Abstract
The new coronavirus disease 2019 (COVID-19) has become a global pandemic leading to over 180 million confirmed cases and nearly 4 million deaths until June 2021, according to the World Health Organization. Since the initial report in December 2019 , COVID-19 has demonstrated a high transmission rate (with an R0 > 2), a diverse set of clinical characteristics (e.g., high rate of hospital and intensive care unit admission rates, multi-organ dysfunction for critically ill patients due to hyperinflammation, thrombosis, etc.), and a tremendous burden on health care systems around the world. To understand the serious and complex diseases and develop effective control, treatment, and prevention strategies, researchers from different disciplines have been making significant efforts from different aspects including epidemiology and public health, biology and genomic medicine, as well as clinical care and patient management. In recent years, artificial intelligence (AI) has been introduced into the healthcare field to aid clinical decision-making for disease diagnosis and treatment such as detecting cancer based on medical images, and has achieved superior performance in multiple data-rich application scenarios. In the COVID-19 pandemic, AI techniques have also been used as a powerful tool to overcome the complex diseases. In this context, the goal of this study is to review existing studies on applications of AI techniques in combating the COVID-19 pandemic. Specifically, these efforts can be grouped into the fields of epidemiology, therapeutics, clinical research, social and behavioral studies and are summarized. Potential challenges, directions, and open questions are discussed accordingly, which may provide new insights into addressing the COVID-19 pandemic and would be helpful for researchers to explore more related topics in the post-pandemic era.
Keywords: COVID-19 pandemic, Artificial intelligence, Electronic health record, Machine learning
Graphical abstract
1. Introduction
The unprecedented outbreak of new coronavirus disease 2019 (COVID-19) has put people around the world at risk. The COVID-19 pandemic in December 2019 has spread throughout the world quickly because of a high transmission rate (with an R0 value bigger than 2) [1]. The scarcity of resources and the worry of overburdened healthcare systems have impelled majority governments to restrict traveling or lockdown cities [2]. The COVID-19 pandemic has caused over 180 million confirmed cases and nearly 4 million deaths until June 2021, according to the World Health Organization [3]. Scientists have identified the genome sequence of the virus and categorized it as a member of the β-CoV genera of the coronavirus family [4], which can attack the human respiratory system, cause fever, cough, and other flu-like symptoms, and further affecting multiple tissues and organ systems [5]. In addition, patients with COVID-19 may rapidly develop serious dysfunctions and even critical illness, leading to a suddenly boosted requirement of hospital beds, mechanical ventilation devices, and critical patient care resources [6]. Therefore, there is an urgent need for new technologies to help clinicians and health care providers to address this pandemic.
Artificial intelligence (AI), advanced by the rapid development of computer hardware and software and mathematics, includes a wide range of techniques that allow computers to think and work like the human brain to support decision making. AI techniques, especially the machine learning (ML) and deep learning (DL), have demonstrated superior performance in many real-world data applications ranging from computer vision to natural language processing. In recent years, AI techniques have also been introduced into the healthcare field and lead to a novel rout to effectively derive knowledge in terms of disease conditions from complex health data to improve human health care, such as clinical decision-making [7], [8]. In COVID-19, the increasing availability of diverse types of data makes it promising to apply AI techniques to assist us to overcome the pandemic [9]. In this context, significant efforts that used AI to address COVID-19 have been drawn from different perspectives, including epidemiology and public health, biology and genomic medicine, as well as clinical care and patient management, etc. In this study, we discussed the applications of AI that mainly focused on ML and DL techniques in COVID-19.
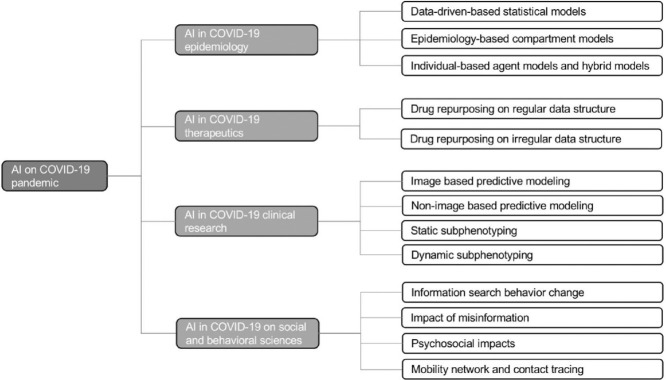
There are several previous studies in terms of using AI to combat COVID-19 [10], [11], [12], [13], [14]. They generally have a specific focus on AI's applications in epidemiology and therapeutics in COVID-19. Islam et al. [11] reviewed 35 studies on the use of AI in COIVD-19 diagnosis, epidemic forecasting, and patient management. Hussain et al. [12] focuses on big data, the Internet of Things (IoT), AI, cloud computing techniques in fighting against the COVID-19 pandemic. In addition, Pham et al. [13], Chen et al. [14], and Nguyen et al. [10] also discussed the use of AI in vaccine and drug development. Compared to the previous studies, we considered a broader spectrum of application areas of AI in fighting the pandemic, including epidemiology, therapeutics, clinical research, social and behavioral studies. In each field, we review existing studies and detail how the AI techniques advanced COVID-19 study, but also discuss unsolved issues and challenges as well as potential opportunities of AI in this field which may provide insights for researchers to bridge the gap between the application of AI and health care in the pandemic. The overall framework of this review is shown in Figure 1 .
Figure 1.
The overall framework of this review. We review four aspects (i.e., epidemiology, therapeutics, clinical research, social and behavioral studies) in terms of applications of AI on COVID-19 pandemic. Also the challenges of each aspect are provided. Finally, the general challenges, directions, and open questions are discussed on model interpretation, model security, model bias, privacy issue and model precision.
References for this Review were obtained through searches of PubMed, Scopus, Google Scholar, and Web of Science for papers. Keywords included “COVID-19” “SARS-CoV-2”, “non-pharmaceutical public health interventions”, “epidemic control”, “drug repositioning”, “drug repurposing”, “network medicine”, “machine learning”, “artificial intelligence”, “convolutional neural networks”, “deep learning”, “subphenotyping”, “misinformation”, “social media”, “health impacts”, “public health”, and “mental health”. The titles and abstracts were furtherly checked for inclusion. Some relevant papers were also collected from the reviews of citations referenced. Most of the reviewed articles were published after June 2020. To clearly summarize these articles, they were grouped into four categories according to the types of applications, including (1) epidemiology, (2) therapeutics, (3) clinical research, and (4) social and behavioral studies.
2. AI in COVID-19 epidemiology
AI models have been involved in the epidemiology studies, mainly focusing on the COVID-19 trend prediction. In particular, the involved AI models include data-driven-based statistical models, epidemiology-based compartment models, and individual-based agent models and hybrid models.
2.1. Data-driven-based statistical models
The data-driven-based statistical models mainly include regression-based parametric or non-parametric models such as Auto-Regressive Integrated Moving Average (ARIMA), Support Vector Regression (SVR), Random Forest (RF), deep learning (DL) model like Recurrent Neural Network (RNN), and so on. For example, Parbat and Chakraborty [15] used the SVR model to predict the COVID-19 trend to the total number of deaths, recovered cases, cumulative number of confirmed cases, and number of daily cases using the Johns Hopkins epidemiological data [16]. The proposed model was efficient and presented higher accuracy than linear or polynomial regression methods. While building a predictive model for COVID-19 trend forecasting, these pure data-driven-based statistical models typically only considered building relationships between a dependent variable such as the number of deaths and independent variables such as the number of days, but did not explicitly consider the epidemiological characteristics of the infectious disease.
2.2. Epidemiology-based compartment models
Compartment models aim to divide entire populations into multiple different compartments (i.e., states) such as susceptible, exposed, infectious and recovered, and then apply ordinary differential equations (ODEs) to model the transitions among these compartments. Two popular compartment models including Susceptible-Infected-Resistant (SIR) [17] and Susceptible-Exposed-Infected-Removed (SEIR) [18] are used to model the spread of infectious disease in terms of multiple previous epidemic outbreaks such as SARS [19] and the ongoing COVID-19 pandemic [20], [21]. Compared to data-driven-based statistical models, the compartment models were built on the well-established mathematical/physical laws, which consider the epidemiological characteristics of infectious disease and there is an assumption for the compartment models that the counts observed from these compartments have the potential to reflect reproduction numbers. Compartment models are still the mainstream approach in epidemiological research of infectious diseases [22]. However, the determination of parameters of the traditional compartment models is difficult and usually relies on predefined hypotheses. The use of AI techniques has shown their strength in estimating the optimal parameters of the compartment models, thus leading to a new way to improve the compartment models in COVID-19 trend prediction [23], [24].
2.3. Individual-based agent models and hybrid models
Recently, several researchers have utilized fine-grained methods to model a population through agent simulation for COVID-19 trend prediction [25]. An individual-based agent model is to simulate a real environment in an abstract representation to estimate the spread of epidemic diseases, which has three main elements including the agent (e.g., person), the factors of each agent (e.g., age), and the links between agents. Rockett et al. [25] used an individual-based agent model to simulate the spread of COVID-19 in an urban area by considering multiple agent factors including age, gender, smoking status, and isolation tendency. They found that the non-pharmaceutical public health interventions, such as staying home, hospital isolation policies, and preventing travel between cities, have contributed to the reduction of the prevalence and the deaths in COVID-19 pandemic.
In addition, some hybrid models such as the combination of mechanistic disease transmission model and a curve-fitting model [26] and the combination of the recurrent neural networks (RNN) model and an improved susceptible-infected (ISI) model [24], have been used in the COVID-19 trend prediction. These hybrid models mainly considered the combination in terms of epidemiology model and ML techniques, which not only capture the epidemiological characteristics of infectious disease but also enhance the ability to build the relationships between input data and output data by a purely data-driven method. The epidemiology model in a hybrid model is usually used to obtain information related to COVID-19 trends such as infection rates, which are utilized as input features for the AI prediction model. The hybrid models have also shown great promise to accelerate the COVID-19 trend prediction.
2.4. Challenge and opportunities
The above summary shows that multiple AI-based epidemiology models have been used to predict the spread of COVID-19 and obtain some promising initial results. However, there remain several challenges and opportunities for the improvement of predictive performance. These include mainly the following:
-
(1)
The spread of infectious disease like COVID-19 is usually complex and influenced by multiple factors such as population density, demographic composition, weather conditions, non-pharmaceutical public health interventions, medical resource disparities, city traffic flows and so on [27], [28], [29], [30], [31]. Researchers need to consider how to combine these factors and set different weights for them. Investigating the impact of individual factors on the spread of the COVID-19 trend is also an interesting topic.
-
(2)
The epidemiology-based compartment models are sensitive to the initial values of model parameters such as infectious population, hospitalized population, and dead population. The determination of initial values of these parameters is usually based on the public reported data (including confirmed cases and recovered cases). However, the reported cases may not be very correct and usually much fewer than their real numbers because of multiple kinds of reasons such as the test capability [32]. Although integrating data-driven machine learning methods can relieve the dependence of initial values and improve the predictive performance, the regular (weekly or daily) updating for AI models to reflect changing dynamics is challenging because of more and more confirmed cases that need more train time.
-
(3)
Several mutations of COVID-19 are more transmissible [33]. The mutated viruses may have higher fatalities, influencing the patterns of the spread of infectious diseases. Incorporating mutation to build predictive models for the COVID-19 trend is important [34] but it is rarely discussed.
-
(4)
Building hybrid models by combining multiple predictive models is a good method for improving the accuracy of predictive models. However, most previous hybrid COVID-19 trend predictive models mainly use the output of one model as the input feature of another model. Building a voting mechanism from many different predictive models would be beneficial for predictive performance.
3. AI in COVID-19 therapeutics: drug discovery
There are two common strategies for the development of drugs to treat diseases including traditional drug development (de novo drug discovery) and drug repurposing [35]. The traditional drug development method starts with building novel chemical compounds based on molecular units and needs multiple steps including preclinical research, safety review, clinical study, FDA review, and FDA postmarket safety monitoring, which usually take more than 10 years and over $ 1 billion to bring a drug to market [36]. Compared to traditional drug development methods, the drug repurposing technique is usually used to identify drugs for emerging and challenging diseases treatment based on approved or investigating existing drugs, which can significantly reduce development timelines and a large number of costs [37]. During the current COVID-19 pandemic, drug repurposing is a very promising approach for discovering effective drugs from existing ones to treat patients with COVID-19 [38]. There are three common strategies for finding drugs in terms of new use by drug repurposing method, including through serendipity, using experimental screening platforms, and computational methods [39]. The serendipity drug repurposing is based on specific pharmacological insights in the lab and clinic. The experimental method based on drug repurposing is usually to bind assays to identify relevant target interactions using some techniques such as affinity chromatography and mass spectrometry, which is costly and time-consuming [40]. A computational method based on drug repurposing is mainly data-driven, which involves systematic analysis on multiple types of large‐scale data such as gene expression, chemical structure, genotype or proteomic data, or electronic health records (EHRs) to acquire meaningful interpretations for repurposing hypotheses [39]. This method provides a great chance for identifying drugs quickly [41].
3.1. Computational drug repurposing
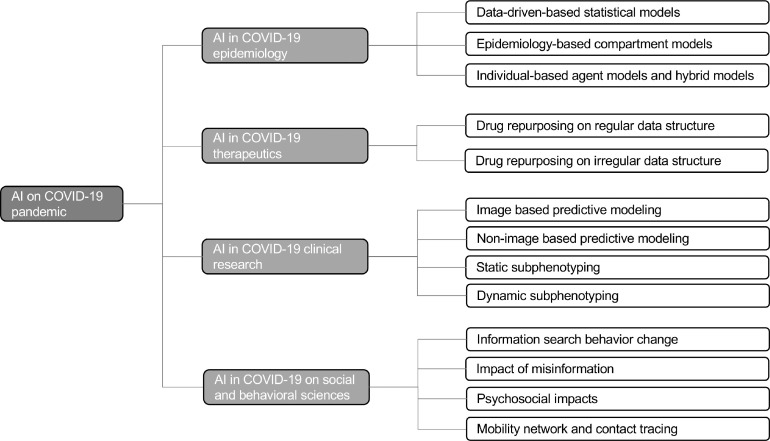
The methods of computational drug discovery can roughly be divided into two categories: Structure-based and ML-based drug discovery. Structure-based drug discovery, one of most popular methods in discovering antiviral drugs, which uses a computational high-throughput ensemble docking technique and obtains the binding affinities by physics-based equations [42]. The ML-based drug discovery attempts to use ML techniques to obtain the representations of drugs or diseases, and then measure the similarities of these entities or build predictive models to obtain the relationships between a drug and disease [35]. During the COVID-19 pandemic, the process of simulations and docking in structure-based drug discovery needs to be refined and reproduced because there are multiple new experimental three-dimensional structures of the S protein and other viral targets [43]. Researchers have started using ML techniques instead of structure-based drug discovery to predict drug binding and find candidate drugs due to the superiority of ML [44]. In MLbased drug repurposing, the representation of the structure of drug and disease is key for training ML models. Drug repurposing using regular and irregular data structure representations is discussed as follows. A general framework of ML-based drug repurposing is demonstrated in Figure 2 .
Figure 2.
A general framework of ML (machine learning) and DL (deep learning) based drug repurposing. FNN: feedforward neural network; CNN: convolutional neural network; RNN: Recurrent neural network.
3.1.1. Drug repurposing on regular data structure
Regular data structures including vector, sequence, and matrix have been used for drug repurposing with different DL architectures [39]. For vector representation of drugs or diseases, a fully connected feedforward neural network (FNN) architecture is usually used to build a predictor or classifier [45]. In FNN, the input variables and output targets are connected by multiple layers with neurons. Each neuron from the preceding layer is connected to all neurons from the subsequent layer, and those connections are assigned different weights, which are trained and optimized through prediction loss and backpropagation. There are several FNN based drug repurposing studies [46], [47], [48] that profile data samples as vector representations. For example, Aliper et al. [46] used vector representations to build transcriptomic profiles for 678 different drugs and then built an FNN model to classify various drugs into therapeutic categories. The FNN model showed better performance compared to other computational methods such as naive Bayes, SVM, and RF. However, if the information of drug or disease is stored in the chemical image, using the FNN method is challenging as it involves a large number of weights in training FNN.
The matrix representation of drugs mainly refers to chemical images, which contain more molecular structure information. In this context, the advanced CNN [49], a preferred DL model specifically designed to obtain insights from those images, could be a promising approach to address the tasks. CNN can build relationships between the pixels in images and final predictive targets by multiple layers of nonlinear transformations [50]. A CNN typically consists of three layers: a convolution layer, a pooling layer, and a fully connected layer. CNN has been applied to explore drug function based on chemical images [51]. For example, Wallach et al. [52] used a CNN to build a predictive architecture, AtomNet, to predict molecular binding affinity to proteins, which obtained an AUC >0.9 on 57.8% of the targets in the DUDE benchmark. Ragoza et al. [53] used CNN to build a protein-ligand scoring system to classify compound poses as binders or non-binders. A grid representation of protein−ligand structures was used as input to the CNN model, which showed better discrimination than AutoDock Vina scoring [54] in terms of pose prediction and virtual screening.
In addition, few studies focused on modeling the molecular sequence of drugs to identify new therapeutic implications. In this context, the recurrent neural networks (RNNs) [55], a kind of DL model for sequence data modeling, are usually involved. In an RNN, a recurrent neuron is used to address each element of a sequence at each timestamp and it integrates the historical information of the current element, which is obtained from the output of the previous timestamp. Several studies used RNN to generate simplified molecular-input line-entry system (SMILES) with desirable properties such as a quantitative estimate of drug-likeness (QED) [56]. By fine tuning of a pre-trained RNN, Olivecrona et al. [57] solved the issue in terms of a combination of handwritten rules for undesirable structure penalties. In addition, RNN architectures have been applied to generate focused molecule libraries for drug discovery by building sequence profiles for molecules based on SMILES codes [58]. Gao et al. [59] designed a hybrid of RNN and graph-based CNN model to identify drug-target interactions based on amino acids sequences and chemical structures.
3.1.2. Drug repurposing on irregular data structure
Irregular data structure-based drug repurposing mainly involves network medicine [60], [61] and graph representation learning [62]. Typically, a biomedical network or biomedical knowledge graph was first built. Then graph-based AI models, such as network embedding or deep graph neural networks, were used to learn low-dimensional representations for nodes and edges while preserving the graph structure. Finally, novel drug implications (e.g., potential drug-disease associations or drug-target interactions) discovery can be done by link prediction based on those representations [38], [63]. For example, Sosa et al. [64] plotted a large and heterogeneous knowledge graph, the Global Network of Biomedical Relationships (GNBR), including drug, disease, and gene (or protein) entities. They used graph embedding techniques to predict the links between drugs and diseases and obtained performance with an AUROC value of 0.89 on a gold-standard test set. Zeng et al. [65] built a COVID-19 knowledge graph, CoV-KGE, to identify drug candidates for treating the SARS-CoV-2 virus from 24 million PubMed research articles (Table 1). Amazon's Amazon Web Services (AWS) computing resources and graph embedding techniques were used on the built knowledge graph that contained 15 million edges, 39 types of relationships among nodes including drugs, diseases, proteins/genes, pathways, and expression, and finally discovered 41 repurposable drugs such as tetrandrine, nadide, estradiol, and so on. Some representative knowledge graph-based studies are shown in Table 2 .
Table 1.
The summary of studies in terms of the applications of AI in epidemiology
| Reference | Task | Data source & size | Model | Result |
|---|---|---|---|---|
| Parbat et al. (May 2020) [15] | Predict the total number of deaths, recovered cases, cumulative number of confirmed cases, and number of daily cases. | Johns Hopkins Github repository (https://github.com/CSSEGISandData/COVID-19) between 01/03/2020–30/04/2020 cases: 35,043 deaths: 1,147 recovered patients: 8,889. |
Support vector regression model | The proposed model was efficient and has higher accuracy (more than 87%) than linear or polynomial regression methods. |
| Zeynep Ceylan (April 2020) [145] | Estimate the prevalence of COVID-19 in Italy, Spain, and France. | The data of COVID-19 collected from the WHO website (https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/) between 21/02/2020–15/04/2020 Italy: mean prevalence case 57,262, mean incidence case 3,009; Spain: mean prevalence case 54,075, mean incidence case 3,521; France: mean prevalence case 30,233, mean incidence case 2,092. |
Auto-Regressive Integrated Moving Average (ARIMA) model | ARIMA (0,2,1), ARIMA (1,2,0), and ARIMA (0,2,1) showed the best prediction performance (more than 82% accuracy) for Italy, Spain, and France, respectively. |
| Benvenuto et al. (February 2020) [146] | Predict the epidemiological trend of the prevalence and incidence of COVID-2019 | the Johns Hopkins epidemiological data (https://gisanddata.maps.arcgis.com/apps/opsdashboard/index.html) |
Auto-Regressive Integrated Moving Average (ARIMA) model | ARIMA (1,0,4) and ARIMA (1,0,3) showed the best performance in terms of determining the prevalence and incidence of COVID-2019, respectively. |
| Rodriguez et al. (September 2020) [147] | Real-time COVID-19 forecasting including incidence and cumulative weekly deaths and Incidence daily hospitalizations. | Johns Hopkins University (JHU) COVID Tracking Project (https://covidtracking.com) |
DeepCOVID including data module, prediction module, and explainability module based on deep learning model | The proposed model was used in CDC COVID-19 Forecast Hub (since April 2020). |
| Singh et al. (September 2020) [148] | Predict the spread of COVID-19 | Data collected from Kaggle website (https://www.kaggle.com/imdevskp/covid19-corona-virus-india-dataset) Data covered 15 States of India. |
Random Forest and Kalman Filter | The proposed model showed good performance in terms of short-term estimation, but not so good for long-term forecasting. |
| Zheng et al. (July 2020) [24] | Predict the development and spread of the COVID-19 | Data collected from the national and provincial health commissions, and dxy.com website (Real-time data API for COVID-19 epidemic) (https://lab.isaaclin.cn/nCoV/zh) | Hybrid AI Model based on susceptible-infected (ISI) model and RNN model | The proposed model acquired the lower mean absolute percentage errors in Wuhan (0.52%), Beijing (0.38%), Shanghai (0.38%), and countrywide (0.86%) for the next 6 days. |
| Huang et al. (May 2021) [22] | Forecast the trend of COVID-19 pandemics under the influence of reopening policies. | Hospitalization and cumulative morality of COVID-19. Houston, Texas, May 1, 2020 – June 29, 2020 |
Risk-stratified SIR-HCD | The proposed model obtained lower mean squared error (MSE) and higher prediction accuracy compared to other models, and supports counterfactual analysis. |
| Liu et al. (May 2021) [149] | Investigate the influence (reproduction number) of non-pharmaceutical public health interventions on COVID-19 epidemics in the United States | COVID Tracking Project (https://covidtracking.com) | A generalized linear model (GLM) | Different NPIs showed different levels of reproduction numbers. The stay-at-home played the most important role and contributed approximately 51% (95% CI: 46%−57%). The gathering ban (more than 50 people) was not very important, which only contributed 7% (2%−11%). |
| Tian et al. (July 2020) [150] | Compare the effect of mild interventions in Shenzhen and countries in the United States | Daily cumulative confirmed cases of COVID-19 in Shenzhen, China and the countries in the United States (https://github.com/CSSEGISandData/COVID-19) | A synthetic control method with a modified selection of control variables and the proposed SIHR model | Implementing the early mild interventions has the potential to subdue the epidemic of COVID-19. |
| Zou et al. (May 2020) [23] | Forecast the spread of COVID-19 | The Johns Hopkins University Center for Systems Science and Engineering; The New York Times data; The data from most states between 03/22/2020 and 05/10/2020. More than 40,000 cases. |
SuEIR model | The proposed model has been adopted by the CDC for COVID-19 death forecasts. |
| Friedman et al. (May 2021) [151] | Predict mortality of patients with COVID-19 | Public data: https://github.com/pyliu47/covidcompare. | SEIR model, Dynamic Growth, SIKJalpha. | Seven predictive models that showed better performance which had a median absolute percent error of 7% to 13% at six weeks. |
| Murray et al. (March 2020) [152] | Predict hospital bed-days, ICU-days, ventilator-days and deaths | Data from local government, national government, and WHO websites were used. | A statistical model based on parametrized Gaussian error function | They forecasted total beds (64,175), ICU beds (17,380), ventilators (19,481), deaths (81,114) at the peak of COVID-19 in the United States between March to June 2020. |
| Hsiang et al. (September 2020) [153] | Investigate the effect (rate of transmission) of non-pharmaceutical public health interventions on COVID-19 epidemics in China, South Korea, Italy, Iran, France and the United States | COVID-19 data collected from government reports, policy briefings and news articles (https://github.com/bolliger32/gpl-covid) | Reduced-form econometric model | The proposed model showed the interventions can reduce the rate of transmission and delay on the order of 61 million confirmed cases across 6 countries. |
| Li et al. (January 2021) [154] | Predict the epidemic trends in terms of future confirmed cases within 7 days | Coronavirus Update (Live): (https://www.worldometers.info/coronavirus/) Coronavirus (COVID-19) Lockdown Tracker Aura Vision. (https://auravision.ai/covid19-lockdown-tracker/) List of countries and dependencies by population: (https://en.wikipedia.org/w/index.php?title=List_of_countries_and_dependencies_by_population&oldid=960653268) |
A transfer learning method called ALeRT-COVID using attention-based RNN architecture | ALeRT-COVID obtained a higher prediction in terms of future confirmed cases |
| Wang et al. (May 2021) [155] | Investigate the impact of the temperature and relative humidity on effective reproductive number in COVID-19 epidemics | Records of 69,498 patients from Chinese National Notifiable Disease Reporting System and 740,843 confirmed cases from COVID-19 database of JHU CSSE (https://github.com/CSSEGISandData/COVID-19/). | Fama-Macbeth Regression [34] | High temperature and humidity can make contributions to the reduction of the transmission of COVID-19. |
| Rockett et al. (July 2020) [25] | Revealing COVID-19 transmission in Australia | Data collected from infected patients during the first 10 weeks of COVID-19 containment in Australia, which reported by New South Wales (NSW) Ministry of Health | Agent-based model | The predictions from ABM were concordant with the local transmission rates. |
| Alzu'bi et al. (December 2020) [25] | Investigate the effect of non-pharmaceutical public health interventions on COVID-19 epidemics | Coronavirus data collected from two urban neighborhoods separated by crossings. 1,000 persons. |
Agent-based model by extending the SIR model | The policies including staying home and hospital isolation policies, and preventing travel between cities made contributions to the reduction of the prevalence and the deaths. |
| Brauer et al. (May 2021) [156] | Estimated global access to handwashing with soap and water | Observational surveys in the context of the Global Burden of Diseases, Injuries, and Risk Factors Study in terms of access to a handwashing station with available soap and water for 1,062 locations from 1990 to 2019. | Spatiotemporal Gaussian process regression modeling | The handwashing access should be considered when building the forecasting models of COVID-19 in terms of low-income counties. |
| Jr et al. (October 2020) [157] | Investigate the effect of social distancing mandates and levels of mask use | COVID-19 case and mortality data from 1 February 2020 to 21 September 2020 in the United States | SEIR model | Keeping universal mask use was enough to relieve the worst effects of epidemic resurgences in multiple states in the United States. Keeping social distancing was helpful for reducing the number of deaths for patients with COVID-19. |
Table 2.
The summary of studies in terms of the applications of AI in drug repurposing
| Reference | Method | Data source & size | Number of identified drug candidates | Identified drug candidates |
|---|---|---|---|---|
| Zhou et al. (March 2020) [158] | Network-based method (drug–target network; human protein–protein interaction network) |
DrugBank database (v4.3), Therapeutic Target Database (TTD), PharmGKB database, ChEMBL (Sv20), BindingDB, and IUPHAR/BPS Guide to PHARMACOLOGY. And other 18 bioinformatics and systems biology databases including 351,444 unique PPIs (edges or links) connecting 17,706 proteins (nodes). |
16 drug candidates and 3 drug combinations |
Candidates: Irbesartan; Toremifene; Camphor; Equilin; Mesalazine; Mercaptopurine; Paroxetine; Sirolimus; Carvedilol; Colchicine; Dactinomycin; Melatonin; Quinacrine; Eplerenone; Emodin; Oxymetholone. Combinations: sirolimus plus dactinomycin, mercaptopurine plus melatonin, and toremifene plus emodin. |
| Zeng et al. (July 2020) [65] | Knowledge-graph and deep learning |
24 million Pubmed research articles. A built knowledge graph contains 15 million edges, 39 types of relationships among nodes including drugs, diseases, proteins/genes, pathways, and expression. |
41 | Tetrandrine, Nadide, Estradiol, and so on (see Table 1 of this reference) |
| Gysi et al. (May 2021) [61] | Network-based method including network proximity, network diffusion, and AI-Net | 21 public databases for compiling protein-protein interactions (PPI) data including 18,505 proteins and 327,924 interactions between them; DrugBank database for obtaining drug-target information including 26,167 interactions between 7,591 drugs and their 4,187 targets. |
4 | Auranofin, Azelastine, Digoxin, and Vinblastine. |
| Wang et al. (May 2021) [159] | Knowledge-graph and deep learning |
25,534 peer-reviewed scientific articles. | 41 | Connecting 41 drugs based on Benazepril, Losartan, and Amodiaquine. |
| Zhang et al. (February 2021) [160] |
Knowledge-graph and deep learning |
PubMed, LitCovid, COVID-19. The built knowledge graph has 131,355 nodes and 2558,935 relations. |
5 | Paclitaxel, SB 203,580, Alpha 2-antiplasmin, Metoclopramide, and Oxymatrine. |
| Gordon et al. (April 2020) [161] | Network-based method | Public sources such as An interactive protein–protein interaction map https://kroganlab.ucsf.edu/network-maps; databases such as ChEMBL [PMID: 27,899,562], ZINC[PMID: 26,479,676] and IUPHAR/BPS Guide to Pharmacology [PMID: 31,691,834]. |
69 | Silmitasertib, Bafilomycin A1, Haloperidol, Loratadine, Entacapone, and so on.(see Supplementary Tables 5 and 6 of this reference) |
| Beck et al. (March 2020) [162] | Knowledge-graph and deep learning |
Drug Target Common (DTC) database and BindingDB database. | 5 | Atazanavir, Remdesivir, Efavirenz, Ritonavir, and Dolutegravir. |
| Mall et al. (July 2020) [163] | Knowledge-graph and deep learning |
MOSES, ChEMBL, UniProt, PubChem and NCBI. | 19 | Remdesivir, lopinavir, Ritonavir, and Hydroxychloroquine (see Table 3 of this reference) |
3.2. Challenge and opportunities
Although computational drug repurposing has shown large potential for identifying effective drug candidates for treating COVID-19 infections, there remain challenges and opportunities for improving the efficiency of discovering drugs. Following are the main challenges and opportunities.
-
(1)
The current dataset for computational drug discovery is very small. Although a gigantic collection of GDB-17 has 166 billion compounds, it is only a tiny fragment of the chemical universe [66]. The ML methods may show poor performance when the model encounters compounds that the molecules have not been seen in train sets. The structure-based drug discovery needs accurate crystal structures to obtain better matching results in terms of proteins with drugs [44]. Building a larger and better dataset that contains more kinds of accurate crystal structures is beneficial for drug discovery, which may need more time, money and expertise.
-
(2)
The Biomedical knowledge graph (BKG)-based approaches for drug development typically rely on the quality of the BKG used. Different resources were used to build the BKGs in different projects, which may hence produce bias during discovering the promising repurposing drug candidates of COVID-19. Efforts such as by Heteionet [67] and our BKG [68] aiming at incorporating and harmonizing data from diverse medical domains and resources to build comprehensive BKGs. However, there is no golden standard to evaluate their quality. This may limit the reliability of the identified therapeutic implications.
-
(3)
Computational data scientists need to work closely with chemists or doctors, which is very crucial for better outcomes. For example, extracting a broad range of properties of molecules based on domain knowledge from chemical experts helps obtain a complete representation of molecules; then feeding them to ML models can improve model performance. Few clinicians and medical school students may need manually reviewed clinical reports to aid model training during BKG building, which may involve bias. More domain experts should work on them and the model developer should iteratively combine feedback from doctors who utilized the developed tool.
4. AI in COVID-19 clinical research
The studies of AI in COVID-19 clinical research can roughly be divided into two types (Table 3 ): the diagnostic and prognostic prediction of COVID-19 and the subphenotyping of COVID-19. For the former, researchers use ML techniques to build classifiers to identify or predict whether patients are suffering from COVID-19 or to assess different levels of severity of COVID-19. For the latter, researchers focus on using clustering methods to identify sub-groups, and further investigate the different characteristics such as hospitalization, intensive services, and death of these sub-groups.
Table 3.
The summary of studies in terms of the applications of AI in clinical research
| Reference | Task | Data source & size | Method | Results |
|---|---|---|---|---|
| Su et al. (March 2021) [164] | Explore albumin level between patients with COVID-19 and patients with sepsis. | 308 patients with COVID-19 and 363 patients with Sepsis | Chow's test, linear mixed-effects models, Fisher's exact test, t-test, and Wilcoxon rank-sum test | Two phases of alterations in albumin levels for patients with COVID-19 were found, which were not presented with patients with sepsis. |
| Liang et al. (May 2021) [85] | Estimate the risk of developing critical illness for patients with COVID-19 | 72 potential predictors were considered from 1,590 patients with COVID-19 in the 575 hospitals of 31 provincial administrative regions in China as of January 31, 2020. | Least Absolute Shrinkage and Selection Operator (LASSO) and Logistic Regression (LR) models | AUC=0.88 (95% CI, 0.84–0.93) on a validation cohort with 710 patients. |
| Burn et al. (October 2020) [165] | Explore the characteristics of patients with COVID-19 and influenza | 34,128 adult patients with COVID-19 and 84,585 patients with influenza (United States: 8,362, South Korea: 7,341, Spain: 18,425) |
Data-driven approach | Compared to patients with influenza, patients with COVID-19 were more male, younger, and with fewer comorbidities and lower medication use. |
| Roth et al. (May 2021) [166] | Investigate the characteristics of patients with COVID-19 in terms of in-hospital mortality in the United States | 20,736 adults with a diagnosis of COVID-19 in the US between March and November 2020. | A multiple mixed-effects logistic regression | The mortality rates for patients with COVID-19 were different between the months of March and April and later months in 2020, which were not fully explained by changes in age, sex, comorbidities, and disease severity. |
| Williams et al. (May 2021) [86] | Predict hospitalization, intensive services, and death for patients with COVID-19 | The cohort for model development has More than 2 million patients diagnosed with influenza or flu-like symptoms any time prior to 2020. The cohort for model validation included 43,061 COVID-19 patients form South Korea, Spain and the United States. |
Data-driven approach | The ranges of AUC on validation in terms of three outcomes including hospitalization, intensive services, and death were 0.73–0.81, 0.73–0.91, and 0.82–0.90, respectively. |
| Liang et al. (July 2020) [90] | Predict the risk of COVID-19 patients developing critical illness | 74 baseline clinical features at admission from 1,590 patients with COVID-19 in the 575 hospitals of 31 provincial administrative regions in China as of January 31, 2020. | Feedforward neural network. | The proposed model was validated on three separate cohorts including 1,393 patients and showed the concordance index of 0.890, 0.852, and 0.967, respectively. |
| Yang et al. (December 2020) [167] | Investigate population drifting in terms of COVID-19 patients | 21 routine blood tests from 5,785 patients in ED of New York Presbyterian Hospital/Weill Cornell Medical Center (NYPH/WCMC) between March 11 and June 30,2020. | Density-based spatial clustering of applications with noise (DBSCAN) and the Unified manifold approximation and projection (UMAP), t-test, Fisher's exact test. | The number of SARS-CoV-2 patients with the COVID-19 HRP became less and less from March to June 2020. |
| Zhang et al. (June 2020) [5] | Diagnose COVID-19 | 532,506 human lung CT scan images from 3,777 patients, China Consortium of Chest CT Image Investigation (CC—CCII) | CNN | Internal validation: Accuracy=92.49%; External validation: Accuracy=90.70%. |
| Wang et al. (May 2020) [75] |
Diagnose COVID-19 | Lung CT images: 5,372 patients from seven cities or provinces in China. | A fully automatic DL model (DenseNet121-FPN) | AUC 0.87 and 0.88 on two validation sets in distinguishing COVID-19 from other pneumonia and AUC 0.86 in distinguishing COVID-19 from viral pneumonia. |
| Ozturk et al. (June 2020) [78] | Diagnose COVID-19 | X-ray images: 127 COVID-19 cases, 500 no-finding, 500 pneumonia. The Cohen JP and the ChestX-ray8 databases |
CNN | An accuracy of 98.08% for classifying COVID-19 and No-findings and 87.02% for classifying COVID-19, No-findings, and Pneumonia. |
| Chen et al. (October 2020) [87] |
Predict the severity of COVID-19 | 52 features from 362 patients with COVID-19 including 214 non-severe and 148 severe cases in China. |
RF | 95% accuracy when considering all features and 99% accuracy when only using top 10 important features selected by Gini impurity. |
| Xu et al. (October 2020) [73] |
Diagnose COVID-19 | 618 CT images in total. 219 samples from 110 patients with COVID-19; 224 samples from 224 patients with IAVP; 175 samples from 175 healthy cases. These samples are from China. |
CNN | Accuracy = 86.7% |
| Avila et al. (June 2020) [88] | Predict COVID-19 | 510 patients including 73 positives for COVID-19 and 437 negatives were from the emergency department of Hospital Israelita Albert Einstein (HIAE, São Paulo, Brazil). |
Gaussian Naïve Bayes (NB) | 100% sensitivity and 22.6% specificity, 76.7% for both sensitivity and specificity, and 0% sensitivity and 100% specificity when prior values were set to 0.9999, 0.2933, 0.001, respectively. |
| An et al. (October 2020) [89] | Predict mortality for patients with COVID-19 | Sociodemographic and medical information from 10,237 patients with COVID-19 in a nationwide Korean cohort. |
LASSO, SVM and RF | The LASSO model obtained best AUC (0.962 (0.945- 0.979)), and identified several significant predictors such as old age and preexisting DM or cancer. |
| Mei et al. (May 2020) [71] | Diagnose COVID-19 | CT scan images and non-image information such as demographic and laboratory tests from 905 patients between 17 January 2020 and 3 March 2020 from 18 medical centers in 13 provinces in China. | CNN+MLP | AUC=0.92 on a test set with 279 patients. |
| Ardakani et al. (June 2020) [74] | Diagnose COVID-19 | 1,020 CT images from 108 patients in Iran University of Medical Sciences (IUMS) hospital. |
CNN (ResNet-101) | AUC = 0.994, Sensitivity = 100%, Specificity = 99.02%, Accuracy = 99.51%. |
| Yang et al. (November 2020) [72] | Predict COVID-19 | Demographic information (i.e., age, sex, race) and 27 routine lab tests from 3,356 SARS-CoV-2 RT-PCR tested patients. These tests were from NYPH/WCM dataset. |
Gradient boosting decision tree (GBDT) | AUC = 0.854 (95% CI: 0.829–0.878). |
| Roy et al. (August 2020) [83] | Diagnose COVID-19 | Italian COVID-19 Lung Ultrasound DataBase: 277 lung ultrasound videos from 35 patients, corresponding to 58,924 images. | Spatial Transformer Networks and CNN | Accurate prediction and localization of COVID-19 imaging biomarkers in three tasks including frame-based classification, video-level grading and pathological artifact segmentation. |
| Narin et al. (May 2020) [76] | Diagnose COVID-19 | 341 images from COVID-19 patients, 2,800 normal chest images, 1,493 viral pneumonia and 2,772 bacterial chest X-ray images | CNN | 96.1%, 99.5%, and 99.7% accuracy on three datasets, respectively. |
| Jain et al. (September 2020) [79] | Diagnose COVID-19 | 1,832 X-ray images strengthened from original 1,215 X-ray images by using data augmentation techniques |
CNN (ResNet-50) | Training-validation-testing: accuracy, recall, and precision were 99.77%, 97.14%, and 97.14%, respectively. 5-fold cross validation: average accuracy, sensitivity, specificity, precision, and F1-score were 98.93%, 98.93%, 98.66%, 96.39%, and 98.15%, respectively. |
| Wang et al. (November 2020) [77] | Diagnose COVID-19 | Two datasets including 1,102 and 625 chest X-ray images, respectively. | CNN and SVM | 99.33%, and 95.02% accuracy on two datasets, respectively. |
| Loey et al. (April 2020) [84] | Detect COVID-19 | 8,100 chest X-ray images strengthened from original 306 chest X-ray images by using data augmentation techniques. | GAN with deep transfer learning | Testing sets: 100% accuracy; Validation set: 99.9% accuracy. |
| Li et al. (September 2020) [100] | Diagnose COVID-19; Identify subphenotypes |
Public dataset: 413 patients with COVID-19 and 1,071 patients with influenza | XGBoost model; a self-organizing map (SOM) |
Sensitivity = 92.5%; Specificity = 97.9%; Identified 4 subphenotypes which showed much difference in terms of gender distribution and levels of CRP and serum immune cells. |
| Zhou et al. (April 2020) [168] | Identify subphenotypes | Mexican Government COVID-19 open data including 778,692 COVID-19 patients. | meta-clustering technique | Identify 3 clusters which showed different recovery rates |
| Su et al. (July 2020) [102] | Identify subphenotypes | NYP-WCMC eligible 318 patients extracted from 1,661 patients with COVID-19 and NYP-LMH eligible 84 patients extracted from 458 patients with COVID-19. | Dynamic time warping and hierarchical agglomerative clustering method | Discovered distinct worsening and recovering subphenotypes within three strata including mild, intermediate, and severe strata. |
| V.Bhavani (December 2020) [103] | Identify subphenotypes | 696 hospitalized patients in University of Chicago Medicine | Group-based trajectory modeling (GBTM) | Discovered 4 subphenotypes which were different in experiencing cytokine storm, coagulopathy, and cardiac and renal injury. |
| Lascarrou et al. (March 2021) [97] | Identify subphenotypes | 416 COVID-19 patients with moderate to severe ARDS at 21 intensive care units in Belgium and France. | Hierarchical clustering method | Identified 3 subphenotypes which have different characteristics on comorbidities, mortality, sex, the duration of symptoms, plateau and driving pressure. |
| Legrand et al. (October 2020) [96] | Identify subphenotypes | 608 patients in at eight teaching hospitals of the Assistance Pub- lique-Hôpitaux de Paris | Consensus cluster analysis method | Identified 3 subphenotypes which are different in terms of a history of chronic hypertension, the presence of fever, respiratory and non-respiratory symptoms, and age. |
| Schinkel et al. (February 2021) [98] | Identify subphenotypes | 2,019 patients collected from COVID Predict project in the Netherlands. | Consensus cluster analysis method | Identified 3 subphenotypes which showed much difference in terms of demographics, comorbidities, and clinical outcomes. |
| Su et al. (July 2021) [99] | Identify subphenotypes | Development cohort with 8,199 patients and internal and external validation cohorts both with 3,519 patients. Those patients were from five major medical centers in New York City (NYC), between March 1 and June 12, 2020. | Data-driven (agglomerative hierarchical clustering model) | Identified 4 subphenotypes which showed much difference in terms of demographics, clinical variables, comorbidities, clinical outcomes, and medication treatments |
4.1. The diagnostic and prognostic prediction in COVID-19
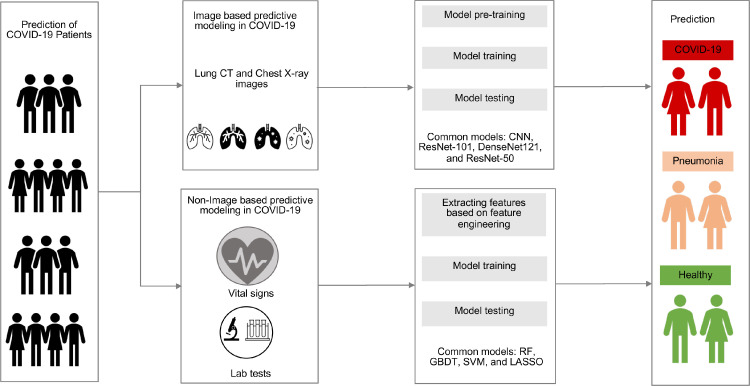
Early and rapid identification of COVID-19 is urgently needed [69], [70], which is important not only for immediate management and treatment of individual patient care but also provides guildance for public health in terms of patient isolation and COVID-19 containment [71]. A COVID-19 virus-specific reverse transcriptase-polymerase chain reaction (RT-PCR) test is widely utilized to detect the COVID-19 virus [72]. However, this test usually takes up to two days to inform final results, and serial tests may be considered to exclude the possibility of false-negative results, which may underestimate the situation of the COVID-19 pandemic, hindering government control in terms of disease transmission and healthcare workforce [71]. Recently, researchers have used data-driven methods to build classifiers with historical medical information for diagnosis or prediction. In particular, researchers have used image or non-image medical information to build classifiers. For image-based studies, they trained classifiers with ML methods using extracted features from medical images, such as human lung CT scan images, chest X-ray, and ultrasound images, or use DL models to build classifiers on raw medical images. For non-image-based studies, they extracted EHR information such as routine lab tests and integrated ML models to train a classifier or build a score system using selected predictors. A general framework of using AI techniques for COVID-19 patients’ prediction is shown in Figure 3 .
Figure 3.
A general framework of using ML (machine learning) and DL (deep learning) techniques in COVID-19 diagnostic and prognostic prediction.
4.1.1. Image-based predictive modeling in COVID-19
Three common types of images including CT images, chest X-rays, and ultrasound images are used to build classifiers to perform COVID-19 diagnosis. With a more accurate tool in CT scans, CT images usually contain more information that is useful for COVID-19 diagnosis [5]. Most previous CT image- based studies mainly use CNN for COVID-19 diagnosis [5, 73-75]. For example, Xu et al. [73] used a CNN architecture to extract lung CT image spatial features from 618 CT images for diagnosing COVID-19, influenza-A viral pneumonia, and healthy cases. Although CT image is a valuable component for COVID-19 diagnosis, CT imaging usually takes more time than X-ray imaging and causes more harm for patients because of more radiation exposure. In addition, compared to CT imaging machines, the equipment for X-ray is cost-effective and easy to operate, which attracted researchers’ attention to COVID-19 diagnosis [76], [77], [78], [79]. For example, Wang et al. [77] built a hybrid model with CNN and SVM for diagnosing COVID-19 on two datasets including 1,102 and 625 chest X-ray images and obtained an accuracy of 99.33% and 95.02% of accuracy, respectively. More recently, clinicians reported that lung ultrasound images can show higher sensitivity than by chest X-rays in diagnosing pneumonia in some cases [80], [81]. Due to the characteristics of a more widely available, lower cost, more safe, and real-time ultrasound imaging technique, using lung ultrasound images for diagnosis of COVID-19 is gaining wide popularity [82], [83]. Roy et al. [83] used lung ultrasound images to predict disease severity using a deep network by integrating spatial transformer networks and CNN, which showed accurate prediction and localization of COVID-19 imaging biomarkers. These previous studies with images mainly use DL techniques to extract spatial information and build classifiers, which need more samples for training classifiers to obtain the best performance. Data augment techniques face the challenge of the lack of medical images for COVID-19 diagnosis [79, 84]. Loey et al. [84] used a generative adversarial network (GAN) with deep transfer learning-based data augmentation techniques to strengthen original 306 chest X-ray images to 8100 images for COVID-19 detection.
4.1.2. Non-image based predictive modeling in COVID-19
Non-image-based classification of COVID-19 focuses on using EHR information to diagnose COVID-19, which consists of two types of studies: score system-based and ML-based COVID-19 diagnosis. For the former, researchers seek to identify important predictors, assign to their scores, sum these scores, and discriminate the severity of disease [85], [86]. For example, Liang et al. [85] built a predictive risk score (COVID-GRAM) system, which included 10 important predictive factors that were screened from 72 potential predictors among epidemiological, clinical, laboratory, and imaging variables, to estimate the risk of developing critical illness for patients with COVID-19 admitted to the hospital. One limitation of these studies is that more professional clinical knowledge or experience is needed for selecting important predictors. Recently, the latter method was widely used for COVID-19 diagnosis [72, 87-89]. Yang et al. [72] built a gradient boosting decision tree (GBDT) model to predict an individual's COVID-19 infection status using three demographic information (i.e., age, sex, race) and 27 routine lab tests, which obtained an AUC of 0.854. With the advance of DL and the availability of EHR information, the DL architectures are gaining more attention for diagnosing COVID-19. Liang et al. [90] built a feedforward neural network-based DL survival model to predict the risk of COVID-19 patients developing critical illness using 74 baseline clinical features at admission from 1,590 patients in 575 medical centers. The proposed model was validated on three separate cohorts including 1,393 patients and showed a high concordance index of 0.890, 0.852, and 0.967.
4.2. The subphenotyping of patients with COVID-19
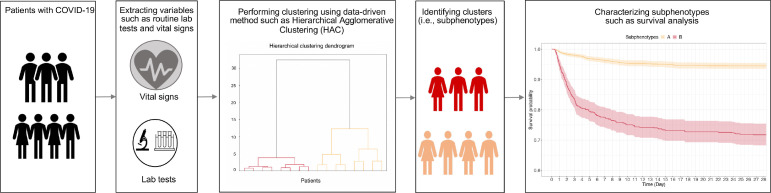
Clinical subphenotyping involves dividing patients who share a phenotype into several clusters [91]. Patients in the same cluster have similar characteristics such as demographics, clinical characteristics, treatments, comorbidities, and outcomes, which differentiate the cluster from other clusters [92]. The identification of subphenotypes helps understand the pathophysiology of critical care syndromes and can lead to personalized treatment and management [93]. Recently, data-driven subphenotyping has been explored for multiple diseases such as sepsis [94], asthma, and allergies [95]. A general framework of using AI techniques for subphenotyping patients is shown in Figure 4 .
Figure 4.
A general framework of using AI techniques for the subphenotyping of patients with COVID-19. SOM: Self-Organizing Map; HAC: Hierarchical Agglomerative Clustering.
The studies of COVID-19 subphenotyping can roughly be divided into two categories: static subphenotyping and dynamic subphenotyping. For the former, the researchers first extract patient clinical variables presenting at admission to the emergency department, hospitalization, or ICU, and then use clustering methods such as hierarchical clustering method, consensus cluster analysis method, and self-organizing map (SOM) to identify clusters, and finally investigate the characteristics such as comorbidities and outcomes of these clusters [96], [97], [98], [99], [100]. For example, Su et al. [99] employed an agglomerative hierarchical clustering model and 30 routinely clinical variables to identify 4 subphenotypes among 8,199 patients with COVID-19 and validated them on internal and external cohorts both with 3,519 patients. There were many differences among discovered subphenotypes in terms of demographics, clinical variables, comorbidities, clinical outcomes, and medication treatments. Li et al. [100] used the SOM method and identified four subphenotypes on 48 clinical variables from 398 patients. These four discovered subphenotypes showed different characteristics. These static variable- based subphenotyping studies mainly identify the short-term subphenotypes, which may ignore the information in terms of the progress of disease and treatment. Although previous studies have discovered several subphenotypes, static assessments of COVID-19 may be incomplete due to the variable presentation to healthcare after developing symptoms and the evolution of organ failure in critical care [101].
For the dynamic subphenotyping, the researchers considered the trajectory of variables during a long- term period such as three days and used trajectory-based clustering methods such as dynamic time warping (DTW) [102] and group-based trajectory modeling (GBTM) to identify clusters [103]. For example, Bhavani et al. [103] used the dynamic trajectories of COVID-19 patient temperature to identify subphenotypes. The differential pattern of temperature change may provide cues to a varied underlying inflammatory response to infection. However, this study only used the trajectory of a single variable, which may ignore the influence of other organ dysfunction. To consider trajectory from multiple organ dysfunction can refine the understanding of the natural history of COVID-19 in response to standard of care treatment and define patterns of disease that may benefit from novel therapeutic strategies [104]. Su et al. [102] used the trajectory of sequential organ failure assessment that described dysfunctions in six organs including respiration, coagulation, liver, cardiovascular, central nervous system, and renal system to identify subphenotypes among the critically ill patients with COVID-19. They discovered distinct worsening and recovering subphenotypes within different baseline severity strata. Compared to baseline severity of illness, demographics and comorbidities, dynamic inflammatory markers and ventilator variables showed significant difference between worsening and recovering subphenotypes. These dynamic variable-based subphenotyping studies consider the longitudinal variable trajectories and have demonstrated great promise to achieve unique insights into the multiorgan dysfunction.
4.3. Challenge and opportunities
Although clinical research including building predictive models and subphenotyping COVID-19 patients has been paid more attention and promising initial results have been obtained, there are some challenges or opportunities. A few are mentioned here below.
-
(1)
Most of the previous clinical research in COVID-19 mainly used structured information such as demographics, lab tests, vital signs, to build the representation of patients for ML modeling. Unstructured information such as clinical notes, the reports of CT scan images may contain more detailed information for COVID-19 diagnosis. For example, Obeid et al. [105] performed text information analysis based on patients’ self-reported symptoms to predict COVID-19 infection risk by a word embedding-based CNN. The unstructured information can be used as complementary information for structured information [106]. Integrating structured and unstructured information can completely represent the patient and improve model performance. How to integrate this information still needs to be investigated by researchers.
-
(2)
For COVID-19 subphenotyping studies, validating the discovered subphenotypes on external sites is very important. However, the distribution difference between derivation cohort and validation cohort such as the size of cohort or heterogeneity of risk factors may generate different subphenotypes. Designing a method to measure the discrepancy of distribution and integrating them into an ML model may make contributions for identifying subphenotypes.
-
(3)
Current static variables-based subphenotyping studies mainly identify subphenotypes for patients at admission to the emergency department or ICU. These discovered subphenotypes may be too early for those patients, which may ignore the progress of COVID-19. Choosing proper time such as the first six hours after admission for subphenotyping patients may be able to avoid premature phenotyping [94].
-
(4)
Although dynamic-based COVID-19 subphenotyping considered the longitudinal trajectories and has the potential to obtain a comprehensive understanding in terms of the natural history of COVID-19, it is still challenging to set a proper time interval for extracting features and building a representation for each patient based on trajectory.
5. AI in COVID-19 on behavioral and social sciences
The outbreak of COVID-19 produced an impact on people's daily behavior. Several specific topics including information search behavior change, the impact of misinformation, psychosocial impacts, mobility network, and contact tracing have been investigated. In particular, for information search behavior change, researchers want to know what kinds of key information would be searched popularly by citizens during the COVID-19 pandemic. For example, Bento et al. [107] investigated information-seeking responses to the first COVID-19 case public announcement in a state. They found more people searched information in terms of “coronavirus”, “coronavirus symptoms”, and “hand sanitizer” after the first case announcement, which increased by about 36% (95% CI: 27% - 44%) on the day immediately and fell back to the baseline level in less than a week or two. The information about community-level policies such as quarantine and personal health strategies such as grocery delivery was not paid more attention, which indicated the study period was relatively early in the epidemic and there were limited elaborate policies from public discourse.
Investigating the information search behavior change can help the government to take proper measures. However, there is a large amount of misinformation in terms of COVID-19, which may mislead people's decisions [108], [109], [110]. Bursztyn et al. [111] discovered the relationship between misinformation and health outcomes based on the two most popular cable news shows (including Hannity and Tucker Carlson Tonight in the United States). An epidemiological model was used to measure the magnitudes in terms of treatment effects, which highlighted the relevance of externalities. Bursztyn et al. reported that misinformation on mass media had significant social consequences. In order to identify low credibility news, Zhou et al. [112] constructed a repository based on 2,029 articles from about 2,000 news publishers and 140,820 tweets, which included multiple types of information on coronavirus, such as textual, visual, temporal, and network information. Several ML methods-based predictive models were built for identifying fake news and obtaining competitive performance.
Investigating social media can find the cues in terms of psychosocial impacts during COVID-19 [113], [114], [115]. Saha et al. [116] discovered the temporal and linguistic changes in symptomatic mental health and support expressions during the COVID-19 pandemic by comparing Twitter streaming posts collected in 2020 and 2019. They found no significant increase in terms of people's mental health symptoms and support expressions during the COVID-19 period. Linguistic analyses showed that people express more concerns in terms of the COVID-19 crisis. Zhang et al. [117] built a fusion classifier that integrated the DL model, psychological text features, and demographic information to investigate the relationships between feature and depression signals. The proposed model demonstrated an accuracy of 78.9% and has been used to analyze the depression level of different groups of people on Twitter in terms of three US states (New York, California, and Florida). These researchers found that people in Florida had a substantially lower level of depression.
In addition, investigating the spread patterns of cases and tracking individuals' movements are useful for controlling the spread of COVID-19. Chang et al. [31] used a metapopulation susceptible-exposed-infectious-removed (SEIR) model based on fine-grained and dynamic mobility networks to investigate the spread of COVID-19 in 10 US metropolitan areas. The built SEIR model can fit a real case trajectory and reveals that setting specific occupancy for different points of interest is more effective than uniformly restricting mobility. With the wide use of smartphones, developing apps can facilitate the tracking of individuals' movements. Ahmed et al. [118] introduced three different kinds of smartphone contact tracing apps based on different ways of using servers and storing data, including centralized, decentralized, and hybrid architecture contact tracing apps. These apps have been used to identify and trace all recent contacts of newly discovered infected individuals.
5.1. Challenge and opportunities
Although more and more researchers have paid attention to behavioral and social sciences during the COVID-19 pandemic, the following challenges or opportunities remain.
-
(1)
The bias of data source: most previous studies used social media data from multiple sources such as different news publishers or Tweet posts. There may be bias for those news publishers. Identifying those biases and integrating them into the model may help in detailed analysis. In addition, there are other types of data such as image and video, which can be integrated with text information to provide more insights for the analysis of COVID-19.
-
(2)
Data privacy for tracing apps: although current apps have used some techniques such as decentralized contact tracing to keep privacy, a fully decentralized architecture has not been proposed [118]. A technique of using a peer-to-peer network may facilitate privacy-preserving information sharing amongst the user devices.
-
(3)
Behavioral changes in different groups such as old and young groups may be different during the COVID-19 pandemic [119]. Interesting findings may show if researchers perform more fine-grained analysis. The government would take proper measures to assist those who may suffer from severe health problems such as depression or anxiety in different groups.
-
(4)
Most previous studies on behavioral changes mainly focused on patients with COVID-19. Currently, with more and more citizens getting vaccinated, investigating the changes in mental health problems after vaccination may be interesting.
6. Discussion: existing challenges and potential future directions
In previous sections, we reviewed studies using AI to address the COVID-19 pandemic from epidemiology, therapeutics, clinical research, social and behavioral aspects, and discussed the potential challenges and opportunities for each kind of application. This section will discuss existing challenges, potential directions, and open questions from a general perspective.
6.1. Model interpretation
Model interpretation is very important in the medical domain because model outputs (e.g., diagnosis) without reasonable reasons make no sense to clinicians [120]. Different types of models may need different types of explainability. Previous models for COVID-19 analysis can be divided into two types: models built on ML and non-ML techniques. For models built on non-ML techniques such as using risk scores, it is not very difficult to explain final results by investigating each risk variable and other related clinical EHR information, which can be seen as intrinsic interpretability [121]. For example, Liang et al. [85] built a predictive risk score (COVID-GRAM) system, which used 10 important factors from the epidemiological, clinical, laboratory, and imaging variables, to estimate the risk of developing critical illness for patients with COVID-19 admitted to the hospital. When interpreting the COVID-GRAM, clinicians only check scores of specific variables among these 10 factors. In addition, clinicians may modify their assessment if they find some values of variables are abnormal. Intrinsically interpretable models based on non-ML techniques can provide better interpretability for clinicians; however, building models based on risk factors is not easy because more professional knowledge is necessary for developers to choose important risk factors.
With a larger amount of EHR, an increasing number of ML models have been used in clinical applications. For models based on ML techniques, the interpretability can be seen as post hoc interpretation [121]. There are two directions for obtaining interpretable ML modeling. One is to derive explainable tools that show the contribution of input features to the final output. Several explainable tools such as local interpretable model-agnostic (LIME) [122] and Shapley Additive Explanation [123] have been developed to determine the feature contributions by assigning importance scores. Adding attention mechanisms to hidden layers in DL models can also contribute to model interpretability [124]. Another key aspect is to interpret complex models based on multiple relatively simple models. For example, outputting the results of each convolutional layer of CNN in identifying specific regions of an image may provide cues for explaining final output results [125]. In addition, considering different levels of explainability in different applications may be sensible [121]. For example, clinicians may be relatively comfortable utilizing black-box models for some specific clinical applications (e.g., image analysis) that clinicians can readily intervene in. On the other hand, applying the black-box model to address unexplored problems may cause less comfort for clinicians.
6.2. Model security
Though ML models have been widely used in COVID-19 related applications, increasing evidence has shown that existing ML models could be fooled by adversarial examples and hence it is hard to obtain desirable performance [126], [127]. Adversarial examples are models’ inputs that are intentionally designed to make a mistake such as misclassification for identifying COVID-19 cases on medical IoT (Internet of Things) devices, which may poison the learning or the inference processes, and further compromising the security of ML models [128]. Recently, adversarial examples have been one of the most popular research topics in ML communities [129]. Although few studies in terms of adversarial examples have been conducted, two directions may be necessary for investigating the detection and defense mechanisms in terms of the COVID-19 DL poisoning process. One is to employ blockchain techniques to address adversarial example attacks on COVID-19 applications. For example, Nassar et al. [130] utilized blockchain to save benign attributes and parameters of each DL model, and furtherly transfer them to explainable AI for high-level users to check whether a particular model is compromised or not. Another is to study transferable adversarial examples [131], which can show better defense mechanisms against inference and model poisoning. Additionally, applying real-world attacks to test DL models and using industry standards such as IBM Adversarial Robustness Toolbox (ART) [132] to estimate and defend DL models against adversarial threats should be encouraged.
6.3. Model bias
AI techniques have become more ubiquitous for users to make or assist decisions in multiple domains such as recruiting (screening job applicants), banking (credit ratings/loan approvals), and judiciary (recidivism risk assessments). However, bias concern has been drawn more attention recently by researchers in terms of whether the learned scoring function in the ML model can make fair decisions in those real-world applications [133]. The bias in ML can be seen as the phenomenon of observing results that the learned model is systematically prejudiced across different groups defined by sensitive variables such as race or gender [134]. The bias may give rise to discrimination for protected groups and lead policymakers to make unfair decisions in real-world applications [135]. Detecting the bias and reducing its likelihood in model design and execution would play more critical roles in creating a fair treatment for specific populations [136].
The bias in ML that may cause discrimination can be roughly divided into three types [137], [138], namely disparate treatment, disparate impact, and disparate mistreatment. To better understand the different types of bias, we take an example (Table 4 ) in terms of a binary classification problem where the ML model learns whether a loan would be returned using n + 1 attributes, of which Q is a sensitive variable such as the user's race. For disparate treatment, it can be detected if the changing of a user's predicted label depends on the changing of the sensitive variable. The above example shows that the learned algorithm predicts positive labels for repaying a loan for the White user population and a negative one for the Black user population. Removing the sensitive variable during model training is a way to avoid the dependence on the sensitive variable. For disparate impact, it can be discovered whether the fraction of positive (negative) labels for the different sensitive groups is different. In terms of the above example, it means more percentage of Black people were classified as defaulters as compared to the White people. Removing the sensitive variable from the dataset is not an excellent way to prevent the disparate impact because other related features such as zip code may cause this issue. Checking the training dataset and making sure there is not much imbalance in terms of positive and negative samples may help prevent disparate impact. For disparate mistreatment, it can be detected if there is a difference in terms of the proportion of accurate labels for different sensitive groups [138]. This bias was found by Propublica in the Northpointe algorithm [139], which misclassified innocent Black defenders as reoffending at twice the rate as White people. Keeping the same percentages of accurate labels for all sensitive groups is useful for rectifying the mistreatment. The objective of ML is to optimize a cost function by minimizing the difference between function outputs and real results. Adding the constraints to the objective function by considering the above-mentioned bias can avoid discrimination. A trade-off between fairness and accuracy should be considered when adding constraints.
Table 4.
An example of a binary classification problem based on machine learning in terms of whether a loan would be returned using n + 1 attributes
| Variable_1 | Variable_2 | … | Variable_n | Q | Label (Y) | |
|---|---|---|---|---|---|---|
| Client_1 | F11 | F12 | … | F1n | Q1 | 1 |
| Client_2 | F21 | F22 | … | F2n | Q2 | 0 |
| … | … | … | … | … | … | … |
| Client_m | Fm1 | Fm2 | … | Fmn | Qm | 1 |
Q is a sensitive variable such as the user's race. Labels “0” and “1” represent “Returned” and “Defaulted”, respectively.
6.4. Privacy issue and model precision
Privacy concerns are very important in applications of ML for healthcare. The “differential privacy” technique has been used to ensure model and data privacy in a single dataset [140], [141]. For example, Chaudhuri et al. [142] proposed differential private approaches to preserve parameters obtained from logistic regression models or support vector machines (SVMs). However, it remains challenging for most AI models to address two issues: (1) more parameters to be safeguarded in DL models; (2) keeping privacy when integrating data from multiple sites. To maintain a balance between privacy and model precision, federated learning (FL) [143], a framework of constructing a central parameter server to train a global model based on the parameters from multiple local sites that store their own sensitive data, has attracted more attention and offer immense promise when integrating fragmented healthcare data from multiple medical sites with privacy-protection. More recently, Swarm Learning (SL) [144], a decentralized ML framework that integrates edge computing, blockchain-based peer-to-peer networking and dynamic central coordinator, has been paid more attention. Warnat-Herresthal et al. [144] used the SL framework to perform predictions in terms of COVID-19, tuberculosis, leukemia and lung pathologies to illustrate the feasibility of SL. Under the SL framework, a shared global model is trained with a dynamic central coordinator that aggregates parameters' from local sites keeping their sensitive data. Blockchain-based peer-to-peer networking is used to keep parameters privacy during data transferring. Thus, data and parameters offer double protection in SL, which can go beyond FL in real-world applications. Although the performance of FL and SL models is usually better than the model trained on single local sites, there exists ample scope for improvement compared to the central model trained by aggregating data from all local sites without any consideration of privacy. How to improve model performance remains an important problem. In addition, most applications using the FL and SW framework mainly focus on disease risk prediction that is relatively simple. Employing these models for more complex applications such as medical treatment and providing medication prescripts may be more worth exploring.
7. Conclusion
In this study, we reviewed existing studies on using AI techniques to deal with COVID-19 pandemic- related problems from four aspects including epidemiology, therapeutics, clinical research, social and behavioral studies. All the results available in that previous literature demonstrated the applicability and great promise of AI in addressing the COVID-19 pandemic. Also, some challenges, directions, and open questions are provided in this review, which may immensely help researchers to explore more related topics in the future.
Conflicts of interest statement
The authors declare no conflicts of interest.
Funding
The research was supported by National Science Foundation (Grant Nos. 1750326 and 2027970), National Institutes of Health (Grant Nos. RF1AG072449 and R01MH124740), as well as Amazon AWS Machine Learning for Research Award and Google Faculty Research Award. The work is also supported by Gates Foundation (Grant No. CORONAVIRUSHUB-D-21-00125).
Author contributions
Fei Wang planned and structured the whole paper. Zhenxing Xu and Chang Su conducted the literature review and drafted the manuscript. Zhenxing Xu, Chang Su, Yunyu Xiao and Fei Wang reviewed and edited the manuscript.
References
- 1.Liu Y, Gayle AA, Wilder-Smith A, et al. The reproductive number of COVID-19 is higher compared to SARS coronavirus. J Travel Med. 2020;27(2) doi: 10.1093/jtm/taaa021. : taaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chamola V, Hassija V, Gupta V, et al. A comprehensive review of the COVID-19 pandemic and the role of IoT, drones, AI, blockchain, and 5G in managing its impact. IEEE Access. 2020;8:90225–90265. doi: 10.1109/ACCESS.2020.2992341. [DOI] [Google Scholar]
- 3.WHO. WHO coronavirus (COVID-19) dashboard. 2021. Available from https://covid19.who.int/.
- 4.Cascella M, Rajnik M, Aleem A, et al. StatPearls. StatPearls Publishing; Treasure Island (FL): 2021. Features, evaluation, and treatment of coronavirus (COVID-19) July 30. [PubMed] [Google Scholar]
- 5.Zhang K., Liu X., Shen J., et al. Clinically applicable AI system for accurate diagnosis, quantitative measurements, and prognosis of covid-19 pneumonia using computed tomography. Cell. 2020;182(5):1360. doi: 10.1016/j.cell.2020.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song CY, Xu J, He JQ, et al. Immune dysfunction following COVID-19, especially in severe patients. Sci Rep. 2020;10(1):15838. doi: 10.1038/s41598-020-72718-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Su C, Xu Z, Pathak J, et al. Deep learning in mental health outcome research: a scoping review. Transl Psychiatry. 2020;10(1):116. doi: 10.1038/s41398-020-0780-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miotto R, Wang F, Wang S, et al. Deep learning for healthcare: review, opportunities and challenges. Brief Bioinform. 2018;19(6):1236–1246. doi: 10.1093/bib/bbx044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sipior JC. Considerations for development and use of AI in response to COVID-19. Int J Inf Manage. 2020;55 doi: 10.1016/j.ijinfomgt.2020.102170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nguyen DC, Ding M, Pathirana PN, et al. Blockchain and AI-based solutions to combat coronavirus (COVID-19)-like epidemics: a survey. IEEE Access. 2020;9:95730–95753. doi: 10.1109/ACCESS.2021.3093633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Islam MN, Inan TT, Rafi S, et al. A survey on the use of AI and ML for fighting the COVID-19 pandemic. arXiv preprint arXiv:2008.07449. 2020. [DOI] [PMC free article] [PubMed]
- 12.Hussain AA, Bouachir O, Al-Turjman F, et al. AI techniques for COVID-19. IEEE Access. 2020;8:128776–128795. doi: 10.1109/ACCESS.2020.3007939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pham QV, Nguyen DC, Huynh-The T, et al. Artificial intelligence (AI) and big data for coronavirus (COVID-19) pandemic: a survey on the state-of-the-arts. arXiv preprint arXiv:210714040. 2021. [DOI] [PMC free article] [PubMed]
- 14.Chen J, Li K, Zhang Z, et al. A survey on applications of artificial intelligence in fighting against covid-19. arXiv preprint arXiv:200702202. 2020.
- 15.Parbat D, Chakraborty M. A python based support vector regression model for prediction of COVID19 cases in India. Chaos Solitons Fractals. 2020;138 doi: 10.1016/j.chaos.2020.109942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020;20(5):533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kermack WO, McKendrick AG. Proceedings of the royal society of London Series A, containing papers of a mathematical and physical character. Vol. 115. 1927. A contribution to the mathematical theory of epidemics; pp. 700–721. [DOI] [Google Scholar]
- 18.Hethcote HW. The mathematics of infectious diseases. SIAM Rev. 2000;42:599–653. doi: 10.1137/S0036144500371907. [DOI] [Google Scholar]
- 19.Fang H, Chen J, Hu J. IEEE Engineering in medicine and biology society conference. Vol. 7. 2005. pp. 7470–7473. [DOI] [PubMed] [Google Scholar]
- 20.Wu JT, Leung K, Leung GM. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020;395(10225):689–697. doi: 10.1016/S0140-6736(20)30260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang B, Wang X, Li Q, et al. Estimation of the Transmission risk of the 2019-nCoV and its implication for public health interventions. J Clin Med. 2020;9(2):462. doi: 10.3390/jcm9020462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang T, Chu Y, Shams S, et al. Population stratification enables modeling effects of reopening policies on mortality and hospitalization rates. J Biomed Inform. 2021;119 doi: 10.1016/j.jbi.2021.103818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zou D, Wang L, Xu P, et al. Epidemic model guided machine learning for COVID-19 forecasts in the United States. medRxiv. 2020 [Google Scholar]
- 24.Zheng N, Du S, Wang J, et al. Predicting COVID-19 in China using hybrid AI model. IEEE Trans Cybern. 2020;50(7):2891–2904. doi: 10.1109/TCYB.2020.2990162. [DOI] [PubMed] [Google Scholar]
- 25.Rockett RJ, Arnott A, Lam C, et al. Revealing COVID-19 transmission in Australia by SARS-CoV-2 genome sequencing and agent-based modeling. Nat Med. 2020;26(9):1398–1404. doi: 10.1038/s41591-020-1000-7. [DOI] [PubMed] [Google Scholar]
- 26.Purkayastha S, Bhattacharyya R, Bhaduri R, et al. A comparison of five epidemiological models for transmission of SARS-CoV-2 in India. BMC Infect Dis. 2021;21(1):533. doi: 10.1186/s12879-021-06077-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kadi N, Khelfaoui M. Population density, a factor in the spread of COVID-19 in Algeria: statistic study. Bull Natl Res Cent. 2020;44(1):138. doi: 10.1186/s42269-020-00393-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pequeno P, Mendel B, Rosa C, et al. Air transportation, population density and temperature predict the spread of COVID-19 in Brazil. PeerJ. 2020;8:e9322. doi: 10.7717/peerj.9322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J, Tang K, Feng K, et al. Impact of temperature and relative humidity on the transmission of COVID-19: a modelling study in China and the United States. BMJ Open. 2021;11(2) doi: 10.1136/bmjopen-2020-043863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Senapati A, Rana S, Das T, et al. Impact of intervention on the spread of COVID-19 in India: a model based study. J Theor Biol. 2021;523 doi: 10.1016/j.jtbi.2021.110711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang S, Pierson E, Koh PW, et al. Mobility network models of COVID-19 explain inequities and inform reopening. Nature. 2021;589(7840):82–87. doi: 10.1038/s41586-020-2923-3. [DOI] [PubMed] [Google Scholar]
- 32.Li R, Pei S, Chen B, et al. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2) Science. 2020;368(6490):489–493. doi: 10.1126/science.abb3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Starr TN, Greaney AJ, Addetia A, et al. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science. 2021;371(6531):850–854. doi: 10.1126/science.abf9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sridhar A, Yağan O, Eletreby R, et al. ICASSP 2021 - 2021 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) IEEE; 2021. pp. 8163–8167. [Google Scholar]
- 35.Ozery-Flato M, Goldschmidt Y, Shaham O, et al. Framework for identifying drug repurposing candidates from observational healthcare data. JAMIA Open. 2020;3(4):536–544. doi: 10.1093/jamiaopen/ooaa048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paranjpe MD, Taubes A, Sirota M. Insights into Computational Drug Repurposing for Neurodegenerative Disease. Trends Pharmacol Sci. 2019;40(8):565–576. doi: 10.1016/j.tips.2019.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ashburn TT, Thor KB. Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov. 2004;3(8):673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 38.Zhou Y, Wang F, Tang J, et al. Artificial intelligence in COVID-19 drug repurposing. Lancet Digit Health. 2020;2(12):e667–e676. doi: 10.1016/S2589-7500(20)30192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pushpakom S, Iorio F, Eyers PA, et al. Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov. 2019;18(1):41–58. doi: 10.1038/nrd.2018.168. [DOI] [PubMed] [Google Scholar]
- 40.Santos R, Ursu O, Gaulton A, et al. A comprehensive map of molecular drug targets. Nat Rev Drug Discov. 2017;16(1):19–34. doi: 10.1038/nrd.2016.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng F, Desai RJ, Handy DE, et al. Network-based approach to prediction and population-based validation of in silico drug repurposing. Nat Commun. 2018;9(1):2691. doi: 10.1038/s41467-018-05116-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Batool M, Ahmad B, Choi S. A structure-based drug discovery paradigm. Int J Mol Sci. 2019;20(11):2783. doi: 10.3390/ijms20112783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parks JM, Smith JC. How to discover antiviral drugs quickly. N Engl J Med. 2020;382(23):2261–2264. doi: 10.1056/NEJMcibr2007042. [DOI] [PubMed] [Google Scholar]
- 44.Mullard A. The drug-maker's guide to the galaxy. Nature. 2017;549(7673):445–447. doi: 10.1038/549445a. [DOI] [PubMed] [Google Scholar]
- 45.Zhang JR, Zhang J, Lok TM, et al. A hybrid particle swarm optimization–back-propagation algorithm for feedforward neural network training. Appl Math Comput. 2007;185:1026–1037. doi: 10.1016/j.amc.2006.07.025. [DOI] [Google Scholar]
- 46.Aliper A, Plis S, Artemov A, et al. Deep Learning Applications for predicting pharmacological properties of drugs and drug repurposing using transcriptomic data. Mol Pharm. 2016;13(7):2524–2530. doi: 10.1021/acs.molpharmaceut.6b00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma J, Sheridan RP, Liaw A, et al. Deep neural nets as a method for quantitative structure-activity relationships. J Chem Inf Model. 2015;55(2):263–274. doi: 10.1021/ci500747n. [DOI] [PubMed] [Google Scholar]
- 48.Mayr A, Klambauer G, Unterthiner T, et al. DeepTox: toxicity prediction using deep learning. Front Environ Sci. 2016;3:80. doi: 10.3389/fenvs.2015.00080. [DOI] [Google Scholar]
- 49.Albawi S, Mohammed TA, Al-Zawi S. The International Conference on Engineering and Technology 2017. IEEE; 2017. pp. 1–6. [Google Scholar]
- 50.Yamashita R, Nishio M, Do RKG, et al. Convolutional neural networks: a overview and application in radiology. Insights Imaging. 2018;9(4):611–629. doi: 10.1007/s13244-018-0639-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meyer JG, Liu S, Miller IJ, et al. Learning drug functions from chemical structures with convolutional neural networks and random forests. J Chem Inf Model. 2019;59(10):4438–4449. doi: 10.1021/acs.jcim.9b00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wallach I, Dzamba M, Heifets A. AtomNet: a deep convolutional neural network for bioactivity prediction in structure-based drug discovery. arXiv preprint arXiv:151002855. 2015.
- 53.Ragoza M, Hochuli J, Idrobo E, et al. Protein-ligand scoring with convolutional neural networks. J Chem Inf Model. 2017;57(4):942–957. doi: 10.1021/acs.jcim.6b00740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mandic D, Chambers J. Wiley; 2001. Recurrent neural networks for prediction: learning algorithms, architectures and stability. [Google Scholar]
- 56.Bickerton GR, Paolini GV, Besnard J, et al. Quantifying the chemical beauty of drugs. Nat Chem. 2012;4(2):90–98. doi: 10.1038/nchem.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Olivecrona M, Blaschke T, Engkvist O, et al. Molecular de-novo design through deep reinforcement learning. J Cheminform. 2017;9(1):48. doi: 10.1186/s13321-017-0235-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Segler MHS, Kogej T, Tyrchan C, et al. Generating Focused molecule libraries for drug discovery with recurrent neural networks. ACS Cent Sci. 2018;4(1):120–131. doi: 10.1021/acscentsci.7b00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gao KY, Fokoue A, Luo H, et al. Interpretable drug target prediction using deep neural representation. IJCAI. 2018:3371–3377. doi: 10.24963/ijcai.2018/468. [DOI] [Google Scholar]
- 60.Barabási AL, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet. 2011;12(1):56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gysi DM, Do Valle Í, Zitnik M, et al. Network medicine framework for identifying. drug-repurposing opportunities for COVID-19. PNASA. 2021;118(19) doi: 10.1073/pnas.2025581118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Su C, Tong J, Zhu Y, et al. Network embedding in biomedical data science. Brief. Bioinform. 2020;21(1):182–197. doi: 10.1093/bib/bby117. [DOI] [PubMed] [Google Scholar]
- 63.Tang J, Qu M, Wang M, et al. Proceedings of the 24th International Conference on World Wide Web. 2015. pp. 1067–1077. [DOI] [Google Scholar]
- 64.Sosa DN, Derry A, Guo M, et al. A literature-based knowledge graph embedding method for identifying drug repurposing opportunities in rare diseases. Pac Symp Biocomput. 2020;25:463–474. [PMC free article] [PubMed] [Google Scholar]
- 65.Zeng X, Song X, Ma T, et al. Repurpose open data to discover therapeutics for COVID-19 using deep learning. J Proteome Res. 2020;19(11):4624–4636. doi: 10.1021/acs.jproteome.0c00316. [DOI] [PubMed] [Google Scholar]
- 66.Mullard A. The drug-maker's guide to the galaxy. Nature. 2017;549(7673):445–447. doi: 10.1038/549445a. [DOI] [PubMed] [Google Scholar]
- 67.Himmelstein DS, Lizee A, Hessler C, et al. Systematic integration of biomedical knowledge prioritizes drugs for repurposing. Elife. 2017;6:e26726. doi: 10.7554/eLife.26726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Su C, Hou Y, Guo W, et al. CBKH: the Cornell biomedical knowledge hub. medRxiv. 2021 [Google Scholar]
- 69.Phan LT, Nguyen TV, Luong QC, et al. Importation and human-to-human transmission of a novel coronavirus in Vietnam. N Engl J Med. 2020;382(9):872–874. doi: 10.1056/NEJMc2001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhu N, Zhang D, Wang W, et al. China novel coronavirus investigating and research team. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mei X, Lee HC, Diao KY, et al. Artificial intelligence-enabled rapid diagnosis of patients with COVID-19. Nat Med. 2020;26(8):1224–1228. doi: 10.1038/s41591-020-0931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang HS, Hou Y, Vasovic LV, et al. Routine Laboratory blood tests predict SARS-CoV-2 infection using machine learning. Clin Chem. 2020;66(11):1396–1404. doi: 10.1093/clinchem/hvaa200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xu X, Jiang X, Ma C, et al. A deep learning system to screen novel coronavirus disease 2019 pneumonia. Engineering (Beijing) 2020;6(10):1122–1129. doi: 10.1016/j.eng.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ardakani AA, Kanafi AR, Acharya UR, et al. Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: results of 10 convolutional neural networks. Comput Biol Med. 2020;121 doi: 10.1016/j.compbiomed.2020.103795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang S, Zha Y, Li W, et al. A fully automatic deep learning system for COVID-19 diagnostic and prognostic analysis. Eur Respir J. 2020;56(2) doi: 10.1183/13993003.00775-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Narin A, Kaya C, Pamuk Z. Automatic detection of coronavirus disease (COVID-19) using X-ray images and deep convolutional neural networks. Pattern Anal Appl. 2021:1–14. doi: 10.1007/s10044-021-00984-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang D, Mo J, Zhou G, et al. An efficient mixture of deep and machine learning models for COVID-19 diagnosis in chest X-ray images. PLoS One. 2020;15(11) doi: 10.1371/journal.pone.0242535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ozturk T, Talo M, Yildirim EA, et al. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput Biol Med. 2020;121 doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jain G, Mittal D, Thakur D, et al. A deep learning approach to detect Covid-19 coronavirus with X-Ray images. Biocybern Biomed Eng. 2020;40(4):1391–1405. doi: 10.1016/j.bbe.2020.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Amatya Y, Rupp J, Russell FM, et al. Diagnostic use of lung ultrasound compared to chest radiograph for suspected pneumonia in a resource-limited setting. Int J Emerg Med. 2018;11(1):8. doi: 10.1186/s12245-018-0170-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hatamabadi H, Shojaee M, Bagheri M, et al. Lung ultrasound findings compared to chest CT scan in patients with COVID-19 associated pneumonia: a pilot study. Adv J Emerg Med. 2020 [Google Scholar]
- 82.Poggiali E, Dacrema A, Bastoni D, et al. Can lung US help critical care clinicians in the early diagnosis of novel coronavirus (COVID-19) pneumonia? Radiology. 2020;295(3):E6. doi: 10.1148/radiol.2020200847. PMC7233381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Roy S, Menapace W, Oei S, et al. Deep learning for classification and localization of COVID-19 markers in point-of-care lung ultrasound. IEEE Trans Med Imaging. 2020;39(8):2676–2687. doi: 10.1109/TMI.2020.2994459. [DOI] [PubMed] [Google Scholar]
- 84.Loey M, Smarandache F, M Khalifa NE. Within the lack of chest COVID-19 X-ray dataset: a novel detection model based on GAN and deep transfer learning. Symmetry (Basel) 2020;12(4):651. doi: 10.3390/SYM12040651. [DOI] [Google Scholar]
- 85.Liang W, Liang H, Ou L, et al. Development and validation of a clinical risk score to predict the occurrence of critical illness in hospitalized patients with COVID-19. JAMA Intern Med. 2020;180(8):1081–1089. doi: 10.1001/jamainternmed.2020.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Williams RD, Markus AF, Yang C, et al. Seek COVER development and validation of a personalized risk calculator for COVID-19 outcomes in an international network. medRxiv. 2020 doi: 10.1186/s12874-022-01505-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen Y, Ouyang L, Bao FS, et al. An interpretable machine learning framework for accurate severe vs non-severe covid-19 clinical type classification. 2020. Available from SSRN 3638427: https://ssrn.com/abstract=3638427 or 10.2139/ssrn.3638427
- 88.Avila E, Kahmann A, Alho C, et al. Hemogram data as a tool for decision-making in COVID-19 management: applications to resource scarcity scenarios. PeerJ. 2020;8:e9482. doi: 10.7717/peerj.9482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.An C, Lim H, Kim DW, et al. Machine learning prediction for mortality of patients diagnosed with COVID-19: a nationwide Korean cohort study. Sci Rep. 2020;10(1):18716. doi: 10.1038/s41598-020-75767-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liang W, Yao J, Chen A, et al. Early triage of critically ill COVID-19 patients using deep learning. Nat Commun. 2020;11(1):3543. doi: 10.1038/s41467-020-17280-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Reddy K, Sinha P, O’Kane CM, et al. Subphenotypes in critical care: translation into clinical practice. Lancet Respir Med. 2020;8(6):631–643. doi: 10.1016/S2213-2600(20)30124-7. [DOI] [PubMed] [Google Scholar]
- 92.Mori M, Krumholz HM, Allore HG. Using Latent Class Analysis to Identify Hidden Clinical Phenotypes. JAMA. 2020;324(7):700–701. doi: 10.1001/jama.2020.2278. [DOI] [PubMed] [Google Scholar]
- 93.Bhavani SV, Carey KA, Gilbert ER, et al. Identifying novel sepsis subphenotypes using temperature trajectories. Am J Respir Crit Care Med. 2019;200(3):327–335. doi: 10.1164/rccm.201806-1197OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Seymour CW, Kennedy JN, Wang S, et al. Derivation, validation, and potential treatment implications of novel clinical phenotypes for sepsis. JAMA. 2019;321(20):2003–2017. doi: 10.1001/jama.2019.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bui DS, Lodge CJ, Perret JL, et al. Trajectories of asthma and allergies from 7 years to 53 years and associations with lung function and extrapulmonary comorbidity profiles: a prospective cohort study. Lancet Respir Med. 2021;9(4):387–396. doi: 10.1016/S2213-2600(20)30413-6. [DOI] [PubMed] [Google Scholar]
- 96.Data Science Collaborative Group. Differences in clinical deterioration among three sub-phenotypes of COVID-19 patients at the time of first positive test: results from a clustering analysis. Intensive Care Med. 2021;47(1):113–115. doi: 10.1007/s00134-020-06236-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lascarrou JB, Gaultier A, Soumagne T, et al. Identifying clinical phenotypes in moderate to severe acute respiratory distress syndrome related to COVID-19: the COVADIS study. Front Med (Lausanne) 2021;8 doi: 10.3389/fmed.2021.632933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.COVID Predict Study Group. Schinkel M, Appelman B, Butler J, , et al. COVID Predict Study Group Association of clinical sub-phenotypes and clinical deterioration in COVID-19: further cluster analyses. Intensive Care Med. 2021;47(4):482–484. doi: 10.1007/s00134-021-06363-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Su C, Zhang Y, Flory JH, et al. Clinical subphenotypes in COVID-19: derivation, validation, prediction, temporal patterns, and interaction with social determinants of health. NPJ Digit Med. 2021;4:1–13. doi: 10.1038/s41746-021-00481-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li WT, Ma J, Shende N, et al. Using machine learning of clinical data to diagnose COVID-19: a systematic review and meta-analysis. BMC Med Inform Decis Mak. 2020;20(1):247. doi: 10.1186/s12911-020-01266-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ferreira FL, Bota DP, Bross A, et al. Serial evaluation of the SOFA score to predict outcome in critically ill patients. JAMA. 2001;286(14):1754–1758. doi: 10.1001/jama.286.14.1754. [DOI] [PubMed] [Google Scholar]
- 102.Su C, Xu Z, Hoffman K, et al. Identifying organ dysfunction trajectory-based subphenotypes in critically ill patients with COVID-19. Sci Rep. 2021;11(1):15872. doi: 10.1038/s41598-021-95431-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bhavani SV, Huang ES, Verhoef PA, et al. Novel temperature trajectory subphenotypes in COVID-19. Chest. 2020;158(6):2436–2439. doi: 10.1016/j.chest.2020.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Vincent JL, de Mendonça A, Cantraine F, et al. Use of the SOFA score to assess the incidence of organ dysfunction/failure in intensive care units: results of a multicenter, prospective study. Working group on “sepsis-related problems” of the European society of intensive care medicine. Crit Care Med. 1998;26(11):1793–1800. doi: 10.1097/00003246-199811000-00016. [DOI] [PubMed] [Google Scholar]
- 105.Obeid JS, Davis M, Turner M, et al. An artificial intelligence approach to COVID-19 infection risk assessment in virtual visits: a case report. J Am Med Inform Assoc. 2020;27(8):1321–1325. doi: 10.1093/jamia/ocaa105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dagliati A, Malovini A, Tibollo V, et al. Health informatics and EHR to support clinical research in the COVID-19 pandemic: an overview. Brief Bioinform. 2021;22(2):812–822. doi: 10.1093/bib/bbaa418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bento AI, Nguyen T, Wing C, et al. Evidence from internet search data shows information-seeking responses to news of local COVID-19 cases. Proc Natl Acad Sci USA. 2020;117(21):11220–11222. doi: 10.1073/pnas.2005335117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tasnim S, Hossain MM, Mazumder H. Impact of Rumors and misinformation on COVID-19 in social media. J Prev Med Public Health. 2020;53(3):171–174. doi: 10.3961/jpmph.20.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cuan-Baltazar JY, Muñoz-Perez MJ, Robledo-Vega C, et al. Misinformation of COVID-19 on the internet: infodemiology study. JMIR Public Health Surveill. 2020;6:e18444. doi: 10.2196/18444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lyu H, Zheng Z, Luo J. Both rates of fake news and fact-based news on twitter negatively correlate with the state-level COVID-19 vaccine uptake. arXiv preprint arXiv:210607435. 2021.
- 111.Bursztyn L, Rao A, Roth CP, et al. Misinformation during a pandemic. Natil Bur Econ Res. 2020 [Google Scholar]
- 112.Zhou X, Mulay A, Ferrara E, et al. International conference on information and knowledge management. 2020. pp. 3205–3212. [DOI] [Google Scholar]
- 113.Pfefferbaum B, North CS. Mental health and the Covid-19 pandemic. N Engl J Med. 2020;383(6):510–512. doi: 10.1056/NEJMp2008017. [DOI] [PubMed] [Google Scholar]
- 114.Usher K, Durkin J, Bhullar N. The COVID-19 pandemic and mental health impacts. Int J Ment Health Nurs. 2020;29(3):315–318. doi: 10.1111/inm.12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cullen W, Gulati G, Kelly BD. Mental health in the COVID-19 pandemic. QJM. 2020;113(5):311–312. doi: 10.1093/qjmed/hcaa110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Saha K, Torous J, Caine ED, et al. Psychosocial Effects of the COVID-19 pandemic: large-scale Quasi-experimental study on social media. J Med Internet Res. 2020;22(11):e22600. doi: 10.2196/22600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhang Y, Lyu H, Liu Y, et al. Monitoring depression trends on twitter during the COVID-19 pandemic: observational study. JMIR Infodemiol. 2021;1:e26769. doi: 10.2196/26769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ahmed N, Michelin RA, Xue W, et al. A survey of COVID-19 contact tracing apps. IEEE Access. 2020;8:134577–134601. doi: 10.1109/ACCESS.2020.3010226. [DOI] [Google Scholar]
- 119.Barber SJ, Kim H. COVID-19 worries and behavior changes in older and younger men and women. J Gerontol B Psychol Sci Soc Sci. 2021;76(2):e17–e23. doi: 10.1093/geronb/gbaa068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hong S, Zhou Y, Shang J, et al. Opportunities and challenges of deep learning methods for electrocardiogram data: a systematic review. Comput Biol Med. 2020;122 doi: 10.1016/j.compbiomed.2020.103801. [DOI] [PubMed] [Google Scholar]
- 121.Wang F, Kaushal R, Khullar D. Should health care demand interpretable artificial intelligence or accept “black box” medicine? Ann Intern Med. 2020;172(1):59–60. doi: 10.7326/M19-2548. [DOI] [PubMed] [Google Scholar]
- 122.Ribeiro MT, Singh S, Guestrin C. Proceedings of the ACM SIGKDD international conference on knowledge discovery and data mining. 2016. pp. 1135–1144. [DOI] [Google Scholar]
- 123.Lundberg SM, Lee SI. A unified approach to interpreting model predictions. Adv Neural Inf Process Syst. 2017;2017:4768–4777. [Google Scholar]
- 124.Hong S, Xiao C, Ma T, et al. Mina: multilevel knowledge-guided attention for modeling electrocardiography signals. arXiv preprint arXiv:190511333. 2019.
- 125.Zhang Q, Zhu SC. Visual interpretability for deep learning: a survey. arXiv preprint arXiv:180200614. 2018.
- 126.Oprea A. Machine learning integrity and privacy in adversarial environments. 2021:1–2.
- 127.Goodfellow IJ, Shlens J, Szegedy C. Explaining and harnessing adversarial examples. arXiv preprint arXiv:14126572. 2014.
- 128.Rahman A, Hossain MS, Alrajeh NA, et al. Adversarial examples–security threats to COVID-19 deep learning systems in medical IoT devices. IEEE Internet Things J. 2020 doi: 10.1109/JIOT.2020.3013710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Finlayson SG, Chung HW, Kohane IS, et al. Adversarial attacks against medical deep learning systems. arXiv preprint arXiv:180405296. 2018.
- 130.Nassar M, Salah K, ur Rehman MH, et al. Blockchain for explainable and trustworthy artificial intelligence. Wiley Interdiscip Rev: Data Min Knowl Discov. 2020;10:e1340. doi: 10.1002/widm.1340. [DOI] [Google Scholar]
- 131.Tramèr F, Papernot N, Goodfellow I, et al. The space of transferable adversarial examples. 2017. arXiv preprint arXiv:170403453.
- 132.Nicolae MI, Sinn M, Tran MN, et al. Adversarial Robustness Toolbox v1.0.0. arXiv preprint arXiv:180701069. 2018.
- 133.Flores AW, Bechtel K, Lowenkamp CT. False positives, false negatives, and false analyses: a rejoinder to machine bias: there’s software used across the country to predict future criminals. and it’s biased against blacks. Fed Probat. 2016;80:38. [Google Scholar]
- 134.Obermeyer Z, Powers B, Vogeli C, et al. Dissecting racial bias in an algorithm used to manage the health of populations. Science. 2019;366(6464):447–453. doi: 10.1126/science.aax2342. [DOI] [PubMed] [Google Scholar]
- 135.Ferrer X, van Nuenen T, Such JM, et al. Bias and discrimination in AI: a cross-disciplinary perspective. IEEE Technol Soc Mag. 2021;40:72–80. doi: 10.1109/MTS.2021.3056293. [DOI] [Google Scholar]
- 136.Sunstein CR. Algorithms, correcting biases. Soc Res (New York) 2019;86(2):499–511. [Google Scholar]
- 137.Criado N, Such JM. OUP; 2019. Digital discrimination algorithmic regulation. [Google Scholar]
- 138.Zafar MB, Valera I, Gomez Rodriguez M, et al. 26th international World Wide Web conference. 2017. pp. 1171–1180. [DOI] [Google Scholar]
- 139.Srinivasan R, Chander A. Biases in AI systems. Commun ACM. 2021;64(8):44–49. doi: 10.1145/3038912.3052660. [DOI] [Google Scholar]
- 140.Dwork C, McSherry F, Nissim K, et al. Joint UNECE/Eurostat work session on statistical data confidentiality. 2011. Differential privacy–a primer for the perplexed. [Google Scholar]
- 141.Leoni D. ACM international conference proceeding series. 2012. pp. 40–52. [DOI] [Google Scholar]
- 142.Chaudhuri K, Monteleoni C, Sarwate AD. Differentially private empirical risk minimization. J Mach Learn Res. 2011;12:1069–1109. [PMC free article] [PubMed] [Google Scholar]
- 143.Xu J, Glicksberg BS, Su C, et al. Federated learning for healthcare informatics. J Healthc Inform Res. 2020:1–19. doi: 10.1007/s41666-020-00082-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Warnat-Herresthal S, Schultze H, Shastry KL, et al. Swarm learning for decentralized and confidential clinical machine learning. Nature. 2021;594(7862):265–270. doi: 10.1038/s41586-021-03583-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ceylan Z. Estimation of COVID-19 prevalence in Italy, Spain, and France. Sci Total Environ. 2020;729 doi: 10.1016/j.scitotenv.2020.138817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Benvenuto D, Giovanetti M, Vassallo L, et al. Application of the ARIMA model on the COVID-2019 epidemic dataset. Data Brief. 2020;29 doi: 10.1016/j.dib.2020.105340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Rodriguez A, Tabassum A, Cui J, et al. Deepcovid: an operational deep learning-driven framework for explainable real-time covid-19 forecasting. medRxiv. 2020 [Google Scholar]
- 148.Singh KK, Kumar S, Dixit P, et al. Kalman filter based short term prediction model for COVID-19 spread. Appl Intell. 2021;51(5):2714–2726. doi: 10.1007/s10489-020-01948-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Liu X, Xu X, Li G, et al. Differential impact of non-pharmaceutical public health interventions on COVID-19 epidemics in the United States. BMC Public Health. 2021;21(1):965. doi: 10.1186/s12889-021-10950-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tian T, Luo W, Tan J, et al. The timing and effectiveness of implementing mild interventions of COVID-19 in large industrial regions via a synthetic control method. Stat Interface. 2021;14(1):3–12. doi: 10.4310/20-SII634. [DOI] [Google Scholar]
- 151.Friedman J., Liu P., Troeger C.E., et al. Predictive performance of international COVID-19 mortality forecasting models. Nat Commun. 2021;12(1):2609. doi: 10.1038/s41467-021-22457-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Murray CJ. Forecasting COVID-19 impact on hospital bed-days, ICU-days, ventilator-days and deaths by US state in the next 4 months. medRxiv. 2020 [Google Scholar]
- 153.Hsiang S, Allen D, Annan-Phan S, et al. Publisher Correction: the effect of large-scale anti-contagion policies on the COVID-19 pandemic. Nature. 2020;585(7824):E7. doi: 10.1038/s41586-020-2691-0. [DOI] [PubMed] [Google Scholar]
- 154.Li Y, Jia W, Wang J, et al. ALeRT-COVID: attentive lockdown-aware transfer learning for predicting COVID-19 pandemics in different countries. J Healthc Inform Res. 2021:1–16. doi: 10.1007/s41666-020-00088-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wang J, Tang K, Feng K, et al. High temperature and high humidity reduce the transmission of COVID-19. arXiv preprint arXiv:200305003. 2020.
- 156.Brauer M, Zhao JT, Bennitt FB, et al. Global Access to handwashing: implications for COVID-19 control in low-income countries. Environ Health Perspect. 2020;128(5):57005. doi: 10.1289/EHP7200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.IHME COVID-19 Forecasting Team. Modeling COVID-19 scenarios for the United States. Nat Med. 2021;27(1):94–105. doi: 10.1038/s41591-020-1132-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhou Y, Hou Y, Shen J, et al. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6:14. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wang Q, Li M, Wang X, et al. COVID-19 literature knowledge graph construction and drug repurposing report generation. arXiv preprint arXiv:200700576. 2020.
- 160.Zhang R, Hristovski D, Schutte D, et al. Drug repurposing for COVID-19 via knowledge graph completion. J Biomed Inform. 2021;115 doi: 10.1016/j.jbi.2021.103696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Gordon DE, Jang GM, Bouhaddou M, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583(7816):459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Beck BR, Shin B, Choi Y, et al. Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model. Comput Struct Biotechnol J. 2020;18:784–790. doi: 10.1016/j.csbj.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Mall R, Elbasir A, Al Meer H, et al. Data-driven drug repurposing for COVID-19. 2020.
- 164.Su C, Hoffman K, Xu Z, et al. Evaluation of albumin kinetics in mechanically ventilated patients with COVID-19 compared to those with sepsis-induced ARDS. medRxiv. 2021 doi: 10.1101/2021.03.16.21253405. [DOI] [Google Scholar]
- 165.Burn E, You SC, Sena AG, et al. Deep phenotyping of 34,128 adult patients hospitalised with COVID-19 in an international network study. Nat Commun. 2020;11(1):5009. doi: 10.1038/s41467-020-18849-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Roth GA, Emmons-Bell S, Alger HM, et al. Trends in patient characteristics and COVID-19 in-hospital mortality in the United States during the COVID-19 pandemic. JAMA Netw Open. 2021;4(5) doi: 10.1001/jamanetworkopen.2021.8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Yang HS, Hou Y, Zhang H, et al. Machine learning analysis highlights the down-trending of the proportion of COVID-19 patients with a distinct laboratory result profile. medRxiv. 2020 [Google Scholar]
- 168.Zhou L, Romero N, Martínez-Miranda J, et al. Subphenotyping of COVID-19 patients at pre-admission towards anticipated severity stratification: an analysis of 778 692 Mexican patients through an age-gender unbiased meta-clustering technique. medRxiv. 2021 [Google Scholar]