Abstract
Viruses are responsible for multiple infections in humans that impose huge health burdens on individuals and populations worldwide. Therefore, numerous diagnostic methods and strategies have been developed for prevention, management, and decreasing the burden of viral diseases, each having its advantages and limitations. Viral infections are commonly detected using serological and nucleic acid-based methods. However, these conventional and clinical approaches have some limitations that can be resolved by implementing other detector devices. Therefore, the search for sensitive, selective, portable, and costless approaches as efficient alternative clinical methods for point of care testing (POCT) analysis has gained much attention in recent years. POCT is one of the ultimate goals in virus detection, and thus, the tests need to be rapid, specific, sensitive, accessible, and user-friendly. In this review, after a brief overview of viruses and their characteristics, the conventional viral detection methods, the clinical approaches, and their advantages and shortcomings are firstly explained. Then, LFA systems working principles, benefits, classification are discussed. Furthermore, the studies regarding designing and employing LFAs in diagnosing different types of viruses, especially SARS-CoV-2 as a main concern worldwide and innovations in the LFAs’ approaches and designs, are comprehensively discussed here. Furthermore, several strategies addressed in some studies for overcoming LFA limitations like low sensitivity are reviewed. Numerous techniques are adopted to increase sensitivity and perform quantitative detection. Employing several visualization methods, using different labeling reporters, integrating LFAs with other detection methods to benefit from both LFA and the integrated detection device advantages, and designing unique membranes to increase reagent reactivity, are some of the approaches that are highlighted.
Keywords: Lateral flow assays, Point of care testing, Viral infections, SARS-CoV-2, HIV, HBV, Influenza, Dengue, Zika
List of abbreviations
- acpcPNA
Pyrrolidinyl peptide nucleic acid
- AIDS
Acquired immunodeficiency syndrome
- AIV
Avian Influenza virus
- anti-FITC
Anti-fluorescein isothiocyanate
- Au NP-DPs
Au NP-detector probes
- Au NPs
Gold nanoparticles
- Au/Pt
Gold-platinum
- AUDG
Antarctic thermolabile uracil-DNA-glycosylase
- BART
Bioluminescent assay in real-time
- BKV
BK virus
- BPE
1,2-Bis(4-pyridyl)ethylene
- BR
Binding ratio
- BRPA
Betaine-assisted recombinase polymerase assay
- BSA
Bovine serum albumin
- CAST
Chinese academy of science test
- cDNA
Complementary DNA
- CDs
Carbon dots
- CDSNs
Carbon dots/SiO2 nanospheres
- CHA
Catalytic hairpin assembly
- CHIKV
Chikungunya virus
- CL
Control line
- CLIA
Chemiluminescence assay
- CMV
Cytomegalovirus
- COPs
Control probes
- CPs
Capture probes
- CRISPR
Clustered regularly interspaced short palindromic repeats
- CuO NPs
Copper oxide nanoparticles
- DENV
Dengue virus
- DIG
Digoxigenin
- DPs
Detector probes
- DRELFA
Dual recognition element LFA
- dsDNA
Double-stranded DNA
- EBV
Epstein-Barr virus
- EBVNA-1
EBV nuclear Ag-1
- eLFA
Electrochemical LFA
- ELFI
Electrochemical lateral flow immunosensor
- ELISA
Enzyme-linked immunosorbent assay
- FCS
Fluorescent CD-based silica
- FITC
Fluorescein isothiocyanate
- FNDPs
Fluorescent nano-diamond particles
- FRET
Fluorescence resonance energy transfer
- GAC
Goat anti-chicken
- GAH
Goat anti-human
- GAM
Goat anti-mouse
- GAR
Goat anti-rabbit
- GNSs
Gold nanostars
- GO
Graphene-oxide
- GPs
Glycoproteins
- HADV
Human adenovirus
- HAV
Hepatitis A virus
- HBB
Human β-globin
- HBeAg
HB e-antigen
- HBsAb
Hepatitis B surface antibody
- HBV
Hepatitis B virus
- HCV
Hepatitis C virus
- HDTMS
Hexadecyltrimethoxysilane
- HIV
Human immunodeficiency virus
- HPV
Human papillomavirus
- HRP
Horseradish peroxidase
- HSV-2
Herpes simplex virus type 2
- IAV
Influenza A virus
- ICC
Immunocytochemistry
- IF
Immunofluorescence
- Ig
Immunoglobulin
- IHC
Immunohistochemistry
- IR
Infrared
- IVB
Influenza B
- LAMP
Loop-mediated isothermal amplification
- LDP NPs
Lanthanide-doped polystyrene NPs
- LFAs
Lateral flow assays
- LFD
Lateral flow dipstick
- LFIA
Lateral flow immunoassay
- LNSM
LFA-NanoSuit method
- LOC
Lab-on-chip
- LOD
Limit of detection
- mAb
Monoclonal antibody
- MAC
IgM Ab capture
- MAH
Mouse anti-human
- MB
Methylene blue
- MBA
4-mercaptobenzoic acid
- MBPs
Multibranched peptides
- MGITC
Malachite green isothiocyanate
- MNPs
Magnetic NPs
- MPXV
Monkeypox virus
- MQDNBs
Magnetic quantum dot nanobeads
- NAA
Nucleic acid amplification
- NASBA
Nucleic acid sequence-based amplification
- NBA
Nile blue A
- NC
Nitrocellulose
- NIR
Near-infrared
- NPs
Nanoparticles
- NSB
Non-specific binding
- NSP1
Nonstructural protein 1
- OHT
Oseltamivir hexylthiol
- OPS
Onset of post-symptoms
- OPV
Orthopox virus
- ORF1ab
open reading frame
- pAbs
Polyclonal antibodies
- PCL
Polycaprolactone
- PCR
Polymerase chain reaction
- PDMS
Polydimethylsiloxane
- PE
Polyethylene
- pGAM
Polyclonal goat anti-mouse
- POCT
Point of care testing
- PSR
Polymerase spiral reaction
- PSR
polymerase spiral reaction
- QDs
quantum dots
- RAM
Rabbit anti-mouse
- r-HIV-1env
Envelope glycoproteins of HIV-1
- r-HIV-2env
Envelope glycoproteins of HIV-2
- RPA
Recombinase polymerase amplification
- RT-LAMP
Reverse transcription-LAMP
- RT-PCR
Reverse transcription polymerase chain reaction
- RT-qPCR
Reverse transcription quantitative polymerase chain reaction
- SAS
Signal amplification system
- SB
Specific binding
- SELEX
Systematic evolution of ligand by exponential enrichment
- SEM
Scanning electron microscope
- SERS
Surface-enhanced Raman scattering
- SFTSV
Severe fever and thrombocytopenia syndrome virus
- SPGE
Screen-printed gold electrode
- ssDNA
Single stranded DNA
- STV
Streptavidin
- TBEV
Tick-borne encephalitis virus
- TCA
Thermal contrast amplification
- TL
Test line
- TP
Treponema pallidum
- UC NP-LFA
Up-conversion NP-based LFA
- UC NPs
Upconversion NPs
- UPT-LFA
Up-converting phosphor technology-based LFA
- VACV
Vaccinia virus
- VLPs
Virus-like particles
- WB
Western blot
- WNV
West Nile virus
- ZKV
Zika virus
1. Introduction
Viruses are infectious entities consisting of one kind of nucleic acid, either DNA or RNA, encapsulated by a protein shell, which their diameters vary from 16 nm to over 300 nm. They sometimes are wrapped in a lipid membrane in some species. Since viruses do not contain ribosomes or any other cell-like organelles, their only means of reproduction are infecting living cells and replicating using host cell ribosomes. Their nucleic acid consists of genes that code structural components, regulatory proteins, and enzymes [1,2]. Viruses evolve through mutations which double-stranded (dsDNA) viruses' mutation rate is as high as 0–7 per nucleotide per year. This rate is higher than in RNA viruses on the order of 3–10 per nucleotide per year [2]. The mutagenic nature of viruses sparked incidents like the sudden emergence of the most recent Covid-19 and gave rise to variants of avian influenza that can be transmitted to humans [3]. Viruses can be also transmitted via aerosol and airborne droplets, fecal-oral, insect transmission, sexual transmission, blood-borne, and other unknown ways and can be entered human body through respiratory, gastrointestinal, and genital tracts, as well as skin, eye, placenta, transplants, and blood transfusion and they can exit the body through entering ways [4]. Currently, 224 virus species are known that can infect humans, every year, three to four new species are discovered, and there is a lot more to be found [5,6]. Occurrences of emerging and re-emerging viral infections and deadly viral diseases have significantly affected human health. They can cause respiratory, intestinal, skin, single organ (e.g., liver, lung, heart, etc.), multi-organ, neurologic, hematologic, immunosuppressive diseases [7]. Furthermore, viruses can alter host cell's genetic material and cause benign and malignant tumors [4]. Due to the urgency and severity of viral infections, accurate, reliable, user-friendly, available, rapid, and low-cost detection methods are necessary [8]. Established approaches such as virus isolation methods like virus culture, nucleic acid-based detection methods like Polymerase chain reaction (PCR), and serologic methods like enzyme-linked immunosorbent assay (ELISA) are widely used in clinical settings. Although these methods are highly sensitive and specific, they require expensive equipment and expert operators and lack the rapidity of newer detection methods [9]. Biosensors are one the alternative methods to conventional methods. A biosensor is a diagnostic device consisting of an electrical transducer that transforms biological responses from a recognition receptor to an electrical signal detected, correlated, and demonstrated by an electrical detector [10,11]. Biosensors can be classified by numerous approaches, considering their electrical transducer, their bio element, and the detected component as the categorization criterion [12,13]. Biosensors, however, have some limitations (e.g., low sensitivity) that can be overcome using nanotechnology [[14], [15], [16]].
Lateral flow assays (LFA) are the most recently used approaches for biosensing and quantifying numerous analytes, including different viruses [17]. The technical principle of LFAs was derived from the latex agglutination test. The first enzyme immunoassay was introduced in the 1960s, with several advantages like rapidity, ease of use, and long shelf life. The LFA principle was first popularized by the human pregnancy test that detected human chorionic gonadotropin in pregnant women urines, and since then, LFAs have been widely used in medicine and other areas like veterinary, agriculture, biowarfare, food, environmental health and safety [18]. LFAs application in medicine are highly versatile and can be used for diagnosis, prognosis, screening, monitoring, and surveillance. Unlike conventional methods, they can be used outside of high-tech laboratories and are applied in hospital wards, clinics, health centers, physicians’ offices, and also can be used by patients themselves for self-diagnosis [17]. This review aims to discuss the recent developments of LFA devices in detection of different viral diseases. Various characteristics of LFA detection approaches used for determination of various kinds of viruses are comprehensively discussed. Additionally, current challenges and problems as well as different strategies considered in the recently introduced LFA-based portable systems for eliminating shortcomings and enhancing their efficiency for the effective determination of different kinds of viral species are highlighted in this review.
2. Clinical and conventional approaches for the recognition of viruses
Since their discovery, scientists have developed multiple methods for virus detection. These methods can be classified into various clinical approaches including serological methods, hybridization methods, methods based on PCR and isothermal amplification methods. Although there are a great variety of detection tests, only some are suitable for clinical use. Here, brief discussions mainly focused on the serological and nucleic acid-based approaches have been provided in the following.
2.1. Serological methods
Serological methods are clinical procedures characterized by detecting immunoglobulins in various samples and consist of multiple tests like immunohistochemistry (IHC), agglutination test, flow cytometry, radioimmunoassay, and ELISA [19,20]. Despite being one of the oldest detection techniques, serological methods are still widely used in clinical recognitions owning to their high sensitivity, specificity, ease of use and cheapness. Based on the manifestation of anti-viral immunoglobulin (Ig) using virus antigen (Ag) in the sample, serological approaches can be employed to diagnose and monitor viral diseases [21]. ELISA is one of the serological methods that use enzyme-labeled antibodies (Abs) to detect viral Ags, making it feasible for detecting Abs in large sample groups [22]. The ELISA method is initiated by coating the plate with specific antigens or antibodies that detect the desired analyte, and then the plate is then washed to remove any unbound substrates. Then animal proteins like ovalbumin, aprotinin, etc., are used to block unbound spots in the container. After another washing step, an enzyme-linked detection antibody is added that binds to the primary antibody. Another washing step is performed, then a substrate/chromophore is added, producing a color detected and measured by the reader to quantify the detected analyte [23]. ELISA method is sensitive, quantitative, and relatively low-cost for significant sample detection. With the emergence of the Zika virus (ZKV) in 2015, several ELISAs were manufactured to detect this virus, and several studies evaluated these methods. In a study, for assessing molecular and serological procedures caused by ZKV infection, the authors used Dengue virus (DENV), ZIKV IgM ELISAs to detect and evaluate these techniques in pre-diagnosed patients by examining IgM cross-reaction among tick-borne encephalitis virus (TBEV), DENV and ZKV. In this study, a specificity around 97.4% was reported for nonstructural protein 1 (NSP1) Ag-based ZKV IgM ELISA (Euroimmun) [24]. In another study, to compare three ZKV IgM screening ELISAs, samples from pre-diagnosed patients in a Colombian hospital were tested with each of these methods. Two of the evaluated ELISAs, the CDC IgM Ab capture (MAC)-ELISA and the In Bios MAC-ELISA, had an agreement above 90% in their overall results; the third evaluated ELISA, Euroimmun ZKV IgM ELISA performance differed from the two others, with an overall agreement of 50% compared to these two ELISAs. However, it wasn't determined whether Euroimmun low agreement with the other two approaches has resulted from its higher specificity to ZKV Ags or it had a lower sensitivity to the tested analyte [25]. In another study, the Euroimmun ZKV IgM ELISA sensitivity was reported to be 29.8% [26]. Agglutination test, immunocytochemistry (ICC), immunohistochemistry (IHC) and immunofluorescence (IF) are considered to be other serological methods based on Abs that are rapid, easy to perform, highly specific and sensitive.
2.2. Nucleic acid-based detection approaches
Polymerase Chain Reaction (PCR) and Reverse transcription PCR (RT-PCR) are the most important nucleic acid-based detection methods and are recognized to be a gold standard for detecting viruses due to their high sensitivity and specificity. On the other hand, real-time PCR is another powerful detection method that is based on the amplification of virus sequences. Continues measurement of fluorescent signals during the amplification reactions allows the detection instrument to quantify the target sequence. This method is not limited by the analyte concentration or other variables (e.g., cycling condition), which results in this method's high sensitivity. Due to these benefits, real-time PCR is considered the gold standard in virus detection [27]. In a study to evaluate two RT-PCRs for diagnosing ZKV, virus RNA was prepared by the Qiagen QIAamp viral RNA mini kit, and DENV and CHIKV were used as the negative control virus pathogens. RT-PCRs performed well and had an overall 96.1% sensitivity and 100% specificity, and showed an excellent agreement (Cohen's kappa = 0.93) [28]. In another investigation, the performance of the triplex RT-PCR was evaluated for the ZKV and it was found that RT-PCR has a sensitivity of 85% for serum components from diagnosed patients. However, the sensitivity is much lower in urine samples. This drop-in sensitivity can be explained by sample collection and processing effect on RT-PCR's sensitivity [29]. Furthermore, a TaqMan MGB real-time PCR assay was developed to detect the Influenza H5N8 subtype. For this approach, two sets of primers were designed for H5 and N8 sequences, and samples were gathered from mice infected with the H5N5 Influenza virus for specificity and sensitivity evaluation. The results were consistent with Influenza A virus (IAV) real-time PCR and indicated that this method is sensitive and specific enough to be used for H5N8 detection [30].
Another nucleic acid-based detection approach is loop-mediated isothermal amplification (LAMP) which is highly specific and sensitive, easy to use, fast, and could be performed within 1 h. LAMP is another nucleic acid-based detection method and unlike PCR, it uses heat to denature dsDNA. LAMP employs four sets of primers to detect six distinct sequences on the target in one-step amplification in an isothermal state to generate 109 copies of DNA, thus increasing its specificity [31]. LAMP's low detection time and cost, high specificity and sensitivity, and being less prone to inhibitors make it an excellent detection tool in clinical settings. It is currently used in some studies for virus recognition. In a study, Gonzalez and coworkers developed a quantitative colorimetric LAMP to detect and amplify SARS-COV-2. LAMP test was coupled with pH indicator (phenol red), and the gene sequences that encoded the nucleocapsid proteins were targeted for detection and amplification. This colorimetric LAMP method performance was as good as reverse transcription quantitative PCR (RT-qPCR), with a sensitivity of 92.85% and specificity of 81.25%. In contrast to RT-qPCR, this approach was more rapid and required small amounts of RNA extract [32]. Moreover, a lab-on-chip (LOC) LAMP-bioluminescent assay in real-time (BART) was designed for detecting viral DNA. A LAMP was coupled with BART to detect and quantify the B19 virus. B19 virus causes fifth disease that is transmitted via respiratory tracts and causes mild rashes in children. The developed LOC was then verified through performance tests. The results were satisfactory for a POCT device and had a significant agreement with the conventional methods [33]. Another investigation describes a multiplex reverse transcription-LAMP (RT-LAMP) which was designed to simultaneously detect human Influenza viruses, coupling RT-LAMP with a colorimetric visualization system to determine results without requiring any extra steps. Multiple primers were designed for various subtypes of Influenza virus, for instance, H1N1, H3N2, H5N1, H5N6, H5N8, and H7N9. This device reached a sensitivity of 92.3% and 98.9% agreement with qRT-PCR [34].
3. Lateral flow assays (LFAs)
Although all of the above-mentioned detection methods have numerous advantages, they, however, have some serious limitations. Unlike other methods that directly detect viral components (Like nucleic acids or nucleoproteins), Ab-based serological methods are not suitable for early detection, and Ab levels are only detectable after infection. Although ELISA is a great tool for clinical settings like hospitals that routinely detect samples in large scales, it's not cost-effective for detecting small numbers of samples. RT-PCR requires high-skilled operators to perform, and is more expensive than other conventional detection methods. In addition, the RT-PCR procedure is very time-consuming, which makes it unsuitable for quick detection in clinical detection. LAMPs have retractions in their primer design and are susceptible to contamination [20,35,36]. Due to such limitations in the current approaches, a rapid, specific, sensitive, user-friendly, and affordable diagnostic method for virus detection is required.
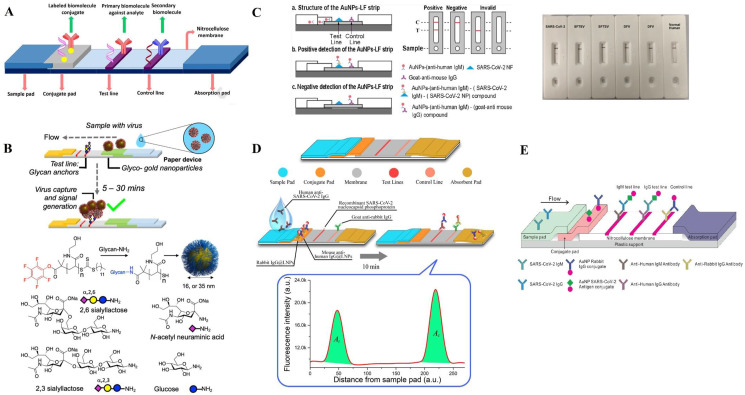
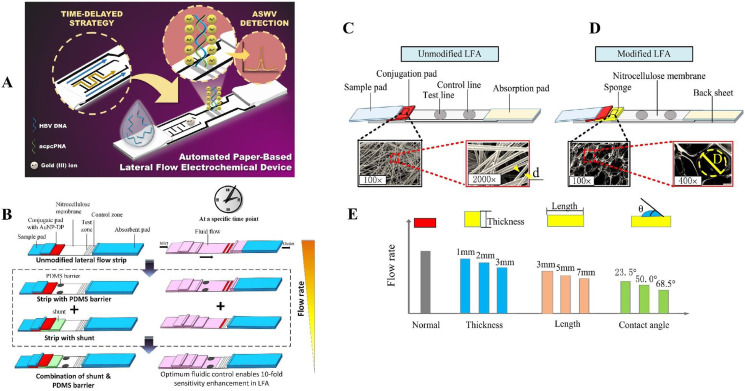
The LFA-based devices may be a good substitute for POCT application in virus detection. LFA is a paper-based biosensor device that can rapidly detect various analytes (e.g., proteins, haptens, nucleic acids, and amplicons) from multiple types of samples (e.g., urine, saliva, sweat, serum, plasma, nasopharyngeal swabs, and stool) within 5–30 min [[37], [38], [39]]. The use of capillary forces in the nitrocellulose (NC) membrane speeds up and facilitates the efficiency of the assay. Furthermore, membrane fiber arrangements can filter out any unwanted analyte in the sample [40]. These tests can be applied in the clinical setting due to their user-friendliness and cheap manufacturing cost and are helpful for onsite decision-making [41]. In addition to these, LFAs have are stable in various environmental conditions and have a long shelf-life, require small sample volume, do not require pre-preparation for fluid samples or washing steps. LFAs mostly require little to no energy, but can be integrated with electronics, have a high potential for commercialization, and be used in various approaches [42]. For these benefits, LFAs are suitable devices for detecting viruses in any sample for clinical practice. LFAs have attracted considerable attention in recent years due to being rapid, cost-effective, user-friendly, and portable [43]. Due to their portability, LFAs can be used in deprived areas, and due to their user-friendliness, they can be performed without expert personnel [38]. LFAs can be classified into lateral flow immunoassay (LFIA), which uses antibodies as recognition elements, and nucleic acid lateral immunoassay (NAFLA), which detects amplicons produced during other nucleic-based amplification devices like PCR. LFIA [38]. Based on their detection approaches, the LFIAs can further be classified into sandwich type and competitive type [44]. As shown in Fig. 1 A, a conventional sandwich LFA strip typically includes some pads such as a sample application pad, conjugate pad as well as NC membrane, and an absorbent pad. Upon introducing sample to the application pad, it will move to the conjugate pad. The conjugate pad contains labeling reporters conjugated to detector probes (which can detect the analyte) to form the reporter probes. The sample is added to the conjugate pad, and the fluid flows toward the NC membrane, which usually contains a test line (TL) and a control line (CL). The sample analyte binds to the detector probe in the conjugate pad or flows through the NC membrane. The TL includes the capture probes (CPs) which can detect the analyte in the immunocomplex. The CL comprises the control probes (COPs) that detect the detector probes. If the TL is detected, then the sample contains the analyte, and the CL should always be detected whether or not the sample contains the analyte. The test is not valid if the CL is not detected. The absorbent pad keeps the flow in the LFA, and the fluid is streamed towards it [42]. In the sandwich format, the analyte binds to both detector probe and capture probe, therefore it is suitable for large molecules. The competitive format is more suitable for small molecule like haptens. Competitive format itself can be divided into two setups. In the first setup, both analyte and the molecule that is conjugated to labeling reporter have a high affinity to bind with Abs on the TL, therefore the analyte compete with the reporter probe. In the second setup, the analyte attaches to the reporter probe, therefore blocks it from attaching to the Abs on the TL. In both formats, with higher concentration of analyte in the sample, the signal intensity on the TL decreases [44].
Fig. 1.
A conventional LFA structure (A). Adapted by permission from Ref. [88]. A design concept for LFA, and the synthesis procedure of Au NPs (B). Adapted by permission form Ref. [45]. The Au NPs LF strip's operating principle and specificity of AuNP-LF strips for SARCS-CoV-2 (C). Adapted by permission from Ref. [46] Structure of a LNP-based LFA applied for SARCS-CoV-2 detection (D). Adapted by permission from Ref. [79]. Schematic depiction of a LFA with Au NPs coated by spike protein from SARS-CoV-2 on the wicking membrane (E). Adapted by permission from Ref. [85].
4. Viral infections determination using LFA sensing platforms
Table 1 summarizes various LFA-based sensing platforms developed in recent years for the determination of various viral infections. Additionally, the main properties of each LFA-based device are provided in this table indicating the interest in clinical assays for sensing different types of viruses using LFA biosensors. In this way, the recent developments regrinding each types of virus are comprehensively highlighted in the following subsections. Thus, various LFAs for detection of different viral infections are comprehensively reviewed taking a deeper look at their design, novelty, performance and analytical figures of merits.
Table 1.
LFA-based sensing assays regarding the determination of different virus species.
| Detected Virus | Detected analyte | Reporter probe | Capture probe | Control Probe | Sensitivity | LOD | Ref. |
|---|---|---|---|---|---|---|---|
| SARS-CoV-2 | Spike protein | Au NPs | SARS-COV-2, S1 spike protein | 2,3′-sialyl lactose-BSA | – | 5 μg | [45] |
| SARS-CoV-2 | Anti-SARS-CoV-2 IgM | Au NPs | SARS-CoV-2 NP | Anti-mouse IgG | 100% | – | [46] |
| SARS-CoV-2 | RNA | Au NPs | Anti-FITC Ab | Biotin | 100% | – | [47] |
| SARS-CoV-2 | Nucleoprotein Ag | Cellulose nanobead | Nucleoprotein Ag | Anti-human IgG antibodies | – | 2 ng | [48] |
| SARS CoV-2 | Anti-SARS-CoV-2 IgM- IgG - IgM | Au-NPs | Protein A & nucleocapsid proteins | Avidin | 94.6% | – | [49] |
| SARS CoV-2 | Anti-SARS-CoV-2 IgA | Au-NPs | Nucleocapsid antigen | staphylococcal protein A | – | – | [50] |
| SARS CoV-2 | Anti-SARS-CoV-2 IgG | Au-NPs | SARS CoV-2 | Anti-mouse IgG | 69.1% | – | [51] |
| SARS CoV-2 | Nucleoprotein and spike protein | Au-NPs | Mouse anti-human IgG and anti-human IgM | mouse anti-tylosin antibody | 96% | – | [52] |
| IAV | Nucleoprotein Ag | Au/Pt NPs conjugated with anti-Influenza A NP | Anti-Influenza A Ab | Anti-mouse IgG | 94% | – | [53] |
| IAV | Nucleoproteins | Ca-coated UC NPs conjugated with anti-AIV NP Ab | Anti-AIV NP Ab | GAM IgG | – | 103.5 EID50/mL | [54] |
| Tamiflu-resistant Influenza virus | Antiviral neuraminidase | Au NPs conjugated with OHT | Anti-AIV nucleoproteins (NP) | Tamiflu resistant NPs | – | 5 × 102 PFU | [55] |
| IAV - IBV | IAV and IBV antigens | Europium | Monoclonal anti-influenza A and monoclonal anti-influenza B mouse IgG | polyclonal mouse IgG | 68.9% | – | [56] |
| HIV, HCV, HAV | Antiviral Abs | Au NPs conjugated with protein A, BSA and proteinticles | Specific proteinticles | GAH Abs | 100% | – | [57] |
| HIV | HIV-1 DNA | Au NPs conjugated with Raman reporter MGITC and DNA detection | Capture DNAs | Control DNAs | – | 0.24 pg/mL | [58] |
| HIV | HIV DNA | QDs conjugated with hairpin DNA | Test-DNA | Control-DNA | – | 0.76 pM | [59] |
| HIV, HCV, HBV | HIV, HCV, HBV Abs | QD nanobeads | HIV-1/2, HCV, HBV, and HBs antigens | GAM IgG | 88.33–93.42% | – | [60] |
| HBV | HBV DNA | detProbe-Au-ssDNA | T-DNA | C-DNA | – | 102 IU/mL | [61] |
| HBV | HBs Ag | UC NPs | AntiHBsAg mAb | RAM IgG | 95.4%, | – | [62] |
| DENV | Anti-Dengue Ab | DYlight-800 | Recombinant Dengue type 1 envelope | GAH IgG | 95% | – | [63] |
| DENV | Dengue NS1 | Magnetic beads | mAbs | GAM Ab | – | 0.1 ng/ml | [64] |
| DENV | Dengue-1 RNA | Au NPs | dDNA | cDNA | – | 1.2 × 104 pfu/mL | [65] |
| HPV | HPV16 E7 and HPV18 L1 | SA-DNPs | Anti-FITC and anti-DIG | Biotin-BSA | – | 5 × 105 copies | [66] |
| TORCH pathogens | IgM Abs | Au/hemin@MOF (metal organic framework) | Torch Ab | GAM IgG Ab | 100% | – | [67] |
| EV | EV glycoprotein | RNs@Au | Anti-Ebola glycoprotein Ab | Anti-human Ab | – | 2 ng/mL | [68] |
| EV | EV glycoprotein | AuNP | EE8 Ab | Anti-mouse immunoglobulin | 85.7% | 2.2 x 104 genome copies/ml | [69] |
| EV | EV glycoprotein | AuNP | mAb 1HK7 - mAb 2HK1 | Goat anti-mouse Ig | 0.23–0.37 ng/mL | [70] | |
| SFTSV | SFTSV NP | Au NPs | mAbs | GAM IgG | – | 1 ng/mL | [71] |
4.1. Sars-Cov-2
In late 2019, Sars-Cov-2 reported from Wuhan city in China has led to a pandemic that is transmitted from person to person and can be presented as mild, moderate, to severe and fulminant disease. Since the discovery, multiple diagnostic methods using various samples (blood, stool, oral swabs, sputum, etc.) have been developed [[72], [73], [74]]. Numerous diagnostic tests like RT-PCR, clustered regularly interspaced short palindromic repeats (CRISPR) are nucleic acid-based, targeting multiple genes. Serological tests are also popular as ELISA, chemiluminescence assay (CLIA), western blot (WB), etc., are commonly used in clinical settings and are the first lines tests at screening these diseases. LFA is a novel method in diagnosing Sars-CoV-2 [75]. In a study, Hoste and coworkers evaluated two serological detection methods, ELISA and LFA, in detecting anti-Sars-CoV-2 nucleocapsid protein antibody. 1065 samples collected from Sars-Cov-2 patients and patients infected with other respiratory viruses were tested on both diagnostic devices. In this study, recombinant N protein was used at the TL, and a monoclonal antibody against BSA was used at the Cl. Black latex beads were conjugated with N protein and BSA to act as reporter probe. Results for LFA and ELISA showed acceptable sensitivity (91.2% and 100%) and specificity (100% and 98.2%), respectively [76].
Catalytic hairpin assembly (CHA) as an application reaction was utilized by Zou and coworkers for designing an isothermal and nonenzymatic signal amplification system coupled with a lateral flow immunoassay (LFIA) strip-based device for signal amplification and sensitive RNA detection of SARS-CoV-2 that takes 90 min for completing diagnostic process. In this study, the hairpins DNA probes H1 and H2 were manufactured and were conjugated with biotin and digoxigenin (DIG). In presence of the target RNA, these primers create hybrids of DIG-biotin double-labeled H1–H2, and the final product is added to LFA. Nanoparticles (NPs) of polyethylene (PE) are conjugated by streptavidin (STV) as a fluorophore. Mouse antidigoxin/DIG monoclonal Abs (mAbs) act as CPs at the TL, and biotin is immobilized at the CL. Samples from 15 Covid patients and 15 healthy individuals were gathered and tested. Using LFA, the detection time decreased from 6–8 h to 90 min. The results showed a limit of detection (LOD) around 2000 copies/mL, and a sensitivity and specificity comparable with RT-PCR and could be an acceptable replacement [77].
In another investigation, Grant and coworkers used commercially available Sars-CoV-2 Abs developed a half-strip LFA (an LFA that doesn't contain sample or conjugate pads and sample and conjugates are premixed in another container) and used an optical reader to measure the LOD. In this design, conjugates of Rockland 200–401–A50 polyclonal antibodies (pAbs) and carboxylic red latex beads (400 nm) were formed to prepare reporter probe. Polystreptavidin was used at CP at the TL and COP was goat anti-chicken (GAC) IgY. Nucleocapsid proteins of SARS-CoV-2 were tested by Genemedi and Genscript half-strips for calculating LOD. The LOD of Geneemedi and Genscript was estimated to be 0.65 and 3.03 ng/mL, respectively [78]. Furthermore, Baker and coworkers designed a LFA detection device using multivalent gold nanoparticles (Au NPs) stabilized in polymer and containing the derivatives of sialic acid with binding ability to SARS-COV-2's spike proteins. In this study, Au NPs were identified as the best target ligand and reporting label. 2,3′-sialyl lactose-bovine serum albumin (BSA) was used as the negative control, for blocking NPs binding (Fig. 1B). The positive control was the immobilized spike protein of recombinant S1 subunit (SARS-COV-2, S1). The device was evaluated and showed that it could detect a concentration of 5 μg of virus-like particles in less than 30 min [45]. Huang and coworkers developed an IgM Ab-based LFA for detecting Sars-Cov-2 onsite. They manufactured a colloidal Au NP strip by coating the SARS-Cov-2 nucleoprotein on the membrane to form the sample capture and conjugated anti-human IgM with Au NPs to produce the detecting reporter (Fig. 1C). The detection device was evaluated and had a sensitivity of 100% and a specificity of 93.3% [46].
In an investigation, for amplifying RNA in the specimens infected by virus, Zhang and coworkers integrated direct RT-LAMP, with LFA to interpret the amplification process by optical means to detect Sars-Cov-2 in less than 40 min. In this study, the anti-fluorescein isothiocyanate (anti-FITC) Ab was used at the TL to act as a CP, and immobilized biotin was used at the CL. STV-coated particles were used as the labeling reporters. LAMP primers were designed to bind and amplify open reading frame (ORF1ab) and nucleoprotein genes. The evaluations demonstrated that this method has no cross-reaction with other viruses and has an accuracy of 100% in detecting the SARS-Cov-2 virus [47]. Additionally, Chen and coworkers developed a LFA anti-SARS-CoV-2 IgG detection device utilizing lanthanide-doped polystyrene NPs (LDP NPs). In this study, at the TL, the CP was recombinant nucleocapsid phosphoprotein of SARS-CoV-2 and at the CL, goat anti-rabbit (GAR) IgG was used (Fig. 1D). The used detector probe was prepared by functionalizing LDP NPs using rabbit IgG and mouse anti-human (MAH) IgG Ab and the whole detection process completed only within 10 min. The device's performance was compared with RT-PCR and 7 positive cases were consistent with LFA's results. One of the samples considered negative with RT-PCR but tested positive with LFA was determined to be SARS-CoV-2 IgG positive, suggesting LFA's higher sensitivity compared with RT-PCR [79].
In a research conducted by Daoud and coworkers, they used two commercially available LFA kits, including the Raybiotech and the Healgen Scientific for SARS-CoV-2 IgM/IgG, and comparatively evaluated the combination of kits and each of them alone. It was reported that the sensitivity of IgM varied between 58.9 and 66.2% and 87.7% for each kit and the combination form of kits, respectively. Still, the IgG sensitivity didn't change while using the combination strategy. Both kits showed high specificity (99.2 and 100%) [80]. Moreover, in another investigation, Azmi and coworkers developed a CRISPR-Cas13a based SARS-CoV-2 RNA detection device coupled with LFA readout integrated with a smartphone application for user-friendliness. The target RNA was amplified using CRISPR-Cas13a and RT-RPA and the final products were added to LFA for visualization. The intensity of the TL was interpreted using Fiji image J software to quantitatively detect amplified RNAs. The device showed a 98% agreement with RT-qPCR results [81].
An LFIA diagnostic test named Chinese academy of science test (CAST) was also introduced by Villarreal and coworkers for detecting IgG and IgM Abs against Sars-CoV-2 and it was applied among the Panamanian blood donors such as healthy volunteers and health care workers. They used samples from symptomatic patients and positive RT-PCR confirmed samples and then tested 19 patients with Covid-19, suspected healthy donors and health workers. The test showed a positive percent of agreement around 97.2% for the detection of both IgG and IgM. Also, this study showed that test sensitivity was associated with the number of days after the onset of symptoms, with a maximum sensitivity detected ≥15 days after the onset of post-symptoms (OPS) [82]. Additionally, Higgins and coworkers examined anti-Sars-CoV-2 Ab levels at various times of OPS via Easy Check Covid-19 IgM/IgG™ LFA. In this research work, 181 patients were confirmed with PCR and also collected samples from 21 donors. The obtained results demonstrated that this test's positivity is at its highest at 31–60 days of OPS. And the percentage results of IgM-/IgG+ is somewhat equal to the outcomes of IgM+/IgG+ at 61–90 days of OPS, resulting from immune response shift from IgM to IgG. This study also evaluated the LFA-based method branded Easy Check Covid-19 IgM/IgG™, demonstrating their high sensitivity at 1–4 days of OPS with a sensitivity of 86% at 5–7 days of OPS [83].
In Broughton and coworkers study, the authors tried to develop SARS-CoV-2 DETECTR, which is a CRISPR-Cas12 based on LFA to detect SARS-CoV-2 RNA extracts. The RNAs were amplified using primers that for target genes corresponding to the envelope and nucleoprotein of SARS-CoV-2 in the RT-LAMP and the final products were added to LFA for visualization. This detection device was evaluated and compared with the real-time PCR assay for SARS-CoV-2 of the U.S. CDC in analyzing samples of patients from infected by virus. Similar results with real-time RT-PCR were obtained for SARS-CoV-2 DETECTR while being rapid (∼30 min), low-cost, and highly specific [84].
In another study, Peng and coworkers used a photon-counting approach integrated with LFA to develop a quantitative and sensitive test. This test utilizes the light scattered by the NC, unconjugated Au NPs, and Au NPs conjugated with SARS-CoV-2 IgG to determine Ab concentration (Fig. 1E) to develop a highly specific, low-cost, rapid, simple, and quantitative test detecting Sars-Cov-2 in clinical samples [85]. Furthermore, Van Elslande and coworkers evaluated 7 LFA-based tests of IgG/IgM Ab against Sars-Cov-2 including Clungene, OrientGene, VivaDiag, StrongStep, Dynamiker, Multi-G and Prima. The authors then compared the results with Euroimmun IgA/IgG ELISA tests in 270 samples. All seven LFA tests had a specificity of over 90% for IgM and over 91% for IgG compared to specificity of 96.1% for ELISA IgG. LFA tests had sensitivities between 92.1% and 100% for IgG at 14–25 OPS compared with 89.5% for ELISA IgG, and IgM results varied between LFA tests [86]. And also, Xiong and coworkers developed a dual-gene Sars-Cov-2 diagnostic device integrating CRISPR/Cas9 with LFA. This test simultaneously discovered the genes associated with the ORF1ab and envelope in Sars-CoV-2 RNAs, and Au NP-DNA probes were used as reporting probes. Evaluations demonstrated that this test reached 97.14% and 100% as positive predictive agreement and negative predictive agreement, respectively when the whole detection process takes under 1 h to complete. Multiple factors can cause single-gene detection methods to have false negatives; therefore, employing dual-gene detection methods can increase test reliability and versatility [87].
In conclusion, these studies demonstrated LFAs' satisfactory performance in detecting SARS-Cov-2 and can be employed for clinical applications. Different biomarkers were used for detecting SARS-CoV-2 in these researches. Multiple approaches, like integrating with other detection methods, using multiplex diagnostic LFA to improve certainty, and using other reading devices, were used to enhance LFAs further.
4.2. Influenza
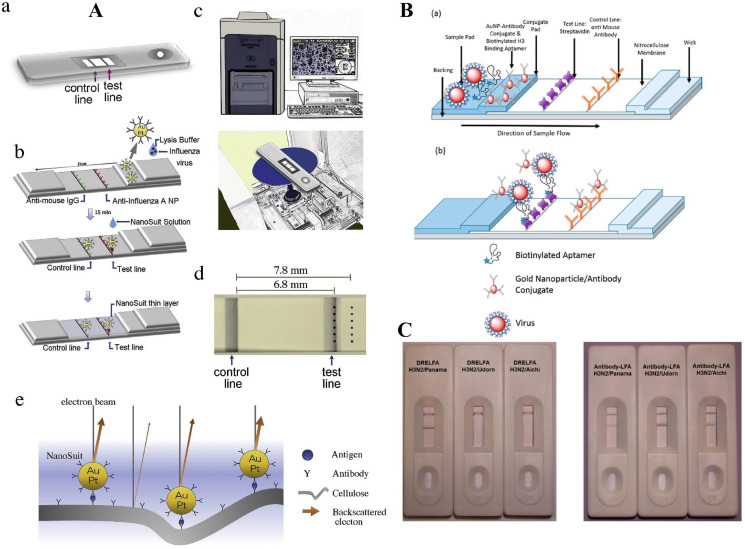
Influenza viruses belong to the Orthomyxoviridae and cause respiratory infections. They can be transmitted by aerosol droplets from person to person or be zoonotic. Influenza causes multiple symptoms like high-grade fever, myalgia, headache, and malaise [89]. Influenza can be diagnosed via serology, Ag detection, nucleic-acid-based detection, and viral culture, RT-PCR being considered the gold standard [90,91]. In the following, various LFA-based sensing platforms regarding Influenza determination are discussed. Kawasaki and coworkers integrated a desktop scanning electron microscope (SEM), NanoSuit approach, and LFA to develop the LFA-NanoSuit method (LNSM) to detect the Influenza virus. In this study, gold-platinum (Au/Pt) NPs were conjugated with anti-Influenza A NP to form the reporter probe. Anti-Influenza A Ab immobilized at the TL operated as a CP, and anti-mouse IgG immobilized at the CL captured remaining NPs (Fig. 2 A). The LFA results can be obtained in various ways, one of which uses the SEM to acquire high-resolution images of Au/Pt NPs to avoid limitations like cellulose swelling. This device was evaluated and reached a sensitivity of 94% and specificity of 100% [53].
Fig. 2.
(A) The TL and CL are shown on the diagnostic kit (a). The Au/Pt-Ab conjugate-linked fast LFA kit is depicted schematically (b). At the TL, the immune complex responds against nucleoprotein of influenza A, whereas at the CL, it reacts with anti-mouse IgG. (top, middle). Formation of a NanoSuit layer after NanoSuit treatment (bottom). SEM (miniscope TM4000plus; top) on a desktop (c). The placement of the kit in the SEM chamber (shown in the bottom). Establishing stations for observing in the TL and also the background (BG) regions, six fields were chosen (d). Au/Pt NPs and cellulose electron backscattering covered by a NanoSuit layer shown in the schematic picture (e). Adapted by permission from Ref. [53]. DREFLA schematic design for Influenza detection. The virus is captured by STV and is visualized by Au NPs (B). Specificity of LFA's DRELFA (C). Adapted by permission from Ref. [92].
In another study, Kim and coworkers developed a near-infrared (NIR) to NIR upconversion NPs (UC NPs)-based LFA detecting Avian Influenza virus (AIV) during 20 min. In this study, Ca-coated UC NPs were conjugated with anti-AIV NP Abs to form the reporter probe. Anti-AIV NP Ab was also immobilized at the TL to act as a CP, and the immobilized goat anti-mouse (GAM) IgG at the CL was used a control probe. The excited UC NP emitted signals that can be read and interpreted by NIR-to-NIR UC NP-based LFA for detecting HPAI H5N6 and LPAI H5N2 viruses. Evaluating this device revealed LOD of 102 and 103.5 EID50/mL for LPAI H5N2 and HPAI H5N6, respectively. This LFA's sensitivity is 10-fold higher than commercial LFAs and unlike them, the detection process isn't disrupted by the opacity and brown color of stool samples [54]. For detecting the H5N2 Influenza virus, Kim and coworkers employed a pair of aptamers to develop an LFA. In this study, systematic evolution of ligand by exponential enrichment (SELEX) and graphene-oxide (GO) were used and an assay based on the fluorescence resonance energy transfer (FRET) mechanism and GO (GO-FRET) were utilized to manufacture virus particle-specific aptamers with capability to bind different parts of virus particles at the same time. Au NPs were used as signaling labels and were conjugated to JH4APT. J3APT was used at the TL as a CP, and poly-A sequence was immobilized at the CL as the COPs. Upon interpreting by ImageJ program and after evaluating the results, the LOD being 1.27 × 105 and 2.09 × 105 EID50/mL was determined for buffer and the duck's feces, respectively [93].
Another LFA approach utilizing surface-enhanced Raman scattering (SERS) phenomenon for simultaneously detecting human adenovirus (HADV) and IAV H1N1 has been designed by Wang and coworkers. Fe3O4@Ag magnetic NPs (MNPs) in this study were coated by Ag, and dye molecules of DTNBs were attached to it. Then HADV Ab or H1N1 Ab was conjugated with it to form the reporter probes. LFA contained two TLs; one used HADV Ab and the other H1N1 Ab, and a CL sprayed with polyclonal goat anti-mouse (pGAM) IgG. The SERS signals emitted from test and CLs then can be read to detect Ags quantitatively. This device was evaluated and showed a LOD of 50 pfu/mL for H1N1 and ten pfu/mL for HADV [94].
In another investigation, a dual recognition element LFA (DRELFA) to highly sensitive detection multiple strains of Influenza have been developed by Le and coworkers. In this study, specific Abs were manufactured and screened using SELEX and were conjugated with biotin. Au NPs were also conjugated with Influenza mAbs, and in the presence of Ags, both the aptamer and the Au NPs would detect the virus and form an immune-complex (Fig. 2B and C). The STV immobilized at the TL would capture the immune-complex, and the Au NPs of the immune-complex would act as reporters. The rest of the Au NPs were captured at the CL by anti-mouse Abs. This device was evaluated and demonstrated a LOD of 2 × 106 virus particles, and the quantitative results, using the TL color intensity, showed a significant correlation with RT-PCR results [92].
Furthermore, Park and coworkers developed a CRISPR-Cas12a-based LFA to detect IAV and Influenza B (IVB). In this research, RNAs were amplified using DNA endonuclease-targeted CRISPR trans reporter (DETECTR) combined with recombinase polymerase amplification (RPA) and LAMP. Primers were manufactured to detect IAV matrix and IBV hemagglutinin genes for amplification. The amplified RNAs were added to an LFA, and the visualization process took about 2 min, with a LOD of 1 × 100 PFUs per reaction [95]. Also, Ma and coworkers integrated RPA and lateral flow dipstick (LFD) to detect IAV H7N9. RPA was used to amplify RNA employing primers that were manufactured to detect H7 and N9 genes. The final amplified RNAs were then added to two LFDs (one for H7 and the other for N9 detection) to be visualized and detected by the naked eye. Both LFDs demonstrated 100% sensitivity and showed no cross-reaction with different Influenza subtypes [96].
Bai and coworkers developed magnetic quantum dot nanobeads (MQDNBs) based LFA for detecting IAV in clinical samples. Magnetic MnFe2O4 nanobeads were conjugated with quantum dots (QDs) and anti-IAV Abs to form the reporter probes. The Abs of IAV sprayed on the TL acted as CPs, and pGAM IgG immobilized at the CL captured remaining MQDNBs. In the presence of Ag, the MQDNB-Ag complexes were arrested at the TL. After excitation, emitted fluorescence signals were collected by the smartphone CMOS image sensor and were analyzed to detect IAV quantitatively. This LFA's LOD was measured to be 22 pfu/mL, and the whole detection process took up about 35 min [97].
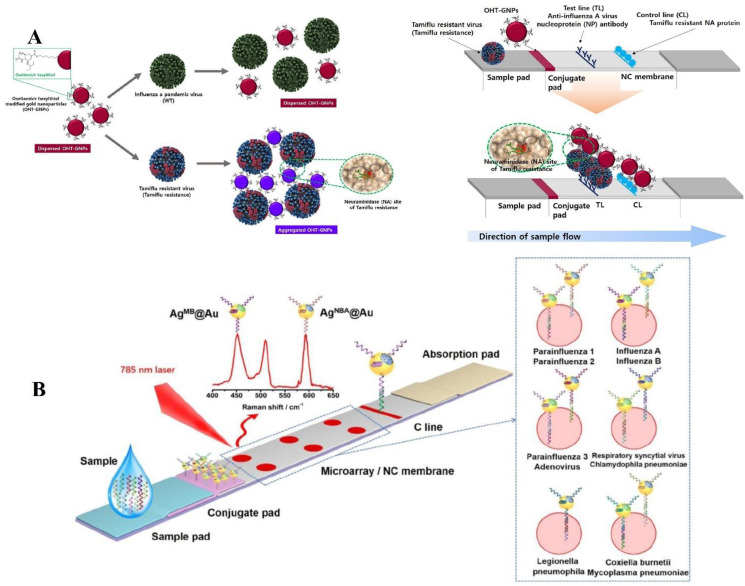
An oseltamivir hexylthiol (OHT)-based LFA to detect Tamiflu-resistant Influenza virus was developed by Hwang and coworkers. OHT has a higher affinity to Tamiflu-resistant Influenza viruses compared with Tamiflu-susceptible ones. In this study, Au NPs were conjugated with OHT and were used as reporter probes. In the TL, anti-AIV nucleoprotein was used as a CP, and Tamiflu resistant NPs at the CL captured OHT-conjugated Au NPs. With the accumulation of Tamiflu-resistant on the TL, a purple line becomes visible to the naked eye (Fig. 3 A). This LFA was evaluated and demonstrated a LOD of 5 × 102 pfu, which is suitable for clinical application [55].
Fig. 3.
Schematic diagram of the colorimetric method for detecting Tamiflu resistance by OHT-modified Au NPs (OHT-Au NPs). Due to the OHT and anti-trimeric fluorescent virus combination, the color of OHT-Au NPs changed from dark red to purple (A). Adapted by permission from Ref. [55]. Schematic diagram of SERS LFM for nucleic acid detection of 11 RTI pathogens with RD-encoded SERS core-shell nano-tags (B). Adapted by permission from Ref. [98].
Sun and coworkers developed an RPA-LFA to detect IAV and IBV simultaneously. In this investigation, primers that detected IAV were conjugated with fluorescein isothiocyanate (FITC) and biotin, and IBV-detecting primers were labeled with DIG and biotin. The Influenza virus TNA was amplified using RPA, and these primers and the final products were added to LFA. Au NPs were conjugated with STV to form the detecting probes. LFA strips consisted of two TLs sprayed with anti-FITC and anti-DIG, and BSA immobilized captured remaining Au NPs at the CL. The formed lines were visible by the naked eye, and a smartphone can capture and interpret the images of the lines to quantitatively detect Influenza RNAs. This device reached sensitivities of 78.57% and 87.50% for Influenza A and B, respectively. The LODs were also determined to be 500 and 50 copies per reaction for Influenza A and B, respectively [99]. In another study, a SERS-based LFA has been developed by Zhang and coworkers to simultaneously detect 11 respiratory tract infection pathogens, including IAV and IBV. In this study, Au NPs were conjugated with Raman Dyes of either Nile blue A (NBA) or methylene blue (MB), and of the 11 pathogens capture nucleic acids (Fig. 3B). The LFA contained 6 T dots, 5 of which included two sets of capture nucleic acids, and one of them had one, and control nucleic acids were immobilized at the CL to capture the remaining Au NPs. A SERS reader can quantitatively detect emitted signals and distinguish between the Raman dyes; therefore, the captured Ags can be distinguished by their specific Raman dyes and their T dot position on the LFA. This LFA was evaluated and showed excellent specificity, and IAV and IBV LOD were determined to be 0.031 pM and 0.035 pM [100].
The design of a carbon nanotag-based LFA for detecting IAV was reported by Wiriyachaiporn and coworkers. In this study, carbon nanostrings were conjugated with BSA and mouse mAb anti-Influenza A nucleoprotein forming detector probe. Mouse anti-Influenza A mAb nucleoprotein was also used at the TL as a CP, and the immobilized GAM IgG captured remaining carbon NPs. The intensity value of the TL was interpreted by Image J program to detect IAV quantitatively. The whole detection process took about 15 min, and LOD was calculated to be 350 TCID50/mL [101]. Also, a horseradish peroxidase (HRP)-based LFA has been introduced by Zhang and coworkers to detect IAV and IBV simultaneously. HRP was conjugated with 11F12 and 10B6 anti-IAV and IBV mAbs to form the detector probes. The LFA strip contained two TLs, with capture Abs immobilized on them, and a CL that used GAM IgG. This device was evaluated and showed sensitivities of 77.5% and 71.2% and specificities of 99.8% and 99.8% for IAV and IBV [100].
These studies showed that LFAs have a satisfactory performance in detecting the influenza virus. Using multiplex LFAs is a great way to diagnose common subtypes of influenza and avoid under treatment. Different approaches like integrating with other detection devices and alternate reading devices were used to improve LFAs performance.
4.3. Human immunodeficiency virus (HIV)
HIV is a member of the Retroviridae family that attacks the host's CD4 + lymphocytes, leading to acquired immunodeficiency syndrome (AIDS) [102]. A third of the people infected with HIV never notice it until it has advanced to a more advanced state and remain an asymptomatic career, which increases the burden of the disease [103]. Therefore, HIV infection should be diagnosed early and needs rapid and cheap POCT for diagnosis and surveillance [104]. Accordingly, Tang and coworkers employed a dialysis method to increase sample concentration in LFA testing to improve its sensitivity. The dialysis device is comprised of a semi-permeable membrane and a glass membrane containing PEG. The LFA device was designed to detect HIV nucleic acid in 15 min, using Au NPs as reporter labels, and conjugated with an HIV detection probe. Evaluations demonstrated that using this method the LFA sensitivity increases 10-fold compared to conventional LFA with a LOD of 0.1 nM [105].
On the other hand, a multiplex LFA to detect HIV, Hepatitis C virus (HCV), and Hepatitis A virus (HAV) simultaneously has been introduced by Lee and coworkers. In this study, diagnostic probes were constructed by conjugating viral Abs to manufacture proteinticles that can attach to multiple Abs to increase sensitivity. Au NPs were conjugated with protein A and BSA to form labeling probes. There were three TLs and one CL on LFA strips; the HIV TL consisted of proteinticles conjugated with the TL of PHIV-1 and PHIV-2, HAV consisted of proteinticles conjugated with PHAV-1, PHAV-2, and PHAV-3, and HCV TL consisted of proteinticles conjugated with PHCV-1 and PHCV-2. The immobilized GAH Abs at the CL captured remaining Au NPs. The formed TLs can be detected by the naked eye or be processed by line analyzer and image analysis software to detect quantitatively. This device was evaluated and reached a sensitivity and specificity of 100% [57]. On the other hand, Calvaria and coworkers worked on a multimodal LFA to detect HIV1 and HIV2 Abs, consisting of only one TL. In this study, AuNPs emitted red lights and were conjugated with gp36 antigen that detects HIV-1 Ab, and gold nano stars emitted blue lights and were conjugated with gp41 antigen that detects HIV-2 Ab. Immobilized gp36 and gp41 were also used at the TL, and avidin was used at the control line. The color of the test line determined the detected HIV subtype, and the evaluations showed that this approach improved LFA's performance [106].
Moreover, Fu and coworkers developed a SERS-based LFA to detect HIV-1 DNA quantitatively. Raman reporter Malachite green isothiocyanate (MGITC) and oligonucleotides as detection DNA to form reporter probes were used. Immobilization of captured DNAs and Control DNAs were performed at the TL and CL of the LFA strip. A Raman instrument reader can be used to interpret Raman intensity to detect HIV-1 DNA quantitatively. This device was evaluated and showed LOD around 0.24 pg/mL, which was 1000-fold greater than the typical LFAs [58]. In another study using QDs, Deng and coworkers developed fluorescence LFA to recognize HIV DNA quantitatively. In this study, QDs conjugated with hairpin DNA were used as reporter probes, t-DNA and C-DNA were immobilized at the TL and CL. Evaluations demonstrated that the LOD was 0.76 pM and was more than satisfactory for clinical application [59].
A magnetic immunochromatography-based LFA to detect HIV-1 and HIV-2 using multibranched peptides (MBPs) has been investigated by Granade and coworkers. Magnetic beads conjugated with protein A were used as reporter probes. Immobilized HIV-1 and HIV-2 MBP Abs were used at the TL, and protein A acted as the COP in the CL. A magnetic reader was used to measure TL activities on minutes 20 and 40 to detect HIV-1 and HIV-2 quantitatively. This LFA's results were concordant with the WB reference test and showed an acceptable reproducibility [107]. In another research, Granade and coworkers developed an LFA to detect recent or long-term HIV-1 simultaneously. In this study, Au NPs conjugated with protein A were used as reporter probes, and protein A was also used as a COP at the CL. Multisubtype gp41 recombinant protein (rIDR-M) was immobilized at two TLs with two different concentrations to differentiate between recent and long-term infections. In recent infection, only the TL with the higher rIDR-M concentration will detect HIV, while both TLs will visualize long-term disease. This device was evaluated and showed 95% agreement with the fabricated assay [108].
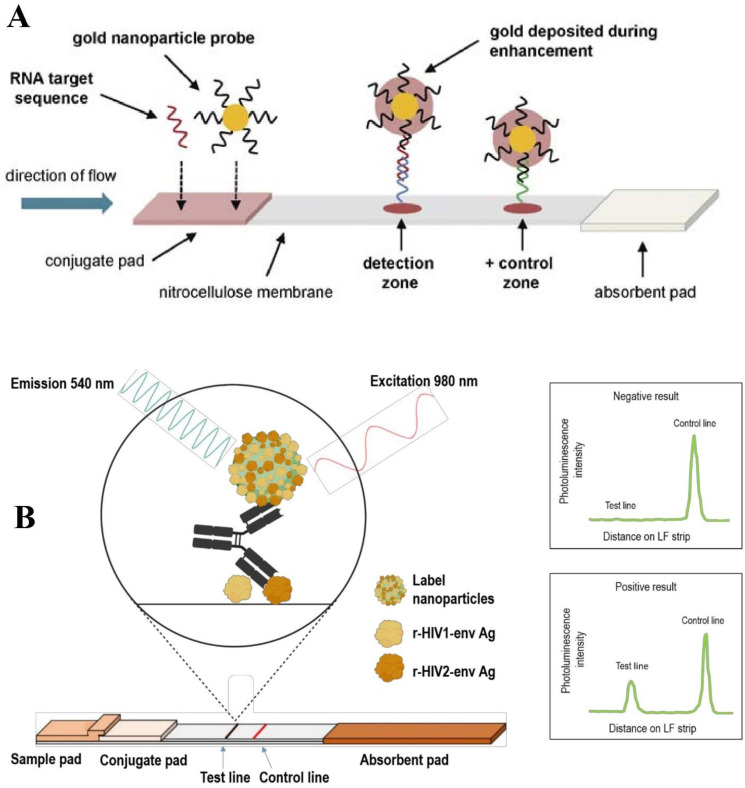
An integrated nucleic acid sequence-based amplification (NASBA) with LFA for quantitatively detecting HIV-1 RNA was designed by Rohrman and coworkers. Au NPs conjugated with detector nucleic acids that bind to the HIV gag gene and form reporter probes (Fig. 4 A). Capture sequences immobilized at the TL act as CPs, and control sequences at the CL bind with the remaining Au NPs. To increase sensitivity, HIV RNAs are amplified by NASBA and are added to LFA. Image analysis was used to interpret TL intensity to detect amplified RNA quantitatively. This sensing assay was evaluated and demonstrated a LOD of 9.5 log10 RNA copies, which is suitable for clinical application [109].
Fig. 4.
Schematic diagram of quantitative LFA for HIV-1 RNA detection (A). Adapted by permission from Ref. [109]. UC NP antiHIV1/2 strip design and dual Ag bridging test principle. Recombinant HIV1 and HIV2 envelope Ags are used as captures on the TL and coupled to the surface of UC NP to detect anti-HIV Abs. UC NP is excited at 980 nm, and the emission peak intensity at 540 nm indicated a negative or positive reading (B). Adapted by permission from Ref. [110].
Bristow and coworkers evaluated the commercial SD BIOLINE HIV/Syphilis Duo test enjoying LFA method for detecting HIV and Syphilis. The participants consisted of 298 GHESKIO clinic attendees who were 18 years or older, and 237 of them were female, of which 49 were pregnant. This LFA reached a sensitivity of 99.2% and a specificity of 97.0%. In pregnant women, the sensitivity was 93.3%, and specificity was 94.1% [111].
Also, thermal contrast amplification (TCA) approach was reported to be useful in improving the LFA detection sensitivity against HIV p24 protein. In this research, Au NPs were conjugated with anti-p24 mAb to form reporter probes. Anti-p24 pAbs were CPs at the TL, and p24 recombinant protein was immobilized at the CL to capture the remaining Au NPs. An infrared (IR) camera was used to measure temperature changes in Au NPs resulted from laser irritations. The IR signals at the CL represented the specific binding (SB), and the emitted IR signals from the TL demonstrated the non-specific binding (NSB). Binding ratio (BR) can be obtained by the BR= SB/NSB, and is an indicator of Au NPs distribution and can be used to detect p24 quantitatively. Evaluations showed this LFA's LOD is 8 pg/mL, which is lower than conventional LFAs and is close to ELISA LOD [112].
UC NP reporters to develop a double Ag LFA introduced by Martinskainen and coworkers luminescent were also used for the detection of Abs against HIV-1 and HIV-2 in 30 min (Fig. 4B). In this study, UC NPs were conjugated with recombinant envelope glycoproteins of HIV-1 (r-HIV-1env) and −2 (r-HIV-2env) to form reporter probes. The Ags corresponding to r-HIV-1env and r-HIV-2env were also immobilized at the TL to act as CPs, and HIV-1 gp41 rabbit pAb was used at the CL to capture the remaining UC NPs. The captured UC NPs at the TL and CL were excited with an IR laser. An Upcon reader device detected the emitted signals from excited UC NPs, and SAS JMP Pro 14 statistics software interpreted the signals to detect Abs of HIV-1 and HIV-2 quantitatively (Fig. 4B). This device was evaluated and reached a sensitivity of 96.6% and a specificity of 98.7% [110].
Another investigation performed by Zhang and coworkers, demonstrated an LFA by fixing cotton threads onto a membrane to detect HIV. In this study, the hydrophilicity of cotton threads was decreased using Hexadecyltrimethoxysilane (HDTMS) and was fixed paper to form the LFA's membrane. Cotton-embedded papers delay fluid flow and therefore increase the reagents' interaction and, in turn, the LFA's sensitivity. Au NPs were conjugated with detector probes, and test probes and COPs were immobilized at the TL and CL. Employing cotton threads increases LFA's sensitivity four-fold compared with conventional LFAs [98].
To detect HIV p24 Ag, A NIR fluorescent microsphere immunochromatography-based LFA was developed by Qi and coworkers. Fluorescent microspheres were conjugated with Dylight 800 and label Abs to form reporter probes. GAM IgG Ab was immobilized at the CL to capture the remaining NPs and HIV-1 P24 Ab was immobilized at the TL to act as a CP. Portable fluorescent scanner KY-100 then detected and interpreted emitted signals from TL and CL to detect HIV p24 Abs quantitatively. This device was evaluated and demonstrated a LOD of 3.4 pg/mL, which is suitable for clinical application [113].
Using multiplex LFAs helps physicians to simultaneously detect various types of HIV and other sexually transmitted viruses. Different methods like using alternate readers and reporter probes and adjusting membranes were used to improve LFAs quality. The alternate readers also helped to detect HIV biomarkers quantitatively.
4.4. Hepatitis B virus (HBV)
HBV as a leading causes of liver disease in the family Hepadnaviridae can be transmitted sexually or via blood transfer. HBV can cause both acute and chronic hepatitis and can lead to liver cancer [114,115]. There are several serum markers for HBV, including HB Abs, HB Ags, HBc Ab, and HBe, each of which is found during different phases of HBV replication [116]. HBV infection can be asymptomatic but can still be transferred, and early diagnosis should be applied using POCTs [117]. By using QDs nanobeads, a multiplex integrated fluorescent LFA to detect infectious pathogens including HIV, HCV, HBV, and Treponema pallidum (TP) was developed Rong and coworkers. In this study, multicolor QDs nanobeads were used as labeling reporters to increase sensitivity and specificity. Different Ags for HIV-1/2, HCV, HBV, and HBs were used both as capture and detection probes. The GAM IgG Ab was used at the CL to detect and capture remaining QD nanobeads. An optical receiver and an Android app were used to receive and interpret fluorescent signals emitted from QD nanobeads at the TL to diagnose and quantify the captured pathogen. This LFA was evaluated, and pathogen detection sensitivities varied between 88.33 and 93.42%, and specificities were between 87.67 and 93.15% [60].
In another investigation, Srsomwat developed an automated electrochemical LFA (eLFA) to detect HBV quantitatively. In this study, pyrrolidinyl peptide nucleic acid (acpcPNA) as an immobilized detector was used at the TL. The used membrane in this device had a unique design, allowing the tested serum to go through a non-delayed channel and the HBV DNA present in the serum to rapidly bind and hybridize with acpcPNA. Then a fluid containing Au3+ (which is used for the metallization process) would go through a delayed channel and reach the TL after the DNA hybridization has occurred. Au3+ binds to hybridized DNA, and after the electrodeposition process, an electrical signal demonstrates HBV DNA detection (Fig. 5 A). The whole detection process takes 7 min and has a LOD of 7.23 pM [118].
Fig. 5.
The unique design of eLFA membrane for delaying the fluid flow (A). Adapted by permission from Ref. [118]. The effect of adding PDMS and shunt to LFA alone and simultaneously on fluid flow rate and sensitivity (B). Adapted by permission from Ref. [119]. The demonstration of unmodified LDA (C) modified LFA (D) and the sponge shunt optimization parameters (E). Adapted by permission from Ref. [120].
Choi and coworkers integrated polydimethylsiloxane (PDMS) barrier and a paper-based shunt with an HBV-detecting LFA device to enhance the assay's sensitivity. Au NPs were used as reporter labels, and STV was used at the TL. Employing a PDMS barrier and a paper-based shunt delays fluid flow, thus, enhances the reagents' interaction chance and eventually leads to signal enhancement. The evaluations demonstrated that this method allows sensitive detection of HBV (∼102 IU/mL) while also retaining the rapidity of LFA tests [119]. Moreover, an LFA to detect HBV DNA was developed by Gao and coworkers. In this study, Au NPs coated with STV and Au NPs labeled probes so-called detProbe-Au-single stranded DNA (ssDNA) were used as amplifying reporters. The synthesized T-DNA and C-DNA at the TL and CL were immobilized (Fig. 5B). Evaluation of such device presented a LOD of 0.01 pM [61]. Another study demonstrated a LFA sensing assay using Au NPs by Kim and coworkers to detect HBsAg. Different sizes of Au NPs and their efficiency in various pH value and Ab concentration was investigated. UV–vis spectroscopy was used to analyze and optimize Au NPs conjugation, and it was established 42.7 nm. Au NPs performed best under multiple circumstances in detecting HBsAg [121].
In addition, Gong and coworkers developed an up-conversion NP-based LFA (UC NP-LFA) procedure for the detection of multiple targets, including HBV. Four different probes including UC NPs and UC NPs-DNA and UC NP-Ab were used in this study. A UC NP-LFA reader calculating the optical density of the TL and background was used to obtain analyte concentration. This LFA was evaluated by testing multiple analytes and correlated with the gold-standard method in testing HBV DNA [122]. Li and coworkers also developed an up-converting phosphor technology-based LFA (UPT-LFA) to rapidly and quantitatively detect HB surface antibody (HBsAb). In this study, up-converting phosphor particles conjugated with HBsAg were used as reporter probes, and HBsAg was immobilized at the TL, and goat anti-HBsAg Ab was loaded at the CL. The UPT-LFA was evaluated and compared with Abbott Axsym AUSAB and conventional ELISA, reaching a good correlation with ELISA, making it a sensitive and rapid quantitative test for detecting HBV [123]. Furthermore, another UC NP-LFA to detect HBsAg has been developed by Martiskainen and coworkers. In this investigation, UCNPs conjugated with anti-HBsAg mAb were considered as reporter probes. Immobilized anti-HBsAg mAb at the TL was used and the CL consisted of rabbit anti-mouse IgG. This UC NP-LFA was evaluated and compared with a conventional LFA. UC NP-LFA reached a sensitivity of 95.4%, while the conventional LFA's sensitivity was 87.7% [62].
An automated fluorescent LFA to detect HBsAg and HCV Ab was also reported by Ryu and coworkers. In this research, fluorescent europium chelates [Eu (III)] conjugated with HBsAg or HCVAg or mouse anti-HBsAg mAbs were used as reporter probes. A fluorescence scanner was used to detect and quantify captured analytes at the TL. The device was evaluated and reached sensitivities of 99.8%, 100%, and 99.8% for HBsAg, anti-HBs, and anti-HCV and specificities of 99.3%, 100%, and 99% for HBsAg, anti-HBs, and anti-HCV tests [55]. On the other hand, Si and coworkers employed a signal amplification system (SAS) to develop an LFA to detect HB e-antigen (HBeAg). In this study, Au NPs conjugated with Ab used as reporter probes and capture Abs labeled with biotin were used as CP. In the presence of HBeAg, the reporter probe and CP would form a dendritic complex. Dendritic complexes would be captured at the TL. The SAS-LFA was evaluated and demonstrated LOD of 9 ng/mL and displayed a complete match with ELISA [124].
Furthermore, Tang et al. demonstrated a hybridization-based LFA by combining a sponge shunt with conventional LFA to detect HBV nucleic acid. Au NPs were used as labeling reporters. A sponge was integrated with an LFA strip to delay fluid flow, increase the interaction time and test sensitivity (Fig. 5C, D and E). The modified LFA was evaluated and reached a LOD of 103 copies/mL, in contrast with conventional LFA, which had 104 copies/mL [120].
An integrated betaine-assisted recombinase polymerase assay (BRPA) with an LFA to detect HBV DNA was introduced by Yi et al. This study showed that in recombinase polymerase assay, nonspecific amplification occurs and the addition of betaine suppresses unwanted nonspecific amplifications. This test was combined with LFA to allow naked-eye detection. The evaluations revealed that 1000 copies of HBV DNA could be detected in a 50 μL mixture using this device with a sensitivity and specificity of 90% and 100%, respectively [125].
An integrated polymerase spiral reaction (PSR) with LFA for sensitive and rapid determination of HBV DNA was developed by Lin and coworkers research group. In this investigation, final products of PSR were labeled with FITC probe and biotin taken to LFA for visualization. STV conjugated Au NPs were used as LFA labeling NPs. Anti-FITC mAb as a capture probe was used at the TL, and biotin-BSA was immobilized at the capture line, and the LFA visualization process takes up to 5 min. This sensing platform was examined and showed a specificity of 100%, which is higher than the qPCR and LAMP assay. LOD was measured 5.4 copies/mL that is ten times more sensitive compared with qPCR and LAMP [126].
LFAs can be more than helpful in developing countries with endemic areas for HBV and lack the expensive conventional devices for HBV detection. Multiplex LFAs are effective in hepatitis viral detection because they also detect various types of hepatitis virus and other sexually transmitted viruses. Integrating LFA with other detection methods, and adjusting the membrane improved sensitivity. These studies also examined alternate readers and reporter probes in quantifying and enhancing detection.
4.5. Dengue virus (DENV)
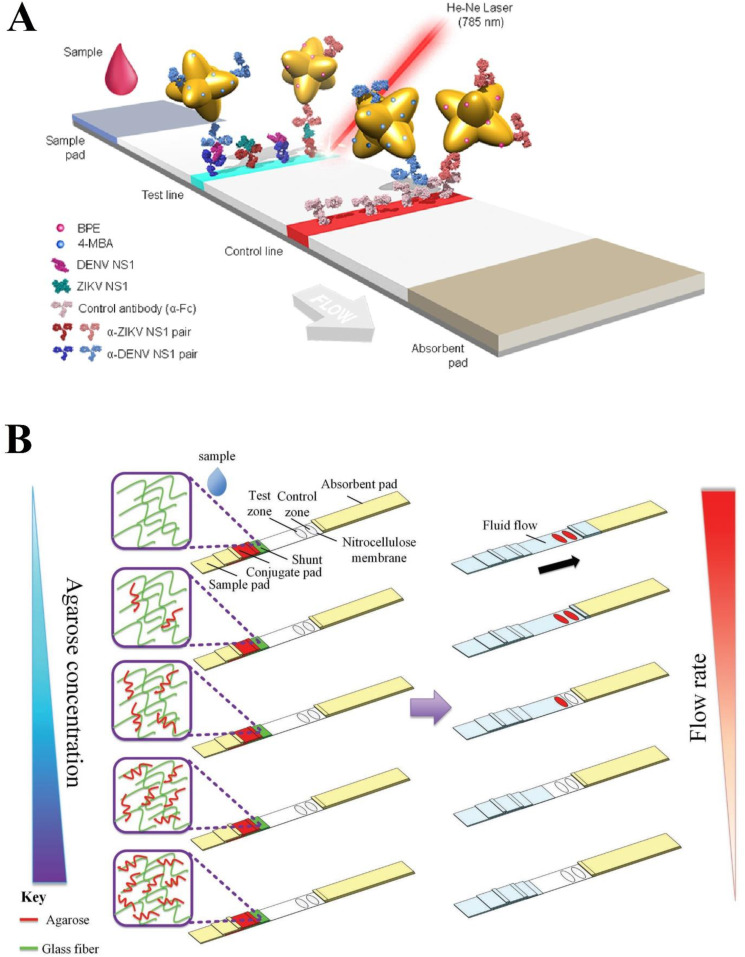
DENV belonging to the family Flaviviridae is transmitted via Aedes mosquitoes. Dengue disease symptoms vary from asymptomatic fever to more severe symptoms like hemorrhagic fever and shock [127,128]. Dengue fever is prevalent in 112 countries, around half of the world population is considered at risk and is estimated to have 390 million incidence cases per year [129,130]. Dengue fever occurs in underdeveloped urban areas and needs a cheap and portable POCT for diagnosis [130]. In the following, some recent advances in LFA sensing platforms are comprehensively explained regarding the DENV. For example, Jeon et al. compared SERS-based LFA strips with conventional LFA to detect mosquito-borne viruses like ZKV and DENV. LFA strips that can resist at high temperatures are needed for these endemic viruses detection in tropical regions. Unlike conventional LFA strips that employ Au NPs, the SERS-based LFA strips in this study used Au NPs conjugated with silica shells as reporter labels to withstand sensitivity loss in higher temperatures. Evaluations demonstrated that conventional LFA strips LOD significantly increase with an increase in temperature, but SERS-based LFA strips LOD value remains the same [131]. Additionally, another SERS-based LFA for detecting and distinguishing the NSP1 of ZKV and DENV was established by Purra et al. In this research, different nanotags were used to differentiate between two viruses. In SERS-encoded gold nanostars, (GNSs), 1,2-bis(4-pyridyl)ethylene (BPE) was conjugated to an anti-ZKV Ab to form GNS-BPE-anti-ZKV, so-called “Z-nanotag”, and 4-mercaptobenzoic acid (MBA) was conjugated to GNSs and anti-DENV Ab to form GNS-MBA-anti-DENV or called “D-nanotag”. The calculated surface enhancement factors could differentiate between nanotags (Fig. 6 A), increasing the specificity of the test while also decreasing the LOD 15- and 7-fold for ZV-NSP1 and DENV-NSP1, respectively [132].
Fig. 6.
Schematic diagram of DENV and ZV multiplexed detection using SERS-based LFA (A). Adapted by permission from Ref. [132]. Schematic design of agarose based-LFA to delay fluid flow for DENV detection (B). Adapted by permission from Ref. [133].
Additionally, Axelrod et al. employed a new approach in LFA to detect and quantify DENV NSP1. Target-specific Ab-conjugated to HRP are present in the conjugation area, and NSP1 molecules are immobilized in the capture area. In negative serums that don't contain NSP1, anti-NSP1-HRPs will be captured by immobilized NSP1 in the capture area, and don't reach the last layer, thus do not generate any signals. However, NSP1 Ags present in positive serums will bind to anti-NSP1-HRPs to form NSP1-specific Ab-HRP immune complex, thus preventing them from being captured by immobilized NSP1s at the capture area, allowing the immune complex to reach the last layer to bind with HRP substrate, generating the signal. This novel LFIA was evaluated and was determined to be five-fold more sensitive than a gold standard ELISA test [134]. Also, Kumar et al. developed an LFA diagnostic test for rapid detection (10 min) of DENV NSP1. In this investigation, Au NPs conjugated with reduced GO (Au NPs-rGO) were used as reporter labels and were conjugated with FITC-NSP1 Abs, increasing the sensitivity and achieving a LOD of 4.9 ng/mL [135]. Tran and coworkers developed a magneto-enzyme LFIA to detect DENV NSP1. In this research, GAM Ab at the CL was used, and at the TL, capture Ab was used. In addition, biotinylated, mAb-conjugated magnetic beads were used as reporter probes. This assay was evaluated and reached a sensitivity and specificity of 100%, demonstrating no cross-reaction with other viruses and LOD between 0.1 and 1 ng/mL applicable for different DENV serotypes [64]. Additionally, a screen-printed gold electrode (SPGE) combination with an LFA strip in the development of an electrochemical lateral flow immunosensor (ELFI), a diagnostic device for DENV was introduced by Sinawang and coworkers. In this device, NSP1 Ab conjugated with immunonanobeads would bind to sample DENV NSP1. The formed immunocomplex would attach with immuno-conjugated electrodes, emitting an electrical signal, allowing a quantitative detection with LOD of 0.5 ng/mL [136].
In another investigation, a NIR fluorescent dye-based LFA detecting anti-DENV1 IgG Abs by Purra et al. was fabricated. This investigation demonstrates that DYlight-800 coated GAH IgG Abs were used as sample capture, and recombinant protein from envelope of DENV1 at the TL was used. This device was evaluated and compared with DENV Duo IgG/IgM cassette and Panbio DENV IgG ELISA. NIR-LFA reached a sensitivity of 95%, which was the same with compared tests. However, testing diluted serum samples, NIR-LFA had a lower LOD compared with these tests [63].
Choi et al. combined agarose hydrogel into the glass fiber shunt to develop a paper-hydrogel hybrid LFA test for detecting the DENV. Using this hybrid allows fluid control while maintaining paper-based advantages, increasing Au NP-detector probes (Au NP-DPs) and DENV RNA interaction (Fig. 6B), thus improving device sensitivity [133].
An LFA for simultaneous detection of DENV and Chikungunya virus (CHIKV) IgM/IgG using two-color detection labels was established by Lee and coworkers. This study used blue labels for anti-IgG and red labels for anti-IgM for both CHIKV and DENV. Two TLs, one consisting of CHIKV Ag and one consisting of DENV Ag, captured virus-specific Abs, while anti-goat IgG was used to capture all Abs in the CL. Captured Abs in the TLs emitted blue or red lights, resulting in a mixture of blue and red light emitted from the TL of virus, captured and calculated to determine IgG and IgM concentration [137].
Yrad and coworkers developed an Au NP-based LFA to detect DENV-1 RNA in less than 20 min. In this investigation, a capture probe specific to DENV-1 (dDNA) was immobilized at the TL to detect DENV-1 RNA and generates a red light, and a control probe (cDNA) was immobilized at the CL to detect dextrin-capped Au NP. The whole detection process took about 20 min, and LOD was calculated to be 1.2 × 104 pfu/mL [65]. Xiong and coworkers integrated reverse transcription recombinase-aided amplification with LFA to detect DENV RNA in blood samples. In this study, primers were designed to detect and amplify DENV RNA and were labeled with FAM and biotin. The amplified nucleic acids were then added to commercially available lateral-flow dipsticks (USTAR, Hangzhou, China) to determine the results. This approach was evaluated and showed a LOD of 10 copies/μL, with no cross-reactivity with other pathogens [138].
LFAs can also help detect DENV in underdeveloped areas. These areas also may have high temperatures that need devices that can withstand environmental inconveniences. These studies examined the impact of using alternate readers and reporter probes in quantifying and improving viral detection. Adjusting the membrane also increased the LFA's sensitivity.
4.6. Human papillomavirus (HPV)
HPV belongs to the Papovaviridae family and is a sexually transmitted virus that causes genital warts [139]. Some subtypes such as 16 and 18 showed high risk and may lead to malignancies [140]. Cervical cancer caused by HPV has a 500,000 incidence rate annually and 250,000 mortalities [141]. Different strategies and approaches are proposed and applied for HPV screening [140]. LFA can be an excellent tool for rapid diagnosis and can be used for screening purposes. For instance, Grant et al. developed a LFA monitoring method to assess HPV vaccination status by detecting anti-HPV16 Abs. In this study, Au NPs conjugated with GAH IgG were used as reporter probes. The LFA had three TLs, spotted with different dilutions of HPV16 L1 VLPs in PBS to discriminate between different serum Ab concentrations, and human IgG was immobilized at the CL. This method was evaluated by testing 28 serum samples, 15 of which received two or more vaccine doses, and 13 received one amount or none. The test could discriminate between these two groups well and could be used two assess patients’ vaccination status [142].
In another investigation, a combination of a miniaturized PCR with LFA to detect HPV DNA has been demonstrated by Liu et al. HPV-nucleic acids were amplified by the IR-COCONUT PCR platform, and the final products were added to LFA for visualization. In this LFA, anti-DIG alkaline phosphatase (AP) conjugate was used as a detector probe, STV was immobilized at the TL and GAR IgG was used at the CL. The LFA visualization process takes up to 25 min, and digital images taken from the TL could be interpreted by computer software to detect PCR products quantitatively [120].
Also, Wang et al. integrated Antarctic thermolabile uracil-DNA-glycosylase (AUDG) with nucleic acid amplification (NAA) techniques and LFA to simultaneously detect HPV16 and HPV18. This research work demonstrates that AUDG-NAAs were used to amplify nucleic acids (HPV16 E7 and HPV18 L1) and labeled with FITC/biotin or DIG/biotin, and the final products were added to LFA for visualization. In this LFA, STV-coated dyed polymer NPs (SA-DNPs) as labeling particles were used and at the first TL, anti-FITC was immobilized. At the second TL, anti-DIG was immobilized. Biotin-BSA forms a firm binding with STV and therefore was used as chromatography control. The final device was evaluated and demonstrated a LOD of 5 × 105 copies and showed no cross-reactivity between HPV16 and HPV18 [66].
Fluorescent probe-based LFA to detect 13 types of HPV simultaneously have been effectively fabricated by Xu and coworkers. This study explains that primers conjugated with fluorophore were used to amplify HPV nucleic acids, and the final products were added to LFA for detection and discrimination. The LFA reader had two-channel color detection; therefore, each line could be conjugated with two different coloring labels, and the probes on the seven lines each were conjugated with FAM or ROX. The probes were designed to detect 13 types of HPV (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68) and capture probe including human β-globin (HBB) was used as a control probe (Fig. 7 A). The LFA visualization process takes up to 30 min. This device was evaluated, and the results showed LODs of 102 copies/μL for HPV type 18 and 10 copies/μL for HPV types 6, 11 and 16. No cross-reactivity was reported, and there was an agreement of 98% with the reverse dot blotting kit (GenoArray) [143].
Fig. 7.
Schematic design of fluorescence LFA for multiplex HPV detection (A). Adapted by permission from Ref. [143]. Schematic illustration of the CuO-based LFSB for HPV detection showing flow direction (B) and the detection procedure (C). Adapted by permission from Ref. [144].
Finally, Yang et al. developed copper oxide nanoparticles (CuO NPs)-based LFSB to detect HPV 16. In this study, CuO NPs conjugated with STV and capture DNA were used as CPs. At the TL and CL, test and control DNAs were immobilized and at the TL or CL, the captured CuO NP-SA-DNAs would form a brown stripe that can be detected qualitatively by the naked eye (Fig. 7B and C), or quantitative detection can be performed by adding washing buffer and using KinBio strip reader software to interpret line color intensity [144].
LFAs are rapid devices that can be great for HPV screening. Using multiplex LFA devices is necessary for screening multiple HPV subtypes. Also, using different readers help to quantify the detection.
4.7. Zika virus (ZKV)
As a member of the Flaviviridae family, ZKV like DENV is commonly transmitted by mosquitos [145]. ZKV infection causes signs and symptoms like fever, arthralgia and myalgia, conjunctivitis, headache, and rash and may cause Guillain-Barré syndrome [146]. Laboratory tests like serological assays, molecular assays, virus isolation, and IHC can be used to detect ZKV [147]. An integrated RT-LAMP with LFA to develop a device for ZKV detection was reported by Lee et al. ZKV RNAs in this research were amplified with RT-LAMP using primers conjugated with DIG, and the final products were added to LFA for visualization. Au NPs conjugated with STV and anti-DIG were used as reporter probes. Amplified ZKV RNAs contained DIG-conjugated primers and were detected by detector probes and were also captured by immobilized anti-DIGs at the TL. Biotin was also affixed in the CL to bind with STV at the Au NPs [148].
In another study, Rong et al. established a smartphone-based LFIA to detect NSP1 in ZKV. In this study, QD microspheres were conjugated with ZKV NSP1Ab and BSA to act as reporter probes. ZKV NSP1 Ab was also immobilized at the TL to capture detected antigen complex, and GAM IgG pAbs at CL was also used to capture QD microspheres. The captured Ag complex at the TL emitted fluorescent can be detected and interpreted by smartphone and its camera, and the whole detection process takes about 20 min. This device was evaluated and showed limited cross-reactivity with similarly structured DENV, and LOD in serum was calculated to be 0.15 ng/mL [149].
Xu et al. developed fluorescent carbon dots (CDs)-based LFA to detect the ZKV. In this investigation, CDs-assembled were loaded into dendritic SiO2 to form fluorescent CD-based silica (FCS) spheres (Fig. 8 A and B). The FCS spheres were conjugated with ZKV-01 Ab and BSA to act as reporter probes. In the presence of ZKV, FCS spheres form a complex, which is captured at the TL by another anti-ZKV Ab, which emits blue light under UV lamp. The remaining FCS spheres migrate to the CL and capture with GAM IgG. This device was evaluated and compared with a conventional Au NP-based LFA. The results showed that this device had a LOD of 10 pg/mL, and had its sensitivity was 100-fold higher than conventional LFA [150].
Fig. 8.
The synthetic processes for CD-embedded SiO2 spheres are depicted in a schematic diagram (A). Schematic illustration for detection of Zika NS1 protein using the iFCS-based LFA (B). Adapted by permission from Ref. [150]. Schematic design of dual-signal readout LFA for EV detection (C). Two-Step Detection of EBV with RNs@Aubased LFA (D). Adapted by permission from Ref. [68].
In another study, polycaprolactone (PCL) electron-coating NC membrane to develop a nucleic acid-based LFA for detecting ZKV was introduced by Yew and coworkers. In this study, Au NPs conjugated with detector probes acted as labeled detectors. Electrospun-coated membrane delays the fluid flow and increases interaction between analyte and reagents. Evaluations demonstrated that this device could detect ZKV cDNA with a LOD of 0.5 nM, and its sensitivity is 10-fold higher than unmodified NC membrane-based LFA [151,152].
4.8. Ebola virus (EV)
EV belongs to the Filoviridae family and causes a hemorrhagic illness called EV disease transmitted via blood and other body fluids [153]. Its signs and symptoms include fever, chills, malaise, anorexia, severe headache, and myalgia [154]. Many laboratory tests are available for EV detection, but due to its infection nature, POCTs are beneficial for early detection and prevention. In this way, Hu et al. developed a rapid, sensitive, and quantitative LFA to detect the EV glycoproteins (GPs). In this study, multifunctional nanosphere (RNs@Au) was used as sample capture. Signal enhancers consisted of RNs@Au with Ab and STV-conjugated on it. These reporters binding with EV glycoprotein were captured by anti-EV glycoprotein at the TL. They emitted two signals, fluorescence signal for quantitative detection interpreted by smartphone and colorimetric signal detectable by the naked eye (Fig. 8C and D). Another set of RNs@Au was conjugated with biotin and developed to bind with STV to amplify signals. This assay allowed the detection of 2 ng/mL glycoprotein by the naked eye in less than 20 min [68]. Additionally, an LFA employing a smartphone reader to detect anti-EV IgG semi-quantitatively was developed by Brangel et al. In this research, anti-human IgG conjugated with Au NPs were used as reporting probes, and recombinant EV proteins were used at the TL. In addition, a smartphone app was developed to acquire semi-quantitative results from the detected IgG at the TL. The device was evaluated and reached a sensitivity of 100% and specificity of 98%, making it a decent test for clinical application [155].
On the other hand, Feuerstein and coworkers developed a NIR light-emitting fluorescent-based LFA for detecting the (GPs of EV. Fluorescent nano-diamond particles (FNDPs) conjugated with anti-EV GPs were used as labeling reporters. In addition, another set of anti-EV recombinant GPs at the TL were immobilized. And, GAH IgG at the CL was immobilized to capture the remaining FNDPs. The captured FNDPs emitted NIR light that could be interpreted by an optoelectronic device [156]. Phan et al. evaluated an LFA for EV detection that developed by the Naval medical research center. Au NPs were used as labeling reporters, and anti-EV Ags were conjugated to them. Polyclonal anti-EV Ag was used at the TL as capture probes, and anti-goat IgG was used to capture remaining reporters at the CL. The LFA was tested and compared with RT-PCR using 290 plasma samples and 237 oral samples. The LFA reached a sensitivity of 87.9% and specificity of 96.8%. EZ1 real-time PCR achieved a sensitivity of 96.7% and specificity of 92.6% [157].
4.9. BK virus (BKV)
BKV infects humans in early childhood and remains there as an asymptomatic infection, and in appropriate circumstances, like immunosuppression, BKV reactivates and begins its pathogenicity process [158]. BKV infection leads to nephropathy; therefore, it is more common in kidney transplant receivers immunocompromised to avoid graft rejection, so BKV detection is crucial in these patients [159,160]. To detect BKV and cytomegalovirus (CMV) in post-transplantation for early infection detection Kaminski et al. developed CRISPR-Cas13-based assay integrated with LFA readout. In this test, BKV DNA and CMV DNA are isolated from urine and blood samples. In addition to this, CXCL9 messenger RNA, a graft rejection marker, is detected in urine samples. This device was integrated with a commercially available LFA for detection optimization, and the final products were added to it. The integrated device was evaluated and showed high sensitivity and specificity for BKV, CMV, and CXCL9 messenger RNA detection [161].
In another study, nucleic acid-based LFA has been established by Huang et al. to detect human polyomavirus BKV in 45 min. In this study, oligonucleotide probes were manufactured and were conjugated to Au NPs to form DPs and CPs that detected different parts of DNA. In the presence of the Ag, the target DNA is captured by both CP and DP. Rabbit polyclonal anti-STV Ab was used as TL and captured the Au NP-DP-target-Au NP-CP immune-complex and formed a red line. Rabbit polyclonal anti-BSA Ab was immobilized at the CL, capturing the remaining Au NPs. The line intensity was interpreted by image software to calculate the quantitative results. The assay LOD was evaluated to be 107 copies/mL, and this test presented no cross-reactivity with other polyomaviruses [162].
4.10. Norovirus
Norovirus belongs to the Caliciviridae family and causes winter vomiting disease [163]. This disease can be asymptomatic or be presented with fever, vomiting, and diarrhea and can be transmitted person-to-person or via food infections [164]. To develop an LFA to detect norovirus, Hagström and coworkers used phage NPs. In this study, Norwalk (norovirus GI.1) virus-like particles (VLPs) were synthetically manufactured and purified. SAM-AviTag M13 phage was cultured and conjugated with BSA and anti-norovirus mAb to be used as reporter probes. Anti-norovirus Abs at the TL were immobilized to detect noroviruses, and anti-M13 Abs were used at the CL to capture remaining M13 phage. This device was evaluated and compared with Au NP-based LFA. The results demonstrated that M13 phage-based LOD is 107 for VLPs per mL, 100-fold lower than Au NP-based LFA [165]. Moreover, Doerflinger and coworkers developed a nanobody-based LFA to detect norovirus. The Nanobody used in this study bound to a region on norovirus capsids and was conjugated with biotin and Au NP. After the addition of norovirus particles, STV on the TL capture immune complexes resulting from nanobodies. This device was evaluated and reached a sensitivity of 80% and specificity of 86% [166].
4.11. Severe fever and thrombocytopenia syndrome virus (SFTSV)
A phlebovirus is known to be the cause of SFTS showing symptoms of gastrointestinal disorders, leukocytopenia, and fever [167]. SFTSV belongs to Bunyaviridae family and can be transmitted via ticks [168]. SFTSV can be diagnosed using real-time PCR, ELISA, and virus isolation, and POCT like LFA can be employed for early detection [169]. In Xu et al. investigation, they developed a fluorescence-based LFA to detect SFTSV. In this study, carbon dots/SiO2 nanospheres (CDSNs) were conjugated with anti-SFTSV mAbs to act as reporter probes. Anti-SFTSV Abs were also immobilized at the TL to capture detected Ags, and GAM IgG Abs at the CL were used to capture remaining CDSNs. This device was evaluated, and the SFTSV detection results were consistent with PCR showing no cross-reactivity with HCG, AFP, CEA, CA125 [170]. In another research, Zuo and coworkers developed an LFA using Au NPs conjugated with anti-SFTSV mAbs as reporter probes. These mAbs at the TL were immobilized to CPs, and GAM IgG at the CL was used for capturing Au NPs. This device was evaluated with a LOD of 1 ng/mL, and the whole detection process took up to 10 min [71].
4.12. LFA sensing assays for determination of other types of viruses
LFA-based devices have also been effectively used in the determination of other kinds of viruses. In an investigation, Li and coworkers constructed an Au NP-based LFA using IgM Abs for detecting four TORCH pathogens including herpes simplex virus type 2 (HSV-2), CMV, rubella virus and toxoplasmosis. In this research, the μ-chain of MAH IgM mAb (anti-human IgM) was conjugated to AuMag to form the reporter probes. TORCH Ags were immobilized at the TL to act as CP, and the GAM IgG Ab at the CL was used as a CP. Serum samples of 162 infected individuals were tested by this device and 100% sensitivity, specificity with no occurrence of cross-reactivity were reported [67]. In another study, Zhu and coworkers developed a fluorescence-based LFA to detect Epstein-Barr virus (EBV) and HPV16. In this study, carboxyl-modified fluorescent microspheres were used as labeling reporters, and NC substrate strips were used as a membrane. BSA and whole human IgG Ab were used as negative and positive controls, respectively. EBV nuclear Ag-1 (EBVNA-1) was immobilized at the TL to capture EBV Ags. This device was again modified to study anti-HPV 16 E7 IgG detection using recombinant HPB16 E7 proteins. The results showed a 100-fold improvement compared with previous studies that used a glass substrate [171].
Additionally, HSV-2 is tried to serologically detect by the development of a LFA device in Goux and coworkers research. In this investigation, persistent luminescent NPs (nanophosphors) of strontium aluminate conjugated with GAH pAbs were used as labeling reporters. At the TL, recombinant gG2 Ag of HSV was immobilized to detect anti-HSV Ab, and GAH IgG capture Ab was used at CL to capture the remaining nanophosphors. Nanophosphors at the TL reacted to light and excited, emitting light that can be detected and interpreted by an iPhone app to quantitatively detect anti-HSV Abs captured at the TL [172].
In an investigation reported by Townsend and coworkers, the authors evaluated commercially available Tetracore Orthopox BioThreat® LFA assay for detecting Orthopoxvirus (OPVs), Vaccinia virus (VACV), and Monkeypox virus (MPXV). In this device, Au NPs conjugated with anti-VACV Abs were used as reporter probes. The anti-VACV Abs were also immobilized at the TL to capture detected OPVs. This assay reached a LOD of 107 pfu/mL and showed a false positive in one out of 11 non-OPV samples; therefore, it is applicable for screening and detecting OPVs [173]. In Rebollo and coworkers study, the authors developed an immune-chromatographic LFA to detect the West Nile virus (WNV). In this study, 1D11 (a mAb that detects WNV's envelope glycoprotein) was conjugated with red carboxyl-latex particles to act as reporter probes, and BSA was conjugated with blue latex particle to serve as a control complex. 1D11 was also immobilized at the TL to capture Ags detected by the detector probe, and the CL consisted of mAbs that captured control-complex proteins. After 10 min, in the presence of WNV, a red line is formed at the TL, informing the test positivity. This test was evaluated and successfully detected different WNV strains with LOD of 105 TCID50/mL and showed no cross-reactivity with other flaviviruses [174].
In recent years, for the effective management of viral diseases, hundreds of POCTs with high sensitivity and specificity have been developed and are commercially available now for detection of most pathogenic viruses such as SARS-Cov-2, HIV, HBV, HCV, Influenza, Ebola, Dengue, HPV, Zika, HCV and etc (Table 2 ).
Table 2.
Some of commercially available LFA strips for fast detection of infectious viruses.
| LFA device | Company name | Detected Virus | Detection time |
|---|---|---|---|
| SARS-Cov-2 Antigen Rapid Test Cassette (swab) | Spring healthcare | SARS-Cov-2 | 15 min |
| E25Bio's SARS-CoV-2 Direct Antigen Rapid Test (DART) | E25bio | SARS-Cov-2 | 15 min |
| Encode SARS-CoV-2 Antigen Rapid Test Device | Zhuhai Encode Medical Engineering Co.,Ltd | SARS-Cov-2 | 20 min |
| SureScreen COVID-19 Rapid Antigen Test Cassette | Surescrenn diagnostics | SARS-Cov-2 | 15 min |
| Statu s™ COVID-19/F lu | Lifesign | SARS-Cov-2/Influenza A and B | 15 min |
| SARS-CoV-2 & Influenza Antigen Combo Rapid Test Kit | MedOmics | SARS-Cov-2/Influenza A and B | 15–20 min |
| SCREEN TEST COVID-19+FLU A/B | ScreenItalia | SARS-Cov-2/Influenza A and B | 15 min |
| SARS-CoV-2 & Influenza A/B & RSV Antigen Combo Test Kit | Shenzhen Microprofit Biotech Co., Ltd | SARS-Cov-2/Influenza A and B/RSV | 15 min |
| QuickProfile COVID-19 ANTIGEN Test | LumiQuick Diagnostics Inc. | SARS-Cov-2 | 15 min |
| quickvue influenza a+b test | Quidell | Influenza A and B | 10 min |
| rapidSTRIPE H1N1 | Analytik Jena | Influenza A H1N1 | 5 min |
| Sofia Influenza A+B FIA | Quidell | Influenza A and B | 15 min |
| Xpect Flu A&B Kit | REMEL INC | Influenza A and B | 15 min |
| OSOM™ Ultra Flu A and B Rapid Test | Sekisui Diagnostics | Influenza A and B | 15 min |
| Influenza A&B test | Nanoentek | Influenza A and B | 15 min |
| OraQuick® In-Home HIV Test | OraSure Technologies | HIV 1 and 2 | 20 min |
| autotest VIH® | AAZ-LMB | HIV 1 and 2 | 15 min |
| BioSURE HIV Self Test | BioSURE United Kingdom Ltd | HIV 1 and 2 | 15 min |
| INSTI HIV Self Test | bioLytical Laboratories | HIV 1 and 2 | 5 min |
| Atomo HIV Self-Test | Atomo Diagnostics | HIV 1 and 2 | 15 min |
| Aware™ HIV-1/2 OMT Oral HIV Self Test | Calypte Biomedical Corporation | HIV 1 and 2 | 20 min |
| Exacto® HIV Screening Test | Biosynex Group | HIV 1 and 2 | 10 min |
| INSTI HIV-1/HIV-2 TEST | Biolytical | HIV 1 and 2 | 1 min |
| DETERMINE HBsAg 2 | Alere Medical Co. Ltd | HBV | 15–30 min |
| iCare Hepatitis B Home Test Kit | iCare | HBV | 15 min |
| SCREEN TEST EPATITE B – HBsAg | ScreenItalia | HBV | 15 min |
| Hepatitis B Rapid diagnostic test | Maccura Biotechnology Co., Ltd. | HBV | 10 min |
| SD BIOLINE HCV | Abbott Laboratories | HCV | 15 min |
| OraQuick® Ebola Rapid Antigen Test | OraSure technologies | Ebola virus | 30 min |
| ReEBOV™ Antigen Rapid TestKit | Corgenix Medical Corporation | Ebola virus | 15 min |
| Panbio™ Dengue Duo Cassette | Abbott Laboratories | Dengue virus | 15 min |
| Dengucheck WB | Adiwara Worldwide | Dengue virus | 15 min |
| Dengue Rapid Test | Maternova inc | Dengue virus | 10 min |
| SD BIOLINE Dengue Duo (Dengue NS1 Ag + IgG/IgM) | Abbott Laboratories | Dengue virus | 15 min |
| OncoE6™ Cervical Test | Arbor Vita Corporation | HPV 16 and 18 | 15 min |
| StrongStep® HPV 16/18 Antigen Rapid Test | Liming bio | HPV 16 and 18 | 15 min |
| AVantage HPV E6 Test | Hologic | HPV 16/18/45/33/58/52 | 15 min |
| Zika SimPlex | E25bio | Zika virus | 15 min |
| One Step Zika IgG/IgM Antibody Test | Maternova inc | Zika virus | 15 min |
| Biopand Zika NS1 Rapid Test | Biopanda reagents | Zika virus | 15 min |
| DPP® Zika IgM Assay USA | ChemBio diagnostics inc. | Zika virus | 15 min |
| Duo HSV-1/2 IgG/IgM Rapid Test CE | CTK Biotech | HSV 1 and 2 | 10 min |
| iCare Herpes-2 Home Test Kit | iCare | HSV-2 | 15 min |
| Herpes Simplex Virus 1 & 2 Blood Test 5 Test Pack | Home health UK | HSV 1 and 2 | 15 mimutes |
| AmpliVue HSV 1 + 2 Assay | Quidell | HSV 1 and 2 | Not mentioned |
5. Conclusions and future outlooks
This review mainly focused on investigating LFAs in virus detection. Conventional LFAs are rapid, user-friendly, and accessible but certain limitations like low sensitivity and showing the results just by an on/off format constrain them for wide application in virus testing. In addition to these, some LFAs suffer from low reproducibility. The final result and detection time depend on various factors like biomolecule affinity, sample nature, fluid viscosity, and surface tension. The analytes must be in solution form, and samples should be fluid; therefore, pretreatment is required for non-fluidic samples, which is time-consuming. Using gold, silver, and other nanoparticles may limit shelf life and increase test costs [175]. However, many LFAs investigated in this study employed novel methods and strategies to overcome these limitations, but other approaches can further improve LFA performance. Immobilizing Abs on the LFA strip can be organized by passive absorption and covalent coupling; the latter conjugates Abs to carboxyl groups on functionalized nanoparticles that improve performance. The LFA strip membrane can also be adjusted to delay fluid flow and increase reagents interactions to ensure that reagents binding is optimal, increase sensitivity and reduce detection time. Using dissolvable barriers between the conjugate pad and test line and between test line and control line has been proven effective in increasing LFA's sensitivity [176,177]. Integrating LFA with other methods can significantly increase its sensitivity. Many techniques like LAMP, PCR, CRISPR-Cas13a, RT-RPA, etc., were used to amplify virus nucleic acids and employed LFA for visualization. With the recent advancements in nanomaterials, many approaches have been suggested to improve LFAs. Two sets of GNPs with two different sizes can be used as reporter probes, as the GNPs with smaller sizes bind to the analytes, and the GNPs with bigger sizes bind to the first group of GNPs [178]. Using alternate labeling reporters, like magnetic particles, quantum dots, upconverting phosphors, europium, latex beads, enzymes, and colloidal carbons could also enhance the signal and increase LFA's sensitivity. Several undesirable interface issues can occur in developing LFAs at molecular levels like in NP-antibody conjugates, immobilized antibodies, and Antibody-biomarker interactions, and with the advancements in nanotechnology, resolving these interface issues can be the next breakthrough in LFA design [179]. Also, the employment of alternative reporters allows novel visualization methods that can be digitized and allows quantitative detection. Visual detection, luminescence detection, magnetic detection, SERS detection, electrochemical detection was used in LFAs, each having its advantages and disadvantages. Quantitative detection can also be obtained using image analysis software that calculated the virus concentration by interpreting the line intensity at the TL. Multiplex detection was also used at these LFAs, some detecting 13 different Ags simultaneously, which can help confirm the differential diagnosis in infectious diseases with similar signs and symptoms. Subsequently, LFAs can be widely used in clinical settings and replace expensive and time-consuming methods with these moderations. Finally, this review finds that LFAs as one of the effective alternative methods in virus detection, mainly due to their rapidity and portability. By further improvements, they can be commonly used in clinical diagnosis.
Declaration of competing interest
The authors have no conflict of interest to declare.
Acknowledgement
We acknowledge the supports from the Immunology Research Center, Tabriz University of Medical Science and University of Tabriz.
References
- 1.Modrow S., Falke D., Truyen U., Schätzl H. Viruses: definition, structure, classification. Molec. Virol. 2013:17–30. doi: 10.1007/978-3-642-20718-1_2. [DOI] [Google Scholar]
- 2.Taylor M.W. What is a virus? Viruses Man: Hist. Interact. 2014:23–40. doi: 10.1007/978-3-319-07758-1_2. [DOI] [Google Scholar]
- 3.Medical importance of viruses [Internet] 2021. https://bio.libretexts.org/@go/page/9870 [cited 2021 Apr 19]. Available from:
- 4.Louten J. Virus transmission and epidemiology. Essent. Human Virol. 2016:71–92. doi: 10.1016/B978-0-12-800947-5.00005-3. [DOI] [Google Scholar]
- 5.Woolhouse M., Scott F., Hudson Z., Howey R., Chase-Topping M. Human viruses: discovery and emergence. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2012;367:2864–2871. doi: 10.1098/rstb.2011.0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walker J.W., Han B.A., Ott I.M., Drake J.M. Transmissibility of emerging viral zoonoses. PLoS One. 2018;13:e0206926. doi: 10.1371/journal.pone.0206926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Payne S. In: Chapter 9 - Viral Pathogenesis. Payne S., editor. Viruses, Academic Press; 2017. pp. 87–95. [Google Scholar]
- 8.Linde A. The importance of specific virus diagnosis and monitoring for antiviral treatment. Antivir. Res. 2001;51:81–94. doi: 10.1016/s0166-3542(01)00129-2. [DOI] [PubMed] [Google Scholar]
- 9.Zhu H., Fohlerová Z., Pekárek J., Basova E., Neužil P. Recent advances in lab-on-a-chip technologies for viral diagnosis. Biosens. Bioelectron. 2020;153:112041. doi: 10.1016/j.bios.2020.112041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hasanzadeh M., Tagi S., Solhi E., Shadjou N., Jouyban A., Mokhtarzadeh A. Immunosensing of breast cancer prognostic marker in adenocarcinoma cell lysates and unprocessed human plasma samples using gold nanostructure coated on organic substrate. Int. J. Biol. Macromol. 2018;118:1082–1089. doi: 10.1016/j.ijbiomac.2018.06.091. [DOI] [PubMed] [Google Scholar]
- 11.Hasanzadeh M., Nahar A.S., Hassanpour S., Shadjou N., Mokhtarzadeh A., Mohammadi J. Proline dehydrogenase-entrapped mesoporous magnetic silica nanomaterial for electrochemical biosensing of L-proline in biological fluids. Enzym. Microb. Technol. 2017;105:64–76. doi: 10.1016/j.enzmictec.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 12.Sohrabi H., Majidi M.R., Arbabzadeh O., Khaaki P., Pourmohammad S., Khataee A., Orooji Y. Recent advances in the highly sensitive determination of zearalenone residues in water and environmental resources with electrochemical biosensors. Environ. Res. 2021:112082. doi: 10.1016/j.envres.2021.112082. [DOI] [PubMed] [Google Scholar]
- 13.Hasanzadeh M., Sadeghi S., Bageri L., Mokhtarzadeh A., Shadjou N., Mahboob S. Poly-dopamine-beta-cyclodextrin: a novel nanobiopolymer towards sensing of some amino acids at physiological pH. Mater. Sci. Eng. C. 2016;69:343–357. doi: 10.1016/j.msec.2016.06.081. [DOI] [PubMed] [Google Scholar]
- 14.Farzin L., Shamsipur M., Samandari L., Sheibani S. HIV biosensors for early diagnosis of infection: the intertwine of nanotechnology with sensing strategies. Talanta. 2020;206:120201. doi: 10.1016/j.talanta.2019.120201. [DOI] [PubMed] [Google Scholar]
- 15.Sohrabi H., Majidi M.R., Asadpour-Zeynali K., Khataee A., Mokhtarzadeh A. Bimetallic Fe/Mn MOFs/MβCD/AuNPs stabilized on MWCNTs for developing a label-free DNA-based genosensing bio-assay applied in the determination of Salmonella typhimurium in milk samples. Chemosphere. 2021:132373. doi: 10.1016/j.chemosphere.2021.132373. [DOI] [PubMed] [Google Scholar]
- 16.Sohrabi H., Khataee A., Ghasemzadeh S., Majidi M.R., Orooji Y. Layer double hydroxides (LDHs)-based electrochemical and optical sensing assessments for quantification and identification of heavy metals in water and environment samples: a review of status and prospects. Trends Environ. Analy. Chem. 2021:e00139. [Google Scholar]
- 17.Di Nardo F., Chiarello M., Cavalera S., Baggiani C., Anfossi L. Ten years of lateral flow immunoassay technique applications: trends, challenges and future perspectives. Sensors. 2021;21:5185. doi: 10.3390/s21155185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Farrell B. Evolution in lateral flow–based immunoassay systems. Lat. Flow Immunoass. 2008:1–33. doi: 10.1007/978-1-59745-240-3_1. [DOI] [Google Scholar]
- 19.Ford P.J. In: Oral Biology: Molecular Techniques and Applications. Seymour G.J., Cullinan M.P., Heng N.C.K., editors. Humana Press; Totowa, NJ: 2010. Immunological techniques: ELISA, flow cytometry, and immunohistochemistry; pp. 327–343. [DOI] [PubMed] [Google Scholar]
- 20.Katsarou K., Bardani E., Kallemi P., Kalantidis K. Viral detection: past, present, and future. BioEssays. 2019;41:1900049. doi: 10.1002/bies.201900049. [DOI] [PubMed] [Google Scholar]
- 21.Vainionpää R., Leinikki P. Diagnostic techniques: serological and molecular approaches. Encyclop. Virol. 2008:29–37. doi: 10.1016/B978-012374410-4.00585-9. [DOI] [Google Scholar]
- 22.Aydin S. A short history, principles, and types of ELISA, and our laboratory experience with peptide/protein analyses using ELISA. Peptides. 2015;72:4–15. doi: 10.1016/j.peptides.2015.04.012. [DOI] [PubMed] [Google Scholar]
- 23.Alhajj F.A.M. StatPearls; StatPearls: 2021. Enzyme Linked Immunosorbeny Assay. [PubMed] [Google Scholar]
- 24.Jääskeläinen A.J., Korhonen E.M., Huhtamo E., Lappalainen M., Vapalahti O., Kallio-Kokko H. Validation of serological and molecular methods for diagnosis of zika virus infections. J. Virol Methods. 2019;263:68–74. doi: 10.1016/j.jviromet.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 25.Granger D., Hilgart H., Misner L., Christensen J., Bistodeau S., Palm J., Strain A.K., Konstantinovski M., Liu D., Tran A., Theel E.S. Serologic testing for zika virus: comparison of three zika virus IgM-screening enzyme-linked immunosorbent assays and initial laboratory experiences. J. Clin. Microbiol. 2017;55:2127–2136. doi: 10.1128/JCM.00580-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huillier A.G., Hamid-Allie A., Kristjanson E., Papageorgiou L., Hung S., Wong C.F., Stein D.R., Olsha R., Goneau L.W., Dimitrova K., Drebot M., Safronetz D., Gubbay J.B. Evaluation of Euroimmun anti-zika virus IgM and IgG enzyme-linked immunosorbent assays for zika virus serologic testing. J. Clin. Microbiol. 2017;55:2462. doi: 10.1128/JCM.00442-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watzinger F., Ebner K., Lion T. Detection and monitoring of virus infections by real-time PCR. Mol. Aspect. Med. 2006;27:254–298. doi: 10.1016/j.mam.2005.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balmaseda A., Zambrana J.V., Collado D., García N., Saborío S., Elizondo D., Mercado J.C., Gonzalez K., Cerpas C., Nuñez A., Corti D., Waggoner J.J., Kuan G., Burger-Calderon R., Harris E. Comparison of four serological methods and two reverse transcription-PCR assays for diagnosis and surveillance of zika virus infection. J. Clin. Microbiol. 2018;56 doi: 10.1128/JCM.01785-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santiago G.A., Vázquez J., Courtney S., Matías K.Y., Andersen L.E., Colón C., Butler A.E., Roulo R., Bowzard J., Villanueva J.M., Muñoz-Jordan J.L. Performance of the Trioplex real-time RT-PCR assay for detection of Zika, dengue, and chikungunya viruses. Nat. Commun. 2018;9:1391. doi: 10.1038/s41467-018-03772-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naranbhai V., Chang C.C., Beltran W.F.G., Miller T.E., Astudillo M.G., Villalba J.A., Yang D., Gelfand J., Bernstein B.E., Feldman J., Hauser B.M., Caradonna T.M., Alter G., Murali M.R., Jasrasaria R., Quinlan J., Xerras D.C., Betancourt J.R., Louis D.N., Schmidt A.G., Lennerz J., Poznansky M.C., Iafrate A.J. High seroprevalence of anti-SARS-CoV-2 antibodies in chelsea, Massachusetts. J. Infect. Dis. 2020;222:1955–1959. doi: 10.1093/infdis/jiaa579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28 doi: 10.1093/nar/28.12.e63. e63-e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.González-González E., Lara-Mayorga I.M., Rodríguez-Sánchez I.P., Zhang Y.S., Martínez-Chapa S.O., Santiago G.T.-d., Alvarez M.M. Colorimetric loop-mediated isothermal amplification (LAMP) for cost-effective and quantitative detection of SARS-CoV-2: the change in color in LAMP-based assays quantitatively correlates with viral copy number. Analy. Meth. 2021;13:169–178. doi: 10.1039/d0ay01658f. [DOI] [PubMed] [Google Scholar]
- 33.Mirasoli M., Bonvicini F., Lovecchio N., Petrucci G., Zangheri M., Calabria D., Costantini F., Roda A., Gallinella G., Caputo D., de Cesare G., Nascetti A. On-chip LAMP-BART reaction for viral DNA real-time bioluminescence detection. Sensor. Actuator. B Chem. 2018;262:1024–1033. [Google Scholar]
- 34.Ahn S.J., Baek Y.H., Lloren K.K.S., Choi W.-S., Jeong J.H., Antigua K.J.C., Kwon H.-i., Park S.-J., Kim E.-H., Kim Y.-i., Si Y.-J., Hong S.B., Shin K.S., Chun S., Choi Y.K., Song M.-S. Rapid and simple colorimetric detection of multiple influenza viruses infecting humans using a reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic platform. BMC Infect. Dis. 2019;19:676. doi: 10.1186/s12879-019-4277-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benzigar M.R., Bhattacharjee R., Baharfar M., Liu G. Current methods for diagnosis of human coronaviruses: pros and cons. Anal. Bioanal. Chem. 2021;413:2311–2330. doi: 10.1007/s00216-020-03046-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sidiq Z., Hanif M., Dwivedi K.K., Chopra K.K. Benefits and limitations of serological assays in COVID-19 infection. Indian J. Tubercul. 2020;67:S163–S166. doi: 10.1016/j.ijtb.2020.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koczula K.M., Gallotta A. Lateral flow assays. Essays Biochem. 2016;60:111–120. doi: 10.1042/EBC20150012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koczula K.M., Gallotta A. Lateral flow assays. Essays Biochem. 2016;60:111–120. doi: 10.1042/EBC20150012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahmoudi T., de la Guardia M., Shirdel B., Mokhtarzadeh A., Baradaran B. Recent advancements in structural improvements of lateral flow assays towards point-of-care testing. Trac. Trends Anal. Chem. 2019;116:13–30. [Google Scholar]
- 40.Peng T., Liu X., Adams L.G., Agarwal G., Akey B., Cirillo J., Deckert V., Delfan S., Fry E., Han Z., Hemmer P., Kattawar G., Kim M., Lee M.-C., Lu C., Mogford J., Nessler R., Neuman B., Nie X., Pan J., Pryor J., Rajil N., Shih Y., Sokolov A., Svidzinsky A., Wang D., Yi Z., Zheltikov A., Scully M. Enhancing sensitivity of lateral flow assay with application to SARS-CoV-2. Appl. Phys. Lett. 2020;117:120601. doi: 10.1063/5.0021842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mahmoudi T., de la Guardia M., Baradaran B. Lateral flow assays towards point-of-care cancer detection: a review of current progress and future trends. Trac. Trends Anal. Chem. 2020;125:115842. [Google Scholar]
- 42.Sajid M., Kawde A.-N., Daud M. Designs, formats and applications of lateral flow assay: a literature review. J. Saudi Chem. Soc. 2015;19:689–705. [Google Scholar]
- 43.Zhao S., Wang S., Zhang S., Liu J., Dong Y. State of the art: lateral flow assay (LFA) biosensor for on-site rapid detection. Chin. Chem. Lett. 2018;29:1567–1577. [Google Scholar]
- 44.Zhang Y., Liu X., Wang L., Yang H., Zhang X., Zhu C., Wang W., Yan L., Li B. Improvement in detection limit for lateral flow assay of biomacromolecules by test-zone pre-enrichment. Sci. Rep. 2020;10:9604. doi: 10.1038/s41598-020-66456-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baker A.N., Richards S.J., Guy C.S., Congdon T.R., Hasan M., Zwetsloot A.J., Gallo A., Lewandowski J.R., Stansfeld P.J., Straube A., Walker M., Chessa S., Pergolizzi G., Dedola S., Field R.A., Gibson M.I. The SARS-COV-2 spike protein binds sialic acids and enables rapid detection in a lateral flow point of care diagnostic device. ACS Cent. Sci. 2020;6:2046–2052. doi: 10.1021/acscentsci.0c00855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang C., Wen T., Shi F.J., Zeng X.Y., Jiao Y.J. Rapid detection of IgM antibodies against the SARS-CoV-2 virus via colloidal gold nanoparticle-based lateral-flow assay. ACS Omega. 2020;5:12550–12556. doi: 10.1021/acsomega.0c01554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang C., Zheng T., Wang H., Chen W., Huang X., Liang J., Qiu L., Han D., Tan W. Rapid one-pot detection of SARS-CoV-2 based on a lateral flow assay in clinical samples. Anal. Chem. 2021;93:3325–3330. doi: 10.1021/acs.analchem.0c05059. [DOI] [PubMed] [Google Scholar]
- 48.Kim H.-Y., Lee J.-H., Kim M.J., Park S.C., Choi M., Lee W., Ku K.B., Kim B.T., Changkyun Park E., Kim H.G., Kim S.I. Development of a SARS-CoV-2-specific biosensor for antigen detection using scFv-Fc fusion proteins. Biosens. Bioelectron. 2021;175:112868. doi: 10.1016/j.bios.2020.112868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cavalera S., Colitti B., Rosati S., Ferrara G., Bertolotti L., Nogarol C., Guiotto C., Cagnazzo C., Denina M., Fagioli F., Di Nardo F., Chiarello M., Baggiani C., Anfossi L. A multi-target lateral flow immunoassay enabling the specific and sensitive detection of total antibodies to SARS COV-2. Talanta. 2021;223:121737. doi: 10.1016/j.talanta.2020.121737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roda A., Cavalera S., Di Nardo F., Calabria D., Rosati S., Simoni P., Colitti B., Baggiani C., Roda M., Anfossi L. Dual lateral flow optical/chemiluminescence immunosensors for the rapid detection of salivary and serum IgA in patients with COVID-19 disease. Biosens. Bioelectron. 2021;172:112765. doi: 10.1016/j.bios.2020.112765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wen T., Huang C., Shi F.-J., Zeng X.-Y., Lu T., jiao Y. Development of a lateral flow immunoassay strip for rapid detection of IgG antibody against SARS-CoV-2 virus. Analyst. 2020;145 doi: 10.1039/d0an00629g. [DOI] [PubMed] [Google Scholar]
- 52.Peng T., Sui Z., Huang Z., Xie J., Wen K., Zhang Y., Huang W., Mi W., Peng K., Dai X., Fang X. Point-of-care test system for detection of immunoglobulin-G and -M against nucleocapsid protein and spike glycoprotein of SARS-CoV-2. Sensor. Actuator. B Chem. 2021;331:129415. doi: 10.1016/j.snb.2020.129415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kawasaki H., Suzuki H., Maekawa M., Hariyama T. Combination of the NanoSuit method and gold/platinum particle-based lateral flow assay for quantitative and highly sensitive diagnosis using a desktop scanning electron microscope. J. Pharmaceut. Biomed. Anal. 2021;196:113924. doi: 10.1016/j.jpba.2021.113924. [DOI] [PubMed] [Google Scholar]
- 54.Kim J., Kwon J.H., Jang J., Lee H., Kim S., Hahn Y.K., Kim S.K., Lee K.H., Lee S., Pyo H., Song C.S., Lee J. Rapid and background-free detection of avian influenza virus in opaque sample using NIR-to-NIR upconversion nanoparticle-based lateral flow immunoassay platform. Biosens. Bioelectron. 2018;112:209–215. doi: 10.1016/j.bios.2018.04.047. [DOI] [PubMed] [Google Scholar]
- 55.Ryu J.H., Kwon M., Moon J.D., Hwang M.W., Lee J.M., Park K.H., Yun S.J., Bae H.J., Choi A., Lee H., Jung B., Jeong J., Han K., Kim Y., Oh E.J. Development of a rapid automated fluorescent lateral flow immunoassay to detect hepatitis B surface antigen (HBsAg), antibody to HBsAg, and antibody to hepatitis C. Ann. Lab. Med. 2018;38:578–584. doi: 10.3343/alm.2018.38.6.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoo S.J., Shim H.S., Yoon S., Moon H.-W. Evaluation of high-throughput digital lateral flow immunoassays for the detection of influenza A/B viruses from clinical swab samples. J. Med. Virol. 2020;92:1040–1046. doi: 10.1002/jmv.25626. [DOI] [PubMed] [Google Scholar]
- 57.Lee J.H., Seo H.S., Kwon J.H., Kim H.T., Kwon K.C., Sim S.J., Cha Y.J., Lee J. Multiplex diagnosis of viral infectious diseases (AIDS, hepatitis C, and hepatitis A) based on point of care lateral flow assay using engineered proteinticles. Biosens. Bioelectron. 2015;69:213–225. doi: 10.1016/j.bios.2015.02.033. [DOI] [PubMed] [Google Scholar]
- 58.Fu X., Cheng Z., Yu J., Choo P., Chen L., Choo J. A SERS-based lateral flow assay biosensor for highly sensitive detection of HIV-1 DNA. Biosens. Bioelectron. 2016;78:530–537. doi: 10.1016/j.bios.2015.11.099. [DOI] [PubMed] [Google Scholar]
- 59.Deng X., Wang C., Gao Y., Li J., Wen W., Zhang X., Wang S. Applying strand displacement amplification to quantum dots-based fluorescent lateral flow assay strips for HIV-DNA detection. Biosens. Bioelectron. 2018;105:211–217. doi: 10.1016/j.bios.2018.01.039. [DOI] [PubMed] [Google Scholar]
- 60.Rong Z., Xiao R., Peng Y., Zhang A., Wei H., Ma Q., Wang D., Wang Q., Bai Z., Wang F., Sun M., Wang S. Integrated fluorescent lateral flow assay platform for point-of-care diagnosis of infectious diseases by using a multichannel test cartridge. Sensor. Actuator. B Chem. 2021;329:129193. [Google Scholar]
- 61.Gao Y., Deng X., Wen W., Zhang X., Wang S. Ultrasensitive paper based nucleic acid detection realized by three-dimensional DNA-AuNPs network amplification. Biosens. Bioelectron. 2017;92:529–535. doi: 10.1016/j.bios.2016.10.068. [DOI] [PubMed] [Google Scholar]
- 62.Martiskainen I., Talha S.M., Vuorenpää K., Salminen T., Juntunen E., Chattopadhyay S., Kumar D., Vuorinen T., Pettersson K., Khanna N., Batra G. Upconverting nanoparticle reporter–based highly sensitive rapid lateral flow immunoassay for hepatitis B virus surface antigen. Anal. Bioanal. Chem. 2021;413:967–978. doi: 10.1007/s00216-020-03055-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen L., Wang H., Guo T., Xiao C., Liu L., Zhang X., Liu B., Li P., Liu A., Li B., Li B., Mao Y. A rapid point-of-care test for dengue virus-1 based on a lateral flow assay with a near-infrared fluorescent dye. J. Immunol. Methods. 2018;456:23–27. doi: 10.1016/j.jim.2018.02.005. [DOI] [PubMed] [Google Scholar]
- 64.Tran T.V., Nguyen B.V., Nguyen T.T.P., Tran T.T., Pham K.G., Le Q.B., Do B.N., Pham H.N., Nguyen C.V., Dinh D.P.H., Ha V.T., Doan T.H.T., Le H.Q. Development of a highly sensitive magneto-enzyme lateral flow immunoassay for dengue NS1 detection. PeerJ. 2019;7:e7779. doi: 10.7717/peerj.7779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yrad F.M., Castañares J.M., Alocilja E.C. Visual detection of dengue-1 RNA using gold nanoparticle-based lateral flow biosensor. Diagnostics (Basel, Switzerland) 2019:9. doi: 10.3390/diagnostics9030074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang Y., Li H., Wang Y., Xu H., Xu J., Ye C. Antarctic thermolabile uracil-DNA-glycosylase-supplemented multiple cross displacement amplification using a label-based nanoparticle lateral flow biosensor for the simultaneous detection of nucleic acid sequences and elimination of carryover contamination. Nano Res. 2018;11:2632–2647. [Google Scholar]
- 67.Xie S., Ye J., Yuan Y., Chai Y., Yuan R. A multifunctional hemin@ metal–organic framework and its application to construct an electrochemical aptasensor for thrombin detection. Nanoscale. 2015;7:18232–18238. doi: 10.1039/c5nr04532k. [DOI] [PubMed] [Google Scholar]
- 68.Hu J., Jiang Y.Z., Wu L.L., Wu Z., Bi Y., Wong G., Qiu X., Chen J., Pang D.W., Zhang Z.L. Dual-signal readout nanospheres for rapid point-of-care detection of Ebola virus glycoprotein. Anal. Chem. 2017;89:13105–13111. doi: 10.1021/acs.analchem.7b02222. [DOI] [PubMed] [Google Scholar]
- 69.Couturier C., Wada A., Louis K., Mistretta M., Beitz B., Povogui M., Ripaux M., Mignon C., Werle B., Lugari A., Pannetier D., Godard S., Bocquin A., Mely S., Béavogui I., Hébélamou J., Leuenberger D., Leissner P., Yamamoto T., Lécine P., Védrine C., Chaix J. Characterization and analytical validation of a new antigenic rapid diagnostic test for Ebola virus disease detection. PLoS Neglected Trop. Dis. 2020;14:e0007965. doi: 10.1371/journal.pntd.0007965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.DeMers H.L., He S., Pandit S.G., Hannah E.E., Zhang Z., Yan F., Green H.R., Reyes D.F., Hau D., McLarty M.E., Altamura L., Taylor-Howell C., Gates-Hollingsworth M.A., Qiu X., AuCoin D.P. Development of an antigen detection assay for early point-of-care diagnosis of Zaire ebolavirus. PLoS Neglected Trop. Dis. 2020;14:e0008817. doi: 10.1371/journal.pntd.0008817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zuo J.Y., Jiao Y.J., Zhu J., Ding S.N. Rapid detection of severe fever with thrombocytopenia syndrome virus via colloidal gold immunochromatography assay. ACS Omega. 2018;3:15399–15406. doi: 10.1021/acsomega.8b02366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu D., Wu T., Liu Q., Yang Z. The SARS-CoV-2 outbreak: what we know. Int. J. Infect. Dis. 2020;94:44–48. doi: 10.1016/j.ijid.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oroojalian F., Haghbin A., Baradaran B., Hemat N., Shahbazi M.-A., Baghi H.B., Mokhtarzadeh A., Hamblin M.R. Novel insights into the treatment of SARS-CoV-2 infection: an overview of current clinical trials. Int. J. Biol. Macromol. 2020;165(part A):18–43. doi: 10.1016/j.ijbiomac.2020.09.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Orooji Y., Sohrabi H., Hemmat N., Oroojalian F., Baradaran B., Mokhtarzadeh A., Mohaghegh M., Karimi-Maleh H. An overview on SARS-CoV-2 (COVID-19) and other human coronaviruses and their detection capability via amplification assay, chemical sensing, biosensing, immunosensing, and clinical assays. Nano-Micro Lett. 2021;13:1–30. doi: 10.1007/s40820-020-00533-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yan Y., Chang L., Wang L. Laboratory testing of SARS-CoV, MERS-CoV, and SARS-CoV-2 (2019-nCoV): current status, challenges, and countermeasures. Rev. Med. Virol. 2020;30:e2106. doi: 10.1002/rmv.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hoste A.C.R., Venteo A., Fresco-Taboada A., Tapia I., Monedero A., López L., Jebbink M.F., Pérez-Ramírez E., Jimenez-Clavero M.A., Almonacid M., Muñoz P., Guinea J., Vela C., van der Hoek L., Rueda P., Sastre P. Two serological approaches for detection of antibodies to SARS-CoV-2 in different scenarios: a screening tool and a point-of-care test. Diagn. Microbiol. Infect. Dis. 2020;98:115167. doi: 10.1016/j.diagmicrobio.2020.115167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zou M., Su F., Zhang R., Jiang X., Xiao H., Yan X., Yang C., Fan X., Wu G. Rapid point-of-care testing for SARS-CoV-2 virus nucleic acid detection by an isothermal and nonenzymatic Signal amplification system coupled with a lateral flow immunoassay strip. Sensor. Actuator. B Chem. 2021;342:129899. doi: 10.1016/j.snb.2021.129899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Grant B.D., Anderson C.E., Williford J.R., Alonzo L.F., Glukhova V.A., Boyle D.S., Weigl B.H., Nichols K.P. SARS-CoV-2 coronavirus nucleocapsid antigen-detecting half-strip lateral flow assay toward the development of point of care tests using commercially available reagents. Anal. Chem. 2020;92:11305–11309. doi: 10.1021/acs.analchem.0c01975. [DOI] [PubMed] [Google Scholar]
- 79.Chen Z., Zhang Z., Zhai X., Li Y., Lin L., Zhao H., Bian L., Li P., Yu L., Wu Y., Lin G. Rapid and sensitive detection of anti-SARS-CoV-2 IgG, using lanthanide-doped nanoparticles-based lateral flow immunoassay. Anal. Chem. 2020;92:7226–7231. doi: 10.1021/acs.analchem.0c00784. [DOI] [PubMed] [Google Scholar]
- 80.Daoud Z., McLeod J., Stockman D.L. Higher sensitivity provided by the combination of two lateral flow immunoassay tests for the detection of COVID-19 immunoglobulins. Front. Cell. Infect. Microbiol. 2020;10 doi: 10.3389/fcimb.2020.00479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Azmi I., Faizan M.I., Kumar R., Raj Yadav S., Chaudhary N., Kumar Singh D., Butola R., Ganotra A., Datt Joshi G., Deep Jhingan G., Iqbal J., Joshi M.C., Ahmad T. A saliva-based RNA extraction-free workflow integrated with Cas13a for SARS-CoV-2 detection. Front. Cell Infect. Microbiol. 2021;11:632646. doi: 10.3389/fcimb.2021.632646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Villarreal A., Rangel G., Zhang X., Wong D., Britton G., Fernandez P.L., Pérez A., Oviedo D., Restrepo C., Carreirra M.B., Sambrano D., Eskildsen G.A., De La Guardia C., Flores-Cuadra J., Carrera J.P., Zaldivar Y., Franco D., López-Vergès S., Zhang D., Fan F., Wang B., Sáez-Llorens X., DeAntonio R., Torres-Atencio I., Blanco I., Subía F.D., Mudarra L., Benzadon A., Valverde W., López L., Hurtado N., Rivas N., Jurado J., Carvallo A., Rodriguez J., Perez Y., Morris J., Luque O., Cortez D., Ortega-Barria E., Kosagisharaf R., Lleonart R., Li C., Goodridge A. Performance of a point of care test for detecting IgM and IgG antibodies against SARS-CoV-2 and seroprevalence in blood donors and health care workers in Panama. Front. Med. 2021;8:616106. doi: 10.3389/fmed.2021.616106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Higgins R.L., Rawlings S.A., Case J., Lee F.Y., Chan C.W., Barrick B., Burger Z.C., Yeo K.J., Marrinucci D. Longitudinal SARS-CoV-2 antibody study using the Easy Check COVID-19 IgM/IgG™ lateral flow assay. PLoS One. 2021;16:e0247797. doi: 10.1371/journal.pone.0247797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Broughton J.P., Deng X., Yu G., Fasching C.L., Singh J., Streithorst J., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.Y., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. Rapid detection of 2019 novel coronavirus SARS-CoV-2 using a CRISPR-based DETECTR lateral flow assay. medRxiv : Preprint Serv. Health Sci. 2020 doi: 10.1101/2020.03.06.20032334. [DOI] [Google Scholar]
- 85.Peng T., Liu X., Adams L.G., Agarwal G., Akey B., Cirillo J., Deckert V., Delfan S., Fry E., Han Z., Hemmer P., Kattawar G., Kim M., Lee M.C., Lu C., Mogford J., Nessler R., Neuman B., Nie X., Pan J., Pryor J., Rajil N., Shih Y., Sokolov A., Svidzinsky A., Wang D., Yi Z., Zheltikov A., Scully M. Enhancing sensitivity of lateral flow assay with application to SARS-CoV-2. Appl. Phys. Lett. 2020;117:120601. doi: 10.1063/5.0021842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Van Elslande J., Houben E., Depypere M., Brackenier A., Desmet S., André E., Van Ranst M., Lagrou K., Vermeersch P. Diagnostic performance of Seven rapid IgG/IgM antibody tests and the Euroimmun IgA/IgG ELISA in COVID-19 patients. Clin. Microbiol. Inf. Offic. Publ. Eur. Soc. Clin. Microbiol. Inf. Dis. 2020;26:1082–1087. doi: 10.1016/j.cmi.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xiong E., Jiang L., Tian T., Hu M., Yue H., Huang M., Lin W., Jiang Y., Zhu D., Zhou X. Simultaneous dual-gene diagnosis of SARS-CoV-2 based on CRISPR/Cas9-Mediated lateral flow assay. Angew. Chem. (Int. Ed. in Engl.) 2021;60:5307–5315. doi: 10.1002/anie.202014506. [DOI] [PubMed] [Google Scholar]
- 88.Kim K., Kashefi-Kheyrabadi L., Joung Y., Kim K., Dang H., Chavan S.G., Lee M.-H., Choo J. Recent advances in sensitive surface-enhanced Raman scattering-based lateral flow assay platforms for point-of-care diagnostics of infectious diseases. Sensor. Actuator. B Chem. 2021;329:129214. doi: 10.1016/j.snb.2020.129214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Moghadami M. A narrative review of influenza: a seasonal and pandemic disease. Iran. J. Med. Sci. 2017;42:2–13. [PMC free article] [PubMed] [Google Scholar]
- 90.Dziąbowska K., Czaczyk E., Nidzworski D. Detection methods of human and animal influenza virus-current trends. Biosensors (Basel) 2018;8:94. doi: 10.3390/bios8040094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hassanpour S., Baradaran B., Hejazi M., Hasanzadeh M., Mokhtarzadeh A., de la Guardia M. Recent trends in rapid detection of influenza infections by bio and nanobiosensor. Trac. Trends Anal. Chem. 2018;98:201–215. [Google Scholar]
- 92.Le T.T., Chang P., Benton D.J., McCauley J.W., Iqbal M., Cass A.E.G. Dual recognition element lateral flow assay toward multiplex strain specific influenza virus detection. Anal. Chem. 2017;89:6781–6786. doi: 10.1021/acs.analchem.7b01149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kim S.H., Lee J., Lee B.H., Song C.-S., Gu M.B. Specific detection of avian influenza H5N2 whole virus particles on lateral flow strips using a pair of sandwich-type aptamers. Biosens. Bioelectron. 2019;134:123–129. doi: 10.1016/j.bios.2019.03.061. [DOI] [PubMed] [Google Scholar]
- 94.Wang C., Wang C., Wang X., Wang K., Zhu Y., Rong Z., Wang W., Xiao R., Wang S. Magnetic SERS strip for sensitive and simultaneous detection of respiratory viruses. ACS Appl. Mater. Interfaces. 2019;11:19495–19505. doi: 10.1021/acsami.9b03920. [DOI] [PubMed] [Google Scholar]
- 95.Park B.J., Park M.S., Lee J.M., Song Y.J. Specific detection of influenza A and B viruses by CRISPR-cas12a-based assay. Biosensors (Basel) 2021;11:88. doi: 10.3390/bios11030088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ma S., Li X., Peng B., Wu W., Wang X., Liu H., Yuan L., Fang S., Lu J. Rapid detection of avian influenza A virus (H7N9) by lateral flow dipstick recombinase polymerase amplification. Biol. Pharm. Bull. 2018;41:1804–1808. doi: 10.1248/bpb.b18-00468. [DOI] [PubMed] [Google Scholar]
- 97.Bai Z., Wei H., Yang X., Zhu Y., Peng Y., Yang J., Wang C., Rong Z., Wang S. Rapid enrichment and ultrasensitive detection of influenza A virus in human specimen using magnetic quantum dot nanobeads based test strips. Sensor. Actuator. B Chem. 2020;325:128780. doi: 10.1016/j.snb.2020.128780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang S.-F., Liu L.-N., Tang R.-H., Liu Z., He X.-C., Qu Z.-G., Li F. Sensitivity enhancement of lateral flow assay by embedding cotton threads in paper. Cellulose. 2019;26:8087–8099. [Google Scholar]
- 99.Sun N., Wang Y., Yao X., Chen F., Gao D., Wang W., Li X. Visual signal generation for the detection of influenza viruses by duplex recombinase polymerase amplification with lateral flow dipsticks. Anal. Bioanal. Chem. 2019;411:3591–3602. doi: 10.1007/s00216-019-01840-z. [DOI] [PubMed] [Google Scholar]
- 100.Zhang D., Huang L., Liu B., Ge Q., Dong J., Zhao X. Rapid and ultrasensitive quantification of multiplex respiratory tract infection pathogen via lateral flow microarray based on SERS nanotags. Theranostics. 2019;9:4849–4859. doi: 10.7150/thno.35824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wiriyachaiporn N., Sirikett H., Maneeprakorn W., Dharakul T. Carbon nanotag based visual detection of influenza A virus by a lateral flow immunoassay. Microchim. Acta. 2017;184:1827–1835. [Google Scholar]
- 102.Douek D.C., Roederer M., Koup R.A. Emerging concepts in the immunopathogenesis of AIDS. Annu. Rev. Med. 2009;60:471–484. doi: 10.1146/annurev.med.60.041807.123549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Antinori A., Coenen T., Costagiola D., Dedes N., Ellefson M., Gatell J., Girardi E., Johnson M., Kirk O., Lundgren J., Mocroft A., D'Arminio Monforte A., Phillips A., Raben D., Rockstroh J., Sabin C., Sönnerborg A., De Wolf F., f.t.E.L.P.C.w. group Late presentation of HIV infection: a consensus definition. HIV Med. 2011;12:61–64. doi: 10.1111/j.1468-1293.2010.00857.x. [DOI] [PubMed] [Google Scholar]
- 104.Boyle D.S., Lehman D.A., Lillis L., Peterson D., Singhal M., Armes N., Parker M., Piepenburg O., Overbaugh J. Rapid detection of HIV-1 proviral DNA for early infant diagnosis using recombinase polymerase amplification. mBio. 2013;4 doi: 10.1128/mBio.00135-13. e00135-00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tang R., Yang H., Choi J.R., Gong Y., Hu J., Feng S., Pingguan-Murphy B., Mei Q., Xu F. Improved sensitivity of lateral flow assay using paper-based sample concentration technique. Talanta. 2016;152:269–276. doi: 10.1016/j.talanta.2016.02.017. [DOI] [PubMed] [Google Scholar]
- 106.Cavalera S., Di Nardo F., Forte L., Marinoni F., Chiarello M., Baggiani C., Anfossi L. Switching from multiplex to multimodal colorimetric lateral flow immunosensor. Sensors. 2020;20:6609. doi: 10.3390/s20226609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Granade T.C., Workman S., Wells S.K., Holder A.N., Owen S.M., Pau C.P. Rapid detection and differentiation of antibodies to HIV-1 and HIV-2 using multivalent antigens and magnetic immunochromatography testing. Clin. Vacc. Immunol. CVI. 2010;17:1034–1039. doi: 10.1128/CVI.00029-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Granade T.C., Nguyen S., Kuehl D.S., Parekh B.S. Development of a novel rapid HIV test for simultaneous detection of recent or long-term HIV type 1 infection using a single testing device. AIDS Res. Hum. Retrovir. 2013;29:61–67. doi: 10.1089/aid.2012.0121. [DOI] [PubMed] [Google Scholar]
- 109.Rohrman B.A., Leautaud V., Molyneux E., Richards-Kortum R.R. A lateral flow assay for quantitative detection of amplified HIV-1 RNA. PLoS One. 2012;7:e45611. doi: 10.1371/journal.pone.0045611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Martiskainen I., Juntunen E., Salminen T., Vuorenpää K., Bayoumy S., Vuorinen T., Khanna N., Pettersson K., Batra G., Talha S.M. Double-antigen lateral flow immunoassay for the detection of anti-HIV-1 and -2 antibodies using upconverting nanoparticle reporters. Sensors (Basel, Switzerland) 2021:21. doi: 10.3390/s21020330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bristow C.C., Severe L., Pape J.W., Javanbakht M., Lee S.J., Comulada W.S., Klausner J.D. Dual rapid lateral flow immunoassay fingerstick wholeblood testing for syphilis and HIV infections is acceptable and accurate, Port-au-Prince, Haiti. BMC Infect. Dis. 2016;16:302. doi: 10.1186/s12879-016-1574-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhan L., Granade T., Liu Y., Wei X., Youngpairoj A., Sullivan V., Johnson J., Bischof J. Development and optimization of thermal contrast amplification lateral flow immunoassays for ultrasensitive HIV p24 protein detection. Microsyst. Nanoeng. 2020;6:54. doi: 10.1038/s41378-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Qi W., Ling H.M., Peng L.L., Jing M., Guang Z.X., Xiang Z.Z., Xi C.Y. A new method for ultra-sensitive P24 antigen assay based on near-infrared fluorescent microsphere immunochromatography. Biomed. Environ. Sci. 2020;33:174. doi: 10.3967/bes2020.024. [DOI] [PubMed] [Google Scholar]
- 114.Raimondo G., Pollicino T., Squadrito G. Clinical virology of hepatitis B virus infection. J. Hepatol. 2003;39:26–30. doi: 10.1016/s0168-8278(03)00135-1. [DOI] [PubMed] [Google Scholar]
- 115.Hassanpour S., Baradaran B., de la Guardia M., Baghbanzadeh A., Mosafer J., Hejazi M., Mokhtarzadeh A., Hasanzadeh M. Diagnosis of hepatitis via nanomaterial-based electrochemical, optical or piezoelectrical biosensors: a review on recent advancements. Microchim. Acta. 2018;185:1–24. doi: 10.1007/s00604-018-3088-8. [DOI] [PubMed] [Google Scholar]
- 116.Peters M.G. Hepatitis B virus infection: what is current and new. Top Antivir. Med. 2019;26:112–116. [PMC free article] [PubMed] [Google Scholar]
- 117.Liang T.J. Hepatitis B: the virus and disease. Hepatology. 2009;49:S13–S21. doi: 10.1002/hep.22881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Srisomwat C., Yakoh A., Chuaypen N., Tangkijvanich P., Vilaivan T., Chailapakul O. Amplification-free DNA sensor for the one-step detection of the hepatitis B virus using an automated paper-based lateral flow electrochemical device. Anal. Chem. 2021;93:2879–2887. doi: 10.1021/acs.analchem.0c04283. [DOI] [PubMed] [Google Scholar]
- 119.Choi J.R., Liu Z., Hu J., Tang R., Gong Y., Feng S., Chai F., Yang H., Qu Z., Pingguan-Murphy B., Xu F. A PDMS-paper hybrid lateral flow assay for highly sensitive point-of-care nucleic acid testing. Anal. Chem. 2016;88 doi: 10.1021/acs.analchem.6b00195. [DOI] [PubMed] [Google Scholar]
- 120.Tang R., Yang H., Gong Y., Liu Z., Li X., Wen T., Qu Z., Zhang S., Mei Q., Xu F. Improved analytical sensitivity of lateral flow assay using sponge for HBV nucleic acid detection. Sci. Rep. 2017;7:1360. doi: 10.1038/s41598-017-01558-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kim D.S., Kim Y.T., Hong S.B., Kim J., Huh N.S., Lee M.-K., Lee S.J., Kim B.I., Kim I.S., Huh Y.S., Choi B.G. Development of lateral flow assay based on size-controlled gold nanoparticles for detection of hepatitis B surface antigen. Sensors (Basel, Switzerland) 2016;16:2154. doi: 10.3390/s16122154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gong Y., Zheng Y., Jin B., You M., Wang J., Li X., Lin M., Xu F., Li F. A portable and universal upconversion nanoparticle-based lateral flow assay platform for point-of-care testing. Talanta. 2019;201:126–133. doi: 10.1016/j.talanta.2019.03.105. [DOI] [PubMed] [Google Scholar]
- 123.Li L., Zhou L., Yu Y., Zhu Z., Lin C., Lu C., Yang R. Development of up-converting phosphor technology-based lateral-flow assay for rapidly quantitative detection of hepatitis B surface antibody. Diagn. Microbiol. Infect. Dis. 2009;63:165–172. doi: 10.1016/j.diagmicrobio.2008.10.020. [DOI] [PubMed] [Google Scholar]
- 124.Si J., Li J., Zhang L., Zhang W., Yao J., Li T., Wang W., Zhu W., Allain J.P., Fu Y., Li C. A signal amplification system on a lateral flow immunoassay detecting for hepatitis e-antigen in human blood samples. J. Med. Virol. 2019;91:1301–1306. doi: 10.1002/jmv.25452. [DOI] [PubMed] [Google Scholar]
- 125.Yi T.-t., Zhang H.-y., Liang H., Gong G.-z., Cai Y. Betaine-assisted recombinase polymerase assay for rapid hepatitis B virus detection. Biotechnol. Appl. Biochem. 2021;68:469–475. doi: 10.1002/bab.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lin L., Guo J., Liu H., Jiang X. Rapid detection of hepatitis B virus in blood samples using a combination of polymerase spiral reaction with nanoparticles lateral-flow biosensor. Front. Molec. Biosci. 2021;7 doi: 10.3389/fmolb.2020.578892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Khetarpal N., Khanna I. Dengue fever: causes, complications, and vaccine strategies. J. Immunol. Res. 2016;2016:6803098. doi: 10.1155/2016/6803098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Eivazzadeh-Keihan R., Pashazadeh-Panahi P., Mahmoudi T., Chenab K.K., Baradaran B., Hashemzaei M., Radinekiyan F., Mokhtarzadeh A., Maleki A. Dengue virus: a review on advances in detection and trends–from conventional methods to novel biosensors. Microchim. Acta. 2019;186:1–24. doi: 10.1007/s00604-019-3420-y. [DOI] [PubMed] [Google Scholar]
- 129.Gurugama P., Garg P., Perera J., Wijewickrama A., Seneviratne S.L. Dengue viral infections. Indian J. Dermatol. 2010;55:68–78. doi: 10.4103/0019-5154.60357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bos S., Gadea G., Despres P. Dengue: a growing threat requiring vaccine development for disease prevention. Pathog. Glob. Health. 2018;112:294–305. doi: 10.1080/20477724.2018.1514136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Jeon J., Lee S.H., Joung Y., Kim K., Choi N., Choo J. Improvement of reproducibility and thermal stability of surface-enhanced Raman scattering-based lateral flow assay strips using silica-encapsulated gold nanoparticles. Sensor. Actuator. B Chem. 2020;321:128521. [Google Scholar]
- 132.Sánchez-Purrà M., Carré-Camps M., de Puig H., Bosch I., Gehrke L., Hamad-Schifferli K. Surface-enhanced Raman spectroscopy-based sandwich immunoassays for multiplexed detection of zika and dengue viral biomarkers. ACS Infect. Dis. 2017;3:767–776. doi: 10.1021/acsinfecdis.7b00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Choi J.R., Yong K.W., Tang R., Gong Y., Wen T., Yang H., Li A., Chia Y.C., Pingguan-Murphy B., Xu F. Lateral flow assay based on paper-hydrogel hybrid material for sensitive point-of-care detection of dengue virus. Adv. Healthc. Mater. 2017;6 doi: 10.1002/adhm.201600920. [DOI] [PubMed] [Google Scholar]
- 134.Axelrod T., Eltzov E., Marks R.S. Capture-layer lateral flow immunoassay: a new platform validated in the detection and quantification of dengue NS1. ACS Omega. 2020;5:10433–10440. doi: 10.1021/acsomega.0c00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kumar S., Bhushan P., Krishna V., Bhattacharya S. Tapered lateral flow immunoassay based point-of-care diagnostic device for ultrasensitive colorimetric detection of dengue NS1. Biomicrofluidics. 2018;12 doi: 10.1063/1.5035113. 034104-034104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sinawang P.D., Rai V., Ionescu R.E., Marks R.S. Electrochemical lateral flow immunosensor for detection and quantification of dengue NS1 protein. Biosens. Bioelectron. 2016;77:400–408. doi: 10.1016/j.bios.2015.09.048. [DOI] [PubMed] [Google Scholar]
- 137.Lee S., Mehta S., Erickson D. Two-color lateral flow assay for multiplex detection of causative agents behind acute febrile Illnesses. Anal. Chem. 2016;88:8359–8363. doi: 10.1021/acs.analchem.6b01828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Xiong Y., Luo Y., Li H., Wu W., Ruan X., Mu X. Rapid visual detection of dengue virus by combining reverse transcription recombinase-aided amplification with lateral-flow dipstick assay. Int. J. Infect. Dis. 2020;95:406–412. doi: 10.1016/j.ijid.2020.03.075. [DOI] [PubMed] [Google Scholar]
- 139.Moens U. Human polyomaviruses and papillomaviruses. Int. J. Mol. Sci. 2018;19:2360. doi: 10.3390/ijms19082360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chan C.K., Aimagambetova G., Ukybassova T., Kongrtay K., Azizan A. Human papillomavirus infection and cervical cancer: epidemiology, screening, and vaccination-review of current perspectives. J. Oncol. 2019;2019 doi: 10.1155/2019/3257939. 3257939-3257939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Braaten K.P., Laufer M.R. Human papillomavirus (HPV), HPV-related disease, and the HPV vaccine. Rev. Obstet. Gynecol. 2008;1:2–10. [PMC free article] [PubMed] [Google Scholar]
- 142.Grant B.D., Smith C.A., Castle P.E., Scheurer M.E., Richards-Kortum R. A paper-based immunoassay to determine HPV vaccination status at the point-of-care. Vaccine. 2016;34:5656–5663. doi: 10.1016/j.vaccine.2016.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Xu Y., Liu Y., Wu Y., Xia X., Liao Y., Li Q. Fluorescent probe-based lateral flow assay for multiplex nucleic acid detection. Anal. Chem. 2014;86:5611–5614. doi: 10.1021/ac5010458. [DOI] [PubMed] [Google Scholar]
- 144.Yang Z., Yi C., Lv S., Sheng Y., Wen W., Zhang X., Wang S. Development of a lateral flow strip biosensor based on copper oxide nanoparticles for rapid and sensitive detection of HPV16 DNA. Sensor. Actuator. B Chem. 2019;285:326–332. [Google Scholar]
- 145.Kazmi S.S., Ali W., Bibi N., Nouroz F. A review on Zika virus outbreak, epidemiology, transmission and infection dynamics. J. Biol. Res.-Thessal. 2020;27:5. doi: 10.1186/s40709-020-00115-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pierson T.C., Diamond M.S. The emergence of Zika virus and its new clinical syndromes. Nature. 2018;560:573–581. doi: 10.1038/s41586-018-0446-y. [DOI] [PubMed] [Google Scholar]
- 147.Noorbakhsh F., Abdolmohammadi K., Fatahi Y., Dalili H., Rasoolinejad M., Rezaei F., Salehi-Vaziri M., Shafiei-Jandaghi N.Z., Gooshki E.S., Zaim M., Nicknam M.H. Zika virus infection, basic and clinical aspects: a review article, Iran. J. Publ. Health. 2019;48:20–31. [PMC free article] [PubMed] [Google Scholar]
- 148.Lee D., Shin Y., Chung S., Hwang K.S., Yoon D.S., Lee J.H. Simple and highly sensitive molecular diagnosis of zika virus by lateral flow assays. Anal. Chem. 2016;88:12272–12278. doi: 10.1021/acs.analchem.6b03460. [DOI] [PubMed] [Google Scholar]
- 149.Rong Z., Wang Q., Sun N., Jia X., Wang K., Xiao R., Wang S. Smartphone-based fluorescent lateral flow immunoassay platform for highly sensitive point-of-care detection of Zika virus nonstructural protein 1. Anal. Chim. Acta. 2019;1055:140–147. doi: 10.1016/j.aca.2018.12.043. [DOI] [PubMed] [Google Scholar]
- 150.Xu L.-D., Du F.-L., Zhu J. Luminous silica colloids with carbon dot incorporation for sensitive immunochromatographic assay of Zika virus. Analyst. 2020;146 doi: 10.1039/d0an02017f. [DOI] [PubMed] [Google Scholar]
- 151.Yew C.-H.T., Azari P., Choi J.R., Li F., Pingguan-Murphy B. Electrospin-coating of nitrocellulose membrane enhances sensitivity in nucleic acid-based lateral flow assay. Anal. Chim. Acta. 2018;1009:81–88. doi: 10.1016/j.aca.2018.01.016. [DOI] [PubMed] [Google Scholar]
- 152.Yew C.H.T., Azari P., Choi J.R., Muhamad F., Pingguan-Murphy B. Electrospun polycaprolactone nanofibers as a reaction membrane for lateral flow assay. Polymers. 2018;10 doi: 10.3390/polym10121387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hasan S., Ahmad S.A., Masood R., Saeed S. Ebola virus: a global public health menace: a narrative review. J. Fam. Med. Prim. Care. 2019;8:2189–2201. doi: 10.4103/jfmpc.jfmpc_297_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kourtis A.P., Appelgren K., Chevalier M.S., McElroy A. Ebola virus disease: focus on children. Pediatr. Infect. Dis. J. 2015;34:893–897. doi: 10.1097/INF.0000000000000707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Brangel P., Sobarzo A., Parolo C., Miller B.S., Howes P.D., Gelkop S., Lutwama J.J., Dye J.M., McKendry R.A., Lobel L., Stevens M.M. A serological point-of-care test for the detection of IgG antibodies against Ebola virus in human survivors. ACS Nano. 2018;12:63–73. doi: 10.1021/acsnano.7b07021. [DOI] [PubMed] [Google Scholar]
- 156.Feuerstein G.Z., Mansfield M.A., Lelkes P.I., Alesci S., Marcinkiewicz C., Butlin N., Sternberg M. The use of near-infrared light-emitting fluorescent nanodiamond particles to detect Ebola virus glycoprotein: technology development and proof of principle. Int. J. Nanomed. 2020;15:7583–7599. doi: 10.2147/IJN.S261952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Phan J.C., Pettitt J., George J.S., Fakoli L.S., 3rd, Taweh F.M., Bateman S.L., Bennett R.S., Norris S.L., Spinnler D.A., Pimentel G., Sahr P.K., Bolay F.K., Schoepp R.J. Lateral flow immunoassays for Ebola virus disease detection in Liberia. J. Infect. Dis. 2016;214:S222–S228. doi: 10.1093/infdis/jiw251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Cohen-Bucay A., Ramirez-Andrade S.E., Gordon C.E., Francis J.M., Chitalia V.C. Advances in BK virus complications in organ transplantation and beyond. Kidney Med. 2020;2:771–786. doi: 10.1016/j.xkme.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Dalianis T., Eriksson B.-M., Felldin M., Friman V., Hammarin A.-L., Herthelius M., Ljungman P., Mölne J., Wennberg L., Swartling L. Management of BK-virus infection – Swedish recommendations. Infect. Dis. 2019;51:479–484. doi: 10.1080/23744235.2019.1595130. [DOI] [PubMed] [Google Scholar]
- 160.Bohl D.L., Brennan D.C. BK virus nephropathy and kidney transplantation. Clin. J. Am. Soc. Nephrol. 2007;2:S36–S46. doi: 10.2215/CJN.00920207. [DOI] [PubMed] [Google Scholar]
- 161.Kaminski M.M., Alcantar M.A., Lape I.T., Greensmith R., Huske A.C., Valeri J.A., Marty F.M., Klämbt V., Azzi J., Akalin E., Riella L.V., Collins J.J. A CRISPR-based assay for the detection of opportunistic infections post-transplantation and for the monitoring of transplant rejection. Nat. Biomed. Eng. 2020;4:601–609. doi: 10.1038/s41551-020-0546-5. [DOI] [PubMed] [Google Scholar]
- 162.Huang Y.H., Yu K.Y., Huang S.P., Chuang H.W., Lin W.Z., Cherng J.H., Hung Y.W., Yeh M.K., Hong P.D., Liu C.C. Development of a nucleic acid lateral flow immunoassay for the detection of human polyomavirus BK. Diagnostics (Basel, Switzerland) 2020:10. doi: 10.3390/diagnostics10060403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Robilotti E., Deresinski S., Pinsky B.A. Norovirus. Clin. Microbiol. Rev. 2015;28:134–164. doi: 10.1128/CMR.00075-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.de Graaf M., van Beek J., Koopmans M.P.G. Human norovirus transmission and evolution in a changing world. Nat. Rev. Microbiol. 2016;14:421–433. doi: 10.1038/nrmicro.2016.48. [DOI] [PubMed] [Google Scholar]
- 165.Hagström A.E.V., Garvey G., Paterson A.S., Dhamane S., Adhikari M., Estes M.K., Strych U., Kourentzi K., Atmar R.L., Willson R.C. Sensitive detection of norovirus using phage nanoparticle reporters in lateral-flow assay. PLoS One. 2015;10 doi: 10.1371/journal.pone.0126571. e0126571-e0126571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Doerflinger S.Y., Tabatabai J., Schnitzler P., Farah C., Rameil S., Sander P., Koromyslova A., Hansman G.S. Development of a nanobody-based lateral flow immunoassay for detection of human norovirus. mSphere. 2016;1 doi: 10.1128/mSphere.00219-16. e00219-00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Casel M.A., Park S.J., Choi Y.K. Severe fever with thrombocytopenia syndrome virus: emerging novel phlebovirus and their control strategy. Exp. Mol. Med. 2021;53:713–722. doi: 10.1038/s12276-021-00610-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hu C., Guo C., Yang Z., Wang L., Hu J., Qin S., Cui N., Peng W., Liu K., Liu W., Cao W. The severe fever with thrombocytopenia syndrome bunyavirus (SFTSV) antibody in a highly endemic region from 2011 to 2013: a comparative serological study. Am. J. Trop. Med. Hyg. 2015;92:479–481. doi: 10.4269/ajtmh.14-0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Park E.-s., Fujita O., Kimura M., Hotta A., Imaoka K., Shimojima M., Saijo M., Maeda K., Morikawa S. Diagnostic system for the detection of severe fever with thrombocytopenia syndrome virus RNA from suspected infected animals. PLoS One. 2021;16:e0238671. doi: 10.1371/journal.pone.0238671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Xu L.D., Zhang Q., Ding S.N., Xu J.J., Chen H.Y. Ultrasensitive detection of severe fever with thrombocytopenia syndrome virus based on immunofluorescent carbon dots/SiO(2) nanosphere-based lateral flow assay. ACS Omega. 2019;4:21431–21438. doi: 10.1021/acsomega.9b03130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zhu M., Obahiagbon U., Anderson K.S., Christen J.B. 2017 IEEE Healthcare Innovations and Point of Care Technologies (HI-POCT) 2017. Highly sensitive fluorescence-based lateral flow platform for point-of-care detection of biomarkers in plasma; pp. 249–252. [Google Scholar]
- 172.Goux H.J., Raja B., Kourentzi K., Trabuco J.R.C., Vu B.V., Paterson A.S., Kirkpatrick A., Townsend B., Lee M., Truong V.T.T., Pedroza C., Willson R.C. Evaluation of a nanophosphor lateral-flow assay for self-testing for herpes simplex virus type 2 seropositivity. PLoS One. 2019;14:e0225365. doi: 10.1371/journal.pone.0225365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Townsend M.B., MacNeil A., Reynolds M.G., Hughes C.M., Olson V.A., Damon I.K., Karem K.L. Evaluation of the Tetracore Orthopox BioThreat® antigen detection assay using laboratory grown orthopoxviruses and rash illness clinical specimens. J. Virol. Methods. 2013;187:37–42. doi: 10.1016/j.jviromet.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Rebollo B., Pérez T., Camuñas A., Pérez-Ramírez E., Llorente F., Sánchez-Seco M.P., Jiménez-Clavero M., Venteo Á. A monoclonal antibody to DIII E protein allowing the differentiation of West Nile virus from other flaviviruses by a lateral flow assay. J. Virol. Methods. 2018;260:41–44. doi: 10.1016/j.jviromet.2018.06.016. [DOI] [PubMed] [Google Scholar]
- 175.Andryukov B.G. Six decades of lateral flow immunoassay: from determining metabolic markers to diagnosing COVID-19. AIMS Microbiol. 2020;6:280–304. doi: 10.3934/microbiol.2020018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Tang R., Alam N., Li M., Xie M., Ni Y. Dissolvable sugar barriers to enhance the sensitivity of nitrocellulose membrane lateral flow assay for COVID-19 nucleic acid. Carbohydr. Polym. 2021;268:118259. doi: 10.1016/j.carbpol.2021.118259. [DOI] [PubMed] [Google Scholar]
- 177.Alam N., Tong L., He Z., Tang R., Ahsan L., Ni Y. Improving the sensitivity of cellulose fiber-based lateral flow assay by incorporating a water-dissolvable polyvinyl alcohol dam. Cellulose. 2021;28:8641–8651. doi: 10.1007/s10570-021-04083-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Napione L. Integrated nanomaterials and nanotechnologies in lateral flow tests for personalized medicine applications. Nanomaterials. 2021;11:2362. doi: 10.3390/nano11092362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.de Puig H., Bosch I., Gehrke L., Hamad-Schifferli K. Challenges of the nano-bio interface in lateral flow and dipstick immunoassays. Trends Biotechnol. 2017;35:1169–1180. doi: 10.1016/j.tibtech.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]