Abstract
The presence and dissemination of carbapenem-resistant Klebsiella pneumoniae (CRKP) often cause life-threatening infections worldwide, but the therapeutic option is limited. In this study, whole-genome sequencing (WGS) was applied to assess the epidemiological characteristics and transmission dynamics of CRKP isolates recovered from two fetal outbreaks of nosocomial infections. Between April 2016 and March 2018, a total of 70 isolates of K. pneumoniae were collected from sterile samples in a tertiary hospital in Hangzhou, China. The minimal inhibitory concentrations (MICs) of 21 antimicrobial agents were determined using the broth microdilution methods. Pulsed-field gel electrophoresis (PFGE) was performed on 47 CRKP isolates, and 16 clonally related isolates were further characterized by Illumina sequencing. In addition, the complete genome sequences of three representative isolates (KP12, KP36, and KP37) were determined by Oxford Nanopore sequencing. The K. pneumoniae isolates were recovered from patients diagnosed with pulmonary infection, cancer, or encephalopathy. For all CRKP isolates, PFGE separated three clusters among all strains. The most predominant PFGE cluster contained 16 isolates collected from patients who shared close hospital units and represented a potential outbreak. All 16 isolates showed an extremely high resistance level (≥87.5%) to 18 antimicrobials tested but remain susceptible to colistin (CST). Multiple antimicrobial resistance and virulence determinants, such as the carbapenem resistance gene bla KPC-2, and genes encoding the virulence factor aerobactin and the regulator of the mucoid phenotype (rmpA and rmpA2), were observed in the 16 CRKP isolates. These isolates belonged to sequence type 11 (ST11) and capsular serotype KL64. A core genome single nucleotide polymorphism (cgSNP)-based phylogenetic analysis indicated that the 16 CRKP isolates could be partitioned into two separate clades (≤15 SNPs), suggesting the two independent transmission scenarios co-occurred. Moreover, a high prevalence of IncFIB/IncHI1B type virulence plasmid with the iroBCDN locus deleted, and an IncFII/IncR type bla KPC-2-bearing plasmid was co-harbored in ST11-KL64 CRKP isolates. In conclusion, our data indicated that the nosocomial dissemination of ST11-KL64 CRKP clone is a potential threat to anti-infective therapy. The development of novel strategies for surveillance, diagnosis, and treatment of this high-risk CRKP clone is urgently needed.
Keywords: Klebsiella pneumoniae, carbapenem resistance, outbreak, whole-genome sequencing, single-nucleotide polymorphisms, genomic surveillance, hypervirulence plasmid
Introduction
Klebsiella pneumoniae, as an increasingly important human pathogen, represents increasingly multidrug-resistance, particularly to carbapenems and the third-generation cephalosporins (Navon-Venezia et al., 2017). Carbapenem-resistant K. pneumoniae (CRKP) is widely reported as a multidrug-resistant bacteria and associated with high morbidity and mortality rates (Wyres et al., 2020a). CRKP can produce carbapenem-hydrolyzing enzymes to hydrolyze carbapenemase and eventually nullify the effectiveness of the last-resort antibiotics. In 1996, the first K. pneumoniae carbapenemase (KPC) enzyme, encoded by the bla KPC gene, was found in K. pneumoniae (Yigit et al., 2001). Subsequently, other carbapenemase genes have emerged, such as bla NDM, bla OXA-48, bla VIM, and bla IMP (Fukigai et al., 2007; Perez-Vazquez et al., 2019; Lu et al., 2020; Nishida et al., 2020). The KPC-producing isolates are always reported to be associated with nosocomial outbreaks worldwide. In China, outbreaks of CRKP isolates mainly carried the bla KPC-2 or bla NDM-1 gene and can be classified as sequence type (ST)11 by multilocus sequence typing (MLST; Gu et al., 2018; Zhang et al., 2020a). Unfortunately, the KPC-producing isolates are resistant to almost all β-lactams and β-lactamase inhibitors, which significantly limits treatment options and eventually leads to high mortality rates, especially among inpatients with prolonged hospitalization (Jiang et al., 2015; Gu et al., 2018; Sui et al., 2018; Zhang et al., 2020a).
Compared with the classic K. pneumoniae (cKP), hypervirulent K. pneumoniae (hvKP) causes more severe infections, such as liver abscesses, endophthalmitis, meningitis, and pneumonia, and displays a higher level of susceptibility to the antimicrobial agents (Wang et al., 2018; Russo and Marr, 2019; Choby et al., 2020). The hvKP is undergoing global dissemination and has been confirmed to be highly associated with several virulence factors as the hallmarks, including the regulator of the mucoid phenotype (encoded by the rmpA gene), the regulator of mucoid phenotype 2 (rmpA2), aerobactin (iucABCD, iutA), salmochelin (iroBCDN), and metabolite transporter (peg-344), which were typically co-located on a classic pLVPK-like virulence plasmid (Wyres et al., 2020b). However, hvKP strains are becoming increasingly resistant to antimicrobials, including carbapenems (Feng et al., 2018; Gu et al., 2018). Moreover, the combination of carbapenem resistance and hypervirulence significantly reduces the efficacy of antimicrobial agents to treat the life-threatening infections caused by carbapenem-resistant hvKP (CR-hvKP). Therefore, they represent an extremely severe health challenge and public concern. Consequently, it is urgent to investigate the genomic characteristics of CRKP to prevent, diagnose, and treat K. pneumoniae infections.
Rapid advances in whole-genome sequencing (WGS) technology and bioinformatics tools can facilitate understanding the spread of K. pneumoniae, including the identification of transmission routes, evolutionary patterns, and antimicrobial resistance mechanisms (Klemm et al., 2018; Schurch et al., 2018). Due to the advances in next-generation sequencing platforms, the cost of WGS continues to decrease. Moreover, WGS can detect minor genomic differences between isolates for its high resolution, limited in traditional molecular typing techniques, such as pulsed-field gel electrophoresis (PFGE) and MLST.
In this study, a total of 70 K. pneumoniae isolates were collected from patients during their hospitalizations in a tertiary hospital in China. The 16 KPC-2-producing CRKP isolates, represented a hospital outbreak based on antimicrobial susceptibility testing and PFGE, were further subjected to WGS analysis. The genomic epidemiological characteristics, the transmission route of this outbreak, and the genetic features of the virulence plasmids and bla KPC-2-bearing plasmids were investigated.
Materials and Methods
Patients and Isolates
Seventy K. pneumoniae isolates, recovered from patients in severe conditions or long-term hospitalization, were collected from a tertiary hospital in Hangzhou, Zhejiang province, China, between April 2016 and March 2018. The quick Sequential Organ Failure Assessment (qSOFA) score and the Confusion, Urea, Respiratory rate, Blood pressure plus age≥65years (CURB-65) score were used to help determine patients with severe illness. An inpatient hospitalization lasting 14 days or more was considered a long hospital stay. All isolates were cultured from the sputum, urine, blood, excreta, catheters, feces, and cerebrospinal fluids specimens. Demographic data, such as gender, age, department of hospitalization, clinical diagnosis, outcome, time of admission, time of discharge, and length of hospital stay, were extracted from the patient administration system. This study was approved by the local Research Ethics Committee of Sir Run Run Shaw Hospital, Zhejiang University School of Medicine. All isolates were generated as part of routine clinical laboratory procedures, and no identifiable patient information was collected.
Bacterial Identification and Antimicrobial Susceptibility
Bacterial identification was performed using VITEK 2 (bioMérieux, Marcy-l’Étoile, France) and Matrix-assisted laser desorption/ionization–time-of-flight mass spectrometry (MALDI-TOF-MS, Bruker, Billerica, MA, United States). Antimicrobial susceptibility testing was carried out for all isolates using the broth microdilution methods. The susceptibility breakpoint was interpreted according to Clinical and Laboratory Standards Institute (CLSI) 2020 guidelines or European Committee on Antibiotic Susceptibility Testing (EUCAST) 10.0 guidelines. The list of tested antimicrobial agents includes the following: aztreonam (ATM), fosfomycin (FOF), ertapenem (ETP), ceftazidime (CAZ), cefepime (FEP), cefoperazone-sulbactam (SCF), cefoxitin (FOX), levofloxacin (LVX), ciprofloxacin (CIP), amikacin (AMK), tetracycline (TET), tigecycline (TGC), minocycline (MH), colistin (CST), cefotaxime (CTX), meropenem (MEM), imipenem (IPM), gentamicin (GEN), trimethoprim-sulfamethoxazole (SXT), piperacillin (PRL), and piperacillin-tazobactam (PRL/TZP). Escherichia coli ATCC 25922 was used as a quality control strain for antimicrobial susceptibility testing.
Pulsed-Field Gel Electrophoresis
All the 47 CRKP isolates were characterized by PFGE. Genomic DNA of isolates was digested overnight with XbaI restriction enzyme (Sangon, Shanghai, China), and the DNA fragments were subjected to electrophoresis in 1% agarose III (Sangon) with a CHEF apparatus (CHEF Mapper XA; Bio-Rad, Hercules, CA, United States). The electrophoresis conditions were 14°C and 6V/cm with alternating pulses at a 120° angle with a 5–35s pulse time gradient for 22h. Salmonella enterica serotype Braenderup H9812 was used as a molecular size marker, covering the fragment ranges generated by K. pneumoniae. The PFGE patterns were analyzed by BioNumerics 7.0 software (Applied Maths, Sint-MartensLatem, Belgium) with the Dice similarity index. Interpretation is based on the criteria proposed by Tenover et al. (1995), that is, two isolates shared no more than three band differences of PFGE patterns are deemed as the same clone.
Whole-Genome Sequencing
Total DNA was extracted from the 16 CRKP isolates using a QIAamp DNA Mini Kit (Qiagen, Valencia, CA, United States) and fragmented by sonication using a Covaris M220 sonicator (Covaris, Woburn, MA, United States). Genomic libraries were prepared with an average insert size of 350bp using a TruSeq DNA Sample Prep kit (Illumina, San Diego, CA, United Staets) and sequenced using the Illumina NovaSeq 6,000 platform (Illumina, San Diego, CA, United States) with the 150bp paired-end protocol. Furthermore, the genomic DNA of three representative isolates (KP12, KP36, and KP37) out of the 16 CRKP isolates was also sequenced on the long-read Oxford Nanopore MinION platform (Nanopore Technologies, Oxford, United Kingdom). The derived short Illumina reads and long MinION reads were assembled using Unicycler v0.4.8 software (Wick et al., 2017).
The genome annotation was performed using the NCBI Prokaryotic Genome Annotation Pipeline (PGAP; Tatusova et al., 2016). Acquired antimicrobial resistance genes and virulence genes were identified using ResFinder 4.1 and VFDB 2019 databases, respectively, with a 90% threshold for gene identification and a 60% minimum length to respective database entries. In silico MLST analysis was performed by BacWGSTdb 2.0 server (Ruan and Feng, 2016; Ruan et al., 2020b; Feng et al., 2021). The type of capsule and lipopolysaccharide serotype of K. pneumoniae was conducted using Kaptive v0.6.1 (Wick et al., 2018).
Single-Nucleotide Polymorphisms and Phylogenetic Analysis
The ST11 K. pneumoniae strain KP58, another ST11 CRKP isolate recovered in Hangzhou, China, was used as a reference sequence to identify genomic variations between 16 CRKP isolates. Bacterial core genome single nucleotide polymorphism (cgSNP) was analyzed using the BacWGSTdb 2.0 server (Ruan and Feng, 2016; Feng et al., 2021). SNPs in the core genome and the removal of recombination regions from SNP alignments were predicted using Snippy v4.4.5. The output was used to construct a phylogenetic tree, with 1,000 bootstraps, under the general time-reversible (GTR) model with RAxML v8.2.12, and also used to identify the pairwise SNP distances using snp-dist v0.6.3 (Stamatakis, 2014). Isolates were considered the same outbreak if the threshold for SNPs distance≤18 (Schurch et al., 2018). Visualization and annotation of the phylogenetic tree and the presence of antimicrobial resistance genes and plasmid-borne virulence genes were performed by the Interactive Tree Of Life (iTOL) v5 webserver (Letunic and Bork, 2021). Sequence comparison of virulence plasmids and bla KPC-2-bearing plasmids was conducted using BLAST Ring Image Generator (BRIG) and Easyfig (Alikhan et al., 2011; Sullivan et al., 2011).
Accession Number
The genome sequences of the 16 CRKP isolates were deposited in the NCBI GenBank database under the BioProject accession numbers PRJNA553055.
Results
Clinical Characteristics of K. pneumoniae Isolates
From April 2016 to March 2018, a total of 70 K. pneumoniae isolates were cultured from 68 inpatients in a tertiary hospital in Hangzhou. The mean age of the inpatients was 70.5±17.3years. Among the 68 inpatients, 47 (69.12%) inpatients were males, and 21 (30.88%) were females. These clinical isolates were cultured from sputum (28/70, 40.00%), urine (20/70, 28.57%), blood (12/70, 17.14%), excreta (7/70, 10.00%), catheters (1/70, 1.43%), feces (1/70, 1.43%), and cerebrospinal fluids (1/70, 1.43%). The inpatients were diagnosed as pulmonary infection (19/68, 27.94%), cancer (9/68, 13.24%), encephalopathy (23/68, 33.82%), upper gastrointestinal bleeding (2/68, 2.94%), abdominal pain (1/68, 1.47%), hydronephrosis (1/68, 1.47%), cardiopulmonary arrest (1/68, 1.47%), chronic sinusitis (1/68, 1.47%), perianal abscess (1/68, 1.47%), diabetes (1/68, 1.47%), and nine (13.24%) inpatients suffered external injury or had a history of surgery. Among the 70 K. pneumoniae isolates, 47 isolates showed a carbapenem resistance phenotype, with a rate of 67.14%.
Comparison of Outbreak Isolates According to PFGE and WGS-Based SNPs
Based on the PFGE typing results of 47 CRKP isolates, three clusters were observed according to the similarity between PFGE banding patterns (Figure 1). The dominant PFGE cluster C contained 16 isolates and represented a putative outbreak in the hospital for similar PFGE patterns (differed by <three bands). The PFGE cluster A and B included two and three isolates, respectively. In contrast, the remaining 26 isolates showed sporadic PFGE patterns.
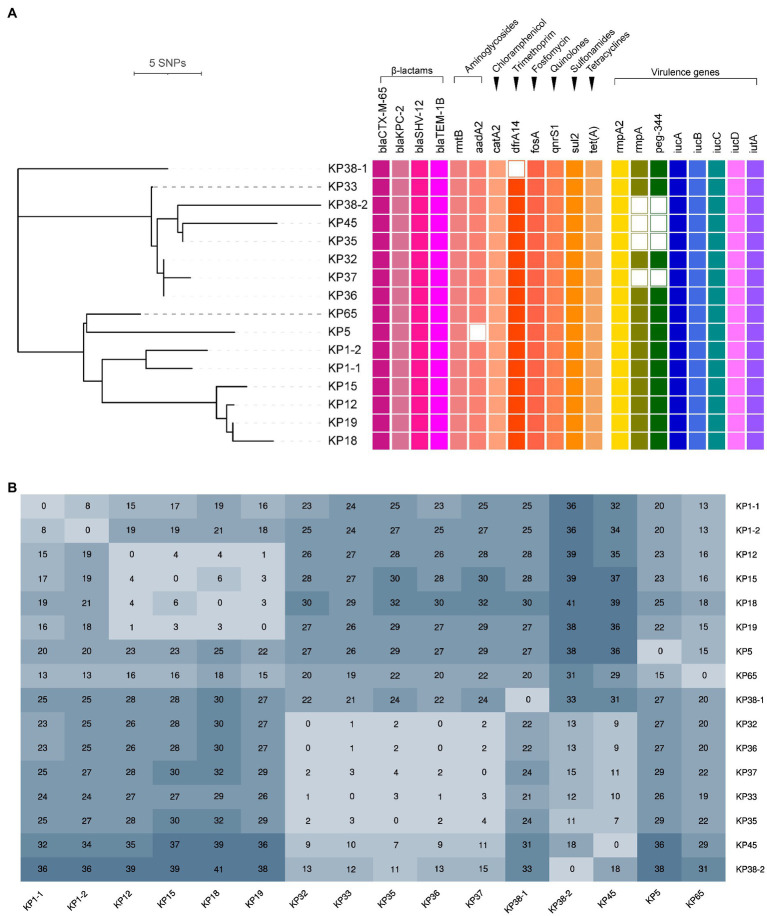
Figure 1.
Clustering of the 16 carbapenem-resistant Klebsiella pneumoniae (CRKP) isolates based on pulsed-field gel electrophoresis (PFGE) profiles. The information of strain ID is listed to the left of the patterns. The three predominant PFGE cluster (A), (B), and (C) are represented by red rectangles.
The draft genomes of the 16 CRKP isolates, presented in PFGE cluster C, were determined by whole-genome sequencing. All the isolates harbored bla KPC-2 gene. According to the MLST scheme of K. pneumoniae, all the 16 KPC-2-producing CRKP isolates were identified to be ST11 and shared the same capsular serotype KL64.
Phylogenetic analysis indicated that all the ST11-KL64 CRKP isolates were partitioned into two clades (Figure 2A). Clade 1 contained seven isolates (KP32, KP33, KP35, KP36, KP37, KP45, and KP38-2), and clade 2 comprised four isolates (KP12, KP15, KP18, and KP19), while the remaining five isolates (KP38-1, KP1-1, KP1-2, KP5, and KP65) were singletons. The number of SNPs separating ST11-KL64 CRKP isolates ranged from 0 to 41 after removing recombination regions (Figure 2B). Concerning the different variants among the 16 isolates, the internal members of each clade were closely related (≤18 SNPs), and a high diversity between the two clades was also observed. The differences in the internal isolates of clade 1 and 2 ranged from 0 to 18 SNPs and 1 to 6 SNPs, respectively, representing two independent outbreaks. It is noteworthy that only eight SNPs were detected between KP1-1 and KP1-2 (recovered from the same patient), suggesting they were the variants of the same clone. In comparison, 33 SNPs were observed between KP38-1 and KP38-2, implying that the patient Pa38 were infected with two different clones of CRKP. The detailed information for SNPs detected in 16 K. pneumoniae strains is shown in Supplementary Table S1.
Figure 2.
Phylogenetic analysis of 16 K. pneumoniae carbapenemase (KPC)-2-producing ST11-KL64 CRKP isolates. (A) Recombination-filtered core genome phylogeny and the distribution of antimicrobial resistance genes and virulence genes in the CRKP isolates. The cell in different colors represents the presence of the gene, while the blank cell represents the absence of the gene. (B) The single nucleotide polymorphisms (SNPs) numbers between each isolate.
Clinical and Microbiological Characteristics of the 16 ST11-KL64 CRKP Isolates
Demographic and clinical characteristics and the timeline of inpatients with 16 KPC-2-producing ST11-KL64 CRKP clinical cultures are shown in Table 1 and Figure 3. The 16 strains were isolated from 14 separate patients admitted into the respiratory (KP1-1, KP1-2, KP5, KP12, KP15, KP18, KP19, and KP65), intensive care unit (ICU; KP32, KP33, KP35, KP36, and KP37), and neurosurgery (KP38-1, KP38-2, and KP45). Isolate KP32, isolated from patient 32 (Pa 32) on May 15, 2017, represented the first case of the first outbreak. In the next 34 days, another four CRKP isolates (KP33, KP35, KP36, and KP37) were cultured from the sputum and blood specimens of four patients in the ICU. About 22 days later, KP38-1 and KP38-2 were isolated from the blood and sputum of the same patient (Pa38) admitted into neurosurgery with a 112-day interval, respectively. KP45 was isolated from Pa45 in the same ward on the same day of KP38-2 isolated. Between November 2017 and January 2018, another seven inpatients were continuously confirmed to have CRKP infection in the respiratory ward. Finally, 10 patients gradually recovered among the 14 patients involved in this outbreak, and four died of CRKP infection.
Table 1.
Characteristics and outcomes of the 14 inpatients involved in the KPC-2-producing ST11-KL64 CRKP isolates outbreaks.
| Patient | Isolate number | Age (years) | Gender | Department | Diagnosis | Outcome | Isolate date | Isolate type |
|---|---|---|---|---|---|---|---|---|
| Pa1 | KP1-1 | 66 | Female | Respiratory | History of pancreatic malignancy | Died | 19/12/2017 | Blood |
| KP1-2 | 66 | Female | Respiratory | History of pancreatic malignancy | Died | 28/12/2017 | Feces | |
| Pa5 | KP5 | 57 | Male | Respiratory | Pulmonary malignancy | Discharged | 20/11/2017 | Blood |
| Pa12 | KP12 | 53 | Male | Respiratory | Pulmonary malignancy | Discharged | 25/12/2017 | Urine |
| Pa15 | KP15 | 91 | Male | Respiratory | Pulmonary infection | Discharged | 13/1/2018 | Urine |
| Pa18 | KP18 | 73 | Male | Respiratory | Pulmonary infection | Discharged | 21/1/2018 | Sputum |
| Pa19 | KP19 | 93 | Female | Respiratory | Pulmonary infection, heart failure | Discharged | 29/1/2018 | Urine |
| Pa32 | KP32 | 61 | Male | ICU | Severe craniocerebral injury | Discharged | 15/5/2017 | Sputum |
| Pa33 | KP33 | 96 | Male | ICU | Pulmonary malignancy, respiratory failure | Died | 25/5/2017 | Sputum |
| Pa35 | KP35 | 60 | Female | ICU | Pulmonary infection, intracranial space-occupied | Discharged | 1/6/2017 | Sputum |
| Pa36 | KP36 | 96 | Male | ICU | Pulmonary malignancy, respiratory failure | Died | 7/6/2017 | Blood |
| Pa37 | KP37 | 36 | Male | ICU | Cardiopulmonary arrest | Discharged | 18/6/2017 | Blood |
| Pa38 | KP38-1 | 92 | Female | Neurosurgery | Aspiration pneumonia, hypertension | Died | 10/7/2017 | Urine |
| KP38-2 | 92 | Female | Neurosurgery | Sequelae of cerebral infarction, Aspiration pneumonia | Died | 30/10/2017 | Blood | |
| Pa45 | KP45 | 83 | Female | Neurosurgery | Post-traumatic syndrome, cerebral hemorrhage | Discharged | 30/10/2017 | Urine |
| Pa65 | KP65 | 73 | Male | Respiratory | Pulmonary infection | Discharged | 14/3/2018 | Excreta |
ICU, intensive care unit.
Figure 3.
The timelines of inpatients harboring KPC-2-producing ST11-KL64 CRKP isolates in the clinical setting. The rectangle on the timeline represents the admitting’s duration of the patients. Different wards are shown in different colors, and the red arrows indicate sampling dates.
Antimicrobial Susceptibility Testing
All 16 ST11-KL64 CRKP isolates were multidrug-resistant (Table 2). According to the antimicrobial susceptibility testing data, all the bla KPC-carrying K. pneumoniae isolates displayed resistance to ATM (MIC≥32mg/L), ETP (MIC≥256mg/L), CAZ (MIC≥128mg/L), FEP (MIC≥256mg/L), SCF (MIC>256mg/L), FOX (MIC≥128mg/L), LVX (MIC≥16mg/L), CIP (MIC≥16mg/L), TET (MIC≥256mg/L), MH (MIC≥16mg/L), CTX (MIC≥64mg/L), MEM (MIC=128mg/L), IPM (MIC≥64mg/L), GEN (MIC≥256mg/L), PRL (MIC>512mg/L), and PRL/TZP (MIC≥512mg/L). Moreover, a high level of resistance (≥87.5%) was observed to AMK (MIC >256mg/L, except for KP5) and SXT (MIC=16mg/L, except for KP38-1 and KP38-2). Additionally, resistance to FOF and TGC was observed in 4 (25.00%) and 3 (18.75%) isolates, respectively, and colistin resistance was detected only in one isolate KP1-2 (6.25%). Compared to the 16 ST11-KL64 CRKP isolates, the remaining 31 CRKP isolates showed relatively more susceptibility to most antimicrobials, especially for tetracycline and minocycline, but higher resistant rates of fosfomycin and trimethoprim-sulfamethoxazole. Most of the remaining 31 CRKP isolates also maintained high resistance (resistance rate>80.65%) to the aztreonam, ertapenem, ceftazidime, cefepime, cefoperazone-sulbactam, cefoxitin, levofloxacin, ciprofloxacin, amikacin, cefotaxime, meropenem, imipenem, gentamicin, trimethoprim-sulfamethoxazole, piperacillin, and piperacillin-tazobactam (Supplementary Table S2).
Table 2.
Minimal inhibitory concentrations (MICs) of the 16 KPC-2-producing ST11-KL64 CRKP isolates to different antimicrobials.
| Isolate | ATMa | FOF | ETP | CAZ | FEP | SCF | FOX | LVX | CIP | AMK | TET | TGC | MH | CST | CTX | MEM | IPM | GEN | SXT | PRL | PRL/TZP |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KP1-1 | 32b | 16 | >256 | >256 | 256 | >256 | 256 | 64 | 128 | >256 | >256 | 8 | 64 | 0.03 | 64 | 128 | 128 | >256 | 16 | >512 | >512 |
| KP1-2 | 32 | 32 | >256 | 256 | 256 | >256 | 128 | 128 | 128 | >256 | >256 | 8 | 64 | 64 | 128 | 128 | 128 | >256 | 16 | >512 | 512 |
| KP5 | 128 | 64 | >256 | >256 | 256 | >256 | >256 | 128 | 128 | 1 | >256 | 4 | 64 | 0.03 | >256 | 128 | 128 | >256 | 16 | >512 | >512 |
| KP12 | 128 | 128 | 256 | >256 | 256 | >256 | 256 | 32 | 64 | >256 | 256 | 2 | 32 | 0.06 | >256 | 128 | 128 | 256 | 16 | >512 | >512 |
| KP15 | 128 | 32 | >256 | >256 | 256 | >256 | >256 | 64 | 128 | >256 | 256 | 4 | 64 | 0.03 | >256 | 128 | 32 | >256 | 16 | >512 | >512 |
| KP18 | 128 | 256 | >256 | >256 | 256 | >256 | >256 | 64 | 64 | >256 | 256 | 4 | 32 | 0.06 | >256 | 128 | 128 | >256 | 16 | >512 | >512 |
| KP19 | 128 | 256 | >256 | >256 | 256 | >256 | 256 | 32 | 64 | >256 | >256 | 4 | 32 | 0.06 | >256 | 128 | 128 | >256 | 16 | >512 | >512 |
| KP32 | 128 | 16 | >256 | >256 | 256 | >256 | >256 | 64 | 128 | >256 | >256 | 4 | 64 | 0.03 | >256 | 128 | 128 | >256 | 16 | >512 | >512 |
| KP33 | 128 | 32 | >256 | >256 | 512 | >256 | >256 | 128 | 128 | >256 | >256 | 4 | 64 | 0.03 | >256 | 128 | 128 | >256 | 16 | >512 | >512 |
| KP35 | 128 | 32 | >256 | >256 | 256 | >256 | >256 | 64 | 128 | >256 | >256 | 4 | 64 | 0.125 | >256 | 128 | 128 | >256 | 16 | >512 | >512 |
| KP36 | 128 | 32 | 256 | >256 | 256 | >256 | 256 | 64 | 128 | >256 | >256 | 4 | 64 | 2 | 256 | 128 | 128 | >256 | 16 | >512 | >512 |
| KP37 | 128 | 32 | >256 | >256 | 256 | >256 | >256 | 128 | 128 | >256 | >256 | 4 | 32 | 0.03 | >256 | 128 | 128 | >256 | 16 | >512 | >512 |
| KP45 | 128 | 32 | 256 | 256 | 256 | >256 | 256 | 16 | 16 | >256 | 256 | 4 | 16 | 0.03 | 256 | 128 | 128 | >256 | 16 | >512 | 512 |
| KP65 | 128 | 64 | >256 | 128 | >256 | >256 | >256 | 128 | 128 | >256 | >256 | 8 | 64 | 0.03 | >256 | 128 | 64 | >256 | 16 | >512 | >512 |
| KP38-1 | 128 | >512 | 512 | 512 | 256 | 512 | 512 | 128 | 128 | 512 | 512 | 2 | 64 | 0.06 | 512 | 128 | 128 | >256 | 1 | >512 | >512 |
| KP38-2 | 128 | >512 | 512 | 512 | 256 | 512 | 512 | 128 | 128 | 512 | 512 | 2 | 64 | 0.06 | 512 | 128 | 128 | >256 | 1 | >512 | >512 |
ATM, aztreonam; FOF, fosfomycin; ETP, ertapenem; CAZ, ceftazidime; FEP, cefepime; SCF, cefoperazone-sulbactam; FOX, cefoxitin; LVX, levofloxacin; CIP, ciprofloxacin; AMK, amikacin; TET, tetracycline; TGC, tigecycline; MH, minocycline; CST, colistin; CTX, cefotaxime; MEM, meropenem; IPM, imipenem; GEN, gentamicin; SXT, trimethoprim-sulfamethoxazole; PRL, piperacillin; and PRL/TZP, piperacillin-tazobactam.
mg/L.
Identification of Antimicrobial Resistance Genes and Virulence Genes
According to the WGS data, antimicrobial resistance genes and virulence genes among the 16 ST11-KL64 CRKP isolates are determined and showed in Figure 2A. All the isolates harbored aminoglycosides (rmtB), chloramphenicol (catA2), fosfomycin (fosA), quinolones (qnrS1), sulfonamides (sul2), tetracyclines [tet(A)], and β-lactams (bla CTX-M-65, bla KPC-2, bla SHV-12, and bla TEM-1B) resistance genes. Aminoglycoside resistance gene aadA2 was observed in 15 CRKP isolates but only absent in isolate KP5. Trimethoprim resistance gene dfrA14 was also found in all CRKP isolates except for KP38-1. Moreover, the analysis of quinolone resistance-determining regions (QRDRs) revealed three amino acid mutations associated with fluoroquinolone resistance, including S83I and D87G in GyrA and S80I in ParC, in all the 16 CRKP isolates. Additionally, all the 16 ST11-KL64 CRKP isolates contained aerobactin (iucABCD and iutA) and hypermucoviscosity (rmpA2) virulence genes, representing a potential CR-hvKP phenotype. The regulator of mucoid phenotype gene (rmpA) and peg-344 were detected in all CRKP isolates except for KP35, KP37, KP38-2, and KP45.
Genetic Features of Virulence Plasmids and bla KPC-2-Bearing Plasmids
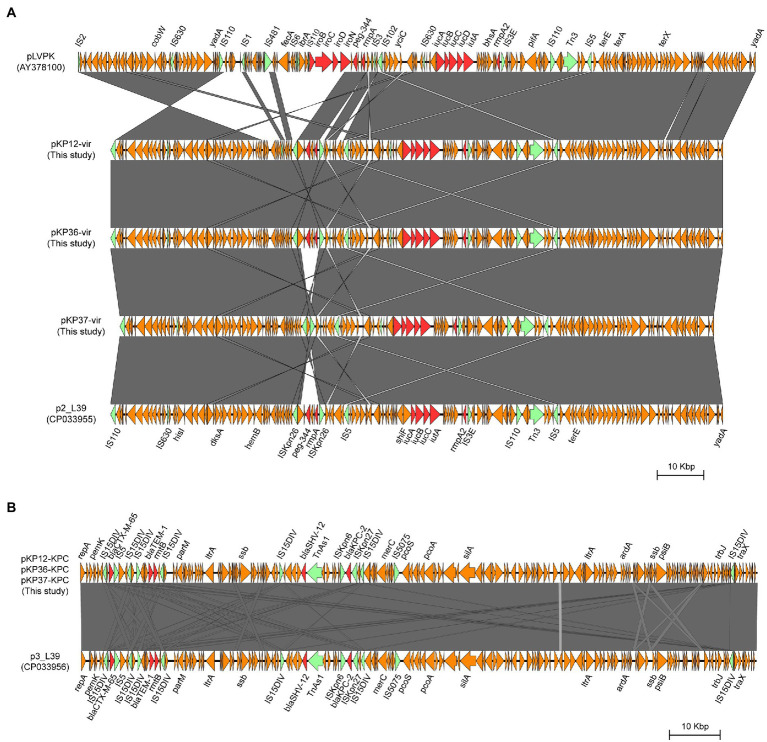
To investigate the genetic features of virulence plasmids and bla KPC-2-bearing plasmids, K. pneumoniae KP12, KP36, and KP37 were further subjected to whole-genome sequencing using the Oxford Nanopore MinION platform. Plasmid sequence analysis confirmed that all the three K. pneumoniae isolates harbored an IncFIB/IncHI1B type virulence plasmid and an IncFII/IncR type bla KPC-2-bearing plasmid (Figure 4). Interestingly, iroBCDN with IS110 transposase on the upstream, located in the same transposon IS3 with rmpA and peg-344 in pLVPK, were absent in KP12, KP36, and KP37 (Figure 5A). It is worth noting that rmpA and peg-344 were located between two ISKpn26 transposons in pKP12-vir and pKP36-vir, but the two virulence genes were further missing in pKP37-vir. According to these results, we hypothesize that IS110 might be responsible for the deletion of iroBCDN, and then forming a new transposon ISKpn26 carrying rmpA and peg-344 from the classical virulence plasmid pLVPK after this losing event.
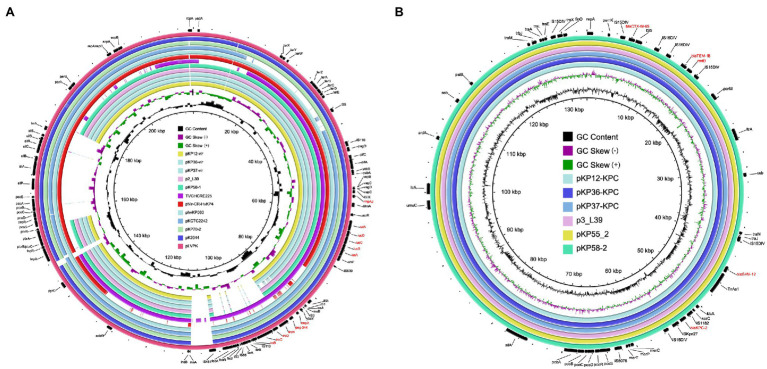
Figure 4.
Genetic comparison of virulence plasmids and bla KPC-2-bearing plasmids recovered from K. pneumoniae KP12, KP36, and KP37 with similar plasmids in the NCBI database, respectively. (A) Alignment of virulence plasmids pKP12-vir, pKP36-vir, and pKP37-vir with p2_L39 (CP033955), pKP58-1 (CP041374), TVGHCRE225 (CP023723), pVir-CR-HvKP4 (MF437313), phvKP060 (CP034776), pKCTC2242 (CP002911), pKP70-2 (MF398271), pK2044 (AP006726), and pLVPK (AY378100). pLVPK was used as the reference plasmid. Virulence genes are highlighted in red. (B) Alignment of bla KPC-2-bearing plasmid pKP12-KPC, pKP36-KPC, and pKP37-KPC with p3_L39 (CP033956), pKP55_2 (CP055296), and pKP58-2 (CP041375). p3_L39 was used as the reference plasmid. Antimicrobial resistance genes are highlighted in red.
Figure 5.
Structural comparison of the genetic context of virulence genes (A) and bla KPC-2 gene (B) in representative plasmids. The arrows represent coding sequences (red arrows, antimicrobial resistance genes or virulence genes; green arrows, mobile elements). Shading denotes the regions with high homology (>95% nucleotide identity).
Plasmids pKP12-KPC, pKP36-KPC, and pKP37-KPC exhibited a high level of homology (99% coverage and 99% identity) with other bla KPC-2-bearing plasmids, i.e., the 133,772-bp IncFII/IncR plasmid p3_L39 recovered from K. pneumoniae strain L39_2 in China. Plasmids pKP12-KPC, pKP36-KPC, and pKP37-KPC also harbored several antimicrobial resistance genes, i.e., bla CTX-M-65, bla SHV-12, bla TEM-1B, and rmtB (Figure 5B). Comparison of genetic surroundings of bla KPC-2 gene indicated that it was flanked by a similar core structure, ISKpn27-bla KPC-2-ISKpn6. Another multi-drug resistance region consisted of bla CTX-M-65, bla TEM-1B, and rmtB genes and five mobile genetic elements on these bla KPC-2-bearing plasmids.
Discussion
The worldwide spread of carbapenem-resistant K. pneumoniae, associated with considerable morbidity and mortality, poses a severe threat to public health. ST11 was found to be the most predominant epidemic clone of CRKP strains in China, which has aroused considerable attention recently due to the scenarios for convergence of resistance and hypervirulence determinants in a single strain (Wyres et al., 2020a). Therefore, surveillance and tracking of high-risk clones of CRKP and understanding their clinical importance are critical. PFGE and MLST, as well-known genotyping methods, can monitor and control the spread of nosocomial pathogens (Singh et al., 2006). However, traditional molecular typing techniques are limited in distinguishing minor genomic differences between closely related strains involved in an outbreak. With the extensive use of next-generation sequencing, WGS analysis easily distinguishes minor differences between clones and is widely used for pathogen identification and tracking the rules underlying pathogen spread (Klemm et al., 2018; Schurch et al., 2018). This study investigated the origin and transmission pattern of KPC-2-producing ST11-KL64 CRKP outbreaks. The genetic features of virulence plasmids and bla KPC-2-bearing plasmids were further investigated by comparative genomic analysis.
During the study period, 47 K. pneumoniae isolates were identified as CRKP, and three dominant PFGE clusters were observed. Among them, 16 CRKP isolates harbored the bla KPC-2 gene, representing a potential clonal spread due to their similar PFGE patterns. In silico MLST analysis identified all the 16 KPC-2-producing CRKP isolates as ST11, which is the most predominant sequence type of KPC-2-producing CRKP in China and has been reported worldwide, including America, Europe, and Asia (Baraniak et al., 2011; Jiang et al., 2015; Zhan et al., 2017; Gu et al., 2018; Ko, 2019; Spencer et al., 2019; Zhou et al., 2020; Jin et al., 2021; Yang et al., 2021). Moreover, all the 16 KPC-2-producing CRKP isolates belonged to the serotype KL64, which differs from those reported previously, such as KL1, KL2, and KL62 (Feng et al., 2018; Gu et al., 2018). However, an infection caused by KPC-2-producing ST11-KL64 CRKP was frequently reported recently (De Campos et al., 2018; Jia et al., 2019; Ruan et al., 2020a; Zhang et al., 2020b; Zhou et al., 2020; Jin et al., 2021; Yang et al., 2021). Zhou et al. (2020) reported that patients infected by ST11-KL64 CRKP had a significantly higher mortality rate than those infected by other CRKP. In this study, four out of 16 patients died of ST11-KL64 CRKP infection. These findings suggested that KL64 clone of CRKP was closely related to high pathogenicity and transmissibility, and continuous monitoring should be taken to prevent further dissemination.
Following cgSNP-based phylogenetic analysis, 16 KPC-2-producing ST11-KL64 CRKP isolates could be divided into two clades, suggesting two independent transmission events. Some patients in the two clades showed overlaps in time frames or hospital stay units that may have contributed to K. pneumoniae dissemination among patients. The index patient of transmission 1 was Pa 32, and the infection was transmitted to Pa 33, Pa 35, Pa 36, and Pa 37 represented in clade 1. Pa 33, Pa 35, Pa 36, and Pa 37 shared months of overlapping ICU stay with Pa 32, and the five isolates from these inpatients were sampled in ICU and displayed considerable genomic similarity. CRKP was detected in the urine and blood culture of Pa 38 and Pa 45 after they were transferred from ICU to neurosurgery. A separate transmission route of KPC-2-producing CRKP infection was identified in Pa 12, Pa 15, Pa 18, and Pa 19. The sampling time of the above four inpatients was close, suggesting that a transmission event occurred within a short period. The spread of ST11 CRKP among different departments or different wards of the same department in the hospital is relatively expected, which has been reported frequently (Jiang et al., 2015; Gu et al., 2018; Sui et al., 2018; Lu et al., 2020; Zhang et al., 2020b). Our data suggested two independent outbreaks of KPC-2-producing ST11-KL64 CRKP isolates in the ICU and the respiratory ward, respectively, between 2016 and 2018. These results confirmed the easy transfer of ST11 CRKP, and practical strategies must be implemented to control the outbreak and avoid nosocomial transmission.
In this study, all the isolates of 16 ST11-KL64 CRKP harbored bla KPC-2 and were multidrug-resistant. In addition to carbapenemases, several extended-spectrum β-lactamase (ESBL) resistance genes, including bla CTX-M-65, bla SHV-12, and bla TEM-1B, were identified in the 16 ST11-KL64 CRKP isolates. This finding indicated that all isolates contained multiple ESBL genes, which is in agreement with the reports on the co-occurrence of bla CTX-M, bla SHV, and bla TEM in K. pneumoniae strains (Baraniak et al., 2011; Jiang et al., 2015; Gu et al., 2018; Sui et al., 2018). Therefore, more attention should be paid to the K. pneumoniae isolates, identified as epidemic ST11 clone and co-harbored carbapenemases and ESBLs in China. Besides the β-lactams resistance genes, other resistance genes that were found to be present in the majority of 16 ST11-KL64 CRKP isolates included rmtB, catA2, fosA, qnrS1, sul2, tet(A), aadA2 (detectable in 15 isolates), and dfrA14 (detectable in 15 isolates), conferring resistance to aminoglycosides, chloramphenicol, fosfomycin, quinolone, sulphonamides, tetracycline, aminoglycoside, and trimethoprim, respectively. Undoubtedly, the presence of resistance genes allows the K. pneumoniae isolates to survive the barrage of antimicrobial agents used in treating infections. Fortunately, tigecycline and colistin were still effective in vitro, which suggested that the above two antimicrobials could be valuable treatment choices against ST11-KL64 CRKP infections.
Several virulence factors have been characterized as contributing to the hypervirulence phenotype of K. pneumoniae, but are not limited to the mucoid regulators rmpA and rmpA2 and the aerobactin synthesis cluster iucABCD/iutA (Russo and Marr, 2019; Choby et al., 2020). The rmpA and rmpA2 genes have both been considered determinants controlling the capsular polysaccharide (CPS) biosynthesis, representing the hypermucoviscous phenotype. The aerobactin has been appreciated as the predominant siderophore system in the hvKP. It has previously been common that ST11 K. pneumoniae is a widely disseminated multidrug-resistant clonal lineage, including carbapenems, but not hypervirulent. However, a fetal outbreak in five patients caused by an ST11 carbapenem-resistant K. pneumoniae strain, which turned into hvKP by acquiring a roughly 170 kb pLVPK-like plasmid, was reported in China (Gu et al., 2018). In this study, all the 16 KPC-2-producing K64-ST11 CRKP isolates harbored hypermucoviscous phenotype regulators and aerobactin synthesis, showed not only resistance to antimicrobials but also hypervirulent phenotype. Alongside the plasmid-borne ESBL and carbapenemase genes, our data revealed a high prevalence of ST11-KL64 CR-hvKPs carrying the IncHI1B/IncFIB virulence plasmids with iro locus deletion. Clinically, the KPC-2-producing ST11-KL64 CRKP isolates carrying the rmpA and rmpA2 genes exhibited enhanced environmental survival and caused more severe infection than classic ST11 KP strains, leading to high mortality (Zhou et al., 2020). This finding is consistent with our study, which showed that all the four dead patients (Pa 1, Pa 33, Pa36, and Pa38) harbored rmpA and rmpA2 genes in ST11-KL64 CRKP isolates. These results revealed the hypervirulence nature of those isolates, and targeted surveillance is urgently needed. Further genomic epidemiology and evolutionary analysis at the national scale are warranted to understand the genetic basis and evolution characteristics of the wide dissemination of carbapenem-resistant hypervirulent ST11-KL64 K. pneumoniae in China.
During the two nosocomial CPKP outbreaks, we had implemented infection prevention and control measures to control the outbreaks in the ICU and respiratory. Several measures were implemented to eliminate the nosocomial CPKP infection. First, we implemented stringent isolation procedures for the CRKP-infected patients and limited the persons who came to contact with these patients. Second, we performed equipment disinfection and periodic environmental cleansing. Third, all the hospital staff contacted with patients carrying CRKP should wear medical gloves and contagion gowns and enforced hand hygiene once the operation was finished. Finally, the ST11-KL64 CRKP nosocomial infection was successfully eliminated and ended in April 2018.
In conclusion, our study identified the emergence of a high-risk clone of KPC-2-producing ST11-KL64 CRKP isolates in a clinical setting. Our results clearly showed that WGS could reveal the two transmission scenarios caused by these CRKP isolates. Due to the acquisition of multiple plasmid-borne antimicrobial resistance and virulence genes, these isolates have presented a significant challenge for public health. The placement of adequate infection control measures is necessary to prevent their further dissemination in nosocomial settings.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
ZR, JZ, and XX designed the experiments. YK, QS, and HC performed the experiments. ZR and YK analyzed the data. YK, ZR, and MD wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (81871696 and 82072342) and the Zhejiang Provincial Medical and Health Science and Technology Plan (2021KY943).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.736896/full#supplementary-material
References
- Alikhan N. F., Petty N. K., Ben Zakour N. L., Beatson S. A. (2011). BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baraniak A., Grabowska A., Izdebski R., Fiett J., Herda M., Bojarska K., et al. (2011). Molecular characteristics of KPC-producing Enterobacteriaceae at the early stage of their dissemination in Poland, 2008-2009. Antimicrob. Agents Chemother. 55, 5493–5499. doi: 10.1128/AAC.05118-11, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choby J. E., Howard-Anderson J., Weiss D. S. (2020). Hypervirulent Klebsiella pneumoniae: clinical and molecular perspectives. J. Intern. Med. 287, 283–300. doi: 10.1111/joim.13007, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Campos T. A., Goncalves L. F., Magalhaes K. G., De Paulo Martins V., Pappas Junior G. J., Peirano G., et al. (2018). A fatal bacteremia caused by hypermucousviscous KPC-2 producing extensively drug-resistant K64-ST11 Klebsiella pneumoniae in Brazil. Front. Med. 5:265. doi: 10.3389/fmed.2018.00265, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Lu Y., Yao Z., Zong Z. (2018). Carbapenem-resistant hypervirulent Klebsiella pneumoniae of sequence type 36. Antimicrob. Agents Chemother. 62, e02644–e02717. doi: 10.1128/AAC.02644-17, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Zou S., Chen H., Yu Y., Ruan Z. (2021). BacWGSTdb 2.0: a one-stop repository for bacterial whole-genome sequence typing and source tracking. Nucleic Acids Res. 49, D644–D650. doi: 10.1093/nar/gkaa821, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukigai S., Alba J., Kimura S., Iida T., Nishikura N., Ishii Y., et al. (2007). Nosocomial outbreak of genetically related IMP-1 beta-lactamase-producing Klebsiella pneumoniae in a general hospital in Japan. Int. J. Antimicrob. Agents 29, 306–310. doi: 10.1016/j.ijantimicag.2006.10.011, PMID: [DOI] [PubMed] [Google Scholar]
- Gu D., Dong N., Zheng Z., Lin D., Huang M., Wang L., et al. (2018). A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect. Dis. 18, 37–46. doi: 10.1016/S1473-3099(17)30489-9, PMID: [DOI] [PubMed] [Google Scholar]
- Jia H., Chen H., Ruan Z. (2019). Unravelling the genome sequence of a pandrug-resistant Klebsiella pneumoniae isolate with sequence type 11 and capsular serotype KL64 from China. J. Glob. Antimicrob. Resist. 19, 40–42. doi: 10.1016/j.jgar.2019.08.013, PMID: [DOI] [PubMed] [Google Scholar]
- Jiang Y., Wei Z., Wang Y., Hua X., Feng Y., Yu Y. (2015). Tracking a hospital outbreak of KPC-producing ST11 Klebsiella pneumoniae with whole genome sequencing. Clin. Microbiol. Infect. 21, 1001–1007. doi: 10.1016/j.cmi.2015.07.001, PMID: [DOI] [PubMed] [Google Scholar]
- Jin X., Chen Q., Shen F., Jiang Y., Wu X., Hua X., et al. (2021). Resistance evolution of hypervirulent carbapenem-resistant Klebsiella pneumoniae ST11 during treatment with tigecycline and polymyxin. Emerg. Microbes Infect. 10, 1129–1136. doi: 10.1080/22221751.2021.1937327, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm E. J., Wong V. K., Dougan G. (2018). Emergence of dominant multidrug-resistant bacterial clades: lessons from history and whole-genome sequencing. Proc. Natl. Acad. Sci. U. S. A. 115, 12872–12877. doi: 10.1073/pnas.1717162115, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko K. S. (2019). Antibiotic-resistant clones in gram-negative pathogens: presence of global clones in Korea. J. Microbiol. 57, 195–202. doi: 10.1007/s12275-019-8491-2, PMID: [DOI] [PubMed] [Google Scholar]
- Letunic I., Bork P. (2021). Interactive tree Of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M. C., Chen Y. T., Tang H. L., Liu Y. Y., Chen B. H., Wang Y. W., et al. (2020). Transmission and evolution of OXA-48-producing Klebsiella pneumoniae ST11 in a single hospital in Taiwan. J. Antimicrob. Chemother. 75, 318–326. doi: 10.1093/jac/dkz431, PMID: [DOI] [PubMed] [Google Scholar]
- Navon-Venezia S., Kondratyeva K., Carattoli A. (2017). Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol. Rev. 41, 252–275. doi: 10.1093/femsre/fux013, PMID: [DOI] [PubMed] [Google Scholar]
- Nishida S., Matsunaga N., Kamimura Y., Ishigaki S., Furukawa T., Ono Y. (2020). Emergence of enterobacter cloacae complex co-producing IMP-10 and CTX-M, and Klebsiella pneumoniae producing VIM-1 in clinical isolates in Japan. Microorganisms 8:1816. doi: 10.3390/microorganisms8111816, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Vazquez M., Sola Campoy P. J., Ortega A., Bautista V., Monzon S., Ruiz-Carrascoso G., et al. (2019). Emergence of NDM-producing Klebsiella pneumoniae and Escherichia coli in Spain: phylogeny, resistome, virulence and plasmids encoding blaNDM-like genes as determined by WGS. J. Antimicrob. Chemother. 74, 3489–3496. doi: 10.1093/jac/dkz366 [DOI] [PubMed] [Google Scholar]
- Ruan Z., Feng Y. (2016). BacWGSTdb, a database for genotyping and source tracking bacterial pathogens. Nucleic Acids Res. 44, D682–D687. doi: 10.1093/nar/gkv1004, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Z., Wu J., Chen H., Draz M. S., Xu J., He F. (2020a). Hybrid genome assembly and annotation of a pandrug-resistant Klebsiella pneumoniae strain using nanopore and illumina sequencing. Infect. Drug Resist. 13, 199–206. doi: 10.2147/IDR.S240404, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Z., Yu Y., Feng Y. (2020b). The global dissemination of bacterial infections necessitates the study of reverse genomic epidemiology. Brief Bioinform. 21, 741–750. doi: 10.1093/bib/bbz010, PMID: [DOI] [PubMed] [Google Scholar]
- Russo T. A., Marr C. M. (2019). Hypervirulent Klebsiella pneumoniae. Clin. Microbiol. Rev. 32, e00001–e00019. doi: 10.1128/CMR.00001-19, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurch A. C., Arredondo-Alonso S., Willems R. J. L., Goering R. V. (2018). Whole genome sequencing options for bacterial strain typing and epidemiologic analysis based on single nucleotide polymorphism versus gene-by-gene-based approaches. Clin. Microbiol. Infect. 24, 350–354. doi: 10.1016/j.cmi.2017.12.016, PMID: [DOI] [PubMed] [Google Scholar]
- Singh A., Goering R. V., Simjee S., Foley S. L., Zervos M. J. (2006). Application of molecular techniques to the study of hospital infection. Clin. Microbiol. Rev. 19, 512–530. doi: 10.1128/CMR.00025-05, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer M. D., Winglee K., Passaretti C., Earl A. M., Manson A. L., Mulder H. P., et al. (2019). Whole genome sequencing detects inter-facility transmission of carbapenem-resistant Klebsiella pneumoniae. J. Infect. 78, 187–199. doi: 10.1016/j.jinf.2018.11.003, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui W., Zhou H., Du P., Wang L., Qin T., Wang M., et al. (2018). Whole genome sequence revealed the fine transmission map of carbapenem-resistant Klebsiella pneumonia isolates within a nosocomial outbreak. Antimicrob. Resist. Infect. Control 7:70. doi: 10.1186/s13756-018-0363-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M. J., Petty N. K., Beatson S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusova T., Dicuccio M., Badretdin A., Chetvernin V., Nawrocki E. P., Zaslavsky L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624. doi: 10.1093/nar/gkw569, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenover F. C., Arbeit R. D., Goering R. V., Mickelsen P. A., Murray B. E., Persing D. H., et al. (1995). Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33, 2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Xie Y., Li G., Liu J., Li X., Tian L., et al. (2018). Whole-genome-sequencing characterization of bloodstream infection-causing hypervirulent Klebsiella pneumoniae of capsular serotype K2 and ST374. Virulence 9, 510–521. doi: 10.1080/21505594.2017.1421894, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick R. R., Heinz E., Holt K. E., Wyres K. L. (2018). Kaptive web: user-friendly capsule and lipopolysaccharide serotype prediction for Klebsiella genomes. J. Clin. Microbiol. 56, e00197–e00218. doi: 10.1128/JCM.00197-18, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick R. R., Judd L. M., Gorrie C. L., Holt K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyres K. L., Lam M. M. C., Holt K. E. (2020a). Population genomics of Klebsiella pneumoniae. Nat. Rev. Microbiol. 18, 344–359. doi: 10.1038/s41579-019-0315-1, PMID: [DOI] [PubMed] [Google Scholar]
- Wyres K. L., Nguyen T. N. T., Lam M. M. C., Judd L. M., Van Vinh Chau N., Dance D. A. B., et al. (2020b). Genomic surveillance for hypervirulence and multi-drug resistance in invasive Klebsiella pneumoniae from south and Southeast Asia. Genome Med. 12:11. doi: 10.1186/s13073-019-0706-y, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Yang Y., Chen G., Lin M., Chen Y., He R., et al. (2021). Molecular characterization of carbapenem-resistant and virulent plasmids in Klebsiella pneumoniae from patients with bloodstream infections in China. Emerg. Microbes Infect. 10, 700–709. doi: 10.1080/22221751.2021.1906163, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yigit H., Queenan A. M., Anderson G. J., Domenech-Sanchez A., Biddle J. W., Steward C. D., et al. (2001). Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45, 1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan L., Wang S., Guo Y., Jin Y., Duan J., Hao Z., et al. (2017). Outbreak by hypermucoviscous Klebsiella pneumoniae ST11 isolates with carbapenem resistance in a tertiary hospital in China. Front. Cell. Infect. Microbiol. 7:182. doi: 10.3389/fcimb.2017.00182, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Jin L., Ouyang P., Wang Q., Wang R., Wang J., et al. (2020b). Evolution of hypervirulence in carbapenem-resistant Klebsiella pneumoniae in China: a multicentre, molecular epidemiological analysis. J. Antimicrob. Chemother. 75, 327–336. doi: 10.1093/jac/dkz446, PMID: [DOI] [PubMed] [Google Scholar]
- Zhang P., Shi Q., Hu H., Hong B., Wu X., Du X., et al. (2020a). Emergence of ceftazidime/avibactam resistance in carbapenem-resistant Klebsiella pneumoniae in China. Clin. Microbiol. Infect. 26, 124.e1–124.e4. doi: 10.1016/j.cmi.2019.08.020, PMID: [DOI] [PubMed] [Google Scholar]
- Zhou K., Xiao T., David S., Wang Q., Zhou Y., Guo L., et al. (2020). Novel subclone of carbapenem-resistant Klebsiella pneumoniae sequence type 11 with enhanced virulence and transmissibility, China. Emerg. Infect. Dis. 26, 289–297. doi: 10.3201/eid2602.190594, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.