Key Words: bioinformatics, blood-brain barrier, circRNA, miRNA, RNA-sequence, tight junction protein, traumatic brain injury, ZO1
Abstract
Circular RNAs (circRNAs) are a new and large group of non-coding RNA molecules that are abundantly expressed in the central nervous system. However, very little is known about their roles in traumatic brain injury. In this study, we firstly screened differentially expressed circRNAs in normal and injured brain tissues of mice after traumatic brain injury. We found that the expression of circLphn3 was substantially decreased in mouse models of traumatic brain injury and in hemin-treated bEnd.3 (mouse brain cell line) cells. After overexpressing circLphn3 in bEnd.3 cells, the expression of the tight junction proteins, ZO-1, ZO-2, and occludin, was upregulated, and the expression of miR-185-5p was decreased. In bEnd.3 cells transfected with miR-185-5p mimics, the expression of ZO-1 was decreased. Dual-luciferase reporter assays showed that circLphn3 bound to miR-185-5p, and that miR-185-5p bound to ZO-1. Additionally, circLphn3 overexpression attenuated the hemin-induced high permeability of the in vitro bEnd.3 cell model of the blood-brain barrier, while miR-185-5p transfection increased the permeability. These findings suggest that circLphn3, as a molecular sponge of miR-185-5p, regulates tight junction proteins’ expression after traumatic brain injury, and it thereby improves the permeability of the blood-brain barrier. This study was approved by the Animal Care and Use Committee of Chongqing Medical University of China (approval No. 2021-177) on March 22, 2021.
Chinese Library Classification No. R453; R741; Q789
Introduction
Long-term pathological changes, including complex cytological reactions, have been shown to be related to traumatic brain injury (TBI) (McGinn and Povlishock, 2016). Impairment of the blood-brain barrier (BBB) is one of the most critical pathological causes of these changes following TBI (Sulhan et al., 2020). Brain microvascular endothelial cells and tight junctions are major factors maintaining the BBB’s integrity (Khellaf et al., 2019). The main functional proteins of the tight junction are zona occludens-1 (ZO-1), ZO-2, ZO-3, and occludin, and their corresponding genes are Tjp1, Tjp2, Tjp3, and Ocln, respectively (Bhowmick et al., 2019). Loss of tight junction proteins (TJPs) can lead to destruction of the BBB and brain edema aggravation (Yuan et al., 2016). So far, there is no effective treatment for BBB damage after TBI. Therefore, new viable treatments are urgently required.
Circular RNAs (circRNAs) are a new type of RNA molecules with a closed loop structure, which is different from linear RNAs (Qu et al., 2015). Across the wide range of eukaryotes, circRNAs are conserved in structure and sequence, and they show cell or tissue-specific expression. CircRNAs are abundant in brain tissues (Mahmoudi et al., 2019). These circRNAs have been shown to be involved in the development, differentiation, and biological function of the nervous system, and to play an important role in the neurological dysfunction and pathological progression after brain injury (Jiang et al., 2020). According to the latest research, there are significant differences in some circRNAs’ expression levels in the injured foci and normal brain tissues after TBI (Han et al., 2017). However, the specific mechanism is still unclear.
In this study, we explored the effects of circRNAs on BBB after TBI. We first established a TBI model by controlling cortical injury in mice, and then we screened the differentially expressed circRNAs in normal and injured brain tissues of mice after TBI by RNA-sequencing (RNA-seq). Our analysis of the sequencing results in terms of expression abundance, expression differences, and strength of RNA-RNA interactions revealed that circLphn3 functions as a molecular sponge of the microRNA (miRNA) miR-185-5p. We also explored its role in the BBB after TBI to reveal potential therapeutic targets for TBI treatment.
Materials and Methods
Animals and TBI model
Many studies have shown that estrogen has protective effects on brain injury (Brotfain et al., 2016; Yilmaz et al., 2019; Zafer et al., 2019; Acosta-Martínez, 2020), hence this study only used male mice. Forty C57BL/6J male mice (18–24 g, 6–8 weeks) were obtained from the Animal Experimental Center of Chongqing Medical University [license No. SCXK (Yu) 2018-0003]. Mice were housed (five per cage) at 24 ± 2°C. All mice were randomly divided into sham and TBI groups. Four mice died after the TBI, and therefore two mice from the sham group were moved to the TBI group after receiving the injury. The mice in each group were used as follows: three for RNA-seq, five for protein extraction, five for RNA extraction, and five for immunofluorescence analysis. A controlled cortical impact was performed on mice to establish the TBI model as previously described (Ma et al., 2019). Briefly, a 5-mm diameter craniotomy was performed on the central aspect of the left parietal bone. Compressed air driven by a flat-tip impactor (Precision Systems and Instrumentation, Fairfax, VA, USA) was positioned to the exposed dura’s surface and set to impact the cortical surface at a velocity of 5.0 m/s, depth of 2.5 mm, and impact duration of 100 ms. Sham animals received the same isoflurane (Macklin, Shanghai, China) anesthesia, skin incision, and craniotomy, but no impact. All animal studies were conducted following the NIH published Care and Use of Laboratory Animals guidelines. Additionally, the study was approved by the Animal Care and Use Committee of Chongqing Medical University (approval No. 2021-177) on March 22, 2021.
RNA-sequencing and bioinformatics analysis
Three days after the operation, mice were anesthetized with 3% isoflurane (Macklin, Shanghai, China) and brains were obtained and frozen in liquid nitrogen for transcriptome sequencing (BGI, Shenzhen, China). The library construction and sequencing were conducted by BGI. Total RNA was extracted from the tissues using Trizol (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. About 60 mg of liquid nitrogen-frozen tissue were ground into powder in a 2-mL tube, and then homogenized for 2 minutes and rested horizontally for 5 minutes. The homogenates were centrifuged for 5 minutes at 12,000 × g at 4°C, and then the supernatant was transferred into a new EP tube with 0.3 mL chloroform/isoamyl alcohol (24:1). The mix was vigorously shaken for 15 seconds, and then centrifuged at 12,000 × g for 10 minutes at 4°C. After centrifugation, the upper aqueous phase containing RNA was transferred into a new tube with an equal volume of isopropyl alcohol, and then centrifuged at 12,000 × g for 20 minutes at 4°C. The RNA pellet was washed twice with 1 mL of 75% ethanol, centrifuged at 12,000 × g for 3 minutes at 4°C to remove residual ethanol, and then air dried for 5–10 minutes. Finally, 25–100 μL of diethyl pyrocarbonate-treated water was added to dissolve the RNA. Subsequently, total RNA was quality checked and quantified using a Nano Drop and Agilent 2100 bioanalyzer (Thermo Fisher Scientific, MA, USA). Oligo(dT)-attached magnetic beads were used to purify the mRNA. Purified mRNA was fragmented into small pieces with fragment buffer at 4°C. Then, first-strand complementary DNA (cDNA) was generated by reverse transcription with random hexamer primers, followed by second-strand cDNA synthesis. Next, A-Tailing Mix and RNA Index Adapters were added for end repair. The cDNA fragments obtained were amplified by PCR, the products were purified using Ampure XP Beads, and then dissolved in EB solution. The products were validated by the Agilent Technologies 2100 bioanalyzer for quality control. The double-stranded PCR products were heat-denatured and circularized by the splint oligo sequence to get the final library. The single-strand circular DNA was the final library. The final library was amplified with phi29 to make a DNA nanoball (DNB), which contained more than 300 copies of one molecule. DNBs were loaded onto the patterned nanoarray and pair end 100-base reads were generated by the BGIseq500 platform (BGI). Three algorithms, RNAhybrid (Krüger and Rehmsmeier, 2006), miRanda (John et al., 2004), and TargetScan (Agarwal et al., 2015), and the Starbase database (Li et al., 2014) were used to predict target genes of miRNAs. The small RNA expression level was calculated by counting the absolute number of molecules using unique molecular identifiers (Kivioja et al., 2011). Subsequently, we used RSEM (Li and Dewey, 2011) to calculate the expression of genes and transcripts (Fragments Per Kilobase per Million Mapped Fragments, FPKM). Differential expression analysis was performed using the DEGseq in the R package (http://www.bioconductor.org/packages/release/bioc/html/DEGseq.html) (Wang et al., 2010). Q ≤ 0.01 and Log2Ratio ≥ 1 as the default threshold were used to judge the significance of expression differences (Additional Table 1).
Additional Table 1.
Softwares for RNA-sequence data analysis
| Software | Source |
|---|---|
| Bowtie2 (2 2.2.9) | http://bowtie-bio.sourceforge.net/index.shtml |
| RNAhybrid | https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid |
| miRanda (3.3a) | http://www.microrna.org/microrna/home.do |
| TargetScan (7.1) | http://www.targetscan.org/vert_72/ |
| starBase | http://starbase.sysu.edu.cn/ |
| DEGseq | http://bioinfo.au.tsinghua.edu.cn/software/degseq/ |
Cell culture and transient transfection
Mouse brain microvascular endothelial cells (bEnd.3, ATCC Cat# CRL-2299, RRID:CVCL_0170; Huzhen Biotech, Shanghai, China) were cultured in high glucose Dulbecco’s modified Eagle’s medium (Gibco, Carlsbad, CA, USA) with 10% fetal bovine serum (PAN-Biotech, Bavaria, Germany) in an incubator at 37°C with 5% CO2, and treated with or without 40 µM hemin (Solarbio, Beijing, China). Cells were transfected with a plasmid and/or miRNA mimics using LipoFiter (HanBio, Shanghai, China) according to the manufacturer’s instructions. Wild type and mutant psiCHECK2-circLphn3 plasmids for the dual-luciferase reporter assay were purchased from HanBio, circLphn3 overexpression vector pUC-circLphn3 (Additional file 1 (83.8KB, pdf) ) was purchased from Cyagen (Guangzhou, China), and mmu-miR-185-5p mimics were purchased from HanBio.
Quantitative real-time PCR
Total RNA of bEnd.3 cells and brain tissues was extracted 3 days after TBI using an RNA Extraction Kit (Beyotime, Shanghai, China) in accordance with the manufacturer’s protocol. RNase R treatment was performed at 37°C for 15–120 minutes with 3 U/mg RNase R (Epicenter, Madison, WI, USA). Using RT Master Mix of a qPCR Kit (MCE, Monmouth Junction, NJ, USA) according to the manufacturer’s instructions, the RNA was reverse transcribed into cDNA. SYBR® Green qPCR Master Mix (MCE) was used as described by the manufacturer to perform quantitative real-time PCR to evaluate the expression of target genes. Analysis of the relative gene expression data was performed using the ΔΔCT method. The primers for mmu-miR-185-5p and U6 were purchased from RiboBio (Guangzhou, China). The PCR primers for circLphn3 (divergent) only amplify circLphn3. These primers were designed by primer-blast (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) according to the circularization junction of circLphn3, which we queried in circbase (http://www.circbase.org/) (Glažar et al., 2014). The sequence of the circularization junction was included in the amplification product. The gene-specific primers are listed in Table 1.
Table 1.
Primer sequences
| Gene | Sequence (5’–3’) |
|---|---|
| circLphn3 (divergent) | F: CAC CAA TTC ACC TCG ACT CTG |
| R: AGG GAC ACA TTC ATA CTG CAC | |
| circLphn3 (convergent) | F: TCT CTC CAC CAA TTC ACC TCG |
| R: ACA CTC CTT TTA GCA GTC CAG G | |
| Lphn3 | F: AGC TTT GAA TGC CTG GAG AGA C |
| R: TCG GTC TTC AAA AGG TTA TCA GC | |
| Tjp1 | F: GTT GTC CCT CCT CTG ATA CCT TC |
| R: TGG CAG TGT CAT TCA CAT CTT TC | |
| Tjp2 | F: ATC TAT GCG GTT CCA ATC AAG |
| R: CCT CTG GTG TCC TGG TAA AGT C | |
| Tjp3 | F: GAA ACG AAG AAA CAG CGA AGA G |
| R: GAC AGA TTA GCA CTG GAC ACC C | |
| Ocln | F: GCT GGA TGA CTA CAG AGA GGA GAG |
| R: TAA GGT TTC CGT CTG TCA TAA TCT C | |
| GAPDH | F: GAC ATC AAG AAG GTG GTG AAG C |
| R: GAA GGT GGA AGA GTG GGA GTT |
circLphn3: Circular Lphn3; F: forward; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; Lphn3: latrophilin 3; Ocln: occludin; R: reverse; Tjp: tight junction protein.
Western blot
The tight junction proteins were examined by western blot analysis. Equal amounts of protein samples extracted from the ipsilesional cortex or bEnd.3 cells were electrophoresed in 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (Beyotime), transferred to polyvinylidene fluoride membranes (Millipore, Burlington, MA, USA), and incubated overnight at 4°C with the following primary antibodies (1:1000): ZO-1 antibody (rabbit, pAb, Cat# 511417, ZEN-BIO, Chengdu, China); ZO-2 (rabbit, pAb, Cat# ab224314) and ZO-3 antibodies (rabbit, pAb, Cat# ab181991, Abcam, Cambridge, UK); Occludin antibody (rabbit, pAb, Cat# GTX114949, GeneTex, Irvine, CA, USA). The bands were detected by an electrochemiluminescence kit (Beyotime) after incubation with a horseradish peroxidase-conjugated secondary antibody (goat anti-rabbit, 1:2000; ZEN-BIO, Cat# 511203) for 1 hour at room temperature. Images were recorded by Fusion FX (VILBER LOURMAT, Marne La Vallée, France) and quantification of band density was conducted using ImageJ software (National Institutes of Health, Bethesda, MD, USA). Relative expression was standardized to the corresponding optical density of β-actin (rabbit, 1:2000; Cat# GTX109639, GeneTex).
Dual-luciferase reporter assay
The target circLphn3 sequences or target mRNA (ZO-1) 3′ untranslated region (UTR) sequences were cloned downstream of hRluc in psiCHECK2 plasmid to construct vectors containing potential miR-185-5p-binding sites. The reporter vectors (HanBio) and miR-185-5p mimics (HanBio) were co-transfected into 293T cells (HanBio) and the renilla luciferase activity (reporter gene) and firefly luciferase activity (internal reference gene) were determined by the Dual-Lumi™ II Luciferase Reporter Gene Assay Kit (BeyoTime). Relative luciferase activity = renilla luciferase activity/firefly luciferase activity.
Monolayer in vitro BBB model and permeability measurement
The in vitro model and the permeability experiments were conducted as described by Watson et al. (2013). Briefly, bEnd.3 cells were seeded in cell inserts with a 1.0-µm transparent polyethylene terephthalate membrane (CORNING Falcon®, Tewksbury, MA, USA) in a 12-well plate. Lucifer yellow (LY; 20 µM, MCE) was added as a tracer for permeability experiments in the inserts of four different experimental groups, and the medium contained 40 µM hemin. At 1, 4, 12, 24, and 36 hours after the LY addition, the medium in the 12-well plate was collected and tested for fluorescence intensity in 96-well plates (200 µL/well). After each collection, the cell inserts were moved into the next well containing the same volume of culture medium as before and the experiment continued for the next period until 36 hours. The fluorescence intensity was measured by a plate reader at a 428 nm excitation wavelength and a 536 nm emission wavelength (Bio-Rad, Richmond, CA, USA). The permeability coefficient was calculated according to the reports of Siflinger-Birnboim et al. (1987) and Deli et al. (2005).
Immunofluorescence assays
The mouse brain was excised and filled with the optimal cutting temperature compound (SAKURA, Torrance, CA, USA), frozen in liquid nitrogen for 30 minutes, and then placed in a –80°C freezer until sectioning. After fixation, samples were permeabilized with 0.5% Triton X-100 in phosphate-buffered saline containing 10% goat serum (Beyotime) at 37°C for 1 hour. Next, the samples were incubated with primary antibodies against ZO-1 (1:100; rabbit, GeneTex) and CD31 (1:100; mouse, Cat# NB100-64796, NOVUS, Centennial, CO, USA) overnight at 4°C, followed by incubation with secondary antibodies labeled with Alexa Fluor® 488 or 594 (1:200; goat anti-mouse Cat# ZF-0512, goat anti-rabbit Cat# ZF-0516, ZSGB-bio, Beijing, China) for 1 hour at 37°C, and counterstaining with 4′,6-diamidino-2-phenylindole (DAPI) (BeyoTime). Cy3-labeled fluorescence in situ hybridization (FISH) probes for circLphn3 were purchased from GeneSeed (Guangzhou, China) and the FISH kit was purchased from RiboBio. FISH was performed according to the manufacturer’s protocol. Briefly, cells were fixed with paraformaldehyde, followed by addition of prehybridization solution and incubation at 37°C for 30 minutes. Then, the cells were incubated with the hybridization solution containing the FISH probes (0.2 µM) overnight in the dark at 37°C. Photomicrographs were taken by a confocal laser scanning microscope (Zeiss LSM800, Braunschweig, Germany).
Cytoplasm/nucleus fraction isolation
The distribution of circLphn3 in the cytoplasm and the nucleus was determined with the Cytoplasmic & Nuclear RNA Purification Kit (NORGEN, Ontario, Canada) following the manufacturer’s protocol. The relative expression levels of circLphn3, a nuclear control (U6), and a cytoplasmic control (GAPDH) were measured by quantitative real-time PCR in RNA samples from the cytoplasm and the nucleus.
Statistical analyses
Statistical analyses and statistical plotting were performed using the GraphPad Prism® 8 (GraphPad, San Diego, CA, USA). All data are presented as the mean ± standard error of the mean (SEM). Two independent groups were analyzed with a two-tailed t-test, while sets of data (> two groups) were analyzed with one-way analysis of variance, followed by Bonferroni multiple comparison post hoc tests. P < 0.05 indicates that the difference is statistically significant.
Results
Differential expression of circRNAs after TBI in mice
To determine the expression status of circRNAs, we selected three pairs of normal brain tissues and TBI tissues and conducted RNA-seq. The results showed 2015 circRNAs with 2-fold differential expression (Q < 0.01, Figure 1A), including 1336 downregulated and 679 upregulated circRNAs. The clustered heat map in Figure 1B shows the top 40 upregulated and downregulated circRNAs. RNAhybrid, miRanda, and TargetScan (Additional Table 1) were subsequently applied to predict the target miRNAs of all 40 differentially expressed circRNA. mmu_circ_0001358 (circLphn3) showed significant expression differences between the TBI and Sham groups, and the prediction showed that many circLphn3-targeted miRNAs were closely related to the TJP genes. In combination with the Starbase database, we found 19 miRNAs (including miR-185-5p) with potential circLphn3-binding sites. Among them, 14 miRNA had potential binding sites for ZO-1 mRNA, 11 miRNAs had binding sites for ZO-2 mRNA, 1 miRNA had binding sites for ZO-3 mRNA, and 8 miRNAs had binding sites for occludin mRNA (Figure 1C). We named mmu_circ_0001358 circLphn3 because of its host gene Lphn3. The average expression (FPKM) of circLphn3 was 1.923 in the Sham group compared with 0.457 in the TBI group, with a –4.2-fold change. Consistently, quantitative real-time PCR indicated that the expression level of circLphn3 in the injured brain tissue was significantly lower than that in the normal brain tissue (P < 0.05, Figure 1D). The in vitro experiment further indicated that the expression of circLphn3 was downregulated after cells were treated with 40 µM hemin for 12 hours (P < 0.05, vs. control group; Figure 1E). Furthermore, immunofluorescence photographs demonstrated colocalization of ZO-1 and CD31, showing loss and disruption of TJPs in brain vascular endothelial cells after TBI (Figure 1F).
Figure 1.
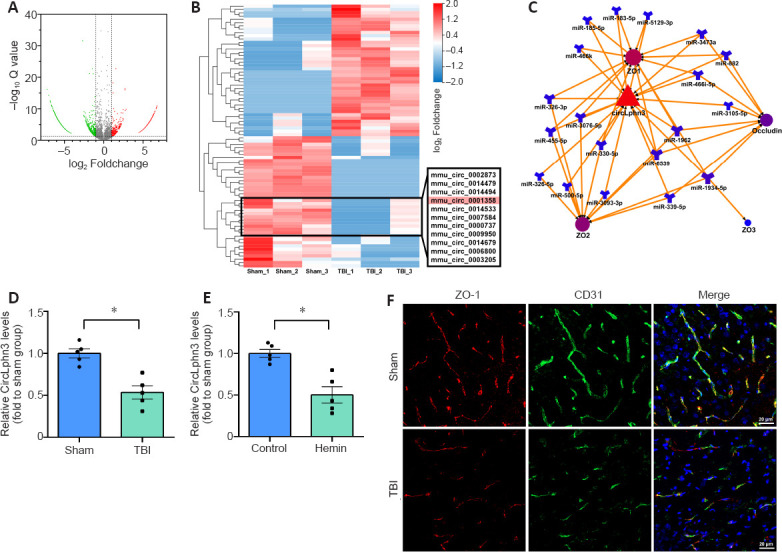
CircRNAs and ZO-1 expression changes after TBI.
(A) Volcano plot of circRNAs. Red spots: upregulated circRNAs (fold change > 2, Q < 0.01); green spots: downregulated circRNAs (fold change < −2, Q < 0.01); gray spots: unchanged circRNAs (−2 < fold change < 2, Q > 0.01). (B) A clustered heat map of the top 40 upregulated and downregulated circRNAs. (C) Schematic illustration of the circLphn3/miRNA/TJPs ceRNA network. MiRNAs in overlapping regions targeted multiple TJPs. (D, E) Quantitative real-time PCR results of circLphn3 expression in the in vivo and in vitro models. CircLphn3 was downregulated after both TBI and hemin treatment. (F) Representative fluorescence images of ZO-1 (red, Alexa Fluor® 594) and CD31 (green, Alexa Fluor® 488) colocalization in the ipsilesional cortex. The expression of ZO-1 was reduced and discontinuous in brain vascular endothelial cells in the TBI group compared with that in the Sham group. Scale bars: 20 µm. All data are presented as mean ± SEM. n = 5 mice per group in vivo, and n = 5 independent experiments in vitro. *P < 0.05 (two-tailed t-test). ceRNA: Competing endogenous RNA; circLphn3: circular Lphn3; CircRNA: circular RNA; Lphn3: latrophilin 3; miRNA: microRNA; TBI: traumatic brain injury; TJPs: tight junction proteins; ZO-1: zona occludens-1.
Identification of circLphn3 as a circRNA in bEnd.3 cells
CircLphn3 was differentially expressed after TBI (Figure 1B). Using a genome browser (UCSC Date, http://genome.ucsc.edu/), we found that circLphn3 (1007 bp) is formed by exons 6–9 of Lphn3 (Figure 2A). Quantitative real-time PCR analysis showed that circLphn3 was amplified by divergent primers with cDNA as the template, but not with genomic DNA (Figure 2B). Additionally, quantitative real-time PCR showed that circLphn3 resisted RNase R, while its host gene, Lphn3 mRNA, was degraded by RNase R (Figure 2C and D). Sanger sequencing of the quantitative real-time PCR products amplified by circLphn3 divergent primers also confirmed a back-spliced junction in circLphn3 (Figure 2A). Moreover, the Cy3-circLphn3 FISH probe (Figure 2E) and cytoplasm/nucleus fraction isolation (Figure 2F) revealed that circLphn3 is primarily expressed in the cytoplasm.
Figure 2.
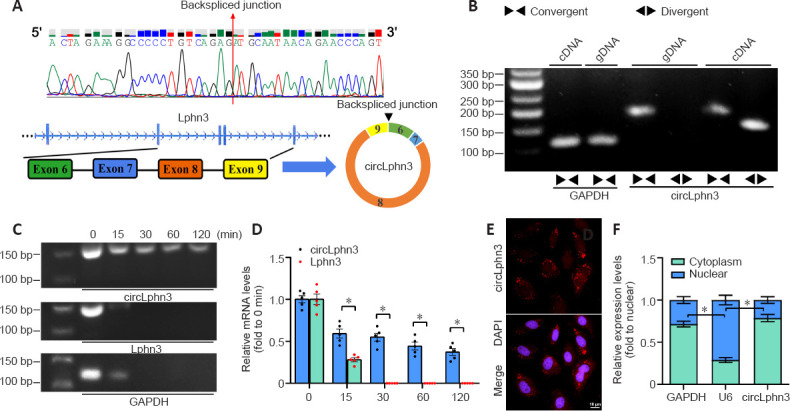
Identification of circLphn3 as a circular RNA.
(A) Sanger sequencing results of quantitative real-time PCR products with circLphn3 primers. Schematic illustration showing the circularization of Lphn3 exons 6–9 that form circLphn3. (B) The expression of circLphn3 was validated in bEnd.3 cells by quantitative real-time PCR and agarose gel electrophoresis. Divergent primers amplified circLphn3 from cDNA but not from genomic DNA. (C) Circular Lphn3 and linear Lphn3 expression levels were examined by quantitative real-time PCR and agarose gel electrophoresis treated with RNase R for 0–120 minutes. Circular Lphn3 is more stable than linear Lphn3. (D) Quantification of circLphn3 and linear Lphn3 levels. (E) Cy3-FISH probe indicated that circLphn3 (red) was enriched in the cytoplasm. Scale bar: 20 µm. (F) CircLphn3 is abundant in the cytoplasm of bEnd.3 cells. U6 and GAPDH were used as positive controls for the nucleus and cytoplasm, respectively. All data are presented as mean ± SEM. The experiments were repeated five times. *P < 0.05. cDNA: Complementary DNA; circLphn3: circular Lphn3; FISH: fluorescence in situ hybridization; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; gDNA: genomic DNA; Lphn3: latrophilin 3.
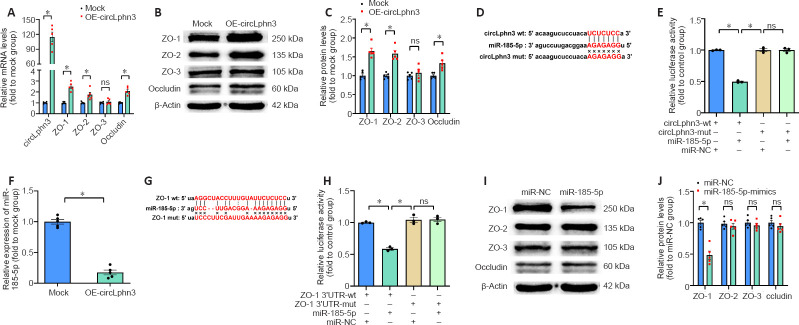
Overexpression of circLphn3 upregulates the expression of tight junction proteins in vitro
To investigate the effect of circLphn3 on the expression of ZO-1, ZO-2, ZO-3, and occludin, we transfected a circLphn3 overexpression plasmid into bEnd.3 cells for 24 hours and examined their mRNA and protein expression by quantitative real-time PCR and western blotting, respectively. The results showed that the mRNA levels of ZO-1, ZO-2, and occludin significantly increased after circLphn3 overexpression compared with those in the control group (P < 0.05; Figure 3A). In contrast, the mRNA levels of ZO-3 did not significantly change (P > 0.05, vs. Mock group; (Figure 3A). Furthermore, circLphn3 overexpression increased the protein expression levels of ZO-1, ZO-2, and occludin (P < 0.05, vs. Mock group), but had no effect on ZO-3 in bEnd.3 cells (P > 0.05, vs. Mock group; Figure 3B and C).
Figure 3.
CircLphn3 functions as a sponge for miR-185-5p and upregulates tight junction genes.
(A, C) CircLphn3 and ZO-1, ZO-2, and occludin mRNA and protein expression were significantly upregulated after circLphn3 overexpression. (B) Representative western blot images of ZO-1, ZO-2, ZO-3, and occludin after circLphn3 overexpression. (D) The predicted binding sites (red bases) of miR-185-5p and circLphn3. (E) Relative luciferase activity in 293T cells transfected with the indicated plasmids. (F) miR-185-5p was strongly downregulated by circLphn3 overexpression. (G) The predicted binding sites of miR-185-5p and ZO-1 mRNA 3′UTR. (H) Dual luciferase reporter gene assay confirmed the binding sites of miR-185-5p and ZO-1 mRNA 3′UTR. (I) Representative western blot images of ZO-1, ZO-2, ZO-3, and occludin after miR-185-5p mimics transfection. (J) ZO-1 was significantly downregulated in bEnd.3 after miR-185-5p mimics transfection. All data are presented as mean ± SEM. The experiments were repeated five times. *P < 0.05. circLphn3: Circular Lphn3; Lphn3: latrophilin 3; mut: mutant; NC: negative control; ns: not significant; OE: overexpression; UTR: untranslated region; wt: wild type; ZO: zona occludens.
circLphn3 binds miR-185-5p
To further verify the targeting of miR-185-5p by circLphn3, we transfected the psiCHECK2-circLphn3-wt or psiCHECK2-circLphn3-mut plasmid and miR-185-5p mimics into 293T cells for the luciferase reporter gene experiment. The results showed that the luciferase activity after co-transfection of miR-185-5p mimics with psiCHECK2-circLphn3-wt was lower than that of the control group (P < 0.05; Figure 3D and E). However, the luciferase activity after co-transfection of miR-185-5p mimics with psiCHECK2-circLphn3-mut was not significantly different from that of the control group (P > 0.05, Figure 3E). Further analysis showed that overexpression of circLphn3 significantly downregulated the expression of miR-185-5p in bEnd.3 cells (P < 0.05, vs. Mock group; Figure 3F). These results indicate that circLphn3 may act as a miR-185-5p sponge to downregulate the expression of miR-185-5p in bEnd.3 cells.
Tjp1 is a target gene of miR-185-5p
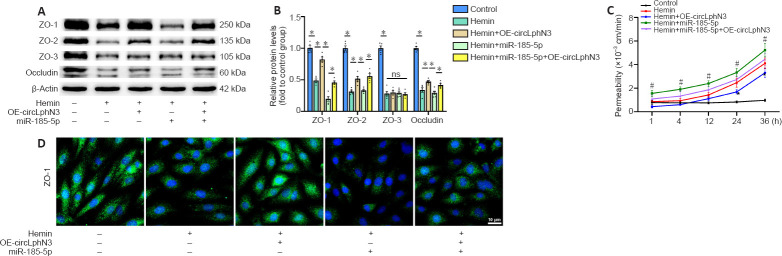
The dual-luciferase reporter system indicated that the luciferase activity after co-transfection of miR-185-5p mimics with psiCHECK2-ZO1 3′UTR-wt was lower than that of the control group (P < 0.05; Figure 3G and H). In contrast, the luciferase activity after co-transfection of miR-185-5p mimics with psiCHECK2-ZO1 3′UTR-mut was not significantly different from that of the control group (P > 0.05; Figure 3H). Namely, miR-185-5p significantly inhibited the luciferase activity of the reporter gene containing the wild type ZO-1 mRNA 3′UTR (P < 0.05, vs. miR-NC group; Figure 3H), but not the mutant isoform (P > 0.05, vs. miR-NC group; Figure 3H). Additionally, our result showed that miR-185-5p mimics reduced the protein level of ZO-1 (P < 0.05, vs. miR-NC group), but had no effect on ZO-2, ZO-3, and occludin protein expression in bEnd.3 cells (P > 0.05, vs. miR-NC group; Figure 3I and J). These data confirmed that Tjp1 is a direct target of miR-185-5p. Furthermore, transfection with miR-185-5p mimics partly reversed the circLphn3-induced upregulation of ZO-1 protein expression (P < 0.05, vs. hemin + OE-circLphn3 group; Figure 4A and B). Importantly, transfection with miR-185-5p mimics reduced the protective effect of circLphn3 on hemin-induced cell permeability (P < 0.05, vs. hemin + OE-circLphn3 group; Figure 4C). These results further confirmed that circLphn3 reduces cell damage by modulating the miR-185-5p/ZO-1 axis, thus suggesting that circLphn3 may be a new target for protecting against subsequent BBB damage.
Figure 4.
CircLphn3 protects BBB from hemin insult.
(A, B) Overexpression of circLphn3 attenuated the hemin-induced downregulation of ZO-1, ZO-2, and occludin, but not ZO-3. miR-185-5p transfection aggravated the ZO-1 decreased expression. (C) In the in vitro monolayer transwell BBB model, circLphn3 overexpression attenuated the hemin-induced high lucifer yellow permeability, while miR-185-5p transfection increased the permeability. (D) Immunofluorescent images of ZO-1 (green, Alexa Fluor® 488) in bEnd.3 cells after different interventions. Overexpression of circLphn3 enhanced ZO-1 expression after hemin treatment, whereas miR-185-5p transfection suppressed the protective effect of circLphn3 on ZO-1. Scale bar: 10 µm. All data are presented as mean ± SEM. The experiments were repeated five times. *P < 0.05; #P < 0.05, vs. hemin group (Bonferroni post hoc test). BBB: blood-brain barrier; circLphn3: circular Lphn3; Lphn3: latrophilin 3; ns: not significant; OE: overexpression; Pe: effective permeability; ZO: zona occludens.
Overexpression of circLphn3 reduces the hemin induced BBB damage in bEnd.3 cells
Next, we examined the function of circLphn3 in bEnd.3 cell dysfunction caused by hemin treatment. Hemin administration for 12 hours significantly decreased the expression of all four TJPs (including ZO-1, ZO-2, ZO-3, and occludin) in bEnd.3 cells (P < 0.05, vs. control group; Figure 4A and B). As shown in Figure 1E, the circLphn3 expression decreased in bEnd.3 cells treated with hemin (40 µM). To further explore the key role of circLphn3 in BBB damage, we established a monolayer bEnd.3 BBB model in the cell inserts for the permeability experiment. Samples were collected from the lower chamber at 1, 4, 12, 24, and 36 hours after hemin addition. The leakage of LY revealed that overexpression of circLphn3 decreased the hemin-induced high LY permeability at 24 and 36 hours (P < 0.05, vs. hemin group), while miR-185-5p transfection increased the permeability from 1 hour (P < 0.05, vs. hemin group; Figure 4C). Finally, ZO-1 immunofluorescence in bEnd.3 cells showed that overexpression of circLphn3 enhanced ZO-1 expression after hemin treatment, whereas miR-185-5p transfection attenuated the protective effect of circLphn3 on ZO-1 expression (Figure 4D).
Discussion
In this study, we used RNA-seq to examine differential circRNA expression in normal and injured brain tissues after TBI. CircRNA biogenesis correlates with the splicing of its host linear pre-mRNA (Chen and Yang, 2015). The changes in cis-elements and trans-factors after TBI may be related to the competition between regulatory canonical splicing and back-splicing of circRNAs. The prediction analysis showed that many circLphn3-targeted miRNAs were closely related to the TJP genes. BBB damage triggered by hemin has previously been reported (Karuppagounder et al., 2016; Min et al., 2017; Zhang et al., 2019). In our study, the expression of circLphn3 in hemin-treated bEnd.3 cells was significantly downregulated, while miR-185-5p expression was upregulated. Overexpression of circLphn3 reduced the hemin-induced cell permeability and enhanced the expression of TJPs. More importantly, we demonstrated for the first time the regulatory role of the circLpnh3/miR-185-5p/ZO-1 axis in hemin-induced cell injury.
In recent years, owing to the wide application of the RNA-seq technology, many circRNAs have been discovered in different species, revealing species and tissue specificities (Li et al., 2018). CircRNAs are abundantly expressed in the mammalian brain and various other tissues (Xu et al., 2017). In this study, the expression level of circLphn3 in the brain tissue after TBI was significantly lower than that in normal brain tissue. Furthermore, we identified that circLphn3 expression was downregulated in hemin-treated cerebral vascular endothelial cells. Further cell experiments showed that circLphn3 overexpression reduced the cell damage and enhanced the expression of tight junction-associated proteins in hemin-treated bEnd.3 cells. On the basis of these findings, we propose that circLphn3 may have an essential role in alleviating TJPs damage in hemin-treated bEnd.3 cells.
By sponging miRNA, circRNAs can regulate transcription, and even influence the pre-mRNA splicing process (Li et al., 2015). We speculated that TJP-related genes are regulated by competitive endogenous RNA (ceRNA). Our study showed that circLphn3 is mainly located in the cytoplasm of cerebral microvascular endothelial cells, and it may have a biological function by adsorbing miRNA. This study explored the protective mechanism of circLphn3 after TBI by specifically binding miR-185-5p. On the basis of prediction algorithms (John et al., 2004; Krüger and Rehmsmeier, 2006; Agarwal et al., 2015) and argonaute2 crosslinking-immunoprecipitation and high-throughput sequencing results in the Starbase database (Li et al., 2014), we hypothesized that circLphn3 may be used as a molecular sponge to adsorb miR-185-5p. Performing a dual-luciferase reporter gene assay, we confirmed that circLphn3 bound to miR-185-5p. Moreover, the miR-185-5p level was significantly reduced after circLphn3 overexpression in bEnd.3. These results confirmed that circLphn3, as a ceRNA, regulates miR-185-5p and participates in regulating TJPs’ expression. However, it is plausible that circLphn3 aslo binds to other miRNAs to mediate its biological function. Our experiments in vascular endothelial cells further confirmed that transfection of miR-185-5p mimics specifically downregulated the expression of ZO-1, but it had no effect on the expression of ZO-2, ZO-3, and occludin. Furthermore, the dual luciferase reporter gene assay confirmed the binding of miR-185-5p and ZO-1 mRNA 3′UTR.
The regulation of circRNA-miRNA-mRNA is a complex regulatory network rather than a linear one (Zhang et al., 2018). In a previous research, Lin et al. (2016) have found that mmu-circRNA-015947 may bind to mmu-miR-188-3p, miR-329-5p, miR-3057-3p, miR-5098, and miR-683, and participate in various pathways including apoptotic, metabolism, and immune pathways, which confirmed that changes in circRNAs are associated with additional miRNA and mRNA-related genes. In our study, circLphn3 overexpression in vascular endothelial cells revealed that the binding of ZO-1 with miR-185-5p and the expression of ZO-2 and occludin were upregulated. Because circLphn3 is a sponge of miRNAs, it may also bind other miRNAs that regulate ZO-2 and occludin. This research suggests that circLphn3, as a molecular sponge of miRNAs, can adsorb multiple miRNAs targeting tight junction-related mRNA genes, and thus may be essential in the regulation of tight junction-related protein expression. Therefore, we believe that our study supports the notion that the regulatory relationship of circRNA-miRNA-mRNA is not linear but a complex regulatory network. However, the network regulation relationship needs further study.
In summary, our study provides novel insight into the function of circLphn3, which is involved in BBB protection after TBI. CircLphn3 binds miR-185-5p and subsequently targets ZO-1. Specific circLphn3 overexpression may be a potential therapeutic target for the treatment of BBB damage. In this study, hemin was used to simulate the damage of cerebral vascular endothelial cells after TBI; however, it lacks the mechanical shear force injury that occurs in TBI. Additionally, the sample size of each group for RNA-seq was only three mice, which may lead to selection bias. Furthermore, the biological effects of overexpression or knockdown of circLphn3 in vivo require further research.
Additional files:
Additional Table 1: Softwares for RNA-sequence data analysis.
Additional file 1 (83.8KB, pdf) : Design and sequence of circLphn3 overexpression plasmid.
Design and sequence of circLphn3 overexpression plasmid
Footnotes
C-Editor: Zhao M; S-Editors: Yu J, Li CH; L-Editors: Yu J, Song LP; T-Editor: Jia Y
Conflicts of interest: The authors declare no conflicts of interest.
Financial support: This study was supported by the National Natural Science Foundation of China, No. 81771355; and the Natural Science Foundation of Chongqing of China, No. CSTC2015jcyjA10096 (both to ZBL). The funding sources had no role in study conception and design, data analysis or interpretation, paper writing or deciding to submit this paper for publication.
Institutional review board statement: The study was approved by Animal Care and Use Committee of Chongqing Medical University (approval No. 2021-177) on March 22, 2021.
Copyright license agreement: The Copyright License Agreement has been signed by all authors before publication.
Data sharing statement: Datasets analyzed during the current study are available from the corresponding author on reasonable request.
Plagiarism check: Checked twice by iThenticate.
Peer review: Externally peer reviewed.
Funding: This study was supported by the National Natural Science Foundation of China, No. 81771355; and the Natural Science Foundation of Chongqing of China, No. CSTC2015jcyjA10096 (both to ZBL).
References
- 1.Acosta-Martínez M. Shaping microglial phenotypes through estrogen receptors: relevance to sex-specific neuroinflammatory responses to brain injury and disease. J Pharmacol Exp Ther. 2020;375:223–236. doi: 10.1124/jpet.119.264598. [DOI] [PubMed] [Google Scholar]
- 2.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhowmick S, D’Mello V, Caruso D, Wallerstein A, Abdul-Muneer PM. Impairment of pericyte-endothelium crosstalk leads to blood-brain barrier dysfunction following traumatic brain injury. Exp Neurol. 2019;317:260–270. doi: 10.1016/j.expneurol.2019.03.014. [DOI] [PubMed] [Google Scholar]
- 4.Brotfain E, Gruenbaum SE, Boyko M, Kutz R, Zlotnik A, Klein M. Neuroprotection by estrogen and progesterone in traumatic brain injury and spinal cord injury. Curr Neuropharmacol. 2016;14:641–653. doi: 10.2174/1570159X14666160309123554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deli MA, Abrahám CS, Kataoka Y, Niwa M. Permeability studies on in vitro blood-brain barrier models: physiology, pathology, and pharmacology. Cell Mol Neurobiol. 2005;25:59–127. doi: 10.1007/s10571-004-1377-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glažar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20:1666–1670. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han D, Li J, Wang H, Su X, Hou J, Gu Y, Qian C, Lin Y, Liu X, Huang M, Li N, Zhou W, Yu Y, Cao X. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151–1164. doi: 10.1002/hep.29270. [DOI] [PubMed] [Google Scholar]
- 9.Jiang S, Zhao G, Lu J, Jiang M, Wu Z, Huang Y, Huang J, Shi J, Jin J, Xu X, Pu X. Silencing of circular RNA ANRIL attenuates oxygen-glucose deprivation and reoxygenation-induced injury in human brain microvascular endothelial cells by sponging miR-622. Biol Res. 2020;53:27. doi: 10.1186/s40659-020-00295-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human microRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karuppagounder SS, Alim I, Khim SJ, Bourassa MW, Sleiman SF, John R, Thinnes CC, Yeh TL, Demetriades M, Neitemeier S, Cruz D, Gazaryan I, Killilea DW, Morgenstern L, Xi G, Keep RF, Schallert T, Tappero RV, Zhong J, Cho S, et al. Therapeutic targeting of oxygen-sensing prolyl hydroxylases abrogates ATF4-dependent neuronal death and improves outcomes after brain hemorrhage in several rodent models. Sci Transl Med. 2016;8:328ra329. doi: 10.1126/scitranslmed.aac6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khellaf A, Khan DZ, Helmy A. Recent advances in traumatic brain injury. J Neurol. 2019;266:2878–2889. doi: 10.1007/s00415-019-09541-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kivioja T, Vähärautio A, Karlsson K, Bonke M, Enge M, Linnarsson S, Taipale J. Counting absolute numbers of molecules using unique molecular identifiers. Nat Methods. 2011;9:72–74. doi: 10.1038/nmeth.1778. [DOI] [PubMed] [Google Scholar]
- 14.Krüger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34:W451–454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2. 0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71:428–442. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 18.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, Zhu P, Chang Z, Wu Q, Zhao Y, Jia Y, Xu P, Liu H, Shan G. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 19.Lin SP, Ye S, Long Y, Fan Y, Mao HF, Chen MT, Ma QJ. Circular RNA expression alterations are involved in OGD/R-induced neuron injury. Biochem Biophys Res Commun. 2016;471:52–56. doi: 10.1016/j.bbrc.2016.01.183. [DOI] [PubMed] [Google Scholar]
- 20.Ma X, Aravind A, Pfister BJ, Chandra N, Haorah J. Animal models of traumatic brain injury and assessment of injury severity. Mol Neurobiol. 2019;56:5332–5345. doi: 10.1007/s12035-018-1454-5. [DOI] [PubMed] [Google Scholar]
- 21.Mahmoudi E, Fitzsimmons C, Geaghan MP, Shannon Weickert C, Atkins JR, Wang X, Cairns MJ. Circular RNA biogenesis is decreased in postmortem cortical gray matter in schizophrenia and may alter the bioavailability of associated miRNA. Neuropsychopharmacology. 2019;44:1043–1054. doi: 10.1038/s41386-019-0348-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGinn MJ, Povlishock JT. Pathophysiology of traumatic brain injury. Neurosurg Clin N Am. 2016;27:397–407. doi: 10.1016/j.nec.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 23.Min H, Choi B, Jang YH, Cho IH, Lee SJ. Heme molecule functions as an endogenous agonist of astrocyte TLR2 to contribute to secondary brain damage after intracerebral hemorrhage. Mol Brain. 2017;10:27. doi: 10.1186/s13041-017-0305-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, Dou K, Li H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 25.Siflinger-Birnboim A, Del Vecchio PJ, Cooper JA, Blumenstock FA, Shepard JM, Malik AB. Molecular sieving characteristics of the cultured endothelial monolayer. J Cell Physiol. 1987;132:111–117. doi: 10.1002/jcp.1041320115. [DOI] [PubMed] [Google Scholar]
- 26.Sulhan S, Lyon KA, Shapiro LA, Huang JH. Neuroinflammation and blood-brain barrier disruption following traumatic brain injury: Pathophysiology and potential therapeutic targets. J Neurosci Res. 2020;98:19–28. doi: 10.1002/jnr.24331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 28.Watson PM, Paterson JC, Thom G, Ginman U, Lundquist S, Webster CI. Modelling the endothelial blood-CNS barriers: a method for the production of robust in vitro models of the rat blood-brain barrier and blood-spinal cord barrier. BMC Neurosci. 2013;14:59. doi: 10.1186/1471-2202-14-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu T, Wu J, Han P, Zhao Z, Song X. Circular RNA expression profiles and features in human tissues: a study using RNA-seq data. BMC Genomics. 2017;18:680. doi: 10.1186/s12864-017-4029-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yilmaz C, Karali K, Fodelianaki G, Gravanis A, Chavakis T, Charalampopoulos I, Alexaki VI. Neurosteroids as regulators of neuroinflammation. Front Neuroendocrinol. 2019;55:100788. doi: 10.1016/j.yfrne.2019.100788. [DOI] [PubMed] [Google Scholar]
- 31.Yuan J, Wang A, He Y, Si Z, Xu S, Zhang S, Wang K, Wang D, Liu Y. Cordycepin attenuates traumatic brain injury-induced impairments of blood-brain barrier integrity in rats. Brain Res Bull. 2016;127:171–176. doi: 10.1016/j.brainresbull.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 32.Zafer D, Aycan N, Ozaydin B, Kemanli P, Ferrazzano P, Levine JE, Cengiz P. Sex differences in hippocampal memory and learning following neonatal brain injury: is there a role for estrogen receptor-α? Neuroendocrinology. 2019;109:249–256. doi: 10.1159/000499661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang F, Zhang R, Zhang X, Wu Y, Li X, Zhang S, Hou W, Ding Y, Tian J, Sun L, Kong X. Comprehensive analysis of circRNA expression pattern and circRNA-miRNA-mRNA network in the pathogenesis of atherosclerosis in rabbits. Aging (Albany NY) 2018;10:2266–2283. doi: 10.18632/aging.101541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Deng H, Hu Y, Pan C, Wu G, Li Q, Tang Z. Adipose-derived mesenchymal stem cells stereotactic transplantation alleviate brain edema from intracerebral hemorrhage. J Cell Biochem. 2019;120:14372–14382. doi: 10.1002/jcb.28693. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Design and sequence of circLphn3 overexpression plasmid