Abstract
MBD1 belongs to a family of mammalian proteins that share a methyl-CpG binding domain. Previous work has shown that MBD1 binds to methylated sites in vivo and in vitro and can repress transcription from methylated templates in transcription extracts and in cultured cells. In the present study we established by several experimental criteria that, contrary to a previous report, MBD1 is not a component of the MeCP1 repressor complex. We identified a powerful transcriptional repression domain (TRD) at the C terminus of MBD1 that can actively repress transcription at a distance. Methylation-dependent repression in vivo depends on the presence of both the TRD and the methyl-CpG binding domain. The mechanism is likely to involve deacetylation, since the deacetylase inhibitor trichostatin A can overcome MBD1-mediated repression. Accordingly, we found that endogenous MBD1 is particularly concentrated at sites of centromeric heterochromatin, where acetylated histone H4 is deficient. Unlike MBD2 and MeCP2, MBD1 is not depleted by antibodies to the histone deacetylase HDAC1. Thus, the deacetylase-dependent pathway by which MBD1 actively silences methylated genes is likely to be different from that utilized by the methylation-dependent repressors MeCP1 and MeCP2.
The mammalian genome is characterized by methylation-free CpG islands interspersed within the methylated bulk chromatin. The great majority of CpGs (about 85% of those outside CpG islands) are in a methylated state in most somatic cell types. The methylated sequences include exons of most genes, as well as intergenic DNA made up of satellites, transposable elements, and single-copy sequences. An established consequence of CpG methylation is transcriptional silencing (reviewed in references 36 and 40). This is most apparent at densely methylated CpG island promoters on the inactive X chromosome (31), at the Xist promoter on the active X chromosome (2, 38) and at many imprinted genes (41, 43). Whether the low-density methylation that affects the bulk of genomic DNA also serves to repress transcription is uncertain (4, 6, 46).
One way of understanding the biological significance of DNA methylation is to characterize fully the mechanisms by which CpG methylation leads to transcriptional silencing. It is known already that the presence of methylated CpG can interfere with binding of some transcription factors to their cognate recognition sites (reviewed in reference 42). Also, DNA methylation can directly influence the translational positioning of a nucleosome at specific DNA sequences in vitro (13) and could lead to masking of essential regulatory elements by nucleosomes. In addition to these direct mechanisms of repression, there is now evidence for indirect repression mechanisms that are mediated by proteins that bind to methylated DNA. Two methylated DNA binding activities have been detected in mammalian cells and shown to repress transcription. MeCP1 (methyl-CpG binding protein 1) exists as large complexes of 400 and 800 kDa and can bind to 12 or more symmetrically methylated CpGs in any sequence context (29). The extent of repression on methylated reporters in living cells correlates strongly with the density of methylation and the in vitro affinity of MeCP1 for the methylated reporters (7, 8). Moreover, cells and extracts deficient in MeCP1 failed to repress methylated reporters (7, 26), further implicating MeCP1 as a methylation-dependent transcriptional repressor.
MeCP2 is distinct from MeCP1 but also has an affinity for methylated DNA. MeCP2 is a chromosomal protein that localizes to methyl-CpG-rich heterochromatin in mouse cells (27, 35). It differs from MeCP1 in that it can bind to a single methylated CpG pair (27, 33) via an 85-amino-acid methyl-CpG binding domain (33) which can bind specifically to a single methyl-CpG pair in vitro. The methyl-CpG binding domain of MeCP2 is required for methyl-CpG binding in vivo, since deletion within this region of an MeCP2-LacZ fusion protein leads to inefficient targeting of the mutant protein to densely methylated heterochromatic foci in mouse cells (35). MeCP2 also interacts with methylated chromatin in vitro (32) and can bind specific methyl-CpG pairs on nucleosomal DNA in a defined system (10). Thus, MeCP2 is an integral component of chromatin that has abundant binding sites in the mammalian genome (32). The likely functional significance of MeCP2 has been revealed by transient cotransfection studies. Fragments of MeCP2 fused to the GAL4 DNA binding domain cause transcriptional silencing of a reporter gene with GAL4 binding sites upstream of the promoter (32). Based on these experiments, a region central to MeCP2 has been defined as the transcriptional repression domain (TRD). This domain can interact with the mSin3A, HDAC1, and HDAC2 corepressor proteins in a glutathione S-transferase (GST) pulldown assay (34). In addition, coimmunoprecipitation assays and biochemical cofractionation assays indicate that MeCP2 is associated with the mSin3 corepressor complex (25, 34). Most importantly, the TRD of both mouse and Xenopus laevis MeCP2 can repress transcription in vivo, and the repression is sensitive to the histone deacetylase inhibitor trichostatin A (TSA). MeCP2 therefore appears to repress transcription by recruiting a histone deacetylase complex that modifies chromatin.
An expressed sequence tag database searching strategy was used to find novel proteins with homology to the methyl-CpG binding domain of MeCP2 (12, 20). The rationale was that this domain is likely to be structurally conserved for recognition of methyl-CpG pairs. A family of novel mammalian proteins with homology to the methyl-CpG binding domain (MBD1 to MBD4) has been identified using this approach. Using bandshift assays, MBD1, MBD2, and MBD4 were shown to bind specifically to a variety of DNA sequences that contained methyl-CpG. Ectopic expression of MBD1, MBD2, and MBD4 in mouse cells resulted in the localization of each protein to 4′,6-diamidino-2-methylindole (DAPI) bright spots, suggesting that they are able to specifically target to methylated DNA in vivo (20). Ectopically expressed MBD2 and MBD4 failed to localize correctly in mouse mutant cells with low levels of genomic methylation, indicating that methylation is an essential determinant for chromosome binding. By the same assay, MBD1 was correctly localized in mutant cells, suggesting that protein factors may also be involved in its localization in vivo. Although MBD2 and MBD3 are related, as revealed by the high level of sequence conservation at the C-terminal region (including the methyl-CpG binding domain), mammalian MBD3 does not bind to methylated DNA specifically either in vitro or in vivo (20, 50). Therefore, among the mammalian methylated DNA binding activities and proteins so far identified, MeCP1, MeCP2, MBD1, MBD2, and MBD4 show specificity for binding CpG only in its methylated state. All but one of these proteins (the exception being MBD4) are now implicated in methylation-directed gene repression (5).
This study concerns MBD1, the largest protein in the MBD family, with an N-terminal methyl-CpG binding domain and two or three cysteine-rich regions (termed the CxxC motifs) that are related to those in DNA methyltransferase protein 1 (DNMT1) and the mammalian trithorax-like protein HRX (see Fig. 1A). A region encompassing the CxxC motif in DNMT1 binds zinc (3, 11). HRX, also known as MLL (mixed-lineage leukemia) or ALL1 (acute lymphoblastic leukemia), is frequently translocated in acute leukemias. A previous study showed that MBD1 is able to repress transcription of a methylated gene in vitro (12). This work indicated that MBD1 is a component of the MeCP1 complex in HeLa cells, since an antibody raised against MBD1 was able to supershift MeCP1 activity (12). Recently it has been shown that another methyl-CpG binding protein, MBD2, is a component of HeLa MeCP1 (37). We report here on functional studies of MBD1. We show that MBD1 can actively repress methylated genes in vivo via a powerful repression domain that depends on deacetylation. Our data rule out an association between MBD1 and MeCP1 of HeLa cells. Like MeCP2, MBD1 appears to be an integral chromosomal protein.
FIG. 1.
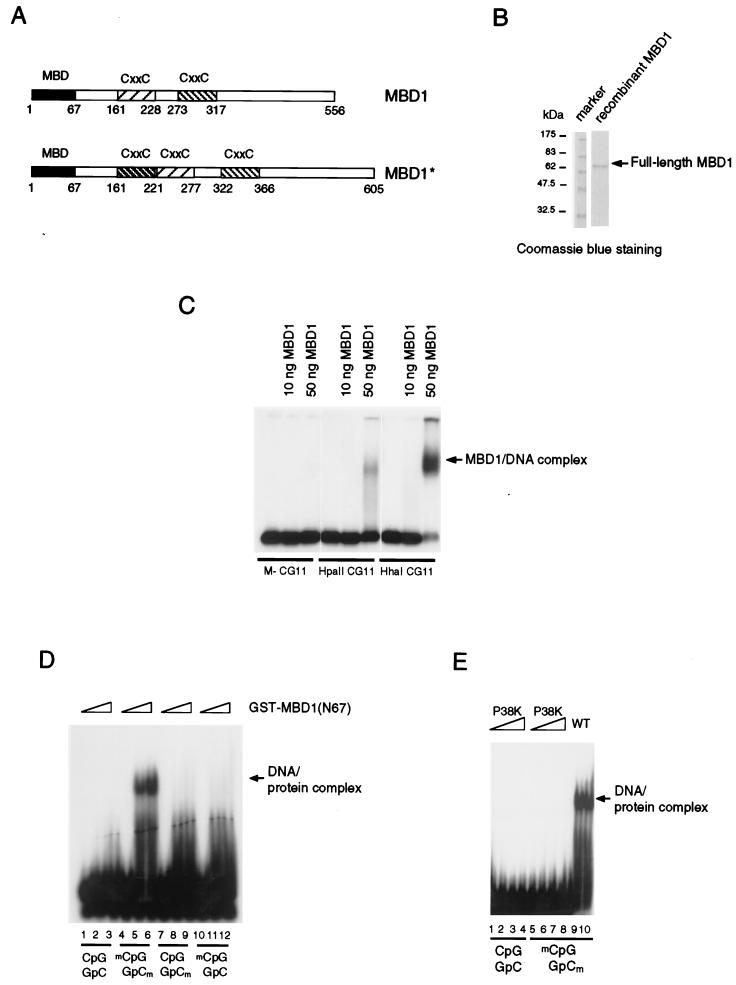
Binding to CpG methylated DNA by the methyl-CpG binding domain of MBD1. (A) Two forms of MBD1 that can be expressed by the human gene (19): MBD1 (accession no. Y10746) and MBD1* (accession no. AAD50371). Both have an N-terminal methyl-CpG binding domain (black box) and two or three CxxC domains (shaded boxes). (B) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of recombinant full-length MBD1 stained with Coomassie blue. (C) Bandshift assays of full-length MBD1 in the presence of differentially methylated CG11 probes. Probes were either nonmethylated (M−), or methylated at 7 HpaII sites (HpaII CG11) or 20 HhaI sites (HhaI CG11) and labeled with 32P. (D) A 67-amino-acid region of the methyl-CpG binding domain fused to GST (N67) binds to symmetrically methylated CpGs but not to nonmethylated or hemimethylated CpGs. (E) Amino acid substitution of the conserved proline 38 residue (P38K) abolishes methyl-CpG binding. WT, wild type.
MATERIALS AND METHODS
Protein expression, production of antibody, immunodepletion assays and Western blotting.
Recombinant proteins were expressed in Escherichia coli as GST fusion or His-tagged proteins. They were purified using glutathione (GSH)-Sepharose (Pharmacia) or Ni-nitrilotriacetic acid superflow (Qiagen) as specified by the manufacturer. Recombinant full-length MBD1 has a C-terminal His tag. Antibodies against MBD1 (S751) were raised against affinity purified GST-MBD1 fusions containing amino acids 351 to 556 of MBD1. For affinity purification of antibody, GST-MBD1 fusion proteins were coupled to Affi-Gel 15 activated matrix (Bio-Rad) as specified by the manufacturer and antibody was purified as described previously (18). The anti-MBD1 serum was passed through a GST-GSH-Sepharose column before being loaded onto a GST-MBD1 column to remove anti-GST antibodies. Immunodepletion experiments and Western blotting were carried out as described previously (37). Anti-MBD2 antibodies (S923 and R593) have been described previously (37). Anti-MTA2, anti-SAP30 and anti-HDAC1 antibodies were a gift from D. Reinberg (50, 51).
Plasmids.
DNA fragments encoding different regions of MBD1 were amplified by PCR with Pfu DNA polymerase (Stratagene). All primers contain either a BglII or BamHI site. After the PCR products were cleaved with appropriate restriction enzymes, they were cloned into pCMV-GAL4 BamHI-digested vector. The cDNA coding for the first 67 amino acids of MBD1 was cloned into the EcoRI site of the pGEX-2TK (Pharmacia) vector. Mutagenesis of the MBD and TRD was carried out by PCR-directed mutagenesis with primers that contained different codons. Full-length MBD1 and fragments thereof were cloned into pCS2+MT vector (which has a cytomegalovirus [CMV] promoter driving MBD1 expression). The resulting constructs expressed proteins with five Myc tags at their N termini. The sequence of the third CxxC domain in MBD1 was isolated by PCR of a HeLa cDNA library with primers that prime bp 486 to 498, encoding the amino acid sequence IAFN (see the published sequence of human MBD1 [12]) and bp 600 to 612, encoding GLRR. The CxxC domain of mouse DNMT1 was amplified with primers corresponding to amino acids 639 and 705 (EKYD and DDDE, respectively [48]). All constructs were verified by DNA sequencing.
Gel filtration chromatography.
HeLa nuclear extract (2.5 mg) prepared by the Dignam method was passed through Superose 6 gel filtration column (HR 10/30; Pharmacia) under conditions described previously (29). The column was calibrated with standard molecular mass markers (blue dextran, 2,000 kDa; thyroglobulin, 669 kDa; apoferritin, 443 kDa; β-amylase, 200 kDa; bovine serum albumin, 66 kDa) under the same conditions for chromotography of nuclear extract (0.15 ml/min). The buffer used was a 50 mM HEPES-KOH (pH 7.9)–250 mM NaCl–1 mM EDTA–0.1% Triton X-100–10% glycerol–10 mM β-mercaptoethanol.
Transfection.
Mouse fibroblast L929 cells were transfected using the DEAE-dextran method as described previously (32). L cells were transfected with 2 μg of reporter plasmid and different amounts of effector plasmids. The total amount of plasmid was normalized to 6 μg by addition of pCMV vector as the carrier. The cells were harvested 60 h after transfection, and β-galactosidase assays were performed as described previously (32). In certain experiments, the transfected cells were treated with 100 ng of TSA per ml for 24 h before harvest. pGL2 luciferase reporter (Promega) was either mock methylated (no enzyme added) or M.HhaI (New England Biolabs) methylated. Complete methylation was checked by restriction digestion with HhaI (New England Biolabs).
Bandshifts.
Bandshift assays were performed as described previously (33) but without addition of unlabeled competitor DNA. The DNA-protein complexes were resolved in a 5% polyacrylamide gel in 0.5× TBE (Tris-borate-EDTA) buffer at 4°C. The CG11 and AB17 probes have been described previously (29, 33).
Immunofluorescence.
Except for minor modifications noted below, double indirect immunolabelling of unfixed metaphase cell preparations of HF19 primary human female fibroblasts was carried out essentially as described previously (22, 24). To the hypotonically swollen HF19 cell suspension, diluted to 0.5 × 105 cells per ml in 75 mM KCl, Tween 20 was added to a final concentration of 0.1%, and then 0.5-ml portions were centrifuged onto glass slides for 10 min at 2,000 rpm by using an Ames Cytotek cytocentrifuge. Slides were incubated for 2 h at room temperature simultaneously with both primary antibodies, sheep anti-MBD1 serum and rabbit anti-acetyl(Lys-12)-histone H4 serum R5/12 (44), diluted 1:200 and 1:100, respectively, in KCM (120 mM KCl, 20 mM NaCl, 10 mM Tris-HCl [pH 7.8], 0.5 mM EDTA, 0.1% Triton X-100) containing 10% normal horse serum. Primary antibody binding was detected by a secondary incubation for 30 min at room temperature with biotin-conjugated donkey anti-sheep immunoglobulin G (Sigma) and fluorescein-isothiocyanate-conjugated donkey anti-rabbit immunoglobulin G (Scottish Antibody Production Unit), both diluted 1:50 in KCM containing 10% horse serum, followed by 30 min at room temperature with tetramethylrhodamine-S-isothiocyanate conjugated Extravidin (Sigma) diluted 1:50 in the same medium. Slides were rinsed after antibody incubations, fixed in formaldehyde, counterstained for DNA with Hoechst 33258, and mounted for fluorescence microscopy by previously described methods. Images were obtained with a Zeiss Axioskop fluorescence microscope fitted with Chroma 83000 triple-band-pass filter set, and a Photometrics CH250 charge-coupled device camera, using Digital Scientific capture and analysis extensions to IPLab Spectrum v3.1 software.
RESULTS
The N-terminal domain of MBD1 is sufficient for binding to methylated DNA.
MBD1 was previously expressed in bacteria and shown to bind to the methylated sequence poly(GAm5C)-poly(GTm5C) but not to its nonmethylated counterpart. Figure 1C shows that full-length MBD1 (Fig. 1B) binds specifically to the 135-bp CG11 duplex probe when methylated at 7 HpaII sites (CCGG) or 20 HhaI sites (GCGC) but not to the unmethylated probe. Since these assays were carried out in the absence of any nonradioactive competitor DNA, it is apparent that MBD1 has no detectable affinity for nonmethylated DNA. The methyl-CpG binding domain-like region in MBD1 (amino acids 7 to 61 [Fig. 1A]) has 43.6% identity to the equivalent region of MeCP2 over a 55-amino-acid sequence. We found that a polypeptide corresponding to the first 67 amino acids of MBD1 fused to GST bound specifically to the 17-mer methylated probe (Fig. 1D) but not to the control nonmethylated probe (probe AB17 [33]). Hemimethylated versions of probes with the same sequence were not bound by the MBD (Fig. 1D). The region of MBD1 that is required for specific interaction with the methyl-CpG pair is therefore smaller than the 85-amino-acid minimal methyl-CpG binding domain of MeCP2 (33). Proline at position 38 is conserved between the MBD of MeCP2 and the MBD-like region of each of the methylated DNA binding proteins (MBD1, MBD2, and MBD4) (20). By using PCR-directed mutagenesis, the proline codon was mutated to encode lysine. The purified GST-MBD1 (amino acids 1 to 67) with this amino acid substitution failed to bind to a methyl-CpG pair as demonstrated in a bandshift assay (Fig. 1E). The result confirms that the N-terminal 67-amino-acid region of MBD1 is responsible for binding to methyl-CpG.
Characterization of an antibody against MBD1.
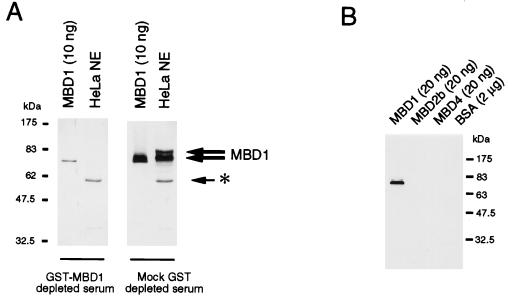
For functional studies of MBD1, an antibody was raised in sheep against a fusion between amino acids 351 to 556 of MBD1 and GST. The C-terminal domain of MBD1 (amino acids 351 to 556) was chosen for the antigen to minimize cross-reactivity, since it does not have significant homology to other MBDs or known proteins. When a Western blot of HeLa nuclear extracts was probed with this antibody (S751), two major bands at 83 and 75 kDa were detected (Fig. 2A). The 75-kDa band has the same mobility as recombinant MBD1 protein, which migrates on sodium dodecyl sulfate-polyacrylamide gel electrophoresis with an apparent size that is larger than its calculated size. Two lines of evidence indicate that the 83- and 75-kDa bands are specific for MBD1. First, the preimmune serum did not detect these proteins (data not shown). Second, the specific antibodies in the immune antiserum can be depleted by preincubation with GST-MBD1 coupled to GSH-Sepharose beads (Fig. 2A). GST alone attached to GSH-Sepharose beads did not deplete the MBD1-specific signal. The two proteins recognized by the antibody are probably splice variants of MBD1 that contain two or three CxxC motifs, since cDNAs encoding two or three CxxC motifs have been found in the mouse (20) and can also be detected in HeLa cells by reverse transcription-PCR (data not shown). The additional CxxC motif of about 68 amino acids is expected to cause an increase of 5 kDa. The S751 immune antiserum is specific for MBD1 and does not cross-react with MBD2b or MBD4 (Fig. 2B). To test for its ability to recognize native MBD1, S751 anti-MBD1 antiserum, together with control preimmune serum, was used to immunoprecipitate nuclear proteins from HeLa nuclear extracts. The presence of MBD1 in the anti-MBD1 immunoprecipitate (Fig. 3A) indicates that the antibody can recognize native MBD1.
FIG. 2.
Characterization of an antibody raised against the C terminus of MBD1. (A) Anti-MBD1 antibody detects two major forms of MBD1 in HeLa nuclear extract (thick arrows; right panel). Adsorption of the antibody with the cognate antigen (GST-MBD1) drastically reduces the reaction of the serum with recombinant MBD1 and prevents the detection of MBD1 in a HeLa nuclear extract (left panel). Mock adsorption with GST does not affect the reactivity of the serum toward MBD1. The asterisk denotes a nonspecific band due to secondary antibody. HeLa NE denotes 30 μg of HeLa nuclear extract. (B) Anti-MBD1 antibody recognizes MBD1 but not MBD2b, MBD4, or bovine serum albumin.
FIG. 3.
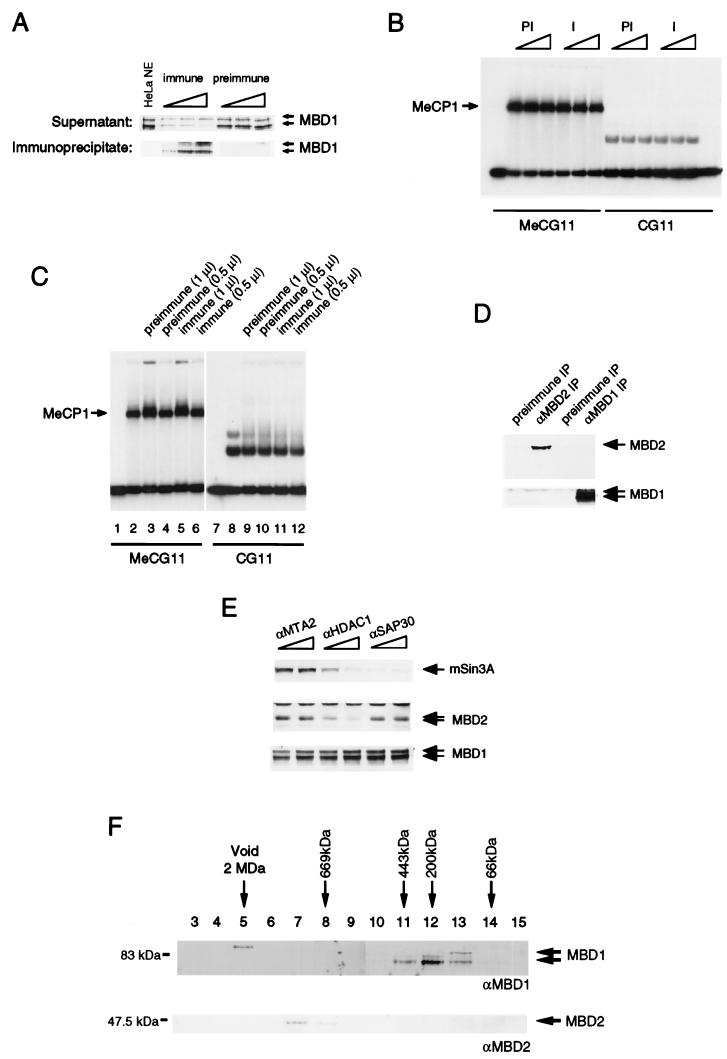
MBD1 is not a component of HeLa MeCP1 activity. (A) Extracts subjected to adsorption by anti-MBD1 antibodies (immune) contained reduced levels of MBD1, whereas preimmune antibodies did not deplete MBD1. Only immunoprecipitates by anti-MBD1 antibodies contained MBD1. We used 1, 5, and 10 μl of sera for the experiments. (B) Both MBD1-immunodepleted extracts (I) and mock-depleted extracts (PI) retained MeCP1 activity, as assayed by the formation of a complex with methylated probe DNA. MeCG11 is the methylated version of CG11. (C) Antibody against MBD1 did not supershift MeCP1 activity. Serum was not added in lanes 2 and 8. Lanes 3, 5, 9, and 11 contain 1 μl of preimmune or immune serum. Half the amount of serum was used in lanes 4, 6, 10, and 12. The complexes formed with the nonmethylated probe are nonspecific complexes not due to MeCP1 (7). (D) MBD1 and MBD2 are not associated in the same complex. Anti-MBD1 antibodies did not immunoprecipitate MBD2, and anti-MBD2 antibodies did not immunoprecipitate MBD1. (E) MBD1 is not associated with the MBD2-HDAC1 MeCP1 complex. Anti-HDAC1 antibodies immunodeplete MBD2 but not MBD1. Anti-MTA2 and anti-SAP30 antibodies were used as controls, since neither MBD1 nor MBD2 associates with these proteins. (F) Apparent molecular masses of MBD1 and MBD2. HeLa nuclear extract was applied to a Superose 6 gel filtration column. Eluted fractions were analyzed by Western blotting to detect MBD1 and MBD2.
MBD1 is not a component of HeLa MeCP1.
The discovery of three novel methyl-CpG binding proteins (MBD1, MBD2, and MBD4) has raised the possibility that these proteins contribute to the MeCP1 activity. A previous antibody raised against full-length MBD1 was apparently able to supershift the MeCP1 activity in a HeLa nuclear extract, suggesting that MBD1 may be a component of MeCP1 (12). However, several lines of evidence based on antibody S751 now preclude MBD1 involvement in MeCP1.
HeLa nuclear extracts were preincubated with both anti-MBD1 and preimmune antibodies coupled on protein G-Sepharose. Anti-MBD1 antibody specifically immunoprecipitated MBD1 and immunodepleted MBD1 from the extracts (Fig. 3A). Bandshift assays using a methylated version of the CG11 probe showed that MeCP1 activity was not affected by this treatment (Fig. 3B). The nonspecific complex on the nonmethylated probe CG11 was not affected by similar treatment (7) (Fig. 3B). Thus, drastic depletion of MBD1 from the extract did not reduce MeCP1 activity. We attempted to “supershift” the MeCP1 complex with methylated DNA by adding anti-MBD1 antibody. Addition of preimmune or immune sera (1 μl) to the extracts slightly altered the mobility of the MeCP1 complex (Fig. 3C, lane 3 and 5) compared to that in extracts with no added serum (lane 2). When half the amount of serum (0.5 μl) was added to the extract, no alteration in mobility was observed. In neither case was there any difference between the effects of preimmune or immune sera on the bandshift complex (Fig. 3C, compare lanes 2, 4, and 6). We conclude that anti-MBD1 was unable to supershift MeCP1 complex, although the same amount of serum could recognize native MBD1 in the extract. Affinity-purified anti-MBD1 antibody also failed to affect the mobility of MeCP1 (data not shown). We suspect that cross-reactivity of the original antibody with unknown components of MeCP1 was responsible for the earlier data (12).
Another methyl-CpG binding protein, MBD2, has recently been conclusively shown to be a component of MeCP1 (37). Antibody against MBD2 was able to both immunodeplete and supershift MeCP1 activity. We investigated whether MBD1 and MBD2 are in the same complex by performing immunoprecipitation assays. We found that MBD1 immunoprecipitates do not contain MBD2 and that MBD2 immunoprecipitates do not contain MBD1 (Fig. 3D). An antibody against HDAC1 depletes both MBD2 and MeCP1 activity from HeLa extracts (37). MBD1 protein, however, was not depleted by anti-HDAC1 antibodies under conditions where MBD2 was specifically depleted by the same antibody (Fig. 3E). Therefore, MBD1 is not associated with the MBD2-HDAC1 complex that is part of MeCP1.
The apparent molecular weights of the MBD1 and MBD2 complexes also differ as shown by gel filtration. A HeLa nuclear extract was passed over a Superose 6 gel filtration column to determine the native size of MBD1. Western blot analysis of the fractions eluted from the column revealed that MBD1 migrates at a molecular mass of 200 to 400 kDa (Fig. 3F). The calculated molecular mass for MBD1 is 66 kDa, and it is therefore possible that MBD1 interacts with itself or other nuclear proteins to form a higher-molecular-mass complex. Alternatively, monomers of MBD1 may adopt a nonglobular conformation with an increased apparent molecular mass in this assay. In contrast, MBD2 (Fig. 3F) comigrates at about 800 kDa under the same conditions. Thus, MBD1 and MeCP1 do not comigrate in a gel filtration assay. This series of experiments argues strongly against involvement of MBD1 in the MBD2 complex of HeLa cells. We cannot exclude the possibility that MBD1 contributes to the MeCP1 activity found in extracts derived from somatic tissues.
MBD1 contains an active repression domain.
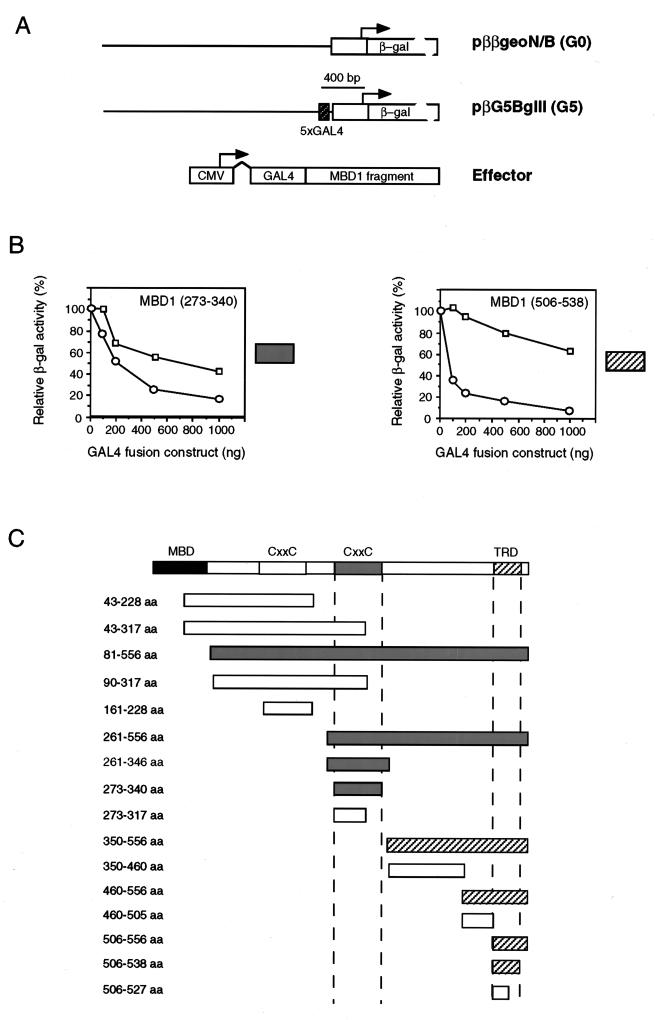
It has previously been shown that MBD1 can repress the expression of methylated genes in transcription extracts (12) and cultured cell lines (16). Is preferential repression of methylated templates by recombinant MBD1 and transiently transfected MBD1 due to steric exclusion of essential regulatory elements (passive repression)? Or does MBD1 repress actively, for example by interference with the initiation complex or recruitment of a corepressor? To answer this question, we fused fragments of the MBD1 coding sequence to DNA encoding a GAL4 DNA binding domain and tested the effect of the expressed fusion proteins on a reporter bearing GAL4 DNA binding sites approximately 80 bp upstream of the human β-actin promoter (Fig. 4A). Individual effector constructs were separately cotransfected into mouse L929 fibroblasts together with a β-galactosidase reporter either with or without five GAL4 binding sites (constructs pββgeoN/B and pβG5BglII [Fig. 4A]). To avoid the mistargeting of the GAL4 fusion proteins to methylated sites in the genome, the first 43 amino acids of MBD1, corresponding to half of the MBD, were omitted from all constructs. Approximately 60 h after transfection, the cells were harvested and extracts were assayed for β-galactosidase activity. For each construct, titration of at least three concentrations of effector was performed. Western blot analyses of the lysates of transfected cells with anti-GAL4 antibody were also carried out to confirm that effectors of the correct size were expressed (data not shown). The results showed that two regions in MBD1 can repress transcription from reporter constructs: the CxxC domain between amino acids 273 to 340 and the C-terminal region spanning amino acids 506 to 538 (Fig. 4B and C). Neither the related N-terminal CxxC domain (amino acids 161 to 228 [Fig. 4B]) nor a third CxxC domain (amino acids 161 to 221), which occurs in the MBD1* splice variant (Fig. 1A), could repress the reporters (data not shown). The CxxC domains of HRX (amino acids 1147 to 1197) and DNA cytosine 5-methyltransferase DNMT1 (amino acids 639 to 705 of mouse DNMT1; accession no. X14805) also show different repression activities, since the HRX domain can specifically repress a reporter with GAL4 binding sites (39, 49) whereas the DNMT1 domain fused to a GAL4 DNA binding domain does not repress (data not shown). The two regions of MBD1 that gave repression differed in that effectors containing the CxxC domain (amino acids 273 to 340) also significantly inhibited the control reporter which lacked GAL4 binding sites, whereas the C-terminal domain (amino acids 506 to 538) showed specificity for repression of the reporter with GAL4 binding sites (Fig. 4B and C). We conclude that inhibition by the CxxC domain can occur without binding to the reporter gene and may be nonspecific. The C-terminal domain, however, has the properties of an active repression domain and is referred to below as the TRD of MBD1.
FIG. 4.
Effect of regions of MBD1 on transcription when tethered to the DNA by a GAL4 DNA binding domain. (A) Map of reporters with five GAL4 binding sites (G5) and no GAL4 binding sites (G0) and of the effectors which express a fusion between the GAL4 binding site and regions of MBD1 downstream. Effector proteins were expressed from a CMV promoter. (B) Examples of the relative transcription levels of G5 (squares) and G0 (circles) reporters in the presence of increasing amounts of different GAL4-MBD1 effectors. The MBD1(273–340) profile was obtained with effector constructs that contained the second CxxC domain, whereas the MBD1(506–538) profile was obtained with effectors containing the C terminus of MBD1. The shaded box next to each panel provides a key to the repression profile obtained with the effector constructs listed in panel C. (C) Summary maps showing regions in MBD1 that can affect the transcription of reporters in the above assay. Repression characteristic of the CxxC domain [see MBD1(273–340) above] was seen with grey constructs, whereas repression characteristic of the C terminus [see MBD1(506–538) above] was seen with diagonally shaded constructs. aa, amino acids.
Characterization of the C-terminal TRD.
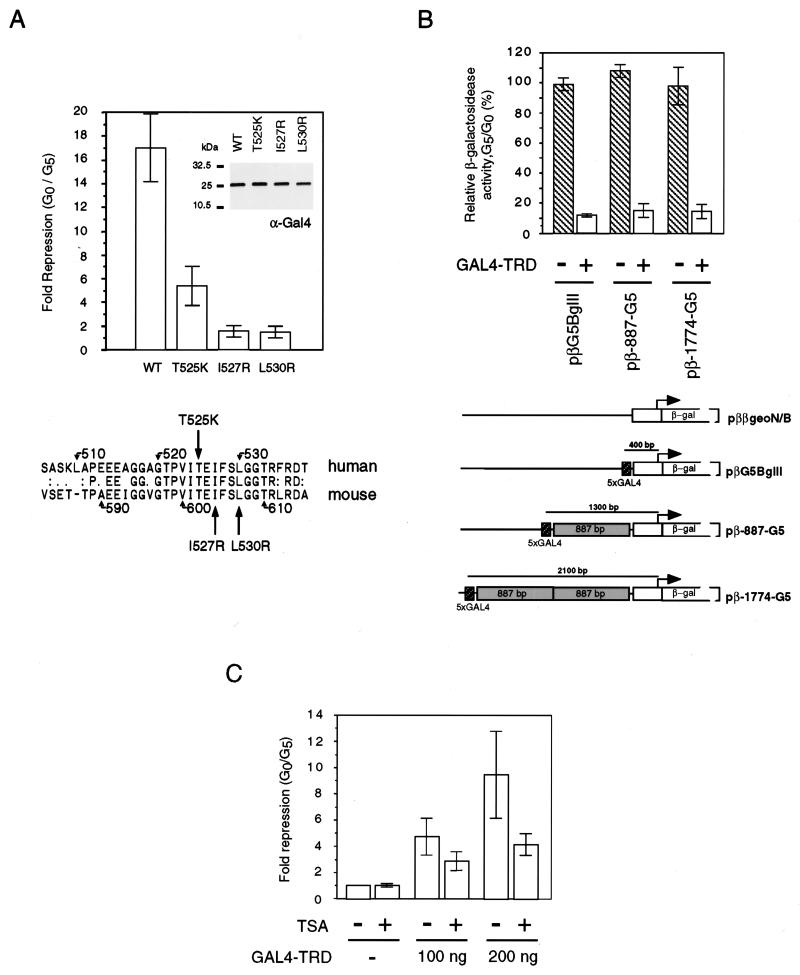
The TRD of MBD1 contains 33% hydrophobic amino acids (alanine, isoleucine, leucine, phenylalanine, and valine) and 15.1% glycine. Comparison between human and mouse MBD1 revealed that the TRD is conserved, with 66.7% identity over a 33-amino-acid region (Fig. 5A). To test the importance of the conserved amino acids, three TRD mutations with single-amino-acid substitutions were generated. Conversion of hydrophobic residues isoleucine-527 and leucine-530 to the basic residue arginine abolished the repression activity of TRD (Fig. 5A). Replacement of polar residue threonine-525 by lysine led to partial loss of repression activity. Western blot analysis using anti-GAL4 antibodies indicated that the expression of these GAL4-TRD mutants was comparable to that of the wild-type TRD fusion protein (Fig. 5A, inset).
FIG. 5.
Repression by the C-terminal TRD acts at a distance but is sensitive to TRD mutations and the presence of TSA. (A) Amino acid sequence conservation of the TRD between human and mouse MBD1s. Mutations of hydrophobic residue isoleucine-527 (I527R) and leucine-530 (L530R) destroys the repression activity of the TRD, but mutation of threonine-525 (T525K) leads to partial loss of repression activity. Inset: wild-type (WT) and mutant GAL4-TRD fusion proteins are expressed at equal levels. (B) GAL4-TRD can repress transcription from a distance. Repression was effective when GAL4 binding sites were placed at 400, 1,300, and 2,100 bp from the transcription start site in the reporter construct. (C) TSA (100 ng/ml) can partially relieve repression by GAL4-TRD. The results shown are based on three independent transfections.
We next asked whether the TRD can repress transcription at a distance. Two reporters (pβ-887-G5 and pβ-1774-G5) with the five GAL4 binding sites located 1.3 and 2.1 kb away from the transcription start site (32) were used to address this question (Fig. 5B). GAL4-TRD repressed the activity of the pβG5BglII reporter in which the GAL4 binding sites were 400 bp upstream of the transcription start site to about 10% of the control level. At the same effector concentration (250 ng), the activity of pββgeoN/B reporter, which lacked GAL4 binding sites, was not affected. GAL4-TRD also efficiently repressed when the GAL4 binding sites were 887 bp (pβ-887-G5) and 1,774 bp (pβ-1774-G5) further upstream than in pβG5BglII (Fig. 5B). The result indicates that the TRD can efficiently mediate repression even when it is more than 2,000 bp distant from the transcription start site. This finding contrasts with the significant reduction with distance in the efficiencies of activation by GAL4-VP16 and of repression by the GAL4-TRD of MeCP2 in the same system (32).
The association between methylated DNA, transcriptional repression, and hypoacetylated histones raised the possibility that MBD1 also represses transcription via deacetylation (14, 25, 34, 36, 37). Repression by GAL4-TRD of MBD1 is sensitive to TSA, suggesting that deacetylation may play a role (Fig. 5C). The relatively nonspecific repression by the CxxC domain was, however, not sensitive to TSA (data not shown). It is unlikely that MBD1 is associated with known histone deacetylase 1 complexes, since MBD1 proteins were not depleted by HDAC1, SAP30, or MTA2 antibodies (Fig. 3E). Furthermore, the apparent molecular weight of MBD1 in extracts was much lower than that of known corepressor complexes (Fig. 3F).
MBD1 repression in vivo depends on the TRD, the methyl-CpG binding domain, and deacetylation.
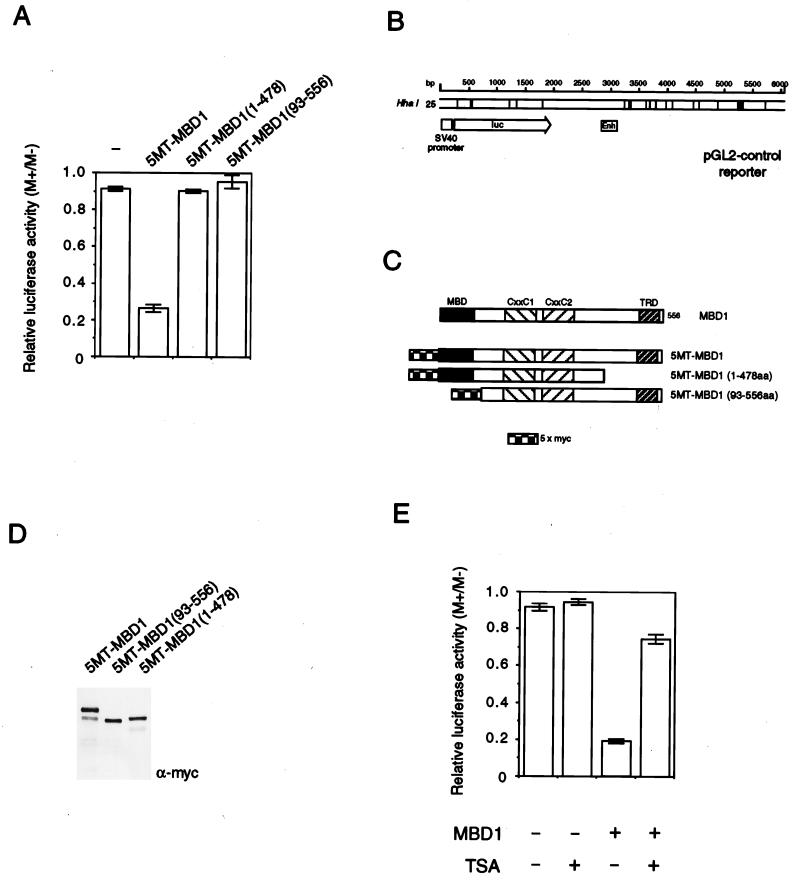
Results with GAL4 fusions to MBD1 fragments indicate that MBD1 contains a TRD whose action may depend on deacetylation. To assess the requirements of methylation-dependent repression in vivo, we expressed MBD1 (or regions thereof [Fig. 6C]) in mouse L929 cells that were cotransfected with a methylated or nonmethylated reporter construct. The reporter consisted of a luciferase coding region plus a simian virus 40 (SV40) promoter and enhancer (Fig. 6B). Methylation of all 25 HhaI sites in the ∼6,000-bp construct had a minimal effect on expression (presumably due to the low density of methylation), but in the presence of intact MBD1, expression was significantly inhibited (Fig. 6A). Repression was abolished by removing the C-terminal TRD (amino acids 479 to 556). Deletion of the methyl-CpG binding domain also abolished repression (Fig. 6A). Comparable expression of the MBD1 proteins was demonstrated by probing Western blots with anti-Myc monoclonal antibody 9e10 (Santa Cruz) to detect the N-terminal Myc epitope tag (Fig. 6D). Nuclear localization of all three MBD1 proteins was verified by immunostaining with anti-Myc antibodies (data not shown). We next asked whether methylation-dependent repression by MBD1 could be reversed by TSA. As shown in Fig. 6E, TSA treatment restored the transcription of the methylated reporter in the presence of MBD1 to over 75% of control levels. These findings confirm that methylated DNA binding and the TRD are required for repression and that deacetylation is an important component of the repression mechanism.
FIG. 6.
Repression of a methylated reporter gene by MBD1 depends on the TRD and methyl-CpG binding domains and is sensitive to TSA. (A) Mouse L929 cells were transfected with an SV40-luciferase reporter (2 μg) that was nonmethylated (M−) or methylated at 25 HhaI (GCGC) sites (M+). Levels of M+ and M− reporter expression (luciferase activity) were normalised to the expression level of cotransfected CMV β-galactosidase control reporter (1 μg). Transcription levels were expressed as the ratio of M+ to M− normalised expression levels. The low density of methylation in the pGL2 control reporter had a negligible effect on transcription in the absence of cotransfected MBD1 but caused repression relative to the nonmethylated reporter in the presence of 0.5 μg of intact MBD1 fused to a Myc epitope tag (5MT-MBD1). N-terminal or C-terminal deletions prevented repression. (B) Diagram of the pGL2 SV40-luciferase reporter, showing the locations of methylated HhaI sites. (C) Diagram of the 5MT-MBD1 proteins used to obtain data shown in panel A. (D) Western blots of proteins extracted from cells transfected by each of the 5MT-MBD1 constructs showing equivalent expression. The blots were probed with anti-Myc monoclonal antibody 9e10. (E) TSA (100 ng/ml) overcomes repression by MBD1 of a methylated reporter gene. The results shown are based on three independent transfections.
MBD1 is a chromosomal protein that is enriched in hypoacetylated constitutive heterochromatin.
Localization of MBD1 has been studied by transient transfection of constructs expressing fusions between MBD1 and green fluorescent protein (16, 20). To avoid possible artifacts associated with overexpression of modified proteins in the presence of endogenous bound protein, we used the anti-MBD1 antibody to study the localization of endogenous MBD1 on metaphase chromosomes of human diploid cells. Unfixed metaphase cell preparations from the primary female fibroblast line HF19 (24) were subjected to immunofluorescent labelling with sheep anti-MBD1 serum S751 (Fig. 7). The major sites of labelling corresponded to centromeric heterochromatin on chromosomes 1, 9, 15, and 16. There was also a concentration in the acrocentric short arms of chromosomes 13, 14, and 15, where rRNA genes are interspersed with highly methylated spacer DNA (9). Nonuniform labelling of euchromatic chromosome arms was also seen (Fig. 7A and B). Affinity-purified MBD1 antibodies gave the same staining pattern as the immune serum, and preimmune serum gave only background staining (data not shown).
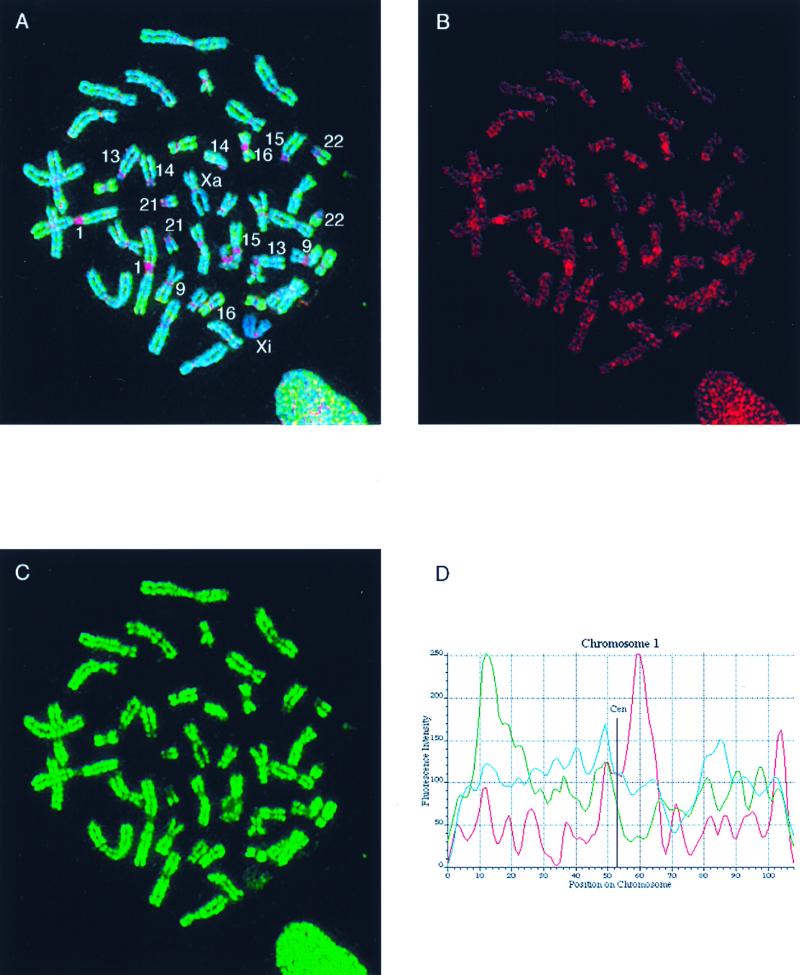
FIG. 7.
Localization of MBD1 on human metaphase chromosomes in relation to the distribution of histone H4 acetylation. (A) Unfixed cytospin HF19 primary female fibroblast metaphase cell, simultaneously immunolabelled with sheep S751 anti-serum (detected in red) and rabbit anti-acetyl(Lys-12)-H4 (detected in green), counterstained for DNA with Hoechst 33258 (blue). Numbered chromosomes were identified from their Hoechst fluorescence and H4-acetylation immunofluorescence patterns. Xa and Xi refer to the active and inactive X chromosomes, respectively. (B) Separated red image showing the MBD1 concentration at centromeres (particularly noticeable in constitutive pericentromeric heterochromatin of chromosomes 1, 9, 15 and 16 [see also panel A]) and weaker, nonuniform labelling of euchromatic chromosome arms. Specific euchromatin localization is evident, since both chromatids exhibit a similar pattern of immunofluorescent bands (this is more apparent in some chromosomes than others). (C) Separated green image showing the histone H4 acetylation profile and identifying hypoacetylated constitutive heterochromatin and the inactive X chromosome (Xi). (D) The intensities of MBD1 and acetylated H4 immunofluorescence (red and green, respectively) and Hoechst 33258 DNA fluorescence (blue) are plotted along the axis of the nonoverlapped chromosome 1 from panel A. (The position along the chromosome axis is given in arbitrary units, starting at the tip of the short arm; fluorescence intensity is likewise given in arbitrary units.) The concentration of MBD1 at the centromere is clearly indicated by the peak in the red trace, in contrast to the very low level of histone H4 acetylation in the same region (green trace). MBD1 fluorescence along the chromosome arms shows much weaker maxima.
The distribution of MBD1 along human metaphase chromosomes was compared with the known distribution of histone H4 acetylation (21, 23, 24) by simultaneous immunofluorescent labelling of fibroblast chromosomes with S751 serum and rabbit anti-acetylated H4 serum R5/12, which recognizes H4 acetylated at lysine-12 and preferentially reacts with multiply acetylated isoforms (24, 44). The results for a typical cell are illustrated in Fig. 7. In contrast to acetylated H4 (Fig. 7A and C, green), which is not detected at centric constitutive heterochromatin (23), MBD1 labelling (Fig. 7A and B, red) is highly concentrated in centromeric domains, particularly on chromosomes 1, 9, 15, and 16. The concentration of MBD1 at the centromere is clearly indicated by the peak of stain intensity in the red trace along chromosome 1, which contrasts with the very low level of histone H4 acetylation (green trace) in the same region (Fig. 7D). The association between regions rich in MBD1 and hypoacetylated chromatin did not apply to the facultative heterochromatin, since the distribution of MBD1 on the inactive X chromosome was not significantly different from that seen on the active X chromosome or the autosomes (Fig. 7A and B).
Anti-MBD1 antiserum also labelled euchromatin at a level significantly above the background labelling observed using preimmune sheep serum (data not shown) although less strongly than it labelled constitutive heterochromatin (Fig. 7A and B). MBD1 fluorescence along the chromosome arms showed much weaker maxima. Although some peaks were common to both the MBD1 distribution and the H4 acetylation profile, in general they showed distinct patterns, which were also different from the G-band like Hoechst 33258 pattern (Fig. 7D). These data suggest that MBD1 is enriched in domains that may not correspond to G- or R-chromosome banding patterns.
DISCUSSION
MBD1 was originally identified by searching the EST database for proteins with homology to the methyl-CpG binding domain of MeCP2. We show here that the N-terminal 67 amino acids of MBD1 are sufficient for binding to a single mCpG pair. This domain is significantly smaller than the minimal methyl-CpG binding domain of MeCP2 (85 amino acids). Amino acid substitution at proline 38, which is conserved among members of the MBD family, abolished the affinity of the MBD1 domain toward the mCpG pair. In a recent nuclear magnetic resonance spectroscopy structure of the methyl-CpG binding domain of MeCP2 (45), proline 38 was found at a sharp turn between two strands of beta sheet. The amino acids around this proline in the MBD of MeCP2 did not undergo a chemical shift upon DNA binding, suggesting that this residue is not in contact with the DNA. Replacement of the proline by lysine is likely to perturb the local structure of the MBD by removing the unique chain-bending property of proline.
Using an antibody specific for MBD1, we investigated the relationship between MBD1 and MeCP1. The antibody binds to native human MBD1 in extracts, but extracts with greatly reduced levels of MBD1 show no loss of MeCP1 activity. In addition, the antibody fails to supershift the MeCP1 complex. Recent studies have demonstrated that another methyl-CpG binding protein, MBD2, is responsible for MeCP1 activity (37). Correspondingly, anti-MBD2 antibodies supershift the MeCP1 complex and can immunodeplete MeCP1 activity from extracts. The distinction between MeCP1 and MBD1 is emphasized by gel filtration chromatography, since MBD2 is part of an approximately 800-kDa complex in HeLa cells whereas native MBD1 migrates at 200 to 400 kDa. Finally, anti-MBD1 antibodies do not immunoprecipitate MBD2, and vice versa. Based on the summation of this work and the MBD2 study (37), we conclude that MeCP1 of HeLa cells does not contain MBD1 protein.
It was shown earlier that recombinant MBD1 can repress transcription in vitro in a methylation-dependent manner (12). While this manuscript was in preparation, Fujita et al. confirmed the in vitro data with additional promoters and showed that human MBD1, when expressed in Drosophila cells, can repress methylated reporter genes specifically (16). Because reporter genes are methylated at many sites within, upstream, and downstream of the transcribed region, it is important to know whether repression is due to passive steric interference with transcription by bound MBD within the gene or within a transcription factor binding site or whether it is due to active interference with the transcription process. Active repression should be apparent even when the repressor is tethered well upstream of the gene region. We used a GAL4 fusion repression assay to identify a TRD within MBD1 that can affect the transcriptional activity of a reporter gene from binding sites at least 2,000 bp pairs upstream of its promoter. We conclude that the TRD represents an active repression domain. A second repression domain, which coincided with one of the CxxC domains, repressed reporters with and without GAL4 binding sites and may therefore represent a nonspecific (or toxic) transcriptional inhibitor.
The specific TRD at the C terminus of MBD1 repressed strongly in the transient-transfection assay. This region was the primary cause of methylation-dependent repression by intact MBD1, since a protein lacking it but having intact CxxC domains did not significantly repress a methylated reporter. Repression by the C-terminal TRD depended on tethering to DNA by either the GAL4 DNA binding domain or the methyl-CpG binding domain. The amino acid sequence of the TRD is conserved between the human and mouse MBD1s. It is enriched in hydrophobic residues, unlike the highly basic TRD of MeCP2 (32). Mutations at conserved residues within the domain abolish repression. TSA treatment relieves the repression by GAL4-TRD and intact MBD1, strongly suggesting that deacetylation is involved in silencing. HDAC1 is unlikely to be responsible, however, since it fails to coimmunoprecipitate with MBD1. MBD1 does not appear to be associated with two other well-studied corepressor complexes, Mi-2/NuRD or Sin3/SAP30, since it does not co-immunoprecipitate with components of these complexes and is considerably smaller than either by gel filtration (Fig. 3). Other members of the mammalian histone deacetylase family have been reported (15, 17, 47) and may conceivably play a role in repression by MBD1.
Previous work showed that overexpressed tagged versions of MBD1 localize preferentially to heterochromatic foci in mouse cells (20) and centromeric foci and chromosome arms in HeLa cells (16). We show here that endogenous MBD1 in diploid human cells localizes to prominent heterochromatic foci close to the centromeres of chromosomes 1, 9, 15, and 16 and more weakly to euchromatic chromosome arms. Regions of pericentromeric heterochromatin that are particularly deficient in acetylated histone H4 tails are enriched in MBD1, although the protein is also found at lower concentrations in euchromatic regions of the genome. The distribution of MBD1 is similar to that of 5-methylcytosine in constitutive heterochromatin, as established using antibodies against the modified base (1, 30). Thus, MBD1 resembles MeCP2 by being stably associated with chromosomes, even during mitosis, when many transcription factors are displaced (28). Although both MeCP2 and MBD1 are chromosome associated, MeCP2 is usually present at low levels in cultured cells. Whether the two proteins normally compete for the same sites or have additional specificities that segregate them (e.g., differing protein-protein interactions) is a topic for future work. More broadly, it will be necessary to dissect the roles of MeCP2, MeCP1 (MBD2), and MBD1 to determine their specific contributions to methylated gene silencing (see reference 5 for a review). Do they back up one another to repress any one promoter? Or are particular genes targeted by one of the three repressors? Answers to these and other pressing questions await the discovery of genes that are specifically targeted for repression in the living organism.
ACKNOWLEDGMENTS
We thank Yi Zhang, Danny Reinberg, and Bryan Turner for antibodies; Xinsheng Nan for plasmids; Harris Morrison for assistance in karyotyping; and Vicky Clark, Joan Davidson, and Aileen Greig for technical assistance.
This work was supported by a Program Grant to A.B. from the Wellcome Trust and by the Medical Research Council (London). H-H.N. is a Darwin Trust Scholar.
REFERENCES
- 1.Barbin A, Montpellier C, Kokalj-Vokac N, Gibaud A, Niveleau A, Malfoy B, Dutrillaux B, Bourgeois C A. New sites of methylcytosine-rich DNA detected on metaphase chromosomes. Hum Genet. 1994;94:684–692. doi: 10.1007/BF00206964. [DOI] [PubMed] [Google Scholar]
- 2.Beard C, Li E, Jaenisch R. Loss of methylation activates Xist in somatic but not in embryonic cells. Genes Dev. 1995;9:2325–2334. doi: 10.1101/gad.9.19.2325. [DOI] [PubMed] [Google Scholar]
- 3.Bestor T H. Activation of mammalian DNA methyltransferase by cleavage of a Zn binding regulatory domain. EMBO J. 1992;11:2611–2617. doi: 10.1002/j.1460-2075.1992.tb05326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bestor T H, Walsh C P, Yoder J A. Does DNA methylation control transposition of selfish elements in the germline. Trends Genet. 1997;13:470–472. doi: 10.1016/s0168-9525(97)01310-3. [DOI] [PubMed] [Google Scholar]
- 5.Bird A, Wolffe A P. Methylation-induced repression—belts, braces and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 6.Bird A P. Gene number, noise reduction and biological complexity. Trends Genet. 1995;11:94–100. doi: 10.1016/S0168-9525(00)89009-5. [DOI] [PubMed] [Google Scholar]
- 7.Boyes J, Bird A. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell. 1991;64:1123–1134. doi: 10.1016/0092-8674(91)90267-3. [DOI] [PubMed] [Google Scholar]
- 8.Boyes J, Bird A. Repression of genes by DNA methylation depends on CpG density and promoter strength: evidence for involvement of a methyl-CpG binding protein. EMBO J. 1992;11:327–333. doi: 10.1002/j.1460-2075.1992.tb05055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brock J R, Bird A. Mosaic methylation of the repeat unit of the human ribosomal RNA genes. Hum Mol Genet. 1997;6:451–456. doi: 10.1093/hmg/6.3.451. [DOI] [PubMed] [Google Scholar]
- 10.Chandler S P, Guschin D, Landsberger N, Wolffe A P. The methyl-CpG binding transcriptional repressor MeCP2 stably associates with nucleosomal DNA. Biochemistry. 1999;38:7008–7018. doi: 10.1021/bi990224y. [DOI] [PubMed] [Google Scholar]
- 11.Chuang L S-H, Ng H-H, Chia J-N, Li B F L. Characterisation of independent DNA and multiple Zn-binding domains at the N terminus of human DNA-(cytosine-5) methyltransferase: modulating the property of a DNA-binding domain by contiguous Zn-binding motifs. J Mol Biol. 1996;257:935–948. doi: 10.1006/jmbi.1996.0213. [DOI] [PubMed] [Google Scholar]
- 12.Cross S H, Meehan R R, Nan X, Bird A. A component of the transcriptional repressor MeCP1 is related to mammalian DNA methyltransferase and trithorax-like protein. Nat Genet. 1997;16:256–259. doi: 10.1038/ng0797-256. [DOI] [PubMed] [Google Scholar]
- 13.Davey C, Pennings S, Allan J. CpG methylation remodels chromatin structure in vitro. J Mol Biol. 1997;267:276–288. doi: 10.1006/jmbi.1997.0899. [DOI] [PubMed] [Google Scholar]
- 14.Eden S, Hashimshony T, Keshet I, Cedar H. DNA methylation models histone acetylation. Nature. 1998;394:842. doi: 10.1038/29680. [DOI] [PubMed] [Google Scholar]
- 15.Emiliani S, Fischle W, Van Lint C, Al-Abed Y, Verdin E. Characterization of a human RPD3 ortholog, HDAC3. Proc Natl Acad Sci USA. 1998;95:2795–2800. doi: 10.1073/pnas.95.6.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujita N, Takebayashi S, Okumura K, Kudo S, Chiba T, Saya H, Nakao M. Methylation-mediated transcriptional silencing in euchromatin by methyl-CpG binding protein MBD1 isoforms. Mol Cell Biol. 1999;19:6415–6426. doi: 10.1128/mcb.19.9.6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grozinger C M, Hassig C A, Schreiber S L. Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proc Natl Acad Sci USA. 1999;96:4868–4873. doi: 10.1073/pnas.96.9.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 19.Hendrich B, Abbott C, McQueen H, Chambers D, Cross S, Bird A. Genomic structure and chromosomal mapping of the murine and human Mbd1, Mbd2, Mbd3, and Mbd4 genes. Mamm Genome. 1999;10:906–912. doi: 10.1007/s003359901112. [DOI] [PubMed] [Google Scholar]
- 20.Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeppesen P. Histone acetylation: a possible mechanism for the inheritance of cell memory at mitosis. Bioessays. 1997;19:67–74. doi: 10.1002/bies.950190111. [DOI] [PubMed] [Google Scholar]
- 22.Jeppesen P. Immunofluorescence techniques applied to mitotic chromosome preparations. Methods Mol Biol. 1994;29:253–285. doi: 10.1385/0-89603-289-2:253. [DOI] [PubMed] [Google Scholar]
- 23.Jeppesen P, Mitchell A, Turner B, Perry P. Antibodies to defined histone epitopes variations in chromatin conformation and underacetylation of centric heterochromatin in human metaphase chromosomes. Chromosoma. 1992;101:322–332. doi: 10.1007/BF00346011. [DOI] [PubMed] [Google Scholar]
- 24.Jeppesen P, Turner B M. The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell. 1993;74:281–289. doi: 10.1016/0092-8674(93)90419-q. [DOI] [PubMed] [Google Scholar]
- 25.Jones P L, Veenstra G J C, Wade P A, Vermaak D, Kass S U, Landsberger N, Strouboulis J, Wolffe A P. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 26.Levine A, Cantoni G L, Razin A. Inhibition of promoter activity by methylation: possible involvement of protein mediators. Proc Natl Acad Sci USA. 1991;88:6515–6518. doi: 10.1073/pnas.88.15.6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewis J D, Meehan R R, Henzel W J, Maurer-Fogy I, Jeppesen P, Klein F, Bird A. Purification, sequence and cellular localisation of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- 28.Martinez-Balbas M, Dey A, Rabindran S K, Ozato K, Wu C. Displacement of sequence-specific transcription factors from mitotic chromatin. Cell. 1995;83:29–38. doi: 10.1016/0092-8674(95)90231-7. [DOI] [PubMed] [Google Scholar]
- 29.Meehan R R, Lewis J D, McKay S, Kleiner E L, Bird A P. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell. 1989;58:499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- 30.Miller O L, Schnoedl W, Allen J, Erlanger B F. Immunofluorescent localization of 5MC. Nature. 1974;251:636–637. doi: 10.1038/251636a0. [DOI] [PubMed] [Google Scholar]
- 31.Mohandas T, Sparkes R S, Shapiro L J. Reactivation of an inactive human X-chromosome: evidence for X-inactivation by DNA methylation. Science. 1981;211:393–396. doi: 10.1126/science.6164095. [DOI] [PubMed] [Google Scholar]
- 32.Nan X, Campoy J, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- 33.Nan X, Meehan R R, Bird A. Dissection of the methyl-CpG binding domain from the chromosomal protein MeCP2. Nucleic Acids Res. 1993;21:4886–4892. doi: 10.1093/nar/21.21.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nan X, Ng H-H, Johnson C A, Laherty C D, Turner B M, Eisenman R N, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 35.Nan X, Tate P, Li E, Bird A P. DNA methylation specifies chromosomal localization of MeCP2. Mol Cell Biol. 1996;16:414–421. doi: 10.1128/mcb.16.1.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ng H-H, Bird A. DNA methylation and chromatin modification. Curr Opin Genet Dev. 1999;9:158–163. doi: 10.1016/s0959-437x(99)80024-0. [DOI] [PubMed] [Google Scholar]
- 37.Ng H-H, Zhang Y, Hendrich B, Johnson C A, Burner B M, Erdjument-Bromage H, Tempst P, Reinberg D, Bird A. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat Genet. 1999;23:58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- 38.Panning B, Jaenisch R. DNA hypomethylation can activate Xist expression and silence X-linked genes. Genes Dev. 1996;10:1991–2002. doi: 10.1101/gad.10.16.1991. [DOI] [PubMed] [Google Scholar]
- 39.Prasad R, Yano T, Sorio C, Nakamura T, Rallapalli R, Gu Y, Leshkowitz D, Croce C M, Canaani E. Domains with transcriptional regulatory activity within the ALL1 and AF4 proteins involved in acute leukemia. Proc Natl Acad Sci USA. 1995;92:12160–12164. doi: 10.1073/pnas.92.26.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Razin A. CpG methylation, chromatin structure and gene silencing—a three-way connection. EMBO J. 1998;17:4905–4908. doi: 10.1093/emboj/17.17.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stöger R, Kubicka P, Liu C-G, Kafri T, Razin A, Cedar H, Barlow D P. Maternal specific methylation of the imprinted mouse Ifg2r locus identifies the expressed locus as carrying the imprinting signal. Cell. 1993;73:61–70. doi: 10.1016/0092-8674(93)90160-r. [DOI] [PubMed] [Google Scholar]
- 42.Tate P H, Bird A. Effects of DNA methylation on DNA-binding proteins and gene expression. Curr Opin Genet Dev. 1993;3:226–231. doi: 10.1016/0959-437x(93)90027-m. [DOI] [PubMed] [Google Scholar]
- 43.Tremblay K D, Duran K L, Bartolomei M S. A 5′ 2-kilobase-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol Cell Biol. 1997;17:4322–4329. doi: 10.1128/mcb.17.8.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turner B M, Fellows G. Specific antibodies reveal ordered and cell cycle-related use of histone H4 acetylation sites in mammalian cells. Eur J Biochem. 1989;79:131–139. doi: 10.1111/j.1432-1033.1989.tb14530.x. [DOI] [PubMed] [Google Scholar]
- 45.Wakefield R I D, Smith B P, Nan X, Free A, Soterious A, Uhrin D, Bird A, Barlow P N. The solution structure of the domain from MeCP2 that binds to methylated DNA. J Mol Biol. 1999;291:1055–1065. doi: 10.1006/jmbi.1999.3023. [DOI] [PubMed] [Google Scholar]
- 46.Walsh C P, Bestor T H. Cytosine methylation and mammalian development. Genes Dev. 1999;13:26–34. doi: 10.1101/gad.13.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang W M, Yao Y L, Sun J M, Davie J R, Seto E. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J Biol Chem. 1997;272:28001–28007. doi: 10.1074/jbc.272.44.28001. [DOI] [PubMed] [Google Scholar]
- 48.Yoder J A, Chui Yen R-W, Vertino P M, Bestor T H, Baylin S B. New 5′ regions of the murine and human genes for DNA (cytosine-5)-methyltransferase. J Biol Chem. 1996;271:31092–31097. doi: 10.1074/jbc.271.49.31092. [DOI] [PubMed] [Google Scholar]
- 49.Zeleznik-Le N J, Harden A M, Rowley J D. 11q23 translocations split the “AT-hook” cruciform DNA-binding region and the transcriptional repression domain from the activation domain of the mixed-lineage leukemia (MLL) gene. Proc Natl Acad Sci USA. 1994;91:10610–10614. doi: 10.1073/pnas.91.22.10610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y, Ng H-H, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NURD subunits reveal a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, Sun Z-W, Iratni R, Erdjument-Bromage H, Tempst P, Hampsey M, Reinberg D. SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol Cell. 1998;1:1021–1031. doi: 10.1016/s1097-2765(00)80102-1. [DOI] [PubMed] [Google Scholar]