Abstract
As our understanding of the human microbiome progresses, so does the need for natural experimental animal models that promote a mechanistic understanding of beneficial microorganism–host interactions. Years of research into the exclusive symbiosis between the Hawaiian bobtail squid, Euprymna scolopes, and the bioluminescent bacterium Vibrio fischeri have permitted a detailed understanding of those bacterial genes underlying signal exchange and rhythmic activities that result in a persistent, beneficial association. as well as glimpses into the evolution of symbiotic competence. Migrating from the ambient seawater to regions deep inside the light-emitting organ of the squid, V. fischeri experiences, recognizes and adjusts to the changing environmental conditions. Here, we review key advances over the last 15 years that are deepening our understanding of these events.
Table of content
In this Review, Visick, Stabb and Ruby describe recent advances in understanding the squid–vibrio symbiosis from the symbiont’s perspective.
[H1] Introduction
Since humans first realized they were affected by the presence of microscopic organisms within their bodies, most of our focus has been directed on those few microbial species that cause disease – who they are, what their impact is and how to eliminate them. Little interest was shown in those bacteria found associated with a healthy body, and which were considered only ‘commensals’, an ecological term the meaning of which specifically connotes their lack of importance to the host. This viewpoint now seems as naïve as a belief in spontaneous generation. The importance of microorganisms to their hosts was well illustrated by the difficulties that pioneering scientists had to overcome to establish the first viable germ-free animals. Moreover, we now recognize that, when compared to pathogens, there are a much greater number and complexity of beneficial microorganisms associated with our bodies, as is the case for virtually all animals and plants. This rapidly expanding realization of the critical importance of the normal microbiota to organismal health has increased our desire to understand the mechanisms underlying the interactions that create and sustain microbial symbioses with eukaryotic partners. In response to such an intellectual frontier, scientists have always turned to model systems in which they could more easily address fundamental questions the answers to which can be broadly applied to many other organisms, including humans. Such ‘simple’ model organisms as Escherichia coli, Caenorhabditis elegans, Drosophila melanogaster, and others, were made ‘famous’ for their basic, and critical, contributions to the fields of biochemistry, genetics and development. Now, as the revolution in microbiome research expands, established models of symbiosis are leading the way, and new models are emerging and being developed1.
One such well-studied animal–bacterial symbiosis is the light-organ association between the squid Euprymna scolopes and its bioluminescent bacterial partner, Vibrio (Aliivibrio) fischeri. This highly species-specific symbiosis begins when a newly hatched juvenile squid emerges into seawater containing planktonic cells of V. fischeri2. These cells aggregate on the surface of the juvenile’s nascent light organ (Fig. 1) and specifically induce host activities, including those that release a chemoattractant from the pores on the surface3. Within 1-3 hours4, the bacteria detach from the aggregate, migrating through surface pores, down a duct, across an antechamber and into the epithelium-lined crypts5 where a few founding cells will grow into a population of several hundred thousand. This dense population induces a visible bioluminescence, which the adult squid uses to avoid predation during their nocturnal activity6. Each morning at dawn, the host expels almost all of the symbiont population into the surrounding seawater2,7; the remaining bacteria repopulate the crypts and are ready to produce light by nightfall. Meanwhile, the vented bacteria cycle through the environment, perhaps in other as yet unknown niches, until they or their progeny colonize the light organs of other E. scolopes hatchlings.
Fig. 1. The juvenile E. scolopes light organ.
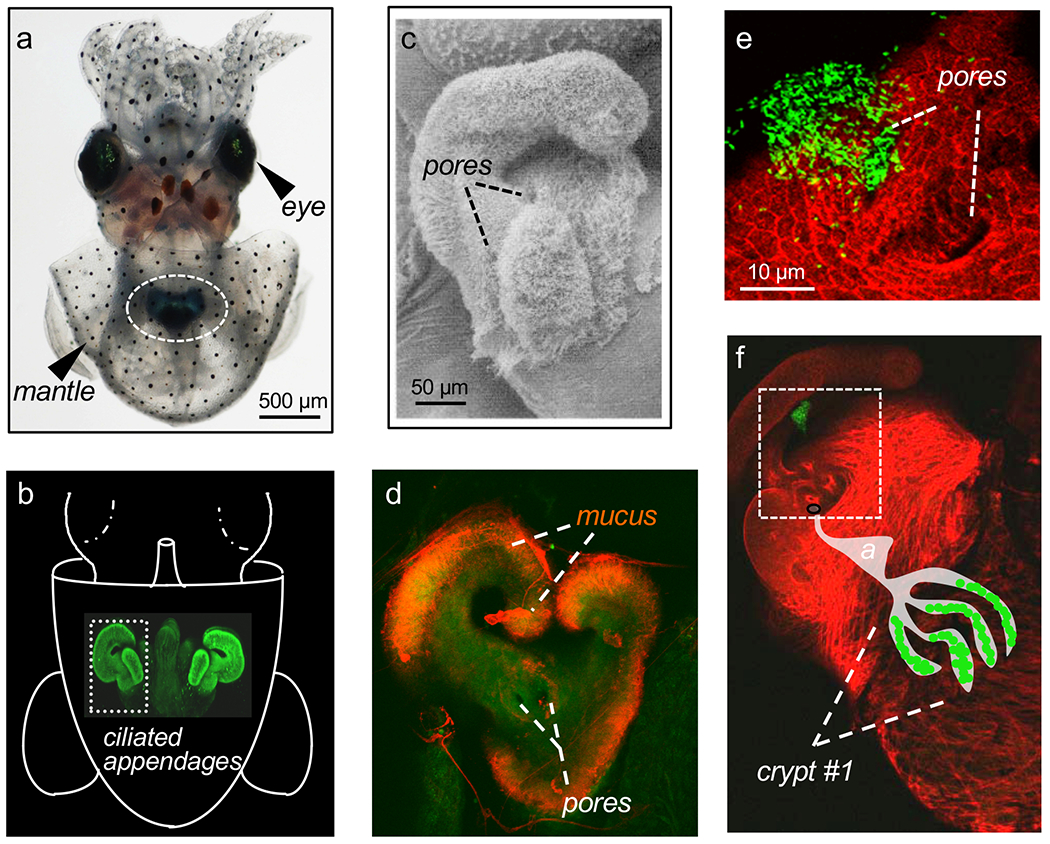
a. The nascent light organ (circle) is located in the mantle cavity of a newly hatched squid. b. Two pairs of ciliated appendages emerge from the outer surface of the light organ, interacting with seawater drawn into the mantle cavity during respiration. c. Three pores are found at the base of each pair of appendages. d. Soon after hatching the appendages begin to produce mucus that covers the ciliated fields. e. Mucus production and the flow fields created by the cilia capture V. fischeri cells (here, labelled with GFP) present in the seawater, and direct them to a zone directly above the pores where they form aggregates. f. After a few hours, the aggregates chemotax to the three pores (square), and migrate into one of three interior ducts, each leading to an antechamber (a), through a bottleneck, and into a crypt. Once in the crypts, the V. fischeri cells proliferate and autoinduce luminescence. This interior pathway to crypt #1 is indicated as a cartoon.
The diurnal rhythmic venting activity of the squid continues as the host develops into maturity, and until its death, which in laboratory aquaria occurs after about 9 months. Throughout the initiation of, accommodation to and persistence in the symbiosis, the species specificity and activity of the bacterial population must be maintained, and the development and immunological response of the host needs to be regulated8-10. To achieve these intricate, intertwined goals, the two partner species have evolved a complex system of physiological and biochemical communication, drawing upon and modifying pre-existing adaptations and capabilities such as chitin-driven fermentation, bacterial chemotaxis to specific nutrients, and the host hemocyanin’s Bohr effect (see companion article by Nyholm and McFall-Ngai 11), to perform new roles3,13,14. Identifying the molecular language of this communication has revealed not only adaptations unique to this association (for example, luminescence), but also modes of signaling (for example, microbe-associated molecular patterns (MAMPs), circadian rhythms, and non-coding RNA)9,15,16 that have later been, or are likely to be, found to occur between mammals and their normal microbiota17,18.
Over a decade ago, a previous contribution to Nature Reviews Microbiology19 focused on the impact that the application of bacterial genetic approaches was having on our understanding of the squid–vibrio association as well as several other emerging models of host–bacteria mutualism. In this Review, we update this analysis with new findings and conclusions about the squid–vibrio system from the symbiont’s perspective. This contribution is preceded by several excellent reviews of the association (see, for example, Refs. 19-23), and accompanies another contribution (see Nyholm & McFall-Ngai11) that describes recent advances in understanding the squid–vibrio symbiosis from the host’s perspective.
[H1] Events at the light organ surface
Initial engagement between the symbiont and its host occurs when ambient seawater containing V. fischeri cells is drawn into the mantle cavity of a juvenile squid (Fig. 1), where the bacteria come into contact with host-derived mucus covering the surface of the light organ. There, the activities of two types of cilia (‘long’ and ‘short’) on the nascent organ combine to direct the flow of bacteria-sized particles in the seawater into stagnant zones above pores in the light organ surface24 (see Nyholm & McFall-Ngai11). Within this zone, the bacteria attach to the cilia25 and form a cohesive cluster known as an aggregate, or symbiotic biofilm26, a critical colonization behavior (for example, Ref. 27). The aggregates can be from 10s to 1000s of bacteria in number, depending on the strain of V. fischeri and its ambient concentration28. Intensive study29,30 has shown that this biofilm, and efficient colonization, depends on an exopolysaccharide (Syp-PS) that is synthesized and exported by proteins encoded in the 18-gene syp locus31,32. Although most biofilm studies have focused on Syp-PS, V. fischeri produces at least one other polysaccharide, cellulose, that contributes to in vitro biofilm formation; however, its influence on the symbiosis is unclear33–36. In addition, other secreted components may contribute to the biofilm matrix, including Bmp proteins37 and outer membrane vesicles (OMVs)38.
Syp-PS production is controlled at the level of transcription by a network of two-component regulators (sensor kinases and response regulators). Sensor kinase proteins sense and relay (via phosphorylation) information about the environment to response regulator proteins that then control bacterial behavior. Because the signal or signals that modulate biofilm formation are generally unknown, for experimental studies biofilm formation is typically induced by overexpression of positive regulators or disruption of negative regulators. For example, overexpression of the sensor kinase RscS is sufficient to induce V. fischeri biofilm formation, contingent on the response regulator SypG, the direct activator of syp transcription (Fig. 2)27,39-42. RscS also depends on the presence of the sensor kinase SypF43. This regulatory scheme is atypical because sensor kinases generally signal directly to a downstream response regulator. Subsequently, it was determined that another sensor kinase, HahK, similarly functions through SypF35. Thus, SypF has an unusual role in integrating sensory information gleaned by itself and other sensor kinases to control SypG and, thus, syp transcription and host colonization35,43,44 (Fig. 2).
Fig. 2. Regulatory pathways controlling production of Syp-PS leading to biofilm formation and colonization.
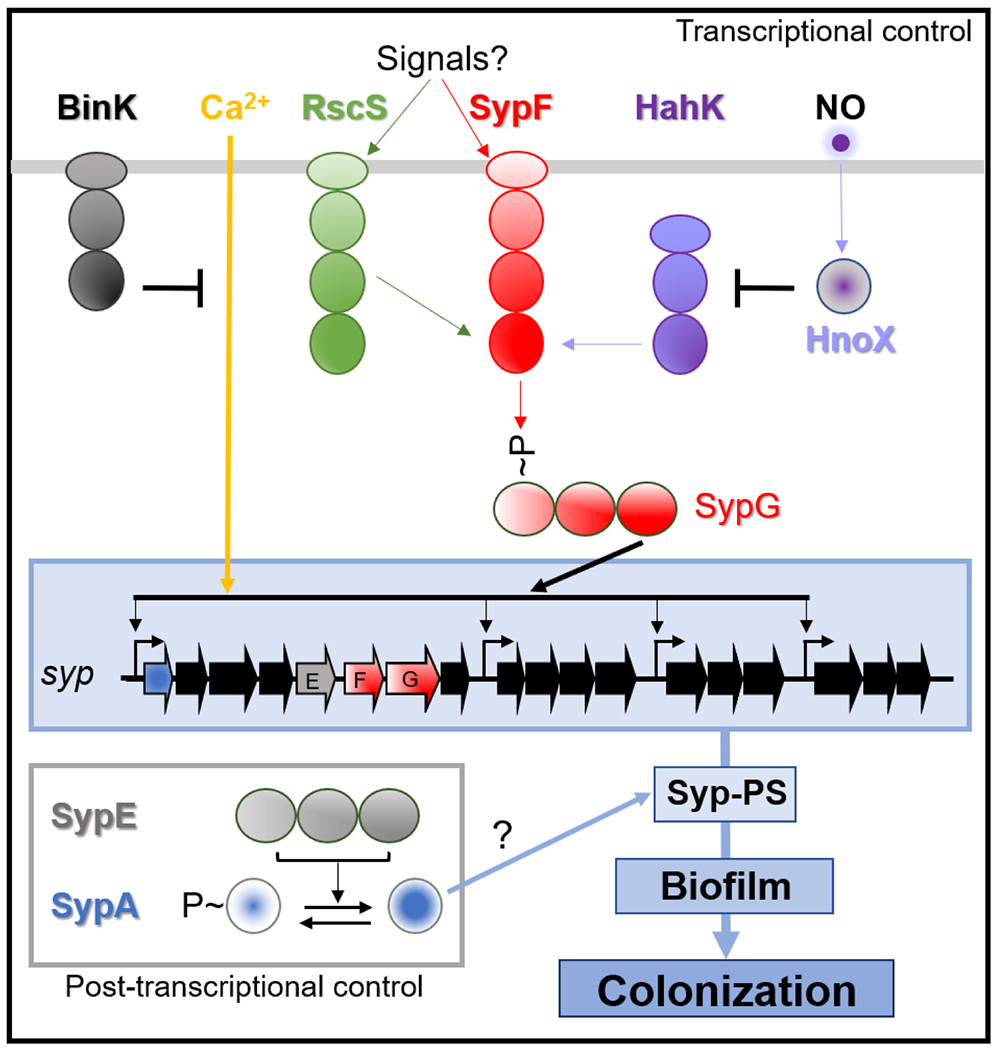
As described in the text, the SKs RscS and HahK function upstream of the SK SypF, which in turn activates, via phosphorylation, the RR SypG. In turn, SypG activates transcription of the syp locus, resulting in production of the structural proteins necessary for Syp-PS production. Calcium is a positive signal inducing syp transcription that is inhibited by the SK BinK. Transcription is also inhibited by nitric oxide (NO) via HahK. Syp-PS production is further regulated post-transcriptionally by the action of the RR SypE, which phosphorylates and dephosphorylates SypA. Unphosphorylated SypA promotes Syp-PS production through an unknown mechanism.
Syp-dependent biofilm formation and symbiotic colonization are inhibited by the response regulator BinK45,46. Loss of binK alone is sufficient to promote syp transcription and syp-dependent biofilm formation in vitro, but only when concentrations of Ca2+ typical in seawater are present35. This combination of factors (lack of BinK and addition of Ca2+) permitted a more natural investigation of the regulation of biofilm formation (without artificial overexpression) and, indeed, facilitated the identification of HahK as a key biofilm regulator35. How BinK and Ca2+ control syp transcription remains unknown. Syp-PS production is also controlled post-transcriptionally, via the response regulator SypE. This response regulator lacks a DNA-binding domain, instead controlling the activity of its target, SypA, via serine phosphorylation/dephosphorylation (Fig. 2)47,48. Unphosphorylated SypA promotes biofilm formation, while phosphorylated SypA is inactive and unable to promote biofilm formation or colonization48. The chaperone protein DnaJ may also function in this pathway: while expressing normal levels of syp transcription, a dnaJ mutant fails to produce Syp-PS or biofilms49. Intriguingly, syp is conserved in most Vibrio species32, and sypA from the pathogens Vibrio vulnificus and Vibrio parahaemolyticus could partially complement a V. fischeri sypA mutant50. Thus, the mechanisms uncovered in the squid symbiosis may have application to understanding biofilm formation and infection by human pathogens as well.
Historically, V. fischeri strain ES114 has been the principal strain used to study the squid–vibrio symbiosis, but new insights are being gleaned with additional isolates (Box 1). For example, the positive regulator rscS, which is required for colonization by ES11451, is either absent or frame-shifted in some colonization-competent isolates, indicating that these strains likely have evolved other mechanisms to induce syp transcription and/or control post-transcriptional processes52. Notably, some isolates, termed ’dominant’, readily out-compete ES114 for colonization53. For at least one, MB13B2, dominance seems to coincide with hyper-production of Syp-PS, as this strain produces substantially larger syp-dependent aggregates, and does so faster than ES11428. MB13B2 contains a frame-shifted rscS allele52 and, thus, understanding how syp regulation is altered in different strains of V. fischeri will be key to determining the diversity of mechanisms by which colonization competence is achieved.
BOX 1: Genomics and molecular genetics.
The availability of the genome for Vibrio fischeri (strain ES114) in 2005 greatly accelerated reverse genetics studies of specific genes, typically chosen through comparisons with those important in pathogens72. The subsequent determination in 2008 of the genome for strain MJ11, a fish light-organ symbiont that fails to colonize squid, permitted a more symbiosis-focused comparison and revealed the absence of a single, key regulator in the fish symbiont40. More recently, the genomes of numerous other V. fischeri symbionts have been reported (see the figure) and contain, in some cases, over 250 kb of additional sequences not found in ES114 (Refs. 52,53,159); the distinct phenotypes of these isolates are prompting investigations of factors that permit superior colonization. Whole-genome sequencing has also facilitated identification of point mutations, ushering in a new era of evolutionary biology. For example, experimental evolution experiments in which the non-colonizing MJ11 acquired the ability to colonize E. scolopes revealed the importance of a negative regulator in preventing colonization46.
At the same time, other advances have facilitated labeling and genetic manipulation of V. fischeri. Study of pES213, a native V. fischeri plasmid, permitted the development of stable, pES213-based expression vectors and reporter plasmids81,161. The application of a hyperactive Tn5 transposon enabled phenotype assessments via generating random mutants162,163. Another major genetic advance came with the finding that ES114 could be induced to take up DNA from the environment, and recombine it into the genome164. This ability to induce DNA uptake, combined with the use of new tools that facilitate mutant construction and complementation, has greatly accelerated the rate at which putative symbiosis factors can be investigated and confirmed through approaches such as ‘backcrossing’165. It has also facilitated deletion mapping of mutant derivatives166. Finally, the ability to interrogate the symbiosis has been advanced by other new approaches, including promoter probe104 and InSeq49 tools.
The geographical and biological sources of the isolates, as well as the number of strains sequenced, are indicated. Almost all the sequences are draft genomes 40,53,72,144,145,159,167,168
* These sequences were deposited into GenBank without an associated publication.
Fig. 4. The rise of V. fischeri genomic information.
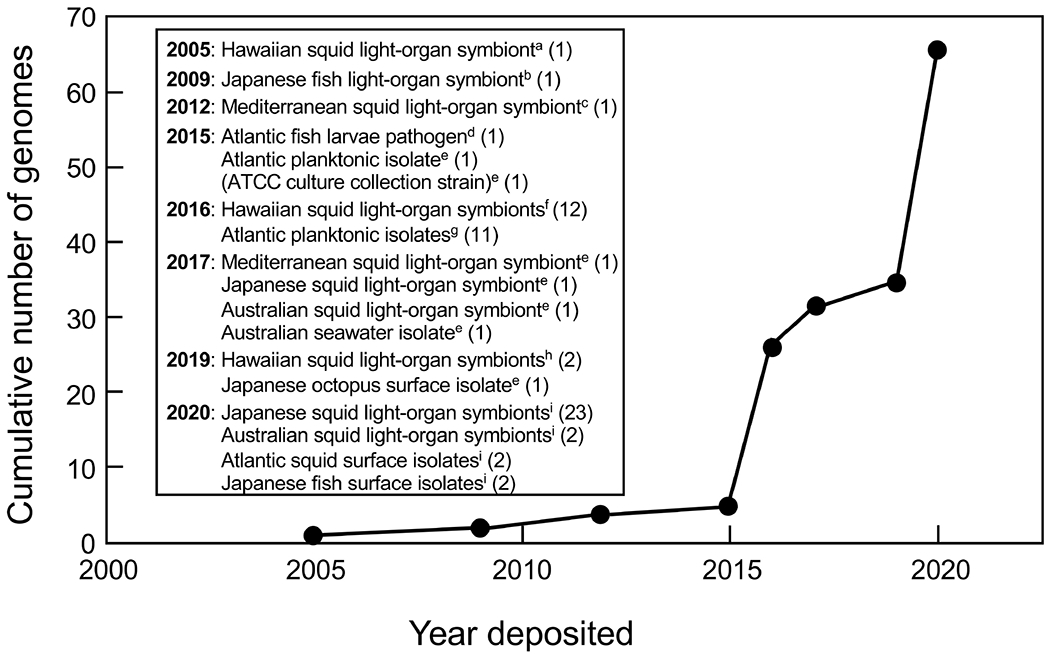
Trajectory of the appearance of V. fischeri genome sequences over the last 15 years, which has seen a rapid increase since 2015. The geographical and biological sources of the isolates, as well as the number of strains sequenced, are indicated. Almost all the sequences are draft genomes. Associated publications are identified by their PMIDs: a15703294; b19182778; c22374964; d26044435; e(none); f27128997; g27653556; h31331977; i32127462.
Despite hyper-aggregation exhibited by both MB13B2 (Ref. 28) and rscS-overproducing ES11427, these (and wild-type) strains ultimately disperse from their symbiotic biofilms to migrate into the light organ. Mechanisms of dispersal remain poorly understood, because it is rare to visualize this transient behavior in squid, it has not been observed in laboratory culture, and it is hard to differentiate mechanisms that promote biofilm formation from those that decrease dispersal, and vice versa. However, recent work has provided the first mechanistic insight by identifying a homolog of the Pseudomonas LapG protease54 as responsible for cleaving a large adhesin from the V. fischeri cell surface, permitting dispersal55. The activity of LapG is controlled by the inner-membrane protein LapD. Loss of LapD results in cells with an increased ability to disperse, a phenomenon that seems to delay symbiotic initiation. Because the lack of LapD does not prevent colonization, it is clear that other dispersal mechanisms must be in place. Additional insights may be gained from conditions or mutations that alter aggregate size (for example, Ref.56). For instance, it is possible that dispersal is facilitated by squid-produced nitric oxide (NO) found on the light organ surface56,57. Consistent with this possibility, addition of a NO synthesis inhibitor causes the formation of larger aggregates57 and could restore aggregation to a mutant deficient for Hmp, which eliminates NO58. NO is recognized and bound by HnoX, which in turn inhibits syp activator HahK. This results in the down-regulation of Syp-PS production (Fig. 2), thereby potentially facilitating dispersal59-60. These observations suggest that other factors49 or biosynthetic capabilities61 are necessary to orchestrate either the production of or a timely exit from the symbiotic aggregate.
[H1] Beyond the light organ surface
[H2] Chemotaxis and motility.
Many horizontally transmitted symbionts are believed to use chemotaxis to find target tissues within their respective hosts62. For instance, the migration of V. fischeri cells from the aggregate to their ultimate destination in the deep crypts depends on bacterial motility and chemotaxis (for example,26,63-67). V. fischeri produces a tuft of sheathed flagella at one pole of the cell, and magnesium in the environment enhances that production68,69. Intriguingly, these flagella are largely lost from symbionts in the crypts, indicating that internal conditions signal the formation of non-motile cells69. A large-scale, unbiased search for factors important for motility yielded both genes previously established as required for flagella biosynthesis and a number of novel genes63. Of note, this work identified VF_1491, which seems to function in chemotaxis, and flagella structural components FlgO, FlgP, and FlgT. These latter proteins are now known to contribute to the scaffolding structure that permits the flagellar motor to produce the high torque necessary to propel migration through high-viscosity environments such as mucus22,70. Confirming previous smaller-scale studies, non-motile mutants were unable to colonize, likely because they could not reach the crypts, whereas mutants with diminished motility exhibited decreased efficiency of colonization63. In addition, mutants defective for chemotaxis regulators CheA and CheZ only colonized ~50% of animals when presented at a dose that enabled full colonization by the wild type, supporting earlier studies (for example,71) that suggested a role for chemotaxis in effective colonization.
Recent work describing the cilia-driven flow of fluids around the light organ surface revealed that these dynamics could cause an accelerated formation of chemical gradients that may permit V. fischeri to chemotax into the light organ24. However, defining chemotactic mechanisms in V. fischeri is particularly challenging because this organism encodes ~43 chemoreceptors63,72. Initial investigations into the roles of 19 of these chemoreceptors revealed the function of only one, VfcA, which recognizes serine as well as certain other amino acids63. Subsequently, VfcB and VfcB2 were shown to recognize short-chain fatty acids such as propionate and butyrate73. However, none of these chemoreceptors seem to be important during the early stages of symbiosis. By contrast, V. fischeri also chemotaxes toward N-acetylneuraminic acid and chitin oligosaccharides, two squid-produced molecules that could promote the recruitment of V. fischeri71,74 (also, see Nyholm & McFall-Ngai11). Indeed, efficient entry depends on the ability to recognize a gradient of chitin oligosaccharides: addition of the disaccharide N, N′ diacetylchitobiose (chitobiose), but not the monosaccharide N-acetylglucosamine, disrupted squid colonization by preventing migration into the duct, while not preventing migration from the aggregate to the pore74. This behavior mimicked that of the colonization-defective cheA mutant. Similarly, efficient colonization could also be blocked by interfering with chitobiose production by the squid enzyme chitotriosidase, present in the ducts and pores3. Thus, it seems that early colonization comprises three stages: aggregate formation; migration to the pore; and movement from the pore into the duct in a manner dependent on the recognition of squid-generated chitin oligosaccharides. To date, the chitobiose chemoreceptor or chemoreceptors remain unknown; however, at least four of the genes encoding methyl-accepting chemotaxis proteins (MCPs) in the V. fischeri genome encode MCPs that recognize N-acetylated sugars like chitobiose75.
Finally, in addition to promoting chemotaxis-based movement, flagellar rotation seems to indirectly trigger the host immune response. Rotation results in the release of lipopolysaccharide molecules from the flagellar sheath and/or outer membrane, which in turn triggers apoptotic cell death in epithelial cells present on the light organ surface76. This apoptosis is part of a program of tissue remodeling that removes colonization-promoting ciliated structures after successful entry by V. fischeri (see below and companion paper) Thus, the role of flagella in symbiosis seems to be multi-factorial22.
[H2] Symbiotic bioluminescence and quorum-dependent regulation.
After colonizing its host, V. fischeri induces the lux operon, which is responsible for bioluminescence (Fig. 3). Once the animal grows large enough to cast a discernable shadow, this light emission is used by the squid to obscure its silhouette in a camouflaging ‘counterillumination’ behavior6, and perhaps other behaviors as well77. Bioluminescence enables V. fischeri to persist in the host78,79; dark mutants persist in the host; mutants deficient in bioluminescence (dark mutants) initially colonize, but within two days their colonization levels begin to diminish relative to the wild type. This initial colonization attenuation seems restricted to the largest, early-developing crypts80, perhaps because lux transcription is delayed in the smaller, later-developing crypts81-83. Although several possibilities have been proposed, the mechanism or mechanisms underlying the contribution of luminescence to colonization persistence remains unknown. Long-term colonization, either with or without a competing wild-type parent, drove a dark mutant to extinction84, perhaps because the host either eliminates the dark cheaters, or restricts their ability to proliferate after the dawn expulsion. These results suggest that luminescence eventually is required in all crypts, and that the light emission of an overwhelming majority of symbionts cannot compensate for the colonization defect of a nearby dark mutant84. The symbiotic fitness advantage to light production is particularly noteworthy given that bioluminescence presents a metabolic fitness cost in culture78, and dark lineages of V. fischeri have arisen in environments that lack hosts with light organs85. Symbiont bioluminescence not only affects the cellular and tissue morphology of the light organ, but also the host transcriptome9, including two transcripts that encode homologs of cryptochromes, light detectors that may permit a response to symbiont-generated light14,79,86-88.
Fig. 3. Genetics and biochemistry of bioluminescence in V. fischeri.
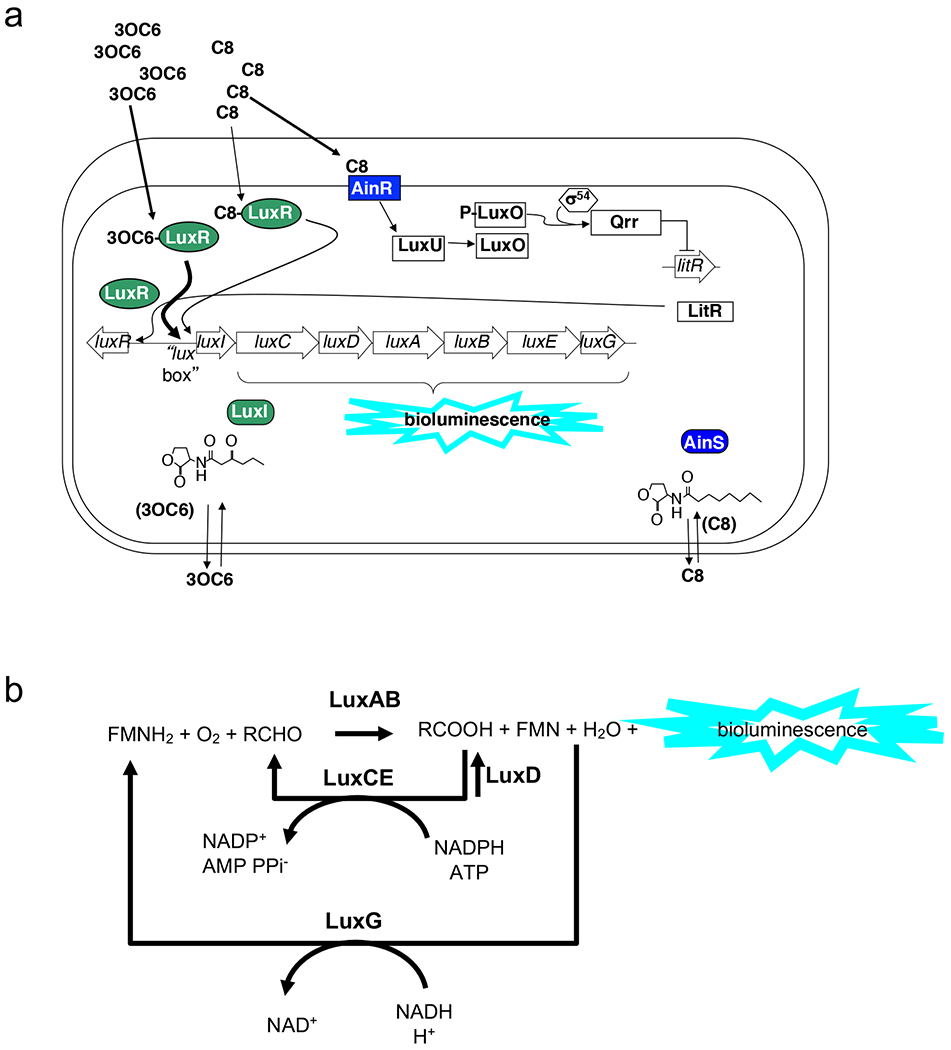
a. Genetics and regulation: large arrows indicate the genes and their orientation at the lux locus. The bioluminescence reaction is driven by products of luxCDABEG, which are co-transcribed with luxI, and this “lux operon” is divergently transcribed from luxR. Two AHL signal molecules, N-3-oxohexanoyl homoserine lactone (3OC6), and N-octanoyl homoserine lactone (C8), produced by LuxI and AinS, respectively, are membrane permeable and can serve as cell-to-cell signals. When 3OC6 binds to LuxR, the activated regulator attaches to a sequence near the promoter of the lux operon called the “lux box”, resulting in an activation of transcription. While C8 is a weak activator of LuxR, it is strongly recognized by its cognate receptor, AinR, and works through a regulatory cascade to relieve the repression of LitR, a master regulator that controls transcription of several genes including LuxR. b. Biochemistry: the LuxAB heterodimer forms the luciferase enzyme, which sequentially binds reduced flavin mononucleotide (FMNH2), O2, and an aliphatic aldehyde (RCHO), and converts these substrates to FMN, water, and the corresponding aliphatic acid (RCOOH), emitting a bluish light (~490 nm wavelength) in the process. LuxC, LuxD, LuxE, and LuxG are responsible for (re)generating the substrates for luciferase.
Bioluminescence is regulated by an archetypic, widespread, transcriptional mechanism controlled by cell–cell signaling and driven by the accumulation of signaling molecules known as pheromones (autoinducers). Only at a high cell density, or a ‘quorum’, do these pheromones reach a sufficient concentration to induce the lux operon. At least three pheromones contribute to induction, although the two acyl-homoserine-lactone (AHL) based systems are the most influential. This interconnected regulatory network has been reviewed extensively23,89,90 (Fig. 3). Briefly, the two AHL-based systems operate sequentially91, with signaling by AinS–AinR triggered at moderate cell densities, and jump-starting the downstream LuxI–LuxR system. LuxR, in turn, provides negative-feedback repression of ainSR92. Consistent with a sentinel role for AinS–AinR, AinR is exquisitely sensitive to its cognate signal, at concentrations <100 pM93. This signaling also influences central metabolism91,94,95 and, thus, exogenous addition of the pheromones, or chemical antagonists, affects not only the initiation of symbiosis, but also the subsequent bioluminescence and symbiotic stability96.
Although a quorum is necessary to stimulate luminescence, it is not sufficient. Regulators of gene expression and positive feedback loops mean that both the environment and the recent history of system induction are also critical for the decision to bioluminesce. This context dependence is evident in V. fischeri strains isolated from squid, which are far more luminescent in the host than in culture, even at similar cell densities97. The combination of environmental regulation and positive feedback could enable V. fischeri to launch population-wide behaviors in response to an environmental cue experienced by only a sub-population98,99. V. fischeri cells in different crypts can communicate using their pheromones, potentially transmitting information about the conditions they experience, despite their physical separation98. Such environmentally responsive regulators that control luminescence include ArcA, CRP, Fur and PhoB, among others98,100-103, and activating conditions and/or signals include oxidative redox, low iron, low phosphate, and the absence of catabolite-repressing sugars (such as glucose). Among these parameters, phosphate availability seems to vary between light organ microenvironments, although this unevenness alone cannot explain the heterogeneity of lux expression in the host104. Although some of these regulatory mechanisms are conserved, the lux locus and its regulation seems to have evolved rapidly among V. fischeri strains105, which suggests distinct selective pressures in different associations.
[H1] Nutritional metabolism
The metabolism of V. fischeri cells in the symbiotic light organ environment has been studied extensively and reviewed elsewhere23,106,107. Bioinformatic analyses illustrate the metabolic diversity of V. fischeri (for example,108), including various catabolic, anabolic, scavenging and energy-generating pathways, and transcriptional profiling shows that V. fischeri actively uses much of this genomic potential in the symbiosis109. Not only is V. fischeri physiology in the light organ complex, but it varies temporally both on a diurnal cycle110 and during maturation of the host106,107,111,112 (see Nyholm & McFall-Ngai11).
Mutant analyses have provided key insights into the nutrients available to symbiotic cells. For example, a pioneering study examining the colonization proficiency of amino acid auxotrophs indicated both that some amino acids were available to symbionts and that de novo amino acid biosynthesis contributed to symbiotic competence7. More recently, mutants with a disrupted tricarboxylic acid cycle (TCA) cycle that required glutamate were found to be proficient in colonization113, suggesting this amino acid or a related compound like glutamine is available in the light organ. Similar mutant analyses have shown that the host provides symbionts with guanine114 and δ-aminolevulinate115, or related compounds, that can overcome these auxotrophies. A recent comprehensive and groundbreaking insertion sequencing (InSeq) study catalogued an expansive number of mutants and their relative abilities to grow in a rich medium or in the light organ, providing a wealth of evidence with respect to what symbionts do or do not have to produce de novo in the light organ49; this study will continue to provide insights that must await additional investigations.
Considerable interest has focused on finding the primary carbon source or sources underpinning symbiont growth, although many questions remain unanswered. N-acetylglucosamine (NAG), and to a lesser extent glucose, have been areas of focus, in part because NAG is a component of chitin, which is the most abundant polysaccharide in the marine environment. Studies of the metabolic regulators CRP116 and NagC111,117 suggest that neither glucose nor NAG is amply available to symbionts initiating colonization, and this interpretation seems corroborated by the poor colonization of a NAG auxotroph115. Interestingly, although the regulatory response of CRP to glucose is consistent with the activity of this regulator in E. coli, the influences of NAG and cellobiose on CRP activity are difficult to square with prevailing models, and may suggest selective pressure to de-couple responses to NAG and glucose20,118. Although NAG seems to have a minor role during initiation of the symbiosis, the host provisions symbionts with chitin, a source of NAG, later during symbiotic development and presumably for most of the duration of this mutualism13 (see below).
Energy generation by the symbionts seems to involve a combination of respiratory and fermentative pathways119,120. Bioluminescence requires oxygen, and light output therefore indicates that the symbiont’s metabolism is not strictly anaerobic; however, luciferase seems to be oxygen-limited at points in the symbiosis, despite its high affinity for this substrate121. These observations and others suggest low-oxygen physiology is important for V. fischeri symbionts12,122. The ArcA–ArcB two-component system, which mediates regulatory changes at the transition from respiration to fermentation, contributes to symbiotic competence100, whereas the anaerobic regulator FNR does not123. V. fischeri possesses three terminal oxidases for aerobic respiration: CydAB, CcoNOQP and the unusual heme-independent NO-resistant AOX, each of which may contribute to symbiotic physiology, although the role of CydAB has been difficult to study owing to its importance in growth ex vivo106,124. Trimethylamine-N-oxide (TMAO) reductases are expressed in the symbiosis, but these anaerobic respiratory chains are not required during initiation of the symbiosis125. Other evidence suggests V. fischeri induces fermentative physiology in the light organ, acidifying the environment and notably producing acetate, which the host itself may use13,110. V. fischeri along with other members of this genus is capable of rapid generation times, which it achieves in the symbiosis69, so production of fermentation acids could indicate a form of overflow metabolism that reflects selection for rapid energy generation over efficient conversion of resources. Thus, the presence of fermentation products need not reflect a lack of electron acceptors to support respiration.
As with many pathogenic bacteria, in host tissues symbiotic V. fischeri cells induce scavenging pathways to obtain essential nutrients such as iron126, phosphate104 and sulfur112. For example, a low-iron response mediated by Fur is induced during colonization, and symbionts use siderophores127,128 and scavenge heme126 to obtain iron. Interestingly, sources of phosphate104 and sulfur112 are non-uniformly distributed across light organ microenvironments, illustrating the heterogenous biogeography of these symbiotic tissues. V. fischeri may obtain these nutrients at least in part from organic biomolecules like glycerophosphate109,110 and cystine7,112 during colonization.
[H1] Responses to the host
One of the key characteristics of bacterial symbioses is that, unlike when they are living outside the host, the surroundings of the microbial partner (that is, host tissues) reciprocally respond to the activities of the symbiont. Furthermore, that response changes depending on the age of the host, health, metabolic state and circadian clock. Because host engagement with the co-evolved mutualistic microbial species, and sometimes even strain, is critical for both partners129,130, there must be effective communication between the partners during the initiation and throughout the persistence of the symbiosis. To achieve this goal in the squid–vibrio association, the host responds to a complex language of chemical signals and cues from V. fischeri87 (see Nyholm & McFall-Ngai11), including OMVs131,132, the composition of which is modified in response to low ambient pH133. Conversely, as in other symbioses134, the squid shapes the microenvironments of its symbiont, which reacts by changing its physiology and behavior.
For example, during the initiation stage of colonization, aggregating V. fischeri cells encounter NO and acidic conditions produced within host mucus3,57. Although the levels of these stresses are insufficient to stop colonization, they do prime V. fischeri to induce resistance responses to withstand not only these conditions but also host-derived antimicrobial proteins135,136 that the symbionts will encounter during their subsequent passage into the crypts59,137,138. In some cases, V. fischeri cells seem uniquely capable of responding to this priming138, providing a possible mechanism that promotes symbiosis specificity. Importantly, transcriptional analysis of symbiont cells immediately after venting from the juvenile host provides evidence of the conditions they were experiencing in the crypts. Specifically, enzymes involved in resisting oxidative stress and antimicrobial peptides are highly upregulated in these cells109, which suggests that the symbionts are under physiological stress139. Having exited the host, the symbionts transition to their planktonic environment by reducing their luminescence and increasing motility, as facilitated by a habitat-transition regulator, HbtR75.
Each day after the morning venting, the remaining symbionts must repopulate the crypts, as well as produce bioluminescence that night. These two energy-intensive activities are supported by nutrients provided by the host. In the developing juvenile host, little is known about the identity of host-derived organic nutrients, although there is evidence from flux-coupling analysis that glycerolipids (perhaps from host membranes) and amino acids and N-acetylneuraminic acid (perhaps from host mucus) are fermented7,109. In addition, the availability of inorganic nutrients104,112, and the resulting nature of population growth82, differ between crypt populations. In contrast to this biogeographic variation within juvenile light organs, the symbionts in adult light organs experience temporally distinct nutritional conditions depending on the time of day110. Specifically, during the night, the host supports and enhances bioluminescence by the provision of chitin oligonucleotides12,13,110. In laboratory studies, catabolism of NAG, the chitin monomer, leads to an increased production of fermentation products, a lowering of ambient pH and a decrease in respiratory oxygen consumption120. Such conditions in the crypt10 would be expected to promote higher levels of luminescence production at night, when the squid needs this activity. Thus, the host controls its symbiont’s bioluminescence by a temporally restricted nutrient availability. How the host constrains any growth in the bacterial population during this period of intense light emission remains an unresolved question.
[H1] Population biology and evolution
It is becoming increasingly clear that strain-level differences (Box 1) have an important role in establishing symbioses19,129,130,140,141. For instance, phenotypically distinct classes of symbiotic V. fischeri isolates exhibited different competitive behaviors during initial colonization of the squid142, and genomic characterization of these strains revealed these classes were phylogenetically consistent, and defined a clade of strains with dominant (‘D’) behavior53. Extensive imaging of these D strains as they initiated light organ colonization showed that they often produced hyperaggregates28, and migrated from their aggregates sooner, reaching the crypts more rapidly4 than non-D strains. Interestingly, the D-strain clade also contains an additional 250 kb of DNA not found in any other V. fischeri53 and these additional 194 ORFs are distributed in 41 loci (‘islets’) that are spread across the two chromosomes, unlike in a single locus as is typical for colonization traits on pathogenicity islands (for example,143). This pattern is similar to the scattering of unique loci of a divergent light organ symbiont from the Mediterranean sepiolid squid Sepiola robusta144. As of yet, none of these genes have been connected to the D-type behavior145. In fact, recent work indicates that strategies for achieving D-type behavior have evolved independently multiple times in V. fischeri145.
Several studies have sought to understand the evolution of symbiosis competence in the highly specific squid–vibrio symbioses, using ecological146, comparative genomic145 or experimental evolution approaches46,147. The latter was designed to uncover specific mutations that increase the effectiveness of poorly colonizing strains of V. fischeri. In one study of two poor colonizers, mutations altering an extracellular polymeric substance (EPS) attenuator, BinK, previously identified in a transposon mutant screen45, readily resulted in mutants with improved ability to initiate colonization46. Besides an increase in the synthesis of Syp-PS, the mutation caused additional altered phenotypes, including reduced pheromone-signal production and decreased luminescence output. Such pleiotropic mutations may produce fitness trade-offs for the bacterium in other environments148, a hypothesis that has not been fully tested. In spite of this potential to increase the effectiveness of poorly colonizing strains, the squid–vibrio symbiosis remains notably species-specific: to date, there have been no effective efforts to convert other Vibrio species into successful colonizers. Thus, with the exception of some strains of the closely related sister species, Vibrio (Aliivibrio) logei149, V. fischeri remains the sole species found colonizing the E. scolopes light organ. Interestingly, like other Vibrio species, V. fischeri has a surprisingly low spontaneous mutation rate that is not uniformly represented around the chromosomes150,151, and identical deletion events resulting in the loss of the entire lux operon85 have apparently occurred across different lineages of this symbiont species. Thus, the population biology of V. fischeri is driven by both known and unknown evolutionary mechanisms.
A single light organ from a field-caught animal typically is colonized by a few of distinct strains152, yet little is known about their distribution among the crypts owing to the complex biogeography of the mature light organ153 and the difficulty of identifying different V. fischeri strains in a naturally colonized adult. An exciting advance in understanding the dynamic nature of symbiont-strain interactions has resulted from directly imaging the juvenile light organ after colonization by pairs of V. fischeri strains. Earlier work using isogenic strains indicated that, although individual crypts are most often colonized by single symbiont cells, ~20% typically develop as co-colonizations142. Colonization of the majority of crypts by only a single cell is believed to occur because of a physical ‘bottleneck’ along the migration pathway that constricts after the first V. fischeri cell enters82. Thus, it was a surprise when co-colonization experiments involving some strains resulted exclusively in singly colonized crypts154, suggesting a competitive incompatibility between certain strains. Subsequent work revealed that some V. fischeri strains encode a second type-6 secretion system (T6SS2) that is lethal to strains that lack this system155, the expression of which is regulated by environmental viscosity156. Carriage of the genes encoding the T6SS2, which includes functionally redundant structural proteins157, apparently constitutes a trade-off for the bacterium, as such strains have not swept through the light organs of squid populations, and still constitute a minority of symbionts; however, when they co-occur in a crypt, these genes are expressed158, and the strain prevails. Interestingly, only some of the dominant-behavior D strains53 carry this lethal T6SS2, which is phylogenetically more widespread among V. fischeri strains155, indicating that additional factors contribute to D-strain dominance. Although this discovery is still a new development in V. fischeri population dynamics, the announcement of the genome sequences of two T6SS-carrying strains159 will facilitate the use of comparative genomics approaches to better understand the origin and function of the mechanisms underlying competitive interactions.
[H1] Concluding comments
Microbial symbionts can affect essentially all aspects of their host’s behavior, life history and evolution and, in turn, adapt to their partner as a changing but predictable habitat. These processes are likely to occur across the extensive array of microbiomes that have become the subject of study in recent decades, including those of humans; however, the complexity of these associations make it difficult to examine individual interactions within a cacophonous consortium. By contrast, a naturally binary symbiosis like the squid–vibrio association has fewer voices in the conversation to decipher, permitting the discovery of novel, but shared, modes and components of a single partner’s reciprocal signaling and response to its host9,14,15. Once identified, these same components can often be recognized within the more complex, consortial symbioses, where they underlie mechanisms of health, resilience and homeostasis. Furthermore, new insights are anticipated from applying conceptual and technological advances to the study of this symbiosis (Box 2). As summarized in this review, the richness of the interaction between V. fischeri and its host is remarkable and continues to provide evidence of the extent to which the two partners have co-evolved, each increasing its fitness as a member of a functioning unit, relative to its success living as an individual organism.
BOX 2: Research areas to watch.
The next decade promises discoveries across a range of research topics, some examples of which are listed below.
Vibrio fischeri isolate ES114 has been a unifying wild-type strain; however, recent studies have shown the power of comparative strain analyses145. V. fischeri populations naturally compete with one another, driving the evolution of distinct genotypes based on fitness tradeoffs that will be identified by strain comparisons and DNA sequencing technology.
Future research will delve into issues of complex biogeography and diurnal variation, once simplified as ‘in the host’, using advances in microscopy such as hybridization chain-reaction fluorescence in situ hybridization (HCR-FISH)75,169, transcriptional reporters and methods such as imaging mass spectrometry that enable the identification of molecules in situ.
New insights into light organ environments enable researchers to better recapitulate relevant conditions ex vivo. No medium can reproduce all host conditions, but a systematic adjustment of media and physio–chemical growth conditions to better reflect light organ environments will give culture-based studies more ‘real-world’ relevance.
Research on symbiotic V. fischeri will benefit from a greater use of computational and bioinformatic approaches. Across biology, mathematical analyses of complex datasets have enabled discoveries in a range of topics, from metabolism to regulatory circuits to the behavior of cells within populations. Research on the light organ symbiosis is generating molecular and imaging data ripe for such deeper analysis.
Results from insertion sequencing (InSeq analysis)49 remain a cornucopia of testable hypotheses, providing insight into the role of every V. fischeri gene with respect to its importance in culture and/or in the symbiosis. This seminal dataset will inform many future studies.
An understanding of ‘global’ symbiont gene expression will benefit from the application of nanoString technology, which allows an accurate determination of the average expression levels for hundreds of genes in as few as 105 symbionts.
Acknowledgements
The authors thank M. McFall-Ngai and S.V. Nyholm for helpful comments about the manuscript. Work in the authors’ laboratories is supported by the US National Institutes of Health (NIH) R37 AI050661 (E.G.R.), R01 GM135254 (E. G. R.), R01 OD011024 (E.G.R.), and R35 GM130355 (K.L.V.); and by the US National Science Foundation (NSF) MCB-1716232 (E.V.S.).
Glossary
- Light organ:
Symbiotic bioluminescence organ inside the squid E. scolopes where V. fischeri resides
- Chemoattractant:
Nutrient or other molecule to which a bacterium migrates, typically using flagella-driven motility
- Mantle cavity:
The region inside E. scolopes where the light organ is located
- Biofilm matrix:
The collection of secreted polysaccharides, proteins, and other substances, produced by and surrounding a group of bacteria, that functions to promote attachment and provide protection
- Oxidative redox:
Chemical conditions that create an oxidative stress on bacteria
- Auxotrophs:
Bacteria that lack the ability to synthesize one or more essential biomolecules from a limited nutrient source
- Cellobiose:
disaccharide composed of glucose
- Siderophores:
Iron-binding molecules secreted by and taken up by bacteria, thereby providing iron to the cell
- Extracellular polymeric substance (EPS):
Polysaccharides, proteins, and other substances produced and secreted by a bacterium, many of which facilitate cell-cell and/or cell-surface attachment, and that contribute to the biofilm matrix
Footnotes
Competing interests
The authors declare no competing interests.
Peer review information
Nature Reviews Microbiology thanks T. Bosch, who co-reviewed with C. Giez, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Citations
- 1.Koch E, & McFall-Ngai M, Model systems for the study of how symbiotic associations between animals and extracellular bacterial partners are established and maintained. Drug Discovery Today: Disease Models 28, 3–12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee KH, & Ruby EG, Effect of the squid host on the abundance and distribution of symbiotic Vibrio fischeri in Nature. Appl. Environ. Microbiol 60, 1565–1571 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kremer N, et al. Initial symbiont contact orchestrates host-organ-wide transcriptional changes that prime tissue colonization. Cell Host Microbe 14, 183–194, doi: 10.1016/j.chom.2013.07.006 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bongrand C, & Ruby EG, Achieving a multi-strain symbiosis: strain behavior and infection dynamics. ISME J. 13, 698–706, doi: 10.1038/s41396-018-0305-8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McFall-Ngai MJ, The importance of microbes in animal development: lessons from the squid-vibrio symbiosis. Annu. Rev. Microbiol 68, 177–194, doi: 10.1146/annurev-micro-091313-103654 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones BW, & Nishiguchi MK, Counterillumination in the Hawaiian bobtail squid, Euprymna scolopes Berry (Mollusca: Cephalopoda). Mar. Biol 144, 1151–1155 (2004). [Google Scholar]
- 7.Graf J, & Ruby EG, Host-derived amino acids support the proliferation of symbiotic bacteria. Proc Natl Acad Sci U S A 95, 1818–1822 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McFall-Ngai MJ, & Ruby EG, Developmental biology in marine invertebrate symbioses. Curr. Opin. Microbiol 3, 603–607 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Moriano-Gutierrez S, et al. The noncoding small RNA SsrA is released by Vibrio fischeri and modulates critical host responses. PLoS Biol. 18, e3000934, doi: 10.1371/journal.pbio.3000934 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwartzman JA, & Ruby EG, Stress as a normal cue in the symbiotic environment. Trends Microbiol. 24, 414–424, doi: 10.1016/j.tim.2016.02.012 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nyholm S, & McFall-Ngai M, A lasting symbiosis: how the Hawaiian bobtail squid finds and keeps its bioluminescent bacterial partner. Nat. Rev. Microbiol doi: 10.1038/s41579-021-00567-y (2021) . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kremer N, et al. The dual nature of haemocyanin in the establishment and persistence of the squid-vibrio symbiosis. Proc. Biol. Sci 281, 20140504, doi: 10.1098/rspb.2014.0504 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwartzman JA, et al. The chemistry of negotiation: rhythmic, glycan-driven acidification in a symbiotic conversation. Proc. Natl. Acad. Sci. U. S. A 112, 566–571, doi: 10.1073/pnas.1418580112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]; In this study, the host’s deliver of chitin-derived GlcNAc is shown to develop 4 weeks post hatch, and this chitin is apparently deliver by hemocytes that lyse in the crypts only at night. A nocturnal acidification of the crypts results, and a model for how this outcome enhances bioluminescence is described.
- 14.Heath-Heckman EA, et al. Bacterial bioluminescence regulates expression of a host cryptochrome gene in the squid-Vibrio symbiosis. mBio 4, doi: 10.1128/mBio.00167-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koropatnick TA, et al. Microbial factor-mediated development in a host-bacterial mutualism. Science 306, 1186–1187 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Bouskra D, et al. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature 456, 507–510, doi: 10.1038/nature07450 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Thaiss CA, et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell 159, 514–529, doi: 10.1016/j.cell.2014.09.048 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Ruby EG, Symbiotic conversations are revealed under genetic interrogation. Nat. Rev. Microbiol 6, 752–762, doi: 10.1038/nrmicro1958 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bongrand C, & Ruby EG, The impact of Vibrio fischeri strain variation on host colonization. Curr. Opin. Microbiol 50, 15–19, doi: 10.1016/j.mib.2019.09.002 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colton DM, & Stabb EV, Rethinking the roles of CRP, cAMP, and sugar-mediated global regulation in the Vibrionaceae. Curr. Genet 62, 39–45, doi: 10.1007/s00294-015-0508-8 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Mandel MJ, & Dunn AK, Impact and Influence of the natural Vibrio-squid symbiosis in understanding bacterial-animal interactions. Front. Microbiol 7, 1982, doi: 10.3389/fmicb.2016.01982 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aschtgen MS, et al. Insights into flagellar function and mechanism from the squid-vibrio symbiosis. NPJ Biofilms Microbiomes 5, 32, doi: 10.1038/s41522-019-0106-5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stabb EV, & Visick KL in The Prokayotes Vol. 1 (eds. Rosenberg E, et al. ) 497–532 (Springer-Verlag, NYC, 2013). [Google Scholar]
- 24.Nawroth JC, et al. Motile cilia create fluid-mechanical microhabitats for the active recruitment of the host microbiome. Proc. Natl. Acad. Sci. U. S. A 114, 9510–9516, doi: 10.1073/pnas.1706926114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; This work provided the first glimpse into the cilia-driven fluid mechanics that position V. fischeri cells to reach and settle in ‘quiet zones’ on the light-organ surface, permitting a selective ‘recruitmen’ of this microbe from the planktonic environment.
- 25.Altura MA, et al. The first engagement of partners in the Euprymna scolopes-Vibrio fischeri symbiosis is a two-step process initiated by a few environmental symbiont cells. Environ. Microbiol 15, 2937–2950, doi: 10.1111/1462-2920.12179 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]; Aggregations of only a few V. fischeri cells were observed to initiate normal host responses, and revealed that aggregation was a two-part process that began with bacterial attachment to the cilia.
- 26.Nyholm SV, Stabb EV, Ruby EG, & McFall-Ngai MJ, Establishment of an animal-bacterial association: recruiting symbiotic vibrios from the environment. Proc. Natl. Acad. Sci. U. S. A 97, 10231–10235 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yip ES, Geszvain K, DeLoney-Marino CR, & Visick KL, The symbiosis regulator RscS controls the syp gene locus, biofilm formation and symbiotic aggregation by Vibrio fischeri. Mol. Microbiol 62, 1586–1600, doi: 10.1111/j.1365-2958.2006.05475.x (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koehler S, et al. The model squid-vibrio symbiosis provides a window into the impact of strain- and species-level differences during the initial stages of symbiont engagement. Environ. Microbiol 21, doi: 10.1111/1462-2920.14392 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morris AR, & Visick KL, Control of biofilm formation and colonization in Vibrio fischeri: a role for partner switching? Environ. Microbiol 12, 2051–2059 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Norsworthy AN, & Visick KL, Gimme shelter: how Vibrio fischeri successfully navigates an animal’s multiple environments. Front. Microbiol 4, 356, doi: 10.3389/fmicb.2013.00356 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shibata S, Yip ES, Quirke KP, Ondrey JM, & Visick KL, Roles of the structural symbiosis polysaccharide (syp) genes in host colonization, biofilm formation, and polysaccharide biosynthesis in Vibrio fischeri. J. Bacteriol 194, 6736–6747, doi: 10.1128/JB.00707-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yip ES, Grublesky BT, Hussa EA, & Visick KL, A novel, conserved cluster of genes promotes symbiotic colonization and sigma-dependent biofilm formation by Vibrio fischeri. Mol. Microbiol 57, 1485–1498, doi: 10.1111/j.1365-2958.2005.04784.x (2005). [DOI] [PubMed] [Google Scholar]
- 33.Bassis CM, & Visick KL, The cyclic-di-GMP phosphodiesterase BinA negatively regulates cellulose-containing biofilms in Vibrio fischeri. J. Bacteriol 192, 1269–1278, doi: 10.1128/JB.01048-09 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chavez-Dozal A, Hogan D, Gorman C, Quintanal-Villalonga A, & Nishiguchi MK, Multiple Vibrio fischeri genes are involved in biofilm formation and host colonization. FEMS Microbiol. Ecol 81, 562–573, doi: 10.1111/j.1574-6941.2012.01386.x (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tischler AH, Lie L, Thompson CM, & Visick KL, Discovery of calcium as a biofilm-promoting signal for Vibrio fischeri reveals new phenotypes and underlying regulatory complexity. J. Bacteriol 200, e00016–18, doi: 10.1128/JB.00016-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]; This article expanded our understanding of the regulatory controls and signals leading to biofilm formation by identifying calcium as a signal that induces a coordinate up-regulation of Syp- and cellulose-dependent biofilm formation and revealing the sensor kinase HahK as a new biofilm regulator.
- 36.Ziemba C, Shabtai Y, Piatkovsky M, & Herzberg M, Cellulose effects on morphology and elasticity of Vibrio fischeri biofilms. NPJ Biofilms Microbiomes 2, 1, doi: 10.1038/s41522-016-0001-2 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ray VA, Driks A, & Visick KL, Identification of a novel matrix protein that promotes biofilm maturation in Vibrio fischeri. J. Bacteriol 197, 518–528, doi: 10.1128/JB.02292-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shibata S, & Visick KL, Sensor kinase RscS induces the production of antigenically distinct outer membrane vesicles That depend on the symbiosis polysaccharide locus in Vibrio fischeri. J. Bacteriol 194, 185–194, doi: 10.1128/JB.05926-11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hussa EA, Darnell CL, & Visick KL, RscS functions upstream of SypG to control the syp locus and biofilm formation in Vibrio fischeri. J. Bacteriol 190, 4576–4583 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mandel MJ, Wollenberg MS, Stabb EV, Visick KL, & Ruby EG, A single regulatory gene is sufficient to alter bacterial host range. Nature 458, 215–218 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ray VA, Eddy JL, Hussa EA, Misale M, & Visick KL, The syp enhancer sequence plays a key role in transcriptional activation by the sigma54-dependent response regulator SypG and in biofilm formation and host colonization by Vibrio fischeri. J. Bacteriol 195, 5402–5412, doi: 10.1128/JB.00689-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Visick KL, & Skoufos LM, Two-component sensor required for normal symbiotic colonization of Euprymna scolopes by Vibrio fischeri. J. Bacteriol 183, 835–842, doi: 10.1128/JB.183.3.835-842.2001 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Norsworthy AN, & Visick KL, Signaling between two interacting sensor kinases promotes biofilms and colonization by a bacterial symbiont. Mol. Microbiol 96, 233–248, doi: 10.1111/mmi.12932 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thompson CM, Marsden AE, Tischler AH, Koo J, & Visick KL, Vibrio fischeri biofilm formation prevented by a trio of regulators. Appl. Environ. Microbiol 84, doi: 10.1128/AEM.01257-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brooks JF 2nd, & Mandel MJ, The histidine kinase BinK Is a negative regulator of biofilm formation and squid colonization. J. Bacteriol 198, 2596–2607, doi: 10.1128/JB.00037-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pankey MS, et al. Host-selected mutations converging on a global regulator drive an adaptive leap by bacteria to symbiosis. eLife 6, doi: 10.7554/eLife.24414 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; Evolutionary pathways that can lead to symbiotic colonization were revealed in this elegant study that followed the serial passage of a non-colonizing strain through many juveniles E. scolopes, resulting in altered, symbiosis-competent strains.
- 47.Morris AR, Darnell CL, & Visick KL, Inactivation of a novel response regulator is necessary for biofilm formation and host colonization by Vibrio fischeri. Mol. Microbiol 82, 114–130, doi: 10.1111/j.1365-2958.2011.07800.x (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morris AR, & Visick KL, The response regulator SypE controls biofilm formation and colonization through phosphorylation of the syp-encoded regulator SypA in Vibrio fischeri. Mol. Microbiol 87, 509–525, doi: 10.1111/mmi.12109 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brooks JF 2nd, et al. Global discovery of colonization determinants in the squid symbiont Vibrio fischeri. Proc. Natl. Acad. Sci. U. S. A 111, 17284–17289, doi: 10.1073/pnas.1415957111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]; This large-scale investigation of colonization factors provides important information on genetic requirements for symbiosis and provides a wealth of data for hypothesis-generation that will foster many subsequent studies.
- 50.Thompson CM, & Visick KL, Assessing the function of STAS domain protein SypA in Vibrio fischeri using a comparative analysis. Front. Microbiol 6, 760, doi: 10.3389/fmicb.2015.00760 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Visick KL, & Skoufos LM, Two-component sensor required for normal symbiotic colonization of Euprymna scolopes by Vibrio fischeri. J. Bacteriol 183, 835–842 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rotman ER, et al. Natural strain variation reveals diverse biofilm regulation in squid-colonizing Vibrio fischeri. J Bacteriol 201, doi: 10.1128/JB.00033-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bongrand C, et al. A genomic comparison of 13 symbiotic Vibrio fischeri isolates from the perspective of their host source and colonization behavior. ISME J. 10, 2907–2917, doi: 10.1038/ismej.2016.69 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study of the genomes and behaviors of a collection of a number of squid symbionts propelled the field from the near-exclusive study of a single isolate, ES114, into new and exciting directions with the genomic sequencing of dominant strains that contain numerous additional genetic sequences and factors.
- 54.Newell PD, Boyd CD, Sondermann H, & O’Toole GA, A c-di-GMP effector system controls cell adhesion by inside-out signaling and surface protein cleavage. PLoS Biol. 9, e1000587, doi: 10.1371/journal.pbio.1000587 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Christensen DG, Marsden AE, Hodge-Hanson K, Essock-Burns T, & Visick KL, LapG mediates biofilm dispersal in Vibrio fischeri by controlling maintenance of the VCBS-containing adhesin LapV. Mol. Microbiol 114, 742–761 doi: 10.1111/mmi.14573 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper addreses a major long-standing question concerning the initiation of the light-organ association; specifically, how do aggregated V. fischeri cells release themselves and migrate into host tissue? One factor may be an adhesin-cleaving protease, which is kept in check by a c-di-GMP-responsive protein, and can promote symbiont dispersal from biofilms.
- 56.Fidopiastis PM, et al. Characterization of a Vibrio fischeri aminopeptidase and evidence for its influence on an early stage of squid colonization. J. Bacteriol 194, 3995–4002, doi: 10.1128/JB.00108-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davidson SK, Koropatnick TA, Kossmehl R, Sycuro L, & McFall-Ngai MJ, NO means ‘yes’ in the squid-vibrio symbiosis: nitric oxide (NO) during the initial stages of a beneficial association. Cellul. Microbiol 6, 1139–1151, doi: 10.1111/j.1462-5822.2004.00429.x (2004). [DOI] [PubMed] [Google Scholar]
- 58.Wang Y, et al. Vibrio fischeri flavohaemoglobin protects against nitric oxide during initiation of the squid-Vibrio symbiosis. Mol. Microbiol 78, 903–915 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stabb EV, Should they stay or should they go? Nitric oxide and the clash of regulators governing Vibrio fischeri biofilm formation. Mol. Microbiol 111, 1–5, doi: 10.1111/mmi.14163 (2019). [DOI] [PubMed] [Google Scholar]
- 60.Thompson CM, Tischler AH, Tarnowski DA, Mandel MJ, & Visick KL, Nitric oxide inhibits biofilm formation by Vibrio fischeri via the nitric oxide sensor HnoX. Mol. Microbiol 111, 187–203, doi: 10.1111/mmi.14147 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; This publication offered insight into the complex role in symbiosis of the squid-produced defense molecule nitric oxide by uncovering its ability to inhibit biofilm formation via the nitric oxide sensor HnoX, a finding that suggests that nitric oxide may influence the location or timing of biofilm formation and/or promote dispersal during symbiotic initiation.
- 61.Singh P, Brooks JF 2nd, Ray VA, Mandel MJ, & Visick KL, CysK plays a role in Biofilm formation and colonization by Vibrio fischeri. Appl. Environ. Microbiol 81, 5223–5234, doi: 10.1128/AEM.00157-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Raina JB, Fernandez V, Lambert B, Stocker R, & Seymour JR, The role of microbial motility and chemotaxis in symbiosis. Nat. Rev. Microbiol 17, 284–294, doi: 10.1038/s41579-019-0182-9 (2019). [DOI] [PubMed] [Google Scholar]
- 63.Brennan CA, DeLoney-Marino CR, & Mandel MJ, Chemoreceptor VfcA mediates amino acid chemotaxis in Vibrio fischeri. Appl. Environ. Microbiol 79, 1889–1896, doi: 10.1128/AEM.03794-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Graf J, Dunlap PV, & Ruby EG, Effect of transposon-induced motility mutations on colonization of the host light organ by Vibrio fischeri. J. Bacteriol 176, 6986–6991 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Millikan DS, & Ruby EG, FlrA, a sigma54-dependent transcriptional activator in Vibrio fischeri, is required for motility and symbiotic light-organ colonization. J. Bacteriol 185, 3547–3557, doi: 10.1128/jb.185.12.3547-3557.2003 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Millikan DS, & Ruby EG, Vibrio fischeri flagellin A is essential for normal motility and for symbiotic competence during initial squid light organ colonization. J. Bacteriol 186, 4315–4325, doi: 10.1128/JB.186.13.4315-4325.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wolfe AJ, Millikan DS, Campbell JM, & Visick KL, Vibrio fischeri sigma54 controls motility, biofilm formation, luminescence, and colonization. Appl. Environ. Microbiol 70, 2520–2524, doi: 10.1128/aem.70.4.2520-2524.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.O’Shea TM, et al. Magnesium promotes flagellation of Vibrio fischeri. J. Bacteriol 187, 2058–2065, doi: 10.1128/JB.187.6.2058-2065.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ruby EG, & Asato LM, Growth and flagellation of Vibrio fischeri during initiation of the sepiolid squid light organ symbiosis. Arch. Microbiol 159, 160–167 (1993). [DOI] [PubMed] [Google Scholar]
- 70.Beeby M, et al. Diverse high-torque bacterial flagellar motors assemble wider stator rings using a conserved protein scaffold. Proc. Natl. Acad. Sci. U. S. A 113, E1917–1926, doi: 10.1073/pnas.1518952113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Deloney-Marino CR, & Visick KL, Role for cheR of Vibrio fischeri in the Vibrio-squid symbiosis. Can. J. Microbiol 58, 29–38, doi: 10.1139/w11-107 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ruby EG, et al. Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc. Natl. Acad. Sci. U. S. A 102, 3004–3009, doi: 10.1073/pnas.0409900102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nikolakakis K, Monfils K, Moriano-Gutierrez S, Brennan CA, & Ruby EG, Characterization of the Vibrio fischeri fatty acid chemoreceptors, VfcB and VfcB2. Appl. Environ. Microbiol 82, 696–704, doi: 10.1128/AEM.02856-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mandel MJ, et al. Squid-derived chitin oligosaccharides are a chemotactic signal during colonization by Vibrio fischeri. Appl. Environ. Microbiol 78, 4620–4626, doi: 10.1128/AEM.00377-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]; While it was long-expected that V. fischeri might sense and be attracted to squid-produced molecules to facilitate directed migration into the light organ crypts, this work was the first to identify squid-produced molecules, chitin oligosaccharides, that function as a chemotactic signal promoting colonization.
- 75.Bennett BD, Essock-Burns T, & Ruby EG, HbtR, a heterofunctional homolog of the virulence regulator TcpP, facilitates the transition between symbiotic and planktonic lifestyles in Vibrio fischeri. mBio 11, doi: 10.1128/mBio.01624-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; Comparisons of V. fischeri with the related pathogen V. cholerae revealed that a regulator conserved among Vibrio spp. plays very different roles in the interactions of these two microorganisms with their respective hosts.
- 76.Brennan CA, et al. A model symbiosis reveals a role for sheathed-flagellum rotation in the release of immunogenic lipopolysaccharide. eLife 3, e01579, doi: 10.7554/eLife.01579 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]; A surprising role for flagellar rotation in the release of LPS molecules that promote squid development was revealed in this work, providing a novel function for the flagellar sheath.
- 77.Stabb EV, & Millikan DS in Defensive mutualism in microbial symbiosis Vol. 27 (eds. White JF, & Torres MS) 85–98 (CRC Press, Boca Raton, 2009). [Google Scholar]
- 78.Bose JL, Rosenberg CS, & Stabb EV, Effects of luxCDABEG induction in Vibrio fischeri: enhancement of symbiotic colonization and conditional attenuation of growth in culture. Arch. Microbiol 190, 169–183, doi: 10.1007/s00203-008-0387-1 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Visick KL, Foster J, Doino J, McFall-Ngai M, & Ruby EG, Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J. Bacteriol 182, 4578–4586 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Verma SC, & Miyashiro T, Niche-specific impact of a symbiotic function on the persistence of microbial symbionts within a natural host. Appl. Environ. Microbiol 82, 5990–5996, doi: 10.1128/AEM.01770-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dunn AK, Millikan DS, Adin DM, Bose JL, & Stabb EV, New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl. Environ. Microbiol 72, 802–810 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Essock-Burns T, Bongrand C, Goldman WE, Ruby EG, & McFall-Ngai MJ, Interactions of symbiotic partners drive the development of a complex biogeography in the squid-vibrio symbiosis. mBio 11, e00853–00820, doi: 10.1128/mBio.00853-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sycuro LK, Ruby EG, & McFall-Ngai M, Confocal microscopy of the light organ crypts in juvenile Euprymna scolopes reveals their morphological complexity and dynamic function in symbiosis. J. Morphol 267, 555–568, doi: 10.1002/jmor.10422 (2006). [DOI] [PubMed] [Google Scholar]
- 84.Koch EJ, Miyashiro T, McFall-Ngai MJ, & Ruby EG, Features governing symbiont persistence in the squid-vibrio association. Mol. Ecol 23, 1624–1634, doi: 10.1111/mec.12474 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wollenberg MS, Preheim SP, Polz MF, & Ruby EG, Polyphyly of non-bioluminescent Vibrio fischeri sharing a lux-locus deletion. Environ. Microbiol 14, 655–668, doi: 10.1111/j.1462-2920.2011.02608.x (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chun CK, et al. Effects of colonization, luminescence, and autoinducer on host transcription during development of the squid-vibrio association. Proc. Natl. Acad. Sci. U. S. A 105, 11323–11328 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McFall-Ngai M, Heath-Heckman EA, Gillette AA, Peyer SM, & Harvie EA, The secret languages of coevolved symbioses: insights from the Euprymna scolopes-Vibrio fischeri symbiosis. Semin. Immunol 24, 3–8, doi: 10.1016/j.smim.2011.11.006 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Moriano-Gutierrez S, et al. Critical symbiont signals drive both local and systemic changes in diel and developmental host gene expression. Proc. Natl. Acad. Sci. U. S. A 116, 7990–7999, doi: 10.1073/pnas.1819897116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Verma SC, & Miyashiro T, Quorum sensing in the squid-Vibrio symbiosis. Int. J. Mol. Sci 14, 16386–16401, doi: 10.3390/ijms140816386 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stabb EV, Schaefer A, Bose JL, & Ruby EG in Chemical communication among bacteria (eds. Winans SC, & Bassler BL) 233–250 (ASM Press, Washington, DC, 2008). [Google Scholar]
- 91.Lupp C, & Ruby EG, Vibrio fischeri uses two quorum-sensing systems for the regulation of early and late colonization factors. J. Bacteriol 187, 3620–3629, doi: 10.1128/JB.187.11.3620-3629.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kimbrough JH, & Stabb EV, Comparative analysis reveals regulatory motifs at the ainS/ainR pheromone-signaling locus of Vibrio fischeri. Sci. Rep 7, 11734, doi: 10.1038/s41598-017-11967-7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kimbrough JH, & Stabb EV, Substrate specificity and function of the pheromone receptor AinR in Vibrio fischeri ES114. J. Bacteriol 195, 5223–5232, doi: 10.1128/JB.00913-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Studer SV, Mandel MJ, & Ruby EG, AinS quorum sensing regulates the Vibrio fischeri acetate switch. J. Bacteriol 190, 5915–5923, doi: 10.1128/JB.00148-08 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cao X, et al. The novel sigma factor-like regulator RpoQ controls luminescence, chitinase activity, and motility in Vibrio fischeri. mBio 3, doi: 10.1128/mBio.00285-11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Studer SV, et al. Non-native acylated homoserine lactones reveal that LuxIR quorum sensing promotes symbiont stability. Environ. Microbiol 16, 2623–2634, doi: 10.1111/1462-2920.12322 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Boettcher KJ, & Ruby EG, Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J. Bacteriol 172, 3701–3706 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Septer AN, & Stabb EV, Coordination of the arc regulatory system and pheromone-mediated positive feedback in controlling the Vibrio fischeri lux operon. PLoS One 7, e49590, doi: 10.1371/journal.pone.0049590 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Stabb EV, Could positive feedback enable bacterial pheromone signaling to coordinate behaviors in response to heterogeneous environmental cues? mBio 9, doi: 10.1128/mBio.00098-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bose JL, et al. Bioluminescence in Vibrio fischeri is controlled by the redox-responsive regulator ArcA. Mol. Microbiol 65, 538–553, doi: 10.1111/j.1365-2958.2007.05809.x (2007). [DOI] [PubMed] [Google Scholar]
- 101.Lyell NL, et al. Cyclic AMP receptor protein regulates pheromone-mediated bioluminescence at multiple levels in Vibrio fischeri ES114. J. Bacteriol 195, 5051–5063, doi: 10.1128/JB.00751-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lyell NL, Dunn AK, Bose JL, & Stabb EV, Bright mutants of Vibrio fischeri ES114 reveal conditions and regulators that control bioluminescence and expression of the lux operon. J. Bacteriol 192, 5103–5114 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Septer AN, Lyell NL, & Stabb EV, The iron-dependent regulator fur controls pheromone signaling systems and luminescence in the squid symbiont Vibrio fischeri ES114. Appl. Environ. Microbiol 79, 1826–1834, doi: 10.1128/AEM.03079-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Stoudenmire JL, et al. An iterative, synthetic approach to engineer a high-performance PhoB-specific reporter. Appl. Environ. Microbiol 84, doi: 10.1128/AEM.00603-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study not only provided a roadmap for synthetic promoter engineering in V. fischeri, but it also uncovered evidence for possible micro-environments present within different crypts of the E. scolopes light organ.
- 105.Bose JL, et al. Contribution of rapid evolution of the luxR-luxI intergenic region to the diverse bioluminescence outputs of Vibrio fischeri strains isolated from different environments. Appl. Environ. Microbiol 77, 2445–2457 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dunn AK, Vibrio fischeri metabolism: symbiosis and beyond. Adv. Microb. Physiol 61, 37 (2012). [DOI] [PubMed] [Google Scholar]
- 107.Schwartzman JA, & Ruby EG, A conserved chemical dialog of mutualism: lessons from squid and vibrio. Microbes Infect. 18, 1–10, doi: 10.1016/j.micinf.2015.08.016 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pan S, et al. Model-enabled gene search (MEGS) allows fast and direct discovery of enzymatic and transport gene functions in the marine bacterium Vibrio fischeri. J. Biol. Chem 292, 10250–10261, doi: 10.1074/jbc.M116.763193 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Thompson LR, et al. Transcriptional characterization of Vibrio fischeri during colonization of juvenile Euprymna scolopes. Environ. Microbiol 19, 1845–1856, doi: 10.1111/1462-2920.13684 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wier AM, et al. Transcriptional patterns in both host and bacterium underlie a daily rhythm of anatomical and metabolic change in a beneficial symbiosis. Proc. Natl. Acad. Sci. U. S. A 107, 2259–2264 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]; In the first dual transcriptional study of an animal host and its symbionts, gene expression in both partners was shown to be regulated over a day/night cycle, revealing a daily remodeling of the crypt epithelial cells, and a night-time provision of chitin to the symbionts.
- 111.Sun Y, Verma SC, Bogale H, & Miyashiro T, NagC represses N-acetyl-glucosamine utilization genes in Vibrio fischeri within the light organ of Euprymna scolopes. Front. Microbiol 6, 741, doi: 10.3389/fmicb.2015.00741 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wasilko NP, et al. Sulfur availability for Vibrio fischeri growth during symbiosis establishment depends on biogeography within the squid light organ. Mol. Microbiol 111, 621–636, doi: 10.1111/mmi.14177 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shed light on both the nutritional adaptability of V. fischeri and the complex biogeography of the light organ by demonstrating that this symbiont uses different sulfur sources within different regions of the light organ.
- 113.Septer AN, et al. Bright luminescence of Vibrio fischeri aconitase mutants reveals a connection between citrate and the Gac/Csr regulatory system. Mol. Microbiol 95, 283–296, doi: 10.1111/mmi.12864 (2015). [DOI] [PubMed] [Google Scholar]
- 114.Lyell NL, & Stabb EV, Symbiotic characterization of Vibrio fischeri ES114 mutants that display enhanced luminescence in culture. Appl. Environ. Microbiol 79, 2480–2483, doi: 10.1128/AEM.03111-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lyell NL, et al. An expanded transposon mutant library reveals that Vibrio fischeri delta-aminolevulinate auxotrophs can colonize Euprymna scolopes. Appl. Environ. Microbiol 83, doi: 10.1128/AEM.02470-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Colton DM, Stoudenmire JL, & Stabb EV, Growth on glucose decreases cAMP-CRP activity while paradoxically increasing intracellular cAMP in the light-organ symbiont Vibrio fischeri. Mol. Microbiol 97, 1114–1127, doi: 10.1111/mmi.13087 (2015). [DOI] [PubMed] [Google Scholar]
- 117.Miyashiro T, et al. The N-acetyl-D-glucosamine repressor NagC of Vibrio fischeri facilitates colonization of Euprymna scolopes. Mol. Microbiol 82, 894–903, doi: 10.1111/j.1365-2958.2011.07858.x (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Adin DM, Visick KL, & Stabb EV, Identification of a cellobiose utilization gene cluster with cryptic beta-galactosidase activity in Vibrio fischeri. Appl. Environ. Microbiol 74, 4059–4069, doi: 10.1128/AEM.00190-08 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dunn AK, Vibrio fischeri metabolism: symbiosis and beyond. Adv. Microb. Physiol 61, 37–68, doi: 10.1016/B978-0-12-394423-8.00002-0 (2012). [DOI] [PubMed] [Google Scholar]
- 120.Pan M, Schwartzman JA, Dunn AK, Lu Z, & Ruby EG, A single host-derived glycan impacts key regulatory nodes of symbiont metabolism in a coevolved mutualism. mBio 6, e00811, doi: 10.1128/mBio.00811-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Boettcher KJ, McFall-Ngai MJ, & Ruby EG, Bioluminescence in the symbiotic squid Euprymna scolopes is controlled by a daily biological rhythm. J. Comp. Physiol 179, 65–73 (1996). [Google Scholar]
- 122.Stabb EV, Shedding light on the bioluminescence “paradox”. ASM News 71, 223–229 (2005). [Google Scholar]
- 123.Septer AN, Bose JL, Dunn AK, & Stabb EV, FNR-mediated regulation of bioluminescence and anaerobic respiration in the light-organ symbiont Vibrio fischeri. FEMS Microbiol. Lett 306, 72–81 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dunn AK, Alternative oxidase activity reduces stress in Vibrio fischeri cells exposed to nitric oxide. J. Bacteriol 200, doi: 10.1128/JB.00797-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dunn AK, & Stabb EV, Genetic analysis of trimethylamine N-oxide reductases in the light organ symbiont Vibrio fischeri ES114. J. Bacteriol 190, 5814–5823 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Septer AN, Wang Y, Ruby EG, Stabb EV, & Dunn AK, The haem-uptake gene cluster in Vibrio fischeri is regulated by Fur and contributes to symbiotic colonization. Environ. Microbiol 13, 2855–2864, doi: 10.1111/j.1462-2920.2011.02558.x (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Graf J, & Ruby EG, Novel effects of a transposon insertion in the Vibrio fischeri glnD gene: defects in iron uptake and symbiotic persistence in addition to nitrogen utilization. Mol. Microbiol 37, 168–179, doi: 10.1046/j.1365-2958.2000.01984.x (2000). [DOI] [PubMed] [Google Scholar]
- 128.Eickhoff MJ, & Bassler BL, Vibrio fischeri siderophore production drives competitive exclusion during dual-species growth. Mol. Microbiol 114, 244–261 doi: 10.1111/mmi.14509 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ferretti P, et al. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe 24, 133–145 e135, doi: 10.1016/j.chom.2018.06.005 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zheng H, et al. Division of labor in honey bee gut microbiota for plant polysaccharide digestion. Proc. Natl. Acad. Sci. U. S. A 116, 25909–25916, doi: 10.1073/pnas.1916224116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Aschtgen MS, et al. Rotation of Vibrio fischeri flagella produces outer membrane vesicles that induce host development. J. Bacteriol 198, 2156–2165, doi: 10.1128/JB.00101-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Aschtgen MS, Wetzel K, Goldman W, McFall-Ngai M, & Ruby E, Vibrio fischeri-derived outer membrane vesicles trigger host development. Cellul. Microbiol 18, 488–499, doi: 10.1111/cmi.12525 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lynch JB, et al. Ambient pH alters the protein content of outer membrane vesicles, driving host development in a beneficial symbiosis. J. Bacteriol 201, doi: 10.1128/JB.00319-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Franzenburg S, et al. Distinct antimicrobial peptide expression determines host species-specific bacterial associations. Proc. Natl. Acad. Sci. U. S. A 110, E3730–3738, doi: 10.1073/pnas.1304960110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chen F, et al. Bactericidal permeability-increasing proteins shape host-microbe interactions mBio 8, e00040–00017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Heath-Heckman EA, et al. Shaping the microenvironment: evidence for the influence of a host galaxin on symbiont acquisition and maintenance in the squid-Vibrio symbiosis. Environ. Microbiol 16, 3669–3682, doi: 10.1111/1462-2920.12496 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wang Y, et al. H-NOX-mediated nitric oxide sensing modulates symbiotic colonization by Vibrio fischeri. Proc. Natl. Acad. Sci. U. S. A 107, 8375–8380 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Schwartzman JA, et al. Acidic pH promotes lipopolysaccharide modification and alters colonization in a bacteria-animal mutualism. Mol. Microbiol 112, 1326–1338, doi: 10.1111/mmi.14365 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Schwartzman JA, & Ruby EG, Stress as a normal cue in the symbiotic environment. Trends Microbiol. 24, 414–424, doi: 10.1016/j.tim.2016.02.012 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kwong WK, & Moran NA, Gut microbial communities of social bees. Nat. Rev. Microbiol 14, 374–384, doi: 10.1038/nrmicro.2016.43 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Murfin KE, et al. Xenorhabdus bovienii strain diversity impacts coevolution and symbiotic maintenance with Steinernema spp. nematode hosts. mBio 6, e00076, doi: 10.1128/mBio.00076-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wollenberg MS, & Ruby EG, Population structure of Vibrio fischeri within the light organs of Euprymna scolopes squid from Two Oahu (Hawaii) populations. Appl. Environ. Microbiol 75, 193–202 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]; The first comparative genome-level study of light organ symbionts both between and within adult squid, suggesting that on average each crypt of an organ is colonized by one or two V. fischeri cells, potentially creating crypt-separated, clonal lineages within a polyclonal organ.
- 143.Tomich M, Planet PJ, & Figurski DH, The tad locus: postcards from the widespread colonization island. Nat. Rev. Microbiol 5, 363–375, doi: 10.1038/nrmicro1636 (2007). [DOI] [PubMed] [Google Scholar]
- 144.Gyllborg MC, Sahl JW, Cronin DC 3rd, Rasko DA, & Mandel MJ, Draft genome sequence of Vibrio fischeri SR5, a strain isolated from the light organ of the Mediterranean squid Sepiola robusta. J. Bacteriol 194, 1639, doi: 10.1128/JB.06825-11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bongrand C, et al. Using colonization assays and comparative genomics to discover symbiosis behaviors and factors in Vibrio fischeri. mBio 11, doi: 10.1128/mBio.03407-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Coryell RL, et al. Phylogeographic patterns in the Philippine archipelago influence symbiont diversity in the bobtail squid-Vibrio mutualism. Ecol. Evol 8, 7421–7435, doi: 10.1002/ece3.4266 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Soto W, Rivera FM, & Nishiguchi MK, Ecological diversification of Vibrio fischeri serially passaged for 500 generations in novel squid host Euprymna tasmanica. Microb. Ecol 67, 700–721, doi: 10.1007/s00248-013-0356-3 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Soto W, Travisano M, Tolleson AR, & Nishiguchi MK, Symbiont evolution during the free-living phase can improve host colonization. Microbiology 165, 174–187, doi: 10.1099/mic.0.000756 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Fidopiastis PM, von Boletzky S, & Ruby EG, A new niche for Vibrio logei, the predominant light organ symbiont of squids in the genus Sepiola. J. Bacteriol 180, 59–64 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Dillon MM, Sung W, Lynch M, & Cooper VS, Periodic variation of mutation rates in bacterial genomes associated with replication timing. mBio 9, doi: 10.1128/mBio.01371-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Dillon MM, Sung W, Sebra R, Lynch M, & Cooper VS, Genome-wide biases in the rate and molecular spectrum of spontaneous mutations in Vibrio cholerae and Vibrio fischeri. Mol. Biol. Evol 34, 93–109, doi: 10.1093/molbev/msw224 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wollenberg MS, & Ruby EG, Phylogeny and fitness of Vibrio fischeri from the light organs of Euprymna scolopes in two Oahu, Hawaii populations. ISME J. 6, 352–362, doi: 10.1038/ismej.2011.92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Koch EJ, et al. The cytokine MIF controls daily rhythms of symbiont nutrition in an animal-bacterial association. Proc. Natl. Acad. Sci. U. S. A 117, 27578–27586, doi: 10.1073/pnas.2016864117 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sun Y, et al. Intraspecific competition impacts Vibrio fischeri strain diversity during initial colonization of the squid light organ. Appl. Environ. Microbiol 82, 3082–3091, doi: 10.1128/AEM.04143-15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Speare L, et al. Bacterial symbionts use a type VI secretion system to eliminate competitors in their natural host. Proc. Natl. Acad. Sci. U. S. A 115, E8528–E8537, doi: 10.1073/pnas.1808302115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]; The finding that V. fischeri engages in biological ‘warfare’ to become the sole colonizer of a given crypt has provided new insight into the dynamics and processes controlling light organ population structure and strain competition in nature.
- 156.Speare L, Smith S, Salvato F, Kleiner M, & Septer AN, Environmental viscosity modulates interbacterial killing during habitat transition. mBio 11, doi: 10.1128/mBio.03060-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Guckes KR, et al. Incompatibility of Vibrio fischeri strains during symbiosis establishment depends on two functionally redundant hcp genes. J. Bacteriol 201, doi: 10.1128/JB.00221-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Guckes KR, Cecere AG, Williams AL, McNeil AE, & Miyashiro T, The bacterial enhancer binding protein VasH promotes expression of a Type VI secretion system in Vibrio fischeri during symbiosis. J. Bacteriol 202, doi: 10.1128/JB.00777-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bultman KM, Cecere AG, Miyashiro T, Septer AN, & Mandel MJ, Draft genome sequences of type VI secretion system-encoding Vibrio fischeri strains FQ-A001 and ES401. Microbiol. Resour. Announc 8, doi: 10.1128/MRA.00385-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Doino JA, & McFall-Ngai MJ, A transient exposure to symbiosis-competent bacteria induces light organ morphogenesis in the host squid. Biol. Bull 189, 347–355, doi: 10.2307/1542152 [DOI] [PubMed] [Google Scholar]
- 161.Dunn AK, Martin MO, & Stabb EV, Characterization of pES213, a small mobilizable plasmid from Vibrio fischeri. Plasmid 54, 114–134, doi: 10.1016/j.plasmid.2005.01.003 (2005). [DOI] [PubMed] [Google Scholar]
- 162.Lyell NL, Dunn AK, Bose JL, Vescovi SL, & Stabb EV, Effective mutagenesis of Vibrio fischeri by using hyperactive mini-Tn5 derivatives. Appl. Environ. Microbiol 74, 7059–7063 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Stoudenmire JL, Black M, Fidopiastis PM, & Stabb EV, Mutagenesis of Vibrio fischeri and other marine bacteria using hyperactive mini-Tn5 derivatives. Methods Mol. Biol 2016, 87–104, doi: 10.1007/978-1-4939-9570-7_9 (2019). [DOI] [PubMed] [Google Scholar]
- 164.Pollack-Berti A, Wollenberg MS, & Ruby EG, Natural transformation of Vibrio fischeri requires tfoX and tfoY. Environ. Microbiol 12, 2302–2311, doi: 10.1111/j.1462-2920.2010.02250.x (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Visick KL, Hodge-Hanson KM, Tischler AH, Bennett AK, & Mastrodomenico V, Tools for rapid genetic engineering of Vibrio fischeri. Appl. Environ. Microbiol 84, doi: 10.1128/AEM.00850-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Brooks JF 2nd, Gyllborg MC, Kocher AA, Markey LE, & Mandel MJ, TfoX-based genetic mapping identifies Vibrio fischeri strain-level differences and reveals a common lineage of laboratory strains. J. Bacteriol 197, 1065–1074, doi: 10.1128/JB.02347-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Califano G, et al. Draft genome sequence of Aliivibrio fischeri strain 5LC, a bacterium retrieved from gilthead seabream (Sparus aurata) larvae reared in aquaculture. Genome Announc 3, e00593–15, doi: 10.1128/genomeA.00593-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hehemann J-H, et al. Adaptive radiation by waves of gene transfer leads to fine-scale resource partitioning in marine microbes. Nat. Commun 7, 12860, doi: 10.1038/ncomms12860 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Nikolakakis K, Lehnert E, McFall-Ngai MJ, & Ruby EG, Use of hybridization chain reaction-fluorescent in situ hybridization to track gene expression by both partners during initiation of symbiosis. Appl. Environ. Microbiol 81, 4728–4735, doi: 10.1128/AEM.00890-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]


