Sarah Budischak, Jasmin Ashraf, and Andrea Graham for valuable advice on the sequencing techniques, protocols, and analyses. We are also grateful to Sara Weinstein, Danny Haelewaters, Anne Pringle, Marie Trest, Rosina Krecek, Vitaliy Kharchenko, and Sonja Matthee for guidance and assistance on the microscopic identification of nematodes and/or the suspected nematophagous fungus.
Footnotes
Note: Supplementary information associated with this article. Nucleotide sequence data reported in this paper are available in the GenBankTM, EMBL and DDJB databases under the accession numbers MZ435497-MZ435587.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijppaw.2021.10.007.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Avramenko R.W., Redman E.M., Lewis R., Bichuette M.A., Palmeira B.M., Yazwinski T.A., Gilleard J.S. The use of nemabiome metabarcoding to explore gastro-intestinal nematode species diversity and anthelmintic treatment effectiveness in beef calves. Int. J. Parasitol. 2017;47:893–902. doi: 10.1016/j.ijpara.2017.06.006. [DOI] [PubMed] [Google Scholar]
- Avramenko R.W., Redman E.M., Lewis R., Yazwinski T.A., Wasmuth J.D., Gilleard J.S. Exploring the gastrointestinal “nemabiome”: deep amplicon sequencing to quantify the species composition of parasitic nematode communities. PLoS One. 2015;10:1–19. doi: 10.1371/journal.pone.0143559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer F., Mercier C., Bonin A., Le Bras Y., Taberlet P., Coissac E. obitools: a unix-inspired software package for DNA metabarcoding. Mol. Ecol. Resour. 2016;16:176–182. doi: 10.1111/1755-0998.12428. [DOI] [PubMed] [Google Scholar]
- Braga F.R., Carvalho R.O., Araújo J.M., Silva A.R., Araújo J.V., Lima W.S., Tavela A.O., Ferreira S.R. Predatory activity of the fungi Duddingtonia flagrans, Monacrosporium thaumasium, Monacrosporium sinense and Arthrobotrys robusta on Angiostrongylus vasorum first-stage larvae. J. Helminthol. 2009;83:303–308. doi: 10.1017/S0022149X09232342. [DOI] [PubMed] [Google Scholar]
- Bredtmann C.M., Krücken J., Kuzmina T., Louro M., Madeira de Carvalho L.M., von Samson-Himmelstjerna G. Nuclear and mitochondrial marker sequences reveal close relationship between Coronocyclus coronatus and a potential Cylicostephanus calicatus cryptic species complex. Infect. Genet. Evol. 2019;75:103956. doi: 10.1016/j.meegid.2019.103956. [DOI] [PubMed] [Google Scholar]
- Bredtmann C.M., Krücken J., Murugaiyan J., Kuzmina T., von Samson-Himmelstjerna G. Nematode species identification—current status, challenges and future perspectives for cyathostomins. Front. Cell. Infect. Microbiol. 2017;7:1–8. doi: 10.3389/fcimb.2017.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu Y., Niu H., Zhang L. Phylogenetic analysis of the genus Cylicocyclus (Nematoda: Strongylidae) based on nuclear ribosomal sequence data. Acta Parasitol. 2013;58:167–173. doi: 10.2478/s11686-013-0124-z. [DOI] [PubMed] [Google Scholar]
- Campbell A.J., Gasser R.B., Chilton N.B. Differences in a ribosomal DNA sequence of Strongylus species allows identification of single eggs. Int. J. Parasitol. 1995;25:359–365. doi: 10.1016/0020-7519(94)00116-6. [DOI] [PubMed] [Google Scholar]
- Canhão-Dias M., Paz-Silva A., Madeira de Carvalho L.M. The efficacy of predatory fungi on the control of gastrointestinal parasites in domestic and wild animals—a systematic review. Vet. Parasitol. 2020;283:109173. doi: 10.1016/j.vetpar.2020.109173. [DOI] [PubMed] [Google Scholar]
- Clarke K.R. Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 1993;18:117–143. [Google Scholar]
- Clauss M. Digestive physiology and feeding behaviour of equids – a comparative approach. Eur. Equine Heal. Nutr. Congr. 2013:25–33. [Google Scholar]
- Dormann C.F., Gruber B., Fründ J. Introducing the bipartite package: analysing ecological networks. R. News. 2008;8:8–11. [Google Scholar]
- Duncan J.L., Pirie H.M. The pathogenesis of single experimental infections with Strongylus vulgaris in foals. Res. Vet. Sci. 1975;18:82–93. [PubMed] [Google Scholar]
- Durette-Desset M.C., Beveridge I., Spratt D.M. The origins and evolutionary expansion of the Strongylida (Nematoda) Int. J. Parasitol. 1994;24:1139–1165. doi: 10.1016/0020-7519(94)90188-0. [DOI] [PubMed] [Google Scholar]
- Dvojnos G., Kharchenko V.A. Nakova Dumka; Kiev: 1994. Strongylids of Domestic and Wild Horses. [In Russian] [Google Scholar]
- Dvojnos G.M. Parazity I Parazitozy Cheloveka I Zhivotnyka. Nakova Dumka; Kiev: 1982. [Systematics and phylogeny of nematodes of the superfamily Strongyloidea Weinland, 1858, parasitic in equines] pp. 106–114. [In Russian] [Google Scholar]
- Edgar R.C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinf. 2004;5:1–19. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felippelli G., Cruz B.C., Gomes L.V.C., Lopes W.D.Z., Teixeira W.F.P., Maciel W.G., Buzzulini C., Bichuette M.A., Campos G.P., Soares V.E., Bergamasco P.L.F., Oliveira G.P. de, Costa A.J. da. Susceptibility of helminth species from horses against different chemical compounds in Brazil. Vet. Parasitol. 2015;212:232–238. doi: 10.1016/j.vetpar.2015.07.041. [DOI] [PubMed] [Google Scholar]
- Gardes M., Bruns T.D. ITS primers with enhanced specificity for basidiomycetes ‐ application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993;2:113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- Gasser R.B., Chilton N.B., Hoste H., Beveridge I. Rapid sequencing of rDNA from single worms and eggs of parasitic helminths. Nucleic Acids Res. 1993;21:2525–2526. doi: 10.1093/nar/21.10.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser R.B., Hung G.-C., Chilton N.B., Beveridge I. Advances in developing molecular-diagnostic tools for strongyloid nematodes of equids: fundamental and applied implications. Mol. Cell. Probes. 2004;18:3–16. doi: 10.1016/j.mcp.2003.10.001. [DOI] [PubMed] [Google Scholar]
- Güiris D.M., Oceguera-Figueroa A., Osorio-Sarabia D., Pérez-Escobar M.E., Nieto-Lopéz M.G., Rojas-Hernández N.M., García-Prieto L. Tziminema unachi n. gen., n. sp. (Nematoda: Strongylidae: Strongylinae) parasite of Baird’s tapir Tapirus bairdii from Mexico. J. Helminthol. 2017;5:1–8. doi: 10.1017/S0022149X17001055. [DOI] [PubMed] [Google Scholar]
- Hung G.-C., Chilton N.B., Beveridge I., Gasser R.B. A molecular systematic framework for equine strongyles based on ribosomal DNA sequence data. Int. J. Parasitol. 2000;30:95–103. doi: 10.1016/s0020-7519(99)00166-6. [DOI] [PubMed] [Google Scholar]
- Hung G.-C., Gasser R.B., Beveridge I., Chilton N.B. Species-specific amplification by PCR of ribosomal DNA from some equine strongyles. Parasitology. 1999;119:69–80. doi: 10.1017/S0031182099004497. [DOI] [PubMed] [Google Scholar]
- Hung G.-C., Jacobs D.E., Krecek R.C., Gasser R.B., Chilton N.B. Strongylus asini (Nematoda, Strongyloidea): genetic relationships with other Strongylus species determined by ribosomal DNA. Int. J. Parasitol. 1996;26:1407–1411. doi: 10.1016/s0020-7519(96)00136-1. [DOI] [PubMed] [Google Scholar]
- Jaccard P. Distribution de la flore alpine dans le bassin des dranses et dans quelques régions voisines. Bull. la Société Vaudoise des Sci. Nat. 1901;37:241–272. [Google Scholar]
- Jónsson H., Schubert M., Seguin-Orlando A., Ginolhac A., Petersen L., Fumagalli M., Albrechtsen A., Petersen B., Korneliussen T.S., Vilstrup J.T., Lear T., Myka J.L., Lundquist J., Miller D.C., Alfarhan A.H., Alquraishi S. a, Al-Rasheid K. a S., Stagegaard J., Strauss G., Bertelsen M.F., Sicheritz-Ponten T., Antczak D.F., Bailey E., Nielsen R., Willerslev E., Orlando L. Speciation with gene flow in equids despite extensive chromosomal plasticity. Proc. Natl. Acad. Sci. Unit. States Am. 2014;111:18655–18660. doi: 10.1073/pnas.1412627111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartzinel T.R., Chen P.A., Coverdale T.C., Erickson D.L., Kress W.J., Kuzmina M.L., Rubenstein D.I., Wang W., Pringle R.M. DNA metabarcoding illuminates dietary niche partitioning by African large herbivores. Proc. Natl. Acad. Sci. Unit. States Am. 2015;112:8019–8024. doi: 10.1073/pnas.1503283112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp S.E., Scialdo-Krecek R.C., Horak I.G., Penzhorn B.L. Helminths and arthropods of black and white rhinoceroses in southern Africa. J. Wildl. Dis. 1997;33:492–502. doi: 10.7589/0090-3558-33.3.492. [DOI] [PubMed] [Google Scholar]
- Krecek R.C., Malan F.S., Reinecke R.K., de Vos V. Nematode parasites from Burchell's zebras in South Africa. J. Wildl. Dis. 1987;23:404–411. doi: 10.7589/0090-3558-23.3.404. [DOI] [PubMed] [Google Scholar]
- Krecek R.C., Reinecke R.K., Malan F.S. Studies on the parasites of zebras. V. Nematodes of the Burchell’s and Hartmann’s mountain zebras from the Etosha National Park, South West Africa/Namibia. Onderstepoort J. Vet. Res. 1987;54:71–78. [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. Mega X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmina T.A., Kharchenko V.A., Zvegintsova N.S., Zhang L., Liu J. Strongylids (Nematoda: Strongylidae) in two zebra species from the “Askania-Nova” Biosphere Reserve, Ukraine: biodiversity and parasite community structure. Helminthologia. 2013;50:172–180. doi: 10.2478/s11687-013-0128-0. [DOI] [Google Scholar]
- Kuzmina T.A., Lyons E.T., Tolliver S.C., Dzeverin I.I., Kharchenko V.A. Fecundity of various species of strongylids (Nematoda: Strongylidae) — parasites of domestic horses. Parasitol. Res. 2012;111:2265–2271. doi: 10.1007/s00436-012-3077-5. [DOI] [PubMed] [Google Scholar]
- Larsen M. Prospects for controlling animal parasitic nematodes by predacious micro fungi. Parasitology. 2000;120:S121–S131. doi: 10.1017/s0031182099005739. [DOI] [PubMed] [Google Scholar]
- Lichtenfels J.R., Kharchenko V.A., Dvojnos G.M. Illustrated identification keys to strongylid parasites (Strongylidae: nematoda) of horses, zebras and asses (Equidae) Vet. Parasitol. 2008;156:4–161. doi: 10.1016/j.vetpar.2008.04.026. [DOI] [PubMed] [Google Scholar]
- Louro M., Kuzmina T.A., Bredtmann C.M., Diekmann I., de Carvalho L.M.M., von Samson-Himmelstjerna G., Krücken J. Genetic variability, cryptic species and phylogenetic relationship of six cyathostomin species based on mitochondrial and nuclear sequences. Sci. Rep. 2021;11:1–10. doi: 10.1038/s41598-021-87500-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love S., Murphy D., Mellor D. Pathogenicity of cyathostome infection. Vet. Parasitol. 1999;85:113–122. doi: 10.1016/S0304-4017(99)00092-8. [DOI] [PubMed] [Google Scholar]
- Madeira F., Park Y.M., Lee J., Buso N., Gur T., Madhusoodanan N., Basutkar P., Tivey A.R.N., Potter S.C., Finn R.D., Lopez R. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47:W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthee S., Krecek R.C., McGeoch M.A. A comparison of the intestinal helminth communities of Equidae in southern Africa. J. Parasitol. 2004;90:1263–1273. doi: 10.1645/GE-3353. [DOI] [PubMed] [Google Scholar]
- McLean E.R., Kinsella J.M., Chiyo P., Obanda V., Moss C., Archie E.A. Genetic identification of five strongyle nematode parasites in wild African elephants (Loxodonta africana) J. Wildl. Dis. 2012;48:707–716. doi: 10.7589/0090-3558-48.3.707. [DOI] [PubMed] [Google Scholar]
- Mercier C., Boyer F., Bonin A., Coissac E. 2013. SUMATRA and SUMACLUST: Fast and Exact Comparison and Clustering of Sequences. [Google Scholar]
- Mitchell C.J., Sullivan C.M.O., Pinloche E., Wilkinson T., Morphew R.M., Mcewan N.R., Group Y.V., Wales S.Y., Sciences L., Campus G., Scotland A.B. Using next-generation sequencing to determine diversity of horse intestinal worms: identifying the equine ‘nemabiome’. J. Equine Sci. 2019;30:1–5. doi: 10.1294/jes.30.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muoria P.K., Muruthi P., Rubenstein D.I., Oguge N.O., Munene E. Cross-sectional survey of gastro-intestinal parasites of Grevy's zebras in southern Samburu, Kenya. Afr. J. Ecol. 2005;43:392–395. doi: 10.1111/j.1469-7998.2006.00139.x. [DOI] [Google Scholar]
- Mwatenga M.S. Kenyatta University; 2017. Gastrointestinal Parasites Infesting Grevy’s Zebra (Equus grevyi) in the Samburu Landscape in Samburu County. [Google Scholar]
- Nielsen M.K., Reinemeyer C.R., Donecker J.M., Leathwick D.M., Marchiondo A.A., Kaplan R.M. Anthelmintic resistance in equine parasites — current evidence and knowledge gaps. Vet. Parasitol. 2014;204:55–63. doi: 10.1016/j.vetpar.2013.11.030. [DOI] [PubMed] [Google Scholar]
- Ogbourne C.P. The prevalence, relative abundance and site distribution of nematodes of the subfamily Cyathostominae in horses killed in Britain. J. Helminthol. 1976;50:203–214. doi: 10.1017/S0022149X00027760. [DOI] [PubMed] [Google Scholar]
- Oksanen J., Blanchet F.G., Friendly M., Kindt R., Legendre P., McGlinn D., Minchin P.R., O'Hara R.B., Simpson G.L., Solymos P., Stevens M.H.H., Szoecs E., Wagner H. 2019. Vegan: Community Ecology Package. [Google Scholar]
- Palomero A.M., Hernández J.A., Cazapal-Monteiro C.F., Balán F.A., Silva M.I., Paz-Silva A., Sánchez-Andrade R., Vázquez M.S.A. Implementation of biological control to the integrated control of strongyle infection among wild captive equids in a zoological park. BioMed Res. Int. 2018;2018:1–7. doi: 10.1155/2018/4267683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pansu J., Guyton J.A., Potter A.B., Atkins J.L., Daskin J.H., Wursten B., Kartzinel T.R., Pringle R.M. Trophic ecology of large herbivores in a reassembling African ecosystem. J. Ecol. 2019;107:1355–1376. doi: 10.1111/1365-2745.13113. [DOI] [Google Scholar]
- Paradis E., Schliep K. Ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 2019;35:526–528. doi: 10.1093/bioinformatics/bty633. [DOI] [PubMed] [Google Scholar]
- Poissant J., Gavriliuc S., Bellaw J., Redman E.M., Avramenko R.W., Robinson D., Workentine M.L., Shury T.K., Jenkins E.J., McLoughlin P.D., Nielsen M.K., Gilleard J.S. A repeatable and quantitative DNA metabarcoding assay to characterize mixed strongyle infections in horses. Int. J. Parasitol. 2021;51:183–192. doi: 10.1016/j.ijpara.2020.09.003. [DOI] [PubMed] [Google Scholar]
- Pringle R.M., Hutchinson M.C. Resolving food-web structure. Annu. Rev. Ecol. Evol. Syst. 2020;12:55–80. [Google Scholar]
- R Core Team . 2019. R: A Language and Environment for Statistical Computing. (Vienna, Austria) [Google Scholar]
- Round M.C. Tech. Comm. Vol. 38. Comm. Bur. Helminthol.; St. Albans, UK: 1968. Check List of the Helminth Parasites of African Mammals. [Google Scholar]
- Rubenstein D.I., Low Mackey B., Davidson Z.D., Kebede F., King S.R.B. Equus grevyi. The IUCN Red List of Threatened Species 2016. 2016 [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Scialdo-Krecek R.C., Reinecke R.K., Malan F.S. Studies on the parasites of zebras. III. Nematodes of the mountain zebra from the farm “Kelpie” and the Namib-Naukluft Park, South West Africa/Namibia. Onderstepoort J. Vet. Res. 1983;50:283–290. [PubMed] [Google Scholar]
- Sim K.A., Hoar B., Kutz S.J., Chilton N.B. Amplification of the second internal transcribed spacer ribosomal DNA of individual trichostrongylid nematode larvae by nested polymerase chain reaction. J. Vet. Diagn. Invest. 2010;22:433–437. doi: 10.1177/104063871002200316. [DOI] [PubMed] [Google Scholar]
- Smith D.P., Peay K.G. Sequence depth, not PCR replication, improves ecological inference from next generation DNA sequencing. PLoS One. 2014;9 doi: 10.1371/journal.pone.0090234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher G., Tamura K., Kumar S. Molecular evolutionary genetics analysis (MEGA) for macOS. Mol. Biol. Evol. 2020;37:1237–1239. doi: 10.1093/molbev/msz312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolliver S., Lyons E., Drudge J. Species of small strongyles and other internal parasites recovered from donkeys at necropsy in Kentucky. Proc. Helminthol. Soc. Wash. 1985;52:260–265. [Google Scholar]
- van Dongen S. Graph clustering via a discrete uncoupling process. SIAM J. Matrix Anal. Appl. 2008;30:121–141. [Google Scholar]
- Yu G., Smith D., Zhu H., Guan Y., Lam T.T.-Y. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 2017;8:28–36. [Google Scholar]
- Zvegintsova N.S., Treus M.J. The investigation of the parasitofauna of zebra in Askania-Nova. Vestn. Zool. 1999;11:98–101. (In Russian) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.