Abstract
Chitosan, a deacetylated form of chitin, is required for the virulence of Cryptococcus neoformans. There are three chitin deacetylase genes (CDA) that are essential for chitosan production, and deletion of all three genes results in the absence of chitosan, loss of virulence, and induction of a protective host response when used as a vaccine. Cda1 plays a major role in deacetylating chitin during pulmonary infection of CBA/J mice. Inoculation with the cda1Δ strain did not lead to a lethal infection. However, the infection was not cleared. The persistence of the fungus in the host suggests that chitin is still being deacetylated by Cda2 and/or Cda3. To test this hypothesis, we subjected strains deleted of two CDA genes to fungal virulence in CBA/J, C57BL/6 and BALB/c and found that cda1Δcda2Δ was avirulent in all mouse lines, as evidenced by its complete clearance. Consistent with the major role of Cda1 in CBA/J, we found that cda2Δcda3Δ was as virulent as its wild-type progenitor KN99. On the other hand, cda1Δcda3Δ displayed virulence comparable to that of cda1Δ. The virulence of each mutant correlates with the amount of chitosan produced when grown under host-mimicking culture conditions. In addition, the avirulence of cda1Δcda2Δ was followed by the induction of a protective immune response in C57BL/6 and CBA/J mice, when a live or heat-killed form of the mutant was used as a vaccine respectively. Taken together, these data imply that, in C. neoformans, coordinated activity of both Cda1 and Cda2 is essential for mediating fungal virulence.
Keywords: Cryptococcus, Chitin deacetylase, Virulence, Fungal vaccine, Cryptococcus Cda
Introduction
Chitin deacetylases (CDA) belong to the CE4 family of carbohydrate esterases and are specific for the deacetylation of chitin as a substrate. Chitin is a homopolymer of N-Acetyl-D-glucosamine residues (GlcNAc) with a β-1,4-linkage, similar to cellulose. It is abundantly present in nature and constitutes an important structural component in fungi, arthropods, and helminths (Muzzarelli, 1997). Upon deacetylation of chitin to chitosan, significant alteration in the physico-chemical properties of the chitin polymer occurs (Tolaimate et al., 2000). The extent and pattern of deacetylation present in a chitosan polymer, as well as its polymer length, influence its biological and immunological properties (Casadidio et al., 2019, Lee et al., 2008). Accordingly, biosynthesis of chitosan plays an important role in the growth of zygomycetous fungi, in the formation of ascospores in S. cerevisiae (Briza et al., 1988, Christodoulidou et al., 1996, Christodoulidou et al., 1999) and influences the outcome of host pathogen interactions for plant and animal pathogens, such as Colletotrichum and Cryptococcus, respectively (Baker et al., 2007, Baker et al., 2011, Blair et al., 2006, Upadhya et al., 2016).
Infections due to C. neoformans are mainly responsible for fungal meningitis, especially in AIDS patients, and are reported to cause over 180,000 annual deaths (Rajasingham et al., 2017)12). In the absence of an effective antifungal therapy and/or vaccine, there is an urgent need to discover new therapeutic targets to develop drugs to treat Cryptococcosis. Understanding the biosynthesis, organization, and function of the components of the fungal cell wall provides one such avenue for developing effective antifungals. Chitin is an important component of fungal cell walls for vegetative growth (Lesage and Bussey, 2006), and we have demonstrated that chitosan is a critical component of the cryptococcal cell wall during mammalian infections (Banks et al., 2005, Upadhya et al., 2018). Previous reports showed that the deacetylation of chitin to chitosan is mediated by three CDAs and is essential for cell wall integrity and virulence (Baker et al., 2007, Baker et al., 2011), and that chitosan-deficient mutants of C. neoformans fail to establish infections when tested in a murine infection model (Baker et al., 2011). This avirulent phenotype was associated with a robust induction of host immune responses, which subsequently led to the clearance of the mutant yeast in the host, and provided protection from a subsequent challenge with wild type KN99 (Upadhya et al., 2016). These studies clearly indicate that the presence of chitosan provides a significant benefit to the yeast cells for survival during infection.
Previously it was reported that, C. neoformans has three chitin deacetylase isozymes and they appeared to be functionally redundant when Cryptococcus was cultured in yeast extract, peptone and dextrose (YPD) medium (Baker et al., 2007). When a cda1Δ strain was tested in a CBA/J mouse model, the chitin deacetylase activity of C. neoformans Cda1 was found to be critical for fungal pathogenesis (Upadhya et al., 2018). In these virulence tests, animals were intranasally infected with 105 colony forming units (CFU) of wild-type or the corresponding individual CDA deletion strains (cda1Δ, cda2Δ or cda3Δ), and their survival was tracked for up to 60 days post infection (DPI). While animals infected with cda2Δ or cda3Δ had median survival times comparable to wild-type KN99, animals infected with cda1Δ showed significant virulence attenuation, with infected animals appearing healthy until the end of the experiment (Upadhya et al., 2018). At this point, however, it was discovered that the animals had a significant fungal burden in their lungs that was nearly equivalent to or slightly higher than the initial inoculum dose. Furthermore, the majority of the animals had detectable fungal burdens in their brains (Upadhya et al., 2018). These findings indicate that, while animals were able to keep the proliferation of cda1Δ in the lungs under control for the duration of the survival experiment, they were unable to eliminate the mutant yeast cells in the lungs or prevent dissemination to the brain. We reasoned that in the absence of Cda1 in the cda1Δ mutant, Cda2 and/or Cda3 could generate enough chitosan to prevent clearance of cda1Δ from the host. As a result, in the current study, we tested the survival and fungal burden of the double CDA deletion strains (cda1Δcda2Δ, cda1Δcda3Δ and cda2Δcda3Δ) using three different inbred mouse lines (CBA/J, BALB/c, and C57BL/6). We discovered that mutant strains of Cryptococcus deleted for both Cda1 and Cda2 (cda1Δcda2Δ) were avirulent in all three mouse lines, with the mutant being completely cleared in each host. We also created a CDA2 reconstituted strain in which a CDA2 gene fragment was re-inserted into the cda1Δcda2Δ mutant's endogenous CDA2 locus. This complemented strain had a lung fungal burden in CBA/J mice similar to the cda1Δ mutant. We show that Cda1 and Cda2 coordinate chitin deacetylation during infection to promote fungal persistence in the lung.
The results of this study also show that the cda1Δ mutant of C. neoformans has a variable ability to cause disease in different laboratory mouse lines. While CBA/J mice were the most resistant to cda1Δ infection, BALB/c mice were highly sensitive, and C57BL/6 mice were moderately sensitive. The virulence of the three CDA double deletion strains correlated with their ability to produce chitosan when grown in Roswell Park Memorial Institute-1640 (RPMI) medium with 10% FBS in the presence of 5% CO2 at 37 °C. We further show that when a used as a vaccine either as live cells or in its heat-killed (HK) form cda1Δcda2Δ conferred robust protection against a subsequent infection with the virulent wild-type KN99 strain. These results shed light on the differential regulation of the three CDA's in chitosan biogenesis during mammalian infection, their impact on fungal virulence and the importance of chitosan-mediated immune responses in controlling C. neoformans infection.
Materials and methods
Strains and media
All strains were derived from C. neoformans serotype A KN99 (KN99) and are listed in Table S1. C. neoformans was grown on YPD (1%yeast, 2% Bacto peptone, 2% dextrose), with solid media containing 2% Bacto agar. Culturing in liquid YPD medium involved inoculating 50 mL of medium with a loop of cells and growing them in a 250 mL flask at 300 rpm at 30 °C. Cells were grown in RPMI as described previously (Upadhya et al., 2018). Briefly, cells were initially grown in YPD culture for 36 h. Cells were collected by centrifugation, washed with PBS and were inoculated to RPMI medium at a cell density of 5 × 105 cells/mL and incubated at 37 °C in the presence of 5% CO2.
Generation of CDA2 complemented strain
To generate a CDA2 reconstituted strain, three DNA fragments were generated by PCR. Fragment A was a 900 bp DNA fragment that was amplified using KN99 genomic DNA as the template and corresponded to a region −2000 to −1100 bp upstream of the ATG of the CDA2 coding region. Fragment B was a 2700 bp fragment of the G418 resistance cassette amplified using pMH12T as a template (Hua et al., 2000). Fragment C was a contiguous 3288 bp genomic DNA fragment of KN99 that included the coding region of CDA2 flanked with 988 bp of its promoter and 694 bp of its terminator regions respectively amplified using KN99 genomic DNA as the template. All three DNA fragments were gel purified and assembled at the NotI restriction site of pGEM®-T Easy (Promega, USA) employing Infusion HD cloning (Takara Bio, USA). Primers were designed to regenerate the NotI site during cloning. After the successful assembly of DNA fragments, the CDA2 reconstitution cassette was released from the vector by NotI digestion and used for transforming cda1Δcda2Δ, isolate A by biolistic transformation. Transformants were initially selected on YPD agar medium with G418 (200 µg/mL). Positive transformants were identified by PCR screening the 5-prime and 3-prime junctions for homologous recombination. During the generation of cda1Δcda2Δ, the CDA2 gene was replaced with a nurseothricin (NAT) selection cassette (Baker et al., 2007). Therefore, in the CDA2 complemented strain, we expected the NAT gene to be replaced by the G418 drug selection cassette. In addition to screening for NAT sensitivity, isolates had their chitosan content measured. One of the isolates that had CDA2 integrated at the endogenous CDA2 locus was designated a CDA2 reconstituted strain (cda1Δcda2Δ::CDA2, JLCN 952) and was used for further experiments.
Cellular chitosan measurement
Cellular chitosan measurement of strains cultured as described above in liquid YPD and RPMI media were performed using the MBTH method as described previously with minor modifications (Upadhya et al., 2018). In brief, C. neoformans cells were collected by centrifugation, washed with PBS and the cell pellets were lyophilized to measure their dry weight. The lyophilized material was suspended in 6 mL of water by vortexing. Potassium hydroxide (KOH) was added to a final concertation of 6% (wt/vol) and the cell suspension was incubated at 80 °C in a bead bath for 60 min with occasional mixing of the cell suspension. The insoluble KOH-treated material was washed four times with PBS, pH 7.4, and finally suspended in PBS to 10 mg/mL (dry weight). The KOH-treated cell material was sonicated into suspension using a probe sonicator (Fisher sonic dismembrator model 300 attached to GraLab model 451 high-accuracy digital electronic timer) for 5 cycles of 60 secs each. A 0.1-mL aliquot of the alkali-treated material was mixed with 0.1 mL of 1 M HCl, followed by the addition of 400 µL of freshly prepared 2.5% (wt/vol) sodium nitrite with vortexing to mix. The deamination and depolymerization reactions were continued for 15 min at 25 °C. Next, 200 µL of 12.5% (wt/vol) ammonium sulfamate was added slowly to neutralize unreacted sodium nitrite. After 5 min of incubation at 25 °C, 200 µL of 0.25% (wt/vol) MBTH prepared in water was added, and the reaction mixture was incubated at 37 °C for 30 min. After the incubation, 200 µL of 0.5% ferric chloride in water (wt/vol) was added, and the contents were mixed and then incubated at 37 °C for 5 min. Finally, the reaction mixture was centrifuged at 12,000×g for 5 min, and the absorbance of the supernatant was measured at 650 nm. Standard curves were prepared from stocks of D-glucosamine hydrochloride in the concentration range of 10 to 100 ηmol. The amount of cellular chitosan was expressed as nanomoles of glucosamine monomer per milligram cells (dry weight).
Survival and fungal burden assays
Virulence and fungal burden assays of CBA/J mice were performed as described previously (Upadhya et al., 2018). The alpha mating type of KN99 was employed as the wild-type control in all animal trials. For animal experiments yeast cells were grown in YPD as described above for 48 h, washed in endotoxin-free PBS, and suspended in 5 mL of the same. The cells were counted with a hemocytometer and diluted to 2 × 106 cells/mL. CBA/J female mice (Jackson Laboratories) were anesthetized with an intraperitoneal injection (200 µL) of ketamine (8 mg/mL)/dexdomitor (0.05 mg/mL) mixture which was reversed by an intraperitoneal injection of atipamezole (200 µL) (0.25 mg/mL). Mice were allowed to inhale 105 CFU of cells in 50 µL, which were dripped into the nares. For virulence assays, the mice were weighed before and during the course of infection. Mice were euthanized by CO2 asphyxiation if they reached 80% of their original body weight. For the determination of CFU, lung or brain from each mouse was placed in two mL of PBS (pH 7), homogenized, serially diluted, plated onto YPD agar supplemented with 100 µg/mL streptomycin and 100 µg/mL ampicillin, and incubated for 2–3 days at 30 °C. The total number of CFU per organ was calculated. Detection limits were 20 CFU/organ. This infection protocol (# 20–0474) was reviewed and approved by the Washington University School of Medicine Animal Care and Use Committee (IACUC).
BALB/c and C57BL/6 mice were from Jackson Laboratories and Charles River Laboratories, respectively. Mice were housed in a pathogen-free environment at the University of Massachusetts Medical School. All experimental procedures were approved by the University of Massachusetts Medical School IACUC (Protocol # A-1802). Mice were infected by the orotracheal infusion of the yeast cells into the lungs with 104 CFU of C. neoformans strain KN99 in 50 µL PBS (Specht et al., 2017). Mice were anesthetized with 2% isoflurane (Piramal Health Care, Andhra Pradesh, India) for inoculation. For fungal burden assays, dissected lungs were homogenized in 4 mL PBS with 100 U/mL penicillin and 100 µg/mL streptomycin and plated on Sabouraud dextrose agar. CFUs were counted following incubation at 30 °C for 2–3 days. Detection limits were 20 CFU/lung.
Statistical analysis
All statistical analyses were performed using GraphPad Prism version 7.03 for Windows, GraphPad Software, La Jolla California USA.
Results
Functional synergy between C. neoformans Cda1 and Cda2 is critical for fungal virulence
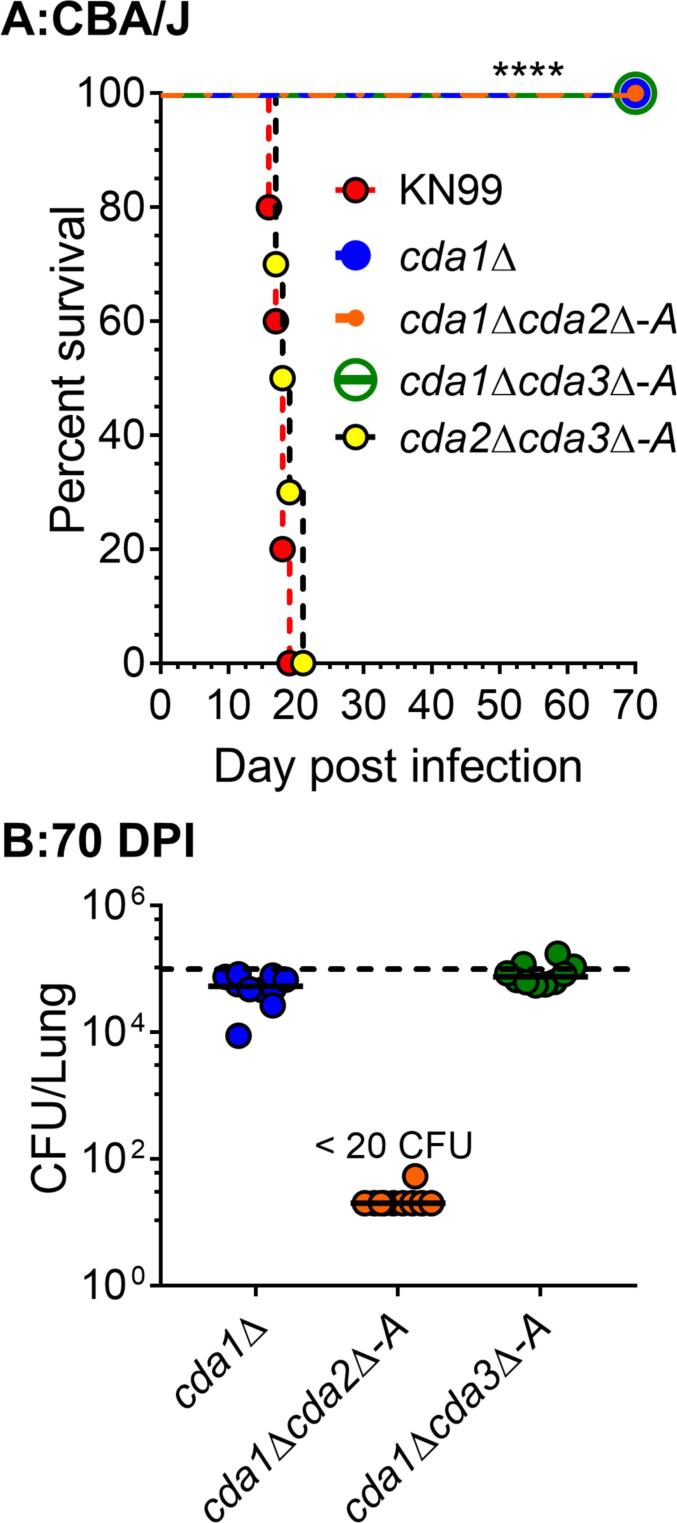
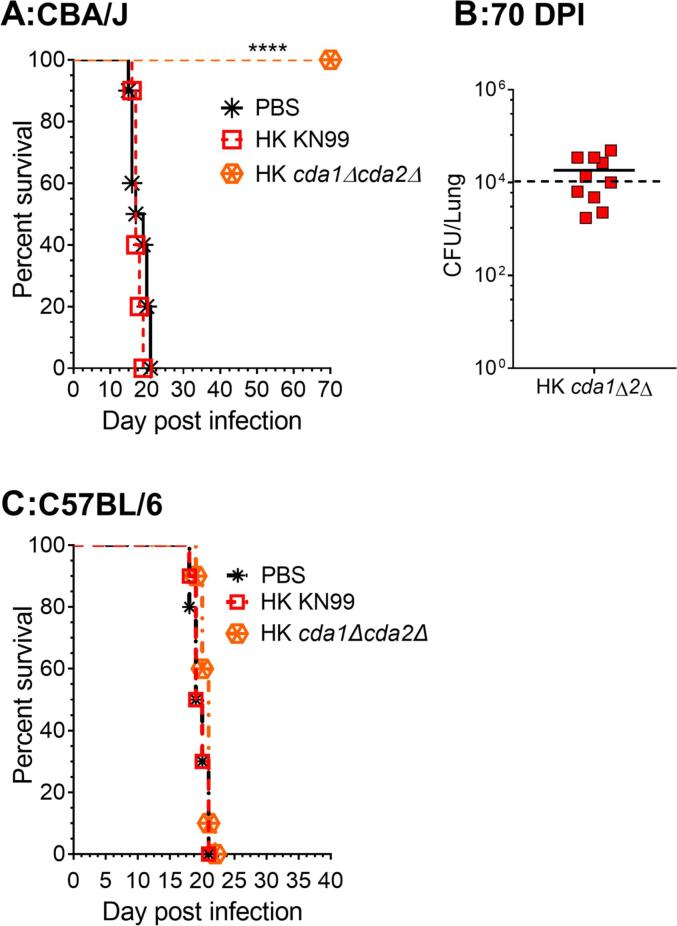
We previously demonstrated that C. neoformans cda1Δ is less virulent in CBA/J mice (Upadhya et al., 2018). However, when the lungs of the surviving mice were examined for fungal burden at 60 DPI, the amount of fungal burden in the lungs and the brains varied among the mice (Upadhya et al., 2018). Some of the animals in the group showed signs of clearance with a several fold decrease in lung CFU compared to the inoculum dose, and no detectable CFU in the brain. Others, on the other hand, had a significant increase in fungal burden in the lungs, with widespread dissemination to the brain (Upadhya et al., 2018). These data imply that, while C. neoformans Cda1 plays a key role in fungal pathogenesis, its deletion is insufficient to induce host-mediated clearance. As a result, we sought to explore how removing the CDA2 or CDA3 genes from the cda1Δ strain would affect its virulence. We infected CBA/J mice with 105 CFU of three different CDA double deletion strains (cda1Δcda2Δ, cda1Δcda3Δ and cda2Δcda3Δ) and tracked their survival for 70 days. For each of the double CDA deletion mutants, two independent isolates were used in the experiment. The survival curves for one set of isolates (isolate A) are depicted in Fig. 1A, while those for the other set of isolates (isolate B) are shown in Fig. S1A. Mice infected with KN99α served as the positive control, while mice infected with cda1Δ were used as the comparison group. Survival data show that double CDA deletion strains cda1Δcda2Δ and cda1Δcda3Δ have an avirulent phenotype similar to cda1Δ, Fig. 1A and Table 1. Infection with a mutant lacking both CDA2 and CDA3 (cda2Δcda3Δ) on the other hand produced a virulent phenotype comparable to KN99, with median survival times of 18.5 and 18 days, respectively (Fig. 1A and Table 1. This is consistent with the wild-type levels of virulence observed upon infecting CBA/J mice with cda2Δ or cda3Δ (single CDA deletion strains) as shown in Fig. 3A and our previously published results (Upadhya et al., 2018). These findings suggest that neither Cda2 nor Cda3 alone, or in combination, are sufficient to cause severe morbidity, lending credence to the role of C. neoformans Cda1 in fungal virulence. When the lungs of cda1Δcda2Δ infected mice were examined for fungal burden at 70 DPI, no CFUs were found (Fig. 1B and Fig. S1B, in contrast to cda1Δ infected mice (Upadhya et al., 2018). This finding suggests that in order to induce an effective clearance from the host, CDA2 must be knocked out in addition to CDA1. In contrast, the lungs of mice infected with cda1Δcda3Δ had varying fungal burdens, with some animals harboring more and others harboring less than the initial inoculum (Fig. 1B and Fig. S1B. The average fungal burden in the lungs of the mice infected with cda1Δcda3Δ was greater than for cda1Δ (Fig. 1B and Fig. S1B, indicating that C. neoformans Cda3 does not contribute to virulence in CBA/J mice by itself or in combination with Cda1 or Cda2.
Fig. 1.
Deletion of C. neoformans CDA2 or CDA3 in conjunction with CDA1 significantly attenuates fungal virulence in CBA/J mice. A. Survival curves of mice infected intranasally with an inoculum equivalent to 105 CFU of each strain (isolate A). Ten mice (6 to 8 weeks old, female) were used for each group and the data is a representative of two independent experiments. ****, p < 0.0001. Statistical analysis of survival rates was determined by log rank (Mantel-Cox) test. B: Fungal burden in survivors' lungs at 70 days’ post infection (DPI). A solid line represents the median CFU per lung for each group. Each datum point represents one mouse's CFU (n = 10). Lower limit of the detection is 20 CFU/lung. The dotted line represents CFU of the inoculum dose.
Table 1.
A summary of the mouse virulence assays performed on various CDA deletion C. neoformans strains in different inbred laboratory mouse lines.
| Yeast strain | Host mouse line | Median survival time (days) |
|---|---|---|
| KN99 | CBA/J | 18 |
| C57BL/6 | 22.5 | |
| BALB/c | 22 | |
| cda1Δ | CBA/J | Avirulent |
| C57BL/6 | 35.5 | |
| BALB/c | 30 | |
| cda2Δ | CBA/J | 17 |
| C57BL/6 | 23.5 | |
| BALB/c | 21.5 | |
| cda3Δ | CBA/J | 17 |
| C57BL/6 | 25 | |
| BALB/c | 26 | |
| cda1Δcda2Δ | CBA/J | Avirulent |
| C57BL/6 | Avirulent | |
| BALB/c | Avirulent | |
| cda1Δcda3Δ | CBA/J | Avirulent |
| C57BL/6 | 51 | |
| BALB/c | 37 | |
| cda2Δcda3Δ | CBA/J | 18.5 |
| C57BL/6 | 34 | |
| BALB/c | 27.5 |
Fig. 3.
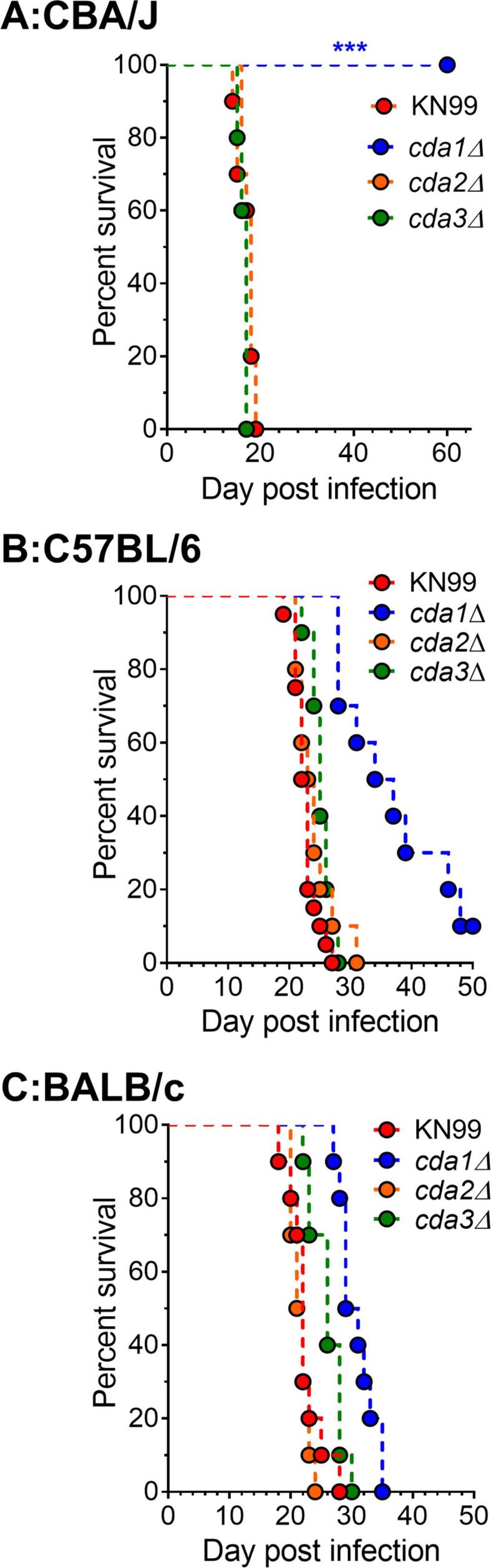
Different inbred mouse lines exhibit varying susceptibility to infection with C. neoformans cda1Δ. A: CBA/J mice were infected intranasally with 105 CFU, B:C57BL/6 mice and C: BALB/c mice were infected orotracheally with 104 CFU of each strain. Ten mice were used to test each strain in each mouse background. ***, p = 0.0002 (determined by log rank [Mantel-Cox] test).
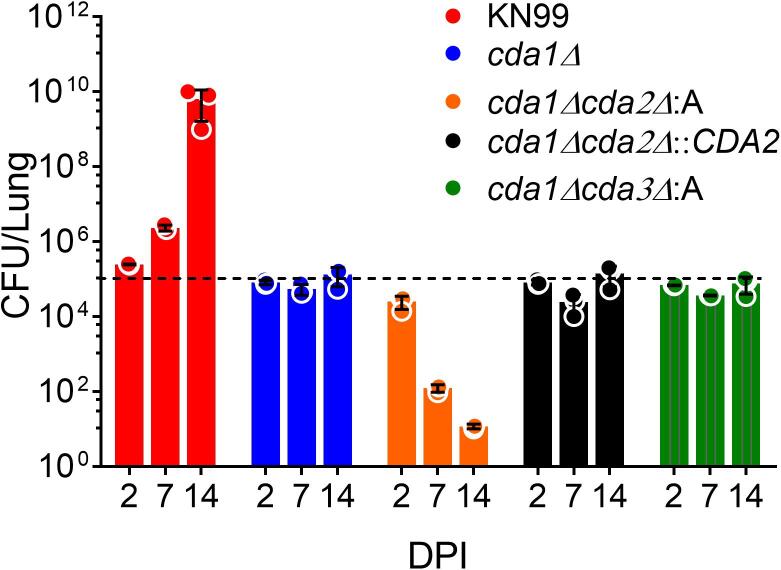
To test the rate of proliferation in the lung, we examined the CFU of infected mice with different CDA deletion strains (cda1Δ, cda1Δcda2Δ, and cda1Δcda3Δ) at different time points, including 2, 7 and 14 DPI. The results shown in Fig. 2 indicate that the cda1Δcda2Δ strain was gradually cleared over 14 days, whereas the cda1Δ and cda1Δcda3Δ strains maintained an average CFU/lung similar to the inoculum, suggesting that there was either no growth or a balance between proliferation and clearance was maintained. To further confirm the importance of Cda2 in the persistence of cda1Δ in the host, we complemented the cda1Δcda2Δ strain with CDA2, which restored the phenotype to that of the cda1Δ strain (Fig. 2). These results suggest that C. neoformans Cda2 rather than Cda3 is required for the persistence of infection in the absence of Cda1.
Fig. 2.
C. neoformans Cda2 contributes to persistence of cda1Δ in the lung. Mean ± SD lung CFU at 2, 7 and 14 DPI of CBA/J mice infected intranasally with an inoculum of 105 CFU (n = 3 mice per group). The data is representative of two biological experiments with three animals in each group, except for the cda1Δcda2Δ::CDA2 mutant, whose data is from a single experiment with three animals. Error bars represent standard errors of the mean. The dotted line represents the CFU/lung of the inoculum dose.
C. neoformans cda1Δcda2Δ is avirulent across different inbred mouse lines
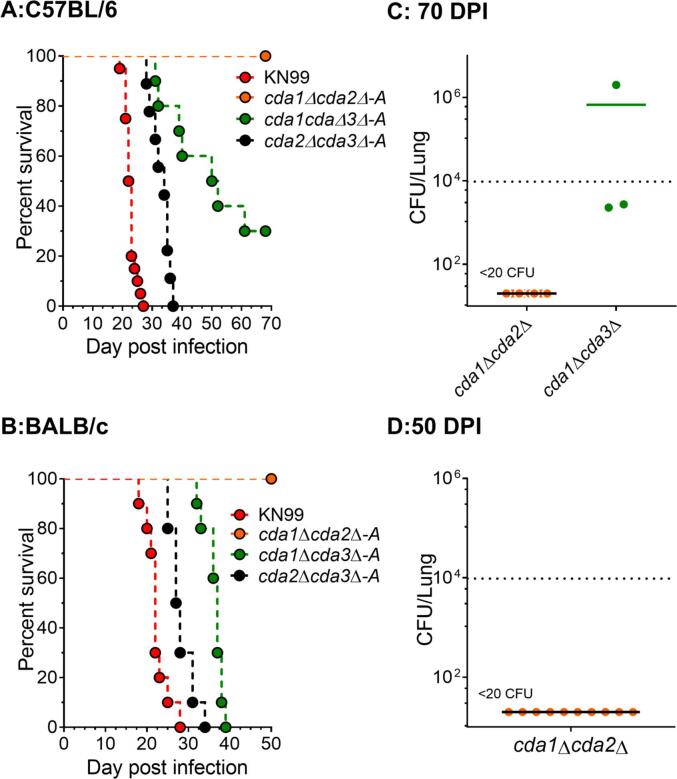
Because different laboratory mouse strains have been reported to have varying degrees of susceptibility to C. neoformans infection, we wanted to see if cda1Δ could cause disease in C57BL/6 and BALB/c mice. We chose inbred mouse lines that are commonly used for immunological studies in different laboratories, and we used a different inoculum (104 CFU), which had been used successfully with these mouse lines (Specht et al., 2017). The results in Fig. 3 and Table 1 show that, in contrast to CBA/J mice, which survived the infection with the cda1Δ strain (as shown in Fig. 3A, both C57B/6 and BALB/c mice were less resistant to infection with this mutant (as shown in Fig. 3B and 3C respectively).
We next determined the virulence of double CDA deletion strains in C57BL/6 and BALB/c mice. We inoculated both of these mouse lines with 104 CFU of the respective double CDA deletion strains by orotracheal inoculation. As shown in Fig. 4A and 4B, cda1Δcda2Δ was avirulent in both C57BL/6 and BALB/c mice when survival was monitored for 70 and 50 DPI respectively. At the end of the experiment, fungal burden analysis on the survivors revealed that both C57BL/6 (Fig. 4C and BALB/c mice (Fig. 4D cleared the mutant from their infected lungs. Unlike in CBA/J mice, where cda2Δcda3Δ was indistinguishable from wild-type in its virulence (Fig. 1A, the virulence of cda2Δcda3Δ was significantly reduced in C57BL/6 and BALB/c when compared to the virulence of wild-type KN99 (Table 1). The reduction in virulence was greater in C57BL/6 mice than in BALB/c mice (Fig. 4 and Table 1). The virulence of cda1Δcda3Δ also followed a similar trend in the three mouse lines, with cda1Δcda3Δ being avirulent in CBA/J mice (Fig. 1A), but attenuated to a greater extent in C57BL/6 mice than in BALB/c mice (Table 1). The virulence of cda1Δcda3Δ appeared to be similar to that of cda1Δ in both C57BL/6 and BALB/c mice (Table 1). Overall, the median survival time of BALB/c was slightly shorter than that of C57BL/6 during infection with various CDA mutants (Table 1).
Fig. 4.
C. neoformans cda1Δcda2Δ is avirulent across different inbred mouse lines: Double CDA deletion strains were tested for their virulence in A: C57BL/6 mice and B: BALB/c mice. Ten mice were infected orotracheally with 104 CFU of each mutant strain and similarly a group of ten mice were infected with wild type KN99. At the conclusion of each survival study lungs were examined for fungal burden. C: Lung CFU for survivors at 70 DPI in C57BL/6 mice, and D: Lung CFU at 50 DPI in BALB/c mice. A solid line represents the median CFU per lung for each group. Each datum point represents one mouse's CFU. The dotted line represents the CFU/lung of the inoculum dose.
The virulence of the CDA mutant strains correlates with their ability to produce chitosan when grown under host-mimicking in vitro tissue culture conditions
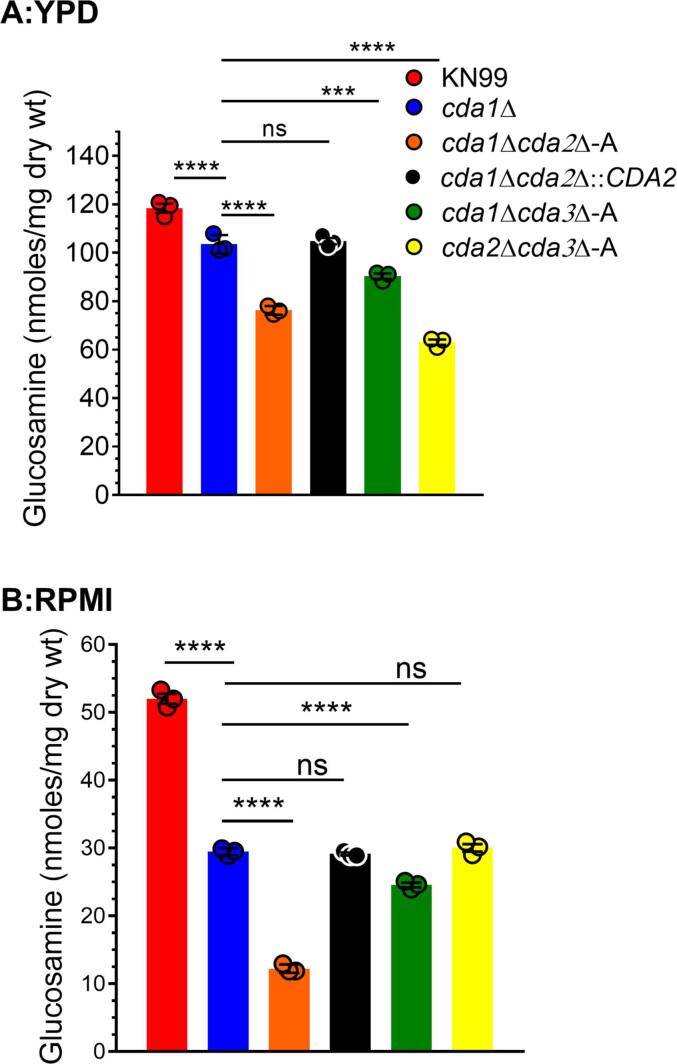
We wanted to see if there was a link between the amount of chitosan produced in the yeast cells during infection and fungal virulence. We previously discovered that the amount of chitosan produced in the lung could be replicated when the strains were grown in conditions such as RPMI containing 10% fetal bovine serum (FBS) at 37 °C and 5% CO2, which mimics the conditions inside the host (Lam et al., 2019, Upadhya et al., 2018). As a result, we looked for differences in the ability of the CDA deletion mutants to synthesize chitosan when grown in either YPD or RPMI containing 10% FBS at 37 °C and 5% CO2. We measured chitosan in two independent isolates of each CDA deletion strain. The results for one group of isolates are shown in Fig. 5, and the results for the other group of independent isolates are shown in Fig. S2. In YPD, all strains, including the wild type KN99, produced more chitosan than in RPMI. The cda1Δcda2Δ and cda2Δcda3Δ made the least chitosan in YPD (Fig. 5A), consistent with the previous findings that Cda2 and Cda3 were responsible for the synthesis of majority of the chitosan when cells were grown in a rich medium (Baker et al., 2007). When cultured in RPMI, the cda1Δcda2Δ strain produced the least chitosan, followed by the cda1Δcda3Δ strain (Fig. 5B). Since these mutants were the least virulent of the three, the amount of chitosan produced in RPMI and virulence correlated. The amount of chitosan produced by cda2Δcda3Δ in RPMI was intermediate between wild type and the other two double mutants (cda1Δcda2Δ and cda1Δcda3Δ; Fig. 5B, and its virulence in BALB/c and C57B/6 were also intermediate (Fig. 4).
Fig. 5.
Deletion of CDA2 in conjunction with CDA1 reduces C. neoformans chitosan production even further. The MBTH assay was used to quantify chitosan extracted from the cell walls of wild-type KN99 and various double CDA deletion strains A: Cells were grown in YPD for 48 h collected, washed and used for the assay. B: Strains were initially grown in YPD for 36 h, collected, washed and inoculated at a cell density of 5 × 105 cells/mL into RPMI medium containing 10% FBS and incubated for 5 days at 37 °C/5% CO2. Cells were collected, washed and used for chitosan measurement. Data is the mean of three biological and two technical replicates (n = 3). Significant differences between the groups were compared by one-way ANOVA followed by Bonferroni’s multiple comparisons test. (****=p < 0.0001 comparing chitosan amount of each strain to cda1Δ). Error bars represent standard errors of the mean.
The reinsertion of the CDA2 gene into the genome of the cda1Δcda2Δ mutant (cda1Δcda2Δ::CDA2) restored the ability to produce chitosan to levels similar to those of the cda1Δ. These results support the notion that deletion of Cda2 in conjunction with Cda1 significantly affects the capacity of the fungal cells to synthesize chitosan, thereby increasing their clearance from the infected lung.
In two different inbred mouse lines, C. neoformans cda1Δcda2Δ confers protection against subsequent KN99 infection
We previously reported that the chitosan-deficient cda1Δcda2Δcda3Δ strain is avirulent due to rapid clearance from the infected lung, and that this clearance was accompanied by the induction of a robust protective immunity against a subsequent infection with the virulent, wild-type KN99 strain (Upadhya et al., 2016). We found that an inoculum dose of 107 CFU of cda1Δcda2Δcda3Δ was minimally required to induce protective immunity and that this protective immunity was also induced when a HK preparation of the mutant was used for vaccination (Upadhya et al., 2016). Therefore, we wanted to test whether vaccinating animals with a HK preparation of an inoculum equivalent to 107 CFU of cda1Δcda2Δ would induce protective immunity. We used intranasal inhalation to vaccinate CBA/J and C57BL/6 mice with a HK preparation of cda1Δcda2Δ equivalent to a dose of 107 CFU. Vaccinated animals were challenged with 50,000 CFU of wild-type KN99 40 days after vaccination. As illustrated in Fig. 6A, vaccination with the HK preparation of cda1Δcda2Δ effectively prevented C. neoformans KN99 infection in CBA/J mice. Surviving animals exhibited varying degrees of clearance of KN99 infection (70-day post infection; Fig. 6B), with the majority of animals having a fungal burden in the lung that was lower than the infection dose of 50,000 CFU. Vaccination with the HK cda1Δcda2Δ failed to induce protective immunity in C57BL/6 mice (Fig. 6C). This is consistent with our previous study where even though HK cda1Δcda2Δcda3Δ induced a robust protective immunity in CBA/J mice, the protection was attenuated in the C57BL/6 mouse line (Upadhya et al., 2016). Other potential vaccine candidates have also reported strain-specific differences in vaccine efficacy against C. neoformans infection (Masso-Silva et al., 2018, Specht et al., 2017, Wang et al., 2019).
Fig. 6.
Vaccination with a heat-killed preparation of cda1Δcda2Δ induces protective immunity against C. neoformans infection A: Mice (female, 6–8 weeks old, CBA/J) C: Mice (female, 6–8 weeks old, C57BL/6) were inoculated intranasally with HK preparations of either KN99 or the cda1Δcda2Δ mutant at a dose equivalent to 107 CFU; PBS inoculated mice served as a negative control. The vaccinated mice were challenged with 50,000 CFU of KN99 after 40 days. Mice were monitored for survival for up to 70 DPI by measuring their body weight. Animals that lost 20% of their body weight during inoculation were deemed morbid and euthanized. The data for each strain (n = 10) are the combined results of two experiments with five animals each. The log rank (Mantel-Cox) test was used to compare survival curves (p < 0.0001 comparing to HK-KN99 vaccinated group. B: Fungal burden in survivors' lungs at 70 DPI. A solid line represents the median CFU per lung for each group. Each datum point represents one mouse's CFU (n = 10). The CFU/lung of the challenge inoculum is indicated by the dotted line.
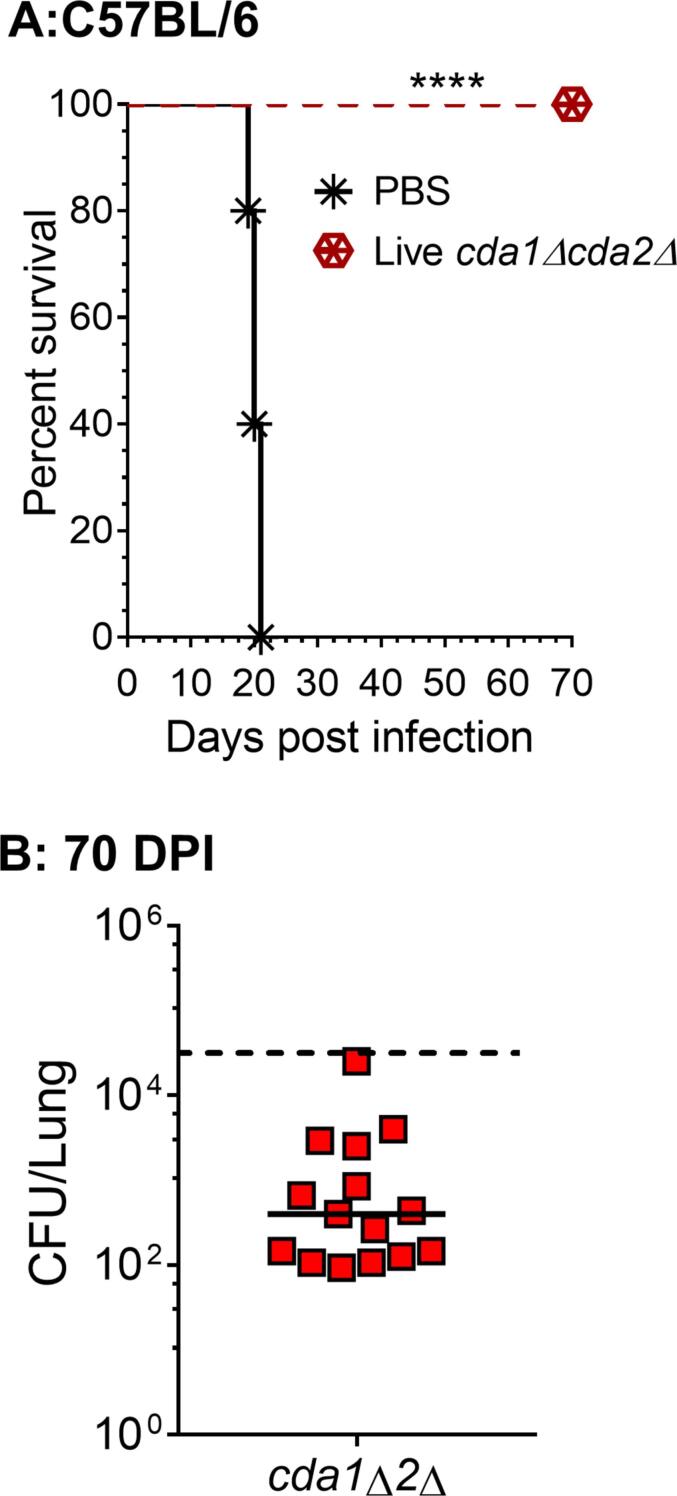
To address the lack of protective immunity with the HK cda1Δcda2Δ vaccine in the C57BL/6 line, we investigated whether a live inoculum of cda1Δcda2Δ would work. For this, we vaccinated C57BL/6 mice with 107 CFU of live cda1Δcda2Δ. After 40 days of vaccination we challenged these animals with 50,000 CFU of wild-type KN99 and monitored their survival as described above. As shown in Fig. 7 all of the vaccinated animals survived infection with wild-type KN99 demonstrating a significant induction of protective immunity in C57BL/6 mice. At the end of the survival experiment (70 DPI), when we tested the lung fungal burden in the survivors, we discovered that all of the animals had a lung fungal burden far less than the initial infection dose of 50,000 CFU/lung (Fig. 7B) indicating that the infection is being cleared effectively by the host.
Fig. 7.
In C57BL/6 mice, vaccination with live cda1Δcda2Δ confers strong protective immunity against C. neoformans infection. A:C57BL/6 mice (female, 6–8 weeks old) were inoculated intranasally with 107 CFU of cda1Δcda2Δ. PBS inoculated mice served as a negative control. The vaccinated mice were challenged with 50,000 CFU of KN99 after 40 days and the survival assay was carried out as described in Fig. 6. The log rank (Mantel-Cox) test was used to compare survival curves (p < 0.0001 comparing to PBS vaccinated group). B: Fungal burden in survivors' lungs at 70 DPI. The data (n = 15) are the combined results of three experiments with five animals each. A solid line represents the median CFU of the group. Each datum point represents one mouse's CFU.
Discussion
In fungi, the chitin polymer provides structural rigidity to the cell wall and to the septum that delineates the junction between hyphal cells and is formed between the mother cell and daughter cell of budding yeast. Fungal chitinases hydrolyze chitin to alter the cell wall during yeast cell separation and hyphal branching, although in Cryptococcus, fungal chitinases are not required for this process, perhaps due to the conversion of much of the chitin to chitosan (Baker et al., 2011). One mechanism that both plants and mammalian systems employ to recognize chitin containing pathogens and to initiate protective resistance mechanisms is through the action of host chitinases (Cord-Landwehr et al., 2016, Lee et al., 2011, Pusztahelyi, 2018, Van Dyken et al., 2017, Van Dyken and Locksley, 2018). To evade this process, plant fungal pathogens have been shown either to produce effector molecules that bind and shield cell wall chitin from being recognized by plant chitinases (Quarantin et al., 2016), or the fungus uses its chitin deacetylases to convert chitin and chitin oligosaccharides to their chitosan counterparts and decrease hydrolysis by chitinases (El Gueddari et al., 2002, Gao et al., 2019, Petutschnig et al., 2010). Our previous reports clearly indicated that chitosan is critical for C. neoformans to sustain growth inside the infected host lung (Baker et al., 2011, Upadhya et al., 2018, Upadhya et al., 2016). However, the mechanisms of clearance of the chitosan-deficient strains are not clearly understood. It may be that the altered cell walls of chitosan deficient mutants lose their integrity inside the lung, possibly through the action of host chitinases.
Chitin is synthesized at the plasma membrane from cytosolic UDP-N-Acetylglucosamine (UDP-GlcNAc) as a homopolymer of β (1 → 4) linked GlcNAc and each GlcNAc can be modified post-synthesis to glucosamine (GlcN) by deacetylation. Renaming of the polymer from chitin to chitosan is generally accepted to occur when the polymer contains more glucosamine subunits than N-acetylglucosamine subunits. However, in our measurements using the biochemical MBTH assay, we can only determine the average amount of glucosamine present in the total chitinous material, not the degree of acetylation or the pattern of deacetylation in a specific polymer molecule. The pattern or degree of deacetylation could be a factor in host recognition or protection from chitinases. Even a small stretch of acetylated subunits could be a substrate for endo-chitinases. The activity and substrate specificity for each of the three Cdas described here has not been determined.
The persistence of the cda1Δ strain in the infected lungs of CBA/J mice indicates that even in the absence of Cda1, sufficient chitosan is being produced to prevent its eradication by the host. The results presented here clearly show that indeed, C. neoformans Cda2 contributes to this chitosan production. The importance of Cda2 is supported by the persistence of the CDA2 reconstituted strain cda1Δcda2Δ::CDA2 in the CBA/J mice (Fig. 2). Interestingly, cda1Δ was more virulent in BALB/c and C57BL/6 mice compared to CBA/J mice. Previously, we have observed a 9-fold and a 2.5-fold upregulation of Cda1 and Cda2 transcript levels, respectively, in wild type yeast cells growing in infected murine lungs compared to their expression levels when being cultured in YPD (Upadhya et al., 2018). Therefore, it is possible that expression of Cda2 is induced to higher levels in a cda1Δ mutant infecting BALB/c and C57BL/6 mice compared to its level in CBA/J mice. An increase in Cda2 could yield more chitosan and increase resistance in the host. Alternatively, BALB/c and C57BL/6 may mount a more substantial host response to a cda1Δ infection, triggering host induced damage.
Irrespective of the mouse line, we found that cda1Δcda2Δ was avirulent and the infection was cleared. This suggests that in the absence of Cda1 and Cda2, C. neoformans Cda3 does not contribute sufficient chitosan to sustain it in the host lung. However, C57BL/6 mice had a survival time of 35.5 days when infected with cda1Δ compared to a survival time of 51 days when infected with cda1Δcda3Δ. This data suggests a potential minor coordinated role for Cda1 and Cda3 in the C57BL/6 mice. These results are in sharp contrast to the virulence phenotype observed by us in the hypervirulent C. gattii R265 strain (Lam et al., 2019). We found that for strain R265, Cda3 alone has an essential role in fungal virulence across all three mouse lines. This suggests that the regulation of Cda1 and Cda3 activity is different in these two pathogenic species of Cryptococcus.
In our previous report on the virulence of cda1Δ in CBA/J mice, we have shown that in the absence of Cda1, the ability of C. neoformans to convert chitin to chitosan in the lung is severely compromised (Upadhya et al., 2018). This was recapitulated in ex vivo experiments when the yeast cells were incubated in a host mimicking conditions of RPMI + 10% FBS, 5% CO2 and at 37 °C (Upadhya et al., 2018). Under these conditions, we found that cda1Δ still had about half of the chitosan as wild type KN99 (Upadhya et al., 2018). Here we show that the deletion of CDA2 in the cda1Δ strain further hampers its ability to synthesize chitosan under host mimicking conditions, reducing the chitosan amount to 23% of levels in KN99 (Fig. 5). The amount of chitosan and the clearance of cda1Δcda2Δ in the host lung imply that chitosan at about a quarter of wild-type levels does not support either the growth or the persistence of the fungus inside the host. We have previously seen that chitosan deficient cda1Δcda2Δcda3Δ was completely cleared from the host within 24 h PI when infected with 105 CFU (Baker et al., 2011). Here we found that at 14 DPI, we were able to recover around 100 CFUs of cda1Δcda2Δ, clearly suggesting that the amount of chitosan present in cda1Δcda2Δ is sufficient to prolong host mediated killing.
At present, we do not know how the three CDAs of Cryptococcus differ enzymatically. We have shown that the removal of Cda2 or Cda3 from the KN99 strain does not affect fungal virulence (Upadhya et al., 2018). In addition, we have shown that the catalytic activity of Cda1 is essential for pathogenesis; the chitin deacetylase activity was eliminated by introducing point mutations in the catalytic site and the strain with the point mutant was avirulent (Upadhya et al., 2018). Multiple factors have been reported to regulate the activity of fungal CDAs (Zhao et al., 2010). Depending on the source of the enzyme, they have strict requirements for pH, temperature and metal ions (Zhao et al., 2010). Importantly, deacetylation reactions of chitin deacetylases are also reported to depend on the degree of polymerization and the position of acetylated sugars in the chitin substrates (Aranda-Martinez et al., 2018, Naqvi et al., 2016, Tsigos et al., 1999, Yamada et al., 2008). Recent heterologous expression and characterization of two chitin deacetylases from the mushroom Coprinopsis cinerea revealed differences in substrate preferences (Wang et al., 20182018). Similarly, concerted action of two chitin deacetylases of the red flour beetle, Tribolium castaneum, has been recently demonstrated to be critical for the higher order organization of the procuticle (Noh et al., 2018). Potentially, C. neoformans may utilize processive deacetylation of chitin during mammalian infection conditions. Cda1 may have a high affinity for chitin, and the partially deacetylated chitin produced by Cda1 may be an ideal substrate for Cda2 and then Cda3, which in turn catalyze further deacetylation of chitin. This highly deacetylated form of chitin promotes fungal survival in the mammalian host. Recent results of recombinant Cda2 and Cda4 activity on various types of chitin/chitosan substrates show that the specific activity of either Cda2 or Cda4 depends on the degree and pattern of acetylation of their oligomeric substrates (Hembach et al., 2020, Hembach et al., 2017).
The protection conferred after vaccination of CBA/J mice with the HK preparation of cda1Δcda2Δ is comparable to that previously reported for the induction of protective immunity with the HK preparation of cda1Δcda2Δcda3Δ (Upadhya et al., 2016). While cda1Δcda2Δcda3Δ was chitosan deficient, cda1Δcda2Δ grown in YPD still harbors a significant amount of chitosan when the heat killed preparation was made for vaccination. This implies that a complete lack of chitosan in the cell wall is not required for an effective vaccine. Despite the fact that HK-cda1Δcda2Δ did not confer protection in C57/BL6, the live form of the cda1Δcda2Δ did. It is possible that the 107 CFU of live-cda1Δcda2Δ takes longer to clear in the host, contributing to more efficient stimulation of the adaptive arm of the immune system and thus conferring robust protective immunity. It will be informative to see if there is a difference in the nature of the protective immunity induced by cda1Δcda2Δ and cda1Δcda2Δcda3Δ, which will be investigated further.
In conclusion, we show that the amount of chitosan in the cell wall of C. neoformans is very dynamic and is coordinately regulated by Cda1 and Cda2 during mammalian infection; Cda3 appears to have a subordinate role. In addition to the transcriptional regulation of individual CDAs, results suggest that there might be differences between Cda1 and Cda2 during infection in their preference for the type of chitin substrate. These results for C. neoformans are in contrast to chitosan regulation in C. gattii, where Cda3 alone plays a critical role in fungal virulence (Lam et al., 2019). Elucidation of the mechanisms of chitosan biosynthesis and its regulation may provide new targets for developing effective antifungals.
CRediT authorship contribution statement
Rajendra Upadhya: Conceptualization, Methodology, Investigation, Validation, Formal analysis, Visualization, Writing – original draft. Woei C. Lam: Conceptualization, Methodology, Investigation, Validation, Formal analysis, Writing – review & editing. Camaron R. Hole: Conceptualization, Methodology, Investigation, Validation, Formal analysis, Writing – review & editing. Danealle Parchment: Investigation, Validation, Formal analysis, Writing – review & editing. Chrono K. Lee: Investigation, Validation, Writing – review & editing. Charles A. Specht: Conceptualization, Methodology, Investigation, Validation, Formal analysis, Visualization, Writing – review & editing, Funding acquisition, Project administration. Stuart M. Levitz: Conceptualization, Methodology, Formal analysis, Writing – review & editing, Funding acquisition, Project administration. Jennifer K. Lodge: Conceptualization, Methodology, Formal analysis, Visualization, Writing – review & editing, Funding acquisition, Project administration.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tcsw.2021.100066.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Aranda-Martinez A., Grifoll-Romero L., Aragunde H., Sancho-Vaello E., Biarnes X., Lopez-Llorca L.V., Planas A. Expression and specificity of a chitin deacetylase from the nematophagous fungus Pochonia chlamydosporia potentially involved in pathogenicity. Sci. Rep. 2018;8:2170. doi: 10.1038/s41598-018-19902-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker L.G., Specht C.A., Donlin M.J., Lodge J.K. Chitosan, the deacetylated form of chitin, is necessary for cell wall integrity in Cryptococcus neoformans. Eukaryot. Cell. 2007;6(5):855–867. doi: 10.1128/EC.00399-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker L.G., Specht C.A., Lodge J.K. Cell wall chitosan is necessary for virulence in the opportunistic pathogen Cryptococcus neoformans. Eukaryot. Cell. 2011;10(9):1264–1268. doi: 10.1128/EC.05138-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks I.R., Specht C.A., Donlin M.J., Gerik K.J., Levitz S.M., Lodge J.K. A chitin synthase and its regulator protein are critical for chitosan production and growth of the fungal pathogen Cryptococcus neoformans. Eukaryot. Cell. 2005;4:1902–1912. doi: 10.1128/EC.4.11.1902-1912.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair D.E., Hekmat O., Schüttelkopf A.W., Shrestha B., Tokuyasu K., Withers S.G., van Aalten D.M.F. Structure and mechanism of chitin deacetylase from the fungal pathogen Colletotrichum lindemuthianum. Biochemistry. 2006;45:9416–9426. doi: 10.1021/bi0606694. [DOI] [PubMed] [Google Scholar]
- Briza P., Ellinger A., Winkler G., Breitenbach M. Chemical composition of the yeast ascospore wall. The second outer layer consists of chitosan. J. Biol. Chem. 1988;263:11569–11574. [PubMed] [Google Scholar]
- Casadidio C., Peregrina D.V., Gigliobianco M.R., Deng S., Censi R., Di Martino P. Chitin and chitosans: characteristics, eco-friendly processes, and applications in cosmetic science. Mar. Drugs. 2019;17 doi: 10.3390/md17060369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christodoulidou A., Bouriotis V., Thireos G. Two sporulation-specific chitin deacetylase-encoding genes are required for the ascospore wall rigidity of Saccharomyces cerevisiae. J. Biol. Chem. 1996;271:31420–31425. doi: 10.1074/jbc.271.49.31420. [DOI] [PubMed] [Google Scholar]
- Christodoulidou A., Briza P., Ellinger A., Bouriotis V. Yeast ascospore wall assembly requires two chitin deacetylase isozymes. FEBS Lett. 1999;460:275–279. doi: 10.1016/s0014-5793(99)01334-4. [DOI] [PubMed] [Google Scholar]
- Cord-Landwehr S., Melcher R.L., Kolkenbrock S., Moerschbacher B.M. A chitin deacetylase from the endophytic fungus Pestalotiopsis sp. efficiently inactivates the elicitor activity of chitin oligomers in rice cells. Sci. Rep. 2016;6:38018. doi: 10.1038/srep38018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Gueddari N.E., Rauchhaus U., Moerschbacher B.M., Deising H.B. Developmentally regulated conversion of surface-exposed chitin to chitosan in cell walls of plant pathogenic fungi. New Phytol. 2002;156:103–112. [Google Scholar]
- Gao F., Zhang B.-S., Zhao J.-H., Huang J.-F., Jia P.-S., Wang S., Zhang J., Zhou J.-M., Guo H.-S. Deacetylation of chitin oligomers increases virulence in soil-borne fungal pathogens. Nat. Plants. 2019;5:1167–1176. doi: 10.1038/s41477-019-0527-4. [DOI] [PubMed] [Google Scholar]
- Hembach L., Cord-Landwehr S., Moerschbacher B.M. Enzymatic production of all fourteen partially acetylated chitosan tetramers using different chitin deacetylases acting in forward or reverse mode. Sci. Rep. 2017;7:17692. doi: 10.1038/s41598-017-17950-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hembach L., Bonin M., Gorzelanny C., Moerschbacher B.M. Unique subsite specificity and potential natural function of a chitosan deacetylase from the human pathogen Cryptococcus neoformans. PNAS. 2020;117:3551–3559. doi: 10.1073/pnas.1915798117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua J., Meyer J.D., Lodge J.K. Development of positive selectable markers for the fungal pathogen Cryptococcus neoformans. Clin. Diagn. Lab. Immunol. 2000;7(1):125–128. doi: 10.1128/cdli.7.1.125-128.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, W.C., Upadhya, R., Specht, C.A., Ragsdale, A.E., Hole, C.R., Levitz, S.M., and Lodge, J.K., 2019. Chitosan Biosynthesis and Virulence in the Human Fungal Pathogen Cryptococcus gattii. mSphere 4 (5), 00644-19. [DOI] [PMC free article] [PubMed]
- Lee C.G., Da Silva C.A., Lee J.-Y., Hartl D., Elias J.A. Chitin regulation of immune responses: an old molecule with new roles. Curr. Opin. Immunol. 2008;20:684–689. doi: 10.1016/j.coi.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.G., Da Silva C.A., Dela Cruz C.S., Ahangari F., Ma B., Kang M.-J., He C.-H., Takyar S., Elias J.A. Role of chitin and chitinase/chitinase-like proteins in inflammation, tissue remodeling, and injury. Annu. Rev. Physiol. 2011;73(1):479–501. doi: 10.1146/annurev-physiol-012110-142250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage G., Bussey H. Cell wall assembly in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2006;70:317–343. doi: 10.1128/MMBR.00038-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masso-Silva, J., Espinosa, V., Liu, T.B., Wang, Y., Xue, C., and Rivera, A., 2018. The F-Box Protein Fbp1 Shapes the Immunogenic Potential of Cryptococcus neoformans. mBio 9. [DOI] [PMC free article] [PubMed]
- Muzzarelli R.A.A. Human enzymatic activities related to the therapeutic administration of chitin derivatives. Cell. Mol. Life Sci. 1997;53:131–140. doi: 10.1007/PL00000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi S., Cord-Landwehr S., Singh R., Bernard F., Kolkenbrock S., El Gueddari N.E., Moerschbacher B.M., Pettinari M.J. A recombinant fungal chitin deacetylase produces fully defined chitosan oligomers with novel patterns of acetylation. Appl. Environ. Microbiol. 2016;82:6645–6655. doi: 10.1128/AEM.01961-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh M.Y., Muthukrishnan S., Kramer K.J., Arakane Y. Group I chitin deacetylases are essential for higher order organization of chitin fibers in beetle cuticle. J. Biol. Chem. 2018;293:6985–6995. doi: 10.1074/jbc.RA117.001454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petutschnig E.K., Jones A.M.E., Serazetdinova L., Lipka U., Lipka V. The lysin motif receptor-like kinase (LysM-RLK) CERK1 is a major chitin-binding protein in Arabidopsis thaliana and subject to chitin-induced phosphorylation. J. Biol. Chem. 2010;285:28902–28911. doi: 10.1074/jbc.M110.116657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusztahelyi T. Chitin and chitin-related compounds in plant-fungal interactions. Mycology. 2018;9:189–201. doi: 10.1080/21501203.2018.1473299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarantin A., Glasenapp A., Schäfer W., Favaron F., Sella L. Involvement of the Fusarium graminearum cerato-platanin proteins in fungal growth and plant infection. Plant Physiol. Biochem. 2016;109:220–229. doi: 10.1016/j.plaphy.2016.10.001. [DOI] [PubMed] [Google Scholar]
- Rajasingham R., Smith R.M., Park B.J., Jarvis J.N., Govender N.P., Chiller T.M., Denning D.W., Loyse A., Boulware D.R. Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. Lancet Infect. Dis. 2017;17:873–881. doi: 10.1016/S1473-3099(17)30243-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Specht, C.A., Lee, C.K., Huang, H., Hester, M.M., Liu, J., Luckie, B.A., Torres Santana, M.A., Mirza, Z., Khoshkenar, P., Abraham, A., et al., 2017. Vaccination with Recombinant Cryptococcus Proteins in Glucan Particles Protects Mice against Cryptococcosis in a Manner Dependent upon Mouse Strain and Cryptococcal Species. mBio 8. [DOI] [PMC free article] [PubMed]
- Tolaimate A., Desbrières J., Rhazi M., Alagui A., Vincendon M., Vottero P. On the influence of deacetylation process on the physicochemical characteristics of chitosan from squid chitin. Polymer. 2000;41:2463–2469. [Google Scholar]
- Tsigos I., Zydowicz N., Martinou A., Domard A., Bouriotis V. Mode of action of chitin deacetylase from Mucor rouxii on N-acetylchitooligosaccharides. Eur. J. Biochem. 1999;261:698–705. doi: 10.1046/j.1432-1327.1999.00311.x. [DOI] [PubMed] [Google Scholar]
- Upadhya R., Lam W.C., Maybruck B., Specht C.A., Levitz S.M., Lodge J.K., Heitman J., Alspaugh J.A., Wormley F. Induction of Protective Immunity to Cryptococcal Infection in Mice by a Heat-Killed, Chitosan-Deficient Strain of Cryptococcus neoformans. mBio. 2016;7(3) doi: 10.1128/mBio.00547-16. e00547-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhya R., Baker L.G., Lam W.C., Specht C.A., Donlin M.J., Lodge J.K. Cryptococcus neoformans Cda1 and Its Chitin Deacetylase Activity Are Required for Fungal Pathogenesis. mBio. 2018;9:e02087–18. doi: 10.1128/mBio.02087-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyken S.J., Locksley R.M. Chitins and chitinase activity in airway diseases. J. Allergy Clin. Immunol. 2018;142:364–369. doi: 10.1016/j.jaci.2018.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyken S.J., Liang H.-E., Naikawadi R.P., Woodruff P.G., Wolters P.J., Erle D.J., Locksley R.M. Spontaneous chitin accumulation in airways and age-related fibrotic lung disease. Cell. 2017;169:497–509.e13. doi: 10.1016/j.cell.2017.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Niu, X., Guo, X., Yu, H., Liu, Z., Zhang, Z., and Yuan, S., 2018. Heterologous expression, characterization and possible functions of the chitin deacetylases, Cda1 and Cda2, from mushroom Coprinopsis cinerea. Glycobiology 28, 318-332. [DOI] [PubMed]
- Wang, Y., Wang, K., Masso-Silva, J.A., Rivera, A., and Xue, C., 2019. A Heat-Killed Cryptococcus Mutant Strain Induces Host Protection against Multiple Invasive Mycoses in a Murine Vaccine Model. mBio, 10(6), e02145-19. [DOI] [PMC free article] [PubMed]
- Yamada, M., Kurano, M., Inatomi, S., Taguchi, G., Okazaki, M., and Shimosaka, M., 2008. Isolation and characterization of a gene coding for chitin deacetylase specifically expressed during fruiting body development in the basidiomycete Flammulina velutipes and its expression in the yeast Pichia pastoris. FEMS Microbiol Lett 289, 130-137. [DOI] [PubMed]
- Zhao Y., Park R.-D., Muzzarelli R.A.A. Chitin deacetylases: properties and applications. Mar. Drugs. 2010;8(1):24–46. doi: 10.3390/md8010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.