Abstract
Mycobacterium tuberculosis complex strains cultured in Denmark have been analyzed by IS6110 restriction fragment length polymorphism (RFLP) on a routine basis from 1992 and onwards. Due to the influx of immigrants with tuberculosis, the number of strains harboring only one to five copies of IS6110 has increased steadily. Since the discriminatory power of IS6110 fingerprinting for such strains is poor, we have performed additional genotyping of all low-copy-number strains by the recently described PCR-based method known as spoligotyping. A total of 311 clinical strains were typed: 14 Mycobacterium bovis BCG, 48 M. bovis, and 249 M. tuberculosis strains. Spoligotyping correctly differentiated M. bovis and M. bovis BCG from M. tuberculosis strains, but it did not differentiate M. bovis from M. bovis BCG. All M. bovis BCG strains exhibited identical spoligotype patterns. The discriminatory power of spoligotyping of low-copy-number M. tuberculosis strains was higher than that of IS6110 fingerprinting. Based on RFLP typing solely, 83% of the low-copy-number M. tuberculosis strains were found to form part of a cluster, and 75% were found to form a cluster on the basis of spoligotyping. When the two techniques were combined, the amount of clustering decreased to 55%. The combination of these two techniques might be valuable in studying the epidemiology of M. tuberculosis strains harboring few copies of the IS6110 element.
Restriction fragment length polymorphism (RFLP) typing performed by using the insertion element IS6110 as a probe has become the most widely used method for strain differentiation of Mycobacterium tuberculosis isolates. The RFLP typing results are stable and reproducible and exhibit a high degree of discrimination. Furthermore, RFLP typing has proven useful for detection of outbreaks (6, 8), control of laboratory cross-contamination (3, 24), and population-based epidemiologic studies of tuberculosis (TB) (1, 23, 30, 31). Due to the development of a standardized methodology (27), it has become possible to exchange DNA fingerprints between laboratories and to establish research collaboration and surveillance efforts within the European Union (28). The detection of clustered strains (i.e., strains exhibiting identical patterns) has been used as a marker of recent transmission (1, 23, 30, 31). However, studies have shown that clustering of strains harboring less than five copies of IS6110 frequently shows discordant results if further typing systems are used (5, 29).
IS6110 RFLP has been used to type TB-associated strains cultured in Denmark from 1992 and onwards (4), and isolates from approximately 93% of the patients have been analyzed. Since the beginning of 1997 the results have been directly used in the control of TB in Denmark; regular reports are supplied to the physicians who use this information in contact tracing and source case identification. Because clustering as an indicator of recent transmission is considered unreliable for strains carrying a low number of IS6110 copies, the results for these strains have not been used for this purpose so far. The number of low-copy-number strains has increased (from 17 in 1992 to 70 in 1997). This can be explained by the increasing percentage of strains originating from immigrants (from 41% of culture-verified cases in 1992 to 68% in 1997). Strains isolated from native Danes almost exclusively exhibit high-copy-number patterns (4). A number of additional typing methods for IS6110 low-copy-number strains have been used (10, 15, 21, 25, 29). One of those methods is the PCR-based spoligotyping based on DNA polymorphism of the direct repeat (DR) region (15). Spoligotyping is a rapid method that allows large numbers of isolates to be handled in a short time. The aim of this study was to evaluate spoligotyping as a method for subtyping the IS6110 low-copy-number M. tuberculosis complex strains cultured in Denmark.
MATERIALS AND METHODS
Bacterial isolates.
TB notifications in Denmark are registered at the Statens Serum Institut in Copenhagen, where the diagnostics of TB in Denmark, Greenland, and the Faeroe Islands are centralized at the Department of Mycobacteriology. In addition, this department serves as a reference laboratory for TB diagnostics in Iceland. Of the notified cases of TB in Denmark, 83 to 91% are culture positive (16–19). Isolates from approximately 93% of the patients with culture-verified cases from 1992 and onwards have been analyzed by RFLP (4, 31). From the beginning of 1995, these analyses have been performed as part of the routine diagnostic procedures. As soon as growth is obtained—usually in the BACTEC culture system (Becton Dickinson and Company, Sparks, Md.)—and species identification by AccuProbe (Gen-Probe Inc., San Diego, Calif.) has revealed the presence of M. tuberculosis complex, the isolate is subcultured in Dubos medium containing Tween 80. Further species identification of Mycobacterium bovis and M. bovis BCG is performed by susceptibility testing (pyrazinamide, thiopheno-2-carboxylic acid hydrazide, and cycloserine), niacin accumulation testing, and evaluation of the ability to reduce nitrate.
RFLP analyses.
After 3 to 4 weeks of growth in Dubos media, the bacteria are harvested by centrifugation and heat killed (90°C for 30 min), and RFLP is performed by using the standardized method (27). In brief, DNA is extracted and digested with PvuII. After electrophoresis on agarose gel the digested DNA is transferred to nylon membranes (Hybond N+; Amersham) and probed with a chemiluminiscence-labelled 245-bp sequence of IS6110. The results are scanned and analyzed by computer by using the Gelcompar software (Applied Maths, Kortrijk, Belgium) as described previously (14).
Spoligotyping.
Strains collected from January 1992 through June 1998 carrying fewer than six copies of IS6110 were selected for spoligotyping. Furthermore, clustered strains (i.e., strains exhibiting 100% identical RFLP patterns) with six copies were also included. From a few strains, purified chromosomal DNA from RFLP typing was still available. For the remaining strains, DNA extraction was performed by mechanical disruption of the cells (either a few colonies from solid media or 100 μl of the frozen culture stock) with glass beads by vortexing (26). Membranes for spoligotyping were obtained from Isogen, Bioscience BV, Utrecht, The Netherlands. The membranes contained oligonucleotides derived from the spacer DNA sequences, interspersing with the directly repeated sequences in the DR region of the M. tuberculosis strain H37RV and M. bovis BCG. The presence or absence of these spacers in M. tuberculosis complex strains can be detected by hybridization following amplification of these spacer regions by primers complementary to the DR. PCR and hybridization were performed as previously described (15). The spoligotyping patterns were scanned and compared by using the Gelcompar program and visually.
RESULTS
A total of 2,745 cases of TB were reported from 1992 to 1997. Of these, 2,336 (85%) were culture positive. From January through June 1998, a total of 280 patients were identified as culture positive for M. tuberculosis complex. Isolates from approximately 93% of the patients with culture-verified cases from January 1992 through June 1998 were previously analyzed by RFLP. For the same period, isolates collected from 213 patients from Greenland, 27 patients from Iceland, and 5 patients from the Faeroe Islands have been analyzed by RFLP.
Bacterial strains with one to five IS6110 copies.
Of 316 M. tuberculosis complex strains with five or fewer IS6110 copies collected from 316 patients and identified, 311 (98%) were available for spoligotyping. The species of the 311 low-copy-number strains are shown in Table 1. Of these strains, 309 originated from patients diagnosed in Denmark (these comprised 13% of RFLP-analyzed isolates). One M. bovis BCG strain originated from a patient in Greenland, and one originated from a patient in the Faeroe Islands. Except for these two, all isolates from Greenland, Iceland, and the Faeroe Islands exhibited RFLP patterns with more than five copies of IS6110. The regions of origin of the patients infected with strains containing few IS6110 copies and of patients with culture-verified TB are shown in Table 2.
TABLE 1.
IS6110 low-copy-number M. tuberculosis complex isolates classified by species and by nationality of patients
| Nationality | No. (%)
|
||
|---|---|---|---|
| M. tuberculosis | M. bovis | M. bovis BCG | |
| Danish | 27 (12) | 41 (84) | 10 (73) |
| Foreigna | 222 (88) | 7 (15) | 4 (27) |
| Total | 249 | 48 | 14b |
Patients of foreign nationality were defined as persons born outside Denmark and their children up to the age of 25 years.
Ten complications that occurred following vaccinations are included. In three other cases BCG was cultured from urine following treatment of bladder cancer with BCG, and in one case disseminated BCG infection occurred following vaccination of a human immunodeficiency virus-infected child.
TABLE 2.
Regions of origin of patients with M. tuberculosis culture-verified TB and with IS6110 low-copy-number M. tuberculosis isolates
| Region or country of origin of patients | No. (%) of patients with culture-verified TBa | No. (%) of low-copy-number isolatesb |
|---|---|---|
| Denmark | 1,035 (44) | 27 (11) |
| Central and East Africa | 565 (24) | 135 (54) |
| West Africa | 25 (1) | 2 (1) |
| North Africa | 27 (1) | 3 (1) |
| Asia | 360 (15.5) | 64 (26) |
| Europe | 173 (7.5) | 11 (4) |
| North America | 63 (3) | 2 (1) |
| South America | 9 (0.5) | 0 |
| Unknown | 79 (3.5) | 5 (2) |
| Total | 2,336 | 249 |
Patients with culture-verified TB identified between 1992 and 1997.
Low-copy-number isolates from 1992 to June 1998.
RFLP typing.
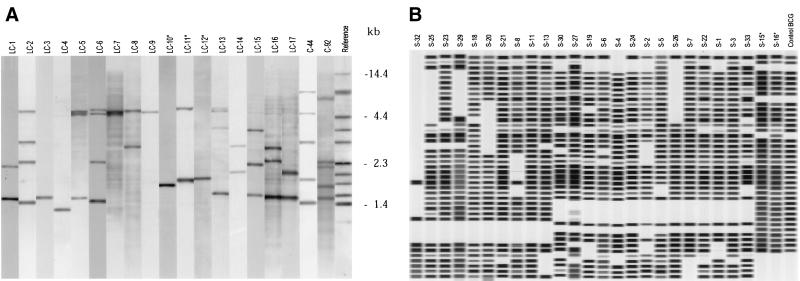
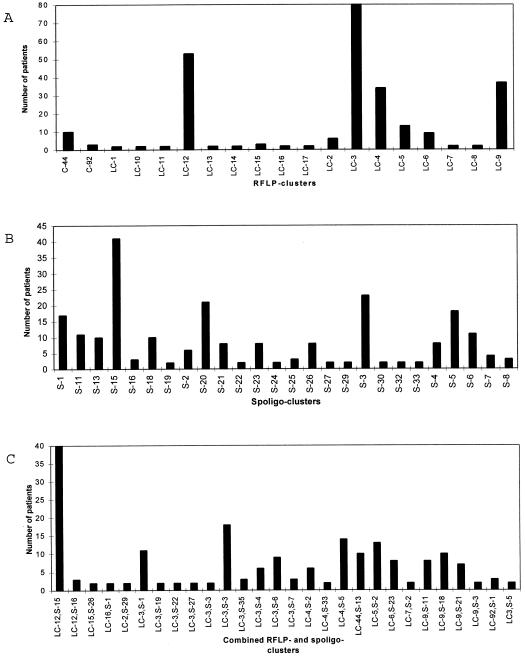
All M. bovis BCG strains exhibited the same, one-band pattern (low-copy-number strain cluster 12 [LC-12], Fig. 1A). Among the 48 M. bovis strains, nine different patterns were observed. Three strains (6%) yielded unique patterns and 45 strains (94%) belonged to one of six different low-copy-number strain clusters, of which three (LC-10 [n = 2], LC-11 [n = 2], and LC-12 [n = 38], Fig. 1A) comprised M. bovis strains or M. bovis BCG strains only. Three other strains belonged to clusters (LC-3 [n = 1], LC-4 [n = 1], and LC-9 [n = 1], Fig. 1A) also comprising M. tuberculosis strains. Among the 249 M. tuberculosis strains, 56 different patterns were observed. Forty strains (16%) were unique, and 209 strains (84%) belonged to one of a total of 16 clusters comprising strains collected from 2 to 80 patients (Fig. 1A and 2A).
FIG. 1.
RFLP patterns and spoligotyping patterns. (A) Computer-generated, normalized image showing representative examples of the RFLP patterns of 19 IS6110 low-copy-number clusters. LC-10, LC-11, and LC-12 comprise M. bovis and M. bovis BCG only. The last lane contains reference strain MT14323. (B) Computer-generated, normalized image showing representative examples of 26 spoligotyping clusters. S-15 and S-16 comprise M. bovis and M. bovis BCG only.
FIG. 2.
(A) Numbers of patients in 19 low-copy-number RFLP clusters. (B) Numbers of patients in 26 spoligotyping clusters. (C) Numbers of patients in 26 combined RFLP and spoligotyping clusters.
Spoligotyping of low-copy-number strains.
All of the M. bovis and M. bovis BCG strains lacked spacers 39 through 43 and contained all spacers from 33 to 38. All 14 M. bovis BCG strains exhibited the same spoligotyping pattern (S-15, Fig. 1B). Among the M. bovis strains, 20 different spoligotyping patterns were observed. Eighteen strains exhibited unique patterns (37%), and 30 strains (63%) belonged to one of two clusters, which comprised 27 and 3 strains (S-15 and S-16, respectively; Fig. 1B).
Among the 249 M. tuberculosis isolates 87 different spoligotyping patterns were observed. Sixty-three (25%) of the 249 isolates exhibited a unique pattern, while 186 (75%) belonged to 1 of 24 different clusters comprising strains from 2 to 23 patients (Fig. 1B and 2B).
Spoligopatterns of RFLP clusters with six IS6110 copies.
Twelve M. tuberculosis isolates each containing six IS6110 copies that belonged to five different clusters consisting of strains from 4, 2, 2, 2, and 2 patients, respectively, were analyzed by spoligotyping. The spoligotyping differentiated the 12 isolates into the same five clusters as did the RFLP analysis. Thus, the results of the spoligotyping were consistent with the results of IS6110 typing, and the ability of spoligotyping to differentiate the isolates was the same as that of RFLP.
Combination of RFLP and spoligotyping for M. tuberculosis strains.
Eleven of the low-copy-number strain clusters identified by RFLP could be further differentiated by spoligotyping, as shown in Table 3, which also shows the differentiation of spoligotyping clusters by RFLP. However, the two five-band clusters both exhibited identical patterns by spoligotyping. Of the 249 M. tuberculosis isolates typed by both RFLP and spoligotyping, 136 (55%) belonged to 1 of 26 combined RFLP and spoligotyping clusters comprising isolates from 2 to 18 patients (Fig. 2C).
TABLE 3.
Polymorphism of RFLP clusters obtained by spoligotyping of strains by number of IS6110 fragments and polymorphism of spoligotyping clusters obtained by RFLP typing
| RFLP low-copy-number cluster | No. of bands | No. of patients | No. of patterns by spoligotyping | Spoligo-typing cluster | No. of patients | No. of patterns by RFLP |
|---|---|---|---|---|---|---|
| LC-1 | 2 | 2 | 2 | S-1 | 17 | 5 |
| LC-2 | 4 | 6 | 3 | S-2 | 6 | 1 |
| LC-3 | 1 | 80 | 34 | S-3 | 23 | 3 |
| LC-4 | 1 | 34 | 16 | S-4 | 8 | 3 |
| LC-5 | 3 | 13 | 1 | S-5 | 18 | 4 |
| LC-6 | 4 | 9 | 2 | S-6 | 11 | 3 |
| LC-7 | 2 | 2 | 1 | S-7 | 4 | 2 |
| LC-8 | 2 | 2 | 2 | S-8 | 3 | 3 |
| LC-9 | 1 | 37 | 15 | S-11 | 11 | 4 |
| LC-10a | 1 | 2 | 2 | S-13 | 10 | 2 |
| LC-11a | 2 | 2 | 2 | S-15a | 41 | 2 |
| LC-12a | 1 | 52 | 12 | S-16a | 3 | 1 |
| LC-13 | 3 | 2 | 2 | S-18 | 10 | 1 |
| LC-14 | 2 | 2 | 2 | S-19 | 2 | 1 |
| LC-15 | 3 | 3 | 2 | S-20 | 21 | 7 |
| LC-16 | 3 | 2 | 1 | S-21 | 8 | 2 |
| LC-17 | 2 | 2 | 2 | S-22 | 2 | 2 |
| C-44 | 5 | 10 | 1 | S-23 | 8 | 1 |
| C-92 | 5 | 3 | 1 | S-24 | 2 | 2 |
| S-25 | 3 | 1 | ||||
| S-26 | 8 | 7 | ||||
| S-27 | 2 | 1 | ||||
| S-29 | 2 | 1 | ||||
| S-30 | 2 | 1 | ||||
| S-32 | 2 | 2 | ||||
| S-33 | 2 | 1 |
Includes exclusively M. bovis or M. bovis BCG strains.
DISCUSSION
The aim of the present study was to evaluate spoligotyping as a technique for further characterizing IS6110 low-copy-number M. tuberculosis complex strains cultured in Denmark. Spoligotyping has been shown to exhibit less discriminatory power than IS6110 RFLP when used for subtyping of high-copy-number strains (9, 12) but has proven useful for subtyping IS6110 low-copy-number strains (12, 15), including M. bovis strains (7, 22). The low-copy-number strains in our study included a mixture of M. tuberculosis, M. bovis, and M. bovis BCG. Our results confirmed those of previous studies showing the ability of spoligotyping to differentiate M. tuberculosis from M. bovis and M. bovis BCG (2, 7, 15). All BCG strains tested gave the same pattern as described previously (i.e., they lacked spacers 39 to 43 plus spacers 3, 9, and 16) (11). This absence of the M. tuberculosis-specific spacers was also observed in 27 of 48 bovine isolates; thus, spoligotyping did not differentiate between M. bovis and M. bovis BCG. However, the differentiation of M. bovis isolates achieved by spoligotyping was superior to the differentiation achieved by RFLP.
A considerable proportion of the patients infected with low-copy-number M. tuberculosis strains originated from Asia and Central and East Africa. Confirming previous observations (11, 15), the polymorphism obtained by spoligotyping of IS6110 low-copy-number M. tuberculosis isolates proved to be superior to the polymorphism obtained by RFLP; 56 patterns were identified among 249 isolates examined by RFLP, whereas 87 patterns were identified by spoligotyping. The RFLP clusters comprising one to four bands could, with few exceptions, be discriminated by spoligotyping. However, the clusters comprising five or six bands gave identical spoligotyping clusters as well. This confirms a previous study, where it was found that IS6110 clusters with fewer than five bands frequently could be further differentiated by a supplementary technique (5).
The percent clustering among TB-associated isolates decreased from 84% when examined with IS6110 fingerprinting alone to 75% when examined with spoligotyping alone and further decreased to 55% when examined with a combination of the two techniques. The number of patients for the combined clusters (2 to 18) was smaller than those for RFLP clusters (2 to 80) and spoligotyping clusters (2 to 23). By a combination of the two techniques, it is thus possible to obtain a cluster percentage close to the one obtained with RFLP of high-copy-number strains for which data are deposited in the Danish database (49%) (4). We did not try to link the patients in the combined clusters epidemiologically, since the majority of the patients were immigrants with noninfectious extrapulmonary TB. However, the combined clusters confirmed several cases of known transmission (data not shown). For example, 10 patients in one of the combined clusters were involved in a known outbreak in an apartment house (20), and two other combined clusters comprised the patients involved in known cases of family transmission. From the names and addresses of the patients in other combined clusters it was possible to detect family transmission (data not shown). Furthermore, the nationalities of the patients in each of the combined clusters were almost exclusively the same (data not shown). These findings suggest that the clustering on the basis of the two typing methods will improve the correlation of DNA fingerprinting results and the epidemiological relatedness of cases encountered in practice.
In our laboratory spoligotyping has, furthermore, become a valuable tool in quality assurance, since it offers a rapid method of typing suspected cases of cross-contamination. During the last few years IS6110 RFLP has revealed that laboratory cross-contamination is a considerable problem in many laboratories (3, 13, 24). However, RFLP typing is hampered by the time delay required to obtain sufficient growth for DNA extraction. This is particularly a problem for specimens suspected of contamination, since these typically have a very small amount of bacterial growth. By spoligotyping it is possible to obtain results within a day after growth has been observed in BACTEC vials.
REFERENCES
- 1.Alland D, Kalkut G E, Moss A R, McAdam R A, Hahn J A, Bosworth W, Drucker E, Bloom B R. Transmission of tuberculosis in New York City, an analysis by DNA fingerprinting and conventional epidemiologic methods. N Engl J Med. 1994;330:1710–1716. doi: 10.1056/NEJM199406163302403. [DOI] [PubMed] [Google Scholar]
- 2.Aranaz A, Liébana E, Mateos A, Dominguez L, Vidal D, Domingo M, Gonzolez O, Rodriguez-Ferri E F, Bunschoten A E, van Embden J D A, Cousins D. Spacer oligonucleotide typing of Mycobacterium bovis strains from cattle and other animals: a tool for studying epidemiology of tuberculosis. J Clin Microbiol. 1996;34:2734–2740. doi: 10.1128/jcm.34.11.2734-2740.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauer J, Thomsen V O, Poulsen S, Andersen Å B. False-positive results from cultures of Mycobacterium tuberculosis due to laboratory cross-contamination confirmed by restriction fragment length polymorphism. J Clin Microbiol. 1997;35:988–991. doi: 10.1128/jcm.35.4.988-991.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauer J, Yang Z, Poulsen S, Andersen Å B. Results from 5 years of nationwide DNA fingerprinting of Mycobacterium tuberculosis complex isolates in a country with a low incidence of M. tuberculosis infection. J Clin Microbiol. 1998;36:305–308. doi: 10.1128/jcm.36.1.305-308.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burman W J, Reves R R, Hawkes A P, Rietmeijer C A, Yang Z, el Hajj H, Bates J H, Cave M D. DNA fingerprinting with two probes decreases clustering of Mycobacterium tuberculosis. Am J Respir Crit Care Med. 1997;155:1140–1146. doi: 10.1164/ajrccm.155.3.9117000. [DOI] [PubMed] [Google Scholar]
- 6.Couldwell D L, Dore G J, Harkness J L, Marriott D J E, Cooper D A, Edwards R, Li Y, Kaldor J M. Nosocomial outbreak of tuberculosis in an outpatient HIV treatment room. AIDS. 1996;10:521–525. doi: 10.1097/00002030-199605000-00011. [DOI] [PubMed] [Google Scholar]
- 7.Cousins D, Williams S, Liébana E, Aranaz A, Bunschoten A, van Embden J, Ellis T. Evaluation of four DNA typing techniques in epidemiological investigations of bovine tuberculosis. J Clin Microbiol. 1997;36:168–178. doi: 10.1128/jcm.36.1.168-178.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daley C L, Small P M, Schecter G F, Schoolnik G K, McAdam R A, Jacobs W R, Jr, Hopewell P C. An outbreak of tuberculosis with accelerated progression among persons infected with the human immunodeficiency virus. An analysis using restriction-fragment-length polymorphisms. N Engl J Med. 1992;326:231–235. doi: 10.1056/NEJM199201233260404. [DOI] [PubMed] [Google Scholar]
- 9.Diaz R, Kremer K, de Haas P E W, Gomez R I, Marrero A, Valdivia J A, van Embden J D A, van Soolingen D. Molecular epidemiology of tuberculosis in Cuba outside of Havana, July 1994–June 1995: utility of spoligotyping versus IS6110 restriction fragment length polymorphism. Int J Tuberc Lung Dis. 1998;2:743–750. [PubMed] [Google Scholar]
- 10.Doveren R F C, Keitzer S T. Studying transmission of tuberculosis with use of DNA fingerprinting. J Infect Dis. 1998;27:412–413. doi: 10.1086/517711. [DOI] [PubMed] [Google Scholar]
- 11.Goguet de la Salmoniére Y, Li H M, Torrea G, Bunschoten A, van Embden J, Gicquel B. Evaluation of spoligotyping in a study of the transmission of Mycobacterium tuberculosis. J Clin Microbiol. 1997;35:2210–2214. doi: 10.1128/jcm.35.9.2210-2214.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goyal M, Saunders N A, van Embden J D A, Young D B, Shaw R J. Differentiation of Mycobacterium tuberculosis isolates by spoligotyping and IS6110 restriction fragment length polymorphism. J Clin Microbiol. 1997;35:647–651. doi: 10.1128/jcm.35.3.647-651.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutiérrez M C, Vincent V, Aubert D, Bizet J, Gaillot O, Lebrun L, Le Pendeven C, Le Pennec M P, Mathieu D, Offredo C, Pangon B, Pierre-Audigier C. Molecular fingerprinting of Mycobacterium tuberculosis and risk factors for tuberculosis transmission in Paris, France, and surrounding areas. J Clin Microbiol. 1998;36:486–492. doi: 10.1128/jcm.36.2.486-492.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heersma H F, Kremer K, van Embden J D A. Computer analysis of IS6110 RFLP patterns of Mycobacterium tuberculosis. Methods Mol Biol. 1998;101:395–422. doi: 10.1385/0-89603-471-2:395. [DOI] [PubMed] [Google Scholar]
- 15.Kammerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van Embden J. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poulsen S, Bennedsen J. Tuberculosis 1994. Copenhagen, Denmark: Statens Serum Institut; 1995. [Google Scholar]
- 17.Poulsen S, Miörner H. Tuberculosis 1995. Copenhagen, Denmark: Statens Serum Institut; 1996. [Google Scholar]
- 18.Poulsen S, Miörner H. Tuberculosis 1996. Copenhagen, Denmark: Statens Serum Institut; 1997. [Google Scholar]
- 19.Poulsen S, Miörner H. Tuberculosis 1997, part I. Copenhagen, Denmark: Statens Serum Institut; 1998. [Google Scholar]
- 20.Poulsen S, Miörner H. Tuberculosis 1997, part II. Copenhagen, Denmark: Statens Serum Institut; 1998. [Google Scholar]
- 21.Prod’hom G, Guilhot C, Gutierrez M C, Varnerot A, Gicquel B, Vincent V. Rapid discrimination of Mycobacterium tuberculosis complex strains by ligation-mediated PCR fingerprint analysis. J Clin Microbiol. 1997;35:3331–3334. doi: 10.1128/jcm.35.12.3331-3334.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roring S, Brittain D, Bunschoten A, Hughes M S, Skuce R A, van Embden J D, Neill S D. Spacer oligotyping of Mycobacterium bovis isolates compared to typing by restriction fragment length polymorphism using PGRS, DR and IS6110 probes. Vet Microbiol. 1998;61:111–120. doi: 10.1016/s0378-1135(98)00178-3. [DOI] [PubMed] [Google Scholar]
- 23.Small P M, Hopewell P C, Singh S P, Paz A, Parsonnet J, Ruston D C, Schecter G F, Daley C L, Schoolnik G K. The epidemiology of tuberculosis in San Francisco, a population-based study using conventional and molecular methods. N Engl J Med. 1994;330:1703–1709. doi: 10.1056/NEJM199406163302402. [DOI] [PubMed] [Google Scholar]
- 24.Small P M, McClenny N B, Singh S P, Schoolnik G K, Tompkins L S, Mickelsen P A. Molecular strain typing of Mycobacterium tuberculosis to confirm cross-contamination in the mycobacteriology laboratory and modification of procedures to minimize occurrence of false-positive cultures. J Clin Microbiol. 1993;31:1677–1682. doi: 10.1128/jcm.31.7.1677-1682.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sola C, Horgen L, Maïsetti J, Devallois A, Goh K S, Rastogi N. Spoligotyping followed by double-repetitive-element PCR as rapid alternative to IS6110 fingerprinting for epidemiological studies of tuberculosis. J Clin Microbiol. 1997;36:1122–1124. doi: 10.1128/jcm.36.4.1122-1124.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Telenti A, Marchesi F, Balz M, Bally F, Böttger E C, Bodmer T. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J Clin Microbiol. 1993;31:175–178. doi: 10.1128/jcm.31.2.175-178.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Embden J D A, Cave M D, Crawford J T, Dale J W, Eisenach K D, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick T M, Small P M. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Embden J D A, van Soolingen D, Heersma H F, De Neeling A J, Jones M E, Steiert M, Grek V, Mooi F R, Verhoef J. Establishment of a European network for the surveillance of Mycobacterium tuberculosis, MRSA and penicillin-resistant pneumococci. J Antimicrob Chemother. 1996;38:905–907. doi: 10.1093/jac/38.5.905. [DOI] [PubMed] [Google Scholar]
- 29.van Soolingen D, de Haas P E W, Hermans P W, Groenen P M, van Embden J D. Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of Mycobacterium tuberculosis. J Clin Microbiol. 1993;31:1987–1995. doi: 10.1128/jcm.31.8.1987-1995.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Z H, de Haas P E W, van Soolingen D, van Embden J D, Andersen Å B. Restriction fragment length polymorphism of Mycobacterium tuberculosis strains isolated from Greenland during 1992: evidence of tuberculosis transmission between Greenland and Denmark. J Clin Microbiol. 1994;32:3018–3025. doi: 10.1128/jcm.32.12.3018-3025.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Z H, de Haas P E W, Wachmann C H, van Soolingen D, van Embden J D, Andersen Å B. Molecular epidemiology of tuberculosis in Denmark in 1992. J Clin Microbiol. 1995;33:2077–2081. doi: 10.1128/jcm.33.8.2077-2081.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]