Abstract
Ophiorrhiza pumila (O. pumila; Op) is a medicinal herbaceous plant, which can accumulate camptothecin (CPT). CPT and its derivatives are widely used as chemotherapeutic drugs for treating malignant tumors. Its biosynthesis pathway has been attracted significant attention. Teosinte branched 1/cycloidea/proliferating cell factors 1/2 (TCP) transcription factors (TFs) regulate a variety of physiological processes, while TCP TFs are involved in the regulation of CPT biosynthesis remain unclear. In this study, a systematic analysis of the TCP TFs family in O. pumila was performed. A total of 16 O. pumila TCP (OpTCP) genes were identified and categorized into two subgroups based on their phylogenetic relationships with those in Arabidopsis thaliana. Tissue-specific expression patterns revealed that nine OpTCP genes showed the highest expression levels in leaves, while the other seven OpTCPs showed a higher expression level in the stems. Co-expression, phylogeny analysis, and dual-luciferase (Dual-LUC) assay revealed that OpTCP15 potentially plays important role in CPT and its precursor biosynthesis. In addition, the subcellular localization experiment of candidate OpTCP genes showed that they are all localized in the nucleus. Our study lays a foundation for further functional characterization of the candidate OpTCP genes involved in CPT biosynthesis regulation and provides new strategies for increasing CPT production.
Keywords: Ophiorrhiza pumila, TCP transcription factors, genome-wide analysis, expression pattern, CPT biosynthesis
Introduction
Ophiorrhiza pumila (O. pumila), belonging to the family Rubiaceae, is an important herbaceous medicinal plant and can accumulate camptothecin (CPT). CPT is a quinoline-type monoterpenoid indole alkaloid and an anticancer compound with potent DNA topoisomerase I inhibitory activity (Johnson et al., 2018). Two semi-synthetic water-soluble CPT analogs known as topotecan and irinotecan have been approved for the treatment of ovarian, colorectal, lung and cervix cancer, and HIV (Liu et al., 2015; Martino et al., 2017). Additionally, a number of other CPT derivatives have shown promising results in preclinical and clinical trials (Xie et al., 2016). Despite CPT with pharmacological relevance, its biosynthesis pathway is complex and partially deciphered (Sadre et al., 2016; Yang et al., 2021). CPT is produced by the iridoid and shikimate pathways, which supply the important precursors, such as secologanin and tryptamine. Subsequently, secologanin and tryptamine are converged into strictosidine, which is finally synthesized into CPT by some unknown catalytic enzymes (Supplementary Figure 1). At present, genomes and transcriptome data of many monoterpenoid indole alkaloid-producing plants, such as O. pumila, Catharanthus roseus, Camptotheca acuminata, and Nothapodytes nimmoniana, provided the foundation for understanding the biosynthesis and regulatory components of plant-specialized metabolites, followed by molecular characterization and functional validation of candidate genes (Kellner et al., 2015; Rather et al., 2018; Kang et al., 2021; Rai et al., 2021). At present, a few genes-encoding key enzymes, such as OpTDC, OpTDC2, OpSTR, OpCPR, OpG10H, OpSLS, OpLAMT, in the CPT biosynthetic pathway and several transcription factors (TFs), such as OpMYB1, OpWRKY1/2/3, and OpERF1/2/3, have been cloned and analyzed in O. pumila (Kai et al., 2015; Rohani et al., 2016; Udomsom et al., 2016; Shi et al., 2020; Hao et al., 2021; Yang et al., 2021; You et al., 2021). For example, OpWRKY2 acted as a positive regulator of CPT biosynthesis by directly binding and activating the gene OpTDC (Hao et al., 2021). Suppression of OpERF2 resulted in reducing expression of genes in the early steps that supplied a precursor for CPT biosynthesis, such as OpTDC, OpG10H, OpSLS, and OpSTR (Udomsom et al., 2016). Nevertheless, limited information is available about the regulatory mechanism of CPT biosynthesis in O. pumila, especially teosinte branched 1/cycloidea/proliferating cell factors 1/2 (TCP) TF.
The TCP family is a group exclusively present in the higher plants (Liu et al., 2019). The name of TCP was derived from three members of this family identified: teosinte branched 1 (TB1) in Zea mays (Doebley et al., 1995), cycloidea (CYC) in Antirrhinum majus (Luo et al., 1996), and proliferating cell factors 1/2 (PCF1/2) in Oryza sativa (Kosugi and Ohashi, 1997; Cubas et al., 1999). These proteins contained a highly conserved 55–59 residue-long basic helix-loop-helix (bHLH) structure at the N-terminus, known as TCP domain (Aggarwal et al., 2010). It was associated with protein nuclear localization, DNA binding, and protein–protein interaction (Cubas et al., 1999). According to the differential features in TCP domains, TCP family members were divided into two subfamilies: class I (also called PCF or TCP-P) and class II (TCP-C) (Martín-Trillo and Cubas, 2010). The most striking difference between these two subfamilies was the deletion of four-amino-acids at the N-terminal, which was exclusive in class I. Class II TCP proteins were further subdivided into the cincinnata (CIN) and cycloidea (CYC)/TB1 subclasses based on the alterations in several amino acids (Howarth and Donoghue, 2006; Horn et al., 2015). Besides the TCP domain, some members within class II had the R domain (18–20 residue arginine-rich motifs) and glutamic acid-cysteine-glutamic acid (ECE) motif (glutamic acid-cysteine-glutamic acid stretch) (Lupas et al., 1991).
Teosinte branched 1/cycloidea/proliferating cell factors 1/2 proteins were involved in diverse physiological and biological processes, such as phytohormone biosynthesis, transport, and signal transduction, leaf development, branching, embryonic growth, floral organ morphogenesis, pollen development, germination, senescence, circadian clock, and cell cycle regulation (Danisman et al., 2012; Manassero et al., 2013). Class I TCP genes, A. thaliana TCP (AtTCP)14 and AtTCP15, played a vital role in seed germination and promoted embryo growth via the gibberellin signaling pathway (Resentini et al., 2015). For CIN subclass TCP genes, repression of AtTCP3, AtTCP4, and AtTCP24 could disturb leaf development (Palatnik et al., 2003; Nag et al., 2009; Sarvepalli and Nath, 2011). Overexpression of CYC-like homolog GhCYC2 caused disk flowers to obtain characteristics typical for ray flowers in Gerbera hybrida (Broholm et al., 2008). Furthermore, several emerging lines of evidence revealed the multi-faceted role of TCP protein in plant specialized metabolism. For example, AtTCP3 interacted with R2R3-MYB proteins to stimulate flavonoid production in Arabidopsis thaliana, while AtTCP15 repressed anthocyanin biosynthesis when the plants were under high light intensity (Li and Zachgo, 2013; Viola et al., 2016). In Lycium ruthenicum, LrTCP4 performed as a positive regulator in kukoamine biosynthesis and other secondary metabolites (Chahel et al., 2019). In Artemisia annua (A. annua), AaTCP14 and AaTCP15 were essential for jasmonate (JA)-induced artemisinin biosynthesis (Ma et al., 2018, 2021).
To date, genome-wide identification of the TCP family members has been identified and characterized in many dicots and monocots plants, such as A. thaliana (Li, 2015), Solanum lycopersicum (Parapunova et al., 2014), Malus domestica (Xu et al., 2014), Citrullus lanatus (Shi et al., 2016), Nicotiana tabacum (Chen et al., 2016), Fragaria vesca (Wei et al., 2016), Glycine max (Feng et al., 2018), Gossypium barbadense (Zheng et al., 2018), Vitis vinifera (Leng et al., 2019), Solanum tuberosum (Bao et al., 2019), Phyllostachys edulis (Liu et al., 2018), Z. mays (Ding et al., 2019), Panicum virgatum (Zheng et al., 2019), Hordeum vulgare subsp. vulgare (Gao et al., 2021), and so on. However, there is no research about the identification and functional characterization of TCP proteins in CPT-producing plants. Here, we first present a detailed and comprehensive analysis of the TCP gene family through the whole O. pumila genome, such as identification of all TCP family members, phylogenetic relationships, conserved motifs, gene structure, cis-elements in gene promoter regions, the expression levels of O. pumila TCP (OpTCP) genes in diverse tissues, co-expression analysis of key enzymes involved in CPT biosynthesis, and subcellular localization, which could provide valuable information for understanding its classification and functions in O. pumila.
Materials and Methods
Identification and Characteristics of Teosinte Branched 1/Cycloidea/Proliferating Cell Factors 1/2 Gene Family in Ophiorrhiza pumila
All annotated protein sequences from O. pumila genome database1 were obtained to comprehensively identify genomic TCP TFs (Rai et al., 2021). Moreover, to avoid missing any OpTCP genes, the Hidden Markov Model (HMM) of the TCP domain (Pfam, PF03634) was applied as a query to blast all TCP-containing sequences against our protein database by HMMER3.2 software.2 Then, all candidate OpTCPs were manually further validate using the online programs of CDD,3 Simple Modular Architecture Research Tool (SMART),4 and Pfam5 confirmed the existence of core domains. Finally, the TCP gene members were identified in O. pumila, after removing incorrect and redundant predicted proteins. The molecular weights (MWs), amino acid lengths, and isoelectric points (pI) of each OpTCP protein were computed by the ExPASy website6 (Gasteiger et al., 2005). The subcellular localization of putative OpTCPs protein was predicted by PSORT7 and pLoc-mPlant.8 In addition, the sequences of the 24 Arabidopsis TCP proteins were downloaded from the Arabidopsis Information Resource (TAIR).9 The sequences of the 23 O. sativa and 18 Coffea canephora TCP proteins were retrieved from PlantTFDB database.10
Chromosomal Localization and Gene Duplication
The physical locations of OpTCP genes on chromosomes were obtained from O. pumila genome database and visualized by TBtools software.11 Multiple collinear scanning toolkits (MCScanX) and the Basic Local Alignment Search Tool (BLASTP) methods were used to analyze gene duplication events. Tandem repeats were identified based on the criteria defined in the previous report (Cheng et al., 2016), in which two or more genes should be located within a 100 kbp window and displayed at least 70% sequence similarity. The synonymous relationship between OpTCP genes and Arabidopsis, rice, and coffee TCP genes was visualized by TBtools software.
Multiple Sequence Alignment and Phylogenetic Analysis
Multi-sequence alignments of all conserved TCP core domains were determined using DNAMAN 6.0 software. The aligned sequences were visualized with WEBLOGO program12 for the conserved amino acid residues analysis. To investigate phylogenetic relationships of OpTCPs and assist their classification, the full-length amino acid sequences of 24 AtTCP, 23 O. sativa TCP (OsTCP), 18 C. canephora TCP (CcTCP), 16 OpTCP, and some functional TCPs were aligned with CLUSTAL software. The protein sequences of those genes involved in the regulation of special secondary metabolism can be found under the following accession numbers: AtTCP3 (At1g53230), AtTCP15 (At1g69690), AaTCP14 (AYF60463.1), AaTCP15 (QKD77227.1), and MdTCP46 (MDP0000319941). The phylogenetic tree was constructed using Neighbor-joining (NJ) method implemented with MEGA 7.0, and its reliability was tested using bootstrapping with 1,000 replicates (Kumar et al., 2016). The display of the phylogenetic tree was performed by Interactive Tree of Life (iTOL).13
Gene Structure and Motif Composition Analysis
The exon-intron structures of OpTCP genes were determined by aligning their genomic sequences with corresponding coding sequences (CDS), while diagrams were visualized with online the program Gene Structure Display Server (GSDS v2.0)14 (Hu et al., 2015). Additionally, the conserved motifs of OpTCP proteins were investigated using Multiple Expectation-Maximization for Motif Elicitation online program (MEME v5.1.1)15 with the following parameters: the number of motifs searched was set as 10; optimum motif width of 6–100 amino acids; and sites of per motif set to ≥2 and ≤600 (Bailey et al., 2009). Subsequently, the TBtools software was used to display and re-edited the gene structure and conserved motif.
Promoter cis-Acting Elements Analysis
To analyze the promoter regions of the OpTCPs and key enzyme genes involved in CPT biosynthesis, genomic DNA sequences in the promoter region (−3,000 to −1 bp) were extracted by the TBtools. Identification of candidate CPT biosynthetic pathway genes in O. pumila by sequence identifies with characterized genes from the prestrictosidine biosynthetic pathway in O. pumila or C. roseus (Supplementary Table 1). These CDS sequences were mapped into the O. pumila genome to find the corresponding promoter regions. Subsequently, the promoter sequences of each OpTCP gene were scanned by Plantpan 3.016 database to predicate and identify cis-acting regulatory elements (Chow et al., 2019). Meanwhile, the identification of TBS elements on the promoter of 15 key enzyme genes in the CPT biosynthetic pathway was scanned with conserved TCP-binding sites by blast methods, such as GGNCCCAC, GGNCC, GCCCR, or G(T/C)GGNCCC (Aggarwal et al., 2010).
Plant Materials, RNA Extraction, and Quantitative Real-Time PCR Analysis
Sterile O. pumila seedlings were cultured on solid B5 medium (pH 5.5) under controlled glasshouse conditions at 25°C and 14 h light/10 h dark photoperiod. The roots, stems, and leaves of 2-month-old O. pumila seedlings were collected to detect the tissue expression pattern of the OpTCP and CPT biosynthetic pathway genes. All of the samples were immediately frozen in liquid nitrogen and then stored at −80°C until used for RNA extraction.
Total RNA was extracted via the RNApure Plant Kit (Tiangen, China). Then cDNA synthesis was performed with PrimeScriptTM II First Strand cDNA Synthesis Kit (Tiangen, China). qRT−PCR was performed by StepOnePlus platform (Bio-Rad, Hercules, CA, United States) with a SYBR Green PCR Master Mix Kit (SYBR® Premix Ex TaqTM, Japan). Transcript abundance was calculated relative to OpUBQ (Ubiquitin) by 2–ΔΔCt method. Primers were listed in Supplementary Table 2. Co-expression analysis of candidate genes was performed by Pearson’s correlation test, and coefficients >0.8 indicated co-expression. Co-expression of candidate genes has been re-visualized as a network figure by Cytoscape_v3.7.2.
Dual-Luciferase Assay
To investigate the ability of OpTCP15 to regulate the expression of CPT biosynthesis pathway genes, the full-length coding sequence of OpTCP15 was amplified and inserted into the pHB-yellow fluorescent protein (YFP) vector (effectors). The promoter regions of Op7DLH and Op8HGO were ligated into the pGreenII0800-LUC vector (reporters). The Renilla luciferase gene driven by the CaMV 35 S promoter was used as an internal control. Empty pHB-YFP was used as the negative control for the effector. Infiltration and detection were performed as described previously (Hao et al., 2021). The ratio of firefly luciferase to Renilla luciferase represents the relative activity of the promoter. All experiments were repeated three times for each combination. All primers used for these constructs are listed in Supplementary Table 2.
Subcellular Localization Assay
To investigate the subcellular localization of candidate OpTCP proteins, the full-length coding sequences were inserted into the pHB-YFP vector. The pHB-YFP (empty vector) was used as the negative control. The plasmids pHB-OpTCPs-YFP and pHB-YFP were transformed into the Agrobacterium tumefaciens strain GV3101, respectively. Then strains GV3101 harboring OpTCPs-YFP and pHB-YFP were transiently infected the epidermal cells of 5-week-old N. benthamiana leaves. YFP fluorescences were analyzed 2 days after infiltration with an LSM880 confocal laser microscope (Carl Zeiss, Germany). Nuclei were stained with 4’ 6-diamidino-2-phenylindole (DAPI, Sigma), and three biological replicates were performed to verify the results (Hao et al., 2021).
Results
Identification of Teosinte Branched 1/Cycloidea/Proliferating Cell Factors 1/2 Family Members in the Ophiorrhiza pumila Genome
To identify TCP genes in O. pumila, an HMM search was conducted using the HMM profiles of TCP domain (PF03634) as queries against the O. pumila genome dataset (see text footnote 1). A total of 16 non-redundant TCP genes were obtained and named as OpTCP1 to OpTCP16 according to their order in the O. pumila genomic sequence (Table 1). Each candidate gene was further analyzed to confirm the integrity of the TCP domain of TCP proteins with the online program of CDD, SMART, and Pfam. Meanwhile, MWs, amino acid lengths, pI, and subcellular location of OpTCP proteins were analyzed (Table 1). The MWs of OpTCP proteins ranged from 18.91 (OpTCP1) to 54.25 (OpTCP13) kDa, with an average of 37.51 kDa. The protein lengths were distributed from 174 (OpTCP1) to 504 (OpTCP13) amino acids, and pI varied from 5.60 (OpTCP11) to 10.01 (OpTCP6). They were all predicted to be located in the nucleus.
TABLE 1.
Detailed information for 16 OpTCP genes in the O. pumila genome.
| ID | Gene name | Type | Chr | Start | Stop | Strand | No. of Exon | CDS (bp) | Protein (aa) | MWs (Da) | pI | Loc |
| Opuchr01_g0001120-1.1 | OpTCP1 | PCF | chr01 | 578509 | 577985 | − | 1 | 525 | 174 | 18907.37 | 9.15 | Nucleus |
| Opuchr04_g0008760-1.1 | OpTCP2 | CYC/TB1 | chr04 | 4881540 | 4880265 | − | 2 | 1,179 | 392 | 44582.07 | 8.36 | Nucleus |
| Opuchr04_g0064620-1.1 | OpTCP3 | CIN | chr04 | 32348228 | 32347020 | − | 1 | 1,209 | 402 | 44514.21 | 7.39 | Nucleus |
| Opuchr05_g0002240-1.1 | OpTCP4 | CIN | chr05 | 1225789 | 1226784 | + | 1 | 996 | 331 | 36943.08 | 6.37 | Nucleus |
| Opuchr05_g0060100-1.1 | OpTCP5 | CIN | chr05 | 27883069 | 27884421 | + | 1 | 1,353 | 450 | 48977.51 | 6.59 | Nucleus |
| Opuchr06_g0114980-1.1 | OpTCP6 | PCF | chr06 | 56622961 | 56623614 | + | 1 | 654 | 217 | 23276.48 | 10.01 | Nucleus |
| Opuchr07_g0000090-1.1 | OpTCP7 | PCF | chr07 | 67153 | 67995 | + | 1 | 843 | 280 | 29639.13 | 8.69 | Nucleus |
| Opuchr07_g0005290-1.1 | OpTCP8 | CIN | chr07 | 2963107 | 2964165 | + | 1 | 1,059 | 352 | 38817.75 | 6.79 | Nucleus |
| Opuchr07_g0006620-1.1 | OpTCP9 | CYC/TB1 | chr07 | 3648191 | 3647069 | − | 2 | 1,203 | 400 | 44972.86 | 9.22 | Nucleus |
| Opuchr07_g0038750-1.1 | OpTCP10 | PCF | chr07 | 19064610 | 19063813 | − | 1 | 798 | 265 | 27475.75 | 9.51 | Nucleus |
| Opuchr09_g0004960-1.1 | OpTCP11 | PCF | chr09 | 3099165 | 3100307 | + | 1 | 1,143 | 380 | 39829.61 | 5.6 | Nucleus |
| Opuchr09_g0016060-1.1 | OpTCP12 | CIN | chr09 | 10055733 | 10054717 | − | 1 | 1,017 | 338 | 37364.12 | 6.26 | Nucleus |
| Opuchr09_g0061970-1.1 | OpTCP13 | CIN | chr09 | 28382461 | 28380947 | − | 1 | 1,515 | 504 | 54252.12 | 7.8 | Nucleus |
| Opuchr10_g0056950-1.1 | OpTCP14 | PCF | chr10 | 28389738 | 28390955 | + | 1 | 1,218 | 405 | 42082.31 | 7.02 | Nucleus |
| Opuchr11_g0082210-1.1 | OpTCP15 | PCF | chr11 | 36501516 | 36502670 | + | 1 | 1,155 | 384 | 41199.34 | 7.86 | Nucleus |
| Opuchr11_g0088190-1.1 | OpTCP16 | PCF | chr11 | 39951958 | 39952734 | + | 1 | 777 | 258 | 27359.43 | 8.57 | Nucleus |
AA, amino acid residues; Chr, chromosome; MW, molecular weight; pI, theoretical isoelectric point; Loc, subcellular location; OpTCP, O. pumila TCP.
Chromosome Localization and Duplication of the OpTCP Gene Family
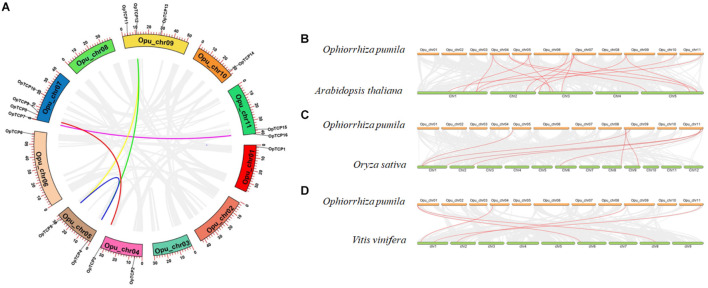
Sixteen OpTCP genes were disproportionately distributed on 8 of 11 O. pumila chromosomes (Figure 1A and Supplementary Figure 2). There was no distribution on Chr 2, 3, and 8. Four OpTCP genes on Chr 7; three OpTCP genes on Chr 9; two on Chr 4, Chr 5, and Chr 11; only one on Chr 1, Chr 6, and Chr 10. The possible relationships with the OpTCP genes and potential gene duplication type, collinear analyses were investigated in O. pumila genome. As illustrated in Figure 1A, seven genes involved in three segmental duplication events, such as Opu_chr05 (OpTCP4, 5)/Opu_chr09 (OpTCP12), Opu_chr04 (OpTCP3)/Opu_chr07 (OpTCP8), and Opu_chr07 (OpTCP7)/Opu_chr11 (OpTCP16). In contrast, no tandem duplication events were observed, suggesting that segmental duplications were the main causes for the amplification of the OpTCP gene family.
FIGURE 1.
The chromosomal location and synteny analysis of OpTCP genes. (A) Circos diagram illustrates the chromosomal locations of OpTCP genes and their synteny. The colored lines display similarity of different genes. (B–D) Synteny analysis of TCP genes between O. pumila and Arabidopsis, and rice, and V. vinifera, respectively. TCP, teosinte branched 1/cycloidea/proliferating cell factors 1/2; OpTCP, O. pumila TCP.
Microsynteny and Evolutionary Patterns of the OpTCP Genes in O. pumila
To further explore the functions and evolutionary relationships of TCP genes, large-scale comparative synteny maps between O. pumila and Arabidopsis, and O. sativa or V. vinifera were analyzed at genome-wide levels (Figures 1B–D). As a result, a total of 15 pairs of syntenic TCP genes were identified between O. pumila and Arabidopsis (Figure 1B and Supplementary Table 3), while 7 and 5 pairs of TCP genes were identified between O. pumila, O. sativa, and V. vinifera (Figures 1C,D and Supplementary Table 3), respectively. Among the synteny events, O. pumila between Arabidopsis and 5 OpTCP genes were found to be associated with two synteny events, such as OpTCP2-AtTCP12/AtTCP18, OpTCP3-AtTCP5/AtTCP17, and OpTCP5-AtTCP3/AtTCP4 (Supplementary Table 3). Interestingly, four of these five genes were in CIN and CYC/TB1subclade, indicating higher conservation of CIN and CYC/TB1 than PIF subclade in the TCP gene family. In addition, the OpTCP15 gene had a homologous relationship in all three plants, indicating that this gene may have an important role in evolution.
Phylogenetic Analysis and Classification of OpTCP Genes
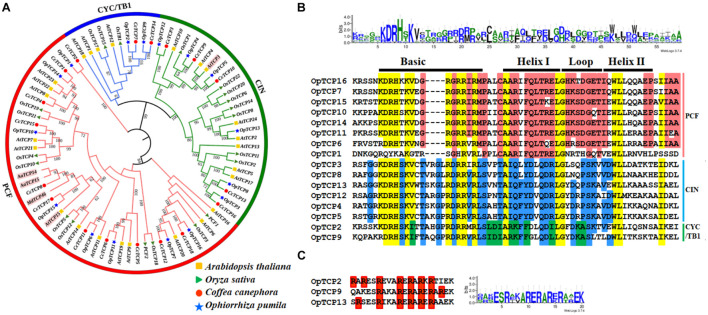
To explore the phylogenetic and evolutionary relationship of the TCP genes in O. pumila and group them with the established subfamilies, an unrooted NJ phylogenetic tree was constructed (Figure 2A). Sixteen OpTCP genes were clustered into classes I and II, each of which contained eight members. Additionally, class II could be further divided into the CIN and CYC/TB1 subgroups, which contained 6 (OpTCP3, OpTCP4, OpTCP5, OpTCP8, OpTCP12, and OpTCP13) and two TCP members (OpTCP2 and OpTCP9), respectively.
FIGURE 2.
Identification of TCP transcription factor genes in O. pumila. (A) Phylogenetic analysis of TCP proteins using the un-rooted Neighbor-Joining (NJ) method with 1,000 bootstrap replicates. Five TCP genes involved in special secondary metabolism are marked with pink shades. (B) The conserved TCP domains analysis using WebLogo and multiple sequence alignments by ClustalW. Each letter represents one amino acid, and the left column corresponds to the name of the gene. The yellow region indicates the highly conserved residues of the OpTCP family members, and the red region indicates residues conserved only in the class I subfamily. The blue region indicates residues conserved in the Class II subfamily. Green indicates the residues conserved in the CYC/TB1 class of proteins. The top black bar represents the conserved domain in TCP proteins. (C) The R domain in Class II members of the OpTCP gene family. TCP, teosinte branched 1/cycloidea/proliferating cell factors 1/2; OpTCP, O. pumila TCP.
Multiple sequences alignments showed that two subgroups were distinguished by a four-amino-acids deletion at the N-terminal of class I (Figure 2B). Additional, three class II proteins, OpTCP2 and OpTCP9 from class II CYC/TB1, and OpTCP13 from CIN also shared an R domain at the C-terminus of the TCP domain (Figure 2C).
Gene Structure and Motif Analysis of OpTCP Genes
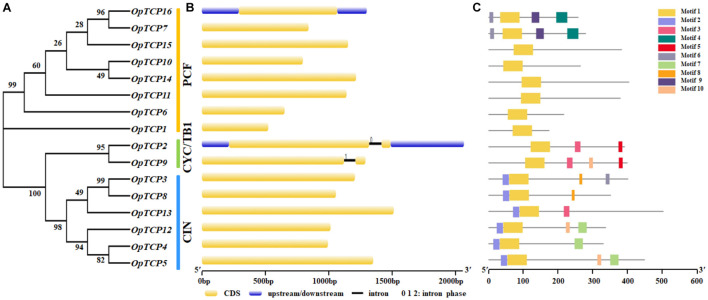
To further understand the pivotal role that exon-intron structural features play in the evolution of O. pumila gene families, the structure of OpTCP genes was obtained through exon-intron organization analysis. The phylogenetic tree (Figure 3A) revealed the most OpTCP proteins in the same group with similar genetic structures, such as the length and number of the exon. As shown in Figure 3B, class I and class II CIN-type TCP genes had only one exon and no intron, while class II CYC/TB1-type TCP genes contained two exons. These results suggested that different OpTCP members tended to share different structure organizations.
FIGURE 3.
Phylogenetic analysis, gene structure, and conserved motifs of TCP family in O. pumila. (A) The conserved TCP domain sequences of OpTCP proteins were constructed a Neighbor-Joining phylogenetic tree and the bootstrap test was performed with 1,000 iterations. (B) Exon-intron structure of the OpTCP genes. Blue indicates untranslated 5′- and 3′-regions, yellow indicates exons; black indicates introns; 0, 1, 2 indicates intron phase. (C) Distribution of conserved motifs of OpTCP proteins. Different motifs are shown by different colors numbered 1–10. TCP, teosinte branched 1/cycloidea/proliferating cell factors 1/2; OpTCP, O. pumila TCP.
The conserved motifs of TCP family proteins in O. pumila were analyzed by MEME online software, and 10 motifs were identified (Supplementary Table 4). These 10 motifs were distributed across different subgroups in the phylogenetic tree (Figure 3C). For OpTCP proteins, motif 1 was broadly distributed in all OpTCP proteins, which was corresponded to TCP domain. The OpTCPs in subfamily CIN had motif 2 and all members of the CYC/TB1 subfamily and OpTCP13 from CIN contained the motif 3 (R domain). Generally, proteins with similar motif compositions were clustered in the same class indicating that members of the same class may have similar functions.
Analysis of cis-Acting Elements
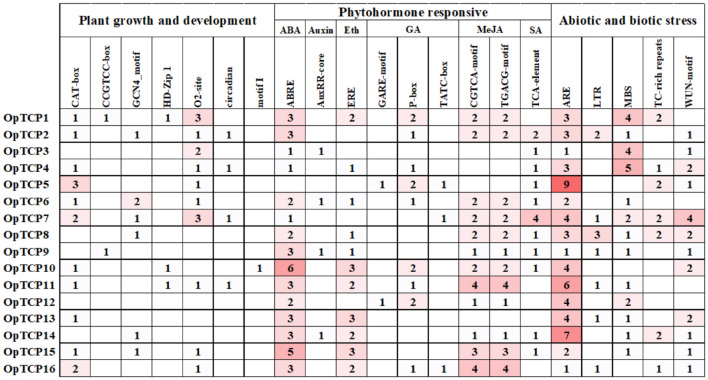
To further investigate the gene function and regulation mechanism of OpTCP genes, the cis-acting elements in promoter sequences were analyzed by Plantpan 3.0 software. As a result, a variety of cis-acting elements involved in plant growth and development, hormone responses, and stress responses were identified. As shown in Figure 4, CAT-box and CCGTCC-box involved in meristem expression were identified in the promoter region of 11 and 2 OpTCP genes, respectively. GCN4_motif was related to endosperm expression in plant growth and development. The zein metabolism regulation element (O2-site) and circadian control element (circadian) were found in 10 and 4 OpTCP genes, respectively. Additionally, leaf development correlated cis-acting regulatory elements (HD-Zip 1) and root-specific (motif I) regulatory elements were also found in the promoter region of the OpTCP genes (Supplementary Table 5).
FIGURE 4.
Promoter cis-regulatory elements analysis of the OpTCP genes. Based on the functional annotation, the cis-acting elements were classified into three major classes: plant growth and development, phytohormone responsive, or abiotic and biotic stresses-related cis-acting elements (detailed results shown in Supplementary Table 5).
In hormone responses, ABA-responsive cis-acting regulatory elements (ABREs) were found in the promoter region of 15 OpTCP genes, except for the OpTCP5 gene. Two MeJA-responsive elements (CGTCA-motif and TGACG-motif) were found in the promoter region of 12 OpTCP genes. In addition, auxin-responsive element (AuxRR-core), ethylene-responsive element (ERE), three gibberellin-responsive elements (GARE-motif, P-box, and TATC-box), and salicylic acid-responsive element (TCA-element) were found in the promoter region of 4, 11, 10, and 11 OpTCP genes, respectively (Supplementary Table 5).
In stress responses, ARE elements essential for the anaerobic induction were found in the promoter region of 16 OpTCP genes. Moreover, low temperature-responsive element (LTR), drought-inducibility element (MBS), TC-rich repeats, and wound-responsive element (WUN-motif) were also found in the promoter region of 7, 13, 7, and 12 OpTCP genes, respectively (Supplementary Table 5).
Expression Patterns of OpTCP Genes in Various Tissues
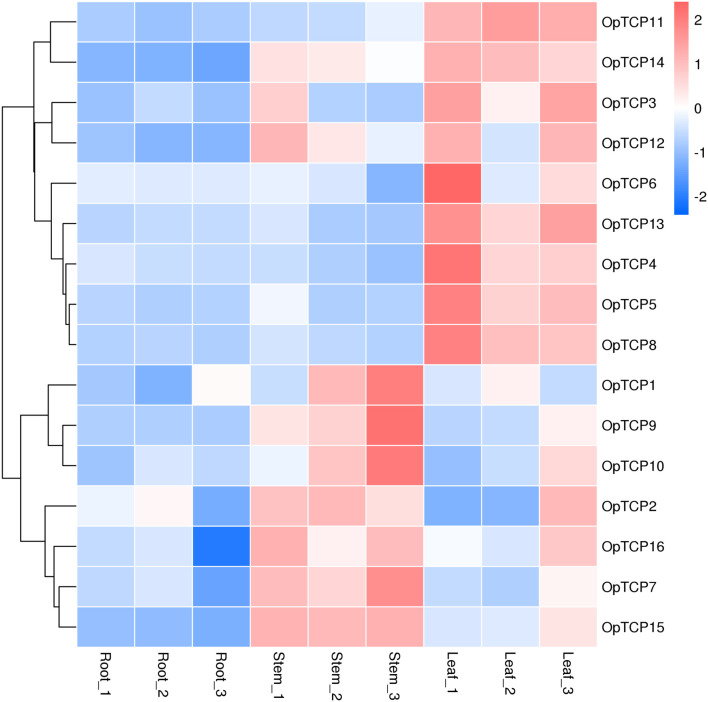
To validate the gene expression profiles, roots, stems, and leaves of O. pumila were collected for RNA extraction and quantitative Real-Time PCR (qRT-PCR) analysis. We performed hierarchical clustering with the expression data and accomplished a heatmap to visualize the expression profiles of the OpTCPs in different tissues (Figure 5). From the heatmap, most of the OpTCP genes preferentially expressed in leaves or stems. For example, 9 OpTCP genes (OpTCP3, 4, 5, 6, 8, 11, 12, 13, and 14) had the highest expression levels in leaves, while the other OpTCPs (OpTCP1, 2, 7, 9, 10, 15, and 16) showed a higher expression level in stems. In addition, OpTCP7, 10, 14, 15, and 16 consistently had high expression level in all tissues, while OpTCP1, 2, 3, 6, and 12 with low expression level.
FIGURE 5.
Expression patterns of OpTCP genes in different tissues. TCP, teosinte branched 1/cycloidea/proliferating cell factors 1/2; OpTCP, O. pumila TCP.
Co-expression Analyses of Candidate Camptothecin Biosynthesis OpTCPs
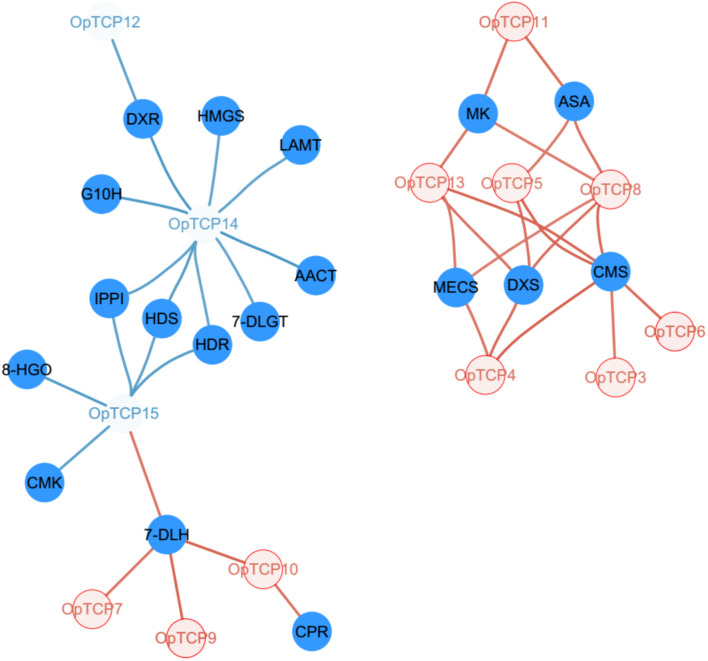
Analysis was conducted to determine the co-expression of all OpTCPs with target genes in CPT biosynthesis pathways. The expression levels of key enzyme genes, such as OpG10H, Op10HGO, Op8HGO, OpSLS1, Op7-DLGT, OpTDC, OpLAMT, OpIS, OpIO, and OpSTR, were extremely higher in roots while Op7-DLH and OpCPR showed higher expression levels in stems (Supplementary Figure 3). The gene co-expression analysis revealed that six OpTCP (OpTCP3, 4, 5, 6, 8, and 13) genes were in strong positive correlations with 2-C-methyl-Derythritol 4-phosphate (MEP) pathway genes (Pearson correlation coefficient r > 0.8 and p < 0.05, Figure 6 and Supplementary Table 6), while OpTCP12, OpTCP14, and OpTCP15 genes were in negative correlations with MEP pathway genes. In addition, OpTCP14 and OpTCP15 were in strong negative correlations with OpG10H, Op7-DLGT, OpLAMT, and Op8HGO (r < −0.8), respectively. Moreover, the Pearson coefficients of four OpTCPs (OpTCP7, 9, 10, and 15) and Op7-DLH were >0.8 (Supplementary Table 6). Overall, this result suggested that 12 of 16 OpTCPs might be associated with CPT and its precursor biosynthesis.
FIGURE 6.
Regulatory network of TCP transcription factors and key CPT-biosynthetic genes. The colored fonts represent TCP TFs and the solid blue circles represent CPT-biosynthetic genes. The edges are drawn when the absolute value of the linear correlation coefficient is >0.8. The blue and red line represents the negative correlation and positive correlation, respectively. TCP, teosinte branched 1/cycloidea/proliferating cell factors 1/2; CPT, camptothecin.
To identify the OpTCP TFs, which potentially involved in the regulation of CPT biosynthesis, we analyzed the phylogenetic relationships between the candidate OpTCPs and functional TCP TFs that regulated specialized metabolite biosynthesis (i.e., AaTCP14, AaTCP15, AtTCP3, AtTCP15, and MdTCP46). These TFs were grouped into two clades in the neighbor-joining tree (Figure 2A). The OpTCP6, OpTCP7, OpTCP10, OpTCP11, OpTCP14, and OpTCP15 proteins clustered with AaTCP14, AaTCP15, AtTCP15, and MdTCP46 in clade I, whereas AtTCP3 and other OpTCPs belonged to clade II. Interestingly, OpTCP5 and OpTCP15 clustered with AtTCP3 and AtTCP15, respectively, and may be most likely participated in regulating CPT and its precursor biosynthesis.
Finally, to identify the cis-elements of TCP TFs, the promoter sequences of the genes encoding enzymes in CPT biosynthesis pathway were analyzed. These results showed that the sequences of GGNCC and GCCCR were identified in the promoter regions of most studied genes (Supplementary Figure 4 and Supplementary Table 7). For example, there were 18, 14, and 9 TBS-binding sites on the promoter sequences of OpSLS, Op8HGO, and Op7DLH genes in CPT biosynthetic pathway, respectively. The finding indicated that the expression of these genes might be regulated by TCP TFs. Overall, OpTCP5 and OpTCP15 were likely to have a functional role in CPT and its precursor biosynthesis.
Dual Luciferase Assay
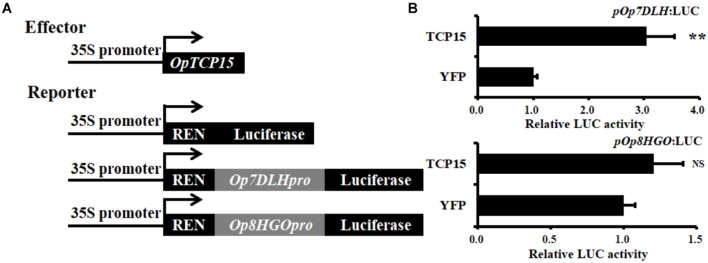
Subsequently, the dual luciferase (Dual-LUC) assay was performed to verify whether OpTCP15 protein affected the transcription of Op7DLH and Op8HGO or not. The results showed that OpTCP15 significantly activated the Op7DLH promoter compared to the YFP control, while the Op8HGO was slightly upregulated (Figure 7). Together, OpTCP15 was likely to have a functional role in CPT and its precursor biosynthesis.
FIGURE 7.
(A) Schematic diagrams of the effector and reporter plasmids used in Dual-LUC assay. (B) Dual-LUC assay in N. benthamiana leaf cells using the constructs shown in panel (A). The relative LUC activity was normalized to that of the reference Renilla (REN) luciferase. Error bars indicate the SD (n = 3). Student’s t-test: **p < 0.01; NS, no significance; Dual-LUC, dual-luciferase assay.
Subcellular Localization Analysis of Selected OpTCPs
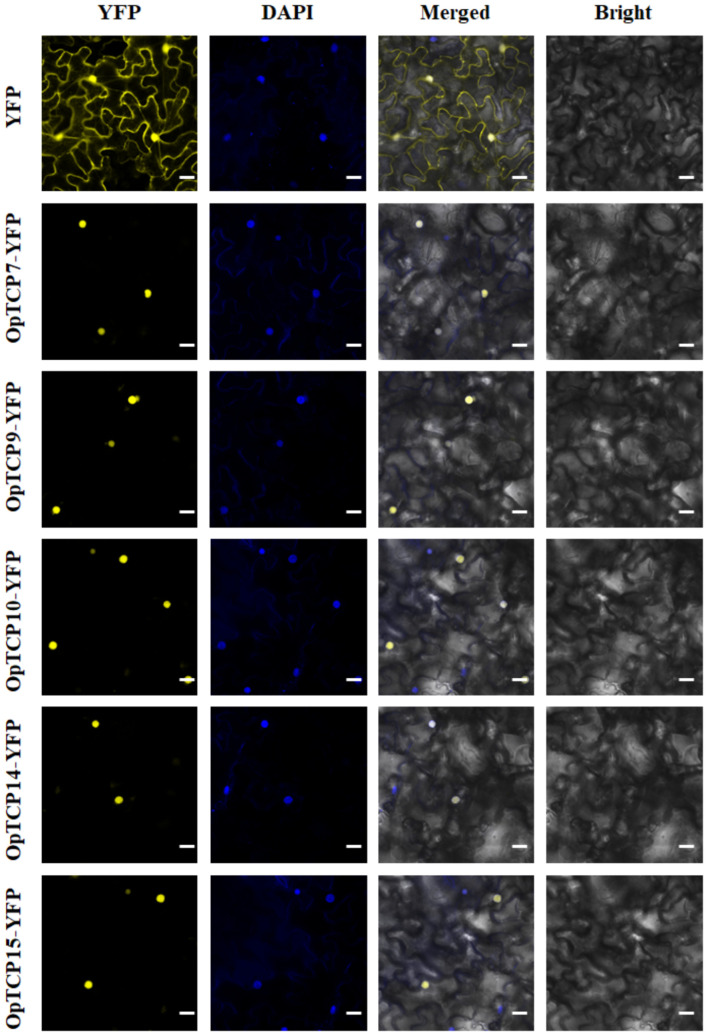
To determine the subcellular localization of five OpTCP genes, which were strongly related to the key enzyme genes of the iridoid pathway, pHB-OpTCP7/9/10/14/15-YFP and pHB-YFP were transiently expressed in N. benthamiana leaves. As shown in Figure 8, the N. benthamiana leaves transformed with pHB-YFP vector displayed fluorescence in nucleus and cytoplasm. In contrast, all fluorescence in cells transformed with pHB-OpTCP7/9/10/14/15-YFP was detected in the nucleus exclusively, suggesting that all five selected OpTCP genes encoded nuclear proteins. It was consistent with their putative role as TFs.
FIGURE 8.
The subcellular localization of the selected five OpTCPs. Scale bars: 20 μm. OpTCP, O. pumila TCP.
Discussion
Evolution and Classification of Teosinte Branched 1/Cycloidea/Proliferating Cell Factors 1/2 Transcription Factor Family
Teosinte branched 1/cycloidea/proliferating cell factors 1/2 genes are a class of terrestrial plant-specific TF, which have been investigated in diverse plants (Liu et al., 2019). However, no systematical study of TCP TFs has been examined in O. pumila. In this study, 16 non-redundant TCP genes were identified and analyzed from the O. pumila genome. Furthermore, a multi-level analysis was performed, such as chromosome location, duplication events, phylogenetic analysis, gene structure, conserved motif, cis-acting elements, expression profiles in different tissues, co-expression analysis of key enzymes in CPT biosynthesis, and subcellular localization of candidate TCPs. Compared with those identified TCP gene family in the higher plant, the number of OpTCP genes was significantly small (Supplementary Table 8). It was consistent with the smaller size of O. pumila genome (439.90 Mb) and did not show signs of any recent whole-genome duplication (WGD) in O. pumila (Rai et al., 2021). It was found that some TCP genes in O. pumila had two counterparts in Arabidopsis and O. sativa (Figures 1B,C), indicating that the deletion of the TCP family in O. pumila. Furthermore, compared with the number of TCP genes in different subgroups (Supplementary Table 8), our results showed that the PCF subfamily had the largest number of genes, accounting for about 50%, while the CYC/TB1 subfamily had the least number of genes, accounting for about 15%. It was in agreement with the CIN clade being larger than the CYC clade in land plants (Liu et al., 2019).
Phylogenetic analysis and sequence alignment showed that OpTCPs in the same group or subgroup shared similar motifs composition and gene structures (Figure 3), which was consistent with the previously described in Arabidopsis (Li, 2015), maize (Ding et al., 2019), potato (Bao et al., 2019), and barley (Gao et al., 2021). For example, motif 2 in N-terminal TCP domain was only present in all CIN subclade, while the conserved R domain (motif 3) was not detected in the PCF subclass (Figure 3C). Additionally, almost all of the OpTCP genes within the same subgroup exhibited similar distribution patterns of exon/intron in terms of exon length and intron number. Taken together, the consistency of the motif compositions and the exon/intron structures of OpTCP genes further supported the close evolutionary relationships.
OpTCP Proteins Play Important Roles in Plant Development and Camptothecin Biosynthesis
Increasing evidence indicated that the main function of class I TCP proteins was to promote cell proliferation in leaves (Li et al., 2005; Resentini et al., 2015), while class II proteins played an important role in preventing tissues overgrowth and cell proliferation (Ori et al., 2007). For instance, AtTCP19 and AtTCP20 were class I TCP proteins that are involved in cell division and affect leaf development (Hervé et al., 2009; Danisman et al., 2013). Furthermore, repression of five CIN-like genes (AtTCP2, 3, 4, 10, and 24) in Arabidopsis disturbed leaf development (Schommer et al., 2008; Nag et al., 2009). TB1 mutant increased the number of lateral branches in Maize (Doebley et al., 1995). According to previous studies, syntenic genes between different species may play similar functions (Angiuoli and Salzberg, 2011). In this study, the two class I clade genes OpTCP7 and OpTCP11 had orthologous genes AtTCP20 and AtTCP19 in Arabidopsis (Figure 1B and Supplementary Table 3), respectively, and might be involved in cell division. OpTCP13 was closely homology with AtTCP2 and highly expressed in O. pumila leaf (Figure 2A). These findings implied that OpTCP13 may be involved in regulating leaf and flower development. OpTCP2, the orthologous gene OsTB1 in O. sativa, was highly expressed in O. pumila stem, which implied that OpTCP2 played a role in regulating the lateral branching. In addition, qRT-PCR results showed that the expression level of OpTCPs in different tissues varied, implying that different OpTCP genes may take part in different organ development.
Interestingly, several TCP proteins were found to regulate special metabolism in plants. In A. annua, AaTCP15 promoted artemisinin production by directly binding to and activating the promoters of ALDH1 and DBR2, in which two genes involved in artemisinin biosynthesis (Ma et al., 2021). In this study, co-expression analysis of all OpTCPs with target genes in the CPT biosynthesis pathways revealed that nine of the 16 TCP genes were in strong correlations with some key enzyme genes in the MEP pathway. Such as the expression patterns of the two genes (OpDXS and OpCMS) in different tissues are consistent with the OpTCP5 gene, and the correlation coefficient was >0.8. Additionally, two TCP genes (OpTCP14 and OpTCP15) were in strong negative correlations with OpG10H, Op7-DLGT, OpLAMT, and Op8HGO (r < −0.8), respectively. Four OpTCPs (OpTCP7, 9, 10, and 15) and Op7-DLH were >0.8 (Supplementary Table 6). Overall, most OpTCPs might be involved in CPT the biosynthesis pathway. Additionally, the phylogenetic tree was constructed using the protein sequences of TCP family genes from A. thaliana, O. sativa, C. canephora, and O. pumila, together with known metabolism-regulating TCPs (AtTCP3, AtTCP15, AaTCP14, AaTCP15, and MdTCP46), to portray their evolutionary relationship. We found that OpTCP15 proteins clustered close with AtTCP15, AaTCP14, AaTCP15, and MdTCP46, while OpTCP5 exhibited a close relationship with AtTCP3 and AtTCP4. Meanwhile, collinearity analyses of TCP genes between O. pumila and Arabidopsis also revealed that OpTCP5 and OpTCP15 have a collinear relationship with AtTCP3, AtTCP15, respectively. In summary, OpTCP5 and OpTCP15 were likely to be involved in the biosynthesis process of CPT in O. pumila. The functions of two genes are worth exploring in the future.
To identify the cis-elements of TCP TFs, the promoter sequences of 15 enzyme-coding genes in CPT biosynthesis were analyzed including three functionally characterized genes (OpSLS, OpTDC, and OpSTR). These results showed that the sequences of GGNCC and GCCCR were identified in the promoter regions of most studied genes (Supplementary Figure 4 and Supplementary Table 7). For example, there were 18, 10, and 7 TBS-binding sites on the promoter sequences of OpSLS, OpSTR, and OpTDC genes, respectively. The finding indicated that TCP TFs may influence the expression of these genes by binding to the TBS binding sites. Furthermore, the results of Dual-LUC assay showed that OpTCP15 significantly activated the transcription of Op7DLH (Figure 7). Investigation on how OpTCP15 is involved in regulating the biosynthesis of CPT needs to be further conducted. Yeast One-Hybrid (Y1H) and electronic mobility shift assays (EMSAs) were performed to examine OpWRKY2 binding W-box in O. pumila (Hao et al., 2021). However, additional methods and techniques are needed to analyze the possible regulatory mechanisms of OpTCP TFs.
Conclusion
This is the first genome-wide study, such as a systematic analysis of the OpTCP gene family in O. pumila. A total of 16 TCP family genes were identified and categorized into two classes based on phylogenetics. Expression patterns of all the 16 OpTCP and central enzyme genes in CPT biosynthetic pathway were investigated. Combining the results of co-expression, phylogeny analysis, and Dual-LUC assay revealed that OpTCP15 potentially participated in the regulation of CPT and its precursor biosynthesis. Additionally, a subcellular localization experiment of five OpTCP genes showed that they were all localized in the nucleus. These results provided a foundation for further functional characterization of the candidate OpTCP genes with the potential to increase CPT production.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
CW analyzed the data. CW and YW wrote the original draft of this manuscript. XH, MS, GK, and Z-GZ revised the manuscript. All authors have read and approved the final version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The handling editor declared a past collaboration with the author GK.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We appreciate the experimental support from the Public Platform of Medical Research Center, Academy of Chinese Medical Science, Zhejiang Chinese Medical University.
Abbreviations
- CPT
camptothecin
- TCP
teosinte branched 1/cycloidea/proliferating cell factors 1/2
- TB1
teosinte branched 1
- CYC
cycloidea
- PCF1/2
proliferating cell factors 1/2
- bHLH
basic helix-loop-helix
- MEP
2-Cmethyl-Derythritol 4-phosphate.
Footnotes
Funding
This work was supported by the National Natural Science Foundation of China (Nos. 31571735, 82073963, 81522049, and 82003889), the Major Science and Technology Projects of Breeding New Varieties of Agriculture in Zhejiang Province (No. 2021C02074), the Zhejiang Provincial Ten Thousands Program for Leading Talents of Science and Technology Innovation (No. 2018R52050), the Zhejiang Provincial Program for the Cultivation of High-level Innovative Health Talents, Zhejiang Provincial Natural Science Foundation of China (LQ21H280004), the Research Project of Zhejiang Chinese Medical University (2021JKZDZC06), and the Opening Project of Zhejiang Provincial Preponderant and Characteristic Subject of Key University (Traditional Chinese Pharmacology), Zhejiang Chinese Medical University (ZYAOX2018009).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.746648/full#supplementary-material
References
- Aggarwal P., Gupta M. D., Joseph A. P., Chatterjee N., Srinivasan N., Nath U. (2010). Identification of specific DNA binding residues in the TCP family of transcription factors in Arabidopsis. Plant Cell 22 1174–1189. 10.1105/tpc.109.066647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angiuoli S. V., Salzberg S. L. (2011). Mugsy: fast multiple alignment of closely related whole genomes. Bioinformatics 27 334–342. 10.1093/bioinformatics/btq665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T. L., Mikael B., Buske F. A., Martin F., Grant C. E., Luca C., et al. (2009). Meme suite: tools for motif discovery and searching. Nucleic Acids Res. 37 W202–W208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S., Zhang Z., Lian Q., Sun Q., Zhang R. (2019). Evolution and expression of genes encoding TCP transcription factors in Solanum tuberosum reveal the involvement of StTCP23 in plant defence. BMC Genet. 20:91. 10.1186/s12863-019-0793-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broholm S. K., Tähtiharju S., Laitinen R. A., Albert V. A., Teeri T. H., Elomaa P. (2008). A TCP domain transcription factor controls flower type specification along the radial axis of the Gerbera (Asteraceae) inflorescence. Proc. Natl. Acad. Sci. U.S.A. 105 9117–9122. 10.1073/pnas.0801359105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahel A. A., Zeng S., Yousaf Z., Liao Y., Yang Z., Wei X., et al. (2019). Plant-specific transcription factor LrTCP4 enhances secondary metabolite biosynthesis in Lycium ruthenicum hairy roots. Plant Cell Tissue Organ Culture 136 323–337. 10.1007/s11240-018-1518-2 [DOI] [Google Scholar]
- Chen L., Chen Y. Q., Ding A. M., Chen H., Xia F., Wang W. F., et al. (2016). Genome-wide analysis of TCP family in tobacco. Genet. Mol. Res. 15:gmr.15027728. 10.4238/gmr.15027728 [DOI] [PubMed] [Google Scholar]
- Cheng Y., Yao Z. P., Ruan M. Y., Ye Q. J., Wang R. Q., Zhou G. Z., et al. (2016). In silico identification and characterization of the WRKY gene superfamily in pepper (Capsicum annuum L.). Genet. Mol. Res. 15 1–12. 10.4238/gmr.15038675 [DOI] [PubMed] [Google Scholar]
- Chow C. N., Lee T. Y., Hung Y. C., Li G. Z., Tseng K. C., Liu Y. H., et al. (2019). PlantPAN3. 0: a new and updated resource for reconstructing transcriptional regulatory networks from ChIP-seq experiments in plants. Nucleic Acids Res. 47 D1155–D1163. 10.1093/nar/gky1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas P., Lauter N., Doebley J., Coen E. (1999). The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 18 215–222. 10.1046/j.1365-313X.1999.00444.x [DOI] [PubMed] [Google Scholar]
- Danisman S., Van der Wal F., Dhondt S., Waites R., de Folter S., Bimbo A., et al. (2012). Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol. 159 1511–1523. 10.1104/pp.112.200303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danisman S., van Dijk A. D. J., Bimbo A., van der Wal F., Hennig L., de Folter S., et al. (2013). Analysis of functional redundancies within the Arabidopsis TCP transcription factor family. J. Exp. Bot. 64 5673–5685. 10.1093/jxb/ert337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S., Cai Z., Du H., Wang H. (2019). Genome-wide analysis of TCP family genes in Zea mays L. identified a role for ZmTCP42 in drought tolerance. Int. J. Mol. Sci. 20:2762. 10.3390/ijms20112762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J., Stec A., Gustus C. (1995). Teosinte branched1 and the origin of maize: evidence for epistasis and the evolution of dominance. Genetics 141 333–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z. J., Xu S. C., Liu N., Zhang G. W., Hu Q. Z., Gong Y. M. (2018). Soybean TCP transcription factors: evolution, classification, protein interaction and stress and hormone responsiveness. Plant Physiol. Biochem. 127 129–142. 10.1016/j.plaphy.2018.03.020 [DOI] [PubMed] [Google Scholar]
- Gao G., Kan J., Jiang C., Ahmar S., Zhang J., Yang P. (2021). Genome-wide diversity analysis of TCP transcription factors revealed cases of selection from wild to cultivated barley. Funct. Integrative Genomics. 21 31–42. 10.1007/s10142-020-00759-4 [DOI] [PubMed] [Google Scholar]
- Gasteiger E., Hoogland C., Gattiker A., Wilkins M. R., Appel R. D., Bairoch A. (2005). Protein Identification and Analysis Tools on the ExPASy Server. The Proteomics Protocols Handbook. Berlin: Springer, 571–607. [Google Scholar]
- Hao X., Xie C., Ruan Q., Zhang X., Wu C., Han B., et al. (2021). The transcription factor OpWRKY2 positively regulates the biosynthesis of the anticancer drug camptothecin in Ophiorrhiza pumila. Horticulture Res. 8 1–14. 10.1038/s41438-020-00437-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervé C., Dabos P., Bardet C., Jauneau A., Auriac M. C., Ramboer A., et al. (2009). In vivo interference with AtTCP20 function induces severe plant growth alterations and deregulates the expression of many genes important for development. Plant Physiol. 149 1462–1477. 10.1104/pp.108.126136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn S., Pabon-Mora N., Theuß V. S., Busch A., Zachgo S. (2015). Analysis of the CYC/TB1 class of TCP transcription factors in basal angiosperms and magnoliids. Plant J. 81 559–571. 10.1111/tpj.12750 [DOI] [PubMed] [Google Scholar]
- Howarth D. G., Donoghue M. J. (2006). Phylogenetic analysis of the “ECE”(CYC/TB1) clade reveals duplications predating the core eudicots. Proc. Natl. Acad. Sci. U.S.A. 103 9101–9106. 10.1073/pnas.0602827103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Jin J., Guo A. Y., Zhang H., Luo J., Gao G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31 1296–1297. 10.1093/bioinformatics/btu817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. J., Rajan R., Baby S. (2018). Secondary metabolites from Ophiorrhiza. Nat. Products J. 8 248–267. 10.2174/2210315508666180515104735 [DOI] [Google Scholar]
- Kai G., Wu C., Gen L., Zhang L., Cui L., Ni X. (2015). Biosynthesis and biotechnological production of anti-cancer drug Camptothecin. Phytochem. Rev. 14 525–539. 10.1007/s11101-015-9405-5 [DOI] [Google Scholar]
- Kang M., Fu R., Zhang P., Lou S., Yang X., Chen Y., et al. (2021). A chromosome-level Camptotheca acuminata genome assembly provides insights into the evolutionary origin of camptothecin biosynthesis. Nat. Commun. 12 1–12. 10.1038/s41467-021-23872-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellner F., Kim J., Clavijo B. J., Hamilton J. P., Childs K. L., Vaillancourt B., et al. (2015). Genome-guided investigation of plant natural product biosynthesis. Plant J. 82 680–692. 10.1111/tpj.12827 [DOI] [PubMed] [Google Scholar]
- Kosugi S., Ohashi Y. (1997). PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell 9 1607–1619. 10.1105/tpc.9.9.1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng X., Wei H., Xu X., Ghuge S. A., Jia D., Liu G., et al. (2019). Genome-wide identification and transcript analysis of TCP transcription factors in grapevine. BMC Genomics 20:786. 10.1186/s12864-019-6159-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Potuschak T., Colón-Carmona A., Gutiérrez R. A., Doerner P. (2005). Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc. Natl. Acad. Sci. U.S.A. 102 12978–12983. 10.1073/pnas.0504039102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. (2015). The Arabidopsis thaliana TCP transcription factors: a broadening horizon beyond development. Plant Signal. Behav. 10:e1044192. 10.1080/15592324.2015.1044192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Zachgo S. (2013). TCP3 interacts with R2R3-MYB proteins, promotes flavonoid biosynthesis and negatively regulates the auxin response in Arabidopsis thaliana. Plant J. 76 901–913. 10.1111/tpj.12348 [DOI] [PubMed] [Google Scholar]
- Liu H. L., Wu M., Li F., Gao Y. M., Chen F., Xiang Y. (2018). TCP transcription factors in moso bamboo (Phyllostachys edulis): genome-wide identification and expression analysis. Front. Plant Sci. 9:1263. 10.3389/fpls.2018.01263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M. M., Wang M. M., Yang J., Wen J., Guo P. C., Wu Y. W., et al. (2019). Evolutionary and comparative expression analyses of TCP transcription factor gene family in land plants. Int. J. Mol. Sci. 20:3591. 10.3390/ijms20143591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. Q., Li W. Q., Morris-Natschke S. L., Qian K., Yang L., Zhu G. X., et al. (2015). Perspectives on biologically active camptothecin derivatives. Med. Res. Rev. 35 753–789. 10.1002/med.21342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D., Carpenter R., Vincent C., Copsey L., Coen E. (1996). Origin of floral asymmetry in Antirrhinum. Nature 383 794–799. 10.1038/383794a0 [DOI] [PubMed] [Google Scholar]
- Lupas A., Van Dyke M., Stock J. (1991). Predicting coiled coils from protein sequences. Science 252 1162–1164. 10.1126/science.252.5009.1162 [DOI] [PubMed] [Google Scholar]
- Ma Y. N., Xu D. B., Li L., Zhang F., Fu X. Q., Shen Q., et al. (2018). Jasmonate promotes artemisinin biosynthesis by activating the TCP14-ORA complex in Artemisia annua. Sci. Adv. 4:eaas9357. 10.1126/sciadv.aas9357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y. N., Xu D. B., Yan X., Wu Z. K., Kayani S. I., Shen Q., et al. (2021). Jasmonate-and abscisic acid-activated AaGSW1-AaTCP15/AaORA transcriptional cascade promotes artemisinin biosynthesis in Artemisia annua. Plant Biotechnol. J. 19 1412–1428. 10.1111/pbi.13561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manassero N. G. U., Viola I. L., Welchen E., Gonzalez D. H. (2013). TCP transcription factors: architectures of plant form. Biomol. Concepts 4 111–127. 10.1515/bmc-2012-0051 [DOI] [PubMed] [Google Scholar]
- Martino E., Della Volpe S., Terribile E., Benetti E., Sakaj M., Centamore A., et al. (2017). The long story of camptothecin: From traditional medicine to drugs. Bioorganic Med. Chem. Lett. 27 701–707. 10.1016/j.bmcl.2016.12.085 [DOI] [PubMed] [Google Scholar]
- Martín-Trillo M., Cubas P. (2010). TCP genes: a family snapshot ten years later. Trends Plant Sci. 15 31–39. 10.1016/j.tplants.2009.11.003 [DOI] [PubMed] [Google Scholar]
- Nag A., King S., Jack T. (2009). miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106 22534–22539. 10.1073/pnas.0908718106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori N., Cohen A. R., Etzioni A., Brand A., Yanai O., Shleizer S., et al. (2007). Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat. Genet. 39 787–791. 10.1038/ng2036 [DOI] [PubMed] [Google Scholar]
- Palatnik J. F., Allen E., Wu X., Schommer C., Schwab R., Carrington J. C., et al. (2003). Control of leaf morphogenesis by microRNAs. Nature 425 257–263. 10.1038/nature01958 [DOI] [PubMed] [Google Scholar]
- Parapunova V., Busscher M., Busscher-Lange J., Lammers M., Karlova R., Bovy A. G., et al. (2014). Identification, cloning and characterization of the tomato TCP transcription factor family. BMC Plant Biol. 14:157. 10.1186/1471-2229-14-157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai A., Hirakawa H., Nakabayashi R., Kikuchi S., Hayashi K., Rai M., et al. (2021). Chromosome-level genome assembly of Ophiorrhiza pumila reveals the evolution of camptothecin biosynthesis. Nat. Commun. 12 1–19. 10.1038/s41467-020-20508-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rather G. A., Sharma A., Pandith S. A., Kaul V., Nandi U., Misra P., et al. (2018). De novo transcriptome analyses reveals putative pathway genes involved in biosynthesis and regulation of camptothecin in Nothapodytes nimmoniana (Graham) Mabb. Plant Mol. Biol. 96 197–215. 10.1007/s11103-017-0690-9 [DOI] [PubMed] [Google Scholar]
- Resentini F., Felipo-Benavent A., Colombo L., Blazquez Rodriguez M. A., Alabadí Diego D., Masiero S. (2015). TCP14 and TCP15 mediate the promotion of seed germination by gibberellins in Arabidopsis thaliana. Mol. Plant 8 482–485. 10.1016/j.molp.2014.11.018 [DOI] [PubMed] [Google Scholar]
- Rohani E. R., Chiba M., Kawaharada M., Asano T., Oshima Y., Mitsuda N., et al. (2016). An MYB transcription factor regulating specialized metabolisms in Ophiorrhiza pumila. Plant Biotechnol. 33 1–9. 10.5511/plantbiotechnology.15.1117a [DOI] [Google Scholar]
- Sadre R., Magallanes-Lundback M., Pradhan S., Salim V., Mesberg A., Jones A. D., et al. (2016). Metabolite diversity in alkaloid biosynthesis: a multilane (diastereomer) highway for camptothecin synthesis in Camptotheca acuminata. Plant Cell 28 1926–1944. 10.1105/tpc.16.00193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarvepalli K., Nath U. (2011). Hyper-activation of the TCP4 transcription factor in Arabidopsis thaliana accelerates multiple aspects of plant maturation. Plant J. 67 595–607. 10.1111/j.1365-313X.2011.04616.x [DOI] [PubMed] [Google Scholar]
- Schommer C., Palatnik J. F., Aggarwal P., Chételat A., Cubas P., Farmer E. E., et al. (2008). Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 6:e230. 10.1371/journal.pbio.0060230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M., Gong H., Cui L., Wang Q., Wang C., Wang Y., et al. (2020). Targeted metabolic engineering of committed steps improves anti-cancer drug camptothecin production in Ophiorrhiza pumila hairy roots. Industrial Crops Products 148:112277. 10.1016/j.indcrop.2020.112277 [DOI] [Google Scholar]
- Shi P., Guy K. M., Wu W., Fang B., Yang J., Zhang M., et al. (2016). Genome-wide identification and expression analysis of the ClTCP transcription factors in Citrullus lanatus. BMC Plant Biol. 16:85. 10.1186/s12870-016-0765-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udomsom N., Rai A., Suzuki H., Okuyama J., Imai R., Mori T., et al. (2016). Function of AP2/ERF transcription factors involved in the regulation of specialized metabolism in Ophiorrhiza pumila revealed by transcriptomics and metabolomics. Front. Plant Sci. 7:1861. 10.3389/fpls.2016.01861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viola I. L., Camoirano A., Gonzalez D. H. (2016). Redox-dependent modulation of anthocyanin biosynthesis by the TCP transcription factor TCP15 during exposure to high light intensity conditions in Arabidopsis. Plant Physiol. 170 74–85. 10.1104/pp.15.01016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., Hu Y., Cui M. Y., Han Y. T., Gao K., Feng J. Y. (2016). Identification and transcript analysis of the TCP transcription factors in the diploid woodland strawberry Fragaria vesca. Front. Plant Sci. 7:1937. 10.3389/fpls.2016.01937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Lin W., Liu H., Deng J., Chen Y., Liu H., et al. (2016). Ultrasound-responsive nanobubbles contained with peptide–camptothecin conjugates for targeted drug delivery. Drug Delivery 23 2756–2764. 10.3109/10717544.2015.1077289 [DOI] [PubMed] [Google Scholar]
- Xu R., Sun P., Jia F., Lu L., Li Y., Zhang S., et al. (2014). Genomewide analysis of TCP transcription factor gene family in Malus domestica. J. Genet. 93 733–746. 10.1007/s12041-014-0446-0 [DOI] [PubMed] [Google Scholar]
- Yang M., Wang Q., Liu Y., Hao X., Wang C., Liang Y., et al. (2021). Divergent camptothecin biosynthetic pathway in Ophiorrhiza pumila. BMC Biol. 19:122. 10.1186/s12915-021-01051-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- You D., Feng Y., Wang C., Sun C., Wang Y., Zhao D., et al. (2021). Cloning, characterization, and enzymatic identification of a new tryptophan decarboxylase from Ophiorrhiza pumila. Biotechnol. Appl. Biochem. 68 381–389. 10.1002/bab.1935 [DOI] [PubMed] [Google Scholar]
- Zheng A., Sun F., Cheng T., Wang Y., Xie K., Zhang C., et al. (2019). Genome-wide identification of members of the TCP gene family in switchgrass (Panicum virgatum L.) and analysis of their expression. Gene 702 89–98. 10.1016/j.gene.2019.03.059 [DOI] [PubMed] [Google Scholar]
- Zheng K., Ni Z., Qu Y., Cai Y., Yang Z., Sun G., et al. (2018). Genome-wide identification and expression analyses of TCP transcription factor genes in Gossypium barbadense. Sci. Rep. 8:14526. 10.1038/s41598-018-32626-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.