Abstract
Identification of a novel mouse nuclear protein termed activator of basal transcription 1 (mABT1) that associates with the TATA-binding protein (TBP) and enhances basal transcription activity of class II promoters is described. We also identify mABT1 homologous counterparts in Caenorhabditis elegans and Saccharomyces cerevisiae and show the homologous yeast gene to be essential for growth. The mABT1 associated with TBP in HeLa nuclear extracts and with purified mouse TBP in vitro. In addition, ectopically expressed mABT1 was coimmunoprecipitated with endogenous TBP in transfected cells. More importantly, mABT1 significantly enhanced transcription from an adenovirus major late promoter in a reconstituted cell-free system. We furthermore demonstrate that mABT1 consistently enhanced transcription from a reporter gene with a minimal core promoter as well as from reporter genes with various enhancer elements in a cotransfection assay. Taken together, these results suggest that mABT1 is a novel TBP-binding protein which can function as a basal transcription activator.
Most genes in eukaryotes show a regulated pattern of expression during the course of development, in the cell cycle, or in response to changes in the cellular environment. The protein-coding genes transcribed by RNA polymerase II (Pol II) are predominantly regulated at the level of transcription (4, 6, 53), and this transcriptional control of RNA Pol II is governed by specific DNA elements and protein factors assembled on these elements. Two types of DNA elements exist: (i) common core promoter elements on which RNA Pol II and general transcription factors (GTFs) such as TFIIA, -B, -D, -E, -F, and -H assemble to form a preinitiation complex and (ii) gene-specific DNA elements that are recognized by regulatory factors (53, 56). According to this scheme, RNA Pol II and cognate GTFs can initiate a low level of intrinsic basal transcription from the core promoter. This basal transcription machinery is an ultimate target of various gene- and cell type-specific regulatory factors, which lend positive and negative signals to modulate transcriptional activity.
The TATA-binding protein (TBP) has been isolated and characterized as a TATA element-binding component of the general transcription factor TFIID (13, 20, 22, 23, 25, 32). TBP is associated with a variety of factors that play important roles in basal or gene-specific regulation of gene expression. For example, TBP interacts with TBP-associated factors (TAFIIs) and forms the TFIID complex, which was initially identified as an essential GTF. Mammalian or Drosophila TFIID can mediate basal and activated transcription in vitro, whereas TBP by itself can mediate only basal transcription, suggesting that mammalian or Drosophila TAFIIs are required for activated transcription (50, 67). While TAFIIs have been proposed to be coactivators that mediate activated transcription, recent studies have shown that TAFIIs have multiple functions including core promoter-selective basal transcription (41, 47, 59), histone acetyltransferase activity (46), and phosphorylation of TFIIF (8). TAFIIs consist of multiproteins ranging in size from 18 to 250 kDa (39, 40). Major TAFIIs have been cloned from yeast, fly, and mammalian cells, and most of the counterparts show remarkable evolutionary conservation. Eleven out of twelve yeast TAFIIs are essential for cell viability (39), indicating the importance of TAFIIs for transcription in eukaryotes.
TBP plays a key role together with TAFIIs in communicating transcriptional regulatory factors and in the basic transcription machinery (67). TBP binds to a variety of factors including c-Fos (45, 52), c-Myc (18, 40), and p53 (58, 64). More recently, other TBP-binding proteins, such as SAGA (3, 10, 55), Mot1 (1, 2, 69), NC2 (17, 26, 33, 42, 44), and NOTs (38), that control class II genes have been found. To account for the diversity of regulation mechanisms of class II genes, it is anticipated that many more factors may be involved in transcription regulation through the TBP.
Here we report cloning and characterization of a novel mouse nuclear protein named activator of basal transcription 1 (mABT1). mABT1 was isolated during the course of yeast two-hybrid screening using the Src homology 2 (SH2) domain of SHD, a previously identified SH2 domain-containing protein (48). The analysis carried out in this study showed that mABT1 associated directly with TBP and activated transcription from an adenovirus major late (AdML) promoter in a cell-free system. Also, expression of mABT1 in mammalian cells was observed to stimulate gene expression regardless of cis-regulatory elements, and we demonstrate that only the core promoter was required for activation. Furthermore, the Saccharomyces cerevisiae yeast counterpart of the mABT1 gene was shown to be essential for growth. These lines of evidence characterizes mABT1 as a novel TBP-binding protein which promotes activation of basal transcription.
MATERIALS AND METHODS
Molecular cloning of mABT1 cDNA.
pB42AD-mABT1, which contains the mouse ABT1 cDNA, was isolated from the Mouse Embryo MATCHMAKER LexA cDNA Library (Clontech) using the SH2 domain of SHD (48). Both strands of the mABT1 cDNA were sequenced with an ABI 377 DNA sequencer (Perkin-Elmer). Human ABT1 cDNA was cloned from the NT2 human teratocarcinoma cell line by rapid amplification of 5′ cDNA ends (5′-RACE) (GIBCO BRL) according to the supplier's instructions. The primer sequences 5′-TGC CTG GAC TAG GCA TTA TCC-3′ and 5′-TTG GAA ATA AAG GCC CTT TCT-3′, used for first-strand cDNA synthesis and PCR, respectively, were obtained from the expressed sequence tags database (dbEST). The PCR product was subcloned in a pT7Blue T-vector (Novagen) and sequenced. The Caenorhabditis elegans ABT1 cDNA was cloned by reverse transcription-PCR (RT-PCR) with primers 5′-TTT GAA TTC ATG GCG CCT ATT CCA AAA AAG-3′ and 5′-GAG AGG ATC CTT ATT TGA AGA TCA TAT TCA TCA ATT C-3′. The PCR product was subcloned in the pT7Blue T-vector and sequenced.
Northern blot analysis.
A mouse multiple-tissue Northern blot (Clontech) was prehybridized and hybridized in ExpressHyb hybridization solution (Clontech) at 65°C with a 32P-labeled mABT1 cDNA probe. The blot was washed twice for 1 h at room temperature in a solution containing 0.3 M NaCl, 0.03 M sodium citrate, and 0.1% sodium dodecyl sulfate (SDS), then washed for 1 h at 65°C in a solution containing 15 mM NaCl, 1.5 mM sodium citrate, 0.1% SDS, and thereafter subjected to autoradiography.
Western blot analysis.
Proteins were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene difluoride membranes (Bio-Rad) with a semidry transfer cell (Bio-Rad). Residual binding sites were blocked by overnight incubation at 4°C in phosphate-buffered saline (PBS; 136.9 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4, 1.5 mM KH2PO4) containing 0.05% Tween 20 and 5% nonfat milk. The blots were incubated for 4 to 16 h at 4°C with primary antibody. Antibody reactions were detected using anti-mouse or anti-rabbit antibody conjugated to horseradish peroxidase (Amersham) and visualized by enhanced Luminol reagent (NEN).
Cell culture and transfections.
COS7 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), penicillin, and streptomycin. The cells were plated approximately 16 h before transfection at a density of 2.0 × 105 cells in a well of 35-mm-diameter multiwell plate (Falcon). Plasmids (2 μg) were preincubated with 13 μl of Lipofectamine (GIBCO BRL) in 200 μl of serum-free DMEM at room temperature for 45 min. The cells were washed once with DMEM. The preincubated mixture was diluted with DMEM to a final volume of 1 ml and added to the cells. The cells were then incubated for 5 h at 37°C, and FBS or bovine serum albumin (BSA) was added to a final concentration of 10 or 1%, respectively. At 24 h after transfection, the cells were washed once with DMEM and then incubated for 24 h in DMEM–10% FBS or in DMEM–1% BSA. At 48 h after transfection, the cells were washed twice with cold PBS and lysed in 150 μl of lysis buffer (25 mM glycylglycine [pH 7.8], 15% glycerol, 8 mM MgSO4, 1 mM EDTA, 1% Triton X-100, 1 mM dithiothreitol [DTT]), followed by incubation for an additional 20 min at 4°C. The cell lysates were transferred to a 1.5-ml tube and centrifuged, and 10 μl of the supernatant was used for luciferase assay.
Plasmids.
pcDNA3-myc was constructed by inserting the annealed primers 5′-A GCT GCC ATG GAA CAA AAA CTC ATC TCA GAA GAG GAT CTG GGA TCC AAG CTT G-3′ and 5′-AA TTC AAG CTT GGA TCC CAG ATC CTC TTC TGA GAT GAG TTT TTG TTC CAT GGC-3′ into the HindIII and EcoRI sites of pcDNA3 (Invitrogen). pcDNA3-mABT1 and pcDNA3-myc-mABT1 were constructed by subcloning the EcoRI-XhoI cDNA fragment of pB42AD-mABT1, which was isolated from the mouse cDNA library, into the EcoRI and XhoI sites of pcDNA3 and pcDNA3-myc, respectively. pFA-CREB, pFA-cFos, pFA-ATF2, pCRE-Luc, pSRE-Luc, pAP1-Luc, and pNF-κB-Luc were purchased from Stratagene. pTATA-Luc was constructed by removing AP-1 binding sequences from plasmid pAP1-Luc. Two oligonucleotides (5′-CGC AAG CTT GCG GAG ACT CTA GAG GG-3′ and 5′-TTC TGC CCG AAC GG-3′) were used for PCR to amplify a sequence containing the TATA box and part of the luciferase coding sequences from pAP1-Luc. The PCR product was digested and replaced with the HindIII-SplI fragment of pAP1-Luc, which contains seven AP-1 binding sites, the TATA box, and the luciferase coding sequence. pEGFP-mABT1 was constructed by subcloning the EcoRI-XhoI fragment of pB42AD-mABT1 into the EcoRI-SalI site of pEGFP-C2 (Clontech). pGEX-mABT1 was constructed by subcloning the EcoRI-XhoI fragment of pB42AD-mABT1 in the EcoRI-XhoI site of pGEX-4T-1 (Pharmacia). pGEX-mABT1 mutant plasmids were constructed by subcloning the EcoRI-XhoI fragment of the PCR products amplified from pcDNA3-myc-mABT1 into the EcoRI-XhoI site of pGEX-4T-1. The oligonucleotide sequences used for amplifying the fragments of mABT1 cDNA were pcDNA3-S (5′-TAA TAC GAC TCA CTA TAG-3′), pcDNA3-AS (5′-ATT TAG GTG ACA CTA TAG-3′), A34 (5′-TTT GAA TTC ATG GCC TGC AGC GCA-3′), A97 (5′-TTT GAA TTC GGA GGA AAG AAG GGA GCT-3′), A39 (5′-TTT CTC GAG GCT GCT GCT GCT GCA GGC-3′), A102 (5′-TTT CTC GAG AGC TCC CTT CTT TCC TCC-3′), and A204 (5′-TTT CTC GAG ATC CCC ATC AGC TGC AAG-3′). Combinations of oligonucleotides for PCR were as follows: A34 and pcDNA3-AS for mABT1(34-269), pcDNA3-S and A39 for mABT1(1-39), pcDNA3-S and A102 for mABT1(1-102), pcDNA3-S and A204 for mABT1(1-204), A34 and A102 for mABT1(34-102), A97 and A204 for mABT1(97-204), and A34 and A204 for mABT1(34-204).
GST fusion protein binding assays.
Escherichia coli JM109 transformed with pGEX plasmids was inoculated in 20 ml of Luria-Bertani medium containing ampicillin (100 μg/ml) and cultured overnight at 37°C. The cultures were diluted 1:10 in 200 ml of the same medium and cultured at 25°C until the optical density at 600 nm reached 0.8; then glutathione S-transferase (GST) fusion proteins were induced by adding isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 0.2 mM. E. coli was collected after 20 h of incubation with IPTG at 25°C and resuspended in 20 ml of PBS containing 0.2 mM phenylmethylsulfonyl fluoride. The E. coli suspension was sonicated followed by incubation in the presence of 1% Triton X-100 for 30 min at 4°C. The suspension was centrifuged, and supernatants were incubated with glutathione-Sepharose 4B (Pharmacia) to immobilize the GST fusion proteins on the Sepharose beads. HeLa nuclear extract was prepared as follows. HeLa cells growing in log phase were collected and washed twice with PBS containing 0.5 mM MgCl2 and twice with buffer A (10 mM Tris-HCl [pH 7.5], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT). The cells resuspended in buffer A were homogenized with a Potter-Elvehjem homogenizer and centrifuged. After removing supernatants, pellets were resuspended in buffer C (20 mM Tris-HCl [pH 7.5], 0.2 mM EDTA, 0.45 M NaCl, 5 mM MgCl2, 0.5 mM DTT, 25% glycerol) and then homogenized with a Potter-Elvehjem homogenizer. The homogenates were transferred to a centrifuge tube and rocked at 4°C for 30 min. The homogenates were then centrifuged, and supernatants were collected as HeLa nuclear extracts. At this point, 4 ml of nuclear extracts was obtained from 5 × 107 cells. The GST fusion protein-Sepharose beads (the amounts of the GST fusion protein and the volume of the Sepharose beads were normalized to approximately 5 μg and 15 μl, respectively) were incubated overnight at 4°C with 200 μl of the HeLa nuclear extracts or approximately 5 ng of recombinant histidine-tagged mouse TBP (His-mTBP) and then washed four times with PBS containing 0.1% Triton X-100. Proteins bound to the Sepharose beads were used for Western blot analysis.
Immunoprecipitation.
COS7 cells (1.2 × 106 cells) were transfected with 12 μg of pcDNA3-myc-mABT1 and 75 μl of Lipofectamine as described above. At 48 h after transfection, cells were lysed with 1 ml of NP-40 buffer (20 mM Tris [pH 8.0], 1 mM EDTA, 150 mM NaCl, 1% NP-40, 10% glycerol) containing aprotinin (10 μg/ml), leupeptin (10 μg/ml), trypsin inhibitor (10 μg/ml), pepstatin A (2 μg/ml), 0.2 mM phenylmethylsulfonyl fluoride, and 1 mM DTT. Lysates were centrifuged, and 400 μl of the supernatants was incubated with an anti-TBP antibody or with a control rabbit immunoglobulin G (IgG) at 4°C for 16 h. Subsequently, 7 μl each of protein A-Sepharose beads and protein G-Sepharose beads were added to the mixture and incubated at 4°C for 2 h to absorb the immunocomplex. The Sepharose beads were washed four times with 1 ml of PBS containing 0.1% Triton X-100, and proteins bound to the Sepharose beads were separated by SDS-PAGE and immunoblotted with an anti-Myc or anti-TBP antibody.
RT-PCR.
Two micrograms of total RNAs isolated from COS7 cells with TRIZOL (GIBCO BRL) was treated with DNase I (GIBCO BRL). Reverse transcription by SuperscriptII (GIBCO BRL) was performed with the RNA samples and random hexamers. The DNA fragment corresponding to the partial sequences of exogenously transfected GAL4-CREB gene was selectively amplified by PCR by using two oligonucleotides (5′-ATT GGC TTC AGT GGA GAC-3′ and 5′-GAA TCA GTT ACA CTA TCC-3′). PCRs were performed for 35 cycles with denaturation at 94°C for 1 min, annealing at 44°C for 1 min, and polymerization at 72°C for 1 min. The PCR products were separated electrophoretically in an agarose gel and stained with ethidium bromide.
mABT1 localization.
pEGFP-mABT1 plasmids were transfected in COS7 cells with Lipofectamine as described above. At 24 h after transfection, localization of the enhanced green fluorescent protein (EGFP)-mABT1 fusion proteins were observed by fluorescence microscopy with an Axiovert 135 microscope (Zeiss).
Disruption of yeast ABT1 gene.
DNA fragments with the yeast ABT1 homologous DNA ends were generated by PCR using the template pFA6a-KanMX4, which contains the known Kanr open reading frame and permits efficient selection of transformants resistant to Geneticin (G418) (68), and the two primers 5′-AGC AAA CAG TTT ACT GCA GCA GAG TGA AGT AAA TTT TTA CGC CGT ACG CTG CAG GTC GAC-3′ and 5′-AGC ATT GGC CAC GGC TTG TTT CCA CAC GAC GTT GTT TAA ATT ATC GAT GAA TTC GAG CTC G-3′. The PCR was carried out with 25 pmol of the primers, 50 ng of template, 250 μM each deoxynucleoside triphosphates, 2 mM MgCl2, 1× KOD polymerase buffer, and 2.5 Unit of KOD polymerase (Toyobo). The program for the PCR consisted of 40 cycles of 15 s at 95°C, 30 s at 60°C, and 60 s at 74°C. S. cerevisiae C110-1 (a/α leu2-3/leu2-3 leu2-112/leu2-112 ura3-52/ura3-52 his6/HIS6) (66) was transformed with the resultant PCR fragment by the lithium acetate method and selected on a YPD plate containing G418 (0.2 mg/ml). G418-resistant clones were isolated, and disruption of the yeast ABT1 gene (YNR054c) was confirmed by PCR. For tetrad analysis, the ABT1-disrupted clones were incubated at 25°C in 1% potassium acetate to form spores; then separated spores were incubated on YPD plates at 30°C for 3 days.
In vitro transcription assay.
The conditions for in vitro transcription assays were as follows. The reaction mixture contained 10 mM HEPES-KOH (pH 7.6), 3% glycerol, 25 mM KCl, 6 mM MgCl2, 620 μM ATP and UTP, 25 μM CTP, 200 μM O-methyl-GTP, 5 μCi of [α-32P]CTP, 800 ng of template (which contains the AdML promoter fused to the G-less cassette [57]), 50 ng of recombinant TFIIB, 120 ng of recombinant TFIIF, 45 ng of recombinant TBP, 0.2 μg of RNA Pol II purified from calf thymus, and different amounts of GST-mABT1 in a total volume of 20 μl. The reaction mixture without nucleotides was incubated on ice for 20 min, and nucleotides were then added to initiate RNA synthesis, which took place at 27°C for 45 min. Synthesized RNA was extracted with phenol-chloroform, precipitated with ethanol, and analyzed on a 5% polyacrylamide–8-M urea gel.
DNA binding assay.
GST-mABT1 or GST protein (approximately 2 μg) was incubated with 15 μl of denatured or native DNA-cellulose (Amersham Pharmacia Biotech) at 4°C for 4 h. The cellulose was washed four times with PBS containing 0.1% Triton X-100. Proteins bound to the cellulose were subjected to SDS-PAGE and then stained with Coomassie brilliant blue.
Nucleotide sequence accession number.
The sequence data for ABT1 have been submitted to the DDBJ/EMBL/GenBank databases with accession no. AB021860 (mouse) and AB027258 (human).
RESULTS
Identification of mouse ABT1 and its homologs in human, yeast, and nematode cells.
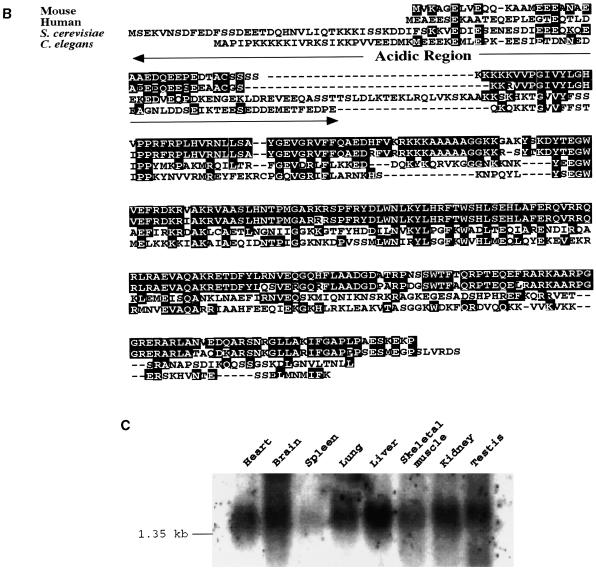
During identification of molecules interacting with SHD by yeast two-hybrid screening, four independent cDNA clones derived from the same gene were cloned from a mouse embryonic cell cDNA library. The representative clone, designated mABT1, has 1,276 bp and a potential open reading frame of 269 amino acids (aa) (Fig. 1A). The sequence around the putative ATG initiation codon at nucleotides 14 to 16 is compatible with the Kozak consensus sequence. The 3′ end is terminated with a poly(A)+ tail preceded by a polyadenylation signal. This clone covers an almost full-length mRNA of 1.4 kb as estimated by Northern blot analysis (see below). A sequence similarity search in the GenBank database was performed using the mABT1 protein sequence as the query, but no similar sequences with known function were found. mABT1 showed similarity only to the hypothetical proteins YNR054c of S. cerevisiae and F57B10.8 of C. elegans (GenBank accession no. P53743 and AF039713, respectively). To determine whether the C. elegans counterpart is actually transcribed, we performed RT-PCR and cloned a 0.8-kb cDNA. Sequence analysis revealed that this cDNA encoded an open reading frame of 268 aa (Fig. 1B) and that it was identical to F57B10.8, which was predicted from the C. elegans genome sequence. According to the sequence information of the 3′ untranslated region of human ABT1 in dbEST, we cloned a human ABT1 cDNA from NT2 human teratocarcinoma cells by 5′-RACE. The human ABT1 cDNA encoded an open reading frame of 272 aa (Fig. 1B). Sequence alignment of the mouse ABT1 protein and its human, S. cerevisiae, and C. elegans counterparts, hABT1, ScABT1, and CeABT1, respectively, is shown in Fig. 1B. The amino acid identities between mABT1 and the three homologs hABT1, ScABT1, and CeABT1 to mABT1 are 75.6, 21.8, and 26.5%, respectively. The homology among these proteins was observed as small conserved stretches distributed throughout the whole sequence. No functional motifs except for two putative nuclear localization signals were found in mABT1. Although ABT1 lacked a typical DNA-binding motif, the N terminus of mABT1 (aa 1 to 39), hABT1 (aa 1 to 39), ScABT1 (aa 1 to 86), and CeABT1 (aa 1 to 75) were rich in glutamic and aspartic acids (30.1, 41.0, 36.0, and 37.3%, respectively), suggesting that ABT1 might be a transcription factor. Northern blot analysis of poly(A)+ RNA from a variety of murine tissues showed that the mRNA of mABT1 was ubiquitously expressed as a transcript of approximately 1.4 kb (Fig. 1C).
FIG. 1.
Structure of mABT1 and expression in mouse. (A) Nucleotide and deduced amino acid sequences of the mouse ABT1 clone. The 269 aa in the open reading frame are represented with one-letter symbols. (B) Alignment of mouse, human, yeast, and nematode ABT1. The hypothetical protein sequences of S. cerevisiae (YNR054), C. elegans (F57B10.8), and human ABT1 exhibit high similarity to the mABT1 sequence. Amino acid residues identical to mABT1 are shown with a black background. Acidic regions of the four ABT1s are indicated with arrows. (C) Northern blot analysis showing the ubiquitous tissue expression of mABT1 mRNA in the mouse. mABT1 mRNA is expressed as an approximately 1.4-kb transcript in all tissues examined.
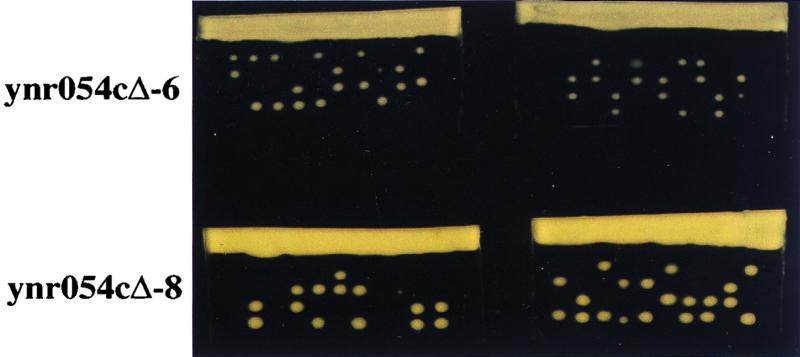
The yeast ABT1 homolog is essential for growth.
Since ABT1 is conserved from yeast to mammalian cells, we anticipate that ABT1 might have an important cellular function. As a step toward obtaining clues for its function, we disrupted the ScABT1 gene (YNR054c). Two yeast clones (ynr054cΔ-6 and ynr054cΔ-8) were generated in which a single copy of the ScABT1 gene was deleted and replaced with a Kanr gene (see Materials and Methods). The consequences of this gene disruption were determined by sporulation and tetrad analysis. Among the 40 tetrads dissected from ynr054cΔ-6 and ynr054cΔ-8, 36 gave rise to only two viable spores that could grow on YPD plates (Fig. 2). All viable progenies were sensitive to G418, indicating that these haploids have intact ScABT1 gene. This result clearly suggests that the yeast ABT1 is an essential gene, and it is therefore appropriate to speculate that the mammalian ABT1 also has an important function.
FIG. 2.
Yeast ABT1 gene is essential for growth. A single copy of the ScABT1 gene in a diploid strain was disrupted, and tetrad analysis were performed. Two independent yeast clones (ynr054Δ-6 and ynr054cΔ-8) were cultured in 1% potassium acetate at 25°C to form spores. Spores derived from approximately 20 tetrads of each clone were separated and incubated on YPD plates at 30°C for 3 days.
mABT1 is localized to the nucleus and nucleolus.
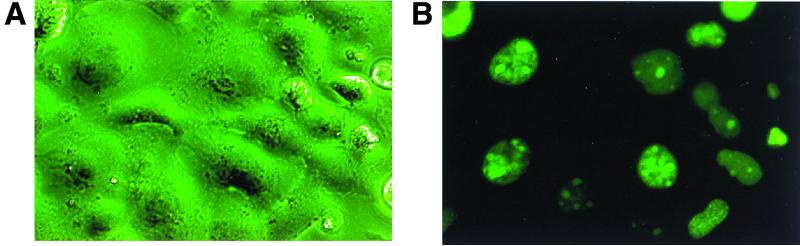
To determine the subcellular localization of mouse ABT1, we transiently expressed in COS7 cells the EGFP-mABT1 fusion protein. While a control EGFP distributed both to the nucleus and the cytoplasm (data not shown), the EGFP-mABT1 fusion protein was confined to the nucleus and in some cases to the nucleolus (Fig. 3). Subcellular localization of hemagglutinin-tagged mABT1 (HA-mABT1) examined by an anti-HA antibody, and immunofluorescence also confirmed nuclear localization of mABT1 (data not shown). Although mABT1 possesses two putative nuclear localization signals, KKKKK (40 to 44) and KRKKK (87 to 91), mABT1 mutants lacking either or both motifs still localized to the nucleus (data not shown). Accordingly, these motifs are not essential for nuclear localization of mABT1. There may be other nuclear localization signals, or mABT1 may passively be transported into the nucleus by diffusion due to its small size.
FIG. 3.
Subcellular localization of mABT1. COS7 cells were transfected with pEGFP-mABT1; after 24 h, cells expressing EGFP proteins were monitored by fluorescence microscopy under phase contrast (A) and dark field (B).
mABT1 interacts with TBP.
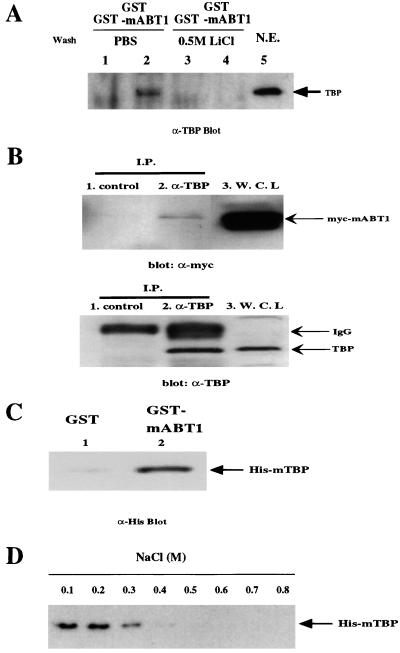
Since an acidic region has been reported to be the transcriptional activation domain in the transcription factors VP16, GAL4, GCN4, and p53 (12, 15, 24, 49, 54, 64), we believe that the function of mABT1 may be related to transcription. Many transcriptional activators and coactivators participate in transcription by interacting with GTFs and especially with TFIID. Thus, we first examined whether mABT1 associates with TBP, one of the major components of TFIID. GST-mABT1 or GST alone was immobilized on glutathione-Sepharose and incubated with nuclear extract prepared from HeLa cells. Glutathione-Sepharose was washed either with PBS containing 0.1% Triton X-100 or with 0.5 M LiCl, and the Sepharose was collected. Proteins bound to GST-mABT1 or to GST were separated by SDS-PAGE and immunoblotted with an anti-TBP antibody (Fig. 4A). A clear band with an approximate molecular mass of 37 kDa corresponding to human TBP was recognized by the anti-TBP antibody when GST-mABT1 but not GST was incubated with the HeLa nuclear extract (Fig. 4A, lanes 2 and 1, respectively). The TBP band disappeared when the Sepharose was washed with 0.5 M LiCl (Fig. 4A, lane 4). The TBP band could also be detected when the Sepharose was washed with NP-40 buffer but not when the Sepharose was washed with the NP-40 buffer containing 0.1% SDS (data not shown). To further confirm the interaction between mABT1 and TBP, we performed an immunoprecipitation assay. Cell lysates from COS7 cells expressing Myc-mABT1 were incubated with an anti-TBP antibody or a control rabbit IgG. Subsequently, the immunocomplex was washed four times with PBS containing 0.1% Triton X-100, separated by SDS-PAGE, and immunoblotted with an anti-Myc antibody or anti-TBP antibody (Fig. 4B). Under these experimental conditions, Myc-mABT1 coimmunoprecipitated with endogenous TBP (Fig. 4B, lane 2), suggesting that mABT1 forms a complex with TBP in transfected cells.
FIG. 4.
mABT1 interacts with TBP. (A) Interaction of GST-mABT1 with TBP in HeLa nuclear extracts (N.E.). The GST-mABT1 fusion protein (lanes 2 and 4) and the GST protein (lanes 1 and 3) expressed in E. coli were immobilized on glutathione-Sepharose beads and incubated with HeLa nuclear extracts. After washing with PBS (136.9 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4, 1.5 mM KH2PO4) containing 0.1% Triton X-100 (lanes 1 and 2) or with 0.5 M LiCl (lanes 3 and 4), proteins bound to the Sepharose beads were subjected to SDS-PAGE and immunodetected with an anti-TBP antibody. HeLa nuclear extracts were used as a positive control (lane 5). The arrow indicates the TBP band, which is 37 kDa. (B) Coimmunoprecipitation of mABT1 and TBP. COS7 cells were transfected with pcDNA3-myc-mABT1, and lysate was prepared. Immunoprecipitations with control rabbit IgG (lane 1) or anti-TBP antibody (lane 2) were performed, and the immunocomplex was analyzed with antibodies against TBP (α-TBP blot) and c-Myc (α-Myc blot). TBP and Myc-mABT1 in the whole cell lysate are shown in lane 3. (C) Interaction of mABT1 with purified mTBP. Recombinant His-mTBP was incubated with GST-mABT1 (lane 2) or GST (lane 1), and proteins bound to the Sepharose beads after washing with PBS containing 0.1% Triton X-100 were subjected to SDS-PAGE and immunodetection with an anti-His antibody. The arrow indicates His-mTBP. (D) Stability of mABT1-TBP. The His-mTBP was incubated with GST-mABT1 and washed with various concentrations of NaCl (0.1 to 0.8 M) containing 0.1% Triton X-100. Then His-mTBP bound to the GST-mABT1 was analyzed as described above.
Next, we examined whether mABT1 interacts directly with TBP. Recombinant His-mTBP produced in E. coli was incubated with GST-mABT1 or GST alone. The Sepharose was washed with PBS, and proteins bound to the Sepharose were analyzed by Western blotting with an anti-His antibody (Fig. 4C). A clear band corresponding to His-mTBP was detected with GST-mABT1 (Fig. 4C, lane 2) but was only weakly detected with GST (Fig. 4C, lane 1). This result indicates that mABT1 and TBP bind directly in vitro. To examine stability of the binding between mABT1 and TBP, we washed the Sepharose with 0.1 to 0.8 M NaCl or 0.5 M guanidium-HCl. His-mTBP and GST-mABT1 was existed as a complex up to 0.3 M NaCl but dissociated at NaCl concentrations of 0.4 or higher (Fig. 4D); 0.5 M guanidinium-HCl also caused the His-mTBP–GST-mABT1 complex to dissociate (data not shown).
Region of mABT1 which binds to TBP.
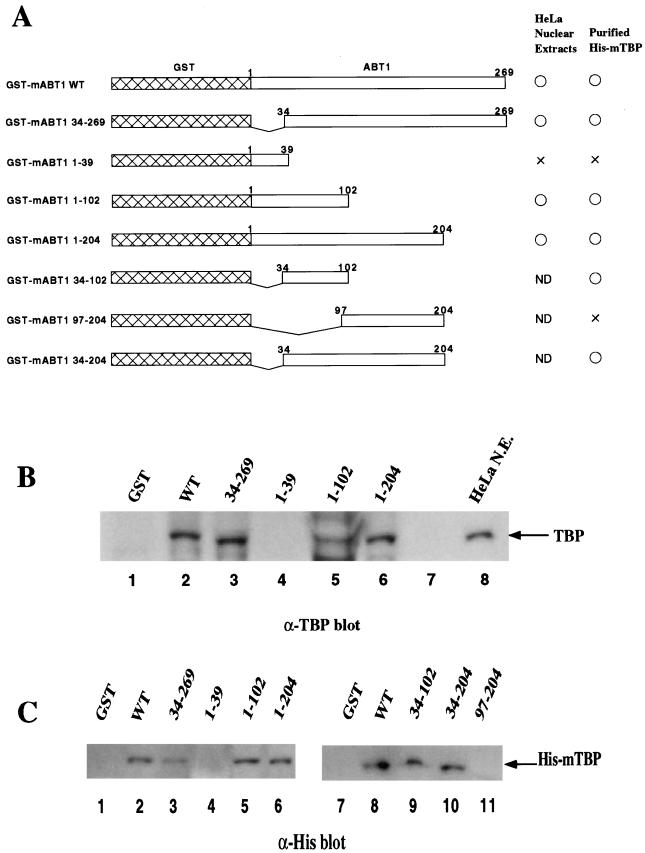
For examination of the TBP-binding region of mABT1, a series of GST-mABT1 fusion proteins constructed from several different regions of mABT1 were generated in E. coli and purified (Fig. 5A). HeLa nuclear extracts or His-mTBP were mixed with these truncated mABT1 fusion proteins, and pull-down experiments were performed. In HeLa nuclear extracts, GST-mABT1(34-269), GST-mABT1(1-102), and GST-mABT1(1-204) were found to interact with TBP (Fig. 5B, lanes 3, 5, and 6, respectively) as did the GST-mABT1 wild type (WT) (Fig. 5B, lane 2). In contrast, GST-mABT1(1-39) (Fig. 5B, lane 4) failed to interact with TBP. These results indicate that the region in mABT1 comprising residues 34 to 102 is necessary for binding to TBP.
FIG. 5.
Delineation of the TBP-binding region of mABT1. (A) Schematic representation of a series of GST-mABT1 mutants and summary of the binding characteristics. Numbers show the sequence position of amino acids in mABT1. Results of the interaction analysis with TBP in HeLa nuclear extracts and purified His-mTBP are summarized to the right (○, interaction; ×, no interaction; ND, not determined). The binding and washing conditions for TBP binding to the GST-mABT1 proteins were the same as in Fig. 4C. (B) Interaction of the GST-mABT1 deletion mutants with TBP in HeLa nuclear extracts. The series of GST-mABT1 deletion mutants were incubated with HeLa nuclear extracts and washed with PBS containing Triton X-100. Proteins bound to the GST fusion proteins were then detected by immunoblotting with an anti-TBP antibody. Lane 1, GST; lane 2, mABT1 WT; lane 3; mABT1(34-269); lane 4, mABT1(1-39); lane 5, mABT1(1-102); lane 6, mABT1(1-204); lane 8, control HeLa nuclear extract (N.E.) (C) The series of GST-mABT1 deletion mutants were incubated with His-mTBP and washed with PBS containing 0.1% Triton X-100. Proteins bound to the GST fusion proteins were detected by immunoblotting with an anti-His antibody. Lanes 1 and 7, GST; lanes 2 and 8, mABT1 WT; lane 3, mABT1(34-269); lane 4, mABT1(1-39); lane 5, mABT1(1-102); lane 6, mABT1(1-204); lane 9, mABT1(34-102); lane 10, mABT1(34-204); lane 11, mABT1(97-204).
As was the case for TBP in HeLa nuclear extracts, purified GST-mABT1 WT (Fig. 5C, lane 2 and 8), GST-mABT1(34-269), GST-mABT1(1-102), and GST-mABT1(1-204) also interacted with His-mTBP (Fig. 5C, lanes 3, 5, and 6, respectively), while GST-mABT1(1-39) (Fig. 5C, lane 4) failed to do so. To further define the region sufficient for complex formation, the GST-mABT1(34-102), GST-mABT1(34-204), and GST-mABT1(97-204) fusion proteins were generated to examine the complex formation. Here, GST-mABT1(34-102) and GST-mABT1(34-204) were observed to interact with His-mTBP as did the WT (Fig. 5C, lanes 9 and 10, respectively). In contrast, GST-mABT1(97-204) did not interact with His-mTBP at all (Fig. 5C, lane 11).
mABT1 stimulates basal transcription in vitro.
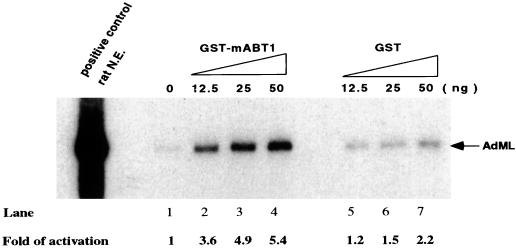
Given that mABT1 directly interacts with TBP, we then examined whether mABT1 will functionally participate in transcription in a reconstituted transcription reaction. We first examined whether mABT1 stimulates basal transcription from the AdML promoter, which contains only the core promoter sequence without any upstream regulatory elements. Addition of 12.5, 25, and 50 ng of GST-mABT1 to the in vitro transcription reaction significantly stimulated transcription levels 3.6-, 4.9-, and 5.4-fold, respectively (Fig. 6, lanes 2 to 4) compared to the control (no addition [Fig. 6, lane 1]). Addition of 12.5, 25, and 50 ng of control GST protein also slightly enhanced transcription levels 1.2-, 1.5-, and 2.2-fold, respectively (Fig. 6, lanes 5 to 7). The stimulatory effect of mABT1 on in vitro transcription was approximately three times higher than that of the GST protein. The transcription enhancement by GST-mABT1 saturated at 50 ng of GST-mABT1, and transcription was rather inhibited at the higher dose of 200 ng of GST-mABT1 (data not shown). These results clearly show that mABT1 can activate basal transcription in vitro.
FIG. 6.
mABT1 stimulates transcription in a reconstituted cell-free system. In vitro transcription from an AdML promoter was performed with addition of increasing amounts of GST-mABT1 or GST. Lane 1, without GST-mABT1; lane 2, 12.5 ng of GST-mABT1; lane 3, 25 ng of GST-mABT1; lane 4, 50 ng of GST-mABT1; lane 5, 12.5 ng of GST; lane 6, 25 ng of GST; lane 7, 50 ng of GST. Nuclear extract (N.E.) prepared from rat liver was used as a positive control. Amounts of transcripts were measured by an image analyzer (BAS 1500; Fuji), and fold activation is indicated at the bottom.
mABT1 stimulates basal transcription in transfected cells.
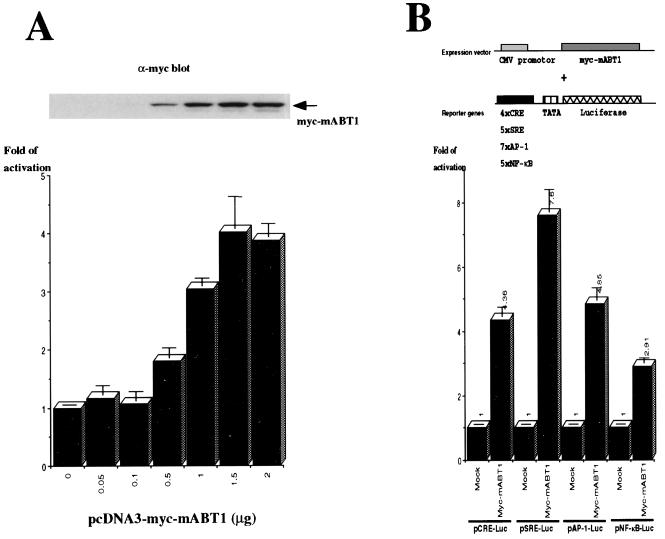
It remains to be shown whether mABT1 can activate basal transcription in the cell. To examine this, the reporter plasmid pTATA-Luc, which consists of a TATA box and a luciferase gene but without any cis-regulatory elements, was cotransfected in COS7 cells with pcDNA3-myc-mABT1, an expression vector of Myc-tagged mABT1. As the expression of mABT1 increased, the luciferase activity from pTATA-Luc was observed to increase up to fourfold in a dose-dependent manner (Fig. 7A). We note here that enhancement of transcription required relatively high expression levels of mABT1. These results convincingly show that overexpression of mABT1 stimulates basal transcription in transfected cells as well as in vitro.
FIG. 7.
mABT1 stimulates transcription in transfected cells. (A) Coexpression of mABT1 stimulates luciferase activity from the reporter plasmid pTATA-Luc. pTATA-Luc (0.1 μg) was cotransfected in COS7 cells with different amounts of pcDNA3-myc-mABT1 (0 to 2 μg). The total amount of plasmids used for each transfection was normalized with pcDNA3-myc. The cells were cultured in DMEM–10% FBS. Following 48 h of incubation, cell lysates were prepared and subjected to immunoblotting and luciferase assays. The expression level of Myc-mABT1 in each sample was detected by immunoblotting with an anti-Myc antibody (inset). (B) Stimulation of luciferase activity from reporter genes with different cis-regulatory elements by mABT1. Structures of the reporter and expression plasmids are shown at the top. One microgram of reporter genes containing CRE, SRE, AP-1, and NF-κB cis-regulatory elements was cotransfected with 1 μg of pcDNA3-myc (Mock) or pcDNA3-myc-mABT1 (Myc-mABT1). The cells were cultured in DMEM–1% BSA; after 48 h of incubation cell lysates were prepared and subjected to luciferase assay. CMV, cytomegalovirus.
We further tested, again by cotransfection, whether mABT1 enhances expression from other reporter genes containing CRE, AP-1, serum response element SRE, and NF-κB regulatory elements. Expression of mABT1 repeatedly increased luciferase activity from these reporter genes, pCRE-Luc (4-fold), pSRE-Luc (7-fold), pAP-1-Luc (5-fold), and pNF-κB-Luc (2.9-fold) (Fig. 7B). The enhancement of transcription from these heterologous promoters was more or less at the same level as that observed for the core promoter (pTATA-Luc; fourfold [Fig. 7A]). This suggests that mABT1 may not act in an enhancer-specific manner but rather acts ubiquitously. This observation is consistent with the idea that mABT1 stimulates basal transcription but does not activate specific enhancer elements. Although Myc-tagged mABT1 was used to monitor expression in these assays, the Myc tag did not contribute to enhance luciferase activity, since tagless mABT1 gave similar results (data not shown).
mABT1 enhances gene expression as monitored by mRNA and protein levels.
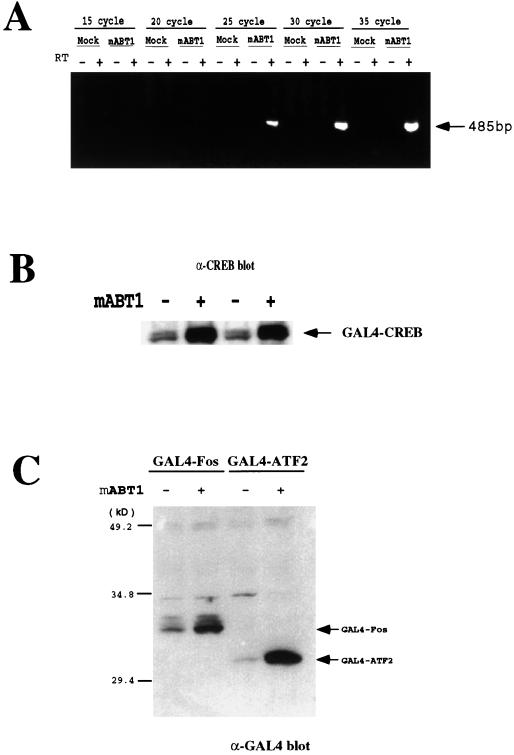
In addition to the measurement of luciferase activity, we confirmed the enhancement of gene expression by monitoring mRNA and protein levels. An expression plasmid for a GAL4-CREB fusion protein was cotransfected into COS7 cells with or without pcDNA3 Myc-mABT1. Total RNA was collected after an incubation period, and RT-PCR analysis was performed to quantify the amount of GAL4-CREB mRNA. As shown in Fig. 8A, the GAL4-CREB transcript was not detected by the RT-PCR in total RNA prepared from the COS7 cells transfected with a GAL4-CREB expression vector and pcDNA3-myc even after 35 cycles under these experimental conditions. In contrast, a GAL4-CREB transcript was readily detected in total RNA prepared from the COS7 cells transfected with the GAL-CREB expression vector and pcDNA3-myc-mABT1 after 25 cycles. We determined the amount of GAL4-CREB protein by Western blotting using an anti-CREB antibody and found that the amount, as well as that of GAL4-CREB mRNA, increased upon cotransfection of mABT1 (Fig. 8B). We further studied whether mABT1 enhances gene expression in other cotransfected plasmids expressing GAL4-Fos or GAL4-ATF2. Western blotting using an anti-GAL4 antibody clearly showed the expression levels of the GAL4-Fos and GAL4-ATF2 chimeric proteins to drastically increase when mABT1 was coexpressed (Fig. 8C).
FIG. 8.
mABT1 enhances mRNA and protein levels from cotransfected genes. (A) COS7 cells were transfected with pFA-CREB with either pcDNA3-myc (as Mock) or pcDNA3-myc-mABT1 (mABT1), and total RNA (1 μg) was subjected to RT-PCR (RT+) or PCR without the RT reaction (RT−). The PCRs were sampled at 15, 20, 25, 30, and 35 cycles, then separated on 1% agarose gel, and stained with ethidium bromide. A PCR product of 485 bp derived from the GAL4-CREB mRNA is indicated with an arrow. (B) Immunoblotting with anti-CREB antibody. Cell lysates prepared from the transfected COS7 cells as mentioned above were subjected to SDS-PAGE, followed by immunoblotting with an anti-CREB antibody. Arrows indicate the GAL4-CREB fusion protein. Two samples of control (−) and mABT1 (+) were independently prepared. (C) Immunoblotting with an anti-GAL4 antibody. COS7 cells were cotransfected with pFA-cFos or pFA-ATF2 with or without pcDNA3-myc-mABT1. Cell lysates were prepared after 48 h and analyzed with an anti-GAL4 antibody. Arrows indicate GAL4-Fos and GAL4-ATF2 fusion proteins detected with the antibody. Lane 1, pFA-cFos plus pcDNA3-myc; lane 2, pFA-cFos plus pcDNA3-myc-mABT1; lane 3, pFA-ATF2 plus pcDNA3-myc; lane 4, pFA-ATF2 plus pcDNA3-myc-mABT1).
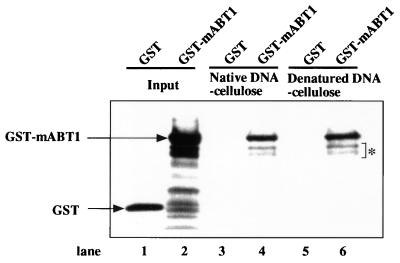
mABT1 interacts with DNA.
To examine whether mABT1 interacts with DNA, GST-mABT1 or control GST protein was incubated with DNA-cellulose. GST-mABT1, but not GST, was captured by both native DNA-cellulose (Fig. 9, lane 4) and denatured DNA-cellulose (Fig. 9, lane 6). The amount of GST-mABT1 protein bound to the cellulose was approximately one-third to one-fifth of the input (Fig. 9, compare lane 2 with lane 4 or 6). These results suggest that mABT1 directly binds to single- and double-stranded DNA. To examine stability of the binding, we washed the complex with 0.1 to 1.0 M NaCl. GST-mABT1 bound to both DNA-celluloses dissociated at 1.0 M NaCl but did not dissociate at 0.8 M NaCl (data not shown). In this assay, some degradation of GST-mABT1 was detected. We do not know whether these degradation products retain DNA-binding ability, since they might be captured by the dimerization of GST (65) between the degradation products and the intact GST-mABT1.
FIG. 9.
mABT1 interacts with DNA. GST or GST-mABT1 (Input; lanes 1 and 2) were incubated with native DNA-cellulose (lanes 3 and 4) or denatured DNA-cellulose (lanes 5 and 6). After washing with PBS containing 0.1% Triton X-100, proteins bound to cellulose were subjected to SDS-PAGE and stained with Coomassie brilliant blue. GST-mABT1, but not GST, was captured by both native and denatured DNA-cellulose (lanes 4 and 6). Degradations of GST-mABT1 are indicated by an asterisk.
DISCUSSION
Recent studies aimed at understanding the molecular mechanism of transcriptional regulation have unraveled a battery of proteins participating in various aspects of the transcriptional event (4, 30, 39, 53, 56, 67). TBP in particular has been recognized as one of the central factors that help multiple proteins to assemble into a large complex leading to transcriptional initiation and regulation (39). To further advance our knowledge, it is important to identify all of the TBP-binding proteins and relevant factors. In the present study, we have described the investigation and resulting characterization of a novel TBP-binding protein, mABT1.
mABT1 is a TBP-binding protein.
ABT1 was found to be ubiquitously expressed in a variety of mouse tissues and conserved from yeast to mammalian cells, suggesting an elementary role of ABT1 in cells. Indeed, the importance of ABT1 was explicitly shown in yeast cells, as the ABT1-deficient cells were unable to grow. We first tested whether mABT1 is involved in transcription, since mABT1 is a nuclear protein and has a characteristic acidic region, which has been described as a transcriptional activation domain in many transcription factors (e.g., VP16, GAL4, GCN4, and p53) (12, 15, 24, 49, 54, 64). Since several transcription factors have been shown to interact with TBP or TFIID, we examined whether mABT1 also interacts with TBP. In fact, mABT1 was found to interact with TBP directly, indicating that mABT1 can be added to the growing list of TBP-binding proteins. Interestingly, mABT1 and TBP could not associate under the high ionic strength of 0.4 M NaCl or 0.5 M guanidine hydrochloride in vitro. Thus, this association appears to be unstable compared with the association between TBP and TAFIIs, which is resistant to 0.5 M guanidine hydrochloride (21, 51). This may be one of the reasons why ABT1 has not previously been identified as a TAFII by conventional immunopurification methods using anti-TBP antibody (9). Although less stable, mABT1-TBP binding could be demonstrated by coimmunoprecipitation in mABT1-transfected cells, indicating that association can occur in the cell.
The fact that TBP can bind so many proteins raises the interesting question of whether this binding occur simultaneously on the same TBP molecule or on different TBP molecules. A quantitative Western blot analysis has shown that the concentration of TBP in the cell is sufficient to permit independent interactions with each of the TBP-binding proteins, including TAFIs, TAFIIs, TAFIIIs, SAGA, Mot1, NC2, and NOTs (39). According to this scheme, mABT1 may interact with a small population of a large global pool of TBP and form a complex with TBP which is not bound to other proteins.
By using a series of mABT1 mutants, we determined that the mABT1 region encompassing residues 34 to 102 is responsible for TBP binding. This domain is well conserved among different species, indicating that the interaction may be an evolutionarily conserved feature of this protein. TBP-binding motifs in several TBP-binding proteins have been reported (29), but we were not able to locate these motifs in the TBP-binding region of mABT1.
mABT1 activates basal transcription.
Since mABT1 binds to TBP, we examined whether mABT1 is involved in transcriptional regulation in a reconstituted cell-free transcription system. mABT1 increased up to threefold in vitro transcription from the AdML promoter in a dose-dependent manner. The level of transcription was inhibited at higher concentrations, possibly due to sequestering of the functional TBP from the promoter DNA. Given that mABT1 binds to TBP, it is likely that functional interaction can occur between mABT1 and the basal transcriptional machinery. The ability of mABT1 to enhance basal transcription appears to render the protein distinct from mammalian TAFIIs (67), which usually do not affect basal transcription but only mediate the response of specific activators. TAFIIs typically act as molecular bridges between specific activators and the general transcription machinery. For example, TAFII110 mediates activation of Sp1 (7, 21, 70), and TAFII40 mediates activation of GAL4-VP16 in cell-free systems (16).
In transfection experiments, we showed that coexpression of mABT1 enhanced transcription from a minimal core promoter and to the same extent from promoters with CRE, SRE, AP-1, and NF-κB cis-regulatory elements. Importantly, mABT1 does not function as a specific regulatory factor for CRE, SRE, AP-1, and NF-κB cis-regulatory elements; only the core promoter is sufficient for this effect. This result is compatible with the notion that mABT1 can activate basal transcription. Again, this feature appears to differentiate mABT1 from TAFIIs, in that TAFII40, TAFII60, TAFII110, and TAFII230 have been shown not to affect basal transcription in cotransfection experiments (11). Taken together, these results show that mABT1 is unique in possessing TBP-binding and basal transcription activities, a feature distinguishing it from typical TAFIIs, which act in a regulator-specific manner.
Recently, a distinct class of factors that activate basal transcription has been identified in yeast. For example, the yeast Tsp1/Sub1 protein, which has sequence similarity to the human coactivator PC4, stimulates basal transcription in vitro (19, 35). Also, the yeast mediator, which consists of approximately 20 polypeptides, interacts with the C-terminal repeat domain of Pol II and stimulates basal transcription in vitro (34, 36). Whether ScABT1 is included in the mediator complex and activates basal transcription remains to be clarified. Tsp1/Sub1 and the mediator have been reported to stimulate activated transcription as well as basal transcription. It will be interesting to determine whether mABT1 stimulates activated transcription. Interestingly, mABT1 can bind to DNA in a sequence-independent manner. This feature of mABT1 suggests that it could be a transcription cofactor, since well-characterized human-positive cofactors such as topoisomerase I (37, 43, 61), topoisomerase II (5), poly(ADP-ribose) polymerase, HMG2 (14, 28, 60, 62, 71, 72), and PC4 (31) are DNA-binding proteins which may affect accessibility to DNA and modulate the activity of RNA Pol II (30). The precise mechanism of how mABT1 exerts its effect on basal transcription remains to be elucidated. Since mABT1 binds to TBP and DNA, we favor the view that mABT1 accelerates or facilitates assembly of the transcriptional initiation complex. However, we cannot totally exclude the possibility that mABT1 affects mRNA stability or transcription elongation. Recently, TBP has been shown to form homodimers and needs to dissociate to acquire TATA element binding and activate transcription. Thus, it is thought that TBP dimerization prevents unregulated gene expression (27, 63). Along the same line, mABT1 might accelerate the dissociation of TBP dimers by binding to TBP and as a result increase the accessibility of TBP to promoter DNA, thereby enhancing basal transcription. To further dissect the molecular mechanism of mABT1 function, it is important to explain which proteins mABT1 interacts with, that is, whether mABT1 binds to GTFs, Pol II, TAFIIs, or other factors included in the preinitiation complex. Yeast two-hybrid screening using mABT1 is under way in our laboratory. mABT1-associated proteins may represent a novel protein-protein complex involved in the regulation of basal transcription. Although mABT1 has an acidic region in its N terminus, the function of this region in transcriptional activation has not been elucidated.
We have provided evidence that mABT1 can act as a regulator of basal transcription for class II genes. However, we still do not know whether mABT1 can act on class I and class III promoters. Since TBP is included in the transcriptional machinery of class I and class III promoters (39), and since mABT1 resides in the nucleolus, these possibilities may be worth testing in the future. Also, it would be interesting to examine if SHD or other SH2 domain-containing proteins regulate mABT1 function.
ACKNOWLEDGMENTS
We thank Y. Makino for the donation of TBP cDNA and for suggestions, and we thank H. Tokumitsu for helpful discussions.
REFERENCES
- 1.Auble D T, Hahn S. An ATP-dependent inhibitor of TBP binding to DNA. Genes Dev. 1993;7:844–856. doi: 10.1101/gad.7.5.844. [DOI] [PubMed] [Google Scholar]
- 2.Auble D T, Hansen K E, Mueller C G, Lane W S, Thorner J, Hahn S. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 1994;8:1920–1934. doi: 10.1101/gad.8.16.1920. [DOI] [PubMed] [Google Scholar]
- 3.Barlev N A, Candau R, Wang L, Darpino P, Silverman N, Berger S L. Characterization of physical interactions of the putative transcriptional adaptor, ADA2, with acidic activation domains and TATA-binding protein. J Biol Chem. 1995;270:19337–19344. doi: 10.1074/jbc.270.33.19337. [DOI] [PubMed] [Google Scholar]
- 4.Björklund S, Kim Y-J. Mediator of transcriptional regulation. Trends Biochem Sci. 1996;21:335–337. doi: 10.1016/s0968-0004(96)10051-7. [DOI] [PubMed] [Google Scholar]
- 5.Brou C, Kuhn A, Staub A, Chaudhary S, Grummt I, Davidson I, Tora L. Sequence-specific transactivators counteract topoisomerase II-mediated inhibition of in vitro transcription by RNA polymerases I and II. Nucleic Acids Res. 1993;21:4011–4018. doi: 10.1093/nar/21.17.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang M, Jaehning J A. A multiplicity of mediators: alternative forms of transcription complexes communicate with transcriptional regulators. Nucleic Acids Res. 1997;25:4861–4865. doi: 10.1093/nar/25.24.4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen J L, Attardi L D, Verrijzer C P, Yokomori K, Tjian R. Assembly of recombinant TFIID reveals differential coactivator requirements for distinct transcriptional activators. Cell. 1994;79:93–105. doi: 10.1016/0092-8674(94)90403-0. [DOI] [PubMed] [Google Scholar]
- 8.Dikstein R, Ruppert S, Tjian R. TAFII250 is a bipartite protein kinase that phosphorylates the basal transcription factor RAP74. Cell. 1996;84:781–790. doi: 10.1016/s0092-8674(00)81055-7. [DOI] [PubMed] [Google Scholar]
- 9.Dynlacht B D, Hoey T, Tjian R. Isolation of coactivators associated with the TATA-binding protein that mediate transcriptional activation. Cell. 1991;66:563–576. doi: 10.1016/0092-8674(81)90019-2. [DOI] [PubMed] [Google Scholar]
- 10.Eisenmann D M, Arndt K M, Ricupero S L, Rooney J W, Winston F. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes Dev. 1992;6:1319–1331. doi: 10.1101/gad.6.7.1319. [DOI] [PubMed] [Google Scholar]
- 11.Farmer G, Colgan J, Nakatani Y, Manley J L, Prives C. Functional interaction between p53, the TATA-binding protein (TBP), and TBP-associated factors in vivo. Mol Cell Biol. 1996;16:4295–4304. doi: 10.1128/mcb.16.8.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedman A D, Triezenberg S J, McKnight S L. Expression of a truncated viral trans-activator selectively impedes lytic infection by its cognate virus. Nature. 1988;335:452–454. doi: 10.1038/335452a0. [DOI] [PubMed] [Google Scholar]
- 13.Gasch A, Hoffmann A, Horikoshi M, Roeder R G, Chua N H. Arabidopsis thaliana contains two genes for TFIID. Nature. 1990;346:390–394. doi: 10.1038/346390a0. [DOI] [PubMed] [Google Scholar]
- 14.Ge H, Roeder R G. The high mobility group protein HMG1 can reversibly inhibit class II gene transcription by interaction with the TATA-binding protein. J Biol Chem. 1994;269:17136–17140. [PubMed] [Google Scholar]
- 15.Gill G, Ptashne M. Mutants of GAL4 protein altered in an activation function. Cell. 1987;51:121–126. doi: 10.1016/0092-8674(87)90016-x. [DOI] [PubMed] [Google Scholar]
- 16.Goodrich J A, Hoey T, Thut C J, Admon A, Tjian R. Drosophila TAFII40 interacts with both a VP16 activation domain and the basal transcription factor TFIIB. Cell. 1993;75:519–530. doi: 10.1016/0092-8674(93)90386-5. [DOI] [PubMed] [Google Scholar]
- 17.Goppelt A, Stelzer G, Lottspeich F, Meisterernst M. A mechanism for repression of class II gene transcription through specific binding of NC2 to TBP-promoter complexes via heterodimeric histone fold domains. EMBO J. 1996;15:3105–3116. [PMC free article] [PubMed] [Google Scholar]
- 18.Hateboer G, Timmers H T, Rustgi A K, Billaud M, van't Veer L J, Bernards R. TATA-binding protein and the retinoblastoma gene product bind to overlapping epitopes on c-Myc and adenovirus E1A protein. Proc Natl Acad Sci USA. 1993;90:8489–8493. doi: 10.1073/pnas.90.18.8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henry N L, Bushnell D A, Kornberg R D. A yeast transcriptional stimulatory protein similar to human PC4. J Biol Chem. 1996;271:21842–21847. doi: 10.1074/jbc.271.36.21842. [DOI] [PubMed] [Google Scholar]
- 20.Hoey T, Dynlacht B D, Peterson M G, Pugh B F, Tjian R. Isolation and characterization of the Drosophila gene encoding the TATA box binding protein, TFIID. Cell. 1990;61:1179–1186. doi: 10.1016/0092-8674(90)90682-5. [DOI] [PubMed] [Google Scholar]
- 21.Hoey T, Weinzierl R O, Gill G, Chen J L, Dynlacht B D, Tjian R. Molecular cloning and functional analysis of Drosophila TAF110 reveal properties expected of coactivators. Cell. 1993;72:247–260. doi: 10.1016/0092-8674(93)90664-c. [DOI] [PubMed] [Google Scholar]
- 22.Hoffmann A, Horikoshi M, Wang C K, Schroeder S, Weil P A, Roeder R G. Cloning of the Schizosaccharomyces pombe TFIID gene reveals a strong conservation of functional domains present in Saccharomyces cerevisiae TFIID. Genes Dev. 1990;4:1141–1148. doi: 10.1101/gad.4.7.1141. [DOI] [PubMed] [Google Scholar]
- 23.Hoffmann A, Sinn E, Yamamoto T, Wang J, Roy A, Horikoshi M, Roeder R G. Highly conserved core domain and unique N terminus with presumptive regulatory motifs in a human TATA factor (TFIID) Nature. 1990;346:387–390. doi: 10.1038/346387a0. [DOI] [PubMed] [Google Scholar]
- 24.Hope I A, Struhl K. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell. 1986;46:885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- 25.Horikoshi M, Wang C K, Fujii H, Cromlish J A, Weil P A, Roeder R G. Cloning and structure of a yeast gene encoding a general transcription initiation factor TFIID that binds to the TATA box. Nature. 1989;341:299–303. doi: 10.1038/341299a0. [DOI] [PubMed] [Google Scholar]
- 26.Inostroza J A, Mermelstein F H, Ha I, Lane W S, Reinberg D. Dr1, a TATA-binding protein-associated phosphoprotein and inhibitor of class II gene transcription. Cell. 1992;70:477–489. doi: 10.1016/0092-8674(92)90172-9. [DOI] [PubMed] [Google Scholar]
- 27.Jackson-Fisher A J, Chitikila C, Mitra M, Pugh B F. A role for TBP dimerization in preventing unregulated gene expression. Mol Cell. 1999;3:717–727. doi: 10.1016/s1097-2765(01)80004-6. [DOI] [PubMed] [Google Scholar]
- 28.Jayaraman L, Moorthy N C, Murthy K G K, Manley J L, Prives C. High mobility group protein-1 (HMG-1) is a unique activator of p53. Genes Dev. 1998;12:462–472. doi: 10.1101/gad.12.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang S-W, Eberhardt N L. TEF-1 transrepression in Be Wo cells is mediated through interactions with the TATA-binding protein, TBP. J Biol Chem. 1996;271:9510–9518. doi: 10.1074/jbc.271.16.9510. [DOI] [PubMed] [Google Scholar]
- 30.Kaiser K, Meisterernst M. The human general co-factors. Trends Biochem Sci. 1996;21:342–345. [PubMed] [Google Scholar]
- 31.Kaiser K, Stelzer G, Meisterernst M. The coactivator p15 (PC4) initiates transcriptional activation during TFIIA-TFIID-promoter complex formation. EMBO J. 1995;14:3520–3527. doi: 10.1002/j.1460-2075.1995.tb07358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kao C C, Lieberman P M, Schmidt M C, Zhou Q, Pei R, Berk A J. Cloning of a transcriptionally active human TATA binding factor. Science. 1990;248:1646–1650. doi: 10.1126/science.2194289. [DOI] [PubMed] [Google Scholar]
- 33.Kim T K, Zhao Y, Ge H, Bernstein R, Roeder R G. TATA-binding protein residues implicated in a functional interplay between negative cofactor NC2 (Dr1) and general factors TFIIA and TFIIB. J Biol Chem. 1995;270:10976–10981. doi: 10.1074/jbc.270.18.10976. [DOI] [PubMed] [Google Scholar]
- 34.Kim Y-J, Björklund S, Li Y, Sayre M H, Kornberg R D. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 35.Knaus R, Pollock R, Guarente L. Yeast SUB1 is a suppressor of TFIIB mutations and has homology to the human co-activator PC4. EMBO J. 1996;15:1933–1940. [PMC free article] [PubMed] [Google Scholar]
- 36.Koleske A J, Young R A. An RNA polymerase II holoenzyme responsive to activators. Nature. 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 37.Kretzschmar M, Meisterernst M, Roeder R G. Identification of human DNA topoisomerase I as a cofactor for activator-dependent transcription by RNA polymerase II. Proc Natl Acad Sci USA. 1993;90:11508–11512. doi: 10.1073/pnas.90.24.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee T I, Wyrick J J, Koh S S, Jennings E G, Gadbois E L, Young R A. Interplay of positive and negative regulators in transcription initiation by RNA polymerase II holoenzyme. Mol Cell Biol. 1998;18:4455–4462. doi: 10.1128/mcb.18.8.4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee T I, Young R A. Regulation of gene expression by TBP-associated proteins. Genes Dev. 1998;12:1398–1408. doi: 10.1101/gad.12.10.1398. [DOI] [PubMed] [Google Scholar]
- 40.Maheswaran S, Lee H, Sonenshein G E. Intracellular association of the protein product of the c-myc oncogene with the TATA-binding protein. Mol Cell Biol. 1994;14:1147–1152. doi: 10.1128/mcb.14.2.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinez E, Ge H, Tao Y, Yuan C-X, Palhan V, Roeder R G. Novel cofactors and TFIIA mediate functional core promoter selectivity by the human TAFII150-containing TFIID complex. Mol Cell Biol. 1998;18:6571–6583. doi: 10.1128/mcb.18.11.6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meisterernst M, Roeder R G. Family of proteins that interact with TFIID and regulate promoter activity. Cell. 1991;67:557–567. doi: 10.1016/0092-8674(91)90530-c. [DOI] [PubMed] [Google Scholar]
- 43.Merino A, Madden K R, Lane W S, Champoux J J, Reinberg D. DNA topoisomerase I is involved in both repression and activation of transcription. Nature. 1993;365:227–232. doi: 10.1038/365227a0. [DOI] [PubMed] [Google Scholar]
- 44.Mermelstein F, Yeung K, Cao J, Inostroza J A, Erdjument-Bromage H, Eagelson K, Landsman D, Levitt P, Tempst P, Reinberg D. Requirement of a corepressor for Dr-1 mediated repression of transcription. Genes Dev. 1996;10:1033–1048. doi: 10.1101/gad.10.8.1033. [DOI] [PubMed] [Google Scholar]
- 45.Metz R, Bannister A J, Sutherland J A, Hagemeier C, O'Rourke E C, Cook A, Bravo R, Kouzarides T. c-Fos-induced activation of a TATA-box-containing promoter involves direct contact with TATA-box-binding protein. Mol Cell Biol. 1994;14:6021–6029. doi: 10.1128/mcb.14.9.6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mizzen C A, Yang X-J, Kokubo T, Brownell J E, Bannister A J, Owen-Hughes T, Workman J, Wang L, Berger S L, Kouzarides T, Nakatani Y, Allis C D. The TAFII250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 47.Moqtaderi Z, Bai Y, Poon D, Weil P A, Struhl K. TBP-associated factors are not generally required for transcriptional activation in yeast. Nature. 1996;383:188–191. doi: 10.1038/383188a0. [DOI] [PubMed] [Google Scholar]
- 48.Oda T, Kujovich J, Reis M, Newman B, Druker B J. Identification and characterization of two novel SH2 domain-containing proteins from a yeast two hybrid screen with the ABL tyrosine kinase. Oncogene. 1997;15:1255–1262. doi: 10.1038/sj.onc.1201299. [DOI] [PubMed] [Google Scholar]
- 49.Oliner J D, Pietenpol J A, Thiagalingam S, Gyuris J, Kinzler K W, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumor suppressor p53. Nature. 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 50.Pugh B F, Tjian R. Mechanism of transcriptional activation by Sp1: evidence for coactivators. Cell. 1990;61:1187–1197. doi: 10.1016/0092-8674(90)90683-6. [DOI] [PubMed] [Google Scholar]
- 51.Pugh B F, Tjian R. Transcription from a TATA-less promoter requires a multisubunit TFIID complex. Genes Dev. 1991;5:1935–1945. doi: 10.1101/gad.5.11.1935. [DOI] [PubMed] [Google Scholar]
- 52.Ransone L J, Kerr L D, Schmitt M J, Wamsley P, Verma I M. The bZIP domains of Fos and Jun mediate a physical association with the TATA box-binding protein. Gene Expr. 1993;3:37–48. [PMC free article] [PubMed] [Google Scholar]
- 53.Roeder R G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 54.Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 55.Saleh A, Lang V, Cook R, Brandl C J. Identification of native complexes containing the yeast coactivator/repressor proteins NGG1/ADA3 and ADA2. J Biol Chem. 1997;272:5571–5578. doi: 10.1074/jbc.272.9.5571. [DOI] [PubMed] [Google Scholar]
- 56.Sauer F, Tjian R. Mechanisms of transcriptional activation: differences and similarities between yeast, Drosophila, and man. Curr Opin Genet Dev. 1997;7:176–181. doi: 10.1016/s0959-437x(97)80126-8. [DOI] [PubMed] [Google Scholar]
- 57.Sawadogo M, Roeder R G. Factors involved in specific transcription by human RNA polymerase II: analysis by a rapid and quantitative in vitro assay. Proc Natl Acad Sci USA. 1985;82:4394–4398. doi: 10.1073/pnas.82.13.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seto E, Usheva A, Zambetti G P, Momand J, Horikoshi N, Weinmann R, Levine A J, Shenk T. Wild-type p53 binds to the TATA-binding protein and represses transcription. Proc Natl Acad Sci USA. 1992;89:12028–12032. doi: 10.1073/pnas.89.24.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shen W-C, Green M R. Yeast TAFII145 functions as a core promoter selectivity factor, not a general coactivator. Cell. 1997;90:615–624. doi: 10.1016/s0092-8674(00)80523-1. [DOI] [PubMed] [Google Scholar]
- 60.Shykind B M, Kim J, Sharp P A. Activation of the TFIID-TFIIA complex with HMG-2. Genes Dev. 1995;9:1354–1365. doi: 10.1101/gad.9.11.1354. [DOI] [PubMed] [Google Scholar]
- 61.Shykind B M, Kim J, Stewart L, Champoux J J, Sharp P A. Topoisomerase I enhances TFIID-TFIIA complex assembly during activation of transcription. Genes Dev. 1997;11:397–407. doi: 10.1101/gad.11.3.397. [DOI] [PubMed] [Google Scholar]
- 62.Stelzer G, Goppelt A, Lottspeich F, Meisterernst M. Repression of basal transcription by HMG2 is counteracted by TFIIH-associated factors in an ATP-dependent process. Mol Cell Biol. 1994;7:4712–4721. doi: 10.1128/mcb.14.7.4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Taggart A K P, Pugh B F. Dimerization of TFIID when not bound to DNA. Science. 1996;272:1331–1333. doi: 10.1126/science.272.5266.1331. [DOI] [PubMed] [Google Scholar]
- 64.Truant R, Xiao H, Ingles C J, Greenblatt J. Direct interaction between the transcriptional activation domain of human p53 and the TATA box-binding protein. J Biol Chem. 1993;268:2284–2287. [PubMed] [Google Scholar]
- 65.Tudyka T, Skerra A. Glutathione S-transferase can be used as a C-terminal, enzymatically active dimerization module for a recombinant protease inhibitor, and functionally secreted into the periplasm of Escherichia coli. Protein Sci. 1997;6:2180–2187. doi: 10.1002/pro.5560061012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Uemura H, Jigami Y. Mutations in GCR1, a transcriptional activator of Saccharomyces cerevisiae glycolytic genes, function as suppressors of gcr2 mutations. Genetics. 1995;139:511–521. doi: 10.1093/genetics/139.2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Verrijzer C P, Tjian R. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem Sci. 1996;21:338–342. [PubMed] [Google Scholar]
- 68.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 69.Wade P A, Jaehning J A. Transcriptional corepression in vitro: a Mot1p-associated form of TATA-binding protein is required for repression by Leu3p. Mol Cell Biol. 1996;16:1641–1648. doi: 10.1128/mcb.16.4.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weinzierl R O, Dynlacht B D, Tjian R. Largest subunit of Drosophila transcription factor IID directs assembly of a complex containing TBP and a coactivator. Nature. 1993;362:511–577. doi: 10.1038/362511a0. [DOI] [PubMed] [Google Scholar]
- 71.Zappavigna V, Falciola M, Citterich M, Mavilio F, Bianchi M E. HMG1 interacts with HOX proteins and enhances DNA binding and transcriptional activation. EMBO J. 1996;15:4981–4991. [PMC free article] [PubMed] [Google Scholar]
- 72.Zwilling S, Koenig H, Wirth T. High mobility group protein 2 functionally interacts with the POU domains of octamer transcription factors. EMBO J. 1995;14:1198–1208. doi: 10.1002/j.1460-2075.1995.tb07103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]