Abstract
Consistent with other members of the family Flaviviridae, hepatitis C virus (HCV) demonstrates a high degree of sequence variation throughout the coding regions of its genome. However, there is a high degree of sequence conservation found within the 5′ untranslated region (UTR) of the genome, making this region a target of choice for most nucleic acid amplification-based detection assays. In this study, the Amplicor HCV test, a commercially available assay which detects the 5′UTR, was used for the detection of HCV RNA in 669 serum samples obtained from a cohort of liver transplantation patients. Amplification products obtained from the HCV-positive cases were subjected to direct sequencing and genotyping based upon seven phylogenetically informative regions within the 5′UTR. Of the 669 specimens, 416 (62.2%) tested positive for the presence of HCV RNA. Of these, 372 (89.4%) specimens were successfully classified into 11 HCV genotypes and subtypes after computer-assisted analysis of the sequence data. Forty-four (10.6%) of the HCV RNA-positive specimens were not classifiable, the majority corresponding to low-titer specimens as determined by the Chiron Quantiplex HCV RNA 2.0 assay. Additional comparative studies targeting the NS-5 region of the viral genome generally confirmed the accuracy and sensitivity of the 5′UTR-based classifications, with the exception of the misclassification of a small number of type 1a cases as type 1b. We conclude that although the high sequence conservation within the 5′UTR results in the misclassification of a small number of HCV subtypes, the overall gains of efficiency, the shorter turnaround time, the inclusion of contamination control measures, and the low rate of test failure compared to that of methods based on the NS-5 gene together constitute significant advantages over other techniques.
Infection with hepatitis C virus (HCV) has been identified as the major cause of posttransfusion non-A, non-B hepatitis (1). HCV has a linear genome approximately 10 kb in length consisting of positive sense single-stranded RNA (ssRNA) (5). Consistent with related members of the family Flaviviridae, HCV demonstrates a high degree of sequence variation throughout its genome; sequence analysis of multiple strains of HCV has demonstrated that the nucleotide sequence can differ by as much as 30% (25). However, the levels of heterogeneity differ considerably among the various regions of the virus. For example, sequence variation ranges from as little as 10% in the 5′ untranslated region (UTR) to as much as 50% or more within the E1 region (2, 4).
Further comparison of published sequence data over the entire HCV genome indicates that the 5′UTR of 324 to 341 nucleotides is the most highly conserved region among HCV strains (3, 14, 23). The high degree of conservation within the 5′UTR has made it the target of choice for most reverse transcriptase (RT)-PCR-based detection assays, including the Amplicor HCV test (Roche Diagnostic Systems, Inc., Branchburg, N.J.) (37). Also, a number of genotyping schemes have been developed which utilize this region and its component phylogenetically informative positions for provisional genotype determination (16, 19, 27, 29–31). Phylogenetic analysis of sequence information obtained from the 5′UTR has been shown to be consistent with that obtained from other regions of the virus, including the core, NS-3, NS-4, and NS-5, thus supporting its use in viral typing (4, 8, 28, 29).
In addition to the determination of the presence or absence of HCV infection, there has also been increased clinical interest in the genotyping of HCV. These genotyping results can be useful in viral transmission studies as well as in epidemiological investigations. Furthermore, in many studies, associations between viral genotype, interferon responsiveness, the progression of disease, and the likelihood of developing hepatocellular carcinoma have been demonstrated (6, 11, 17, 18, 21, 22, 34, 38, 39). Thus, although the basis for these associations is poorly understood, HCV genotyping is being performed increasingly in clinical studies of HCV patients. The use of a single amplification reaction for HCV detection with the potential for genotype determination from these amplification products is the most efficient means of genotyping.
In this study, we utilized a commercially available HCV amplification assay, the Amplicor HCV test, to determine the presence or absence of HCV RNA in a large patient cohort from which 669 serum samples were tested. HCV genotyping of all HCV RNA-positive specimens was attempted by direct sequencing of the amplification products generated in the detection assay. In addition to these methods, a subset of the HCV RNA-positive specimens were also subjected to amplification and sequence analysis of a 222-bp region within the NS-5 gene as a means of validating results obtained from the 5′UTR (26). Viral load determination was also performed on all HCV RNA-positive samples as a part of this study.
MATERIALS AND METHODS
Specimens.
The samples analyzed in this study were obtained from the National Institute of Diabetes and Digestive and Kidney Diseases Liver Transplantation Database (NIDDK LTD), a prospective cohort study of patients evaluated for liver transplantation at the following three large U.S. transplant centers: Mayo Clinic Foundation, Rochester, Minn.; University of Nebraska Medical Center, Omaha; and the University of California, San Francisco (35). The comprehensive results of that study are presented elsewhere by Everhart et al. (10).
A total of 669 frozen serum samples were analyzed as part of our comprehensive HCV study performed for the NIDDK LTD. This sample set consisted of donor as well as pre- and postorthotopic liver transplant serum samples selected by the NIDDK LTD for analysis and provided to our laboratory in a blind fashion. In an effort to minimize sample degradation from multiple freeze-thaw cycles, all samples were thawed on receipt, aliquoted, and stored at −70°C prior to testing (13).
RNA extraction. (i) 5′UTR extractions.
Aliquots, each containing 100 μl of serum, were thawed and extracted by using a guanidine thiocyanate lysis protocol with reagents supplied with the Amplicor HCV test kit. RNA was resuspended in 100 μl of sample diluent in a modification of the Amplicor protocol (33). Extracts were maintained at room temperature (<3 h) until amplification reactions were assembled and extracts were added. The positive and negative controls supplied with the Amplicor kit were processed in parallel with each batch of samples in a dedicated extraction area free of amplification products.
(ii) NS-5 extractions.
Independent 100-μl serum aliquots were thawed and extracted by using 1 ml of RNAzol B reagent (Leedo Medical Laboratories, Houston, Tex.) per specimen. Sample disruption was followed by isopropanol precipitation of the aqueous phase with 2 μl of pellet paint (Novagen, Madison, Wis.), an ethanol wash step and, finally, resuspension in 25 μl of RNase-free water containing 40 U of recombinant RNasin (rRNasin) (Promega, Madison, Wis.). Extracts were held at 4°C (<3 h) until amplification reactions were assembled. Appropriate positive and negative controls were also processed in parallel with each batch of samples extracted by this method. These extractions were also performed in a dedicated extraction area free of amplification products.
RT-PCR amplification. (i) 5′UTR amplification.
Nucleotide primer sequences specific for a 244-bp target located within the 5′UTR of the virus were used to generate amplification products for detection and direct sequence analysis. Combined RT-PCR amplifications were performed by using reagents and protocols supplied with the Amplicor HCV test kit. Reaction mixtures were assembled in an amplicon-free work area prior to thermal cycling on a GeneAmp 9600 thermal cycler (PE Applied Biosystems, Foster City, Calif.).
(ii) NS-5 amplification.
Nucleotide primers capable of amplifying a 401-bp target thought to be highly conserved among various HCV strains were used to generate amplification products from the NS-5 region for sequence analysis and genotyping (9). Amplifications were performed by using a combined reverse transcription-PCR format consisting of a 20-μl reverse transcription reaction mixture and direct addition of 80 μl of PCR mix, resulting in a final volume of 100 μl. Reverse transcription was performed in a 20-μl reaction mixture consisting of 10 μl of master mix (3.0 mM MgCl2, 1× reverse transcription buffer [Promega], 1.0 mM [each] deoxynucleoside triphosphate [Roche Molecular Biochemicals, Inc., Indianapolis, Ind.], 2.5 μM antisense primer [5′GGC GGA ATT CCT GGT CAT AGC CTC CGT GAA 3′], 20 U of rRNasin [Promega], and 15 U of avian myeloblastosis virus RT [Promega]) and 3 drops of sterile mineral oil, followed by the addition of 10 μl of RNA extract. Reverse transcription reactions were set up and performed in an amplicon-free work area under the following conditions: 42°C for 60 min, 99°C for 5 min, and 5°C for 5 min. Amplification of HCV cDNA was accomplished through the addition of 80 μl of a PCR master mix containing 0.81 mM MgCl2, 1× PCR buffer II (PE Applied Biosystems), 13% glycerol, 250 μM dUTP (Roche Molecular Biochemicals, Inc.), 0.625 μM sense primer (5′ TGG GGA TCC CGT ATG ATA CCC GCT GCT TTG A 3′), and 2.5 U of AmpliTaq DNA polymerase (PE Applied Biosystems). The preparation and addition of this second master mix was also performed in an amplicon-free work area in order to minimize possible carryover contamination. A partial dUTP substitution was performed in these amplification reactions to generate products susceptible to degradation by the enzyme uracil N-glycosylase yet allow direct sequencing of reaction products. In this study, preamplification enzymatic degradation was not performed, since no contamination events were detected. Thermal cycling was performed on a model 480 DNA thermal cycler (PE Applied Biosystems) under the following conditions: 94°C for 5 min, 50 cycles of 94°C for 1 min, and 58°C for 1 min, followed by a final extension step at 72°C for 5 min.
Detection. (i) 5′UTR detection.
After completion of thermal cycling, 50 μl of each 100-μl reaction mixture was quickly transferred to a second tube containing 50 μl of chloroform and mixed. This treatment ensures complete inactivation of uracil N-glycosylase in these aliquots. The aqueous phase was removed from each tube and stored at −20°C until preparation for direct sequencing could be completed. The remaining 50 μl of each reaction mixture was denatured in preparation for detection according to the Amplicor protocol by adding 50 μl of the denaturation solution provided in the kit. Note that 50 μl of denaturation solution was used rather than 100 μl, to compensate for the removal of 50 μl of each reaction mixture for direct sequence analysis. Detection was completed without further modification of the Amplicor protocol.
(ii) NS-5 detection.
Upon completion of thermal cycling, 10-μl aliquots of each reaction mixture were screened by ethidium bromide agarose gel electrophoresis (2% SeaKem agarose; FMC Bioproducts, Rockland, Maine) to determine suitability for sequence analysis. Only those reaction mixtures demonstrating the presence of the appropriate size amplification product (401 bp) were subjected to further analysis. Previous experience in our laboratory demonstrated that the minimum amount of DNA required to produce a visible band by ethidium bromide staining approximates the quantity required to generate direct sequencing products, thus supporting the use of this screening method.
Direct sequencing. (i) 5′UTR sequencing.
Frozen aliquots of all Amplicor-positive reaction mixtures were thawed and processed in batches. Direct sequencing was performed as follows. Six microliters of crude PCR product was digested for 30 min at 37°C by using 1 U of shrimp alkaline phosphatase (Amersham Life Sciences, Inc., Cleveland, Ohio) and 10 U of Exonuclease I (Amersham Life Sciences, Inc.), followed by heat inactivation at 80°C for 15 min (15, 36). This treatment was performed to eliminate excess deoxyribonucleotides and amplification primers prior to the dye terminator cycle sequencing. Cycle sequencing reactions were completed with the addition of 2 μl (3.2 pmol) of internal antisense sequencing primer (5′ AGT ACC ACA AGG CCT TTC 3′ or 5′ ACC ACA AGG CCT TTC GC 3′), 2 μl of RNase-free water, and 8 μl of dye terminator premix included in the dye terminator cycle sequencing ready reaction kit (PE Applied Biosystems). Thermal cycling was performed on a GeneAmp 9600 thermal cycler (PE Applied Biosystems) by using the following protocol: 96°C for 10 s, 50°C for 5 s, and 60°C for 4 min, for a total of 25 cycles, followed by a 4°C soak. Completed reaction mixtures were passed through a CentriSep gel filtration spin column (Princeton Separations, Adelphia, N.J.) to remove excess dye terminators. Purified reaction mixtures were vacuum dried and reconstituted in 25 μl of template suppression reagent (PE Applied Biosystems) prior to analysis on an ABI Prism 310 genetic analyzer (PE Applied Biosystems), the sequencing platform routinely used in our clinical laboratory.
(ii) NS-5 sequencing.
Aliquots of all reaction mixtures demonstrating a band of the appropriate size were further analyzed by direct sequencing. Crude PCR products were processed in batches by using the same methods as previously described for 5′UTR products. However, reactions were set up by using the sense-specific amplification primer (5′ TGG GGA TCC CGT ATG ATA CCC GCT GCT TTG A 3′) as the sequencing primer. Analysis of these reactions was performed with an ABI 377 genetic analyzer (PE Applied Biosystems), the sequencing platform routinely used in our research laboratories.
Sequence analysis. (i) 5′UTR analysis.
The resulting sequence data were analyzed with the aid of ABI Sequence Navigator software version 1.0.1. All ambiguous base calls were reviewed, and the overall quality of the sequence information was assessed. Once these steps were completed, multiple sequence alignments were generated by using the Sequence Navigator software. Genotype determinations were based upon the comparison of sequence information from seven distinct informative regions found within the 5′UTR to a previously published database (31, 32). The informative regions include R1 (nucleotides −240 to −233), R2 (nucleotides −167 to −155), R3 (nucleotides −147 to −142), R4 (nucleotides −138 to −132), R5 (nucleotides −128 to −118), R6 (nucleotides −100 to −92), and R7 (nucleotides −81 to −70). Unique sequence patterns located within these informative regions were used to assign either a genotype and subtype or a genotype without a subtype designation if the subtype was not clearly distinguishable. Assignments were based upon the methods and limitations described by Stuyver et al. (31). The following genotype and subtype designations were used in this study: 1a, 1b, 1, 2a/2c, 2b, 2, 3a, 3b, 3, 4a, 4b, 4c/4d, 4e, 4f, 4h, 4, 5a, 5, 6a, and 6.
(ii) NS-5 analysis.
The resulting sequence data from the NS-5 region were reviewed for overall quality prior to comparison with previously described consensus reference sequences (26). These 222-bp consensus sequences are representative of each of the following genotypes: 1a, 1b, 1c, 2a, 2b, 2c, 3a, 3b, 4a, 5a, and 6a. Sequence analysis was performed with the aid of the FASTA algorithm of the Wisconsin package (12), and provisional genotype assignments were based upon percent identity scores generated by using this program. The percent identity scores used to define a specific subtype match were 88 to 100%. Scores used for the differentiation of subtypes within a single genotype ranged from 74 to 86%, while the scores used to distinguish between unique genotypes ranged from 56 to 72% (26). This provisional classification scheme has been validated by formal phylogenetic analysis of multiple HCV strains (26).
Phylogenetic analysis.
Phylogenetic analysis of a 196-bp segment (nucleotides −256 to −70) of the 5′UTR was performed on a subset of the sequences generated in this study. Included in this analysis were 16 prototype sequences obtained from GenBank. The prototype sequences used are presented here along with their designations and GenBank accession numbers as follows: 1a, HCV1 (M62321); 1b, HCVJ (D90208); 2a, HC J6 (D00944); 2b, HC J8 (D01221); 2c, HC J5 (D10075); 3a, Th85 (D14307); 3b, HPCENCR (D11443); 3c, NE048 (D16612); 4a, Z4 (M84848); 4b, Z1 (M84845); 4c, Z6 (M84862); 4d, DK13 (M84832); 4e, Z5 (M84828); 4f, Z8 (M84829); 5a, SA1 (M84860); and 6a, HK2 (M84827). The complete 5′UTR sequence of the 4h prototype (GB438) was not available from GenBank and therefore was not included in this analysis.
The analysis of these sequences was performed using the Molecular Evolutionary Genetics Analysis (MEGA) computer program, version 1.01 (20). Both the unweighted pair group method with arithmetic means (UPGMA) described by Sneath and Sokal (29a) and the neighbor-joining method of Saitou and Nei (24a) were used to construct trees depicting the relationships between sequences generated from the 5′UTR and the prototype sequences. In each case, the Jukes-Cantor algorithm was used to estimate the number of nucleotide substitutions between sequences. The branching of both trees was tested by bootstrap analysis (500 replicates).
Viral quantitation.
Independent aliquots of all serum specimens demonstrating a positive Amplicor HCV result were thawed, and viral load determinations were performed by using the Quantiplex HCV RNA 2.0 assay (Chiron Diagnostics, Emeryville, Calif.) according to the manufacturer’s directions.
RESULTS
Direct sequence analysis of the HCV 5′UTR and correlation with viral titer.
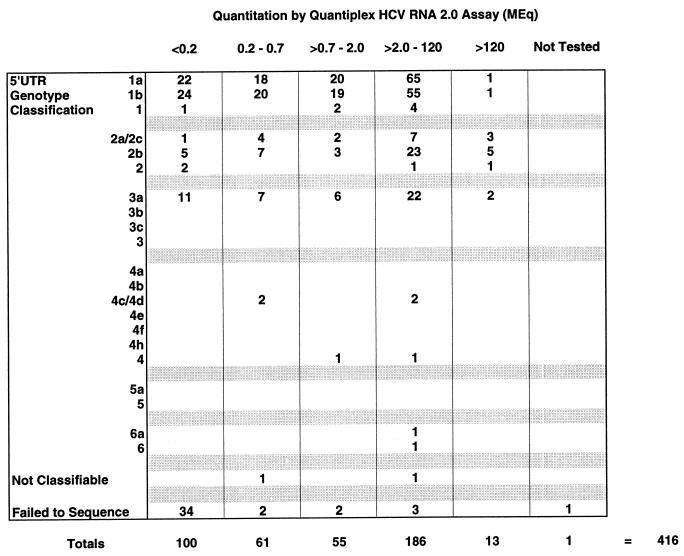
A total of 416 of 669 (62.2%) samples tested positive for the presence of HCV RNA by the Amplicor HCV test. Performance of the PCR test relative to clinical and serologic criteria has been described elsewhere by Everhart et al. (10). The amplification products from all positive reactions were subjected to direct sequencing without further exclusions. Of these, 372 (89.4%) were successfully sequenced and classified into 11 distinct types and subtypes. A total of 44 samples (10.6%) were not classifiable due to the absence of usable sequence information (42 samples) or to an inability to assign a genotype based upon the available sequence information (2 samples). Thirty-four of 44 (77%) of the failed sequencing reactions were disproportionally clustered among those specimens with viral loads of <0.2 Meq/ml as determined by the Quantiplex HCV RNA 2.0 assay (Fig. 1). Finally, there were an additional seven failed sequencing reactions with viral loads of >0.2 Meq and one in which viral load testing was not performed due to insufficient sample volume (Fig. 1).
FIG. 1.
Comparison of 5′UTR genotype classifications and viral titers as determined by the Chiron Quantiplex HCV RNA 2.0 assay. The results of the direct sequencing and genotype classification are indicated on the left, while viral quantitations are expressed in Meq/ml (1 Meq = 106 viral equivalents) across the top of the chart. Quantitative results were grouped into five distinct ranges as indicated, and the specimen totals for each quantitative range are indicated under the appropriate columns. The quantitative range of the Quantiplex HCV RNA 2.0 assay extends from 0.2 to 120 Meq/ml.
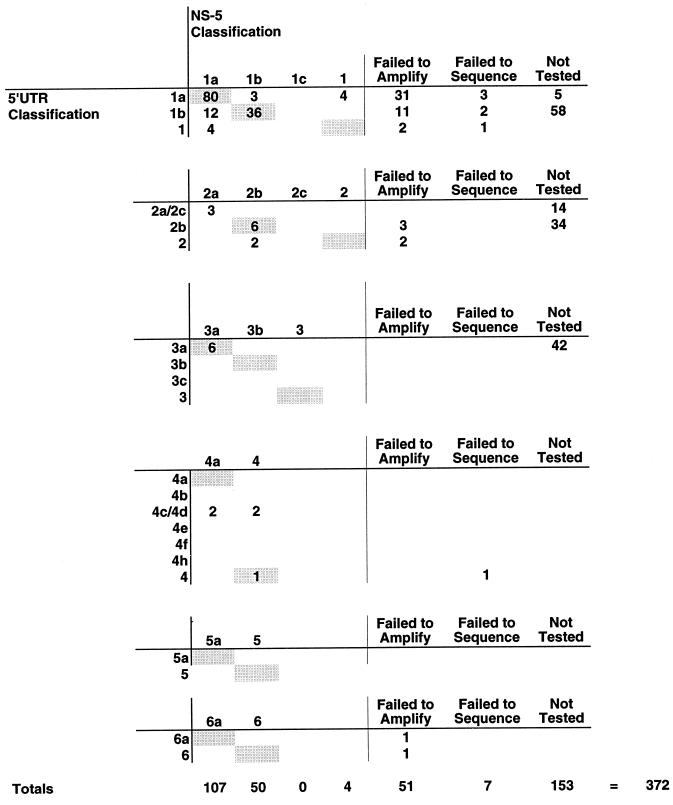
Comparison of 5′UTR and NS-5 regions for HCV genotyping.
Of the 372 samples with 5′UTR genotyping results, 219 (59%) were also characterized by amplification and sequence analysis of the NS-5 region. The results of this testing were compared to the genotyping results obtained from the 5′UTR (Fig. 2). Among these 219 samples, 58 (26.5%) failed to generate an NS-5 amplification product suitable for sequence analysis. In 51 (23.3%) of the cases, no NS-5 amplification products were detectable; 7 (3.2%) samples failed to generate usable sequence information despite the presence of detectable quantities of amplified product. For the 161 remaining specimens, comparable results were obtained by sequence analysis of both regions of the viral genome, despite the presence of a much larger number of phylogenetically informative positions in the NS-5 region. However, there were several notable exceptions (Fig. 2). Among those samples classified as genotype 1a by 5′UTR analysis, 80 of 87 (92%) were in agreement with the NS-5 classification, 3 of 87 (3.4%) were discordant, and 4 of 87 (4.6%) were incompletely identified by NS-5 analysis. Comparison of samples classified as genotype 1b by 5′UTR analysis indicated that 36 of 48 (75%) of the samples were in agreement with the NS-5 classification, while 12 of 48 (25%) were discordant; all of the discrepant specimens were classified as 1b by 5′UTR analysis but as type 1a by NS-5 classification. Finally, among those samples classified as genotype 4c/4d by the 5′UTR analysis, 2 of 4 were discordant, while the other two samples were incompletely identified by NS-5 analysis.
FIG. 2.
Comparison of 5′UTR and NS-5 genotyping. The 5′UTR genotyping classifications are indicated on the left, while the NS-5 genotyping classifications are presented horizontally above each section. The number of specimens which failed to amplify by the NS-5 protocol or failed to generate sequencing data are also indicated along with those not tested by the NS-5 protocol. The totals for each column are indicated.
Phylogenetic analysis of the 5′UTR from a subset of specimens.
Representative 5′UTR sequences obtained from 77 specimens were selected for phylogenetic analysis. These sequences were chosen from the data presented in Fig. 2 and divided into two groups (type 1 versus non-type 1) prior to the analyses. The first analytical set consisted of 47 sequences which included (i) 12 representative sequences from the 80 sequences concordantly identified as genotype 1a by both methods, (ii) 3 sequences identified discordantly as genotype 1a (via 5′UTR) versus 1b (via NS-5), (iii) 12 sequences identified discordantly as genotype 1b (via 5′UTR) versus 1a (via NS-5), (iv) 12 representative sequences from a group of 36 identified as genotype 1b by both methods, and (v) 8 sequences which could not be subtyped by one or both methods used. Prototype sequences for genotypes 1a and 1b were also included in these analyses. The second analytical set consisted of 120 sequences identified as a genotype other than type 1 by the 5′UTR classification method. This subset of sequences was further restricted to those obtained from specimens which were also analyzed by the NS-5 methods (Fig. 2). A total of 30 sequences were included in this group, including 8 sequences which could not be definitively classified by NS-5 amplification or sequencing. The latter were included in the analysis because of the relatively small number of sequences available from these genotype groups. Prototype sequences of genotypes 2a, 2b, 2c, 3a, 3b, 3c, 4a, 4b, 4c, 4d, 4e, 4f, 5a, and 6a were included in the phylogenetic analysis of the second set.
Both UPGMA and neighbor-joining methods were used to analyze these sets of sequences. In the analysis of the second analytical set (non-type 1 sequences), the sequences cosegregated with the type strains by both analytical methods; statistical support of the branching order was provided by bootstrap analysis in which viral types and, to a lesser extent, subtypes were supported in >50% of bootstrap replicates (data not shown). In contrast, similar analysis of type 1 sequences provided generally good separation of subtypes 1a and 1b (with the exceptions previously noted), but none of the bootstrap analyses reached the 50% level of support (data not shown). This was the result of the small number of phylogenetically informative positions found within the 5′UTR. While the bootstrap analyses do not strongly support the branching of these phylogenetic trees, the cosegregation of sequences along with well-characterized prototype sequences does provide support of genotype assignment based upon the 5′UTR, and these results were usually corroborated by the NS-5 sequence analysis.
Nucleotide sequence accession numbers.
The sequences obtained from these 77 samples have been deposited into GenBank under accession no. AF141981 to AF142057.
DISCUSSION
Our experience with direct sequence analysis of the Amplicor HCV PCR product indicates that this approach is an efficient means of genotyping HCV. It does not require an additional specimen-processing step and utilizes products obtained from a single, nonnested amplification reaction, thus eliminating the delays and expense involved in performing additional amplifications. In addition, the direct sequencing of amplification products provides more detailed sequence information than genotyping assays based upon either hybridization or restriction analysis. This additional information could prove to be quite useful in the detection of novel viral types, several of which may have been already observed in this and other studies (31).
Using this method, we were able to successfully sequence and classify 89.4% of those 416 specimens demonstrating a positive HCV RNA test result. Of those 44 of 416 (10.6%) HCV RNA-positive specimens which could not be classified based on the 5′UTR, the majority corresponded to low-titer specimens as determined by the Chiron Quantiplex HCV RNA 2.0 assay (Fig. 2). Despite the fact that there were detectable levels of amplification products found in these reactions, these results suggest the lack of sufficient quantities of amplified material as the primary cause of direct sequencing failure. In turn, the most likely explanation for lack of amplification products is limited quantities of viral RNA present in the original specimen. We could have performed nested amplification to generate sufficient product for sequencing, but this procedure would add delays, expense, and the risk of carryover contamination to the procedure (24). Ultimately, we decided against further amplification, since the sensitivity of the procedure already exceeded that of the NS-5 sequencing protocol (see below).
We attempted to minimize the occurrence of failed or uninterpretable sequencing reactions through the use of the same amplification target for both detection and genotyping. Compared to the current 5′UTR protocol, NS-5 amplification, analysis, and sequencing demonstrated a high failure rate. Failure rates for NS-5 amplification and genotyping were found to be 26.5% higher than those established for the 5′UTR, a total of 58 of 219 specimens which were tested by both methods. This discrepancy is likely due in large part to a reduction in amplification efficiency resulting from differential primer binding efficiencies. Although the primers utilized in the NS-5 amplifications are thought to bind highly conserved sequences (9, 26), primer-target mismatching within this region is still likely because of inherently high sequence variability. In our experience, PCR amplifications targeting the 5′UTR are consistently more likely to detect HCV, consistent with the data presented in this study. Viral quantitation data also suggest a correlation between low viral titer and failure to amplify or generate usable sequence information from the NS-5 region (data not shown).
Disadvantages of genotyping schemes based on the 5′UTR may also ironically be related to its high sequence conservation. Although amplification and genotyping of HCV based upon the 5′UTR appears to be quite sensitive and efficient, the higher level of conservation found within this region can make discrimination of all genotypes and subtypes difficult (25, 29, 32). In fact, genotypes 7, 8, and 9 cannot reliably be distinguished from genotype 1 based upon the 5′UTR and would therefore be misclassified as genotype 1 if this region alone was used for classification (31). In other situations, subtype distinctions cannot be made; such is the case between subtypes 2a and 2c. There are also examples in which only one or two minor nucleotide changes distinguish unique subtypes. An example of this situation is illustrated by the minor differences seen in the 5′UTR sequences of subtypes 1a and 1b. A single base change at position −99 (A/G) is the only change within the 5′UTR differentiating these two subtypes, thus leading to poor statistical support for their discrimination by phylogenetic analysis of this region alone (data not shown). In addition, based on analysis of other regions, it has been shown that this nucleotide difference is not absolute, and interpretations based upon this nucleotide position (−99) can lead to errors in the determination of viral subtype (25, 29, 31, 32). The occurrence of both type 1a and 1b variants was supported by the results of this study. We found that 3 of 87 (3.4%) of the subtype 1a specimens were actually subtyped as 1b by NS-5 sequence analysis, while 12 of 48 (25%) of the subtype 1b specimens were shown to contain subtype 1a sequences by NS-5 analysis. Another possible explanation for these results could have been preferential amplification of different subtypes from a mixed population. However, the clarity of the NS-5 sequencing data obtained from these samples suggests the presence of single viral subtypes (data not shown). Finally, although other studies have suggested that viral recombination appears to be a rare occurrence in HCV (4, 8, 28, 29), further sequencing studies of these isolates could provide evidence for recombination in these specimens.
Despite the inability to completely resolve all existing HCV genotypes and subtypes, the use of this 5′UTR genotyping method provided a sensitive and efficient means of HCV genotyping in a clinical setting, especially in light of studies which show that the clinically relevant distinction is between genotype 1 and non-type 1 (7). The use of a commercially available 5′UTR assay designed for the detection of HCV in clinical specimens provides the user with a sensitive, standardized amplification protocol specifically designed for large-volume testing and rapid turnaround time. In addition to these features, this method utilizes uracil N-glycosylase as an amplicon carryover prevention method. Together with the use of dedicated work areas and a nested sequencing primer, this method provides a reliable means of carryover control in a high-volume testing facility and still provides an acceptable template for direct sequence analysis. For these reasons, in addition to those mentioned above, we have adopted the 5′UTR direct sequencing approach to HCV genotyping as our routine diagnostic procedure.
ACKNOWLEDGMENTS
We thank Theresa Hoff for her assistance in manuscript preparation and all of the members of our laboratory who made this work possible. We also thank the NIDDK LTD for their assistance. Members who contributed to this study included the following: from the Department of Epidemiology, University of Pittsburgh Graduate School of Public Health, Pittsburgh, Pa., Katherine M. Detre, A. Jake Demetris, Steven H. Belle, Yuling L. Wei, Manuel Lombardero, Eric Seaberg, Sharon Lawlor, Heather Eng, Jacqueline Haber, Shannon FitzGerald, and Gerald L. Swanson; from the Departments of Medicine and Surgery, Mayo Clinic Foundation, Russell H. Wiesner, Ruud A. F. Krom, Michael K. Porayko, and Lori Schwerman; from the Departments of Medicine and Surgery, the University of Nebraska Medical Center, Omaha, Rowen K. Zetterman, Byers W. Shaw, Jr., and Karen Taylor; from the Departments of Medicine and Surgery, University of California, San Francisco, Nancy L. Ascher, John R. Lake, and Cherie Bremer-Kamp; and from the Division of Digestive Diseases and Nutrition, NIDDK, National Institutes of Health, Bethesda, Md., James Everhart and Jay H. Hoofnagle.
The LTD is supported by contracts no. N01-DK-0-2251, N01-DK-0-2252, N01-DK-0-2253, and N01-DK-0-2254 from the NIDDK.
REFERENCES
- 1.Alter H J, Purcell R H, Shih J W, Melpolder J C, Houghton M, Choo Q L, Kuo G. Detection of antibody to hepatitis C virus in prospectively followed transfusion recipients with acute and chronic non-A, non-B hepatitis. N Engl J Med. 1989;321:1494–1500. doi: 10.1056/NEJM198911303212202. [DOI] [PubMed] [Google Scholar]
- 2.Bukh J, Purcell R H, Miller R H. At least 12 genotypes of hepatitis C virus predicted by sequence analysis of the putative E1 gene of isolates collected worldwide. Proc Natl Acad Sci USA. 1993;90:8234–8238. doi: 10.1073/pnas.90.17.8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cha T A, Kolberg J, Irvine B, Stempien M, Beall E, Yano M, Choo Q L, Houghton M, Kuo G, Han J H, et al. Use of a signature nucleotide sequence of hepatitis C virus for detection of viral RNA in human serum and plasma. J Clin Microbiol. 1991;29:2528–2534. doi: 10.1128/jcm.29.11.2528-2534.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan S W, McOmish F, Holmes E C, Dow B, Peutherer J F, Follett E, Yap P L, Simmonds P. Analysis of a new hepatitis C virus type and its phylogenetic relationship to existing variants. J Gen Virol. 1992;73:1131–1141. doi: 10.1099/0022-1317-73-5-1131. [DOI] [PubMed] [Google Scholar]
- 5.Choo Q L, Kuo G, Weiner A J, Overby L R, Bradley D W, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 6.Clarysse C, Van den Eynde C, Nevens F, Portier C, Aelbrecht N, Fevery J, Yap S H. Genotype, serum level of HCV-RNA and response to interferon-alpha treatment in patients with chronic hepatitis C. Neth J Med. 1995;47:265–271. doi: 10.1016/0300-2977(95)00101-8. [DOI] [PubMed] [Google Scholar]
- 7.Davis G L, Esteban-Mur R, Rustgi V, Hoefs J, Gordon S C, Trepo C, Shiffman M L, Zeuzem S, Craxi A, Ling M H, Albrecht J for The International Hepatitis Interventional Therapy Group. Interferon alfa-2b alone or in combination with ribavirin for the treatment of relapse of chronic hepatitis C. N Engl J Med. 1998;339:1493–1499. doi: 10.1056/NEJM199811193392102. [DOI] [PubMed] [Google Scholar]
- 8.Dixit V, Quan S, Martin P, Larson D, Brezina M, DiNello R, Sra K, Lau J Y, Chien D, Kolberg J, et al. Evaluation of a novel serotyping system for hepatitis C virus: strong correlation with standard genotyping methodologies. J Clin Microbiol. 1995;33:2978–2983. doi: 10.1128/jcm.33.11.2978-2983.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enomoto N, Takada A, Nakao T, Date T. There are two major types of hepatitis C virus in Japan. Biochem Biophys Res Commun. 1990;170:1021–1025. doi: 10.1016/0006-291x(90)90494-8. [DOI] [PubMed] [Google Scholar]
- 10.Everhart J, Wei Y, Eng H, Charlton M, Persing D, Wiesner R, Germer J, Lake J, Zetterman R, Hoofnagle J. Recurrent and new hepatitis C virus infection after liver transplantation. Hepatology. 1999;29:1220–1226. doi: 10.1002/hep.510290412. [DOI] [PubMed] [Google Scholar]
- 11.Gayowski T, Singh N, Marino I R, Vargas H, Wagener M, Wannstedt C, Morelli F, Laskus T, Fung J J, Rakela J, Starzl T E. Hepatitis C virus genotypes in liver transplant recipients: impact on posttransplant recurrence, infections, response to interferon-alpha therapy and outcome. Transplantation. 1997;64:422–426. doi: 10.1097/00007890-199708150-00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Genetics Computer Group. Program manual for the Wisconsin package. 8th ed. Madison, Wis: Genetics Computer Group; 1994. [Google Scholar]
- 13.Halfon P, Khiri H, Gerolami V, Bourliere M, Feryn J M, Reynier P, Gauthier A, Cartouzou G. Impact of various handling and storage conditions on quantitative detection of hepatitis C virus RNA. J Hepatol. 1996;25:307–311. doi: 10.1016/s0168-8278(96)80116-4. [DOI] [PubMed] [Google Scholar]
- 14.Han J H, Shyamala V, Richman K H, Brauer M J, Irvine B, Urdea M S, Tekamp-Olson P, Kuo G, Choo Q L, Houghton M. Characterization of the terminal regions of hepatitis C viral RNA: identification of conserved sequences in the 5′ untranslated region and poly(A) tails at the 3′ end. Proc Natl Acad Sci USA. 1991;88:1711–1715. doi: 10.1073/pnas.88.5.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanke M, Wink M. Direct DNA sequencing of PCR-amplified vector inserts following enzymatic degradation of primer and dNTPs. BioTechniques. 1994;17:858–860. . (Erratum, 18:636, 1995.) [PubMed] [Google Scholar]
- 16.Holland J, Bastian I, Ratcliff R M, Beers M Y, Hahesy P, Harley H, Shaw D R, Higgins G D. Hepatitis C genotyping by direct sequencing of the product from the Roche AMPLICOR test: methodology and application to a South Australian population. Pathology. 1998;30:192–195. doi: 10.1080/00313029800169226. [DOI] [PubMed] [Google Scholar]
- 17.Hopf U, Berg T, Konig V, Kuther S, Heuft H G, Lobeck H. Treatment of chronic hepatitis C with interferon alpha: long-term follow-up and prognostic relevance of HCV genotypes. J Hepatol. 1996;24(Suppl. 2):67–73. [PubMed] [Google Scholar]
- 18.Isaacson A H, Davis G L, Lau J Y. Should we test hepatitis C virus genotype and viraemia level in patients with chronic hepatitis C? J Viral Hepatitis. 1997;4:285–292. doi: 10.1046/j.1365-2893.1997.00065.x. [DOI] [PubMed] [Google Scholar]
- 19.Kleter G E, van Doorn L J, Brouwer J T, Schalm S W, Heijtink R A, Quint W G. Sequence analysis of the 5′ untranslated region in isolates of at least four genotypes of hepatitis C virus in The Netherlands. J Clin Microbiol. 1994;32:306–310. doi: 10.1128/jcm.32.2.306-310.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar S, Tamura K, Nei M. MEGA: molecular evolutionary genetic analysis, version 1.01. University Park, Pa: Pennsylvania State University; 1993. [Google Scholar]
- 21.Martinot-Peignoux M, Marcellin P, Pouteau M, Castelnau C, Boyer N, Poliquin M, Degott C, Descombes I, Le Breton V, Milotova V, et al. Pretreatment serum hepatitis C virus RNA levels and hepatitis C virus genotype are the main and independent prognostic factors of sustained response to interferon alpha therapy in chronic hepatitis C. Hepatology. 1995;22(Suppl. 4, part 1):1050–1056. [PubMed] [Google Scholar]
- 22.Mizokami M, Orito E, Gibo Y, Suzuki K, Ohba K, Ohno T, Lau J Y. Genotype, serum level of hepatitis C virus RNA and liver histology as predictors of response to interferon-alpha 2a therapy in Japanese patients with chronic hepatitis C. Liver. 1996;16:23–27. doi: 10.1111/j.1600-0676.1996.tb00699.x. [DOI] [PubMed] [Google Scholar]
- 23.Okamoto H, Okada S, Sugiyama Y, Yotsumoto S, Tanaka T, Yoshizawa H, Tsuda F, Miyakawa Y, Mayumi M. The 5′-terminal sequence of the hepatitis C virus genome. Jpn J Exp Med. 1990;60:167–177. [PubMed] [Google Scholar]
- 24.Persing D H. In vitro nucleic acid amplification techniques. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology principles and applications. 1st ed. Washington, D.C: American Society for Microbiology; 1993. pp. 51–87. [Google Scholar]
- 24a.Saiton N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 25.Simmonds P. Variability of hepatitis C virus. Hepatology. 1995;21:570–583. doi: 10.1002/hep.1840210243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simmonds P, Holmes E C, Cha T A, Chan S W, McOmish F, Irvine B, Beall E, Yap P L, Kolberg J, Urdea M S. Classification of hepatitis C virus into six major genotypes and a series of subtypes by phylogenetic analysis of the NS-5 region. J Gen Virol. 1993;74:2391–2399. doi: 10.1099/0022-1317-74-11-2391. [DOI] [PubMed] [Google Scholar]
- 27.Simmonds P, McOmish F, Yap P L, Chan S W, Lin C K, Dusheiko G, Saeed A A, Holmes E C. Sequence variability in the 5′ non-coding region of hepatitis C virus: identification of a new virus type and restrictions on sequence diversity. J Gen Virol. 1993;74:661–668. doi: 10.1099/0022-1317-74-4-661. [DOI] [PubMed] [Google Scholar]
- 28.Simmonds P, Smith D B, McOmish F, Yap P L, Kolberg J, Urdea M S, Holmes E C. Identification of genotypes of hepatitis C virus by sequence comparisons in the core, E1 and NS-5 regions. J Gen Virol. 1994;75:1053–1061. doi: 10.1099/0022-1317-75-5-1053. [DOI] [PubMed] [Google Scholar]
- 29.Smith D B, Mellor J, Jarvis L M, Davidson F, Kolberg J, Urdea M, Yap P L, Simmonds P The International HCV Collaborative Study Group. Variation of the hepatitis C virus 5′ non-coding region: implications for secondary structure, virus detection and typing. J Gen Virol. 1995;76:1749–1761. doi: 10.1099/0022-1317-76-7-1749. [DOI] [PubMed] [Google Scholar]
- 29a.Sneath P H A, Sokal R R. Numerical taxonomy. San Francisco, Calif: Freeman; 1973. [Google Scholar]
- 30.Stuyver L, Rossau R, Wyseur A, Duhamel M, Vanderborght B, Van Heuverswyn H, Maertens G. Typing of hepatitis C virus isolates and characterization of new subtypes using a line probe assay. J Gen Virol. 1993;74:1093–1102. doi: 10.1099/0022-1317-74-6-1093. [DOI] [PubMed] [Google Scholar]
- 31.Stuyver L, Wyseur A, van Arnhem W, Hernandez F, Maertens G. Second-generation line probe assay for hepatitis C virus genotyping. J Clin Microbiol. 1996;34:2259–2266. doi: 10.1128/jcm.34.9.2259-2266.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stuyver L, Wyseur A, van Arnhem W, Lunel F, Laurent-Puig P, Pawlotsky J M, Kleter B, Bassit L, Nkengasong J, van Doorn L J, et al. Hepatitis C virus genotyping by means of 5′-UR/core line probe assays and molecular analysis of untypeable samples. Virus Res. 1995;38:137–157. doi: 10.1016/0168-1702(95)00052-r. [DOI] [PubMed] [Google Scholar]
- 33.Thorvilson J, Keach N, Mitchell P, Germer J, Persing D. Abstracts of the 96th General Meeting of the American Society for Microbiology, 1996. Washington, D.C: American Society for Microbiology; 1996. In-house developed reverse transcription-polymerase chain reaction (RT-PCR) assay vs. modified Roche Amplicor RT-PCR assay for Hepatitis C Virus (HCV) RNA detection, abstr. C141. [Google Scholar]
- 34.Tsubota A, Chayama K, Ikeda K, Yasuji A, Koida I, Saitoh S, Hashimoto M, Iwasaki S, Kobayashi M, Hiromitsu K. Factors predictive of response to interferon-alpha therapy in hepatitis C virus infection. Hepatology. 1994;19:1088–1094. [PubMed] [Google Scholar]
- 35.Wei Y L, Detre K M, Everhart J E. The NIDDK liver transplantation database. Liver Transplant Surgery. 1997;3:10–22. doi: 10.1002/lt.500030102. [DOI] [PubMed] [Google Scholar]
- 36.Werle E, Schneider C, Renner M, Volker M, Fiehn W. Convenient single-step, one tube purification of PCR products for direct sequencing. Nucleic Acids Res. 1994;22:4354–4355. doi: 10.1093/nar/22.20.4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young K K, Resnick R M, Myers T W. Detection of hepatitis C virus RNA by a combined reverse transcription-polymerase chain reaction assay. J Clin Microbiol. 1993;31:882–886. doi: 10.1128/jcm.31.4.882-886.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zein N N, Persing D H. Hepatitis C genotypes: current trends and future implications. Mayo Clin Proc. 1996;71:458–462. doi: 10.4065/71.5.458. [DOI] [PubMed] [Google Scholar]
- 39.Zein N N, Rakela J, Krawitt E L, Reddy K R, Tominaga T, Persing D H. Hepatitis C virus genotypes in the United States: epidemiology, pathogenicity, and response to interferon therapy. Collaborative Study Group. Ann Intern Med. 1996;125:634–639. doi: 10.7326/0003-4819-125-8-199610150-00002. [DOI] [PubMed] [Google Scholar]