Abstract
Introduction
Data extraction from electronic health record (EHR) systems occurs through manual abstraction, automated extraction, or a combination of both. While each method has its strengths and weaknesses, both are necessary for retrospective observational research as well as sudden clinical events, like the COVID-19 pandemic. Assessing the strengths, weaknesses, and potentials of these methods is important to continue to understand optimal approaches to extracting clinical data. We set out to assess automated and manual techniques for collecting medication use data in patients with COVID-19 to inform future observational studies that extract data from the electronic health record (EHR).
Materials and methods
For 4,123 COVID-positive patients hospitalized and/or seen in the emergency department at an academic medical center between 03/03/2020 and 05/15/2020, we compared medication use data of 25 medications or drug classes collected through manual abstraction and automated extraction from the EHR. Quantitatively, we assessed concordance using Cohen’s kappa to measure interrater reliability, and qualitatively, we audited observed discrepancies to determine causes of inconsistencies.
Results
For the 16 inpatient medications, 11 (69%) demonstrated moderate or better agreement; 7 of those demonstrated strong or almost perfect agreement. For 9 outpatient medications, 3 (33%) demonstrated moderate agreement, but none achieved strong or almost perfect agreement. We audited 12% of all discrepancies (716/5,790) and, in those audited, observed three principal categories of error: human error in manual abstraction (26%), errors in the extract-transform-load (ETL) or mapping of the automated extraction (41%), and abstraction-query mismatch (33%).
Conclusion
Our findings suggest many inpatient medications can be collected reliably through automated extraction, especially when abstraction instructions are designed with data architecture in mind. We discuss quality issues, concerns, and improvements for institutions to consider when crafting an approach. During crises, institutions must decide how to allocate limited resources. We show that automated extraction of medications is feasible and make recommendations on how to improve future iterations.
Keywords: Electronic health record, Chart review, COVID-19, Research data repositories, Data quality
Abbreviations: EHR, electronic health record; IDR, institutional data repository; ED, emergency department; CDM, common data model; OMOP, observational medical outcomes partnership; SQL, structured query language; NDF-RT, National Drug File-Reference Terminology; ATC, Anatomical Therapeutic Chemical; κ, Cohen’s kappa; PABAK, prevalence-adjusted bias-adjusted kappa; PI, prevalence index; ETL, extract-transform-load
1. Introduction
Data collection from electronic health record (EHR) systems may be conducted through manual abstraction, automated extraction, or a combination of both. Manual abstraction, which involves trained personnel reviewing patient charts and completing case report forms, is often considered the gold standard for retrospective observational research. Many variables require manual adjudication by clinically trained personnel, depending on the complexity of institutional workflows, clinical questions, medical record structure, etc. [1], [2], [3], [4], [5], [6], [7], [8], [9], [10]. Notably, Flatiron Health, with its data set for analytics generated through manual review, sold for nearly $2 billion to pharmaceutical company Roche, demonstrating the significant value of manually abstracted data [11]. Although considered the gold standard, manual abstraction has limitations, as human reviewers are not infallible and can be less accurate in certain cases [12], [13], [14], [15], [16], [17]. Importantly, manual abstraction consumes significant time for clinically trained personnel who are needed for patient care and other capacities, especially during times of crisis as occurred in the COVID-19 pandemic.
To address these challenges, studies have demonstrated that automated data extraction from the EHR, which involves direct database queries, can produce data sets of similar quality to manual abstraction for certain variables while saving time for study teams and reducing error [16], [18], [19], [20], [21], [22], [23], [24]. Even so, automated extraction is similarly susceptible to data quality issues relating to high complexity or fragmentation of data across many EHR systems [25], [26], [27], [28]. Manual abstraction and automated extraction both ultimately depend on the EHR, which is not an objective, canonical source of truth but rather an artifact with its own bias, inaccuracies, and subjectivity [29], [30], [31], [32], [33], [34], [35]. While previous work has explored these concepts, optimal approaches for acquiring data from EHR systems for research are unknown.
Alongside other academic medical centers, our institution, Weill Cornell Medicine, deployed informatics to support COVID-19 pandemic response efforts [36], [37], [38], [39], [40], [41], [42], [43], [44], [45], [46], [47], [48]. This included systematic data collection from the EHR, which at our institution occurred through a combination of manual abstraction and automated extraction. Prior investigations have compared manual to automated data collection techniques in conditions other than COVID-19, described informatics resources specific to COVID-19[47], [48], [49], and evaluated the performance of automated extraction in COVID-19 [47], [48], [49]. Such evaluations of automated extraction have included problem lists as well as natural language processing extracting signs and symptoms [50], [51], [52], [53]. To the best of our knowledge, no studies have compared manual to automated data acquisition of medications in COVID-19 data. Medication use data is critical to studying new diseases, especially concerning their risk factors and outcomes following certain treatments. For example, ACE inhibitors were debated early in the pandemic due to concerns about their role in exacerbating disease [54]. We sought to quantitatively assess the concordance between manual abstraction and automated extraction of EHR data for inpatient and outpatient medications using Cohen’s kappa while also qualitatively reviewing instances of discordance to understand sources of error, similar to previous work [15], [16], [17], [18], [23]. The COVID-19 pandemic uniquely fueled parallel database creation through both manual and automated methods, given the dire need for information. In turn, this enabled us to compare these methods in ways not done previously, given the number of patients included, number of medications included, and the parallel creation of the databases. Through our comparison and suggestions, we hope to support institutions in improving data collection methods and in the allocation of resources in future efforts, whether related to COVID or other clinical scenarios [49], [55].
2. Materials and methods
2.1. Setting
This retrospective observational study occurred at Weill Cornell Medicine (WCM), the biomedical research and education unit of Cornell University, which has 1,600 attending physicians with faculty appointments in the WCM Physician Organization and admitting privileges to NewYork-Presbyterian. Affiliated facilities included NewYork-Presbyterian/Weill Cornell Medical Center (NYP/WCMC), an 862-bed teaching hospital; NewYork-Presbyterian Hospital/Lower Manhattan Hospital (NYP/LMH), a 180-bed community hospital; and NewYork-Presbyterian/Queens (NYP/Q), a 535-bed community teaching hospital. For inpatient and emergency settings, clinicians used the Allscripts Sunrise Clinical Manager EHR system. For outpatient settings, NYP/WCMC and NYP/LMH clinicians used the Epic EHR system while NYP/Q clinicians used the Athenahealth EHR system. The study period was 03/03/2020 (date of first COVID-positive admission to a WCM campus) to 05/15/2020. The WCM Institutional Review Board approved this study (#20-03021681).
2.2. System description
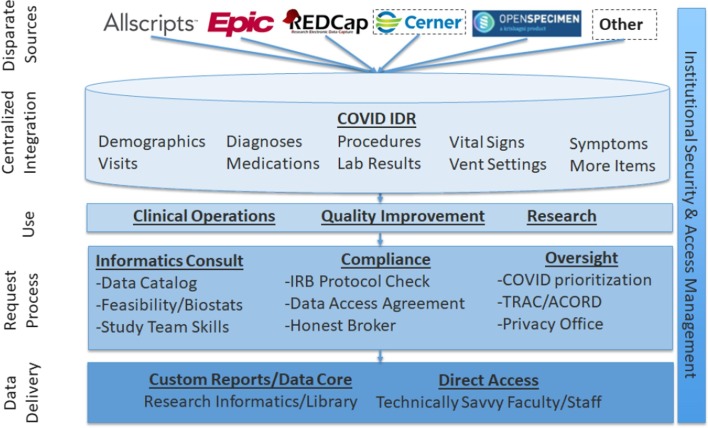
To support institutional pandemic response efforts, we created the COVID Institutional Data Repository (IDR), comprised of data retrieved through manual abstraction and automated extraction from EHR systems. The COVID IDR used existing institutional infrastructure for secondary use of EHR data, including Microsoft SQL Server-based pipelines for data acquisition from ambulatory and inpatient EHR systems and research systems, described in prior work [56]. As illustrated in Fig. 1 , the IDR includes many clinical domains and was designed to support diverse use cases, including clinical operations, quality improvement, and research with processes to determine access and oversee regulatory approval [56]. Data collected through manual abstraction and automated extraction in the IDR are ultimately derived from the EHR, which, as demonstrated by Hripcsak et al, constitutes an imperfect proxy for the true underlying patient state [30]. Inpatient and outpatient medication data examined in this study were derived from a few different sources. Inpatient medication data was derived from the medication administration record present in Allscripts SCM, the electronic health record system in use at the time of the pandemic. Outpatient medication data was derived from a combination of free text mentions in clinical notes, “historical medication” orders entered into the EHR as the result of medication reconciliation at the time of admission, and prescriptions entered into the EHR either at discharge or at an ambulatory care visit.
Fig. 1.
COVID Institutional Data Repository – architectural overview.
2.2.1. Manual abstraction
A team of clinicians (PG, JC, HL, GW, MA, MS) identified data elements in the EHR and created a REDCap case report form [57]. The team provided training to furloughed medical students and other clinicians (the abstractors) on abstraction methods [58]. A daily query identified patients based on the inclusion criteria (admitted to or seen in the emergency department (ED) at NYP/WCMC, NYP/LMH, or NYP/Q AND positive RT-PCR for SARS-CoV-2). Abstractors followed these patients (n = 4,123) through their entire hospitalization until discharge, including any subsequent encounters for any of these same patients who presented to the ED again or were readmitted (n = 4,414 visits). The case report form included 14 sections: patient information, comorbidities, symptoms, home (outpatient) medications, ED course, mechanical ventilation, ICU stay, discharge, imaging, disposition, complications, testing, inpatient medications, and survey status. Reviewers relied principally on the inpatient EHR. For portions of the case report form regarding medications, abstractors answered a mix of binary (“yes” or “no”) and check box (“check all that apply”) questions. Medications were abstracted at the drug class level (e.g. Statins) or individual level (e.g. Hydroxychloroquine). To determine inpatient medication exposure, abstractors used structured order entry and medication administration record data to determine whether a patient received a given medication. To determine outpatient medication exposure (those drugs prescribed prior to the hospitalization), manual abstractors relied on outpatient medication orders from the ambulatory EHR system, mentions of drug exposure in clinical notes, and “historical medication” orders entered into the inpatient order-entry system as a result of medication reconciliation after admission. The case report form listed examples of medications from drug classes for abstractors to reference. As a quality check prior to initial publication of registry data, a second abstractor reviewed 10% of records, calculating mean Cohen’s kappas of 0.92 and 0.94 for categorical and continuous variables, respectively. Table 2, Table 3 in Appendix A show the results from this secondary extraction in the “Validation” columns. The entirety of these methods has been described previously [36]. For the manual abstraction data dictionary, see Appendix B.
Table 2.
Prevalence of each medication/drug class in the manual abstraction and automated extraction data. Data show the number of visits (out of the total 4,414) in which each medication was administered (inpatient medications) or an active prescription at the time of hospitalization (outpatient medications).
| Number of visits in which medication was administered during hospitalization |
Number of visits in which medication was actively prescribed at the time of hospitalization |
||||
|---|---|---|---|---|---|
| Inpatient | Manual | Automated | Outpatient | Manual | Automated |
| Hydroxychloroquine | 2656 | 2576 | Statin | 1323 | 1425 |
| Antibiotics | 2513 | 2138 | NSAID | 1021 | 1270 |
| Statins | 1115 | 986 | ACEi/ARB | 1090 | 1180 |
| NSAID | 878 | 698 | PPI | 624 | 619 |
| Vasopressors | 747 | 646 | Steroids | 360 | 595 |
| Diuretics | 706 | 275 | Immunosuppressives | 279 | 305 |
| Steroids | 697 | 880 | Hydroxychloroquine | 83 | 126 |
| ACEi/ARB | 334 | 267 | Antivirals | 69 | 148 |
| Remdesivir | 188 | 0 | Oseltamivir | 50 | 24 |
| Tocilizumab | 171 | 168 | |||
| Oseltamivir | 75 | 77 | |||
| Inotropes | 62 | 49 | |||
| Sarilumab | 27 | 0 | |||
| IVIG | 21 | 19 | |||
| Lopinavir/Ritonavir | 1 | 5 | |||
| Protease Inhibitors | 0 | 0 | |||
Table 3.
Classifications of discrepancies between manual and automated detection of medications. Issues were generally categorized into one of 3 groups: human error, ETL or mapping error, and abstraction-query mismatch. Descriptions and examples of each issue are provided.
| Common Issues | Description | Example |
|---|---|---|
| Human Error | ||
| Abstractors overlook desired information (false negative) | In 16% of discrepancies, the manual abstractors overlooked a medication that should have been included based on instructions | Patient received hydroxychloroquine during admission, but was categorized by the manual abstractor as not having received hydroxychloroquine |
| Abstractors include inappropriate information (false positive) | In 10% of discrepancies, complex drug classes or questions led manual reviewers to classify patients as having been exposed to a medication when they were not | Patient classified by manual abstractor as exposed to NSAIDs despite only receiving acetaminophen (a non-NSAID drug) during hospitalization |
| ETL/Mapping Error | ||
| Missing data leading to query error | In 31% of discrepancies, data missing in the EHR led to the query incorrectly categorizing patients as having continued exposure to a given drug | Outpatient medications were only included by manual abstractors if the patient was exposed based on admission documentation, but many orders in the outpatient EHR lack end dates, requiring further work for appropriate automated calculation |
| Local errors | In 5% of discrepancies, issues with missing reference terminology in source systems caused failure to detect some medications during automated extraction of data | Remdesivir and sarilumab were not coded to RxNorm vocabulary due to investigational status and exposures to these drugs were not detected in the automated extracted data |
| Patient identifier inconsistency | In 4% of discrepancies, patient identifiers were either missing or incorrect, leading to discrepancies in specific drug exposures | Two patients shared the same enterprise master patient index, resulting in conflation of their data |
| Cross institutional differences | In 1% of discrepancies, data were not mapped correctly between differing hospital campuses, which led to incorrectly classified drug classes in the data extracted by automated methods | The formulary from one hospital was mapped to the formulary from another, yielding incorrect classes for some drugs |
| Abstraction-Query Mismatch | ||
| Mismatch between query and data format | In 30% of discrepancies, for inpatient medications where duration of administration was important, the query overlooked medications that were ordered daily as opposed to ordered continuously as order duration was used to measure duration of administration. | Diuretics were commonly ordered as single doses each day, thus although a patient could receive diuretics for consecutive days, the query only detected doses as having a 24-hour duration when instructions for manual abstraction asked for a minimum 48-hour duration. |
| Complex instructions/confounding medications | In 3% of discrepancies, lack of clarity in some special instructions for specific medication categories created challenges in developing the query | Manual abstractors were instructed to only capture protease inhibitor exposure if the drug was part of an HIV regimen – the automated extraction method and query did not take this into account. Confounding medication names also led to inappropriate inclusion. |
2.2.2. Automated extraction
Automated extraction captured data for all patients tested for SARS-CoV-2 and/or diagnosed with COVID-19 as documented by EHR systems in the study period. Data were transferred from their underlying raw format (vendor-specific proprietary EHR data models) and loaded into a Microsoft SQL Server database designed with a custom schema. Data were stored in tables corresponding to clinical domains (Appendix C). Instead of using an existing common data model (CDM) such as the Observational Medical Outcomes Partnership (OMOP) or PCORnet CDM, we used a simplified format based on OMOP to include data elements not always assigned reference terminology in EHR source data and to present data in keeping with clinical preconceptions (e.g. separating in-hospital medication administration from outpatient prescriptions to preserve their distinct provenance and usage rather than combining both into a single table)[59], [60].
2.3. Research methods of comparison
First, we characterized both constituent elements of the IDR: its manually abstracted and automatically extracted components. For both data sets, we determined how many patients were included, tallied total observations, and determined basic demographic characteristics. Second, for all patient visits (as some patients had multiple hospitalizations) with data collected through both manual abstraction and automated extraction, we quantitatively assessed agreement between the methods using Cohen’s kappa. Third, we audited a subset of discrepancies between the results of the automated and manual processes for each medication or drug class to determine the underlying error. Of note, in this comparison of manual abstraction and automated extraction, we did not assume either to be the gold standard, instead seeking objective strengths and weaknesses of each approach and the concordance between their data. Past studies have used this approach to compare strengths and weaknesses of manual and automated data collection [15], [16], [17], [18], [23]. Individual medications and drug classes were studied to better understand both common and specific causes of discrepancies within the two methods and allow for more targeted recommendations for improvement.
2.3.1. Data transformation
Because the data set formats differed, both required transformation before comparison. For example, medication data from manual abstraction were stored as dichotomous variables on a per-patient basis, while automated extraction stored them on a per-order basis. In order to compare, we developed Structured Query Language (SQL) queries with outputs displaying the presence or absence of agreement between the two methods (example of query output displayed in Appendix A, Table 1). SQL code for queries is available on Github (https://github.com/wcmc-research-informatics/covid_comparison). Queries were designed to align with the instructions given to manual abstractors. For example, they did not use RxNorm-derived definitions of drug classes, such as the National Drug File-Reference Terminology (NDF-RT) or Anatomical Therapeutic Chemical (ATC) hierarchies, instead using generic names from the manual abstractors’ instructions and clinical discretion of members of the research team (AY, WG, PG, JC) who participated in the manual abstraction. Queries identified inpatient medications from the automated extraction database if the date of administration fell within the dates of a given hospitalization. Queries identified outpatient medications based on whether the medication was actively prescribed for a patient at the time of admission to the hospital. It is important to note that a closed system to guarantee medication administration does not exist in the outpatient setting as it does in the inpatient setting.
Table 1.
Characteristics of patients included in the COVID-19 manual abstraction effort and the IDR.
| Patients with manually abstracted data (N = 4123) | Patients with only automated extracted data (N = 20821) | Patients with data through manual abstraction, automated extraction, or both (N = 24944) | |
|---|---|---|---|
| Race/Ethnicity | |||
| Asian, non-Hispanic | 640 (15.5%) | 1835 (8.8%) | 2475 (9.9%) |
| Black, non-Hispanic | 433 (10.5%) | 2966 (14.2%) | 3399 (13.6%) |
| White, non-Hispanic | 896 (21.7%) | 6732 (32.3%) | 7628 (30.6%) |
| Other non-Hispanic | 434 (10.5%) | 1718 (8.3%) | 2152 (8.6%) |
| Hispanic/Latino | 1213 (29.4%) | 4375 (21.0%) | 5588 (22.4%) |
| Unknown | 507 (12.3%) | 3195 (15.3%) | 3702 (14.8%) |
| Sex | |||
| Female | 1711 (41.5%) | 12,907 (62.0%) | 14,618 (58.6%) |
| Male | 2397 (58.1%) | 7892 (37.9%) | 10,289 (41.2%) |
| Other | 1 (0.0%) | 5 (0.0%) | 6 (0.0%) |
| Missing | 14 (0.3%) | 17 (0.1%) | 31 (0.1%) |
| Age | |||
| 0–18 | 16 (0.4%) | 791 (3.8%) | 807 (3.2%) |
| 18–35 | 245 (5.9%) | 4292 (20.6%) | 4537 (18.2%) |
| 35–50 | 571 (13.8%) | 5752 (27.6%) | 6323 (25.3%) |
| 50–65 | 1129 (27.4%) | 4438 (21.3%) | 5567 (22.3%) |
| 65–89 | 1785 (43.3%) | 4764 (22.9%) | 6549 (26.3%) |
| 89+ | 363 (8.8%) | 767 (3.7%) | 1130 (4.5%) |
| Missing | 14 (0.3%) | 17 (0.1%) | 31 (0.1%) |
2.3.2. Measuring agreement between manual abstraction and automated extraction
For each medication or drug class in the query (both inpatient and outpatient), we calculated Cohen’s kappa (κ), a statistic commonly used to measure interrater reliability, to quantify the agreement between data obtained through manual abstraction and automated extraction. We used the scale developed by McHugh to determine the strength of agreement based on the following thresholds: “almost perfect” (κ > 0.9), “strong” (0.9 > κ > 0.8), “moderate” (0.8 > κ > 0.6), “weak” (0.6 > κ > 0.4), “minimal” (0.4 > κ > 0.2), and “none” (0.2 > κ > 0.0) [61]. 95% CIs were calculated for each medication. To assess whether having previous records in our system improved data quality, we calculated κ values based on whether patients had EHR documentation of a prior outpatient, inpatient, or emergency visit to our healthcare system. To account for different prevalence in medications (i.e. some had very low prevalence) in the data set, we calculated the prevalence-adjusted bias-adjusted kappa (PABAK) and prevalence index (PI) [62].
2.3.3. Explaining discrepancies
For each medication or drug class, we randomly audited 10% of the identified discrepancies or 20 discrepancies, whichever was greater. Two members of the research team (AY and WG) adjudicated discrepancies in the results of the manual abstraction and automated extraction to determine the cause of error and determine the correct output. In order to do this, they reviewed both information from inpatient and outpatient EHRs as well as the manual abstraction and automated extraction data. ES adjudicated cases of disagreement between AY and WG. We classified discrepancies into three principal categories based on whether each was attributable to error in the manual abstraction, in the automated extraction, or to errors that could not be attributed to either method specifically. Respectively, these categories are: 1) human error in manual abstraction, 2) automated extraction error in the extract-transform-load (ETL) or mapping process, or 3) unattributable error due to abstraction-query mismatch between the instructions supplied to manual abstractors and the extraction query that flattened automated data for comparison. Although designed closely, the manual abstraction process was designed first, with instructions that did not necessarily consider the logic of automated extraction. We calculated descriptive statistics of the distribution of error types across these three categories.
3. Results
During the study period, manual abstraction yielded data for 4,123 patients while automated extraction collected data for 24,944 patients, including the 4,123 patients from manual abstraction. Table 1 describes the distribution of patient characteristics collected through manual abstraction and automated extraction.
All 25 medications (16 inpatient and 9 outpatient) in the manual abstraction process were included. The 4,123 patients identified in the manual abstraction process had a total of 4,414 visits, as some patients had multiple hospitalizations during the study period. Based on the manual abstraction, the percent of visits receiving a certain medication or drug class ranged from 0% to 60% (e.g. protease inhibitors in 0 of 4,414 visits; hydroxychloroquine in 2,656 of 4,414 visits). Prevalence of each medication or drug class in the manual abstraction and automated extraction can be seen in Table 2. For all counts, percentages, and a more detailed breakdown of the data, see Appendix A, Table 2, Table 3.
3.1. Measuring agreement between manual abstraction and automated extraction
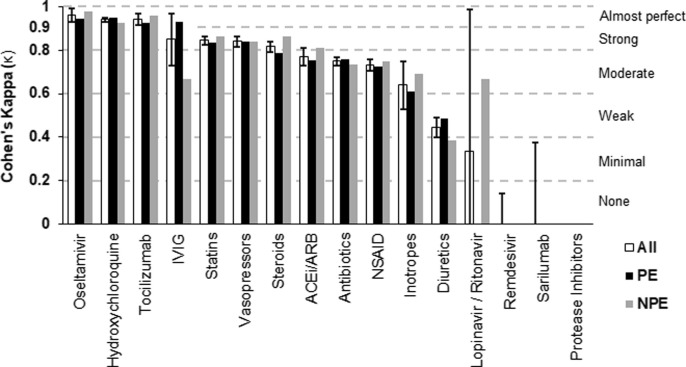
For inpatient medications, we compared data from 16 different medications or drug classes and report the resulting Cohen’s kappa (κ) values for each category in Fig. 2 . To see the data used to calculate κ values, see Appendix A, Table 2. Based on McHugh’s benchmark, agreement between manual abstraction and automated extraction was almost perfect for 3 (19%) inpatient medications, strong for 4 (25%), moderate for 4 (25%), weak for 1 (6%), and minimal or none for 4 (25%). The median κ for inpatient medications was 0.75.
Fig. 2.
Interrater reliability (Cohen’s kappa) between manual and automated methods of detecting inpatient medications. Data for each medication category are shown as “All” (includes all 4,123 patients; with 95% CIs overlaid), and then as two separate groups (PE: prior exposure to our health system; NPE: no prior exposure). ACEi/ARB: ACE Inhibitors and Angiotensin II Receptor blockers; NSAID: Non-steroidal anti-inflammatory drugs; IVIG: Intravenous Immunoglobulin.
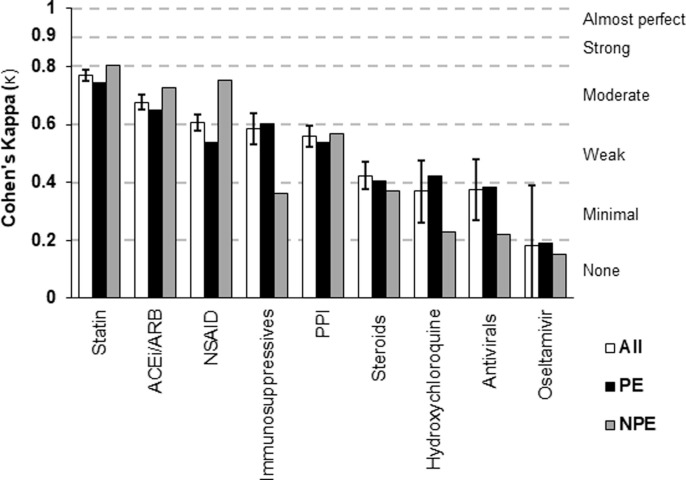
For outpatient medications, we compared data from 9 different medications or drug classes and report the resulting Cohen’s kappa (κ) values in Fig. 3 . To view the data used to calculate the κ values, see Appendix A, Table 3. Based on McHugh’s benchmark, agreement between manual abstraction and automated extraction was moderate for 3 (33%) outpatient medications, weak for 3 (33%), minimal for 2 (22%), and none for 1 (11%). The median κ for outpatient medications was 0.56.
Fig. 3.
Interrater reliability (Cohen’s kappa) between manual and automated methods of detecting outpatient medications. Data for each medication category are shown as “All” (includes all 4,123 patients; with 95% CIs overlaid), and then as two separate groups (PE: prior exposure to our health system; NPE: no prior exposure). ACEi/ARB: ACE Inhibitors and Angiotensin II Receptor Blockers; NSAID: Non-steroidal anti-inflammatory drugs; PPI: Proton Pump Inhibitor.
3.2. Classifying discrepancies
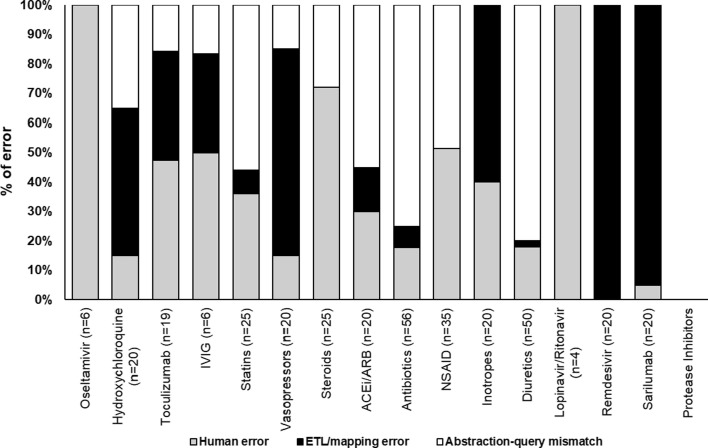
We audited 716 discrepancies, representing 12.37% of the 5,790 total discrepancies detected. For inpatient medications, we audited 13.2% (346/2,621) of all discrepancies and found that in 31% (107/346) the automated extraction was correct and there was human error in the manual abstraction, in 27% (94/346) ETL or mapping error led to error in the automated extraction, and in 42% (145/346) abstraction-query mismatch occurred where the logic in the automated extraction did not match that of the manual abstraction. Fig. 4 shows the breakdown of errors by individual inpatient medications and drug classes.
Fig. 4.
Proportion of inpatient errors as attributable to the 3 major categories of error: human error, ETL or mapping error, and abstraction-query mismatch. The number in parentheses represents the total number of discrepancies audited for a given medication. ACEi/ARB: ACE Inhibitors and Angiotensin II Receptor Blockers; NSAID: Non-steroidal anti-inflammatory drugs; IVIG: Intravenous Immunoglobulin.
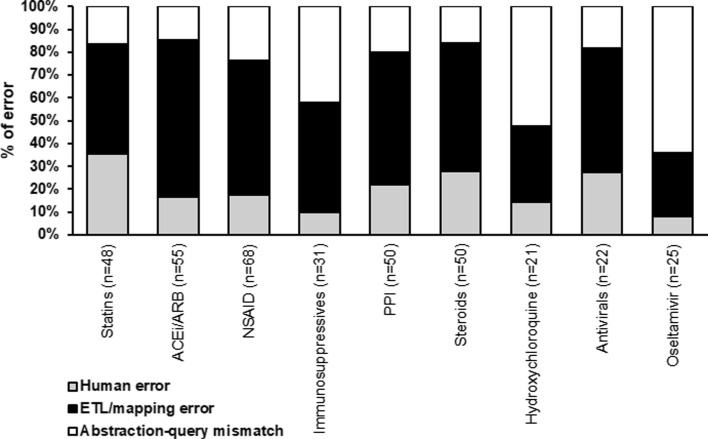
For outpatient medications, we audited 11.68% (370/3,169) of all discrepancies and found that 21% (77/370) were due to human error in the manual abstraction, 54% (199/370) to ETL or mapping error in the automated extraction, and 25% (94/370) to abstraction-query mismatch where errors could not be attributed to either manual abstraction or automated extraction. For a breakdown of errors by individual inpatient medications and drug classes, see Fig. 5 .
Fig. 5.
Proportion of outpatient errors as attributable to the 3 major categories of error: human error, ETL or mapping error, and abstraction-query mismatch. The number in parentheses represents the total number of discrepancies audited for a given medication. ACEi/ARB: ACE Inhibitors and Angiotensin II Receptor Blockers; NSAID: Non-steroidal anti-inflammatory drugs; PPI: Proton Pump Inhibitor.
In addition to identifying the 3 primary underlying causes of error (i.e. human, ETL/mapping, or abstraction-query mismatch), we identified 8 sub-categories to further characterize the observed discrepancies. The largest of these 8 causes were missing data (31%), predominantly in outpatient medication data; mismatches between the query and the data format (30%); and human errors in manual abstraction (26%), including both missed and inappropriately included information. These issues, along with descriptions and examples, are listed in Table 3.
4. Discussion
4.1. Findings
In a comparison of data collected through manual abstraction and automated extraction for COVID-19 patients at the height of the pandemic, we observed that automated extraction performed equal to manual abstraction for many inpatient medications and poorly for most outpatient medications. This suggests that future efforts to collect inpatient medication data need not rely on manual abstraction, allowing institutions to direct valuable human resources toward other needs.
For inpatient medications, 44% (7/16) medications or drug classes reached strong agreement or higher, many of which were the more prevalent medications (e.g. statins, hydrochlorothiazide, etc.). The 25% (4/16) of inpatient medications with minimal to no agreement (Cohen’s kappa < 0.4) were due to formulary related errors in the ETL/mapping of the automated extraction (remdesivir, sarilumab), or infrequent clinical usage (protease inhibitors, lopinavir/ritonavir) making kappa incalculable. Query related challenges in matching time-specific manual extraction instructions affected diuretics, NSAIDs, statins, antibiotics, and ACEi/ARBs. Of the 9 outpatient medications and drug classes, 33% (3/9) reached moderate agreement, while none achieved strong or almost perfect agreement. Based on our audit, these outcomes were driven by poor data quality in the EHR. Outpatient medications are not recorded with the same rigor of inpatient medications given the inability to truly confirm if a patient is taking a prescription. Additionally, outpatient medications often lack “end dates” in the EHR as medications commonly go unreconciled or have undefined order lengths. Thus, the query categorized these as “active” (the alternative being broad under-detection of home medications), even if the medication was neither included in admission documentation nor recorded by manual abstraction.
Interrater reliability was not consistently increased or decreased in patients with previous visits to our health system for inpatient or outpatient medications (i.e. patients with a previous visit did not consistently have a higher kappa value). This variation likely stems from a double-edged benefit: more data enriches the EHR, but entries of previous medications that have poor/missing documentation lead to erroneous detection.
In considering data sources for retrospective observational research, manual abstraction is often considered the gold standard. Although manual abstractors can navigate interface errors, read free text, and generally benefit from use of clinical judgement to interpret data, they are a limited resource and susceptible to human error [12], [13], [14]. Automated abstraction, while theoretically capable of flawlessly mirroring data in the EHR, is subject to several prominent issues impacting the utility and fitness of the data for secondary use [25], [26], [29], [30], [31], [32]. Because we observe errors in both methods, our findings suggest there is not a one-size-fits-all solution to generating research data sets using the EHR and that the conceptualization of “data quality” should be expanded in these contexts to better consider the provenance of the data in question and the nature of the downstream use case. Inpatient medications were thoroughly documented in the EHR, and automated extraction techniques performed well, suggesting manual efforts could target other areas such as outpatient medications or other domains requiring interpretation of context or setting such as provider notes. We believe that many observed issues could be improved in future work by designing studies to account for automated extraction logic. In Table 4 , we suggest ways to address common errors presented in Table 3.
Table 4.
Recommendations for avoiding discrepancies between manual and automated data extraction. Issues were generally categorized into one of 3 groups: human error, ETL or mapping error, and abstraction-query mismatch. Suggestions for future implementations are provided for each issue.
| Common Issues | Suggestions for improvement |
|---|---|
| Human Error | |
| Manual abstractors overlook desired information. | Provide instructions with as complete a list as possible of inclusion examples. Rapid extraction efforts limited the time available for data collectors to meticulously consider all possible responses. |
| Manual abstractors include inappropriate information. (false positive) | Explain complex drug classes, confusing content, and any foreseeable misconceptions (with examples), otherwise reported data will reflect this confusion. |
| ETL/Mapping Error | |
| Missing data | Increase use of health information exchanges (HIEs) and foster adoption of HIE data into research repositories. Encourage more robust data capture within the EHR (e.g. recording end dates of medication orders). Incorporate, where possible, medication fill data into research repositories. |
| Local errors | Actively surveil and address mapping issues by hardcoding placeholder reference terminology (e.g. RxNorm code 2284718) in ETL code for investigational agents (e.g. remdesivir). |
| Patient identifier inconsistency | Work with existing health information management teams within the clinical informatics domain to address observed issues in identity management. |
| Cross institutional differences | Ensure that differences between EHR systems at subsites of large hospital systems are properly addressed before incorporating their data. Implement “sanity checks” on mappings to identify errors before pushing new data. |
| Abstraction-Query Mismatch | |
| Mismatch between query and data format | Develop imputation logic for extending repeated standing orders into continuous drug exposure variable. Develop abstraction instructions that either consider the logic of automated processes or clarify inclusion criteria. |
| Complex instructions | Minimize conditional chart review instructions where possible and ensure contingent logic is necessary to yield clinically meaningful data. Minimize ambiguity by providing lists of acceptable responses and examples of chart review in ambiguous conditions. |
4.2. Relevance to previous work
While previous work on COVID-19 data has evaluated the quality of extraction for problem lists and natural language processing for signs and symptoms [50], [51], [52], [53], to our knowledge, our work is the first evaluating manual abstraction and automated extraction of EHR medication data relevant to COVID-19. Previous studies comparing manual abstraction and automated extraction have usually reviewed fewer patients or focused on theoretical principles [16], [18], [19], [20], [21], [22], [23], [24], [25]. To support future studies of this kind—particularly recent needs to create a widely-available data set of COVID-19 cases such as N3C—we hope this work provides a roadmap and highlights new variables eligible for automated extraction with high accuracy (i.e. inpatient medications), allowing valuable clinical resources needed for manual abstraction to be redirected toward other domains [47], [48], [49], [63]. Similarly, automated extraction methods as demonstrated here can be the foundation for more closely adhering to certain best practice methods for data quality standards and assessment [64], [65].
The current study demonstrates the viability of automated extraction of many inpatient medications or drug classes from the medical record, with outpatient medications showing weaker results. Institutions with large databases should be capable of employing similar methods for automated data extraction. Given the size and thorough auditing, this study demonstrates the importance of avoiding a ‘one-size-fits-all’ mentality in data extraction. Instead, it is important to be rigorous about data sources and transparent about the methods used, accepting that different errors will occur and different resources will be required in manual abstraction and automated extraction. It is also important to consider that refinement of automated techniques can systematically improve accuracy, while manual abstraction can face the challenges of staff turnover, training, and ongoing quality improvement. Lastly, automated approaches allow access to significant amounts of data for analysis. Manual data abstraction consisted of mainly binary variables to describe the parameters of each of the 4,414 patient visits, including 15 comorbidities and 25 medications. Over the same period of time, automated extraction holistically captured 3,997,956 diagnoses, 758,448 prescriptions, and 22,533,020 laboratory values from 2,036,243 EHR encounters, including data on a host of comorbidities and prescriptions not captured in the manual abstraction effort. As of April of 2021, over 318,000 patients have had automated data extracted, with 53 users executing approximately 26,000 queries.
4.3. Limitations
This study has certain limitations. Although the findings and implications of this work are more broadly applicable, both abstraction methods were tailored to our hospital system. Other hospital systems should create queries specific to their own data architecture. Second, although the audit process was extensive, the process focused on discrepancies rather than a complete random audit of all results. Certain errors may have existed among agreeing results (e.g. manual and automated approaches may have both detected a medication that a patient didn’t receive or vice versa). The Cohen’s kappa calculation is unaffected by this and there may be other errors that were not characterized. Third, systematic data quality issues may have affected certain patient populations, such as reduced clinical resources in certain hospitals during the peak of the crisis. Although extracting across 3 different hospitals is an overall strength of the study, it also introduces variation in the staff, procedures, and patient populations. Future work hopes to deliver similar analyses for other data points such as comorbidities, outcomes, etc. Future work will also explore some concepts that are outside of the scope of this current work, including both further pursuit of data quality improvement topics in Table 4 as well as better characterization of the timeline and context in which these errors in the data occur.
5. Conclusion
COVID-19 has changed the landscape of healthcare, creating opportunities to improve data infrastructures. The current study assessed agreement on outpatient and inpatient medication exposure between data collected through manual abstraction and automated extraction, exploring underlying causes of discrepancies and offering ways to avoid them. It demonstrates that automated collection of medication data is feasible and, for many inpatient medications, could save time and resources required for manual abstraction. As with many institutions during this pandemic, institutions must make tough decisions about where to allocate resources. This work outlines quality issues for institutions to be aware of and improvements that could be made.
Summary Table
Already known on this topic:
-
•
Data extraction from electronic health record (EHR) systems occurs through manual abstraction, automated extraction, or a combination of both, with each process having its strengths and weaknesses depending on setting and data type.
-
•
Prior investigations have compared manual to automated data collection techniques in conditions other than COVID-19, described informatics resources specific to COVID-19 and evaluated the performance of automated extraction in COVID-19, including problem lists and natural language processing to extract signs and symptoms.
What this study adds:
-
•
Automated extraction performed equal to manual abstraction for many inpatient medications and poorly for most outpatient medications, suggesting future efforts to collect inpatient medication data need not rely on manual abstraction and allowing institutions to direct valuable human resources toward other needs.
-
•
Both automated extraction and manual abstraction have strengths and weaknesses that must be considered in any data extraction effort. Institutions can now be more aware of the potential trade-offs and areas of improvement.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
The authors would like to acknowledge the large institutional efforts that enabled this work and the individuals who advised, shaped, and supported this manuscript.
Weill Cornell COVID-19 Data Abstraction Consortial Authors:
Zara Adamou, BAa, Bryan Ang, MDa, Elena Beideck, MDa, Orrin S. Belden, MD MBAa, Anthony F. Blackburn, MDa, Joshua Bliss, PharmD MDa, Kimberly A. Bogardus, MDa, Chelsea D. Boydstun, MDa, Clare A. Burchenal, MDa, Eric T. Caliendo, MDa, John K. Chae, BAa, David L. Chang, MDa, Frank R. Chen, MDa, Kenny Chen, MDa, Andrew Cho, MD PhDa, Alice Chung, MDa, Marcos A. Davila III, MCSb, Alisha N. Dua, M.Resa, Andrew Eidelberg, MDa, Rahmi S. Elahjji, MDa, Mahmoud Eljalby, MDa, Emily R. Eruysal, MDa, Kimberly N. Forlenza, MDa, Rana K. Fowlkes, MDa, Rachel L. Friedlander, MDa, Xiaobo Fuld, MSb, Gary F. George, BSa, Shannon M. Glynn, MDa, Leora Haber, MDa, Janice Havasy, BSa, Alex L. Huang, MDa, Hao Huang, BSa, Jennifer Huang, MDa, Sonia Iosima, MD, Mitali Kini, MDa, Rohini V. Kopparam, MDa, Jerry Y. Lee, BAa, Aretina K. Leung, MDa, Bethina Liu, MDa, Charalambia Louka, MDa, Brienne C. Lubor, MDa, Dianne Lumaquin, BSa, Matthew L. Magruder, MDa, Ruth Moges, MDa, Prithvi M. Mohan, MDa, Max F. Morin, MDa, Sophie Mou, MDa, Joel Jose Q. Nario, MDa, Yuna Oh, BSa, Emma M. Schatoff, MD PhDa, Pooja D. Shah, MDa, Sachin P. Shah, BAa, Daniel A. Skaf, MDa, Ahmed Toure, MDa, Camila M. Villasante, BAa, Gal Wald, MDa, Samuel C. Williams, BAa, Ashley Wu, MDa, Lisa Zhang, BAa.
aWeill Cornell Medical College, Weill Cornell Medicine, New York, NY.
bInformation Technologies & Services Department, Weill Cornell Medicine, New York, NY.
Author Contribution:
Please see Appendix D for details of contributions.
Funding Statement:
Research reported in this publication was supported by the National Center For Advancing Translational Sciences of the National Institutes of Health under Award Number UL1TR002384. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijmedinf.2021.104622.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Vassar M., Matthew H. The retrospective chart review: important methodological considerations. J. Educ. Eval. Health Prof. 2013;10:12. doi: 10.3352/jeehp.2013.10.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu L., Ashton C.M. Chart Review. Eval. Health Prof. 1997;20(2):146–163. doi: 10.1177/016327879702000203. [DOI] [PubMed] [Google Scholar]
- 3.Cassidy L.D., Marsh G.M., Holleran M.K., Ruhl L.S. Methodology to improve data quality from chart review in the managed care setting. Am. J. Manag. Care. 2002;8:787–793. http://www.ncbi.nlm.nih.gov/pubmed/12234019 [PubMed] [Google Scholar]
- 4.Engel L., Henderson C., Fergenbaum J., Colantonio A. Medical Record Review Conduction Model for Improving Interrater Reliability of Abstracting Medical-Related Information. Eval. Health Prof. 2009;32(3):281–298. doi: 10.1177/0163278709338561. [DOI] [PubMed] [Google Scholar]
- 5.Nahm M. Springer; London: 2012. Data Quality in Clinical Research, in; pp. 175–201. [Google Scholar]
- 6.Pan L., Fergusson D., Schweitzer I., Hebert P.C. Ensuring high accuracy of data abstracted from patient charts: the use of a standardized medical record as a training tool. J. Clin. Epidemiol. 2005;58:918–923. doi: 10.1016/j.jclinepi.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 7.Kreimeyer K., Foster M., Pandey A., Arya N., Halford G., Jones S.F., Forshee R., Walderhaug M., Botsis T. Natural language processing systems for capturing and standardizing unstructured clinical information: A systematic review. J. Biomed. Inform. 2017;73:14–29. doi: 10.1016/j.jbi.2017.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uzuner O., Bodnari A., Shen S., Forbush T., Pestian J., South B.R. Evaluating the state of the art in coreference resolution for electronic medical records. J. Am. Med. Informatics Assoc. 2012;19(5):786–791. doi: 10.1136/amiajnl-2011-000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uzuner Ö., South B.R., Shen S., DuVall S.L. i2b2/VA challenge on concepts, assertions, and relations in clinical text. J. Am. Med. Informatics Assoc. 2010;18(2011):552–556. doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koleck T.A., Dreisbach C., Bourne P.E., Bakken S. Natural language processing of symptoms documented in free-text narratives of electronic health records: a systematic review. J. Am. Med. Informatics Assoc. 2019;26:364–379. doi: 10.1093/jamia/ocy173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D. Shaywitz, The Deeply Human Core of Roche’s $2.1 Billion Tech Acquisition -- And Why It Made It, Forbes. (2018) 1–6. https://www.forbes.com/sites/davidshaywitz/2018/02/18/the-deeply-human-core-of-roches-2-1b-tech-acquisition-and-why-they-did-it/?sh=4964c72329c2 (accessed March 15, 2021).
- 12.Zozus M.N., Pieper C., Johnson C.M., Johnson T.R., Franklin A., Smith J., Zhang J. Factors Affecting Accuracy of Data Abstracted from Medical Records. PLoS One. 2015;10 doi: 10.1371/journal.pone.0138649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng J.E., Anoushiravani A.A., Tesoriero P.J., Ani L., Meftah M., Schwarzkopf R., Leucht P. Transcription Error Rates in Retrospective Chart Reviews. Orthopedics. 2020;43:e404–e408. doi: 10.3928/01477447-20200619-10. [DOI] [PubMed] [Google Scholar]
- 14.Nordo A.H., Eisenstein E.L., Hawley J., Vadakkeveedu S., Pressley M., Pennock J., Sanderson I. A comparative effectiveness study of eSource used for data capture for a clinical research registry. Int. J. Med. Inform. 2017;103:89–94. doi: 10.1016/j.ijmedinf.2017.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campion T.R., May A.K., Waitman L.R., Ozdas A., Gadd C.S. Effects of blood glucose transcription mismatches on a computer-based intensive insulin therapy protocol. Intensive Care Med. 2010;36(9):1566–1570. doi: 10.1007/s00134-010-1868-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brundin-Mather R., Soo A., Zuege D.J., Niven D.J., Fiest K., Doig C.J., Zygun D., Boyd J.M., Parsons Leigh J., Bagshaw S.M., Stelfox H.T. Secondary EMR data for quality improvement and research: A comparison of manual and electronic data collection from an integrated critical care electronic medical record system. J. Crit. Care. 2018;47:295–301. doi: 10.1016/j.jcrc.2018.07.021. [DOI] [PubMed] [Google Scholar]
- 17.Vawdrey D.K., Gardner R.M., Evans R.S., Orme J.F., Clemmer T.P., Greenway L., Drews F.A. Assessing Data Quality in Manual Entry of Ventilator Settings. J. Am. Med. Informatics Assoc. 2007;14(3):295–303. doi: 10.1197/jamia.M2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newgard C.D., Zive D., Jui J., Weathers C., Daya M. Electronic Versus Manual Data Processing: Evaluating the Use of Electronic Health Records in Out-of-hospital Clinical Research. Acad. Emerg. Med. 2012;19:217–227. doi: 10.1111/j.1553-2712.2011.01275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karanevich A.G., Weisbrod L.J., Jawdat O., Barohn R.J., Gajewski B.J., He J., Statland J.M. Using automated electronic medical record data extraction to model ALS survival and progression. BMC Neurol. 2018;18:205. doi: 10.1186/s12883-018-1208-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Byrne M.D., Jordan T.R., Welle T. Comparison of manual versus automated data collection method for an evidence-based nursing practice study. Appl. Clin. Inform. 2013;04(01):61–74. doi: 10.4338/ACI-2012-09-RA-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haerian K., Varn D., Vaidya S., Ena L., Chase H.S., Friedman C. Detection of Pharmacovigilance-Related Adverse Events Using Electronic Health Records and Automated Methods. Clin. Pharmacol. Ther. 2012;92(2):228–234. doi: 10.1038/clpt.2012.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tinoco A., Evans R.S., Staes C.J., Lloyd J.F., Rothschild J.M., Haug P.J. Comparison of computerized surveillance and manual chart review for adverse events. J. Am. Med. Informatics Assoc. 2011;18(4):491–497. doi: 10.1136/amiajnl-2011-000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knake L.A., Ahuja M., McDonald E.L., Ryckman K.K., Weathers N., Burstain T., Dagle J.M., Murray J.C., Nadkarni P. Quality of EHR data extractions for studies of preterm birth in a tertiary care center: guidelines for obtaining reliable data. BMC Pediatr. 2016;16:59. doi: 10.1186/s12887-016-0592-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.J.L. Warner, M.A. Levy, M.N. Neuss, J.L. Warner, M.A. Levy, M.N. Neuss, ReCAP: Feasibility and Accuracy of Extracting Cancer Stage Information From Narrative Electronic Health Record Data., J. Oncol. Pract. 12 (2016) 157–8; e169-7. 10.1200/JOP.2015.004622. [DOI] [PubMed]
- 25.Hersh W.R., Weiner M.G., Embi P.J., Logan J.R., Payne P.R.O., Bernstam E.V., Lehmann H.P., Hripcsak G., Hartzog T.H., Cimino J.J., Saltz J.H. Caveats for the use of operational electronic health record data in comparative effectiveness research. Med. Care. 2013;51:S30–S37. doi: 10.1097/MLR.0b013e31829b1dbd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.W.R. Hersh, J. Cimino, P.R.O. Payne, P. Embi, J. Logan, M. Weiner, E. V Bernstam, H. Lehmann, G. Hripcsak, T. Hartzog, J. Saltz, Recommendations for the use of operational electronic health record data in comparative effectiveness research., EGEMS (Washington, DC). 1 (2013) 1018. 10.13063/2327-9214.1018. [DOI] [PMC free article] [PubMed]
- 27.M.G. Kahn, T.J. Callahan, J. Barnard, A.E. Bauck, J. Brown, B.N. Davidson, H. Estiri, C. Goerg, E. Holve, S.G. Johnson, S.-T. Liaw, M. Hamilton-Lopez, D. Meeker, T.C. Ong, P. Ryan, N. Shang, N.G. Weiskopf, C. Weng, M.N. Zozus, L. Schilling, A Harmonized Data Quality Assessment Terminology and Framework for the Secondary Use of Electronic Health Record Data., EGEMS (Washington, DC). 4 (2016) 1244. 10.13063/2327-9214.1244. [DOI] [PMC free article] [PubMed]
- 28.Wei W.-Q., Leibson C.L., Ransom J.E., Kho A.N., Caraballo P.J., Chai H.S., Yawn B.P., Pacheco J.A., Chute C.G. Impact of data fragmentation across healthcare centers on the accuracy of a high-throughput clinical phenotyping algorithm for specifying subjects with type 2 diabetes mellitus. J. Am. Med. Informatics Assoc. 2012;19(2):219–224. doi: 10.1136/amiajnl-2011-000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Botsis T., Hartvigsen G., Chen F., Weng C. Secondary Use of EHR: Data Quality Issues and Informatics Opportunities. Summit on Translat. Bioinforma. 2010;2010:1–5. http://www.ncbi.nlm.nih.gov/pubmed/21347133 [PMC free article] [PubMed] [Google Scholar]
- 30.Hripcsak G., Albers D.J. Next-generation phenotyping of electronic health records. J. Am. Med. Inform. Assoc. 2013;20(1):117–121. doi: 10.1136/amiajnl-2012-001145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prokosch H.U., Ganslandt T. Perspectives for Medical Informatics, Methods. Inf. Med. 2009;48:38–44. doi: 10.3414/ME9132. [DOI] [PubMed] [Google Scholar]
- 32.Weiskopf N.G., Rusanov A., Weng C. Sick patients have more data: the non-random completeness of electronic health records. AMIA Annu. Symp. Proceedings. AMIA Symp. 2013;2013:1472–1477. http://www.ncbi.nlm.nih.gov/pubmed/24551421 [PMC free article] [PubMed] [Google Scholar]
- 33.Weiskopf N.G., Hripcsak G., Swaminathan S., Weng C. Defining and measuring completeness of electronic health records for secondary use. J. Biomed. Inform. 2013;46(5):830–836. doi: 10.1016/j.jbi.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weiskopf N.G., Cohen A.M., Hannan J., Jarmon T., Dorr D.A. Towards augmenting structured EHR data: a comparison of manual chart review and patient self-report. AMIA Annu. Symp. Proceedings. AMIA Symp. 2019;2019:903–912. http://www.ncbi.nlm.nih.gov/pubmed/32308887 [PMC free article] [PubMed] [Google Scholar]
- 35.Kern L.M., Malhotra S., Barrón Y., Quaresimo J., Dhopeshwarkar R., Pichardo M., Edwards A.M., Kaushal R. Accuracy of Electronically Reported “Meaningful Use” Clinical Quality Measures. Ann. Intern. Med. 2013;158:77. doi: 10.7326/0003-4819-158-2-201301150-00001. [DOI] [PubMed] [Google Scholar]
- 36.Goyal P., Choi J.J., Pinheiro L.C., Schenck E.J., Chen R., Jabri A., Satlin M.J., Campion T.R., Nahid M., Ringel J.B., Hoffman K.L., Alshak M.N., Li H.A., Wehmeyer G.T., Rajan M., Reshetnyak E., Hupert N., Horn E.M., Martinez F.J., Gulick R.M., Safford M.M. Clinical Characteristics of Covid-19 in New York City. N. Engl. J. Med. 2020;382(24):2372–2374. doi: 10.1056/NEJMc2010419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richardson S., Hirsch J.S., Narasimhan M., Crawford J.M., McGinn T., Davidson K.W., Barnaby D.P., Becker L.B., Chelico J.D., Cohen S.L., Cookingham J., Coppa K., Diefenbach M.A., Dominello A.J., Duer-Hefele J., Falzon L., Gitlin J., Hajizadeh N., Harvin T.G., Hirschwerk D.A., Kim E.J., Kozel Z.M., Marrast L.M., Mogavero J.N., Osorio G.A., Qiu M., Zanos T.P. Presenting Characteristics, Comorbidities, and Outcomes Among 5700 Patients Hospitalized With COVID-19 in the New York City Area. JAMA. 2020;323:2052. doi: 10.1001/jama.2020.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hsu H., Greenwald P.W., Laghezza M.R., Steel P., Trepp R., Sharma R. Clinical informatics during the COVID-19 pandemic: Lessons learned and implications for emergency department and inpatient operations. J. Am. Med. Informatics Assoc. 2021;28:879–889. doi: 10.1093/jamia/ocaa311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Helmer T.T., Lewis A.A., McEver M., Delacqua F., Pastern C.L., Kennedy N., Edwards T.L., Woodward B.O., Harris P.A. Creating and implementing a COVID-19 recruitment Data Mart. J. Biomed. Inform. 2021;117 doi: 10.1016/j.jbi.2021.103765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Y. Huang, X. Li, G.-Q. Zhang, ELII: A Novel Inverted Index for Fast Temporal Query, with Application to a Large Covid-19 EHR Dataset, J. Biomed. Inform. (2021) 103744. 10.1016/j.jbi.2021.103744. [DOI] [PMC free article] [PubMed]
- 41.Y. Du, L. Tu, P. Zhu, M. Mu, R. Wang, P. Yang, X. Wang, C. Hu, R. Ping, P. Hu, T. Li, F. Cao, C. Chang, Q. Hu, Y. Jin, G. Xu, Clinical Features of 85 Fatal Cases of COVID-19 from Wuhan. A Retrospective Observational Study., Am. J. Respir. Crit. Care Med. 201 (2020) 1372–1379. 10.1164/rccm.202003-0543OC. [DOI] [PMC free article] [PubMed]
- 42.Li Y., Li M., Wang M., Zhou Y., Chang J., Xian Y., Wang D., Mao L., Jin H., Hu B. Acute cerebrovascular disease following COVID-19: a single center, retrospective, observational study. Stroke Vasc. Neurol. 2020;5:279–284. doi: 10.1136/svn-2020-000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., Guan L., Wei Y., Li H., Wu X., Xu J., Tu S., Zhang Y., Chen H., Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang X., Yu Y., Xu J., Shu H., Xia J., Liu H., Wu Y., Zhang L., Yu Z., Fang M., Yu T., Wang Y., Pan S., Zou X., Yuan S., Shang Y. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Price-Haywood Eboni G., Burton Jeffrey, Fort Daniel, Seoane Leonardo. Hospitalization and Mortality among Black Patients and White Patients with Covid-19. N. Engl. J. Med. 2020;382(26):2534–2543. doi: 10.1056/NEJMsa2011686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosenberg E.S., Dufort E.M., Udo T., Wilberschied L.A., Kumar J., Tesoriero J., Weinberg P., Kirkwood J., Muse A., DeHovitz J., Blog D.S., Hutton B., Holtgrave D.R., Zucker H.A. Association of Treatment With Hydroxychloroquine or Azithromycin With In-Hospital Mortality in Patients With COVID-19 in New York State. JAMA. 2020;323:2493. doi: 10.1001/jama.2020.8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reeves J.J., Hollandsworth H.M., Torriani F.J., Taplitz R., Abeles S., Tai-Seale M., Millen M., Clay B.J., Longhurst C.A. Rapid response to COVID-19: health informatics support for outbreak management in an academic health system. J. Am. Med. Informatics Assoc. 2020;27:853–859. doi: 10.1093/jamia/ocaa037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stefan M.S., Salvador D., Lagu T.K. Pandemic as a Catalyst for Rapid Implementation. Am. J. Med. Qual. 2021;36:60–62. doi: 10.1177/1062860620961171. [DOI] [PubMed] [Google Scholar]
- 49.Madhavan S., Bastarache L., Brown J.S., Butte A.J., Dorr D.A., Embi P.J., Friedman C.P., Johnson K.B., Moore J.H., Kohane I.S., Payne P.R.O., Tenenbaum J.D., Weiner M.G., Wilcox A.B., Ohno-Machado L. Use of electronic health records to support a public health response to the COVID-19 pandemic in the United States: a perspective from 15 academic medical centers. J. Am. Med. Informatics Assoc. 2021;28:393–401. doi: 10.1093/jamia/ocaa287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Poulos J., Zhu L., Shah A.D. Data gaps in electronic health record (EHR) systems: An audit of problem list completeness during the COVID-19 pandemic. Int. J. Med. Inform. 2021;150 doi: 10.1016/j.ijmedinf.2021.104452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang J., Abu-el-Rub N., Gray J., Pham H.A., Zhou Y., Manion F.J., Liu M., Song X., Xu H., Rouhizadeh M., Zhang Y. COVID-19 SignSym: a fast adaptation of a general clinical NLP tool to identify and normalize COVID-19 signs and symptoms to OMOP common data model. J. Am. Med. Informatics Assoc. 2021 doi: 10.1093/jamia/ocab015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lybarger K., Ostendorf M., Thompson M., Yetisgen M. Extracting COVID-19 diagnoses and symptoms from clinical text: A new annotated corpus and neural event extraction framework. J. Biomed. Inform. 2021;117 doi: 10.1016/j.jbi.2021.103761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao J., Grabowska M.E., Kerchberger V.E., Smith J.C., Eken H.N., Feng Q., Peterson J.F., Trent Rosenbloom S., Johnson K.B., Wei W.-Q. ConceptWAS: A high-throughput method for early identification of COVID-19 presenting symptoms and characteristics from clinical notes. J. Biomed Inform. 2021;117 doi: 10.1016/j.jbi.2021.103748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reynolds Harmony R., Adhikari Samrachana, Pulgarin Claudia, Troxel Andrea B., Iturrate Eduardo, Johnson Stephen B., Hausvater Anaïs, Newman Jonathan D., Berger Jeffrey S., Bangalore Sripal, Katz Stuart D., Fishman Glenn I., Kunichoff Dennis, Chen Yu, Ogedegbe Gbenga, Hochman Judith S. Renin–Angiotensin–Aldosterone System Inhibitors and Risk of Covid-19. N. Engl. J. Med. 2020;382(25):2441–2448. doi: 10.1056/NEJMoa2008975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bakken S. Biomedical and health informatics approaches remain essential for addressing the COVID-19 pandemic. J. Am. Med. Informatics Assoc. 2021;28:425–426. doi: 10.1093/jamia/ocab007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sholle E.T., Kabariti J., Johnson S.B., Leonard J.P., Pathak J., Varughese V.I., Cole C.L., Campion T.R. Secondary Use of Patients’ Electronic Records (SUPER): An Approach for Meeting Specific Data Needs of Clinical and Translational Researchers. AMIA Annu. Symp. Proceedings. AMIA Symp. 2017;2017:1581–1588. http://www.ncbi.nlm.nih.gov/pubmed/29854228 [PMC free article] [PubMed] [Google Scholar]
- 57.Harris Paul A., Taylor Robert, Thielke Robert, Payne Jonathon, Gonzalez Nathaniel, Conde Jose G. Research electronic data capture (REDCap)—A metadata-driven methodology and workflow process for providing translational research informatics support. J. Biomed. Inform. 2009;42(2):377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alshak M.N., Li H.A., Wehmeyer G.T. Medical Students as Essential Frontline Researchers During the COVID-19 Pandemic. Acad. Med. Publish Ah. 2021 doi: 10.1097/ACM.0000000000004056. [DOI] [PubMed] [Google Scholar]
- 59.Fleurence R.L., Curtis L.H., Califf R.M., Platt R., Selby J.V., Brown J.S. Launching PCORnet, a national patient-centered clinical research network. J. Am. Med. Informatics Assoc. 2014;21(4):578–582. doi: 10.1136/amiajnl-2014-002747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stang P.E., Ryan P.B., Racoosin J.A., Overhage J.M., Hartzema A.G., Reich C., Welebob E., Scarnecchia T., Woodcock J. Advancing the Science for Active Surveillance: Rationale and Design for the Observational Medical Outcomes Partnership. Ann. Intern. Med. 2010;153:600. doi: 10.7326/0003-4819-153-9-201011020-00010. [DOI] [PubMed] [Google Scholar]
- 61.McHugh M.L. Interrater reliability: the kappa statistic., Biochem. Medica. 2012;22:276–282. http://www.ncbi.nlm.nih.gov/pubmed/23092060 [PMC free article] [PubMed] [Google Scholar]
- 62.Flight Laura, Julious Steven A. The disagreeable behaviour of the kappa statistic. Pharm. Stat. 2015;14(1):74–78. doi: 10.1002/pst.1659. [DOI] [PubMed] [Google Scholar]
- 63.M.A. Haendel, C.G. Chute, T.D. Bennett, D.A. Eichmann, J. Guinney, W.A. Kibbe, P.R.O. Payne, E.R. Pfaff, P.N. Robinson, J.H. Saltz, H. Spratt, C. Suver, J. Wilbanks, A.B. Wilcox, A.E. Williams, C. Wu, C. Blacketer, R.L. Bradford, J.J. Cimino, M. Clark, E.W. Colmenares, P.A. Francis, D. Gabriel, A. Graves, R. Hemadri, S.S. Hong, G. Hripscak, D. Jiao, J.G. Klann, K. Kostka, A.M. Lee, H.P. Lehmann, L. Lingrey, R.T. Miller, M. Morris, S.N. Murphy, K. Natarajan, M.B. Palchuk, U. Sheikh, H. Solbrig, S. Visweswaran, A. Walden, K.M. Walters, G.M. Weber, X.T. Zhang, R.L. Zhu, B. Amor, A.T. Girvin, A. Manna, N. Qureshi, M.G. Kurilla, S.G. Michael, L.M. Portilla, J.L. Rutter, C.P. Austin, K.R. Gersing, The National COVID Cohort Collaborative (N3C): Rationale, design, infrastructure, and deployment, J. Am. Med. Informatics Assoc. 28 (2021) 427–443. 10.1093/jamia/ocaa196. [DOI] [PMC free article] [PubMed]
- 64.Weiskopf N.G., Weng C. Methods and dimensions of electronic health record data quality assessment: enabling reuse for clinical research. J. Am. Med. Inform. Assoc. 2013;20(1):144–151. doi: 10.1136/amiajnl-2011-000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.N.G. Weiskopf, S. Bakken, G. Hripcsak, C. Weng, A Data Quality Assessment Guideline for Electronic Health Record Data Reuse., EGEMS (Washington, DC). 5 (2017) 14. 10.5334/egems.218. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.