Abstract
To clarify the effect of retinoid X receptor-α/γ (RXR-α/γ) genes functional genetic variants (RXR-α rs4842194 G>A, RXR-γ rs100537 A>G and rs2134095 T>C) on the risk of gestational diabetes mellitus (GDM), a case–control study with 573 GDM patients and 740 pregnant women with normal glucose tolerance was performed in Guangxi area of China. An odds ratio (OR) with its corresponding 95% confidence interval (CI) was used to assess the strengths of the association between genetic variation and GDM. After adjustment of age and pre-BMI, the logistic regression analysis showed that the rs2134095 was significantly associated with GDM risk (CC vs. TT/TC: adjusted OR = 0.71, 95% CI = 0.56–0.90) in all subjects, and this result remained highly significant after Bonferroni’s correction for multiple testing (P=0.004). The stratified analysis showed that rs2134095 was significantly associated with the risk of GDM among age > 30 years (adjusted OR = 0.61, 95% CI = 0.39–0.97), BMI > 22 kg/m2 (adjusted OR = 0.46, 95% CI = 0.30–0.70), systolic blood pressure (SBP) > 120 mmHg (adjusted OR = 1.96, 95% CI = 1.14–3.36), glycosylated hemoglobin A1c (HbA1c) < 6.5% (adjusted OR = 1.41, 95% CI = 1.11–1.78), TG ≤ 1.7 mmol/l (adjusted OR = 2.57, 95% CI = 1.45–4.53), TC ≤ 5.18 mmol/l (adjusted OR = 1.58, 95% CI = 1.13–2.22), high-density lipoprotein cholesterol (HDL-c) ≤ 1.5 mmol/l (adjusted OR = 1.70, 95% CI = 1.16–2.49) and low-density lipoprotein cholesterol (LDL-c) > 3.12 mmol/l (adjusted OR = 1.47, 95% CI = 1.08–2.00) subjects, under the recessive genetic model. We also found that rs2134095 interacted with age (Pinteraction=0.039), pre-BMI (Pinteraction=0.040) and TG (Pinteraction=0.025) influencing individual’s genetic susceptibility to GDM. The rs2134095 T>C is significantly associated with the risk of GDM by effect of a single locus and/or complex joint gene–gene and gene–environment interactions. Larger sample-size and different population studies are required to confirm the findings.
Keywords: association study, gestational diabetes mellitus, retinoid X receptor, single nucleotide polymorphisms, susceptibility
Introduction
Gestational diabetes mellitus (GDM) is defined as different degrees of impaired glucose tolerance that is first recognized during pregnancy, often manifested as insufficient islet secretion and insulin resistance, which seriously threatens maternal and child health [1]. Worldwide, it affects approximately 2–20% of all pregnancies [2]. Clinical and epidemiological studies have demonstrated that GDM is associated with maternal and perinatal complications, such as fetal macrosomia, pre-eclampsia, preterm birth, spontaneous abortion, respiratory distress syndrome, small for gestational age (SGA), large for gestational age (LGA), shoulder dystocia, hypoglcemia, polycythemia etc [3–5]. As yet, the main risk factors are known as older age at pregnancy, obesity, family history of type 2 diabetes (T2DM) and past history of GDM, previous poor obstetric history and genetic factors [6–10]. So far, the pathogenesis of GDM is not clear.
GDM was thought to have similar pathophysiological mechanisms with T2DM, such as insulin resistance, β-cell dysfunction and defects in insulin action [11]. Retinoic acid, as a product of retinol catabolism, can maintain the activity of vitamin A by activating retinoic acid receptor (RAR) and retinoid acid X receptor (RXR). Studies have confirmed that retinoid is highly associated with diabetes, islet function, and glucose and lipid metabolism by regulating the transcription of downstream target genes [12–14]. The RXRs are nuclear receptors that mediate the biological effects of retinoids by their involvement in retinoic acid-mediated genes activation. It functions as transcription factors by binding as heterodimers to the specific sequences in the promoter of target genes. In addition, RXRs are members of the vitamin D (VD) pathway that regulate the transcription of vitamin D receptor (VDR) and helps to repair islet β cells, improve insulin resistance and promote the glucose and lipid metabolism [15–18]. It is reasonable to believe that RXRs play a key role in the pathogenesis of GDM.
Single nucleotide polymorphism (SNP) mainly refer to the polymorphism of DNA sequence caused by the conversion or transversion of a single nucleotide, as the most common genetic variation in humans, accounting for more than 90% of all known variants. It has been confirmed that some variants located in the gene may directly affect the protein structure or expression level, and may represent some factors in the genetic mechanism of GDM [19]. A genome-wide association study (GWAS) from South Korea showed that T2DM-associated genetic variants were also associated with GDM risk, such as SLC30A8 rs3802177, CDKAL1 rs10440833, CDKN2A/2B rs10965250 etc [20]. In addition, studies have shown that the genetic variation rs10830963 of MTNR1B gene is associated with the pathogenesis of T2DM by affecting the expression of MTNR1B [21], and carrying the MTNR1B rs10830963G allele significantly increased the odds of antenatal insulin therapy (AIT) in pregnant women of GDM with pre-pregnancy BMI > 29 kg/m2 [22]. The above mentioned evidence revealed that inherited genetic factors are also contributed to the genesis and development of GDM.
The chromosomal 1q21-24 is identified as the susceptible region of T2DM, and the retinoid X receptor-γ (RXR-γ) rs10918169, apolipoprotein A2 (APOA2) rs6413453, calsequestrin 1 (CASQ1) rs617698 and dual specificity phosphatase 12 (DUSP12) rs1503814 are screened as the susceptible genes and loci of T2DM belonging to this region [23]. Shi et al.’s study indicated that the allele frequencies of rs4240711, rs4842194 and rs3132291 variants in RXR-α gene were statistically different between metabolic syndrome (MetS) and normal controls in a Chinese Han population, and the genotypes of studied variants were associated with the decreased risk of MetS [24]. A collection of studies showed that the variants of RXR-α and RXR-γ genes were associated with the risk of T2DM [25–27].
GDM may have a similar genetic basis to T2DM and are considered to be an early stage of T2DM [28]. We propose the hypothesis that the functional genetic variations of RXR-α/γ genes may affect transcription or post-transcriptional regulation of genes and change the susceptibility of pregnant women to GDM. Hence, we investigated the relationship between the functional variations of candidate RXR-α/γ genes, and the risk of GDM in pregnancy women in Guangxi area by using a molecular epidemiological case–control study.
Materials and methods
Study subjects
All 1313 subjects were recruited from the Affiliated Hospital of Guilin Medical University, from September 2014 to June 2016. The project has been approved by the ethics committee of Guilin Medical University, and all subjects signed informed consent. All pregnant women were routinely offered a 75-g oral glucose tolerance test (OGTT) at 24–28 weeks and were evaluated according to the International Association of the Diabetes and Pregnancy Study Groups criteria (IADPSG). The GDM cases were identified when one or more values of glucose concentration were above cut-off points (≥5.10, ≥10.0 and ≥8.5 mmol/l at 0, 60 and 120 min, respectively). If a pregnant woman who had been previously diagnosed with endocrine diseases, serious systemic diseases, history of pre-pregnancy type 1 or type 2 diabetes and other pregnancy complications or long-term using of glucose metabolism affecting drugs, was excluded. In the present study, 573 GDM cases and 740 pregnant women with normal glucose tolerance at the same period were included.
Research methods
Clinical and biochemical data
According to the study purpose, clinical indicators included age, height, weight, systolic blood pressure (SBP) and diastolic blood pressure (DBP) were collected. Meanwhile, body mass index before gestation (pre-BMI) was calculated as body weight (kg) divided by the square of height (m2). Five milliliter of peripheral venous blood from each subject was collected for detection of biochemical indicators. Biochemical data consisted of results of OGTT and glycosylated hemoglobin A1c (HbA1c).
Genomic DNA extraction
The genomic DNA was extracted from EDTA-treated whole blood using a DNA extraction kit (Aidlab Biotechnologies Co., Ltd, China) and stored in a refrigerator at −20°C prior to polymerase chain reaction (PCR).
SNP selection
The potential functional SNPs of candidate RXR-α and RXR-γ genes were screened by using NCBI dbSNP database (http://www.ncbi.nlm.nih.gov/projects/SNP), SNPinfo Web Server (http://snpinfo.niehs.nih.gov/) and SHEsis online tool (http://analysis.bio-x.cn/) [29], and should meet the following criteria: (1) SNPs with minor allele frequency (MAF) reported in HapMap based Han Chinese in Beijing (CHB) was greater than 0.05; (2) located at the regulatory region of studied genes (i.e., the 5′ near gene, 5′ untranslated regions [UTRs], 3′ UTR, 3′ near gene or splice sites); (3) might affect the microRNA–mRNA binding sites (MMBSs) activity, transcription factor-binding sites (TFBSs) activity in the putative promoter region or the post-transcriptional splicing process; (4) the linkage disequilibrium (LD) coefficient D′ or r2 < 0.8 between candidate SNPs.
According to the selection strategy, three SNPs including rs4842194 G>A in the RXR-α gene, and rs100537 A>G and rs2134095 T>C in the RXR-γ gene were identified for further study. Among them, rs4842194 G>A located at the 3′-UTR region of RXR-α gene may affect the MMBS activity. The SNP rs100537 A>G located in the 5′ near gene may affect the TFBS function, and rs2134095 T>C in the splice sites may affect the post-transcriptional splicing process of RXR-γ gene.
Genotyping
The PCR and restriction fragment length polymorphism (RFLP) methods were used to analyze the genotypes of the studied loci. The specific primers were designed and synthesized according to the sequence of candidate SNPs by Suzhou Hongxun Biotechnology Co., Ltd, China, in Table 1.
Table 1. The information of primer sequences for selected SNPs.
| Primer name | Primer sequence | Amplified length | Restriction enzyme |
|---|---|---|---|
| rs4842194-F | 5′-CCGTCCTGAGGCAGAGTGTTG-3′ | 299 bp | Pst I |
| rs4842194-R | 5′-TCCCAAGAAGCAGCCGACC-3′ | ||
| rs100537-F | 5′-CCTGGCACGGTGACTTGA-3′ | 667 bp | Msp I |
| rs100537-R | 5′-TGGGAAATACGAATGACTGGAT-3′ | ||
| rs2134095-F | 5′-CTTGCTGTGCCTGTTG-3′ | 525 bp | Hinf I |
| rs2134095-R | 5′-GTGGAAGAGGGCTGAA-3′ |
Abbreviations: F, the forward primer; R, the reverse primer.
Each 20-μl PCR mixture contained genomic DNA 1 μl (100–200 ng), 2× M5 Taq HiFi PCR mix 10 μl, primer F (5 μM) 1 μl, primer R (5 μM) 1 μl and ddH2O 7 μl. Reaction procedures were as follows: pre-degenerated at 94°C for 4 min, denatured at 94°C for 30 s, annealed at 56–60°C for 25 s, extended at 72°C for 25 s, followed by 30 cycles and with a final extension at 72°C for 7 min. The restriction enzyme digestion gave rise to RFLP of the PCR products. Each digestion reaction mixture with PCR products 8 μl, FastDigest Green Buffer (10×) 1 μl, restriction enzyme 1 unite and ddH2O 6 μl was incubated in water at 37°C for 2 h, and then separated by 2.0% agarose gel electrophoresis.
Statistical analysis
The chi-square (χ2) test was used to assess the differences of selected demographic data, environmental exposure factors and genotypes of studied variants between cases and controls. A χ2 goodness-of-fit test was performed to determine whether the distribution about genotypes of SNPs in control group conformed to Hardy–Weinberg equilibrium (HWE). Unconditional logistic regression analysis was used to estimate the associations between the genotypes of SNPs and the risk of GDM by computing the odds ratios (ORs) and their 95% confidence intervals (CIs). In the present study, the two-sided test was used, and P≤0.05 was considered statistically significant. Bonferroni’s correction of the significance level (0.05/3 = 0.017) according to the number of comparisons was applied to account for multiple testing. The statistical analysis was performed with SPSS 17.0 (SPSS, Chicago, IL, U.S.A.). In addition, false positive report probability (FPRP) was estimated to assess the robustness of findings statistically significant association by using the method described by Wacholder et al. [30]. The FPRP threshold was set to 0.2, and the prior probability was set to 0.1 to detect the noteworthiness for OR of 1.5 or 0.67, with an α level equal to the observed P-value.
The multiple dimension reduction (MDR) software (version 3.0.2) was used to investigate the joint effect of SNPs. A 10-fold cross-validation and 1000-fold permutation testing were adopted under the null hypothesis of no association. The cross-validation consistency (CVC) and the testing balanced accuracy was help to identify the best model among all possibilities considered [31,32].
Functional prediction of studied variants
The online bioinformatics tool SNPinfo Web Server (https://snpinfo.niehs.nih.gov/cgi-bin/snpinfo/snpfunc.cgi) and Human Splicing Finder 3.1 [33] were conducted to investigate the putative functional effect of the variants and the gene alternative splicing regulatory.
Results
Characteristics of research objects
In the study, 1313 pregnant women in Guilin were recruited, including 573 patients in the GDM case group, with an average age of 31.48 ± 4.74 years; 740 patients in the normal pregnancy group, with an average age of 28.91 ± 4.18 years. The comparison of general data showed that age, pre-pregnancy BMI, SBP, DBP, 0/60/120 min OGTT and HbA1c in the cases were much higher than that of controls (P<0.05) as shown in Table 2.
Table 2. Demographic and clinical characteristics in GDM cases and controls ().
| Variables | Cases (n= 573) | Controls (n=740) | t | P-values |
|---|---|---|---|---|
| Age (years) | 31.48 ± 4.74 | 28.91 ± 4.18 | 10.25 | <0.001 |
| Pre-pregnancy BMI (kg/m2) | 23.14 ± 3.63 | 21.66 ± 3.06 | 7.83 | <0.001 |
| SBP (mmHg) | 111.52 ± 10.60 | 109.28 ± 9.48 | 3.98 | <0.001 |
| DBP (mmHg) | 71.42 ± 7.88 | 69.96 ± 6.73 | 4.37 | <0.001 |
| OGTT 0 min glucose (mmol/l) | 5.23 ± 1.32 | 4.41 ± 0.36 | 14.36 | <0.001 |
| OGTT 60 min glucose (mmol/l) | 9.76 ± 2.23 | 7.00 ± 1.43 | 25.80 | <0.001 |
| OGTT 120 min glucose (mmol/l) | 8.31 ± 2.15 | 6.10 ± 1.10 | 22.39 | <0.001 |
| HbA1c (%) | 5.44 ± 0.68 | 4.93 ± 0.70 | 13.17 | <0.001 |
| Triglyceride (mmol/l) | 2.67 ± 1.20 | 2.37 ± 1.03 | 4.90 | <0.001 |
| Total cholesterol (mmol/l) | 5.37 ± 1.16 | 5.24 ± 1.07 | 2.18 | 0.029 |
| HDL-c (mmol/l) | 1.61 ± 0.37 | 1.66 ± 0.41 | 2.09 | 0.036 |
| LDL-c (mmol/l) | 3.49 ± 1.03 | 3.35 ± 1.04 | 2.29 | 0.022 |
Abbreviations: HDL-c, high -ensity lipoprotein cholesterol; LDL-c, low-density lipoprotein cholesterol.
Relationship between studied variants and GDM risk
The genotypes of candidate SNPs were detected by a PCR-RFLP method. The results showed that the genotype frequencies of rs4842194 G>A, rs100537 A>G and rs2134095 T>C were in agreement with HWE in the controls (P>0.05), and the P-values were 0.727, 0.668 and 0.418, respectively. The D′ and r2 values were calculated to evaluate the LD of studied SNPs from their genotyping data by using the SHEsis online tool, and showed that there was no significant degree of LD detected among three SNPs (data not shown). A multivariable logistic regression analysis adjusted for age and BMI showed that the rs2134095 T>C in RXR-γ gene was significantly associated with the risk of GDM, individuals with CT genotype were 1.32-times higher than those with TT genotype (adjusted OR = 1.32, 95% CI = 1.01–1.73, P=0.043). However, the significance did not exist after Bonferroni’s correction. Further, under recessive genetic model, the CC genotype of rs2134095 could significantly reduce the risk of GDM compared with TT/CT genotypes (adjusted OR = 0.71, 95% CI = 0.56–0.90, P=0.004). We failed to find a significant association between the RXR-α rs4842194 G>A and RXR-γ rs100537 A>G and susceptibility to GDM in present study (P>0.05) as shown in Table 3.
Table 3. Association between genotypes of selected SNPs and GDM susceptibility.
| Gene | Genotype | Case | Control | P 1 | Crude OR (95% CI) | P 2 | Adjusted OR (95% CI) | P 3 |
|---|---|---|---|---|---|---|---|---|
| RXR-α | rs4842194 | |||||||
| GG | 235 | 327 | 0.326 | 1 | 1 | |||
| GA | 263 | 333 | 1.10 (0.87–1.39) | 0.427 | 1.05 (0.82–1.35) | 0.68 | ||
| AA | 75 | 80 | 1.31 (0.91–1.86) | 0.144 | 1.31 (0.90–1.91) | 0.164 | ||
| AA/GA | 338 | 413 | 0.249 | 1.14 (0.91–1.42) | 0.249 | 1.10 (0.87–1.39) | 0.418 | |
| GG/GA | 498 | 660 | 0.227 | 1 | 1 | |||
| AA | 75 | 80 | 1.24 (0.89–1.74) | 0.205 | 1.27 (0.89–1.82) | 0.183 | ||
| RXR-γ | rs100537 | |||||||
| AA | 48 | 55 | 0.199 | 1 | 1 | |||
| AG | 257 | 302 | 0.98 (0.64–1.49) | 0.907 | 0.87 (0.56–1.36) | 0.546 | ||
| GG | 268 | 383 | 0.80 (0.53–1.22) | 0.3 | 0.78 (0.50–1.21) | 0.268 | ||
| GG/AG | 525 | 685 | 0.536 | 0.88 (0.59–1.32) | 0.528 | 0.82 (0.54–1.26) | 0.364 | |
| AA/AG | 305 | 357 | 0.075 | 1 | ||||
| GG | 268 | 383 | 0.82 (0.66–1.02) | 0.073 | 0.88 (0.70–1.10) | 0.259 | ||
| rs2134095 | ||||||||
| TT | 158 | 219 | 0.004 | 1 | 1 | |||
| TC | 324 | 357 | 1.26 (0.98–1.62) | 0.076 | 1.32 (1.01–1.73) | 0.043 | ||
| CC | 91 | 164 | 0.77 (0.55–1.07) | 0.117 | 0.85 (0.60–1.20) | 0.363 | ||
| CC/TC | 415 | 521 | 0.425 | 1.10 (0.87–1.41) | 0.422 | 1.17 (0.91–1.52) | 0.221 | |
| TT/TC | 482 | 576 | 0.003 | 1 | ||||
| CC | 91 | 164 | 0.72 (0.58–0.89) | 0.003 | 0.71 (0.56–0.90) | 0.004 |
The significance level after Bonferroni's correction for multiple testing is 0.017. Bold values indicate that the difference is statistically significant.
Two-sided χ2 test for genotypes’ distributions between cases and controls.
Unconditional logistic regression analysis.
Adjusted for age, pre-BMI in logistic regression models.
We also estimated the rs2134095 T>C on GDM risk with a stratified analysis under the recessive genetic model. The results showed that compared with TT/CT genotypes, the rs2134095 CC genotype could significantly reduce the risk of GDM in age > 30 years old subjects (adjusted OR = 0.61, 95% CI = 0.39–0.97, P=0.037) and in BMI > 22 kg/m2 subgroup (adjusted OR = 0.46, 95% CI = 0.30–0.70, P<0.001), and increase the GDM risk in SBP > 120 mmHg people (adjusted OR = 1.96, 95% CI = 1.14–3.36, P<0.015), DBP ≤ 80 mmHg people (adjusted OR = 1.33, 95% CI = 1.04–1.71, P<0.023), DBP > 80 mmHg people (adjusted OR = 2.33, 95% CI = 1.15–4.75, P<0.020), HbA1c < 6.5% subgroup (adjusted OR = 1.41, 95% CI = 1.11–1.78, P<0.004), TG ≤ 1.7 mmol/l (adjusted OR = 2.57, 95% CI = 1.45–4.53, P<0.001), TC ≤ 5.18 mmol/l subgroup (adjusted OR = 1.58, 95% CI = 1.13–2.22, P<0.007), high-density lipoprotein cholesterol (HDL-c) ≤ 1.5 mmol/l subjects (adjusted OR = 1.70, 95% CI = 1.16–2.49, P<0.006) and LDL-c > 3.12 mmol/l subjects (adjusted OR = 1.47, 95% CI = 1.08–2.00, P<0.013). We also found that rs2134095 interacted with age (Pinteraction=0.039), pre-BMI (Pinteraction=0.040) and TG (Pinteraction=0.025) influencing individual’s susceptibility to GDM in the recessive genetic model as shown in Table 4.
Table 4. Stratification analysis for associations between RXR-γ rs2134095 T>C and GDM risk.
| Variables | TT/TC (case/control) |
CC (case/ control) | Crude OR (95%CI) | P1 | Adjusted OR (95% CI) | P2 | P3 |
|---|---|---|---|---|---|---|---|
| Age (years) | 0.039 | ||||||
| ≤30 | 210/395 | 49/117 | 0.79 (0.54–1.14) | 0.210 | 0.78 (0.53–1.14) | 0.202 | |
| >30 | 272/181 | 42/47 | 0.60 (0.38–0.94) | 0.026 | 0.61 (0.39–0.97) | 0.037 | |
| Pre-BMI (kg/m2) | 0.040 | ||||||
| ≤22 | 187/361 | 48/90 | 1.03 (0.70–1.52) | 0.884 | 1.06 (0.701.59) | 0.787 | |
| >22 | 295/215 | 43/74 | 0.42 (0.28–0.64) | <0.001 | 0.46 (0.30–0.70) | <0.001 | |
| SBP (mmHg) | |||||||
| ≤120 | 206/324 | 243/299 | 1.28 (1.00–1.63) | 0.048 | 1.29 (1.00–1.67) | 0.055 | 0.165 |
| >120 | 43/59 | 81/58 | 1.92 (1.14–3.22) | 0.014 | 1.96 (1.14–3.36) | 0.015 | |
| DBP (mmHg) | 0.134 | ||||||
| ≤80 | 214/347 | 271/333 | 1.32 (1.04–1.67) | 0.020 | 1.33 (1.04–1.71) | 0.023 | |
| >80 | 35/36 | 53/24 | 2.27 (1.16–4.44) | 0.016 | 2.33 (1.15–4.75) | 0.020 | |
| HbA1c (%) | |||||||
| ≤6.5 | 238/382 | 309/357 | 1.39 (1.12–1.74) | 0.004 | 1.41 (1.11–1.78) | 0.004 | - |
| >6.5 | 11/1 | 15/0 | - | - | - | - | |
| TG (mmol/L) | |||||||
| ≤1.7 | 30/106 | 55/81 | 2.40 (1.41–4.08) | 0.001 | 2.57 (1.45–4.53) | 0.001 | 0.025 |
| >1.7 | 219/277 | 269/276 | 1.23 (0.97–1.57) | 0.093 | 1.22 (0.94–1.58) | 0.133 | |
| TC (mmol/l) | 0.761 | ||||||
| ≤5.18 | 118/208 | 142/175 | 1.43 (1.04–1.96) | 0.027 | 1.58 (1.13–2.22) | 0.007 | |
| >5.18 | 131/175 | 182/182 | 1.34 (0.98–1.81) | 0.063 | 1.21 (0.87–1.67) | 0.256 | |
| HDL-c (mmol/l) | 0.554 | ||||||
| ≤1.5 | 96/147 | 128/129 | 1.52 (1.07–2.17) | 0.021 | 1.70 (1.16–2.49) | 0.006 | |
| >1.5 | 153/236 | 196/228 | 1.33 (1.00–1.75) | 0.047 | 1.27 (0.95–1.70) | 0.112 | |
| LDL-c (mmol/l) | 0.670 | ||||||
| ≤3.12 | 105/168 | 125/152 | 1.32 (0.94–1.85) | 0.113 | 1.29 (0.90–1.85) | 0.160 | |
| >3.12 | 144/215 | 199/205 | 1.45 (1.09–1.93) | 0.011 | 1.47 (1.08–2.00) | 0.013 |
Unconditional logistic regression analysis.
Adjusted for age, pre-BMI in logistic regression models.
Test for multiplicative interaction obtained from logistic regression models.
FPRP analysis
The FPRP test was adopted to assess the noteworthiness of the observed significant associations between the studied rs2134095 T>C SNP and the risk of GDM, and the prior probability of 0.1 and a relatively stringent FPRP cut-off value of 0.2 were set. The FPRP values were 0.050, 0.031, 0.049, 0.149, 0.176 and 0.178 for the associations of rs2134095 and GDM risk in all subjects, and subgroups of BMI > 22 kg/m2, HbA1c ≤ 6.5%, TC ≤ 5.18 mmol/l, HDL-c ≤ 1.5 mmol/l and LDL-c > 3.12 mmol/l, respectively, under the recessive genetic model (CC vs. TT/TC). It suggested that the above associations were noteworthy findings as presented in the present study as shown in Table 5.
Table 5. FPRP analysis for the significant associations of the rs2134095 T>C and GDM risk.
| Comparisons | Adjusted OR (95% CI) | Prior probability | |||||
|---|---|---|---|---|---|---|---|
| 0.25 | 0.1 | 0.01 | 0.001 | 0.0001 | 0.00001 | ||
| TC vs. TT | 1.32 (1.01–1.73) | 0.136 | 0.321 | 0.839 | 0.981 | 0.998 | 1.000 |
| CC vs. TT/TC | 0.71 (0.56–0.90) | 0.017 | 0.050 | 0.367 | 0.854 | 0.983 | 0.998 |
| Subgroup | |||||||
| Age > 30 years | 0.61 (0.39–0.97) | 0.238 | 0.484 | 0.912 | 0.990 | 0.999 | 1.000 |
| BMI > 22 kg/m2 | 0.46 (0.30–0.70) | 0.010 | 0.031 | 0.259 | 0.779 | 0.972 | 0.997 |
| SBP > 120 mmHg | 1.96 (1.14–3.36) | 0.210 | 0.444 | 0.898 | 0.989 | 0.999 | 1.000 |
| DBP (mmHg) | |||||||
| ≤80 | 1.33 (1.04–1.71) | 0.078 | 0.203 | 0.737 | 0.966 | 0.966 | 1.000 |
| >80 | 2.33 (1.15–4.75) | 0.347 | 0.614 | 0.946 | 0.994 | 0.999 | 1.000 |
| HbA1c ≤ 6.5% | 1.41 (1.11–1.78) | 0.017 | 0.049 | 0.360 | 0.850 | 0.983 | 0.998 |
| TG ≤ 1.7 mmol/l | 2.57 (1.45–4.53) | 0.092 | 0.234 | 0.771 | 0.971 | 0.997 | 1.000 |
| TC ≤ 5.18 mmol/l | 1.58 (1.13–2.22) | 0.055 | 0.149 | 0.658 | 0.951 | 0.995 | 0.999 |
| HDL-c ≤ 1.5 mmol/l | 1.70 (1.16–2.49) | 0.066 | 0.176 | 0.701 | 0.960 | 0.996 | 1.000 |
| LDL-c > 3.12 mmol/l | 1.47 (1.08–2.00) | 0.067 | 0.178 | 0.705 | 0.960 | 0.996 | 1.000 |
Bold values indicate that the difference is statistically significant at the test level of α=0.2.
High-order interaction with GDM risk by MDR analysis
The results of MDR analysis indicated that rs2134095 was the best one-factor model for GDM with a maximum CVC of 10/10, a testing balanced accuracy of 0.531, and a P-value of statistical test of 0.003. More interestingly, the best interaction model to predict GDM risk for the present study population was a two-factor model of rs2134095 and rs100537 with a maximum CVC of 10/10, a testing balanced accuracy of 0.552, and a P-value <0.001 (Table 6).
Table 6. MDR analysis for the GDM risk prediction.
| Best model | Training balanced accuracy | Testing balanced accuracy | CVC | λ² | P |
|---|---|---|---|---|---|
| 1 | 0.542 | 0.531 | 10/10 | 8.91 | 0.003 |
| 1, 2 | 0.555 | 0.552 | 10/10 | 17.62 | <0.001 |
| 1, 2, 3 | 0.574 | 0.540 | 10/10 | 27.04 | <0.001 |
Labels: 1. rs2134095; 2. rs100537; 3. rs4842194.
P-value for 1000-fold permutation test.
The best model was selected as the one in boldface with the maximum prediction precision and CVC.
Functional prediction of rs2134095
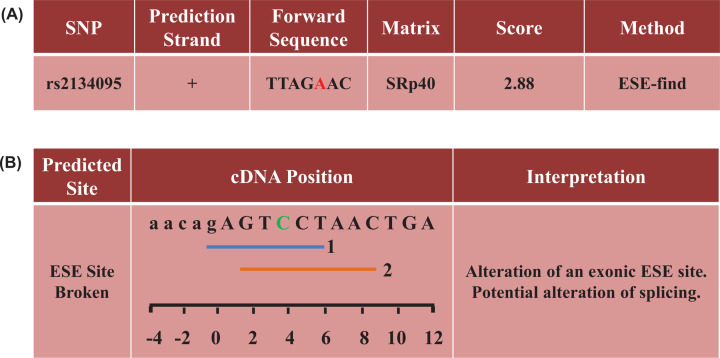
Based on the positive findings identified above, the RXR-γ rs2134095 was further explored of a putative alteration in regulation of gene splicing. It seems that the variant has an impact in splicing regulation according to SNP Function Prediction of SNPinfo Web Server and Human Splicing Finder 3.1 tool. The variant is located in an acceptor splicing site, and alteration of the C→T allele of rs2134095 leads to the disruption of an exonic splicing enhancer (ESE) SRp40 (Figure 1). It is biologically reasonable to hypothesize that the variant has an effect on the splicing of the RXR-γ mRNA, thus altering the function of the gene product.
Figure 1. Predicted the function of rs2134095 on RXR-γ post-transcriptional splicing sites.
(A) Predicted by SNPinfo Web Server (base complementary pairing principle: A=T, G≡C). (B) Predicted by Human Splicing Finder 3.1 tool.
Discussion
GDM is characterized by β-cell dysfunction, insulin secretory defect and peripheral insulin resistance. It is likely to bring adverse consequences for both mothers and their offspring, for instance, pregnant women with GDM are often at a relatively higher risk of T2DM, obesity, dyslipidemia, preterm birth, spontaneous abortion, respiratory distress syndrome of newborn etc. Studies showed that the RXR-α/γ proteins perform the function of transcription factors by binding as heterodimers to the specific sequences in the promoter of target genes of VD and retinoid pathways, which are closely related to diabetes, islet function, and glucose and lipid metabolism. We believe that it is necessary to explore the relationship between RXR-α/γ and GDM. GWASs and meta-analysis have shown that association genetic loci could provide insight into pathogenetic mechanisms underlying the GDM [20,34–36]. To date, there have been studies on multiple genes and their SNPs of VD pathway with the susceptibility to T2DM and GDM [26,37–39]. Studies have shown that the variants in RXR-α/γ genes might be associated with the risk of T2DM and metabolic syndrome [24–26]. We genotyped three potential functional SNPs in RXR-α/γ genes explore the pathogenesis of GDM.
In the single locus analysis, we found that the RXR-γ rs2134095 T>C was significantly associated with the risk of GDM under the recessive genetic model in a Southern Chinese population. In the stratified analysis, the reduced risk of GDM associated with rs2134095 SNP was more pronounced among the elderly (>30 years old) and relatively higher pre-BMI (>22 kg/m2) subjects carrying the CC genotype in the recessive genetic model. Further, the interaction between rs2134095 and age and pre-BMI was detected. The above findings suggest that not only a single environmental or genetic risk factor but also their interaction might affect the individual suffering from GDM under different etiologies. Additionally, recent studies have confirmed that the variants located in RXR-γ were significantly associated with the lipid metabolism of chronic metabolic diseases. These variants are closely related to abnormal lipid metabolism by affecting the levels of TG, LDL-c, apolipoprotein B etc [40–42]. In the hierarchical analysis of this study, higher risk effects in HbA1c (≤6.5%), TG (≤1.7 mmol/l), TC (≤5.18 mmol/l) and LDL-c (>3.12 mmol/l) are observed in rs2134095CC carrier subjects. These findings might be helpful to clarify the genetic component of RXR-γ and lipid profile.
Very interesting, results from the MDR analysis also consistently identified RXR-γ rs2134095 T>C as the main single susceptibility locus contributed to the risk of GDM, and some complex gene–gene interaction effects exist among the RXR-γ SNPs. It showed that a two-factor model including the combination of rs2134095 and rs100537, is the best model to predict GDM risk in the study population. These results suggest that RXR-γ gene variants may have a joint effect on the genesis of GDM.
These above findings seem to be biologically plausible. Study confirmed that gene variants might change the splicing pattern of pre-mRNA leading to a wrong transcript product and ultimately influence the gene expression or function [43–45]. According to the online prediction database ‘snpinfo web server’, the SNP rs2134095 may play the role of exonic splicing enhancer (ESE) or silencer (ESS) performing a function of RXR-γ gene post transcriptional splicing. Grave et al. used a general linear model to evaluate the VD pathway gene–gene interaction and detected a significant effect of the interaction between RXR-γ rs2134095 and GC rs7041 on low-density lipoprotein cholesterol (LDL-c) levels. The further functional analysis indicated that the studied variants were involved in the regulation of the gene expression [42]. Another study of Sentinelli et al. found that the ‘at-risk’ haplotype of RXR-γ gene variants (rs1128977, rs2651860, rs2134095, rs283696 and rs10918169) were associated with the risk of familial combined hyperlipidemia (FCHL) and a higher LDL-c level [40]. It is speculated that the rs2134095 T>C may affect the function of RXR-γ and alter the activation of retinoid and VD pathway genes, which are closely related to diabetes, islet function, and glucose and lipid metabolism. In addition, maternal plasma levels of vitamin A and vitamin D reduce during pregnancy, thus exacerbating the functional impairment of above pathways caused by rs2134095. The above is likely to explain the mechanism of the changes in the susceptibility of pregnant individual to GDM.
The FPRP analysis is an effective method to verify the noteworthiness of significant associations [46]. In the present study, a relatively stringent FPRP threshold of 0.2 was set. The FPRP values of the observed significant associations between rs2134095 T>C and the risk GDM is much lower than the preset threshold 0.2 in BMI > 22 kg/m2, HbA1c ≤ 6.5%, TC ≤ 5.18 mmol/l, HDL-c ≤ 1.5 mmol/l and LDL-c > 3.12 mmol/l subgroups under the recessive genetic model. It suggests that the positive findings are probability authentic and reliable. Meanwhile, the detected FPRP values in some comparisons much greater than 0.2 indicated that these significant findings might be chance observations. Therefore, the conclusion obtained here must be viewed as preliminary and needs to be verified.
To our knowledge, it was the first time to explore the relationship between the studied functional variants of RXR-α/γ genes and the risk of GDM, and some etiological clues of GDM are suggested from the perspective of genetics. The strengths of our study include its good design, relatively large sample size and multiple analysis methods. However, some limitations in this study should not be neglected. First, designed as a hospital-based case–control study, it may be still subject to inherent biases, like selection bias and recall bias. Second, although relatively large samples were recruited, however, the lower minor allele frequency of some tested SNPs might still limit the statistical power to detect significant association in some subgroups. Third, some positive correlations between rs2134095 T>C and GDM risk were observed, however, the specific mechanisms underlying the effect of the above-mentioned positive findings should be investigated.
Conclusion
In summary, the present study support that the RXR-γ gene rs2134095 T>C variant is significant associated with the increased risk of GDM by effect of a single locus and/or complex joint gene–gene and gene–environment interactions. Furthermore, larger sample-size and different population studies are required to confirm our findings.
Abbreviations
- BMI
body mass index
- CI
confidence interval
- CVC
cross-validation consistency
- ESE
exonic splicing enhancer
- FPRP
false positive report probability
- GDM
gestational diabetes mellitus
- GWAS
genome-wide association study
- HbA1c
glycosylated hemoglobin A1c
- HDL-c
high-density lipoprotein cholesterol
- HWE
Hardy–Weinberg equilibrium
- LD
linkage disequilibrium
- LDL-c
low-density lipoprotein cholesterol
- MDR
multiple dimension reduction
- MMBS
microRNA–mRNA binding site
- OGTT
oral glucose tolerance test
- OR
odds ratio
- PCR
polymerase chain reaction
- RFLP
restriction fragment length polymorphism
- RXRα/γ
retinoid X receptor -α/γ
- SBP
systolic blood pressure
- SNP
single nucleotide polymorphism
- TC
total cholesterol
- TFBS
transcription factor-binding site
- TG
triglyceride
- T2DM
type 2 diabetes
- UTR
untranslated region
- VD
vitamin D
Contributor Information
Xiang-yuan Yu, Email: 112013033@glmc.edu.cn.
Da-bin Liu, Email: 13489997701@139.com.
Data Availability
The data used to support the findings of the present study are included within the article.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by the Guangxi Young and Middle-Aged Teachers’ Basic Ability Improvement Project [grant number 2020KY12028]; the Guangxi ‘Bagui Scholars’ Reproductive and Maternal and Child Health Project, National Natural Science Foundation of China [grant number 81760614]; the Key Research and Development Program [grant number 2018AB62004]; and the College Students’ Innovation Project of Guangxi, China [grant numbers 201710601103, 201810601031].
CRediT Author Contribution
Xiang-yuan Yu: Resources, Software, Validation, Writing—review & editing. Li-ping Song: Formal analysis, Validation. Hui-ting Zheng: Data curation, Formal analysis. Shu-dan Wei: Formal analysis, Investigation. Xiao-lan Wen: Data curation, Investigation. Bo Huang: Data curation, Validation, Writing—review & editing. Da-bin Liu: Conceptualization, Writing—review & editing.
References
- 1.American Diabetes Association . (2004) Gestational diabetes mellitus. Diabetes Care 27, S88–S90 10.2337/diacare.27.2007.S88 [DOI] [PubMed] [Google Scholar]
- 2.Ben-Haroush A., Yogev Y. and Hod M. (2004) Epidemiology of gestational diabetes mellitus and its association with Type 2 diabetes. Diabet. Med. 21, 103–113 10.1046/j.1464-5491.2003.00985.x [DOI] [PubMed] [Google Scholar]
- 3.Farrar D., Simmonds M., Bryant M.et al. (2016) Hyperglycaemia and risk of adverse perinatal outcomes: systematic review and meta-analysis. BMJ 354, i4694 10.1136/bmj.i4694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coustan D.R. (2013) Pregnancy outcomes in women with and without gestational diabetes mellitus according to the international association of the diabetes and pregnancy study groups criteria. Obstet. Gynecol. 121, 377 10.1097/AOG.0b013e318280e05d [DOI] [PubMed] [Google Scholar]
- 5.Li M., Rahman M.L., Wu J.et al. (2020) Genetic factors and risk of type 2 diabetes among women with a history of gestational diabetes: findings from two independent populations. BMJ Open Diabetes Res. Care 8, e000850 10.1136/bmjdrc-2019-000850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leng J., Shao P., Zhang C.et al. (2015) Prevalence of gestational diabetes mellitus and its risk factors in Chinese pregnant women: a prospective population-based study in Tianjin, China. PLoS ONE 10, e0121029 10.1371/journal.pone.0121029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu L., Han L., Zhan Y.et al. (2018) Prevalence of gestational diabetes mellitus and associated risk factors in pregnant Chinese women: a cross-sectional study in Huangdao, Qingdao, China. Asia Pac. J. Clin. Nutr. 27, 383–388 [DOI] [PubMed] [Google Scholar]
- 8.Chu S.Y., Callaghan W.M., Kim S.Y.et al. (2007) Maternal obesity and risk of gestational diabetes mellitus. Diabetes Care 30, 2070–2076 10.2337/dc06-2559a [DOI] [PubMed] [Google Scholar]
- 9.Rhee S.Y., Kim J.Y., Woo J.T.et al. (2010) Familial clustering of type 2 diabetes in Korean women with gestational diabetes mellitus. Korean J. Intern. Med. 25, 269–272 10.3904/kjim.2010.25.3.269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang C., Bao W., Rong Y.et al. (2013) Genetic variants and the risk of gestational diabetes mellitus: a systematic review. Hum. Reprod. Update 19, 376–390 10.1093/humupd/dmt013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J.Y., Cheong H.S., Park B.L.et al. (2011) Melatonin receptor 1 B polymorphisms associated with the risk of gestational diabetes mellitus. BMC Med. Genet. 12, 82 10.1186/1471-2350-12-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou Y., Wang H., Zhou J.et al. (2021) Vitamin A and its multi-effects on pancreas: recent advances and prospects. Front. Endocrinol. (Lausanne) 12, 620941 10.3389/fendo.2021.620941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao S., Li R., Li Y.et al. (2012) Roles of vitamin A status and retinoids in glucose and fatty acid metabolism. Biochem. Cell Biol. 2, 142–152 10.1139/o11-079 [DOI] [PubMed] [Google Scholar]
- 14.Trasino S.E. and Gudas L.J. (2015) Vitamin A: a missing link in diabetes? Diabetes Manag. (Lond.) 5, 359–367 10.2217/dmt.15.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haddad Kashani H., Seyed Hosseini E., Nikzad H.et al. (2018) The effects of vitamin D supplementation on signaling pathway of inflammation and oxidative stress in diabetic hemodialysis: a randomized, double-blind, placebo-controlled trial. Front. Pharmacol. 9, 50 10.3389/fphar.2018.00050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui J., Chen H. and Huang B.R. (2011) The recent progress in the studies of vitamin D receptor. Sheng Li Ke Xue Jin Zhan 42, 95–99 [PubMed] [Google Scholar]
- 17.Liu K.N., Meng H.X. and Hou J.X. (2018) Influence of vitamin D receptor FokI polymorphism on expression of CYP24A1 in periodontal cells. Beijing Da Xue Xue Bao 50, 13–19 [PubMed] [Google Scholar]
- 18.Liu Y., He Y., Wang Q.et al. (2019) Vitamin D3 supplementation improves testicular function in diabetic rats through peroxisome proliferator-activated receptor-gamma/transforming growth factor-beta 1/nuclear factor-kappa B. J. Diabetes Investig. 10, 261–271 10.1111/jdi.12886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radha V., Kanthimathi S., Anjana R.M.et al. (2016) Genetics of gestational diabetes mellitus. J. Pak. Med. Assoc. 66, S11–S14 [PubMed] [Google Scholar]
- 20.Kwak S.H., Kim S.H., Cho Y.M.et al. (2012) A genome-wide association study of gestational diabetes mellitus in Korean women. Diabetes 61, 531–541 10.2337/db11-1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaulton K.J., Ferreira T., Lee Y.et al. (2015) Genetic fine mapping and genomic annotation defines causal mechanisms at type 2 diabetes susceptibility loci. Nat. Genet. 12, 1415–1425 10.1038/ng.3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Firneisz G., Rosta K., Al-Aissa Z.et al. (2018) The MTNR1B rs10830963 variant in interaction with pre-pregnancy BMI is a pharmacogenetic marker for the initiation of antenatal insulin therapy in gestational diabetes mellitus. Int. J. Mol. Sci. 12, 3734 10.3390/ijms19123734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hasstedt S.J., Chu W.S., Das S.K.et al. (2008) Type 2 diabetes susceptibility genes on chromosome 1q21-24. Ann. Hum. Genet. 72, 163–169 10.1111/j.1469-1809.2007.00416.x [DOI] [PubMed] [Google Scholar]
- 24.Shi H., Yu X., Li Q.et al. (2012) Association between PPAR-gamma and RXR-alpha gene polymorphism and metabolic syndrome risk: a case-control study of a Chinese Han population. Arch. Med. Res. 43, 233–242 10.1016/j.arcmed.2012.03.006 [DOI] [PubMed] [Google Scholar]
- 25.Lu Y., Ye X., Cao Y.et al. (2011) Genetic variants in peroxisome proliferator-activated receptor-gamma and retinoid X receptor-alpha gene and type 2 diabetes risk: a case-control study of a Chinese Han population. Diabetes Technol. Ther. 13, 157–164 10.1089/dia.2010.0122 [DOI] [PubMed] [Google Scholar]
- 26.Du J., Shi H., Lu Y.et al. (2011) Tagging single nucleotide polymorphisms in the PPAR-gamma and RXR-alpha gene and type 2 diabetes risk: a case-control study of a Chinese Han population. J. Biomed. Res. 25, 33–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi H., Lu Y., Du J.et al. (2012) Application of back propagation artificial neural network on genetic variants in adiponectin ADIPOQ, peroxisome proliferator-activated receptor-gamma, and retinoid X receptor-alpha genes and type 2 diabetes risk in a Chinese Han population. Diabetes Technol. Ther. 14, 293–300 10.1089/dia.2011.0071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lowe W.L. Jr, Scholtens D.M.et al. (2016) Genetics of gestational diabetes mellitus and maternal metabolism. Curr. Diabetes Rep. 16, 15 10.1007/s11892-015-0709-z [DOI] [PubMed] [Google Scholar]
- 29.Shi Y.Y. and He L. (2005) SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2, 97–98 [DOI] [PubMed] [Google Scholar]
- 30.Wacholder S., Chanock S., Garcia-Closas M.et al. (2004) Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J. Natl. Cancer Inst. 96, 434–442 10.1093/jnci/djh075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He J., Wang M.Y., Qiu L.X.et al. (2013) Genetic variations of mTORC1 genes and risk of gastric cancer in an Eastern Chinese population. Mol. Carcinog. 52, E70–E79 10.1002/mc.22013 [DOI] [PubMed] [Google Scholar]
- 32.Hahn L.W., Ritchie M.D. and Moore J.H. (2003) Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics 19, 376–382 10.1093/bioinformatics/btf869 [DOI] [PubMed] [Google Scholar]
- 33.Desmet F.O., Hamroun D., Lalande M.et al. (2009) Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 37, e67 10.1093/nar/gkp215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu N.N., Zhao D., Ma W.et al. (2021) A genome-wide association study of gestational diabetes mellitus in Chinese women. J. Mater. Fetal Neonatal Med. 34, 1557–1564 [DOI] [PubMed] [Google Scholar]
- 35.Huang B., Wang Y.K., Qin L.Y.et al. (2019) A functional polymorphism rs10830963 in melatonin receptor 1B associated with the risk of gestational diabetes mellitus. Biosci. Rep. 39, BSR20190744 10.1042/BSR20190744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo F., Long W., Zhou W.et al. (2018) FTO, GCKR, CDKAL1 and CDKN2A/B gene polymorphisms and the risk of gestational diabetes mellitus: a meta-analysis. Arch. Gynecol. Obstet. 298, 705–715 10.1007/s00404-018-4857-7 [DOI] [PubMed] [Google Scholar]
- 37.Shi A., Wen J., Liu G.et al. (2016) Genetic variants in vitamin D signaling pathways and risk of gestational diabetes mellitus. Oncotarget 7, 67788–67795 10.18632/oncotarget.11984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siqueira T.W., Araujo Junior E., Mattar R.et al. (2019) Assessment of polymorphism of the VDR gene and serum vitamin D values in gestational diabetes mellitus. Revis. Bras. Ginecol. E Obstet. 41, 425–431 10.1055/s-0039-1693678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.El-Beshbishy H.A., Tawfeek M.A., Taha I.M.et al. (2015) Association of vitamin D receptor gene BsmI (A>G) and FokI (C>T) polymorphism in gestational diabetes among Saudi Women. Pak. J. Med. Sci. 31, 1328–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang H., Chu W., Hemphill C.et al. (2002) Mutation screening and association of human retinoid X receptor gamma variation with lipid levels in familial type 2 diabetes. Mol. Genet. Metab. 76, 14–22 10.1016/S1096-7192(02)00016-1 [DOI] [PubMed] [Google Scholar]
- 41.Sentinelli F., Minicocci I., Montali A.et al. (2013) Association of RXR-gamma gene variants with familial combined hyperlipidemia: genotype and haplotype analysis. J. Lipids 2013, 517943 10.1155/2013/517943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grave N., Tovo-Rodrigues L., da Silveira J.et al. (2016) A vitamin D pathway gene-gene interaction affects low-density lipoprotein cholesterol levels. J. Nutr. Biochem. 38, 12–17 10.1016/j.jnutbio.2016.08.002 [DOI] [PubMed] [Google Scholar]
- 43.Thi Tran H.T., Takeshima Y., Surono A., Yagi M.et al. (2005) A G-to-A transition at the fifth position of intron-32 of the dystrophin gene inactivates a splice-donor site both in vivo and in vitro. Mol. Genet. Metab. 85, 213–219 10.1016/j.ymgme.2005.03.006 [DOI] [PubMed] [Google Scholar]
- 44.Xie M., Zhang J., Singan V.R.et al. (2020) Identification of functional single nucleotide polymorphism of Populus trichocarpa PtrEPSP-TF and determination of its transcriptional effect. Plant Direct 4, e00178 10.1002/pld3.178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moorehead A., Hanna R., Heroux D.et al. (2020) A thymic stromal lymphopoietin polymorphism may provide protection from asthma by altering gene expression. Clin. Exp. Allergy 50, 471– 478 10.1111/cea.13568 [DOI] [PubMed] [Google Scholar]
- 46.Tong X., Ma Y., Deng H.et al. (2016) The SDF-1 rs1801157 polymorphism is associated with cancer risk: an update pooled analysis and FPRP test of 17,876 participants. Sci. Rep. 6, 27466 10.1038/srep27466 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of the present study are included within the article.