Abstract
The recognition of a new yeast, Candida auris, in 2009 in East Asia, and its rapid global spread, was a reminder of the threats posed by multidrug-resistant fungal pathogens. C. auris had likely remained unrecognized for a long time as accurate tests were not available. The laboratory community responded to the C. auris challenge by publishing 35 new or revised diagnostic methods between 2014 and early 2021. The commercial sector also modified existing diagnostic devices. These C. auris diagnostic tests run the gamut from traditional culture-based differential and selective media, biochemical assimilations, and rapid protein profiles, as well as culture-independent DNA-based diagnostics. We provide an overview of these developments, especially the tests with validation data that were subsequently adopted for common use. We share a workflow developed in our laboratory to process over 37,000 C. auris surveillance samples and 5,000 C. auris isolates from the outbreak in the New York metropolitan area. Our preview covers new devices and diagnostic approaches on the horizon based on microfluidics, optics, and nanotechnology. Frontline laboratories need rapid, cheap, stable, and easy-to-implement tests to improve C. auris diagnosis, surveillance, patient isolation, admission screening, and environmental control. Among the urgent needs is a lateral flow assay or similar device for presumptive C. auris identification. All laboratories will benefit from devices that allow rapid antifungal susceptibility testing, including detection of mutations conferring drug resistance. Hopefully, multiplex test panels are on the horizon for synergy of C. auris testing with ongoing surveillance of other healthcare-associated infections. C. auris genome analysis has a proven role for outbreak investigations, and diagnostic laboratories need quick access to regional and national genome analysis networks.
Keywords: PCR, real-time PCR, MALDI-TOF MS, LAMP, biosensor, laboratory-developed tests, clinical, surveillance, healthcare-associated infections
Candida auris, a Novel Pathogen
Clinical laboratories made significant progress in the identification of fungal pathogens with the introduction of ribosomal RNA gene sequencing and matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (White et al., 1990; Qian et al., 2008; Marklein et al., 2009). These DNA- and protein-based approaches enhanced the capacity of many frontline laboratories to recognize new pathogens, which were previously the province of highly specialized centers of excellence (Sullivan et al., 1995; Alcoba-Florez et al., 2005; Sugita et al., 2006; Jensen and Arendrup, 2011; Cendejas-Bueno et al., 2012; Castanheira et al., 2013; Eddouzi et al., 2013; Tsang et al., 2014). This scenario played out perfectly when Candida auris was recognized as a new yeast species by Satoh et al. (2009). The authors studied a single yeast isolate from the external ear canal discharge of an elderly patient at a Tokyo metropolitan hospital using biochemical tests and ribosomal RNA gene sequences (ITS and D1-D2) to delineate novel C. auris formally (Satoh et al., 2009). Around the same timeframe, Kim et al. (2009) reported 15 isolates of a novel Candida species, now confirmed as C. auris, from ear canals of chronic otitis media patients in South Korea. These isolates were susceptible to relatively high concentrations of amphotericin B and fluconazole. A subsequent study from South Korea by Lee et al. (2011) established C. auris as a causal agent of fatal fungemia with intrinsic and acquired resistance to fluconazole. The two teams highlighted misidentification of C. auris by commercial systems. They also emphasized the value of internal transcribed spacer (ITS) sequencing as a confirmatory test (Kim et al., 2009; Lee et al., 2011). Teun Boekhout’s group reclassified Candida haemulonii species complex to recognize C. haemulonii, C. haemulonii var. vulnera, Candida pseudohaemulonii, Candida duobushaemulonii, and C. auris (Cendejas-Bueno et al., 2012). Notably, these investigators reported that MALDI-TOF MS was as good as the ITS sequencing for identification purposes (Cendejas-Bueno et al., 2012). Thus, within a short span of 4 years, a rare group of yeast species with intrinsic antifungal resistance was characterized for clinical significance, and the MALDI-TOF MS test was prescribed for rapid laboratory identification.
Candida auris Internal Transcribed Spacer (ITS) and Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry
Subsequent notable developments in the C. auris saga involved reports from outside East Asia. Sarma et al. (2013) described two C. auris isolates from candidemia cases from a hospital in North India. These isolates were amphotericin B- and fluconazole-resistant. The tally was most likely higher than reported, as only two of 15 isolates tentatively identified as C. haemulonii using a commercial yeast kit were sent out for re-identification by ITS sequencing (Sarma et al., 2013). Later, Chowdhury and coworkers described a series of C. auris cases from North and South India (Chowdhary et al., 2013, 2014). The authors suggested a clonal population based on amplicon typing and noted nucleotide variability in ITS gene sequences from East Asian C. auris isolates (Chowdhary et al., 2013, 2014). The C. auris isolates in these series were resistant to fluconazole, voriconazole, caspofungin, and flucytosine (Chowdhary et al., 2014). An additional case of fluconazole-resistant C. auris candidemia was reported from Kuwait in 2014, attesting to broader distribution in South Asia and Middle East (Emara et al., 2015). Also, in 2014, four cases of fluconazole-resistant C. auris candidemia were described from South Africa, and the investigators reported nucleotide variability in ITS sequences (Magobo et al., 2014). Thus, multiple reports documented widespread misidentification of C. auris by available specialized culture media, biochemical test kits, or commercial systems, highlighting the need for ITS sequencing (Chowdhary et al., 2013, 2014; Sarma et al., 2013; Magobo et al., 2014; Won et al., 2014; Emara et al., 2015). Using a more extensive collection of 102 C. auris isolates, the Chowdhury group also affirmed an earlier report by Cendejas-Bueno et al. (2012) on the suitability of MALDI-TOF MS as a more facile approach for C. auris confirmation (Kathuria et al., 2015; Prakash et al., 2016). In a preliminary study, VITEK MS instrument was found to be efficient in identification of eight of twelve reference strains of C. auris (Girard et al., 2016). Ghosh et al. (2015) and Bao et al. (2018) created main spectrum projections (MSP) and an in-house database (CMdb), respectively, to demonstrate it was possible to identify C. auris using a commercial Bruker Biotyper MALDI-TOF MS system. Other investigators also generated in-house databases to improve their Bruker MALDI-TOF MS reference library (Ceballos-Garzon et al., 2020). Of note, the US Food and Drug Administration (FDA) approved the BRUKER MALDI Biotyper CA system (April 20, 2018) and the bioMérieux Vitek MS (December 21, 2018) for C. auris identification (Zhu et al., 2020).
Candida auris PCR and Loop-Mediated Isothermal Amplification
Polymerase chain reaction (PCR) in many configurations remains the technology of choice for newly described laboratory tests for C. auris (Table 1). Among the most straightforward applications, a duplex one-tube ITS-PCR assay was developed to speciate C. auris from C. haemulonii by the size selection of amplicons (Theill et al., 2018). Another direct PCR application targeted glycosylphosphatidylinositol (GPI) protein-encoding genes to speciate C. auris and 18 other Candida species by gel visualization of different size amplicons (Ruiz-Gaitan et al., 2018). The Boekhout group described novel tetraplex PCR and 21-Multiplex PCR for C. auris and other closely related and other yeast species, and extensive testing with 405 reference strains and 804 clinical strains of yeasts from three different countries (Arastehfar et al., 2018, 2019a,b, 2019c). Prospective and retrospective validations were performed in hospitals that appear not to have C. auris (Arastehfar et al., 2018, 2019a). Although targeted for resource-poor settings, direct PCR methods appear not to be widely adopted either due to the potential for contamination, the difficulties of recording minor variations in the gel bands or the unavailability of validation data needed for a laboratory-developed test (LDT) (Bockstahler, 1994; Millar et al., 2002; Hoorfar et al., 2004). A commercial multiplex nucleic acid test panel that includes C. auris, received FDA approval on March 18, 2020; we did not find any independent evaluation of this product in peer-reviewed publication (BioFire® Blood Culture Identification 2 (BCID2) Panel, FilmArray® 2.0 or FilmArray® Torch systems, bioMérieux, Inc., Durham, NC, United States).
TABLE 1.
Candida auris culture-independent DNA-based identification and confirmation methods.
| Method | Target | C. auris (No. isolates) | Closely-Related Yeasts (No. isolates) | Other Fungi (No. isolates) | Clinical specimen (No.) | Sensitivity (%) | Specificity (%) | LOD (CFU/reaction) | References |
| PCR | |||||||||
| GPI | 139 | 18 | ND (Not Done) | ND | 98 | 100 | ND | Ruiz-Gaitan et al., 2018 | |
| ITS2 | 20 | 30 | ND | ND | 100 | 100 | ND | Theill et al., 2018 | |
| Multiplex PCR | |||||||||
| 26S | 138 | 34 | 9 | Mouse blood and tissue (21) | 100 | 100 | ND | Arastehfar et al., 2018 | |
| 26S | 3 | 297 | ND | ND | 100 | 100 | ND | Arastehfar et al., 2019a | |
| 26S | 35 | 1149 | 47 | ND | 100 | 100 | ND | Arastehfar et al., 2019c | |
| Real-time PCR | |||||||||
| ITS2 | 44 | 92 | 5 | ND | 100 | 100 | 10 | Kordalewska et al., 2017 | |
| ITS2 | 17 | 40 | 31 | Swab (365) | 89 | 99 | 1 | Leach et al., 2018 | |
| 58 | Sponge (258) | 100 | 89 | 1 | Leach et al., 2018 | ||||
| ITS2 | 47 | Swab (110) | 96 | 92 | 1 | Leach et al., 2019 | |||
| ITS2 | 73 | Swab (247) | 93.6 | 97.2 | 1 | Ahmad et al., 2019 | |||
| ITS1/2 | 10 | 103 | 13 | Simulated sputum, Urine, Wound swabs, and Serum (11) | 100 | 100 | 1 | Lima et al., 2019 | |
| ITS2 | 32 | 18 | 54 | Simulated urine, Blood, and Swab (30) | 93.3–100 | 96 | 4–54 | Walchak et al., 2020 | |
| GPI | 155 | 18 | ND | Simulated serum (1) | 100 | 100 | 5 | Alvarado et al., 2021 | |
| GPI | 8 | 62 | 2,123 | Stool and water samples (2073) | 100 | 100 | 13 | Ibrahim et al., 2021a | |
| ITS | 4 | 113 | 8 | ND | 100 | 100 | 1 | Jafarian et al., 2020 | |
| LAMP | Pyruvate synthase | 20 | 32 | 13 | ND | 100 | 100 | 20 | Yamamoto et al., 2018 |
| Commercial | |||||||||
| Real-time, AurisID | 72 | 40 | ND | Swab (113) | 96.6 | 100 | 1 | Mulet Bayona et al., 2021 | |
| Real-time, AurisID | 29 | 13 | ND | Simulated serum (2) | 100 | 0 | 1 | Sattler et al., 2021 | |
| Real-time, Fungiplex Candida Auris RUO | 29 | 13 | ND | Simulated serum (2) | 100 | 0 | 9 | Sattler et al., 2021 | |
| qPCR, GPSTM MONODOSE dtec | 117 | 25 | ND | ND | 100 | 100 | 5 | Martinez-Murcia et al., 2018 | |
| qPCR, SYBR green | ITS2 | 48 | ND | 55 | ND | 93 | 96 | 4 | Sexton et al., 2018b |
| T2Dx | 50 | 43 | ND | ND | 89 | 98 | 5 | Sexton et al., 2018a | |
| GenMark Dx ePlex | 49 | 700 | 70 | Blood (141) | 100 | 100 | ND | Zhang et al., 2020 | |
| Biosensor | GPI | 31 | 13 | ND | Simulated serum (22) | 85 | 100 | 6 | Pla et al., 2021 |
Yamamoto et al. (2018) developed a loop-mediated isothermal amplification (LAMP)-based approach to detect C. auris. LAMP forward/reverse primers targeting C. auris the pyruvate: ferredoxin oxidoreductase domain from uncharacterized protein (NCBI CJI97_003625, Gene ID 40028770) were tested in PCR at 56°C for 90 min and the product visualized with a turbidimeter (Yamamoto et al., 2018). The assay had an excellent limit of detection (LOD) (20 copies/reaction) and specificity (100%) with a large panel of C. auris and other fungi, positive C. auris identification from a clinical swab, and simulated environmental specimens (Yamamoto et al., 2018). Surprisingly, C. auris LAMP assay has not seen broader adaptation, perhaps because test kit manufacturers appear to be unenthused about LAMP technology (Cantera et al., 2019).
Candida auris Real-Time PCR
Kordalewska et al. (2017) described a real-time PCR assay for C. auris (Table 1). The new assay targeted Candida ribosomal genes for specific primers used either for direct PCR and detection of variable amplicon sizes or for real-time PCR and melting curve analysis of the double-strand-specific dye SYBR® Green I (Wittwer et al., 2013). The investigators reported 100% accuracy (100% sensitivity and specificity) for C. auris from closely related species based on a proficiency panel of 44. C. auris and 97 other yeast isolates (Kordalewska et al., 2017). Further evaluation and modification of the assay by the Perlin laboratory and CDC investigators allowed direct detection of C. auris from patient swabs with 93% sensitivity and 96% specificity (Sexton et al., 2018b). This milestone marked the availability of a rapid C. auris real-time PCR test for surveillance purposes with the accompanying validation data required of an LDT. Within 3 months of the publication of the new real-time assay, one of us (SC) led a team that developed and validated a TaqMan-based real-time PCR assay targeting the ribosomal ITS2 of C. auris (Leach et al., 2018). The validation study comprised 623 surveillance samples, including 365 patient swabs and 258 environmental sponges. We found 49 swabs and 58 sponge samples positive, with 89 and 100% clinical sensitivity vis-a-vis culture-positive results (Leach et al., 2018). The distinguishing features of our assay vis-à-vis earlier publication from Perlin laboratory were the use of TaqMan probe chemistry, higher sensitivity (with an LOD of 1 C. auris CFU/PCR), the inclusion of all known clades of C. auris as reported by whole-genome sequencing, and direct utilization of the test for detection of C. auris from large numbers of surveillance samples (Kordalewska et al., 2017; Leach et al., 2018; Sexton et al., 2018b). We further expanded our manual assay to an automated sample-to-result real-time C. auris PCR assay using the BD Max open system (Leach et al., 2019). The new assay, with culture as gold standard, yielded 96% clinical sensitivity, and 94% clinical accuracy with 110 patient surveillance samples (Leach et al., 2019). The new assay appeared promising for broader adaptability and availability of high throughput surveillance testing (Leach et al., 2019). CDC investigators adapted our manual assay to an even higher throughput platform by automating the extraction steps and achieved diagnostic sensitivity and specificity of 93.6 and 97.2%, respectively (Ahmad et al., 2019). Both manual and semi-automated C. auris assays developed by our group were adopted by other laboratories, including the CDC Antifungal Resistance Lab Network (personal communications) (Caceres et al., 2019; Malczynski et al., 2020; Pacilli et al., 2020; CDC, 2021a). A few months before submission/publication of our BD Max assay, Lima et al. (2019) described a BD Max assay for C. auris. The authors designed primers to target many fungal ribosomal genes and tested a collection of fungi and bacteria and 50 contrived clinical specimens to report 100% clinical sensitivity and specificity (Lima et al., 2019). The authors indicated that their primer-design strategy was superior to other reported assays, and the new assay was widely applicable. However, only 10 C. auris isolates and no C. auris positive clinical or surveillance samples were tested during the validation steps (Lima et al., 2019). Further information is awaited about the performance and adoption of the assay in other laboratories.
In 2020–2021, several additional laboratories described real-time PCR assays for C. auris (Walchak et al., 2020; Alvarado et al., 2021; Ibrahim et al., 2021a; Table 1). Commercial primers and probes, melting curve analysis, and validation with limited C. auris isolates and contrived clinical samples allowed ≤100 CFU/reaction sensitivity from blood and urine (Walchak et al., 2020). A similar melting curve approach using primers against GPI-modified protein-encoding genes allowed specific identification and detection of C. auris with an LOD of 5 CFU/reaction for isolates, 20 CFU/reaction from spiked blood and serum (Alvarado et al., 2021). A TaqMan-chemistry assay with an ITS2-specific probe achieved LOD 1 CFU/reaction when four C. auris isolates and other fungal and bacterial strains were tested (Jafarian et al., 2020). Another study utilizing a GPI-target probe with TaqMan-chemistry and simulated samples achieved an LOD of 13 C. auris CFU/qPCR reaction (Ibrahim et al., 2021a). The number of publications suggests real-time PCR is a preferred approach for rapid identification of C. auris from clinical, surveillance, and environmental samples. However, in the absence of head-to-head comparisons, it is not clear if SYBR® Green I or TaqMan chemistry, or a particular real-time machine, are preferable for C. auris real-time PCR assays. A few studies not involving C. auris describe the relative merits of various real-time probes. Moreover, a process was published to compare in-development real-time platforms; we refer the readers to these studies to make an informed choice about primers, probes, and a platform suitable for their laboratory needs (Van Poucke et al., 2012; Polinski et al., 2013; Ahrberg and Neužil, 2015; Cantera et al., 2019).
Candida auris Real-Time PCR Kits
AurisID® is a commercial kit with ready-made reagents for qPCR to identify C. auris from fungal culture (OLM Diagnostics, Newcastle upon Tyne, England) (Table 1). A retrospective evaluation with C. auris positive swab samples reported 96.6% sensitivity (Mulet Bayona et al., 2021). In a second study, AurisID® detected C. auris with an LOD of 1 genome copy/reaction but gave false positives with high DNA amounts of the C. haemulonii, C. duobushaemulonii, and C. pseudohaemulonii (Sattler et al., 2021). Fungiplex® Candida Auris is a real-time, research use only (RUO) PCR assay for the rapid detection of C. auris in hospital hygiene applications including ready-made reagents for qPCR (Bruker Daltonics GmbH & Co. KG. Bremen, Germany). An independent evaluation reported the Fungiplex® Candida Auris kit LOD to be 9 copies/reaction, much lower than AurisID® (LOD = one copy/reaction) and 100% specificity for five C. auris isolates tested (Sattler et al., 2021). The investigators also tested Fungiplex® Candida Auris in “off-label use” with blood samples spiked with two C. auris isolates to obtain ∼45 viable CFU/reaction (Sattler et al., 2021). All three commercial ready-to-use real-time PCR reagent kits do not disclose primer and probe details, which may or may not impact their future use as new C. auris clades are discovered. None of these kits have current regulatory approval for routine use in diagnostic laboratories.
Differential and Selective Media
Differential culture media continue to be widely used in busy clinical laboratories. CHROMagarTM Candida Plus is a new chromogenic differential medium (Table 2). A comparative evaluation with HiCrome C. auris MDR selective agar, CandiSelect, CHROMagarTM Candida, and Chromatic Candida commercial media revealed C. auris colonies develop a species-specific coloration, as do closely related pathogenic species C. pseudohaemulonii and Candida vulturna (de Jong et al., 2021). In a similarly designed laboratory evaluation, CHROMagarTM Candida Plus agar was judged to be an excellent alternative to conventional mycological media for the screening of patients with C. auris, as only Candida diddensiae yielded a similar coloration (Borman et al., 2021). An earlier evaluation of CHROMagarTM Candida Plus found 100% sensitivity and specificity for C. auris when 14 surveillance samples were tested (Mulet Bayona et al., 2020). Even in an off-label modification, CHROMagar Candida was reported to offer differentiation between C. auris and C. haemulonii complex (Kumar et al., 2017). Thus, CHROMagar Candida Plus agar, and possibly other commercial differential media, hold promise for presumptive identification of C. auris when the laboratory confirms the identification with MALDI-TOF MS or another confirmatory test as per the manufacturer’s recommendation. Welsh et al. (2017) described the first selective medium when they reported C. auris growth at an elevated temperature (40°C) and salinity (10% wt/vol) in the Sabouraud or yeast nitrogen base broths with dulcitol or mannitol as the carbon source. The high salt medium had an excellent performance in the selective enrichment of C. auris cells from patient and environmental surveillance samples (Zhu et al., 2020; Sexton et al., 2021). A modified Selective Auris Medium (SAM) was recently described with YPD agar comprising 12.5% NaCl and 9 mM ferrous sulfate and incubation at 42°C (Das et al., 2021). Another variation of selective medium described by Walsh is termed SCA (specific C. auris) medium, incorporating crystal violet to prevent the growth of Candida tropicalis (Ibrahim et al., 2021b). Both SAM and SCA are reported to improve the original C. auris selective medium, but independent confirmations are not yet available.
TABLE 2.
Candida auris identification by mass spectrometry and other biochemical methods.
| Method | C. auris (No. isolates) | Closely-Related Yeasts (No. isolates) | Other Yeasts (No. isolates) | Clinical specimen (No.) | Sensitivity (%) | Specificity (%) | References |
| Mass spectrometry | |||||||
| Bruker | 90 | 12 | ND | ND | 100 | 100 | Kathuria et al., 2015 |
| Bruker | 82 | 11 | ND | ND | 100 | 100 | Girard et al., 2016 |
| Bruker, CMdb database | 33 | 62 | 16 | ND | 100 | 100 | Bao et al., 2018 |
| MALDI | 3 | 298 | ND | ND | 100 | 100 | Arastehfar et al., 2019a |
| MALDI, PXD016387 database | 300 | 8 | ND | ND | 100 | 100 | Ceballos-Garzon et al., 2020 |
| Autof MS 1000 | 2 | 1216 | ND | ND | 100 | 100 | Yi et al., 2021 |
| Vitek MS | 2 | 1216 | ND | ND | 100 | 100 | Yi et al., 2021 |
| Biotyper | 61 | ND | ND | ND | 75.4–83.6 | ND | Kwon et al., 2019 |
| Vitek MS | 61 | ND | ND | ND | 93.4–96.7 | ND | Kwon et al., 2019 |
| Differential media | |||||||
| CHROMagarTM with Pal’s medium | 15 | 13 | ND | ND | 100 | 100 | Kumar et al., 2017 |
| CHROMagarTM Candida Plus | 37 | 58 | ND | swab (23) | 100 | 100 | Mulet Bayona et al., 2020 |
| CHROMagarTM Candida Plus | 10 | 52 | ND | ND | ND | 98 | Borman et al., 2021 |
| CHROMagarTM Candida Plus | 9 | 35 | ND | ND | 90 | 100 | de Jong et al., 2021 |
| Selective media | |||||||
| Selective Auris Medium | 133 | 446 | ND | Bactec blood culture broth (40) | 100 | 100 | Das et al., 2021 |
| Specific C. auris (SCA) Medium | 7 | 128 | 50 | stool (200) | 100 | 100 | Ibrahim et al., 2021b |
In laboratories without access to ITS sequencing and MALDI-TOF MS, it is convenient to use manual or automated biochemical panels and systems to confirm the identification of yeast isolated from culture. Earlier reports and recent re-evaluations found incomplete or incorrect identifications of C. auris by the API ID 32 C system (version 4.0 database), AuxaColorTM 2 (Bio-Rad Laboratories, Marnes-la-Coquette, France), Vitek 2 (bioMérieux, Marcy l’Étoile, France), BD Phoenix (BD Diagnostics, Sparks, MD, United States), and RapID Yeast Plus (Remel, Thermo Fisher Scientific, Lenexa, KS, United States) (Won et al., 2014; Kathuria et al., 2015; Kim et al., 2016; Ruiz Gaitan et al., 2017; Iguchi et al., 2018; Sharp et al., 2018; Snayd et al., 2018; Ambaraghassi et al., 2019; Tan et al., 2019; CDC, 2021b; Du et al., 2021; Table 2). A recent report on API ID 32 C concluded that C. auris could be identified if the percentage of positive reactions is registered in the database or calculated manually (Du et al., 2021). It appears these panels and systems need further refinements and evaluation to assess their utility for the identification of rare yeasts, including C. auris (Du et al., 2021). It might also be prudent for the end users to promptly install the latest product updates from the manufacturers and review any new information on approvals and evaluations.
Candida auris Antifungal Resistance Testing
Antifungal susceptibility testing (AFST) of C. auris is performed with CLSI or EUCAST methods and commercial devices (Chowdhary et al., 2014, 2018; Arendrup et al., 2017; Bidaud et al., 2019; Escandon et al., 2019; Kwon et al., 2019; O’Brien et al., 2020; Zhu et al., 2020; CDC, 2021a). Several new drugs in development were also tested for their efficacy against C. auris using standard AFST (Arendrup et al., 2018; Ghannoum et al., 2020). A few studies describe antifungal combination testing for C. auris using LDTs (Fakhim et al., 2017; Bidaud et al., 2019; O’Brien et al., 2020). The literature on C. auris antifungal testing is voluminous, and discussion of these publications is not possible here. The focus is on the evaluation of established or new laboratory tests. Nevertheless, it is incumbent upon laboratories to perform both identification and antifungal testing for C. auris as part of their diagnostic service. The drug-resistance pattern in C. auris remains variable. Also, regional patterns have been observed for fluconazole and other triazoles, echinocandins, and amphotericin B (Chowdhary et al., 2018; Escandon et al., 2019). There is emerging evidence about the utility of susceptibility testing in selecting appropriate antifungal drugs in the management of patients (Alatoom et al., 2018; Chen et al., 2018; Dewaele et al., 2020). Lessons learned from C. auris outbreak investigations included a vital role for antifungal susceptibility data for surveillance purposes, including monitoring emerging drug resistance patterns in the community (Mulet Bayona et al., 2020; Ostrowsky et al., 2020; Zhu et al., 2020; Tian et al., 2021). AFST test innovations for C. auris remain sparse, with just one report describing same-day identification and echinocandin-resistance testing using MALDI-TOF MS (Vatanshenassan et al., 2019). Further confirmations and correlation of the new test approach with standard AFST are not yet available. The phenotype based CLSI, EUCAST, and commercial AFST tests are slow. Ideally, phenotype tests should be preceded by rapid DNA tests for drug-resistant C. auris, ensuring timely treatment of infected patients and effective pathogen control measures. In promising developments, Perlin laboratory described a molecular beacon-based platform for detecting FKS1 (echinocandins) and ERG11 (azoles) mutations in C. auris isolates and patient swab samples (Hou et al., 2019; Kordalewska et al., 2019). The molecular tests for drug resistance are desirable as surrogates for standard AFST tests standalone or multiplexed with rapid ID tests for C. auris. Molecular platforms offer flexibility to target new mutations encountered in drug-resistant C. auris.
Candida auris Diagnostic Algorithm
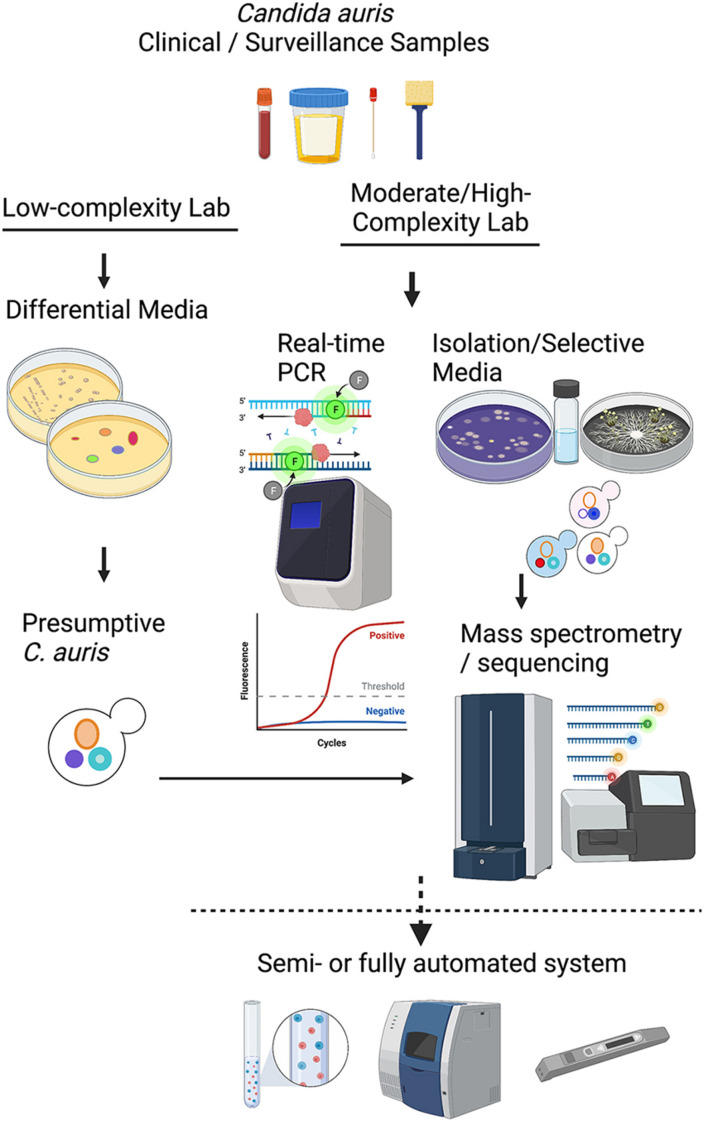
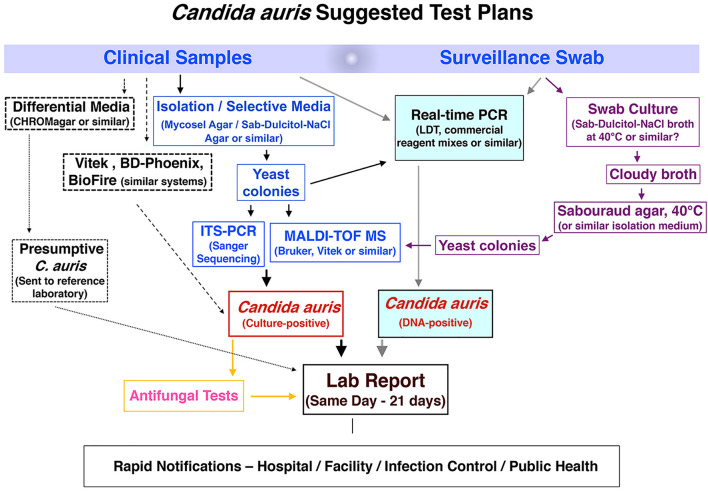
Candida auris colonization and infection among hospitalized patients and long-term care residents have become a global problem, with localized outbreaks reported from several countries. Several laboratory modes of operation are possible regarding pathogen isolation, rapid identification, susceptibility testing, and genotyping. Figure 1 provides a conceptual illustration of testing method differences between low-complexity and moderate- to high-complexity laboratories. A significant challenge for specialized diagnostic mycology laboratories concerns supporting C. auris surveillance and outbreak investigations. Such a service is available in large bacteriology laboratories but seldom needed for fungal pathogens. Consequently, many clinical recommendations and guidelines were published for laboratories that work with or want to expand their services for C. auris (Lockhart et al., 2017; Tsay et al., 2018; Caceres et al., 2019; Kenters et al., 2019). In the United States, the latest versions of the CLSI M-54 document and the Manual of Clinical Microbiology, peer-recommended guidelines for diagnostic microbiology laboratories, do not have C. auris-specific guidelines for isolation and identification (Carroll et al., 2019; CLSI, 2021). Early in 2017, we devised an algorithm which has been modified regularly since then to cope with an unprecedented volume of surveillance and clinical samples of C. auris. Alternate laboratory approaches are shown with dotted lines. We share the scheme hoping it will help other laboratories to modify their workflows to suit local needs (Figure 2).
FIGURE 1.
A conceptual illustration of testing method differences between low-complexity and moderate- to high-complexity laboratories.
FIGURE 2.
Workflow algorithm for high-volume surveillance and clinical sample testing of C. auris. Dotted lines indicate alternate laboratory approaches.
Candida auris Diagnostics on the Horizon
Among commercial products likely to be available soon, T2MR (T2 magnetic resonance) is a portable system that detects candidemia by measuring signals from PCR-amplified Candida nanoparticle clusters in the blood directly within 3 h (Neely et al., 2013; Mylonakis et al., 2015). In 2018, CDC investigators evaluated a T2Cauris panel (T2 Biosystems, Lexington, MA, United States) with axilla/groin swab samples and reported 89% sensitivity and 98% specificity for detecting C. auris (Sexton et al., 2018a; Table 2). The T2Biosystems® website states that T2Cauris is not cleared for diagnostic testing and is available for research use only (RUO). MONODOSE dtec-qPCR C. auris is a commercial ready-to-use qPCR kit for pathogen detection (Genetic PCR SolutionsTM, Alicante, Spain). The manufacturer completed a validation study with academic partners according to UNE-EN ISO/IEC 17025:2005 standard (Martinez-Murcia et al., 2018). The investigators reported MONODOSE dtec-qPCR C. auris passed validation in two independent laboratories and is ready to undergo clinical evaluation (Martinez-Murcia et al., 2018). Among new sample-to-answer systems, ePlex BCID-FP is an investigational use only proprietary reagent test panel for 15 Candida species used with a proprietary ePlex System (GenMark Diagnostics, Inc., Carlsbad, CA, United States). A multi-laboratory evaluation with 3 C. auris isolates and 49 contrived blood samples obtained 100% sensitivity and specificity (Zhang et al., 2020). Another sample-to-answer approach based on an oligonucleotide-functionalized gated nanosystem for C. auris achieved 85% sensitivity and 100% specificity in limited testing with 22 blood samples (Pla et al., 2021). Mass spectrometry systems from manufacturers other than Bruker are also available and in use in clinical laboratories. A minimal evaluation with two C. auris isolates reported equal efficacy of Vitek MS and Autof MS 1000 systems (Yi et al., 2021). It is safe to say that any MALDI-TOF MS machine will provide identification of C. auris provided enough representative isolates encompassing all known clades are included in the database to train the decision algorithm (van Belkum et al., 2017). Limited but promising results are also being published on sample processing and machine learning applications to MALDI platforms, which will benefit C. auris diagnostics eventually (Muthu et al., 2018; Weis et al., 2020; De Bruyne et al., 2021).
Candida auris Biosafety, Disinfectants, and Advisories
Candida auris poses unique occupational risks for laboratory personnel due to potential exposure to multidrug-resistant organisms. The reported persistence of C. auris on inanimate objects requires focused efforts at environmental decontamination. No C. auris-specific guidelines are available in the latest edition of BMBL Biosafety in Microbiological and Biomedical Laboratories (DHH, 2020). CDC and many state and local jurisdictions in the United States have issued C. auris advisories for healthcare professionals. The guidance information is regularly updated online for easy consultation. The United States EPA’s (Environmental Protection Agency) List P includes 23 disinfectant products with claims about their ability to kill C. auris (EPA, 2021). We follow institutional biosafety guidelines for BSL 2 laboratories. Our additional precautions include frequent changes of hand gloves and disposable lab apparel. The laboratory work surfaces are decontaminated pre- and post-procedure with freshly prepared 10% bleach solution, followed by 70% ethanol. We secure fungal cultures in secondary containers for transfer and incubation within the laboratory. Periodic sampling of space and instruments are done by C. auris real-time PCR and culture to check for any inadvertent fungal contamination.
Candida auris Test Wish List
The availability of rapid C. auris DNA tests onsite remain severely restricted, especially in resource-poor settings. The results are available only after 24–72 h, assuming samples are sent out for reference testing. DNA tests are challenging for many frontline laboratories and long-term care facilities due to a lack of equipment and trained personnel. MALDI-TOF MS assays work best with isolated C. auris colonies, but the time required delays diagnosis by 3–7 days on average (Zhu et al., 2020). Frontline laboratories need rapid and facile onsite testing of C. auris to inform their efforts at identification, surveillance, patient isolation, admission screening, and environmental control (Durante et al., 2018; Wang et al., 2020). Thus, there is an unmet, urgent need for simple C. auris tests, especially for surveillance samples. Lateral flow assays (LFAs), also known as lateral flow immunoassay or immunochromatographic assay, could be a good choice as they are rapid, cheap, stable, and easy-to-implement for presumptive identification of microbes (Koczula and Gallotta, 2016; Boutal et al., 2018). At the other end of the service spectrum, many hospital laboratories already use multiplex test panels for Clostridioides difficile and carbapenem-resistant Enterobacterales (CRE), and synergy of such panels with C. auris testing would be welcome (Binnicker, 2015; Crobach et al., 2016; Kost et al., 2017; McDonald et al., 2018; Bogaerts et al., 2020; CDC, 2021c). Current AFST services are inadequate as turnaround time is woefully inadequate, and new test formats/devices are needed for faster reporting and AFST-directed treatment of C. auris infections. Laboratory surveillance is crucial for monitoring and control of C. auris outbreaks. C. auris genome analysis has a proven role in the control of hospital outbreaks by pinpointing the common source (Eyre et al., 2018; Theodoropoulos et al., 2020). However, fungal genome sequencing and analysis remain beyond the capabilities of most diagnostic laboratories. Therefore, local outbreak investigations require diagnostic laboratories to quickly access regional and national collaborative networks with standardized C. auris sequencing tools (Ladner et al., 2019; Mintzer et al., 2019; NIHR Global Health Research Unit on Genomic Surveillance of AMR, 2020).
Author Contributions
ED: draft preparation, review and editing, visualization. SC: supervision, conceptualization, and review and editing. VC: supervision, conceptualization, draft preparation, and review and editing, visualization. All authors approved the final version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank YanChun Zhu, Lynn Leach, and Brittany O’Brien for their contributions to methods development, evaluations, and discussions about C. auris testing. Kimberly McClive-Reed is thanked for editorial comments.
Funding
Supported in part by a Centers for Disease Control and Prevention-Antibiotic Resistance Lab Network grant (NU50CK000423) and a grant (1R21AI156573-01A1) from the National Institutes of Health. The contents are solely the responsibility of the authors and do not necessarily represent the official views of the Centers for Disease Control and Prevention or the Department of Health and Human Services.
References
- Ahmad A., Spencer J. E., Lockhart S. R., Singleton S., Petway D. J., Bagarozzi D. A., et al. (2019). A high-throughput and rapid method for accurate identification of emerging multidrug−resistant Candida auris. Mycoses 62 513–518. 10.1111/myc.12907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrberg C. D., Neužil P. (2015). Doubling Throughput of a Real-Time PCR. Sci. Rep. 5:12595. 10.1038/srep12595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alatoom A., Sartawi M., Lawlor K., AbdelWareth L., Thomsen J., Nusair A., et al. (2018). Persistent candidemia despite appropriate fungal therapy: First case of Candida auris from the United Arab Emirates. Int. J. Infect. Dis. 70 36–37. 10.1016/j.ijid.2018.02.005 [DOI] [PubMed] [Google Scholar]
- Alcoba-Florez J., Mendez-Alvarez S., Cano J., Guarro J., Perez-Roth E., del Pilar Arevalo M. (2005). Phenotypic and molecular characterization of Candida nivariensis sp. nov., a possible new opportunistic fungus. J. Clin. Microbiol. 43 4107–4111. 10.1128/JCM.43.8.4107-4111.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarado M., Bartolome Alvarez J., Lockhart S. R., Valentin E., Ruiz-Gaitan A. C., Eraso E., et al. (2021). Identification of Candida auris and related species by multiplex PCR based on unique GPI protein-encoding genes. Mycoses 64 194–202. 10.1111/myc.13204 [DOI] [PubMed] [Google Scholar]
- Ambaraghassi G., Dufresne P. J., Dufresne S. F., Vallieres E., Munoz J. F., Cuomo C. A., et al. (2019). Identification of Candida auris by Use of the Updated Vitek 2 Yeast Identification System, Version 8.01: a Multilaboratory Evaluation Study. J. Clin. Microbiol. 57 e884–e819. 10.1128/JCM.00884-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arastehfar A., Daneshnia F., Kord M., Roudbary M., Zarrinfar H., Fang W., et al. (2019a). Comparison of 21-Plex PCR and API 20C AUX, MALDI-TOF MS, and rDNA sequencing for a wide range of clinically isolated yeast species: Improved identification by combining 21-Plex PCR and API 20C AUX as an alternative strategy for developing countries. Front. Cell. Infect. Microb. 9:21. 10.3389/fcimb.2019.00021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arastehfar A., Fang W., Badali H., Vaezi A., Jiang W., Liao W., et al. (2018). Low-Cost Tetraplex PCR for the Global Spreading Multi-Drug Resistant Fungus, Candida auris and Its Phylogenetic Relatives. Front. Microbiol. 9:1119. 10.3389/fmicb.2018.01119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arastehfar A., Fang W., Daneshnia F., Al-Hatmi A. M., Liao W., Pan W., et al. (2019b). Novel multiplex real-time quantitative PCR detecting system approach for direct detection of Candida auris and its relatives in spiked serum samples. Future Microbiol. 14 33–45. 10.2217/fmb-2018-0227 [DOI] [PubMed] [Google Scholar]
- Arastehfar A., Fang W., Pan W., Lackner M., Liao W., Badiee P., et al. (2019c). YEAST PANEL multiplex PCR for identification of clinically important yeast species: stepwise diagnostic strategy, useful for developing countries. Diagn. Microbiol. Infect. Dis. 93 112–119. 10.1016/j.diagmicrobio.2018.09.007 [DOI] [PubMed] [Google Scholar]
- Arendrup M. C., Chowdhary A., Astvad K. M. T., Jorgensen K. M. (2018). APX001A In Vitro Activity against Contemporary Blood Isolates and Candida auris Determined by the EUCAST Reference Method. Antimicrob. Agents Chemother. 62 e1225–e1218. 10.1128/AAC.01225-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendrup M. C., Prakash A., Meletiadis J., Sharma C., Chowdhary A. (2017). Comparison of EUCAST and CLSI reference microdilution MICs of eight antifungal compounds for Candida auris and associated tentative epidemiological cutoff values. Antimicrob. Chemother. 61 e417–e485. 10.1128/AAC.00485-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao J. R., Master R. N., Azad K. N., Schwab D. A., Clark R. B., Jones R. S., et al. (2018). Rapid, Accurate Identification of Candida auris by Using a Novel Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry (MALDI-TOF MS) Database (Library). J. Clin. Microbiol. 56 e1700–e1717. 10.1128/JCM.01700-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidaud A., Botterel F., Chowdhary A., Dannaoui E. (2019). In vitro antifungal combination of flucytosine with amphotericin B, voriconazole, or micafungin against Candida auris shows no antagonism. Antimicrob. Agents Chemother. 2019:63. 10.1128/AAC.01393-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binnicker M. J. (2015). Multiplex Molecular Panels for Diagnosis of Gastrointestinal Infection: Performance, Result Interpretation, and Cost-Effectiveness. J. Clin. Microbiol. 53 3723–3728. 10.1128/JCM.02103-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockstahler L. E. (1994). Overview of international PCR standardization efforts. PCR Methods Appl. 3 263–267. 10.1101/gr.3.5.263 [DOI] [PubMed] [Google Scholar]
- Bogaerts P., Berger A. S., Evrard S., Huang T. D. (2020). Comparison of two multiplex immunochromatographic assays for the rapid detection of major carbapenemases in Enterobacterales. J. Antimicrob. Chemother. 75 1491–1494. 10.1093/jac/dkaa043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borman A. M., Fraser M., Johnson E. M. (2021). CHROMagarTM Candida Plus: A novel chromogenic agar that permits the rapid identification of Candida auris. Med. Mycol. 59 253–258. 10.1093/mmy/myaa049 [DOI] [PubMed] [Google Scholar]
- Boutal H., Vogel A., Bernabeu S., Devilliers K., Creton E., Cotellon G., et al. (2018). A multiplex lateral flow immunoassay for the rapid identification of NDM-, KPC-, IMP- and VIM-type and OXA-48-like carbapenemase-producing Enterobacteriaceae. J. Antimicrob. Chemother. 73 909–915. 10.1093/jac/dkx521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres D. H., Forsberg K., Welsh R. M., Sexton D. J., Lockhart S. R., Jackson B. R., et al. (2019). Candida auris: A Review of Recommendations for Detection and Control in Healthcare Settings. J. Fungi. 5:111. 10.3390/jof5040111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantera J. L., White H., Diaz M. H., Beall S. G., Winchell J. M., Lillis L., et al. (2019). Assessment of eight nucleic acid amplification technologies for potential use to detect infectious agents in low-resource settings. PLoS One 14:e0215756. 10.1371/journal.pone.0215756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll K. C., Pfaller M. A., Landry M. L., McAdam A. J., Patel R., Richter S. S., et al. (2019). Manual of Clinical Microbiology. Hoboken, NJ: John Wiley & Sons, Inc. 10.1128/9781555819842 [DOI] [Google Scholar]
- Castanheira M., Woosley L. N., Diekema D. J., Jones R. N., Pfaller M. A. (2013). Candida guilliermondii and other species of Candida misidentified as Candida famata: assessment by vitek 2, DNA sequencing analysis, and matrix-assisted laser desorption ionization-time of flight mass spectrometry in two global antifungal surveillance programs. J. Clin. Microbiol. 51 117–124. 10.1128/JCM.01686-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC (2021a). How to Participate in CDC’s AR Lab Network: Lab Testing. Centers for Disease Control and Prevention NCfEaZIDN. Atlanta, GA: Division of Healthcare Quality Promotion (DHQP). [Google Scholar]
- CDC (2021b). Identification of Candida auris. Centers for Disease Control and Prevention NCfEaZIDN. Atlanta, GA: Division of Healthcare Quality Promotion (DHQP). [Google Scholar]
- CDC (2021c). Laboratory Resources- Healthcare-associated Infection. Centers for Disease Control and Prevention NCfEaZIDN. Atlanta, GA: Division of Healthcare Quality Promotion (DHQP). [Google Scholar]
- Ceballos-Garzon A., Amado D., Velez N., Jimenez A. M., Rodriguez C., Parra-Giraldo C. M. (2020). Development and Validation of an in-House Library of Colombian Candida auris Strains with MALDI-TOF MS to Improve Yeast Identification. J. Fungi. 6:72. 10.3390/jof6020072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cendejas-Bueno E., Kolecka A., Alastruey-Izquierdo A., Theelen B., Groenewald M., Kostrzewa M., et al. (2012). Reclassification of the Candida haemulonii complex as Candida haemulonii (C. haemulonii group I), C. duobushaemulonii sp. nov. (C. haemulonii group II), and C. haemulonii var. vulnera var. nov.: three multiresistant human pathogenic yeasts. J. Clin. Microb. 50 3641–3651. 10.1128/JCM.02248-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Zhao J., Han L., Qi L., Fan W., Liu J., et al. (2018). Emergency of fungemia cases caused by fluconazole-resistant Candida auris in Beijing. China. J. Infect. 77 561–571. 10.1016/j.jinf.2018.09.002 [DOI] [PubMed] [Google Scholar]
- Chowdhary A., Anil Kumar V., Sharma C., Prakash A., Agarwal K., Babu R., et al. (2014). Multidrug-resistant endemic clonal strain of Candida auris in India. Eur. J. Clin. Microbiol. Infect. Dis. 33 919–926. 10.1007/s10096-013-2027-1 [DOI] [PubMed] [Google Scholar]
- Chowdhary A., Prakash A., Sharma C., Kordalewska M., Kumar A., Sarma S., et al. (2018). A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009–17) in India: role of the ERG11 and FKS1 genes in azole and echinocandin resistance. J. Antimicrob. Chemother. 73 891–899. 10.1093/jac/dkx480 [DOI] [PubMed] [Google Scholar]
- Chowdhary A., Sharma C., Duggal S., Agarwal K., Prakash A., Singh P. K., et al. (2013). New clonal strain of Candida auris, Delhi, India. Emerg. Infect. Dis. 19 1670–1673. 10.3201/eid1910.130393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI (2021). CLSI. Principles and procedures for detection and culture of fungi in clinical specimens. 2nd ed. CLSI guideline M54. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Crobach M. J., Planche T., Eckert C., Barbut F., Terveer E. M., Dekkers O. M., et al. (2016). European Society of Clinical Microbiology and Infectious Diseases: update of the diagnostic guidance document for Clostridium difficile infection. Clin. Microbiol. Infect. 22(Suppl. 4), S63–S81. 10.1016/j.cmi.2016.03.010 [DOI] [PubMed] [Google Scholar]
- Das S., Singh S., Tawde Y., Chakrabarti A., Rudramurthy S. M., Kaur H., et al. (2021). A Selective Medium for Isolation and Detection of Candida auris, an Emerging Pathogen. J. Clin. Microbiol. 59 e326–e320. 10.1128/JCM.00326-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bruyne S., Speeckaert M. M., Van Biesen W., Delanghe J. R. (2021). Recent evolutions of machine learning applications in clinical laboratory medicine. Crit. Rev. Clin. Lab. Sci. 58 131–152. 10.1080/10408363.2020.1828811 [DOI] [PubMed] [Google Scholar]
- de Jong A. W., Dieleman C., Carbia M., Mohd Tap R., Hagen F. (2021). Performance of Two Novel Chromogenic Media for the Identification of Multidrug-Resistant Candida auris Compared with Other Commercially Available Formulations. J. Clin. Microbiol. 59 e3220–e3220. 10.1128/JCM.03220-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewaele K., Frans J., Smismans A., Ho E., Tollens T., Lagrou K. (2020). First case of Candida auris infection in Belgium in a surgical patient from Kuwait. Acta Clin. Belg. 75 221–228. 10.1080/17843286.2018.1555114 [DOI] [PubMed] [Google Scholar]
- DHH (2020). Biosafety in Microbiological and Biomedical Laboratories. Services. U.S. Department of Health and Human Services. [Google Scholar]
- Du M., Hu W., Tamura T., Alshahni M. M., Satoh K., Yamanishi C., et al. (2021). Investigation of the Physiological, Biochemical and Antifungal Susceptibility Properties of Candida auris. Mycopathologia 186 189–198. 10.1007/s11046-020-00526-w [DOI] [PubMed] [Google Scholar]
- Durante A. J., Maloney M. H., Leung V. H., Razeq J. H., Banach D. B. (2018). Challenges in identifying Candida auris in hospital clinical laboratories: a need for hospital and public health laboratory collaboration in rapid identification of an emerging pathogen. Infect. Control. Hosp. Epidemiol. 39 1015–1016. 10.1017/ice.2018.133 [DOI] [PubMed] [Google Scholar]
- Eddouzi J., Hofstetter V., Groenewald M., Manai M., Sanglard D. (2013). Characterization of a new clinical yeast species, Candida tunisiensis sp. nov., isolated from a strain collection from Tunisian hospitals. J. Clin. Microbiol. 51 31–39. 10.1128/JCM.01627-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emara M., Ahmad S., Khan Z., Joseph L., Al-Obaid Im, Purohit P., et al. (2015). Candida auris candidemia in Kuwait, 2014. Emerg. Infect. Dis. 21:1091. 10.3201/eid2106.150270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EPA (2021). List P: Antimicrobial Products Registered with EPA for Claims Against Candida Auris. Washington, DC: United States Environmental Protection Agency. [Google Scholar]
- Escandon P., Chow N. A., Caceres D. H., Gade L., Berkow E. L., Armstrong P., et al. (2019). Molecular epidemiology of Candida auris in Colombia reveals a highly related, countrywide colonization with regional patterns in amphotericin B resistance. Clin. Infect. Dis. 68 15–21. 10.1093/cid/ciy411 [DOI] [PubMed] [Google Scholar]
- Eyre D. W., Sheppard A. E., Madder H., Moir I., Moroney R., Quan T. P., et al. (2018). A Candida auris outbreak and its control in an intensive care setting. New Engl. J. Med. 379 1322–1331. 10.1056/NEJMoa1714373 [DOI] [PubMed] [Google Scholar]
- Fakhim H., Chowdhary A., Prakash A., Vaezi A., Dannaoui E., Meis J. F., et al. (2017). In vitro interactions of echinocandins with triazoles against multidrug-resistant Candida auris. Antimicrob. Agents Chemother. 2017:61. 10.1128/AAC.01056-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghannoum M., Arendrup M. C., Chaturvedi V. P., Lockhart S. R., McCormick T. S., Chaturvedi S., et al. (2020). Ibrexafungerp: A Novel Oral Triterpenoid Antifungal in Development for the Treatment of Candida auris Infections. Antibiotics 2020:9. 10.3390/antibiotics9090539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A. K., Paul S., Sood P., Rudramurthy S. M., Rajbanshi A., Jillwin T. J., et al. (2015). Matrix-assisted laser desorption ionization time-of-flight mass spectrometry for the rapid identification of yeasts causing bloodstream infections. Clin. Microbiol. Infect. 21 372–378. 10.1016/j.cmi.2014.11.009 [DOI] [PubMed] [Google Scholar]
- Girard V., Mailler S., Chetry M., Vidal C., Durand G., van Belkum A., et al. (2016). Identification and typing of the emerging pathogen Candida auris by matrix-assisted laser desorption ionisation time of flight mass spectrometry. Mycoses 59 535–538. 10.1111/myc.12519 [DOI] [PubMed] [Google Scholar]
- Hoorfar J., Wolffs P., Rådström P. (2004). Diagnostic PCR: validation and sample preparation are two sides of the same coin. Apmis 112 808–814. 10.1111/j.1600-0463.2004.apm11211-1207.x [DOI] [PubMed] [Google Scholar]
- Hou X., Lee A., Jimenez-Ortigosa C., Kordalewska M., Perlin D. S., Zhao Y. (2019). Rapid detection of ERG11-associated azole resistance and FKS-associated echinocandin resistance in Candida auris. Antimicrob. Agents Chemother. 63:e01811-18. 10.1128/AAC.01811-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim A., Baron S. A., Yousfi H., Hadjadj L., Lalaoui R., Morand S., et al. (2021a). Development and standardization of a specific real-time PCR assay for the rapid detection of Candida auris. Eur. J. Clin. Microb. Infect. Dis. 2021:41768. 10.1007/s10096-021-04176-8 [DOI] [PubMed] [Google Scholar]
- Ibrahim A., Peyclit L., Abdallah R., Khelaifia S., Chamieh A., Rolain J. M., et al. (2021b). SCA Medium: A New Culture Medium for the Isolation of All Candida auris Clades. J. Fungi. 2021:7. 10.20944/preprints202104.0658.v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iguchi S., Mizushima R., Kamada K., Itakura Y., Yoshida A., Uzawa Y., et al. (2018). The Second Candida auris Isolate from Aural Discharge in Japan. JPN J. Infect. Dis. 71 174–175. 10.7883/yoken.JJID.2017.466 [DOI] [PubMed] [Google Scholar]
- Jafarian H., Khodadadi H., Badiee P. (2020). Development a hydrolysis probe-based quantitative PCR assay for the specific detection and quantification of Candida auris. Curr. Med. Mycol. 6 50–56. 10.18502/cmm.6.3.4665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R. H., Arendrup M. C. (2011). Candida palmioleophila: characterization of a previously overlooked pathogen and its unique susceptibility profile in comparison with five related species. J. Clin. Microbiol. 49 549–556. 10.1128/JCM.02071-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathuria S., Singh P. K., Sharma C., Prakash A., Masih A., Kumar A., et al. (2015). Multidrug-resistant Candida auris misidentified as Candida haemulonii: characterization by matrix-assisted laser desorption ionization–time of flight mass spectrometry and DNA sequencing and its antifungal susceptibility profile variability by Vitek 2, CLSI broth microdilution, and Etest method. J. Clin. Microbiol. 53 1823–1830. 10.1128/JCM.00367-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenters N., Kiernan M., Chowdhary A., Denning D. W., Pemán J., Saris K., et al. (2019). Control of Candida auris in healthcare institutions: Outcome of an International Society for Antimicrobial Chemotherapy expert meeting. Int. J. Antimicrob. Agents 54 400–406. 10.1016/j.ijantimicag.2019.08.013 [DOI] [PubMed] [Google Scholar]
- Kim G. Y., Jeon J. S., Kim J. K. (2016). Isolation Frequency Characteristics of Candida Species from Clinical Specimens. Mycobiology 44 99–104. 10.5941/MYCO.2016.44.2.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. N., Shin J. H., Sung H., Lee K., Kim E. C., Ryoo N., et al. (2009). Candida haemulonii and closely related species at 5 university hospitals in Korea: identification, antifungal susceptibility, and clinical features. Clin. Infect. Dis. 48 e57–e61. 10.1086/597108 [DOI] [PubMed] [Google Scholar]
- Koczula K. M., Gallotta A. (2016). Lateral flow assays. Essays Biochem. 60 111–120. 10.1042/EBC20150012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordalewska M., Lee A., Zhao Y., Perlin D. S. (2019). Detection of Candida auris Antifungal Drug Resistance Markers Directly from Clinical Skin Swabs. Antimicrob. Agents Chemother. 63 e1754–e1719. 10.1128/AAC.01754-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordalewska M., Zhao Y., Lockhart S. R., Chowdhary A., Berrio I., Perlin D. S. (2017). Rapid and accurate molecular identification of the emerging multidrug-resistant pathogen Candida auris. J. Clin. Microb. 55 2445–2452. 10.1128/JCM.00630-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost K., Yi J., Rogers B., Jerris R. (2017). Comparison of clinical methods for detecting carbapenem-resistant Enterobacteriaceae. Pract. Lab. Med. 8 18–25. 10.1016/j.plabm.2017.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Sachu A., Mohan K., Vinod V., Dinesh K., Karim S. (2017). Simple low cost differentiation of Candida auris from Candida haemulonii complex using CHROMagar Candida medium supplemented with Pal’s medium. Rev. Iberoam. Micol. 34 109–111. 10.1016/j.riam.2016.11.004 [DOI] [PubMed] [Google Scholar]
- Kwon Y. J., Shin J. H., Byun S. A., Choi M. J., Won E. J., Lee D., et al. (2019). Candida auris Clinical Isolates from South Korea: Identification. Antifung. Suscept. Genot. J. Clin. Microbiol. 57 e1618–e1624. 10.1128/JCM.01624-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladner J. T., Grubaugh N. D., Pybus O. G., Andersen K. G. (2019). Precision epidemiology for infectious disease control. Nat. Med. 25 206–211. 10.1038/s41591-019-0345-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach L., Russell A., Zhu Y., Chaturvedi S., Chaturvedi V. (2019). A rapid and automated sample-to-result Candida auris real-time PCR assay for high-throughput testing of surveillance samples with the BD Max Open system. J. Clin. Microb. 57 e619–e630. 10.1128/JCM.00630-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach L., Zhu Y., Chaturvedi S. (2018). Development and validation of a real-time PCR assay for rapid detection of Candida auris from surveillance samples. J. Clin. Microb. 56 e1217–e1223. 10.1128/JCM.01223-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W. G., Shin J. H., Uh Y., Kang M. G., Kim S. H., Park K. H., et al. (2011). First three reported cases of nosocomial fungemia caused by Candida auris. J. Clin. Microbiol. 49 3139–3142. 10.1128/JCM.00319-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima A., Widen R., Vestal G., Uy D., Silbert S. (2019). A TaqMan Probe-Based Real-Time PCR Assay for the Rapid Identification of the Emerging Multidrug-Resistant Pathogen Candida auris on the BD Max System. J. Clin. Microbiol. 57 e1604–e1618. 10.1128/JCM.01604-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart S. R., Etienne K. A., Vallabhaneni S., Farooqi J., Chowdhary A., Govender N. P., et al. (2017). Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin. Infect. Dis. 64 134–140. 10.1093/cid/ciw691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magobo R. E., Corcoran C., Seetharam S., Govender N. P. (2014). Candida auris–associated candidemia, South Africa. Emerg. Infect. Dis. 20:1250. 10.3201/eid2007.131765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malczynski M., Dowllow N., Rezaeian S., Rios J., Dirnberger L., Zembower J. A., et al. (2020). Optimizing a real-time PCR assay for rapid detection of Candida auris in nasal and axillary/groin samples. J. Med. Microb. 69 824–829. 10.1099/jmm.0.001207 [DOI] [PubMed] [Google Scholar]
- Marklein G., Josten M., Klanke U., Muller E., Horre R., Maier T., et al. (2009). Matrix-assisted laser desorption ionization-time of flight mass spectrometry for fast and reliable identification of clinical yeast isolates. J. Clin. Microbiol. 47 2912–2917. 10.1128/JCM.00389-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Murcia A., Navarro A., Bru G., Chowdhary A., Hagen F., Meis J. F. (2018). Internal validation of GPS() MONODOSE CanAur dtec-qPCR kit following the UNE/EN ISO/IEC 17025:2005 for detection of the emerging yeast Candida auris. Mycoses 61 877–884. 10.1111/myc.12834 [DOI] [PubMed] [Google Scholar]
- McDonald L. C., Gerding D. N., Johnson S., Bakken J. S., Carroll K. C., Coffin S. E., et al. (2018). Clinical Practice Guidelines for Clostridium difficile Infection in Adults and Children: 2017 Update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin. Infect. Dis. 66 e1–e48. 10.1093/cid/ciy149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar B. C., Xu J., Moore J. E. (2002). Risk assessment models and contamination management: implications for broad-range ribosomal DNA PCR as a diagnostic tool in medical bacteriology. J. Clin. Microbiol. 40 1575–1580. 10.1128/JCM.40.5.1575-1580.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintzer V., Moran-Gilad J., Simon-Tuval T. (2019). Operational models and criteria for incorporating microbial whole genome sequencing in hospital microbiology - A systematic literature review. Clin. Microbiol. Infect. 25 1086–1095. 10.1016/j.cmi.2019.04.019 [DOI] [PubMed] [Google Scholar]
- Mulet Bayona J. V., Salvador García C., Tormo Palop N., Gimeno Cardona C. (2020). Evaluation of a novel chromogenic medium for Candida spp. identification and comparison with CHROMagarTM Candida for the detection of Candida auris in surveillance samples. Diagn. Microbiol. Infect. Dis. 98:115168. 10.1016/j.diagmicrobio.2020.115168 [DOI] [PubMed] [Google Scholar]
- Mulet Bayona J. V., Salvador García C., Tormo Palop N., Gimeno Cardona C. (2021). Validation and implementation of a commercial real-time PCR assay for direct detection of Candida auris from surveillance samples. Mycoses 64 612–615. 10.1111/myc.13250 [DOI] [PubMed] [Google Scholar]
- Muthu M., Chun S., Wu H. F., Duncan M. W., Gopal J. (2018). The ongoing evolution of laser desorption/ionization mass spectrometry: Some observations on current trends and future directions. J. Mass Spectr. 53 525–540. 10.1002/jms.4083 [DOI] [PubMed] [Google Scholar]
- Mylonakis E., Clancy C. J., Ostrosky-Zeichner L., Garey K. W., Alangaden G. J., Vazquez J. A., et al. (2015). T2 magnetic resonance assay for the rapid diagnosis of candidemia in whole blood: a clinical trial. Clin. Infect. Dis. 60 892–899. 10.1093/cid/ciu959 [DOI] [PubMed] [Google Scholar]
- Neely L. A., Audeh M., Phung N. A., Min M., Suchocki A., Plourde D., et al. (2013). T2 magnetic resonance enables nanoparticle-mediated rapid detection of candidemia in whole blood. Sci. Transl. Med. 5:182ra54. 10.1126/scitranslmed.3005377 [DOI] [PubMed] [Google Scholar]
- NIHR Global Health Research Unit on Genomic Surveillance of AMR (2020). Whole-genome sequencing as part of national and international surveillance programmes for antimicrobial resistance: a roadmap. BMJ Global Health 5:e002244. 10.1136/bmjgh-2019-002244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien B., Chaturvedi S., Chaturvedi V. (2020). In vitro evaluation of antifungal drug combinations against multidrug-resistant Candida auris isolates from New York outbreak. Antimicrob. Agents Chemother. 64 e2195–e2119. 10.1128/AAC.02195-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowsky B., Greenko J., Adams E., Quinn M., O’Brien B., Chaturvedi V., et al. (2020). Candida auris Isolates Resistant to Three Classes of Antifungal Medications - New York, 2019. MMWR Morb. Mort. Weekly Rep. 69 6–9. 10.15585/mmwr.mm6901a2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacilli M., Kerins J. L., Clegg W. J., Walblay K. A., Adil H., Kemble S. K., et al. (2020). Regional emergence of Candida auris in Chicago and lessons learned from intensive follow-up at 1 ventilator-capable skilled nursing facility. Clin. Infect. Dis. 71 e718–e725. 10.1093/cid/ciaa435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pla L., Santiago-Felipe S., Tormo-Mas M. A., Ruiz-Gaitan A., Peman J., Valentin E., et al. (2021). Oligonucleotide-capped nanoporous anodic alumina biosensor as diagnostic tool for rapid and accurate detection of Candida auris in clinical samples. Emerg. Microbes Infect. 10 407–415. 10.1080/22221751.2020.1870411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polinski M., Hamilton D. B., Nowak B., Bridle A. (2013). SYBR, TaqMan, or both: highly sensitive, non-invasive detection of Cardicola blood fluke species in Southern Bluefin Tuna (Thunnus maccoyii). Mol. Biochem. Parasitol. 191 7–15. 10.1016/j.molbiopara.2013.07.002 [DOI] [PubMed] [Google Scholar]
- Prakash A., Sharma C., Singh A., Kumar Singh P., Kumar A., Hagen F. (2016). Evidence of genotypic diversity among Candida auris isolates by multilocus sequence typing, matrix-assisted laser desorption ionization time-of-flight mass spectrometry and amplified fragment length polymorphism. Clin. Microbiol. Infect. 2016:22. 10.1016/j.cmi.2015.10.022 [DOI] [PubMed] [Google Scholar]
- Qian J., Cutler J. E., Cole R. B., Cai Y. (2008). MALDI-TOF mass signatures for differentiation of yeast species, strain grouping and monitoring of morphogenesis markers. Anal. Bioanal. Chem. 392 439–449. 10.1007/s00216-008-2288-1 [DOI] [PubMed] [Google Scholar]
- Ruiz Gaitan A. C., Moret A., Lopez Hontangas J. L., Molina J. M., Aleixandre Lopez A. I., Cabezas A. H., et al. (2017). Nosocomial fungemia by Candida auris: First four reported cases in continental Europe. Rev. Iberoam. Micol. 34 23–27. 10.1016/j.riam.2016.11.002 [DOI] [PubMed] [Google Scholar]
- Ruiz-Gaitan A. C., Fernandez-Pereira J., Valentin E., Tormo-Mas M. A., Eraso E., Peman J., et al. (2018). Molecular identification of Candida auris by PCR amplification of species-specific GPI protein-encoding genes. Int. J. Med. Microbiol. 308 812–818. 10.1016/j.ijmm.2018.06.014 [DOI] [PubMed] [Google Scholar]
- Sarma S., Kumar N., Sharma S., Govil D., Ali T., Mehta Y., et al. (2013). Candidemia caused by amphotericin B and fluconazole resistant Candida auris. Indian J. Med. Microbiol. 31 90–91. 10.4103/0255-0857.108746 [DOI] [PubMed] [Google Scholar]
- Satoh K., Makimura K., Hasumi Y., Nishiyama Y., Uchida K., Yamaguchi H. (2009). Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol. Immunol. 53 41–44. 10.1111/j.1348-0421.2008.00083.x [DOI] [PubMed] [Google Scholar]
- Sattler J., Noster J., Brunke A., Plum G., Wiegel P., Kurzai O., et al. (2021). Comparison of Two Commercially Available qPCR Kits for the Detection of Candida auris. J. Fungi. 7:154. 10.3390/jof7020154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton D. J., Bentz M. L., Welsh R. M., Derado G., Furin W., Rose L. J., et al. (2021). Positive correlation between Candida auris skin-colonization burden and environmental contamination at a ventilator-capable skilled nursing facility in Chicago. Clin. Infect. Dis. 2021:327. 10.1093/cid/ciab327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton D. J., Kordalewska M., Bentz M. L., Welsh R. M., Perlin D. S., Litvintseva A. P. (2018b). Direct detection of emergent fungal pathogen Candida auris in clinical skin swabs by SYBR Green-based quantitative PCR assay. J. Clin. Microbiol. 56 e1318–e1337. 10.1128/JCM.01337-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton D. J., Bentz M. L., Welsh R. M., Litvintseva A. P. (2018a). Evaluation of a new T2 Magnetic Resonance assay for rapid detection of emergent fungal pathogen Candida auris on clinical skin swab samples. Mycoses 61 786–790. 10.1111/myc.12817 [DOI] [PubMed] [Google Scholar]
- Sharp A., Borman A. M., Perera N., Randle M., Braham S., Taori S., et al. (2018). Assessing routine diagnostic methods for detecting Candida auris in England. J. Infect. 77 448–454. 10.1016/j.jinf.2018.07.012 [DOI] [PubMed] [Google Scholar]
- Snayd M., Dias F., Ryan R. W., Clout D., Banach D. B. (2018). Misidentification of Candida auris by RapID Yeast Plus, a Commercial, Biochemical Enzyme-Based Manual Rapid Identification System. J. Clin. Microbiol. 56 e80–e18. 10.1128/JCM.00080-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita T., Takashima M., Poonwan N., Mekha N. (2006). Candida pseudohaemulonii Sp. Nov., an amphotericin B-and azole-resistant yeast species, isolated from the blood of a patient from Thailand. Microbiol. Immunol. 50 469–473. 10.1111/j.1348-0421.2006.tb03816.x [DOI] [PubMed] [Google Scholar]
- Sullivan D. J., Westerneng T. J., Haynes K. A., Bennett D. E., Coleman D. C. (1995). Candida dubliniensis sp. nov.: phenotypic and molecular characterization of a novel species associated with oral candidosis in HIV-infected individuals. Microbiology 141(Pt 7), 1507–1521. 10.1099/13500872-141-7-1507 [DOI] [PubMed] [Google Scholar]
- Tan Y. E., Teo J. Q., Rahman N. B. A., Ng O. T., Kalisvar M., Tan A. L., et al. (2019). Candida auris in Singapore: Genomic epidemiology, antifungal drug resistance, and identification using the updated 8.01 VITEK(®)2 system. Int. J. Antimicrob. Agents 54 709–715. 10.1016/j.ijantimicag.2019.09.016 [DOI] [PubMed] [Google Scholar]
- Theill L., Dudiuk C., Morales-Lopez S., Berrio I., Rodriguez J. Y., Marin A., et al. (2018). Single-tube classical PCR for Candida auris and Candida haemulonii identification. Rev. Iberoam Micol. 35 110–112. 10.1016/j.riam.2018.01.003 [DOI] [PubMed] [Google Scholar]
- Theodoropoulos N. M., Bolstorff B., Bozorgzadeh A., Brandeburg C., Cumming M., Daly J. S., et al. (2020). Candida auris outbreak involving liver transplant recipients in a surgical intensive care unit. Am. J. Transplant. 20 3673–3679. 10.1111/ajt.16144 [DOI] [PubMed] [Google Scholar]
- Tian S., Bing J., Chu Y., Chen J., Cheng S., Wang Q., et al. (2021). Genomic epidemiology of Candida auris in a general hospital in Shenyang, China: a three-year surveillance study. Emerg. Microbes Infect. 10 1088–1096. 10.1080/22221751.2021.1934557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang C. C., Chan J. F., Ip P. P., Ngan A. H., Chen J. H., Lau S. K., et al. (2014). Subcutaneous phaeohyphomycotic nodule due to Phialemoniopsis hongkongensis sp. nov. J. Clin. Microbiol. 52 3280–3289. 10.1128/JCM.01592-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsay S., Kallen A., Jackson B. R., Chiller T. M., Vallabhaneni S. (2018). Approach to the Investigation and Management of Patients With Candida auris, an Emerging Multidrug-Resistant Yeast. Clin. Infect. Dis. 66 306–311. 10.1093/cid/cix744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Belkum A., Welker M., Pincus D., Charrier J. P., Girard V. (2017). Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry in Clinical Microbiology: What Are the Current Issues? Ann. Lab. Med. 37 475–483. 10.3343/alm.2017.37.6.475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Poucke M., Van Zeveren A., Peelman L. J. (2012). [Letter to the editor] Combined FAM-labeled TaqMan probe detection and SYBR green I melting curve analysis in multiprobe qPCR genotyping assays. Biotechniques 52 81–86. 10.2144/000113808 [DOI] [PubMed] [Google Scholar]
- Vatanshenassan M., Boekhout T., Meis J. F., Berman J., Chowdhary A., Ben-Ami R., et al. (2019). Candida auris Identification and Rapid Antifungal Susceptibility Testing Against Echinocandins by MALDI-TOF MS. Front. Cell. Infect. Microbiol. 9:20. 10.3389/fcimb.2019.00020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walchak R. C., Buckwalter S. P., Zinsmaster N. M., Henn K. M., Johnson K. M., Koelsch J. M., et al. (2020). Candida auris Direct Detection from Surveillance Swabs, Blood, and Urine Using a Laboratory-Developed PCR Method. J. Fungi. 6:224. 10.3390/jof6040224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T. Z., White K. N., Scarr J. V., Simon M. S., Calfee D. P. (2020). Preparing your healthcare facility for the new fungus among us: An infection preventionist’s guide to Candida auris. Am. J. Infect. Control. 48 825–827. 10.1016/j.ajic.2020.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis C. V., Jutzeler C. R., Borgwardt K. (2020). Machine learning for microbial identification and antimicrobial susceptibility testing on MALDI-TOF mass spectra: a systematic review. Clin. Microbiol. Infect. 26 1310–1317. 10.1016/j.cmi.2020.03.014 [DOI] [PubMed] [Google Scholar]
- Welsh R. M., Bentz M. L., Shams A., Houston H., Lyons A., Rose L. J., et al. (2017). Survival, Persistence, and Isolation of the Emerging Multidrug-Resistant Pathogenic Yeast Candida auris on a Plastic Health Care Surface. J. Clin. Microbiol. 55 2996–3005. 10.1128/JCM.00921-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T. J., Bruns T., Lee S., Taylor J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, diagnostics and forensics,” in PCR Protocols:A guide to methods and applications, eds Innis M. A., Gelfand D. H., Sninsky J. J., White T. J. (San Diego: Academic Press; ). 10.1016/B978-0-12-372180-8.50042-1 [DOI] [Google Scholar]
- Wittwer C. T., Herrmann M. G., Moss A. A., Rasmussen R. P. (2013). Continuous fluorescence monitoring of rapid cycle DNA amplification. 1997. Biotechniques 54 314–320. 10.2144/000114043 [DOI] [PubMed] [Google Scholar]
- Won E. J., Shin J. H., Kim M.-N., Choi M. J., Joo M. Y., Kee S. J., et al. (2014). Evaluation of the BD Phoenix system for identification of a wide spectrum of clinically important yeast species: a comparison with Vitek 2-YST. Diagnost. Microbiol. Infect. Dis. 79 477–480. 10.1016/j.diagmicrobio.2014.05.011 [DOI] [PubMed] [Google Scholar]
- Yamamoto M., Alshahni M. M., Tamura T., Satoh K., Iguchi S., Kikuchi K., et al. (2018). Rapid Detection of Candida auris Based on Loop-Mediated Isothermal Amplification (LAMP). J. Clin. Microbiol. 56 e518–e591. 10.1128/JCM.00591-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Q., Xiao M., Fan X., Zhang G., Yang Y., Zhang J. J., et al. (2021). Evaluation of Autof MS 1000 and Vitek MS MALDI-TOF MS System in Identification of Closely-Related Yeasts Causing Invasive Fungal Diseases. Front. Cell Infect. Microbiol. 11:628828. 10.3389/fcimb.2021.628828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. X., Carroll K. C., Lewis S., Totten M., Mead P., Samuel L., et al. (2020). Multicenter Evaluation of a PCR-Based Digital Microfluidics and Electrochemical Detection System for the Rapid Identification of 15 Fungal Pathogens Directly from Positive Blood Cultures. J. Clin. Microbiol. 2020:58. 10.1128/JCM.02096-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., O’Brien B., Leach L., Clarke A., Bates M., Adams E., et al. (2020). Laboratory analysis of an outbreak of Candida auris in New York from 2016 to 2018: Impact and lessons learned. J. Clin. Microb. 58 e1503–e1519. 10.1128/JCM.01503-19 [DOI] [PMC free article] [PubMed] [Google Scholar]