Summary
Greater understanding of the spatial relationships between members of the human gut microbiota and available nutrients is needed to gain deeper insights about community dynamics and expressed functions. Therefore, we generated a panel of artificial food particles, each type composed of microscopic paramagnetic beads coated with a fluorescent barcode and one of 60 different dietary or host glycan preparations. Analysis of 160 Bacteroides and Parabacteroides strains disclosed diverse strain-specific and glycan-specific binding phenotypes. We identified carbohydrate structures that correlated with binding by specific bacterial strains in vitro, and noted strain-specific differences in catabolism of glycans that mediate adhesion. Mixed in vitro cultures revealed that these adhesion phenotypes are maintained in more complex communities. Additionally, orally administering glycan-beads to gnotobiotic mice confirmed specificity in glycan binding. This approach should facilitate analyses of how strains occupying the same physical niche interact, and advance development of synbiotics, more nutritious foods, and microbiota-based diagnostics.
Keywords: bacterial adhesion, glycan recognition and utilization, imaging microbiota spatial organization, multiplex bead-based phenotypic screens, retrievable artificial food particles, gnotobiotic mice
Graphical Abstract
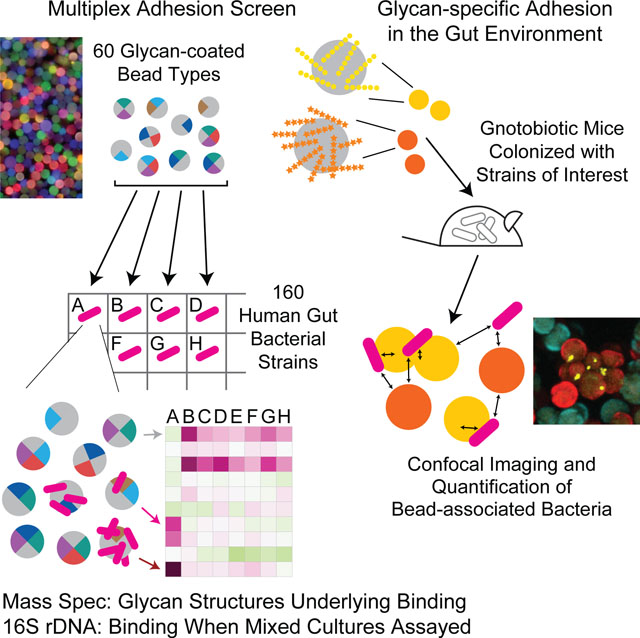
eTOC Blurb
Using fluorescently-labeled, microscopic glass beads containing different bound glycans, Patnode et al. examine carbohydrate-dependent adhesion of gut bacterial strains. Strain-specific binding correlated with linkage composition of dietary glycans and adhesion was observed in mixed in vitro cultures and gnotobiotic mice. The method has basic scientific plus diagnostic and therapeutic applications.
Introduction
The human gut is populated by microbial communities that exhibit a remarkable degree of strain-level diversity (Human Microbiome Project, 2012). While this diversity is currently being described at a genomic level (Karcher et al., 2020; Vatanen et al., 2019; Yaffe and Relman, 2020), characterizing its functional significance is very challenging (Yang et al., 2020). One testable hypothesis is that an aspect of this diversity is related to the capacity of organisms to physically interact with and utilize various components of the diets consumed by their hosts. The ability of members of the human gut microbiota to forge competitive or cooperative relationships with one another is influenced by their physical proximity. The spatial arrangement of microbial cells affects diffusion of the products of their metabolism to neighbors that have the capacity to further transform these products for their benefit and/or the benefit of other community members and their host (Rakoff-Nahoum et al., 2016). Spatial relationships also profoundly impact contact-dependent ‘interference competition’ in a variety of ecosystems (Cordero and Datta, 2016; Ting et al., 2020). Moreover, ‘exclusion competition’ between species depends on their ability to gain access to nutrient depots that are heterogeneously distributed in the gut (Earle et al., 2015).
Glycan-mediated adhesion is one of the primary means by which bacteria increase their access to nutrients in diverse ecosystems. Adhesion to chitin is prevalent among marine microbes that degrade this polysaccharide (Sun et al., 2015), as is adhesion to cellulose among cellulolytic species in the rumen (Miron et al., 2001). Pathogenic microbes adhere to host cells to enable access to nutrients that these cells provide (Poole et al., 2018); in many cases the adhesins involved bind to specific host glycan structures (Kalas et al., 2018; Le Guennec et al., 2020; Skoog et al., 2017). Fibers in human diets contain polysaccharides embedded within plant cell wall fragments; their surfaces represent potential sites of attachment that could allow gut microbes and their syntrophic partners to co-metabolize fiber components (Macfarlane and Macfarlane, 2006). However, little is known about how these microbes control their localization relative to ingested fiber in vivo or whether they interact with the surfaces of fibers (or other particulate material derived from ingested foods) stochastically as a result of turbulent peristaltic flow within the gut. If binding does occur, what factors influence the duration of affiliation with particles? Are glycan targets of bacterial binding also substrates for bacterial metabolism or do they represent ‘landing platforms’ so that other particle-associated nutrient depots can be mined?
In the present study, we use a generalizable and scalable approach to uncover differences in the adhesion of human gut-derived Bacteroides and Parabacteroides strains to distinct classes of dietary polysaccharides in vitro and in vivo.
Results
Multiplex screening reveals strain-specific adhesion to dietary fiber polysaccharides
To determine whether adhesion controls gut bacterial proximity to dietary nutrient sources, we first carried out a screen using a collection of 160 human gut-derived Bacteroides and Parabacteroides strains belonging to 13 different species (Table S1A). Many members of the phylum Bacteroidetes are highly adapted for harvesting dietary fiber glycans; their genomes contain expanded repertoires of genes involved in glycan sensing, binding, import and metabolism (Martens et al., 2014). The screen employed a library of microscopic paramagnetic glass beads, each coated with one of 60 distinct types of glycan preparations, including algal polysaccharides, mammalian glycans, and plant hemicelluloses and pectins, many of which are known substrates for gut microbial metabolic processing (Desai et al., 2016). This collection also contained heterogeneous, food-grade fiber preparations isolated from fruits, vegetables, grains, and legumes (Table S1B). Each bead type was labeled with a unique combination of fluorophores (up to six, allowing 64 combinations), enabling its identification in a pooled mixture of all bead types. The bead library also contained four sets of control (“empty”) beads with attached fluorophores but no glycan, to account for potential non-specific adhesion to the glass surface of the beads (Mihajlovic et al., 2019) (Table S1C).
Figure 1A describes the screening procedure. A monoculture of each bacterial strain was grown in a multi-well format, then labeled with a fluorescent nucleic acid-binding dye (Syto-82). Each strain was subsequently pelleted by centrifugation and the cells were incubated with an aliquot of the 60-member glycan-bead library. Incubations were performed at 4 °C for 30 minutes to minimize potential microbial degradation of glycans on the bead surface. Beads were purified magnetically, and the gating strategy shown in Table S1C was employed to identify each bead-type by flow cytometry. The extent of bacterial binding was quantified by measuring the mean intensity of Syto-82 fluorescence for each bead type in each bacterial sample. We established assay background by measuring the fluorescence associated with aliquots of the bead library introduced into wells lacking bacteria (n=8 aliquots); based on the results we selected a positive fluorescence threshold of three standard deviations above the mean background fluorescence for each bead type in these wells, and considered all signals above this threshold in the remaining wells as indicative of bacterial binding. We then defined glycan-dependent binding as adhesion >4-fold above the signal observed on empty beads with no glycan coating. The presence of bacterial cells bound to beads identified in this way was confirmed by confocal microscopic examination of samples of beads (Video S1).
Figure 1: Multiplex screening of human gut Bacteroides and Parabacteroides species/strains for their ability to bind to fluorescent, barcoded, paramagnetic microscopic glycan-coated beads.
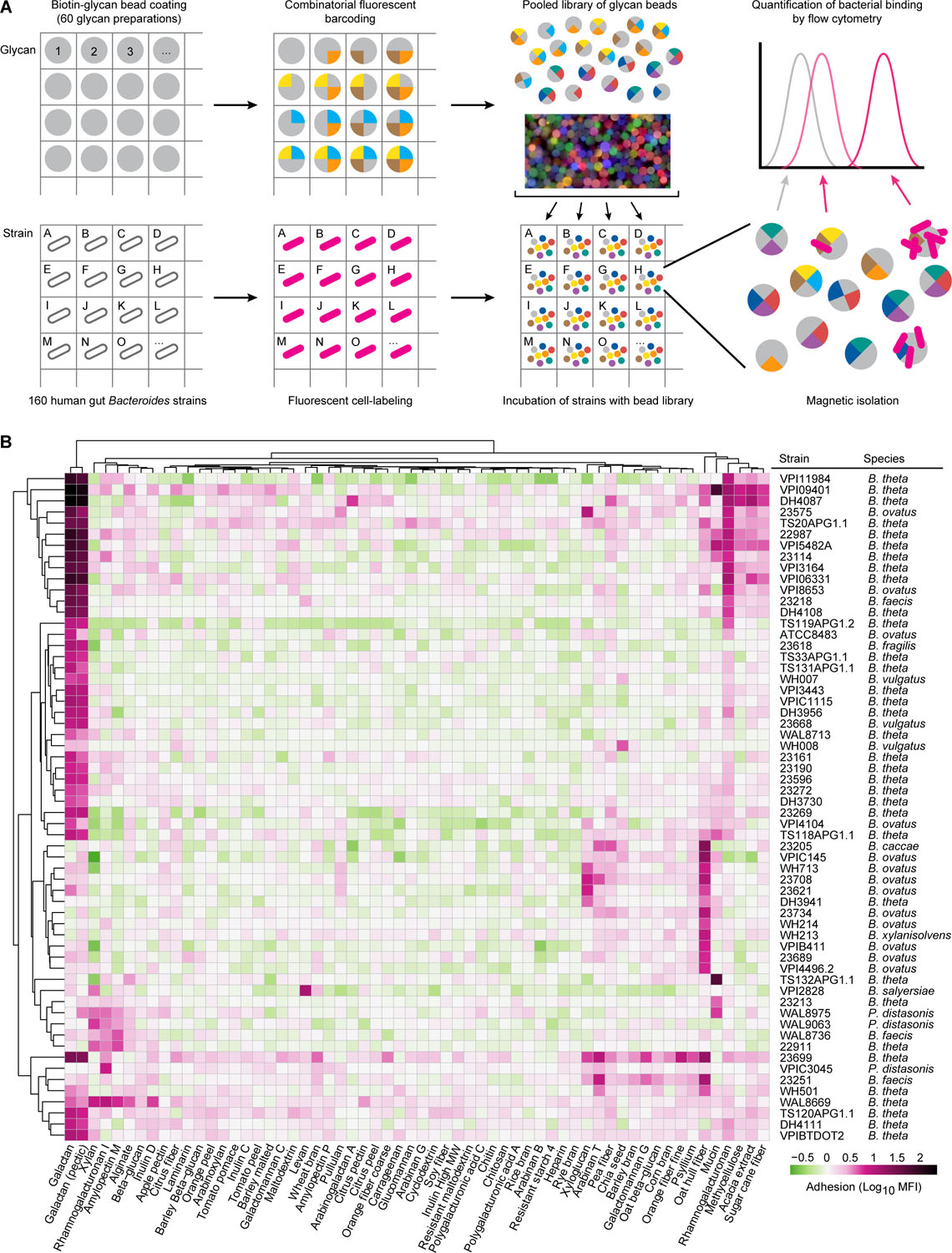
(A) Schematic design of how the polysaccharide bead library was generated and used to simultaneously characterize the glycan binding phenotypes of multiple bacterial strains. The matrix depicts how a given bacterial strain was added to a given well of a multi-well plate, labeled with Syto-82 and incubated with the 60-member library of fluorescently barcoded beads (depicted as a cartoon as well as by a photograph of labeled beads). Beads were recovered from each monoculture based on their magnetism and then subjected to flow cytometry according to the gating strategy depicted in Table S1C to identify bead types with bound bacteria. (B) Quantification of bacterial adhesion for all strains that exhibited glycan-dependent binding (>4-fold above the average level of fluorescence of “empty” beads not coated with glycans). The log10 geometric mean fluorescent intensity in the Syto-82 channel for each population of beads in each well relative to empty beads is indicated by the color bar. Columns denote the carbohydrate preparation coated on each bead type. Bacterial strains incubated with beads are listed in the rows along with their species classification. Binding profiles for a given bead type and strain are hierarchically clustered using Ward’s minimum variance (Ward, 1963). See also Table S1, S2.
A total of 60 of the 160 Bacteroides and Parabacteroides strains exhibited glycan-specific binding to one or more glycan-coated bead types (Figure 1B). Adherent strains of B. thetaiotaomicron, B. ovatus, B. faecis, B. vulgatus, P. distasonis, B. caccae, B. salyersiae, B. xylanisolvens, and B. fragilis were identified. Six of the strains (B. thetaiotaomicron 23165 and 23584; B. fragilis 23212, 23618, 23576, and VPI-BV8526) bound to all bead types, including control beads not coated with glycans (Table S2). The adherence of these strains likely involves binding to streptavidin, biotin, or features inherent to the bead surface. Bacterial cells were added in ~80,000-fold excess relative to beads and as anticipated, binding phenotypes were not significantly correlated with the number of cells per well for any bead type (t-test for Pearson correlation, minimum p-value = 0.12; Table S1, Table S2). (Note that validation experiments indicated that a 3-fold reduction in cell density, the maximum difference between isolates in our screen, corresponded to a 1.24-fold reduction in bead fluorescence).
The screen revealed pronounced strain-to-strain variation in binding phenotypes (Figure 1B). For each strain that exhibited glycan-specific binding, there were other strains belonging to the same species that did not (Table S2). For example, B. salyersiae strain VPI-2828 exhibited preferential binding to levan beads, while B. salyersiae 23369 did not. Different strains of B. thetaiotaomicron adhered to either galactan (DH4108) or mucin (TS132APG1.1), while representatives of B. ovatus bound with high specificity to either oat hull fiber (VPI-4496.2) or galactan and xyloglucan (23575). (Figure 1B). Among P. distasonis strains, one bound rhamnogalacturonan I (VPI-C3045) and another bound xylan (WAL9063). These results provided evidence that diverse dietary glycans serve as sites of attachment for gut bacterial species, and that one manifestation of functional heterogeneity between Bacteroides as well as Parabacteroides strains is their capacity to gain physical proximity to carbohydrate resources present in their gut habitat.
Mass spectrometric identification of glycan structures correlated with bacterial binding
To characterize molecular features associated with glycan-specific binding by human gut Bacteroides and Parabacteroides, we performed monosaccharide and glycosyl linkage analysis on 50 of the food-grade fiber preparations and purified polysaccharides used in the adhesion screen (Table S3A, S3B). Although food-grade fiber preparations were heterogeneous with respect to their linkage composition, our analyses confirmed similarities between samples from related sources (e.g., brans and beta-glucans; peels from citrus fruits).
To understand whether particular glycan structures were associated with binding phenotypes, we calculated Spearman’s rank correlation coefficients for the adhesion of all 160 strains tested versus each carbohydrate linkage across all polysaccharides (Figure 2A; Table S4). Statistically significant correlations were observed between linkages and binding (p<0.05; t-test). Moreover, the glycan linkages correlated with binding were distinct from strain to strain within a given species. For example, binding of B. thetaiotaomicron strains 23161 and 23584 was strongly correlated with 3,6-mannose and 3,4-glucose, respectively. Adhesion of B. ovatus strain 23621 significantly correlated with the abundance of 2-rhamnose in polysaccharides, whereas B. ovatus strain VPI-C145 binding was correlated with 5-linked arabinofuranose (Figure 2B). These data illustrate how the assay platform can disclose strain-specific differences in binding that are associated with specific carbohydrate structures.
Figure 2: Glycosyl linkage analysis highlights structural features correlated with strain-specific adhesion and strain-specific catabolism.
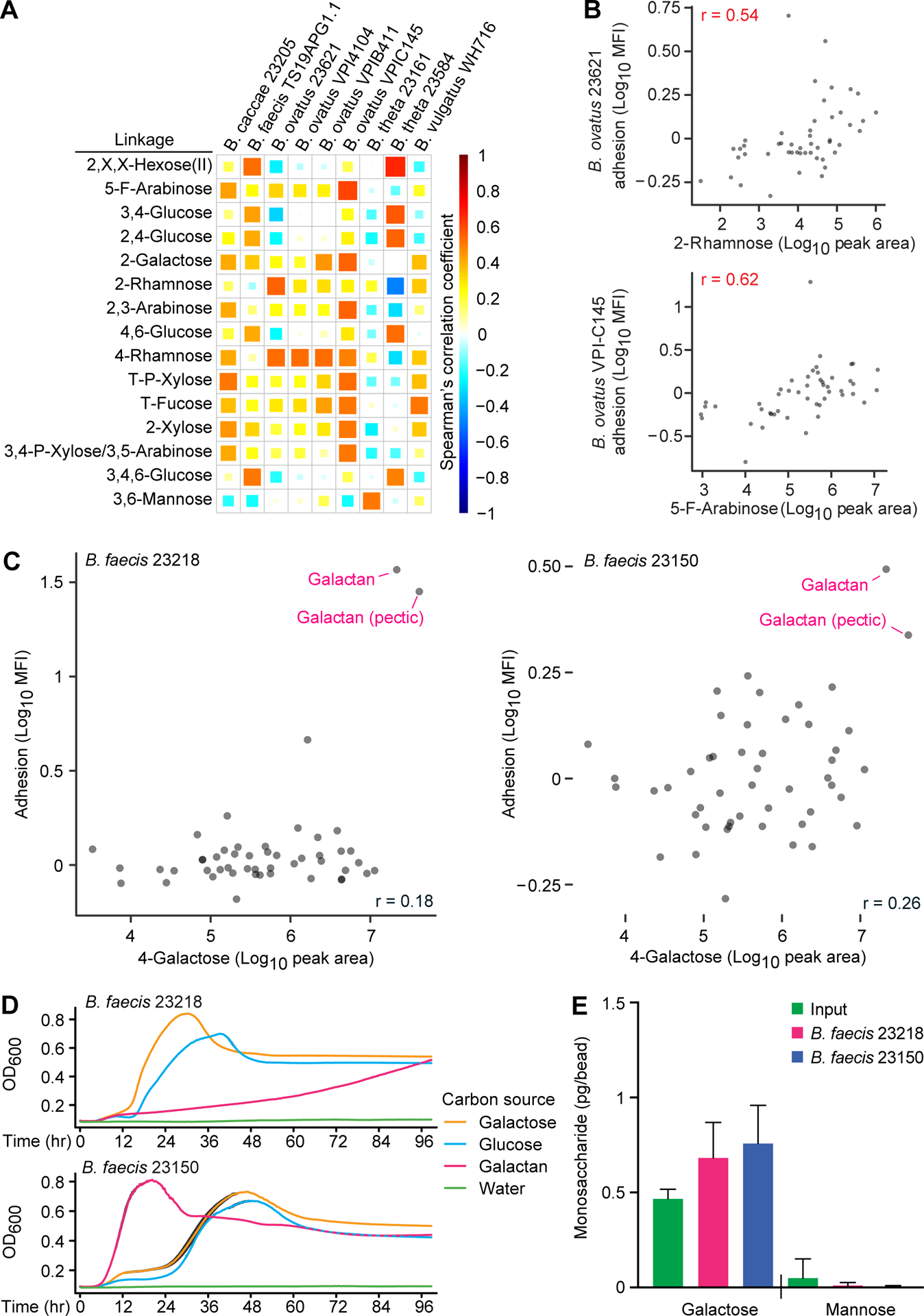
(A) Spearman’s rank correlation coefficients generated using results of mass spectrometry-based linkage analysis of 50 glycan preparations. The magnitude of the correlation between the adhesion phenotype of each organism (columns) and each glycosyl linkage (rows) is indicated by the color intensity and the completeness with which each cell in the matrix is filled (the direction of the correlation is indicated by color hue). Shown are the subset of linkages and strains tested for which one or more correlation coefficients was >0.5. Linkages are described as ring position followed by monosaccharide (T, terminal; X, single linkage of undetermined position; P-Xylose, xylopyranose; roman numerals designate distinct incompletely defined analytes). Strains belonging to the same species are grouped. (B) Plots showing correlations between adhesion phenotypes of two B. ovatus strains (23621, top; VPI-C145, bottom) and two glycosyl linkages (2-rhamnose, top; 5-F-arabinose, bottom). Spearman’s rank correlation coefficients are shown in red. (C) Adhesion values for B. faecis 23218 (left) and B. faecis 23150 (right) versus abundance of 4-linked galactose across 50 glycan preparations. The two purified galactans are highlighted with red arrows. Spearman’s rank correlation coefficients are shown. (D) Growth of B. faecis strains in minimal medium supplemented with the indicated carbon sources. Optical density (600nm) measured over 96 hours is shown as the mean value ± sd of quadruplicate cultures. (E) Quantification of bound galactan on beads before and after in vitro incubation at 4 °C with the indicated strains, as defined by GC-MS measurements of the mass of monosaccharide liberated by acid hydrolysis (each circle represents results obtained with a separate monoculture; mean values + sd are plotted). Values obtained with the different strains were not significantly different (p>0.05; t-test). See also Table S2–S5.
Assessing strain-specific differences in utilization of glycans that mediate adhesion
Purified polysaccharides, such as the two galactan preparations, were less complex in composition and less similar to any of the other preparations. Accordingly, some strains bound these polysaccharides with high specificity, including B. faecis 23218. We confirmed that galactan was predominantly composed of 4-linked galactose and terminal galactose (77.7% and 10.5% of detected carbohydrates, respectively), as was pectic galactan (70.5% and 12.1% respectively; Table S3). Our initial screen disclosed that B. faecis 23150 also bound most strongly to β-1,4-galactans (Figure 2C). Follow-up tests of these two strains’ adhesion phenotypes, performed in triplicate against the panel of 60 glycan-bead types, not only verified their binding specificity (Table S2C, S2D) but also established that the magnitude of their binding was significantly different; this difference remained when the adhesion assay was conducted at 37 °C, as opposed to 4 °C (mean fluorescence intensity ± sd; 97.7 ± 9.7 versus 32.2 ± 0.8 for strains 23218 and 23150, respectively; n=3 replicate determinations/strain; p<0.05; t-test).
To determine whether galactan metabolism by these two B. faecis strains was distinct, we cultured each strain in minimal medium with soluble galactan, galactose, or glucose as the sole carbon source. Figure 2D shows that strain 23150 displayed significantly more rapid growth in the presence of galactan compared to strain 23218 (reference control, glucose; n=3–4 replicates/condition; p<0.001, t-test). More efficient galactan degradation might eliminate the number of binding sites present on particulate galactan, reducing the ability of bacterial cells to adhere. To test whether catabolism of galactan by strain 23150 might account for its reduced adhesion, we directly measured galactose on beads before and after the adhesion assay by performing gas chromatography-mass spectrometry (GC-MS) of the quantity of monosaccharides released from beads by acid hydrolysis. We did not detect a significant difference in the amount of galactan remaining on beads exposed to these two strains during the adhesion assay which was conducted for 30 minutes at 4 °C (p>0.5, t-test; Figure 2E). In addition, we did not detect a significant difference in staining of galactan beads with a galactan-specific antibody before versus after carrying out the adhesion assay with either B. faecis strain (p>0.5, t-test). Together, our results indicate that the different binding phenotypes of these strains could not be attributed to differences in removal of galactan from the beads, at least as judged by these two detection methods.
Bacteroides possess multiple polysaccharide utilization loci (PULs) each containing various carbohydrate active enzymes (CAZymes) that allow them to sense, import, and metabolize glycan structures present in the gut. We sequenced the genomes of these two B. faecis strains and compared all of their predicted PULs to those present in B. thetaiotaomicron VPI-5482 (see Table S5), for which patterns of PUL gene expression in response to different glycans have been extensively characterized both in vitro and in vivo (Martens et al., 2011). We found that B. faecis 23218 and 23150 both contain a PUL (23218_2032 to 23218_2038 and 23150_4035 to 23150_4041) with high similarity to a PUL in the type-strain genome (BT_4667 to BT_4673) involved in the degradation of galactan (Martens et al., 2011; Luis et al., 2018). Notably, the PUL in B. faecis 23218 encodes a truncated hybrid two-component system (HTCS) that lacks amino acids 1 to 327 of the N-terminal, periplasmic sensor domain, as compared to the full length HTCS in B. faecis 23150 and B. thetaiotaomicron VPI-5482 (Table S5). The development of genetic tools applicable to these strains of B. faecis may reveal whether this or other genomic features provide a basis for their discrepant adhesion and galactan utilization phenotypes.
16S rDNA-based and confocal image-based analyses of the specificity of bacterial binding to dietary glycans
We developed a DNA sequence-based readout of bacterial adhesion in order to quantify the binding of co-existing species/strains to different glycan-bead types (Figure 3A). We first incubated beads coated with either galactan or arabinoxylan with either B. faecis 23218 or B. ovatus 23708 that had been grown in monoculture. The separate samples of beads were then isolated, washed free of non-adherent cells, combined with a defined quantity of E. coli as a normalization control and DNA was isolated from the washed beads. Variable region 4 of 16S rDNA genes present in each sample was amplified by PCR and the resulting amplicons were sequenced. By quantifying the number of Bacteroides reads relative to E. coli reads, we documented significantly greater binding of B. faecis cells to galactan beads than arabinoxylan beads (0.31% versus 0.007% of the input respectively; n=3 independent adhesion assays/strain/bead type; p<0.05; t-test; Figure 3B). As predicted from our screen, B. ovatus 23708 showed negligible binding to both bead types (0.015% of the input recovered on galactan-beads and 0.009% on arabinoxylan-beads). We then combined B. faecis 23218 and B. ovatus 23708 cells in a 1:1 ratio and incubated the mixture with galactan or arabinoxylan beads. V4–16S rDNA analysis revealed that galactan bead samples contained significantly more of the galactan-binding strain B. faecis 23218 (0.28% of the input cells) than the control strain B. ovatus 23708 (0.016% of the input cells; p<0.005, t-test; Figure 3C). Arabinoxylan bead samples contained 0.009% and 0.006% of the input cells for these two strains, respectively.
Figure 3: 16S rDNA-based and confocal image-based analyses of the specificity of bacterial binding to dietary glycans.
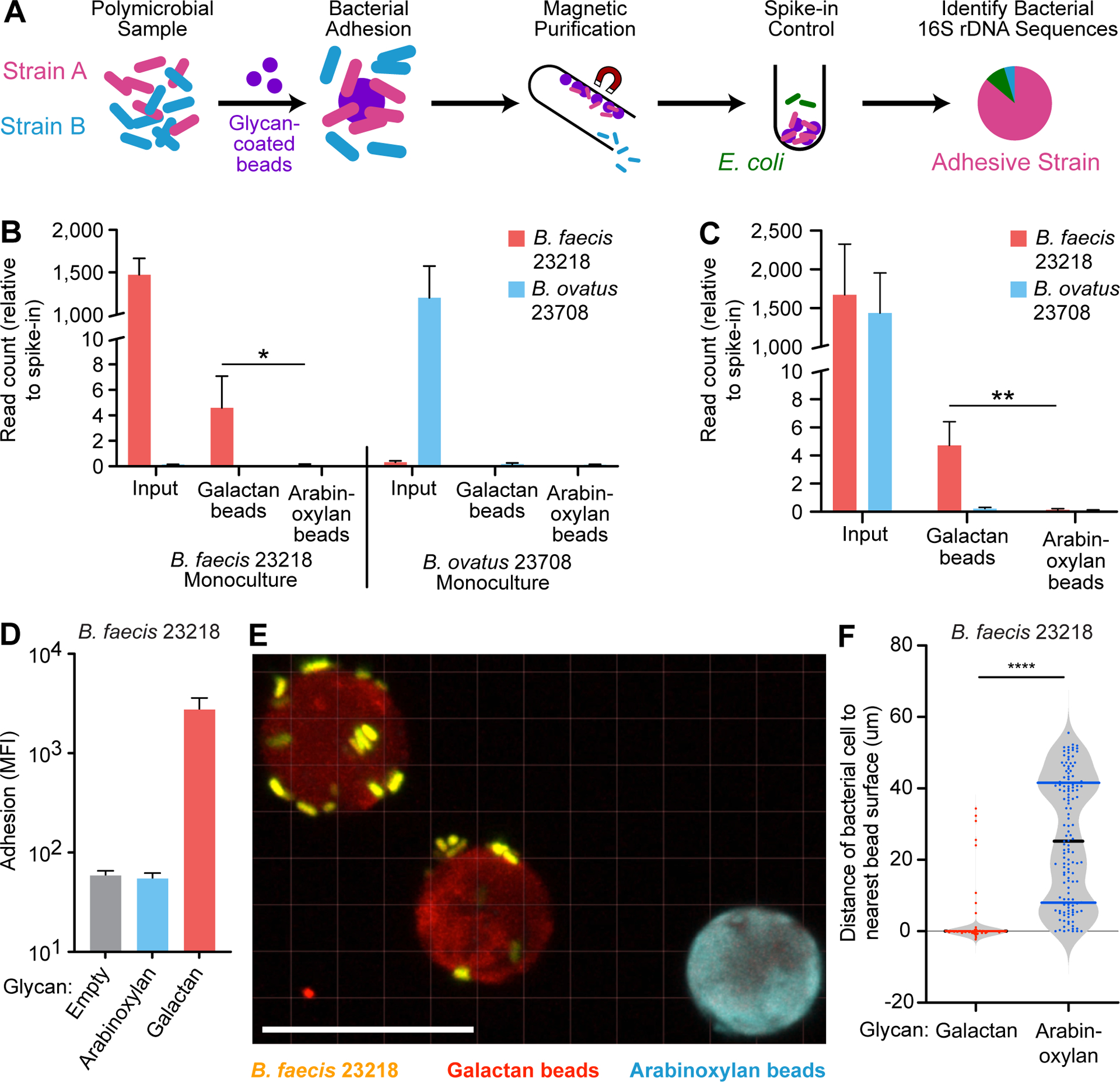
(A) Schematic depicting the implementation of beads to identify adhesive species in a mixed microbial culture using 16S rDNA sequencing. (B) Quantification of B. faecis 23218 and B. ovatus 23708 associated with galactan or arabinoxylan beads after their incubation with the indicated strains grown in monoculture. The Y-axis denotes V4–16S rDNA read count relative to E. coli spike-in control read count for each sample (n=3 separate adhesion assays; mean + sd is plotted for each group; *, p<0.05; t-test). (C) Quantification of B. faecis 23218 and B. ovatus 23708 associated with galactan or arabinoxylan beads after incubation with the both strains in a 1:1 mixed culture. The Y-axis denotes read count relative to E. coli spike-in control read count for each sample (n=4 separate adhesion assays; mean + sd is plotted for each group; **, p<0.005; t-test). (D) Fluorescence measurement of adhesion of B. faecis 23218, grown and assayed by flow cytometry in triplicate, incubated with a mixture of three bead types. Log10 geometric mean fluorescent intensity in the Syto-82 channel for each bead type in each sample is indicated on the y-axis (mean + sd). (E) Confocal image of galactan beads (red) and arabinoxylan beads (blue) incubated with B. faecis 23218 cells (yellow) in vitro. Scale bar, 20μm. (F) Quantification of distances from bacterial isosurfaces to the nearest isosurface of each bead type from the experiment shown in panel B. Each dot represents a bacterial isosurface (n=143 isosurfaces scored; median and quartiles are shown in the violin plots). ****, p<0.001 (Mann-Whitney U test).
To assess whether glycan-specific adhesion to artificial food particles could be quantified microscopically, we incubated B. faecis 23218 with galactan and arabinoxylan beads and verified bacterial adhesion by FACS (Figure 3D). Confocal microscopy disclosed individual bacterial cells, oriented parallel to and ≤0.5 μm from the surfaces of galactan but not arabinoxylan beads (Figure 3E; Video S1). Isosurfaces (i.e., computer-generated, 3-dimenional representations of the volume of fluorescent signal exceeding a defined grey-value threshold) of the distinct bead types and bacterial cells were created. The distance from each bacterial isosurface to the nearest bead (of each type) was then measured. This in vitro analysis established that there were statistically significant differences in the proximity of bacterial cells to galactan beads compared to arabinoxylan-coated control beads (Figure 3C). Taken together, the sequencing and imaging data validate the binding specificity (at the level of organism and glycan) detected in our screen and provide evidence that that the assay technology can be applied to polymicrobial communities.
Glycan-specific binding in vivo
A number of factors might modulate adhesion in vivo that are not represented in our in vitro assay; the intestine not only contains polysaccharides from diet, but also glycans produced by cell lineages represented in its mucosa, present in exfoliated epithelial cells, and generated by microbiota members themselves (e.g., capsular polysaccharides). Therefore, we used gnotobiotic mice to test whether the carbohydrate-specific adhesion phenotypes we uncovered in vitro could be recapitulated in an in vivo context. Adult C57Bl/6J germ-free mice were fed a standard chow diet rich in diverse plant polysaccharides and monocolonized with B. faecis strain 23218 or 23150. Beads coated with galactan or arabinoxylan, each labeled with a distinct fluorophore, were combined, sterilized with 70% ethanol, and orally administered to the monocolonized mice (Figure 4A). Beads were recovered from cecal contents 4.5 hours after gavage, washed, and embedded in agar on an imaging slide. Qualitative assessments based on confocal microscopy disclosed specific binding of each of the strains to galactan- compared to arabinoxylan-beads (n=4 animals/strain; Figure 4B,C; Video S2,S3). A noticeable difference between our in vitro and in vivo assay results was the predominance of aggregates composed of multiple galactan-coated beads with bacterial cells surrounded by neighboring beads with their surfaces closely approximated, suggesting that the bacteria themselves served to agglutinate the individual particles in vivo. As shown in Figure 4B,C, the vast majority of bacteria were present in these aggregates of galactan-beads. In contrast, in vitro the vast majority of galactan-beads with bound bacteria were present as singletons [86.5 ± 3.4% (mean ± sd) based on the FACS gating described in Table S1C]; aggregation, when encountered, was most often manifest as bead pairs rather than large agglomerations.
Figure 4: Adhesion of bacteria to glycan-beads in gnotobiotic mice.
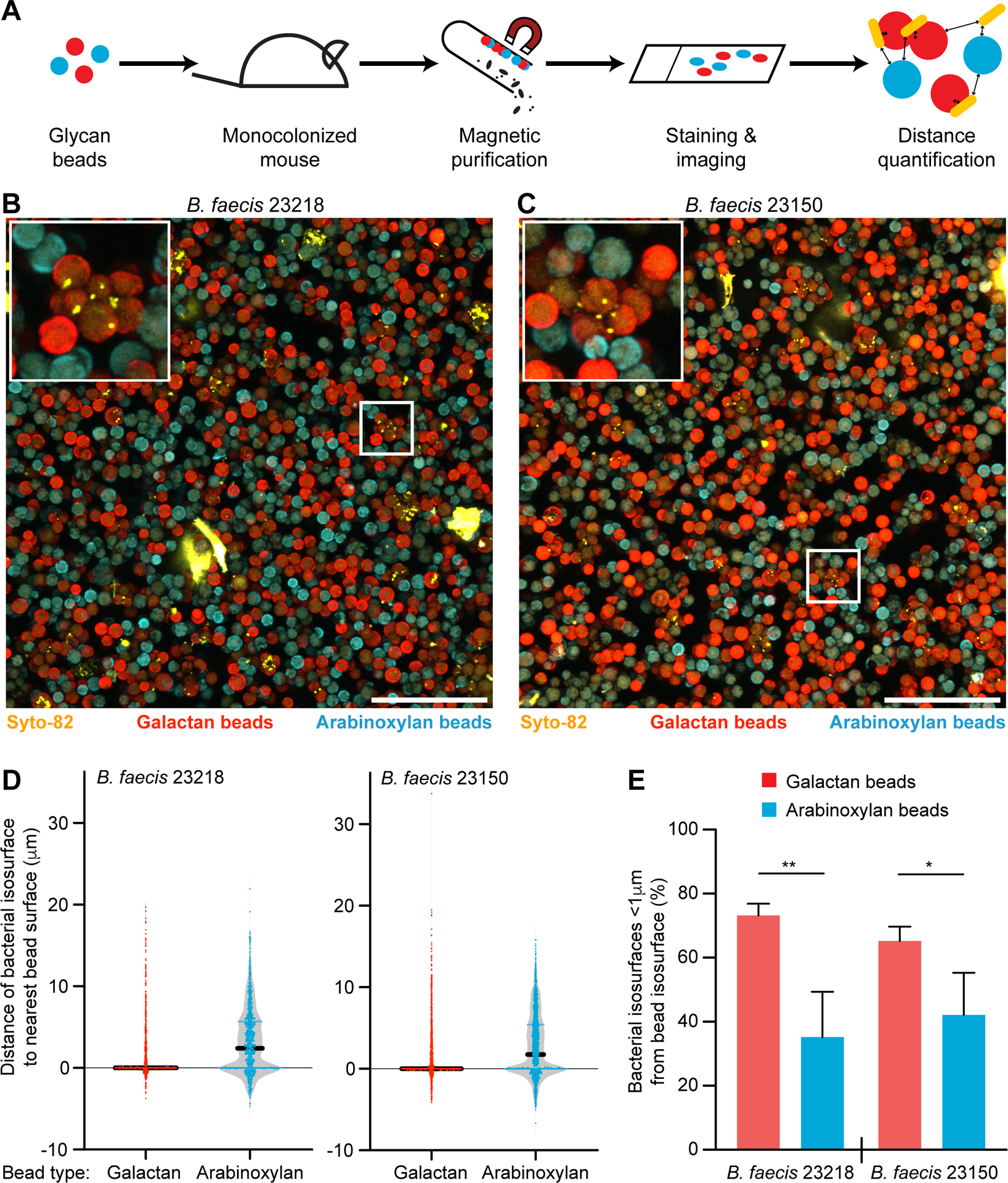
(A) Schematic depicting the oral gavage, isolation, quantitative imaging of beads administered to monocolonized gnotobiotic mice to test for in vivo bacterial adhesion to distinct dietary glycans. (B,C) Beads were recovered from cecal contents 4.5 hours after their administration to gnotobiotic mice monocolonized with either B. faecis 23218 or B. faecis 23150. Recovered beads were embedded in agarose, tiled confocal z-stacks were acquired, and maximum intensity projections were generated. Syto-82 labeled bacterial cells (yellow), arabinoxylan beads (cyan), and galactan beads (red) are shown. Scale bar, 100 μm. Insets depict aggregates of beads with bound bacterial cells. (D) Representative plots (one of four mice analyzed per group) show the shortest distances between each bacterial isosurface and the nearest bead surface. Distances are negative if the bacterial surface lies completely within the borders of the bead surface. Dots represent bacterial isosurfaces (n=473–1780 bacteria quantified per mouse) (E) The percentage of bacterial isosurfaces within 1 μm of each bead type isosurface is plotted (n=4 mice/strain; bars show mean + sd; *, p<0.05; **, p<0.01; t-test).
Quantification of these confocal microscopy imaging data sets revealed that bacterial isosurfaces were significantly further from arabinoxylan beads compared to galactan beads (Figure 4D). Operationally defining ‘associated bacteria’ as those located <1 μm from a bead isosurface allowed us to conclude that the percentage of bacteria associated with galactan beads was significantly higher than the percentage associated with arabinoxylan beads for both B. faecis strains (p<0.01, t-test; Figure 4E). Although the aggregation phenomenon prevented us from accurately determining in vivo whether there were statistically significant differences in bacterial load per galactan bead between strains, aggregation was glycan-dependent; there were small but significant differences between the average volumes of galactan bead versus arabinoxylan bead isosurfaces produced in vivo (2750 ± 386 versus 2316 ± 324 μm3; mean ± 95%CI; p<0.0001; Mann-Whitney U test).
Discussion
The library of artificial food particles described in this study, composed of multi-label microscopic paramagnetic beads each coated with a different glycan preparation, provides a quantitative functional readout of strain-level diversity. The observed variation in binding specificities within and across Bacteroides and Parabacteroides species and strains has a number of implications. A microbial community that contains strains with diverse binding specificities for polysaccharides would be expected to have greater capacity to gain access to the nutrient resources contained in fibers whose varied physical-chemical and compositional properties reflect differences in their sources and differences in the food processing methods used to incorporate them into contemporary diets. The fact that adhesion is not necessarily linked to degradation of soluble glycan provides one explanation for the evolution of adhesion phenotypes, namely, the capacity to bind to one component of a plant cell wall/fiber to gain access other component; it also has implications regarding how syntrophic (nutrient sharing) relationships can be established and sustained among bacterial strains/species that have evolved the capacity to cohabitate on the surfaces of food fragments or host-derived structures (e.g., mucus or mucus fragments released into the lumen; exfoliated cells or cell surfaces). The glycan-specificity we observed may also indicate that binding of members of Bacteroides and Parabacteroides is mediated by bacterial cell surface lectins, including, for example, lectins incorporated into pili that extend beyond polysaccharide capsule (Berne et al., 2018; Xu et al., 2016).
The bead-based approach described has a number of potential applications. For example, our in vitro screening method enables the selection of strains with adhesion phenotypes that are robust to changes in various factors (i.e., nutrient availability, pH, growth phase). These strains can then be administered as candidate probiotics with defined glycan binding specificity to gnotobiotic mice colonized with defined consortia of human gut microbes, together with artificial particles containing a glycan that they recognize. In addition, that bead-type could be used to engineer recruitment of other organisms to a microcosm where syntrophic partnerships that yield desired metabolic outputs can be forged. Glycan-coated beads could also be exploited (i) to intentionally foster competition between microbes when both competitors adhere to the same carbohydrate or to two distinct diet- or host-derived carbohydrates affixed to the same bead surface, or (ii) as tests of “parasitic adhesion” not to bead-associated glycans, but rather to the other microbes that bind to a certain bead-linked glycan (i.e., a strategy for glycan foraging that exploits the binding of other microbiota members). This approach has implications for development of pre-, pro-, and synbiotics as well as providing insights about mechanisms underlying bacterial pathogenesis. The micron scale biogeography of organisms, and the metabolic consequences of such physical relationships, could be assessed within the gut via in situ microscopy or imaging mass spectrometry (Rath et al., 2012) using beads as a well-defined spatial landmark. This application may be useful to food scientists who wish to identify combinations of polysaccharides and ways for embedding them in a common matrix so that together they can be more efficiently processed by a consumer’s microbiota.
Bead libraries could be added to human fecal samples to define functional diversity as it relates to diet, health status or other parameters. The results could have diagnostic and therapeutic implications. For example, what are the effects of prolonged consumption of a fiber-poor diet on glycan-mediated adhesion phenotypes? To what extent is adhesive capacity lost and how can it be rescued? Glycan binding patterns generated by the type of bead-binding assays described in this report could provide a way to characterize healthy community development in infants and children, serve as a biomarker for predicting risk for or the early advent of dysbioses, or provide a metric for quantifying the recovery from community perturbations, such as those prompted by diarrheal diseases or oral antibiotics, as a function of time or a therapeutic invention. Finally, looking beyond human communities, combining this bead-coating chemistry (which is not restricted to glycans) with 16S rDNA sequencing approaches may be useful in mining communities from various host species and environmental ecosystems for previously uncharacterized, uncultured microbes that adhere to diverse biomolecules of interest.
STAR Methods
Resource Availability
Lead Contact –
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jeffrey I. Gordon (jgordon@wustl.edu).
Materials Availability –
This study did not generate new unique reagents.
Data and Code Availability –
Full-length 16S rRNA sequences from the 160 Bacteroides and Parabacteroides strains, 16S rDNA sequences from polymicrobial experiments, and shotgun sequencing datasets generated from the genomes of Bacteroides faecis strains 23218 and 23150 have been deposited, in raw format prior to post-processing and data analysis, in the European Nucleotide Archive under study accession PRJEB39728.
Experimental Model and Subject Details
Gnotobiotic Mice –
All experiments involving mice were carried out in accordance with protocols approved by the Animal Studies Committee of Washington University in St. Louis. For in vivo bacterial adhesion assays, germ-free male C57BL/6J mice (10–16-weeks-old) were singly-housed in cages located within flexible plastic isolators. Cages contained paper houses for environmental enrichment. Animals consumed a standard rodent chow diet (Teklad 2018S) for the duration of the experiment and were maintained on a strict light cycle (lights on at 0600 h, off at 1900 h). On experimental day 0, aliquots of bacterial cultures were thawed, the outer surface of their tubes were sterilized with Clidox (Pharmacal) and the tubes were introduced into gnotobiotic isolators. The cell suspension was administered through a plastic tipped oral gavage needle. Pre-colonization fecal samples were collected and assayed by culture and culture-independent assays to verify the germ-free status of the mice.
Human gut bacterial strains –
Bacterial strains (Table S1A) were clonally arrayed in a 96-well format, frozen in TYGS plus 15% glycerol (Martens et al., 2008) and maintained at −80 °C until use. Strains were derived from existing collections of Bacteroides and Parabacteroides isolates (Johnson et al., 1978; Shoemaker et al., 2001; Snydman et al., 2007; Peterson et al., 2015). The identities of each of the strains used in adhesion assays was determined by near-full length (V1-V9) 16S rRNA gene sequencing (Genewiz).
Method Details
Generation of a 60-member glycan-bead library
Biotinylated glycans were generated as described previously (Patnode et al., 2019). Briefly, glycan preparations listed in Table S1B were suspended in water at a concentration of 20 mg/mL, sonicated, heated to 100 °C for one minute, and then centrifuged at 24,000 × g for 10 minutes at room temperature. The resulting solutions were diluted to 5 mg/mL in water and arrayed in a 96-well plate format (Applied Biosystems). TFPA-PEG3-biotin (Thermo Scientific), dissolved in DMSO (10 mg/mL) was added to the glycan solutions at a ratio of 1:5 (v/v). Samples were subjected to UV irradiation for 10 minutes (UV-B 306 nm, 7844 mJ total), and then diluted 1:4 to facilitate desalting (96-well ZEBA plates with a 7kD molecular weight cut-off; Thermo Scientific). Biotinylated glycans (5 μL aliquots) were incubated with 3×105 paramagnetic, ~15 μm-diameter streptavidin-coated silica beads (LSKMAGT, Millipore Sigma) for 24 hours at room temperature. Beads were washed three times with 180 μL HNTB buffer [10mM HEPES, 150mM NaCl, 0.05% Tween-20, 0.1% bovine serum albumin (MilliporeSigma, catalog number A2058)] using a magnetic stand. Beads were subsequently incubated for 30 minutes at room temperature with 5 μg/mL streptavidin-fluorophore mixtures in HNTB according to the layout in Table S1C. The process of washing, biotin-glycan incubation, washing, and streptavidin incubation was repeated for two additional cycles. Equal volumes of uniquely labeled bead types were then pooled into a single mixture. The pooled bead library was assessed using an Aria III cell sorter (BD Biosciences) to confirm detection of non-overlapping fluorescent populations.
Bacterial adhesion screening assay
For adhesion experiments, strains were grown at 37 °C in TYGS medium in an anaerobic chamber (atmosphere; 75% N2, 20% CO2, 5% H2) to an optical density at 600 nm of 0.80 ± 0.15 (mean ± sd). Bacterial cells were pelleted from cultures by centrifugation at 3,000 × g for 5 minutes. Cell pellets were immediately resuspended in HNTB containing 5mM Syto-82 dye (Invitrogen) for 5 minutes at room temperature and washed twice by centrifugation and addition of 200 μL HNTB. Washed cells were centrifuged 3,000 × g for 5 minutes, the supernatant was removed, a 5 μL aliquot of the pooled bead library was added to each cell pellet and the solution was mixed by repeated pipetting; the average number and standard deviation of beads per well was 9,443 ± 1,032. The suspensions were incubated in the dark at 4 °C with constant rotation for 30 minutes; beads were then collected using a magnetic rack and washed three times with 200 μL HNTB (with minimal agitation). Beads were run on a FACSAriaIII, identified based on fluorescence (see Table S1C), and Syto-82 fluorescence was measured (585/42nm bandpass filter) for each bead type. Samples of the glycan bead library were incubated with 5mM Syto-82 in the absence of bacteria to establish background dye binding.
Glycosyl linkage analysis
A stock solution of each polysaccharide was prepared in water at a concentration of 10 μg/μL. Homogenization of sample was accomplished by bead blasting for 3 minutes using 1.4 mm-diameter stainless steel magnetic beads and a Next Advance Bullet Blender Storm 24 (Next Advance, Troy, NY). The resulting sample solutions were incubated at 100 °C for 1 h followed by a second round of bead blasting. A 5 μL aliquot of the homogenized sample was plated onto a 96-well plate and dried by vacuum centrifugation.
Methylation of polysaccharides was adapted from previously described methods (Galermo et al., 2018). In short, free hydroxyl groups on the polysaccharides were methylated by reacting the samples with saturated NaOH and iodomethane in DMSO. Dichloromethane extractions and water washes were performed to isolate the methylated polysaccharides and remove residual NaOH and DMSO. The methylated polysaccharides were dried to completeness by vacuum centrifugation and subjected to acid hydrolysis in 4 M trifluoroacetic acid at 100 °C for 1 h. Samples were dried by vacuum centrifugation and labeled with 1-phenyl-3-methyl-5-pyrazolone (PMP). The partially methylated, PMP-labeled compounds were separated and analyzed on an Agilent 1290 Infinity II UHPLC system coupled to an Agilent 6495A triple quadrupole mass spectrometer operated in MRM mode. Acquired UPLC-QqQ-MS data were retrieved using Agilent MassHunter Workstation Data Acquisition software (Version B.06.01) and analyzed with Agilent MassHunter Quantitative Analysis software (Version B.06.00).
16S rDNA-based quantification of bead-bound bacterial strains
Strains grown in monoculture were aliquoted into a deep-well culture plate either alone or mixed in a 1:1 ratio. The cells were exposed to beads coated with dietary glycans; the beads were then collected and washed exactly as described for the multiplex screening assay outlined above. Ten microliters of an OD600 0.035 culture of E. coli TSDC17.2–1.2 (Patnode et al., 2019) was added to each sample as a spike-in normalization control. DNA was then isolated from each sample by first bead-beating the sample with 0.15mm-diameter zirconium oxide beads and a 5mm-diameter steel ball in 2X buffer A (200 mM NaCl, 200 mM Tris, 20 mM EDTA), followed by extraction in phenol:chloroform:isoamyl alcohol, and further purification (QiaQuick 96 purification kit). PCR amplification of the V4 region of bacterial 16S rRNA genes was performed as described (Bokulich et al., 2013). Amplicons with sample-specific barcodes were pooled for multiplex sequencing using an Illumina MiSeq instrument. Reads were demultiplexed and reads sharing ≥97% nucleotide sequence identity [97% ID operational taxonomic units (OTUs)], that mapped to a reference OTU in the GreenGenes 16S rRNA gene database (McDonald et al., 2012) were assigned to that OTU.
Measurements of bead-based glycan degradation by bound bacterial strains
For quantification of polysaccharide on the bead surface after adhesion, beads were incubated with bacteria at 4 °C as described above, then collected and washed in a solution containing 1% SDS, 6M urea and HNTB for 10 minutes at room temperature to remove bound bacteria and exogenous components. Beads were then washed three times with 200 μL HNTB. Hybridoma culture supernatant (anti-galactan monoclonal CCRC-133, CarboSource) was subsequently incubated with the beads for 30 minutes at room temperature. Beads were washed and incubated with 1μg/mL R-phycoerythrin conjugated goat anti-mouse IgG (Jackson Immunoresearch) for 15 minutes. Samples were analyzed on an FACSAria III cell sorter (BD Biosciences).
The mass of polysaccharide on glycan coated beads was quantified using monosaccharide analysis as described previously (Patnode et al., 2019). Briefly, an aliquot of each sample was taken to verify the number of beads, and the beads were subjected to acid hydrolysis (2M trifluoroacetic acid) in 300 μL glass vials (ThermoFisher; catalog number C4008–632C). Vials were crimped with Teflon-lined silicone caps (ThermoFisher) and incubated at 100 °C with rocking for 1 hour. Samples were subsequently dried in a SpeedVac for 24 hours, incubated with 30 μL O-methoxyamine (15mg/mL pyridine) for 24 h at 37 °C, followed by addition of 30 μL MSTFA/TMCS [N-Methyl-N-trimethylsilyltrifluoroacetamide/2,2,2-trifluoro-N-methyl-N-(trimethylsilyl)-acetamide, chlorotrimethylsilane] (ThermoFisher) and incubation for 1 h at 70 °C. Heptane (30 μL) was added before loading the samples for injection onto an Agilent model 7890B gas chromatography system coupled to an Agilent model 5977B MS detector. The mass of each monosaccharide detected in each sample of beads was determined using standard curves generated with purified monosaccharides. This mass was then divided by the final count of beads in each sample to produce a measurement of mass of recoverable monosaccharide per bead.
Bacterial adhesion assays in the intestines of gnotobiotic mice
Wheat arabinoxylan and lupin seed galactan (P-WAXYL, P-GALLU; Megazyme) were biotinylated as described above and incubated with paramagnetic glass beads, along with biotinylated fluorophores at a concentration of 50ng/mL (PF-505, PF-633; Promokine). Beads were incubated with 70% ethanol for 1 minute in a biosafety cabinet, then washed three times with 1 mL of sterile HNTB using a magnetic stand. The bead types were combined, diluted, and aliquoted (107 beads/650μL HNTB) into sterile Eppendorf microcentrifuge tubes. Tubes containing beads were introduced into gnotobiotic isolators and the beads were administered by oral gavage (600μL per mouse). Animals were sacrificed 4.5 hours after gavage and cecal contents were vortexed and filtered through nylon mesh (100 μm pore-diameter). The resulting suspension of luminal contents was layered over sterile Percoll Plus (GE Health Care) and centrifuged at 500 × g for 5 minutes. Beads were collected from underneath the Percoll layer and washed four times using a magnetic stand, each time with 1 mL of fresh HNTB.
Beads from each mouse were suspended in low melting temperature agarose and added to a multi-well imaging slide (Ibidi). Tiled confocal z-stacks were acquired on a Zeiss 880 laser confocal microscope to a depth of 40μm. Isosurfaces (computer-generated representations of a specified gray value range in the data set) were created for each channel, and distances between surfaces were calculated using Imaris software (Bitplane).
Bacterial growth assays
Frozen stocks of each strain (TYGs/15% glycerol) were streaked on BHI blood agar plates and single colonies were used to inoculate TYGs medium (Martens et al., 2008). The concentration of cells was normalized based on OD600 and aliquots were then added to minimal medium (Martens et al., 2008) or minimal medium supplemented with 5mg/mL carbohydrate. Cultures were grown at 37 °C in an anaerobic chamber (atmosphere; 75% N2, 20% CO2, 5% H2) with OD600 measurements acquired with an automated plate reader (BioTek) every 30 minutes for 96 hours (n=3–4 monocultures of each strain analyzed in parallel).
Genome sequencing and annotation
Bacterial cultures were centrifuged at 18,000 × g for 5 minutes to pellet cells. DNA was isolated by bead-beating with 0.15 mm-diameter zirconium oxide beads and a 4 mm-diameter steel ball in 2X buffer A (200 mM NaCl, 200 mM Tris, 20 mM EDTA), followed by extraction in phenol:chloroform:isoamyl alcohol (25:24:1), and further purification (QiaQuick 96 purification kit; Qiagen). Sequencing libraries were generated by tagmentation using the Nextera DNA Library Prep Kit (Illumina) and combinations of custom barcoded primers (Adey et al., 2010). Libraries were sequenced on an Illumina NextSeq instrument (paired-end 150 nt reads) and genomes were assembled using SPAdes (Bankevich et al., 2012). Open reading frames were annotated with Prokka (Seemann, 2014). Genes encoding carbohydrate-active enzymes were annotated using the CAZy database through manual curation (Lombard et al., 2014); PULs and clusters of CAZymes were predicted and assigned sequential ID numbers using the algorithms from PULDB (Terrapon et al., 2018).
Quantification and Statistical Analysis
For statistical tests comparing the binding of different bacterial strains to 60 glycan bead types, three independent bacterial cultures per strain were used, and differences were calculated using Students t-test with FDR p-value correction. For analysis of correlations between bacterial binding and carbohydrate linkages, a Spearman’s rank correlation coefficient was calculated for each strain versus each linkage across all analyzed glycan preparations. When a linkage was not detected in a glycan sample, an estimate of the limit of detection (lowest detected value across all polysaccharide samples divided by 5) was used as a pseudocount. Images of bacteria binding to beads in vitro were collected for 13 subsamples of the bead population and isosurfaces were generated for both bead types in parallel using identical parameters for each subsample. For in vivo quantification of bacteria binding to beads, nine fields of view were analyzed for each monocolonized mouse (n=4 mice per colonization condition). Isosurfaces for bacterial cells were generated using identical parameters across all images, and image processing (channel subtraction) was used to compensate bead fluorescence and generate isosurfaces for each bead type. The shortest distance between each bacterial isosurface and the nearest bead isosurface of each type was calculated (Imaris, Bitplane). Bead isosurfaces with a bacterial isosurface within 1μm were considered to have bacteria associated with their surface. Among all bacteria isosurfaces scored, the percentage associated with beads was calculated and differences between bead types were determined using t-tests. All tests were two-tailed with a p-value cutoff of 0.05. Mean values and standard deviations are shown in the figures.
Supplementary Material
Table S1 (related to Figure 1): Bacterial strains, glycans, and fluorophores used in adhesion screen. (A) Bacterial strains. (B) Glycan preparations. (C) Fluorophore combinations.
Table S2 (related to Figure 1): Bacterial adhesion data for the bead-based in vitro screen and subsequent validation. (A) Geometric mean of Syto-82 fluorescence intensity for each bead type in each culture well. (B) Bead count for each bead type in each culture well. (C) Geometric mean of Syto-82 fluorescence intensity for each bead type in each culture well containing replicate cultures of B. faecis strains 23218 and 23150. (D) Bead count for each bead type in each culture well containing replicate cultures of B. faecis strains 23218 and 23150.
Table S3 (related to Figure 2): Monosaccharide and linkage analysis for glycan preparations used in the bead-based adhesion screen. (A) Monosaccharides detected in glycan preparations. (B) Glycosidic linkages detected in glycan preparations.
Table S4 (related to Figure 2): Correlation coefficients between binding of each bacterial strain to tested glycan preparations and the abundance of each carbohydrate linkage across all glycan preparations.
Table S5 (related to Figure 2): Comparison of carbohydrate-active enzymes (CAZymes) and polysaccharide utilization loci (PULs) identified in the genomes of B. faecis 23218 and B. faecis 23150 and the reference type strain B. thetaiotaomicron VPI-5482.
Video S1 (related to Figure 1): Confocal images of Bacteroides adhesion to glycan-coated beads. Rotations (360 degrees) of reconstructed z-stacks showing galactan-coated beads (red), arabinoxylan-coated beads (cyan), and B. faecis 23218 cells (green).
Video S2 (related to Figure 4): Confocal images of beads orally administered and recovered from gnotobiotic mouse cecal contents after monocolonization with B. faecis 23218. Rotations (360 degrees) of reconstructed z-stacks showing galactan-coated beads (red), arabinoxylan-coated beads (cyan), and B. faecis 23218 cells (yellow). Raw fluorescence data are shown followed by the corresponding computed isosurfaces.
Video S3 (related to Figure 4): Confocal images of beads orally administered and recovered from gnotobiotic mouse cecal contents after monocolonization with B. faecis 23150. Rotations (360 degrees) of reconstructed z-stacks showing galactan-coated beads (red), arabinoxylan-coated beads (cyan), and B. faecis 23150 cells (yellow). The raw fluorescence data are shown followed by the corresponding computed isosurfaces.
Highlights.
Library of glass beads with unique multi-fluorescent labels and glycans was created
Imaging and 16S rRNA in vitro assays assessed gut bacterial strain glycan binding
Strain-specific binding correlated with linkage composition of dietary glycans
Administering glycan-beads to gnotobiotic mice confirms specificity of binding
Acknowledgements
We thank Maria Karlsson, David O’Donnell, Justin Serugo, Marty Meier, Sabrina Wagoner, and J. Hoisington-Lopez, MariaLynn Crosby, and Darryl Wesener for their invaluable technical assistance. This work was supported by grants from the NIH (DK70977, DK78669, DK107158, and DK124445). J.I.G. is the recipient of a Thought Leader Award from Agilent Technologies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
J.I.G. is a co-founder of Matatu, Inc., a company characterizing the role of diet-by-microbiota interactions in animal health. C.B.L. is co-founder of Evolve Biosciences.
References
- Adey A, Morrison HG, Asan, Xun X, Kitzman JO, Turner EH, Stackhouse B, MacKenzie AP, Caruccio NC, Zhang X, et al. (2010). Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol 11, R119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19, 455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berne C, Ellison CK, Ducret A, and Brun YV (2018). Bacterial adhesion at the single-cell level. Nature Rev Microbiol 16, 616–627. [DOI] [PubMed] [Google Scholar]
- Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, and Caporaso JG (2013). Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nature Methods 10, 57–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero OX, and Datta MS (2016). Microbial interactions and community assembly at microscales. Curr Opin Microbiol 31, 227–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, Pudlo NA, Kitamoto S, Terrapon N, Muller A, et al. (2016). A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell 167, 1339–1353 e1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earle KA, Billings G, Sigal M, Lichtman JS, Hansson GC, Elias JE, Amieva MR, Huang KC, and Sonnenburg JL (2015). Quantitative Imaging of Gut Microbiota Spatial Organization. Cell Host & Microbe 18, 478–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galermo AG, Nandita E, Barboza M, Amicucci MJ, Vo TT, and Lebrilla CB (2018). Liquid Chromatography-Tandem Mass Spectrometry Approach for Determining Glycosidic Linkages. Anal Chem 90, 13073–13080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project, C. (2012). Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JL, and Ault DA (1978). Taxonomy of Bacteroides .2. Correlation of Phenotypic Characteristics with Deoxyribonucleic-Acid Homology Groupings for Bacteroides-Fragilis and Other Saccharolytic Bacteroides Species. Int J Syst Bacteriol 28, 257–268. [Google Scholar]
- Kalas V, Hibbing ME, Maddirala AR, Chugani R, Pinkner JS, Mydock-McGrane LK, Conover MS, Janetka JW, and Hultgren SJ (2018). Structure-based discovery of glycomimetic FmlH ligands as inhibitors of bacterial adhesion during urinary tract infection. Proc Natl Acad Sci USA 115, E2819–E2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karcher N, Pasolli E, Asnicar F, Huang KD, Tett A, Manara S, Armanini F, Bain D, Duncan SH, Louis P, et al. (2020). Analysis of 1321 Eubacterium rectale genomes from metagenomes uncovers complex phylogeographic population structure and subspecies functional adaptations. Genome Biol 21, 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guennec L, Virion Z, Bouzinba-Segard H, Robbe-Masselot C, Leonard R, Nassif X, Bourdoulous S, and Coureuil M (2020). Receptor recognition by meningococcal type IV pili relies on a specific complex N-glycan. Proc Natl Acad Sci USA 117, 2606–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, and Henrissat B (2014). The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42, D490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luis AS, Briggs J, Zhang X, Farnell B, Ndeh D, Labourel A, Basle A, Cartmell A, Terrapon N, Stott K, et al. (2018). Dietary pectic glycans are degraded by coordinated enzyme pathways in human colonic Bacteroides. Nat Microbiol 3, 210–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane S, and Macfarlane GT (2006). Composition and metabolic activities of bacterial biofilms colonizing food residues in the human gut. Appl. Environ Microbiol 72, 6204–6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Chiang HC, and Gordon JI (2008). Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host & Microbe 4, 447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Kelly AG, Tauzin AS, and Brumer H (2014). The devil lies in the details: how variations in polysaccharide fine-structure impact the physiology and evolution of gut microbes. J Mol Biol 426, 3851–3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Lowe EC, Chiang H, Pudlo NA, Wu M, McNulty NP, Abbott DW, Henrissat B, Gilbert HJ, Bolam DN, et al. (2011). Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biology 9, e1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNulty NP, Wu M, Erickson AR, Pan C, Erickson BK, Martens EC, Pudlo NA, Muegge BD, Henrissat B, Hettich RL, et al. (2013). Effects of diet on resource utilization by a model human gut microbiota containing Bacteroides cellulosilyticus WH2, a symbiont with an extensive glycobiome. PLoS Biology 11, e1001637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihajlovic J, Bechon N, Ivanova C, Chain F, Almeida A, Langella P, Beloin C, and Ghigo JM (2019). A Putative Type V Pilus Contributes to Bacteroides thetaiotaomicron Biofilm Formation Capacity. J Bacteriol 201, e00650–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miron J, Ben-Ghedalia D, and Morrison M (2001). Invited review: adhesion mechanisms of rumen cellulolytic bacteria. J Dairy Sci 84, 1294–1309. [DOI] [PubMed] [Google Scholar]
- Patnode ML, Beller ZW, Han ND, Cheng J, Peters SL, Terrapon N, Henrissat B, Le Gall S, Saulnier L, Hayashi DK, et al. (2019). Interspecies Competition Impacts Targeted Manipulation of Human Gut Bacteria by Fiber-Derived Glycans. Cell 179, 59–73 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson DA, Planer JD, Guruge JL, Xue L, Downey-Virgin W, Goodman AL, Seedorf H, and Gordon JI (2015). Characterizing the interactions between a naturally primed immunoglobulin A and its conserved Bacteroides thetaiotaomicron species-specific epitope in gnotobiotic mice. J Biol Chem 290, 12630–12649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole J, Day CJ, von Itzstein M, Paton JC, and Jennings MP (2018). Glycointeractions in bacterial pathogenesis. Nature Rev Microbiol 16, 440–452. [DOI] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Foster KR, and Comstock LE (2016). The evolution of cooperation within the gut microbiota. Nature 533, 255–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath CM, Alexandrov T, Higginbottom SK, Song J, Milla ME, Fischbach MA, Sonnenburg JL, and Dorrestein PC (2012). Molecular analysis of model gut microbiotas by imaging mass spectrometry and nanodesorption electrospray ionization reveals dietary metabolite transformations. Anal Chem 84, 9259–9267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. [DOI] [PubMed] [Google Scholar]
- Shoemaker NB, Vlamakis H, Hayes K, and Salyers AA (2001). Evidence for extensive resistance gene transfer among Bacteroides spp. and among Bacteroides and other genera in the human colon. Appl Environ Microbiol 67, 561–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoog EC, Padra M, Aberg A, Gideonsson P, Obi I, Quintana-Hayashi MP, Arnqvist A, and Linden SK (2017). BabA dependent binding of Helicobacter pylori to human gastric mucins cause aggregation that inhibits proliferation and is regulated via ArsS. Sci Rep 7, 40656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snydman DR, Jacobus NV, McDermott LA, Ruthazer R, Golan Y, Goldstein EJ, Finegold SM, Harrell LJ, Hecht DW, Jenkins SG, et al. (2007). National survey on the susceptibility of Bacteroides fragilis group: report and analysis of trends in the United States from 1997 to 2004. Antimicrobial Agents Chemotherapy 51, 1649–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Tay QX, Kjelleberg S, Rice SA, and McDougald D (2015). Quorum sensing-regulated chitin metabolism provides grazing resistance to Vibrio cholerae biofilms. ISME J 9, 1812–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrapon N, Lombard V, Drula E, Lapebie P, Al-Masaudi S, Gilbert HJ, and Henrissat B (2018). PULDB: the expanded database of Polysaccharide Utilization Loci. Nucleic Acids Res 46, D677–D683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting SY, Martinez-Garcia E, Huang S, Bertolli SK, Kelly KA, Cutler KJ, Su ED, Zhi H, Tang Q, Radey MC, et al. (2020). Targeted Depletion of Bacteria from Mixed Populations by Programmable Adhesion with Antagonistic Competitor Cells. Cell Host & Microbe 28, 313–321 e316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatanen T, Plichta DR, Somani J, Munch PC, Arthur TD, Hall AB, Rudolf S, Oakeley EJ, Ke X, Young RA, et al. (2019). Genomic variation and strain-specific functional adaptation in the human gut microbiome during early life. Nat Microbiol 4, 470–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JH (1963). Hierarchical Grouping to Optimize an Objective Function. J Am Statistical Assoc 58, 236–244. [Google Scholar]
- Wu M, McNulty NP, Rodionov DA, Khoroshkin MS, Griffin NW, Cheng J, Latreille P, Kerstetter RA, Terrapon N, Henrissat B, et al. (2015). Genetic determinants of in vivo fitness and diet responsiveness in multiple human gut Bacteroides. Science 350, aac5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Shoji M, Shibata S, Naito M, Sato K, Elsliger MA, Grant JC, Axelrod HL, Chiu HJ, Farr CL, et al. (2016). A Distinct Type of Pilus from the Human Microbiome. Cell 165, 690–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe E, and Relman DA (2020). Tracking microbial evolution in the human gut using Hi-C reveals extensive horizontal gene transfer, persistence and adaptation. Nat Microbiol 5, 343–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Mogno I, Contijoch EJ, Borgerding JN, Aggarwala V, Li Z, Siu S, Grasset EK, Helmus DS, Dubinsky MC, et al. (2020). Fecal IgA Levels Are Determined by Strain-Level Differences in Bacteroides ovatus and Are Modifiable by Gut Microbiota Manipulation. Cell Host & Microbe 27, 467–475 e466. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 (related to Figure 1): Bacterial strains, glycans, and fluorophores used in adhesion screen. (A) Bacterial strains. (B) Glycan preparations. (C) Fluorophore combinations.
Table S2 (related to Figure 1): Bacterial adhesion data for the bead-based in vitro screen and subsequent validation. (A) Geometric mean of Syto-82 fluorescence intensity for each bead type in each culture well. (B) Bead count for each bead type in each culture well. (C) Geometric mean of Syto-82 fluorescence intensity for each bead type in each culture well containing replicate cultures of B. faecis strains 23218 and 23150. (D) Bead count for each bead type in each culture well containing replicate cultures of B. faecis strains 23218 and 23150.
Table S3 (related to Figure 2): Monosaccharide and linkage analysis for glycan preparations used in the bead-based adhesion screen. (A) Monosaccharides detected in glycan preparations. (B) Glycosidic linkages detected in glycan preparations.
Table S4 (related to Figure 2): Correlation coefficients between binding of each bacterial strain to tested glycan preparations and the abundance of each carbohydrate linkage across all glycan preparations.
Table S5 (related to Figure 2): Comparison of carbohydrate-active enzymes (CAZymes) and polysaccharide utilization loci (PULs) identified in the genomes of B. faecis 23218 and B. faecis 23150 and the reference type strain B. thetaiotaomicron VPI-5482.
Video S1 (related to Figure 1): Confocal images of Bacteroides adhesion to glycan-coated beads. Rotations (360 degrees) of reconstructed z-stacks showing galactan-coated beads (red), arabinoxylan-coated beads (cyan), and B. faecis 23218 cells (green).
Video S2 (related to Figure 4): Confocal images of beads orally administered and recovered from gnotobiotic mouse cecal contents after monocolonization with B. faecis 23218. Rotations (360 degrees) of reconstructed z-stacks showing galactan-coated beads (red), arabinoxylan-coated beads (cyan), and B. faecis 23218 cells (yellow). Raw fluorescence data are shown followed by the corresponding computed isosurfaces.
Video S3 (related to Figure 4): Confocal images of beads orally administered and recovered from gnotobiotic mouse cecal contents after monocolonization with B. faecis 23150. Rotations (360 degrees) of reconstructed z-stacks showing galactan-coated beads (red), arabinoxylan-coated beads (cyan), and B. faecis 23150 cells (yellow). The raw fluorescence data are shown followed by the corresponding computed isosurfaces.
Data Availability Statement
Full-length 16S rRNA sequences from the 160 Bacteroides and Parabacteroides strains, 16S rDNA sequences from polymicrobial experiments, and shotgun sequencing datasets generated from the genomes of Bacteroides faecis strains 23218 and 23150 have been deposited, in raw format prior to post-processing and data analysis, in the European Nucleotide Archive under study accession PRJEB39728.


