Abstract
High-risk neuroblastoma (HR-NB) is branded with hematogenous metastasis, relapses, and dismal long-term survival. Intensification of consolidation therapy with tandem/triple autologous stem-cell (SC) rescue (with bone-marrow [BM]/ peripheral-blood [PB] CD34+ selection) after myeloablative chemotherapy has improved long-term survival. However, the benefit is limited by the indication of NB cells in CD34+ PBSCs, CD34-expression in NB cells, and the risk of reinfusing NB-cancer stem cells (NB-CSCs) that could lead to post-transplant relapse. We investigated the association of CD34 surface expression (92 patients) with NB evolution/clinical outcomes. CD34 gene-level status in NB was assessed through RNA-Seq data mining (18 cohorts, n=3,324). Genetic landscape of CD34-expressing NB-CSCs (CD133+CD34+) was compared with CD34− CSCs (CD133+CD34−). RNA-seq data revealed equivocal association patterns of CD34 expression with patient survival. Our IHC-data revealed definite, but rare (Mean=0.73%; Range 0.00-7.87%; Median=0.20%) CD34 positivity in NB. CD34+ significantly associated with MYCN amplification (p=0.003), advanced disease stage (P=0.016), and progressive disease (PD, p<0.0009) after clinical therapy. A general high-is-worse tendency was observed in patients with relapsed disease. High CD34+ correlated with poor survival in patients with N-MYC-amplified HR-NB. Gene expression analysis of CD34+-NB-CSCs identified significant up- (4,631) and down-modulation (4,678) of genes compared with NB-CSCs that lack CD34. IPA recognized the modulation of crucial signaling elements (EMT, stemness-maintenance, differentiation, inflammation, clonal-expansion, drag-resistance, metastasis) that orchestrate NB disease evolution in CD34+-CSCs compared with CD34−CSCs. While the function of CD34 in NB evolution requires further in-depth investigation, careful consideration should be exercised for autologous stem-cell rescue with CD34+ selection in NB patients.
Keywords: Neuroblastoma, CD34, Cancer stem cells, Stem cell rescue, Disease evolution, Tumor progression
Graphical abstract
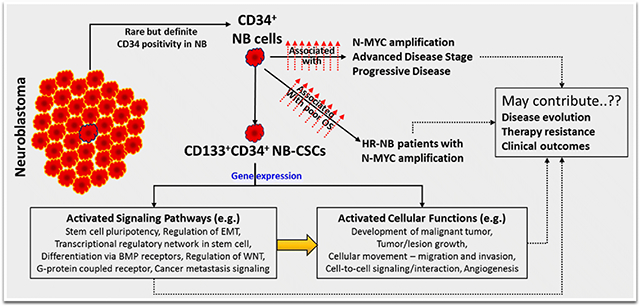
INTRODUCTION
Neuroblastoma (NB), the most common cancer at infancy, (Gurney., Smith. & Ross., 1999; Marc et al., 1999) profoundly contributes to childhood cancer deaths (American-Cancer-Society., 2013; Morgenstern, Baruchel & Irwin, 2013; Smith et al., 2010). Clinically, NB progression is branded with hematogeneous metastasis and frequent relapses, with a rapidly decreasing timeline (Santana et al., 2008; Simon et al., 2011). The current intensive multi-modal clinical therapy (IMCT) for high-risk NB comprises induction phase with alternating regimens of high-dose chemotherapeutic drugs (cyclophosphamide, cisplatin, vincristine, doxorubicin, etoposide, topotecan) and load reduction surgery; consolidation phase with more intensive chemotherapy (carboplatin, etoposide, topotecan, bisulfan and melphalan, thiotepa), along with radiotherapy and stem cell transplant (autologous bone marrow transplantation, ABMT; peripheral blood stem cell reinfusion); and maintenance phase with 13-cis-retinoic acid treatment, immunotherapy (dinutuximab), and immune-activating cytokine (GM-CSF, IL-2) treatment (www.cancer.org/cancer/neuroblastoma/treating.html; www.childrensoncologygroup.org/index.php/in-treatment-for-neuroblastoma). Intensification of consolidation therapy with autologous stem-cell rescue after myeloablative chemotherapy with/without radiotherapy has resulted in relatively improved patient survival (Frappaz et al., 2000; Matthay et al., 1999). Further, for the treatment of high-risk NB, studies have indicated the benefit of tandem or triple autologous stem-cell rescue (in combination with the high-dose therapy) with encouraging long-term survival and, slashed CNS relapse and secondary malignancies (George et al., 2006; Kletzel et al., 2002). However, the benefit of such a major therapeutic advancement is limited by the risk of reinfusing NB cancer cells that could lead to and/or attributable to the post-transplant relapse (Moss et al., 1994; Rill et al., 1994). Such limitation is realized both in hematopoietic stem cell and in peripheral blood stem cells (PBSCs) transplantations (Moss et al., 1994; Rill et al., 1994). In light of sidestepping graft contamination, many ex vivo purging techniques, including negative selection with tumor cell killing toxic concentrations of chemotherapy, monoclonal antibodies targeting neuroblasts, transgenic virus directed enzyme-prodrug therapy, and positive selection of hematopoietic stem cells using specific anti-CD34+ monoclonal antibodies, have been adopted (Marabelle et al., 2011). The feasibility of CD34+ cell immunoselection, its purging efficacy against contaminating neuroblasts, the ability of such CD34+ grafts in reconstitution and maintenance of hematopoiesis, and its benefit in long-term clinical outcomes have been extensively investigated (Berger et al., 1997; Kanold et al., 2003; Marabelle et al., 2011).
As the part of current multi-modal therapy, CD34+ bone marrow or peripheral blood stem cell selection is widely used with increasing frequency for transplantation in patients with NB (Illhardt et al., 2018; Ning et al., 2016; Salazar-Riojas et al., 2015; Seif et al., 2013; Veljkovic et al., 2013). Development of rapid isolation approaches for CD34+ hematopoietic stem cell selection accentuates its clinical utility (Fritsch et al., 1993; Lebkowski et al., 1992; Salazar-Riojas et al., 2015; Veljkovic et al., 2013). However, studies indicated the detection of NB cells in CD34+ selected peripheral blood stem cells (Lode et al., 1997). In addition, researchers recognized the surface membrane expression of CD34 in NB cells and further identified relatively stronger CD34 expression in clonogenic, less differentiated, non-adherent NB cells when compared with the adherent NB cells (Hafer et al., 1999; Leung et al., 1998; Tchirkov et al., 1998; Voigt et al., 1997). These observations indicate that caution is warranted for transplants obtained by CD34+ stem cell selection, at least for the NB setting. However, the criticality of CD34+ NB cell contamination in tumor relapse after autologous stem cell transplantation remains debatable with conflicting results (Choi et al., 2005; Corrias et al., 2006; Hafer et al., 1999; Handgretinger et al., 2003; Hansford et al., 2007; Leung et al., 1998; Lode et al., 1997; Tchirkov et al., 1998). Although heightened expression of CD34 was indicated in clonally active, less differentiated, non-adherent neuroblastoma cells (Hafer et al., 1999; Leung et al., 1998; Tchirkov et al., 1998; Voigt et al., 1997), the physiognomies of CD34+ NB clones and their relevance/capabilities in disease initiation and/or evolution remain unexplored.
Our recent investigation agrees with previous studies (Hafer et al., 1999; Leung et al., 1998; Tchirkov et al., 1998; Voigt et al., 1997), and recognized the existence and/or selection of CD34+ clones in a metastatic site-derived neuroblastoma cancer stem cell (NB-CSC) population (Khan et al., 2015b). Hence, hypothetically, CD34+ NB-CSC clones may harbor elite genetic rearrangements and could thereby contribute to disease evolution. Thus far, the availability and magnification of CD34 surface expression in human NB and its relevance in disease progression and/or clinical outcomes is not realized. Likewise, the genetic and molecular physiognomies of the CD34-expressing NB cancer stem cells (NB-CSCs) and their probable significance in NB disease pathogenesis are largely unknown. Therefore, we aimed to investigate (i) the surface expression blueprint of CD34 positivity in human NB and its significance in clinical outcomes, (ii) association of CD34 with disease evolution (progression, dissemination, and therapy resistance), and (iii) the gene expression rearrangements in CD34-expressing NB-CSCs (CD34+NB-CSCs) compared with NB-CSCs that lack CD34 (CD34−CD133+-NB-CSCs) and their association with disease evolution.
MATERIALS AND METHODS
CD34 immunohistochemistry (IHC) in human NB:
We examined 119 specimens from 92 cases of human NB collected from our institutional (University of Oklahoma Health Sciences Center, OUHSC) pediatrics pathology collection. Cooperative Human Tissue Network (CHTN), and the Oregon Health and Science University Biospecimen core. Table 1 lists patient demographics. All protocols were approved by the OUHSC Institutional Review Board (IRB), with permission for the research use of de-identified specimens. All experiments were performed in accordance with OUHSC IRB guidelines and regulations for the protection of human subjects. Hematoxylin-Eosin (H&E)-stained sections were examined. Only the cases with sufficient percent tumor (with minimal necrosis) and adequate tumor volume for tissue microarray (TMA) construction were included. TMA construction and CD34 IHC was performed in the Tissue Pathology Core of the Stephenson Cancer Center, as described in our earlier studies. (Khan et al., 2015a; Khan et al., 2015b; Somasundaram et al., 2019) The cores (1 mm, in triplicate) from 10% NBF-fixed and paraffin-embedded (FFPE) tissue blocks were randomly arrayed with no predictability in the recipient paraffin block. Four-μm-thick sections from the TMAs were baked for IHC. Immunohistochemical staining was performed using a Leica BOND-III fully automated staining system® (Leica Biosystem Inc., Buffalo Grove, IL). Deparaffinized TMA sections were rehydrated, neutralized (0.25 M Tris-HCl), blocked (0.5% BSA-2% FBS), and labeled with 1 μg purified anti-human mouse monoclonal CD34 antibody (Biolegend, San Diego, CA) under humidified conditions. Appropriate negative (no primary antibody) controls were examined in parallel. The slides were micro-digitally scanned using an Aperio Scanscope (Aperio Technologies, Inc., Buffalo Grove, IL) slide scanner, and the virtual slide was constructed with digital histology. This process allows for the assembly of tissue collections in a TMA with variable magnifications. The digital images were then group analyzed with appropriate algorithms for membrane-specific CD34 positivity using Aperio integrated Spectrum (Aperio) software. To avoid equivocal representations, we considered only strong surface expression positivity for analysis, and presented as percent total cells. Group-wise comparisons and survival analysis (log rank tests, Kaplan-Meier plots) were performed using GraphPad Prism (version 8.3.1, GraphPad Software, San Diego, CA).
Table 1.
Demographics of neuroblastoma patients.
| Total cases | (n=92) | |||
|---|---|---|---|---|
| Total specimens | (n=119) | |||
| Gender | Male (n = 51) | Female (n = 41) | ||
| Age | <2Y or =2Y (n = 47) | >2Y (n = 45) | ||
| N-Myc Status | Amplified (n = 15) | Not-amplified (n = 72) | ||
| INSS Stage | I (n = 21) | II (n = 12) | III (n = 16) | IV (n = 43) |
| Tumor site | Primary (n = 47) | Metastatic (n = 43) | Unknown (n = 29) | |
| Relapse | No Relapse (n = 53) | Relapsed (n = 20) | Unknown (n = 19) | |
| Survival | Live (n = 55) | Dead (n = 29) | Unknown (n = 8) | |
| Disease status | Diagnosis (n = 51) | PD-IMCT (n = 47) | Dx & PD from same case (n = 9) | |
Cell culture and whole genome gene expression:
The human NB (SH-SY5Y) cell line obtained from ATCC (Manassas, VA) was cultured and maintained as described earlier (Khan et al., 2015b; Veeraraghavan et al., 2011). NB-CSCs (unsorted, CD133+CD34−, CD133+CD34+) derived and characterized earlier(Khan et al., 2015a; Khan et al., 2015b; Pandian et al., 2015) were cultured and maintained in stem cell medium (DMEM:F12 with 1% N2 Supplement, 2% B27 supplement, 20 ng/ml hPDGF, 100 ng/ml EGF, and 1% antibiotic-antimycotic). Whole genome gene expression analysis was performed in SH-SY5Y, unsorted NB-CSCs, CD133+CD34− NB-CSCs, and CD133+CD34+ NB-CSCs using the Human V2 SurePrint G3 Unrestricted GE 8x60K gene expression microarray (Agilent Technologies, Inc., Santa Clara, CA). In brief, RNA from NB-CSCs was extracted using RNeasy mini spin columns (RNeasy Mini Kit, Qiagen, German town, MD) following the manufacturer’s instructions and quality/quantity controlled using a bioanalyzer (Agilent Technologies). 100 ng of each RNA sample was used for the linear T7-based amplification step. Cy3-labelled cRNA was generated from total RNA using the One Color Low Input Quick Amp Labeling (Agilent Technologies) following the manufacturer’s protocol. Equal amounts of purified Cy3-labelled cRNA were hybridized according to the Agilent microarray processing protocol (Gene Expression Hybridization Kit, Agilent Technologies) for 17 h at 65°C to Agilent SurePrint G3 Human GE 8 × 60K V2 microarrays. The microarrays were washed using the Gene Expression Wash Buffer Kit (Agilent) and scanned using a high-resolution microarray scanner (G2600D, Agilent Technologies, Palo Alto, USA). The data were processed using Agilent Feature Extraction software (version 11.0.1) and analyzed using the Agilent GeneSpring GX version 12.5 software (Agilent). All data were background-corrected, normalized between arrays (quantile-normalization), and transformed to log2-scale. After normalization, filtering low-quality probes was based on quality flags set by the FES. The processed and filtered data were then subjected to between sample comparisons using GraphPad Prism.
Functional annotation of modulated genes in CD34+ NB-CSCs with Ingenuity pathway analysis:
To investigate the functional relevance of modulated genes in CD34 surface-expressing NB-CSCs, we utilized ingenuity pathway analysis (IPA). Only the genes that displayed significant (≥2 fold) up and down modulation were included for the analysis. Gene probe ID with corresponding expression levels were first annotated. The downstream analysis was restricted only to those mapped (defined or predicted data availability) by the IPA. Core analysis was performed with criteria including direct relationship with causal path scoring for networks and upstream regulator analysis and experimentally observed confidence level. Significant associations of the genes with diseases, molecular and cellular functions, and canonical pathways and networks were examined.
In silico data mining to assess the association of CD34 transcription with clinical outcomes:
We examined the correlation of CD34 transcription with overall survival, relapse-free survival, disease stage, prognosis, and patient status. For this, we used the R2: a web-based microarray analysis and visualization platform (http://r2.amc.nl. by Dr. Koster, Department of Oncogenomics, Academic Medical Center, Amsterdam, Netherlands) that allows us to correlate select gene expression profile with clinical outcomes for various patient cohorts that were submitted by individual investigators. Moreover, R2 is the only application that holds the comprehensive collection of NB studies with RNA-seq or microarray data. Eighteen individual studies (combined n=3,324) were examined for the association of CD34 with disease progression and clinical outcomes.
RESULTS
CD34 transcription in NB displays dubious association with disease evolution and clinical outcomes:
Eighteen patient cohorts were independently assayed for CD34 correlation with advanced disease stage (INSS) and clinical outcomes (overall survival, OS; Relapse-free survival, RFS) in overall, N-MYC-amplified, or N-MYC non-amplified patient subsets (Table 2). A direct correlation of CD34 with advanced disease stage was observed in overall (GSE27608, n=47; GSE54720, n=23; TARGET161, n=161 [Fig S1a]). N-MYC-amplified (TARGET161, n = 161; TARGET249, n = 249; GSE54720, n = 23; GSE85047, n=283) and in N-MYC non-amplified (GSE45547, n = 649) patient cohorts. An independent study (GSE12460, n = 64) showed an inverse relationship of CD34 levels with disease progression. While some studies (GSE16476, n=88; GSE3446, n=102; GSE85047, n=283) showed positive association with poor OS and RFS (Fig S1b–d), others (GSE62564, n=498; NB251, n=251; GSE45547, n=649) displayed inverse correlation with poor OS and RFS. A number of other independent studies (e.g. TARGET 249; n=249; GSE49710, n=498) exhibited MYCN status-specific mixed associations with OS or RFS. A handful of studies displayed no significant association (direct or inverse) of CD34 expression with disease progression and clinical outcomes (Table 2). Overall, these observations from in silico data mining suggest no agreement across NB patient cohorts. However, these findings could indicate that fluctuations in CD34 transcription may effect tumor evolution and poor clinical outcomes (Fig S1).
Table 2.
In silico analysis of gene expression profiles in NB patients from various independent studies showing differential association of CD34 gene levels with NB advanced disease stage and clinical outcomes. The results of these gene expression (RNA sequencing, microarray) studies did not conclusively agree and affirm beneficial or detrimental effects of CD34 expression in NB. Momentous differences were observed in terms of patient cohorts, platform used, MYCN status, and disease status.
| Geo ID | n | OS All |
OS MA |
OS MN |
RFS All |
RFS MA |
RFS MN |
ADSAll | ADSMA | ADSMN |
|---|---|---|---|---|---|---|---|---|---|---|
| GSE16476 | 88 | HW | X | HW | HW | X | HW | X | X | X |
| GSE3446 | 102 | HW | ||||||||
| TARGET161 | 161 | D | D | D | ||||||
| TARGET249 | 249 | LW | HW | LW | HW | HW | LW | X | D | X |
| GSE27608 | 47 | D | ||||||||
| GSE73517 | 105 | X | X | X | ||||||
| NBpreter24 | 24 | X | X | X | ||||||
| GSE49710 | 498 | LW | HW | LW | LW | HW | LW | X | X | X |
| GSE62564 | 498 | LW | LW | LW | LW | X | X | |||
| NB251 | 251 | LW | LW | LW | LW | LW | LW | X | X | X |
| GSE3960 | 101 | X | X | |||||||
| GSE54720 | 23 | D | D | X | ||||||
| GSE13136 | 30 | X | X | |||||||
| GSE45547 | 649 | LW | LW | LW | LW | LW | LW | X | X | D |
| GSE19274 | 100 | X | X | X | ||||||
| GSE16237 | 51 | X | X | X | ||||||
| GSE12460 | 64 | I | X | I | ||||||
| GSE85047 | 283 | HW | HW | HW | HW | LW | LW | X | D | X |
n = number of patients; OS = overall survival; RFS = Relapse-free survival; MA = N-MYC amplified; MN = N-MYC non-amplified; ADS = Advanced disease stage; HW = High is Worse; LW = Low is Worse; D = Direct association; I = Inverse Association; X = no association. Note: Fields left blank had no data available or computed.
CD34 surface-expressing cells are sparse in neuroblastoma:
IHC assessment of the levels of CD34 revealed a definite surface expression of CD34 in human NB (Fig 1a). We used custom-archived TMAs constructed with a manifold of tumors in primary/metastatic sites from patients during diagnosis/progressive disease after IMCT. Three randomly positioned independent cores were examined for each specimen. To avoid any inconclusive representations, we considered only strong surface expression positivity. Although the presence of CD34 surface-expressing cells was observed within most of the NB tissues, their occurrence remained in low to very-low percentile. For the 119 NB specimens analyzed, CD34 IHC analysis revealed that expression ranged from a minimum of 0.00% to a maximum of 7.87%, with an average of 0.73% and a median of 0.20%.
Figure 1. Availability of CD34-expressing cells with advanced disease in neuroblastoma patients:
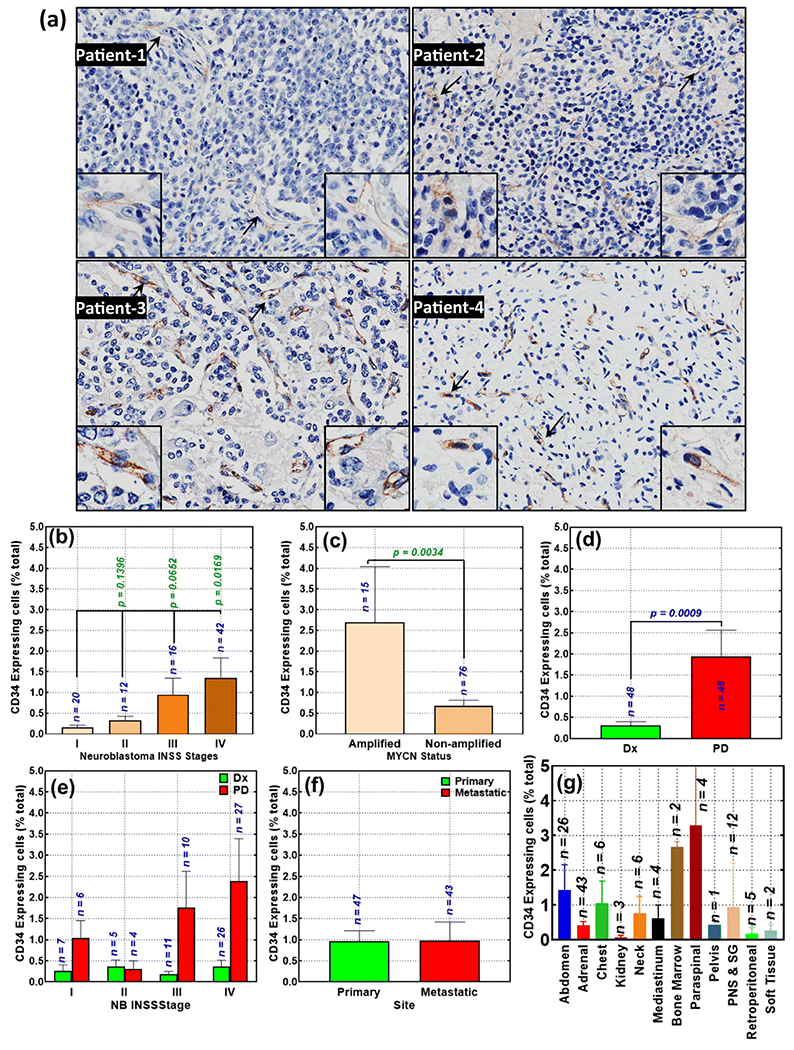
(a) Representative microphotographs showing surface expression of CD34 in human NB [Magnification 20x; Insert, 40x], Histograms constructed from Aperio TMA image analysis of CD34 strong positivity identified (b) significant association of CD34 expression with advanced disease stage; (c) profound correlation of high CD34 expression with N-MYC amplification compared with N-MYC non-amplified subset; (d) acquired gain of CD34-expressing cells in progressive tumors after intensive multimodal clinical therapy (PD, progressive disease) compared with the disease at diagnosis (Dx); (e) acquired gain of CD34-expressing cells in progressive tumors after IMCT in patients presenting with various stages of disease; (f) no significant association with primary or metastatic disease; and (g) the levels of CD34-expressing cells in NB tumors from various recovery sites, irrespective of the primary or metastatic status. Group-wise comparisons were performed with GraphPad Prism. P values of less than 0.05 were considered significant.
CD34 surface expression is associated with NB disease progression:
We sought to define and typify the association of CD34 to progressive NB. First, we examined the INSS stage-dependent fluctuations of CD34. Patients who presented with stage 1 (n=20) disease showed marginal (mean 0.17 ±0.04) levels of CD34 surface expression (Fig 1b). We observed increasing levels of CD34 expression in stage 2 (n=12, 0.33±0.96), stage 3 (n=16, 0.95±0.39), and stage 4 (n=42, 1.36±0.48) NB. Group-wise comparison analysis showed significantly (P<0,016) higher levels of CD34 in high-risk (stage 4) NB than in clinically stable (stage 1) disease (Fig 1b).
We also assessed CD34 surface expression variation in relation to N-MYC status. N-MYC amplification and its function in NB disease evolution has been extensively realized and serves as a dominant variable for the prediction of clinical outcomes in NB. Our IHC analysis revealed significantly (P=0,0034) high incidence of CD34 surface-expressing cells in the N-MYC-amplified subset (n=15) compared with the N-MYC non-amplified (n=76) group (Fig 1c).
Then, we investigated whether the increase in the availability of CD34 surface-expressing cells is an ongoing acquisition process in cells surviving therapy. For this, disease at diagnosis was compared to the progressive disease after IMCT. We observed a significant (P=0.0009) increase in CD34 surface-expressing cells in progressive disease after IMCT compared with disease at diagnosis (Fig 1d). Our analysis on acquired gain, revealed profound acquisition of CD34-expressing cells in progressive disease (compared with diagnosis) for the patients with stages 1, 3, and 4 (Fig 1e). We did not see similar pattern in stage 2 disease, however could speculate that this could be due to the limitations in the n for comparison.
We examined whether there was a difference in availability of CD34 surface-expressing cells in primary (n=47) and disseminated tumors (n=43). There was no significant difference between the primary and metastatic tumors (Fig 1f). Then, we investigated whether there was a difference in the availability of CD34-expressing cells in relation to the site of tumor tissue, irrespective of primary/metastatic status. Compared with adrenal tumors (generally considered a primary site for NB), we observed high CD34 levels in tumors of abdomen, bone marrow, and paraspinal tumors; and relatively high CD34 levels in chest, neck, mediastinum, sympathetic ganglion ,and peripheral nerve structure tumors (Fig 1g). Tumors recovered from kidney, retroperitoneal, and soft tissues harbored few CD34+ cells compared with adrenal tumors. Together, these observations strongly suggest that CD34-expressing cells are associated with advanced disease stage, N-MYC amplification, and disease evolution after IMCT.
CD34-expressing cells and NB clinical outcomes:
We performed survival analysis to assess the influence of the CD34 surface-expressing cells on NB patient outcomes. Since the availability of CD34 surface-expressing cells in NB is minimal, the survival analysis was computed for the presence and absence of CD34 surface-expressing cells. CD34 positivity above 0.2% (median value) was considered as presence and anything less than 0.2% was considered absence. Of the 84 patients with available survival data, 41 were negative CD34-expressing cells and 43 were positive for CD34-expressing cells. The presence of CD34+ NB cells had no significant association with poor (Fig 2a) and relapse-free survival (RFS; Fig 2b) in this cohort of NB patients. However, in patients with relapsed disease, we found a profound (HR = 2.248) association of CD34+ cells presence with poor survival (Fig 2c). Within the N-MYC amplified subset, we found a robust (HR=2.067) association of CD34+ cells presence with OS, although this association was not significant (Fig 2d). There was no association of CD34+ cell presence with RFS in this subset (Fig 2e). However, in patients with relapsed disease, we observed a complete decline in patient survival with the presence of CD34+ cells (Fig 2f). Interestingly, in the N-MYC non-amplified patient subset, though not significant, CD34+ cell presence showed inverse association with OS (HR=2.325; Fig 2g) and RFS (HR=2.187; Fig 2h). In patients with relapsed disease, survival analysis revealed a sharp drop in patients’ survival with the presence of CD34+ cells (Fig 2i).
Figure 2. Presence of CD34+ expressing cells in NB clinical outcomes:
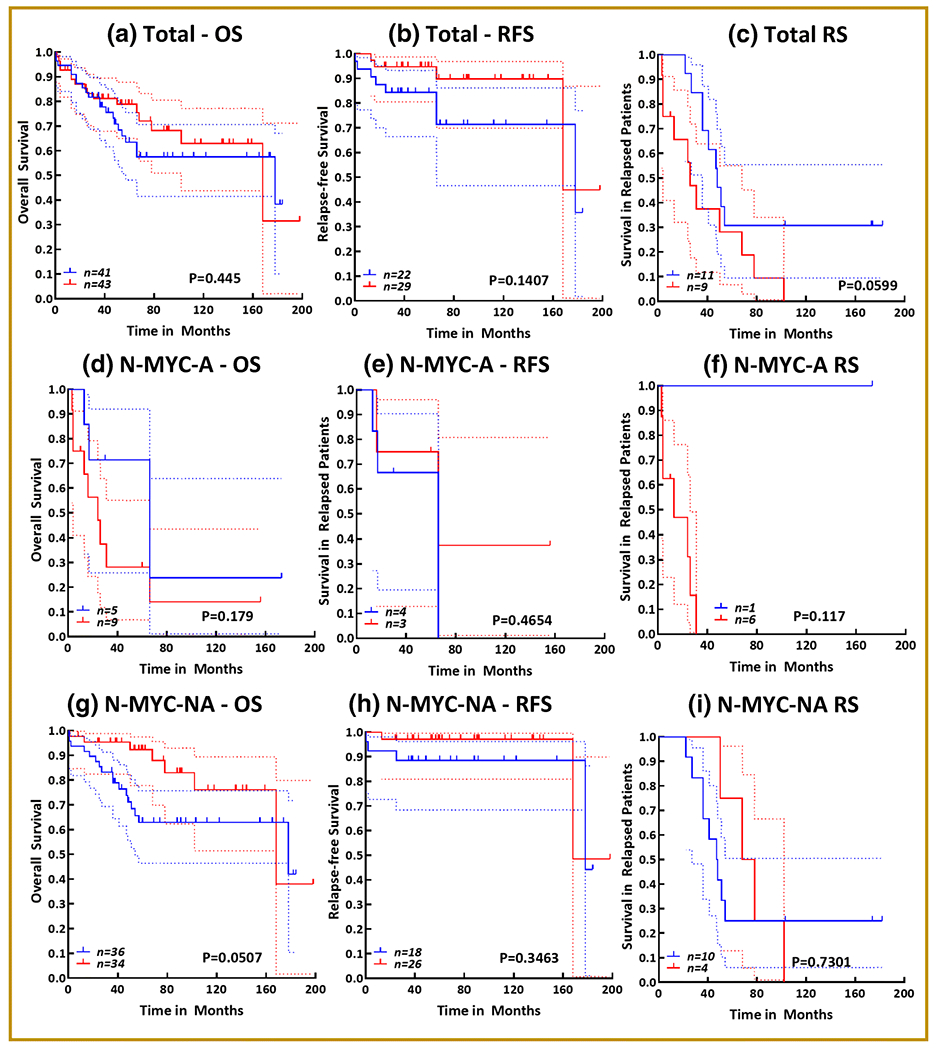
Kaplan-Meier curves showing the association of the presence of CD34+ cells with (a) overall survival, (b) relapse-free survival, and (c) survival in patients with relapsed disease in a cohort of 84 NB patients. Kaplan-Meier curves showing the association of the presence of CD34+ cells with (d) overall survival, (e) relapse-free survival, and (f) survival in patients with relapsed disease in a cohort of 14 N-MYC amplified NB patient subset. Kaplan-Meier curves showing the association of the presence of CD34+ cells with (g) overall survival, (h) relapse-free survival, and (i) survival in patients with relapsed disease in a cohort of 70 N-MYC non-amplified NB patient subset. All survival analysis (log rank Mantle cox test and hazard ratio) and Kaplan-Meier curves were performed in GraphPad Prism. Blue = absence of CD34+ cells; Red = presence of CD34+ cells; dotted lines = 95% confidence interval.
In high-risk progressive (stage 4) disease, presence of CD34+ cells had no significant association with OS (Fig 3a) and RFS (Fig 3b). Although a high HR (HR = 1.679) indicating a positive linear correlation of CD34+ cell presence to poor survival in patients with relapsed disease (Fig 3c). High-risk patients with N-MYC amplification showed a significant (P=0.0097; HR = 4.883) correlation of CD34+ cell presence with poor OS (Fig 3d). However, in high-risk patients with N-MYC non-amplified status, there was no association of CD34 with poor OS (Fig 3e) and RFS (Fig 3f). Although there was no significant relationship between CD34+ cell presence to poor survival in patients with relapsed disease, the trend of linear association between them was noted in this subset (Fig 3g). Together, these data strongly suggest a significant relationship between the presence of CD34+ cells and poor survival in NB patients, particularly in N-MYC amplified high-risk patients and NB patients who present with relapsed disease after IMCT.
Figure 3. Association of CD34+-expressing cells with clinical outcomes in high-risk NB patients:
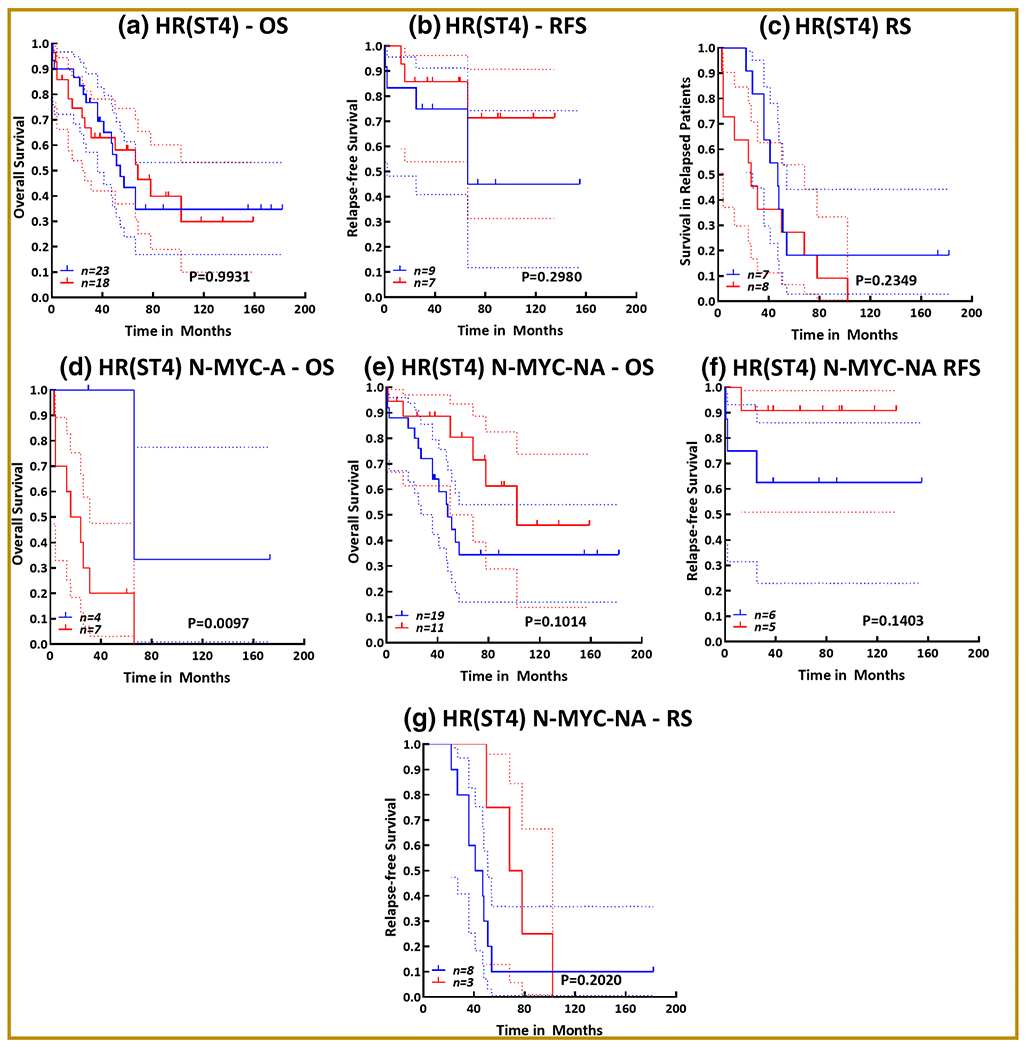
Kaplan-Meier curves showing the association of the presence of CD34+ cells with (a) overall survival, (b) relapse-free survival, and (c) survival in patients with relapsed disease in a cohort of 41 high-risk (stage 4) NB patients. Kaplan-Meier curves showing the association of the presence of CD34+ cells with (d) overall survival in a cohort of 11 N-MYC amplified high-risk patient subset. Kaplan-Meier curves showing the association of the presence of CD34+ cells with (e) overall survival, (f) relapse-free survival, and (g) survival in patients with relapsed disease in a cohort of 30 N-MYC non-amplified high-risk (stage 4) NB patient subset. All survival analysis (log rank Mantle cox test and hazard ratio) and Kaplan-Meier curves were performed in GraphPad Prism. Blue = absence of CD34+ cells; Red = presence of CD34+ cells; dotted lines = 95% confidence interval.
Genetic landscape of CD34+ cancer stem Cells (CSCs):
To define the genetic blueprint of CD34-expressing NB CSCs, we performed whole genome gene expression analysis. For this, NB CSCs, including unsorted (SM-CSC-L), CD133+CD34− (SM-CSC-L-CD133+), and CD133+CD34+ (SM-CSC-L-CD34+) derived and characterized from our earlier studies,(Khan et al., 2015a; Khan et al., 2015b; Pandian et al., 2015) were screened with Agilent SurePrint G3 Human GE 8 × 60K V2 microarrays. Human NB SH-SY5Y cells were used as controls. For better comparative understanding, we performed three-step analysis: 1) we defined the CSCs genetic makeup by comparing the CSC-like (unsorted) and CSCs (CD133+, CD133+CD34+) to the non-CSC tumor populations (SH-SY5Y); 2) we defined the genetic makeup of designated CSCs (CD133+, CD133+CD34+) compared with unsorted CSC-like populations; and 3) we defined the additive gene modulations in CD34+ NB CSCs. For the exploration of additive gene modulations, it is essential to determine the differences in CD34-negative and CD34-positive conditions in clearly defined CSCs. Thus, we used CD133, the well-documented and extensively recognized NB-CSC marker (reviewed in detail elsewhere(Aravindan et al., 2019a)), to designate NB CSCs. Table 3 presents the upmodulations and downmodulations for each case and for each comparison individually. A two-step significance criterion (significant, ≥2 fold and highly significant, ≥10 fold) was adopted for each comparison.
Table 3.
Comparison analysis from whole genome gene expression profile showing genes that are upmodulated and downmodulated in NB-CSCs (SM-CSC-L, SM-CSC-L-CD133+, vs. SM-CSC-L-CD34+) compared with non-CSC tumor cells (SH-SY5Y); in designated CSCs (SM-CSC-L-CD133+, vs. SM-CSC-L-CD34+) compared with unsorted NB-CSCs (SM-CSC-L; and in CD133+CD34+ (SM-CSC-L-CD34+) CSCs compared with CD133+CD34− (SM-CSC-L-CD133+) CSCs. Outcomes of the crisscross analysis are presented in overall, significant (≥2 fold), and highly significant (≥10 fold) settings. CSCs = Cancer Stem Cells; NB = Neuroblastoma
| Group | Comparisons | Total | Overall Up | ≥2 Fold Up | ≥10 Fold Up | Overall Down | ≥2 Fold Down | ≥10 Fold Down |
|---|---|---|---|---|---|---|---|---|
| A | SH-SY5Y vs. SM-CSC-L | 50,737 | 30,131 | 3,255 | 270 | 20,468 | 3,142 | 249 |
| B | SH-SY5Y vs. SM-CSC-L-CD133+ | 50,737 | 29,778 | 2,905 | 164 | 20,821 | 3,391 | 214 |
| C | SH-SY5Y vs. SM-CSC-L-CD34+ | 50,737 | 30,127 | 4,902 | 406 | 20,472 | 5,082 | 738 |
| D | SM-CSC-L vs. SM-CSC-L-CD133+ | 50,737 | 20,133 | 2,215 | 72 | 29,466 | 2,818 | 70 |
| E | SM-CSC-L vs. SM-CSC-L-CD34+ | 50,737 | 22,016 | 4,846 | 290 | 28,583 | 5,314 | 659 |
| F | SM-CSC-L-CD133+ vs. SM-CSC-L-CD34+ | 50,737 | 22,715 | 4,631 | 274 | 27,884 | 4,678 | 572 |
| Common Up | Magnified | Reverted to Up | Common Down | Magnified | Reverted to Down | |||
| G | A vs. B (only ≥10 Fold) | 163 | 56 | 1 | 196 | 145 | 18 | |
| H | A vs. C (only ≥10 Fold) | 366 | 317 | 40 | 542 | 711 | 196 | |
| Overall Up | ≥2 Fold Up | ≥10 Fold Up | Overall Down | ≥2 Fold Down | ≥10 Fold Down | |||
| I | D vs. E (Common) | 14,271 | 1,152 | 49 | 21,721 | 1,060 | 4 |
The gene modulation in SM-CSC-L, SM-CSC-L-CD133+, and SM-CSC-L-CD34+ were compared with the gene profile of non-tumor (SH-SY5Y) NB cells. Each CSC type investigated displayed a robust activation (upmodulation) of genes (Fig 4–ai). Crisscross comparisons between the CSCs upmodulated genes identified ‘CSC type-specific’ and ‘shared’ gene signatures. Interestingly, 17,868 transcripts showed CSC-type independent upregulation (Fig 4–ai). Applying stringent criteria (≥2 fold), we observed a unique gene signature with CSC type-specific (CSC-L, 882; CD133+, 581; CD34+, 2582), shared (CSC-L & CD133+, 581; CSC-L & CD34+, 521; CD133+ & CD34+, 472), and CSC type-independent (1,327) profiles (Fig 4–aii). Further, focusing on highly significant upregulation, we observed that 85, 13, and 243 genes are specifically activated in CSC-L, CD133+, and CD34+ CSCs, respectively. Thirty-nine genes were shared between CSC-L and CD133+, 51 genes were shared between CSC-L and CD34+, and 17 genes were shared between CD133+ and CD34+ (Fig 4–aiii). Ninety-five genes showed CSC type-independent significant activation compared with non-CSC tumor cells. Similarly, CSCs also displayed distinct downmodulation signatures and identified ‘CSC type-specific’, ‘shared’, and ‘independent’ (9,064) gene signatures (Fig 4–aiv).
Figure 4. Genetic landscape of NB-CSCs:
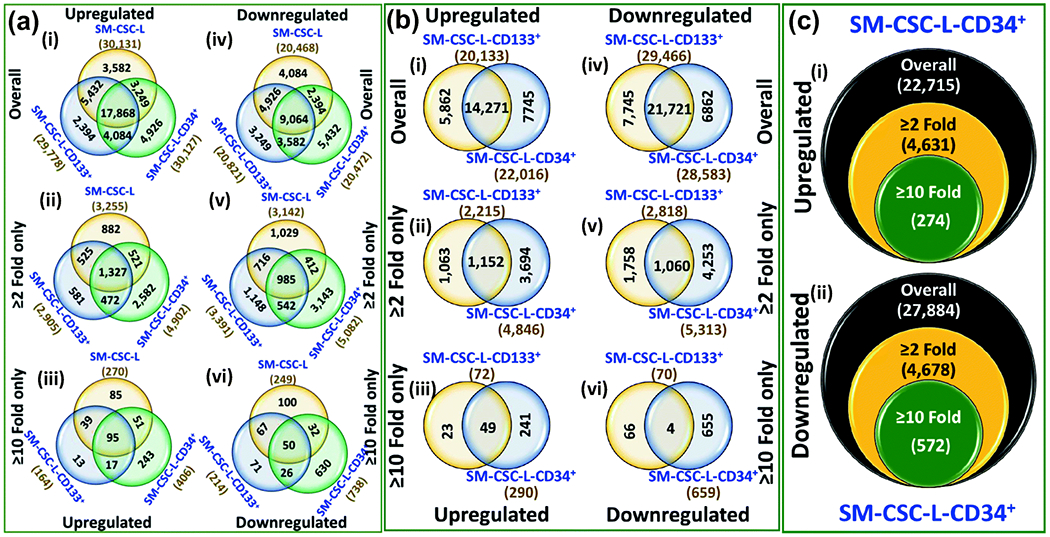
(a) Venn diagrams of gene comparison analysis from whole genome gene expression profile showing genes that are upmodulated and downmodulated in NB-CSCs (SM-CSC-L, SM-CSC-L-CD133+, vs. SM-CSC-L-CD34+) compared with non-CSC tumor cells (SH-SY5Y). (b) Venn diagrams of gene comparison analysis from whole genome gene expression profile showing genes that are upmodulated and downmodulated in designated NB-CSCs (SM-CSC-L-CD 133+, vs. SM-CSC-L-CD34+) compared with unsorted CSCs (SM-CSC-L). (c) Venn diagrams of gene comparison analysis from whole genome gene expression profile showing genes that are upmodulated and downmodulated in CD133+CD34+ (SM-CSC-L-CD34+) CSCs compared with CD133+CD34− (SM-CSC-L-CD133+) CSCs. Venn diagrams constructed for overall modulated, significantly modulated (≥2 fold) .and highly significant (≥10 fold) are arranged vertically. Numbers outside the circle are the total number of genes that are upregulated or downregulated in any given CSCs; numbers inside the circle are specific to the particular CSC; numbers in overlapping regions between two circles are the genes commonly modulated between those specific CSCs; numbers in the center are the commonly modulated genes.
With stringent criteria (≥2 fold), we observed a CSC type-specific (CSC-L, 1,029; CD133+, 1,148; CD34+, 3,143), shared (CSC-L & CD133+, 716; CSC-L & CD34+, 412; CD133+ & CD34+, 542), and CSC type-independent (985) downmodulation (Fig 4–av). Computing for highly significant (≥10 Fold) downregulated genes, we observed that 100, 71, and 630 genes are specifically suppressed in CSC-L, CD133+, and CD34+ CSCs, respectively. Sixty-seven genes were shared between CSC-L and CD133+, 32 genes were shared between CSC-L and CD34+, and 26 genes were shared between CD133+ and CD34+ (Fig 4–avi). Fifty genes showed CSC type-independent suppression compared with non-CSC tumor cells.
Together, these gene profiles clearly indicate the gene fluctuations in NB-CSCs compared with non-CSC tumor cells. The results identified the unsorted, CD133+, and CD34+CSCs genetic blueprints, clone-dependent differences, and the common NB-CSC designation signature. More importantly, these signature profiles (both upmodulation and downmodulation) under general, significant, and highly significant stratifications clearly indicate a unique and magnified SM-CSC-L-CD34+ clone-specific signature compared with SM-CSC-L or SM-CSC-L-CD133+.
Next, we compared the genetic profiles of the designated NB CSCs (CD133+ and CD133+CD34+) with that of the general population of unsorted CSC-like cells (SM-CSC-L). Compared with unsorted CSCs, CD133+ CSCs displayed upmodulation of 20,133 genes, of which 5,862 are specific to this population. Likewise, CD133+CD34+ CSC clones exhibited an upregulation of 22,016 genes, within which 7,745 are specific to this type. With stringent criteria in place, 2,215 and 4,846 genes were significantly upregulated in CD133+and CD133+CD34+ CSCs, respectively (Fig 4–bii). Of these, 1,063 are unique to CD133+, 3,694 are unique to CD133+CD34+, and another 1,152 genes were commonly upregulated between these CSCs. With ≥10 fold significance in place, we observed an activation of 72 and 290 genes in CD133+and CD133+CD34+ CSCs, respectively (Fig 4–biii). With 49 of these genes shared between both designations, 23 genes were uniquely activated in CD133+CSCs and 241 were specific to CD133+CD34+ CSCs. Compared with the unsorted population, a large number of genes are downregulated in these designated CSCs, with 7,745 genes specific to CD133+ CSCs, 6,862 genes unique to CD133+CD34+ CSCs, and 21,721 genes that are commonly downmodulated (Fig 4–biii). Of this number, 2,818 and 5,313 genes are significantly (≤2 fold) downmodulated in CD133+ and CD133+CD34+ CSCs, respectively; these genes include a shared downregulation of 1,060 genes (Fig 4–bv). More importantly, 70 and 659 genes were completely (<10 fold) inhibited in CD133+ and CD133+CD34+ CSCs, with only four genes commonly downregulated between both groups (Fig 4–bvi). These data identified genetic signatures (specific, shared) of the designated (CD133+ and CD133+CD34+) NB-CSCs compared with the unsorted CSC population. Interestingly, within this comparison, we observed a heightened number of molecules that were upmodulated and downmodulated in CD133+CD34+ CSCs compared with CD133+ CSCs.
We compared the gene expression profiles between CD133+CD34− (SM-CSC-L-CD133) and CD133+CD34+ (SM-CSC-L-CD34+) NB-CSCs. Compared with CD34− NB-CSCs, CD34+ CSCs displayed upmodulation of 22,715 genes, of which 4,631 genes were significantly (≥2 fold) upregulated. Two hundred seventy-four genes showed highly significant (≥10 fold) activation (Fig 4–ci). Conversely, we found 27,884 genes were downregulated in CD34+ CSCs when compared with CD34− CSCs. While 4,678 of these genes showed significant downmodulation 572 genes were completely (310 fold) suppressed (Fig 4–cii). These results show that CD34+ NB-CSCs have additive gene modulations that warrant investigation to define their function in disease evolution and clinical outcomes.
Since this was the prime focus of this study, the significantly upregulated (4,631) and downregulated (4,678) genes in CD34+ NB-CSCs were subjected to IPA analysis. Of the 9,309 inputs, IPA was able to map 7,602 for core analysis. Confidence was set for ‘experimentally observed’ interactions and predictions were excluded for this analysis. IPA comparison analysis identified >500 canonical pathways (Table S1) in which these molecules have critical (statistically significant) and functional roles. Their statistical significance indicates the probability of association of the molecules modulated in CD34+ NB-CSCs with the documented canonical pathways. Furthermore, constructing an overlapping network with the top 100 canonical signaling pathways including the ‘direct relationship’ and excluding any indirect interactions (Fig 5) revealed a tightly regulated signaling. To shed some light on the characteristics of CD34+ CSCs in regards to epithelial-to mesenchymal transition, stemness maintenance, self-renewal, clonal selection, and NB disease evolution, a few examples of the activated signaling pathways (stem cell pluripotency, transcriptional regulatory network in stem cell, regulation of EMT, differentiation via BMP receptors, regulation of WNT, G-protein coupled receptor and cancer metastasis signaling) and involved molecules from this tightly regulated overlapping network are highlighted in Fig 5. In addition to their defined roles and association in canonical signaling, we also evaluated their intricate roles in molecular and cellular functions. The genes modulated in CD34+ CSCs are critically involved in more than 500 annotated cellular functions, which clearly showed their all-inclusive role in the orchestration of disease progression and evolution (Table S2). The annotated cellular functions and diseases that have high relevance to NB progression and evolution include: development of malignant tumor, tumor/lesion growth, cellular movement – migration and invasion, cell-to-cell signaling/interaction, angiogenesis, and secondary tumor. Together, the gene expression data identified the unique and additive gene modulations in CD34+ NB-CSCs, and the IPA analyses indicated the functional relevance of such candidates in the translation of stemness maintenance, clonal selection and expansion, tumor progression, dissemination, and evolution.
Figure 5. Relevance of CD34+ NB-CSC gene signature in stemness maintenance and tumor evolution:
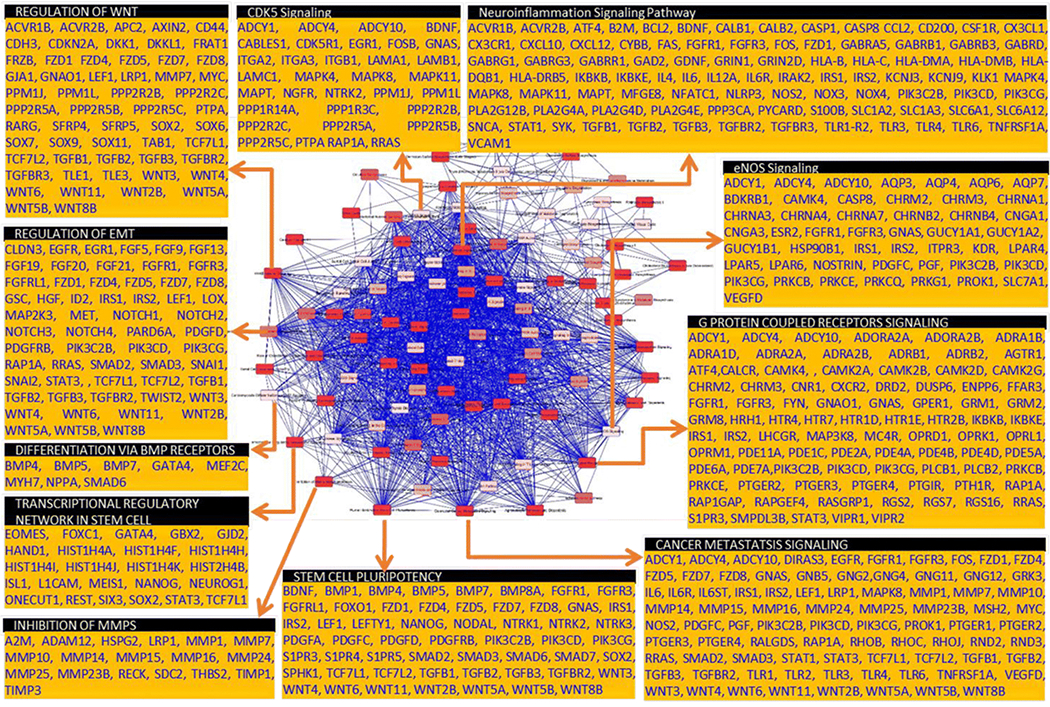
Overlay of top 100 statistically significant canonical pathways identified by IPA analysis with direct relationship showing tightly inter-regulated signaling events that drive stemness maintenance, self-renewal capacity, EMT, clonal expansion, tumor progression, and evolution. Pull out boxes for some of the canonical pathways showing the list of candidate genes involved in these pathways that are modulated in the CD34+NB-CSCs.
DISCUSSION
In a cohort of NB patients in the present study, we showed the definite but rare presence of CD34 surface-expressing cells within NB. CD34 surface expression in general is thought to be confined to lymphohematopoietic cells, capillary endothelial cells, and brain cells (Civin, 1995). Early reviews speculated that, other than the malignancies of vascular origin (e.g., Kaposi’s sarcoma), CD34 is negative in solid tumors, including NB (Krause et al., 1996). However, studies in other tumor systems indicated that tumor cells are still detectable after CD34 enrichment, and CD34 positivity exists in tumor cells (Leung et al., 1998; Mapara et al., 1997; Verbeek et al., 1995). Zintl and colleagues showed CD34 surface expression in primary and permanent NB cell lines (Voigt et al., 1997). Later, the same group validated the surface expression of CD34 in NB cells, and further indicated that CD34 mRNA expression in NB cells may or may not correspond to surface protein expression (Hafer et al., 1999). However, to our knowledge, this study is the first attempt to identity the positivity of CD34 surface expression in NB tumors. Our results showed the presence of CD34 surface-expressing cells within NB, but such positive cells are rare (mean of 0.73% of total cell population). This key finding significantly addresses two contradictory claims: 1) that CD34 surface expression is negative in solid tumors, including NB, and 2) that NB cells exhibit surface expression of CD34. Our findings of rarity in CD34 surface-expressing cells (considering only the strong positivity) within NB suggest that the threshold for positivity could be the deciding factor in no-to-low frequency graft contamination in the clinics. If no threshold for positivity is applied, graft contamination might be overestimated.
Though it is relatively sparse, their definite existence with the NB population, we examined the relevance of CD34 surface expressing cells frequency to the NB disease evolution and clinical outcomes. For the first time, the results of this study demonstrated a linear relationship of the increasing number of CD34 surface expressing NB cells to the advanced disease stage. Significantly high number of CD34+ NB cells in tumors of high-risk (stage 4) patients indicate its possible influence in disease progression. The amplification of the N-MYC gene that codes for an oncogenic transcription factor was observed in approximately 20% of all NB, and is considered to be a molecular marker to identity high-risk patients (Ambros et al., 2009). N-MYC amplification predicting poor prognosis in NB has been extensively documented, and also serves as a tool for better patient stratification for therapy (Ahmed et al., 2017; Yue et al., 2017; Zhang et al., 2017). Results from our comparison analysis between N-MYC amplified and non-amplified statuses identified significantly more CD34 surface-expressing cells in the N-MYC amplified subset. Considering the criticality of MYCN status in poor NB prognosis, one could easily attribute the increased CD34+ positivity to high-risk status and disease progression. More importantly, patients with disease that does not respond to the front-line therapy and progresses are of major concern (Cho et al., 2018; Choi et al., 2005; Modak et al., 2018; Peinemann et al., 2017; Zhou et al., 2015). The cells that survive first-line IMCT may undergo continuous acquisition of molecular and genetic rearrangements and contribute to disease evolution. Our results comparing the disease at diagnosis to the progressive disease after IMCT showed a significant increase in the number of CD34 surface-expressing cells in progressive disease without any sample number bias. Such a finding relates to the acquisition or clonal expansion of CSCs in resistant populations that orchestrate disease evolution. We recently reviewed the role of CSCs in therapy resistance and NB disease evolution (Aravindan et al., 2019a). Interestingly, our findings also showed fluctuations in CD34-surface expressing cells in relation to the site of tumor tissue. Compared to the adrenal tumors, CD34-levels were profoundly high in tumors of bone marrow and paraspinal tumors. With the documented evidence of high numbers of CSCs in NB spheres from bone marrow (Bahmad et al., 2019; Veschi, Verona & Thiele, 2019), we could speculate that the relative high-levels of CD34 in these tumors could be linear to the selection and/or enrichment CSCs in these tissues. Together, these data strongly suggest that there is a correlation between the increasing number of CD34 surface-expressing cells and disease progression, therapy resistance, and disease evolution. Although we observed this linear relationship, functional studies are still needed to confirm the direct role of CD34-expressing cells in NB progression.
With such a low incidence, we did not expect that we would be able to predict any statistically definable relationship with clinical outcomes. However, to our surprise, we observed a tendency of high number of CD34+ cells → low survival in the patients with relapsed disease. This pattern was visible when survival curves were computed for the overall cohort, N-MYC-amplified or non-amplified, and high-risk subsets. More importantly, in N-MYC amplified high-risk (stage 4) subset, our results for the first time identified a significant correlation with poor survival. This finding is significant and indicates that CD34 may serve as a prognostic indicator for this highly progressive, hard to treat subset.
The recurrent clinical course of NB, as well as their self-renewal ability and uninhibited growth pattern despite IMCT, suggest the existence and/or de novo evolution of NB-CSCs (Abdullah & Chow, 2013; Alisi et al., 2013). CSCs are known to harbor critical genetic pro-oncogenic mutations that instigate clonal selection and expansion, promoting disease progression and evolution (Hansford et al., 2007; Mahller et al., 2009; Ross, Biedler & Spengler, 2003). CSCs display high resistance to IMCT, which is attributed to cancer recurrence, metastatic growth, and poor clinical outcomes (Peiris-Pages et al., 2016; Prieto-Vila et al., 2017; Zhu & Fan, 2018). Accordingly, recent investigations are focused on characterizing NB-CSCs on genetic, molecular, and functional levels. In our recent review, we have extensively discussed the roles of NB-CSCs in therapy resistance (Aravindan et al., 2019a). Of the many surface markers indicated for NB-CSCs, CD133 (Prominin-1) has been widely accepted as the marker for CSCs(Li, 2013). CD133 levels were associated with the gain in 16p13 (a genetic profile different from that of non-stem tumor cells), associated with chemoresistance (Sartelet et al., 2012), and inversely correlated with OS of NB patients (Tong et al., 2008).
For the first time, our characterization of CD133+CD34+ NB-CSCs in the present study revealed a unique genetic profile of these NB-CSCs compared with CD133+CD34− NB-CSCs. The difference in the genetic makeup clearly identified activated stemness maintenance, self-renewal, clonal selection and expansion, tumor growth and metastasis, and other key functions (Figure 5) in CD133+CD34+ NB-CSCs that could correspond to tumor progression, disease evolution, and poor clinical outcomes. Our recent reviews extensively documented (Aravindan et al., 2019a; Aravindan et al., 2019b) the direct functional significance of the candidate genes and/or the signaling pathways (identified by IPA in this study) in driving NB stemness maintenance, self-renewal capacity, EMT, clonal expansion, tumor progression, and evolution. We have unveiled the complete variation, significant fluctuations, and the lead candidates with a stratified approach. Further, our step-wise experimental design to display the genetic blueprint of NB-CSCs compared with non-stem cancer cells, surface marker designated NB-CSCs compared with undesignated CSCs, and CD133+CD34+ NB-CSCs compared with CD133+CD34− clearly identified the evolution of NB-CSCs. Hierarchically, CD34+ NB-CSCs exhibited heightened pro-tumorigenic and tumor evolution genetic imprint, which indicates an aggressive phenotype. Consistently, studies have recognized relatively stronger CD34 expression in clonogenic, less differentiated, non-adherent NB cells when compared with the adherent NB cells (Hafer et al., 1999; Leung et al., 1998; Tchirkov et al., 1998; Voigt et al., 1997). The unique, yet tumor evolution-dedicated, genetic signature warrants in-depth functional characterization of CD34+ CSCs in NB and in other tumor systems.
The authors acknowledge that the genetic characterization of the circulating CD133+CD34+ NB-CSCs would have more direct relevance to clinical translation. In addition, we recognize that appropriate pre-clinical animal model studies are warranted to underscore the function of CD34+ CSCs in tumor development, metastasis, therapy resistance, and recurrence. Experimental approaches to address these critical questions are currently under development and/or in the pipeline in our laboratory.
CONCLUSIONS
The results presented here identify for the first time the levels of hematopoietic surface antigen CD34 in NB cells; their likely relevance in disease progression, evolution, and clinical outcomes; and the genetic blueprint of CD34-expressing NB-CSCs. The key findings include: (i) the existence of CD34 positivity of tumor cells in a cohort of NB patients; (ii) the sparse prevalence of CD34 surface-expressing cells; (iii) presence of CD34-expressing cells are significantly associated with advanced disease stage, N-MYC amplification, and disease evolution (progressive disease after IMCT); (iv) that presence of CD34-expressing cells show the tendency to associate with poor survival in patients who present with relapsed disease; (v) that high CD34 is significantly associated with the survival of high-risk patients with N-MYC amplification; (vi) the genetic blueprint of NB-CSCs (compared with non-stem tumor cells), surface marker designated NB-CSCs (compared with undesignated CSCs), and the thus-far unrealized genetic profile of CD133+CD34+ CSCs (compared with CD133+CD34−); and characterization of the activated pathways in CD133+CD34+ CSCs corresponding to tumor evolution. Together, the results indicate that CD34 positivity in NB could correspond to advanced disease stage and disease evolution to progressive disease despite IMCT, and could relate to the poor survival in N-MYC-amplified high-risk patients. In-depth genetic characterization of circulating CD34-expressing cells and preclinical assessment of CD34 function in tumor development and evolution are warranted and are currently under development in our laboratory. Additional studies with bigger cohorts of NB patients are warranted to define the role of CD34 in NB evolution and for any clinical applicability.
Supplementary Material
Figure S1. Association of CD34 transcription with NB disease evolution and poor clinical outcomes: (a) Box-whiskers plot with circles showing high levels of CD34 in high-risk aggressive metastatic stage 4 disease in a cohort of 161 patients. (b) Correlation of CD34 expression with NB clinical outcomes in a cohort of 283 patients. Kaplan-Meier curves showing decreased (b-i) overall survival (OS) and (b-ii) progression-free survival in patients with high CD34 expression compared with low CD34 expression. (c) Kaplan-Meier curve showing a pronounced decrease in relapse-free survival in N-MYC non-amplified subset (n=102) of NB patients with high CD34 expression. (d) Kaplan-Meier curves from an independent cohort of 88 NB patients showing significant decrease in (d-i) OS and (d-ii) relapse-free survival (RFS) with high levels of CD34 expression compared with patients who presented with low CD34. Within the same population, patients who presented with high-risk aggressive disease (INSS stage 4) displayed poor (d-iii) OS and (d-iv) RFS.
Highlights.
A small fraction of NB cells displays CD34 surface expression.
Presence of CD34-expressing cells associated with advanced disease stage, N-MYC amplification, and disease evolution and, associated with the poor survival of high-risk patients with N-MYC amplification.
Comparative gene expression profile of CD34+ NB-CSCs recognized activated signaling pathways and cellular functions those correspond to disease evolution.
ACKNOWLEDGEMENTS
The authors acknowledge the NB specimen providers: Department of Pathology, University of Oklahoma Health Sciences Center; Cooperative Human Tissue Network (CHTN), which is funded by the National Cancer Institute (NCI), and; Oregon Health and Science University Biospecimen core. The authors acknowledge the Stephenson Cancer Center (SCC) Cancer Tissue pathology core for all TMA and IHC services and the SCC Cancer Functional Genomics core for help with whole genome gene expression. The authors also acknowledge the OUHSC Staff Editor (Ms. Kathy Kyler) for the help in critically reviewing this manuscript.
Funding: The National Institutes of Health, NIH-P20GM103639, and Oklahoma Center for the Advancement of Science and Technology, OCAST-HR19-04.
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Conflicts of interest: All other authors have nothing to disclose.
Data Sharing Statement: No additional data are available.
Institutional Review Board Statement: The study was reviewed and approved by the Institutional Review Board at the University of Oklahoma Health Sciences Center (OUHSC), with permission for the research use of de-identified specimens. All experiments were performed in accordance with OUHSC IRB guidelines and regulations for the protection of human subjects.
REFERENCES
- Abdullah LN, Chow EK (2013). Mechanisms of chemoresistance in cancer stem cells. Clin Transl Med 2:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed AA, Zhang L, Reddivalla N, Hetherington M (2017). Neuroblastoma in children: Update on clinicopathologic and genetic prognostic factors. Pediatr Hematol Oncol 34:165–185 [DOI] [PubMed] [Google Scholar]
- Alisi A, Cho WC, Locatelli F, Fruci D (2013). Multidrug resistance and cancer stem cells in neuroblastoma and hepatoblastoma. Int J Mol Sci 14:24706–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros PF, Ambros IM, Brodeur GM, Haber M, Khan J, Nakagawara A, Schleiermacher G, Speleman F, Spitz R, London WB, Cohn SL, Pearson AD, Maris JM (2009). International consensus for neuroblastoma molecular diagnostics: report from the International Neuroblastoma Risk Group (INRG) Biology Committee. Br J Cancer 100:1471–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- American-Cancer-Society. (2013). Cancer Facts & Figures Atlanta: American Cancer Society 1–64 [Google Scholar]
- Aravindan N, Jain D, Somasundaram DB, Herman TS, Aravindan S (2019a). Cancer stem cells in neuroblastoma therapy resistance. Cancer Drug Resist 2:948–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravindan N, Subramanian K, Somasundaram DB, Herman TS, Aravindan S (2019b). MicroRNAs in neuroblastoma tumorigenesis, therapy resistance, and disease evolution. Cancer Drug Resist 2:1086–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahmad HF, Chamaa F, Assi S, Chalhoub RM, Abou-Antoun T, Abou-Kheir W (2019). Cancer Stem Cells in Neuroblastoma: Expanding the Therapeutic Frontier. Front Mol Neurosci 12:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger M, Kanold J, Rapatel C, deLumley L, Lutz P, Plantaz D, Vannier JP, Mechinaud F, Bergeron C, Favrot M, Gembara P, Halle-Hauss P, Bonhomme J, Travade P, Demeocq F (1997). Feasibility of a PB CD34+ cell transplantation procedure using standard leukapheresis products in very small children. Bone Marrow Transplant 20:191–8 [DOI] [PubMed] [Google Scholar]
- Cho HW, Lee JW, Ma Y, Yoo KH, Sung KW, Koo HH (2018). Treatment Outcomes in Children and Adolescents with Relapsed or Progressed Solid Tumors: a 20-year, Single-Center Study. J Korean Med Sci 33:e260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HS, Koh SH, Park ES, Shin HY, Ahn HS (2005). CNS recurrence following CD34+ peripheral blood stem cell transplantation in stage 4 neuroblastoma. Pediatr Blood Cancer 45:68–71 [DOI] [PubMed] [Google Scholar]
- Civin CI (1995). Purification and expansion of haemopoietic stem cells. Schlossmann SF et al. (Eds.), Leucocyte Typing V. White Cell Differentiation Antigens 1:869–871 [Google Scholar]
- Corrias MV, Haupt R, Carlini B, Parodi S, Rivabella L, Garaventa A, Pistoia V, Dallorso S (2006). Peripheral blood stem cell tumor cell contamination and survival of neuroblastoma patients. Clin Cancer Res 12:5680–5 [DOI] [PubMed] [Google Scholar]
- Frappaz D, Michon J, Coze C, Berger C, Plouvier E, Lasset C, Bernard JL, Stephan JL, Bouffet E, Buclon M, Combaret V, Fourquet A, Philip T, Zucker JM (2000). LMCE3 treatment strategy: results in 99 consecutively diagnosed stage 4 neuroblastomas in children older than 1 year at diagnosis. J Clin Oncol 18:468–76 [DOI] [PubMed] [Google Scholar]
- Fritsch G, Buchinger P, Printz D, Fink FM, Mann G, Peters C, Wagner T, Adler A, Gadner H (1993). Rapid discrimination of early CD34+ myeloid progenitors using CD45-RA analysis. Blood 81:2301–9 [PubMed] [Google Scholar]
- George RE, Li S, Medeiros-Nancarrow C, Neuberg D, Marcus K, Shamberger RC, Pulsipher M, Grupp SA, Diller L (2006). High-risk neuroblastoma treated with tandem autologous peripheral-blood stem cell-supported transplantation: long-term survival update. J Clin Oncol 24:2891–6 [DOI] [PubMed] [Google Scholar]
- Gurney. JG, Smith. MA, Ross. JA (1999). Cancer Among Infants. Cancer Incidence and Survival among Children and Adolescents: United States SEER Program 1975–1995, National Cancer Institute, Bethesda, MD: NIH Pub. No. 99-4649 149-156 [Google Scholar]
- Hafer R, Voigt A, Gruhn B, Zintl F (1999). Neuroblastoma cells can express the hematopoietic progenitor cell antigen CD34 as detected at surface protein and mRNA level. J Neuroimmunol 96:201–6 [DOI] [PubMed] [Google Scholar]
- Handgretinger R, Leung W, Ihm K, Lang P, Klingebiel T, Niethammer D (2003). Tumour cell contamination of autologous stem cells grafts in high-risk neuroblastoma: the good news? Br J Cancer 88:1874–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansford LM, McKee AE, Zhang L, George RE, Gerstle JT, Thorner PS, Smith KM, Look AT, Yeger H, Miller FD, Irwin MS, Thiele CJ, Kaplan DR (2007). Neuroblastoma cells isolated from bone marrow metastases contain a naturally enriched tumor-initiating cell. Cancer Res 67:11234–43 [DOI] [PubMed] [Google Scholar]
- Illhardt T, Toporski J, Feuchtinger T, Turkiewicz D, Teltschik HM, Ebinger M, Schwarze CP, Holzer U, Lode HN, Albert MH, Gruhn B, Urban C, Dykes JH, Teuffel O, Schumm M, Handgretinger R, Lang P (2018). Haploidentical Stem Cell Transplantation for Refractory/Relapsed Neuroblastoma. Biol Blood Marrow Transplant 24:1005–1012 [DOI] [PubMed] [Google Scholar]
- Kanold J, Halle P, Tchirkov A, Berger M, Giarratana MC, Kobari L, Boiret N, Paillard C, Demeocq F, Douay L (2003). Ex vivo expansion of autologous PB CD34+ cells provides a purging effect in children with neuroblastoma. Bone Marrow Transplant 32:485–8 [DOI] [PubMed] [Google Scholar]
- Khan FH, Pandian V, Ramraj S, Natarajan M, Aravindan S, Herman TS, Aravindan N (2015a). Acquired genetic alterations in tumor cells dictate the development of high-risk neuroblastoma and clinical outcomes. BMC Cancer 15:514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan FH, Pandian V, Ramraj SK, Aravindan S, Natarajan M, Azadi S, Herman TS, Aravindan N (2015b). RD3 loss dictates high-risk aggressive neuroblastoma and poor clinical outcomes. Oncotarget 6:36522–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kletzel M, Katzenstein HM, Haut PR, Yu AL, Morgan E, Reynolds M, Geissler G, Marymount MH, Liu D, Kalapurakal JA, Shore RM, Bardo DM, Schmoldt J, Rademaker AW, Cohn SL (2002). Treatment of high-risk neuroblastoma with triple-tandem high-dose therapy and stem-cell rescue: results of the Chicago Pilot II Study. J Clin Oncol 20:2284–92 [DOI] [PubMed] [Google Scholar]
- Krause DS, Fackler MJ, Civin CI, May WS (1996). CD34: structure, biology, and clinical utility. Blood 87:1–13 [PubMed] [Google Scholar]
- Lebkowski JS, Schain LR, Okrongly D, Levinsky R, Harvey MJ, Okarma TB (1992). Rapid isolation of human CD34 hematopoietic stem cells--purging of human tumor cells. Transplantation 53:1011–9 [DOI] [PubMed] [Google Scholar]
- Leung W, Chen AR, Klann RC, Moss TJ, Davis JM, Noga SJ, Cohen KJ, Friedman AD, Small D, Schwartz CL, Borowitz MJ, Wharam MD, Paidas CN, Long CA, Karandish S, McMannis JD, Kastan MB, Civin CI (1998). Frequent detection of tumor cells in hematopoietic grafts in neuroblastoma and Ewing’s sarcoma. Bone Marrow Transplant 22:971–9 [DOI] [PubMed] [Google Scholar]
- Li Z (2013). CD133: a stem cell biomarker and beyond. Exp Hematol Oncol 2:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lode HN, Handgretinger R, Schuermann U, Seitz G, Klingebiel T, Niethammer D, Beck J (1997). Detection of neuroblastoma cells in CD34+ selected peripheral stem cells using a combination of tyrosine hydroxylase nested RT-PCR and anti-ganglioside GD2 immunocytochemistry. Eur J Cancer 33:2024–30 [DOI] [PubMed] [Google Scholar]
- Mahller YY, Williams JP, Baird WH, Mitton B, Grossheim J, Saeki Y, Cancelas JA, Ratner N, Cripe TP (2009). Neuroblastoma cell lines contain pluripotent tumor initiating cells that are susceptible to a targeted oncolytic virus. PLoS One 4:e4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapara MY, Korner IJ, Hildebrandt M, Bargou R, Krahl D, Reichardt P, Dorken B (1997). Monitoring of tumor cell purging after highly efficient immunomagnetic selection of CD34 cells from leukapheresis products in breast cancer patients: comparison of immunocytochemical tumor cell staining and reverse transcriptase-polymerase chain reaction. Blood 89:337–44 [PubMed] [Google Scholar]
- Marabelle A, Merlin E, Halle P, Paillard C, Berger M, Tchirkov A, Rousseau R, Leverger G, Piguet C, Stephan JL, Demeocq F, Kanold J (2011). CD34+ immunoselection of autologous grafts for the treatment of high-risk neuroblastoma. Pediatr Blood Cancer 56:134–42 [DOI] [PubMed] [Google Scholar]
- Marc TG, Gurney. JG, Smith. MA, Olshan. AF (1999). Sympathetic Nervous System Tumors. Cancer Incidence and Survival among Children and Adolescents: United States SEER Program 1975–1995, National Cancer Institute, Bethesda, MD: NIH Pub. No. 99-4649:65-72 [Google Scholar]
- Matthay KK, Villablanca JG, Seeger RC, Stram DO, Harris RE, Ramsay NK, Swift P, Shimada H, Black CT, Brodeur GM, Gerbing RB, Reynolds CP (1999). Treatment of high-risk neuroblastoma with intensive chemotherapy, radiotherapy, autologous bone marrow transplantation, and 13-cis-retinoic acid. Children’s Cancer Group. N Engl J Med 341:1165–73 [DOI] [PubMed] [Google Scholar]
- Modak S, Le Luduec JB, Cheung IY, Goldman DA, Ostrovnaya I, Doubrovina E, Basu E, Kushner BH, Kramer K, Roberts SS, O’Reilly RJ, Cheung NV, Hsu KC (2018). Adoptive immunotherapy with haploidentical natural killer cells and Anti-GD2 monoclonal antibody m3F8 for resistant neuroblastoma: Results of a phase I study. Oncoimmunology 7:e1461305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern DA, Baruchel S, Irwin MS (2013). Current and future strategies for relapsed neuroblastoma: challenges on the road to precision therapy. J Pediatr Hematol Oncol 35:337–47 [DOI] [PubMed] [Google Scholar]
- Moss TJ, Cairo M, Santana VM, Weinthal J, Hurvitz C, Bostrom B (1994). Clonogenicity of circulating neuroblastoma cells: implications regarding peripheral blood stem cell transplantation. Blood 83:3085–9 [PubMed] [Google Scholar]
- Ning B, Cheuk DK, Chiang AK, Lee PP, Ha SY, Chan GC (2016). Autologous cord blood transplantation for metastatic neuroblastoma. Pediatr Transplant 20:290–6 [DOI] [PubMed] [Google Scholar]
- Pandian V, Ramraj S, Khan FH, Azim T, Aravindan N (2015). Metastatic neuroblastoma cancer stem cells exhibit flexible plasticity and adaptive stemness signaling. Stem Cell Res Ther 6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinemann F, van Dalen EC, Enk H, Berthold F (2017). Retinoic acid postconsolidation therapy for high-risk neuroblastoma patients treated with autologous haematopoietic stem cell transplantation. Cochrane Database Syst Rev 8:CD010685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris-Pages M, Martinez-Outschoorn UE, Pestell RG, Sotgia F, Lisanti MP (2016). Cancer stem cell metabolism. Breast Cancer Res 18:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto-Vila M, Takahashi RU, Usuba W, Kohama I, Ochiya T (2017). Drug Resistance Driven by Cancer Stem Cells and Their Niche. Int J Mol Sci 18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rill DR, Santana VM, Roberts WM, Nilson T, Bowman LC, Krance RA, Heslop HE, Moen RC, Ihle JN, Brenner MK (1994). Direct demonstration that autologous bone marrow transplantation for solid tumors can return a multiplicity of tumorigenic cells. Blood 84:380–3 [PubMed] [Google Scholar]
- Ross RA, Biedler JL, Spengler BA (2003). A role for distinct cell types in determining malignancy in human neuroblastoma cell lines and tumors. Cancer Lett 197:35–9 [DOI] [PubMed] [Google Scholar]
- Salazar-Riojas R, Garcia-Lozano JA, Valdes-Galvan M, Martinez-Gonzalez O, Cantu-Rodriguez OG, Gonzalez-Llano O, Gomez-De Leon A, Jaime-Perez JC, Gomez-Almaguer D, Gutierrez-Aguirre CH (2015). Effective collection of peripheral blood stem cells in children weighing 20 kilogram or less in a single large-volume apheresis procedure. J Clin Apher 30:281–7 [DOI] [PubMed] [Google Scholar]
- Santana VM, Furman WL, McGregor LM, Billups CA (2008). Disease control intervals in high-risk neuroblastoma. Cancer 112:2796–801 [DOI] [PubMed] [Google Scholar]
- Sartelet H, Imbriglio T, Nyalendo C, Haddad E, Annabi B, Duval M, Fetni R, Victor K, Alexendrov L, Sinnett D, Fabre M, Vassal G (2012). CD133 expression is associated with poor outcome in neuroblastoma via chemoresistance mediated by the AKT pathway. Histopathology 60:1144–55 [DOI] [PubMed] [Google Scholar]
- Seif AE, Naranjo A, Baker DL, Bunin NJ, Kletzel M, Kretschmar CS, Maris JM, McGrady PW, von Allmen D, Cohn SL, London WB, Park JR, Diller LR, Grupp SA (2013). A pilot study of tandem high-dose chemotherapy with stem cell rescue as consolidation for high-risk neuroblastoma: Children’s Oncology Group study ANBL00P1. Bone Marrow Transplant 48:947–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon T, Berthold F, Borkhardt A, Kremens B, De Carolis B, Hero B (2011). Treatment and outcomes of patients with relapsed, high-risk neuroblastoma: results of German trials. Pediatr Blood Cancer 56:578–83 [DOI] [PubMed] [Google Scholar]
- Smith MA, Seibel NL, Altekruse SF, Ries LA, Melbert DL, O’Leary M, Smith FO, Reaman GH (2010). Outcomes for children and adolescents with cancer: challenges for the twenty-first century. J Clin Oncol 28:2625–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somasundaram DB, Subramanian K, Aravindan S, Yu Z, Natarajan M, Herman T, Aravindan N (2019). De novo regulation of RD3 synthesis in residual neuroblastoma cells after intensive multi-modal clinical therapy harmonizes disease evolution. Sci Rep 9:11766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchirkov A, Kanold J, Giollant M, Halle-Haus P, Berger M, Rapatel C, Lutz P, Bergeron C, Plantaz D, Vannier JP, Stephan JL, Favrot M, Bordigoni P, Malet P, Briancon G, Demeocq F (1998). Molecular monitoring of tumor cell contamination in leukapheresis products from stage IV neuroblastoma patients before and after positive CD34 selection. Med Pediatr Oncol 30:228–32 [DOI] [PubMed] [Google Scholar]
- Tong QS, Zheng LD, Tang ST, Ruan QL, Liu Y, Li SW, Jiang GS, Cai JB (2008). Expression and clinical significance of stem cell marker CD133 in human neuroblastoma. World J Pediatr 4:58–62 [DOI] [PubMed] [Google Scholar]
- Veeraraghavan J, Natarajan M, Aravindan S, Herman TS, Aravindan N (2011). Radiation-triggered tumor necrosis factor (TNF) alpha-NFkappaB cross-signaling favors survival advantage in human neuroblastoma cells. J Biol Chem 286:21588–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veljkovic D, Nonkovic OS, Radonjic Z, Kuzmanovic M, Zecevic Z (2013). Bone marrow processing for transplantation using Cobe Spectra cell separator. Transfus Apher Sci 48:359–63 [DOI] [PubMed] [Google Scholar]
- Verbeek W, Pies A, Humpe A, Grove D, Troff C, Kunze E, Hiddemann W, Wormann B (1995). Mobilization of CD34-positive tumour cells in a patient with testicular mixed germ cell tumour. Br J Haematol 90:947–50 [DOI] [PubMed] [Google Scholar]
- Veschi V, Verona F, Thiele CJ (2019). Cancer Stem Cells and Neuroblastoma: Characteristics and Therapeutic Targeting Options. Front Endocrinol (Lausanne) 10:782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt A, Hafer R, Gruhn B, Zintl F (1997). Expression of CD34 and other haematopoietic antigens on neuroblastoma cells: consequences for autologous bone marrow and peripheral blood stem cell transplantation. J Neuroimmunol 78:117–26 [DOI] [PubMed] [Google Scholar]
- Yue ZX, Huang C, Gao C, Xing TY, Liu SG, Li XJ, Zhao Q, Wang XS, Zhao W, Jin M, Ma XL (2017). MYCN amplification predicts poor prognosis based on interphase fluorescence in situ hybridization analysis of bone marrow cells in bone marrow metastases of neuroblastoma. Cancer Cell Int 17:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Wu X, Basu M, Dong C, Zheng P, Liu Y, Sandler AD (2017). MYCN Amplification Is Associated with Repressed Cellular Immunity in Neuroblastoma: An In Silico Immunological Analysis of TARGET Database. Front Immunol 8:1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou MJ, Doral MY, DuBois SG, Villablanca JG, Yanik GA, Matthay KK (2015). Different outcomes for relapsed versus refractory neuroblastoma after therapy with (131)I-metaiodobenzylguanidine ((131)I-MIBG). Eur J Cancer 51:2465–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P, Fan Z (2018). Cancer stem cells and tumorigenesis. Biophys Rep 4:178–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Association of CD34 transcription with NB disease evolution and poor clinical outcomes: (a) Box-whiskers plot with circles showing high levels of CD34 in high-risk aggressive metastatic stage 4 disease in a cohort of 161 patients. (b) Correlation of CD34 expression with NB clinical outcomes in a cohort of 283 patients. Kaplan-Meier curves showing decreased (b-i) overall survival (OS) and (b-ii) progression-free survival in patients with high CD34 expression compared with low CD34 expression. (c) Kaplan-Meier curve showing a pronounced decrease in relapse-free survival in N-MYC non-amplified subset (n=102) of NB patients with high CD34 expression. (d) Kaplan-Meier curves from an independent cohort of 88 NB patients showing significant decrease in (d-i) OS and (d-ii) relapse-free survival (RFS) with high levels of CD34 expression compared with patients who presented with low CD34. Within the same population, patients who presented with high-risk aggressive disease (INSS stage 4) displayed poor (d-iii) OS and (d-iv) RFS.


