Abstract
Exceptional efforts have been undertaken to shed light into the biology of adaptive immune responses to SARS-CoV-2. T cells occupy a central role in adaptive immunity to mediate helper functions to different arms of the immune system and are fundamental to mediate protection, control, and clearance of most viral infections. Even though many questions remain unsolved, there is a growing literature linking specific T cell characteristics to differential COVID-19 severity and vaccine outcome. In this review, we summarize our current understanding of CD4+ and CD8+ T cell responses in acute and convalescent COVID-19. Further, we discuss the T cell literature coupled to pre-existing immunity and vaccines and highlight the need to look beyond blood to fully understand how T cells function in the tissue space.
Keywords: SARS-CoV-2, COVID-19, Memory T cells, Vaccine
1. Introduction
With an evolving pathogen like SARS-CoV-2, it remains absolutely necessary to gain a better understanding of the precise mechanisms that enable immune protection of SARS-CoV-2 and future viral variants. In this context, the adaptive immune system occupies a key role by re-recognizing viral antigens and rapidly block or generate control to limit disease progression. Adaptive immunity consists of two major components: B cells and T cells. In simplified terms, B cells produce antibodies and T cells provide helper functions to other immune cells and directly eliminate virus-infected cells. The current literature is replete with studies, from both natural infection and vaccination, showing that neutralizing antibodies serve as a key effector correlate, appropriately, of COVID-19 protection (reviewed in [1]). However, current SARS-CoV-2 variants of concern (e.g. Delta variant) partly subvert neutralizing antibodies [2], emphasizing that more studies are needed to grasp the full picture of SARS-CoV-2-specific adaptive immunity. T cells are harder to examine and therefore fewer studies, in relative terms, have focused on this arm of adaptive immunity. Nevertheless, T cells serve as the critical node to generate both effective humoral- and cellular immunity; without T cells the host loses the capacity to control normally harmless infections and succumbs, as demonstrated in both inherent and acquired immunodeficiencies (e.g. AIDS).
T cells recognize epitopes from a wide range of viral antigens through a combined array of T cell receptors (TCRs) that generates a repertoire of 25 million unique clonotypes [3]. This tremendous diversity provides T cells an advantage to recognize for example viral variants of respiratory viruses, such as SARS-CoV-2. T cells are broadly divided up into two subsets: The first one, T helper cells (CD4+ T cells), orchestrate different arms of the immune system and are critical in the defense against both extracellular and intracellular pathogens. Following viral infections, naïve CD4+ T cells differentiate mainly into two subsets, known as T helper 1 (Th1) and T follicular helper (Tfh) cells (reviewed in [4]). Th1 promote cell-mediated immune responses by activating other immune cells to control viral spread, while Tfh provide B cell help by mediating somatic hypermutations and affinity maturation of germinal center reactions, leading to the generation of high-affinity antibodies capable of neutralizing viruses. The second arm, known as cytotoxic (CD8+) T cells, are important to generate control and clear most viral infections (reviewed in [5]). Following antigen clearance, long-lived memory CD8+ T cells are formed to mediate an effective secondary response against future viral infections (e.g. Influenza A). In the setting of chronic infections, CD8+ T cells are instead known to develop into an exhausted trajectory where both dysfunctional and more functional subsets exist to generate durable control of persistent viral infections (e.g. HIV, HCV, HBV). Independent of their trajectories, most still view non-naïve CD8+ T cells simply as killer T cells with a main function to eliminate virus-infected cells. However, newer studies suggest a greater extent of CD8+ T cell diversity in humans [6], where these cells can function also as innate-like sensors in tissues to promote recruitment of other immune cells during pathogen invasion [7]. Nevertheless, based on an extensive literature it is appropriate to state that both CD4+ and CD8+ T cells are critical to generate control and provide helper mechanisms to increase resistance to viral infection and re-infection.
Studies of adaptive immunity to SARS-CoV-2 are starting to shed light into the correlates of T cell responses to differential COVID-19 progression and vaccine outcome. In this review, we provide an update of our basic knowledge of T cell characteristics and function to SARS-CoV-2 in acute and convalescent infection and discuss how cross-reactive, vaccine-induced, and tissue-resident T cells may impact protection and disease progression.
2. T cell responses in the acute phase
COVID-19 is a disease of many different outcomes. Certain individuals remain asymptomatic, while others experience severe and critical symptoms, including respiratory failure, septic shock, multiorgan failure and, ultimately, fatal outcome. It is now well established that severe infection leads to a dysregulated and hyper-activated immune response, similar to “cytokine storm”, creating a misfire of the antiviral response [8,9]. Elevated cytokine and chemokine signatures of IL-6, IL-8, IL-10, CXCL10 among others are usually manifestations of severe COVID-19 [10,11], while the early antiviral misfire is partly mediated by a defective type I IFN response in severe and fatal COVID-19 [12,13]. Another prominent hallmark of SARS-CoV-2 infection is lymphopenia, a general phenomenon of COVID-19 progression, involving major alterations in the B cell composition and a profound impact on the T cell lineage [14,15]. The decline of T cells in the circulation combined with increased immune activation profiles are probably direct consequences of an emerging antiviral T cell response re-distributing to the respiratory tract.
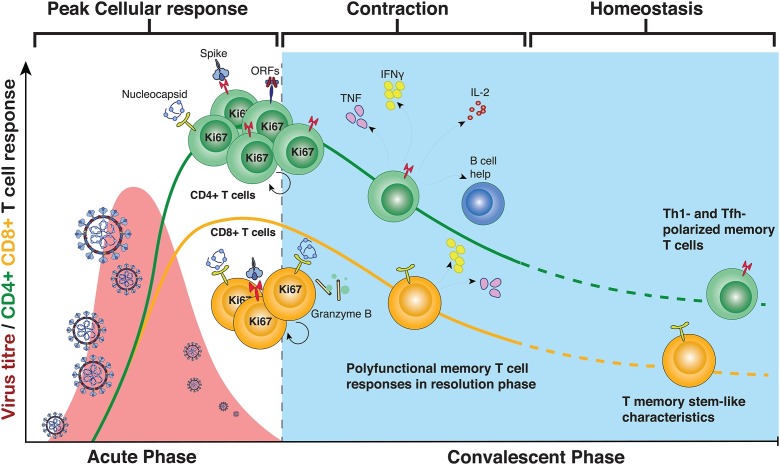
SARS-CoV-2-specific T cells are observed in most donors during acute infection [12,16,17] (Fig. 1 ). The majority of SARS-CoV-2-specific CD4+ T cells exhibit a CCR7+CD45RA− (central memory; 50–60 %) and CCR7−CD45RA− (effector memory; 25–40 %) during acute and over the course of infection. Instead, SARS-CoV-2-specific CD8+ T cells are preferentially categorized into an effector memory phenotype during acute infection (40–60 %), that slowly declines with time and is replenished with the other circulating memory subsets during the course of convalescence [18,19]. Both the CD4+ and CD8+ T cell response is commonly directed against multiple different SARS-CoV-2 antigens, with strong responses particularly observed against spike and nucleocapsid [[18], [19], [20]]. Similar to other acute viral infections [21,22], most SARS-CoV-2-specific CD4+ and CD8+ T cells display a Th1 and effector profile, respectively, with increased cytotoxic capacity, elevated levels of immune activation markers (Ki-67, CD38, HLA-DR, CD69) and lower Th2 and Th17-mediated responses [17,23,24]. Many studies have also observed an upregulation of inhibitory receptors (PD-1, TIM-3, CD39, among others) during acute infection, initially suggesting that the early T cell response in acute disease was dysfunctional and overactivated, possibly contributing to the cytokine storm [15,25,26]. However, it has now become more apparent that the upregulation of inhibitory receptors are likely an activation-induced phenomenon rather than due to exhaustion, as SARS-CoV-2-specific T cells expressing inhibitory receptors are fully functional [27]. It has also now been shown that the source of pro-inflammatory cytokine flares contributing to immunopathology does not originate from T cells, but are most likely derived from the myeloid cell lineage [28]. There have been initial concerns of pro-inflammatory cytokines promoting deviation from the Th1 trajectory of the T cells into a pathogenic Th2 or Th17 fate, however no such phenomenon has been reported thus far – at least not in circulation [29].
Fig. 1.
Phenotype and kinetics of SARS-CoV-2-specific T cell responses after natural infection.
SARS-CoV-2-specific T cells exhibit many specific attributes that are associated with more or less severe COVID-19. For instance, detection of an early CD4+ T cell response show a stronger correlation to less severe COVID-19 than antibody and CD8+ T cell responses [16]. Likewise, early induction of a functional (IFN-γ-producing) CD4+ T cell response are found much earlier in patients with mild disease and correlates with viral clearance [20]. These data go in line with recent preliminary data suggesting that very rapid induction of CD8+ T cells could be a cause of asymptomatic disease [30]. A new study found that severe COVID-19 is not preferentially associated with the quantity of the CD4+ T cell response, but rather poor polyfunctionality and proliferative capacity, as well as enhanced immune activation [31]. An increased immune activation profile has been distinguished in numerous studies, and together with differential Th2, Th17 or Tfh responses to SARS-CoV-2 been associated with distinct disease outcomes [32,33]. Collectively, the current human data indicate that early induction of functional SARS-CoV-2-specific T cells correlate with viral control and mild disease, whereas delayed T cell activation leads to poor functional responses and clearance of the virus and, ultimately, severe and fatal COVID-19.
Human studies are pivotal to establish immune correlates to disease severity, but do not offer always the same mechanistic insight as animal studies. Early reports using transgenic mice models provided evidence that T cells are also critical for viral clearance and disease resolution after SARS-CoV-2 infection [34]. Recent depletion studies in similar murine models have demonstrated that CD8+ T cells serve as a fundamental adaptive component of early SARS-CoV-2 control and clearance, while CD4+ T cells are necessary to induce antibody responses [35]. Much of these data are supported by studies in non-human primates, showing that CD8+ T cell depletion in animals leads to partial abrogation of protection against SARS-CoV-2 rechallenge [36], and that CD4+ T cell responses serve as a strong immune correlate of vaccine efficacy and neutralizing antibody levels [37]. In contrast, a new study on non-human primates surprisingly found that T cells are not critical for antibody class switching, development of immunological memory, and protection from secondary SARS-CoV-2 infection [38]. These differential outcomes could be due to differences of animal handling and experimental setups, which highlights the needs to conduct more thorough animal studies to gain a full picture of the immune correlates of protection.
3. T cell responses in the convalescent phase
Most convalescent patients display a durable antibody and SARS-CoV-2-specific memory T cell response after COVID-19 [39]. SARS-CoV-2-specific CD4+ and CD8+ T cells are detected ex vivo in 100 % and 70 % of patients with previous COVID-19 respectively [40] – although more recent studies using barcoded tetramers indicate that CD8+ T cell responses are detectable at low levels in all patients [24]. After the rapid expansion of CD4+ and CD8+ T cells during the acute phase, both subsets decay with a half-life of approximately 200 days [18]. Similar to antibody levels, the frequencies of SARS-CoV-2-specific T cells stabilize over time, which is associated with an increased fraction of memory stem-like T cells, regardless of disease severity [19]. This subset is known to express TCF-1 as well as other memory stem cell-like markers (such as CD45RA, CD95) in the absence of inhibitory receptors, distinguishing them from previously described precursor exhausted T cells [41]. During the first wave of the SARS-CoV-2 pandemic, several studies simultaneously reported on the presence of polyfunctional SARS-CoV-2-specific CD4+ and CD8+ T cells displaying specificity both to structural [23,42] and non-structural proteins [43,44]. Both CD4+ and CD8+ T cells retain their polyfunctional characteristics for >180 days. Whereas CD4+ T cells exhibit equal functional responses against both structural and accessory SARS-CoV-2 proteins, CD8+ T cells show a preferential recognition of the nucleocapsid protein [18,19].
Convalescent SARS-CoV-2-specific T cell responses are usually present at somewhat lower frequencies in people with previous asymptomatic COVID-19 [45]. These individuals also exhibit discordant T cell and neutralizing antibody responses to some degree [46]. Despite these facts, several studies have shown that asymptomatic disease also induces polyfunctional T cell responses with coordinated secretion of proinflammatory and regulatory (IL-10) cytokines (IL-6, IL-1β) [47,48]. In addition to asymptomatic patients, highly-exposed seronegative individuals show a readily detectable response that is significantly stronger than in healthy and unexposed donors [23,49,50]. Polyfunctional T cell responses in highly-exposed seronegative individuals has also been described in the context of HIV [51], which correlated to people resistant to infections [52]. Whether this type of T cell responses in the absence of antibodies will be able to protect from SARS-CoV-2 infection or severe disease remains to be determined.
A long-standing question has been whether the adaptive immune response following natural infection will provide protection from re-infection. A recent study, published before the spread of the Delta variant, indicated that protection against repeat infection is around 80 % [53]. T cell responses are maintained at stable levels and capable of inducing robust anamnestic responses without any decline in quality to a period of up to 10 months [[18], [19], [20],39,45,54]. In addition, stable frequencies of spike-IgG+ memory B cells and long-lived plasmablasts have been detected from recovered patients, indicative of durable humoral immunity despite low antibody titers [18,55]. With incidents of new viral variants continuing to emerge [56], it remains likely that many will come in contact with distinct SARS-CoV-2 strains again. However, given that SARS-CoV-1-specific T cells can be detected almost 20 years after the 2003 outbreak, even in the absence of detectable antibodies [44,57], it remains likely that coordinated memory B and T cells will form an efficient response to provide protection from recurrent severe COVID-19.
4. Cross-reactive T cells to SARS-CoV-2
The recognition of multiple peptide-MHC ligands by one unique TCR provides the host with an advantage to mount a functional immune response against an immense plethora of antigens, exceeding the number of naïve T cells by far [58]. TCR cross-reactivity is preferentially a consequence of peptide sequence similarities between closely related pathogens, such as different influenza virus strains or flaviviruses. However, the extreme flexibility of the TCR, or specifically focused engagement of the TCR on a few residues within a peptide sequence, allows a promiscuous binding capacity of TCRs between unrelated pathogens and antigens also without sequence homology [[59], [60], [61]].
Since the outbreak of COVID-19, multiple studies have detected T cell responses in the peripheral blood of unexposed individuals that cross-recognize several structural and non-structural SARS-CoV-2 proteins [17,23,40,43,44,[62], [63], [64], [65], [66], [67], [68]]. Cross-reactive SARS-CoV-2-specific memory CD4+ T cells are readily detectable ex vivo in approximately 20–50 % of unexposed people and detectable in almost all individuals after in vitro expansion [62], whereas cross-reactive SARS-CoV-2-specific memory CD8+ T cells are less commonly detected in peripheral blood [40]. Given their sequence homology, it has been proposed that previous infections with endemic common cold human coronaviruses (HCoVs) such as HCoV-OC43, HCoV-229E, HCoV-HKU1, and HCoV-NL63 induce SARS-CoV-2-cross-reactive T cell immunity. Indeed, expanded HCoV-specific CD4+ and CD8+ T cells from unexposed donors show some level of reactivity to SARS-CoV-2 antigens and vice versa [43,62,64,[66], [67], [68], [69]], and shared clonotypes can be detected between HCoV- and SARS-CoV-2-specific T cells in uninfected individuals [66,68,69]. These cross-reactive CD4+ and CD8+ T cell are generally directed against highly conserved epitopes among coronaviruses [66,[63], [64], [65], [66], [67], [68],70], further supporting the concept that SARS-CoV-2-cross-reactive-T cell responses are preferentially mediated by HCoVs.
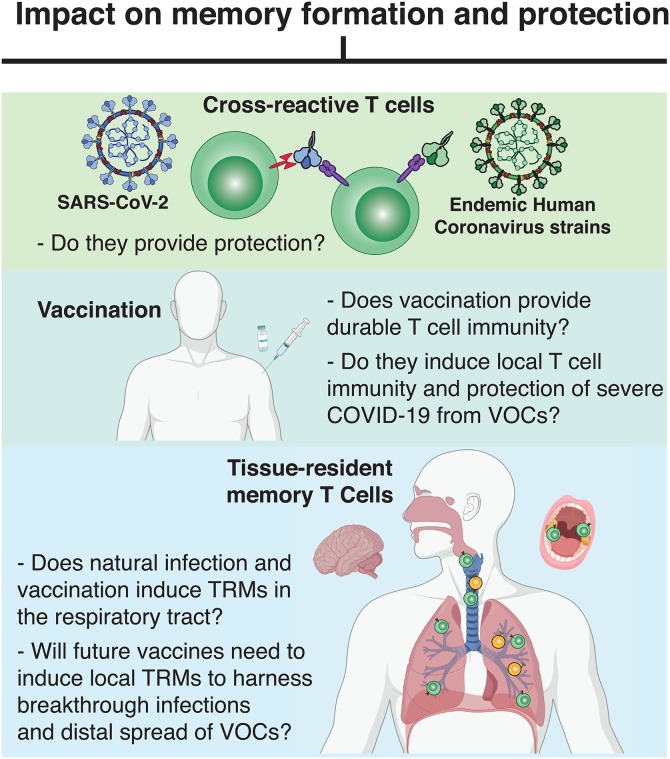
SARS-CoV-2 infection can cause a wide array of clinical outcome ranging from asymptomatic to severe infection and death. Whether pre-existing SARS-CoV-2-cross-reactive T cells have any impact on disease outcome remains debated [71] (Fig. 2 ). We know from other viral infections that cross-reactive immunity may be a double-edged sword: A recall of pre-existing immune cells could induce an early efficient antiviral immune response [[72], [73], [74]], but potentially also contribute to infection-related immunopathology [60]. In addition, cross-reactive memory T cells, which generally show lower affinity and functional capacity [62,69], might outcompete and prevent the activation of naïve T cells – a phenomenon also called “original antigenic sin” [75] – and therefore inhibit the generation of a primary high-affinity T cell response to SARS-CoV-2. The antigenic sin concept, however, does not recapitulate studies showing that recent HCoV infection is associated with milder COVID-19 disease progression and reduced mortality [76]. Furthermore, sampling of peripheral blood from individuals prior to and after SARS-CoV-2 infection demonstrate that pre-existing CD4+ and CD8+ T cells can be recalled and expanded after infection [66,68], but do not seem to preclude the generation of a highly polyclonal primary T cell response after infection [66]. An early activation of SARS-CoV-2-specific CD4+ and CD8+ T cells has been associated with fast viral clearance and a milder clinical disease outcome [20,30], but whether an early T cell response stems from the activation of pre-existing T cell responses or rapid priming of naïve responses has remained unclear. A new study suggest, however, that persons with cross-reactive CD4+ T cell responses demonstrate a higher frequency of SARS-CoV-2-spike-specific CD4+ T cells and neutralizing antibody responses post infection [67]. A similar effect can be seen after mRNA COVID-19 vaccination, where increased SARS-CoV-2-spike-specific CD4+ T cell and antibody responses are observed in individuals with detectable pre-existing CD4+ T cell responses ex vivo [67,77]. Although pre-existing CD8+ T cell responses are less well studied, a recent preliminary study shows that CD8+ T cell responses directed against a highly conserved region of coronavirus polymerase can be detected in uninfected individuals and are expanded in SARS-CoV-2-exposed health care workers that did not subsequently become infected [68]. These T cell responses target a protein that is expressed early during the viral life cycle, which could potentially induce abortive replication and prevent infection [68]. Additionally, coronavirus vaccination provides heterologous protection against other coronaviruses including SARS-CoV-2 in mice [121]
Fig. 2.
Major unknowns within the SARS-CoV-2 T cell field. Which impact will interventions and T cell subsets have on memory formation and COVID-19 protection?
Together these findings highlight the importance of an effective early T cell response to dampen SARS-CoV-2 infection, which may be augmented by pre-existing immunity for infection. Further studies with larger cohorts of longitudinal samples collected prior to and after SARS-CoV-2 infection are necessary to better decipher the impact of pre-existing SARS-CoV-2 immunity on the outcome of COVID-19 and vaccine-generated immunity.
5. T cell immunity after SARS-CoV-2 vaccination
The unprecedented effort in SARS-CoV-2 vaccine development has led to the approval of several vaccine regimens for emergency use in numerous countries [54]. Current efforts are ongoing for their worldwide administration to reach wide-spread immunity against SARS-CoV-2 to prevent infection, severe COVID-19 and halt the pandemic. To date, there are around 20 approved vaccines in at least one country and more than 100 are in phase II or III clinical trials [78]. We will in this review focus on vaccine-induced immunity of the widely distributed mRNA-based BNT162b2 (Pfizer/BioNTech) and mRNA-1273 (Moderna) vaccines, as well as the adenoviral-vector-based ChAdOx1 (Oxford-AstraZeneca) and Ad26.COV2.S (Johnson & Johnson) vaccines [[79], [80], [81], [82]]. All these vaccine regimens encode for the full-length spike protein to induce a potent spike-specific antibody response. Recent studies by Khoury et al. and Earle et al. have shown a strong association between vaccine efficacy and neutralizing antibody levels after two vaccine doses, indicating that vaccine-induced humoral immunity is a key correlate of protection [83,84]. However, the onset of mRNA vaccine protection has been observed as early as 12 days after vaccination, when neutralizing antibody levels are still undetectable, implicating that other immune components may induce early COVID-19 protection [85]. Furthermore, T cell responses are a necessity to generate humoral immune responses in murine models to SARS-CoV-2 [35], and possess a critical role in vaccine-induced immunity for respiratory infections in mice [86,87]. Potent antibody responses rely on Tfh help [88], and SARS-CoV-2-specific CD4+ and CD8+ T cell responses are central for viral control during natural infection [35,89]. Importantly, depletion of CD8+ T cell responses allows SARS-CoV-2 replication in the upper respiratory tract of rechallenged non-human primates [36], and COVID-19-recovered patients with impaired or non-existent B cell responses show a strong SARS-CoV-2-specific CD8+ T cell response [89], thus highlighting the need of vaccines to induce complimentary humoral and cellular anti-SARS-CoV-2 immunity for protection.
Early immunogenicity and follow-up studies demonstrate that BNT162b2, mRNA-1273, ChAdOx1 and Ad26.COV2.S vaccination induces CD4+ and CD8+ T cell responses [[90], [91], [92], [93], [94]]. T cell immunity is generated after the prime dose prior to the mobilization of neutralizing antibody responses and thus may contribute together with Fc-mediated antibody responses to the early protective effect of vaccination [95,96]. T cell responses further increase after the second booster dose in two-dose regimen vaccines [[90], [91], [92]], and correlate with SARS-CoV-2 neutralizing antibody responses in some studies [90,96]. Vaccine-induced spike-specific CD4+ T cell responses are polarized towards a Th1- and Tfh- but not Th2-profile [90,[94], [95], [96], [97]], similar to responses induced during natural infection [16,23,42,43]. Similarly, vaccination-induced CD8+ T cell responses show comparable functional capacity compared to responses induced by natural infection, mainly characterized by the expression of IFN-γ [[90], [91], [92],[94], [95], [96]] and the degranulation marker CD107a [[94], [95], [96]]. However, some studies have implicated that CD8+ T cell responses during SARS-CoV-2 infection demonstrate a more early differentiated profile [95,98], in comparison to vaccine-specific CD8+ T cells after BNT162b vaccination [95]. Thus, further longitudinal studies are necessary to determine possible differential long-term memory generation, maintenance, and longevity of vaccine- and infection-induced SARS-CoV-2-specific T cell responses (Fig. 2).
Due to rare cases of vaccine-induced thrombotic thrombocytopenia after ChAdOx1 vaccination [99], some countries opted to recommend mRNA-based vaccines as boost dose for individuals who had already received a ChAdOx1 prime. Despite the potent induction of humoral and cellular immunity using standard homologous vaccination (prime and boost with same vaccine regimen), vaccine-induced immunity is further increased after heterologous ChAdOx1/BNT162b2 vaccination [92]. Vaccinees demonstrate increased spike-specific antibody titers and neutralization against SARS-CoV-2 including variants of concern (VOC) Alpha, Beta and Gamma, and mount stronger spike-specific CD4+ and CD8+ T cell immunity compared to individuals who received ChAdOx1 vaccine for both doses [92]. Although these results are promising, large efficacy studies of heterologous vaccine regimens are still lacking.
A very potent immune response against SARS-CoV-2 can also be induced through so-called “hybrid immunity”, a combination of naturally generated immunity through infection and vaccine-induced immunity [88]. Several recent studies compared vaccine-induced immunity in naïve versus previously infected individuals. One dose to SARS-CoV-2 convalescent individuals induced antibody titers and neutralization capacity against several SARS-CoV-2 VOCs that was significantly higher compared to naïve individuals who received both vaccine doses by boosting infection-induced immunity [100,101]. Similarly, one vaccine dose to previously infected individuals seems sufficient to raise SARS-CoV-2-specific CD4+ T cell responses to the level in fully vaccinated naïve individuals [100,102]. While the second dose to convalescent individuals does not boost spike-specific CD4+ immunity [100,102], CD8+ T cell responses are further increased [102]. Whether heterologous immunization or hybrid immunity induce a preserved response and of distinct phenotypic or functional T cell profiles compared to standard vaccination remains to be determined.
Concerns of current vaccine actions include existing and future VOCs, which might escape from vaccine-induced immunity. Immunogenicity studies demonstrate that although one dose for convalescent and two vaccine doses for SARS-CoV-2-naïve individuals induce a potent antibody response, binding affinity as well as neutralization and Fc functionality is decreased against some spike-variants when compared to the wild-type strain [[100], [101], [102]]. Although single sequence-specific T cell response against variants decrease dependent on HLA polymorphism [101], the overall T cell response against VOCs seems to be less affected [56,93,101,102]. These data suggest that current VOCs do not fully escape adaptive immunity and that T cells may prevent severe disease progression despite limited protection from VOC infections due to inefficient spike-specific humoral immunity. The emergence of vaccine-escaping SARS-CoV-2 variants could further be limited by including additional viral antigens for vaccination, such as the highly conserved nucleocapsid protein, which is likely more resistant to immune selection pressure. Indeed, nucleocapsid vaccination in mice elicited T cell immunity that mediated protection from SARS-CoV-2 infection [103]. Combined spike and nucleocapsid-adenoviral vaccines furthermore decreased SARS-CoV-2 replication not only in lung, but also brain of challenged mice [104]. Although local tissue immunity in the brain was not assessed in this study, protection was likely mediated through vaccine-induced T cells.
Vaccine-induced neutralization titers decline over time with a similar half-life than infection-induced humoral responses [83,105,106], which has sparked a debate about the necessity of a third booster immunization. However, initial decay rates typically slow down within the first years and long-lived memory B and T cell responses may persist for years and likely protect at least from severe COVID-19. Indeed, the CD8+ T cell response induced after yellow-fever vaccination initially declined with a half-life of around 123 days within the first year [107], which is similar to T cell decay rates after SARS-CoV-2 infection [18,39]. However, vaccination generates a fraction of long-lived memory T cells, which are still detectable in peripheral blood for decades after yellow-fever vaccination [107], or up to 60 years after smallpox vaccination [108]. Altogether, a two-dose vaccination regimen will likely induce a long-lasting humoral- and cell-mediated immune response that protects most from severe illness. However, in the short term, a third booster dose would probably be needed in elderly patients and certain risk groups, including certain immunocompromised individuals who generate a poor adaptive immune response [109,110].
6. Memory T cells in tissues
As highlighted before, T cell responses have been demonstrated to be a crucial factor for full control and clearance of SARS-CoV-2 infection [35,36]. SARS-CoV-2-specific blood memory CD4+ and CD8+ T cells persists for more than 8 months post infection [18,39]. Yet, most studies have relied solely on the analysis of peripheral blood samples and do not take into consideration “tissue-specific immunity”, specifically in the upper and lower respiratory tract. One specific T cell population therefore missed in these analyses are tissue-resident memory T cells (TRMs). These cells were identified about a decade ago and are generated preferentially at the site of initial infection, reside solely in tissues, do not recirculate to peripheral blood, but can give rise to peripheral blood effector memory T cells [123], and are crucial mediators of protection from secondary viral exposure [7]. In the context of respiratory viral infections, TRMs provide rapid viral containment at secondary exposure by directly killing infected cells and releasing cytokines and chemokines to recruit other immune cells to the viral entry site in the upper and lower respiratory tract. Under certain circumstances, TRMs might even provide heterologous immune pressure to different coronaviruses. Studies in mouse models have demonstrated that resident memory T cell immunity can provide heterologous protection from severe disease, e.g. SARS-CoV and MERS [72,87,111]. Studies from our own group demonstrate the localization of SARS-CoV-2-cross-reactive CD4+ and/or CD8+ T cells with TRM phenotype in about 50 % of oropharyngeal lymphoid tissue samples collected from children and adults prior to the pandemic [120]. However, whether pre-existing TRMs can affect disease outcome during primary SARS-CoV-2 infection or following SARS-CoV-2 vaccination remains to be determined (Fig. 2).
SARS-CoV-2 infection induces a virus-specific T cell immunity that can be detected at least 8 months after infection in peripheral blood, but analyses on tissue samples are scarce. Recently, studies on lung and nasal tissue obtained from SARS-CoV-2 convalescent individuals have found TRM responses with functional reactivity against several SARS-CoV-2 proteins that persist for at least 2–10 months after infection [112,113]. In addition to lung and lung-associated lymph nodes, SARS-CoV-2-specific CD4+ and CD8+ T cells can also be found in other organs from convalescent individuals [122]. Virus-specific TRMs located in the upper respiratory tract are known to be long-lived [114], but further studies are required to determine the longevity of SARS-CoV-2-specific TRMs in the lower respiratory tract, given that studies in mice have demonstrate their gradual loss in the lung [115].
Similar to studies on natural SARS-CoV-2 infection, analyses of vaccine-induced tissue immunity have been rare due to limited access. A preliminary study by Ssemaganda et al. reports the expansion of TRMs in the nasal mucosa after mRNA vaccination [116]. However, while intramuscular injection – the standard route for current SARS-CoV-2 vaccine regimens – induces a systemic immune response, induction of TRM in the respiratory tract was inefficient in mice and did not fully abrogate lung SARS-CoV-2 replication [117]. This is consistent with other vaccine studies demonstrating inefficient TRM induction with conventional systemic routes [111,118]. In contrast, intranasal delivery of a ChAd-SARS-CoV-2-S vaccine induced both systemic and localized immune responses in the lung including vaccine-specific IgA and CD8+ TRM responses, which correlated with reduced viral replication and shedding [117]. Seven of the 100 SARS-CoV-2 vaccines currently in clinical trials are delivered intranasally [119]. It therefore remains to be seen whether this delivery route shows higher vaccine efficacy in clinical trials and whether they will induce long-term systemic and mucosal immunity.
7. Conclusions
SARS-CoV-2 will be nearly impossible to eradicate in any near future. As such, it has never been more important than now to understand how different immune cell components provide protective immune responses to SARS-CoV-2 infection and disease severity. Many studies emerging in the animal field show that T cells together with antibodies are essential to provide full protection. However, many unknowns remain, such as whether rapid induction of adaptive immune responses to SARS-CoV-2 and less severe COVID-19 manifestations are driven by pre-existing T cells. If a pre-existing response might tweak the general immune response, is that orchestrated by TRMs present within the upper respiratory tract? Furthermore, there is an increasing concern that current vaccine regimens do not protect as well from new VOCs, including the Delta variant. If breakthrough infections occur, will that then cause long COVID, and would it therefore be necessary to include other antigens (e.g. nucleocapsid) and new administration routes (e.g. nasal vaccination) to promote resident memory cells in future vaccines to hinder spread to distal sites? Thus, additional studies are needed to fully understand how T cells and antibodies together orchestrate long-lasting natural and vaccine-mediated immunity to SARS-CoV-2.
Declaration of Competing Interest
M.B. is a consultant for Oxford Immunotec. All other authors report no competing interest.
Acknowledgements
We would like to express gratitude to our funders. J.N. was supported by an EMBO Postdoctoral Fellowship (ALTF 1062-2020). T.S. was supported by the Swedish Research Council and the Swedish Childhood Cancer Fund. M.B. was supported by the Swedish Research Council, the Karolinska Institutet, the Swedish Society of Medicine, the Swedish Cancer Society, the Swedish Childhood Cancer Fund, the Åke Wibergs Stiftelse, and the Jonas Söderquist Stiftelse. Figures were conducted using Biorender.
References
- 1.Khoury D.S., Wheatley A.K., Ramuta M.D., Reynaldi A., Cromer D., Subbarao K., O’Connor D.H., Kent S.J., Davenport M.P. Measuring immunity to SARS-CoV-2 infection: comparing assays and animal models. Nat. Rev. Immunol. 2020;20:727–738. doi: 10.1038/s41577-020-00471-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., Planchais C., Porrot F., Robillard N., Puech J., Prot M., Gallais F., Gantner P., Velay A., Le Guen J., Kassis-Chikhani N., Edriss D., Belec L., Seve A., Courtellemont L., Péré H., Hocqueloux L., Fafi-Kremer S., Prazuck T., Mouquet H., Bruel T., Simon-Lorière E., Rey F.A., Schwartz O. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021 doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 3.Arstila T.P., Casrouge A., Baron V., Even J., Kanellopoulos J., Kourilsky P. A direct estimate of the human alphabeta T cell receptor diversity. Science. 1999;286:958–961. doi: 10.1126/science.286.5441.958. [DOI] [PubMed] [Google Scholar]
- 4.Vella L.A., Herati R.S., Wherry E.J. CD4+ T cell differentiation in chronic viral infections: the Tfh perspective. Trends Mol. Med. 2017;23:1072–1087. doi: 10.1016/j.molmed.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wherry E.J., Ahmed R. Memory CD8 T-cell differentiation during viral infection. J. Virol. 2004;78:5535–5545. doi: 10.1128/JVI.78.11.5535-5545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buggert M., Vella L.A., Nguyen S., Wu V.H., Chen Z., Sekine T., Perez-Potti A., Maldini C.R., Manne S., Darko S., Ransier A., Kuri-Cervantes L., Japp A.S., Brody I.B., Ivarsson M.A., Gorin J.-B., Rivera-Ballesteros O., Hertwig L., Antel J.P., Johnson M.E., Okoye A., Picker L., Vahedi G., Sparrelid E., Llewellyn-Lacey S., Gostick E., Sandberg J.K., Björkström N., Bar-Or A., Dori Y., Naji A., Canaday D.H., Laufer T.M., Wells A.D., Price D.A., Frank I., Douek D.C., Wherry E.J., Itkin M.G., Betts M.R. The identity of human tissue-emigrant CD8+ T cells. Cell. 2020;183:1946–1961. doi: 10.1016/j.cell.2020.11.019. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schenkel J.M., Masopust D. Tissue-resident memory T cells. Immunity. 2014;41:886–897. doi: 10.1016/j.immuni.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanco-Melo D., Nilsson-Payant B.E., Liu W.-C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., Wang T.T., Schwartz R.E., Lim J.K., Albrecht R.A., tenOever B.R. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045. doi: 10.1016/j.cell.2020.04.026. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lucas C., Wong P., Klein J., Castro T.B.R., Silva J., Sundaram M., Ellingson M.K., Mao T., Oh J.E., Israelow B., Takahashi T., Tokuyama M., Lu P., Venkataraman A., Park A., Mohanty S., Wang H., Wyllie A.L., Vogels C.B.F., Earnest R., Lapidus S., Ott I.M., Moore A.J., Muenker M.C., Fournier J.B., Campbell M., Odio C.D., Casanovas-Massana A., Herbst R., Shaw A.C., Medzhitov R., Schulz W.L., Grubaugh N.D., Cruz C.D., Farhadian S., Ko A.I., Omer S.B., Iwasaki A. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laing A.G., Lorenc A., del Molino del Barrio I., Das A., Fish M., Monin L., Muñoz-Ruiz M., McKenzie D.R., Hayday T.S., Francos-Quijorna I., Kamdar S., Joseph M., Davies D., Davis R., Jennings A., Zlatareva I., Vantourout P., Wu Y., Sofra V., Cano F., Greco M., Theodoridis E., Freedman J.D., Gee S., Chan J.N.E., Ryan S., Bugallo-Blanco E., Peterson P., Kisand K., Haljasmägi L., Chadli L., Moingeon P., Martinez L., Merrick B., Bisnauthsing K., Brooks K., Ibrahim M.A.A., Mason J., Lopez Gomez F., Babalola K., Abdul-Jawad S., Cason J., Mant C., Seow J., Graham C., Doores K.J., Di Rosa F., Edgeworth J., Shankar-Hari M., Hayday A.C. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat. Med. 2020;26:1623–1635. doi: 10.1038/s41591-020-1038-6. [DOI] [PubMed] [Google Scholar]
- 11.Chua R.L., Lukassen S., Trump S., Hennig B.P., Wendisch D., Pott F., Debnath O., Thürmann L., Kurth F., Völker M.T., Kazmierski J., Timmermann B., Twardziok S., Schneider S., Machleidt F., Müller-Redetzky H., Maier M., Krannich A., Schmidt S., Balzer F., Liebig J., Loske J., Suttorp N., Eils J., Ishaque N., Liebert U.G., von Kalle C., Hocke A., Witzenrath M., Goffinet C., Drosten C., Laudi S., Lehmann I., Conrad C., Sander L.-E., Eils R. COVID-19 severity correlates with airway epithelium–immune cell interactions identified by single-cell analysis. Nat. Biotechnol. 2020;38:970–979. doi: 10.1038/s41587-020-0602-4. [DOI] [PubMed] [Google Scholar]
- 12.Bastard P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.-H., Zhang Y., Dorgham K., Philippot Q., Rosain J., Béziat V., Manry J., Shaw E., Haljasmägi L., Peterson P., Lorenzo L., Bizien L., Trouillet-Assant S., Dobbs K., de Jesus A.A., Belot A., Kallaste A., Catherinot E., Tandjaoui-Lambiotte Y., Le Pen J., Kerner G., Bigio B., Seeleuthner Y., Yang R., Bolze A., Spaan A.N., Delmonte O.M., Abers M.S., Aiuti A., Casari G., Lampasona V., Piemonti L., Ciceri F., Bilguvar K., Lifton R.P., Vasse M., Smadja D.M., Migaud M., Hadjadj J., Terrier B., Duffy D., Quintana-Murci L., van de Beek D., Roussel L., Vinh D.C., Tangye S.G., Haerynck F., Dalmau D., Martinez-Picado J., Brodin P., Nussenzweig M.C., Boisson-Dupuis S., Rodríguez-Gallego C., Vogt G., Mogensen T.H., Oler A.J., Gu J., Burbelo P.D., Cohen J.I., Biondi A., Bettini L.R., D’Angio M., Bonfanti P., Rossignol P., Mayaux J., Rieux-Laucat F., Husebye E.S., Fusco F., Ursini M.V., Imberti L., Sottini A., Paghera S., Quiros-Roldan E., Rossi C., Castagnoli R., Montagna D., Licari A., Marseglia G.L., Duval X., Ghosn J., HGID Lab, NIAID-USUHS Immune Response to COVID Group, COVID Clinicians, COVID-STORM Clinicians, Imagine COVID Group, French COVID Cohort Study Group, Milieu Intérieur Consortium, CoV-Contact Cohort, Amsterdam UMC Covid-19 Biobank, COVID Human Genetic Effort, Tsang J.S., Goldbach-Mansky R., Kisand K., Lionakis M.S., Puel A., Zhang S.-Y., Holland S.M., Gorochov G., Jouanguy E., Rice C.M., Cobat A., Notarangelo L.D., Abel L., Su H.C., Casanova J.-L. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370:eabd4585. doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Q., Bastard P., Liu Z., Pen J.L., Moncada-Velez M., Chen J., Ogishi M., Sabli I.K.D., Hodeib S., Korol C., Rosain J., Bilguvar K., Ye J., Bolze A., Bigio B., Yang R., Arias A.A., Zhou Q., Zhang Y., Onodi F., Korniotis S., Karpf L., Philippot Q., Chbihi M., Bonnet-Madin L., Dorgham K., Smith N., Schneider W.M., Razooky B.S., Hoffmann H.-H., Michailidis E., Moens L., Han J.E., Lorenzo L., Bizien L., Meade P., Neehus A.-L., Ugurbil A.C., Corneau A., Kerner G., Zhang P., Rapaport F., Seeleuthner Y., Manry J., Masson C., Schmitt Y., Schlüter A., Voyer T.L., Khan T., Li J., Fellay J., Roussel L., Shahrooei M., Alosaimi M.F., Mansouri D., Al-Saud H., Al-Mulla F., Almourfi F., Al-Muhsen S.Z., Alsohime F., Turki S.A., Hasanato R., van de Beek D., Biondi A., Bettini L.R., D’Angio’ M., Bonfanti P., Imberti L., Sottini A., Paghera S., Quiros-Roldan E., Rossi C., Oler A.J., Tompkins M.F., Alba C., Vandernoot I., Goffard J.-C., Smits G., Migeotte I., Haerynck F., Soler-Palacin P., Martin-Nalda A., Colobran R., Morange P.-E., Keles S., Çölkesen F., Ozcelik T., Yasar K.K., Senoglu S., Karabela Ş.N., Rodríguez-Gallego C., Novelli G., Hraiech S., Tandjaoui-Lambiotte Y., Duval X., Laouénan C., Clinicians C.-S., Clinicians C., I.C. Group, F.C.C.S. Group, Cohort C.-C., A.U.C.-19 Biobank, Effort C.H.G., N.-U.C.I. Group, Snow A.L., Dalgard C.L., Milner J.D., Vinh D.C., Mogensen T.H., Marr N., Spaan A.N., Boisson B., Boisson-Dupuis S., Bustamante J., Puel A., Ciancanelli M.J., Meyts I., Maniatis T., Soumelis V., Amara A., Nussenzweig M., García-Sastre A., Krammer F., Pujol A., Duffy D., Lifton R.P., Zhang S.-Y., Gorochov G., Béziat V., Jouanguy E., Sancho-Shimizu V., Rice C.M., Abel L., Notarangelo L.D., Cobat A., Su H.C., Casanova J.-L. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370 doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Cheng L., Li J., Wang X., Wang F., Liu L., Amit I., Zhang S., Zhang Z. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 15.Mathew D., Giles J.R., Baxter A.E., Oldridge D.A., Greenplate A.R., Wu J.E., Alanio C., Kuri-Cervantes L., Pampena M.B., D’Andrea K., Manne S., Chen Z., Huang Y.J., Reilly J.P., Weisman A.R., Ittner C.A.G., Kuthuru O., Dougherty J., Nzingha K., Han N., Kim J., Pattekar A., Goodwin E.C., Anderson E.M., Weirick M.E., Gouma S., Arevalo C.P., Bolton M.J., Chen F., Lacey S.F., Ramage H., Cherry S., Hensley S.E., Apostolidis S.A., Huang A.C., Vella L.A., Unit T.Up.C.P., Betts M.R., Meyer N.J., Wherry E.J. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369 doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rydyznski Moderbacher C., Ramirez S.I., Dan J.M., Grifoni A., Hastie K.M., Weiskopf D., Belanger S., Abbott R.K., Kim C., Choi J., Kato Y., Crotty E.G., Kim C., Rawlings S.A., Mateus J., Tse L.P.V., Frazier A., Baric R., Peters B., Greenbaum J., Ollmann Saphire E., Smith D.M., Sette A., Crotty S. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183:996–1012. doi: 10.1016/j.cell.2020.09.038. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weiskopf D., Schmitz K.S., Raadsen M.P., Grifoni A., Okba N.M.A., Endeman H., van den Akker J.P.C., Molenkamp R., Koopmans M.P.G., van Gorp E.C.M., Haagmans B.L., de Swart R.L., Sette A., de Vries R.D. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.abd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen K.W., Linderman S.L., Moodie Z., Czartoski J., Lai L., Mantus G., Norwood C., Nyhoff L.E., Edara V.V., Floyd K., De Rosa S.C., Ahmed H., Whaley R., Patel S.N., Prigmore B., Lemos M.P., Davis C.W., Furth S., O’Keefe J.B., Gharpure M.P., Gunisetty S., Stephens K., Antia R., Zarnitsyna V.I., Stephens D.S., Edupuganti S., Rouphael N., Anderson E.J., Mehta A.K., Wrammert J., Suthar M.S., Ahmed R., McElrath M.J. Longitudinal analysis shows durable and broad immune memory after SARS-CoV-2 infection with persisting antibody responses and memory B and T cells. Cell Rep. Med. 2021;2 doi: 10.1016/j.xcrm.2021.100354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jung J.H., Rha M.-S., Sa M., Choi H.K., Jeon J.H., Seok H., Park D.W., Park S.-H., Jeong H.W., Choi W.S., Shin E.-C. SARS-CoV-2-specific T cell memory is sustained in COVID-19 convalescent patients for 10 months with successful development of stem cell-like memory T cells. Nat. Commun. 2021;12:4043. doi: 10.1038/s41467-021-24377-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan A.T., Linster M., Tan C.W., Le Bert N., Chia W.N., Kunasegaran K., Zhuang Y., Tham C.Y.L., Chia A., Smith G.J.D., Young B., Kalimuddin S., Low J.G.H., Lye D., Wang L.-F., Bertoletti A. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep. 2021;34 doi: 10.1016/j.celrep.2021.108728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buggert M., Nguyen S., de Oca G.S.-M., Bengsch B., Darko S., Ransier A., Roberts E.R., del Alcazar D., Brody I.B., Vella L.A., Beura L., Wijeyesinghe S., Herati R.S., Estrada P.M.D.R., Ablanedo-Terrazas Y., Kuri-Cervantes L., Japp A.S., Manne S., Vartanian S., Huffman A., Sandberg J.K., Gostick E., Nadolski G., Silvestri G., Canaday D.H., Price D.A., Petrovas C., Su L.F., Vahedi G., Dori Y., Frank I., Itkin M.G., Wherry E.J., Deeks S.G., Naji A., Reyes-Terán G., Masopust D., Douek D.C., Betts M.R. Identification and characterization of HIV-specific resident memory CD8+ T cells in human lymphoid tissue. Sci. Immunol. 2018;3 doi: 10.1126/sciimmunol.aar4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller J.D., van der Most R.G., Akondy R.S., Glidewell J.T., Albott S., Masopust D., Murali-Krishna K., Mahar P.L., Edupuganti S., Lalor S., Germon S., Del Rio C., Mulligan M.J., Staprans S.I., Altman J.D., Feinberg M.B., Ahmed R. Human effector and memory CD8+ T cell responses to smallpox and yellow fever vaccines. Immunity. 2008;28:710–722. doi: 10.1016/j.immuni.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 23.Sekine T., Perez-Potti A., Rivera-Ballesteros O., Strålin K., Gorin J.-B., Olsson A., Llewellyn-Lacey S., Kamal H., Bogdanovic G., Muschiol S., Wullimann D.J., Kammann T., Emgård J., Parrot T., Folkesson E., Rooyackers O., Eriksson L.I., Henter J.-I., Sönnerborg A., Allander T., Albert J., Nielsen M., Klingström J., Gredmark-Russ S., Björkström N.K., Sandberg J.K., Price D.A., Ljunggren H.-G., Aleman S., Buggert M. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183:158–168. doi: 10.1016/j.cell.2020.08.017. e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saini S.K., Hersby D.S., Tamhane T., Povlsen H.R., Hernandez S.P.A., Nielsen M., Gang A.O., Hadrup S.R. SARS-CoV-2 genome-wide T cell epitope mapping reveals immunodominance and substantial CD8+ T cell activation in COVID-19 patients. Sci. Immunol. 2021;6 doi: 10.1126/sciimmunol.abf7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng H.-Y., Zhang M., Yang C.-X., Zhang N., Wang X.-C., Yang X.-P., Dong X.-Q., Zheng Y.-T. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell. Mol. Immunol. 2020;17:541–543. doi: 10.1038/s41423-020-0401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song J.-W., Zhang C., Fan X., Meng F.-P., Xu Z., Xia P., Cao W.-J., Yang T., Dai X.-P., Wang S.-Y., Xu R.-N., Jiang T.-J., Li W.-G., Zhang D.-W., Zhao P., Shi M., Agrati C., Ippolito G., Maeurer M., Zumla A., Wang F.-S., Zhang J.-Y. Immunological and inflammatory profiles in mild and severe cases of COVID-19. Nat. Commun. 2020;11:3410. doi: 10.1038/s41467-020-17240-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rha M.-S., Jeong H.W., Ko J.-H., Choi S.J., Seo I.-H., Lee J.S., Sa M., Kim A.R., Joo E.-J., Ahn J.Y., Kim J.H., Song K.-H., Kim E.S., Oh D.H., Ahn M.Y., Choi H.K., Jeon J.H., Choi J.-P., Kim H.B., Kim Y.K., Park S.-H., Choi W.S., Choi J.Y., Peck K.R., Shin E.-C. PD-1-expressing SARS-CoV-2-specific CD8+ t cells are not exhausted, but functional in patients with COVID-19. Immunity. 2021;54:44–52. doi: 10.1016/j.immuni.2020.12.002. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou R., To K.K.-W., Wong Y.-C., Liu L., Zhou B., Li X., Huang H., Mo Y., Luk T.-Y., Lau T.T.-K., Yeung P., Chan W.-M., Wu A.K.-L., Lung K.-C., Tsang O.T.-Y., Leung W.-S., Hung I.F.-N., Yuen K.-Y., Chen Z. Acute SARS-CoV-2 infection impairs dendritic cell and T cell responses. Immunity. 2020;53:864–877. doi: 10.1016/j.immuni.2020.07.026. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cohen K.W., Linderman S.L., Moodie Z., Czartoski J., Lai L., Mantus G., Norwood C., Nyhoff L.E., Edara V.V., Floyd K., De Rosa S.C., Ahmed H., Whaley R., Patel S.N., Prigmore B., Lemos M.P., Davis C.W., Furth S., O’Keefe J.B., Gharpure M.P., Gunisetty S., Stephens K., Antia R., Zarnitsyna V.I., Stephens D.S., Edupuganti S., Rouphael N., Anderson E.J., Mehta A.K., Wrammert J., Suthar M.S., Ahmed R., McElrath M.J. Longitudinal analysis shows durable and broad immune memory after SARS-CoV-2 infection with persisting antibody responses and memory B and T cells. Cell Rep. Med. 2021;2:100354. doi: 10.1016/j.xcrm.2021.100354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chandran A., Rosenheim J., Nageswaran G., Swadling L., Pollara G., Gupta R.K., Guerra-Assuncao J.A., Woolston A., Ronel T., Pade C., Gibbons J., Estrada B.S.-M.D.D., de Massy M.R., Whelan M., Semper A., Brooks T., Altmann D.M., Boyton R.J., McKnight Á., Manisty C., Treibel T.A., Moon J., Tomlinson G.S., Maini M.K., Chain B.M., Noursadeghi M., Covid. Investigators Non-severe SARS-CoV-2 infection is characterised by very early T cell proliferation independent of type 1 interferon responses and distinct from other acute respiratory viruses. MedRxiv. 2021 doi: 10.1101/2021.03.30.21254540. 2021.03.30.21254540. [DOI] [Google Scholar]
- 31.Riou C., du Bruyn E., Stek C., Daroowala R., Goliath R.T., Abrahams F., Said-Hartley Q., Allwood B.W., Hsiao N.-Y., Wilkinson K.A., Arlehamn C.S.L., Sette A., Wasserman S., Wilkinson R.J., HIATUS consortium Relationship of SARS-CoV-2-specific CD4 response to COVID-19 severity and impact of HIV-1 and tuberculosis coinfection. J. Clin. Invest. 2021;131:149125. doi: 10.1172/JCI149125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Juno J.A., Tan H.-X., Lee W.S., Reynaldi A., Kelly H.G., Wragg K., Esterbauer R., Kent H.E., Batten C.J., Mordant F.L., Gherardin N.A., Pymm P., Dietrich M.H., Scott N.E., Tham W.-H., Godfrey D.I., Subbarao K., Davenport M.P., Kent S.J., Wheatley A.K. Humoral and circulating follicular helper T cell responses in recovered patients with COVID-19. Nat. Med. 2020;26:1428–1434. doi: 10.1038/s41591-020-0995-0. [DOI] [PubMed] [Google Scholar]
- 33.Shahbaz S., Xu L., Sligl W., Osman M., Bozorgmehr N., Mashhouri S., Redmond D., Perez Rosero E., Walker J., Elahi S. The quality of SARS-CoV-2-specific T cell functions differs in patients with mild/moderate versus severe disease, and T cells expressing coinhibitory receptors are highly activated. J. Immunol. 2021;207:1099–1111. doi: 10.4049/jimmunol.2100446. [DOI] [PubMed] [Google Scholar]
- 34.Sun J., Zhuang Z., Zheng J., Li K., Wong R.L.-Y., Liu D., Huang J., He J., Zhu A., Zhao J., Li X., Xi Y., Chen R., Alshukairi A.N., Chen Z., Zhang Z., Chen C., Huang X., Li F., Lai X., Chen D., Wen L., Zhuo J., Zhang Y., Wang Y., Huang S., Dai J., Shi Y., Zheng K., Leidinger M.R., Chen J., Li Y., Zhong N., Meyerholz D.K., McCray P.B., Perlman S., Zhao J. Generation of a broadly useful model for COVID-19 pathogenesis, vaccination, and treatment. Cell. 2020;182:734–743.e5. doi: 10.1016/j.cell.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Israelow B., Mao T., Klein J., Song E., Menasche B., Omer S.B., Iwasaki A. Adaptive immune determinants of viral clearance and protection in mouse models of SARS-CoV-2. Sci. Immunol. 2021;6 doi: 10.1126/sciimmunol.abl4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McMahan K., Yu J., Mercado N.B., Loos C., Tostanoski L.H., Chandrashekar A., Liu J., Peter L., Atyeo C., Zhu A., Bondzie E.A., Dagotto G., Gebre M.S., Jacob-Dolan C., Li Z., Nampanya F., Patel S., Pessaint L., Van Ry A., Blade K., Yalley-Ogunro J., Cabus M., Brown R., Cook A., Teow E., Andersen H., Lewis M.G., Lauffenburger D.A., Alter G., Barouch D.H. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature. 2021;590:630–634. doi: 10.1038/s41586-020-03041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Corbett K.S., Nason M.C., Flach B., Gagne M., O’Connell S., Johnston T.S., Shah S.N., Edara V.V., Floyd K., Lai L., McDanal C., Francica J.R., Flynn B., Wu K., Choi A., Koch M., Abiona O.M., Werner A.P., Moliva J.I., Andrew S.F., Donaldson M.M., Fintzi J., Flebbe D.R., Lamb E., Noe A.T., Nurmukhambetova S.T., Provost S.J., Cook A., Dodson A., Faudree A., Greenhouse J., Kar S., Pessaint L., Porto M., Steingrebe K., Valentin D., Zouantcha S., Bock K.W., Minai M., Nagata B.M., van de Wetering R., Boyoglu-Barnum S., Leung K., Shi W., Yang E.S., Zhang Y., Todd J.-P.M., Wang L., Alvarado G.S., Andersen H., Foulds K.E., Edwards D.K., Mascola J.R., Moore I.N., Lewis M.G., Carfi A., Montefiori D., Suthar M.S., McDermott A., Roederer M., Sullivan N.J., Douek D.C., Graham B.S., Seder R.A. Immune correlates of protection by mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. Science. 2021 doi: 10.1126/science.abj0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hasenkrug K.J., Feldmann F., Myers L., Santiago M.L., Guo K., Barrett B.S., Mickens K.L., Carmody A., Okumura A., Rao D., Collins M.M., Messer R.J., Lovaglio J., Shaia C., Rosenke R., van Doremalen N., Clancy C., Saturday G., Hanley P., Smith B.J., Meade-White K., Shupert W.L., Hawman D.W., Feldmann H. Recovery from acute SARS-CoV-2 infection and development of anamnestic immune responses in T cell-depleted Rhesus macaques. MBio. 2021 doi: 10.1128/mBio.01503-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dan J.M., Mateus J., Kato Y., Hastie K.M., Yu E.D., Faliti C.E., Grifoni A., Ramirez S.I., Haupt S., Frazier A., Nakao C., Rayaprolu V., Rawlings S.A., Peters B., Krammer F., Simon V., Saphire E.O., Smith D.M., Weiskopf D., Sette A., Crotty S. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371 doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., Marrama D., de Silva A.M., Frazier A., Carlin A.F., Greenbaum J.A., Peters B., Krammer F., Smith D.M., Crotty S., Sette A. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501. doi: 10.1016/j.cell.2020.05.015. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Galletti G., De Simone G., Mazza E.M.C., Puccio S., Mezzanotte C., Bi T.M., Davydov A.N., Metsger M., Scamardella E., Alvisi G., De Paoli F., Zanon V., Scarpa A., Camisa B., Colombo F.S., Anselmo A., Peano C., Polletti S., Mavilio D., Gattinoni L., Boi S.K., Youngblood B.A., Jones R.E., Baird D.M., Gostick E., Llewellyn-Lacey S., Ladell K., Price D.A., Chudakov D.M., Newell E.W., Casucci M., Lugli E. Two subsets of stem-like CD8+ memory T cell progenitors with distinct fate commitments in humans. Nat. Immunol. 2020;21:1552–1562. doi: 10.1038/s41590-020-0791-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peng Y., Mentzer A.J., Liu G., Yao X., Yin Z., Dong D., Dejnirattisai W., Rostron T., Supasa P., Liu C., López-Camacho C., Slon-Campos J., Zhao Y., Stuart D.I., Paesen G.C., Grimes J.M., Antson A.A., Bayfield O.W., Hawkins D.E.D.P., Ker D.-S., Wang B., Turtle L., Subramaniam K., Thomson P., Zhang P., Dold C., Ratcliff J., Simmonds P., de Silva T., Sopp P., Wellington D., Rajapaksa U., Chen Y.-L., Salio M., Napolitani G., Paes W., Borrow P., Kessler B.M., Fry J.W., Schwabe N.F., Semple M.G., Baillie J.K., Moore S.C., Openshaw P.J.M., Ansari M.A., Dunachie S., Barnes E., Frater J., Kerr G., Goulder P., Lockett T., Levin R., Zhang Y., Jing R., Ho L.-P., Cornall R.J., Conlon C.P., Klenerman P., Screaton G.R., Mongkolsapaya J., McMichael A., Knight J.C., Ogg G., Dong T. Broad and strong memory CD4 + and CD8 + T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 2020;21:1336–1345. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Braun J., Loyal L., Frentsch M., Wendisch D., Georg P., Kurth F., Hippenstiel S., Dingeldey M., Kruse B., Fauchere F., Baysal E., Mangold M., Henze L., Lauster R., Mall M.A., Beyer K., Röhmel J., Voigt S., Schmitz J., Miltenyi S., Demuth I., Müller M.A., Hocke A., Witzenrath M., Suttorp N., Kern F., Reimer U., Wenschuh H., Drosten C., Corman V.M., Giesecke-Thiel C., Sander L.E., Thiel A. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587:270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- 44.Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., Chng M.H.Y., Lin M., Tan N., Linster M., Chia W.N., Chen M.I.-C., Wang L.-F., Ooi E.E., Kalimuddin S., Tambyah P.A., Low J.G.-H., Tan Y.-J., Bertoletti A. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 45.Zuo J., Dowell A.C., Pearce H., Verma K., Long H.M., Begum J., Aiano F., Amin-Chowdhury Z., Hoschler K., Brooks T., Taylor S., Hewson J., Hallis B., Stapley L., Borrow R., Linley E., Ahmad S., Parker B., Horsley A., Amirthalingam G., Brown K., Ramsay M.E., Ladhani S., Moss P. Robust SARS-CoV-2-specific T cell immunity is maintained at 6 months following primary infection. Nat. Immunol. 2021;22:620–626. doi: 10.1038/s41590-021-00902-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reynolds C.J., Swadling L., Gibbons J.M., Pade C., Jensen M.P., Diniz M.O., Schmidt N.M., Butler D.K., Amin O.E., Bailey S.N.L., Murray S.M., Pieper F.P., Taylor S., Jones J., Jones M., Lee W.-Y.J., Rosenheim J., Chandran A., Joy G., Di Genova C., Temperton N., Lambourne J., Cutino-Moguel T., Andiapen M., Fontana M., Smit A., Semper A., O’Brien B., Chain B., Brooks T., Manisty C., Treibel T., Moon J.C., COVIDsortium investigators, Noursadeghi M., COVIDsortium immune correlates network, Altmann D.M., Maini M.K., McKnight Á., Boyton R.J. Discordant neutralizing antibody and T cell responses in asymptomatic and mild SARS-CoV-2 infection. Sci. Immunol. 2020;5:eabf3698. doi: 10.1126/sciimmunol.abf3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Long Q.-X., Tang X.-J., Shi Q.-L., Li Q., Deng H.-J., Yuan J., Hu J.-L., Xu W., Zhang Y., Lv F.-J., Su K., Zhang F., Gong J., Wu B., Liu X.-M., Li J.-J., Qiu J.-F., Chen J., Huang A.-L. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020;26:1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- 48.Le Bert N., Clapham H.E., Tan A.T., Chia W.N., Tham C.Y.L., Lim J.M., Kunasegaran K., Tan L.W.L., Dutertre C.-A., Shankar N., Lim J.M.E., Sun L.J., Zahari M., Tun Z.M., Kumar V., Lim B.L., Lim S.H., Chia A., Tan Y.-J., Tambyah P.A., Kalimuddin S., Lye D., Low J.G.H., Wang L.-F., Wan W.Y., Hsu L.Y., Bertoletti A., Tam C.C. Highly functional virus-specific cellular immune response in asymptomatic SARS-CoV-2 infection. J. Exp. Med. 2021;218 doi: 10.1084/jem.20202617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Z., Yang X., Zhong J., Zhou Y., Tang Z., Zhou H., He J., Mei X., Tang Y., Lin B., Chen Z., McCluskey J., Yang J., Corbett A.J., Ran P. Exposure to SARS-CoV-2 generates T-cell memory in the absence of a detectable viral infection. Nat. Commun. 2021;12:1724. doi: 10.1038/s41467-021-22036-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ogbe A., Kronsteiner B., Skelly D.T., Pace M., Brown A., Adland E., Adair K., Akhter H.D., Ali M., Ali S.-E., Angyal A., Ansari M.A., Arancibia-Cárcamo C.V., Brown H., Chinnakannan S., Conlon C., de Lara C., de Silva T., Dold C., Dong T., Donnison T., Eyre D., Flaxman A., Fletcher H., Gardner J., Grist J.T., Hackstein C.-P., Jaruthamsophon K., Jeffery K., Lambe T., Lee L., Li W., Lim N., Matthews P.C., Mentzer A.J., Moore S.C., Naisbitt D.J., Ogese M., Ogg G., Openshaw P., Pirmohamed M., Pollard A.J., Ramamurthy N., Rongkard P., Rowland-Jones S., Sampson O., Screaton G., Sette A., Stafford L., Thompson C., Thomson P.J., Thwaites R., Vieira V., Weiskopf D., Zacharopoulou P., Oxford Immunology Network Covid-19 Response T Cell Consortium, Oxford Protective T Cell Immunology for COVID-19 (OPTIC) Clinical Team, Turtle L., Klenerman P., Goulder P., Frater J., Barnes E., Dunachie S. T cell assays differentiate clinical and subclinical SARS-CoV-2 infections from cross-reactive antiviral responses. Nat. Commun. 2021;12:2055. doi: 10.1038/s41467-021-21856-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rowland-Jones S., Sutton J., Ariyoshi K., Dong T., Gotch F., McAdam S., Whitby D., Sabally S., Gallimore A., Corrah T. HIV-specific cytotoxic T-cells in HIV-exposed but uninfected Gambian women. Nat. Med. 1995;1:59–64. doi: 10.1038/nm0195-59. [DOI] [PubMed] [Google Scholar]
- 52.Munusamy Ponnan S., Thiruvengadam K., Kathirvel S., Shankar J., Rajaraman A., Mathaiyan M., Dinesha T.R., Poongulali S., Saravanan S., Murugavel K.G., Swaminathan S., Tripathy S.P., Neogi U., Velu V., Hanna L.E. Elevated numbers of HIV-specific poly-functional CD8+ T cells with stem cell-like and follicular homing phenotypes in HIV-exposed seronegative individuals. Front. Immunol. 2021;12:638144. doi: 10.3389/fimmu.2021.638144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hansen C.H., Michlmayr D., Gubbels S.M., Mølbak K., Ethelberg S. Assessment of protection against reinfection with SARS-CoV-2 among 4 million PCR-tested individuals in Denmark in 2020: a population-level observational study. Lancet. 2021;397:1204–1212. doi: 10.1016/S0140-6736(21)00575-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carvalho T., Krammer F., Iwasaki A. The first 12 months of COVID-19: a timeline of immunological insights. Nat. Rev. Immunol. 2021;21:245–256. doi: 10.1038/s41577-021-00522-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vaisman-Mentesh A., Dror Y., Tur-Kaspa R., Markovitch D., Kournos T., Dicker D., Wine Y. MedRxiv; 2020. SARS-CoV-2 Specific Memory B Cells Frequency in Recovered Patient Remains Stable While Antibodies Decay Over Time. [Google Scholar]
- 56.Tarke A., Sidney J., Methot N., Yu E.D., Zhang Y., Dan J.M., Goodwin B., Rubiro P., Sutherland A., Wang E., Frazier A., Ramirez S.I., Rawlings S.A., Smith D.M., da Silva Antunes R., Peters B., Scheuermann R.H., Weiskopf D., Crotty S., Grifoni A., Sette A. Impact of SARS-CoV-2 variants on the total CD4+ and CD8+ T cell reactivity in infected or vaccinated individuals. Cell Rep Med. 2021;2:100355. doi: 10.1016/j.xcrm.2021.100355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ng O.-W., Chia A., Tan A.T., Jadi R.S., Leong H.N., Bertoletti A., Tan Y.-J. Memory T cell responses targeting the SARS coronavirus persist up to 11 years post-infection. Vaccine. 2016;34:2008–2014. doi: 10.1016/j.vaccine.2016.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sewell A.K. Why must T cells be cross-reactive? Nat. Rev. Immunol. 2012;12:669–677. doi: 10.1038/nri3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nilges K., Höhn H., Pilch H., Neukirch C., Freitag K., Talbot P.J., Maeurer M.J. Human papillomavirus type 16 E7 peptide-directed CD8+ T cells from patients with cervical cancer are cross-reactive with the coronavirus NS2 protein. J. Virol. 2003;77:5464–5474. doi: 10.1128/JVI.77.9.5464-5474.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Clute S.C., Watkin L.B., Cornberg M., Naumov Y.N., Sullivan J.L., Luzuriaga K., Welsh R.M., Selin L.K. Cross-reactive influenza virus-specific CD8+ T cells contribute to lymphoproliferation in Epstein-Barr virus-associated infectious mononucleosis. J. Clin. Invest. 2005;115:3602–3612. doi: 10.1172/JCI25078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Acierno P.M., Newton D.A., Brown E.A., Maes L.A., Baatz J.E., Gattoni-Celli S. Cross-reactivity between HLA-A2-restricted FLU-M1:58–66 and HIV p17 GAG:77–85 epitopes in HIV-infected and uninfected individuals. J. Transl. Med. 2003;1:3. doi: 10.1186/1479-5876-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bacher P., Rosati E., Esser D., Martini G.R., Saggau C., Schiminsky E., Dargvainiene J., Schröder I., Wieters I., Khodamoradi Y., Eberhardt F., Vehreschild M.J.G.T., Neb H., Sonntagbauer M., Conrad C., Tran F., Rosenstiel P., Markewitz R., Wandinger K.-P., Augustin M., Rybniker J., Kochanek M., Leypoldt F., Cornely O.A., Koehler P., Franke A., Scheffold A. Low-avidity CD4+ T cell responses to SARS-CoV-2 in unexposed individuals and humans with severe COVID-19. Immunity. 2020;53:1258–1271. doi: 10.1016/j.immuni.2020.11.016. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nelde A., Bilich T., Heitmann J.S., Maringer Y., Salih H.R., Roerden M., Lübke M., Bauer J., Rieth J., Wacker M., Peter A., Hörber S., Traenkle B., Kaiser P.D., Rothbauer U., Becker M., Junker D., Krause G., Strengert M., Schneiderhan-Marra N., Templin M.F., Joos T.O., Kowalewski D.J., Stos-Zweifel V., Fehr M., Rabsteyn A., Mirakaj V., Karbach J., Jäger E., Graf M., Gruber L.-C., Rachfalski D., Preuß B., Hagelstein I., Märklin M., Bakchoul T., Gouttefangeas C., Kohlbacher O., Klein R., Stevanović S., Rammensee H.-G., Walz J.S. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat. Immunol. 2021;22:74–85. doi: 10.1038/s41590-020-00808-x. [DOI] [PubMed] [Google Scholar]
- 64.Mateus J., Grifoni A., Tarke A., Sidney J., Ramirez S.I., Dan J.M., Burger Z.C., Rawlings S.A., Smith D.M., Phillips E., Mallal S., Lammers M., Rubiro P., Quiambao L., Sutherland A., Yu E.D., da R., Antunes S., Greenbaum J., Frazier A., Markmann A.J., Premkumar L., de Silva A., Peters B., Crotty S., Sette A., Weiskopf D. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370:89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Meckiff B.J., Ramírez-Suástegui C., Fajardo V., Chee S.J., Kusnadi A., Simon H., Eschweiler S., Grifoni A., Pelosi E., Weiskopf D., Sette A., Ay F., Seumois G., Ottensmeier C.H., Vijayanand P. Imbalance of regulatory and cytotoxic SARS-CoV-2-reactive CD4+ T cells in COVID-19. Cell. 2020;183:1340–1353. doi: 10.1016/j.cell.2020.10.001. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Low J.S., Vaqueirinho D., Mele F., Foglierini M., Jerak J., Perotti M., Jarrossay D., Jovic S., Perez L., Cacciatore R., Terrot T., Pellanda A.F., Biggiogero M., Garzoni C., Ferrari P., Ceschi A., Lanzavecchia A., Sallusto F., Cassotta A. Clonal analysis of immunodominance and cross-reactivity of the CD4 T cell response to SARS-CoV-2. Science. 2021;372:1336–1341. doi: 10.1126/science.abg8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Loyal L., Braun J., Henze L., Kruse B., Dingeldey M., Reimer U., Kern F., Schwarz T., Mangold M., Unger C., Dörfler F., Kadler S., Rosowski J., Gürcan K., Uyar-Aydin Z., Frentsch M., Kurth F., Schnatbaum K., Eckey M., Hippenstiel S., Hocke A., Müller M.A., Sawitzki B., Miltenyi S., Paul F., Mall M.A., Wenschuh H., Voigt S., Drosten C., Lauster R., Lachman N., Sander L.-E., Corman V.M., Röhmel J., Meyer-Arndt L., Thiel A., Giesecke-Thiel C., the Charité Corona Cross Study Group Cross-reactive CD4 + T cells enhance SARS-CoV-2 immune responses upon infection and vaccination. Allergy Immunol. 2021 doi: 10.1101/2021.04.01.21252379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Swadling L., Diniz M.O., Schmidt N.M., Amin O.E., Chandran A., Shaw E., Pade C., Gibbons J.M., Le Bert N., Tan A.T., Jeffery-Smith A., Tan C., Tham C.Y.L., Kucyowicz S., Aidoo-Micah G., Rosenheim J., Davies J., Jensen M.P., Joy G., McCoy L.E., Valdes A.M., van Dorp L., Altmann D.M., Boyton R.J., Manisty C., Treibel T.A., Moon J.C., COVIDsortium investigators, Balloux F., McKnight A., Noursadeghi M., Bertoletti A., Maini M.K. MedRxiv; 2021. Pre-existing Polymerase-specific T Cells Expand in Abortive Seronegative SARS-CoV-2 Infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dykema A.G., Zhang B., Woldemeskel B.A., Garliss C.C., Cheung L.S., Choudhury D., Zhang J., Aparicio L., Bom S., Rashid R., Caushi J.X., Hsiue E.H.-C., Cascino K., Thompson E.A., Kwaa A.K., Singh D., Thapa S., Ordonez A.A., Pekosz A., D’Alessio F.R., Powell J.D., Yegnasubramanian S., Zhou S., Pardoll D.M., Ji H., Cox A.L., Blankson J.N., Smith K.N. Functional characterization of CD4+ T cell receptors crossreactive for SARS-CoV-2 and endemic coronaviruses. J. Clin. Invest. 2021;131 doi: 10.1172/JCI146922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schulien I., Kemming J., Oberhardt V., Wild K., Seidel L.M., Killmer S., Sagar, Daul F., Salvat Lago M., Decker A., Luxenburger H., Binder B., Bettinger D., Sogukpinar O., Rieg S., Panning M., Huzly D., Schwemmle M., Kochs G., Waller C.F., Nieters A., Duerschmied D., Emmerich F., Mei H.E., Schulz A.R., Llewellyn-Lacey S., Price D.A., Boettler T., Bengsch B., Thimme R., Hofmann M., Neumann-Haefelin C. Characterization of pre-existing and induced SARS-CoV-2-specific CD8 + T cells. Nat. Med. 2021;27:78–85. doi: 10.1038/s41591-020-01143-2. [DOI] [PubMed] [Google Scholar]
- 71.Lipsitch M., Grad Y.H., Sette A., Crotty S. Cross-reactive memory T cells and herd immunity to SARS-CoV-2. Nat. Rev. Immunol. 2020;20:709–713. doi: 10.1038/s41577-020-00460-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao J., Zhao J., Mangalam A.K., Channappanavar R., Fett C., Meyerholz D.K., Agnihothram S., Baric R.S., David C.S., Perlman S. Airway memory CD4+ T cells mediate protective immunity against emerging respiratory coronaviruses. Immunity. 2016;44:1379–1391. doi: 10.1016/j.immuni.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Koutsakos M., Illing P.T., Nguyen T.H.O., Mifsud N.A., Crawford J.C., Rizzetto S., Eltahla A.A., Clemens E.B., Sant S., Chua B.Y., Wong C.Y., Allen E.K., Teng D., Dash P., Boyd D.F., Grzelak L., Zeng W., Hurt A.C., Barr I., Rockman S., Jackson D.C., Kotsimbos T.C., Cheng A.C., Richards M., Westall G.P., Loudovaris T., Mannering S.I., Elliott M., Tangye S.G., Wakim L.M., Rossjohn J., Vijaykrishna D., Luciani F., Thomas P.G., Gras S., Purcell A.W., Kedzierska K. Human CD8 + T cell cross-reactivity across influenza A, B and C viruses. Nat. Immunol. 2019;20:613–625. doi: 10.1038/s41590-019-0320-6. [DOI] [PubMed] [Google Scholar]
- 74.Chen H.D., Fraire A.E., Joris I., Brehm M.A., Welsh R.M., Selin L.K. Memory CD8+ T cells in heterologous antiviral immunity and immunopathology in the lung. Nat. Immunol. 2001;2:1067–1076. doi: 10.1038/ni727. [DOI] [PubMed] [Google Scholar]
- 75.Klenerman P., Zinkernagel R.M. Original antigenic sin impairs cytotoxic T lymphocyte responses to viruses bearing variant epitopes. Nature. 1998;394:482–485. doi: 10.1038/28860. [DOI] [PubMed] [Google Scholar]
- 76.Sagar M., Reifler K., Rossi M., Miller N.S., Sinha P., White L.F., Mizgerd J.P. Recent endemic coronavirus infection is associated with less-severe COVID-19. J. Clin. Invest. 2021;131 doi: 10.1172/JCI143380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mateus J., Dan J.M., Zhang Z., Moderbacher C.R., Lammers M., Goodwin B., Sette A., Crotty S., Weiskopf D. Low dose mRNA-1273 COVID-19 vaccine generates durable T cell memory and antibodies enhanced by pre-existing crossreactive T cell memory. Science. 2021;374(6566) doi: 10.1126/science.abj9853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.2021. COVID-19 Vaccine Tracker and Landscape. (n.d.). https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines (accessed August 13, 2021) [Google Scholar]
- 79.Voysey M., Clemens S.A.C., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K., Angus B., Baillie V.L., Barnabas S.L., Bhorat Q.E., Bibi S., Briner C., Cicconi P., Collins A.M., Colin-Jones R., Cutland C.L., Darton T.C., Dheda K., Duncan C.J.A., Emary K.R.W., Ewer K.J., Fairlie L., Faust S.N., Feng S., Ferreira D.M., Finn A., Goodman A.L., Green C.M., Green C.A., Heath P.T., Hill C., Hill H., Hirsch I., Hodgson S.H.C., Izu A., Jackson S., Jenkin D., Joe C.C.D., Kerridge S., Koen A., Kwatra G., Lazarus R., Lawrie A.M., Lelliott A., Libri V., Lillie P.J., Mallory R., Mendes A.V.A., Milan E.P., Minassian A.M., McGregor A., Morrison H., Mujadidi Y.F., Nana A., O’Reilly P.J., Padayachee S.D., Pittella A., Plested E., Pollock K.M., Ramasamy M.N., Rhead S., Schwarzbold A.V., Singh N., Smith A., Song R., Snape M.D., Sprinz E., Sutherland R.K., Tarrant R., Thomson E.C., Török M.E., Toshner M., Turner D.P.J., Vekemans J., Villafana T.L., Watson M.E.E., Williams C.J., Douglas A.D., Hill A.V.S., Lambe T., Gilbert S.C., Pollard A.J., Aban M., Abayomi F., Abeyskera K., Aboagye J., Adam M., Adams K., Adamson J., Adelaja Y.A., Adewetan G., Adlou S., Ahmed K., Akhalwaya Y., Akhalwaya S., Alcock A., Ali A., Allen E.R., Allen L., Almeida T.C.D.S.C., Alves M.P.S., Amorim F., Andritsou F., Anslow R., Appleby M., Arbe-Barnes E.H., Ariaans M.P., Arns B., Arruda L., Azi P., Azi L., Babbage G., Bailey C., Baker K.F., Baker M., Baker N., Baker P., Baldwin L., Baleanu I., Bandeira D., Bara A., Barbosa M.A.S., Barker D., Barlow G.D., Barnes E., Barr A.S., Barrett J.R., Barrett J., Bates L., Batten A., Beadon K., Beales E., Beckley R., Belij-Rammerstorfer S., Bell J., Bellamy D., Bellei N., Belton S., Berg A., Bermejo L., Berrie E., Berry L., Berzenyi D., Beveridge A., Bewley K.R., Bexhell H., Bhikha S., Bhorat A.E., Bhorat Z.E., Bijker E., Birch G., Birch S., Bird A., Bird O., Bisnauthsing K., Bittaye M., Blackstone K., Blackwell L., Bletchly H., Blundell C.L., Blundell S.R., Bodalia P., Boettger B.C., Bolam E., Boland E., Bormans D., Borthwick N., Bowring F., Boyd A., Bradley P., Brenner T., Brown P., Brown C., Brown-O’Sullivan C., Bruce S., Brunt E., Buchan R., Budd W., Bulbulia Y.A., Bull M., Burbage J., Burhan H., Burn A., Buttigieg K.R., Byard N., Puig I.C., Calderon G., Calvert A., Camara S., Cao M., Cappuccini F., Cardoso J.R., Carr M., Carroll M.W., Carson-Stevens A., Carvalho Yde M., Carvalho J.A.M., Casey H.R., Cashen P., Castro T., Castro L.C., Cathie K., Cavey A., Cerbino-Neto J., Chadwick J., Chapman D., Charlton S., Chelysheva I., Chester O., Chita S., Cho J.-S., Cifuentes L., Clark E., Clark M., Clarke A., Clutterbuck E.A., Collins S.L.K., Conlon C.P., Connarty S., Coombes N., Cooper C., Cooper R., Cornelissen L., Corrah T., Cosgrove C., Cox T., Crocker W.E.M., Crosbie S., Cullen L., Cullen D., Cunha D.R.M.F., Cunningham C., Cuthbertson F.C., Guarda S.N.F.D., da Silva L.P., Damratoski B.E., Danos Z., Dantas M.T.D.C., Darroch P., Datoo M.S., Datta C., Davids M., Davies S.L., Davies H., Davis E., Davis J., Davis J., Nobrega M.M.D.D., Kalid L.M.D.O., Dearlove D., Demissie T., Desai A., Marco S.D., Maso C.D., Dinelli M.I.S., Dinesh T., Docksey C., Dold C., Dong T., Donnellan F.R., Santos T.D., dos Santos T.G., Santos E.P.D., Douglas N., Downing C., Drake J., Drake-Brockman R., Driver K., Drury R., Dunachie S.J., Durham B.S., Dutra L., Easom N.J.W., van Eck S., Edwards M., Edwards N.J., Muhanna O.M.E., Elias S.C., Elmore M., English M., Esmail A., Essack Y.M., Farmer E., Farooq M., Farrar M., Farrugia L., Faulkner B., Fedosyuk S., Felle S., Feng S., Silva C.F.D., Field S., Fisher R., Flaxman A., Fletcher J., Fofie H., Fok H., Ford K.J., Fowler J., Fraiman P.H.A., Francis E., Franco M.M., Frater J., Freire M.S.M., Fry S.H., Fudge S., Furze J., Fuskova M., Galian-Rubio P., Galiza E., Garlant H., Gavrila M., Geddes A., Gibbons K.A., Gilbride C., Gill H., Glynn S., Godwin K., Gokani K., Goldoni U.C., Goncalves M., Gonzalez I.G.S., Goodwin J., Goondiwala A., Gordon-Quayle K., Gorini G., Grab J., Gracie L., Greenland M., Greenwood N., Greffrath J., Groenewald M.M., Grossi L., Gupta G., Hackett M., Hallis B., Hamaluba M., Hamilton E., Hamlyn J., Hammersley D., Hanrath A.T., Hanumunthadu B., Harris S.A., Harris C., Harris T., Harrison T.D., Harrison D., Hart T.C., Hartnell B., Hassan S., Haughney J., Hawkins S., Hay J., Head I., Henry J., Herrera M.H., Hettle D.B., Hill J., Hodges G., Horne E., Hou M.M., Houlihan C., Howe E., Howell N., Humphreys J., Humphries H.E., Hurley K., Huson C., Hyder-Wright A., Hyams C., Ikram S., Ishwarbhai A., Ivan M., Iveson P., Iyer V., Jackson F., Jager J.D., Jaumdally S., Jeffers H., Jesudason N., Jones B., Jones K., Jones E., Jones C., Jorge M.R., Jose A., Joshi A., Júnior E.A.M.S., Kadziola J., Kailath R., Kana F., Karampatsas K., Kasanyinga M., Keen J., Kelly E.J., Kelly D.M., Kelly D., Kelly S., Kerr D., Kfouri R. de Á., Khan L., Khozoee B., Kidd S., Killen A., Kinch J., Kinch P., King L.D.W., King T.B., Kingham L., Klenerman P., Knapper F., Knight J.C., Knott D., Koleva S., Lang M., Lang G., Larkworthy C.W., Larwood J.P.J., Law R., Lazarus E.M., Leach A., Lees E.A., Lemm N.-M., Lessa A., Leung S., Li Y., Lias A.M., Liatsikos K., Linder A., Lipworth S., Liu S., Liu X., Lloyd A., Lloyd S., Loew L., Ramon R.L., Lora L., Lowthorpe V., Luz K., MacDonald J.C., MacGregor G., Madhavan M., Mainwaring D.O., Makambwa E., Makinson R., Malahleha M., Malamatsho R., Mallett G., Mansatta K., Maoko T., Mapetla K., Marchevsky N.G., Marinou S., Marlow E., Marques G.N., Marriott P., Marshall R.P., Marshall J.L., Martins F.J., Masenya M., Masilela M., Masters S.K., Mathew M., Matlebjane H., Matshidiso K., Mazur O., Mazzella A., McCaughan H., McEwan J., McGlashan J., McInroy L., McIntyre Z., McLenaghan D., McRobert N., McSwiggan S., Megson C., Mehdipour S., Meijs W., Mendonça R.N.Á., Mentzer A.J., Mirtorabi N., Mitton C., Mnyakeni S., Moghaddas F., Molapo K., Moloi M., Moore M., Moraes-Pinto M.I., Moran M., Morey E., Morgans R., Morris S., Morris S., Morris H.C., Morselli F., Morshead G., Morter R., Mottal L., Moultrie A., Moya N., Mpelembue M., Msomi S., Mugodi Y., Mukhopadhyay E., Muller J., Munro A., Munro C., Murphy S., Mweu P., Myasaki C.H., Naik G., Naker K., Nastouli E., Nazir A., Ndlovu B., Neffa F., Njenga C., Noal H., Noé A., Novaes G., Nugent F.L., Nunes G., O’Brien K., O’Connor D., Odam M., Oelofse S., Oguti B., Olchawski V., Oldfield N.J., Oliveira M.G., Oliveira C., Oosthuizen A., O’Reilly P., Osborne P., Owen D.R.J., Owen L., Owens D., Owino N., Pacurar M., Paiva B.V.B., Palhares E.M.F., Palmer S., Parkinson S., Parracho H.M.R.T., Parsons K., Patel D., Patel B., Patel F., Patel K., Patrick-Smith M., Payne R.O., Peng Y., Penn E.J., Pennington A., Alvarez M.P.P., Perring J., Perry N., Perumal R., Petkar S., Philip T., Phillips D.J., Phillips J., Phohu M.K., Pickup L., Pieterse S., Piper J., Pipini D., Plank M., Plessis J.D., Pollard S., Pooley J., Pooran A., Poulton I., Powers C., Presa F.B., Price D.A., Price V., Primeira M., Proud P.C., Provstgaard-Morys S., Pueschel S., Pulido D., Quaid S., Rabara R., Radford A., Radia K., Rajapaska D., Rajeswaran T., Ramos A.S.F., Lopez F.R., Rampling T., Rand J., Ratcliffe H., Rawlinson T., Rea D., Rees B., Reiné J., Resuello-Dauti M., Pabon E.R., Ribiero C.M., Ricamara M., Richter A., Ritchie N., Ritchie A.J., Robbins A.J., Roberts H., Robinson R.E., Robinson H., Rocchetti T.T., Rocha B.P., Roche S., Rollier C., Rose L., Russell A.L.R., Rossouw L., Royal S., Rudiansyah I., Ruiz S., Saich S., Sala C., Sale J., Salman A.M., Salvador N., Salvador S., Sampaio M., Samson A.D., Sanchez-Gonzalez A., Sanders H., Sanders K., Santos E., Guerra M.F.S.S., Satti I., Saunders J.E., Saunders C., Sayed A., van der Loeff I.S., Schmid A.B., Schofield E., Screaton G., Seddiqi S., Segireddy R.R., Senger R., Serrano S., Shah R., Shaik I., Sharpe H.E., Sharrocks K., Shaw R., Shea A., Shepherd A., Shepherd J.G., Shiham F., Sidhom E., Silk S.E., Moraes A.C. da S., Silva-Junior G., Silva-Reyes L., Silveira A.D., Silveira M.B.V., Sinha J., Skelly D.T., Smith D.C., Smith N., Smith H.E., Smith D.J., Smith C.C., Soares A., Soares T., Solórzano C., Sorio G.L., Sorley K., Sosa-Rodriguez T., Souza C.M.C.D.L., Souza B.S.D.F., Souza A.R., Spencer A.J., Spina F., Spoors L., Stafford L., Stamford I., Starinskij I., Stein R., Steven J., Stockdale L., Stockwell L.V., Strickland L.H., Stuart A.C., Sturdy A., Sutton N., Szigeti A., Tahiri-Alaoui A., Tanner R., Taoushanis C., Tarr A.W., Taylor K., Taylor U., Taylor I.J., Taylor J., Naude R. te W., Themistocleous Y., Themistocleous A., Thomas M., Thomas K., Thomas T.M., Thombrayil A., Thompson F., Thompson A., Thompson K., Thompson A., Thomson J., Thornton-Jones V., Tighe P.J., Tinoco L.A., Tiongson G., Tladinyane B., Tomasicchio M., Tomic A., Tonks S., Towner J., Tran N., Tree J., Trillana G., Trinham C., Trivett R., Truby A., Tsheko B.L., Turabi A., Turner R., Turner C., Ulaszewska M., Underwood B.R., Varughese R., Verbart D., Verheul M., Vichos I., Vieira T., Waddington C.S., Walker L., Wallis E., Wand M., Warbick D., Wardell T., Warimwe G., Warren S.C., Watkins B., Watson E., Webb S., Webb-Bridges A., Webster A., Welch J., Wells J., West A., White C., White R., Williams P., Williams R.L., Winslow R., Woodyer M., Worth A.T., Wright D., Wroblewska M., Yao A., Zimmer R., Zizi D., Zuidewind P. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397:99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Baden L.R., Sahly H.M.E., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., McGettigan J., Khetan S., Segall N., Solis J., Brosz A., Fierro C., Schwartz H., Neuzil K., Corey L., Gilbert P., Janes H., Follmann D., Marovich M., Mascola J., Polakowski L., Ledgerwood J., Graham B.S., Bennett H., Pajon R., Knightly C., Leav B., Deng W., Zhou H., Han S., Ivarsson M., Miller J., Zaks T. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Marc G.P., Moreira E.D., Zerbini C., Bailey R., Swanson K.A., Roychoudhury S., Koury K., Li P., Kalina W.V., Cooper D., Robert J., Frenck W., Hammitt L.L., Türeci Ö., Nell H., Schaefer A., Ünal S., Tresnan D.B., Mather S., Dormitzer P.R., Şahin U., Jansen K.U., Gruber W.C. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sadoff J., Gray G., Vandebosch A., Cárdenas V., Shukarev G., Grinsztejn B., Goepfert P.A., Truyers C., Fennema H., Spiessens B., Offergeld K., Scheper G., Taylor K.L., Robb M.L., Treanor J., Barouch D.H., Stoddard J., Ryser M.F., Marovich M.A., Neuzil K.M., Corey L., Cauwenberghs N., Tanner T., Hardt K., Ruiz-Guiñazú J., Le Gars M., Schuitemaker H., Van Hoof J., Struyf F., Douoguih M. Safety and efficacy of single-dose Ad26.COV2.S vaccine against Covid-19. N. Engl. J. Med. 2021;384:2187–2201. doi: 10.1056/NEJMoa2101544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., Subbarao K., Kent S.J., Triccas J.A., Davenport M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- 84.Earle K.A., Ambrosino D.M., Fiore-Gartland A., Goldblatt D., Gilbert P.B., Siber G.R., Dull P., Plotkin S.A. Evidence for antibody as a protective correlate for COVID-19 vaccines. Vaccine. 2021;39:4423–4428. doi: 10.1016/j.vaccine.2021.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kalimuddin S., Tham C.Y.L., Qui M., de Alwis R., Sim J.X.Y., Lim J.M.E., Tan H.-C., Syenina A., Zhang S.L., Le Bert N., Tan A.T., Leong Y.S., Yee J.X., Ong E.Z., Ooi E.E., Bertoletti A., Low J.G. Early T cell and binding antibody responses are associated with COVID-19 RNA vaccine efficacy onset. Med (N Y) 2021;2:682–688. doi: 10.1016/j.medj.2021.04.003. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Slütter B., Pewe L.L., Kaech S.M., Harty J.T. Lung airway-surveilling CXCR3hi memory CD8+ T cells are critical for protection against influenza a virus. Immunity. 2013;39:939–948. doi: 10.1016/j.immuni.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Channappanavar R., Fett C., Zhao J., Meyerholz D.K., Perlman S. Virus-specific memory CD8 T cells provide substantial protection from lethal severe acute respiratory syndrome coronavirus infection. J. Virol. 2014;88:11034–11044. doi: 10.1128/JVI.01505-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Crotty S. Hybrid immunity. Science. 2021;372:1392–1393. doi: 10.1126/science.abj2258. [DOI] [Google Scholar]
- 89.Bange E.M., Han N.A., Wileyto P., Kim J.Y., Gouma S., Robinson J., Greenplate A.R., Hwee M.A., Porterfield F., Owoyemi O., Naik K., Zheng C., Galantino M., Weisman A.R., Ittner C.A.G., Kugler E.M., Baxter A.E., Oniyide O., Agyekum R.S., Dunn T.G., Jones T.K., Giannini H.M., Weirick M.E., McAllister C.M., Babady N.E., Kumar A., Widman A.J., DeWolf S., Boutemine S.R., Roberts C., Budzik K.R., Tollett S., Wright C., Perloff T., Sun L., Mathew D., Giles J.R., Oldridge D.A., Wu J.E., Alanio C., Adamski S., Garfall A.L., Vella L.A., Kerr S.J., Cohen J.V., Oyer R.A., Massa R., Maillard I.P., Maxwell K.N., Reilly J.P., Maslak P.G., Vonderheide R.H., Wolchok J.D., Hensley S.E., Wherry E.J., Meyer N.J., DeMichele A.M., Vardhana S.A., Mamtani R., Huang A.C. CD8+ T cells contribute to survival in patients with COVID-19 and hematologic cancer. Nat. Med. 2021;27:1280–1289. doi: 10.1038/s41591-021-01386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sahin U., Muik A., Derhovanessian E., Vogler I., Kranz L.M., Vormehr M., Baum A., Pascal K., Quandt J., Maurus D., Brachtendorf S., Lörks V., Sikorski J., Hilker R., Becker D., Eller A.-K., Grützner J., Boesler C., Rosenbaum C., Kühnle M.-C., Luxemburger U., Kemmer-Brück A., Langer D., Bexon M., Bolte S., Karikó K., Palanche T., Fischer B., Schultz A., Shi P.-Y., Fontes-Garfias C., Perez J.L., Swanson K.A., Loschko J., Scully I.L., Cutler M., Kalina W., Kyratsous C.A., Cooper D., Dormitzer P.R., Jansen K.U., Türeci Ö. COVID-19 vaccine BNT162b1 elicits human antibody and T H 1 T cell responses. Nature. 2020;586:594–599. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- 91.Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., McDermott A., Flach B., Doria-Rose N.A., Corbett K.S., Morabito K.M., O’Dell S., Schmidt S.D., Swanson P.A., Padilla M., Mascola J.R., Neuzil K.M., Bennett H., Sun W., Peters E., Makowski M., Albert J., Cross K., Buchanan W., Pikaart-Tautges R., Ledgerwood J.E., Graham B.S., Beigel J.H. An mRNA vaccine against SARS-CoV-2 — preliminary report. N. Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Barros-Martins J., Hammerschmidt S.I., Cossmann A., Odak I., Stankov M.V., Morillas Ramos G., Dopfer-Jablonka A., Heidemann A., Ritter C., Friedrichsen M., Schultze-Florey C., Ravens I., Willenzon S., Bubke A., Ristenpart J., Janssen A., Ssebyatika G., Bernhardt G., Münch J., Hoffmann M., Pöhlmann S., Krey T., Bošnjak B., Förster R., Behrens G.M.N. Immune responses against SARS-CoV-2 variants after heterologous and homologous ChAdOx1 nCoV-19/BNT162b2 vaccination. Nat. Med. 2021:1–5. doi: 10.1038/s41591-021-01449-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Alter G., Yu J., Liu J., Chandrashekar A., Borducchi E.N., Tostanoski L.H., McMahan K., Jacob-Dolan C., Martinez D.R., Chang A., Anioke T., Lifton M., Nkolola J., Stephenson K.E., Atyeo C., Shin S., Fields P., Kaplan I., Robins H., Amanat F., Krammer F., Baric R.S., Le Gars M., Sadoff J., de Groot A.M., Heerwegh D., Struyf F., Douoguih M., van Hoof J., Schuitemaker H., Barouch D.H. Immunogenicity of Ad26.COV2.S vaccine against SARS-CoV-2 variants in humans. Nature. 2021:1–5. doi: 10.1038/s41586-021-03681-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ewer K.J., Barrett J.R., Belij-Rammerstorfer S., Sharpe H., Makinson R., Morter R., Flaxman A., Wright D., Bellamy D., Bittaye M., Dold C., Provine N.M., Aboagye J., Fowler J., Silk S.E., Alderson J., Aley P.K., Angus B., Berrie E., Bibi S., Cicconi P., Clutterbuck E.A., Chelysheva I., Folegatti P.M., Fuskova M., Green C.M., Jenkin D., Kerridge S., Lawrie A., Minassian A.M., Moore M., Mujadidi Y., Plested E., Poulton I., Ramasamy M.N., Robinson H., Song R., Snape M.D., Tarrant R., Voysey M., Watson M.E.E., Douglas A.D., Hill A.V.S., Gilbert S.C., Pollard A.J., Lambe T. T cell and antibody responses induced by a single dose of ChAdOx1 nCoV-19 (AZD1222) vaccine in a phase 1/2 clinical trial. Nat. Med. 2021;27:270–278. doi: 10.1038/s41591-020-01194-5. [DOI] [PubMed] [Google Scholar]
- 95.Oberhardt V., Luxenburger H., Kemming J., Schulien I., Ciminski K., Giese S., Csernalabics B., Lang-Meli J., Janowska I., Staniek J., Wild K., Basho K., Marinescu M.S., Fuchs J., Topfstedt F., Janda A., Sogukpinar O., Hilger H., Stete K., Emmerich F., Bengsch B., Waller C.F., Rieg S., Sagar, Boettler T., Zoldan K., Kochs G., Schwemmle M., Rizzi M., Thimme R., Neumann-Haefelin C., Hofmann M. Rapid and stable mobilization of CD8+ T cells by SARS-CoV-2 mRNA vaccine. Nature. 2021:1–9.. doi: 10.1038/s41586-021-03841-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tauzin A., Nayrac M., Benlarbi M., Gong S.Y., Gasser R., Beaudoin-Bussières G., Brassard N., Laumaea A., Vézina D., Prévost J., Anand S.P., Bourassa C., Gendron-Lepage G., Medjahed H., Goyette G., Niessl J., Tastet O., Gokool L., Morrisseau C., Arlotto P., Stamatatos L., McGuire A.T., Larochelle C., Uchil P., Lu M., Mothes W., Serres G.D., Moreira S., Roger M., Richard J., Martel-Laferrière V., Duerr R., Tremblay C., Kaufmann D.E., Finzi A. A single dose of the SARS-CoV-2 vaccine BNT162b2 elicits Fc-mediated antibody effector functions and T-cell responses. Cell Host Microbe. 2021;0 doi: 10.1016/j.chom.2021.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Painter M.M., Mathew D., Goel R.R., Apostolidis S.A., Pattekar A., Kuthuru O., Baxter A.E., Herati R.S., Oldridge D.A., Gouma S., Hicks P., Dysinger S., Lundgreen K.A., Kuri-Cervantes L., Adamski S., Hicks A., Korte S., Giles J.R., Weirick M.E., McAllister C.M., Dougherty J., Long S., D’Andrea K., Hamilton J.T., Betts M.R., Bates P., Hensley S.E., Grifoni A., Weiskopf D., Sette A., Greenplate A.R., Wherry E.J. Rapid induction of antigen-specific CD4+ T cells is associated with coordinated humoral and cellular immune responses to SARS-CoV-2 mRNA vaccination. Immunity. 2021;54 doi: 10.1016/j.immuni.2021.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Habel J.R., Nguyen T.H.O., van de Sandt C.E., Juno J.A., Chaurasia P., Wragg K., Koutsakos M., Hensen L., Jia X., Chua B., Zhang W., Tan H.-X., Flanagan K.L., Doolan D.L., Torresi J., Chen W., Wakim L.M., Cheng A.C., Doherty P.C., Petersen J., Rossjohn J., Wheatley A.K., Kent S.J., Rowntree L.C., Kedzierska K. Suboptimal SARS-CoV-2−specific CD8+ T cell response associated with the prominent HLA-A*02:01 phenotype. PNAS. 2020;117:24384–24391. doi: 10.1073/pnas.2015486117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Greinacher A., Thiele T., Warkentin T.E., Weisser K., Kyrle P.A., Eichinger S. Thrombotic thrombocytopenia after ChAdOx1 nCov-19 vaccination. N. Engl. J. Med. 2021;384:2092–2101. doi: 10.1056/NEJMoa2104840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Stamatatos L., Czartoski J., Wan Y.-H., Homad L.J., Rubin V., Glantz H., Neradilek M., Seydoux E., Jennewein M.F., MacCamy A.J., Feng J., Mize G., Rosa S.C.D., Finzi A., Lemos M.P., Cohen K.W., Moodie Z., McElrath M.J., McGuire A.T. mRNA vaccination boosts cross-variant neutralizing antibodies elicited by SARS-CoV-2 infection. Science. 2021;372:1413–1418. doi: 10.1126/science.abg9175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Reynolds C.J., Pade C., Gibbons J.M., Butler D.K., Otter A.D., Menacho K., Fontana M., Smit A., Sackville-West J.E., Cutino-Moguel T., Maini M.K., Chain B., Noursadeghi M., U.Covid.I.C. Network, Brooks T., Semper A., Manisty C., Treibel T.A., Moon J.C., U. Covid. Investigators, Valdes A.M., McKnight Á., Altmann D.M., Boyton R. Prior SARS-CoV-2 infection rescues B and T cell responses to variants after first vaccine dose. Science. 2021;372:1418–1423. doi: 10.1126/science.abh1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Geers D., Shamier M.C., Bogers S., den Hartog G., Gommers L., Nieuwkoop N.N., Schmitz K.S., Rijsbergen L.C., van Osch J.A.T., Dijkhuizen E., Smits G., Comvalius A., van Mourik D., Caniels T.G., van Gils M.J., Sanders R.W., Munnink B.B.O., Molenkamp R., de Jager H.J., Haagmans B.L., de Swart R.L., Koopmans M.P.G., van Binnendijk R.S., de Vries R.D., GeurtsvanKessel C.H. SARS-CoV-2 variants of concern partially escape humoral but not T-cell responses in COVID-19 convalescent donors and vaccinees. Sci. Immunol. 2021;6 doi: 10.1126/sciimmunol.abj1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Matchett W.E., Joag V., Stolley J.M., Shepherd F.K., Quarnstrom C.F., Mickelson C.K., Wijeyesinghe S., Soerens A.G., Becker S., Thiede J.M., Weyu E., O’Flanagan S.D., Walter J.A., Vu M.N., Menachery V.D., Bold T.D., Vezys V., Jenkins M.K., Langlois R.A., Masopust D. Nucleocapsid vaccine elicits spike-independent SARS-CoV-2 protective immunity. J. Immunol. 2021 doi: 10.4049/jimmunol.2100421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Class J., Dangi T., Richner J.M., Penaloza-MacMaster P. Combining spike- and nucleocapsid-based vaccines improves distal control of SARS-CoV-2. Cell Reports. 2021;36(10) doi: 10.1016/j.celrep.2021.109664. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Doria-Rose N., Suthar M.S., Makowski M., O’Connell S., McDermott A.B., Flach B., Ledgerwood J.E., Mascola J.R., Graham B.S., Lin B.C., O’Dell S., Schmidt S.D., Widge A.T., Edara V.-V., Anderson E.J., Lai L., Floyd K., Rouphael N.G., Zarnitsyna V., Roberts P.C., Makhene M., Buchanan W., Luke C.J., Beigel J.H., Jackson L.A., Neuzil K.M., Bennett H., Leav B., Albert J., Kunwar P. Antibody persistence through 6 months after the second dose of mRNA-1273 vaccine for Covid-19. N. Engl. J. Med. 2021;384:2259–2261. doi: 10.1056/NEJMc2103916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ibarrondo F.J., Hofmann C., Fulcher J.A., Goodman-Meza D., Mu W., Hausner M.A., Ali A., Balamurugan A., Taus E., Elliott J., Krogstad P., Tobin N.H., Ferbas K.G., Kitchen S.G., Aldrovandi G.M., Rimoin A.W., Yang O.O. Primary, recall, and decay kinetics of SARS-CoV-2 vaccine antibody responses. ACS Nano. 2021;15:11180–11191. doi: 10.1021/acsnano.1c03972. [DOI] [PubMed] [Google Scholar]
- 107.Akondy R.S., Fitch M., Edupuganti S., Yang S., Kissick H.T., Li K.W., Youngblood B.A., Abdelsamed H.A., McGuire D.J., Cohen K.W., Alexe G., Nagar S., McCausland M.M., Gupta S., Tata P., Haining W.N., McElrath M.J., Zhang D., Hu B., Greenleaf W.J., Goronzy J.J., Mulligan M.J., Hellerstein M., Ahmed R. Origin and differentiation of human memory CD8 T cells after vaccination. Nature. 2017;552:362–367. doi: 10.1038/nature24633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hammarlund E., Lewis M.W., Hansen S.G., Strelow L.I., Nelson J.A., Sexton G.J., Hanifin J.M., Slifka M.K. Duration of antiviral immunity after smallpox vaccination. Nat. Med. 2003;9:1131–1137. doi: 10.1038/nm917. [DOI] [PubMed] [Google Scholar]
- 109.Prendecki M., Clarke C., Edwards H., McIntyre S., Mortimer P., Gleeson S., Martin P., Thomson T., Randell P., Shah A., Singanayagam A., Lightstone L., Cox A., Kelleher P., Willicombe M., McAdoo S.P. Humoral and T-cell responses to SARS-CoV-2 vaccination in patients receiving immunosuppression. Ann. Rheum. Dis. 2021 doi: 10.1136/annrheumdis-2021-220626. annrheumdis-2021-220626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bertrand D., Hamzaoui M., Lemée V., Lamulle J., Hanoy M., Laurent C., Lebourg L., Etienne I., Lemoine M., Le Roy F., Nezam D., Plantier J.-C., Boyer O., Guerrot D., Candon S. Antibody and T cell response to SARS-CoV-2 messenger RNA BNT162b2 vaccine in kidney transplant recipients and hemodialysis patients. J. Am. Soc. Nephrol. 2021 doi: 10.1681/ASN.2021040480. ASN.2021040480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wu T., Hu Y., Lee Y.-T., Bouchard K.R., Benechet A., Khanna K., Cauley L.S. Lung-resident memory CD8 T cells (TRM) are indispensable for optimal cross-protection against pulmonary virus infection. J. Leukoc. Biol. 2014;95:215–224. doi: 10.1189/jlb.0313180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Grau-Expósito J., Sánchez-Gaona N., Massana N., Suppi M., Astorga-Gamaza A., Perea D., Rosado J., Falcó A., Kirkegaard C., Torrella A., Planas B., Navarro J., Suanzes P., Álvarez-Sierra D., Ayora A., Sansano I., Esperalba J., Andrés C., Antón A., Ramón y Cajal S., Almirante B., Pujol-Borrell R., Falcó V., Burgos J., Buzón M.J., Genescà M. Peripheral and lung resident memory T cell responses against SARS-CoV-2. Nat. Commun. 2021;12:3010. doi: 10.1038/s41467-021-23333-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Roukens A.H.E., König M., Dalebout T., Tak T., Azimi S., Kruize Y., Pothast C.R., Hagedoorn R.S., Arbous S.M., Zhang J.L.H., Verheij M., Prins C., van der Does A.M., Hiemstra P.S., de Vries J.J.C., Janse J.J., Roestenberg M., Myeni S.K., Kikkert M., Heemskerk M.H.M., Yazdanbakhsh M., Smits H.H., Jochems S.P., B.-C. Group Prolonged activation of nasal immune cell populations and development of tissue-resident SARS-CoV-2 specific CD8 T cell responses following COVID-19. MedRxiv. 2021 doi: 10.1101/2021.04.19.21255727. 2021.04.19.21255727. [DOI] [PubMed] [Google Scholar]
- 114.Pizzolla A., Nguyen T.H.O., Smith J.M., Brooks A.G., Kedzieska K., Heath W.R., Reading P.C., Wakim L.M. Resident memory CD8+ T cells in the upper respiratory tract prevent pulmonary influenza virus infection. Sci. Immunol. 2017;2 doi: 10.1126/sciimmunol.aam6970. [DOI] [PubMed] [Google Scholar]
- 115.Slütter B., Braeckel-Budimir N.V., Abboud G., Varga S.M., Salek-Ardakani S., Harty J.T. Dynamics of influenza-induced lung-resident memory T cells underlie waning heterosubtypic immunity. Sci. Immunol. 2017;2 doi: 10.1126/sciimmunol.aag2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ssemaganda A., Nguyen H.M., Nuhu F., Jahan N., Card C.M., Kiazyk S., Severini G., Keynan Y., Su R.-C., Ji H., Abrenica B., McLaren P.J., Ball T.B., Bullard J., Caeseele P.V., Stein D., McKinnon L.R. Expansion of tissue-resident CD8+ T cells and CD4+ Th17 cells in the nasal mucosa following mRNA COVID-19 vaccination. BioRxiv. 2021 doi: 10.1101/2021.05.07.442971. 2021.05.07.442971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hassan A.O., Kafai N.M., Dmitriev I.P., Fox J.M., Smith B.K., Harvey I.B., Chen R.E., Winkler E.S., Wessel A.W., Case J.B., Kashentseva E., McCune B.T., Bailey A.L., Zhao H., VanBlargan L.A., Dai Y.-N., Ma M., Adams L.J., Shrihari S., Danis J.E., Gralinski L.E., Hou Y.J., Schäfer A., Kim A.S., Keeler S.P., Weiskopf D., Baric R.S., Holtzman M.J., Fremont D.H., Curiel D.T., Diamond M.S. A single-dose intranasal ChAd vaccine protects upper and lower respiratory tracts against SARS-CoV-2. Cell. 2020;183:169–184. doi: 10.1016/j.cell.2020.08.026. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Takamura S., Roberts A.D., Jelley-Gibbs D.M., Wittmer S.T., Kohlmeier J.E., Woodland D.L. The route of priming influences the ability of respiratory virus–specific memory CD8+ T cells to be activated by residual antigen. J. Exp. Med. 2010;207:1153–1160. doi: 10.1084/jem.20090283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lund F.E., Randall T.D. Scent of a vaccine. Science. 2021;373:397–399. doi: 10.1126/science.abg9857. [DOI] [PubMed] [Google Scholar]
- 120.Niessl J., Sekine T., Lange J., Konya V., Forkel M., Maric J., Rao A., Mazzurana L., Kokkinou E., Weigel W., Llewellyn-Lacey S., Hodcroft E.B., Karlsson A.C., Fehrm J., Sundman J., Price D.A., Mjösberg J., Friberg D., Buggert M. Identification of resident memory CD8+ T cells with functional specificity for SARS-CoV-2 in unexposed oropharyngeal lymphoid tissue. Sci. Immunol. 2021 doi: 10.1126/sciimmunol.abk0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dangi T., Palacio N., Sanchez S., Park M., Class J., Visvabharathy L., Ciucci T., Koralnik I.J., Richner J.M., Penaloza-MacMaster P. Cross-protective immunity following coronavirus vaccination and coronavirus infection. J. Clin. Invest. 2021 doi: 10.1172/JCI151969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Poon M.M.L., Rybkina K., Kato Y., Kubota M., Matsumoto R., Bloom N.I., Zhang Z., Hastie K.M., Grifoni A., Weiskopf D., Wells S.B., Ural B.B., Lam N., Szabo P.A., Dogra P., Lee Y.S., Gray J.I., Bradley M.C., Brusko M.A., Brusko T.M., Saphire E.O., Connors T.J., Sette A., Crotty S., Farber D.L. SARS-CoV-2 infection generates tissue-localized immunological memory in humans. Sci. Immunol. 2021 doi: 10.1126/sciimmunol.abl9105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wijeyesinghe S., Beura L.K., Pierson M.J., Stolley J.M., Adam O.A., Ruscher R., Steinert E.M., Rosato P.C., Vezys V., Masopust D. Expansible residence decentralizes immune homeostasis. Nature. 2021;592 doi: 10.1038/s41586-021-03351-3. [DOI] [PMC free article] [PubMed] [Google Scholar]