Abstract
Background
Wildtype mice are not susceptible to SARS-CoV-2 infection. Emerging SARS-CoV-2 variants, including B.1.1.7, B.1.351, P.1, and P.3, contain mutations in spike that has been suggested to associate with an increased recognition of mouse ACE2, raising the postulation that these SARS-CoV-2 variants may have evolved to expand species tropism to wildtype mouse and potentially other murines. Our study evaluated this possibility with substantial public health importance.
Methods
We investigated the capacity of wildtype (WT) SARS-CoV-2 and SARS-CoV-2 variants in infecting mice (Mus musculus) and rats (Rattus norvegicus) under in vitro and in vivo settings. Susceptibility to infection was evaluated with RT-qPCR, plaque assays, immunohistological stainings, and neutralization assays.
Findings
Our results reveal that B.1.1.7 and other N501Y-carrying variants but not WT SARS-CoV-2 can infect wildtype mice. High viral genome copies and high infectious virus particle titres are recovered from the nasal turbinate and lung of B.1.1.7-inocluated mice for 4-to-7 days post infection. In agreement with these observations, robust expression of viral nucleocapsid protein and histopathological changes are detected from the nasal turbinate and lung of B.1.1.7-inocluated mice but not that of the WT SARS-CoV-2-inoculated mice. Similarly, B.1.1.7 readily infects wildtype rats with production of infectious virus particles.
Interpretation
Our study provides direct evidence that the SARS-CoV-2 variant, B.1.1.7, as well as other N501Y-carrying variants including B.1.351 and P.3, has gained the capability to expand species tropism to murines and public health measures including stringent murine control should be implemented to facilitate the control of the ongoing pandemic.
Funding
A full list of funding bodies that contributed to this study can be found in the Acknowledgements section.
Keywords: SARS-CoV-2 variants, mouse, rat, murine, susceptibility, N501Y
Research in context.
Evidence before this study
We searched PubMed on July 14, 2021, with no starting date limitations, using the terms “SARS-CoV-2” and “B.1.1.7 or B.1.351 or P.1 or P.3” and “mouse or rat” and “susceptibility or infection” for articles in English. Our search did not reveal any report that performed comprehensive side-by-side investigations of the susceptibility of SARS-CoV-2 N501Y-carrying variants in murines in both in vitro and in vivo settings.
Added value of this study
Our study reveals that B.1.1.7 and other variants carrying the N501Y mutation but not WT SARS-CoV-2 can infect wildtype mice. High viral genome copies and high infectious virus particle titres are recovered from the nasal turbinate and lung of N501Y-carrying variants-inoculated mice for 4-to-7 days post infection. Robust expression of viral nucleocapsid protein and histopathological changes are detected from the nasal turbinate and lung of B.1.1.7-inocluated mice but not that of the WT SARS-CoV-2-inoculated mice. Similarly, B.1.1.7 readily infects wildtype rats with production of infectious virus particles. These findings provide direct evidence that B.1.1.7 and other N501Y-carrying variants have gained the capability to expand species tropism to murine species.
Implications of all the available evidence
Due to the abundance of murines and their proximate habitats with humans, humans-to- murines and murines-to-humans infection events may be inevitable. Our findings suggest that public health control measures including stringent murine control should be implemented to facilitate the control of the ongoing pandemic. Considering the high susceptibility of mice to N501Y-carrying variants and the endogenous ACE2 expression in wildtype mice in comparison to the hACE2-transgenic mice, the N501Y-carrying variant infection model may represent a physiologically-relevant model for SARS-CoV-2 studies on pathogenesis, immunology, and antivirals. In addition, infection of N501Y-carrying variants is compatible with all existing knock-out and knock-in mouse models that will greatly facilitate further functional studies on host genes and pathways.
Alt-text: Unlabelled box
1. Introduction
Coronaviruses have repeatedly crossed species barriers to cause human infection [1]. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a novel lineage B human-pathogenic betacoronavirus that was first identified in December 2019 [2]. SARS-CoV-2 is highly transmissible and has rapidly spread worldwide to cause more than 197 million cases of Coronavirus Disease 2019 (COVID-19) with over 4.0 million deaths as of 31st July, 2021 [3, 4]. Same as SARS-CoV-1 and human coronavirus NL63 (HCoV-NL63), SARS-CoV-2 also uses angiotensin-converting enzyme II (ACE2) as the cellular receptor for entry [5]. Importantly, SARS-CoV-2 variants of concern (VOC) with amino acid substitutions in the spike protein continue to emerge as the pandemic expands and are potentially associated with increased transmissibility and/or virulence [6].
Mammals, especially bats and rodents, are considered as major animal reservoirs of alphacoronaviruses and betacoronaviruses [1, 7]. The global distribution, rapid growth rate, high population densities, and close peridomestic relationship with humans of synanthropic murine species like rats and mice give them important roles in the transmission cycle of various zoonotic pathogens [[8], [9], [10]]. We and others have previously shown that murine cells are not susceptible to SARS-CoV-2 infection in vitro and mice are not infectable by SARS-CoV-2 in vivo [11, 12], which is due to the amino acid substitutions present at the ACE2-spike interacting surface on mouse ACE2 (mACE2) compared with human ACE2 (hACE2) that render SARS-CoV-2 spike unable to utilize mACE2 for cell entry [13, 14]. Interestingly, emerging SARS-CoV-2 variants, including B.1.1.7 [6], B.1.351 [15], P.1 [16], and P.3 [17], all contain the N501Y mutation in spike that has been suggested to be associated with mouse adaptation [18], raising the postulation that these emerging SARS-CoV-2 variants may have evolved to expand species tropism to mouse and possibly other murines. To evaluate this possibility with major public health significance, we evaluated the capacity of SARS-CoV-2 wildtype (WT), B.1.1.7, and additional N501Y-carrying variants to infect mice (Mus musculus). Our results demonstrate that while wildtype mice are not permissive to WT SARS-CoV-2, B.1.1.7 and N501Y-carrying variants efficiently infect wildtype mice with production of infectious virus particles in the nasal turbinate and lung. Similarly, B.1.1.7 but not WT SARS-CoV-2 readily infects rats (Rattus norvegicus). These findings indicate that B.1.1.7 and other N501Y-carrying variants have gained the capacity to expand species tropism to murine species and that public health control measures including stringent murine control should be implemented to facilitate the control of the ongoing pandemic.
2. Methods
2.1. Viruses and biosafety
All virus strains used in this study were shown in Figure S1. SARS-CoV-2 wildtype virus HKU-001a (GenBank: MT230904) was a clinical isolate as previously described [11]. SARS-CoV-2 B.1 (GISAID: EPI_ISL_497840), B.1.1.7 (GISAID: EPI_ISL_1273444), B.1.1.28.3 (P.3) (GISAID: EPI_ISL_1660475), B.1.351 (GISAID: EPI_ISL_2423556) and B.1.617.2 (GISAID: EPI_ISL_3221329) were isolated from laboratory-confirmed COVID-19 patients in Hong Kong. In vitro and in vivo experiments involving infectious wild type SARS-CoV-2, B.1, B.1.1.7, B.1.351, P.3, and B.1.617.2 were performed in Biosafety Level 3 laboratory and strictly followed the approved standard operation procedures.
2.2. SARS-CoV-2-Spike-pseudovirus entry assay
The SARS-CoV-2-WT-S and SARS-CoV-2-N501Y-S pseudoviruses were generated in 293T cells according to a previously established protocol [19, 20]. In brief, 293T cells were transfected with SARS-CoV-2-WT-S or SARS-CoV-2-N501Y-S plasmids with Lipofectamine 3000 (Thermo Fisher Scientific). At 24h post transfection, the cells were infected with VSV-ΔG-firefly pseudotyped with VSV-G. The cells were washed extensively after 2h and fresh culture medium was added. The generated SARS-CoV-2-WT-S-, or SARS-CoV-2-N501Y-S-pseudoviruses were harvested at 16h post inoculation. For pseudovirus entry assays, target cells were inoculated with pseudoviruses for 24h, before washed and lysed for detection of luciferase signal with a luciferase assay system (Promega). MLV-based pseudoviruses carrying full-length spike from B.1, B.1.1.7, B.1.351, P.1 and B.1.617.2 were obtained from Codex BioSolutions, Inc. For pseudovirus entry assays, mouse ACE2, rat ACE2, or human ACE2 were transfected into BHK21 cells. At 24h post transfection, transfected cells were inoculated with pseudoviruses for 48h before washed and lysed for measuring luciferase activity with a firefly luciferase assay system (Promega). The raw data for pseudovirus entry is provided in Table S1.
2.3. In vivo virus challenge in mice
The use of animals was approved by the Committee on the Use of Live Animals in Teaching and Research of The University of Hong Kong (CULATR). Protocols were approved under CULATR #5440-20 and #5193-19 for the experiments regarding to mice and rats, respectively. Heterogenous K18-hACE2 C57BL/6J mice (2B6.Cg-Tg(K18-ACE2)2Prlmn/J) were obtained from The Jackson Laboratory. For virus challenge in mice, 6- to 8- week-old female C57BL/6J mice or K18-hACE2 transgenic mice were anaesthetized with ketamine and xylazine, followed by intranasal inoculation with 20 µl/mouse of WT, B.1, B.1.1.7, B.1.351, P.3 or B.1.617.2 SARS-CoV-2 at 4 × 103 PFU/mouse as we previously described [21]. Remdesivir treatment was dosed at 15 mg per kg via the intraperitoneal route as we previously reported [22]. Briefly, remdesivir was prepared as 100 mg/ml stock in DMSO and further diluted using 12% SBE-β-CD before intraperitoneal injection. Mice receiving 2% DMSO in 12% SBE-β-CD were used as mock-treatment controls. Mice were treated once prior to infection, followed by treatment on a daily basis until sacrifice.
2.4. In vivo virus challenge in rats
For SARS-CoV-2 infection in rats, 6-week-old female Sprague Dawley rats were anaesthetized with ketamine and xylazine, followed by intranasal inoculation with 4 × 105 PFU/rat of WT or B.1.1.7 SARS-CoV-2 diluted in 50µl PBS. On day 2-, 4- and 7-day post virus challenge, animals were euthanized for harvesting organs for viral load titration and histology staining.
2.5. RNA extraction and quantitative RT-PCR
RNA extraction and RT-qPCR were performed as we previously described [11]. SARS-CoV-2 genome copies were quantified with sequence-specific probe and primers targeting the RNA-dependent RNA polymerase [11]. Subgenomic viral RNA was detected using primer and probe targeting the E gene as described previously [23].
2.6. Infectious virus titration by plaque assays
Organs harvested from virus-inoculated animals were homogenized in DMEM with Tissue Lyzer II, followed by centrifugation and titration. To titrate the infectious virus titre, tissue homogenates were 10-fold serially-diluted with DMEM and applied to monolayered VeroE6 cells for 2h. After inoculation, cells were washed once before overlaid with 1% low-melting agarose containing 1% FBS. Cells were further incubated for 72 h and fixed with 0.5% crystal violet for visualizing plaque formation [24].
2.7. Histology and immunohistochemistry staining
Animal tissues were harvested and fixed with 10% neutral-buffered formalin. Nasal turbinates were decalcified with 10% formic acid for 7 days before being processed with the TP1020 Leica semi-enclosed benchtop tissue processor. IHC was performed with the DAB (3,3’-diaminobenzidine) substrate kit (Vector Laboratories) as we previously described [25]. To detect the viral antigen, in-house mouse monoclonal biotinylated anti-SARS-CoV-2 nucleocapsid protein antibody was used, followed by colour development with the DAB substrate kit. The nuclei were detected with haematoxylin before the tissue sections was mounted with the VectaMount permanent mounting medium (Vector Laboratories). For H&E staining, tissue sections were stained with Gill's haematoxylin and eosin-Y. Images were acquired with the Olympus BX53 light microscope. Five mice or three rats were sampled each group (as specified in the figure legends) and four to six sections from each animal were used for histology analysis.
2.8. Immunofluorescence staining
Nucleocapsid of SARS-CoV-2 was detected by the in-house rabbit polyclonal anti-SARS-CoV-2 nucleocapsid antibody as we previously reported [26]. Nuclei were stained with the DAPI dye (Thermo Fisher Scientific) before the tissue sections being mounted with the Diamond Prolong Antifade mounting buffer (Thermo Fisher Scientific). Images were acquired with the Olympus BX53 light microscope.
2.9. Neutralizing antibody assay
Mice serum was obtained immediately before virus rechallenge and 14 days post rechallenge. Sera were heat-inactivated for 30 min at 56 °C before use. Two-fold serially diluted sera was incubated with 100 TCID50 WT SARS-CoV-2 for 1 h at 37 °C. After incubation, the virus-sera mixture was inoculated in VeroE6 cells and further incubated for 72 h for cytopathic effect (CPE) development. The neutralization titre was determined by observing the highest dilution without CPE.
3.10. ELISA
96-well plates were coated with 100 ng/well His-tagged SARS-CoV-2 receptor binding domain (RBD) or nucleocapsid protein (N) and incubated at 4 °C overnight. Plates were washed with 0.05% Tween-20 in PBS (PBST) once and blocked for 1 h at room temperature. Mouse sera were 4-fold diluted starting from 1:40 and incubated with the protein-coated plates for 1 h at 37 °C. Uninfected mouse serum and previously in-house raised anti-RBD/anti-N mouse sera were included as negative and positive control, respectively. Plates were washed for three times with PBST and incubated with horseradish peroxidase-conjugated goat anti-mouse IgG antibody (Thermo Fisher Scientific) for 1 h at room temperature. After incubation, plates were washed for six times with PBST followed by adding 3,3′,5,5′-tetramethylbenzidine (TMB) solution (Thermo Fisher Scientific) for colour development. Plate development was terminated by adding equal volume of 0.3 N H2SO4. Absorbance at 450nm was measured with a Thermo Scientific Multiskan FC Microplate Photometer.
3.11. Multiple sequence alignment and docking
Sequence alignment and docking were performed as we previously described [27]. ACE2 protein sequences of human (Q9BYF1), mouse (Q8R0I0), and rat (Q5EGZ1) were retrieved from Uniprot. Multiple sequence alignment was constructed with Muscle. The crystal structure of wildtype SARS-CoV-2 spike RBD and human ACE2 complex (code: 6M0J) was retrieved from the Protein Data Bank. Mouse and rat ACE2 structures were generated by mutating the interface residues within 4.0Å of RBD according to the multiple sequence alignment. The complex of ACE2 and N501Y RBD was built by superimposition of N501Y RBD (code: 7NEG) with wildtype RBD. All ACE2-RBD complexes were relaxed and locally refined with Rosetta Relax and Docking protocols. Interface binding energies were estimated with InterfaceAnalyzer application.
3.12. Ethics
The use of all animals in this study was approved by the Committee on the Use of Live Animals in Teaching and Research of The University of Hong Kong.
3.13. Statistical analysis
Data analysis were performed with Graphpad prism 8.0. Statistical comparison between different groups was performed by one-way ANOVA, two-way ANOVA, or Student's t-test. Differences were considered statistically significant when p<0.05.
3.14. Role of the funding source
All funders had no role in study design, data collection, data analyses, interpretation, or writing of the study.
4. Results
4.1. The SARS-CoV-2 N501Y-carrying variants gained the capability to utilize mouse and rat ACE2 in mediating virus entry
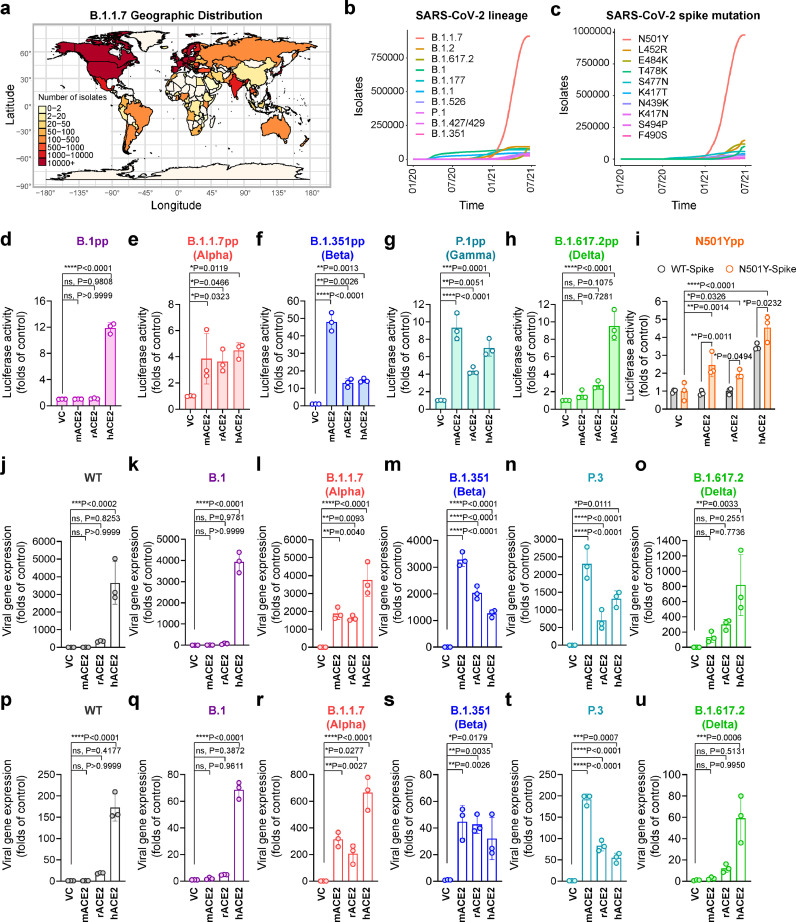
B.1.1.7 emerged in late 2020 and quickly became a dominant SARS-CoV-2 variant among recently reported SARS-CoV-2 isolates in countries on different continents (Fig. 1a-b). In parallel with the surge of B.1.1.7, the N501Y mutation in spike, which is one of the B.1.1.7-defining mutations, has emerged as the most predominant spike mutation since late 2020 (Fig. 1c). This is associated with critical public health importance since N501Y has been implicated in an increased recognition of mouse ACE2 [18], hinting that the naturally-emerging N501Y-carrying SARS-CoV-2 variants, including B.1.1.7, B.1.351, P.1, and P.3, may have evolved to infect rodents. To evaluate this possibility, we assessed pseudovirus entry of all four SARS-CoV-2 VOCs by far, including B.1.1.7 (Alpha), B.1.351 (Beta), P.1 (Gamma) and B.1.617.2 (Delta), in the non-permissive BHK21 cells with mouse ACE2 (mACE2), rat ACE2 (rACE2), or human ACE2 (hACE2) overexpression, with B.1 pseudovirus serving as a control (Fig. 1d-h). Overall, pseudovirus entry to both mACE2 and rACE2 was enabled by full-length spike of Alpha, Beta, and Gamma variants that carried the N501Y mutation (Fig. 1e-g), but not by the full-length spike of B.1 or Delta variant (Fig. 1d and h). Interestingly, unlike the B.1.1.7 pseudoviruses, both B.1.351 and P.1 pseudoviruses appeared to preferentially utilize mACE2 over hACE2 (Fig. 1e-g), suggesting the K417N/T and E484K mutation in the RBD of these variants might further facilitate mouse adaptation in addition to the N501Y substitution. To further confirm the N501Y mutation was sufficient to mediate virus entry to murine cells, we generated pseudovirus expressing spike with the single N501Y amino acid substitution. Our results demonstrated that BHK21 cells with mACE2 or rACE2 overexpression were permissive to the entry of SARS-CoV-2-N501Y-spike pseudoviruses, but not SARS-CoV-2-WT-spike pseudoviruses. In contrast, both pseudoviruses efficiently utilized hACE2 for entry (Fig. 1i). Next, we evaluated the capacity of SARS-CoV-2 N501Y variants (Fig. S1) to infect BHK21 cells expressing mACE2, rACE2, or hACE2. Wildtype SARS-CoV-2 HKU001a from lineage A (SARS-CoV-2 WT) and two other lineage B variants, B.1 and B.1.617.2, which do not carry the N501Y mutation, were included as controls. Our results demonstrated that SARS-CoV-2 WT, B.1 and B.1.617.2 did not infect or replicate in BHK21 cells overexpressing mACE2 or rACE2 (Fig. 1j,k,o,p,q,u). In stark contrast, B.1.1.7, B.1.351, and P.3, which carry N501Y, efficiently infected and replicated in mACE2- or rACE2-expressing BHK21 cells (Fig. 1l-n,r-t). Together, these findings suggest that the recently emerged N501Y-carrying SARS-CoV-2 variants, including B.1.1.7, B.1.351, P.1, and P.3, can utilize murine ACE2 as entry receptor and may infect murine species without the need of further adaptation.
Figure 1.
SARS-CoV-2 B.1.1.7 and other N501Y-carrying variants efficiently utilize murine ACE2 for virus entry. (a) Geographic distribution of the B.1.1.7 variant. Heatmap of the geographic distribution of B.1.1.7 isolates retrieved from the GISAID EpiCoV database as of 28th June, 2021. (b) Top 10 prevalent lineages of SARS-CoV-2 and (c) Top 10 amino acid mutations in the spike of SARS-CoV-2 variants retrieved from the GISAID EpiCoV database as of July 21st, 2021. (d-i) BHK21 cells transfected with empty vector, mouse ACE2, rat ACE2 or human ACE2 were inoculated with SARS-CoV-2-S-pseudoviruses with (d) B.1, (e) B.1.1.7, (f) B.1.351, (g) P.1, (h) B.1.617.2 or (i) WT and N501Y spike. Pseudovirus entry was determined at 48h post inoculation (n = 3). (j-u) BHK21 cells overexpressing empty vector, mouse ACE, rat ACE2 or human ACE2 were infected with (k,q) B.1, (l,r) B.1.1.7, (m,s) B.1.351, (n,t) P.3, (o,u) B.1.617.2 or (j,p) WT SARS-CoV-2. Relative viral gene expression in (j-o) cell lysates and (p-u) supernatants were determined by RT-qPCR (n = 3). Data represents mean ± SD from the indicated number of biological repeats. Statistical differences were determined with two-way analysis of variance (ANOVA) in (i) and one-way ANOVA in (d-h, j-u). * represented P < 0.05; ** represented P < 0.01; *** represented P < 0.001; **** represented P < 0.0001. pp, pseudovirus particles; WT, wildtype SARS-CoV-2; VC, vector control.
4.2. SARS-CoV-2 N501Y-carrying variants efficiently infect mice
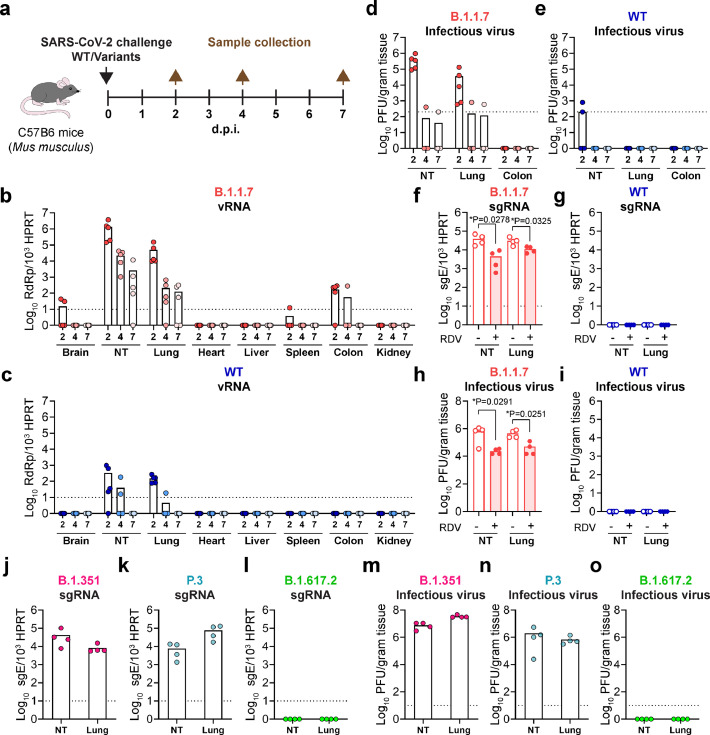
To directly address the question of whether these emerging SARS-CoV-2 variants have evolved to gain the capacity to cross species barrier to murine species, we intranasally challenged 6- to 8-week-old C57B6 mice (Mus musculus) with WT SARS-CoV-2, B.1.1.7, or other N501Y-carrying variants and collected tissues for virological and histological assessment (Fig. 2a). Importantly, in B.1.1.7-inoculated mice, virus gene copies were detected in brain, nasal turbinate, lung, spleen, and colon tissues (Fig. 2b). In particular, robust virus replication was detected in the nasal turbinate and lung tissues, which were 4044 and 330 folds higher than that of WT SARS-CoV-2-inoculated mice at 2 d.p.i., respectively. Viral gene copies were cleared from WT SARS-CoV-2-inoculated mice at 7 d.p.i., but remained readily detectable from the nasal turbinate and lung of B.1.1.7-incolated mice (Fig. 2b,c). High-titre infectious virus particles were recovered from the nasal turbinate and lung of B.1.1.7-inocluated mice at 2 d.p.i., and remained detectable in one out of five mice at 4 d.pi. and 7 d.p.i, respectively (Fig. 2d). In comparison, infectious viral titres were only marginally detected in the nasal turbinate of mice inoculated with WT SARS-CoV-2 at 2 d.p.i. but not at 4 d.p.i. and 7 d.p.i. (Fig. 2e). To exclude the possibility that the viral RNA and infectious viral titres detected at early time points were remnants of the inoculum, we additionally quantify the subgenomic RNA (sgRNA), which was an intermediate generated during active coronavirus replication. We detected high copy number of the sgRNA from mice infected with B.1.1.7 but not WT SARS-CoV-2, suggesting active virus replication took place in B.1.1.7-infected but not WT SARS-CoV-2-infected mice (Fig. 2f-i). In addition, viral burdens could be reduced by remdesivir treatment in both the nasal turbinate and lung of mice infected with B.1.1.7 (Fig 2f-i), representing the inhibition of active virus replication. Moreover, we similarly detected high sgRNA level and high-titre infectious viral particles in the nasal turbinate and lung of mice inoculated with B.1.351 and P.3 but not B.1.617.2 (Fig. 2j-o). Since B.1.1.7, B.1.351, and P.3 all shares both the D614G and N501Y mutation in spike, we included B.1 as a control virus, which carries the D614G but not the N501Y mutation, to exclude the possibility that D614G conferred infectivity to mice. Our results showed that permissibility to mouse cells was not due to the D614G mutation as B.1 did not replicate in the nasal turbinate and lung of the inoculated mice (Fig. S2). Together, these findings indicate that the SARS-CoV-2 variant B.1.1.7 and other N501Y-carrying variants can infect mice with robust virus shedding from the nasal turbinate.
Figure 2.
B.1.1.7 and other N501Y-carrying variants expanded tropism to infect Mus musculus. (a) Schematic illustration of the in vivo infection experiment in C57B6 mice. (b-e) 6-to-8-week-old C57B6 mice were intranasally inoculated with 4 × 103 PFU B.1.1.7 or WT SARS-CoV-2. Mice were euthanized on 2, 4 or 7 d.p.i. and tissues were harvested for viral titre quantification. Viral gene copies were measured from the brain, nasal turbinate, lung, heart, liver, spleen, colon and kidney of mice infected with (b) B.1.1.7 and (c) WT SARS-CoV-2. Infectious viral titre in the nasal turbinate, lung, and colon of mice inoculated with (d) B.1.1.7 and (e) WT SARS-CoV-2 was determined with plaque assays. (f-i) 6-to-8-week-old C57B6 mice were intranasally inoculated with 4 × 103 PFU B.1.1.7 or WT SARS-CoV-2 with or without 15 mg/kg remdesivir treatment. Nasal turbinate and lung of the infected mice were harvested on 2 d.p.i. for (f and g) viral envelope subgenomic RNA quantification with RT-qPCR and (h and i) infectious viral particle titration with plaque assays. (j-o) 6-to-8-week-old C57B6 mice were intranasally inoculated with 4 × 103 PFU B.1.351, P.3, and B.1.617.2. Nasal turbinate and lung of the infected mice were harvested on 2 d.p.i. for (j-l) viral envelope subgenomic RNA quantification with RT-qPCR and (m-o) infectious viral particle titration with plaque assays. Statistical differences were determined with two-tailed Student's t-test in (f) and (h). * represented P < 0.05. WT, wildtype SARS-CoV-2; NT, nasal turbinate.
4.3. Mice (Mus musculus) develop pathological changes after primary B.1.1.7 inoculation
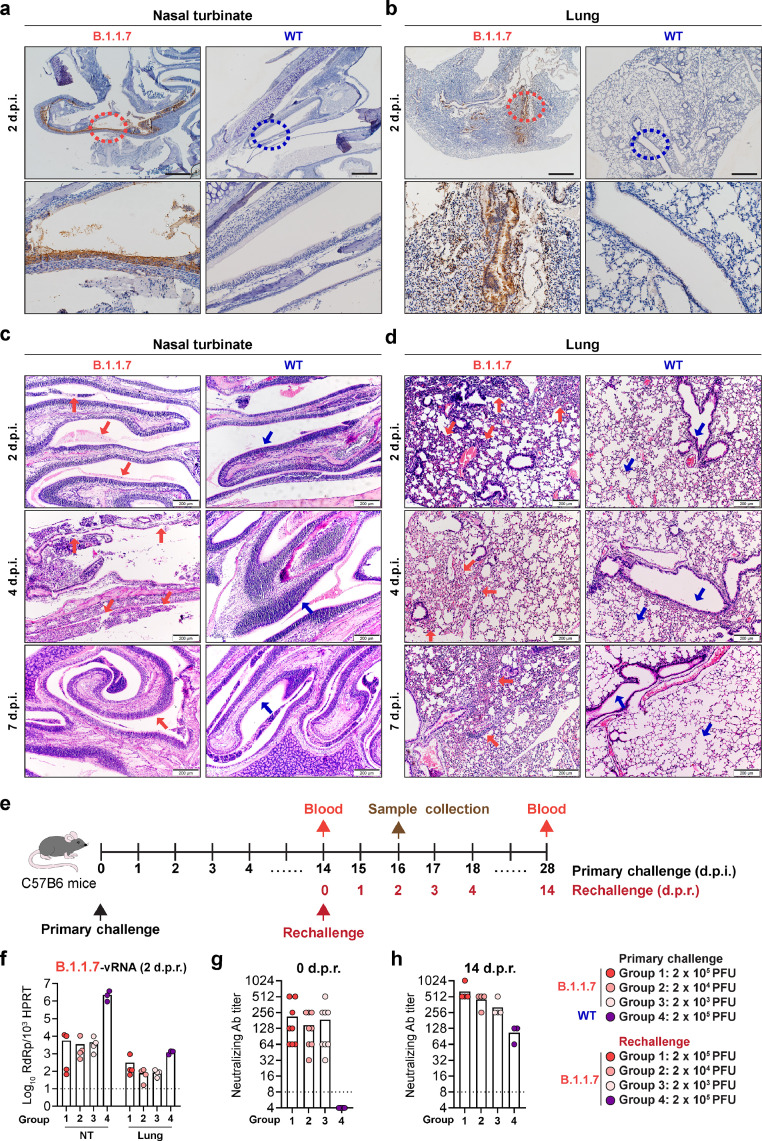
Next, we evaluated viral antigen expression and virus-induced pathology in the respiratory tract of mice upon B.1.1.7 or WT SARS-CoV-2 inoculation. As demonstrated in Figure 3a and 3b, abundant viral nucleocapsid (N) protein expression was detected in mice inoculated with B.1.1.7 but not WT SARS-CoV-2. In the nasal turbinate, robust viral N protein expression was detected in the epithelial lining at 2 d.p.i. (Fig. 3a) and remained detectable at 4 d.p.i. (Fig. S3). In the lung, abundant N protein was detected in the bronchioles and alveoli (Fig. 3b). Viral N protein in the lung of B.1.1.7-inoculated mice was most readily detected at 2 d.p.i. and remained detectable at 7 d.p.i. (Fig. S3). In contrast to B.1.1.7-inoculated mice, no viral N protein could be detected in the nasal turbinate and lung of WT SARS-CoV-2-incoluated mice (Fig. 3a,b and Fig. S3). Histological examination of B.1.1.7-inoculated mice showed epithelial damage with secretion and cell debris in the nasal turbinate (Fig. 3c, 2 d.p.i., red arrows) at 2 d.p.i.. At 4 d.p.i., more severe submucosal infiltration and loss of epithelial integrity were observed in multiple locations (Fig. 3c, 4 d.p.i., red arrows). At 7 d.p.i., intact nasal turbinate epithelium with resolved inflammatory infiltration was observed in the B.1.1.7-inoculated mice, indicating regeneration of the epithelial lining after virus infection (Fig. 3c, 7 d.p.i., red arrow). In mice inoculated with WT SARS-CoV-2, the nasal turbinate epithelium remained intact from 2 d.p.i. to 7 d.p.i. (Fig. 3c, blue arrows). In line with the histopathological findings of the nasal turbinate, the lung of B.1.1.7-inoculated mice showed alveolar wall congestion and inflammatory infiltration (Fig. 3d, 2 d.p.i, red arrows) accompanied by alveolar haemorrhage (Fig. 3d, 4 d.p.i, red arrows). Perivascular infiltration and alveolar septal thickening in the lung of B.1.1.7-inoculated mice remained evident at 7 d.p.i (Fig. 3d, 7 d.p.i, red arrows). In contrast, the lung of WT SARS-CoV-2-inoculated mice showed no histopathological changes at all examined time points (Fig. 3d, blue arrows). Despite the histopathological changes found in B.1.1.7-inoculated mice, no significant body weight loss was observed (Fig. S4). To evaluate the transmission from B.1.1.7-infected mice to naïve mice, we co-housed B.1.1.7-infected mice with naïve mice. Although B.1.1.7 replicated robustly along the airway of the directly inoculated mice, viral RNA was undetectable in the contact mice (Fig. S5). In agreement with this result, sera reactivity to the receptor binding domain (RBD) of the SARS-CoV-2 spike were strong in the index mice with direct exposure to B.1.1.7 infection, yet none of the contact mice had significant increase in the sera reactivity compared to the negative control (Fig. S5), suggesting mice-to-mice transmission did not occur in our experimental setup. Moreover, we rechallenged B.1.1.7- or WT SARS-CoV-2-incoluated mice at 14 d.p.i. with B.1.1.7 and harvested their nasal turbinate, lung, and blood samples at 14 days post rechallenge (d.p.r.). Our results showed that mice previously inoculated with B.1.1.7 but not WT SARS-CoV-2 were partially protected from virus rechallenge as evidenced by the lower virus gene copy detected in the nasal turbinate and lung (Fig. 3f). In agreement with the observed partial protection, we detected the presence of serum SARS-CoV-2-specific neutralizing antibodies at 14 days post primary virus challenge in B.1.1.7-incoluated but not WT SARS-CoV-2-inoculated mice (Fig. 3g). The level of neutralizing antibody was further enhanced upon rechallenge, in a virus inoculum-dependent manner (Fig. 3h). Taken together, our results demonstrate that the B.1.1.7 variant has expanded species tropism to mice with high infectious virus titre shedding from their nasal turbinate, but with no evidence of mice-to-mice transmission.
Figure 3.
Mice develop pathological changes along the respiratory tract after primary B.1.1.7 infection. (a-d) C57B6 mice intranasally inoculated with B.1.1.7 or WT SARS-CoV-2 were euthanized for harvesting nasal turbinate and lung tissues on 2, 4 and 7 d.p.i. (n = 5). Representative images of immunohistochemistry staining for the detection of nucleocapsid protein (brown) in the (a) nasal turbinates and (b) lung on 2 d.p.i.. Insets were shown below for the circled areas. Scale bar, 500 µm. (c) Representative images of haematoxylin and eosin (H&E) staining of mouse nasal turbinates harvested on 2, 4 and 7 d.p.i.. Pathological changes in the B.1.1.7-inoculated mice on 2 d.p.i. and 4 d.p.i. were pointed out with red arrows. On 7 d.p.i., the integrity of the nasal cavity epithelium was restored (red arrow). Intact nasal epithelium layers of the WT SARS-CoV-2-inoculated mice were shown for comparison (blue arrows). (d) Representative images of H&E staining of mouse lungs harvested on 2, 4 and 7 d.p.i.. In the lungs of B.1.1.7-inoculated mice, alveolar wall congestion and alveolar infiltrations were detected on 2 d.p.i. (red arrows). Alveolar haemorrhage was developed on 4 d.p.i. (red arrows). Alveolar septa thickening remained evident on 7 d.p.i.. Normal structure of bronchioles and alveoli in the lung of the WT SARS-CoV-2-inoculated mice were indicated by blue arrows for comparison. Scale bar, 200 µm. (e-h) C57B6 mice were intranasally inoculated with 2 × 105 PFU WT SARS-CoV-2 or B.1.1.7 at 2 × 103, 2 × 104, or 2 × 105 PFU, followed by rechallenge on 14 d.p.i. with B.1.1.7 at the indicated titres. (e) Schematic illustration of the rechallenge experiment. (f) Viral titre in nasal turbinate and lung of the infected mice on day 2 post rechallenge quantified with RT-qPCR (n = 4 for group 1-3, n = 3 for group 4). (g) Neutralizing antibody titre in mouse sera harvested on 0 day post rechallenge (d.p.r.) (n = 8 for group 1-3, n = 3 for group 4) and (h) 14 d.p.r. (n = 4 for group 1-3, n = 3 for group 4). Dashed line indicates the detection limit of each assay. Images in (a-d) are representative images from five mice. Four to six sections were taken from each mouse for histology and immunochemistry analysis. NT, nasal turbinate.
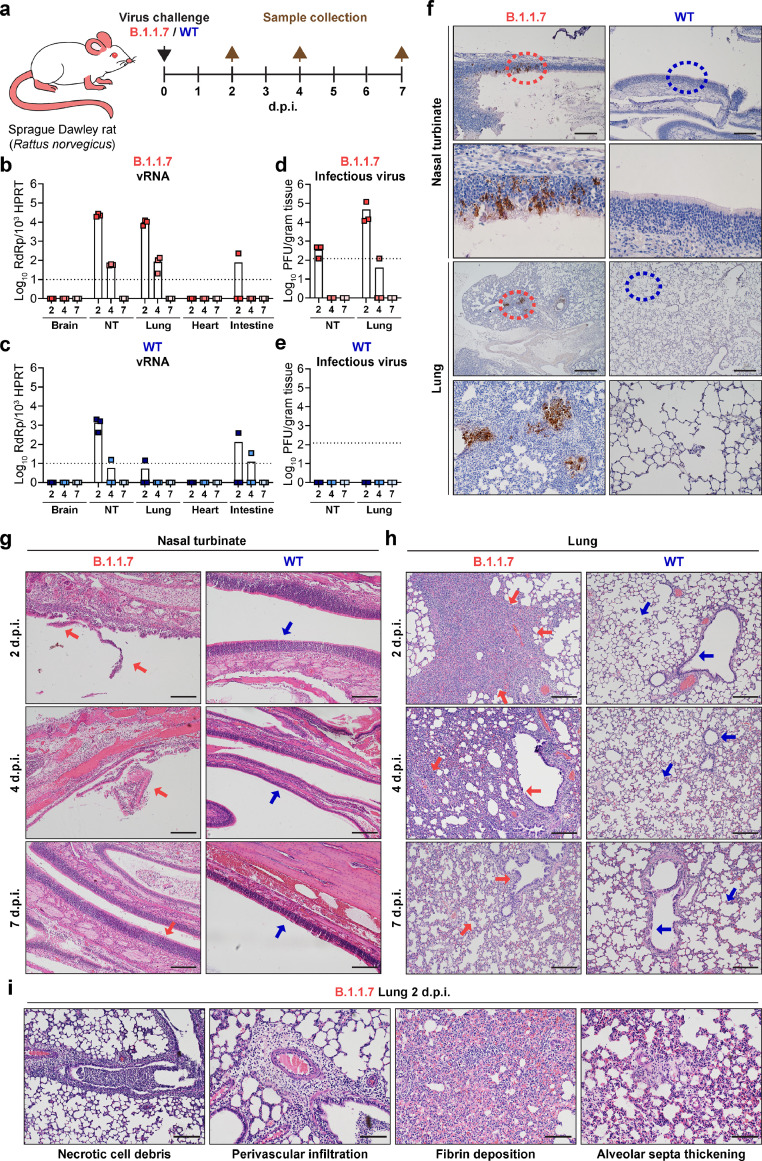
Figure 4.
B.1.1.7 infects and causes pathological damages in rats (Rattus norvegicus). (a) Schematic illustration of in vivo infection experiment in Sprague Dawley rats. 6-week-old Sprague Dawley rats were intranasally inoculated with B.1.1.7 or WT SARS-CoV-2. On 2, 4 and 7 d.p.i., rats were euthanized for harvesting tissues for virological assessment and histopathological analysis (n = 3). (b and c) Viral titre quantified with RT-qPCR in the brain, nasal turbinate, lung, heart and intestine of the inoculated rats. NT, nasal turbinate. (d and e) Infectious viral titre quantified with plaque assays in the nasal turbinate and lung of the inoculated rats. (f) Immunohistochemistry staining for the detection of nucleocapsid protein expression (brown) in the nasal turbinate and lung of the inoculated rats on 2 d.p.i. Insets were shown below for the circled areas. Scale bar, 200 µm for nasal turbinate; 500 µm for lung. (g-i) Histopathological analysis for nasal turbinate and lung harvested on 2, 4, and 7 d.p.i.. (g) Representative H&E images showing pathological changes in the nasal cavity of B.1.1.7-inocuated rats, including epithelial damage and loss of mucosal integrity (2 d.p.i., red arrow). On 4 d.p.i., more severe epithelial sloughing and submucosal infiltrations were detected (4 d.p.i., red arrow). On 7 d.p.i., the epithelial lining was largely restored (7 d.p.i., red arrow). In contrast, the structure of the nasal turbinate in WT SARS-CoV-2-inoculated rats remained undisrupted throughout the course of experiment. (h) Representative H&E images showing severe alveoli consolidation (2 d.p.i., red arrows), epithelium disruption and alveoli infiltrations (4 d.p.i., red arrows) in the lungs of B.1.1.7-inoculated rats. Acute lung injuries were largely repaired by 7 d.p.i. and normal structure of the alveoli and lung were indicated by red arrows. No pathological change was observed in either alveoli or bronchioles of WT SARS-CoV-2-inoculated rats (2, 4, and 7 d.p.i., blue arrows). Scale bar, 200 µm. (i) Typical pulmonary lesions detected in the lungs of B.1.1.7-inoculated rats on 2 d.p.i.. Scale bar, 100 µm. Images in (f-i) are representative images from three rats. Four to six sections were taken from each rat for histology and immunochemistry analysis.
4.4. B.1.1.7 infects and causes pathological damages in rats (Rattus norvegicus)
Street rats (Rattus norvegicus, hereafter referred to as rat) live in close proximity to humans and play key roles in disseminating zoonotic diseases. Rat ACE2 (rACE2) is similar to mACE2 in amino acid sequences (Fig. S6) and supported B.1.1.7 entry in vitro (Fig. 1e,l,r). To evaluate whether rats are susceptible to B.1.1.7 infection, we challenged 6-week-old Sprague Dawley rats (Rattus norvegicus) intranasally with B.1.1.7 or WT SARS-CoV-2 (Fig. 4a). Similar to B.1.1.7 infection in mice, the viral gene copies of the inoculated rats peaked at 2 d.p.i. and were 17.0- and 1831.7-folds higher in the nasal turbinate and lung, respectively, than that of WT SARS-CoV-2-inoculated rats (Fig. 4b,c). Importantly, infectious virus titres were recovered from the nasal turbinate and lung of B.1.1.7-inoculated but not WT SARS-CoV-2-inoculated rats, indicating that rats were permissive to B.1.1.7 but not WT SARS-CoV-2 (Fig. 4d,e). In line with the infectious virus titre results, immunohistochemistry staining revealed that viral N protein was more frequently detected in the lung than the nasal turbinate of B.1.1.7-inoculated rats (Fig. 4f), while viral N protein was absent in the lung and nasal turbinate of WT SARS-CoV-2-inoculated rats (Fig. 4f).
In the nasal turbinate of B.1.1.7-inoculated rats, histological examination showed loss of mucosal integrity and epithelial damage at 2 d.p.i. (Fig. 4g, 2 d.p.i., red arrows). More severe epithelial sloughing and submucosal infiltration were detected at 4 d.p.i. (Fig. 4g, 4 d.p.i., red arrow), which were largely regenerated at 7 d.p.i. (Fig. 4g, 7 d.p.i., red arrow). In contrast, the epithelial lining in the nasal turbinate of WT SARS-CoV-2-inoculated rats remained intact at all time points examined (Fig. 4g, blue arrows). In the lung of B.1.1.7-inoculated rats, moderate to severe multi-focal consolidation in the alveoli was observed at 2 d.p.i.. (Fig. 4h, 2 d.p.i., red arrows). Moreover, necrotic cell debris in the bronchioles, perivascular cuffing caused by inflammatory infiltration, fibrin deposition in the interstitium, and alveolar septa thickening were also observed (Fig. 4i). Alveolar congestion, epithelial disruption in the bronchioles, and alveoli infiltration remained evident at 4 d.p.i. (Fig. 4h, 4 d.p.i., red arrows). Upon viral clearance at 7 d.p.i., these lung pathologies were largely resolved (Fig. 4h, 7 d.p.i., red arrows). In contrast to B.1.1.7-inocluated rats, no pathological change was observed in the lung of WT SARS-CoV-2-inoculated rats at any examined time points (Fig. 4h, blue arrows). These findings reveal that the SARS-CoV-2 variant, B.1.1.7, has evolved to expand species tropism beyond WT SARS-CoV-2, and can infect wildtype murines including rats.
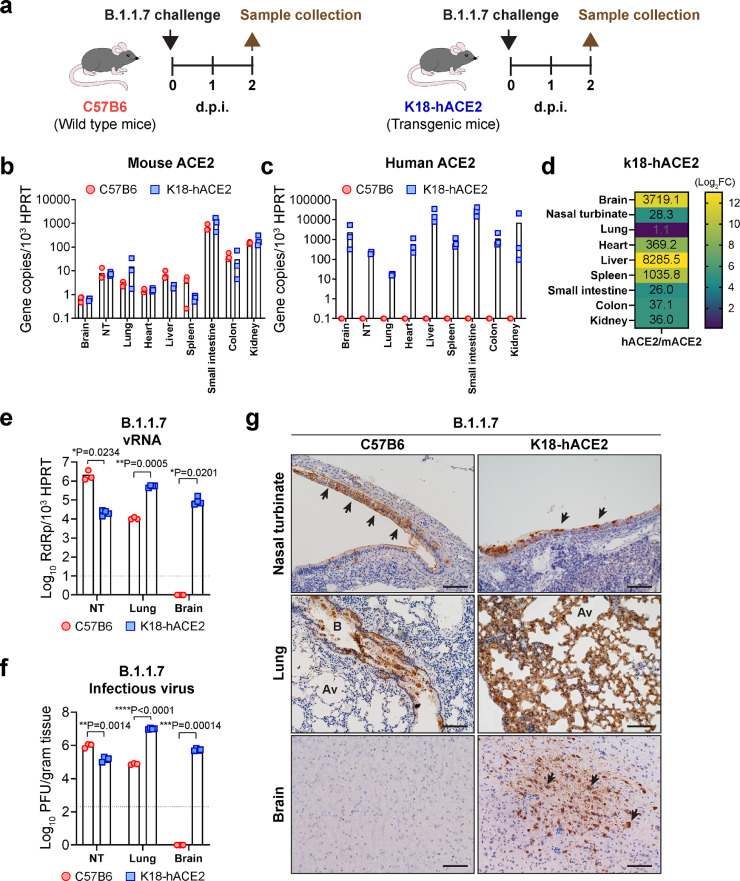
4.5. Differential SARS-CoV-2 tropism in K18-hACE2 and wildtype mice
Since wildtype mice are not susceptible to WT SARS-CoV-2 infection (12), the K18-hACE2 transgenic mice are utilized as an animal model for SARS-CoV-2 infection [28]. Our current findings reveal that B.1.1.7 and other N501Y-carrying variants can naturally infect mice, which may serve as a new animal model for SARS-CoV-2 infection that does not require prior virus adaptation or introduction of human ACE2 to the mice. To compare the two infection models (Fig. 5a), we first characterized the physiological distribution of ACE2 in the two mouse models. Our data showed that the K18-hACE2 and C57B6 mice expressed comparable levels of mACE2. Mouse ACE2 expression in the small intestine was the highest, followed by kidney, colon, lung, and nasal turbinate (Fig. 5b), which resembled the natural ACE2 distribution in human [29, 30]. In addition to mACE2, K18-hACE2 mice express high level of hACE2 at all evaluated organs (Fig. 5c). The hACE2 expression level was 3719.1-, 28.3-, 1.1-, 369.2-, 8285.5-, 1035.8, 26.0-, 37.1-, and 36.0-folds higher than that of the mACE2 expression in brain, nasal turbinate, lung, heart, liver, spleen, small intestine, colon, and kidney of K18-hACE2 mice, respectively (Fig. 5d).
Figure 5.
Differential SARS-CoV-2 tropism in K18-hACE2 and wild type mice. (a) Schematic illustration of comparative study of SARS-CoV-2 B.1.1.7 tropism in C57B6 and K18-hACE2 mice. (b) Mouse ACE2 expression quantified with RT-qPCR. (c) Human ACE2 expression quantified with RT-qPCR. (d) Quantification of the relative expression ratio between human and mouse ACE2 in the K18-hACE2 transgenic mouse. Fold changes in different organs were indicated with numbers. (e) Viral gene copy in nasal turbinate, lung and brain of the B.1.1.7-inoculated C57B6 and K18-hACE2 mice quantified with RT-qPCR. (f) Infectious viral titres in nasal turbinate, lung, and brain of B.1.1.7-inoculated C57B6 and K18-hACE2 mice quantified with plaque assays. (g) Representative immunohistochemistry images of nasal turbinate, lung, and brain of B.1.1.7-inoculated C57B6 and K18-hACE2 mice (n = 3). Presence of SARS-CoV-2 nucleocapsid protein were shown in brown and indicated by black arrows. Scale bar, 100 µm. Images in (g) are representative images from three mice. Four to six sections were taken from each mouse for histology and immunochemistry analysis. Statistical differences were determined with two-tailed Student's t-test in (d) and (e). * represented P < 0.05; ** represented P < 0.01; *** represented P < 0.001; **** represented P < 0.0001. Av, alveoli; B, bronchioles. NT, nasal turbinates; SI, small intestine; FC, fold change.
Next, we in parallel inoculated 6- to 8-week-old C57B6 and K18-hACE2 mice with B.1.1.7 and evaluated virus replication at 2 d.p.i.. Interestingly, our results indicated that B.1.1.7 replication was more robust in the nasal turbinate of C57B6 mice than that of the K18-hACE2 mice [(vRNA; C57B6: 2.15 × 106 vs K18-hACE2: 2.17 × 104; p=0.0234) and (PFU; C57B6: 9.87 × 105 vs K18-hACE2: 1.50 × 105; p=0.0014]. In contrast, B.1.1.7 replication was more robust in the lungs of K18-hACE2 mice than that of the C57B6 mice [(vRNA; C57B6: 1.08 × 104/103HPRT vs K18-hACE2: 5.36 × 106/103HPRT; p=0.0005) and (PFU; C57B6: 7.67 × 104/gram tissue vs K18-hACE2: 1.02 × 107/gram tissue; p<0.0001] (Fig. 5e,f). These findings were validated by immunohistochemistry staining, which consistently detected more abundant viral N protein expression in the nasal turbinate of C57B6 mice than K18-ACE2 mice. Similarly, viral N protein was most frequently and abundantly detected in the lung of K18-hACE2 mice than C57B6 mice (Fig. 5g). In addition to the nasal turbinate and lung, high levels of infectious virus titre and viral antigen expression were detected in the brain of K18-hACE2 mice, which is suggested to cause fatal outcome in the infected animals [31]. In contrast, the brain of C57B6 mice was free of either infectious virus particles or viral antigen expression (Fig. 5f,g). In addition to evaluating B.1.1.7 infection in C57B6 and K18-hACE2 mice, we additionally compared the infection of B.1.1.7 and WT SARS-CoV-2 in K18-hACE2 mice. Our results indicated that the B.1.1.7 and WT SARS-CoV-2 have similar replication efficiency in the nasal turbinate and lung of K18-hACE2 mice (Fig. S7). Taken together, the B.1.1.7-inoculated C57B6 mouse model may represent a more physiologically relevant model for studying SARS-CoV-2 pathogenesis and therapeutics evaluation due to its native ACE2 distribution profile, while the K18-hACE2 mouse model will be useful for studying the central nervous system pathologies of SARS-CoV-2 infection.
5. Discussion
In summary, we demonstrate that B.1.1.7 and other N501Y-carrying SARS-CoV-2 variants have evolved to gain the capability to cross species barrier to infect wildtype murines including mice and rats, which are not considered permissive to WT SARS-CoV-2. Mice and rats reside in close proximity to humans, including densely populated areas with circulating B.1.1.7 and/or other SARS-CoV-2 variants. The acquired susceptibility of these murines to SARS-CoV-2 variants has substantially increased the risk of SARS-CoV-2 infection in the murine population, which may result in a zoonotic reservoir of SARS-CoV-2. Substantial evidence has been raised to demonstrate the increased binding between N501Y mutant spike and mACE2 as well as the role of N501Y in mouse-adapted or genetically-engineered SARS-CoV-2 strains in mouse infection [32]. However, direct experimental evidence on whether authentic N501Y VOCs and other emerging variants can infect mice or other murine is still largely lacking. Our results indicate that B.1.1.7-infected mice and rats can shed infectious virus particles in the respiratory tract for approximately 4 to 7 days after infection, which may contaminate the environment and become a persistent source of human infection. Our results corroborated with the recent findings reported by Montagutelli X et al. that B.1.351 was capable of infecting wildtype mice [33]. We have extended these findings to demonstrate that among the four VOCs, only the N501Y-carrying variants infect wildtype mice. Furthermore, we demonstrated that in addition to wildtype Mus musculus, Rattus norvegicus is also susceptible to the N501Y-carrying mutant B.1.1.7.
The N501Y mutation contributed to an over 40-fold increase in pseudovirus entry in mACE2 overexpressed cells [34]. Apart from the N501Y mutation, several amino acid substitutions including K417N, E484K, Q493H/K, and Q498H were also suggested to be critical for SARS-CoV-2 adaptation in murine species [[34], [35], [36]]. This is congruous with our observation that B.1.351 replicated to a higher level than B.1.1.7 and P.3 in wildtype mice since B.1.351 carries K417N, E484K in addition to N501Y.
Our current data demonstrated B.1.1.7 infection caused significant pathological changes in the airway of both wildtype mice and rats at 2 d.p.i. although infectious viral progenies was largely cleared by the host immune response at 4 d.p.i.. These findings agreed with the existing data published with mouse-adapted SARS-CoV-2 infection in young wildtype mice [18, 37, 38]. Aging is reported to associate with increased incidence of comorbidities, impaired T cell responses, aberrant cytokine/chemokine production [[39], [40], [41]], which may results in more severe outcomes during viral infections. Therefore, infection of N501Y-carrying variants in aged mice may represent a more severe disease model with enhanced pathological tissue damage upon virus infection and ultimately succumbed to high-titre inoculum [37, 38, 42].
Human ACE2 transgenic mice was previously employed for SARS-CoV-1 study and later demonstrated to be a useful tool in investigating SARS-CoV-2 pathogenesis [12, 43, 44]. Different from the non-lethal infection in young wildtype mice, B.1.1.7 and B.1.351 caused lethal outcomes in hACE2 transgenic mice accompanied with high tittered viral burdens as well as extensive tissue damage throughout the whole course of infection [45, 46]. However, as revealed in this study, the high level of human ACE2 expression in the hACE2 transgenic mice, especially in extrapulmonary organs such as the brain, liver, and spleen, may potentially alter the tissue tropism of SARS-CoV-2, which may complicate the interpretation of research findings in a physiological-relevant context. Another rodent infection model commonly used for SARS-CoV-2 studies is the Golden Syrian hamster model. Similar to our current wildtype mouse model, hamsters support robust virus replication in the airway, develop significant tissue damage at the infection sites, but the animals eventually recover from viral infection without therapeutic interventions [27]. In addition, the hamster model allows transmission studies since hamster-to-hamster transmission by close contact is highly efficient [27, 47].
Interspecies spill-over of SARS-CoV-2 is not uncommon. While SARS-CoV-2 is presumed to have originated from an animal reservoir, humans are also known to infect other animals with SARS-CoV-2, including cats, dogs, and even tigers in zoos [[48], [49], [50]]. A previous study demonstrated that bidirectional SARS-CoV-2 transmission could occur between humans and minks [51]. Although our current data showed no evidence of mice-to-mice transmission among B.1.1.7-infected wildtype mice, the possibility of murine species serving as a reservoir for SARS-CoV-2 should not be completely excluded, since SARS-CoV-2 transmission was recently documented among hamsters [47], deer mice [52] and hACE2 transgenic mice [53]. Importantly, SARS-CoV-2 adaptation in mice occurs rapidly, with mouse-adapting mutations developing within 1-to-10 passages [18, 37, 54, 55]. These mutations will result in even more robust SARS-CoV-2 replication in rodents and may further facilitate virus dissemination. As demonstrated in the current study, the evolving SARS-CoV-2 variants with the N501Y mutation in spike can now naturally infect mice and rats, and potentially other rodent species. Considering the abundance of these animals and their proximate habitats with humans, humans-to-rodents and rodents-to-humans transmissions may be inevitable. Public health measures including stringent rodent control and surveillance on SARS-CoV-2 infection in the rodent populations especially in densely populated regions should be implemented to facilitate the control of the pandemic.
Contributors
HS, JF-WC, K-YY, and HC had roles in the study design, data collection, data analysis, data interpretation, and writing of the manuscript. TT-TY, CY, J-CH, LW, BH, DY, YW, YH, XH, YC, CC-SC, VK-MP, LL, R-QZ, W-MC, JDI, AW-HC, Y-FH, J-PC, K-HC, JZ, SS, B-ZZ, SY, AJZ, J-DH, and KK-WT had roles in the experiments, data collection, data analysis, and data interpretation. All authors reviewed and approved the final version of the manuscript.
Materials and correspondence
Correspondence and material requests should be addressed to Dr. Hin Chu, Prof. Kwok-Yung Yuen, or Dr. Jasper Fuk-Woo Chan.
Data Sharing
The data that support the findings of this study are available from the corresponding authors upon reasonable request.
Declaration of Competing Interest
Authors declare that they have no competing interests.
Acknowledgments
This work was partly supported by funding from the Health and Medical Research Fund (CID-HKU1-5 and COVID1903010-14), the Food and Health Bureau, The Government of the Hong Kong Special Administrative Region; Innovation and Technology Fund (ITF), the Government of the Hong Kong Special Administrative Region; the Consultancy Service for Enhancing Laboratory Surveillance of Emerging Infectious Diseases and Research Capability on Antimicrobial Resistance for Department of Health of the Hong Kong Special Administrative Region Government; the National Program on Key Research Project of China (grant no. 2020YFA0707500 and 2020YFA0707504); Sanming Project of Medicine in Shenzhen, China (SZSM201911014); the High Level-Hospital Program, Health Commission of Guangdong Province, China; and the research project of Hainan academician innovation platform (YSPTZX202004); the donations of the Shaw Foundation of Hong Kong, the Richard Yu and Carol Yu, Michael Seak-Kan Tong, May Tam Mak Mei Yin, Lee Wan Keung Charity Foundation Limited, Hong Kong Sanatorium & Hospital, Hui Ming, Hui Hoy and Chow Sin Lan Charity Fund Limited, Chan Yin Chuen Memorial Charitable Foundation, Marina Man-Wai Lee, the Hong Kong Hainan Commercial Association South China Microbiology Research Fund, the Jessie & George Ho Charitable Foundation, Perfect Shape Medical Limited, Kai Chong Tong, Tse Kam Ming Laurence, Foo Oi Foundation Limited, Betty Hing-Chu Lee, Ping Cham So, and Lo Ying Shek Chi Wai Foundation. The funding sources had no role in the study design, data collection, analysis, interpretation, or writing of the report.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103643.
Contributor Information
Jasper Fuk-Woo Chan, Email: jfwchan@hku.hk.
Kwok-Yung Yuen, Email: kyyuen@hku.hk.
Hin Chu, Email: hinchu@hku.hk.
Appendix. Supplementary materials
References
- 1.Chan JF, To KK, Tse H, Jin DY, Yuen KY. Interspecies transmission and emergence of novel viruses: lessons from bats and birds. Trends Microbiol. 2013;21(10):544–555. doi: 10.1016/j.tim.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan JF, Yuan S, Kok KH, To KK, Chu H, Yang J. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395(10223):514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO. 2021. https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---3-august-2021.
- 5.Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leung K, Shum MH, Leung GM, Lam TT, Wu JT. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Euro Surveill. 2021;26(1) doi: 10.2807/1560-7917.ES.2020.26.1.2002106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lau SK, Woo PC, Li KS, Tsang AK, Fan RY, Luk HK. Discovery of a novel coronavirus, China Rattus coronavirus HKU24, from Norway rats supports the murine origin of Betacoronavirus 1 and has implications for the ancestor of Betacoronavirus lineage A. J Virol. 2015;89(6):3076–3092. doi: 10.1128/JVI.02420-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Firth C, Bhat M, Firth MA, Williams SH, Frye MJ, Simmonds P. Detection of zoonotic pathogens and characterization of novel viruses carried by commensal Rattus norvegicus in New York City. mBio. 2014;5(5):e01933. doi: 10.1128/mBio.01933-14. -14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McFarlane R, Sleigh A, McMichael T. Synanthropy of wild mammals as a determinant of emerging infectious diseases in the Asian-Australasian region. Ecohealth. 2012;9(1):24–35. doi: 10.1007/s10393-012-0763-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sridhar S, Yip CC, Wu S, Chew NF, Leung KH, Chan JF. Transmission of Rat Hepatitis E Virus Infection to Humans in Hong Kong: A Clinical and Epidemiological Analysis. Hepatology. 2021;73(1):10–22. doi: 10.1002/hep.31138. [DOI] [PubMed] [Google Scholar]
- 11.Chu H, Chan JF, Yuen TT, Shuai H, Yuan S, Wang Y. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet Microbe. 2020;1(1):e14–e23. doi: 10.1016/S2666-5247(20)30004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bao L, Deng W, Huang B, Gao H, Liu J, Ren L. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583(7818):830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- 13.Zhai X, Sun J, Yan Z, Zhang J, Zhao J, Zhao Z. Comparison of Severe Acute Respiratory Syndrome Coronavirus 2 Spike Protein Binding to ACE2 Receptors from Human, Pets, Farm Animals, and Putative Intermediate Hosts. J Virol. 2020;94(15) doi: 10.1128/JVI.00831-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao X, Chen D, Szabla R, Zheng M, Li G, Du P. Broad and Differential Animal Angiotensin-Converting Enzyme 2 Receptor Usage by SARS-CoV-2. J Virol. 2020;94(18) doi: 10.1128/JVI.00940-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv. 2020 [Google Scholar]
- 16.Voloch CM, da Silva Francisco R, Jr., de Almeida LGP, Cardoso CC, Brustolini OJ, Gerber AL. Genomic characterization of a novel SARS-CoV-2 lineage from Rio de Janeiro. Brazil. J Virol. 2021 doi: 10.1128/JVI.00119-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tablizo F, Kim K, Lapid C, Castro M, Yangzon M, Maralit B. Genome sequencing and analysis of an emergent SARS-CoV-2 variant characterized by multiple spike protein mutations detected from the Central Visayas Region of the Philippines. medRxiv. 2021 [Google Scholar]
- 18.Gu H, Chen Q, Yang G, He L, Fan H, Deng YQ. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science. 2020;369(6511):1603–1607. doi: 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whitt MA. Generation of VSV pseudotypes using recombinant DeltaG-VSV for studies on virus entry, identification of entry inhibitors, and immune responses to vaccines. J Virol Methods. 2010;169(2):365–374. doi: 10.1016/j.jviromet.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chu H, Hu B, Huang X, Chai Y, Zhou D, Wang Y. Host and viral determinants for efficient SARS-CoV-2 infection of the human lung. Nat Commun. 2021;12(1):134. doi: 10.1038/s41467-020-20457-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chu H, Shuai H, Hou Y, Zhang X, Wen L, Huang X. Targeting highly pathogenic coronavirus-induced apoptosis reduces viral pathogenesis and disease severity. Sci Adv. 2021;7(25) doi: 10.1126/sciadv.abf8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan S, Yin X, Meng X, Chan JF, Ye ZW, Riva L. Clofazimine broadly inhibits coronaviruses including SARS-CoV-2. Nature. 2021;593(7859):418–423. doi: 10.1038/s41586-021-03431-4. [DOI] [PubMed] [Google Scholar]
- 23.Wolfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Muller MA. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 24.Chu H, Chan JF, Wang Y, Yuen TT, Chai Y, Hou Y. Comparative Replication and Immune Activation Profiles of SARS-CoV-2 and SARS-CoV in Human Lungs: An Ex Vivo Study With Implications for the Pathogenesis of COVID-19. Clin Infect Dis. 2020;71(6):1400–1409. doi: 10.1093/cid/ciaa410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chu H, Chan JF, Wang Y, Yuen TT, Chai Y, Shuai H. SARS-CoV-2 Induces a More Robust Innate Immune Response and Replicates Less Efficiently Than SARS-CoV in the Human Intestines: An Ex Vivo Study With Implications on Pathogenesis of COVID-19. Cell Mol Gastroenterol Hepatol. 2021;11(3):771–781. doi: 10.1016/j.jcmgh.2020.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shuai H, Chu H, Hou Y, Yang D, Wang Y, Hu B. Differential immune activation profile of SARS-CoV-2 and SARS-CoV infection in human lung and intestinal cells: Implications for treatment with IFN-beta and IFN inducer. J Infect. 2020;81(4):e1–e10. doi: 10.1016/j.jinf.2020.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan JF, Zhang AJ, Yuan S, Poon VK, Chan CC, Lee AC. Simulation of the Clinical and Pathological Manifestations of Coronavirus Disease 2019 (COVID-19) in a Golden Syrian Hamster Model: Implications for Disease Pathogenesis and Transmissibility. Clin Infect Dis. 2020;71(9):2428–2446. doi: 10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng J, Wong LR, Li K, Verma AK, Ortiz ME, Wohlford-Lenane C. COVID-19 treatments and pathogenesis including anosmia in K18-hACE2 mice. Nature. 2021;589(7843):603–607. doi: 10.1038/s41586-020-2943-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203(2):631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hikmet F, Mear L, Edvinsson A, Micke P, Uhlen M, Lindskog C. The protein expression profile of ACE2 in human tissues. Mol Syst Biol. 2020;16(7):e9610. doi: 10.15252/msb.20209610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCray PB, Jr., Pewe L, Wohlford-Lenane C, Hickey M, Manzel L, Shi L. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J Virol. 2007;81(2):813–821. doi: 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang H, Zhu Y, Niu Z, Zhou L, Sun Q. SARS-CoV-2 N501Y variants of concern and their potential transmission by mouse. Cell Death Differ. 2021;28(10):2840–2842. doi: 10.1038/s41418-021-00846-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Montagutelli X, Prot M, Levillayer L, Salazar E, Jouvion G, Conquet L. The B1.351 and P.1 variants extend SARS-CoV-2 host range to mice. bioRxiv. 2021 [Google Scholar]
- 34.Wang R, Zhang Q, Ge J, Ren W, Zhang R, Lan J. Analysis of SARS-CoV-2 variant mutations reveals neutralization escape mechanisms and the ability to use ACE2 receptors from additional species. Immunity. 2021;54(7):1611–1621. doi: 10.1016/j.immuni.2021.06.003. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Q, Nie J, Wu J, Zhang L, Ding R, Wang H. SARS-CoV-2 501Y.V2 variants lack higher infectivity but do have immune escape. Cell. 2021;184(9):2362–2371. doi: 10.1016/j.cell.2021.02.042. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang K, Zhang Y, Hui X, Zhao Y, Gong W, Wang T. Q493K and Q498H substitutions in Spike promote adaptation of SARS-CoV-2 in mice. EBioMedicine. 2021;67 doi: 10.1016/j.ebiom.2021.103381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leist SR, Dinnon KH, 3rd Schafer A, Tse LV, Okuda K, Hou YJ. A Mouse-Adapted SARS-CoV-2 Induces Acute Lung Injury and Mortality in Standard Laboratory Mice. Cell. 2020;183(4):1070–1085. doi: 10.1016/j.cell.2020.09.050. e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun S, Gu H, Cao L, Chen Q, Yang G, Li R. Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2. bioRxiv. 2021 doi: 10.1038/s41467-021-25903-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toapanta FR, Ross TM. Impaired immune responses in the lungs of aged mice following influenza infection. Respir Res. 2009;10:112. doi: 10.1186/1465-9921-10-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao J, Zhao J, Legge K, Perlman S. Age-related increases in PGD(2) expression impair respiratory DC migration, resulting in diminished T cell responses upon respiratory virus infection in mice. J Clin Invest. 2011;121(12):4921–4930. doi: 10.1172/JCI59777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Driscoll M, Ribeiro Dos Santos G, Wang L, Cummings DAT, Azman AS, Paireau J. Age-specific mortality and immunity patterns of SARS-CoV-2. Nature. 2021;590(7844):140–145. doi: 10.1038/s41586-020-2918-0. [DOI] [PubMed] [Google Scholar]
- 42.Rathnasinghe R, Jangra S, Cupic A, Martinez-Romero C, Mulder LCF, Kehrer T. The N501Y mutation in SARS-CoV-2 spike leads to morbidity in obese and aged mice and is neutralized by convalescent and post-vaccination human sera. medRxiv. 2021 [Google Scholar]
- 43.Jiang RD, Liu MQ, Chen Y, Shan C, Zhou YW, Shen XR. Pathogenesis of SARS-CoV-2 in Transgenic Mice Expressing Human Angiotensin-Converting Enzyme 2. Cell. 2020;182(1):50–58. doi: 10.1016/j.cell.2020.05.027. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winkler ES, Bailey AL, Kafai NM, Nair S, McCune BT, Yu J. SARS-CoV-2 infection in the lungs of human ACE2 transgenic mice causes severe inflammation, immune cell infiltration, and compromised respiratory function. bioRxiv. 2020 [Google Scholar]
- 45.Horspool AM, Ye C, Wong TY, Russ BP, Lee KS, Winters MT. SARS-CoV-2 B.1.1.7 and B.1.351 variants of concern induce lethal disease in K18-hACE2 transgenic mice despite convalescent plasma therapy. bioRxiv. 2021 [Google Scholar]
- 46.Radvak P, Kwon H, Kosikova M, Ortega-Rodriguez U, Xiang R, Phue J. B.1.1.7 and B.1.351 variants are highly virulent in K18-ACE2 transgenic mice and show different pathogenic patterns from early SARS-CoV-2 strains. bioRxiv. 2021 doi: 10.1038/s41467-021-26803-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chan JF, Yuan S, Zhang AJ, Poon VK, Chan CC, Lee AC. Surgical Mask Partition Reduces the Risk of Noncontact Transmission in a Golden Syrian Hamster Model for Coronavirus Disease 2019 (COVID-19) Clin Infect Dis. 2020;71(16):2139–2149. doi: 10.1093/cid/ciaa644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garigliany M, Van Laere AS, Clercx C, Giet D, Escriou N, Huon C. SARS-CoV-2 Natural Transmission from Human to Cat, Belgium, March 2020. Emerg Infect Dis. 2020;26(12):3069–3071. doi: 10.3201/eid2612.202223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sit THC, Brackman CJ, Ip SM, Tam KWS, Law PYT, To EMW. Infection of dogs with SARS-CoV-2. Nature. 2020;586(7831):776–778. doi: 10.1038/s41586-020-2334-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McAloose D, Laverack M, Wang L, Killian ML, Caserta LC, Yuan F. From People to Panthera: Natural SARS-CoV-2 Infection in Tigers and Lions at the Bronx Zoo. mBio. 2020;11(5) doi: 10.1128/mBio.02220-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oude Munnink BB, Sikkema RS, Nieuwenhuijse DF, Molenaar RJ, Munger E, Molenkamp R. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science. 2021;371(6525):172–177. doi: 10.1126/science.abe5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Griffin BD, Chan M, Tailor N, Mendoza EJ, Leung A, Warner BM. SARS-CoV-2 infection and transmission in the North American deer mouse. Nat Commun. 2021;12(1):3612. doi: 10.1038/s41467-021-23848-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bao L, Gao H, Deng W, Lv Q, Yu H, Liu M. Transmission of Severe Acute Respiratory Syndrome Coronavirus 2 via Close Contact and Respiratory Droplets Among Human Angiotensin-Converting Enzyme 2 Mice. J Infect Dis. 2020;222(4):551–555. doi: 10.1093/infdis/jiaa281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Y, Huang K, Wang T, Deng F, Gong W, Hui X. SARS-CoV-2 rapidly adapts in aged BALB/c mice and induces typical pneumonia. J Virol. 2021 doi: 10.1128/JVI.02477-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang J, Shuai L, Wang C, Liu R, He X, Zhang X. Mouse-adapted SARS-CoV-2 replicates efficiently in the upper and lower respiratory tract of BALB/c and C57BL/6J mice. Protein Cell. 2020;11(10):776–782. doi: 10.1007/s13238-020-00767-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.