Abstract
Cholangiocarcinomas (CCAs) are diverse biliary epithelial tumours involving the intrahepatic, perihilar and distal parts of the biliary tree. The three entirely variable entities have distinct epidemiology, molecular characteristics, prognosis and strategy for clinical management. However, many cholangiocarcinoma tumor-cells appear to be resistant to current chemotherapeutic agents. The role of autophagy and the therapeutic value of autophagy-based therapy are largely unknown in CCA. The multistep nature of autophagy offers a plethora of regulation points, which are prone to be deregulated and cause different human diseases, including cancer. However, it offers multiple targetable points for designing novel therapeutic strategies. Tumor cells have evolved to use autophagy as an adaptive mechanism for survival under stressful conditions such as energy imbalance and hypoxic region of tumors within the tumor microenvironment, but also to increase invasiveness and resistance to chemotherapy. The purpose of this review is to summarize the current knowledge regarding the interplay between autophagy and cholangiocarcinogenesis, together with some preclinical studies with agents that modulate autophagy in order to induce tumor cell death. Altogether, a combinatorial strategy, which comprises the current anti-cancer agents and autophagy modulators, would represent a positive CCA patient approach.
Keywords: Autophagy, Autophagy modulators, Chemotherapy, Cholangiocarcinoma
Core Tip: The significant role of autophagy in maintaining the energy balance of cancer cells in tumorigenesis remains controversial. A grown body of research data suggests that autophagy is a promising target for several cancer types, including cholangiocarcinomas (CCAs). A novel therapeutic approach which could involve autophagy manipulation plus chemotherapeutic agents may open a new field for more beneficial therapeutic strategies for patients with CCA.
INTRODUCTION
Cholangiocarcinoma (CCA) constitutes a highly malignant group of epithelial tumours, originated in the biliary tree, consisting of three heterogeneous entities based on their anatomical occurrence: (1) The intrahepatic (ICC), 10% of primary liver malignancies, the second most common after hepatocellular carcinoma (HCC); (2) Perihilar (PCC), the most frequent type of CCA ( 50%-60%); and (3) Distal (DCC) which comprises the 20%-30% of all CCA[1-5]. A rare mixed type of CCA and HCC is hepatocellular (CHC-CCA), arising from transdifferentiated hepatocytes[6,7]. CCA is a rare gastrointestinal cancer (3%), however, it exhibits a noticeably increased incidence in the last decades in Western countries (0.3-6 per 10people). It is characterized by a late diagnostic type which contributes to a high mortality rate (1-6 per 105 people) and a worrisome prognosis[8,9]. The highest incidence of CCA is reported in Southeast Asia, especially in Northeast Thailand (85 per 105 people) based on Age-standardized global incidence rates[10]. Except for the geographical variations that imply an interactive relationship between genetic and local environmental risk factors, CCA exhibits gender disparity with a slight male predominance (1.5 fold higher), mostly in the 5th decade of life[4,5,10], as well as racial variation based on karyotyping studies[11,12]. In endemic areas, a well–documented risk factor is the contamination with liver fluke, larvae of Opisthorchis viverrini, and Clonorchissinesisvia food consumption, and occupational exposure aflatoxins, asbestos and plutonium manual-labor and industrial work[13,14]. The majority of CCA cases in the Western world are not related to any obvious predisposing factor[13,15,16]. However, primary sclerosing cholangitis (PSC) is the most reported risk factor[17]. Pathologies related to chronic biliary inflammation account for risk factors like hepatobiliary lithiasis, chronic pancreatitis, fibropolycystic liver disease, non-alcoholic fatty liver disease (NAFLD), cirrhosis, as well as, Hepatitis B and C, viral infections, which are strongly associated with iCCA occurrence[18]. Metabolic diseases like diabetes mellitus type2 (T2DM), obesity, hypertension, as well as other inflammatory diseases may also contribute to the disease[19,20].
Cholangiocarcinogenesis is a multistep event, resulted from deregulated signaling pathways and genomic aberrations[2,21]. Chronic biliary inflammation leads to the proinflammatory cytokine overexpression, like interleukin-6 (IL6), which has the role of growth factor in CCA[22,23]. FGFR gene fusion with MGEA5, TACC3, BICC1, PPHLN1 and ROS is reported, consisting of therapeutic targets[10,24-26]. A variety of mutations have been reported, like KRAS, TP53, RNF43, ROBO2, CDKN2A MLL3, SMAD4, ARID1A, and a recently reported in IDH, which also composes a druggable target[10,27]. KRAS and Tp53 mutations are associated with an aggressive behaviour of tumours and poor prognosis [E], while the latter is frequently coexisting with viral hepatitis B inflection[28,29]. Extrahepatic CCA, are frequently associated with ERBBE, ELF3 mutations and PRKACA-PRKACB fusions, while iCCA with IDH1/2, BRAF, ARID1A and FGFR gene fusions[30]. Epigenetic and microRNAs deregulation, are also reported. The former is frequently resulted by the mutation of MLLE, ARIDA1A and IDH[4,12,31], involved in chromatin remodeling and DNA methylation[3,32,33]. microRNAs up or down-regulation is closely involved in cell cycle function, including autophagy, as well as in invasion, metastasis and chemoresistance[34,35], while they constitute biomarkers for survival and prognosis prediction, especially miR-10b, miR-22 and miR-551b[10,36-38].
Autophagy is a multiphasic, homeostatic, self-degenerative cellular mechanism by which non-functional, clustered or mutant proteins and impaired organelles such as Endoplasmic reticulum, peroxisomes or mitochondria, are insulated into vesicles, which are further fused with lysosomes for the degeneration process[39]. Autophagy appears to have a dual role in cancer, either promotes or suppress carcinogenesis. This peculiar capacity has created new therapeutic strategies for cancer via interfering in autophagy steps[40]. Despite the fact that autophagy’s regulatory mechanism on tumors is still examined, many studies demonstrate propitious results of its therapeutic potency, especially in combination with other chemotherapeutic agents[41].
Based on several preclinical studies, disturbances in autophagy regulation are closely related to carcinogenesis in cholangiocytes, as well as with metastasis and dismal outcomes, while it can act as a potent anti-cancer drug target[42].
This review gathers information from the current clinical and preclinical research data, about autophagy modulation in CCA and the therapeutic strategies for this highly invasive malignancy.
MAIN ROLE OF AUTOPHAGY IN CANCER BIOLOGY
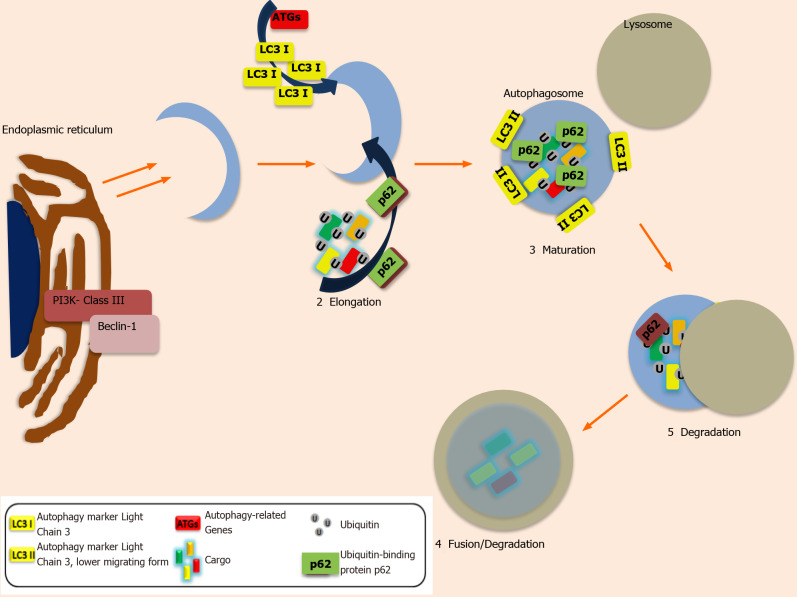
Autophagy (previously described as Macroautophagy) ensures cellular survival under stressful conditions[40]. Other less described entities of autophagy are: Microautophagy, which includes engulfment of intracellular components via the invagination of cell-membrane and fusion with lysosomes, as well as chaperon-mediated autophagy direct translocation of the targeted protein towards lysosomes for the degradation process[39]. In Figure 1, the main steps of autophagy are presented.
Figure 1.
The stages of autophagosome formation. The autophagy process includes five distinct steps: initiation, elongation, maturation, fusion and degradation. In the first step or initiation (1), the double-membrane structure, the Phagophore, is formed after activation of PI3K-classIII – Beclin-1 complex in endoplasmic reticulum. Elongation (2) is the second step where the new-formed phagophore begin to enclose Ubiquitin-labeled cytosolic cargos such as proteins. A plethora of proteins such as LC3 (LC3-I is conjugated to phosphatidylethanolamine to form LC3-phosphatidylethanolamine conjugate or LC3-II, responsible for the autophagosomal membrane structure), Tags (Autophagy-related genes) and p62 (an adaptor protein responsible for the docking of cargoes) have a key role in the Maturation (3) step where the Autophagosome has already formed. In the fourth step or Fusion/degradation (4) step, the Autophagosome is fused with a Lysosome in order to create the autolysosomes wherein the degradation step (5) the cytosolic cargos are digested from lysosomal enzymes.
Despite the fact that it is a physiological mechanism, it has a dual role (as it was mentioned before), either as a tumour suppressor or promoter of tumorigenesis and metastasis[43,44]. This complex procedure includes a series of steps in order to allow the engulfment of the cellular organelles by vesicles, the formation and the expansion of phagophore, the maturation into autophagosome and the fusion of the latter with the lysosome, with the formation of autolysosome, which is responsible for the degradation and recycling of the organelles[39]. The first step of the mechanism (induction) is initiated, by the inactivation of mammalian target of rapamycin (mTOR), allowing the activation of Unc-51-like kinase1 complex (ULK1) and the cargo selection and engulfment by vacuoles. The second step (nucleation), includes the activation and phosphorylation of activated class III PI3K complex by ULK1, with the formation of PI3K -Beclin-1 complex[39]. In the third step, phagophore starts to expand via membrane elongation, which includes two conjugations of ATG5–ATG12 complex with ATG16 and the LC3I (soluble-form) to lipid phosphatidylethanolamine (PE), with the former recruiting more cargo for the phagophore expansion and the latter resulting into LC3II (lipid form), which locates in autophagosome-membrane for the binding of degradation-products[45].The fourth step includes the formation of autolysosome (fusion of the autophagosome with lysosome) and the fifth, the degradation of cargo and the recycling of the products[39,46]. Autophagy is closely related to the tumour microenvironment (TME), exhibiting a protective role via the degradation of damaged cargo and the inhibition of tumour growth in early malignant stages[45], like damaged mitochondria, a major source of mutagenic reactive-oxygen species (ROS)[47-49]. In the late stages, it is used by the cancer cells for their adaptation and survival, in extreme micro-environmental conditions, like hypoxia and starvation, having the role of tumour growth promoter[42,45,50]. All the steps can be targeted in anti-cancer therapy via the induction of an autophagiccytoprotective mechanism, which can further reduce the chemo-resistance and induce cancer cell death[40,51].
ROLE OF AUTOPHAGY IN CHOLANGIOCARCINOMA
CCA is a highly diversified group of malignancies that exhibit various risk factors and an aberrant epigenetic and genetic landscape[2]. The well-established therapeutic strategies include surgical tumor resection, chemotherapy regimens, as well as locoregional therapies. Only a limited portion of patients (1/3) are eligible for tumor resection at the diagnostic time, which are further receive adjuvant chemotherapy, including either gemcitabine, cisplatin, or 5-fluorouracil (5-FU). However, they cannot put a halt to tumor recurrence and resistance. Due to the highly aggressive behavior of these malignancies, the majority of patients are diagnosed when the metastasis already occurs, or the resection is unfeasible. Palliative care is reserved for these cases, including the combination of chemotherapeutic agents as gemcitabine and cisplatin, which nevertheless exhibit limited benefits[52,53]. In unsuccessful treatment cases with the former combination, another regimen is reserved, based on fluoropyrimidine[2].
Genetic and epigenetic information, as well as the knowledge of the molecular pathways in CCA, which contribute to tumor resistance, relapse, as well as metastatic behavior, open up more therapeutic approaches via the usage of molecular agents, although with moderate overall survival enhancement[1,54,55]. Genomic profiling of iCCA sub-classifies it into: (i) inflammatory and (ii) proliferative classes. In the former, the activation of inflammatory pathways mainly occurs, while on the latter, the activation of oncogenes demonstrates a more worrisome prognosis[12].
The heterogeneity of CCAs subtypes is also demonstrated by Next-generation sequencing analysis, which indicates different genetic mutations based on CCA’s anatomical location. (iCCA vs extrahepatic: pCCA and dCCA). RAS mutation is more frequently exhibited in CCA, particularly in dCCA[56], however, there is a subclass of CCA, without exhibiting it. There is also emerging evidence of gene FGFR2 fusions involvement in cholangiocarcinogenesis, based on exome sequencing analysis[57]. Aberrations are also identified in the epigenetic level of gene regulation, such as histone modification, DNA hypermethylation and microRNAs (miRNAs) dysregulation, all implied in CCA Tumorigenesis[34].
As alluded to previously, a better understanding of the molecular, genomic and epigenetic affected pathways driving to CCA development and progression could give rise to new and improved generations of therapeutic approaches based on patient-stratification. Major factors implicated in CCA establishment are chronic biliary inflammation, ductal obstruction with cholestasis and bile duct injury[58]. As a consequence of chronic inflammation, proinflammatory cytokines’ overexpression occurs (TNF, IL-6, endotoxins). Persistent secretion of IL-6 by inflamed cholangiocytes and immune cells contributes to cancer establishment and progress. IL-6 oversecretion, induces nitric oxide production (via nitric oxide synthase), which is implied in DNA oxidation and damage[59], as well as stimulates the secretion of cyclic oxygenase (COX)-2-mediated prostaglandin, which promotes angiogenesis and disrupts the programmed cell death[60].
Autophagy has a crucial role in inflammation; however, their correlation is still being researched[44]. A large number of signaling pathways are involved in chronic inflammation, during cancer establishment, which influences the process of autophagy. An example of this interconnection is the persistent overexpression of IL6 by lung cells, as a response to arsenic exposure, that down-regulates autophagy and promotes malignant transformation[61]. IL-6 up-regulation influences the STAT3 signalling pathway via the inhibition of the Beclin1-Bcl2 complex, which further enables an IL-6-dependent transformation. On the contrary, Beclin1 over-stimulation enables the blockage of this transformation[61]. The above interrelation between IL-6-dependent transformation and autophagy during tumorigenesis could open up treatment opportunities in inflammatory-type iCCA. Additionally, many studies demonstrate the correlation of different pro-inflammatory signaling pathways with autophagy and stress[42].
Based on studies, many genetic mutations have been reported, implied in CCA. Harboring mutant KRAS has been identified in 40% of CCAs, particularly in dCCA with dismal outcomes[56]. Moreover, it is also related to lymphatic dissemination, lower long-term OS and higher grade, as was demonstrated in a study with a limited number of patients with iCCA and mutant KRAS gene (7.4%)[62]. In addition, based on an animal model study, concomitant mutations of KRAS and P53 are related to worse overall survival and malignant transformation in murine[63], while they constitute the most frequently reported genetic modifications[56,64]. The iCCA in murine demonstrates similar morphopathological characteristics with humans and presents an upregulation of autophagy mechanism, contributing to tumor development. The utilization of chloroquine (CQ) ceased tumor growth via the inhibition of the mechanism, resulting in the accumulation of LC3-II[42]. Human iCCA, with KRAS and P53 alterations, exposed as well increased mechanism of autophagy, compared with iCCA without them and the tumor progression was similarly inhibited via the use of CQ[42,63].
The development of KRAS axis inhibitors, such as selumetinib, opens up new therapeutic strategies, which potentially could be enhanced via the addition of autophagy modulators[1,65]. Mutations in MET lead to STAT modulation, Akt/PI3K and MAPK signaling pathways and are associated with aggressiveness, higher tumor stage, and reduced survival[66,67].
Furthermore, c-MET inhibition is related to an increased level of autophagy, as it was demonstrated in lung cancer[68]. Similarly, mutant EGFR and ERBB genes are associated as well with poor outcome and invasiveness[69,70]. In many cancers, treated with inhibitors of tyrosine kinase, autophagy acts as a tumor suppressor[71]. The combination of autophagy and tyrosine kinase inhibitors could potentially improve the treatment results. Moreover, the fusion of FGFR2 genes is demonstrated in CCA[72], and they are correlated with decreased autophagy levels, leading to tumorigenesis. Inhibition of the above gene induces autophagy as a tumor suppressor mechanism in breast and lung malignancies, and its effect can be enhanced with the combination of autophagy inhibitors[73,74]. All the above data support that the combination of these inhibitors could potentially increase the therapeutic potential in CCA. Alteration in the SMAD4 gene is mainly identified in dCCA[75] and in pancreatic malignancy, in which increased autophagy is associated with resistance to radiotherapy[76]. Similarly, inhibition of autophagy could also be beneficial to this type of cancer.
In the initial phase of cholangiocarcinogenesis, Adenomatous Polyposis Coli (APC) mutation has been also reported[77], with the altered mechanism of autophagy[78] and during the establishment of cancer models[79]. Aberrations in the epigenetic level, such as histone modification, DNA hypermethylation, and miRNAs deregulation, are crucial for CCA establishment and development[80] while modulating the autophagy process[81], as well. The expression and the characteristics of the cilium are influenced by the increased expression of histone deacetylase 6 (HDAC6), which reduces its length and increases its proliferation. Inhibition of HDAC6 is correlated with reduced tumor progression and restoration of cilia[82,83]. Suppression of autophagy contributes to the effects of HDAC6 inhibition in many cancers such as neuroblastoma, colorectal and multiple myeloma[84].
Aggressiveness and dismal outcome of iCCA, are also reported in cases of modified HDAC1expression[85]. Significant autophagy regulators are the methylations of histone, which decelerate it[86]. Inactivation of tumor suppressors, caused by DNA methylation, is reported in cholangiocarcinogenesis. DNA hypermethylation of IDH1/2, is identified in some iCCA cases (10%), which leads to deregulation of cellular functions, such as their differentiation[87,88]. Mutation of IDH, identified in gliomas, demonstrates the interconnection of autophagy suppression and methylations of histone[81,86], which open up therapeutic opportunities via autophagy inhibitors[89]. Deregulation of many non-coding RNA sequences, such as miR-21, miR-29, miR-141 and others, present either up or down-regulation and they constitute biomarkers for tumor progression, invasion, cancer cell-death and chemoresistance in CCA[90,91]. Autophagy and its components, such as autophagy-associated proteins (ATG4, ATG9), beclin1, LC3 and ULK2, are also modulated via miRNAs[92,93]. Induction of autophagy, via the action of miR-124, resulted in an altered STAT3 signaling pathway, as it was reported[94].
Autophagy modulators in combination with immunotherapy, targeted therapies and chemotherapy are positioning as a promising strategy to increase therapeutic benefits for cancer patients. Current treatment options for patients with CCA are limited to chemotherapy, thus, combinatorial scheme including autophagy modulators could offer an opportunity to increase survival of patients with CCA. Autophagy inhibition such as Hydroxy-chloroquine (HCQ) alters the mechanism of resistance and could potentially decrease CCA metastatic potential; therefore, clinical results of this study would be of great help for further design of novel therapeutic approaches involving autophagy inhibitors in CCA. Recent studies revealed the potential of the well-known autophagy marker, Beclin-1, as a prognostic factor in different cancers including CCA. It has emphasized the necessity to combine Beclin-1 expression with other autophagy-related proteins such as Bcl-2 family proteins Bcl-xL and BNIP3, HIF-1α, PI3KC3 or ATGs to increase its clinical value for patients with CCA.
TARGETING AUTOPHAGY—A PUTATIVE THERAPEUTIC OPTION
Autophagy activators and cancer therapy
Many studies demonstrate the correlation of autophagy mechanism with the microenvironment of tumors and the antitumor immune response, in many cancers, including CRC. Major histocompatibility complex (MCH) I/II Ag presentation is closely regulated by autophagy mechanism, as well as the cellular apoptosis. The multi-roles of autophagy gave the opportunity for the development of antitumor agents that induce this mechanism. Notable activators are Rapamycin and its analog-like, deforolimus, rapalogs like temsirolimus and everolimus and mTOR inhibitors, which activate the mechanism of autophagy[95].
More particularly, it is demonstrated that therapy with Rapamycin intensifies radiotherapy effects on A549 malignant lung cells via autophagy activation and by expressing a dilatory effect on genome damage repairing[96]. Rapalogs, like everolimus, have been indicated that suppress the progression and the growth of malignant endometrial cells, especially when Paclitaxel is added to the therapeutic scheme[97,98]. Both of the above autophagy activators can be added to anti-cancer therapeutic strategies, with another kind of antitumor medication. However, their use in clinical practice should be further examined[97].
Furthermore, it is reported that another anti-proliferative agent, that inducts autophagy mechanism is the well-known metformin, which directs inhibition of autophagy, or via blocking beclin-1. Moreover, it is reported that metformin induces autophagy mechanism in the case of adenocarcinoma in the lung, as well as cell apoptosis via increasing tumor necrosis factor (TNF), the so-called TNF-Related-Apoptosis-Inducing Ligand (TRAIL), apoptosis[99]. In breast cancer therapy, without BRCA1 mutation, metformin is combined with another autophagy inhibitor, spautin-1, which sensitizes these tumors, for the mitochondrial-targeted disruptors. In this case, the combination of an autophagy activator and inhibitor, like metformin and spautin-1, responsively can modify the function of mitochondria differently, resulting in reducing the cell life span[100].
Induction of autophagy can be achieved via another agent, like Obatoclax, commonly reported in Hematologic malignant diseases[101]. This agent aims at the Bcl-2 protein family, which is closely associated with cell-apoptosis at the mitochondrion, while is also influencing autophagosome membranes vianecrosome congregation, resulting in necroptosis[45,102].
Alkaloids are identified as another group of autophagy inducers in malignancies[103]. Some of them are liensinine, isoliensinine and cepharanthine[48], which target AMPK phosphorylation and mTOR blockage. These agents have been utilized in cases of MEFs, in which we are presenting resistance in the cell-apoptosis mechanism[102].
In addition to the well-established antioxidant function of omega-3polyunsaturated fatty acids (ω-3 PUFAs)[104], it has been shown that these safenatural compounds can induce 15-hydroxyprostaglandin dehydrogenase(15-PGDH) leading to inactivation of prostaglandin E2 (PGE2) that is knownto drive human cholangiocarcinoma[105]. The latter, combined with the fact that ω-3 PUFAs induce autophagy-mediated cell death in cancer cells support the use of ω-3 PUFAs as non-toxic adjuvant therapeutic agents for the treatment of human cholangiocarcinoma[106].
Autophagy inhibitors and cancer therapy
A wide range of studies about autophagy and its influence on the efficacy of other cancer treatments, such as chemotherapy, radiotherapy, or immunotherapy, has been reported in the last years[107]. These studies focused on this mechanism, used by cancer cells for their energy, metabolic regulation and survival[40,108]. The dual role of autophagy, either as tumor promoter, or tumor suppressor, opened up new opportunities for anti-cancer treatment via autophagy- inhibitors.
The most widely known inhibitors are Chloroquine (CQ) and hydroxychloroquine (HCQ), which impede the fusion of autophagosomes with the lysosomes. Their efficiency as anti-cancer therapy has been evaluated in a variety of malignancies[43]. However, their clinical significance as monotherapy was limited due to their non-persistent inhibition[109]. The combination of other cancer therapies demonstrated better therapeutic results[41,110], such as the combination of HCQ with gemcitabine in the case of pancreatic adenocarcinoma, which resulted in a significant reduction of tumor marker 19-9 (60%)[111].
Moreover, the combination of immunotherapy and autophagy inhibitors, such as CQ with IL-2, has been proven beneficial with reduced toxicity, such as in animal-model studies of murine with hepatic metastasis. Furthermore, it was demonstrated that this dual therapeutic strategy, has a better survival rate in the long term as well as a better response by immune cells[107]. However, the response to CQ derivatives, including HCQ is variable, due to the lack of specificity, which leads to the interaction with other medical substances and the modification of tumor properties, like pH[109,112]. Additionally, the efficacy of autophagy inhibition by the above agents, cannot be evaluated due to the absence of biomarkers, which is a significant limitation in the clinical practice. This is the reason that new inhibitors with higher specificity have been developed[41,107].
There are some new, efficacious inhibitors, such as Lys05, also described as dimeric chloroquine, which is well–tolerated and exhibits a strong antitumor action via the modification of lysosome enzymes[112]. Another one is SAR405, an inhibitor of kinase, which is more specific and targets Vps18 and Vps34 vacuole proteins, which have a crucial role in the initiation of autophagy-mechanism. More particularly, the initiation step is regulated by Beclin-1 and Vps34, whereas Vps34 suppression, results in the impairment of lysosomal and vesicular transport[113]. Initiation-step can also be targeted, via the use of Beclin-1 inhibitors, which suppress the tumor progression, intensify the antitumor activity of Natural Killer (NK) cells and induce CCL5 cytokine overexpression by cancer cells, a condition that influences the transporting of NK cells towards the malignant tumors[107].
Based on studies in various malignancies, the inhibition of ULK1 (Unc-51 Like kinase-1) by SBI-0206965, has great antitumor potential due to its higher selectivity, resulting from the suppression of ULK1-phosphorylations[114]. Some other agents, are DCMI including desmethylclomipramine, verteporfin and clomipramine, impeding the fusion of autophagosome with lysosomes or acidification lysosomes[115], whereas the addition of DCMI to doxorubicin, in vitro, demonstrated higher effectiveness of the latter[116]. Moreover, spautin-1, is another effective inhibitor, which impedes the initiation step of autophagy, by suppressing the crucial for the process ubiquitin-specific peptidases USP13, USP10, as well as Beclin-1, which is deubiquitinated in Vps34 complex[99].
The microenvironment of tumors, is closely related to the autophagy mechanism, as well as with the antitumor immune response. According to this fact, the inhibition of autophagy could have a negative impact on the adaptive immune response against malignant tumors. However, Starobinetset al[117] in 2016 confuted this hypothesis by proving that inhibition of autophagy does not have an adverse impact on the adaptive anti-cancer immunity in melanoma and breast cancers. For this reason, inhibitors of autophagy can be combined with another chemotherapeutic agent without negatively influencing the antitumor response of T cells towards malignant tumors[117].
Herein, we provide two summarized tables about small agents that inhibit or activate autophagy. Autophagy manipulation is already used in research to develop putative chemotherapeutic strategies with a plethora of agents for different types of cancer (Tables 1 and 2).
Table 1.
Small molecules able to induce autophagic activity
|
Agents
|
Mechanism of action
|
| GDC-0941 | Inhibitor of class I PI3K |
| GDC-0980 | Dual inhibitor of PI3K and mTORC1 |
| Everolimus | mTORC1 inhibitor |
| Temsirolimus | mTORC1 inhibitor |
| Rapamycin | mTORC1 inhibitor |
| Tat–beclin 1 peptide | Releases beclin-1 into cytoplasm-regulate autophagosome formation |
| Metformin | AMPK activator |
| Fluspirilene | Antagonists of L-type Ca2+ channels |
| Loperamide | Antagonists of L-type Ca2+ channels |
| Amiodarone | Antagonists of L-type Ca2+ channels |
| Isoliensinine | Natural alkaloid |
| Cepharanthine | Natural alkaloid |
mTORC1: Mammalian target of rapamycin complex 1; AMPK: 5’ AMP-activated protein kinase; PI3K: Phosphatidylinositol 3-kinases; AKT: Protein kinase B; Beclin-1: The mammalian ortholog of the yeast autophagy-related gene 6 (Atg6).
Table 2.
Small molecules able to inhibit autophagic activity
|
Agents
|
Mechanism of action
|
| 3-Methyladenine (3-MA) | Inhibitor of class III PI3K |
| LY294002 | PI3K inhibitor |
| Wortmannin | PI3K inhibitor |
| SB202190 | Cross-inhibition of the PI3K/mTOR and MAPKs pathway |
| MHY1485 | Activator of mTOR |
| Azithromycin | Inhibitor of v-ATPase, inhibition of lysosomal acidification |
| Bafilomycin A1 | Inhibitor of v-ATPase, inhibition of lysosomal acidification |
| Concanamycin A | Inhibitor of v-ATPase, inhibition of lysosomal acidification |
| Chloroquine (CQ) | Autophagosome-lysosome fusion |
| Hydroxy-chloroquine (HCQ) | Autophagosome-lysosome fusion |
| Clomipramine | Alter acidification of lysosomes |
| Verteporfin | Alter acidification of lysosomes |
| Paclitaxel | Microtubule stabilizer- inhibit phosphorylation of VPS34 at T159 |
| Spain-1 | Inhibits the activity of ubiquitin-specific peptidases, USP10 and USP13 |
| Monensin | Inhibit autophagosome-lysosome fusion |
PI3K: Phosphatidylinositol 3-kinases; mTORC1: Mammalian target of rapamycin complex 1; AMPK: 5’ AMP-activated protein kinase;VPS: Vacuolar protein sorting; ATG: Autophagy-related proteins; USP: Ubiquitin-specific protease.
CONCLUSION
It is a well-established knowledge that autophagy’s prominent role is strongly correlated with the degradation of dysfunctional cellular proteins and organelles. A plethora of studies in the field of cancer research and autophagy highlights the controversial role of this mechanism either as tumor suppressor or promoter mechanism in different types of cancer, including CCA. Several in vitro and in vivo studies in CCAs have associated autophagy with cholangiocarcinogenesis development and progression. Furthermore, autophagy markers such as Beclin-1 and LC3 and/or autophagy-associated proteins appeared to associate with a different CCAs stage through miRNAs expression. Current treatment options for CCA are limited to chemotherapy with limited efficacy on CCA patients. Agents that modulate autophagy in different steps in combination with the currents chemotherapeutic drugs are proposed as a promising therapeutic strategy in order to increase the beneficial effect of the therapeutic expectancy of cancer patients.
Footnotes
Conflict-of-interest statement: The authors declare no conflict of interests for this article.
Manuscript source: Invited manuscript
Peer-review started: February 21, 2021
First decision: April 19, 2021
Article in press: August 3, 2021
Specialty type: Oncology
Country/Territory of origin: Greece
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Abdel Moneim AE, Li Y S-Editor: Ma YJ L-Editor: Filipodia P-Editor: Yuan YY
Contributor Information
Evangelos Koustas, Department of Biological Chemistry, Medical School, National and Kapodistrian University of Athens, Athens 11527, Greece. vang.koustas@gmail.com.
Eleni-Myrto Trifylli, Department of Biological Chemistry, Medical School, National and Kapodistrian University of Athens, Athens 11527, Greece.
Panagiotis Sarantis, Department of Biological Chemistry, Medical School, National and Kapodistrian University of Athens, Athens 11527, Greece.
Athanasios G Papavassiliou, Department of Biological Chemistry, Medical School, National and Kapodistrian University of Athens, Athens 11527, Greece.
Michalis V Karamouzis, Department of Biological Chemistry, Medical School, National and Kapodistrian University of Athens, Athens 11527, Greece.
References
- 1.Banales JM, Cardinale V, Carpino G, Marzioni M, Andersen JB, Invernizzi P, Lind GE, Folseraas T, Forbes SJ, Fouassier L, Geier A, Calvisi DF, Mertens JC, Trauner M, Benedetti A, Maroni L, Vaquero J, Macias RI, Raggi C, Perugorria MJ, Gaudio E, Boberg KM, Marin JJ, Alvaro D. Expert consensus document: Cholangiocarcinoma: current knowledge and future perspectives consensus statement from the European Network for the Study of Cholangiocarcinoma (ENS-CCA) Nat Rev Gastroenterol Hepatol. 2016;13:261–280. doi: 10.1038/nrgastro.2016.51. [DOI] [PubMed] [Google Scholar]
- 2.Rizvi S, Khan SA, Hallemeier CL, Kelley RK, Gores GJ. Cholangiocarcinoma - evolving concepts and therapeutic strategies. Nat Rev Clin Oncol. 2018;15:95–111. doi: 10.1038/nrclinonc.2017.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cholangiocarcinoma Working Group. Italian Clinical Practice Guidelines on Cholangiocarcinoma - Part I: Classification, diagnosis and staging. Dig Liver Dis. 2020;52:1282–1293. doi: 10.1016/j.dld.2020.06.045. [DOI] [PubMed] [Google Scholar]
- 4.Lendvai G, Szekerczés T, Illyés I, Dóra R, Kontsek E, Gógl A, Kiss A, Werling K, Kovalszky I, Schaff Z, Borka K. Cholangiocarcinoma: Classification, Histopathology and Molecular Carcinogenesis. Pathol Oncol Res. 2020;26:3–15. doi: 10.1007/s12253-018-0491-8. [DOI] [PubMed] [Google Scholar]
- 5.Khan SA, Tavolari S, Brandi G. Cholangiocarcinoma: Epidemiology and risk factors. Liver Int. 2019;39 Suppl 1:19–31. doi: 10.1111/liv.14095. [DOI] [PubMed] [Google Scholar]
- 6.Munoz-Garrido P, Rodrigues PM. The jigsaw of dual hepatocellular-intrahepatic cholangiocarcinoma tumours. Nat Rev Gastroenterol Hepatol. 2019;16:653–655. doi: 10.1038/s41575-019-0185-z. [DOI] [PubMed] [Google Scholar]
- 7.Xue R, Chen L, Zhang C, Fujita M, Li R, Yan SM, Ong CK, Liao X, Gao Q, Sasagawa S, Li Y, Wang J, Guo H, Huang QT, Zhong Q, Tan J, Qi L, Gong W, Hong Z, Li M, Zhao J, Peng T, Lu Y, Lim KHT, Boot A, Ono A, Chayama K, Zhang Z, Rozen SG, Teh BT, Wang XW, Nakagawa H, Zeng MS, Bai F, Zhang N. Genomic and Transcriptomic Profiling of Combined Hepatocellular and Intrahepatic Cholangiocarcinoma Reveals Distinct Molecular Subtypes. Cancer Cell. 2019;35:932–947.e8. doi: 10.1016/j.ccell.2019.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bertuccio P, Malvezzi M, Carioli G, Hashim D, Boffetta P, El-Serag HB, La Vecchia C, Negri E. Global trends in mortality from intrahepatic and extrahepatic cholangiocarcinoma. J Hepatol. 2019;71:104–114. doi: 10.1016/j.jhep.2019.03.013. [DOI] [PubMed] [Google Scholar]
- 9.Maemura K, Natsugoe S, Takao S. Molecular mechanism of cholangiocarcinoma carcinogenesis. J Hepatobiliary Pancreat Sci. 2014;21:754–760. doi: 10.1002/jhbp.126. [DOI] [PubMed] [Google Scholar]
- 10.Banales JM, Marin JJG, Lamarca A, Rodrigues PM, Khan SA, Roberts LR, Cardinale V, Carpino G, Andersen JB, Braconi C, Calvisi DF, Perugorria MJ, Fabris L, Boulter L, Macias RIR, Gaudio E, Alvaro D, Gradilone SA, Strazzabosco M, Marzioni M, Coulouarn C, Fouassier L, Raggi C, Invernizzi P, Mertens JC, Moncsek A, Rizvi S, Heimbach J, Koerkamp BG, Bruix J, Forner A, Bridgewater J, Valle JW, Gores GJ. Cholangiocarcinoma 2020: the next horizon in mechanisms and management. Nat Rev Gastroenterol Hepatol. 2020;17:557–588. doi: 10.1038/s41575-020-0310-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koo SH, Ihm CH, Kwon KC, Park JW, Kim JM, Kong G. Genetic alterations in hepatocellular carcinoma and intrahepatic cholangiocarcinoma. Cancer Genet Cytogenet. 2001;130:22–28. doi: 10.1016/s0165-4608(01)00460-5. [DOI] [PubMed] [Google Scholar]
- 12.Sia D, Hoshida Y, Villanueva A, Roayaie S, Ferrer J, Tabak B, Peix J, Sole M, Tovar V, Alsinet C, Cornella H, Klotzle B, Fan JB, Cotsoglou C, Thung SN, Fuster J, Waxman S, Garcia-Valdecasas JC, Bruix J, Schwartz ME, Beroukhim R, Mazzaferro V, Llovet JM. Integrative molecular analysis of intrahepatic cholangiocarcinoma reveals 2 classes that have different outcomes. Gastroenterology. 2013;144:829–840. doi: 10.1053/j.gastro.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suk WA, Bhudhisawasdi V, Ruchirawat M. The Curious Case of Cholangiocarcinoma: Opportunities for Environmental Health Scientists to Learn about a Complex Disease. J Environ Public Health. 2018;2018:2606973. doi: 10.1155/2018/2606973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilcox BA, Echaubard P. Balancing biomedical and ecological perspectives in research framing of liver fluke and cholangiocarcinoma in NE Thailand. Parasitol Int. 2017;66:372–377. doi: 10.1016/j.parint.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 15.Fiorino S, Bacchi-Reggiani L, de Biase D, Fornelli A, Masetti M, Tura A, Grizzi F, Zanello M, Mastrangelo L, Lombardi R, Acquaviva G, di Tommaso L, Bondi A, Visani M, Sabbatani S, Pontoriero L, Fabbri C, Cuppini A, Pession A, Jovine E. Possible association between hepatitis C virus and malignancies different from hepatocellular carcinoma: A systematic review. World J Gastroenterol. 2015;21:12896–12953. doi: 10.3748/wjg.v21.i45.12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petrick JL, Yang B, Altekruse SF, Van Dyke AL, Koshiol J, Graubard BI, McGlynn KA. Risk factors for intrahepatic and extrahepatic cholangiocarcinoma in the United States: A population-based study in SEER-Medicare. PLoS One. 2017;12:e0186643. doi: 10.1371/journal.pone.0186643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fung BM, Lindor KD, Tabibian JH. Cancer risk in primary sclerosing cholangitis: Epidemiology, prevention, and surveillance strategies. World J Gastroenterol. 2019;25:659–671. doi: 10.3748/wjg.v25.i6.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsumoto K, Onoyama T, Kawata S, Takeda Y, Harada K, Ikebuchi Y, Ueki M, Miura N, Yashima K, Koda M, Sakamoto T, Endo M, Horie Y, Murawaki Y. Hepatitis B and C virus infection is a risk factor for the development of cholangiocarcinoma. Intern Med. 2014;53:651–654. doi: 10.2169/internalmedicine.53.1410. [DOI] [PubMed] [Google Scholar]
- 19.Kongpetch S, Jusakul A, Ong CK, Lim WK, Rozen SG, Tan P, Teh BT. Pathogenesis of cholangiocarcinoma: From genetics to signalling pathways. Best Pract Res Clin Gastroenterol. 2015;29:233–244. doi: 10.1016/j.bpg.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 20.Jing W, Jin G, Zhou X, Zhou Y, Zhang Y, Shao C, Liu R, Hu X. Diabetes mellitus and increased risk of cholangiocarcinoma: a meta-analysis. Eur J Cancer Prev. 2012;21:24–31. doi: 10.1097/CEJ.0b013e3283481d89. [DOI] [PubMed] [Google Scholar]
- 21.Ong CK, Subimerb C, Pairojkul C, Wongkham S, Cutcutache I, Yu W, McPherson JR, Allen GE, Ng CC, Wong BH, Myint SS, Rajasegaran V, Heng HL, Gan A, Zang ZJ, Wu Y, Wu J, Lee MH, Huang D, Ong P, Chan-on W, Cao Y, Qian CN, Lim KH, Ooi A, Dykema K, Furge K, Kukongviriyapan V, Sripa B, Wongkham C, Yongvanit P, Futreal PA, Bhudhisawasdi V, Rozen S, Tan P, Teh BT. Exome sequencing of liver fluke-associated cholangiocarcinoma. Nat Genet. 2012;44:690–693. doi: 10.1038/ng.2273. [DOI] [PubMed] [Google Scholar]
- 22.Isomoto H, Mott JL, Kobayashi S, Werneburg NW, Bronk SF, Haan S, Gores GJ. Sustained IL-6/STAT-3 signaling in cholangiocarcinoma cells due to SOCS-3 epigenetic silencing. Gastroenterology. 2007;132:384–396. doi: 10.1053/j.gastro.2006.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmoldt A, Benthe HF, Haberland G. Digitoxin metabolism by rat liver microsomes. Biochem Pharmacol. 1975;24:1639–1641. [PubMed] [Google Scholar]
- 24.Charest A, Lane K, McMahon K, Park J, Preisinger E, Conroy H, Housman D. Fusion of FIG to the receptor tyrosine kinase ROS in a glioblastoma with an interstitial del(6)(q21q21) Genes Chromosomes Cancer. 2003;37:58–71. doi: 10.1002/gcc.10207. [DOI] [PubMed] [Google Scholar]
- 25.Borad MJ, Champion MD, Egan JB, Liang WS, Fonseca R, Bryce AH, McCullough AE, Barrett MT, Hunt K, Patel MD, Young SW, Collins JM, Silva AC, Condjella RM, Block M, McWilliams RR, Lazaridis KN, Klee EW, Bible KC, Harris P, Oliver GR, Bhavsar JD, Nair AA, Middha S, Asmann Y, Kocher JP, Schahl K, Kipp BR, Barr Fritcher EG, Baker A, Aldrich J, Kurdoglu A, Izatt T, Christoforides A, Cherni I, Nasser S, Reiman R, Phillips L, McDonald J, Adkins J, Mastrian SD, Placek P, Watanabe AT, Lobello J, Han H, Von Hoff D, Craig DW, Stewart AK, Carpten JD. Integrated genomic characterization reveals novel, therapeutically relevant drug targets in FGFR and EGFR pathways in sporadic intrahepatic cholangiocarcinoma. PLoS Genet. 2014;10:e1004135. doi: 10.1371/journal.pgen.1004135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Luca A, Esposito Abate R, Rachiglio AM, Maiello MR, Esposito C, Schettino C, Izzo F, Nasti G, Normanno N. FGFR Fusions in Cancer: From Diagnostic Approaches to Therapeutic Intervention. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21186856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abou-Alfa GK, Mercade TM, Javle M, Kelley RK, Lubner S, Adeva J, Cleary JM, Catenacci DV, Borad MJ, Bridgewater JA, Harris WP, Murphy AG, Oh DY, Whisenant J, Wu B, Jiang L, Gliser C, Pandya SS, Valle JW, Zhu AX. LBA10_PR - ClarIDHy: A global, phase III, randomized, double-blind study of ivosidenib (IVO) vs placebo in patients with advanced cholangiocarcinoma (CC) with an isocitrate dehydrogenase 1 (IDH1) mutation. Ann Oncol. 2019;30:872–873. [Google Scholar]
- 28.Nepal C, O'Rourke CJ, Oliveira DVNP, Taranta A, Shema S, Gautam P, Calderaro J, Barbour A, Raggi C, Wennerberg K, Wang XW, Lautem A, Roberts LR, Andersen JB. Genomic perturbations reveal distinct regulatory networks in intrahepatic cholangiocarcinoma. Hepatology. 2018;68:949–963. doi: 10.1002/hep.29764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zou S, Li J, Zhou H, Frech C, Jiang X, Chu JS, Zhao X, Li Y, Li Q, Wang H, Hu J, Kong G, Wu M, Ding C, Chen N, Hu H. Mutational landscape of intrahepatic cholangiocarcinoma. Nat Commun. 2014;5:5696. doi: 10.1038/ncomms6696. [DOI] [PubMed] [Google Scholar]
- 30.Gingras MC, Covington KR, Chang DK, Donehower LA, Gill AJ, Ittmann MM, Creighton CJ, Johns AL, Shinbrot E, Dewal N, Fisher WE Australian Pancreatic Cancer Genome Initiative, Pilarsky C, Grützmann R, Overman MJ, Jamieson NB, Van Buren G 2nd, Drummond J, Walker K, Hampton OA, Xi L, Muzny DM, Doddapaneni H, Lee SL, Bellair M, Hu J, Han Y, Dinh HH, Dahdouli M, Samra JS, Bailey P, Waddell N, Pearson JV, Harliwong I, Wang H, Aust D, Oien KA, Hruban RH, Hodges SE, McElhany A, Saengboonmee C, Duthie FR, Grimmond SM, Biankin AV, Wheeler DA, Gibbs RA. Ampullary Cancers Harbor ELF3 Tumor Suppressor Gene Mutations and Exhibit Frequent WNT Dysregulation. Cell Rep. 2016;14:907–919. doi: 10.1016/j.celrep.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bridgewater J, Galle PR, Khan SA, Llovet JM, Park JW, Patel T, Pawlik TM, Gores GJ. Guidelines for the diagnosis and management of intrahepatic cholangiocarcinoma. J Hepatol. 2014;60:1268–1289. doi: 10.1016/j.jhep.2014.01.021. [DOI] [PubMed] [Google Scholar]
- 32.Jiao Y, Pawlik TM, Anders RA, Selaru FM, Streppel MM, Lucas DJ, Niknafs N, Guthrie VB, Maitra A, Argani P, Offerhaus GJA, Roa JC, Roberts LR, Gores GJ, Popescu I, Alexandrescu ST, Dima S, Fassan M, Simbolo M, Mafficini A, Capelli P, Lawlor RT, Ruzzenente A, Guglielmi A, Tortora G, de Braud F, Scarpa A, Jarnagin W, Klimstra D, Karchin R, Velculescu VE, Hruban RH, Vogelstein B, Kinzler KW, Papadopoulos N, Wood LD. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat Genet. 2013;45:1470–1473. doi: 10.1038/ng.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lazaridis KN, LaRusso NF. Primary Sclerosing Cholangitis. N Engl J Med. 2016;375:1161–1170. doi: 10.1056/NEJMra1506330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu HT, Gao P. The roles of microRNAs related with progression and metastasis in human cancers. Tumour Biol. 2016 doi: 10.1007/s13277-016-5436-9. [DOI] [PubMed] [Google Scholar]
- 35.Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduct Target Ther. 2016;1:15004. doi: 10.1038/sigtrans.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cao J, Sun L, Li J, Zhou C, Cheng L, Chen K, Yan B, Qian W, Ma Q, Duan W. A novel threemiRNA signature predicts survival in cholangiocarcinoma based on RNA-Seq data. Oncol Rep. 2018;40:1422–1434. doi: 10.3892/or.2018.6534. [DOI] [PubMed] [Google Scholar]
- 37.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 38.Zhong XY, Yu JH, Zhang WG, Wang ZD, Dong Q, Tai S, Cui YF, Li H. MicroRNA-421 functions as an oncogenic miRNA in biliary tract cancer through down-regulating farnesoid X receptor expression. Gene. 2012;493:44–51. doi: 10.1016/j.gene.2011.11.028. [DOI] [PubMed] [Google Scholar]
- 39.Koustas E, Karamouzis MV, Mihailidou C, Schizas D, Papavassiliou AG. Co-targeting of EGFR and autophagy signaling is an emerging treatment strategy in metastatic colorectal cancer. Cancer Lett. 2017;396:94–102. doi: 10.1016/j.canlet.2017.03.023. [DOI] [PubMed] [Google Scholar]
- 40.Yun CW, Lee SH. The Roles of Autophagy in Cancer. Int J Mol Sci. 2018;19 doi: 10.3390/ijms19113466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer. 2017;17:528–542. doi: 10.1038/nrc.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aredia F, Giansanti V, Mazzini G, Savio M, Ortiz LM, Jaadane I, Zaffaroni N, Forlino A, Torriglia A, Scovassi AI. Multiple effects of the Na(+)/H (+) antiporter inhibitor HMA on cancer cells. Apoptosis. 2013;18:1586–1598. doi: 10.1007/s10495-013-0898-3. [DOI] [PubMed] [Google Scholar]
- 43.Rosenfeldt MT, Ryan KM. The multiple roles of autophagy in cancer. Carcinogenesis. 2011;32:955–963. doi: 10.1093/carcin/bgr031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gewirtz DA. The four faces of autophagy: implications for cancer therapy. Cancer Res. 2014;74:647–651. doi: 10.1158/0008-5472.CAN-13-2966. [DOI] [PubMed] [Google Scholar]
- 45.Koustas E, Sarantis P, Kyriakopoulou G, Papavassiliou AG, Karamouzis MV. The Interplay of Autophagy and Tumor Microenvironment in Colorectal Cancer-Ways of Enhancing Immunotherapy Action. Cancers (Basel) 2019;11 doi: 10.3390/cancers11040533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perez-Montoyo H. Therapeutic Potential of Autophagy Modulation in Cholangiocarcinoma. Cells. 2020;9 doi: 10.3390/cells9030614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moloney JN, Cotter TG. ROS signalling in the biology of cancer. Semin Cell Dev Biol. 2018;80:50–64. doi: 10.1016/j.semcdb.2017.05.023. [DOI] [PubMed] [Google Scholar]
- 48.Ávalos Y, Canales J, Bravo-Sagua R, Criollo A, Lavandero S, Quest AF. Tumor suppression and promotion by autophagy. Biomed Res Int. 2014;2014:603980. doi: 10.1155/2014/603980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Filomeni G, De Zio D, Cecconi F. Oxidative stress and autophagy: the clash between damage and metabolic needs. Cell Death Differ. 2015;22:377–388. doi: 10.1038/cdd.2014.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koustas E, Sarantis P, Papavassiliou AG, Karamouzis MV. Upgraded role of autophagy in colorectal carcinomas. World J Gastrointest Oncol. 2018;10:367–369. doi: 10.4251/wjgo.v10.i11.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.White E, DiPaola RS. The double-edged sword of autophagy modulation in cancer. Clin Cancer Res. 2009;15:5308–5316. doi: 10.1158/1078-0432.CCR-07-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Valle J, Wasan H, Palmer DH, Cunningham D, Anthoney A, Maraveyas A, Madhusudan S, Iveson T, Hughes S, Pereira SP, Roughton M, Bridgewater J ABC-02 Trial Investigators. Cisplatin plus gemcitabine vs gemcitabine for biliary tract cancer. N Engl J Med. 2010;362:1273–1281. doi: 10.1056/NEJMoa0908721. [DOI] [PubMed] [Google Scholar]
- 53.Valle JW, Furuse J, Jitlal M, Beare S, Mizuno N, Wasan H, Bridgewater J, Okusaka T. Cisplatin and gemcitabine for advanced biliary tract cancer: a meta-analysis of two randomised trials. Ann Oncol. 2014;25:391–398. doi: 10.1093/annonc/mdt540. [DOI] [PubMed] [Google Scholar]
- 54.Benavides M, Antón A, Gallego J, Gómez MA, Jiménez-Gordo A, La Casta A, Laquente B, Macarulla T, Rodríguez-Mowbray JR, Maurel J. Biliary tract cancers: SEOM clinical guidelines. Clin Transl Oncol. 2015;17:982–987. doi: 10.1007/s12094-015-1436-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simile MM, Bagella P, Vidili G, Spanu A, Manetti R, Seddaiu MA, Babudieri S, Madeddu G, Serra PA, Altana M, Paliogiannis P. Targeted Therapies in Cholangiocarcinoma: Emerging Evidence from Clinical Trials. Medicina (Kaunas) 2019;55 doi: 10.3390/medicina55020042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Simbolo M, Fassan M, Ruzzenente A, Mafficini A, Wood LD, Corbo V, Melisi D, Malleo G, Vicentini C, Malpeli G, Antonello D, Sperandio N, Capelli P, Tomezzoli A, Iacono C, Lawlor RT, Bassi C, Hruban RH, Guglielmi A, Tortora G, de Braud F, Scarpa A. Multigene mutational profiling of cholangiocarcinomas identifies actionable molecular subgroups. Oncotarget. 2014;5:2839–2852. doi: 10.18632/oncotarget.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arai Y, Totoki Y, Hosoda F, Shirota T, Hama N, Nakamura H, Ojima H, Furuta K, Shimada K, Okusaka T, Kosuge T, Shibata T. Fibroblast growth factor receptor 2 tyrosine kinase fusions define a unique molecular subtype of cholangiocarcinoma. Hepatology. 2014;59:1427–1434. doi: 10.1002/hep.26890. [DOI] [PubMed] [Google Scholar]
- 58.Cheng Z, Lei Z, Shen F. Coming of a precision era of the staging systems for intrahepatic cholangiocarcinoma? Cancer Lett. 2019;460:10–17. doi: 10.1016/j.canlet.2019.114426. [DOI] [PubMed] [Google Scholar]
- 59.Landskron G, De la Fuente M, Thuwajit P, Thuwajit C, Hermoso MA. Chronic inflammation and cytokines in the tumor microenvironment. J Immunol Res. 2014;2014:149185. doi: 10.1155/2014/149185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nzeako UC, Guicciardi ME, Yoon JH, Bronk SF, Gores GJ. COX-2 inhibits Fas-mediated apoptosis in cholangiocarcinoma cells. Hepatology. 2002;35:552–559. doi: 10.1053/jhep.2002.31774. [DOI] [PubMed] [Google Scholar]
- 61.Qi Y, Zhang M, Li H, Frank JA, Dai L, Liu H, Zhang Z, Wang C, Chen G. Autophagy inhibition by sustained overproduction of IL6 contributes to arsenic carcinogenesis. Cancer Res. 2014;74:3740–3752. doi: 10.1158/0008-5472.CAN-13-3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moeini A, Sia D, Bardeesy N, Mazzaferro V, Llovet JM. Molecular Pathogenesis and Targeted Therapies for Intrahepatic Cholangiocarcinoma. Clin Cancer Res. 2016;22:291–300. doi: 10.1158/1078-0432.CCR-14-3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.O'Dell MR, Huang JL, Whitney-Miller CL, Deshpande V, Rothberg P, Grose V, Rossi RM, Zhu AX, Land H, Bardeesy N, Hezel AF. Kras(G12D) and p53 mutation cause primary intrahepatic cholangiocarcinoma. Cancer Res. 2012;72:1557–1567. doi: 10.1158/0008-5472.CAN-11-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Labib PL, Goodchild G, Pereira SP. Molecular Pathogenesis of Cholangiocarcinoma. BMC Cancer. 2019;19:185. doi: 10.1186/s12885-019-5391-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mertens JC, Rizvi S, Gores GJ. Targeting cholangiocarcinoma. Biochim Biophys Acta Mol Basis Dis. 2018;1864:1454–1460. doi: 10.1016/j.bbadis.2017.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Socoteanu MP, Mott F, Alpini G, Frankel AE. c-Met targeted therapy of cholangiocarcinoma. World J Gastroenterol. 2008;14:2990–2994. doi: 10.3748/wjg.14.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pu XH, Yue S, Wu HY, Yang J, Fan XS, Fu Y, Ye Q, Chen J. C-MET in intrahepatic cholangiocarcinoma: High-Frequency amplification predicts protein expression and a unique molecular subtype. Pathol Res Pract. 2020;216:152857. doi: 10.1016/j.prp.2020.152857. [DOI] [PubMed] [Google Scholar]
- 68.Liu Y, Liu JH, Chai K, Tashiro S, Onodera S, Ikejima T. Inhibition of c-Met promoted apoptosis, autophagy and loss of the mitochondrial transmembrane potential in oridonin-induced A549 lung cancer cells. J Pharm Pharmacol. 2013;65:1622–1642. doi: 10.1111/jphp.12140. [DOI] [PubMed] [Google Scholar]
- 69.Xu L, Hausmann M, Dietmaier W, Kellermeier S, Pesch T, Stieber-Gunckel M, Lippert E, Klebl F, Rogler G. Expression of growth factor receptors and targeting of EGFR in cholangiocarcinoma cell lines. BMC Cancer. 2010;10:302. doi: 10.1186/1471-2407-10-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lai GH, Zhang Z, Shen XN, Ward DJ, Dewitt JL, Holt SE, Rozich RA, Hixson DC, Sirica AE. erbB-2/neu transformed rat cholangiocytes recapitulate key cellular and molecular features of human bile duct cancer. Gastroenterology. 2005;129:2047–2057. doi: 10.1053/j.gastro.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 71.Tanaka H, Hino H, Moriya S, Kazama H, Miyazaki M, Takano N, Hiramoto M, Tsukahara K, Miyazawa K. Comparison of autophagy inducibility in various tyrosine kinase inhibitors and their enhanced cytotoxicity via inhibition of autophagy in cancer cells in combined treatment with azithromycin. Biochem Biophys Rep. 2020;22:100750. doi: 10.1016/j.bbrep.2020.100750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu YM, Su F, Kalyana-Sundaram S, Khazanov N, Ateeq B, Cao X, Lonigro RJ, Vats P, Wang R, Lin SF, Cheng AJ, Kunju LP, Siddiqui J, Tomlins SA, Wyngaard P, Sadis S, Roychowdhury S, Hussain MH, Feng FY, Zalupski MM, Talpaz M, Pienta KJ, Rhodes DR, Robinson DR, Chinnaiyan AM. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov. 2013;3:636–647. doi: 10.1158/2159-8290.CD-13-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.van der Wekken AJ, Saber A, Hiltermann TJ, Kok K, van den Berg A, Groen HJ. Resistance mechanisms after tyrosine kinase inhibitors afatinib and crizotinib in non-small cell lung cancer, a review of the literature. Crit Rev Oncol Hematol. 2016;100:107–116. doi: 10.1016/j.critrevonc.2016.01.024. [DOI] [PubMed] [Google Scholar]
- 74.Chen Y, Xie X, Li X, Wang P, Jing Q, Yue J, Liu Y, Cheng Z, Li J, Song H, Li G, Liu R, Wang J. FGFR antagonist induces protective autophagy in FGFR1-amplified breast cancer cell. Biochem Biophys Res Commun. 2016;474:1–7. doi: 10.1016/j.bbrc.2016.03.017. [DOI] [PubMed] [Google Scholar]
- 75.Kang YK, Kim WH, Jang JJ. Expression of G1-S modulators (p53, p16, p27, cyclin D1, Rb) and Smad4/Dpc4 in intrahepatic cholangiocarcinoma. Hum Pathol. 2002;33:877–883. doi: 10.1053/hupa.2002.127444. [DOI] [PubMed] [Google Scholar]
- 76.Wang F, Xia X, Yang C, Shen J, Mai J, Kim HC, Kirui D, Kang Y, Fleming JB, Koay EJ, Mitra S, Ferrari M, Shen H. SMAD4 Gene Mutation Renders Pancreatic Cancer Resistance to Radiotherapy through Promotion of Autophagy. Clin Cancer Res. 2018;24:3176–3185. doi: 10.1158/1078-0432.CCR-17-3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cong WM, Bakker A, Swalsky PA, Raja S, Woods J, Thomas S, Demetris AJ, Finkelstein SD. Multiple genetic alterations involved in the tumorigenesis of human cholangiocarcinoma: a molecular genetic and clinicopathological study. J Cancer Res Clin Oncol. 2001;127:187–192. doi: 10.1007/s004320000194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sasaki M, Nitta T, Sato Y, Nakanuma Y. Autophagy may occur at an early stage of cholangiocarcinogenesis via biliary intraepithelial neoplasia. Hum Pathol. 2015;46:202–209. doi: 10.1016/j.humpath.2014.09.016. [DOI] [PubMed] [Google Scholar]
- 79.Qu X, Sheng J, Shen L, Su J, Xu Y, Xie Q, Wu Y, Zhang X, Sun L. Autophagy inhibitor chloroquine increases sensitivity to cisplatin in QBC939 cholangiocarcinoma cells by mitochondrial ROS. PLoS One. 2017;12:e0173712. doi: 10.1371/journal.pone.0173712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Isomoto H. Epigenetic alterations associated with cholangiocarcinoma (review) Oncol Rep. 2009;22:227–232. [PubMed] [Google Scholar]
- 81.Peixoto P, Grandvallet C, Feugeas JP, Guittaut M, Hervouet E. Epigenetic Control of Autophagy in Cancer Cells: A Key Process for Cancer-Related Phenotypes. Cells. 2019;8 doi: 10.3390/cells8121656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gradilone SA, Radtke BN, Bogert PS, Huang BQ, Gajdos GB, LaRusso NF. HDAC6 inhibition restores ciliary expression and decreases tumor growth. Cancer Res. 2013;73:2259–2270. doi: 10.1158/0008-5472.CAN-12-2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gradilone SA, Habringer S, Masyuk TV, Howard BN, Masyuk AI, Larusso NF. HDAC6 is overexpressed in cystic cholangiocytes and its inhibition reduces cystogenesis. Am J Pathol. 2014;184:600–608. doi: 10.1016/j.ajpath.2013.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kaliszczak M, van Hechanova E, Li Y, Alsadah H, Parzych K, Auner HW, Aboagye EO. The HDAC6 inhibitor C1A modulates autophagy substrates in diverse cancer cells and induces cell death. Br J Cancer. 2018;119:1278–1287. doi: 10.1038/s41416-018-0232-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pant K, Peixoto E, Richard S, Gradilone SA. Role of Histone Deacetylases in Carcinogenesis: Potential Role in Cholangiocarcinoma. Cells. 2020;9 doi: 10.3390/cells9030780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hu LF. Epigenetic Regulation of Autophagy. Adv Exp Med Biol. 2019;1206:221–236. doi: 10.1007/978-981-15-0602-4_11. [DOI] [PubMed] [Google Scholar]
- 87.Borger DR, Tanabe KK, Fan KC, Lopez HU, Fantin VR, Straley KS, Schenkein DP, Hezel AF, Ancukiewicz M, Liebman HM, Kwak EL, Clark JW, Ryan DP, Deshpande V, Dias-Santagata D, Ellisen LW, Zhu AX, Iafrate AJ. Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist. 2012;17:72–79. doi: 10.1634/theoncologist.2011-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang P, Dong Q, Zhang C, Kuan PF, Liu Y, Jeck WR, Andersen JB, Jiang W, Savich GL, Tan TX, Auman JT, Hoskins JM, Misher AD, Moser CD, Yourstone SM, Kim JW, Cibulskis K, Getz G, Hunt HV, Thorgeirsson SS, Roberts LR, Ye D, Guan KL, Xiong Y, Qin LX, Chiang DY. Mutations in isocitrate dehydrogenase 1 and 2 occur frequently in intrahepatic cholangiocarcinomas and share hypermethylation targets with glioblastomas. Oncogene. 2013;32:3091–3100. doi: 10.1038/onc.2012.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Trejo-Solís C, Serrano-Garcia N, Escamilla-Ramírez Á, Castillo-Rodríguez RA, Jimenez-Farfan D, Palencia G, Calvillo M, Alvarez-Lemus MA, Flores-Nájera A, Cruz-Salgado A, Sotelo J. Autophagic and Apoptotic Pathways as Targets for Chemotherapy in Glioblastoma. Int J Mol Sci. 2018;19 doi: 10.3390/ijms19123773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li Z, Shen J, Chan MT, Wu WK. The role of microRNAs in intrahepatic cholangiocarcinoma. J Cell Mol Med. 2017;21:177–184. doi: 10.1111/jcmm.12951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Puik JR, Meijer LL, Le Large TY, Prado MM, Frampton AE, Kazemier G, Giovannetti E. miRNA profiling for diagnosis, prognosis and stratification of cancer treatment in cholangiocarcinoma. Pharmacogenomics. 2017;18:1343–1358. doi: 10.2217/pgs-2017-0010. [DOI] [PubMed] [Google Scholar]
- 92.Gozuacik D, Akkoc Y, Ozturk DG, Kocak M. Autophagy-Regulating microRNAs and Cancer. Front Oncol. 2017;7:65. doi: 10.3389/fonc.2017.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Frankel LB, Lund AH. MicroRNA regulation of autophagy. Carcinogenesis. 2012;33:2018–2025. doi: 10.1093/carcin/bgs266. [DOI] [PubMed] [Google Scholar]
- 94.Ma J, Weng L, Wang Z, Jia Y, Liu B, Wu S, Cao Y, Sun X, Yin X, Shang M, Mao A. MiR-124 induces autophagy-related cell death in cholangiocarcinoma cells through direct targeting of the EZH2-STAT3 signaling axis. Exp Cell Res. 2018;366:103–113. doi: 10.1016/j.yexcr.2018.02.037. [DOI] [PubMed] [Google Scholar]
- 95.Onorati AV, Dyczynski M, Ojha R, Amaravadi RK. Targeting autophagy in cancer. Cancer. 2018;124:3307–3318. doi: 10.1002/cncr.31335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang H, Li D, Li X, Ou X, Liu S, Zhang Y, Ding J, Xie B. Mammalian target of rapamycin inhibitor RAD001 sensitizes endometrial cancer cells to paclitaxel-induced apoptosis via the induction of autophagy. Oncol Lett. 2016;12:5029–5035. doi: 10.3892/ol.2016.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Byun S, Lee E, Lee KW. Therapeutic Implications of Autophagy Inducers in Immunological Disorders, Infection, and Cancer. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18091959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Koustas E, Papavassiliou AG, Karamouzis MV. The role of autophagy in the treatment of BRAF mutant colorectal carcinomas differs based on microsatellite instability status. PLoS One. 2018;13:e0207227. doi: 10.1371/journal.pone.0207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu J, Xia H, Kim M, Xu L, Li Y, Zhang L, Cai Y, Norberg HV, Zhang T, Furuya T, Jin M, Zhu Z, Wang H, Yu J, Hao Y, Choi A, Ke H, Ma D, Yuan J. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell. 2011;147:223–234. doi: 10.1016/j.cell.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yeo SK, Paul R, Haas M, Wang C, Guan JL. Improved efficacy of mitochondrial disrupting agents upon inhibition of autophagy in a mouse model of BRCA1-deficient breast cancer. Autophagy. 2018;14:1214–1225. doi: 10.1080/15548627.2018.1460010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Merino D, Kelly GL, Lessene G, Wei AH, Roberts AW, Strasser A. BH3-Mimetic Drugs: Blazing the Trail for New Cancer Medicines. Cancer Cell. 2018;34:879–891. doi: 10.1016/j.ccell.2018.11.004. [DOI] [PubMed] [Google Scholar]
- 102.Yang ZJ, Chee CE, Huang S, Sinicrope FA. The role of autophagy in cancer: therapeutic implications. Mol Cancer Ther. 2011;10:1533–1541. doi: 10.1158/1535-7163.MCT-11-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Law BY, Chan WK, Xu SW, Wang JR, Bai LP, Liu L, Wong VK. Natural small-molecule enhancers of autophagy induce autophagic cell death in apoptosis-defective cells. Sci Rep. 2014;4:5510. doi: 10.1038/srep05510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Siasos G, Tousoulis D, Oikonomou E, Zaromitidou M, Verveniotis A, Plastiras A, Kioufis S, Maniatis K, Miliou A, Siasou Z, Stefanadis C, Papavassiliou AG. Effects of Ω-3 fatty acids on endothelial function, arterial wall properties, inflammatory and fibrinolytic status in smokers: a cross over study. Int J Cardiol. 2013;166:340–346. doi: 10.1016/j.ijcard.2011.10.081. [DOI] [PubMed] [Google Scholar]
- 105.Yao L, Han C, Song K, Zhang J, Lim K, Wu T. Omega-3 Polyunsaturated Fatty Acids Upregulate 15-PGDH Expression in Cholangiocarcinoma Cells by Inhibiting miR-26a/b Expression. Cancer Res. 2015;75:1388–1398. doi: 10.1158/0008-5472.CAN-14-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kim S, Jing K, Shin S, Jeong S, Han SH, Oh H, Yoo YS, Han J, Jeon YJ, Heo JY, Kweon GR, Park SK, Park JI, Wu T, Lim K. ω3-polyunsaturated fatty acids induce cell death through apoptosis and autophagy in glioblastoma cells: In vitro and in vivo. Oncol Rep. 2018;39:239–246. doi: 10.3892/or.2017.6101. [DOI] [PubMed] [Google Scholar]
- 107.Janji B, Berchem G, Chouaib S. Targeting Autophagy in the Tumor Microenvironment: New Challenges and Opportunities for Regulating Tumor Immunity. Front Immunol. 2018;9:887. doi: 10.3389/fimmu.2018.00887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Qian HR, Shi ZQ, Zhu HP, Gu LH, Wang XF, Yang Y. Interplay between apoptosis and autophagy in colorectal cancer. Oncotarget. 2017;8:62759–62768. doi: 10.18632/oncotarget.18663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rosenfeld MR, Ye X, Supko JG, Desideri S, Grossman SA, Brem S, Mikkelson T, Wang D, Chang YC, Hu J, McAfee Q, Fisher J, Troxel AB, Piao S, Heitjan DF, Tan KS, Pontiggia L, O'Dwyer PJ, Davis LE, Amaravadi RK. A phase I/II trial of hydroxychloroquine in conjunction with radiation therapy and concurrent and adjuvant temozolomide in patients with newly diagnosed glioblastoma multiforme. Autophagy. 2014;10:1359–1368. doi: 10.4161/auto.28984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Goulielmaki M, Koustas E, Moysidou E, Vlassi M, Sasazuki T, Shirasawa S, Zografos G, Oikonomou E, Pintzas A. BRAF associated autophagy exploitation: BRAF and autophagy inhibitors synergise to efficiently overcome resistance of BRAF mutant colorectal cancer cells. Oncotarget. 2016;7:9188–9221. doi: 10.18632/oncotarget.6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Boone BA, Bahary N, Zureikat AH, Moser AJ, Normolle DP, Wu WC, Singhi AD, Bao P, Bartlett DL, Liotta LA, Espina V, Loughran P, Lotze MT, Zeh HJ 3rd. Safety and Biologic Response of Pre-operative Autophagy Inhibition in Combination with Gemcitabine in Patients with Pancreatic Adenocarcinoma. Ann Surg Oncol. 2015;22:4402–4410. doi: 10.1245/s10434-015-4566-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Amaravadi RK, Winkler JD. Lys05: a new lysosomal autophagy inhibitor. Autophagy. 2012;8:1383–1384. doi: 10.4161/auto.20958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ronan B, Flamand O, Vescovi L, Dureuil C, Durand L, Fassy F, Bachelot MF, Lamberton A, Mathieu M, Bertrand T, Marquette JP, El-Ahmad Y, Filoche-Romme B, Schio L, Garcia-Echeverria C, Goulaouic H, Pasquier B. A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy. Nat Chem Biol. 2014;10:1013–1019. doi: 10.1038/nchembio.1681. [DOI] [PubMed] [Google Scholar]
- 114.Egan DF, Chun MG, Vamos M, Zou H, Rong J, Miller CJ, Lou HJ, Raveendra-Panickar D, Yang CC, Sheffler DJ, Teriete P, Asara JM, Turk BE, Cosford ND, Shaw RJ. Small Molecule Inhibition of the Autophagy Kinase ULK1 and Identification of ULK1 Substrates. Mol Cell. 2015;59:285–297. doi: 10.1016/j.molcel.2015.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Vakifahmetoglu-Norberg H, Xia HG, Yuan J. Pharmacologic agents targeting autophagy. J Clin Invest. 2015;125:5–13. doi: 10.1172/JCI73937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rossi M, Munarriz ER, Bartesaghi S, Milanese M, Dinsdale D, Guerra-Martin MA, Bampton ET, Glynn P, Bonanno G, Knight RA, Nicotera P, Melino G. Desmethylclomipramine induces the accumulation of autophagy markers by blocking autophagic flux. J Cell Sci. 2009;122:3330–3339. doi: 10.1242/jcs.048181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Starobinets H, Ye J, Broz M, Barry K, Goldsmith J, Marsh T, Rostker F, Krummel M, Debnath J. Antitumor adaptive immunity remains intact following inhibition of autophagy and antimalarial treatment. J Clin Invest. 2016;126:4417–4429. doi: 10.1172/JCI85705. [DOI] [PMC free article] [PubMed] [Google Scholar]